ABSTRACT
Grasslands converted to agricultural land use can be reestablished by sowing seeds of native species and temporal dynamics of diversity under altered climate can inform community assembly in the context of global change. We quantified three aspects of diversity (species richness, phylogenetic diversity, and functional diversity) in restored prairie plots sown with different ecotypes of two dominant grass species and manipulated rainfall to understand the relative importance of abiotic filtering and population source of dominant species on community assembly. We also evaluated the contributions of intra‐ and interspecific variations in functional traits across plots sown with different ecotypes of dominant species. Since the fourth year of community establishment, species richness decreased over time as dominant species gradually established. Phylogenetic and functional diversity was unaffected by the ecotypic sources of dominant species during restoration. Experimental drought did not affect species richness, phylogenetic, or functional diversity. Community structure in the grasslands was mainly shaped by intraspecific, within‐population variation in the dominant species rather than by differences in traits among species. Our results showed that intraspecific biotic interactions contribute more than environmental filtering to community assembly in a tallgrass‐dominated prairie ecosystem.
Keywords: biological filter, community assembly, grassland, intraspecific trait variation, long term, multitrait space
In restored prairie plots with different ecotypes of dominant grass species and manipulated rainfall, we examined the effects of abiotic filtering and population source on community assembly, finding that species richness declined over time as dominant species established, while phylogenetic and functional diversity remained unaffected. Our results indicated that intraspecific, within‐population variation in dominant species primarily shapes community structure, highlighting the greater role of biotic interactions over environmental filtering in prairie ecosystems.
1. Introduction
Understanding factors influencing temporal dynamics of biodiversity during restoration is one of the key foci of ecological research (Pavoine and Bonsall 2011; Gibson et al. 2012; Baer, Gibson, and Johnson 2019). Traditional diversity indices measure community members as evolutionarily independent and ecologically equivalent but lack adequate details about how species are related and assembled in specific patterns (Arnan, Cerda, and Retana 2015). Therefore, additional diversity methods have been explored to provide important information about evolutionary history and trait patterns of communities: phylogenetic diversity measures the assembled evolution and history of species in a community, while functional diversity quantifies the states of morphological, physiological, and phenological traits affecting species' fitness (Webb et al. 2002; Petchey and Gaston 2006). Past efforts to substitute one diversity pattern with another have brought about criticism of the proxy diversity measures. For instance, using richness instead of trait‐based metrics oversimplifies diversity, ignores ecological redundancy, and misguides conservation efforts (Chave, Chust, and Thébaud 2007; Losos 2008). Kraft et al. (2007) found that local phylogenetic overdispersion reflects trait overdispersion only if traits are highly conserved. Kluge and Kessler (2011) observed no phylogenetic diversity pattern along elevation, despite varying in functional diversity. Spasojevic and Suding (2012) found no correlation between phylogenetic and functional diversity along resource–stress gradients, and E‐Vojtkó et al. (2023) noted that phylogenetic diversity rarely represents functional diversity in temperate vegetation.
Grassland biodiversity patterns vary across different scales and metrics such as species richness, evolutionary history, and functional traits of plant species (Khalil et al. 2018). Recent studies of grassland assembly have changed the emphasis from the straightforward measurement of species diversity to more process‐centered indicators, including assessing evolutionary‐ and trait‐based assembly drivers and determinants (Webb et al. 2002; Hardy and Senterre 2007; Pavoine, Baguette, and Bonsall 2010; Khalil et al. 2018; Jones, Barber, and Gibson 2019). Mechanistic studies on assembly drivers in tallgrass prairie, focusing on environmental factors like rainfall (Johnson et al. 2015; Knapp et al. 2024; Mount et al. 2024) and biotic drivers such as locally adapted seed sources of Andropogon gerardi (Galliart et al. 2020; Ren et al. 2023) and Sorghastrum nutans (Wilson et al. 2016; Vogel et al. 2018), can guide management decisions to avoid undesirable restoration outcomes (Baer, Gibson, and Johnson 2019; Jones, Barber, and Gibson 2019).
Knowledge of multidimensional diversity patterns of restored grasslands can serve as a critical tool in steering community assembly over time, thereby mitigating loss of biodiversity. In long‐term restoration efforts of North American grasslands, species losses have been observed (McLachlan and Knispel 2005; Twidwell et al. 2012; Young et al. 2015; McKone, Williams, and Beck 2021). However, functional and phylogenetic relationships in grasslands might not be necessarily connected to species richness (Belinchon, Hemrova, and Munzbergova 2019). Grassland functional and phylogenetic diversity could be expected to persist despite a decrease in species richness (Belinchon, Hemrova, and Munzbergova 2019). Historical climate changes could have driven greater evolutionary similarity within grassland communities (Li, Miller, and Harrison 2019; Harrison, Spasojevic, and Li 2020; Luong, Holl, and Loik 2021). Midolo, Kuss, and Wellstein (2021) further showed that increasing drought can reinforce trait similarities, such as seed mass and specific leaf area, linked to water availability in grasslands.
Recent trait‐based community studies on grassland restoration have highlighted the significant role of intraspecific trait variation (ITV). This variability is crucial for fostering species richness (Crawford et al. 2019) and maintaining diversity of functional traits (He et al. 2021). It also plays a key role in shaping competitive interactions (Fajardo and Siefert 2018; Carmona et al. 2019) and preserving genetic diversity (Zeldin et al. 2020). Additionally, ITV contributes to increasing adaptability (Lanuza et al. 2020) and elevating ecosystem stability (Lambert, Baer, and Gibson 2011). ITV can be influenced by phenotypic plasticity, environmental contexts, and evolutionary processes (Messier, McGill, and Lechowicz 2010; Violle et al. 2012). Plant species often show a high ITV in functional traits due to plasticity and heritable genetic variation (Violle et al. 2012; Siefert et al. 2015). ITV is considered a component of “internal filtering” to influence community assembly through biotic interactions, for example, competition or commensalism (Crawford et al. 2019). Yet, the exact mechanism by which ITV influences community assembly remains a subject of debate. On one hand, coexistence theory (Chesson 2000) states that ITV‐induced niche overlap exacerbates the dominance of the better competitor. In contrast, “individual variation” theories (Violle et al. 2012) declare that community assembly arising from ITV are challenging to model accurately while vital to the maintenance of diversity (Clark 2010).
While ITV is a major “internal filter,” climate stressors such as variation in rainfall are crucial “external filters” affecting grassland assembly (Funk 2021). Hallett et al. (2019) found that greater rainfall variability enhanced species coexistence in Californian grasslands. Manning and Baer (2018) noted that interannual rainfall variations influenced community assembly and ecosystem functioning in restored tallgrass prairie. Atkinson et al. (2023) highlighted that variation in rainfall during establishment significantly impacted trait diversity and biodiversity success in restored grasslands.
To better understand the relative importance of ITV as an internal filter and manipulated rainfall as an external filter on community assembly in restored grassland, we asked the following questions: (1) Does a manipulated rainfall × ITV interaction influence grassland diversity over time? (2) Does intraspecific trait variation influence grassland community assembly similarly to interspecific trait variation? We use a long‐term field experiment that contained plots established with different ecotypes of two dominant grass species and the same eight nondominant species with and without rainfall reduction treatments to test two hypotheses. First, we hypothesized that experimental drought would act as an environmental filter and decrease species richness while increasing evolutionary similarity and trait similarity compared to an ambient treatment. Previous studies showed that severe experimental drought can significantly decrease species diversity and exacerbate shifts in grassland community structure due to the local extinction of subordinate species (Tilman and Haddi, 1992; Smith et al. 2020; Knapp et al. 2024). Second, we hypothesized that intraspecific trait variation (ITV) serves as a more influential biotic filter than interspecific trait variations in shaping a restored grassland. For example, studies have indicated that ITV could have a predominant influence on grassland assembly, such as in mesic meadows (Volf et al. 2016), semiarid grasslands (Zhang et al. 2019), and urban–rural grassland gradients (Cochard et al. 2019).
2. Materials and Methods
2.1. Study Site and Seed Sources
A common garden consisting of plots sown with different ecotypes of dominant grass species was established in the spring of 2009 in Carbondale, Illinois (37°41′47.0″ N, 89°14′19.2″ W). This site receives an average annual rainfall of 1198 mm and the average mean temperature is 13.5°C (Galliart et al. 2020; NCEI, 2019). The site was under agricultural cultivation prior to common garden establishment and characterized as silt loam soils (Mendola et al. 2015).
Big bluestem (A. gerardi) and Indiangrass (S. nutans) were chosen as the dominant species for our tallgrass prairie restoration experiment as these C4 grasses dominate large areas of native tallgrass prairie (Risser et al. 1981). In the fall of 2008, seeds of A. gerardi and S. nutans were collected from three regions along a rainfall gradient from southern Illinois (WET; mean annual rainfall [MAP] = 1097 mm) to eastern (MESIC; mean annual rainfall [MAP] = 849 mm) and central (DRY; mean annual rainfall [MAP] = 654 mm) Kansas (Johnson et al. 2015; Wilson et al. 2016). Genetic, phenotypic, and chemical variations confirmed the identity of A. gerardi ecotypes in the Great Plains (Gibson et al. 2013; Caudle et al. 2014; Gray et al. 2014; Johnson et al. 2015; Galliart et al. 2019, 2020). The mechanism behind the regional differences among S. nutans seed sources remained unknown, though ecotypes of S. nutans were anticipated to match the pattern of A. gerardi in the Great Plains (McMillan, 1959; Gustafson et al. 2014; Wilson et al. 2016).
Along with the two dominant species, seeds of eight subordinate species were also sown into the common garden plots (Table 1). The nondominant species sown included Canada wild rye (Elymus canadensis), butterfly milkweed (Asclepias tuberosa), partridge pea (Chamaecrista fasciculata), purple prairie clover (Dalea purpurea), wild bergamot (Monarda fistulosa), stiff goldenrod (Solidago rigida), foxglove beardtongue (Penstemon digitalis), and wild petunia (Ruellia humilis). The subordinate species were included to foster competitive dynamics akin to those found in prairie restorations (Johnson et al. 2015). Seeds of the eight subordinate species were purchased from a commercial provider (Ion Exchange, Inc.) volunteer species were allowed to establish in the plots, with no weeding done to preserve the natural species composition.
TABLE 1.
