Abstract
Reduction of the survival of motor neurons (SMN) protein levels causes the motor neuron degenerative disease spinal muscular atrophy, the severity of which correlates with the extent of reduction in SMN. SMN, together with Gemins 2 to 7, forms a complex that functions in the assembly of small nuclear ribonucleoprotein particles (snRNPs). Complete depletion of the SMN complex from cell extracts abolishes snRNP assembly, the formation of heptameric Sm cores on snRNAs. However, what effect, if any, reduction of SMN protein levels, as occurs in spinal muscular atrophy patients, has on the capacity of cells to produce snRNPs is not known. To address this, we developed a sensitive and quantitative assay for snRNP assembly, the formation of high-salt- and heparin-resistant stable Sm cores, that is strictly dependent on the SMN complex. We show that the extent of Sm core assembly is directly proportional to the amount of SMN protein in cell extracts. Consistent with this, pulse-labeling experiments demonstrate a significant reduction in the rate of snRNP biogenesis in low-SMN cells. Furthermore, extracts of cells from spinal muscular atrophy patients have a lower capacity for snRNP assembly that corresponds directly to the reduced amount of SMN. Thus, SMN determines the capacity for snRNP biogenesis, and our findings provide evidence for a measurable deficiency in a biochemical activity in cells from patients with spinal muscular atrophy.
The process of pre-mRNA splicing is carried out by a macromolecular complex, the spliceosome, the major components of which are the U1, U2, U5, and U4/U6 small nuclear ribonucleoprotein particles (snRNPs) (18, 34, 47). Each of the snRNPs (except for U6) is composed of one snRNA molecule, a set of seven common proteins, and several proteins that are specific to individual snRNAs (18, 27, 28, 47). SnRNP biogenesis begins with the transcription of the snRNAs in the nucleus followed by their nuclear export to the cytoplasm, where the major assembly process of the snRNPs takes place. The common proteins, called Sm proteins, B/B′, D1, D2, D3, E, F, and G, are arranged into a stable heptameric ring, the Sm core, on a uridine-rich sequence motif, the Sm site, of the snRNAs (1, 2, 19, 41). The assembly of Sm cores is required for the subsequent modification of the 7-methyl guanosine cap of snRNAs into a 2,2,7-trimethyl guanosine cap as well as for the stability and function of the snRNPs (30, 38). Properly assembled and modified snRNPs are then imported into the nucleus, where additional snRNP-specific proteins associate to form fully functional snRNPs (10, 11, 13, 30, 31, 47).
Earlier studies have shown that snRNP assembly readily occurs in vitro with purified total snRNP proteins (TPs) and snRNAs in an ATP-independent manner and without requirement for non-snRNP proteins (39, 40, 43). However, reconstitution of snRNPs in extracts from Xenopus laevis eggs and mammalian cells requires ATP (21, 32, 33, 37, 44), suggesting that snRNP assembly might be regulated by additional factors in vivo. Studies on a macromolecular complex containing the survival of motor neurons (SMN) protein indicated that the SMN complex is required for the ATP-dependent snRNP assembly (3, 9, 32, 33, 36, 37, 49). SMN is the protein product of the gene responsible for spinal muscular atrophy (SMA), a common and often fatal genetic disorder in which motor neurons in the spinal cord degenerate (6, 8, 15, 22). Based on the age of onset and the severity of the disease, SMA is clinically classified into three types: the severe type I, the moderate type II, and the mild type III. Studies on SMA patient-derived cell lines have shown that the severity of SMA clinical phenotypes is closely linked to the degree of reduction of SMN protein levels (7, 23).
Immunodepletion or antibody inhibition of the SMN complex in vitro demonstrated that the SMN complex is required for snRNP assembly (32, 33, 37). However, how much the SMN protein as well as individual Gemins contribute to snRNP assembly and what happens in SMA patients' cells, where the amount of SMN protein is reduced to various degrees, have not been determined. Current methods using gel mobility shift assay to monitor snRNP assembly are not suitable for quantitative analysis, due to the heterodisperse migration of large RNP complexes on native gels. To assess the relationship between the amount of SMN and the activity of Sm core assembly in cells and to facilitate further studies on the mechanism of SMN complex function, we developed a sensitive and quantitative assay for snRNP assembly.
The assay is based on the isolation of high-salt- and heparin-resistant Sm cores formed on biotin-labeled snRNAs with magnetic beads bearing anti-Sm antibodies. The amount of snRNPs assembled in the reaction is then determined by luminescence detection of the biotin molecules on the snRNAs. Importantly, we show that the extent of assembly is directly dependent on the amount of the SMN complex in cell extracts and that extracts from cells of SMA patients have a lower capacity for snRNP assembly, proportional to the reduction of SMN protein in these cells. In vivo pulse-labeling experiments demonstrate that the rate of biogenesis of the major snRNPs is strongly reduced in cells with low SMN. These findings provide evidence for a deficiency in a specific biochemical activity in SMA patients and present a powerful method for studying the activity of the SMN complex.
MATERIALS AND METHODS
In vitro transcription of RNAs.
Plasmids for in vitro transcription of snRNAs were described elsewhere (10, 13, 16, 30). For radiolabeling of RNAs, in vitro transcription was carried out in the presence of [32P]UTP as previously described (48). Biotin-labeled RNAs were produced according to the suggestions of the manufacturer (Ambion) with the modification that 5 mM biotin-UTP (Roche) and 2.5 mM UTP were present. All of the labeled RNAs were purified by electrophoresis on 7 M urea-6% polyacrylamide gels, precipitated with ethanol, and resuspended in nuclease-free water. The concentrations of the biotin-labeled RNAs were determined by absorbance at 260 nm.
Cell lines and cell culture maintenance.
