Abstract
Numerous studies indirectly implicate Rac GTPases in cancer. To investigate if Rac3 contributes to normal or malignant cell function, we generated rac3 null mutants through gene targeting. These mice were viable, fertile, and lacked an obvious external phenotype. This shows Rac3 function is dispensable for embryonic development. Bcr/Abl is a deregulated tyrosine kinase that causes chronic myelogenous leukemia and Ph-positive acute lymphoblastic leukemia in humans. Vav1, a hematopoiesis-specific exchange factor for Rac, was constitutively tyrosine phosphorylated in primary lymphomas from Bcr/Abl P190 transgenic mice, suggesting inappropriate Rac activation. rac3 is expressed in these malignant hematopoietic cells. Using lysates from BCR/ABL transgenic mice that express or lack rac3, we detected the presence of activated Rac3 but not Rac1 or Rac2 in the malignant precursor B-lineage lymphoblasts. In addition, in female P190 BCR/ABL transgenic mice, lack of rac3 was associated with a longer average survival. These data are the first to directly show a stimulatory role for Rac in leukemia in vivo. Moreover, our data suggest that interference with Rac3 activity, for example, by using geranyl-geranyltransferase inhibitors, may provide a positive clinical benefit for patients with Ph-positive acute lymphoblastic leukemia.
Rac1 and -2 are distinct members of the Rho family of small GTPases that regulate a very large number of cellular functions (12, 13, 43). We identified Rac3, the last member of the human Rac family, by cloning it as a cDNA from a chronic myelogenous leukemia cell line (10). Rac1, -2, and -3 share a high degree of amino acid sequence similarity, with the largest degree of divergence found at their C-terminal ends (10). Biochemically, Rac1 and Rac3 are also closely related (12). Rac GTPases cycle between two distinct conformations that are dictated by their binding to GTP or GDP. RacGTP represents the active conformation of the protein. Guanine nucleotide exchange factors such as the hematopoiesis-specific protein Vav1 stimulate the generation of activated Rac. Vav1 guanine nucleotide exchange factor activity, in turn, is stimulated by tyrosine phosphorylation (5, 14).
Functions ascribed to Rac include those that are abnormal in many types of cancer, such as regulation of programmed cell death and cell proliferation. Of note, some Rac guanine nucleotide exchange factors such as Tiam1 and Vav were originally identified as oncogenes (11, 24). Rac1 was shown to be needed to transduce Ras transforming signals in transfected cells (25, 34, 37, 39). In addition, Rac3, similar to Rac1, cooperates with Raf in a transformation assay of NIH 3T3 fibroblasts (22). Constitutively active Rac3 stimulated DNA synthesis in breast cancer cells (32). When constitutively active Rac3 was targeted to mammary epithelium in transgenic mice, involution was found to be incomplete and older female mice developed benign mammary gland lesions (27). These and other data (21, 26, 30, 35, 47) suggest that activated Racs may contribute to cancer development or progression.
To explore the different roles of Rac1 and Rac2 in vivo, conditional and standard null mutant mice have been generated. These studies have unveiled interesting distinct functions as well as overlapping activities for these highly related GTPases (7, 9, 38, 42, 46). We similarly sought to investigate the function of Rac3 in normal and neoplastic cells. In the current study, we report the generation of a mutant mouse that lacks any rac3 function, and using these mice, we have specifically evaluated the contribution of Rac3 to the class of acute lymphoblastic leukemia generated by the Bcr/Abl oncogene (8).
MATERIALS AND METHODS
Generation of rac3 null mutant mice.
We cloned 22.5 kb of DNA containing the complete mouse genomic rac3 locus (located on murine chromosome 11), including flanking sequences, in five overlapping phage genomic clones, using a human RAC3 cDNA probe from the 3′ untranslated region. No other highly homologous sequences were isolated using this probe. The mouse rac3 gene contains six exons and the cDNA-homologous region is dispersed over about 2.4 kb. Sequencing of the exons confirmed the identity of the locus (mouse genomic rac3 locus, NT_036064).
A targeting vector was made in which a 1.8-kb SstII-Eco47III fragment was replaced by a phosphoglycerol kinase (PGK)-neomycin resistance cassette. The SstII site is located in the rac3 promoter region, and the Eco47III site is present just 3′ to the stop codon within the 3′ untranslated region of rac3. This cassette replaced exons 1 to 5 and part of exon 6 of mouse rac3. The final targeting vector included 7.2 kb of 5′ homology and 7.1 kb of 3′ homology to endogenous murine rac3. A herpes simplex virus thymidine kinase cassette was included as a negative selection marker. The targeting vector was linearized using XbaI. Homologous recombination was obtained essentially as described (23).
We obtained three homologous recombinants, R63, R92, and R107. R107 and R63 gave germ line transmission in chimeric mice. The targeted loci of R107 and R63 were mapped in detail. One of these showed evidence of gene rearrangement 3′ in the rac3 genomic locus and caused embryonic lethality before embryonic day 9.5 (E9.5) in a 129Sv mouse background. The second, correctly recombined allele was also used to make chimeric mice and to obtain germ line transmission. This null mutant allele was bred into the C57BL/6J background until F9. Genotyping was performed using either Southern blots with an internal probe or PCR. Forward primer F6 from rac3 exon 6 (GGT TCC GTC AAG TAC CTG GA) was used in combination with a reverse primer from the 3′ untranslated region (GGG ACA TCA CAC AGC TCA ACA CGA) to detect the wild-type allele as a 200-bp product. To detect the targeted allele, we used a NeoP primer from the PGK-neo cassette (AGC TCA TTC CTC CCA CTC ATG) and the AS1 reverse primer, which generate a 300-bp product. A multiplex PCR was performed using all three primers and an annealing temperature of 58°C.
RT-PCR and quantitative real-time PCR.
Total RNAs from bone marrow-derived macrophages of a rac3−/− and a rac3+/+ mice were isolated using Trizol (Invitrogen). RNA was also isolated from cultured P190 BCR/ABL rac3−/− and rac3+/+ lymphoblasts. RNAs were subjected to reverse transcription (RT)-PCR for rac3. As control, the same RNAs were subjected to RT-PCR for β-actin, rac1, and rac2. The sequences of the primers and annealing temperature are summarized in Table 1. For quantitative real-time RT-PCR, RNAs were treated with DNase I (Invitrogen, amplification grade). First-strand synthesis on 1 μg total, DNase I-treated RNA was performed using SuperScript and random primers in a 20-μl volume. Real-time PCRs were on 1 μl of cDNA with Sybergreen (Invitrogen). Amplification reactions were monitored with a SmartCycler (Cepheid). An average Ct value was calculated from triplicate samples.
