Abstract
Chromatin modification complexes are key gene regulatory factors which posttranslationally modify the histone component of chromatin with epigenetic marks. To address what features of chromatin modification complexes are responsible for the specific recognition of nucleosomes compared to naked histones, we have performed a functional dissection of the Esa1-containing Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex. Our studies define the Piccolo determinants sufficient to assemble its three subunits into a complex as well as Piccolo determinants sufficient to specifically acetylate a chromatin template. We find that the conserved Enhancer of Polycomb A (EPcA) homology region of the Epl1 component and the N-terminal 165 amino acids of the Yng2 component of Piccolo are sufficient with Esa1 to specifically act on nucleosomes. We also find that the Esa1 chromodomain plays a critical role in Piccolo's ability to distinguish between histones and nucleosomes. In particular, specific point mutations in the chromodomain putative hydrophobic cage which strongly hinder growth in yeast greatly reduce histone acetyltransferase activity on nucleosome substrates, independent of histone methylation or other modifications. However, the chromodomain is not required for Piccolo to bind to nucleosomes, suggesting a role for the chromodomain in a catalysis step after nucleosome binding.
Transcriptional regulation in a eukaryotic nucleus requires cellular activities that recognize and act on chromatin. Such activities include chromatin modification enzymes, which tag nucleosomes with posttranslational modifications (18), and chromatin remodeling enzymes, which render the constituent DNA in nucleosomes accessible to other transcription factors by disrupting or remodeling chromatin (4). One of the best-characterized posttranslational chromatin modifications is histone acetylation, which is generally associated with an open chromatin or activated transcriptional state (7). Despite the characterization of many histone acetyltransferase (HAT) complexes and catalytic subunits, it is not clear how HAT complexes specifically recognize a chromatin substrate versus naked histones, even though this is a fundamental property of many chromatin modification complexes.
Several distinct nuclear HAT complexes have been isolated from the budding yeast Saccharomyces cerevisiae, but only the NuA4 (nucleosome acetylating H4) complex is essential for cell viability (7). The 1.3-MDa NuA4 complex contains at least 12 polypeptides (2, 13) including the essential Esa1 (essential Sas-related acetyltransferase 1) subunit (8, 29). Although Esa1 is the catalytic subunit of the NuA4 complex, it acetylates only naked histones on its own and is unable to acetylated NuA4's physiological substrate of nucleosomes (2, 6). This inadequacy of the catalytic subunit to recognize or act on nucleosomes is a common feature of many HAT enzyme complexes (33). For example, yeast Gcn5 can acetylate histones on its own but additionally requires Ada2 and Ada3 to acetylate nucleosomes efficiently (3), and four separate yeast Gcn5-containing complexes (SAGA, SLIK/SALSA, ADA, and HAT A2) which acetylate nucleosomes also include Ada2 and Ada3 subunits (7).
Like Gcn5, Esa1 can also function in multiple complexes. Fractionation of yeast extracts showed the presence of Esa1 in the NuA4 complex as well as a smaller complex termed Piccolo NuA4, or Piccolo for short (6). Both NuA4 and Piccolo exhibit strong HAT activity towards nucleosomes, but strikingly, only Piccolo acetylates a nucleosome substrate in preference to naked histones. Combined biochemical and genetic data indicate that the Piccolo complex is responsible for global acetylation in yeast in contrast to activator-directed acetylation at transcription promoters by the NuA4 coactivator complex (6).
The Piccolo complex is an attractive chromatin modification enzyme for study because it is a relatively compact enzyme able to recognize and acetylate nucleosomes and because its three subunits Epl1, Yng2, and Esa1 boast rich genetic and biochemical backgrounds. Epl1 (Enhancer of Polycomb-Like 1) is the yeast homolog of the Drosophila Enhancer of Polycomb, E(Pc), isolated as a gene that enhanced Polycomb group mutations and suppressed position-effect variegation in Drosophila through some undefined mechanism involving chromatin (28, 30). E(Pc) homologs found in organisms such as yeast, worms, and mammals each contain a common 280-residue N-terminal Enhancer of Polycomb A (EPcA) domain. Yng2 shares significant sequence homology to the p33Ing1 human tumor suppressor candidate involved in cell proliferation and apoptosis (9, 14, 20, 25), particularly a common C-terminal plant homeodomain (PHD) finger domain, although weaker sequence homology also exists in the N-terminal regions. Finally, the catalytic subunit Esa1 contains the MYST histone acetyltransferase domain (33) and a 60-residue chromodomain found in many chromatin modification and remodeling proteins (19). The function of the chromodomain may be protein specific since the chromodomains of heterochromatin proteins HP1 and Polycomb bind to histone tails containing methylated lysines (11, 16, 17, 23, 24), but the chromodomain of the dosage compensation protein MOF binds to RNA (1).
Since relatively little information is available to explain how HAT enzymes recognize a chromatin template, we have examined the determinants of Piccolo necessary to act on a chromatin template compared to naked histones. We find that the conserved EPcA domain and chromodomain are critical for Piccolo to acetylate nucleosomes. Our results also suggest that a putative hydrophobic cage on the chromodomain surface is necessary for Piccolo to specifically act on nucleosomes after binding of this substrate. Surprisingly, chromodomain function in chromatin acetylation by Piccolo is independent of histone methylation, indicating a new distinct role of the Esa1 chromodomain.
