Abstract
Cellular differentiation is governed by changes in gene expression, but at the same time, a cell's identity needs to be maintained through multiple cell divisions during maturation. In myeloid cell lines, retinoids induce gene expression and a well-characterized two-step lineage-specific differentiation. To identify mechanisms that contribute to cellular transcriptional memory, we analyzed the epigenetic changes taking place on regulatory regions of tissue transglutaminase, a gene whose expression is tightly linked to retinoid-induced differentiation. Here we report that the induction of an intermediary or “primed” state of myeloid differentiation is associated with increased H4 arginine 3 and decreased H3 lysine 4 methylation. These modifications occur before transcription and appear to prime the chromatin for subsequent hormone-regulated transcription. Moreover, inhibition of methyltransferase activity, preacetylation, or activation of the enzyme PAD4 attenuated retinoid-regulated gene expression, while overexpression of PRMT1, a methyltransferase, enhanced retinoid responsiveness. Taken together, our results suggest that H4 arginine 3 methylation is a bona fide positive epigenetic marker and regulator of transcriptional responsiveness as well as a signal integration mechanism during cell differentiation and, as such, may provide epigenetic memory.
The HL-60 cell line is a well-characterized M2 myeloid leukemia cell line (6) that can be induced to undergo differentiation along different pathways, myeloid versus monocytic, in response to a variety of physiological and pharmacological stimuli. The process of myeloid differentiation itself involves two distinct and sequential steps. The first is an identifiable intermediate state termed the precommitment or primed state, and the second is a series of late events that lead to the onset of lineage-specific terminal differentiation (55-58). Primed cells are characterized by altered nuclear structure and feature retention of phenotype, a form of cellular memory that can last for several cell cycles. Most significantly, primed cells require only an abbreviated exposure to an appropriate inducer, such as all-trans-retinoic acid, for onset of G1/0-specific growth arrest and phenotypic differentiation (55). For example, a short exposure (16 to 24 h) to dimethyl sulfoxide (DMSO) or vitamin D induces a state of precommitment in HL-60 cells. Subsequent exposure of these precommitted cells to another inducer, such as all-trans-retinoic acid, results in the onset of growth arrest and differentiation that is much more rapid (<24 h) than for cells that have not been subject to precommitment (>48 h). In this example, DMSO exposure results in the acquisition of a precommitment memory, a state which can be sustained for more than one cell cycle (55). These experimental observations have been explained by suggesting a two-step model for induction of terminal differentiation in the cells. In this model, early events preceding precommitment regulate growth arrest, and late events that occur after precommitment regulate the choice of a specific differentiation lineage. Interestingly, both events occur while the cells are still proliferating (57). Very little is known about the molecular characteristics of the precommitment or primed state, and in particular, very little is known about how these molecular changes impact the regulation of gene expression.
One mechanism that is an obvious candidate to explain the generation of a primed state of transcriptional memory is the alteration in chromatin structure. The structure of the chromatin plays a fundamental role in regulating gene expression by controlling the access of transcription factors to the regulatory regions of genes. Two classes of enzymes are known to play a role in regulating chromatin structure. The ATP-dependent chromatin-remodeling enzymes act by the sliding of DNA and the triggering of conformational changes of the nucleosomes (20). The other class of chromatin-modifying enzymes is responsible for the posttranslational modification of histone tails. According to the “histone code” hypothesis, the covalent modifications of the histone tails are responsible for the sustained maintenance and modulation of patterns of gene expression (25, 45, 49, 50). Modified histone tails have been reported to form binding sites of specific classes of proteins: bromodomain-containing proteins bind to histone tails with acetylated lysine residues, while chromodomain-containing proteins bind to methylated histone lysine residues. It is thought that these histone binding proteins form a functional link between the covalently modified histone tails and the effectors of transcription initiation (32).
To investigate the molecular mechanisms that may be associated with the priming of HL-60 cells and the establishment of a precommitted state, we have characterized the covalent modification of the tails of histones specifically associated with the promoter of tissue transglutaminase, a gene that undergoes marked up-regulation during retinoid-induced myeloid cell differentiation.
We used chromatin immunoprecipitation combined with real-time PCR to quantify the level of histone acetylation and methylation on the nucleosomes associated with the proximal regions of the tissue transglutaminase promoter. We found that acetylation of histone 4 (H4) is correlated with increases in gene expression. Changes in methylation on histone H3 lysine 4 (H3K4) and histone H4 arginine 3 (H4R3) are linked to the presence of the primed state of differentiation and increased hormonal responsiveness. In an attempt to establish more-mechanistic links between H4R3 methylation and retinoid responsiveness, we evaluated the role of an enzyme pair shown to be responsible for the methylation and demethylation (citrullination) of H4R3 (53). We found that inhibition of methylation by preacetylation or activation of PAD4 by Ca2+ ionophores attenuated retinoid responsiveness and overexpression of PRMT1 enhanced it.
MATERIALS AND METHODS
Cells and materials.
HL-60/CDM-1 cells, a kind gift of Diane Lucas (Walter Reed Army Medical Center, Washington, D.C.), were cultured in suspension in RPMI 1640 medium supplemented with ITS (I1884; Sigma) using standard cell culture conditions. Cells were treated with 9-cis retinoic acid at a concentration of 1 μM dissolved in ethanol-DMSO. Priming of cells was done with 1.25% DMSO for 16 h. Blocking of methyltransferases was achieved by treatment with 10 μM adenosine dialdehyde (ADOX, A7154;Sigma) for the same period. After pretreatment, medium was replaced with fresh medium and retinoic treatment was carried out. If not specified, all materials were purchased from Sigma. Antibodies for flow cytometric analysis, if not otherwise mentioned, were purchased from DAKO A/S, as were the appropriate control antibodies.
Plasmids.
Full-length wild-type pcDNA3.1 human PRMT1 was cloned from the pGEX-HRMT1L2 (v2) (43) by PCR introducing a BamHI site at the 5′ end and an EcoRI site at the 3′ end of the HMRT1L2 cDNA (deleting the stop at the same time). Subsequently, the HMRT1L2 cDNA was subcloned with BamHI and EcoRI into pcDNA3.1-B (Invitrogen), generating a C-terminal myc/His tag. The point mutations S69A, G70A, and T71A were introduced into pcDNA3.1 human PRMT1 via site-directed mutagenesis using the QuikChange kit (Stratagene) to give rise to the pcDNA3.1 PRMT1 full-length catalytic mutant. All constructs were verified by DNA sequencing. Mammalian expression vectors for PAD4 and the PAD4 C645S mutant were received from Yanming Wang and described previously (53). All other plasmids used were described previously (4).
Transfection.
HL-60 cells were transfected with the AMAXA electroporator system using 1 μg of plasmid for 2 million cells according to the manufacturer's instructions (electroporation solution V and program V01). 293T cells were transfected with jetPEI reagent (Qbiogene) according to the manufacturer's instructions. The βRARE thymidine kinase Luc reporter plasmid was cotransfected with full-length pCMV retinoic acid receptor α (RARα), full-length pCMV retinoic X receptor α (RXRα), and the β-galactosidase plasmid. Wild-type PRMT1, mutant PRMT1, wild-type PAD4, and mutant PAD4 were added to the reporter plasmids.
Extraction of total RNA.
Total RNA was extracted by using Trizol reagent (Invitrogen) according to the manufacturer's instructions.
Real-time QPCR.
Real-time quantitative PCR (QPCR) was carried out on ABI7700 and ABI7900 real-time sequence detection systems. QPCR measurements were performed as described previously (51). For the TGM2 promoter assays, the reactions were carried out similarly without the reverse transcription step. For standard calibration, DNA from the bacterial artificial chromosome clone RP5-1054A22 was used which contained the whole promoter of TGM2 and was received from the Sanger Centre, Clone Resources Group, Hinxton, United Kingdom.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was carried out as described by Kuo and Allis (29) with modifications. Briefly, cells were fixed with 1% formaldehyde for 10 min at room temperature. Fixation was stopped by adding chilled glycine to a final concentration of 150 mM. Cells were scraped and washed twice with ice-cold phosphate-buffered saline (PBS) that contained proteinase inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/μl aprotinin, and 1 μg/μl pepstatin A). Nuclei were prepared by incubation for 10 min on ice in a buffer containing 5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 8, 85 mM KCl, 0.5% NP-40, and proteinase inhibitors. After centrifugation at 3,000 × g for 10 min at 4°C, nuclei were resuspended in sonication buffer (1% sodium dodecyl sulfate [SDS], 0.1 M NaHCO3, and proteinase inhibitors), lysed on ice for 10 min, and sonicated on ice to an average fragment size of 300 base pairs. Cell debris was pelleted twice by centrifugation at 10,000 × g for 30 min at 4°C in a bench-top centrifuge. Soluble chromatin was aliquoted, frozen in liquid nitrogen, and stored at −70°C. For immunoprecipitation, chromatin was diluted 10-fold in an immunoprecipitation (IP) buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.1, 16.7 mM NaCl, and proteinase inhibitors). One milliliter of diluted chromatin was precleared twice with 40 μl and blocked with protein A-Sepharose beads. Immunoprecipitation was carried out with specific antibodies purchased from Upstate Biotech and Abcam against modified histones as follows: from Upstate, anti-acetyl H4 (2 μl/IP, catalog no. 06-866), anti-dimethyl H4 Arg 3 (6 μl/IP, catalog no. 07-213), anti-dimethyl H3 Lys 4 (5 μl/IP, catalog no. 07-030), anti-dimethyl H3 Lys 9 (5 μl/IP, catalog no. 07-212); from Abcam, H4 methyl R3 antibody (5 μl/IP, catalog no. ab5823), pan dimethyl arginine (5 μl/IP, catalog no. 413-200). Incubation with the antibodies was carried out overnight on a rotating plate at 4°C. Complexes were collected with 40 μl blocked protein A-agarose (catalog no. 16-157; Upstate). An aliquot of the no-antibody control supernatant was used to measure and calculate the input DNA. Beads were pelleted and washed twice with each of the following buffers: buffer A (low salt) (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, 150 mM NaCl, and propidium iodide [PI]), buffer B (high salt) (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8.1, 500 mM NaCl, and PI), buffer C (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris, pH 8.1, and PI), and TE buffer (10 mM Tris, 10 mM EDTA, pH 8, and PI). Immunoprecipitated nucleosomes were eluted twice from beads with elution buffer (1% SDS, 0.1 M NaHCO3), and eluates were combined. Cross-links were reversed by incubation for 6 h at 65°C after the addition of 20 μl 5 M NaCl. The eluate was combined with 10 μl of 0.5 M EDTA, 20 μl 1 M Tris, pH 6.5, and 2 μg proteinase K and incubated for 1 h at 45°C. DNA was recovered after phenol-chloroform extraction and ethanol precipitation using 20 μg of glycogen as a carrier. DNA was resuspended in 50 μl of 50 ng/μl yeast tRNA (catalog no. 15401-011; Invitrogen). Two microliters of this solution was used for real-time QPCR in a 25-μl reaction mixture. All measurements were done in triplicate. All chromatin results were verified from independent chromatin preps.