TABLE 1.
Primers used for quantitative real-time and conventional RT-PCR
| Target | Primer | Sequence (5′ to 3′) | Tm (°C) |
|---|---|---|---|
| β-Actin | Forward | GAG AGG GAA ATC GTG CGT GAC A | 60 |
| Reverse | GTT GGC ATA GAG GTC TTT ACG GA | ||
| rac1 | Forward | TGG TAT CCT GAA GTG CGA CA | 60 |
| Reverse | CTT GAG TCC TCG CTG TGT GA | ||
| rac2 | Forward | GTG GCG TTC TTT CCC AGT TA | 57 |
| Reverse | CGA GAG AGG TGT CAG GAA GG | ||
| rac3 | Forward | CGA TTG AAC GGC TGC GGG AC | 60 |
| Reverse | TCC AGG TAC TTG ACG GAA CC |
Vav immunoprecipitation.
Mouse tumor lysates were precleared by incubation with 50 μl of protein A-agarose beads for 1 h at 4°C. Two mg of precleared lysate in a volume of 500 μl was incubated with 500 μl of radioimmunoprecipitation assay (RIPA) buffer B (phosphate-buffered saline containing 1% NP-40, 0.25% deoxycholic acid, 0.1% sodium dodecyl sulfate, 10 mM sodium fluoride, 10 mM pyrophosphate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 0.5 mM orthovanadate, and 25 μM phenylarsine oxide) and 2 μg anti-Vav antibodies (C-14; Santa Cruz Biotechnology). The membranes were incubated with antiphosphotyrosine antibodies (PY20-HRPO, Transduction Laboratories). The membranes were stripped with a Re-blot kit (Chemicon) and reprobed with anti-Vav antibodies.
P190 rac−/− and rac3+/+ mice.
BCR/ABL P190 transgenic mice have been described previously (44). To obtain matched mice to follow leukemogenesis, these P190 doubly transgenic mice (on a mixed C57BL × CBA background) were mated with rac3−/− male 7 (f7 on a C57Bl background). Resultant P190 single transgenic, rac3−/+ mice were interbred to obtain the cohort of mice studied here. We initially obtained 28 rac3−/− and 42 rac3+/+ P190 transgenics; of these, 20 rac3−/− (9 females, 11 males) and 38 rac3+/+ (18 females, 20 males) were eventually included in the study. At the end of the study all remaining animals were sacrificed and evaluated for lymphomas. For survival analysis, a log rank sum test was performed and a Kaplan-Meier survival curve was obtained with SPSS 2.0 statistics software.
Assays for activated Rac.
Protein lysates from lymphomas were prepared in modified RIPA buffer (10 mM sodium phosphate, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin). Because of the very limited amount of lysate that can be obtained from bone marrow, which precludes the possibility of repeating experiments, lymphomas were primarily used as a more abundant source of leukemic cells. To confirm results, all pull-down reactions, Western blotting, and immunoprecipitations were performed at least twice, with similar results, using the same samples.
Lysates were cleared by centrifugation and the protein concentration was measured by a BCA assay (Pierce). To assay GTP-bound Rac, affinity precipitation with glutathione S-transferase (GST)-Pak-RBD was performed as described by Benard et al. (3) with slight modifications. The cleared lymphoma lysates (2 mg, in a volume of 200 μl) were mixed with GST-Pak RBD (20 μg) and reaction buffer (200 μl, 25 mM Tris, pH 7.5, 40 mM NaCl, 30 mM MgCl2, 0.5% Nonidet P-40, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM orthovanadate) and then incubated for 1 h at 4°C. Glutathione-agarose beads (50% vol/vol in reaction buffer, 50 μl) were added to the mixture and incubated for 1 h at 4°C. These glutathione-agarose beads were precipitated and washed twice with reaction buffer without inhibitors and twice with the same buffer without Nonidet P-40 and inhibitors. The proteins precipitated with the beads and 20 μg of the lymphoma lysates were applied to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to Hybond P membranes (Pharmacia). The membranes were probed for Rac proteins. Details of the antibodies used for each experiment are indicated in the figures. Either CH21 anti-Rac3 antipeptide antibodies (10) or pAb106 anti-Rac3 antibodies (4) were used to detect Rac3. The CH21 antibody is not suitable for immunoprecipitation, whereas pAb106 can be used to immunoprecipitate total Rac3 from lysates (4). Anti-Rac1 monoclonal antibodies (BD Transduction Labs) and anti-Rac2 antipeptide polyclonal antibodies (Santa Cruz) were used to detect Rac1 and Rac2, respectively.
Culture and assays on P190 rac3−/− and rac3+/+ lymphoblasts, plasmids, and transfections.
Primary lymphoma cells from P190 rac3−/− and rac3+/+ mice were isolated and cultured on mitotically inactivated mouse embryonic fibroblast feeder layers as described previously (33). After cells were treated with 20 μM GGTI-298 (Calbiochem) for 24 h, a viable cell count was performed using trypan blue. Bone marrow-derived macrophages from rac3−/− and rac3+/+ mice were isolated essentially as described by Stanley (41). Tissue culture reagents were from Life Technologies (Gaithersburg, MD). To construct plasmids expressing constitutively active Rac, pcDNA 3.1 was digested with HindIII and EcoRI, which removes the 6xHis/Xpress tag. V12 Rac1, -2, or -3 was subcloned as HindIII-EcoRI fragments into this vector.
Flow cytometry analysis of B-lineage cells in blood, bone marrow, and spleen of rac3−/− and rac3+/+ mice.
The peripheral blood, spleen, and bone marrow of two rac3−/− and two rac3+/+ mice of around 14 months of age were analyzed using anti-CD43-fluorescein isothiocyanate, anti-CD24-R-phycoerythrin, anti-CD45R-allophycocyanin, and anti-IgM-PerCP-Cy5.5 (PharminGen BD) on a FACScan (Beckton Dickinson). These cell surface markers were used by Hardy and colleagues to subclassify early B-lineage cells in the bone marrow (15, 28). We treated the samples with rat anti-mouse FcBlock (anti-CD16/CD32, Pharmingen BD) according to the supplier's instructions to avoid nonspecific binding to Fc receptors.
RESULTS
rac3 null mutant phenotype.