MATERIALS AND METHODS
Coexpression and purification of Piccolo complexes.
Deletions of Piccolo subunits were designed after considering sequence homologies and secondary structure predictions using the PredictProtein server (26) (http://www.predictprotein.org/). Deletions and point mutations of Piccolo subunits were prepared using standard molecular biology techniques, verified by sequencing through entire coding regions, and subcloned into the pST44 polycistronic expression vector (32) before expression in Escherichia coli BL21(DE3)pLysS cells. The coexpressed proteins were purified by Talon (Clontech) cobalt affinity chromatography in P100 buffer (50 mM sodium phosphate, pH 7.5, 100 mM NaCl, 5 mM 2-mercaptoethanol, 1 mM benzamidine) with additional 100 mM imidazole for elution. yEpl1 and Yng2 truncations were almost completely insoluble when expressed on their own in E. coli but become predominantly soluble when coexpressed with Esa1.
HAT and ELISA assays.
HAT assays were performed minimally three times for each sample using previously described procedures except that 0.125 μCi of [3H]acetyl-coenzyme A was used per reaction (15). HAT assays were normalized by the amount of histones, whether provided as naked histones or nucleosomes. Native chicken histones and oligonucleosomes were isolated from chicken erythrocytes (15, 21). Recombinant Xenopus core histones were expressed, purified, reconstituted with mouse mammary tumor virus long terminal repeat NucB DNA into nucleosome core particles as described previously (12, 21). Recombinant yeast core histones (27) were similarly reconstituted into nucleosome core particles. Both Xenopus and yeast recombinant nucleosome core particles were purified by anion exchange Source Q (Amersham) chromatography.
Appropriate dilutions of each Piccolo complex were prepared to ensure the measured activity remained in the linear range of the HAT assay. The samples were normalized by Esa1 content by enzyme-linked immunosorbent assay (ELISA) using anti-His polyclonal rabbit antibodies (Santa Cruz) or affinity-purified anti-Esa1 polyclonal rabbit antibodies prepared against recombinant full-length Esa1 (this study) because normalization by Coomassie blue staining of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels proved to be insufficiently precise. Piccolo v14 and 15 histone and nucleosome HAT activities were normalized to Piccolo v7 by Esa1 content via visual inspection of Western blots using anti-His antibodies because ELISAs using anti-His or anti-Esa1 antibodies produced results clearly inconsistent with the Western blot and Coomassie blue-stained SDS-PAGE gels. All other Piccolo constructs produced self-consistent ELISA, Western, and Coomassie blue quantitation of Esa1.
Plasmid shuffle.
Esa1 chromodomain mutations were transferred from a pBluescript plasmid to a yeast vector by amplification with the following primers containing BamHI sites; For 5′ TATAAGGATCCTCCCATGACG GAAAAGAAGAACCTGGTATTG-3′, and Rev 5′-GCGGGATCCTTACCAGGCAAAGCGTAACTGAGAGGC-3′. BamHI sites are indicated in italics, and stop codon is underlined. The product was digested with BamHI and ligated into a BamHI-digested pBFG6 plasmid, a 2μm plasmid providing the phosphoglycerol kinase promoter, six N-terminal hemagglutinin (HA) epitopes and the LEU2 selection marker (25). Each plasmid was verified by sequencing and transformed into QY118 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 esa1Δ::KanMX pLP795 (ESA1 ARS/CEN URA3) (8) by standard protocols. Resulting clones were selected on SC medium lacking leucine and uracil. Good in vivo expression of each Esa1 mutant protein was confirmed by Western blot on whole-cell extracts (with anti-HA antibodies).
Plasmid shuffling experiments for viability tests were performed following standard procedure. Yeast containing wild-type and mutant versions of ESA1 on URA3 and LEU2 plasmids, respectively, were grown overnight in medium lacking leucine and with uracil, diluted to an optical density of 0.25, and grown for 90 min at 30°C. Tenfold serial dilutions in water were spotted on YPD and HC lacking leucine 0.1% 5′-fluoroorotic acid plates (to chase the wild-type ESA1/URA3 plasmid) and grown at 30°C for 2 to 4 days.
Piccolo binding of nucleosomes via size-exclusion chromatography.
To preparatively purify Piccolo v7, v17, v69, v72, and v74, soluble extract from 6 liters of BL21(DE3)pLysS cells expressing the appropriate Piccolo complex were purified successively by Talon cobalt affinity, Source Q anion-exchange, Source S cation-exchange, and Source ISO hydrophobic interaction chromatography (all Source resins from Amersham). The Piccolo/nucleosome core particle complex prepared by mixing Piccolo and recombinant Xenopus nucleosome core particles were fractionated by Superdex HR 200 (Amersham) size exclusion chromatography in 20 mM Tris-Cl, pH 8.0, 50 mM KCl, 5 mM 2-mercaptoethanol at 0.4 ml/min flow rate.
Piccolo binding of nucleosomes via StrepTactin pulldown.