Plant species and seeding density used for common garden establishment (Johnson et al. 2015).
| Planted species | Family | Source | Seeding rate (seeds m−2) | |
|---|---|---|---|---|
| Grasses | Andropogon gerardi | Poaceae | Local a | 270 |
| Sorghastrum nutans | Poaceae | Local b | 70 | |
| Elymus canadensis | Poaceae | Commercial c | 30 | |
| Forbs | Asclepias tuberosa | Apocynaceae | Commercial c | 30 |
| Chamaecrista fasciculata | Fabaceae | Commercial c | 30 | |
| Dalea purpurea | Fabaceae | Commercial c , d | 30 | |
| Monarda fistulosa | Lamiaceae | Commercial c | 30 | |
| Solidago rigida | Asteraceae | Commercial c | 30 | |
| Penstemon digitalis | Plantaginaceae | Commercial c | 30 | |
| Ruellia humilis | Acanthaceae | Commercial c | 30 | |
| Total seeds (m−2) | 580 | |||
Seeds were collected from multiple remnants.
Seeds were collected from one remnant prairie within the native habitat for each ecotype (e.g., DRY ecotype in Hays, KS, USA, 38°51′13.2″ N, 99°19′08.6″ W; MESIC ecotype in Manhattan, KS, USA, 39°08′22.3″ N, 96°38′23.3″ W; and WET ecotype in Carbondale, IL, USA, 37°41′47.0″ N, 89°14′19.2″ W).
Seeds purchased from Ion Exchange Inc. Harpers Ferry, IA, USA.
Dalea purpurea was initially sown for the experiment but was absent in field surveys from 2012 to 2019.
2.2. Common Garden Establishment
The long‐term experiment contained a randomized complete block design. Four blocks contained three 4 × 4 m plots randomly assigned to be sown with one of three ecotypes (WET, MESIC, or DRY) of the dominant species (A. gerardi and S. nutans). The sown seed density of A. gerardi was 270 live seeds m−2 and S. nutans was at a density of 70 live seeds m−2 (live seed percentage was determined by the Kansas Seed Crop Improvement Center, Manhattan, Kansas, USA). Seeds of each subordinate species were added at a rate of 30 live seeds m−2 (Johnson et al. 2015). For a single plot, the total live seed density was 580 seeds m−2, as suggested for grassland restoration (Packard and Mutel 2005). The seed was mixed with damp sand, hand broadcast into plots, and raked into the soil (Johnson et al. 2015). The buffer zones between plots were seeded with little bluestem (Schizachyrium scoparium) and sideoats grama (Bouteloua curtipendula) supplied by a commercial seed company Ion Exchange Inc. (Johnson et al. 2015). Prescribed burning was applied in the site after the end of the growing season each year, starting from the fall of 2009 (Wilson et al. 2016).
In 2011, rainfall reduction shelters were installed according to a split‐plot design. Rainout shelters were placed in over half of each plot sown with a single ecotype of the dominant species. The shelters were designed to intercept 50% of ambient rainfall (Yahdjian and Sala 2002) and reduced rainfall by 34%–38%, based on measurements of rainfall and intercepted water collected in the site from June to September 2012 (Wilson et al. 2016). The size of shelter frames was 2.4 × 2.5 m, and each roof was constructed of clear acrylic, V‐shaped plates (0.13‐m wide and 2‐m long) spaced 20 cm apart. The roof was angled at a 20° slope to direct rain into a gutter on the low side, guiding it away from the plots (Yahdjian and Sala 2002). Shelters were placed in the field to cover a 2 × 2 m area of each plot. To minimize shading and warming greenhouse effects, we maintained a 150‐cm gap between the lower roof edge of the rainout shelters and the ground, preventing interference with the plant canopy (Kreyling et al. 2017). Kramer et al. (2018) found that the rainout shelters had little effect on the morphological traits of dominant species A. gerardi in the North American Tallgrass Prairie. All the experimental drought shelters were erected close to the beginning of the growing season, early June of each year, when a quarter of the mean annual cumulative temperature had elapsed (Johnson et al. 2015).
2.3. Plant Surveys
We identified and visually estimated percent cover of each species rooted in each of four 1 m2 (1 × 1 m) quadrats in each single plot (two quadrats in each of the ambient and reduced rainfall treatment). Plant surveys were conducted in late summer each survey year. Field surveys were conducted in 2012, 2014, 2018, and 2019.
2.4. Phylogeny and Functional Traits
The taxonomic name of each plant species was standardized with the Taxonomic Name Resolution System (TNRS) implemented in the R package “taxize” (version 0.9.98; Chamberlain and Szocs, 2013). We employed the largest fossil‐dated mega‐phylogeny for spermatophytes, GBOTB, comprising 79,881 taxa, as the basis to construct a phylogeny for plant species in our common garden site (Smith and Brown 2018). At the species level, 91 species from 32 families in the restoration experiment were identified in the latest mega‐phylogeny. Phylogeny in the site was performed using phylo.maker function in the “V.PhyloMaker” package version 0.1.0 (Jin and Qian 2019). We added sago palm (Cycas revoluta) as the outgroup. We employed scenario three and “build.nodes.1” in V.PhyloMaker. We eventually pruned the mega‐phylogeny to maintain only the plant species in the experiment.
We measured functional traits from 10 individuals for the three ecotypes of A. gerardi and S. nutans in late August 2019. We followed a standardized protocol to measure dominant species' traits (Cornelissen et al. 2003) of specific leaf area (cm g−1), height (cm), leaf nitrogen (N) content (mg g−1), leaf area (cm2), and seed mass (mg). These functional traits were selected to describe either interspecific or intraspecific competition relevant to nutrient and light uptake (Violle et al. 2012; Swenson et al. 2012; Lasky et al. 2014) and are considered advantageous compared to discrete traits since continuous traits can account for quantitative modeling and forecast plant functions (Swenson and Weiser 2010). We quantified specific leaf area (cm g−1) by dividing leaf fresh area by dry leaf mass. We acquired fresh leaf area (cm2) with an LI‐3000C leaf area meter (Licor, Lincoln, Nebraska, USA). We measured leaf dry mass (g) following oven drying at 45°C for 3 days (Khalil et al. 2018). We estimated leaf nitrogen content (mg g−1) with a Thermo Scientific Flash 2000 CNHSO Elemental Analyzer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). We calculated the seed mass (mg) by weighing 1000 seeds. We measured functional traits of 39 subordinate and volunteer species collected from the same field in 2015 (Agronomy Research Center SIU, Carbondale, IL, USA; 37°41′47.0″ N, 89°14′19.2″ W). The traits were obtained from either 20 replicates (canopy % cover ≥ 10) or five replicates (canopy % cover < 10; Khalil et al. 2018). We utilized functional traits of the remaining 50 volunteer species available from the TRY plant trait database (version 5.0) if traits were unavailable at the time of surveys (Maitner et al. 2018; Kattge et al. 2020; Appendix S1). To ensure the quality of trait observations, we followed the standard data cleaning protocol for TRY database (Augustine et al. 2024) to retain continuous trait data that met the following criteria: (1) marked by TRY database as unduplicated (unique in the database), (2) represented as a mean or single observation (e.g., excluding minimum and maximum values), and (3) not marked as an outlier by TRY database (e.g., within three standard deviations of the species trait mean).
2.5. Data Analysis
We assessed species richness in each plot by averaging the abundance of individual species within each subplot. We estimated phylogenetic or functional mean pairwise distance by the standardized effect size (sesmpd) using the “picante” (version 1.8.2) R package (Kembel et al. 2010). Phylogenetic diversity (PDsesmpd) and functional diversity (FDsesmpd) are abundance‐weighted metrics calculated as:
where Meanobs is the observed mean pairwise distance, Meanrand is the mean of random mean pairwise distance, and SDrand is the standard deviation of the random mean pairwise distance (Swenson 2014). Random communities were produced by random shuffling of taxa labels across the branching diagram's tips 999 times (Swenson 2014). Positive values of sesmpd suggest a high degree of trait or evolutionary dissimilarity, while negative values imply a low degree of trait or evolutionary dissimilarity. To ensure comparability across traits and mitigate biases from varying scales or units, we standardized functional traits to have a mean of zero and a standard deviation of one, and quantified sesmpd using the Gower distance (Swenson 2014). Values of sesmpd were quantified based on species abundance in plots, that is, they were abundance weighted by using relative percentage cover of each species (Webb et al. 2002; Kembel et al. 2010). We used a repeated measures generalized linear mixed model (GLMM) with a Poisson distribution and a log link function to analyze the discrete response variable of species richness (number of species). Experimental drought, ecotype, year, and their interactions were included in the model as the fixed factors. We utilized a repeated measures linear mixed model (LMM) to examine the effects of experimental drought, ecotype, year, and their interactions on the continuous response variables PDsesmpd or FDsesmpd. Block and plot (as repeated measures) were treated as random factors in GLMM and LMM. For post hoc evaluation, we applied Tukey's multiple comparison test. We used Cohen's d to estimate effect size to show the magnitude of temporal change in PDsesmpd or FDsesmpd. An effect size of 0.7 means the mean response of 1 year is 0.7 standard deviations different from another year. Temporal differences are considered trivial (0 < d ≤ 0.2), small (0.2 < d ≤ 0.5), moderate (0.5 < d ≤ 0.8), and strong (d > 0.8; Cohen 1992). We used the R (version 4.0.2) packages, including “lme4” (version 1.1.23), “multcomp” (version 1.4.13), and “emmeans” (version 1.5.0) for the models (Hothorn, Bretz, and Westfall 2008; Bates et al. 2015; Lenth 2016; R Core Team 2020; Appendix S2).
To evaluate the effect of internal (ITV) and external (rainfall) filters in determining grassland community assembly, we assessed trait statistics (T‐statistics) to estimate where functional traits were most significant with different ecotypes of the dominant species (Violle et al. 2012). Functional traits of a community were represented from each ecotype (WET, MESIC, or DRY) of the dominant species A. gerardi (Gray et al. 2014; Galliart et al. 2020) and S. nutans (Khalil, Gibson, and Baer 2019). Three components of T‐statistics were summarized to partition phenotypic variance in traits into three organizational levels: (i) is the ratio of trait variance within ecotype (e.g., ITV within a WET ecotype) to total trait variance within a plot (e.g., trait variation of all individuals within a plot sown with the WET ecotype). The component serves as a measure of internal filtering, aiming to assess the role of ITV in shaping community assembly, highlighting that the two individuals are members from the same population and can show more similar trait values than two individuals selected randomly from a plot (Taudiere and Violle 2016). (ii) is the ratio of trait variance within a plot (e.g., WET‐ecotype plot) to trait variance of the whole common garden experiment (e.g., trait variance of all individuals across plots sown with WET, MESIC, and DRY ecotypes of dominant species). Thus, can be interpreted as a measure of external filtering (e.g., controlled by manipulated rainfall) while accounting for trait variation of individuals (Jordani et al. 2019). (iii) is the ratio of trait variance within a plot (e.g., WET‐ecotype plot) relative to the total trait variance in the common garden experiment as a quantity of the power of external filtering without taking intraspecific variation into account. ITV among three ecotypes were summarized in a principal component analysis (PCA). Ellipses with a 68% probability (i.e., the proportion of samples within one standard deviation) were added around points from each ecotype for both dominant grasses to visualize the degree of intraspecific trait variability (Vu 2011). We computed PCA with the prcomp function in “ggbiplot” (version 0.55) package in R software (version 4.0.2; R Core Team 2020).