Cell cycle analysis.
Cell cycle analysis was performed as follows: cells were washed in PBS and fixed in 70% ethanol overnight. Briefly, cells were washed in PBS and fixed in 70% ethanol overnight. Fixed cells were then washed twice and resuspended in PI working solution (50 μg of propidium iodide, 20 μg/ml RNase, and 0.5% Tween-20 in PBS). After a 15-min incubation at 37°C, cells were analyzed on a Coulter flow cytometer and data were analyzed with WinMDI software.
Intracellular staining and flow cytometry.
Cells were fixed (2% paraformaldehyde, 0.1% sodium azide, 1% heat-inactivated filtered fetal bovine serum) at room temperature for 20 min. After washing, cells were permeabilized with buffer P (PBS, 2% fetal bovine serum, 0.2% sodium azide, 0.5% saponin) for 10 min on ice. Immunostaining was carried out with a mouse monoclonal antibody which specifically recognizes the TGM2 protein (a kind gift of Laszlo Fesus, University of Debrecen) and an isotype control antibody. As a secondary antibody, fluorescein isothiocyanate-conjugated anti-mouse antibody was used. All staining procedures were done in saponin-containing buffers. Flow cytometry analysis was done in buffer P as described above.
Microarray analysis of gene expression patterns.
Total RNA was extracted as described above. RNA was labeled with Cy3 and Cy5 and hybridized to a microarray containing 3,200 human genes as described previously (42). Data processing and statistical analysis were done as described previously (41).
Protein expression pattern analysis.
Protein expression pattern analysis was performed as described previously (31). Briefly, 150 μg of total protein was run on parallel gels. After isoelectric focusing between pH 3 and 10, samples were run on an 11% SDS gel. Gels were silver stained and analyzed as described previously (31) using Phoretix 5.01 software (NonLinear Dynamics Ltd., Newcastle upon Tyne, United Kingdom).
Quantification of DNase I sensitivity.
Quantification of DNase I sensitivity was performed as described previously (33) with modifications. Briefly, cells were washed with ice-cold PBS resuspended in lysis buffer (50 mM Tris-Cl pH 7.9, 100 KCl, 5 mM MgCl2, 0.05% saponin, 50% glycerol, 200 mM β-mercaptoethanol) and incubated on ice for 10 min. Nuclei were recovered by centrifugation at 1,300 × g for 15 min at 4°C and resuspended in buffer A (50 mM Tris-Cl, pH 7.9, 100 mM NaCl, 3 mM MgCl2 1 mM dithiothreitol, and proteinase inhibitors). After centrifugation at 1,300 × g for 15 min at 4°C, nuclei were resuspended in buffer A and divided into several aliquots. After this step, nuclei were treated with different concentrations of DNase I for 20 min at 37°C. The reaction was stopped with a 1/10 volume of 0.5 M EDTA. After RNase and proteinase K digestion, DNA was extracted with phenol-chloroform and precipitated with absolute ethanol. Extracted DNA was treated with EcoRI, purified with PCR purification columns (QIAGEN), and measured by QPCR for the specific promoter regions. Values were normalized to the total DNA concentration in each sample as measured with a photometer (A260 and A280).
RESULTS
Retinoid regulation of tissue transglutaminase gene expression in naive and primed myeloid leukemia cells.
The expression of tissue transglutaminase type 2 (TGM2) is very tightly regulated in myeloid leukemia cells. In HL-60 cells, in the absence of exposure to retinoids, the level of TGM2 mRNA is below the limits of detection of a sensitive real-time reverse transcription (RT)-QPCR assay (less than 10 copies per nanogram of total RNA). Exposure of cells to either natural or synthetic retinoid receptor agonists increases transglutaminase gene expression markedly (17). Priming of the cells by pretreatment with differentiating agents such as vitamin D or the polar-planar solvent DMSO increases retinoid-induced TGM2 expression. This experimental system (Fig. 1A) has allowed us to study the effects of priming on the expression of a specific gene, the induction of which is increased by the process of precommitment. We will refer to the unprimed and uncommitted cells as “naive” and those that advanced to precommitment as “primed.”
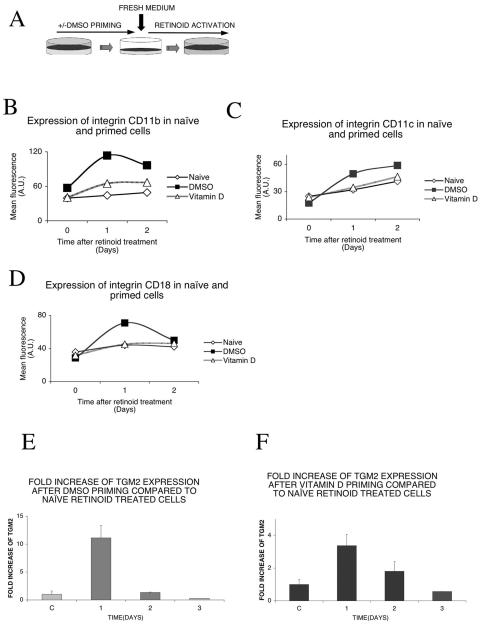
FIG. 1.
Priming enhances retinoid response in HL-60 cells. (A) Experiment outline. HL-60/CDM-1 cells were primed with 1.25% DMSO overnight. The control (naive) cells received no priming. After overnight incubation, cells were washed, resuspended in fresh medium, and treated with 1 μM 9-cis retinoic acid. (B) Priming with DMSO or vitamin D enhances 9-cis retinoic acid-dependent expression of integrin CD11b. Cells were primed with 1.25% DMSO or 100 nM vitamin D. After overnight incubation and washing out of the priming agent, cells were treated with 1 μM 9-cis retinoic acid and analyzed on a flow cytometer at the indicated time points as described in Materials and Methods. Isotype controls had no change (not shown). (C) Priming with DMSO or, to a lesser extent, with vitamin D enhances 9-cis retinoic acid-dependent expression of integrin CD11c. Cells were primed with 1.25% DMSO or 100 nM vitamin D. After overnight incubation and washing out of the priming agent, cells were treated with 1 μM 9-cis retinoic acid and analyzed with a flow cytometer at the indicated time points as described in Materials and Methods. Isotype controls had no change (not shown). (D) Priming with DMSO but not with vitamin D enhances 9-cis retinoic acid-dependent expression of integrin CD18. Cells were primed with 1.25% DMSO or 100 nM vitamin D. After overnight incubation and washing out of the priming agent, cells were treated with 1 μM 9-cis retinoic acid and analyzed with a flow cytometer at the indicated time points as described in Materials and Methods. Isotype controls had no change (not shown). (E) Relative increase of TGM2 mRNA induction compared to that of naive, retinoid-treated cells. Assessment of the maintenance in time of the priming effect: TGM2 mRNA was measured by real-time QPCR after priming with 1.25% DMSO and compared to naive, retinoid-treated cells. After overnight incubation and washing out of the priming agent, cells were treated with 1 μM 9-cis retinoic acid at different time points after the priming, as indicated. The retinoid treatment was 12 h long in all cases. Values are the means of the results from three independent QPCR measurements ± standard deviations. (F) Relative increase of TGM2 induction compared to that of naive, retinoid-treated cells. Assessment of the maintenance in time of the priming effect: TGM2 mRNA was measured by real-time QPCR after priming with 100 nM vitamin D and compared to naive, retinoid-treated cells. After overnight incubation and washing out of the priming agent, cells were treated with 1 μM 9-cis retinoic acid at different time points after the priming, as indicated. The retinoid treatment was 12 h long in all cases. Values are the means of the results from three independent QPCR measurements ± standard deviations.
Priming enhances retinoid response in HL-60 cells.
To study the effect of priming on the differentiation of HL-60 cells, we analyzed the expression of a set of differentiation-linked surface markers. Cells were primed with DMSO or vitamin D. The priming agent was washed out, and cells were treated with 9-cis retinoic acid. The cell surface expression of integrin molecules CD18 or integrin β2 and its heterodimeric partners CD11b and CD11c or integrin alpha M and integrin alpha X showed an increased expression after retinoid treatment in primed cells if compared to naive cells (Fig. 1B, C, and D). An increased expression could be detected in the case of all three markers with DMSO and for CD11b with vitamin D priming.
The priming effect of both DMSO and vitamin D pretreatment was detectable on the retinoid response of tissue transglutaminase type 2, a molecular marker of myeloid cell differentiation and a direct target of retinoid receptors (37). As seen on Fig. 1E, priming with DMSO produced a marked increase in the expression of TGM2. To determine whether priming conferred some sort of transcriptional memory, we analyzed the expression of tissue transglutaminase type 2 after the priming agent was washed out and the cells were treated with 9-cis retinoic acid after 1, 2, or 3 days. The priming effect of DMSO proved to be transient and declined rapidly (Fig. 1E). Similarly, in the case of vitamin D priming, an increase in the expression of TGM2 was detected but with a smaller amplitude (Fig. 1F). This effect decreased in 2 days to the level of naive cells. These experiments showed that the priming effect is transient, lasting for 24 to 48 h. Since both of these agents, DMSO and vitamin D, produced similar effects, we decided to carry out our experiments with the more potent priming agent available, DMSO.