Piccolo v78 (Epl1Δ3, C-terminal Strep II peptide-tagged Yng2, Esa1) and Piccolo (Epl1Δ3, C-terminal Strep II-tagged Yng2, Esa1Δ3) were coexpressed and purified by Talon metal affinity chromatography as described above. These Strep-tagged Piccolo complexes were incubated with Strep-Tactin resin (IBA GmbH) in TG50 buffer (50 mM Tris-Cl, pH 8.0, 50 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% Tween 20, and 10 mM 2-mercaptoethanol) for 1 h at 4°C, washed three times with 20 resin volumes of TG50 buffer, and then incubated with recombinant Xenopus nucleosome core particles. After 1 h incubation at 4°C, the supernatant was stored as the unbound fraction, and the resin was washed three times with 20 resin volumes of TG50 buffer. Equivalent volumes of input nucleosome core particles, unbound (supernatant), and bound fractions were fractionated by SDS-PAGE and detected by Western blotting using anti-H3 antibodies (Abcam).
RESULTS
EPcA homology in Epl1 is sufficient for nucleosomal HAT activity of Piccolo.
We have previously shown that the N-terminal half of Epl1, specifically Epl1 residues 1 to 485, and full-length Yng2 and Esa1 are sufficient for Piccolo's strong nucleosomal HAT activity in vivo and in vitro (6). Here, we define the functionally important regions of each subunit by assaying Piccolo containing deletions or mutations in the three subunits. Use of a polycistronic expression system (31) modified to permit subcloning of individual genes in any order made it possible to rapidly prepare 28 variant Piccolo complexes for this study. Piccolo deletion complexes expressed and reconstituted in E. coli were then partially purified using an engineered hexahistidine tag on the Esa1 subunit (Fig. 1a).
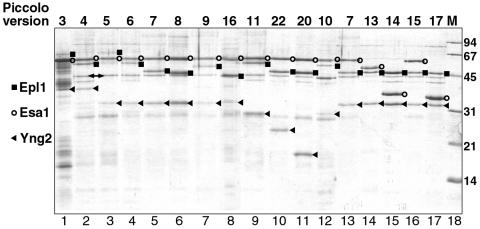
FIG. 1.
Representative selection of Piccolo deletion complexes used for HAT assays. The Coomassie-stained SDS-PAGE gel shows recombinant Piccolo complexes coexpressed in E. coli and purified by Talon chromatography. Constituent polypeptides in each deletion are described in Fig. 2. Piccolo v3 in lane 1 was additionally purified by Source Q and S chromatography. Epl1, Yng2, and Esa1 polypeptides for each deletion are indicated with a black square, black triangle, and open circle, respectively, to the right of the appropriate band. An E. coli contaminant with apparent size of 43 kDa that copurifies over Talon chromatography is labeled with a double-headed arrow between lanes 2 and 3. Molecular size standards are shown in lane 18 with corresponding sizes to the right (in kilodaltons).
Complex formation was judged by the copurification of untagged subunits (Epl1 and Yng2) with the tagged subunit (Esa1). The substoichiometric amounts of Epl1 for Piccolo v5 in lane 3 reflects partial degradation of the Epl1Δ1 component (data not shown).
To compare the activities of the deletion variants, we calculate a preference for nucleosomes ratio defined as HAT activity on nucleosome substrate divided by the HAT activity on naked histone substrate. This ratio has several advantages since it provides a measure of the ability of Piccolo to specifically recognize and acetylate a nucleosome substrate, the ratio greatly reduces the effect of variations in specific activity between samples, and the ratio automatically normalizes for the amount of Esa1 between different Piccolo deletions. To further facilitate the comparison between different Piccolo deletion constructs, we express the preference for nucleosomes ratio relative to Piccolo v7, where Piccolo v7 is set to 1.0. The HAT activities of Piccolo v5, v7, and v20 additionally purified by anion- and cation-exchange chromatography were very similar to what we observed for the respective partially purified complexes shown in Fig. 1, suggesting that the single-step affinity-purified Piccolo complexes are sufficiently pure for HAT activity measurements (data not shown).
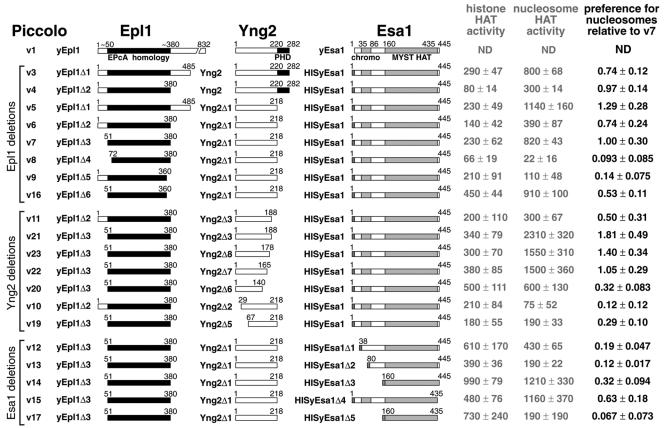
Our deletions of Epl1 demonstrate the importance of the EPcA homology domain in Epl1 for Piccolo function (Fig. 2). Since removing the Yng2 PHD domain did not decrease the preference of the Piccolo complex to acetylate nucleosomes (compare Piccolo v3 and v5) but produced more homogeneous protein complexes, we examined Epl1 deletions in the context of Yng2(1-218). Piccolo v4, v5, and v6, which all contain the EPcA homology, retain strong nucleosomal HAT activity, as does Piccolo v7, which contains just the EPcA homology for its Epl1 component. Removing just the respective N-terminal and C-terminal 20 residues from the EPcA homology in Piccolo v8 and v9 dramatically reduces the preference for nucleosomes to 10 to 15% compared to Piccolo v7 (also compare Piccolo v6 and v9 in Fig. 2). Interestingly, Piccolo v16, which contains the minimal EPcA homology lacking its C-terminal 20 residues, still maintains approximately half of its preference for nucleosomes.