We utilized standardized effect sizes (SES) of T‐statistic values to test the deviation of observed trait distributions from randomization (n = 999 permutations). SES was calculated:
where and are respectively the mean and the standard deviation of the randomized values of trait and is the observed value of trait. SES estimates the number of standard deviations which differentiate the observed trait values from the average values of the simulated communities (Gotelli and McCabe 2002). A negative SES value indicates the T‐statistic value lower than random expectation, representing the overlap of trait distribution less than expected value by chance (Jordani et al. 2019). By contrast, a positive SES value suggests the T‐statistic value higher than random expectation, showing trait distribution overlapped more than null expectation. The trait analysis was performed in R version 4.0.2 (R Core Team 2020; Appendix S2), using tstats function in the “cati” (version 0.99.4) package for the T‐statistics (Taudiere and Violle 2016).
3. Results
3.1. Impact of Rainfall × ITV on Grassland Diversity
To address the question, “(1) Does a manipulated rainfall × ITV interact to influence grassland diversity overtime?,” we surveyed 91 species comprising 32 plant families. The most abundant species were among five angiosperm families including Asteraceae (n = 21 species), Poaceae (n = 19 species), Fabaceae (n = 8 species), Brassicaceae (n = 4 species), and Convolvulaceae (n = 3 species). Species richness showed no response to the experimental drought (χ2 < 0.01, df = 1, p = 0.98), or interactions with drought (ecotype × experimental drought: χ2 = 2.07, df = 2, p = 0.36; year × experimental drought: χ2 = 0.58, df = 3, p = 0.90). There was an ecotype × year interaction on richness (χ 2 = 15.44, df = 6, p = 0.02; Figure 1a). Species richness in local WET ecotype plots declined during the first two surveyed years from 2012 to 2014, though there were no differences in the WET‐ecotype plots in the following years (Figure 1a: blue line). Species richness in nonlocal DRY‐ecotype plots in 2012 was higher than all the plots in the later years (Figure 1a).
FIGURE 1.
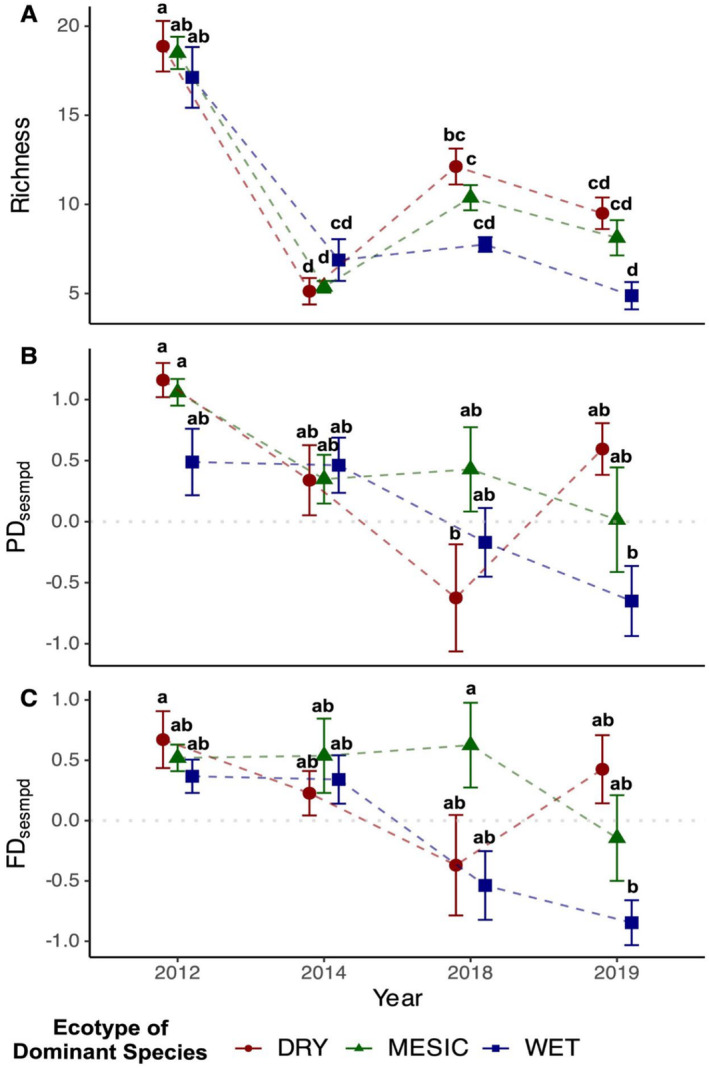
Results showing the interactive effect of dominant grass ecotype (DRY, MESIC, or WET) and year (2012, 2014, 2018, or 2019) on (A) species richness (number of species m−2), (B) phylogenetic diversity (PDsesmpd), and (C) functional diversity (FDsesmpd). Values of metrics from the same year are horizontally jittered to aid visualization. Above data points and error bars sharing the same letter indicate nonsignificant differences (p > 0.05).
Overall phylogenetic diversity showed a structural shift of species from a distant evolutionary relationship in 2012 (PDsesmpd = 0.90 ± 0.12) to a random evolutionary relationship in 2019 (PDsesmpd = −0.01 ± 0.21), accompanied by a Cohen's d effect size of 1.13, indicating a substantial decline in PDsesmpd values from early year 2012 to later year 2019. Phylogenetic diversity showed no response to experimental drought (F 1,63 = 0.61, p = 0.44), ecotype × experimental drought (F 2,63 = 0.20, p = 0.82), or year × experimental drought (F 3,63 = 0.24, p = 0.87) interactions. There was an ecotype × year effect (F 6,63 = 2.43, p = 0.04) on phylogenetic diversity (PDsesmpd; Figure 1b) resulting from differences between 2012 and 2019. Specifically, species in local WET‐ecotype plots in 2019 were more closely related evolutionarily than species in nonlocal MESIC‐ or DRY‐ecotype plots in 2012 (Figure 1b). Species in nonlocal DRY‐ecotype plots in 2018 were also more closely related evolutionarily than species in non‐local MESIC‐ or DRY‐ecotype plots in 2012. In contrast, no ecotype effect occurred between 2014 and 2018.
Overall functional diversity also showed a trait compositional shift for co‐occurring species from high dissimilarity in early year 2012 (FDsesmpd = 0.52 ± 0.10) to a random relationship of trait pattern in later year 2019 (FDsesmpd = −0.18 ± 0.19), accompanied by a Cohen's d effect size of 0.92, indicating a substantial decrease in FDsesmpd values from 2012 to 2019. Besides, functional diversity showed no response to experimental drought alone (F 1,63 = 1.22, p = 0.27), or ecotype × experimental drought (F 2,63 = 0.01, p = 0.988), or year × experimental drought (F 3,63 = 0.05, p = 0.98) interactions. There was an ecotype × year effect (F 6,63 = 2.33, p = 0.04) on functional diversity (FDsesmpd; Figure 1c). Species in local WET‐ecotype plots in 2019 showed higher trait similarity than species in nonlocal MESIC‐ecotype plots in 2018 and DRY‐ecotype plots in 2012 (Figure 1c).
3.2. Intraspecific Versus Interspecific Trait Variation in Grassland Assembly
To address the question, “(2) Does intraspecific trait variation influence grassland community assembly similarly to interspecific trait variation?,” we first summarized the trait values (mean ± standard error) for the three ecotypes of the dominant species (both A. gerardi and S. nutans; Table 2) to assess ITV among the dominant grasses. In local WET‐ecotype plots, A. gerardi exhibited heights 32% and 42% greater than those in nonlocal MESIC‐ or DRY‐ecotype plots, respectively, while S. nutans displayed heights 24% and 23% higher in local WET‐ecotype plots compared to nonlocal MESIC‐ or DRY‐ecotype plots. The leaf area of A. gerardi in WET‐ or DRY‐ecotype plots increased by 33% and 28%, respectively, compared to MESIC‐ecotype plots, with no difference observed among S. nutans ecotypes. In local WET‐ecotype plots, A. gerardi exhibited 15% and 11% higher seed mass compared to nonlocal MESIC‐ or DRY‐ecotype plots, while in MESIC‐ecotype plots, S. nutans displayed 20% and 26% greater seed mass compared to WET‐ or DRY‐ecotype plots, respectively. The specific leaf area of both species did not differ across ecotypes. Finally, in WET‐ or MESIC‐ecotype plots, S. nutans leaf nitrogen content was 58% and 44% higher than in DRY‐ecotype plots, respectively, with no difference observed among A. gerardi ecotypes.
TABLE 2.
Trait measurements (mean ± standard error) of ecotypes (WET, MESIC, or DRY) of each dominant grass species ( Andropogon gerardi or Sorghastrum nutans ).
| Functional trait | Andropogon gerardi (n = 10 per ecotype) | Sorghastrum nutans (n = 10 per ecotype) | ||||
|---|---|---|---|---|---|---|
| WET | MESIC | DRY | WET | MESIC | DRY | |
| Height (cm) | 286.7 ± 2.8 a | 217.4 ± 3.23 b | 201.3 ± 4.47 c | 257.0 ± 7.23 a | 206.9 ± 5.57 b | 208.5 ± 4.02 b |
| Leaf area (cm2) | 37.1 ± 1.28 a | 27.9 ± 1.38 b | 35.7 ± 2.49 a | 30.3 ± 2.17 a | 28.8 ± 2.24 a | 28.8 ± 2.27 a |
| Seed mass (mg) | 3.1 ± 0.06 a | 2.7 ± 0.11 b | 2.8 ± 0.06 b | 2.0 ± 0.05 b | 2.4 ± 0.04 a | 1.9 ± 0.03 b |
| Specific leaf area (cm2 g−1) | 170.0 ± 6.62 a | 149.2 ± 20.43 a | 210.1 ± 21.95 a | 142.7 ± 12.61 a | 149.2 ± 21.13 a | 137.6 ± 6.92 a |
| Leaf N content (mg g−1) | 11.2 ± 0.56 a | 10.6 ± 0.78 a | 11.7 ± 0.78 a | 11.4 ± 0.54 a | 10.4 ± 0.56 a | 7.2 ± 0.53 b |
Note: Sample sizes (n) refer to the number of individual plants from which traits were measured. (mean ± standard error) followed by identical letters were not significantly different from each other (experiment‐wide α = 0.05, Tukey adjusted).
