Abstract
Esf2p is the Saccharomyces cerevisiae homolog of mouse ABT1, a protein previously identified as a putative partner of the TATA-element binding protein. However, large-scale studies have indicated that Esf2p is primarily localized to the nucleolus and that it physically associates with pre-rRNA processing factors. Here, we show that Esf2p-depleted cells are defective for pre-rRNA processing at the early nucleolar cleavage sites A0 through A2 and consequently are inhibited for 18S rRNA synthesis. Esf2p was stably associated with the 5′ external transcribed spacer (ETS) and the box C+D snoRNA U3, as well as additional box C+D snoRNAs and proteins enriched within the small-subunit (SSU) processome/90S preribosomes. Esf2p colocalized on glycerol gradients with 90S preribosomes and slower migrating particles containing 5′ ETS fragments. Strikingly, upon Esf2p depletion, chromatin spreads revealed that SSU processome assembly and compaction are inhibited and glycerol gradient analysis showed that U3 remains associated within 90S preribosomes. This suggests that in the absence of proper SSU processome assembly, early pre-rRNA processing is inhibited and U3 is not properly released from the 35S pre-rRNAs. The identification of ABT1 in a large-scale analysis of the human nucleolar proteome indicates that its role may also be conserved in mammals.
Ribosome biogenesis is a complicated pathway that requires in eukaryotes an excess of 200 protein trans-acting factors and just as many small nucleolar RNAs (snoRNAs) (27, 28, 30, 38). The process is initiated and largely takes place in the nucleolus, a dedicated subcellular compartment. RNA polymerase I (Pol I) transcription generates a large precursor (35S in Saccharomyces cerevisiae) encoding three out of the four mature rRNAs (5.8S, 18S, and 25S rRNAs). Mature sequences are released from noncoding spacer regions (5′ and 3′ ETS and ITS1 and ITS2), following a relatively well-established processing pathway involving both endoribonucleolytic and exoribonucleolytic digestions (Fig. 1A) (reviewed in references 30 and 45). 5S rRNA is encoded by a Pol III transcript and matured independently.
FIG. 1.
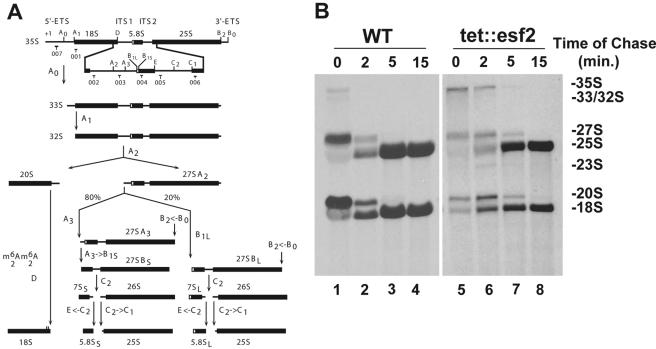
Esf2p is required for normal pre-rRNA processing. (A) Yeast pre-rRNA processing pathway. The 18S, 5.8S, and 25S rRNAs are encoded within a single large RNA Pol I transcript (35S). Mature sequences are interspersed with noncoding external (5′ and 3′) as well as internal (1 and 2) transcribed spacers. For a complete description of the pathway, see the text and reference 30. Cleavage sites (A0 to E) and the oligonucleotide probes used (001 through 007) are indicated. (B) Pulse-chase analysis. tet::esf2 and wild-type isogenic cells were grown in synthetic medium containing doxycycline for 24 h, pulsed with methyl-3H, and chased with an excess of cold methionine and samples collected at the times indicated. Total RNA was extracted and fractionated on denaturing agarose gels. Equal counts were loaded (20,000 cpm); the left exposure was for 3 days, the right for 18 days.
Most ribosome biogenesis factors are localized to the nucleolus, are essential for cell viability, and are components of large preribosomal protein complexes. Consequently, proteomic analyses have resulted in the identification of many potential new rRNA processing factors (RRPs) (reviewed in references 10 and 19), and the use of affinity purification has delineated distinct and presumably successive preribosomal complexes (reviewed in references 10, 12, and 46). Among these is the small-subunit (SSU) processome (3, 4, 9, 14, 35), a U3-containing ribonucleoprotein complex primarily involved in the three initial pre-rRNA processing reactions (cleavage at sites A0 through A2) (see Fig. 1A) and therefore required for 18S rRNA synthesis.
As 35S pre-rRNA molecules emerge from the Pol I transcription machinery, 5′ pre-rRNA termini are bound by U3 and early RRPs, generating the so-called “terminal knobs.” These are visualized by electron microscopy on chromatin spreads at the ends of the rRNA gene “Christmas tree” branches (9, 11, 35). In yeast, initial knobs condense around pre-18S rRNA sequences into larger structures that correspond to the SSU processome/90S preribosomes. SSU processomes are first detected in a loose configuration and, as pre-18S rRNA sequences undergo additional folding, are further compacted into tight structures (35). Yeast chromatin spreads also reveal a significant level of cotranscriptional cleavage at site A2, resulting in the SSU processomes being cleaved from the transcripts (35). Compaction of the knobs and cotranscriptional cleavage follow strict kinetics under optimal growth conditions, with knob compaction occurring roughly once transcription reaches the middle third of the rRNA gene and cotranscriptional cleavage soon after (35). Note that the 90S preribosomes, formally based on the 35S pre-rRNAs, and the SSU processomes, resulting from the assembly of U3 and pre-18S rRNA sequences, are biochemically similar and may be functionally indistinguishable. However, results of affinity purifications with individual tagged protein subunits vary considerably and indicate that this large RNP is dynamic and heterogeneous in composition and that its internal architecture is complex and modular (9, 18, 29, 36, 41).
Many of the U3-associated proteins (UTPs) remain associated with 5′ ETS fragments following cleavages at sites A0 through A2 (29, 36). In total cellular extracts, U3 is detected in three pools as follows: (i) as free small nucleolar RNPs (snoRNPs), (ii) engaged through Watson-Crick base pair interactions with the 35S pre-rRNA within the SSU processome/90S preribosomes, and (iii) within the cleaved 5′ ETS-based RNP (5, 20, 48). It is not known how the U3 RNP is recruited to nucleolar preribosomes or whether or how any of its constituents are recycled following cleavages at sites A0 through A2.
Ynr054cp is a protein that several genomic and proteomic analyses have identified as a potential RRP factor (see below); however, the mouse homolog was previously characterized as ABT1 (activator of basal transcription) and was reported to bind directly to TATA binding protein (34). Here, we present several lines of evidence that Ynr054cp is a bona fide component of the SSU processome. Both biochemical and phenotypic analyses indicate a role in early nucleolar cleavage sites A0 through A2, required for 18S rRNA synthesis, a stable association with U3 and 5′ ETS fragments, and a requirement for terminal knob formation and SSU processome assembly. We have named the gene ESF2 (for eighteen S factor 2) following our previous nomenclature for a gene with similar genetic and biochemical properties (ESF1 [36]).