FIG. 2.
Deletion analysis of Piccolo subunits. The chart shows the identity of subunits present in each Piccolo deletion construct, with evolutionarily conserved regions (EPcA, PHD, and chromodomain), structurally determined regions (Esa1 HAT domain), and hexahistidine tags shown as black and shaded regions. Naked histone and nucleosome HAT activities were normalized by ELISA-determined Esa1 content using anti-Esa1 antibodies. The preference for nucleosome ratio shown on the right side is the ratio of naked histone HAT activity to nucleosome HAT activity, with Piccolo v7 set to 1.00.
In general, the evolutionarily nonconserved N-terminal 50 amino acids of yEpl1 inhibit the nucleosomal preference (compare Piccolo v6 and v7, v9 and 16, and v11 and v21 in Fig. 2). Taken together, these results show that the yeast 280-residue EPcA homology near the N terminus of each Drosophila E(Pc) homolog is sufficient to form a Piccolo complex with Yng2 and Esa1 and is important for Piccolo's ability to recognize and acetylate nucleosomes.
Piccolo's ability to acetylate nucleosomes requires Yng2 N-terminal sequences but not the C-terminal PHD domain.
We find that removing the highly conserved C-terminal PHD finger does not significantly affect recombinant Piccolo complex formation or the nucleosome preference of Piccolo's HAT activity (Fig. 2, compare v3, v4, and v5). This observation is consistent with previous studies which showed that PHD region is not necessary for the HAT activity of Piccolo (20, 25). Our results further establish that the C-terminal 40% of Yng2 does not appear to play a significant role for complex formation or ability to acetylate nucleosomes since Piccolo v7, v21, v23, and v22, which all contain at least Yng2(1-165) retain both functions. However, Piccolo v20 containing Yng2(1-140) possesses significantly less preference for nucleosomes, suggesting that some aspect of Yng2 residues 140-165 is involved in recognition and/or acetylation of nucleosomes.
In contrast to the PHD finger, the N-terminal half of Yng2 bears less sequence similarity with its p33Ing1 homologs, but is critical for Piccolo's nucleosomal HAT activity. Deletion of the first 28 or 66 amino acids of Yng2 is sufficient to reduce Piccolo's preference for nucleosomes by at least a factor of three (Fig. 2, compare v7 with v10 and v19).
Esa1 chromodomain is essential for Piccolo nucleosomal HAT activity.
The 445-residue Esa1 contains a chromodomain near its N terminus and a MYST catalytic core which occupies the C-terminal two-thirds of the protein. Since the catalytic core possesses a well-defined structure (34), we chose not to analyze deletions within residues 160 to 435. However, we did examine the role of the chromodomain since this region was not necessary for naked histone HAT activity in vitro and yet essential for Esa1 function in vivo (34). We find that deleting just 3 residues from the N terminus of the Esa1 chromodomain (Piccolo v12) causes a fivefold decrease in Piccolo preference for nucleosomes, a similar effect observed when most of the chromodomain is removed in Piccolo v13 (Fig. 2). This suggests that the integrity of the chromodomain is critical for Esa1 function in the Piccolo complex. It is conceivable that misfolding of the chromodomain which may occur when it is partially deleted accounts for the lower nucleosomal HAT activity of Piccolo v12 and v13 compared to Piccolo v14, where the chromodomain is completely deleted.
The C-terminal 10 residues of Esa1 (residues 436 to 445) also appears to play a role in nucleosomal HAT activity. Deleting these 10 amino acids from full-length Esa1 reduces the preference for nucleosome ratio by nearly 40% in Piccolo v15 compared to v7 (Fig. 2). Furthermore, removing the same 10 C-terminal amino acids in the context of the catalytic core in Piccolo v17 reduces the preference for nucleosomes to less than 10% of v7. Our results therefore show that the Esa1 catalytic core defined structurally as Esa1(160-435) is capable of forming a ternary complex with Epl1 and Yng2 but possesses substantially less activity on nucleosome substrates compared to full-length Esa1, which additionally contains the chromodomain.
Putative hydrophobic cage in Esa1 chromodomain is critical for both Piccolo function in vitro and cell viability.
The crystal structure of the HP1 chromodomain bound to an H3 peptide containing a trimethylated lysine residue shows that the H3 peptide forms an extended chain that completes a beta-sandwich created by the HP1 chromodomain (16, 24). Furthermore, the trimethylated lysine binds tightly in a hydrophobic cage formed by aromatic chromodomain residues Y24, Y48, and W45 (Fig. 3a). Since not all these hydrophobic cage residues are conserved between HP1 and Esa1, Esa1 chromodomain is not expected to bind methylated lysine residues. In fact, Jacobs et al. have demonstrated that Esa1 chromodomain binds unmodified H3 tail, binds more weakly to methylated H3 tail, and binds very poorly to unmodified or acetylated H4 tails (17). In the context of nucleosomes, Piccolo acetylates histone H4 tails preferentially to H2A tails without significant activity on H3 tails (6). Given our results showed that the Esa1 chromodomain plays a major role in Piccolo's ability to acetylate nucleosomes, we asked if mutating the potential chromodomain hydrophobic cage residues would affect Piccolo's nucleosomal HAT activity.