The maintenance of the S5 cell line, which is a chicken DT40 cell line with targeted disruption of the SMN gene, has been described previously (45). Epstein-Barr virus-transformed lymphoblast cell lines derived from a 6-month-old SMA type I patient (GM10684) and an age- and gender-matched individual with a syndrome unrelated to SMA as a control (GM12497) were obtained from Coriell Cell Repositories and maintained in RPMI 1640 medium (Gibco BRL) containing 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Gibco BRL). Primary fibroblast cell lines from four SMA type I patients (GM00232, GM09677, GM03813, and GM03815), one heterozygous carrier (GM03814), and two apparently healthy controls (GM08333 and GM00498) were also obtained from Coriell Cell Repositories. They were maintained in minimal essential medium (Gibco BRL) containing 15% fetal bovine serum, 2 mM l-glutamine, and 1% penicillin-streptomycin.
Preparation of cytoplasmic extracts from cultured cells.
For each sample in the assays shown, the same number (≈4 × 107) of cells were harvested and washed twice with phosphate-buffered saline. For the SMA fibroblast cell lines, cells were used at an early and identical passage stage. Cytoplasmic extracts competent for snRNP assembly were prepared as described previously (37). The protein concentrations of the various extracts were determined using the Bio-Rad protein assay (Bio-Rad). All of the extracts were adjusted to the same final protein concentration (≈15 mg/ml for extracts prepared from HeLa cells, S5 cells, or SMA lymphoblastoid cells, and 3 mg/ml for extracts of SMA fibroblast cells) and were quickly frozen in liquid nitrogen and stored in aliquots at −80°C.
Assay for in vitro assembly of snRNPs.
For in vitro Sm core assembly on 32P-labeled snRNAs in cytoplasmic extracts, the reactions were carried out at 30°C for 1 h using standard assembly reaction conditions (37). Subsequently, half of the reaction mixtures was loaded onto native gels for electrophoretic mobility shift assays as described previously (37). The other half of the samples was immunoprecipitated with the anti-Sm monoclonal antibody Y12 (24), and the immunoprecipitated RNAs were isolated and analyzed by electrophoresis on 7 M urea-8% polyacrylamide gels.
For quantitative in vitro assembly assays on magnetic beads, cytoplasmic extracts were prepared and used for assembly on biotin-labeled snRNAs using the standard reconstitution conditions in 96-well plates. Following the reactions, Y12 antibodies immobilized onto the magnetizable Dynabeads protein A (Dynal Biotech ASA, Oslo, Norway) in 100 μl of RSB-500 buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 2.5 mM MgCl2) containing 2 mg/ml heparin, 0.1% NP-40, and 0.2 U/μl RNasin RNase inhibitor (Promega) were added to each well. Immunoprecipitations in the 96-well plates were carried out with gentle mixing at 750 rpm in a Thermomixer (Eppendorf, Germany) at 30°C for 1 h. The plates were subsequently transferred to a Kingfisher 96 magnetic particle processor (Thermo Labsystems, Vantaa, Finland) for automatic washing of the Dynabeads in each well with 200 μl of wash buffer (RSB-500, 0.1% NP-40) five times. After the last wash, the beads that were bound with Y12-immunoprecipitated snRNPs were then resuspended in 120 μl of wash buffer containing 0.08 μg/ml horseradish peroxidase-conjugated NeutrAvidin (Pierce). Following incubation at 30°C for 1 h with gentle mixing, the beads in each well were again washed five times by the Kingfisher 96 magnetic particle processor and finally resuspended in 150 μl of SuperSignal enzyme-linked immunosorbent assay (ELISA) Femto substrate working solution (Pierce). The plates were transferred to a Wallac Victor2 multilabel plate reader (Perkin-Elmer) for luminescence measurements at 495 nm. The resulting data were analyzed with Microsoft Excel.
Antibodies and quantitative immunoblotting.
The anti-SMN (62E7), anti-Gemin2 (2E17), anti-Gemin3 (12H12), anti-Gemin4 (17D10), anti-Gemin5 (10G11), anti-Y14 (1F12), and anti-Sm (Y12) monoclonal antibodies have been described previously (4, 5, 12, 20, 24-26). The anti-mouse immunoglobulin G secondary antibody labeled with IRDye800 (Rockland) was used at 1:5,000. Proteins from 20 μg of cytoplasmic extracts were separated on NuPAGE 4 to 12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes. Quantitative immunoblotting was performed as suggested by the manufacturer (Li-Cor). The membranes were scanned on an Odyssey infrared imaging system (Li-Cor), and the intensity of the protein bands was analyzed using the software provided by the manufacturer.
Pulse-label measurements of the rate of snRNP biogenesis in cells.
Three days after chicken S5 cells were cultured in medium containing 10 or 18 ng/ml tetracycline, equal numbers of cells (107) were pulse-labeled with 25 μCi/ml [3H]uridine (Amersham) for one hour. Total RNAs were isolated from 10% of the labeled cells using TRIzol reagent (Invitrogen). Total cell extracts from the remaining cells were subjected to immunoprecipitation by Y12. The immunoprecipitated RNAs were isolated by proteinase K treatment followed by phenol-chloroform extraction and ethanol precipitation. Both total RNAs and Y12-immunoprecipitated RNAs were analyzed by electrophoresis on 7 M urea-8% polyacrylamide gels. Gels were then treated with Amplify solution (Amersham) and dried for autoradiography.
Analysis of steady-state snRNP levels in cells.
To determine the overall amount of snRNPs at steady state in cells, Y12 immunoprecipitations were performed using cell extracts from 107 S5 cells that were grown in the presence of either 10 or 18 ng/ml tetracycline. Y12 immunoprecipitated RNAs were isolated and radioactively labeled at the 3′ end with [α-32P]pCp (Perkin-Elmer) and T4 RNA ligase (New England Biolabs). The labeled RNAs were resolved by electrophoresis on 7 M urea/8% polyacrylamide gels and analyzed by autoradiography.