MATERIALS AND METHODS
Bioinformatics.
ESF2 is YNR054C. Multiple alignments were performed with Clustal W, and setup was performed with Blosum62. The physical interaction network was visualized with Osprey (http://biodata.mshri.on.ca/osprey/servlet/Index) and edited manually. The set of interactions reported in the manuscript was used as a backbone and complemented with interactions from high-throughput data sets (15, 23, 26, 29), as well as specific data sets for Esf2p (22), Esf1p (36), and Pwp2p (8). For clarity, nodes with fewer than two connections are not represented in Fig. 4D.
FIG. 4.
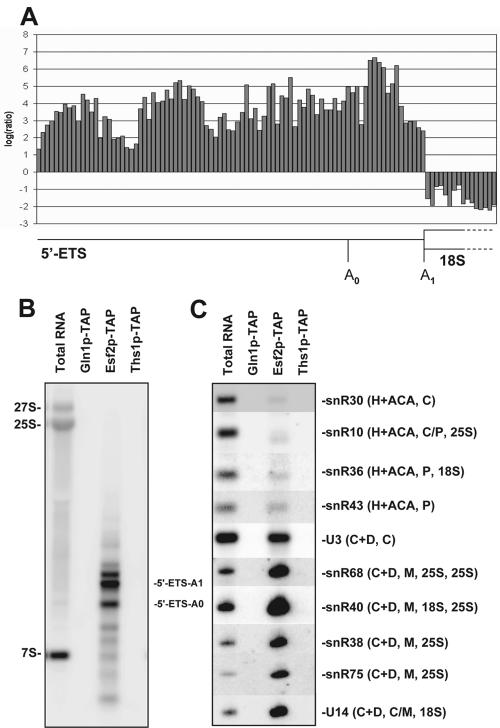
Esf2p is stably associated with 5′ ETS fragments, the box C+D snoRNA U3 and several other box C+D snoRNAs. (A) Ratios (Esf2p-TAP purification RNA versus total RNA) from oligonucleotide microarray probes spaced every five bases along the 5′ ETS fragments and the 5′ end of the 18S rRNA. Tandem affinity-purified fractions equivalent to 1.3 liters of cells were phenol extracted and subjected to microarray analysis or Northern blotting (see Materials and Methods). Microarray data are listed on our website (http://hugheslab.med.utoronto.ca/Hoang). The salt concentration used in washes was 150 mM. (B) Northern blot confirming that the Esf2p-TAP purification contains cleaved 5′ ETS. The bands detected are virtually identical to those detected in association with other UTPs (29, 36). Gln1p-TAP and Ths1p-TAP are included as negative controls for specificity; Gln1p is a metabolic enzyme, and Ths1p binds threonyl-tRNAs (25). The oligonucleotide probes used were EC2 and 5′ ETS-A0 (see Materials and Methods). (C) Northern blots confirming that the Esf2p-TAP purification contains U3 and a subset of other box C+D snoRNAs but not box H+ACA snoRNAs. The snoRNAs involved in cleavages (C) and/or modification (P, pseudouridylation; M, methylation), as well as known target substrates for modification (18S and/or 25S rRNA), are indicated. The snoRNA probes used are listed in Materials and Methods.
Yeast strains.
The tet::esf2 strain is TH_5627 and the isogenic wild type is R1158 (32). The strains expressing Esf2p tagged with tandem affinity purification (Esf2p-TAP), Gln1p-TAP, and Ths1p-TAP were obtained from the University of California—San Francisco collection (16). The untagged control strain is BY4741 (17). For Esf2p-eGFP strains, a PCR cassette for carboxyl-terminal tagging was generated with plasmid pKT128 or pKT209 and integrated at the ESF2 locus, according to reference 42, generating strains YDL892 (ESF2-eGFP-CaURA3) and YDL893 (ESF2-eGFP-SpHIS5), respectively. The tet::esf2 UTP18-TAP strain (YDL902) was generated by the integration of a PCR UTP18-TAP-HIS3MX6 cassette at the UTP18 locus in strain TH_5627.
RNA extraction and Northern blotting.
For the depletion of Esf2p protein, the tet::esf2 strain (and the isogenic wild-type control strain R1158) was exposed to 10 μg/ml doxycycline (DOX) (Sigma) for a total of up to 24 h before harvesting for RNA extraction. For Fig. 2, strains were grown in complete yeast extract-peptone-dextrose (YPD) medium. Identical results were obtained when cells were grown in synthetic dextrose (SD) medium (data not shown). RNA extraction and Northern blotting were performed as described previously (7).
FIG. 2.
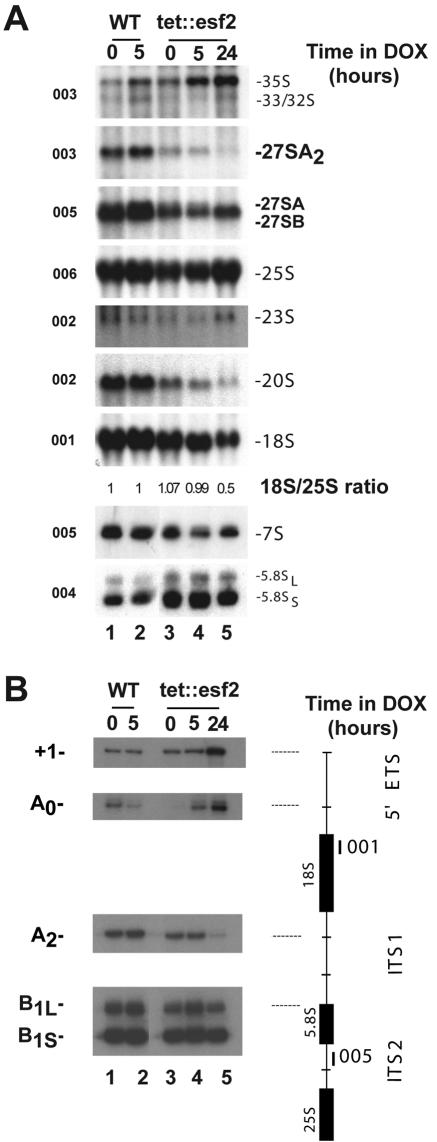
Esf2p is required for nucleolar pre-rRNA processing at cleavage sites A0 through A2. (A) Northern blot analysis. tet::esf2 and wild-type isogenic cells were grown in complete medium and maintained in exponential phase by dilution with fresh medium. Samples were collected in the absence of doxycycline (0-h time point) and following the addition of the antibiotic for 5 and 24 h. Total RNA was extracted, separated on agarose/formaldehyde gels, and processed for Northern blot hybridization. The oligonucleotide probes used (Fig. 1A) are indicated. The 18S-to-25S ratio was established by phosphorimager quantitation (Typhoon 9200/ImageQuantTL v2003.03; GE Healthcare). Note that for tet::esf2 strains, the pre-rRNA processing defect in the absence of doxycycline is already detected (0-h time point), suggesting that the level of ESF2 mRNA is already reduced by the Tet transcriptional fusion in the absence of doxycycline-mediated repression L, Long; S, short. (B) Primer extension analysis. Total RNA from Esf2p-depleted and isogenic control cells was also used as a template in primer extension experiments with oligonucleotides 001 and 005.