To evaluate the effect of internal and external filters in determining community assembly, we assessed the departure of observed estimates of T‐statistics from randomized values for five functional traits (Figure 2), including leaf area, seed mass, leaf N content, height, and specific leaf area. The result was mostly consistent across different traits. values were calculated to measure internal filtering in grassland assembly. The mean estimates of were significantly less than expected by chance for all the traits, except specific leaf area, which was the only trait showing mean value within null expectation (Figure 3: red markings). Hence, within each community (planted with one of the three dominant species ecotypes WET, MESIC, or DRY), there was minimal overlap among species in terms of trait distributions when considering intraspecific variations. In other words, two individuals from a dominant species ecotype exhibited more similar trait values than two individuals selected randomly from the same community. In contrast, the values used for measuring external filtering did not deviate from null expectations on average for most communities (Figure 2: purple markings). Thus, two individuals randomly selected from a community planted with a particular ecotype were not necessarily more comparable or more distinctive than two individuals randomly selected from the entire common garden experiment (Jordani et al. 2019). In the same way, values quantified the overlap among community trait distributions within the whole common garden without considering ITVs. The lack of departure for values from null expectations showed that there was limited overlap in trait distributions between communities when focusing on population‐level trait estimates (Jordani et al. 2019, Figure 2: green markings).
FIGURE 2.
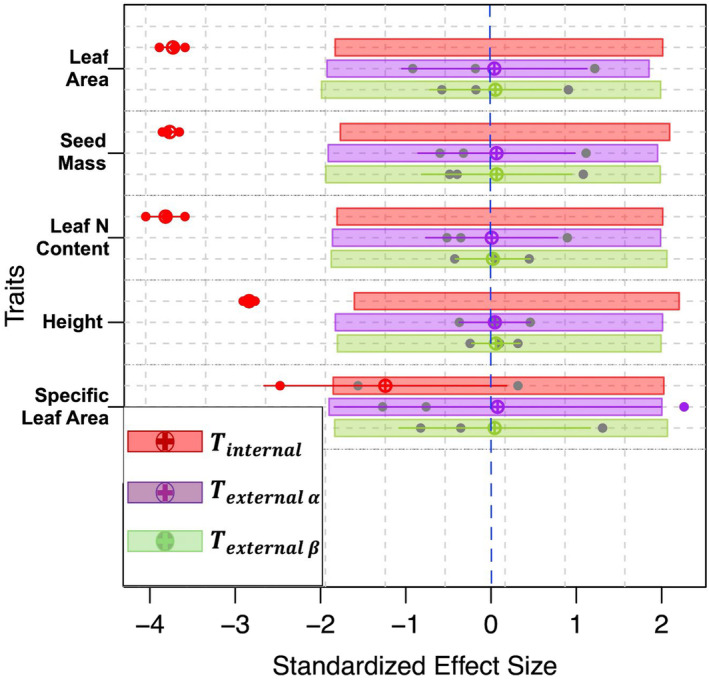
Standardized effect size (SES) of T‐statistics for the five traits: leaf area (cm2), seed mass (mg), leaf N content (mg g−1), height (cm), and specific leaf area (cm2 g−1) collected from the common garden experiment. The horizontal axis (SES) was employed to quantify the magnitude of changes, enabling comparison across distinct trait measures. Colored dots represent the SES value for plots planted with one dominant species ecotype (e.g., DRY, MESIC, or WET) when different from the null model. Tinternal = the ratio of trait variance within ecotype (e.g., intraspecific variation of WET ecotype for dominant species) relative to total trait variance within the plot (e.g., including both intraspecific and interspecific variations); T external α = the ratio of trait variance within a plot relative to trait variance of all plots in the common garden experiment; and T external β = the ratio of trait variance within a plot relative to trait variance of all plots in the common garden experiment, excluding intraspecific trait variation. The crossed circles and the segments represent the mean and standard deviation of the SES values for a given T‐statistic and a given trait. For a given T‐statistic, the mean SES (crossed circle) is significantly different from the null distribution if not embedded within the colored bar (e.g., ). The more the SES value departs from the null model, the stronger the filtering effect.
FIGURE 3.
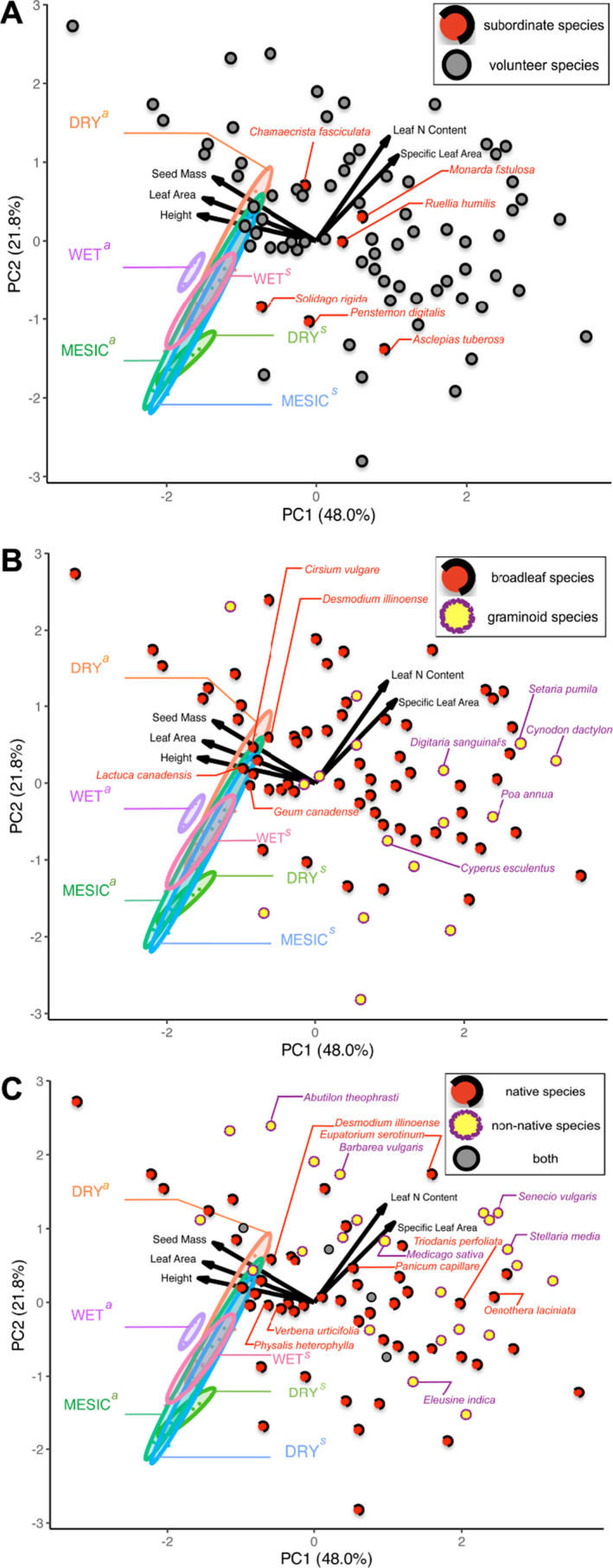
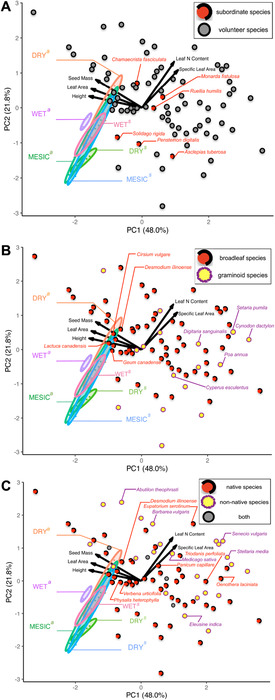
Principal components analysis (PCA) summarizing ITVs among ecotypes (DRY, MESIC, or WET) of dominant species (a = Andropogon gerardi ; s = Sorghastrum nutans ) in multivariate trait space. The ellipses are 68% data ellipses for A. gerardi and S. nutans . Dots represent the positions of nondominant species within PCA trait space grouped by (A) their roles in communities: red dots with dashed circles = subordinate species sown with A. gerardi and S. nutans in 2008 (Table 1); gray dots with solid circles = volunteer species, (B) their morphological features: red dots with dashed circles = broadleaf species; yellow dots with rough circles = graminoid species, and (C) their nativeness: red dots with dashed circles = native species; yellow dots with rough circles = non‐native species; and gray dots with solid circles = species can be both native and non‐native. Information on whether a species is native or non‐native to Illinois, USA was obtained from the US Department of Agriculture (USDA) PLANTS Database (https://plants.usda.gov). Representative species are labeled with their scientific names: subordinate (red) in A, broadleaf (red) or graminoid (purple) in B, and native (red) or non‐native (purple) in C. The solid arrowed lines show the direction and loadings of the traits including height, leaf area, seed mass, leaf N content, and specific leaf area.
Overall, the PCA (Figure 3a) revealed that the ellipse of local A. gerardi WET‐ecotype was much smaller compared with the ellipses of nonlocal MESIC‐ or DRY‐ecotype of A. gerardi. In contrast, the ellipses among ecotypes of S. nutans mostly overlapped (e.g., MESIC and DRY, MESIC and WET). Our result showed that the multidimensional trait overlaps of dominant species was shaped by interspecific (e.g., between A. gerardi and S. nutans) and ITVs (e.g., among ecotypes) of functional traits. Many broadleaf species, including common thistle (Cirsium vulgare), Illinois ticktrefoil (Desmodium illinoense), white avens (Geum canadense), and tall lettuce (Lactuca canadensis), exhibited a multitrait space close to dominant A. gerardi or S. nutans in the PCA (Figure 3b; nondominant species represented as colored dots instead of name/code labels due to significant label overlap in multitrait PCA space). In contrast, graminoids such as Bermuda grass (Cynodon dactylon), yellow nutsedge (Cyperus esculentus), large crabgrass (Digitaria sanguinalis), annual bluegrass (Poa annua), and yellow foxtail (Setaria pumila) occupied a multitrait space opposite to the two dominant species (Figure 3b). Native broadleaf species (Figure 3c) such as late boneset (Eupatorium serotinum), cutleaf evening primrose (Oenothera laciniata), and clasping bellflower (Triodanis perfoliata) also occupied a trait space opposite to the two dominant species in the PCA. Other natives such as Illinois ticktrefoil (D. illinoense), clammy groundcherry (Physalis heterophylla), and white vervain (Verbena urticifolia) were close to the dominant species. Most non‐native species, particularly agricultural weeds such as velvetleaf (Abutilon theophrasti), goosegrass (Eleusine indica), groundsel (Senecio vulgaris), and chickweed (Stellaria media), exhibited a multitrait space distinct from the dominant species (Figure 3c). Furthermore, height, leaf area, and seed mass showed high negative loadings on PC1 axis, while leaf nitrogen content and specific leaf area showed relatively high positive loadings on both axes.