RESULTS
Development of a quantitative assay for snRNP assembly.
Assembled Sm cores comprised of seven-membered rings of Sm proteins, unlike the complexes of individual Sm proteins or a subset of Sm proteins with RNA, are extremely sturdy and resist dissociation even in the presence of high salt, heparin, and urea (14, 16, 40). This property provides the basis for a gel mobility shift assay to assess Sm core formation. In these assays, 32P-labeled snRNAs are incubated with purified total snRNP proteins or a cell fraction containing Sm proteins, such as extracts from HeLa cells or Xenopus laevis eggs, and the stable Sm cores, resolved by electrophoresis from free RNAs, are visualized by their characteristic mobility shift after autoradiography of the native polyacrylamide gels (21, 32, 33, 37, 39, 44).
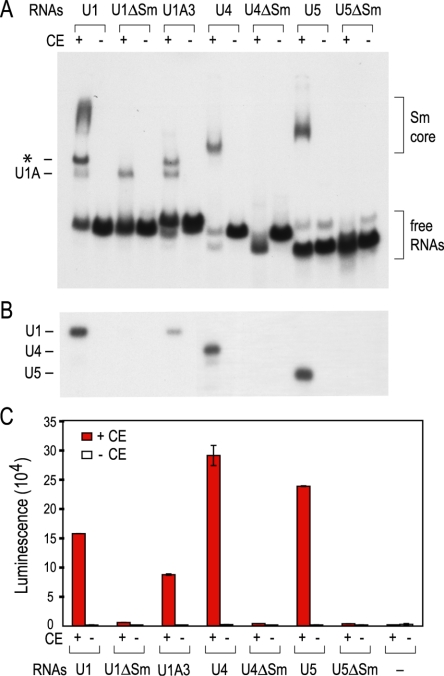
As has been shown previously, using this gel shift assay, Sm cores formed on Sm site-containing U1, U4, and U5 snRNAs when incubated in HeLa cytoplasmic extracts (Fig. 1A). Sm core assembly was strictly dependent on Sm proteins supplied by the extracts (Fig. 1A, +CE) and on the presence of an Sm site on the snRNAs (Fig. 1A, ΔSm). Sm cores assembled only inefficiently on U1A3 (Fig. 1A), a U1 snRNA mutant that has three nucleotide substitutions in the loop of stem-loop 1 and is impaired in binding to the SMN complex (48). However, due to the heterodisperse migration of large RNP complexes on native gels, the gel shift assay is not a sensitive and objective method to accurately quantitate Sm core assembly.
FIG. 1.
Analysis of in vitro-assembled snRNPs by the mobility gel shift assay and by the magnetic beads assay. (A) [32P]UTP-labeled U1, U1ΔSm, U1A3, U4, U4ΔSm, U5, or U5ΔSm snRNA was mixed with HeLa cytoplasmic extracts (CE) containing 25 μg total proteins (+ lanes) or with buffer only (− lanes) for in vitro assembly of Sm cores. The reaction mixtures were analyzed by electrophoresis on 6% native polyacrylamide gels. The brackets on the right indicate the positions of assembled Sm cores and the free RNAs. The complex that results from the binding of the U1-specific protein U1A to stem-loop 2 of the U1 snRNA is marked on the left. The band indicated by an asterisk is likely to be U1A/Sm core complexes. (B) The same reaction mixtures as used in panel A were immunoprecipitated by Y12 bound to protein A-Sepharose beads. RNAs were isolated from the bound fractions and analyzed by electrophoresis on 7 M urea-8% polyacrylamide gels. The RNAs that migrate at different positions on the gel are indicated on the left. (C) SnRNAs were produced by in vitro transcription in the presence of biotin-UTP instead of [32P]UTP, and similar assembly reactions as in A were carried out with these biotinylated RNAs or without any RNA using either HeLa cytoplasmic extracts (+ lanes) or buffer (− lanes). The amount of the Sm cores assembled on these RNAs was assessed by the magnetic beads assay as depicted in Fig. 2. The error bars, which for some data points are too small to be seen on the figure, represent standard deviations from three independent experiments.
To further confirm the identity of the assembled complexes and to assess snRNP assembly more accurately, the assembly reaction mixtures were subjected to immunoprecipitations with the anti-Sm antibody Y12 (24) under stringent conditions such that only snRNAs that have assembled high-salt- and heparin-resistant Sm cores could be immunoprecipitated. Following Y12 immunoprecipitation, 32P-labeled snRNAs on which Sm cores formed were eluted and analyzed by urea-polyacrylamide gel electrophoresis (14, 16, 37). Similar to the gel shift assay, Sm cores assembled on the wild-type snRNAs but not on the respective ΔSm mutants (Fig. 1B). A weak signal was detected for the U1A3 mutant, demonstrating more clearly than on the gel shift assay a low level of Sm core formation on this mutant (Fig. 1B).
The level of snRNP assembly can be estimated by autoradiography on a phosphorimager. While the 32P-labeled RNA bands obtained after immunoprecipitation with anti-Sm antibodies are more suitable to estimate assembled Sm cores, this approach, using radiolabeled RNAs, still has considerable shortcomings, such as the short half-life of the 32P-labeled RNAs and the laborious handling of the hazardous materials.