To provide a baseline for our further studies we determined the time course of mRNA induction. Exposure of “naive” HL-60 cells to retinoids (9-cis retinoic acid) results in a very rapid (<2 h) increase in transglutaminase gene expression (Fig. 2A) that reached a plateau after 12 h (data not shown). This induction is strikingly enhanced (approximately 100-fold) upon DMSO priming (Fig. 2A and B). DMSO alone does not increase TGM2 expression (Fig. 2A and B). It was apparent by comparing the induction in the naive and primed cells that, while the magnitude of the induction was very different (Fig. 2B), the kinetics was very similar whether or not the cells had been primed (Fig. 2A). Thus, priming resulted in a state characterized by greater induction of the target gene without an alteration in the time course of the transcriptional response.
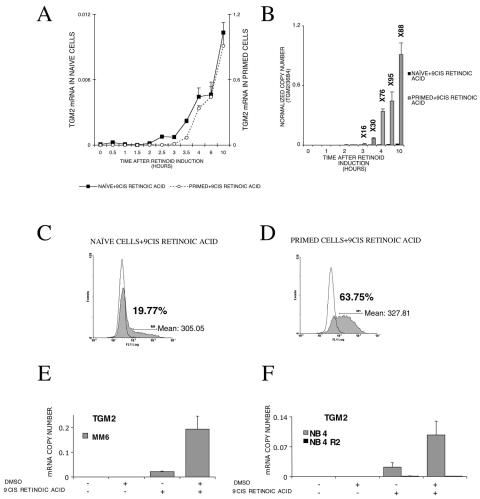
FIG. 2.
TGM2 expression is enhanced by DMSO priming at both the RNA and protein levels, and this effect is not cell type specific. (A) 9-cis retinoic acid induction of TGM2 mRNA as measured by real-time QPCR. The expression of TGM2 mRNA in primed and naive cells is shown on the same graph with two different y axes with a difference of 2 orders of magnitude of the scales. Transcript copy numbers were normalized to 36B4 transcript levels. Values are the means of the results from three independent QPCR measurements ± standard deviations. (B) Relative difference in gene expression in naive and primed cells. Fold changes are indicated above the bars. Copy numbers of TGM2 mRNA are determined by real-time QPCR and normalized to 36B4 transcript levels. Values are the means of the results from three independent QPCR measurements ± standard deviations. (C) Intracellular immunostaining and flow cytometric analysis of retinoid-treated HL-60 cells as described in Materials and Methods. The expression of TGM2 protein in naive cells (shaded) and its isotype control (white) after retinoid induction. The percentage of cells expressing TGM2 protein is shown on the graph along with the mean fluorescence intensity of the positive cells. Values are expressed in arbitrary units (AU). (D) Intracellular immunostaining and flow cytometric analysis of retinoid-treated HL-60 cells as described in Materials and Methods. TGM2 expression in DMSO-primed cells (shaded) along with its isotype control (white). The percentage of cells expressing TGM2 is shown on the graph along with the mean fluorescence intensity of the positive cells, expressed in arbitrary units (AU). At least three independent determinations have been carried out. Data from a representative experiment are shown. (E) Effect of priming upon TGM2 induction by retinoids in MonoMac6 (MM6) cells. TGM2 copy numbers were normalized to cyclophilin transcript levels. Values are the means of the results from three independent QPCR measurements ± standard deviations. (F) TGM2 induction in NB4 and RARα mutant NB4R2 cells in naive and primed states. The mRNA copy numbers were normalized to cyclophilin transcript levels. Values are the means of the results from three parallel QPCR measurements ± standard deviations, and results were confirmed from at least three independent biological samples. +, present; −, absent.
A key question is whether increased induction of transglutaminase expression in the primed cells is a result of a higher mRNA expression level in each individual cell or is due to an increase in the fraction of the cells responding to the inducer (retinoid). Since it has already been demonstrated that there is a correlation between TGM2 protein and mRNA levels in HL-60 cells (9, 10, 14-16), we addressed the issue of the induction of the enzyme by using a coupled immunohistochemical/flow cytometric analysis to evaluate the levels of TGM2 in individual cells prior to and following retinoid treatment. Using these techniques, the basal level of TGM2 was undetectable in both untreated HL-60 cells and HL-60 cells treated with DMSO alone (data not shown). In naive cells (as shown in Fig. 2C) 9-cis retinoic acid induced detectable levels of TGM2 in 19.7% of the cells. The level of TGM2 was normally distributed in the population of cells with a mean fluorescence intensity of 305.05 arbitrary units (AU). When DMSO-primed cells were treated with 9-cis retinoic acid, a much greater fraction of the population responded to the retinoid stimulus than did the naive, retinoid-treated cells. Among primed cells, 63.7% had detectable levels of TGM2 protein (Fig. 2D) and this also showed a normal distribution. The mean fluorescence intensity in primed, retinoid-treated cells was not different (327.8 AU) from the level of fluorescence intensity in retinoid-treated naive cells (305.0 AU). The mean fluorescence intensity of the entire cell population increased from 39.6 to 179.43 AU after DMSO priming and retinoid induction. The most likely explanation for these findings is that the maximal level of TGM2 expression by individual cells did not change after DMSO priming, but more cells gained competence to respond to retinoids with increased TGM2 expression. It appears that priming is likely to lower the threshold for induction of gene expression and differentiation, resulting in a larger portion of cells able to respond. This is consistent with our previous findings shown in Fig. 2A, namely that priming affects the amplitude but not the dynamics of the response to retinoic acid.
Role of receptor levels and cell cycle distribution of cells in enhanced retinoid response.
To investigate possible explanations for the increased frequency of response by primed cells, we tested two obvious hypotheses. The first is that the effect of priming is to increase the level of expression of retinoid receptors in individual cells. To address this hypothesis, we measured the levels of transcripts of RARα, -β, and -γ and RXRα and γ in naive and DMSO-primed HL-60 cells. There was no significant change in the expression level of RARα and there was only a slight increase (less than twofold, as measured by real-time RT-QPCR assay) in the level of RXRα mRNA as the result of DMSO priming (data not shown). There was also no change in the levels of RARβ and RXRγ mRNA (data not shown). The second is that differentiation is linked to cell cycle arrest. We considered it possible that the change in the fraction of cells responding to retinoids following DMSO priming was linked to alterations in the distribution of cells in different stages of the cell cycle. It has previously been shown that priming HL-60 cells with DMSO for a period shorter than 24 h does not significantly change its cell cycle distribution (58), but to be sure that something different was not happening in our cell population, we compared the distribution of cells in naive and DMSO-primed populations. Our results confirmed the previously reported findings, DMSO priming had only a minor effect on the distribution of cells: there was a slight increase in the number of cells in G1 (51% to 63%) that was compensated for by slight decreases in the fraction of cells in the S and M phases (from 27% to 20% and from 22% to 17%, respectively) (data not shown).
To test whether the effect of priming on retinoid-regulated gene expression was restricted to HL-60 cells, we carried out comparable studies in a human macrophage-like cell line (monocytic leukemia cell line, FAB M5/MonoMac6 or MM6) and in the well-characterized NB4 and NB4R2 promyelocytic leukemia (FAB M3) cell lines. The two NB4 cell lines also allowed us to address the issue of retinoid receptor dependence. As shown in Fig. 2E, we have found that retinoid-induced expression of TGM2 was increased in DMSO-primed MM6 cells compared to unprimed cells. In the NB4 cells, DMSO priming also increased retinoid-induced TGM2 expression (Fig. 2F). The NB4R2 cells have a mutation in the ligand binding domain of RARα that disrupts retinoid signaling (18). In this cell line, TGM2 could not be induced after retinoid treatment either in naive or in DMSO-primed cells (Fig. 2F). These findings demonstrated that the effects of priming on retinoid-regulated gene expression are not unique to HL-60 cells but represent a generalized phenomenon at least among human myeloid leukemia cell lines.
Alterations in chromatin modifications in naive and primed cells upon retinoid treatment.
A precommitment state of HL-60 cells can be generated by short treatment (<24 h) with several differentiating agents (57). This transient primed state is not associated with growth arrest but is characterized by lack of lineage commitment, by altered nuclear structure, and by the retention of a cellular memory that lasts for at least three rounds of cell division (55, 56). Changes in chromatin structure are major contributors to the regulation of transcriptional activity that can, through epigenetic modifications, provide transcriptional memory. We therefore investigated the question of whether DMSO priming had any demonstrable effects on either the DNA or histone components of chromatin associated with retinoid-regulated genes such as tissue transglutaminase. Previous studies have identified an 1,800-bp fragment in the 5′-flanking DNA of the human tissue transglutaminase gene as containing the core promoter and the HR1 enhancer (37). We therefore focused our attention on the effects of DMSO priming on the chromatin and covalent modification of histones on this key regulatory sequence.
To find out whether priming is producing changes on the chromatin level at the regulatory regions of this gene, we performed a DNase I hypersensitivity analysis of the promoter of TGM2. The promoter of RARβ that is not inducible in this cell line by retinoid treatment and priming did not change this property of the gene (data not shown); DNase I hypersensitivity was not induced by priming (Fig. 3A). On the other hand, as shown on Fig. 3B, the core promoter of the TGM2 gene became more sensitive to DNase I solely by DMSO priming, suggesting that priming induces changes at the chromatin level of TGM2.
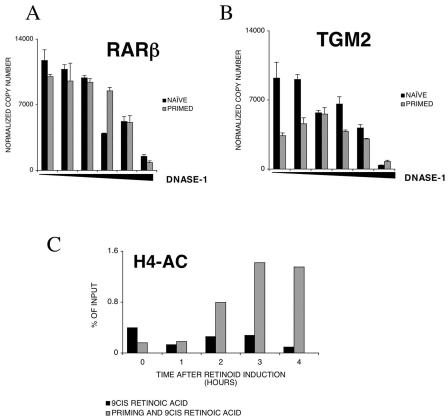
FIG. 3.