To generate a mouse lacking rac3, we deleted the entire rac3 coding region using a replacement strategy (Fig. 1A). After obtaining rac3−/+ genotypes, we bred the rac3 null allele into an inbred C57BL/6J background and then mated rac3 heterozygotes. Animals were genotyped using Southern blotting and PCR (Fig. 1B and 1C). rac3−/− mice were obtained at the expected Mendelian frequency. The mice lacking rac3 were initially smaller at birth than heterozygote or wild-type littermates, but were otherwise fully viable and fertile. On a mixed genetic background, no size differences were observed between newborn wild-types and null mutants.
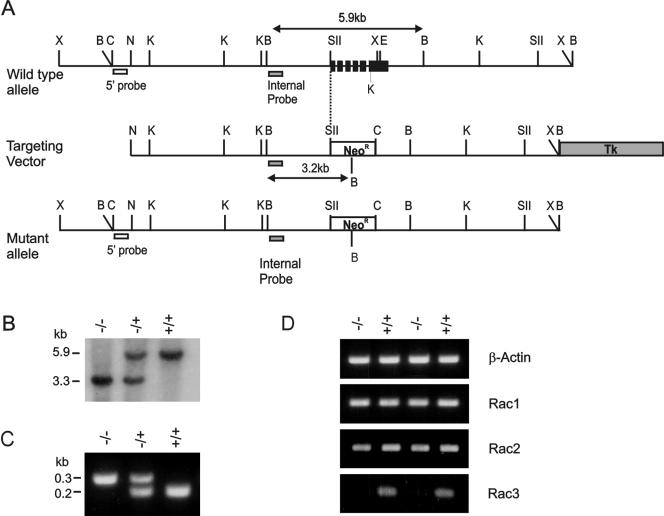
FIG. 1.
Generation of mice deficient in rac3. (A) Schematic illustration of the wild-type rac3 genomic locus (top line), the targeting vector (middle line), and the correctly targeted allele (bottom line). The targeting vector was designed to replace a 1.8-kb SstII-Eco47III fragment with a PGK-neomycin resistance cassette. The grey box in the targeting vector indicates the location of the herpes simplex virus thymidine kinase gene and plasmid sequences. The open box shows the location of the PGK-neo cassette. We used an 0.4-kb ClaI-NruI fragment from outside the targeting vector as an external probe, indicated as 5′ probe beneath the maps. The approximate locations of the six exons of mouse rac3 are indicated in the wild-type allele as black boxes. Restriction enzymes: B = BamHI; C = ClaI; E = Eco47III; K = KpnI; N = Nru I; SII = SstII; Xb = XbaI. (B and C) Genotyping was done using Southern blot analysis on BamHI-digested genomic DNA from the F9 progeny hybridized to the internal probe indicated in panel A (B) or using PCR (C). (D) RT-PCR on two different sets of rac3−/− and rac3+/+ bone marrow-derived macrophages shows that no rac3 mRNA is made in rac3−/− samples. The primers used are indicated in Table 1.
We found no evidence for a critical role of Rac3 in tissues that, in humans, express relatively the highest amount of RAC3, such as brain, heart, placenta, and pancreas (10). Immunohistochemistry showed a particularly high concentration of Rac3 in the deep cerebellar nuclei and in the pons in the mouse (4), and the chicken homolog of Rac3 was isolated from brain (29). However, rac3−/− mice have no external phenotype that would suggest cerebellar developmental defects, and null mutant rac3 females are able to produce progeny, indicating that maternal placental formation is normal. Animals appear to have a normal life span, which is not consistent with severe cardiac or pancreatic functional defects.
Analysis of RNA isolated from bone marrow-derived macrophages using RT-PCR confirmed that no rac3 was present in RNA from rac3−/− mice, whereas it was clearly present in wild-type RNA (Fig. 1D, bottom panel). Similar results were obtained with testis and spleen RNA (not shown). No compensatory increase in either rac1 or rac2 mRNA was evident in the bone marrow-derived macrophages of rac3−/− mice using RT-PCR (Fig. 1D), which was confirmed using real-time quantitative RT-PCR. The Ct values for expression in bone marrow-derived macrophages were 20.21 ± 0.44 for β-actin, 23.94 ± 0.48 for rac1, 23.46 ± 0.47 for rac2, and 28.26 ± 0.46 for rac3 (n = 4 for β-actin, rac1, and rac2; n = 2 for rac3).
An examination of steady-state hematopoiesis of the peripheral blood, bone marrow, and spleen at 3 months of age using fluorescence-activated cell sorting analysis and Gr-1, Mac-1, B220, and Thy-1.2 did not reveal consistent differences (n = 4). We also performed more detailed analysis for possible differences in the early B-lineage using fluorescence-activated cell sorting and antibodies against CD43, CD24 (heat-stable antigen), CD45R/B220, and surface immunoglobulin M as described (15, 28) on peripheral blood, bone marrow, and spleen in two pairs of matched female rac3−/− and rac3+/+ mice at 14 months of age. We detected no genotype-associated differences in the blood and spleen samples. However, within the CD45R+ population in the bone marrow, there were slightly more (mature) CD43− cells (36% versus 27.5%) and less (immature) CD43+ cells (64% versus 72.5%) in the rac3−/− samples.
Rac has been typically associated with regulation of cytoskeletal reorganization (13). However, we did not detect any obvious abnormalities in actin cytoskeletal organization, proliferation in the presence of CSF-1, or haptotactic motility on fibronectin with or without CSF-1 in bone marrow-derived macrophages of rac3−/− mice or in the actin cytoskeleton of P190 BCR/ABL rac3−/− and rac3+/+ lymphoblasts (see below) cultured in the presence of interleukin-3 on mouse embryonic fibroblasts or attached to poly-l-lysine-coated slides (not shown). We conclude that Rac3 does not play an indispensable role in embryonic development, nor is loss of Rac3 associated with overt phenotypic defects in the adult.
Bcr/Abl lymphoblasts contain tyrosine-phosphorylated Vav.
We next wished to examine a possible contribution of Rac3 in cancer. Previous data including the tyrosine phosphorylation of Vav1, a hematopoiesis-specific exchange factor for Rac, suggested that a Rac may be involved in the leukemia generated by the BCR/ABL oncogene (2, 16, 31, 40). This oncogene is the cause of human chronic myelogenous leukemia and a subset of acute lymphoblastic leukemia. It encodes a deregulated tyrosine kinase (8, 18).