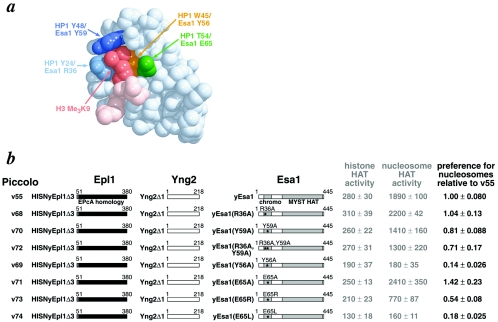
FIG. 3.
Piccolo variants containing Esa1 chromodomain mutations. (a) Space-filling representation of H3 peptide containing trimethylated K9 bound to Drosophila HP1 chromodomain. The HP1 chromodomain is shown in light blue, while the H3 peptide backbone and the trimethylated K9 are shown in pink and maroon, respectively. The residues that create or line the hydrophobic cage are displayed as follows: Y24 in light blue, Y48 in dark blue, W45 in yellow, and T54 in green. The corresponding Esa1 residues are also provided. Figure prepared using MidasPlus molecular graphics software (10) and PDB coordinates 1KNE (16). (b) Effect of Esa1 chromodomain point mutations on Piccolo HAT activity. Piccolo v55 and v68 to v74 each contain an N-terminal hexahistidine-tobacco etch virus nuclear inclusion a (NIa) protease site tag (HISN) on the Epl1 component, and all complexes were expressed and purified to similar levels. The preference for nucleosome ratio shown on the right side is the ratio of nucleosomal HAT activity to naked histone HAT activity, with Piccolo v55 set to 1.00 for all eight Piccolo variants.
Our results suggest that the putative Esa1 chromodomain hydrophobic cage plays a significant role in Piccolo's ability to acetylate nucleosomes, but in a different manner than the HP1 chromodomain. The Esa1 chromodomain single point mutation Y56A (Esa1 Y56 corresponds to HP1 W45) alone caused a sevenfold reduction in Piccolo's preference for nucleosomes (Fig. 3b, v69). However, removing either or both of the side chains of R36 and Y59 (corresponding to HP1 Y24 and Y48) did not affect Piccolo complex formation, naked histone, or nucleosomal HAT activity (Fig. 3b, v68, v70, and v72). In contrast, the corresponding alanine substitution mutations in HP1 reduced its binding to K9 trimethylated H3 peptide at least 20-fold (16).
We also examined Esa1 E65 mutations because our model building suggested the possibility that an unmodified positive charged lysine residue might contact this acidic residue. We find that removing the side chain actually increased Piccolo's preference for nucleosome by 40% (Fig. 3b, v71), while changing the ionic charge with the E65R mutation decreased this ability by almost 50% (Fig. 3b, v73). We observe an even more dramatic effect when the negatively charge glutamic acid is replaced with a hydrophobic leucine residue: the E65L mutation causes a sixfold reduction in nucleosome preference, a similar deleterious effect as the Y56A mutation (Fig. 3b, v74).
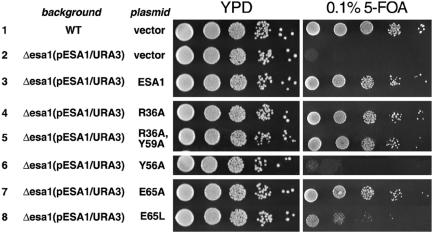
Since Esa1 is an essential yeast gene, we have used a URA3-based plasmid shuffle system to examine the effect of these Esa1 chromodomain mutations in vivo. We find a strong correlation between the in vitro nucleosomal HAT activity of Piccolo and the viability of yeast cells (Fig. 4). The single R36A and double R36A Y59A mutations possess nearly wild-type nucleosomal HAT activity (104% and 71%, respectively, compared to wild-type v55), and yeast cells with these mutations grow at the same rate as the positive control (Fig. 4, lanes 3 to 5). Similarly, yeast cells with the E65A mutation, which results in slightly higher than wild-type nucleosomal HAT activity, do not display any growth defect (Fig. 4, lane 7). However, the E65L mutation associated with a sixfold reduction in nucleosomal HAT activity causes severe growth defects in yeast, while cells with the Y56A mutation which caused a sevenfold decrease in HAT activity in vitro are not viable (Fig. 4, lanes 8 and 6). Western blots confirm that cellular Esa1 levels are similar for each mutant, indicating that the growth defects do not result from instability of mutant Esa1 proteins in the yeast cells (data not shown). These results strongly suggest that the Esa1 chromodomain putative hydrophobic cage plays an critical role in yeast related to Piccolo's ability to acetylate nucleosomes.
FIG. 4.