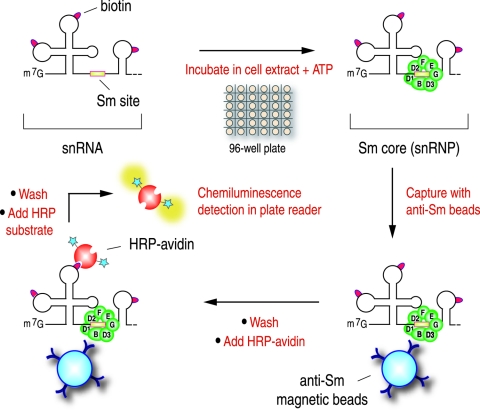
To facilitate the study of the snRNP assembly process, we developed an assay that allows quantitative measurement of Sm core formation (Fig. 2). Instead of labeling RNAs with [32P]UTP, snRNAs were prepared by in vitro transcription in the presence of biotin-UTP. Following in vitro assembly reactions with the biotin-labeled RNAs, immunoprecipitations of the Sm cores were carried out under stringent conditions, including high salt (500 mM NaCl) and heparin (2 mg/ml). The immunoprecipitations were carried out with Y12 antibodies immobilized on magnetic beads in a 96-well plate format, which allows automatic cycles of washing and mixing of the beads on a robotic manifold. Subsequently, horseradish peroxidase-conjugated avidin, which binds tightly to biotin, was used to recognize the biotinylated RNAs in the Y12 immunoprecipitated Sm cores. This step also serves to amplify the signals for the luminescence measurement of the horseradish peroxidase activity on an automatic plate reader.
FIG. 2.
Schematic depiction of the magnetic bead assay procedure for the detection of in vitro-assembled snRNPs.
The results obtained using this assay system (Fig. 1C) strongly resemble those of Fig. 1A and 1B. The signal indicating the amount of assembly on U1, U4, and U5 is more than 100-fold above background (Fig. 1C, -RNA, -CE). Readings on all ΔSm mutants were close to background, demonstrating the strict dependence of assembly on an Sm site. In this assay, the assembly of Sm cores on U1A3 is more significant than what was observed by the gel shift assay or Y12 immunoprecipitation with 32P-labeled snRNAs. Since U1A3 binds to the SMN complex with lower affinity than wild-type U1 (data not shown) and Sm cores assemble on U1A3 with slower kinetics in vivo in Xenopus oocytes (48), the difference might be due to the higher concentration of U1A3 RNA and longer reaction time used in this assay. As indicated by the error bars in Fig. 1C, the experimental variation in independent experiments is typically less than 5% of the signal. This assay for snRNP assembly is more sensitive, much less labor-intensive than previous methods, and amendable to high-throughput automation.
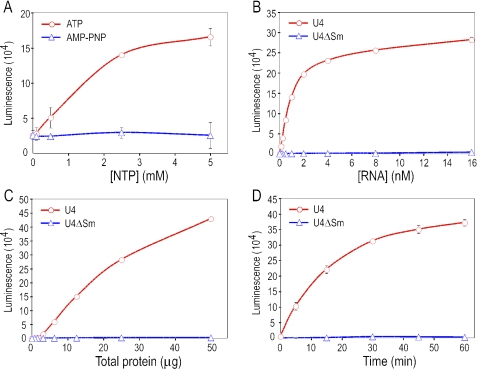
While the assembly of Sm cores from TPs is ATP independent, energy is required for this process to occur in cell extracts (21, 32, 33, 37, 44). As shown in Fig. 3A, Sm core assembly in extracts requires the addition of exogenous ATP and does not occur with adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP), a nonhydrolysable ATP analog, suggesting that ATP hydrolysis is required for assembly. To optimize the assay conditions, we examined the effect of several parameters on the Sm core assembly. Assembly on wild-type snRNA depends on RNA concentration, the amount of cytoplasmic extracts, and the reaction time (Fig. 3B to D). In contrast, Sm cores do not form on ΔSm mutant snRNAs even with excess RNAs, extracts, and reaction time (Fig. 3B to D). Profiles similar to those presented in Fig. 3 for U4 were obtained for U1 and U5 (data not shown). Since assembly activity corresponds linearly to the amount of extract (Fig. 3C) and plateaus at higher RNA concentration and longer reaction time (Fig. 3B and 3D), we set the parameters used for our standard assembly reaction at 5 nM biotinylated snRNA, cytoplasmic extracts containing 25 μg total proteins, and a reaction time of 60 min.
FIG. 3.
Specific parameters that affect snRNP assembly in vitro. In vitro assembly reactions were performed using standard conditions except one of the following parameters. Sm core assembly on U4 (A, B, C, and D) and U4ΔSm (B, C, and D) was examined by the magnetic beads assay and graphed against ATP or AMP-PNP concentration (A), the snRNA concentration (B), the amount of HeLa cytoplasmic extract (C), or the assembly reaction time (D). Error bars, which are too small to be visible for some of the data points, represent standard deviations from two independent experiments.
SMN protein determines the capacity for snRNP assembly.
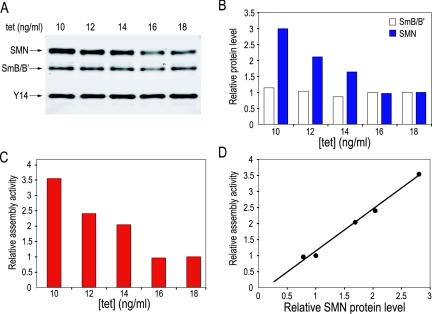
It has been previously shown that the assembly of Sm cores is abolished when SMN complexes are depleted in vitro from cell extracts (32, 37). Since the assembly activity correlates with the amount of cytoplasmic extract (Fig. 3C), we asked whether it is the amount of SMN in the extract that determines snRNP assembly capacity. To address this, SMN expression was reduced in HeLa cells by RNA interference (RNAi) and this resulted in a drastic reduction in the assembly activity of the extracts prepared from these cells (Feng and Dreyfuss, personal communication). However, RNAi cannot be regulated to obtain a desired level of reduction in the amount of the protein. We therefore used the S5 cell line, derived from chicken DT40 cells, in which the endogenous SMN gene is disrupted by homologous recombination and SMN protein is exogenously expressed from a tetracycline-repressible promoter (45). This allows SMN expression to be tightly controlled to various levels by adjusting the concentration of tetracycline in the culture media (45). Since the growth of S5 cells is arrested after 72 h in the presence of 18 ng/ml tetracycline and higher concentrations of tetracycline lead to cell death (45), we analyzed S5 cells which were normally maintained in 10 ng/ml tetracycline and then were split into culture media containing 10, 12, 14, 16, or 18 ng/ml tetracycline for 3 days. Equal numbers of cells (≈4 × 107) were harvested for each of the tetracycline concentrations and used to prepare cytoplasmic extracts.