Tyrosine phosphorylation of Vav is known to activate its exchange factor activity for Rac (5, 14). Since tyrosine-phosphorylated Vav was detected in two myeloid (chronic myelogenous leukemia) cell lines (2), we first examined if Vav was possibly also tyrosine phosphorylated in malignant primary precursor B lymphoblasts from BCR/ABL transgenic mice. These mice, which are transgenic for the P190 form of Bcr/Abl, model the human acute lymphoblastic leukemia that is generated by Bcr/Abl (17, 44).
We immunoprecipitated Vav from lysates prepared from a number of different primary lymphomas, including two from BCR/ABL P190 transgenic mice and three from mice that were not BCR/ABL transgenic. Tyrosine phosphorylation of Vav was detected by immunoblotting the precipitates with antiphosphotyrosine antibodies. Interestingly, as shown in Fig. 2A (top panel), tyrosine-phosphorylated Vav was detected at the highest level in the two P190 Bcr/Abl lymphomas, whereas the other three samples contained much lower phosphorylated Vav. The same filter was stripped and reprobed with anti-Vav antibodies to show approximately equal loading of all samples (Fig. 2A, bottom panel). Tyrosine-phosphorylated Vav was also detected in four other independent P190 lymphoma samples (not shown). These experiments demonstrated for the first time that tyrosine-phosphorylated Vav is present in primary lymphoblastic leukemic cells. Because tyrosine phosphorylation can activate the guanine nucleotide exchange factor function of Vav, this suggests that these primary lymphomas may also contain one or more Racs that are constitutively activated.
FIG. 2.
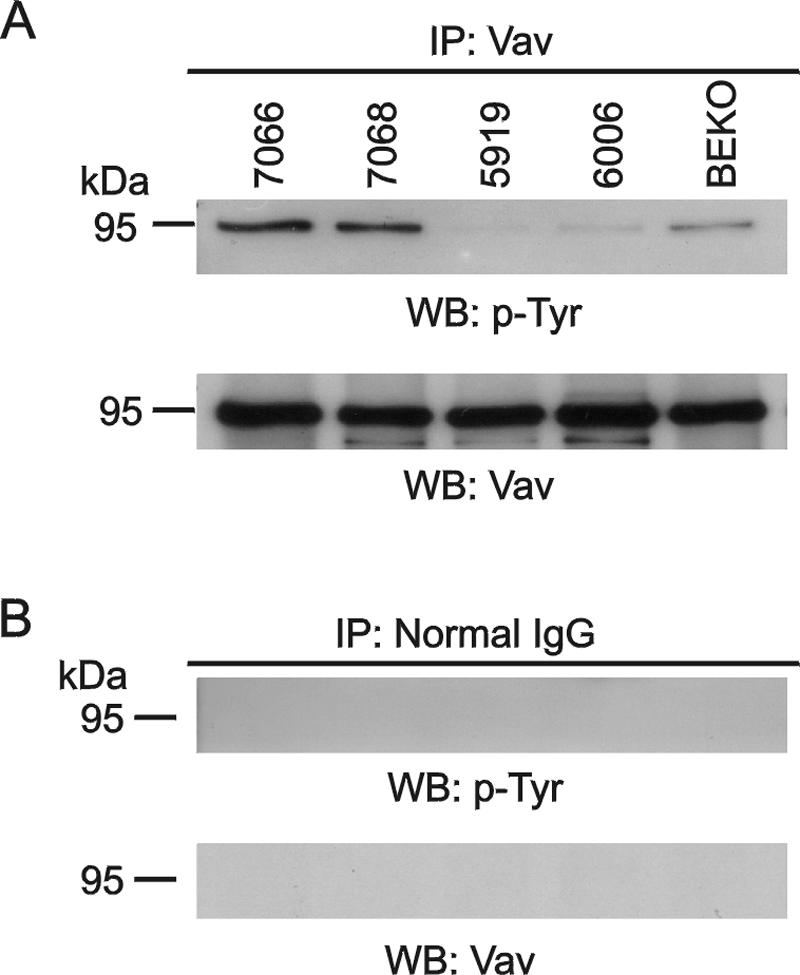
Vav is tyrosine phosphorylated in primary P190 lymphoblastic leukemia cells. (A) Vav was immunoprecipitated (IP) with anti-Vav antibodies from lymphoma lysates of mice 7066 (P190 BCR/ABL rac3+/+); 7068 (P190 BCR/ABL rac3−/−); 5919 (wild type), 6006 (10xCRKL transgenic); and BEKO (bcr−/−). The samples were separated on a 7.5% SDS-polyacrylamide gel, and the presence of phosphorylated Vav (top) and total Vav (bottom) was investigated with antiphosphotyrosine antibodies (PY-20) or anti-Vav antibodies (C-14). (B) Control immunoprecipitation with immunoglobulin G. The Western blot (WB) was reacted with the antibodies shown.
Lymphoblastic leukemia cells from P190 rac3−/− and rac3+/+ mice.
We then generated a cohort of matched P190 BCR/ABL rac3−/− and P190 BCR/ABL rac3+/+ mice through breeding and isolated malignant leukemia/lymphoma cells from these animals.
We used RT-PCR to check for rac expression in these lymphoblasts (Fig. 3A). Real-time quantitative RT-PCR analysis of two independently isolated cultures of P190 rac3−/− and rac3+/+ lymphoblasts for all three Racs yielded average Ct values of 18.9, 21.5, 20.8, and 31.3 for β-actin, rac1, rac2, and rac3, respectively. No differences in rac mRNA expression were found in cultured lymphoblasts from females and males (not shown). We additionally performed real-time quantitative RT-PCR to confirm that lack of rac3 was not compensated for by increased expression of rac1 and/or rac2 mRNA levels in the rac3−/− precursor B-lineage lymphomas (not shown). These experiments showed that rac3 was present, but at lower levels than rac1 and rac2, and confirmed lack of significant differences in rac1 or rac2 levels in cells with or without rac3.