Specific mutations in Esa1 chromodomain severely affect growth of yeast cells. Yeast strains deleted for Esa1 and containing a wild-type Esa1 gene on a low-copy-number URA3 plasmid were transformed with a LEU2 plasmid expressing wild-type Esa1 or Esa1 containing the indicated point mutations in the chromodomain. The left panels show growth of 10-fold serial dilutions on YPD rich medium, while the right panels show similar growth on 0.1% fluoroorotic acid plates. Rows 6 and 8 document the severe growth defects of yeast cells expressing the Y56A and E65L Esa1 chromodomain mutations.
Histone modifications are not necessary for specific effects of Piccolo chromodomain mutations.
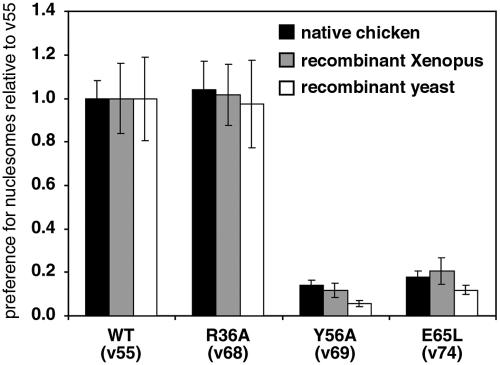
Since one established role for the chromodomain is to bind a modified histone tail, we have examined the activity of the Piccolo containing Esa1 chromodomain mutations on native and recombinant nucleosome substrates. In particular, we have compared native chicken blood nucleosome substrates (as used in Fig. 2 and 3) with recombinant Xenopus and recombinant yeast nucleosomes. Since the recombinant Xenopus and yeast nucleosomes contain core histones expressed in E. coli, these histones are not expected to contain posttranslational modifications.
We find that the Piccolo containing the Esa1 R36A chromodomain mutation does not affect the preference for nucleosome, consistent with our HAT results using chicken nucleosomes and the in vivo complementation studies (Fig. 5, v68). Furthermore, the Y56A and E65L mutations, which severely affected yeast viability and HAT activity using native chicken nucleosome substrates, also severely reduced HAT activity using recombinant yeast nucleosomes (Fig. 5, v69 and 74). Essentially the same results were obtained when recombinant Xenopus nucleosome substrates were used. These results indicate that the chromodomain's critical role in Piccolo acetylation of nucleosomes does not require posttranslational modifications, since similar effects on HAT activity of specific mutations are detected with both native substrates with posttranslational modifications and recombinant substrates without such modifications. In particular, our results show that the Esa1 chromodomain does not bind a methylated lysine for Piccolo acetylation function.
FIG. 5.
Specific effects of chromodomain mutations do not require histone modifications. Piccolo v55, v68, v69, and v74 were assayed using native chicken (black bars), recombinant Xenopus (grey bars), and recombinant yeast (white bars) nucleosomes. Equivalent amounts of nucleosome substrates determined by Coomassie-stained SDS-PAGE gels of the histone proteins were used in the assays. The preference for nucleosome ratio shown on the right side is the ratio of nucleosomal HAT activity to naked histone HAT activity, with Piccolo v55 set to 1.00.
Esa1 chromodomain is not required for Piccolo to bind to nucleosomes.
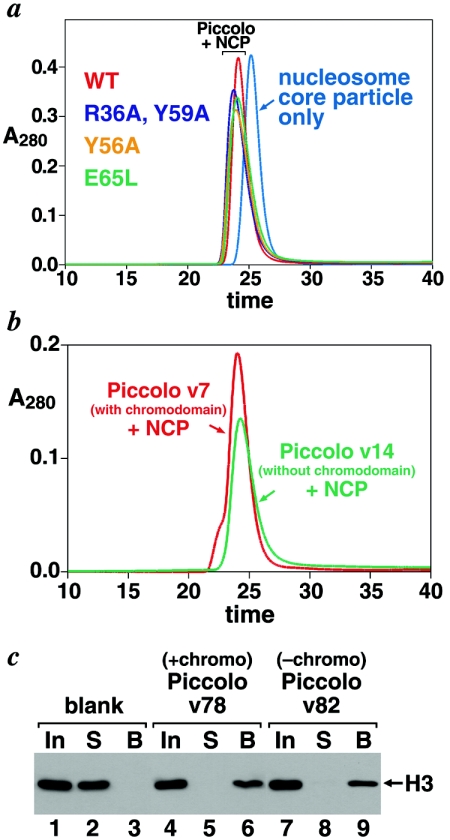
For Piccolo to acetylate nucleosomes, it must presumably first bind to its nucleosome substrate. In fact, a stable Piccolo/nucleosome core particle complex is observed when we incubate Piccolo complex with recombinant nucleosome core particles and analyze the mixture by gel filtration chromatography (Fig. 6a). If the Esa1 chromodomain is required for binding to nucleosomes, we would expect that the Y56A and E65L chromodomain mutations, which greatly affected nucleosomal HAT activity and cell viability, would adversely affect Piccolo/nucleosome complex formation. However, no noticeable change in Piccolo/nucleosome complex formation is detected by gel filtration for either the Y56A or the E65L mutation, showing that Piccolo complexes containing these mutations still form stable complexes with nucleosomes. We have also examined Piccolo/nucleosome complex formation using Piccolo v17, which lacks the entire chromodomain. We find that Piccolo v17 binds to nucleosomes similarly to Piccolo v7, which contains full-length Esa1 (Fig. 6b).
FIG. 6.