For Fig. 3, oligonucleotides 001 through 006 are identical to those described in reference 7; oligonucleotide probe 007 is ETS-A0 (see below). For Fig. 5B and C, the oligonucleotide probes used and their sequences were as follows: EC2, 5′-TCCAATGAAAAGGCCAGCAATTTCAAGTTAACTCCAAAGAGTATCACTCAC-3′; 5′ ETS-A0, 5′-GGAAATGCTCTCTGTTCAAAAAGCTTTTACACTCTTGACCAGCGCACTCC-3′; U3, 5′-CCCTATCCCTTCAAAAAAGAAGTACATAGGATGGGTCAAGATCATCGCGC-3′; U14, 5′-GCGGTCACCGAGAGTACTAACGA-3′; snR10, 5′-CACATTCTTCATGGGTCAAGAACGCCCCGG-3′; snR30, 5′-TCCATATATATCATGGCAACAGCCCCCGAA-3′; snR40, 5′-TGGGTATACTTAATCCTTCATAGGACACCT-3′; snR68, 5′-ACAGCCCCCGTCAATACGATAACGCAGTAA-3′; snR36, 5′-GTCATCCAGCTCAAGATCGTAATATTG-3′; snR43, 5′-CGAGACGCCGTCTACGGTTGTATC-3′; snR38, 5′-ACAAATATCAACATATGAGAGGTTACCTATTATTACCCATTCAGACAGGG-3′; snR75, 5′-CATGAATGGTAATTTTAATAGTTGTGTCTTCACGAATGATCAGACTCGTC-3′; and tRNATrp, AACCTGCAACCCTTCGA.
FIG. 3.
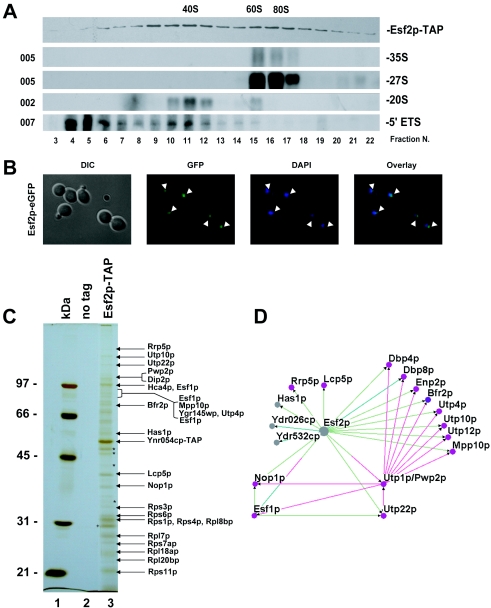
Esf2p, a component of 90S preribosomes and 5′ ETS-based RNPs. (A) Glycerol gradient. Total cellular extracts from a strain expressing a functional Esf2p-TAP construct were layered on 10 to 30% glycerol gradients and resolved by ultracentrifugation. Twenty-four fractions were collected manually and analyzed by Western and Northern blotting for protein and RNA compositions. The peroxidase antiperoxidase antibody (Sigma) that recognizes the protein A moiety of the tandem affinity purification tag and specific oligonucleotides (to the left) (Fig. 1A) were used in the hybridizations. (B) GFP fluorescence. Esf2p-eGFP-expressing cells were grown to mid-log phase in complete medium and the fusion wasdetected directly in live cells by use of a Zeiss Axioskop2 Plus microscope equipped with a Plan Neofluar 100×/1.30 objective. Acquisition was performed with an AxiocamHRm camera and the native Axiovision4 software from Zeiss (release 4.1). Cells were counterstained with DAPI that labels the bulk of the DNA; a differential interference contrast (DIC) is provided. The strain presented here is YDL892; the same result was obtained with strain YDL893. (C) MALDI-TOF mass spectrometry analysis. Total cellular extracts, as shown in panel A, were affinity purified according to the tandem affinity purification protocol. Pellet fractions were resolved by SDS-PAGE and analyzed by MALDI. Major copurifying bands are annotated. As a control, a wild-type isogenic strain (no tag) was used. Molecular weight markers (MW) are on the left. The asterisks represent degradation products of Esf2p. The cross corresponds to tobacco etch virus protease, which was used in the purification procedure. (D) Esf2p-centric view of preribosome assembly. This diagram was compiled with the Osprey software from several high-throughput data sets, as well as specific analysis (see Materials and Methods). Interactions are color coded as follows: light green, affinity purification (this work); turquoise, two-hybrid interaction (22); and pink, affinity purification (15, 23).
FIG. 5.
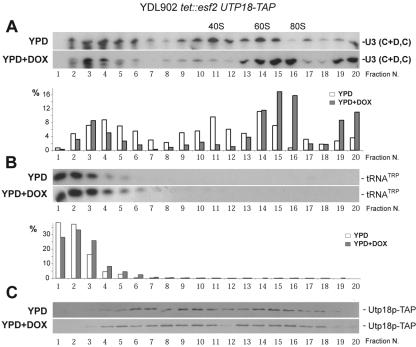
Esf2p is required for the efficient cycling of U3 and other snoRNAs. Total cellular extracts from a tet::esf2 UTP18-TAP strain grown to mid-log phase in complete medium in the absence of doxycycline (YPD) and following its addition for 24 h (YPD+DOX) were analyzed by a 10 to 30% glycerol gradient in 200 mM salt. Identical amounts of total proteins were loaded on each gradient. Total RNA was extracted from 20 fractions and processed for Northern blotting with a probe specific to U3 (A) and mature tRNATrp (B). The detected signal in each fraction was quantitated with a Typhoon 9200 and the ImageQuanTL V2003.3 native software (GE Healthcare). Note that it is unclear why the level of the large particles pelleted at the bottom of the tubes (fractions 19 and 20) is slightly more elevated in the YPD+DOX condition. (C) Total proteins were extracted from all fractions, resolved by PAGE, and analyzed by Western blotting for Utp18p-TAP detection with the PAP antibody (see Fig. 3).
Primer extension was as described previously (7).
Pulse-chase labeling.
R1158 (wild type) and the tet::esf2 mutant were transformed with pRS411 (a plasmid containing MET15) and grown to 0.8 × 107 cells/ml over a period of 24 h in SD-Met medium containing 10 μg/ml doxycycline. Pulse-chase labeling, use of gels, and autoradiography were performed as described previously (36).
Affinity purification.