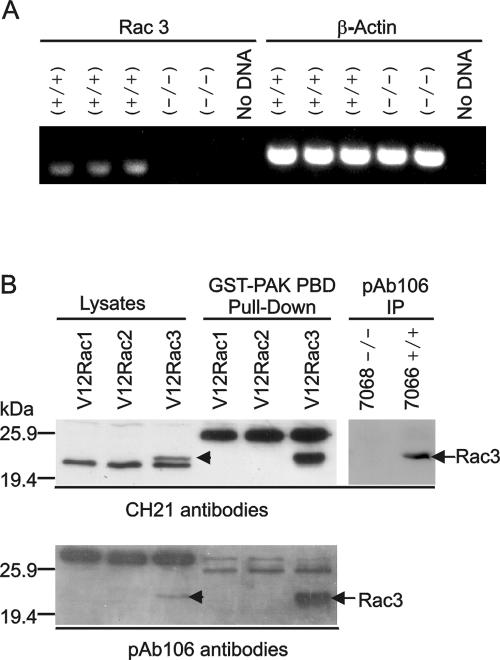
FIG. 3.
Detection of Rac3 expression. (A) RT-PCR of rac3 mRNA in leukemic precursor B cells. The primers used are indicated above the lanes. rac3+/+ and rac3−/− indicate samples from P190 rac3+/+ and P190 rac3−/− mice, respectively. (B) Western blot analysis using two different Rac3 antibodies (as indicated beneath the panels) to detect Rac3. CHO cells were transfected with constitutively active (V12) Rac1, Rac2, or Rac3, and activated Rac was detected using GST-Pak. Total Rac3 protein was also immunoprecipitated from lymphoma lysates of a rac3−/− and a rac3+/+ mouse using pAb106 antibodies. The location of Rac3 protein is indicated.
The absolute expression levels of rac provide only very limited information on their possible impact on a cell, because Rac proteins can be present in an active, GTP-bound conformation or in an inactive, GDP-bound state. Since low levels of active (GTP-bound) Rac may be more significant than high levels of inactive (GDP-bound) Rac, it is important to measure if the cells contain GTP-bound Rac. We addressed this using a GST-Pak pull-down assay which, from the total pool of Rac, affinity purifies only activated GTP-bound Rac.
The ability to distinguish Rac3 from the more abundant Rac1 and Rac2 depends on the availability of Rac3 antibodies that do not cross-react with Rac1 and Rac2. The amino acid sequences of human Rac1, -2, and -3 are nearly identical, but their very C-terminal ends exhibit the most significant divergence. Therefore, we reevaluated antipeptide antibodies raised against the Rac3 C terminus (3, 10). In CHO cells transfected with constitutively active Rac1, -2, or -3, CH21 antibodies reacted with Rac3 but not Rac1 or Rac2 (Fig. 3B, lysate, top panel). The CH21 antibodies did not cross-react with Cdc42 (not shown). However, they did react with an unidentified protein that migrates slightly faster than Rac proteins and which is present in many primary lysates, including those derived from rac3−/− cells (also see Fig. 4A, panel with total lysates).
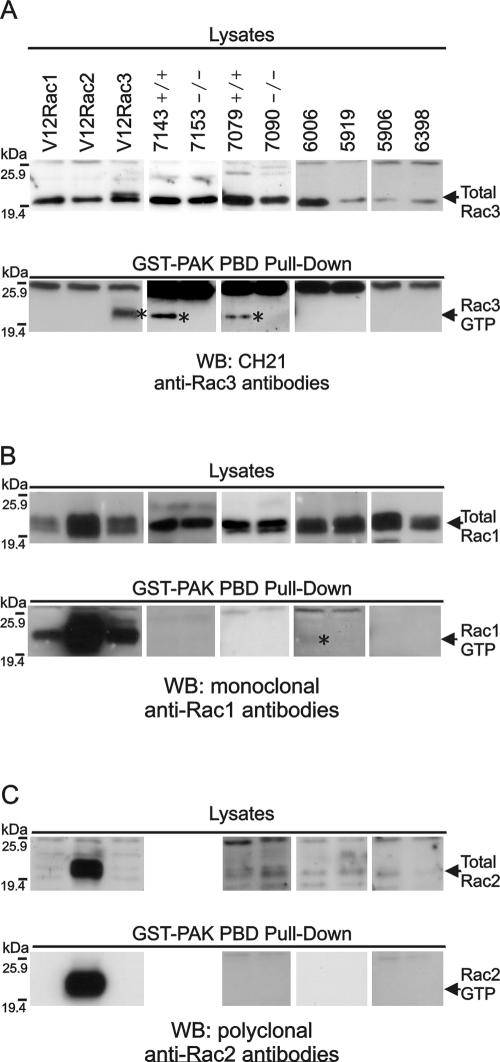
FIG. 4.
Analysis of Rac activation in primary lymphomas, for activated Rac3 using anti-CH21 antibodies (A); for activated Rac1 using monoclonal anti-Rac1 (Transduction Laboratories) antibodies (B); and for activated Rac2 using anti-Rac2 (Santa Cruz) antibodies (C). Top panels, lysates for total Rac levels; bottom panels, GST-Pak-RBD pull-down reactions for GTP-bound Rac. COS cells transfected with V12Rac1, Rac2, or Rac3 are shown in the three left lanes as positive methodological controls. All samples were lymphomas (unless otherwise indicated) and include 7143 (P190 BCR/ABL rac3+/+); 7153 (P190 BCR/ABL rac3−/−); 7079 (P190 BCR/ABL rac3+/+); 7090 (P190 BCR/ABL rac3−/−); 6006 (10xCRKL transgenic); 5919 (wild type); 5906 (wild type, benign mammary gland tumor); and 6398 (10xCRKL transgenic). Different exposures of the same blot are shown for lysates and GST-Pak-RBD pull-downs. The position of activated GTP-bound Rac (RacGTP) is indicated with an asterisk.
Experiments with pAb106, a second set of independently generated Rac3 antibodies, confirmed that the CH21 antiserum did detect Rac3 but not Rac1 or Rac2 (Fig. 3B, lower panel). Immunoprecipitation of Rac3 protein from P190 rac3+/+ and rac3−/− lymphoblastic leukemia lysates with the pAb106 antiserum, followed by a Western blot using the CH21 antibodies, also confirmed the ability of the CH21 antibodies to specifically detect Rac3 in the wild-type cells (Fig. 3B, pAb106 IP, top panel). Affinity purification of activated Rac using GST-Pak-RBD further showed that the CH21 antibodies very strongly detected activated Rac3 (Fig. 3B, GST-Pak pull-down). Importantly, the unknown cross-reacting protein did not bind to GST-Pak-RBD.
Bcr/Abl-expressing lymphoblastic leukemia/lymphoma cells contain activated Rac3.