4. Discussion
Manipulated rainfall did not affect plant diversity in our grassland community common garden experiment, indicating that developing tallgrass prairie may be resilient to less precipitation in the initial decade of restoration. Our first hypothesis that drought lowers species richness and increases evolutionary and trait similarity was not supported because there were no observed effects of the experimental drought treatment or its interaction with ecotype or year on species richness, phylogenetic (PD‐), and functional diversity (FD‐sesmpd). Yue et al. (2020) also reported a similar result following a meta‐analysis of experimental rainfall manipulations, discovering no overall treatment effect on plant diversity on all levels. Moreover, Komatsu et al. (2019) synthesized studies on manipulating precipitation either experimentally increased or reduced and found no effect of drought on taxonomic diversity. Our study site in southern Illinois, USA is located on the mesic edge of North American Tallgrass Prairie. Korell et al. (2021) studied 74 rainfall control experiments and found those plant communities in relatively wetter regions are often less sensitive to predicted variations in rainfall than water‐limited ecosystems. Although experimental drought might be expected to function as an abiotic filter by decreasing the possibility that certain plant species with lower drought tolerance will establish, there can be no correlation between grassland phylogenetic diversity and manipulated rainfall (Barber et al. 2017). This result supports a previous study that grassland responses to rainfall were not phylogenetically conserved (Bennett and Cahill 2013; Luong, Holl, and Loik 2021). Moreover, our result showed no effect of drought on plant functional diversity. It reinforced that trends in grassland functional diversity were not necessarily linked to loss of species during restoration, and variability of functional diversity is less prone to be only shaped by experimental treatments (Miller et al. 2019; Zuo et al. 2021; Karimi et al. 2022). Although our findings indicated no impact from the rainout shelter roofs, we cannot completely rule out the possibility of unwanted side effects on the microenvironment.
Species richness decreased significantly during the early years of assembly, coinciding with the increasing dominance of sown grasses (Ren et al. 2023). Similar declines in richness have been observed during the initial years of restoration in other grasslands. For example, the increased density of dominant species could lead to richness losses (Keddy et al. 2006; Avolio et al. 2019). Declines in richness may also result from decreased financial and labor resources postplanting, as long‐term maintenances are essential for high species diversity (Luong, Holl, and Loik 2021; Luong, Press, and Holl 2023). Contrary to the trend of decreasing richness, ecotypic effects on phylogenetic and functional diversity were generally minimal. However, our findings suggest that communities with locally adapted WET ecotypes, benefiting from a home‐site advantage, displayed a trajectory of increasing evolutionary and trait similarity over time, contrasting with communities hosting nonlocal ecotypes (Johnson et al. 2015; Mendola et al. 2015; Wilson et al. 2016). Previous studies showed that genetic differences shaped the competitive traits of the dominant grasses, causing A. gerardi from wet regions to display greater canopy cover, leaf count, stem diameter, and maximum leaf width compared to those from xeric areas (Kramer et al. 2018; Galliart et al. 2020).
Despite rejecting our first hypothesis regarding experimental drought's influence on biodiversity, we observed ecotype × year effect on functional (FD‐) and phylogenetic diversity (PD‐sesmpd) in the experiment. Previous studies showed that changes in the ITV of dominant grasses could alter grassland functional or phylogenetic diversity by impacting nondominant species (Gustafson et al. 2014; Khalil, Gibson, and Baer 2019). For example, cultivars impacted grassland phylogenetic diversity more than noncultivar population sources of a dominant grass species (Khalil, Gibson, and Baer 2017) by reducing the abundance of an evolutionarily distinct community of less closely related subordinate species. Khalil, Gibson, and Baer (2017) showed diversity patterns varied among metrics: phylogenetic and functional diversity were maintained at constant levels while taxonomic diversity declined during restoration. Purschke et al. (2013) also found contrasting changes in taxonomic, functional, and phylogenetic diversity within a chronosequence during a long‐term seminatural grassland succession. Understanding the role of intraspecific variation during grassland restoration is essential to inform predictions of how temperate grassland ecosystems will respond to global climate change (Baer, Gibson, and Johnson 2019). Furthermore, understanding the relative importance of evolutionary history and environmental conditions on the dominant grasses is necessary to determine the best sources of seed materials for prairie restoration and forecast ecosystem response to biotic or environmental assembly drivers (Mendola et al. 2015).
Our trait‐based analysis indicated variability in the grassland communities was principally due to within‐population trait variation resulting from ecotypic differentiation in the dominant species rather than differences between species in a community. This result supports our second hypothesis that an internal biotic filter plays a key role in grassland assembly with a less important external environmental filter. The ITV of dominant species as a primary internal filter significantly influences restoration efforts, ecosystem functions, environmental filtering, and species coexistence (Laughlin et al. 2012; Hart, Schreiber, and Levine 2016). Overall, the global pattern of plant traits showed that ITV constituted 32% of trait variation between communities and 25% within communities (Siefert et al. 2015). Although patterns of trait variation within species might seem idiosyncratic, the inter‐ and intraspecific variations of functional traits can be interpreted by environmental context, functional tactics, and evolutionary history (Sandel et al. 2021). Nevertheless, a limitation of trait‐based analyses is its reliance on empirical correlations and null models (Swenson 2014). Consequently, our analyses on functional traits did not encompass all life stages of dominant species and their interactions with all possible volunteer species, which could connect grassland function and phylogeny to the demography of dominant species (Enquist et al. 2015).
We found little support for an influence of an external filter on the communities planted with different dominant grass ecotypes. Similarly, Khalil, Gibson, and Baer (2019) showed that ITV as an internal filter was predominant among functional traits rather than trait variation among species as the external filter in restored grassland in southern Illinois. Fang et al. (2019) found that ITV analyses showed the importance of limiting similarity in driving community assembly at an early stage of succession. We also observed a strong internal filter effect on most functional traits, including height, seed mass, leaf area, and leaf N content in grassland communities, implying the internal filter with a low overlap in trait distributions. This less‐than‐random (negative SES value) trait overlap indicated that ITV among dominant species in our study, was largely driving the restored grassland assembly process (Khalil, Gibson, and Baer 2019). Likewise, Crawford et al. (2019) found that intraspecific variation played pivotal roles in grassland assembly processes. The PCA result revealed that the local WET‐ecotype of dominant species had a lower ITV compared to nonlocal ecotypes. Non‐native volunteer species such as rocketcress (Barbarea vulgaris), alfalfa (Medicago sativa), and native volunteer species such as witchgrass (Panicum capillare), which were exclusive to local WET‐ecotype plots, displayed traits such as higher leaf nitrogen content and specific leaf area than the dominant species A. gerardi. The differences in traits between dominant and nondominant species suggest that many volunteer species might occupy niches that are different from those of the dominant species due to limiting similarity.
In general, our results highlight the importance of integrating interspecific and intraspecific trait variabilities. We focused on functional traits to comprehend better how trait variability is coupled with species coexistence (Jordani et al. 2019). Future empirical and experimental studies should investigate ongoing theoretical research on ITV, such as the eco‐evolutionary theory of community structure (Wickman, Koffel, and Klausmeier 2023) and the niche packing hypothesis (Violle et al. 2012). These investigations are necessary to examine the distinctive origins of variability in plant traits and how they contribute to community assembly in restored grasslands. Embracing diverse practice and management strategies is crucial for enhancing ecological restoration efforts. These strategies may include selecting locally adapted seed sources or vegetative propagules to promote survival and growth, conducting long‐term monitoring to anticipate future challenges, and identifying native populations that thrive under controlled conditions, such as manipulated rainfall, to foster establishment in naturally variable environments.
Author Contributions
Zhe Ren: conceptualization (lead), data curation (equal), formal analysis (lead), investigation (equal), methodology (equal), project administration (lead), resources (equal), software (lead), visualization (lead), writing – original draft (lead), writing – review and editing (equal). Sara G. Baer: investigation (equal), project administration (equal), resources (equal), writing – review and editing (equal). Loretta C. Johnson: funding acquisition (equal), project administration (equal), resources (equal), writing – review and editing (equal). Matthew B. Galliart: methodology (equal), project administration (equal), resources (equal), writing – review and editing (equal). David J. Gibson: data curation (equal), funding acquisition (equal), investigation (equal), methodology (equal), project administration (supporting), resources (equal), supervision (supporting), writing – review and editing (equal).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research Badges
This article has earned an Open Data badge for making publicly available the digitally‐shareable data necessary to reproduce the reported results. The data is available at (insert provided URL from Open Research Disclosure Form).
Supporting information
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.
Acknowledgments
We extend our appreciation to the collaborators at the Southern Illinois University and Kansas State University, particularly Mohammed I. Khalil, Drew A. Scott, Laurel R. Wilson, Xian Liu, and Saroj Thapa for the efforts in maintaining the common garden experiment and the contributions to the long‐term ecological restoration survey. This project received support from the U.S. Department of Agriculture Abiotic Stress Program (grant no. 2008‐35001‐04545) and the National Science Foundation, with grants awarded to D.J.G. (grant no. DUE‐1758497 and DUE‐1949969).
Funding: This work was supported by U.S. Department of Agriculture (2008‐35001‐04545). National Science Foundation (DUE‐1758497, DUE‐1949969).
Data Availability Statement
The data and codes used in this study are available with the corresponding Supporting Information.