Four liters of tandem affinity purification-tagged cells (or untagged negative control cells) was grown in YPD to 1.5 × 107 cells/ml and harvested by brief centrifugation. The cell pellets were washed twice with cold water and frozen with liquid nitrogen. Frozen cell pellets were broken with dry ice in a small coffee grinder (Krups), and 10 ml IPP150 buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Triton X-100) plus 1 mM dithiothreitol and protease inhibitors were added to the lysed cells. The broken cells were subjected to centrifugation at 13,000 rpm in an SS-34 rotor (20,200 × g) for 1 h at 4°C. The lysates were mixed with 200 μl immunoglobulin G-agarose beads for 2 h at 4°C. After five washes with 0.5 ml IPP150 buffer, the eluates from immunoglobulin G columns were further purified with calmodulin as previously described (29). The RNA associated with the complex was extracted with phenol-chloroform, and RNase-free DNase I was used to remove DNA in the samples as described previously (29). The purified proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gels containing 10% polyacrylamide, visualized by silver staining, and identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry.
For Northern and microarray analysis of RNA in protein complexes purified by tandem affinity purification, affinity-purified fractions equivalent to 1.3 liters of cells were extracted with phenol, ethanol precipitated, and treated with DNase I as described previously (29). Array design, array hybridizations, and data processing followed reference 49. Briefly, the raw log ratios for each oligonucleotide were calculated from the median intensity at each spot, and the data were normalized by applying a lowest smoother to the ratios of each experiment over intensity. RNA species with high ratios over multiple oligonucleotides detecting the same transcript were taken as the strongest candidates for bona fide enrichment in the Esf2p-TAP purification. Probe sequences and all data are compiled at our website (http://hugheslab.med.utoronto.ca/Hoang).
Glycerol gradients.
For Fig. 3, lysis in buffer A200 (20 mM Tris-HCl, pH 8.0, 200 mM K acetate, 5 mM Mg acetate, 0.2% Triton X-100, 1 mM dithiothreitol) was performed, and the gradients were prepared in 60 mM salt as described in reference 6. For Fig. 5, both cell lysis and the gradients were prepared in A200.
GFP fluorescence.
Esf2p-enhanced green fluorescent protein (eGFP)-expressing cells were grown to mid-log phase in complete medium. The fusion protein was detected directly in live cells with a Zeiss Axioskop2 Plus microscope equipped with a Plan Neofluar 100×/1.30 objective. Acquisition was performed with an AxiocamHRm camera and the native Axiovision4 software from Zeiss (release 4.1). Cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole), which labels the bulk of the DNA.
Chromatin spreads were performed as described in reference 35. Semiquantitative analysis (Table 1) was established by visual inspection of printed micrographs and evaluated with Student's t test.
TABLE 1.
Average numbers of SSU processomes and cleaved transcripts in Esf2p depletion and control conditions
| Condition (no. of genes analyzed) | Avg value (range)b
|
||||
|---|---|---|---|---|---|
| No. of SSU processomes/gene | Position of first SSU processomec | No. of transcripts in last half of gene | No. of cleaved transcripts | % of cleaved transcripts in second half of gene | |
| Control (11) | 9.6 (3-18) | 52.3 (40-65) | 28 (17-40) | 8.1 (4-20) | 29 (14-50) |
| Depletion time (h)a | |||||
| 10 (15) | 9.7 (5-16) | 48 (40-75) | 20.6 (12-31) | 7.4 (1-19) | 36 (6-76) |
| 18 (17) | 7.9 (0-26) | 53.8 (20-75) | 24 (13-39) | 2.1 (0-5) | 8.6 (0-21) |
| 24 (15) | 6.1 (1-18) | 64 (50-100) | 18.2 (10-23) | 1.7 (0-4) | 9.5 (0-27) |
Depleted strains were grown in 10 μg/ml doxycycline for the number of hours indicated.
The percentage of cleaved transcripts at each time point was evaluated with Student's t test. Results for the control and 10-hour depletion were not significantly different from each other, and neither were those for the 18-hour and 24-hour depletions. All other paired combinations were significantly different, with P values of 0.0002 or lower.
Percent distance along gene from 5′.
RESULTS
Bioinformatic analyses of large-scale data suggest that Esf2p is a preribosomal trans-acting factor.
Our analysis of Esf2p was prompted by the results of several large-scale studies that, taken together, suggested that this protein may play a role in ribosome biogenesis. First, Esf2p was found to be physically associated with Utp1p/Pwp2p, a core component of the SSU processome (9), in the tandem affinity purification data of Gavin et al. (15). With over 20 listed interactions with known UTPs, Utp1p is the most highly connected constituent of the SSU processome. In a separate study (23), Esf2p was found to be in physical association with Esf1p, an 18S rRNA biogenesis factor also associated with some components of the SSU processome (36). A large-scale GFP localization study (24) placed Esf2p in the nucleolus and, to some minor extent, in the cytoplasm. The ESF2 gene is required for cell viability (34), which is characteristic of many ribosome biogenesis factors, and the mRNA expression profile of a tetracycline (Tet)-regulatable promoter allele of ESF2 correlated most highly with Tet-regulatable promoter mutants in other genes encoding ribosome biogenesis factors (32).
Esf2p in both plants and mammals is conserved at the primary sequence level (data not shown), and the only characterized homolog of Esf2p is encoded by ABT1 (see Discussion), which appears to be single copy in all fully sequenced mammalian genomes. Although sequence similarity is high throughout most of the alignment, there are some differences between the sequences from different organisms; for example, yeast Esf2p has a 43-amino-acid N-terminal extension and a 20-amino-acid insertion (data not shown). SMART (31), pfam (2), and prosite (25) domain searches detected a noncanonical RNA recognition motif in most of these sequences, supporting a role in RNA metabolism. Taken together, these pieces of evidence motivated us to explore in more detail the potential role of Esf2p in ribosome biogenesis.
Esf2p is required for early nucleolar pre-rRNA processing reactions.
Since Esf2p is an essential gene (34), our functional analysis used a Tet-regulated allele (tet::esf2) (32, 37) in which ESF2 was placed under the control of a Tet-regulatable promoter, allowing specific transcriptional shutoff upon the addition of doxycycline to the growth medium (see Materials and Methods). In the case of ESF2, the depletion by the Tet transcriptional fusion was incomplete, as judged by growth defects (doubling times of 140, 166, and 260 min at the 0-, 10-, and 20-h time points of depletion, respectively). After the strains had been in doxycycline for 24 h, the doubling time had reached a steady value (260 min) corresponding to about twice the doubling time of the wild-type strain (120 min). This level of repression was, however, sufficient to cause not only a growth inhibition but, in addition, specific defects in ribosome synthesis and SSU processome metabolism (see below).