Priming produces changes at the chromatin level on the promoter of TGM2. (A) DNase I sensitivity of the RARβ promoter in naive and primed cells. DNase I hypersensitivity was measured as described in Materials and Methods. Values are the means of three independent QPCR measurements ± standard deviations of a representative experiment. (B) DNase I sensitivity of the TGM2 promoter in naive and primed cells. DNase I hypersensitivity was measured as described in Materials and Methods. Values are the means of three independent QPCR measurements ± standard deviations of a representative experiment. (C) Chromatin immunoprecipitation analysis of HR1 enhancer element of TGM2. H4 acetylation was measured as described in Materials and Methods. The acetylation level is expressed as a percentage of input DNA. All no-antibody controls were lower than 0.2% of input DNA. Values are the means of three independent QPCR measurements of a representative experiment. Chromatin immunoprecipitation results were confirmed in at least three independent chromatin preps.
The next step in our studies was to characterize the effects of DMSO priming on the posttranslational modifications of histones associated with regions of the tissue transglutaminase gene promoter. We have used chromatin immunoprecipitation in combination with QPCR (real-time QPCR with TaqMan probes) to obtain accurate quantitation of the level of posttranslational modification of specific histone tails in chromatin isolated from naive and DMSO-primed HL-60 cells. We designed five promoter-specific probe sets spanning the 1,800-bp fragment from the HR1 enhancer to the core promoter (Fig. 4). The AG- and TG-rich tandem repeat regions embedded in the promoter were not covered in this analysis.
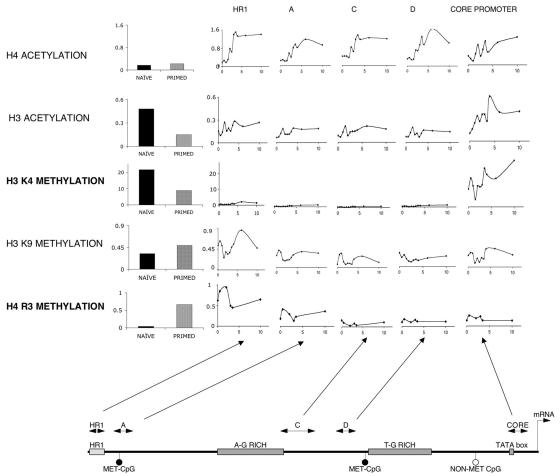
FIG. 4.
Detailed map of histone tail modifications on the promoter/enhancer of TGM2. The bar graphs on the left show the effect of priming on the histone tails bound to the enhancer region HR1 (except H3K4, which is shown on the core promoter). The line graphs show the changes in histone tail modifications along the 1.8-kb studied fragment of the promoter/enhancer in primed cells after 9-cis retinoic acid treatment for the indicated period (in hours). Copy numbers are expressed as percentages of input. All no-antibody controls were lower than 0.2% of input DNA. Values are the means of three independent QPCR measurements of a representative experiment. Chromatin immunoprecipitation results were confirmed in at least three independent chromatin preps. The scheme of the 1.8-kb human TGM2 promoter and the location of the various QPCR assays are shown at the bottom.
We first examined the effect of retinoid treatment on H4 acetylation in naive and DMSO-primed cells. As shown on Fig. 3C, in naive cells, retinoid treatment produced little change in the level of acetylation of H4 histones associated with the HR1 region of the transglutaminase promoter. In primed cells, on the other hand, retinoid treatment for 2 h results in increased levels of H4 acetylation. There is a similar and significant increase in H3 acetylation on the core promoter region in retinoid-treated primed cells and no detectable change in the level of acetylation of histones in this enhancer region HR1 in the retinoid-treated naive cells (data not shown). These findings demonstrate that after retinoid induction of transcription, histones associated with both the HR1 enhancer and the core promoter of TGM2 gene become acetylated in primed cells but do so to a lesser extent in naive cells. Due to the fact that this method is not suitable for analysis of individual cells, we can only state that, after retinoid treatment of DMSO-primed HL-60 cells, the acetylation level of the TGM2 promoter of the entire cell population is much higher than in naive retinoid-treated cells. The level of acetylation of the TGM2 promoter in naive cells after retinoid treatment is below the levels of detection by this method. Whether there is a small fraction of cells in which this region is acetylated or not cannot be assessed.
Based on these findings, we carried out a more comprehensive analysis of the effects of DMSO priming and retinoid treatment of primed cells on histone modifications (acetylation of H3 and H4 and methylation of H3K4, H3K9, and H4R3) (Fig. 4). The upper two panels of Fig. 4 show the changes in histone acetylation that occurred in response to retinoid treatment. In the case of H4 acetylation, retinoid treatment of primed cells results in a significant and uniform increase in the level of acetylation of this histone H4 in all five regions of the promoter. This increase in acetylation starts within 2 h of the addition of the retinoid and reaches a plateau in 6 to 8 h. Retinoid treatment also results in increased acetylation of H3, but unlike H4 acetylation, this effect is not global but is restricted to histones associated with the core promoter.
The lower three panels of Fig. 4 profile the effects of retinoid treatment on the pattern of histone tail methylation. In the case of H3K4, DMSO priming results in a marked decrease in the level of methylation of histones on the core promoter region, as seen in the left bar graph of Fig. 4 (for other regions, H3K4 levels were lower by at least 1 order of magnitude [data not shown]). DMSO priming does not have any marked effect on the K9 methylation of H3 in any region of the TGM2 promoter. Retinoid treatment does induce transient changes in the methylation of this histone in most regions of the promoter, and these changes are most prominent in histones associated with the distal enhancer (HR1) region of the promoter. While DMSO priming decreased methylation of the H3 histone side chains (K4), it increases the level of methylation on the H4 histone (R3). The effect is selective, being most marked on histones associated with the distal regions of the promoter, particularly the HR1 enhancer, and less marked for the histones associated with the proximal regions and the core of the promoter. Retinoid treatment induces a rapid increase in H4R3 methylation that peaks at about 2 h and then rapidly returns to the baseline.
These results demonstrate that the priming of HL-60 cells results in a coordinated set of histone modifications that are likely to be linked to alterations in chromatin structure. Among the most prominent of these effects are the suppression of H3K4 methylation at the core promoter and the increased methylation of the R3 residue of H4 on histones associated with the distal regulatory regions of the promoter. Furthermore, although DMSO priming itself has little effect on either H3 or H4 acetylation, retinoid treatment results in a marked and generalized increase in H4 acetylation and more localized increase in H3 acetylation at the core promoter region.
Role of H4R3 methylation and H3K4 demethylation.
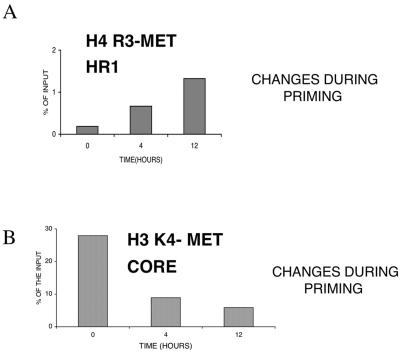
The comparison of histone modifications between naive and DMSO-primed HL-60 cells indicated that reciprocal changes in side chain modifications might be linked to the altered activation of gene expression associated with the precommitment process. To better understand the types of processes that might be involved in the observed alterations in histone methylation, we examined the time course for the changes in H4R3 and H3K4 methylation that followed the initiation of priming with DMSO. Naive cells were treated with DMSO for 4 and 12 h, and the chromatin immunoprecipitation (ChIP) analysis was carried out to determine the levels of H4R3 methylation of histones bound to the HR1 enhancer and H3K4 methylation of histones bound to the core promoter (Fig. 5). There is a striking similarity between the reciprocal changes of H4R3 and H3K4 methylation. Both are substantially changed within 4 h of the initiation of DMSO priming, and these changes remained until the end of the monitored period.
FIG. 5.
Histone tail modifications during priming. (A) H4 arginine 3 methylation levels detected by chromatin immunoprecipitation on the enhancer element (HR1) during priming with DMSO. (B) H3 lysine 4 methylation levels detected by chromatin immunoprecipitation on the core promoter during priming with DMSO. Values are the means of three independent QPCR measurements of a representative experiment. Chromatin immunoprecipitation results were confirmed in at least three independent chromatin preps.
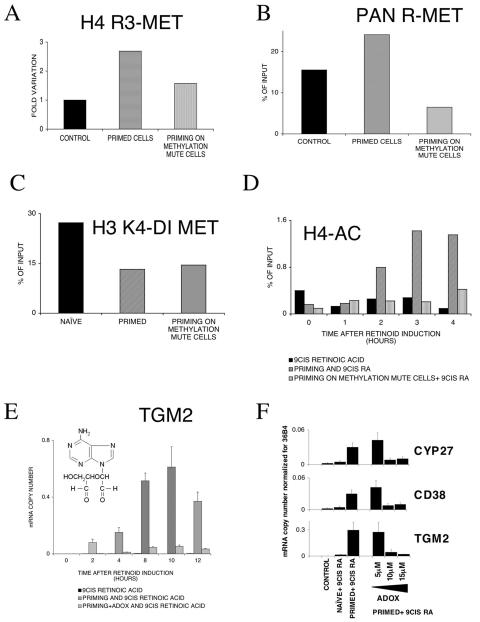
To test whether there is a functional link between the changes in H4R3 methylation induced by priming and altered gene expression, we used ADOX, an inhibitor of methyltransferases (38, 48) to suppress methylation. Cotreatment of HL-60 cells with ADOX and DMSO eliminated H4R3 methylation (Fig. 6A) and also reduced arginine methylation in general on the studied enhancer element, as measured by ChIP QPCR analyses with an anti-Pan-methylated arginine antibody (Fig. 6B). The decrease in H3K4 methylation induced by DMSO priming was not blocked with ADOX (Fig. 6C). Inhibition of methylation with ADOX also blocked retinoid-induced acetylation of H4 histones (Fig. 6D). In parallel with the inhibition of H4R3 methylation and H4 acetylation, there was marked, but not complete, inhibition in the retinoic acid-induced expression of tissue transglutaminase (Fig. 6E). It is important to note that simultaneous treatment of HL-60 cells with ADOX and DMSO reduced but did not completely block the induction of the transglutaminase gene. Collectively, these data suggest that the inhibition of methyltransferases by ADOX leads to the inhibition of H4 arginine 3 methylation and a concomitant decrease in retinoid-induced transglutaminase promoter activation. We speculate that the decrease in acetylation of H4 histones associated with the transglutaminase promoter is the consequence of the decrease in the retinoid-dependent activation of transcription of this gene. Moreover, the results we have obtained point to a strong correlation between H4 arginine 3 methylation and the precommitment of HL-60 cells to terminal differentiation.