The expression level of Y14, a component of the exon-junction complex (20) and unrelated to the snRNP assembly process, remained largely unchanged at various tetracycline concentrations (Fig. 4A) and was therefore used as an internal control to determine the relative protein levels of SMN and Sm proteins B/B′. As shown in Fig. 4B, the level of the SMN protein decreased with increasing tetracycline concentration. Cells cultured at 18 ng/ml tetracycline contained about 30% SMN compared to those at 10 ng/ml tetracycline. Unlike the reduction of SMN, SmB/B′ protein levels were not significantly changed.
FIG. 4.
Reduced snRNP assembly correlates with decreased SMN protein levels in extracts from chicken S5 cells. (A) Cytoplasmic extracts were prepared from S5 cells cultured at 10, 12, 14, 16, or 18 ng/ml tetracycline. SMN, SmB/B′, and Y14 proteins in each of these extracts containing 20 μg total proteins were detected by quantitative Li-Cor Western blotting analysis. (B) The intensity of each protein band in panel A was analyzed using the Odyssey infrared imaging system. SMN and SmB/B′ proteins levels in extracts of various tetracycline concentrations were normalized using the Y14 signals as an internal control. The relative SMN and SmB/B′ protein levels at 18 ng/ml tetracycline were set to 1. The relative protein levels at other tetracycline concentrations were calculated as ratios to those at 18 ng/ml tetracycline. (C) SnRNP assembly capacities of the same extracts as in A on U4 snRNA were examined by the magnetic bead assay. The relative assembly activity of extract at 18 ng/ml tetracycline was set to 1. The relative assembly activities of the other extracts were calculated as ratios to that at 18 ng/ml tetracycline. (D) The relative assembly activity at each tetracycline concentration as in panel C was plotted on the y axis against the corresponding relative SMN protein level as in panel B on the x axis. The data points fitted best to a linear graph (R2 = 0.9911).
We next employed the magnetic beads assay to quantitate the assembly activity of these cell extracts. The capacity for assembly of Sm cores decreased with the reduction in SMN protein (Fig. 4C). When the relative assembly activity at each tetracycline concentration was plotted against the corresponding relative SMN protein level, the data points fitted best to a linear graph with a significant R2 value (R2 = 0.9911), indicating a direct correlation between the SMN protein level and assembly activity (Fig. 4D). These results indicate that the SMN protein determines the capacity of Sm core assembly in cell extracts even if they contain similar, nonlimiting amounts of Sm proteins.
Cells with low SMN accumulate snRNPs more slowly.
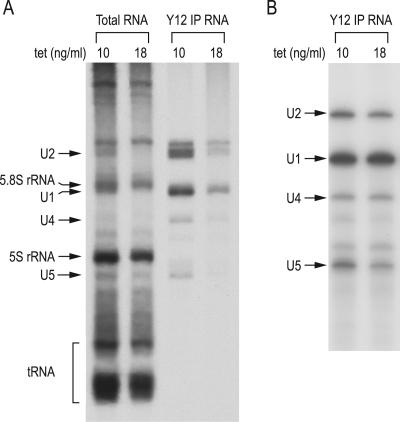
To determine if the reduced snRNP assembly capacity observed in extracts of cells with low SMN is reflected in vivo, we measured the rate of snRNP biogenesis in S5 cells cultured in the presence of either 10 ng/ml tetracycline (normal SMN) or 18 ng/ml tetracycline (low SMN) by pulse-labeling. Pulse-labeling of these cells showed that the rate of accumulation of the major snRNAs and snRNPs, determined by one hour of [3H]uridine incorporation into the major Sm core-containing snRNAs, particularly U1 and U2, was drastically reduced (Fig. 5A, Y12 IP RNA). Albeit to a lesser extent, the total amount of labeled U1 and U2 snRNAs, as well as other RNAs (e.g., 5S and tRNA) produced in the same time interval was also reduced (Fig. 5A, Total RNA).
FIG. 5.
Cells with low SMN accumulate snRNPs more slowly. (A) To determine the rate of snRNP biogenesis in vivo, S5 cells cultured at 10 or 18 ng/ml tetracycline were pulse-labeled with 25 μCi/ml [3H]uridine for one hour. Total RNAs from 10% of the labeled cells and snRNAs isolated by immunoprecipitations with Y12 from the remaining cells were analyzed on 7 M urea-8% polyacrylamide gel. Known RNAs are indicated on the left. (B) To measure the level of snRNPs in vivo at steady state, Y12 immunoprecipitations were performed on S5 cells cultured at 10 or 18 ng/ml tetracycline. The immunoprecipitated RNAs were isolated, radioactively labeled at the 3′ end, and analyzed on 7 M urea-8% polyacrylamide gel. Known RNAs are indicated on the left.