To investigate a possible involvement of Esf2p in RNA Pol I transcription and/or pre-rRNA processing, tet::esf2 and wild-type isogenic cells were grown in synthetic (SD-Met) medium to mid-log phase in the presence of doxycycline for 24 h, pulse-labeled with 250 μCi tritiated methyl-methionine for 2 min and chased with an excess of cold methionine. Samples were collected at 0, 2, 5, and 15 min of chase. Total RNA was extracted, separated on denaturing agarose gels, and transferred to nylon membranes for autoradiography (Fig. 1B).
In wild-type yeast cells, the primary Pol I transcript, 35S pre-rRNA, is initially cleaved in the 5′ external transcribed spacer (5′ ETS) at sites A0 and A1 and within the internal transcribed spacer 1 (ITS1) at site A2 (Fig. 1A), generating successively the 33S and 32S pre-rRNAs. Cleavage at sites A0 through A2 occurs very rapidly following transcription, and consequently, only low levels of the primary transcript and the 33S/32S precursors are normally detected in wild-type cells (Fig. 1B).
Upon depletion of Esf2p, the 35S pre-rRNA and all other precursor and mature forms were observed. However, the 35S and, to some lesser extent, the 33S/32S precursors were mildly accumulated and chased to 27SA and 20S pre-rRNAs with slowed kinetics. This indicates that the early nucleolar processing events (cleavage at processing sites A0 through A2) are inhibited. Consistent with this, the aberrant 23S RNA that results from direct cleavage of the 35S pre-rRNA at site A3 in the ITS1, in the absence of cleavage at sites A0 through A2, was accumulated. In addition, the maturation of the 20S pre-rRNA to 18S rRNA appeared substantially slowed, resulting in a decrease in the production of mature 18S rRNA. In contrast, 27S processing was not severely perturbed and 25S rRNA synthesis was only marginally affected.
Inhibition of cleavage at sites A0 through A2 was confirmed by Northern blotting and primer extension experiments (Fig. 2A and B, respectively). tet::esf2 and isogenic wild-type cells were grown in complete medium to mid-log phase, and cells were collected at regular intervals following the addition of doxycycline (Fig. 2 shows analyses of samples under conditions of 5 and 24 h of depletion in the presence of doxycycline). Total RNA was extracted, and processed for either Northern blotting or primer extension analysis (Fig. 2A and B, respectively).
Hybridization with a collection of oligonucleotide probes specific to subsets of pre-rRNAs and mature rRNAs (Fig. 1A) confirmed our initial observations of a significant delay in the initial processing at sites A0 and A1 (elevated amounts of 35S and 33/32S) and inhibition of cleavage at A2 (substantial reduction in 27SA2 and 20S). The reduction in 20S pre-rRNA accumulation led to an inhibition in 18S rRNA synthesis relative to 25S that was estimated by phosphorimager quantitation to be about 50% after 24 h of depletion (Fig. 2A). The mild accumulation of the aberrant 23S RNA is consistent with inhibition of cleavage at sites A0 through A2. The pathways of 5.8S and 25S rRNA synthesis were not significantly affected (essentially equal levels of 27SB, 25S, 7S, and two forms of 5.8S were observed).
For the primer extension analysis, cDNAs were extended across the 5′ ETS and the ITS1 from oligonucleotides 001 and 005, respectively, in order to detect the transcription start site (+1), as well as cleavage sites A0, A2, A3, B1L, and B1S (Fig. 2B). Delayed cleavage at sites A0 and A1 was confirmed by an increased signal at +1 and A0. The reduction in the steady-state level of 27SA2 observed by Northern blot hybridization was corroborated by a strong reduction in the signal at A2 in the primer extension assay (Fig. 2B). Processing at sites A3, B1L, and B1S, which initiate the synthesis of the 5.8S and 25S rRNAs, was not affected (Fig. 2B and data not shown).
Finally, the steady-state level of the snoRNAs known to be required for pre-rRNA processing at sites A0 through A2 (i.e., U3, snR10, and snR30) was tested and found to be unaffected (data not shown).
In conclusion, Esf2p is required for early nucleolar pre-rRNA processing at sites A0 through A2. Inhibition of cleavage at these sites results in a decrease of about 50% in the production of the 18S rRNA, the RNA component of the small ribosomal subunit. However, Esf2p is not required for the accumulation of snoRNAs required for processing at these sites.
Esf2p is stably associated with U3-specific proteins within 90S preribosomes and 5′ ETS-containing RNPs.
The requirement for Esf2p in cleavage at sites A0 through A2 suggested that the protein is acting at an early stage in preribosome assembly. To establish whether Esf2p is a component of preribosomal particles, in particular, of the SSU processome/90S preribosomes, the sedimentation profile of a functional epitope-tagged version of the protein was established on glycerol gradients. For this, we employed a strain in which Esf2p was fused at its carboxyl-terminal end to the tandem affinity purification tag and expressed under its own promoter from its chromosomal locus (16).
Total extracts from yeast cells expressing an Esf2p-TAP construct were layered and fractionated on 10 to 30% glycerol gradients. Following ultracentrifugation, total RNA and total proteins were extracted from all 24 fractions and processed for either Western or Northern blotting analysis (Fig. 3A). We concluded that Esf2p showed a bipartite distribution. The protein colocalized with fast-migrating particles that correspond to 90S preribosomes (fractions 15 through 17 and upwards), as well as with slower-migrating particles (fractions 9 through 12), that largely cosediment with 5′ ETS fragments. Note that the initial assignment of 90S by Trapman et al. (43) corresponded to gradient analyses performed at a low salt concentration (10 mM) and that large preribosomes are known to sediment significantly slower at the higher salt concentration used here. Importantly, Esf2p was not significantly enriched in fractions corresponding to pre-40S subunits (20S pre-rRNA peaks at fraction 11) indicating that the protein is likely not a pre-40S component, and presumably cycles off from the preribosomes following cleavage at site A2. This is supported by a previous proteomic analysis of the pre-40S complex, which did not detect Esf2p (41, 44).
We therefore reinvestigated the subcellular localization of Esf2p (18) in a freshly made Esf2p-eGFP strain (see Materials and Methods). In our hands, the protein was unambiguously restricted to the nucleolus (Fig. 3B), unlike in a previous study, in which it was additionally detected in the cytoplasm (24).
To establish which proteins associate with Esf2p, total extract from Esf2p-TAP-expressing cells was affinity purified by use of the tandem affinity purification protocol (see Materials and Methods) (39). The affinity-purified fraction was characterized by SDS-PAGE and MALDI-TOF mass spectrometry analysis (Fig. 3C). Esf2p was stably associated with at least 14 known RRPs, as well as with at least 10 ribosomal proteins, constituents of both the small and large subunits. Many of the RRPs found in association with Esf2p-TAP have been annotated as UTPs and described as core components of the U3 RNPs; however, we know now that 90S preribosomes and SSU processomes are similar entities (see the introduction) (9, 35). Our preparation also contained Esf1p, a protein with a similar spectrum of physical interactions, which we have previously proposed as an SSU accessory factor (36). Based on these novel interactions and the results of several high-throughput data sets, as well as specific analysis, we assembled a high-confidence network of physical associations centered on Esf2p (see Materials and Methods) (Fig. 3D).