References
- Arnan, X. , Cerda X., and Retana J.. 2015. “Partitioning the Impact of Environment and Spatial Structure on Alpha and Beta Components of Taxonomic, Functional, and Phylogenetic Diversity in European Ants.” Peer J 3: e1241. 10.7717/peerj.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, J. , Groves A. M., Towers I. R., Catano C. P., and Brudvig L. A.. 2023. “Trait‐Mediated Community Assembly During Experimental Grassland Restoration Is Altered by Planting Year Rainfall.” Journal of Applied Ecology 60, no. 8: 1587–1596. 10.1111/1365-2664.14430. [DOI] [Google Scholar]
- Augustine, S. P. , Bailey‐Marren I., Charton K. T., Kiel N. G., and Peyton M. S.. 2024. “Improper Data Practices Erode the Quality of Global Ecological Databases and Impede the Progress of Ecological Research.” Global Change Biology 30, no. 1: e17116. 10.1111/gcb.17116. [DOI] [PubMed] [Google Scholar]
- Avolio, M. L. , Forrestel E. J., Chang C. C., La Pierre K. J., Burghardt K. T., and Smith M. D.. 2019. “Demystifying Dominant Species.” New Phytologist 223, no. 3: 1106–1126. 10.1111/nph.15789. [DOI] [PubMed] [Google Scholar]
- Baer, S. G. , Gibson D. J., and Johnson L. C.. 2019. “Restoring Grassland in the Context of Climate Change.” In Grasslands and Climate Change, edited by Gibson D. J. and Newman J. A., 310–322. Cambridge, United Kingdom: Cambridge University Press. 10.1017/9781108163941. [DOI] [Google Scholar]
- Barber, N. A. , Jones H. P., Duvall M. R., Wysocki W. P., Hansen M. J., and Gibson D. J.. 2017. “Phylogenetic Diversity Is Maintained Despite Richness Losses Over Time in Restored Tallgrass Prairie Plant Communities.” Journal of Applied Ecology 54, no. 1: 137–144. 10.1111/1365-2664.12639. [DOI] [Google Scholar]
- Bates, D. , Kliegl R., Vasishth S., and Baayen H.. 2015. “Parsimonious Mixed Models.” arXiv 1: 04967. [Google Scholar]
- Belinchon, R. , Hemrova L., and Munzbergova Z.. 2019. “Abiotic, Present‐Day and Historical Effects on Species, Functional and Phylogenetic Diversity in Dry Grasslands of Different Age.” PLoS One 14, no. 10: e0223826. 10.1371/journal.pone.0223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J. A. , and Cahill J. F.. 2013. “Conservatism of Responses to Environmental Change Is Rare Under Natural Conditions in a Native Grassland.” Perspectives in Plant Ecology, Evolution and Systematics 15, no. 6: 328–337. 10.1016/j.ppees.2013.10.001. [DOI] [Google Scholar]
- Carmona, C. P. , de Bello F., Azcárate F. M., Mason N. W. H., and Peco B.. 2019. “Trait Hierarchies and Intraspecific Variability Drive Competitive Interactions in Mediterranean Annual Plants.” Journal of Ecology 107, no. 5: 2078–2089. 10.1111/1365-2745.13248. [DOI] [Google Scholar]
- Caudle, K. L. , Johnson L. C., Baer S. G., and Maricle B. R.. 2014. “A Comparison of Seasonal Foliar Chlorophyll Change Among Ecotypes and Cultivars of Andropogon gerardii (Poaceae) by Using Nondestructive and Destructive Methods.” Photosynthetica 52, no. 4: 511–518. 10.1007/s11099-014-0057-2. [DOI] [Google Scholar]
- Chamberlain, S. A. , and Szöcs E.. 2013. “Taxize: Taxonomic Search and Retrieval in R.” F1000Research 2: 191. 10.12688/f1000research.2-191.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chave, J. , Chust G., and Thébaud C.. 2007. “The Importance of Phylogenetic Structure in Biodiversity Studies.” In Scaling Biodiversity, edited by Storch D., Marquet D., and Brown J., 150–167. Cambridge, United Kingdom: Cambridge University Press. 10.1017/CBO9780511814938.010. [DOI] [Google Scholar]
- Chesson, P. 2000. “General Theory of Competitive Coexistence in Spatially‐Varying Environments.” Theoretical Population Biology 58, no. 3: 211–237. 10.1006/tpbi.2000.1486. [DOI] [PubMed] [Google Scholar]
- Clark, J. S. 2010. “Individuals and the Variation Needed for High Species Diversity in Forest Trees.” Science 327, no. 5969: 1129–1132. 10.1126/science.1183506. [DOI] [PubMed] [Google Scholar]
- Cochard, A. , Pithon J., Braud F., Beaujouan V., Bulot A., and Daniel H.. 2019. “Intraspecific Trait Variation in Grassland Plant Communities Along Urban‐Rural Gradients.” Urban Ecosystems 22, no. 3: 583–591. 10.1007/s11252-019-0827-5. [DOI] [Google Scholar]
- Cohen, J. 1992. “A Power Primer.” Psychological Bulletin 112, no. 1: 155–159. 10.1037/0033-2909.112.1.155. [DOI] [PubMed] [Google Scholar]
- Cornelissen, J. H. C. , Lavorel S., Garnier E., et al. 2003. “A Handbook of Protocols for Standardised and Easy Measurement of Plant Functional Traits Worldwide.” Australian Journal of Botany 51, no. 4: 335–380. 10.1071/Bt02124. [DOI] [Google Scholar]
- Crawford, M. , Jeltsch F., May F., Grimm V., and Schlagel U. E.. 2019. “Intraspecific Trait Variation Increases Species Diversity in a Trait‐Based Grassland Model.” Oikos 128, no. 3: 441–455. 10.1111/oik.05567. [DOI] [Google Scholar]
- Enquist, B. J. , Norberg J., Bonser S. P., et al. 2015. “Scaling From Traits to Ecosystems: Developing a General Trait Driver Theory via Integrating Trait‐Based and Metabolic Scaling Theories.” Advances in Ecological Research 52: 249–318. 10.1016/bs.aecr.2015.02.001. [DOI] [Google Scholar]
- E‐Vojtkó, A. , De Bello F., Lososová Z., and Götzenberger L.. 2023. “Phylogenetic Diversity Is a Weak Proxy for Functional Diversity but They Are Complementary in Explaining Community Assembly Patterns in Temperate Vegetation.” Journal of Ecology 111, no. 10: 2218–2230. 10.1111/1365-2745.14171. [DOI] [Google Scholar]
- Fajardo, A. , and Siefert A.. 2018. “Intraspecific Trait Variation and the Leaf Economics Spectrum Across Resource Gradients and Levels of Organization.” Ecology 99, no. 5: 1024–1030. 10.1002/ecy.2194. [DOI] [PubMed] [Google Scholar]
- Fang, S. , Cadotte M. W., Yuan Z., et al. 2019. “Intraspecific Trait Variation Improves the Detection of Deterministic Community Assembly Processes in Early Successional Forests, But Not in Late Successional Forests.” Journal of Plant Ecology 12, no. 4: 593–602. 10.1093/jpe/rty053. [DOI] [Google Scholar]
- Funk, J. L. 2021. “Revising the Trait‐Based Filtering Framework to Include Interacting Filters: Lessons From Grassland Restoration.” Journal of Ecology 109, no. 10: 3466–3472. 10.1111/1365-2745.13763. [DOI] [Google Scholar]
- Galliart, M. , Bello N., Knapp M., et al. 2019. “Local Adaptation, Genetic Divergence, and Experimental Selection in a Foundation Grass Across the US Great Plains' Climate Gradient.” Global Change Biology 25, no. 3: 850–868. 10.1111/gcb.14534. [DOI] [PubMed] [Google Scholar]
- Galliart, M. , Sabates S., Tetreault H., et al. 2020. “Adaptive Genetic Potential and Plasticity of Trait Variation in the Foundation Prairie Grass Andropogon gerardii Across the US Great Plains' Climate Gradient: Implications for Climate Change and Restoration.” Evolutionary Applications 13, no. 9: 2333–2356. 10.1111/eva.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D. J. , Allstadt A. J., Baer S. G., and Geisler M.. 2012. “Effects of Foundation Species Genotypic Diversity on Subordinate Species Richness in an Assembling Community.” Oikos 121, no. 4: 496–507. 10.1111/j.1600-0706.2011.19447.x. [DOI] [Google Scholar]
- Gibson, D. J. , Sendor G., Donatelli J., Baer S. G., and Johnson L.. 2013. “Fitness Among Population Sources of a Dominant Species ( Andropogon gerardii Vitman) Used in Prairie Restoration.” Journal of the Torrey Botanical Society 140, no. 3: 269–279. 10.3159/Torrey-D-12-00063.1. [DOI] [Google Scholar]
- Gotelli, N. J. , and McCabe D. J.. 2002. “Species Co‐Occurrence: A Meta‐Analysis of J. M. Diamond's Assembly Rules Model.” Ecology 83, no. 8: 2091–2096. 10.2307/3072040. [DOI] [Google Scholar]
- Gray, M. M. , St Amand P., Bello N. M., et al. 2014. “Ecotypes of an Ecologically Dominant Prairie Grass ( Andropogon gerardii) Exhibit Genetic Divergence Across the U.S. Midwest grasslands' Environmental Gradient.” Molecular Ecology 23, no. 24: 6011–6028. 10.1111/mec.12993. [DOI] [PubMed] [Google Scholar]
- Gustafson, D. J. , Major C., Jones D., Synovec J., Baer S. G., and Gibson D. J.. 2014. “Genetic Sorting of Subordinate Species in Grassland Modulated by Intraspecific Variation in Dominant Species.” PLoS One 9, no. 3: e91511. 10.1371/journal.pone.0091511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett, L. M. , Shoemaker L. G., White C. T., and Suding K. N.. 2019. “Rainfall Variability Maintains Grass‐Forb Species Coexistence.” Ecology Letters 22, no. 10: 1658–1667. 10.1111/ele.13341. [DOI] [PubMed] [Google Scholar]
- Hardy, O. J. , and Senterre B.. 2007. “Characterizing the Phylogenetic Structure of Communities by an Additive Partitioning of Phylogenetic Diversity.” Journal of Ecology 95, no. 3: 493–506. 10.1111/j.1365-2745.2007.01222.x. [DOI] [Google Scholar]
- Harrison, S. , Spasojevic M. J., and Li D. J.. 2020. “Climate and Plant Community Diversity in Space and Time.” Proceedings of the National Academy of Sciences 117, no. 9: 4464–4470. 10.1073/pnas.1921724117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, S. P. , Schreiber S. J., and Levine J. M.. 2016. “How Variation Between Individuals Affects Species Coexistence.” Ecology Letters 19, no. 8: 825–838. 10.1111/ele.12618. [DOI] [PubMed] [Google Scholar]
- He, D. , Biswas S. R., Xu M. S., Yang T. H., You W. H., and Yan E. R.. 2021. “The Importance of Intraspecific Trait Variability in Promoting Functional Niche Dimensionality.” Ecography 44, no. 3: 380–390. 10.1111/ecog.05254. [DOI] [Google Scholar]
- Hothorn, T. , Bretz F., and Westfall P.. 2008. “Simultaneous Inference in General Parametric Models.” Biometrical Journal 50, no. 3: 346–363. 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Jin, Y. , and Qian H.. 2019. “V.PhyloMaker: An R Package That Can Generate Very Large Phylogenies for Vascular Plants.” Ecography 42, no. 8: 1353–1359. 10.1111/ecog.04434. [DOI] [Google Scholar]