FIG. 6.
Epigenetic changes on TGM2 promoter and mRNA expression upon priming in methylation mute cells (methyltransferases were blocked with 10 μM ADOX for 16 h as described in Materials and Methods). Values are the means of three parallel QPCR measurements ± standard deviations, and results were confirmed from at least three independent biological samples. All chromatin immunoprecipitation values are the means of three independent QPCR measurements of a representative experiment. Chromatin immunoprecipitation results were confirmed in at least three independent chromatin preps. (A) H4 arginine 3 methylation levels determined by chromatin immunoprecipitation on the HR1 enhancer element. (B) Arginine methylation levels determined by chromatin immunoprecipitation on the HR1 enhancer element with an anti-Pan methyl arginine antibody. (C) H3 lysine 4 methylation studied by chromatin immunoprecipitation on the core promoter. (D) Chromatin immunoprecipitation analysis of H4 acetylation levels in naive, primed, and primed methylation mute cells on the enhancer element HR1. (E) TGM2 mRNA levels normalized to 36B4 copy numbers in naive, primed, and primed methylation mute cells (the structure of ADOX is shown in the inset). (F) TGM2, CD38, and CYP27 transcript levels after blocking methylation with ADOX in naive and primed cells. ADOX was used in increasing concentrations as shown. The mRNA copy numbers were normalized to 36B4 transcript levels. Values are the means of three parallel QPCR measurements ± standard deviations, and results were confirmed from at least three independent biological samples.
There were two main concerns with the results obtained in the previous studies and these pertained to how widespread the priming effect was (i.e., was it limited to the tissue transglutaminase gene or were other retinoid-regulated genes also involved?) and whether DMSO induced a general induction in gene expression rather than affecting only a subset of genes during priming. To address the issue of whether DMSO priming was limited to only one gene, we compared the induction of tissue transglutaminase to the induction of two other genes (CD38 and CYP27) regulated by retinoids in myeloid leukemia cells (27, 47). The expression of both CD38 and CYP27 was increased by 9-cis retinoic acid, and this induction is much greater in cells that had been primed by pretreatment with DMSO (Fig. 6F). Cotreatment of the cells with different concentrations of ADOX and DMSO resulted in a dose-dependent suppression of retinoid-induced expression of both of these genes.
To address the issue of a potential general effect on transcription by DMSO and ADOX on primed gene expression, we used a general expression profiling approach. HL-60 cells were primed with DMSOADOX, or DMSO and ADOX for 16 h and subsequently treated with 9-cis retinoic acid for 6 h. RNA was then prepared from these cells, and global expression profiles were determined using a 3,200-feature human cDNA microarray. There were approximately the same number of genes (104 for DMSO and 111 for ADOX) induced >1.5-fold in the DMSO- and ADOX-treated cultures (http://www.biochem.dote.hu/nagylab/suppdata.htm). These results confirmed that treatment of HL-60 cells with ADOX does not result in a generalized perturbation of the preexisting patterns of gene expression.
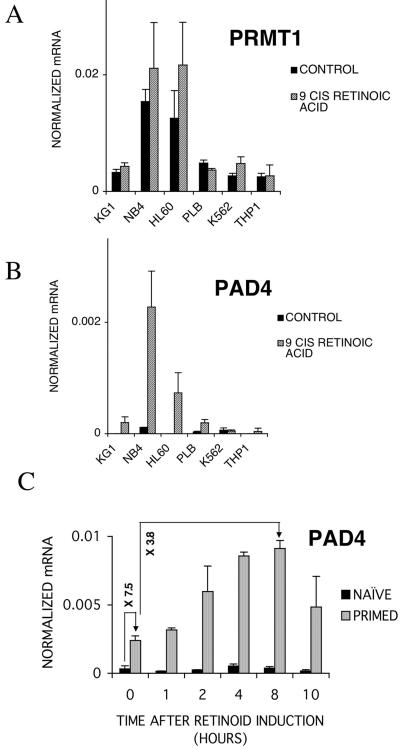
To address the question of the more general effects of DMSO priming and ADOX on retinoid-regulated gene expression, we compared the profiles of retinoid-induced genes in RNA from control naive cells and retinoid-treated cells that had been primed with either DMSO alone or DMSO and ADOX. We detected 38 genes with expression that was upregulated more than 1.5-fold by the treatment of naive HL-60 cells with retinoids for 6 h. Exposure of similar cells to retinoids following DMSO priming resulted in a substantially larger pool of genes with expression that was changed (for 104 genes, the expression was increased more than 1.5-fold compared to the DMSO-primed cells). Of the 75 retinoid-induced genes with expression which was selectively increased in DMSO-primed cells, the induction of 62 was blocked by coadministration of DMSO plus ADOX (http://www.biochem.dote.hu/nagylab/suppdata.htm). These experiments suggested that neither DMSO nor ADOX had a widespread effect on transcription in these cells. Since methylation is involved not only in RNA processing but also in protein synthesis, we asked whether a general inhibitor of methyltransferases like ADOX would produce a major rearrangement in the proteins of the studied cells. For this, we analyzed, with two-dimensional electrophoresis and silver staining, the proteins of HL-60 cells and the ways ADOX is changing this protein expression pattern. We could not detect any substantial change in the protein composition of HL-60 cells after ADOX treatment with this method. In fact, there was less than 5% change in the number of spots detected. These data strongly suggest that ADOX is not a general inhibitor of RNA and protein synthesis under the conditions used (http://www.biochem.dote.hu/nagylab/suppdata.htm). To gain a more mechanistic insight into this process and also to take advantage of recent developments in the field, we have evaluated the role of the enzymes proposed to be responsible for H4R3 methylation. These are PRMT1, a methyltransferase, and PAD4, a peptidyl arginine deiminase recently identified as the enzyme responsible for methyl arginine's conversion into citrulline and thereby removing the methyl group (13, 23, 53). We used gene-specific TaqMan assays and carried out real-time RT-QPCR analysis to determine the expression level of PRMT1 and PAD4 in the myeloid cell lines used in our studies in the absence and presence of 9-cis retinoic acid. As shown in Fig. 7A, the expression level of PRMT1 was determined. All cell lines expressed an appreciable level of the PRMT1 mRNA, and retinoid treatment did not appear to significantly change the expression level. PAD4 is expressed at low or not detectable levels in the six cell lines examined but induced to high levels upon 2 days of retinoid treatment in NB4 and HL-60 cells and to a lesser degree in KG1 and PLB cells (Fig. 7B). Finally, we examined the expression level of PAD4 during priming and the subsequent retinoid response in HL-60 cells. As shown in Fig. 7C, PAD4 is induced by priming itself and further induced during retinoid treatment.
FIG. 7.
Analysis of PRMT1 and PAD4 mRNA levels in different cell lines upon retinoid treatment. (A) PRMT1 mRNA copy numbers were normalized to cyclophilin transcript levels. Values are the means of three independent QPCR measurements ± standard deviations. (B) PAD4 mRNA copy numbers were normalized to cyclophilin transcript levels. Values are the means of three independent QPCR measurements ± standard deviations. (C) mRNA levels of PAD4 in 9-cis retinoic acid-treated naive and DMSO-primed HL-60 cells. Relative increases of mRNA levels during priming or retinoid treatment of primed cells are shown on the arrows. PAD4 mRNA copy numbers were normalized to cyclophilin transcript levels. Values are the means of three independent QPCR measurements ± standard deviations.