In this network, Esf2p appears connected to at least three putative RNA helicases (Has1p, Dbp4p/Hca4p, and Dbp8p; see Discussion), as well as to several uncharacterized open reading frames. Ydr026cp is a nonessential nucleolar protein; ydr026cΔKanMx cells showed no growth defect (tested at 18°C, 23°C, 30°C, and 37°C) and showed no SSU rRNA synthesis defect (data not shown). We also note the conspicuous presence of Nop1p and Utp1p/Pwp2p (see below and Discussion).
Esf2p is stably associated with 5′ ETS fragments, the box C+D snoRNA U3 and several box C+D snoRNAs enriched in 90S preribosomes.
To determine whether Esf2p stably associates with RNAs, we phenol extracted a fraction of the tandem affinity-purified Esf2p complex, labeled the extracted material with fluors, and hybridized it to a microarray containing 21,939 oligonucleotides that tile every five bases of all known yeast noncoding RNAs in the antisense orientation. The copurifying RNA was compared against total cellular RNA by using a two-color (i.e., red/green) system such that spots with the highest red/green ratios should represent the probes with the highest enrichment in the Esf2p-containing complex(es). The most striking result was that virtually all of the probes complementary to the 5′ ETS had high ratios (Fig. 4A); the entire 5′ ETS sequence appears to be associated with Esf2p, whereas the adjacent 18S sequences are underrepresented (the means of all RNAs on the array are zero due to normalization). We confirmed by Northern blotting that this corresponds to the cleaved 5′ ETS fragment(s) (Fig. 4B) as previously observed for other components of the SSU processome (29, 36).
The array data also suggested that Esf2p is associated with the U3 snoRNA, as well as with several other box C+D snoRNAs that are expected to be enriched within the SSU processome/90S preribosomes (http://hugheslab.med.utoronto.ca/Hoang). We confirmed several of these interactions by Northern blotting (Fig. 4C). The enrichment of box C+D snoRNAs is consistent with the fact that the protein purification contains Nop1p (Fig. 3D), which is a component of all box C+D snoRNPs. It is likely that these interactions are indirectly mediated through Nop1p. Other core components of box C+D snoRNPs were not detected; however, as in other affinity purifications of large preribosomal particles (e.g., references 9, 18, 21, 33, 40, and 41) it is possible that not all associated proteins were identified, presumably due to either fragmentation of the complex during purification or the protein being associated with multiple dynamic complexes in vivo (Fig. 3A).
We conclude that Esf2p is a novel U3-associated protein. Esf2p is unlikely to be a core constituent of the U3 RNP, since it was not present in the originally described SSU processome (9) and it is also not a component of the UTP “A,” “B,” or “C” subcomplexes (29). Rather, it presumably interacts only transiently with the SSU processome, a notion that is also supported by the fact that many of the bands in Fig. 3C appear substoichiometric.
Upon depletion of Esf2p, U3 and other snoRNAs remain associated within 90S preribosomes.
Upon fractionation in glycerol gradients, U3 is detected in three distinctive peaks that correspond to a free pool (also referred to as monosome particles or 12S), a peak that comigrates with the 5′ ETS at about 40S, and a fraction that localizes with the 35S pre-rRNA at about 60S to 80S (5, 20, 48; see below).
To test whether this distribution is affected upon Esf2p depletion, total extracts from cells grown to mid-log phase in either YPD or YPD and doxycycline for 24 h were layered on 10 to 30% glycerol gradients under stringent salt conditions (200 mM) and separated by ultracentrifugation; 20 fractions were collected and analyzed by Northern blotting and Western hybridizations (Fig. 5). Under wild-type conditions (YPD), U3 showed the expected tripartite distribution, with fractions 2 through 6 corresponding to the monosomes, fractions 9 through 12 to the 5′ ETS-based particles, and fractions 13 through 15 to the large 90S preribosomes. Upon Esf2p depletion (YPD and DOX), the fraction of U3 that comigrates with the large preribosomes was significantly enriched (Fig. 5A, compare fractions 15 and 16 under both YPD and YPD plus DOX conditions and see quantitation in the lower panel), while the fraction associated with the 5′ ETS fragments was proportionally reduced. The pools of monosomes were not affected. The detected signal in each fraction was quantitated by phosphorimager counting and represented graphically as a percentage of the total input (Fig. 5A, lower panel). Fraction 15 showed an approximately twofold increase; fraction 16 was increased by a factor of 22. As a loading control, the membrane was probed for a mature tRNA species (tRNATrp); a signal similar in strength was detected across the top fractions of the gradient for this RNA (Fig. 5B). The experiment was repeated for U14, a box C+D snoRNA involved in pre-rRNA methylation and processing, as well as for snR3, a box H+ACA required for pseudouridylation. The distribution of these two snoRNAs was affected in a fashion similar to that described for U3 (http://www.ulb.ac.be/sciences/rna). Note that this experiment was carried out both with a tet::esf2 strain (data not shown) and with a tet::esf2 UTP18-TAP strain (Fig. 5), with identical results.
In conclusion, the fraction of U3 that comigrates with the 35S pre-rRNA increases substantially upon Esf2p depletion, while the amount detected in association with the 5′ ETS, resulting from early pre-rRNA processing cleavages, proportionally decreases. This is consistent with the biochemical phenotype described above, i.e., inhibition of pre-rRNA processing at sites A0 through A2.
The tet::esf2 UTP18-TAP strain was further used to characterize the protein composition of the preribosomes isolated in the presence and absence of Esf2p. For this, we first established the distribution of Utp18p-TAP in a glycerol gradient by Western blotting, in both the presence and absence of Esf2p (Fig. 5C). Utp18p-TAP was similarly distributed across the gradient under both conditions. Preribosomes were specified selected from Utp18p-TAP in cells grown in YPD and in YPD plus DOX for 24 h, and the proteins and RNAs isolated were characterized by mass spectrometry and microarray analysis; however, no significant qualitative differences were unveiled (http://hugheslab.med.utoronto.ca/Hoang).
We conclude that the overall protein and RNA composition of the SSU processome is not affected upon Esf2p depletion.
Esf2p is required for SSU processome assembly and compaction.