- Johnson, L. C. , Olsen J. T., Tetreault H., et al. 2015. “Intraspecific Variation of a Dominant Grass and Local Adaptation in Reciprocal Garden Communities Along a US Great Plains' Precipitation Gradient: Implications for Grassland Restoration With Climate Change.” Evolutionary Applications 8, no. 7: 705–723. 10.1111/eva.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, H. P. , Barber N. A., and Gibson D. J.. 2019. “Is Phylogenetic and Functional Trait Diversity a Driver or a Consequence of Grassland Community Assembly?” Journal of Ecology 107, no. 5: 2027–2032. 10.1111/1365-2745.13260. [DOI] [Google Scholar]
- Jordani, M. , Mouquet N., Casatti L., Menin M., de Cerqueira Rossa‐Feres D., and Albert C. H.. 2019. “Intraspecific and Interspecific Trait Variability in Tadpole Meta‐Communities From the Brazilian Atlantic Rainforest.” Ecology and Evolution 9, no. 7: 4025–4037. 10.1002/ece3.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, N. , Larkin D. J., Glasenhardt M. C., et al. 2022. “Selection on Convergent Functional Traits Drives Compositional Divergence in Early Succession of a Tallgrass Prairie Restoration Experiment.” Journal of Ecology 110, no. 2: 415–429. 10.1111/1365-2745.13808. [DOI] [Google Scholar]
- Kattge, J. , Bonisch G., Diaz S., et al. 2020. “TRY Plant Trait Database—Enhanced Coverage and Open Access.” Global Change Biology 26, no. 1: 119–188. 10.1111/gcb.14904. [DOI] [PubMed] [Google Scholar]
- Keddy, P. A. , Smith L., Campbell D. R., Clark M., and Montz G.. 2006. “Patterns of Herbaceous Plant Diversity in Southeastern Louisiana Pine Savannas.” Applied Vegetation Science 9, no. 1: 17–26. 10.1111/j.1654-109X.2006.tb00652.x. [DOI] [Google Scholar]
- Kembel, S. W. , Cowan P. D., Helmus M. R., et al. 2010. “Picante: R Tools for Integrating Phylogenies and Ecology.” Bioinformatics 26, no. 11: 1463–1464. 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Khalil, M. I. , Gibson D. J., and Baer S. G.. 2017. “Phylogenetic Diversity Reveals Hidden Patterns Related to Population Source and Species Pools During Restoration.” Journal of Applied Ecology 54, no. 3: 1010. 10.1111/1365-2664.12896. [DOI] [Google Scholar]
- Khalil, M. I. , Gibson D. J., and Baer S. G.. 2019. “Functional Response of Subordinate Species to Intraspecific Trait Variability Within Dominant Species.” Journal of Ecology 107, no. 5: 2040–2053. 10.1111/1365-2745.13249. [DOI] [Google Scholar]
- Khalil, M. I. , Gibson D. J., Baer S. G., and Willand J. E.. 2018. “Functional Diversity Is More Sensitive to Biotic Filters Than Phylogenetic Diversity During Community Assembly.” Ecosphere 9, no. 3: ecs2.2164. 10.1002/ecs2.2164. [DOI] [Google Scholar]
- Kluge, J. , and Kessler M.. 2011. “Phylogenetic Diversity, Trait Diversity and Niches: Species Assembly of Ferns Along a Tropical Elevational Gradient.” Journal of Biogeography 38, no. 2: 394–405. 10.1111/j.1365-2699.2010.02433.x. [DOI] [Google Scholar]
- Knapp, A. K. , Condon K. V., Folks C. C., et al. 2024. “Field Experiments Have Enhanced Our Understanding of Drought Impacts on Terrestrial Ecosystems—But Where Do We Go From Here?” Functional Ecology 38, no. 1: 76–97. 10.1111/1365-2435.14460. [DOI] [Google Scholar]
- Komatsu, K. J. , Avolio M. L., Lemoine N. P., et al. 2019. “Global Change Effects on Plant Communities Are Magnified by Time and the Number of Global Change Factors Imposed.” Proceedings of the National Academy of Sciences 116, no. 36: 17867–17873. 10.1073/pnas.1819027116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korell, L. , Auge H., Chase J. M., Harpole W. S., and Knight T. M.. 2021. “Responses of Plant Diversity to Precipitation Change Are Strongest at Local Spatial Scales and in Drylands.” Nature Communications 12, no. 1: 2489. 10.1038/s41467-021-22766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, N. J. B. , Cornwell W. K., Webb C. O., and Ackerly D. D.. 2007. “Trait Evolution, Community Assembly, and the Phylogenetic Structure of Ecological Communities.” American Naturalist 170, no. 2: 271–283. 10.1086/519400. [DOI] [PubMed] [Google Scholar]
- Kramer, D. L. , Maricle K. L., Hilt C. J., et al. 2018. “Drought Tolerance in Ecotypes of Big Bluestem ( Andropogon gerardii) Relates to Above‐Ground Surface Area: Results From a Common Garden Experiment.” Flora 246: 52–60. 10.1016/j.flora.2018.07.005. [DOI] [Google Scholar]
- Kreyling, J. , Khan M. A. S. A., Sultana F., et al. 2017. “Drought Effects in Climate Change Manipulation Experiments: Quantifying the Influence of Ambient Weather Conditions and Rain‐Out Shelter Artifacts.” Ecosystems 20, no. 2: 301–315. 10.1007/s10021-016-0025-8. [DOI] [Google Scholar]
- Lambert, A. M. , Baer S. G., and Gibson D. J.. 2011. “Intraspecific Variation in Ecophysiology of Three Dominant Prairie Grasses Used in Restoration: Cultivar Versus Non‐cultivar Population Sources.” Restoration Ecology 19: 43–52. 10.1111/j.1526-100X.2010.00673.x. [DOI] [Google Scholar]
- Lanuza, O. R. , Espelta J. M., Peñuelas J., and Peguero G.. 2020. “Assessing Intraspecific Trait Variability During Seedling Establishment to Improve Restoration of Tropical Dry Forests.” Ecosphere 11, no. 2: ecs2.3052. 10.1002/ecs2.3052. [DOI] [Google Scholar]
- Lasky, J. R. , Uriarte M., Boukili V. K., and Chazdon R. L.. 2014. “Trait‐Mediated Assembly Processes Predict Successional Changes in Community Diversity of Tropical Forests.” Proceedings of the National Academy of Sciences 111, no. 15: 5616–5621. 10.1073/pnas.1319342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin, D. C. , Joshi C., van Bodegom P. M., Bastow Z. A., and Fule P. Z.. 2012. “A Predictive Model of Community Assembly That Incorporates Intraspecific Trait Variation.” Ecology Letters 15, no. 11: 1291–1299. 10.1111/j.1461-0248.2012.01852.x. [DOI] [PubMed] [Google Scholar]
- Lenth, R. V. 2016. “Least‐Squares Means: The R Package Lsmeans.” Journal of Statistical Software 69, no. 1: 1–33. 10.18637/jss.v069.i01. [DOI] [Google Scholar]
- Li, D. , Miller J. E. D., and Harrison S.. 2019. “Climate Drives Loss of Phylogenetic Diversity in a Grassland Community.” Proceedings of the National Academy of Sciences 116, no. 40: 19989–19994. 10.1073/pnas.1912247116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos, J. B. 2008. “Phylogenetic Niche Conservatism, Phylogenetic Signal and the Relationship Between Phylogenetic Relatedness and Ecological Similarity Among Species.” Ecology Letters 11, no. 10: 995–1003. 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- Luong, J. C. , Holl K. D., and Loik M. E.. 2021. “Leaf Traits and Phylogeny Explain Plant Survival and Community Dynamics in Response to Extreme Drought in a Restored Coastal Grassland.” Journal of Applied Ecology 58, no. 8: 1670–1680. 10.1111/1365-2664.13909. [DOI] [Google Scholar]
- Luong, J. C. , Press D. M., and Holl K. D.. 2023. “Lessons Learned From an Interdisciplinary Evaluation of Long‐Term Restoration Outcomes on 37 Restored Coastal Grasslands in California.” Biological Conservation 280: 109956. 10.1016/j.biocon.2023.109956. [DOI] [Google Scholar]
- Maitner, B. S. , Boyle B., Casler N., et al. 2018. “The BIEN R Package: A Tool to Access the Botanical Information and Ecology Network (BIEN) Database.” Methods in Ecology and Evolution 9, no. 2: 373–379. 10.1111/2041-210x.12861. [DOI] [Google Scholar]
- Manning, G. C. , and Baer S. G.. 2018. “Interannual Variability in Climate Effects on Community Assembly and Ecosystem Functioning in Restored Prairie.” Ecosphere 9, no. 6: ecs2.2327. 10.1002/ecs2.2327. [DOI] [Google Scholar]
- McKone, M. J. , Williams E. W., and Beck J. J.. 2021. “Evidence for Local‐Scale Community Assembly Processes From Long‐Term Observations of Biodiversity in a Grassland Chronosequence.” Journal of Vegetation Science 32, no. 4: e13065. 10.1111/jvs.13065. [DOI] [Google Scholar]
- McLachlan, S. M. , and Knispel A. L.. 2005. “Assessment of Long‐Term Tallgrass Prairie Restoration in Manitoba, Canada.” Biological Conservation 124, no. 1: 75–88. 10.1016/j.biocon.2005.01.014. [DOI] [Google Scholar]
- Mcmillan, C. 1959. “The Role of Ecotypic Variation in the Distribution of the Central Grassland of North America.” Ecological Monographs 29, no. 4: 285–308. 10.2307/1942132. [DOI] [Google Scholar]
- Mendola, M. L. , Baer S. G., Johnson L. C., and Maricle B. R.. 2015. “The Role of Ecotypic Variation and the Environment on Biomass and Nitrogen in a Dominant Prairie Grass.” Ecology 96, no. 9: 2433–2445. 10.1890/14-1492.1. [DOI] [PubMed] [Google Scholar]
- Messier, J. , McGill B. J., and Lechowicz M. J.. 2010. “How Do Traits Vary Across Ecological Scales? A Case for Trait‐Based Ecology.” Ecology Letters 13, no. 7: 838–848. 10.1111/j.1461-0248.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- Midolo, G. , Kuss P., and Wellstein C.. 2021. “Land Use and Water Availability Drive Community‐Level Plant Functional Diversity of Grasslands Along a Temperature Gradient in the Swiss Alps.” Science of the Total Environment 764: 142888. 10.1016/j.scitotenv.2020.142888. [DOI] [PubMed] [Google Scholar]
- Miller, J. E. D. , Li D., LaForgia M., Harrison S., and Jones H.. 2019. “Functional Diversity Is a Passenger but Not Driver of Drought‐Related Plant Diversity Losses in Annual Grasslands.” Journal of Ecology 107, no. 5: 2033–2039. 10.1111/1365-2745.13244. [DOI] [Google Scholar]
- Mount, H. E. , Smith M. D., Knapp A. K., et al. 2024. “Drought‐Tolerant Grassland Species Are Generally More Resistant to Competition.” Journal of Ecology 112, no. 2: 416–426. 10.1111/1365-2745.14243. [DOI] [Google Scholar]
- NCEI . 2019. “U.S. Climate Normals (Version 1.0.1).” https://www.ncei.noaa.gov/products/land‐based‐station/us‐climate‐normals/.