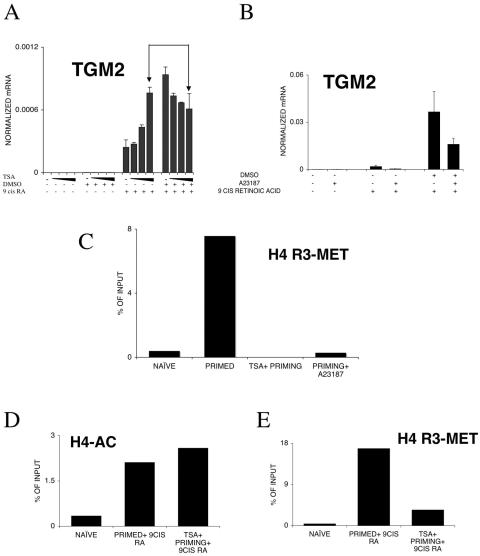
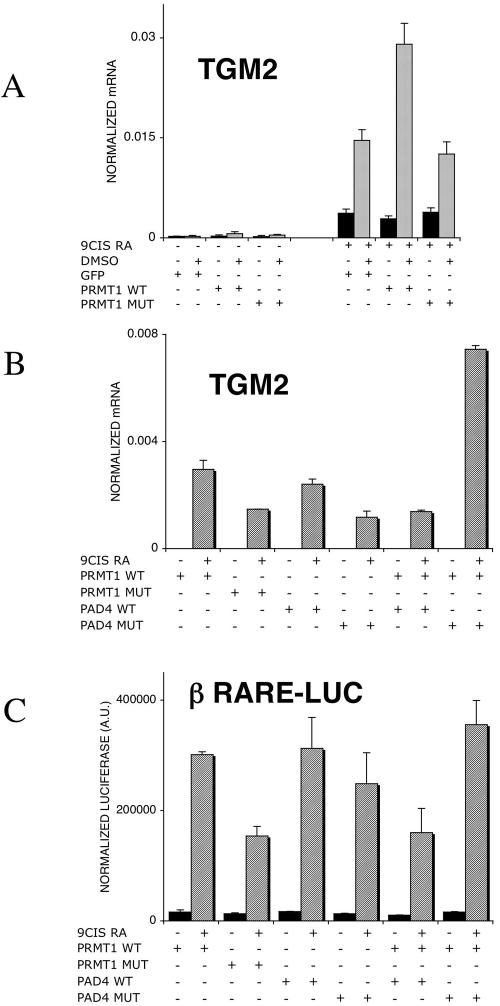
These data established that both PRMT1 and PAD4 are present in HL-60 cells and, while PRMT1's expression level appears to be constant, PAD4 is dynamically changing during priming and retinoid stimulation, in agreement with previous findings (39). If one assumes that H4R3 methylation is the cause of the priming effect, a few predictions can be put forward and tested. One is that preacetylation of chromatin interferes with H4R3 methylation (52); therefore, the priming is likely to be attenuated. We tested this by using trichostatin A (TSA), a histone deacetylase inhibitor. TSA alone had no effect on TGM2 expression (Fig. 8A). Increasing the amount of TSA potentiated the retinoid's effect as anticipated and as was shown by us previously (36). Importantly, the DMSO priming effect was completely abolished if cells were pretreated with TSA, suggesting that acetylation of histones interfering with H4R3 methylation eliminates the priming effect. These findings confirm previous reports on the interference of acetylation and H4R3 methylation (52). Another prediction is that activation of PAD4, the enzyme converting methyl arginine to citrulline, also attenuates the priming effect. To test this prediction, we used calcium ionophores to activate PAD4 in HL-60 cells similar to that reported by Wang and coworkers (53). Cells, after priming and prior to retinoid induction, were exposed to a short 15-min treatment of 1 μM A23187 calcium ionophore. After this treatment, cells were washed extensively and treated with 9-cis retinoic acid. The presence of A23187 reduced retinoid responsiveness and priming, as shown in Fig. 8B. The interpretation of these data is corroborated by chromatin immunoprecipitation results. As shown in Fig. 8C, both TSA pretreatment and PAD4 activation prevented/eliminated H4R3 methylation, respectively. Moreover, comparison of H4 acetylation and H4R3 methylation revealed that TSA treatment enhanced acetylation while preventing H4R3 methylation (Fig. 8D and E). The third prediction we tested was that the increased level of PRMT1, the methylase responsible for H4R3 methylation, would lead to increased priming. On one hand, as shown in Fig. 9A, in the absence of retinoid treatment, transfection of PRMT1 does not induce gene expression. On the other hand, it can further induce the priming effect. It is noteworthy that under the conditions used, increased PRMT1 expression does not substitute for priming. Finally, we have evaluated the combined effect of transfected wild-type or mutant PRMT1 and PAD4 on the retinoid-regulated expression of TGM2 and also that of a retinoid-inducible reporter gene. As shown in Fig. 9B and C, PRMT1's enzymatic activity is required for its coactivator activity. PAD4 does not act as a corepressor, but its enzymatically inactive mutant synergizes with PRMT1 in enhancing transcription (Fig. 9B). These data, if put together, make it very likely that the priming effect involves H4R3 methylation and that the level of H3R4 methylation is regulated by the activity of PRMT1 and PAD4.
FIG. 8.
Modulation of TGM2 expression levels in HL-60 cells. (A) TSA pretreatment for 1 h of HL-60 cells enhances the retinoid response in naive cells (50, 100, and 150 nM TSA was used). TSA pretreatment for 1 h of HL-60 cells before DMSO priming reduces the effect of priming on TGM2 induction (50, 100, and 150 nM TSA was used). Arrows indicate the effect of pretreatment with 150 nM TSA on primed versus naive retinoid-treated cells. mRNA copy numbers were normalized to 36B4 transcript levels. Values are the means of three independent QPCR measurements ± standard deviations. (B) Activation of PAD4 after priming by a 15-min treatment with 1 μM calcium ionophore A23187 reduces the priming effect. mRNA copy numbers were normalized to cyclophilin transcript levels. Values are the means of three independent QPCR measurements ± standard deviations. (C) H4R3 methylation levels are changed during priming and modulated by pretreatment with 100 nM TSA for 1 hour or a 15-min treatment with 1 μM calcium ionophore A23187. H4R3 methylation was determined by chromatin immunoprecipitation on the HR1 enhancer element. Copy numbers are expressed as percentages of the input. (D) H4 acetylation levels in primed, retinoid-treated cells and TSA-pretreated, primed retinoid-treated cells. Cells were pretreated with 100 nM TSA for 1 hour, and H4 acetylation levels were determined by chromatin immunoprecipitation on the HR1 enhancer element. Copy numbers are expressed as percentages of the input. (E) H4R3 methylation levels in primed, retinoid-treated cells and TSA-pretreated, primed retinoid-treated cells. Cells were pretreated with 100 nM TSA for 1 hour, and H4R3 methylation levels were determined by chromatin immunoprecipitation on the HR1 enhancer element. Copy numbers are expressed as percentages of the input. The values of the no-antibody controls were subtracted. Values are the means of three independent QPCR measurements of a representative experiment. Chromatin immunoprecipitation results were confirmed in two independent chromatin preps with two independent immunoprecipitations, respectively. +, present; −, absent.
FIG. 9.
Effect of PRMT1 and PAD4 on retinoid-regulated gene expression. (A) Transfection of PRMT1 expression vector into HL-60 cells increases the TGM2 induction after retinoid treatment in primed cells, while the catalytic mutant has no effect on retinoid responsiveness. Cells were treated with 1 μM 9-cis retinoic acid for 10 h. mRNA copy numbers were normalized to cyclophilin transcript levels. Values are the means of three independent QPCR measurements ± standard deviations. (B) Cotransfection of PAD4 and PRMT1 expression vectors and their catalytic mutants into HL-60 cells modulates TGM2 induction. Cells were treated with 1 μM 9-cis retinoic acid for 24 h. mRNA copy numbers were normalized for cyclophilin transcript levels. Values are the means of two independent biological replicates and three independent QPCR measurements for each sample ± standard deviations. (C) Cotransfection of PAD4 and PRMT1 expression vectors and their catalytic mutants into 293T cells modulates the luciferase expression on a βRARE tkLuc reporter system. Values were normalized to measured β-galactosidase values and expressed in arbitrary units (AU) as described in Materials and Methods. Values are the mean of three independent biological replicates ± standard deviations. +, present; −, absent.
DISCUSSION
The profile of gene expression in cells, both differentiated and in their precursors, is often viewed from the perspective of a static “snapshot” rather than as parts of a dynamic process akin to the many individual frames that when combined constitute a movie. For cells undergoing differentiation, the phenotypic identity of a cell, as defined by its distinctive pattern of gene expression, has to be maintained through multiple cycles of DNA replication, chromatin assembly, and repackaging. It has been suggested that there has to be some sort of cellular memory that provides a cell with an epigenetically coded identity that can be preserved during differentiation (50). One form of cellular memory, provided by gene “silencing,” has been studied in detail (22, 40). However, the mechanisms that allow for either persistent activation of gene expression or a presensitization to expression of specific genes has not been well characterized. An understanding of these mechanisms will contribute a significant new level of insight to the regulation of gene expression.
Epigenetic changes that affect the structure of chromatin associated with regulated genes and gene networks are obvious candidates to serve as components of the postulated cellular memory of gene expression. Methylation of CpG islands and deacetylation of lysines have been clearly linked to the induction of a silenced state of chromatin (26). On the other hand, acetylation of histones H3 and H4 have been linked to the activation of gene expression (1). In the studies reported here, we have set out to address a complex issue, namely to investigate the changes in epigenetic markers that are linked to the regulation of gene expression during retinoid-induced myeloid cell differentiation. We have chosen to work with the HL-60 cell line because previous work from several laboratories has established that differentiation of these cells involves a two-step process that can be separated pharmacologically (11, 55-57). The first step, termed precommitment or priming, involves a persistent state of presensitization that constitutes a well-defined model of cellular memory. The second step, which is also well characterized, involves selective activation of gene expression by inducers of terminal differentiation for this lineage, such as retinoids. We have then used the promoter of a well-characterized retinoid-regulated gene in this pathway, tissue transglutaminase, a key marker of retinoid response, to investigate the role of epigenetic modification of chromatin components in the establishment of the precommitted state of differentiation of these cells. We have carried out a detailed analysis of the covalent modifications of histones bound to different regions of the transglutaminase promoter during three distinct states of differentiation: the naive state that occurs prior to the initiation of differentiation, the primed or precommitted state that is induced by brief exposure of the naive cells to vitamin D or DMSO, and the differentiated state that occurs following the addition of a retinoid to the primed cells. In characterizing the precommitted state, we found that it was a “threshold phenomenon.” The increased induction of gene expression in primed versus naive cells was not due to an increase in the transcriptional activation of individual cells but rather to an increase in the fraction of the population of cells that was able to respond to retinoids with activation of gene expression (Fig. 2). This result is entirely consistent with a model that suggests that the development of epigenetic memory entails a persistent “marking” of those cells that have been exposed to the memory inducer (in this case, vitamin D or DMSO).
The analysis of histone tail modifications during both priming and transcriptional activation revealed distinct mechanisms that mark both of these processes. Priming itself appears to be linked to major changes in histone side chain methylation. In particular, methylation of K4 on histone H3 (H3K4) associated with the core promoter is rapidly decreased after the initiation of priming (Fig. 4 and 5). This decrease in H3K4 methylation level was not blocked by the methyltransferase inhibitor ADOX (Fig. 6C), suggesting that a non-methyltransferase-dependent pathway might be responsible for this effect. This observation is in agreement with the recent identification of the enzymes responsible for demethylation (44). According to Shi et al., demethylation is mediated by LSD1, a member of the amine oxidase enzyme family. Priming increases the methylation of arginine on histone H4 (H4R3) specifically on histones associated with a prominent enhancer element in the transglutaminase promoter. Arginine methylation of histones seems particularly important to the induction of the primed state, since its elimination with a pharmacologic inhibitor of methyltransferases resulted in the loss of priming for the induction of tissue transglutaminase and several other retinoid-regulated genes (Fig. 6).
Cross talk between enhancer and core promoter.