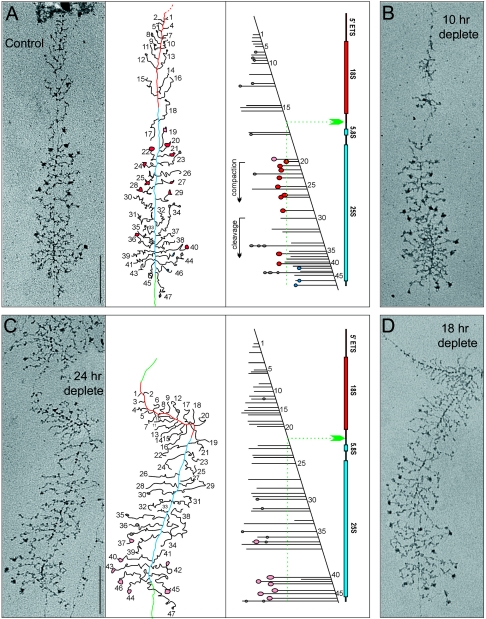
To test whether the SSU processome assembly kinetics and/or its internal architecture is dependent upon Esf2p, chromatin spreads were performed with the tet::esf2 strain grown to mid-log phase in YPD and at different time points of depletion in doxycycline (10, 18, and 24 h) (Fig. 6).
FIG. 6.
Esf2p is required for terminal knob deposition and SSU processome assembly. Yeast rRNA genes are shown as visualized by chromatin spread. Cells were grown to mid-log phase in YPD (control) or YPD plus DOX (10-h, 18-h, and 24-h depletion time points) and spreads made according to reference 35. For the control and 24-h depletion time points (panels A and C), interpretive tracing of the genes and transcript mapping are provided. DNA is color coded as follows: the 5′ end to A2 is red, A2 to the 3′ end is blue, and the intergene spacer is green. Particles that appear on the transcripts are shown on the tracing as follows: gray particles correspond to the initial small 5′ terminal knobs, pink to the newly formed (loose) large SSU processomes, red to the mature (tight) SSU processomes, and blue to pre-large-subunit knobs that form at the 5′ end of cleaved transcripts.
In the control condition (YPD), terminal knob deposition, as well as SSU processome assembly and compaction, occurred with the proper timing and a normal level of cotranscriptional cleavage was observed (Fig. 6A, gene mapping cartoon; see Table 1 for a semiquantitative analysis). Following 24 h of depletion (Fig. 6C), many fewer knobs were formed, and their appearance was severely delayed. In addition, very few SSU processomes were assembled, and those detected were all in the loose conformation. Finally, no significant level of cotranscriptional cleavage was observed (Fig. 6C and Table 1). The results of the 10-h depletion were similar to those of the control, while the 18-h and 24-h depletion results were similar to each other (Fig. 6B and D and Table 1).
We conclude that in the absence of Esf2p, terminal knob deposition and SSU processome assembly upon nascent rRNA are kinetically delayed and that once assembled, the SSU processomes fail to rearrange into a compact structure. This is fully compatible with the pre-rRNA processing phenotype and presumably underlies the inability of U3 to cycle off from the 90S preribosomes as part of a cleaved 5′ ETS RNP.
DISCUSSION
Esf2p is a novel accessory component of the SSU processome.
Esf2p was previously described as the yeast homolog of ABT1, a mouse protein implicated in general transcription by virtue of its ability to associate with the TATA binding protein and to stimulate mildly the transcription of reporter genes (34). However, accumulating evidence, including protein-protein interactions and subcellular localization, pointed to a role for Esf2p in ribosome biogenesis. Our results show unequivocally that yeast Esf2p is required for normal pre-rRNA processing, as well as for SSU processome assembly and function. The tet::esf2 mutant accumulates specific aberrant 18S rRNA precursors, and the proteins and RNAs contained in our Esf2p affinity purifications are consistent with a function closely related to that of the SSU processome. The distribution of Esf2p on glycerol gradients is also consistent with a function that involves its association with large preribosomal particles. Finally, the requirement for Esf2p in proper U3 sedimentation profile, as well as in terminal knob deposition and SSU processome assembly, clearly demonstrates that Esf2p is closely linked to the function of U3. Note that it is the kinetics of formation of the SSU processome, as well as its internal organization (transition from loose to tight configuration is defective), rather than its gross composition, that is affected.
While we cannot rule out a role for Esf2p in basal Pol II transcription, neither we nor others have confirmed the physical association of Esf2p with yeast TATA binding protein (15, 23). Since most RRPs are essential for cell viability, it is conceivable that ribosome biogenesis is the essential function of Esf2p. This is supported by the fact that the most highly conserved region of Esf2p appears to be a noncanonical RNA recognition motif, which is one of the most common RNA binding motifs.
We also note that human ABT1 was identified as a component of the human nucleolar proteome in an independent study (1). This is consistent with a role for mammalian ABT1 in ribosome biogenesis. The fact that ABT1 appears to be constitutively expressed across mammalian tissues (34) is also consistent with such a role.
What is the mechanistic role of Esf2p in ribosome biogenesis?
Our data point to a role for Esf2p in early ribosome biogenesis, specifically at the SSU processome-mediated cleavage sites A0, A1, and A2. This is supported by phenotypic analysis, by the proteins with which Esf2p associates, and by its stable physical association with U3 as well as with the 5′ ETS.
Although Esf2p physically associates with Nop1p and several box C+D snoRNAs, the fact that our pulse-chase analysis with methionine (Fig. 1B) did not display any gross alterations demonstrates that rRNA methylation is not greatly inhibited. Esf2p is therefore apparently not critical for the function of the snoRNAs in RNA modification. Other core box C+D snoRNPs were not detected in our Esf2p-TAP purification; this could be due to the technical difficulty of comprehensive peptide identification in a large and dynamic complex. However, another possible explanation is that the steady-state levels of the different box C+D snoRNP core proteins within the pre-rRNPs isolated from Esf2p are different from those previously described for isolated box C+D snoRNPs (13, 47).
The strong interaction between Esf2p and 5′ ETS fragments suggests that the Esf2 protein, like several previously inspected U3 components (29, 36), cycles off from the 90S preribosomes following cleavage at site A2. This predicts that Esf2p would not follow pre-40S subunits to the cytoplasm, and indeed, the protein has been neither copurified with the recently described SSU RRP complex (41, 44) nor detected in the cytoplasm, in our hands (Fig. 3B). The presence of several snoRNAs in our tandem affinity purifications (which largely select for the 5′ ETS-containing particles) suggests that their release, following early pre-rRNA cleavages, may also be Esf2p dependent and thus coupled to the release of the 5′ ETS. The identification of several helicases in Esf2p-TAP preparations also supports a role in recycling snoRNPs and/or the SSU processome. It is also intriguing that our 5′ ETS-based rRNPs are specifically enriched for box C+D snoRNAs and not for H+ACA components, since 2′-O methylation and pseudouridine formation are not known to operate mechanistically on distinct kinetic timelines; together, these observations suggest that Esf2p could play a role in the dissociation of box C+D snoRNPs other than U3. Indeed, the distribution on glycerol gradients of U3 and other snoRNAs was dramatically affected upon Esf2p depletion, with the notable accumulation of these RNA species in fractions cosedimenting with 90S preribosomes (Fig. 5). The snoRNAs involved were from both box C+D and box H+ACA families, were involved in RNA modification and/or processing, and were not all found in stable association with Esf2p.