- Packard, S. , and Mutel C. F.. 2005. The Tallgrass Restoration Handbook: For Prairies, Savannas, and Woodlands. Washington, DC: Island Press. [Google Scholar]
- Pavoine, S. , Baguette M., and Bonsall M. B.. 2010. “Decomposition of Trait Diversity Among the Nodes of a Phylogenetic Tree.” Ecological Monographs 80, no. 3: 485–507. 10.1890/09-1290.1. [DOI] [Google Scholar]
- Pavoine, S. , and Bonsall M. B.. 2011. “Measuring Biodiversity to Explain Community Assembly: A Unified Approach.” Biological Reviews 86, no. 4: 792–812. 10.1111/j.1469-185X.2010.00171.x. [DOI] [PubMed] [Google Scholar]
- Petchey, O. L. , and Gaston K. J.. 2006. “Functional Diversity: Back to Basics and Looking Forward.” Ecology Letters 9, no. 6: 741–758. 10.1111/j.1461-0248.2006.00924.x. [DOI] [PubMed] [Google Scholar]
- Purschke, O. , Schmid B. C., Sykes M. T., et al. 2013. “Contrasting Changes in Taxonomic, Phylogenetic and Functional Diversity During a Long‐Term Succession: Insights Into Assembly Processes.” Journal of Ecology 101, no. 4: 857–866. 10.1111/1365-2745.12098. [DOI] [Google Scholar]
- R Core Team . 2020. R: A Language and Environment for Statistical Computing. R Version 4.0.2. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ren, Z. , Baer S. G., Johnson L. C., Galliart M. B., Wilson L. R., and Gibson D. J.. 2023. “The Role of Dominant Prairie Species Ecotypes on Plant Diversity Patterns of Restored Grasslands Across a Rainfall Gradient in the US Great Plains.” Applied Vegetation Science 26, no. 2: e012725. 10.1111/avsc.12725. [DOI] [Google Scholar]
- Risser, P. G. , Birney E., Blocker H., May S., Parton W. J., and Wiens J. A.. 1981. “Grasslands Across North America.” In The True Prairie Ecosystem, 9–18. Stroudsburg, PA: Hutchinson Ross Publishing Company. [Google Scholar]
- Sandel, B. , Pavelka C., Hayashi T., et al. 2021. “Predicting Intraspecific Trait Variation Among California's Grasses.” Journal of Ecology 109, no. 7: 2662–2677. 10.1111/1365-2745.13673. [DOI] [Google Scholar]
- Siefert, A. , Violle C., Chalmandrier L., et al. 2015. “A Global Meta‐Analysis of the Relative Extent of Intraspecific Trait Variation in Plant Communities.” Ecology Letters 18, no. 12: 1406–1419. 10.1111/ele.12508. [DOI] [PubMed] [Google Scholar]
- Smith, S. A. , and Brown J. W.. 2018. “Constructing a Broadly Inclusive Seed Plant Phylogeny.” American Journal of Botany 105, no. 3: 302–314. 10.1002/ajb2.1019. [DOI] [PubMed] [Google Scholar]
- Smith, M. D. , Koerner S. E., Knapp A. K., et al. 2020. “Mass Ratio Effects Underlie Ecosystem Responses to Environmental Change.” Journal of Ecology 108, no. 3: 855–864. 10.1111/1365-2745.13330. [DOI] [Google Scholar]
- Spasojevic, M. J. , and Suding K. N.. 2012. “Inferring Community Assembly Mechanisms From Functional Diversity Patterns: The Importance of Multiple Assembly Processes.” Journal of Ecology 100, no. 3: 652–661. 10.1111/j.1365-2745.2011.01945.x. [DOI] [Google Scholar]
- Swenson, N. G. 2014. “Functional Diversity.” In Functional and Phylogenetic Ecology in R. New York, NY: Springer. 10.1007/978-1-4614-9542-0_4. [DOI] [Google Scholar]
- Swenson, N. G. , Enquist B. J., Pither J., et al. 2012. “The Biogeography and Filtering of Woody Plant Functional Diversity in North and South America.” Global Ecology and Biogeography 21, no. 8: 798–808. 10.1111/j.1466-8238.2011.00727.x. [DOI] [Google Scholar]
- Swenson, N. G. , and Weiser M. D.. 2010. “Plant Geography Upon the Basis of Functional Traits: An Example From Eastern North American Trees.” Ecology 91, no. 8: 2234–2241. 10.1890/09-1743.1. [DOI] [PubMed] [Google Scholar]
- Taudiere, A. , and Violle C.. 2016. “Cati: An R Package Using Functional Traits to Detect and Quantify Multi‐Level Community Assembly Processes.” Ecography 39, no. 7: 699–708. 10.1111/ecog.01433. [DOI] [Google Scholar]
- Tilman, D. , and El Haddi A.. 1992. “Drought and Biodiversity in Grasslands.” Oecologia 89, no. 2: 257–264. 10.1007/BF00317226. [DOI] [PubMed] [Google Scholar]
- Twidwell, D. , Rogers W. E., McMahon E. A., Thomas B. R., Kreuter U. P., and Blankenship T. L.. 2012. “Prescribed Extreme Fire Effects on Richness and Invasion in Coastal Prairie.” Invasive Plant Science and Management 5, no. 3: 330–340. 10.1614/IPSM-D-12-00017.1. [DOI] [Google Scholar]
- Violle, C. , Enquist B. J., McGill B. J., et al. 2012. “The Return of the Variance: Intraspecific Variability in Community Ecology.” Trends in Ecology & Evolution 27, no. 4: 244–252. 10.1016/j.tree.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Vogel, K. P. , Johnson K. D., Carlson I. T., and Schmer M. R.. 2018. “Big Bluestem and Indiangrass From Remnant Prairies: Plant Biomass and Adaptation.” Crop Science 58, no. 2: 728–738. 10.2135/cropsci2017.09.0572. [DOI] [Google Scholar]
- Volf, M. , Redmond C., Albert A. J., et al. 2016. “Effects of Long‐ and Short‐Term Management on the Functional Structure of Meadows Through Species Turnover and Intraspecific Trait Variability.” Oecologia 180, no. 4: 941–950. 10.1007/s00442-016-3548-y. [DOI] [PubMed] [Google Scholar]
- Vu, V. Q. 2011. “Ggbiplot: A ggplot2 Based Biplot.” https://github.com/friendly/ggbiplot.
- Webb, C. O. , Ackerly D. D., McPeek M. A., and Donoghue M. J.. 2002. “Phylogenies and Community Ecology.” Annual Review of Ecology and Systematics 33: 475–505. 10.1146/annurev.ecolsys.33.010802.150448. [DOI] [Google Scholar]
- Wickman, J. , Koffel T., and Klausmeier C. A.. 2023. “A Theoretical Framework for Trait‐Based Eco‐Evolutionary Dynamics: Population Structure, Intraspecific Variation, and Community Assembly.” American Naturalist 201, no. 4: 501–522. 10.1086/723406. [DOI] [PubMed] [Google Scholar]
- Wilson, L. R. , Gibson D. J., Baer S. G., and Johnson L. C.. 2016. “Plant Community Response to Regional Sources of Dominant Grasses in Grasslands Restored Across a Longitudinal Gradient.” Ecosphere 7, no. 4: ecs2.1329. 10.1002/ecs2.1329. [DOI] [Google Scholar]
- Yahdjian, L. , and Sala O. E.. 2002. “A Rainout Shelter Design for Intercepting Different Amounts of Rainfall.” Oecologia 133, no. 2: 95–101. 10.1007/s00442-002-1024-3. [DOI] [PubMed] [Google Scholar]
- Young, D. J. , Porensky L. M., Wolf K. M., Fick S. E., and Young T. P.. 2015. “Burning Reveals Cryptic Plant Diversity and Promotes Coexistence in a California Prairie Restoration Experiment.” Ecosphere 6, no. 5: 1–11. 10.1890/ES14-00303.1. [DOI] [Google Scholar]
- Yue, K. , Jarvie S., Senior A. M., et al. 2020. “Changes in Plant Diversity and Its Relationship With Productivity in Response to Nitrogen Addition, Warming and Increased Rainfall.” Oikos 129, no. 7: 939–952. 10.1111/oik.07006. [DOI] [Google Scholar]
- Zeldin, J. , Lichtenberger T. M., Foxx A. J., Williams E. W., and Kramer A. T.. 2020. “Intraspecific Functional Trait Structure of Restoration‐Relevant Species: Implications for Restoration Seed Sourcing.” Journal of Applied Ecology 57, no. 5: 864–874. 10.1111/1365-2664.13603. [DOI] [Google Scholar]
- Zhang, J. , Zuo X., Lv P., Zhao S., and Zhao X.. 2019. “Plant Functional Trait Response to Habitat Change and Grazing in a Semiarid Grassland: Unravelling Species Turnover and Intraspecific Variation Effects.” Polish Journal of Ecology 67, no. 1: 62–74. 10.3161/15052249PJE2019.67.1.005. [DOI] [Google Scholar]
- Zuo, X. , Zhao S., Cheng H., et al. 2021. “Functional Diversity Response to Geographic and Experimental Precipitation Gradients Varies With Plant Community Type.” Functional Ecology 35, no. 9: 2119–2132. 10.1111/1365-2435.13875. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.
Data Availability Statement
The data and codes used in this study are available with the corresponding Supporting Information.