There is an apparent discrepancy between the epigenetic changes occurring on the HR1 enhancer and the core promoter, as revealed by our studies. While histone methylation of H4R3 on the enhancer element appears to play an important role in the establishment of the precommitted state, histone acetylation seems to contribute to the activation of transcription. Retinoid-activated transcription of the primed cells resulted in a marked increase in H4 acetylation, particularly in regions of the enhancer element, and also a lesser increase in H3 acetylation of histones associated with the core promoter. There could be numerous reasons for this discrepancy. For example, acetylation and arginine methylation were shown by An and colleagues to be detectable on a larger promoter fragments than the regions in the proximity of acetyltransferases or methyltransferases (2). The activation of transcription of the transglutaminase gene in addition to the changes in acetylation results in increased H3 methylation (H3K4) of the core promoter and, to a lesser and more transient extent, H3 methylation (H3K9) of the enhancer. H3K4 methylation was reported as being localized to core promoters and transcribed regions of active genes if studied by the ChIP technique on larger genomic regions (5). If studied on individual promoters, the location of increased H3K4 methylation seems also to correlate with core promoters (2, 24). We have also found a clear and marked difference in the H3K4 levels (more than 50 times higher levels on the core promoter than on the enhancer HR1). Taken together, these results suggest a cross talk between the promoter and enhancer of TGM2, with the activation of distinct enzymatic pathways both during priming and retinoid activation. This so far unsubstantiated cross talk suggests a physical interaction between these regulatory DNA elements, an interaction that needs to be investigated further.
Nuclear receptor complexes induce histone tail modifications.
The epigenetic changes associated with both the priming and transcriptional activation that occur during retinoid-induced differentiation are clearly linked to the activity of retinoid receptors. In the absence of a ligand, RAR-RXR heterodimers, the mediators of the effects of retinoids on myeloid cell differentiation, are believed to bind to their cognate response elements and repress transcription. Liganding of these receptors results both in the loss of this repressive effect and in the induction of transcriptional activity. While little detail is known about the molecular steps involved in the activation of transcription by retinoid receptors, significantly more is known about the activity of other members of the nuclear receptor superfamily. A general model of transcriptional activation, developed primarily from studies on estrogen and glucocorticoid receptor-regulated genes (3, 34) suggests that liganding of the receptors results in a sequential recruitment of proteins involved in transcriptional activation. According to the present concept, the sequential recruitment of cofactors may be different from gene to gene (reviewed by M. P. Cosma) (12) but, in all cases, results in an orderly process of covalent modifications of the tails of histone proteins associated with the promoter and enhancer elements of the target gene. Our data on the effects of retinoids on the covalent modifications of histone proteins associated with the transglutaminase promoter are in agreement with this general model. We have demonstrated that induced transcription of the transglutaminase gene in these myeloid cells is paralleled by a wave of histone tail acetylation that is most likely due to the recruitment of histone acetyltransferase (HAT)-containing complexes to the promoter. Our studies have demonstrated, however, that activation of transcription is not only linked to histone acetylation but also to some very selective effects on histone methylation. H3K4 methylation has been characterized in several systems and has generally been found to oppose H4K9 methylation and to mark areas of increased gene expression (19, 21). We have found that this methylation pattern is concentrated in the core promoter and is reduced by priming and increased by retinoid treatment. These data are in agreement with a purported role of H3K4 as a marker of positive gene expression.
A role for histone 4 arginine 3 methylation in nuclear receptor signaling.
H4 arginine 3 methylation is one of the least-characterized histone tail modifications. Arginine methylation on both H4 and H3 tails has been shown to be related to nuclear receptor coactivation in several experimental systems. The family of p160 transcriptional coactivators (e.g., SRC1, GRIP1, ACTR) binds two members of the arginine methyltransferase family, PRMT1 and CARM1 (also called PRMT4). Both these transferases have been implicated in the activation of nuclear receptor-dependent genes (7, 46). The arginine methyltransferases modify histone tails and arginine side chains on other proteins as well (e.g., CBP, STAT) (8, 35, 54). The PRMT1 enzyme has been shown in cotransfection studies to be a cofactor of nuclear receptor-activated gene expression (28). In vitro studies have demonstrated that PRMT1 methylates H4 arginine 3 and that, once methylated, H4 is a better substrate for HAT-s and vice versa, once acetylated, the histone tails lose their ability to become methylated by PRMT1 (52). The other factor implicated in the regulation of arginine methylation is the enzyme called peptidyl arginine deiminase, or PAD4. In HL-60 cells, PAD4 was shown to be regulated by DMSO, vitamin D, and retinoic acid (39). All of these agents have the potential to prime HL-60 cells (56, 57). Recently, this gene was found to be responsible for the removal of the methyl mark on H4R3 by converting it to a citrullinated histone H4 (53). These observations provide a plausible link between our observations that the precommitment state of HL-60 cells involves increased H4R3 methylation and increased retinoid-activated gene expression. In parallel with the increase in H4R3 methylation, the gene responsible for the removal of the methyl mark is also induced. Our transfection data (Fig. 9) also suggest that PAD4 may not act as a transcriptional repressor per se, but some of its activity it is required along with PRMT1 for synergistic/cooperative activation of retinoid-mediated transcription. This activity does not require but is inhibited by its peptidyl arginine deiminase activity. The complex role of PAD4 in the regulation of transcription is further supported by recent work of Lee and colleagues (30). According to their data, demethylimination of p300 by PAD4 is changing its activity to an activated or even hyperactive form. This change in p300 activity is modulated via the regulation of p300-GRIP1 interaction. We propose that priming-induced modifications of H4 arginine 3 could increase retinoid-induced gene activation by potentiating HAT activity on histones. This in turn would result in increased histone acetylation and increased transcriptional activation. Our data also suggest that the full activation can be achieved in two distinct ways. The first is by switching off histone deacetylase activity with a histone deacetylase inhibitor such as TSA. In this case, the response to a retinoid signal will increase. The other way is by transforming the histone tails to become better substrates of HAT-s. Arginine methylation on H4R3 is making such a change. The two mechanisms are mutually exclusive and antagonistic. By this, the two parallel pathways provide a signal integration mechanism for the cell.
The most detailed characterization of the dynamics of nuclear receptor-mediated transcriptional activation has been carried out by Metivier and colleagues (34). By analyzing the estrogen receptor-induced transcription on the pS2 gene, they described the ordered and sequential recruitment of specific cofactors and the resultant ordered sequence of histone tail modifications. According to their data, in the first, “unproductive,” cycle, binding of PRMT1 to the receptor complex and subsequent H4R3 methylation occur. In the second, “productive,” cycle, recruitment of PRMT1 is followed by binding of HAT-s and extensive acetylation of the histones H3 and H4. We propose that the two phases of gene activation proposed by Metivier and colleagues (34) may be dissociated in HL-60 cell differentiation. Our results suggest that the transition to the precommitted state (i.e., the priming of the cells) induces an increase in H4R3 methylation but no change in H4 acetylation and no activation of transcription. This first cycle of HL-60 differentiation does not require liganding of the nuclear receptors, it occurs via an independent and uncharacterized vitamin D- or DMSO-dependent step. This step provides a further signal integration node for the cell by integrating two independent signals in time and space.
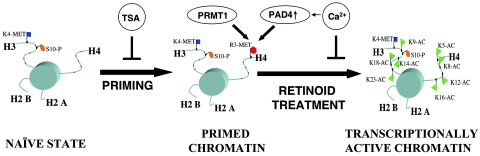
We propose a model for the epigenetic regulation of retinoid response and differentiation competence in myeloid leukemia cells (Fig. 10). In this study, we were able to dissect some of the epigenetic changes taking place on a retinoid-regulated gene. We found that H4R3 methylation takes place prior to gene activation and is a hallmark of the primed cell state. This modification represents a transcriptionally silent (unproductive) but primed state (34) which marks key histones and makes them better substrates for receptor-bound acetyltransferases (HAT-s). We propose that this mechanism accounts for the increased susceptibility of the cell to respond to terminal differentiating agents, such as a retinoid, with increased gene expression and an increased potential for phenotypic differentiation. Preacetylated histones are refractory to this mechanism. This model is consistent with the proposal that histone tail modifications function as the physical mediators of cellular memory. By providing docking sites for transcription factors and marking histones for subsequent covalent modifications, these methylation reactions serve as silent switches of gene expression. Our findings suggest an active and physiological role for arginine 3 methylation on H4 tails in retinoid response and provide a model amenable to further investigation and potentially to pharmacological exploitation.
FIG. 10.
Proposed model of gene expression modulation by changes in H4 arginine 3 methylation. PRMT1 and PAD4 regulate arginine 3 methylation. The naive state is characterized by lack of H4 arginine methylation on the enhancer region. Priming results in increased H4 R3 methylation, while retinoid treatment leads to decreased H4R3 methylation and to the indicated changes in lysine acetylation. Preacetylation of histone tails prior to priming by TSA is likely to reduce the affinity of histone tails toward PRMT1 and, by this, prevents the enhanced retinoid responsiveness caused by priming. The activation of PAD4 after priming by calcium ionophore treatment leads to the removal of the methyl mark from H4R3 and abolition of the enhanced retinoid responsiveness caused by priming.
Acknowledgments
We thank Ibolya Furtos and Marta Beladi for excellent technical assistance and members of the Nagy laboratory and Laszlo Tora, Mate Demeny, Zsuzsanna Nagy, and Beata Scholtz for suggestions and comments on the manuscript.
This work was supported by grant FP5-RTN from the EU (to L.N.), FIRCA award 5 RO3 TW 01146-02 (to P.J.A.D and L.N.), and Hungarian Scientific Research Fund (OTKA) T034434 (to L.N.). L.N. is an International Scholar of HHMI and an EMBO Young Investigator and holds a Wellcome Trust Senior Research Fellowship in Biomedical Sciences in Central Europe. B.L.B. is a Young Researcher of the EU NUC REC NET (an EU FP5 training network).