The observation that Esf2p is required for proper terminal knob deposition and SSU processome assembly indicates that the protein is required for initial preribosomal assembly. Although we cannot formally state at this stage whether the assembly defect is primary or not, this inhibition alone could easily account for the pre-rRNA processing defect reported as well as for the retention of U3 within 90S preribosomes (Fig. 5).
Alternatively, it could be that Esf2p is primarily required to recycle U3 (i.e., to dissociate U3 from the 35S pre-rRNA) and that, in the absence of U3 release, early pre-rRNA processing and SSU processome formation are inhibited. Esf2p may be assisted in its functions by some of its associated partners, that include several putative RNA helicases (Fig. 3D). Dissociation of the SSU processome from large nucleolar preribosomes is expected to involve a substantial remodeling of the overall structure that may underlie the effects observed for the other snoRNA species tested.
As far as U3 is concerned, a nearly reciprocal phenomenon has recently been described for Utp1p/Pwp2p: the depletion of Utp1p led to the exclusive detection of U3 in fractions corresponding to free snoRNPs, indicating that in this case, the Utp1 protein (and presumably associated partners) is involved in loading the SSU processome onto the 90S preribosomes (8). We note that Esf2p and Utp1p stably associate with each other (Fig. 4). It will be of particular interest to determine whether Esf2p and Utp1p have antagonistic roles and/or whether one is epistatic to the other. It will also be of interest to determine whether mutations of other components of the SSU processome can be discriminated functionally, for example, whether other members of the highly stable UTP B subcomplex, of which Utp1p appears to be a member (8, 25), have phenotypes similar to those of proteins observed after Utp1p depletion.
Acknowledgments
We thank Dawn Richards and Victoria Canadienne (Affinium Pharmaceuticals) for protein identification by MALDI mass spectrometry and Mouna Derfoufi (Institut de Biologie et de Médecine Moléculaires, Université Libre de Bruxelles) for expert technical support.
This work was supported by the National Science Foundation (grant MCB-9513589) (Beyer), the Canadian Institutes for Health Research and CFI (Hughes), Genome Canada (Greenblatt), Banque Nationale de Belgique, Defay, European Molecular Biology Organization, Fonds National de la Recherche Scientifique, International Brachet Stiftüng, Université Libre de Bruxelles, Van Buuren and Xenophilia (Lafontaine). S. Hiley is supported by a Canadian Institutes for Health Research postdoctoral fellowship; T. Hoang is supported by a Ph.D. fellowship from the Ministry of Education, Hanoï; and E. Vanrobays was the recipient of a European Molecular Biology Organization long-term fellowship (ALTF728-2002).
REFERENCES
- 1.Andersen, J. S., C. E. Lyon, A. H. Fox, A. K. Leung, Y. W. Lam, H. Steen, M. Mann, and A. I. Lamond. 2002. Directed proteomic analysis of the human nucleolus. Curr. Biol. 12:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32(Database issue):D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, K. A., and S. J. Baserga. 2004. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell 15:5038-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, K. A., J. E. Gallagher, B. M. Mitchell, S. Granneman, and S. J. Baserga. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 3:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billy, E., T. Wegierski, F. Nasr, and W. Filipowicz. 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 19:2115-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousquet-Antonelli, C., E. Vanrobays, J. P. Gelugne, M. Caizergues-Ferrer, and Y. Henry. 2000. Rrp8p is a yeast nucleolar protein functionally linked to Gar1p and involved in pre-rRNA cleavage at site A2. RNA 6:826-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colau, G., M. Thiry, V. Leduc, R. Bordonné, and D. L. J. Lafontaine. 2004. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology. Mol. Cell. Biol. 24:7976-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dosil, M., and X. R. Bustelo. 2004. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90S pre-ribosomal particle. J. Biol. Chem. 279:37385-37397. [DOI] [PubMed] [Google Scholar]
- 9.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 11.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 13.Galardi, S., A. Fatica, A. Bachi, A. Scaloni, C. Presutti, and I. Bozzoni. 2002. Purified box C/D snoRNPs are able to reproduce site-specific 2′-O-methylation of target RNA in vitro. Mol. Cell. Biol. 22:6663-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 16.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 17.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 18.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 19.Granneman, S., and S. J. Baserga. 2003. Probing the yeast proteome for RNA-processing factors. Genome Biol. 4:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granneman, S., J. E. Gallagher, J. Vogelzangs, W. Horstman, W. J. van Venrooij, S. J. Baserga, and G. J. Pruijn. 2003. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 31:1877-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 22.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 23.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 24.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 25.Hulo, N., C. J. Sigrist, V. Le Saux, P. S. Langendijk-Genevaux, L. Bordoli, A. Gattiker, E. De Castro, P. Bucher, and A. Bairoch. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32(Database issue):D134-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Kressler, D., P. Linder, and J. de La Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 30.Lafontaine, D. L. J. 2004. Eukaryotic ribosome synthesis, p. 107-143. In K. Nierhaus (ed.), Protein synthesis and ribosome structure. Wiley-InterScience, New York, N.Y.
- 31.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32(Database issue):D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 118:31-44. [DOI] [PubMed] [Google Scholar]
- 33.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oda, T., K. Kayukawa, H. Hagiwara, H. T. Yudate, Y. Masuho, Y. Murakami, T. A. Tamura, and M. A. Muramatsu. 2000. A novel TATA-binding protein-binding protein, ABT1, activates basal transcription and has a yeast homolog that is essential for growth. Mol. Cell. Biol. 20:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osheim, Y. N., S. L. French, K. M. Keck, E. A. Champion, K. Spasov, F. Dragon, S. J. Baserga, and A. L. Beyer. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16:943-954. [DOI] [PubMed] [Google Scholar]
- 36.Peng, W. T., N. J. Krogan, D. P. Richards, J. F. Greenblatt, and T. R. Hughes. 2004. ESF1 is required for 18S rRNA synthesis in Saccharomyces cerevisiae. Nucleic Acids Res. 32:1993-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 38.Raué, H. A. 2003. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine, p. 199-222. In M. J. O. Olson (ed.), The nucleolus. Kluwer Academic/Plenum, New York, N.Y.
- 39.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 40.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661-670. [DOI] [PubMed] [Google Scholar]
- 43.Trapman, J., J. Retel, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 44.Vanrobays, E., J.-P. Gélugne, M. Caizergues-Ferrer, and D. L. J. Lafontaine. 2004. Dim2p, a KH-domain protein required for small ribosomal subunit synthesis. RNA 10:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 46.Warner, J. R. 2001. Nascent ribosomes. Cell 107:133-136. [DOI] [PubMed] [Google Scholar]
- 47.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Luhrmann. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 48.Wegierski, T., E. Billy, F. Nasr, and W. Filipowicz. 2001. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA 7:1254-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xing, F., S. L. Hiley, T. R. Hughes, and E. M. Phizicky. 2004. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 279:17850-17860. [DOI] [PubMed] [Google Scholar]