It is possible that low-SMN cells also have reduced transcription rates and/or that RNAs, especially snRNAs, that are not assembled are rapidly degraded (29). The possibility that the rate of transcription is reduced is consistent with previous observations suggesting a role for the SMN complex in the formation of transcription factories (35). We conclude that SMN is a critical factor in determining the rate of production of snRNPs in cells. We note that the overall steady-state amount of the major snRNAs and snRNPs, determined by 3′-end labeling of the Y12 immunoprecipitated RNAs (Fig. 5B), was not significantly different in normal- and low-SMN cells. However, the low-SMN cells have a much slower growth rate than the normal-SMN cells, possibly to allow these cells to accumulate the necessary amount of RNPs, which now occurs at a slower rate.
Deficiency in snRNP assembly in cell extracts of SMA patients.
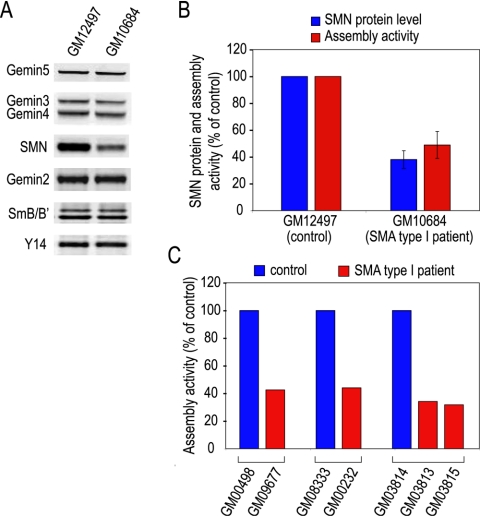
In light of the findings described above that SMN protein levels parallel the capacity of snRNP assembly in HeLa cells and in chicken S5 cells, we asked whether cells from SMA patients were also deficient in snRNP assembly activity. We first examined a lymphoblastoid cell line derived from a type I SMA patient (GM10684), using a similarly established cell line from an age- and gender-matched individual as a control (GM12497). Immunoblotting showed that SMN in the patient cells was considerably reduced, whereas the levels of Gemins 2 to 5 and SmB/B′ were similar to the control (Fig. 6A). Compared to the control, the snRNP assembly activity of the patient cell extract was 48% ± 10% in three independent experiments. This assembly capacity correlates well with the SMN level, which was 38% ± 6.5% of the control (Fig. 6B), demonstrating a biochemical deficiency in the cells of an SMA patient.
FIG. 6.
Cells of SMA patients are deficient in their capacity for snRNP assembly. (A) Cytoplasmic extracts were prepared from two lymphoblast cell lines. One was established from an SMA type I patient (GM10684) and the other from an age- and gender-matched individual (GM12497) as a control. SMN, Gemins 2 to 5, and SmB/B′ proteins were detected by quantitative Li-Cor Western blotting analysis, using Y14 as a loading control. (B) The intensity of each of the protein bands in panel A was analyzed using the Odyssey infrared imaging system. The SMN protein level in each of the cell extract was normalized using Y14 signal as an internal control. The relative level of SMN protein in the cell extract of an SMA patient (GM10684) was calculated as the percentage of that of the control (GM12497). Similarly, the relative assembly activity in cell extracts of the patient (GM10684) was calculated as a percentage of the control (GM12497). (C) Cytoplasmic extracts from SMA patient fibroblast cell lines were assayed by the magnetic bead assay. SnRNP assembly activities in cell extracts of the controls (GM00498, GM08333, and GM03814) were set to be 100%. Assembly activities in cell extracts of the SMA type I patients (GM09677, GM00232, GM03813, and GM03815) were expressed as percentages of their respective controls.
We further studied several available sets of primary fibroblast cell lines derived from type I SMA patients or heterozygous carriers, as well as unaffected individuals (Table 1). The full-length SMN transcript levels and SMN protein levels in patient cells (GM09677, GM00232, and GM03813) were estimated to be about 40 to 50% of those of the respective controls (GM00498, GM08333, and GM03814) (42). However, we observed that the overall SMN protein levels in these fibroblast cells are very low and passage dependent (data not shown). Consequently, the assembly activities in these cells were considerably lower than those in the lymphoblastoid cells.
TABLE 1.
Fibroblast cell lines derived from SMA patients and control individualsa
| Parameter | GM00498 | GM09677 | GM08333 | GM00232 | GM03814 | GM03813 | GM03815 |
|---|---|---|---|---|---|---|---|
| Phenotype | Unaffected | SMA | Unaffected | SMA | Unaffected | SMA | SMA |
| Age (yr) | 3 | 2 | 5 | 7 | N/A | 3 | N/A |
| Gender | Male | Male | Male | Male | Female | Male | Male |
| Relationship | N/A | N/A | N/A | N/A | Mother | Son | Son |
SMA, spinal muscular atrophy; N/A, not available.
The assembly assay revealed that extracts from the patient fibroblast cells had approximately 40% of the assembly activity of the corresponding controls (Fig. 6C), consistent with the degree of reduction of SMN protein in these patient cells. Unexpectedly, GM03815 cells, described in the Coriell Cell Repository as having been obtained from the heterozygous carrier father of the donor of the GM03813 cells, also showed a similar level of deficiency in snRNP assembly (Fig. 6C). Consistent with this, genetic linkage analysis indicated that GM03815 is, in fact, a male sibling of GM03813 (R. Wilson, personal communication). Indeed, another affected son was listed in the family tree. These observations demonstrate that as a result of reduced levels of SMN protein expression, cells of SMA patients are deficient in snRNP assembly in vitro and suggest that the snRNP assembly assay can be used as a potential diagnostic tool for SMA.