To investigate possible Rac activation in primary P190 Bcr/Abl lymphoblasts, we prepared lysates from a number of different lymphomas and from a benign mammary tumor. Two lymphomas were from P190 BCR/ABL rac3−/− and two from P190 BCR/ABL rac3+/+ mice. Mesenteric lymph node lymphoma lysates were also prepared from a wild-type mouse and from two different mice that were transgenic for a DNA construct encoding the small adapter protein Crkl (19). We performed GST-Pak pull-down reactions, which precipitate all three Racs in their GTP-bound conformation, followed by Western blotting to detect specific Racs. As a positive methodological control, COS-1 cells were transfected with constitutively active V12 Rac1, Rac2, or Rac3.
Using the GST pull-down technique, we could clearly detect activated V12Rac3 (Fig. 4A, bottom panel) as well as activated V12Rac1 and 2 (Fig. 4B and C, bottom panels) in the COS cells. Interestingly, GST-Pak pull-down reactions of the lymphoma lysates followed by immunoblotting with the anti-Rac3 antibodies revealed that activated Rac3 could only be detected in the rac3+/+ P190 BCR/ABL lymphoma samples (Fig. 4A, compare pull-down of the 7143 and 7079 rac3+/+ samples to that of the 7153 and 7090 rac3−/− samples). We did not detect activated Rac3 in any of the other lymphoma samples.
We reprobed the membranes using Rac1 monoclonal antibodies. In concordance with observations made by others (4), these antibodies reacted relatively poorly with Rac3. They detected total Rac2 quite strongly (Fig. 4B, lysates). Interestingly, in the GST-Pak pull-down, no activated Rac1 or -2 was detected with these antibodies in the lymphoma lysates of either rac3−/− or rac3+/+ mice (Fig. 4B) whereas total Rac was clearly detectable. Also, in the GST-Pak pull-down, the same antibodies detected V12Rac1 and V12Rac2. One of the non-Bcr/Abl lymphoma lysates, 6006, showed a low level of activated Rac1. Since no activated Rac2 or Rac3 was detected in the same sample, we infer that this is Rac1. Using Rac2-specific antibodies (Santa Cruz) we also failed to detect activated Rac2 in the P190 rac3−/− or rac3+/+ lymphoma lysates (Fig. 4C), although the antibodies strongly detected V12Rac2. Taken together, these results clearly demonstrate that the pre-B lymphoblastic leukemia cells that express Bcr/Abl P190 contain activated Rac3.
Survival of P190 rac3−/− and P190 rac3+/+ mice.
After a latency period, P190 BCR/ABL transgenic mice typically develop a highly malignant type of cancer that is characterized by very rapidly proliferating cells (17, 44). To date, we have not identified any factor(s) that will significantly affect the growth of the lymphoblasts once the cancer has progressed. Even the Bcr/Abl tyrosine kinase inhibitor imatinib does not ultimately affect their proliferation (Mishra et al., in preparation).
To investigate whether lack of Rac3 affects the leukemia caused by Bcr/Abl P190 in vivo, we generated a cohort of 58 matched P190 BCR/ABL rac3−/− and P190 BCR/ABL rac3+/+ mice through breeding. These mice were followed for the development of leukemia/lymphoma for up to 2 years. When the mice became moribund, they were sacrificed and necroscopies were performed. Fluorescence-activated cell sorting analysis on seven rac3−/− and three rac3+/+ mice showed that most mice developed B-lineage lymphoblastic leukemia. All three rac3+/+ samples consisted mainly of B220+ cells, with a small population of B220+, Thy-1+ cells. Four of the rac3−/− samples contained mainly B220+ cells; the other three were more heterogeneous, including a higher percentage of B220+ Thy-1+ cells, B220lo cells, or cells that also stained for myeloid markers. In comparison with similar analyses on other lymphomas from wild-type P190 BCR/ABL transgenic mice, these results are fairly typical (not shown). Overall, terminal white blood cells in both wild-type and null mutants ranged widely, from around 2 × 106 to 150 × 106/liter. The number and the anatomical location of lymphomas in individual animals ranged from 1 to 12 in different individual animals. The size of the lymphomas in the rac3−/− and rac3+/+ mice was overall similar. Peritoneal exudates were found in both rac3−/− and rac3+/+ mice.
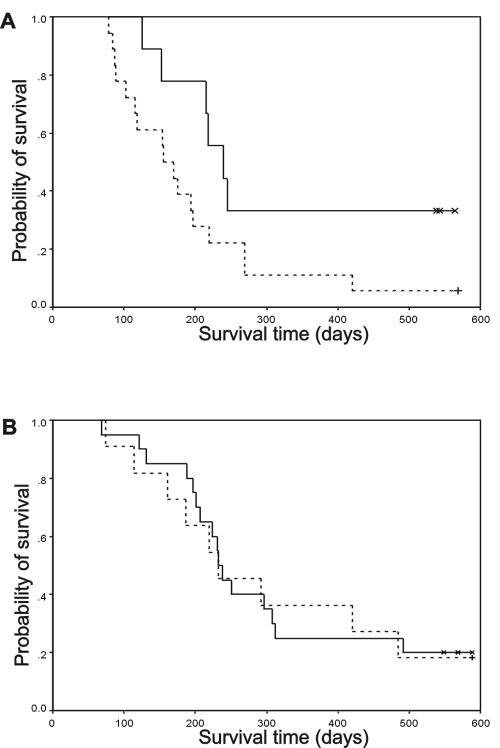
We compared the cumulative survival of male and female animals separately, since in our mouse model, female P190 transgenics develop leukemia/lymphoma more rapidly than males (45). In a pairwise comparison, no statistically significant difference was found between survival of wild-type and null mutant males (P = 0.87) (Fig. 5B). However, the mean survival of females lacking Rac3 was 321 ± 59 days, whereas this was 193 ± 29 days for mice that expressed Rac3 (P = 0.06) (Fig. 5A).
FIG. 5.
Kaplan-Meier survival curve of P190 BCR/ABL transgenic mice in the presence and absence of rac3. (A) A genetically matched set of P190 BCR/ABL rac3+/+ female mice (dashed line, n = 18) were compared to P190 BCR/ABL rac3−/− female mice (solid line, n = 9). (B) A matched set of P190 BCR/ABL rac3+/+ male mice (dashed line, n = 20) were compared to P190 BCR/ABL rac3−/− male mice (solid line, n = 11). Censored data are marked (asterisks and cross).
Dependence of B-lymphoblastic leukemia cells on rac3.