Esa1 chromodomain is not necessary for Piccolo to bind nucleosomes. (a) Gel filtration chromatograms of recombinant nucleosome core particles alone (cyan) and nucleosome core particles incubated with Piccolo v7 (red), Piccolo v72 (blue), Piccolo v69 (yellow), and Piccolo v74 (green). All Piccolo samples form stable complexes with nucleosomes, as shown by the appearance of a larger peak, distinct from the nucleosome peak and from the Piccolo-only peak at approximately 27 min (data not shown). SDS-PAGE gels of the peaks confirm the assignment of the gel filtration peaks. For example, fractions for the Piccolo/nucleosome peaks show all three Piccolo subunits and all four nucleosome histone subunits (data not shown). (b) Gel filtration chromatograms of recombinant nucleosome core particles (NCP) incubated with Piccolo v7 (red), which contains full-length Esa1, or Piccolo v14 (green), which lacks the Esa1 chromodomain. Both samples form the Piccolo/nucleosome complex, establishing that the chromodomain is not required for stable binding of Piccolo to nucleosomes. (c) Piccolo/nucleosome pulldown experiment confirms that the Esa1 chromodomain is not necessary for binding to nucleosomes. Blank (lanes 1 to 3), Piccolo v78 (lanes 4 to 6), or v82 (lanes 7 to 9) was immobilized on Strep-Tactin beads via their Strep-tagged Yng2 subunits and incubated with nucleosome core particles, and unbound and bound fractions were analyzed by Western blots to detect the histone H3 component of nucleosomes. Input samples are shown in lanes 1, 4, and 7; unbound (supernatant) fractions in lanes 2, 5, and 8; and bound fractions in lanes 3, 6, and 9.
To independently verify that Piccolo binds to nucleosomes and that the Esa1 chromodomain is not necessary for this interaction, we employed a pulldown assay using epitope-tagged Piccolo. Specifically, Piccolo v78, which contains Epl1Δ3, C-terminally Strep II-tagged full-length Yng2, and full-length Esa1 was bound to Strep-Tactin resin via the Strep tag, and the immobilized Piccolo complex was incubated with nucleosome core particles. The unbound and bound fractions were analyzed after washing by Western blotting using anti-H3 antibodies. Figure 6c shows that nucleosome core particles alone do not bind to the Strep-Tactin resin (lanes 1 to 3), whereas nucleosomes do bind to Piccolo v78 immobilized on the same resin (lanes 4 to 6). Furthermore, similar binding is detected when Piccolo v82, equivalent to Piccolo v78 without the Esa1 chromodomain, is bound to the Strep-Tactin resin (lanes 7 to 9). Thus, we conclude that the chromodomain is not required for Piccolo to bind to nucleosomes but may instead play a role in a catalysis step after binding.
DISCUSSION
We have exploited a new E. coli modular polycistronic expression system to coexpress and to functionally dissect the three-protein yeast Piccolo NuA4 complex. This complex represents the catalytic core of the 1.3-MDa NuA4 histone acetyltransferase coactivator but also appears to be a bona fide HAT complex on its own, responsible for the global H2A and H4 acetylation of histones in yeast (6). Our biochemical analysis of the variant Piccolo complexes defines the regions of the component proteins sufficient for Piccolo assembly as well as regions necessary to recognize and acetylate nucleosomes.
We find that the EPcA homology region of Epl1 is sufficient for Piccolo complex formation with Yng2 and Esa1. In contrast, the well-conserved PHD finger of Yng2 is dispensable for Piccolo complex formation, as are the N-terminal 66 residues. We also find that the HAT catalytic domain of Esa1 is sufficient for Piccolo complex assembly. However, the requirements for the preferential acetylation of nucleosomes are more stringent. The N-terminal 20 amino acids of the EPcA homology region of Epl1 are necessary for nucleosome HAT activity, while some aspect of the Yng2 N-terminal 28 residues is necessary for Piccolo's robust ability to acetylate nucleosomes even though they are not necessary for Piccolo complex formation. Our results also show that the chromodomain of Esa1 near its N terminus plays a critical role in Piccolo's preference for nucleosomes, although Piccolo can assemble in the absence of the chromodomain. Similar results attest to a significant role for the very C-terminal 10 amino acids of the Esa1 protein in Piccolo's ability to acetylate a nucleosome substrate.
Although this deletion analysis defines the regions of Epl1, Yng2, and Esa1 that assemble Piccolo and confer upon Piccolo its nucleosome acetylation function, sequence homologies for these regions unfortunately do not provide insight into the mechanisms for the assembly and catalytic functions. However, point mutations introduced into the Esa1 chromodomain do shed light on possible mechanisms for the catalytic function. Our results show that specific mutations in the putative hydrophobic cage of Esa1 affect Piccolo's ability to acetylate nucleosomes but not in a way that could be predicted based on the HP1 chromodomain hydrophobic cage's binding to a methylated lysine histone peptide. This suggests that Piccolo does use the Esa1 chromodomain hydrophobic cage in its catalytic mechanism but not necessarily the same way HP1 binds H3 peptides through methylated lysine.