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M., C. Baumann, S. John, D. A. Walker, M. Vigneron, J. G. McNally, and G. L. Hager. 2002. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 3:1188-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benko, S., J. D. Love, M. Beladi, J. W. Schwabe, and L. Nagy. 2003. Molecular determinants of the balance between co-repressor and co-activator recruitment to the retinoic acid receptor. J. Biol. Chem. 278:43797-43806. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., M. Kamal, K. Lindblad-Toh, S. Bekiranov, D. K. Bailey, D. J. Huebert, S. McMahon, E. K. Karlsson, E. J. Kulbokas III, T. R. Gingeras, S. L. Schreiber, and E. S. Lander. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169-181. [DOI] [PubMed] [Google Scholar]
- 6.Chen, A., J. D. Licht, Y. Wu, N. Hellinger, W. Scher, and S. Waxman. 1994. Retinoic acid is required for and potentiates differentiation of acute promyelocytic leukemia cells by nonretinoid agents. Blood 84:2122-2129. [PubMed] [Google Scholar]
- 7.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 8.Chevillard-Briet, M., D. Trouche, and L. Vandel. 2002. Control of CBP co-activating activity by arginine methylation. EMBO J. 21:5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiocca, E. A., P. J. Davies, and J. P. Stein. 1988. The molecular basis of retinoic acid action. Transcriptional regulation of tissue transglutaminase gene expression in macrophages. J. Biol. Chem. 263:11584-11589. [PubMed] [Google Scholar]
- 10.Chiocca, E. A., P. J. Davies, and J. P. Stein. 1989. Regulation of tissue transglutaminase gene expression as a molecular model for retinoid effects on proliferation and differentiation. J. Cell. Biochem. 39:293-304. [DOI] [PubMed] [Google Scholar]
- 11.Collins, S. J., K. A. Robertson, and L. Mueller. 1990. Retinoic acid-induced granulocytic differentiation of HL-60 myeloid leukemia cells is mediated directly through the retinoic acid receptor (RAR-alpha). Mol. Cell. Biol. 10:2154-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 13.Cuthbert, G. L., S. Daujat, A. W. Snowden, H. Erdjument-Bromage, T. Hagiwara, M. Yamada, R. Schneider, P. D. Gregory, P. Tempst, A. J. Bannister, and T. Kouzarides. 2004. Histone deimination antagonizes arginine methylation. Cell 118:545-553. [DOI] [PubMed] [Google Scholar]
- 14.Davies, P. J., J. P. Basilion, E. A. Chiocca, J. Johnson, S. Poddar, and J. P. Stein. 1988. Retinoids as generalized regulators of cellular growth and differentiation. Am. J. Med. Sci. 296:164-170. [DOI] [PubMed] [Google Scholar]
- 15.Davies, P. J., E. A. Chiocca, J. P. Basilion, S. Poddar, and J. P. Stein. 1988. Transglutaminases and their regulation: implications for polyamine metabolism. Adv. Exp. Med. Biol. 250:391-401. [DOI] [PubMed] [Google Scholar]
- 16.Davies, P. J., E. A. Chiocca, and J. P. Stein. 1988. Retinoid-regulated expression of tissue transglutaminase in normal and leukemic myeloid cells. Adv. Exp. Med. Biol. 231:63-71. [DOI] [PubMed] [Google Scholar]
- 17.Davies, P. J., M. P. Murtaugh, W. T. Moore, Jr., G. S. Johnson, and D. Lucas. 1985. Retinoic acid-induced expression of tissue transglutaminase in human promyelocytic leukemia (HL-60) cells. J. Biol. Chem. 260:5166-5174. [PubMed] [Google Scholar]
- 18.Duprez, E., G. Benoit, M. Flexor, J. R. Lillehaug, and M. Lanotte. 2000. A mutated PML/RARA found in the retinoid maturation resistant NB4 subclone, NB4-R2, blocks RARA and wild-type PML/RARA transcriptional activities. Leukemia 14:255-261. [DOI] [PubMed] [Google Scholar]
- 19.Elgin, S. C., and S. I. Grewal. 2003. Heterochromatin: silence is golden. Curr. Biol. 13:R895-R898. [DOI] [PubMed] [Google Scholar]
- 20.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-R197. [DOI] [PubMed] [Google Scholar]
- 21.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 22.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 23.Hagiwara, T., K. Nakashima, H. Hirano, T. Senshu, and M. Yamada. 2002. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem. Biophys. Res. Commun. 290:979-983. [DOI] [PubMed] [Google Scholar]
- 24.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 26.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 27.Kishimoto, H., S. Hoshino, M. Ohori, K. Kontani, H. Nishina, M. Suzawa, S. Kato, and T. Katada. 1998. Molecular mechanism of human CD38 gene expression by retinoic acid. Identification of retinoic acid response element in the first intron. J. Biol. Chem. 273:15429-15434. [DOI] [PubMed] [Google Scholar]
- 28.Koh, S. S., D. Chen, Y. H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 29.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 30.Lee, Y. H., S. A. Coonrod, W. L. Kraus, M. A. Jelinek, and M. R. Stallcup. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. USA 102:3611-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madi, A., S. Mikkat, B. Ringel, H. J. Thiesen, and M. O. Glocker. 2003. Profiling stage-dependent changes of protein expression in Caenorhabditis elegans by mass spectrometric proteome analysis leads to the identification of stage-specific marker proteins. Electrophoresis 24:1809-1817. [DOI] [PubMed] [Google Scholar]
- 32.Marmorstein, R. 2001. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2:422-432. [DOI] [PubMed] [Google Scholar]
- 33.McArthur, M., S. Gerum, and G. Stamatoyannopoulos. 2001. Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNaseI-hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol. 313:27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 35.Mowen, K. A., J. Tang, W. Zhu, B. T. Schurter, K. Shuai, H. R. Herschman, and M. David. 2001. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell 104:731-741. [DOI] [PubMed] [Google Scholar]
- 36.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 37.Nagy, L., M. Saydak, N. Shipley, S. Lu, J. P. Basilion, Z. H. Yan, P. Syka, R. A. Chandraratna, J. P. Stein, R. A. Heyman, and P. J. Davies. 1996. Identification and characterization of a versatile retinoid response element (retinoic acid receptor response element-retinoid X receptor response element) in the mouse tissue transglutaminase gene promoter. J. Biol. Chem. 271:4355-4365. [DOI] [PubMed] [Google Scholar]
- 38.Najbauer, J., B. A. Johnson, A. L. Young, and D. W. Aswad. 1993. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J. Biol. Chem. 268:10501-10509. [PubMed] [Google Scholar]
- 39.Nakashima, K., T. Hagiwara, A. Ishigami, S. Nagata, H. Asaga, M. Kuramoto, T. Senshu, and M. Yamada. 1999. Molecular characterization of peptidylarginine deiminase in HL-60 cells induced by retinoic acid and 1alpha,25-dihydroxyvitamin D(3). J. Biol. Chem. 274:27786-27792. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto, I., A. P. Otte, C. D. Allis, D. Reinberg, and E. Heard. 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303:644-649. [DOI] [PubMed] [Google Scholar]
- 41.Onody, A., A. Zvara, L. Hackler, Jr., L. Vigh, P. Ferdinandy, and L. G. Puskas. 2003. Effect of classic preconditioning on the gene expression pattern of rat hearts: a DNA microarray study. FEBS Lett. 536:35-40. [DOI] [PubMed] [Google Scholar]
- 42.Puskas, L. G., K. Kitajka, C. Nyakas, G. Barcelo-Coblijn, and T. Farkas. 2003. Short-term administration of omega 3 fatty acids from fish oil results in increased transthyretin transcription in old rat hippocampus. Proc. Natl. Acad. Sci. USA 100:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott, H. S., S. E. Antonarakis, M. D. Lalioti, C. Rossier, P. A. Silver, and M. F. Henry. 1998. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2). Genomics 48:330-340. [DOI] [PubMed] [Google Scholar]
- 44.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, and R. A. Casero. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 45.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 46.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 47.Szanto, A., S. Benko, I. Szatmari, B. L. Balint, I. Furtos, R. Ruhl, S. Molnar, L. Csiba, R. Garuti, S. Calandra, H. Larsson, U. Diczfalusy, and L. Nagy. 2004. Transcriptional regulation of human CYP27 integrates retinoid, peroxisome proliferator-activated receptor, and liver X receptor signaling in macrophages. Mol. Cell. Biol. 24:8154-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang, J., A. Frankel, R. J. Cook, S. Kim, W. K. Paik, K. R. Williams, S. Clarke, and H. R. Herschman. 2000. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275:7723-7730. [DOI] [PubMed] [Google Scholar]
- 49.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 50.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 51.Uray, I. P., J. H. Connelly, V. Thomazy, G. L. Shipley, W. K. Vaughn, O. H. Frazier, H. Taegtmeyer, and P. J. Davies. 2002. Left ventricular unloading alters receptor tyrosine kinase expression in the failing human heart. J. Heart Lung Transplant. 21:771-782. [DOI] [PubMed] [Google Scholar]
- 52.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Y., J. Wysocka, J. Sayegh, Y. H. Lee, J. R. Perlin, L. Leonelli, L. S. Sonbuchner, C. H. McDonald, R. G. Cook, Y. Dou, R. G. Roeder, S. Clarke, M. R. Stallcup, C. D. Allis, and S. A. Coonrod. 2004. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306:279-283. [DOI] [PubMed] [Google Scholar]
- 54.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 55.Yen, A. 1985. Control of HL-60 myeloid differentiation. Evidence of uncoupled growth and differentiation control, S-phase specificity, and two-step regulation. Exp. Cell Res. 156:198-212. [DOI] [PubMed] [Google Scholar]
- 56.Yen, A., D. Brown, and J. Fishbaugh. 1987. Precommitment states induced during HL-60 myeloid differentiation: possible similarities of retinoic acid- and DMSO-induced early events. Exp. Cell Res. 173:80-84. [DOI] [PubMed] [Google Scholar]
- 57.Yen, A., M. Forbes, G. DeGala, and J. Fishbaugh. 1987. Control of HL-60 cell differentiation lineage specificity, a late event occurring after precommitment. Cancer Res. 47:129-134. [PubMed] [Google Scholar]
- 58.Yen, A., and R. Sturgill. 1998. Hypophosphorylation of the RB protein in S and G2 as well as G1 during growth arrest. Exp. Cell Res. 241:324-331. [DOI] [PubMed] [Google Scholar]