To investigate the importance of these small GTPases to the proliferation of the malignant lymphoblasts, we compared the viability of P190 rac3−/− and rac3+/+ lymphoblasts in the presence of a geranyl-geranyltransferase inhibitor (GGTI). GGTIs block the geranyl-geranylation of Rho family GTPases, including Rac (22). Since, to initiate a biological response, Rac needs to localize to membranes via its prenylated C-terminal end, treatment with GGTIs results in a nonfunctional Rac. As shown in Fig. 6, GGTI-298 had a differential effect on the B-lineage lymphoblasts, with P190 rac3+/+ cells being clearly more sensitive to the GGTI than P190 rac3−/− cells. This suggests that P190 BCR/ABL rac3+/+ lymphoblasts react to the removal of Rac3 with increased cell death because they depend upon the presence of Rac3 function for survival. We speculate that in the P190 BCR/ABL rac3−/− lymphoblasts, the lack of Rac3 has been compensated for, over time, by other pathways involving proteins less sensitive to geranyl-geranyltransferase inhibitors.
FIG. 6.
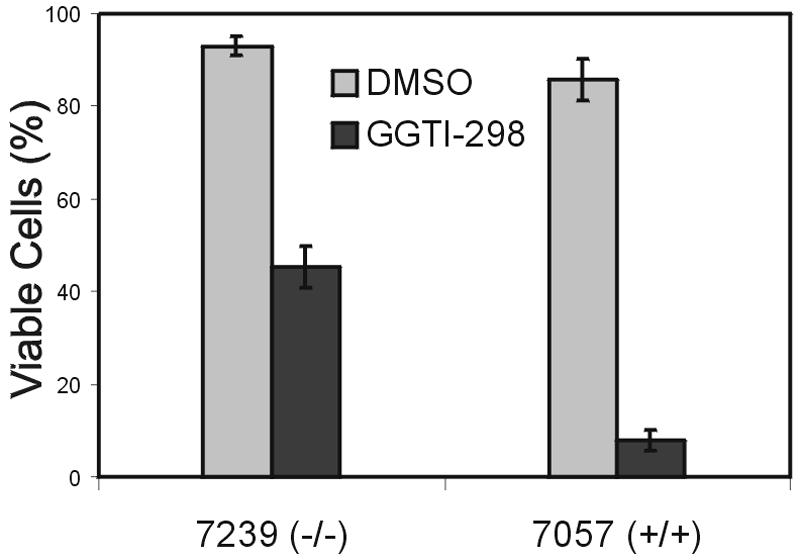
P190 BCR/ABL precursor B lymphoblasts expressing Rac3 are more sensitive to GGTI treatment than cells lacking Rac3. Lymphoblasts grown on mitotically inactivated murine embryonic fibroblast feeder layers were treated for 24 h with 20 μM GGTI-298 in dimethyl sulfoxide or dimethyl sulfoxide alone, after which the percentage of viable cells was determined. The result shown is one of two independently performed experiments with similar results. A second set of rac3−/− and rac3+/+ lymphoblasts yielded a qualitatively similar result. Bars indicate the standard error of the mean.
DISCUSSION
The mammalian genome contains three RAC genes, RAC1, -2, and -3, which are expressed in different tissues at different levels. Based on the phenotype of the total rac1 knockout, which is lethal at the stage of gastrulation, Rac1 has an important function during development (42). Both rac2 (38) and rac3 (the current study) appear to be dispensable for embryogenesis and for overall viability in postnatal life. Rac2 and Rac3 differ, however, in that the former is only expressed in hematopoietic cells and is relatively abundant there, whereas expression of the latter is more widespread but at a lower level of abundance.
Rac1 and Rac2 have been characterized in most detail with respect to expression and function in hematopoietic cell types. For example, both Rac1 and Rac2 are abundant in neutrophils, but rac2 null mutant neutrophils have markedly impaired production of reactive oxygen species though the NADPH oxidase, whereas neutrophils lacking rac1 from a conditional rac1 null mutant are normal (9). This indicates that Racs that are coexpressed may have distinct functions in the same cell. We speculate that Rac3 also has a very specialized function that, to date, we have not been able to precisely identify.
A number of earlier studies had suggested that Rac is involved in leukemias caused by the deregulated tyrosine kinase Bcr/Abl. The activity of Vav as an exchange factor towards the small GTPase Rac is regulated by tyrosine phosphorylation (1). Matsuguchi et al. (31) reported that Vav1 became constitutively tyrosine phosphorylated in MO7e cells transfected by Bcr/Abl. Skorski et al. (40) transfected 32D cells with a dominant negative Rac and provided evidence that Bcr/Abl-mediated leukemogenesis requires the activity of Rac. Bassermann et al. (2) also reported the tyrosine phosphorylation of Vav1 by Bcr/Abl, showed increased levels of active Rac in cells transiently transfected with Bcr/Abl and Rac, and demonstrated a direct complex between Bcr/Abl and Vav in two chronic myelogenous leukemia cell lines. Harnois et al. (16) further showed activation of Rac in BaF3 cells transfected with Bcr/Abl P210 and P190.
Our results show for the first time that Vav is constitutively tyrosine phosphorylated in pro/pre-B lymphoblasts isolated directly from lymphomas of mice transgenic for BCR/ABL P190, indicating that such cells may indeed contain one or more activated Rac family members. However, we cannot deduce from our experiments if the observed tyrosine phosphorylation of Vav1 is the result of a direct tyrosine phosphorylation of Vav1 by Bcr/Abl or if it is indirect. Since Bcr/Abl is known to activate other hematopoietic tyrosine kinases, such as Lck, which use Vav1 as a substrate in vivo, it is possible that Bcr/Abl activates a Src family kinase that activates Vav1 (6, 20, 36).
Rac3 expression has also been reported in hematopoietic cells. We originally isolated human Rac3 from a chronic myelogenous leukemia cell line, and Northern blot analysis showed RAC3 mRNA in human B-lineage lymphocytic leukemia and promyelocytic HL-60 cell lines (10). Gu et al. (9) reported that Rac3 mRNA was present in both primitive and differentiated murine hematopoietic cells using RT-PCR, and Zhao et al. (48) also reported RAC3 mRNA in human monocytes. In the current study we detected rac3 in malignant precursor B-lineage lymphoid cells. These cells also expressed rac1 and rac2 mRNAs. Since the activation state of Racs may be functionally more significant than their expression levels, we investigated these cells directly for endogenous levels of activated Rac3, Rac2, and Rac1. Surprisingly, we could only positively identify activated, GTP-bound Rac3 in the Bcr/Abl P190-expressing leukemic cells. However, since the detection limits of such assays are determined by the number of cells used for the pull-down assay, we cannot exclude the possibility that these cells do contain activated Rac1 and Rac2 that was not detectable under the assay conditions used.