Since the Esa1 chromodomain can bind unmodified histone tails (17), we considered the possibility that the chromodomain is required for Piccolo to bind to nucleosomes, but our results using chromodomain point mutations or removing the entire chromodomain show that Piccolo does not require the chromodomain to form a stable complex with nucleosome core particles. Since we used recombinant nucleosomes containing E. coli-expressed histones for our binding studies, our results also suggest that Piccolo does not require histone modifications to form a stable complex with nucleosomes. The specific effects of the chromodomain point mutations on Piccolo HAT activity also do not require histone modifications, since similar effects are observed whether nucleosomes used were isolated from natural sources or produced recombinantly.
Our data suggests the possibility that the Esa1 chromodomain is involved in some catalysis event after Piccolo binds to nucleosomes. For example, since nucleosomal histone tails are likely to be associated with DNA (22), Piccolo may use the Esa1 chromodomain to pry histone tails away from the DNA onto the Esa1 MYST HAT domain. One possible mechanism for Piccolo to separate histone tails from DNA might be for the Esa1 chromodomain to bind directly to histone tails. This would be consistent with the observation that recombinant Esa1 chromodomain can bind unmodified histone tails, although that study found the Esa1 chromodomain interacted with unmodified histone H3 tail and not the H4 tail acetylated by Piccolo (17). An alternative but not mutually exclusive model is that the chromodomain binds nucleic acid and displaces histone tails by competing for the histone tail's DNA binding site on the nucleosome. Several studies have shown that various chromodomains can bind to nucleic acid: the dosage compensation histone acetyltransferase MOF binds to noncoding RNA in Drosophila melanogaster (1) and the Mi-2 chromodomain binding of DNA is involved in the Mi-2 complex's ATP-dependent nucleosome remodeling activity (5).
Our deletion and mutational analysis of Piccolo have defined the regions of this histone modification complex which allow the specific recognition of its physiological nucleosome substrate and refined our ideas of how the Esa1 chromodomain functions in nucleosome acetylation. It will be important now to investigate precisely how Piccolo binds its nucleosome substrate, including the roles of individual histone tails for this process.
Acknowledgments
We dedicate this manuscript to Bob Simpson, whose pioneering chromatin studies inspired us and whose support and friendship we cherished.
We are grateful to the Tan Lab and the entire Penn State gene regulation community for support and fruitful discussions. We also thank Tim Richmond and Brad Cairns for recombinant Xenopus and yeast histone expression plasmids, respectively, and Marianne Potvin, Andreanne Auger, and Tim Kelly for technical help.
I.F. is supported by a Canada Graduate studentship. S.T. is a Pew Scholar in the Biomedical Sciences. This work was supported by grant GM-60489 from the National Institutes of Health to S.T. and by grants from the Canadian Institutes of Health Research to J.C.
REFERENCES
- 1.Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405-409. [DOI] [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Bouazoune, K., A. Mitterweger, G. Langst, A. Imhof, A. Akhtar, P. B. Becker, and A. Brehm. 2002. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21:2430-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley, S. Allard, J. Savard, W. S. Lane, S. Tan, and J. Cote. 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, X., Y. Hara, and K. Riabowol. 2002. Different HATS of the ING1 gene family. Trends Cell Biol. 12:532-538. [DOI] [PubMed] [Google Scholar]
- 10.Ferrin, T. E., C. C. Huang, L. E. Jarvis, and R. Langridge. 1988. The MIDAS display system. J. Mol. Graphics 6:13-27. [Google Scholar]
- 11.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flaus, A., and T. J. Richmond. 1998. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 275:427-441. [DOI] [PubMed] [Google Scholar]
- 13.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 14.Garkavtsev, I., I. A. Grigorian, V. S. Ossovskaya, M. V. Chernov, P. M. Chumakov, and A. V. Gudkov. 1998. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391:295-298. [DOI] [PubMed] [Google Scholar]
- 15.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 19.Jones, D. O., I. G. Cowell, and P. B. Singh. 2000. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22:124-137. [DOI] [PubMed] [Google Scholar]
- 20.Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol, and D. Young. 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119:1-16. [DOI] [PubMed] [Google Scholar]
- 22.Luger, K., and T. J. Richmond. 1998. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8:140-146. [DOI] [PubMed] [Google Scholar]
- 23.Min, J., Y. Zhang, and R. M. Xu. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 25.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Cote. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rost, B., and J. Liu. 2003. The PredictProtein server. Nucleic Acids Res. 31:3300-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha, A., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinclair, D. A., N. J. Clegg, J. Antonchuk, T. A. Milne, K. Stankunas, C. Ruse, T. A. Grigliatti, J. A. Kassis, and H. W. Brock. 1998. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics 148:211-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stankunas, K., J. Berger, C. Ruse, D. A. Sinclair, F. Randazzo, and H. W. Brock. 1998. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development 125:4055-4066. [DOI] [PubMed] [Google Scholar]
- 31.Tan, S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21:224-234. [DOI] [PubMed] [Google Scholar]
- 32.Tan, S., R. C. Kern, and W. Selleck. 2005. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40:385-395. [DOI] [PubMed] [Google Scholar]
- 33.Utley, R. T., and J. Cote. 2003. The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 274:203-236. [DOI] [PubMed] [Google Scholar]
- 34.Yan, Y., N. A. Barlev, R. H. Haley, S. L. Berger, and R. Marmorstein. 2000. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6:1195-1205. [DOI] [PubMed] [Google Scholar]