DISCUSSION
Previous experiments have demonstrated that the SMN complex is required for snRNP assembly. Specifically, complete or nearly complete removal or inhibition of the SMN complex results in the inhibition of Sm core assembly in vitro (32, 37). Here, using an in vitro assay we developed for the quantitative measurement of the Sm core assembly process in cell extracts, we show that there is a linear correlation between the amount of SMN present in the cell extract and the amount of Sm cores that can be formed on specific RNA substrates, suggesting that the amount of SMN determines the capacity for Sm core assembly. SMA results from a reduction in the amount of the full-length SMN protein (6, 8, 15, 22). Studies on a collection of SMA patient cell lines revealed that SMN expression is more reduced in the severe form (type I) than the mild form (type III) of the disease, demonstrating a direct correlation between the degree of reduction of SMN protein levels in SMA patients and the severity of their clinical phenotypes (7, 23). However, an understanding of the molecular consequences of the reduced levels of SMN in patients' cells has been lacking.
Our finding that snRNP assembly is impaired in cells of SMA patients provides the first direct evidence for a biochemical deficiency, namely, reduced capacity to assemble Sm cores, in SMA patients. Due to a limited availability of SMA patient cells, our studies focused on the severe type I SMA patients. However, the correlation we find is strong, exhibiting direct proportionality between the amount of the SMN protein and assembly capacity in vitro. Consistent with the reduced activity observed in extracts of cells with low SMN, the rate of production of snRNPs in these cells is profoundly reduced.
Although the overall amount of the major snRNPs in the same cells is not reduced, the strong deficiency in their rate of accumulation could be detrimental to cells in several ways. For dividing cells, biosynthetic capacity, such as the overall translation rate, has been linked to the control of cell growth and division (17, 46). Similarly, the slower rate of snRNP accumulation could cause a delay in cell cycle progression. This is indeed the case for S5 cells where the growth rate slows down proportionally to the reduction in SMN (45). In nondividing cells, such as motor neurons, an unmet demand for timely snRNP production at a particular point in the growth and development of the cell could have severe consequences for the cell. It could lead to a deficit in functions that depend on an adequate amount of the major snRNPs (e.g., a general decrease in pre-mRNA splicing or an altered processing pattern of pre-mRNAs that are required by that cell), or to a deficit in a specific snRNP (e.g., an RNA of a lower abundance or one that has a lower affinity for the SMN complex). It is also possible that reduced amounts of the SMN complex result in some loss of the regulation of Sm core assembly, leading to loss of fidelity of Sm core assembly, such that Sm cores assemble on RNAs that are not supposed to receive them. This could be harmful to cells, as it may interfere with the normal functions of these RNAs or cause them to aggregate. We note that the experiments we carried out here examined only the formation of the major snRNPs, and it is therefore possible that the formation of minor snRNPs, motor neuron-specific snRNPs, or other RNAs that require the SMN complex for RNP assembly is strongly affected by the reduction in SMN.
The method we describe here is sensitive, quantitative, easy to set up, and readily suitable for laboratory automation. Unlike the methods commonly used so far, such as RNP gel shift and anti-Sm immunoprecipitation followed by gel electrophoresis, this assay does not require radioactive labeling of the RNAs, nor does it require gel electrophoresis. The biotinylated RNA substrates used for the assay can be prepared ahead of time and stored for many months, allowing off-the-shelf and long-term use. The assay should facilitate detailed studies of the mechanisms of action of the SMN complex and on the process of Sm core assembly. Combined with other approaches, such as RNA interference to systematically reduce protein expression, it should now be possible to determine the roles of the individual Gemins and other components of interest in the assembly reaction. The assay can also be used on extracts from patient cells. The sensitivity of the assay and its colinearity with the amount of SMN make it suitable as an additional measure for the characterization of SMA and as a means to identify potential modifiers, both genetic and pharmacological, of the disease phenotype.
Acknowledgments
We are grateful to Joan A. Steitz for providing the Y12 monoclonal antibody. We thank Stephen A. Liebhaber, Tracey J. Golembe, and the members of our laboratory for stimulating discussions and helpful comments on the manuscript. We are also grateful to Gina Daly for secretarial assistance.
This work was supported by the Association Française Contre les Myopathies (AFM) and by a grant from the National Institutes of Health. G.D. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Achsel, T., H. Stark, and R. Luhrmann. 2001. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. USA 98:3685-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branlant, C., A. Krol, J. P. Ebel, E. Lazar, B. Haendler, and M. Jacob. 1982. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1:1259-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buhler, D., V. Raker, R. Luhrmann, and U. Fischer. 1999. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 8:2351-2357. [DOI] [PubMed] [Google Scholar]
- 4.Charroux, B., L. Pellizzoni, R. A. Perkinson, A. Shevchenko, M. Mann, and G. Dreyfuss. 1999. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147:1181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charroux, B., L. Pellizzoni, R. A. Perkinson, J. Yong, A. Shevchenko, M. Mann, and G. Dreyfuss. 2000. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 148:1177-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cifuentes-Diaz, C., T. Frugier, and J. Melki. 2002. Spinal muscular atrophy. Semin. Pediatr. Neurol. 9:145-150. [DOI] [PubMed] [Google Scholar]
- 7.Coovert, D. D., T. T. Le, P. E. McAndrew, J. Strasswimmer, T. O. Crawford, J. R. Mendell, S. E. Coulson, E. J. Androphy, T. W. Prior, and A. H. Burghes. 1997. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 6:1205-1214. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, T. O., and C. A. Pardo. 1996. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 3:97-110. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, U., Q. Liu, and G. Dreyfuss. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90:1023-1029. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, U., and R. Luhrmann. 1990. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science 249:786-790. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, U., V. Sumpter, M. Sekine, T. Satoh, and R. Luhrmann. 1993. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 12:573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubitz, A. K., Z. Mourelatos, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277:5631-5636. [DOI] [PubMed] [Google Scholar]
- 13.Hamm, J., E. Darzynkiewicz, S. M. Tahara, and I. W. Mattaj. 1990. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell 62:569-577. [DOI] [PubMed] [Google Scholar]
- 14.Hamm, J., M. Kazmaier, and I. W. Mattaj. 1987. In vitro assembly of U1 snRNPs. EMBO J. 6:3479-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iannaccone, S. T., S. A. Smith, and L. R. Simard. 2004. Spinal muscular atrophy. Curr. Neurol. Neurosci. Rep. 4:74-80. [DOI] [PubMed] [Google Scholar]
- 16.Jarmolowski, A., and I. W. Mattaj. 1993. The determinants for Sm protein binding to Xenopus U1 and U5 snRNAs are complex and non-identical. EMBO J. 12:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorgensen, P., and M. Tyers. 2004. How cells coordinate growth and division. Curr. Biol. 14:R1014-1027. [DOI] [PubMed] [Google Scholar]
- 18.Kambach, C., S. Walke, and K. Nagai. 1999. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9:222-230. [DOI] [PubMed] [Google Scholar]
- 19.Kambach, C., S. Walke, R. Young, J. M. Avis, E. de la Fortelle, V. A. Raker, R. Luhrmann, J. Li, and K. Nagai. 1999. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell 96:375-387. [DOI] [PubMed] [Google Scholar]
- 20.Kim, V. N., J. Yong, N. Kataoka, L. Abel, M. D. Diem, and G. Dreyfuss. 2001. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20:2062-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinschmidt, A. M., J. R. Patton, and T. Pederson. 1989. U2 small nuclear RNP assembly in vitro. Nucleic Acids Res. 17:4817-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefebvre, S., L. Burglen, S. Reboullet, O. Clermont, P. Burlet, L. Viollet, B. Benichou, C. Cruaud, P. Millasseau, M. Zeviani, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155-165. [DOI] [PubMed] [Google Scholar]
- 23.Lefebvre, S., P. Burlet, Q. Liu, S. Bertrandy, O. Clermont, A. Munnich, G. Dreyfuss, and J. Melki. 1997. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16:265-269. [DOI] [PubMed] [Google Scholar]
- 24.Lerner, E. A., M. R. Lerner, C. A. Janeway, Jr., and J. A. Steitz. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 78:2737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Q., and G. Dreyfuss. 1996. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15:3555-3565. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Q., U. Fischer, F. Wang, and G. Dreyfuss. 1997. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90:1013-1021. [DOI] [PubMed] [Google Scholar]
- 27.Luhrmann, R. 1990. Functions of U-snRNPs. Mol. Biol. Rep. 14:183-192. [DOI] [PubMed] [Google Scholar]
- 28.Luhrmann, R., B. Kastner, and M. Bach. 1990. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta 1087:265-292. [DOI] [PubMed] [Google Scholar]
- 29.Mangin, M., M. Ares, Jr., and A. M. Weiner. 1985. U1 small nuclear RNA genes are subject to dosage compensation in mouse cells. Science 229:272-275. [DOI] [PubMed] [Google Scholar]
- 30.Mattaj, I. W. 1986. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46:905-911. [DOI] [PubMed] [Google Scholar]
- 31.Mattaj, I. W., W. Boelens, E. Izaurralde, A. Jarmolowski, and C. Kambach. 1993. Nucleocytoplasmic transport and snRNP assembly. Mol. Biol. Rep. 18:79-83. [DOI] [PubMed] [Google Scholar]
- 32.Meister, G., D. Buhler, R. Pillai, F. Lottspeich, and U. Fischer. 2001. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3:945-949. [DOI] [PubMed] [Google Scholar]
- 33.Meister, G., and U. Fischer. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21:5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? BioEssays 25:1147-1149. [DOI] [PubMed] [Google Scholar]
- 35.Pellizzoni, L., B. Charroux, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 152:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellizzoni, L., N. Kataoka, B. Charroux, and G. Dreyfuss. 1998. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95:615-624. [DOI] [PubMed] [Google Scholar]
- 37.Pellizzoni, L., J. Yong, and G. Dreyfuss. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298:1775-1779. [DOI] [PubMed] [Google Scholar]
- 38.Plessel, G., U. Fischer, and R. Luhrmann. 1994. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol. 14:4160-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raker, V. A., K. Hartmuth, B. Kastner, and R. Luhrmann. 1999. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 19:6554-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raker, V. A., G. Plessel, and R. Luhrmann. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15:2256-2269. [PMC free article] [PubMed] [Google Scholar]
- 41.Stark, H., P. Dube, R. Luhrmann, and B. Kastner. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409:539-542. [DOI] [PubMed] [Google Scholar]
- 42.Sumner, C. J., T. N. Huynh, J. A. Markowitz, J. S. Perhac, B. Hill, D. D. Coovert, K. Schussler, X. Chen, J. Jarecki, A. H. Burghes, J. P. Taylor, and K. H. Fischbeck. 2003. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann. Neurol. 54:647-654. [DOI] [PubMed] [Google Scholar]
- 43.Sumpter, V., A. Kahrs, U. Fischer, U. Kornstadt, and R. Luhrmann. 1992. In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol. Biol. Rep. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 44.Temsamani, J., M. Rhoadhouse, and T. Pederson. 1991. The U2 small nuclear ribonucleoprotein particle associates with nuclear factors in a pre-mRNA independent reaction. J. Biol. Chem. 266:20356-20362. [PubMed] [Google Scholar]
- 45.Wang, J., and G. Dreyfuss. 2001. A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem. 276:9599-9605. [DOI] [PubMed] [Google Scholar]
- 46.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 47.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 48.Yong, J., L. Pellizzoni, and G. Dreyfuss. 2002. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 21:1188-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yong, J., L. Wan, and G. Dreyfuss. 2004. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 14:226-232. [DOI] [PubMed] [Google Scholar]