Walmsley et al. (46) investigated the effect of ablation of Rac1 and Rac2 on B-cell development and signaling using rac2 null mutants and targeted deletion of rac1 in B-lineage (CD19+) cells. Double null mutants had profound abnormalities in more mature B-cell stages but no changes in pro-B, pre-B, or immature B cells, demonstrating that neither Rac1 nor Rac2 is required for the generation or expansion of early B-lineage progenitors. Interestingly, in the BCR/ABL P190 transgenic mice, leukemia arises mainly in the early B-lineage compartment, at the pre- or pro-B-cell stage of development (17, 44). Preliminary analysis of rac3−/− and rac3+/+ bone marrow did suggest that there may be a difference in the composition of the pre/pro-B cell population. However, additional experiments with mice of different ages and including males are needed to substantiate this. Based on the finding that lack of rac3 in BCR/ABL P190 female transgenics was associated with a delay in the average age at death and on the presence of Rac3GTP in the lymphomas, we conclude that of the three Racs, Rac3 has a specific involvement in Bcr/Abl P190-caused acute lymphoblastic leukemia/lymphoma.
Based on the increasing knowledge of the signal transduction pathways that are affected by Bcr/Abl and the proteins that interact with this deregulated tyrosine kinase, small-molecule inhibitors are emerging as potential alternative drugs to eradicate Ph-positive acute lymphoblastic leukemia and chronic myelogenous leukemia. Joyce and Cox (22) compared the transforming activity of Rac1 and Rac3 in NIH 3T3 cells and the effect of GGTIs on this activity. They concluded that Rac1 and Rac3 are potential physiological targets of GGTIs. Our study strongly supports the concept that interference with Rac3 function through the use of GGTIs may have a beneficial therapeutic effect on Ph-positive acute lymphoblastic leukemias. Therefore, therapies based on blocking Rac function using inhibitors of Rac could have application in the treatment of advanced Ph-positive leukemias.
Acknowledgments
We thank Michael Anderson and his lab for help, advice, and access to real-time PCR, Aljona Antipova for involvement in cloning murine rac3 genomic DNA, Jess Cunnick for help with optimizing lymphoma culture and imaging of the cells, Hyungtaek Lee for help with evaluation of rac3−/− and rac3+/+ mice injected with LPS, Dinithi Silva for assistance with construction of the targeting vector, Ana Romero for help with isolation of mouse tail DNAs, Donna Foster for excellent care of the mice, and Bianca Hemmeryckx for preparation of some of the lymphoma lysates. Gary Bokoch and Ulla Knaus (Scripps Research Institute, La Jolla, CA) are acknowledged for GST-Pak-RBD.
This work was supported by PHS grants CA50248 and CA90321 (N.H.), HL060231 and HL071945 (J.G.), the Kenneth T and Eileen L Norris Foundation (B.Z. and J.G.), and the T. J. Martell Foundation (J.G. and N.H.).
REFERENCES
- 1.Aghazadeh, B., W. E. Lowry, X. Y. Huang, and M. K. Rosen. 2000. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102:625-633. [DOI] [PubMed] [Google Scholar]
- 2.Bassermann, F., T. Jahn, C. Miething, P. Seipel, R. Y. Bai, S. Coutinho, V. L. Tybulewicz, C. Peschel, and J. Duyster. 2002. Association of Bcr-Abl with the proto-oncogene Vav is implicated in activation of the Rac-1 pathway. J. Biol. Chem. 277:12437-12445. [DOI] [PubMed] [Google Scholar]
- 3.Benard, V., B. P. Bohl, and G. M. Bokoch. 1999. Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 274:13198-13204. [DOI] [PubMed] [Google Scholar]
- 4.Bolis, A., S. Corbetta, A. Cioce, and I. de Curtis. 2003. Differential distribution of Rac1 and Rac3 GTPases in the developing mouse brain: implications for a role of Rac3 in Purkinje cell differentiation. Eur. J. Neurosci. 18:2417-2424. [DOI] [PubMed] [Google Scholar]
- 5.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the Vav proto-oncogene product. Nature 385:169-172. [DOI] [PubMed] [Google Scholar]
- 6.Danhauser-Riedl, S., M. Warmuth, B. J. Druker, B. Emmerich, and M. Hallek. 1996. Activation of Src kinases p53/56lyn and p59hck by p210bcr/abl in myeloid cells. Cancer Res. 56:3589-3596. [PubMed] [Google Scholar]
- 7.Glogauer, M., C. C. Marchal, F. Zhu, A. Worku, B. E. Clausen, I. Foerster, P. Marks, G. P. Downey, M. Dinauer, and D. J. Kwiatkowski. 2003. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J. Immunol. 170:5652-5657. [DOI] [PubMed] [Google Scholar]
- 8.Groffen, J., and Heisterkamp, N. 1997. The chimeric BCR-ABL gene. Baillieres Clin. Haematol. 10:187-201. [DOI] [PubMed] [Google Scholar]
- 9.Gu, Y., M. D. Filippi, J. A. Cancelas, J. E. Siefring, E. P. Williams, A. C. Jasti, C. E. Harris, A. W. Lee, R. Prabhakar, S. J. Atkinson, D. J. Kwiatkowski, and D. A. Williams. 2003. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science 302:445-449. [DOI] [PubMed] [Google Scholar]
- 10.Haataja, L., J. Groffen, and N. Heisterkamp. 1997. Characterization of RAC3, a novel member of the Rho family. J. Biol. Chem. 272:20384-23388. [DOI] [PubMed] [Google Scholar]
- 11.Habets, G. G., R. A. van der Kammen, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, A. Hagemeijer, and J. G. Collard. 1995. The invasion-inducing TIAM1 gene maps to human chromosome band 21q22 and mouse chromosome 16. Cytogenet. Cell. Genet. 70:48-51. [DOI] [PubMed] [Google Scholar]
- 12.Haeusler, L. C., L. Blumenstein, P. Stege, R. Dvorsky, and M. R. Ahmadian. 2003. Comparative functional analysis of the Rac GTPases. FEBS Lett. 555:556-560. [DOI] [PubMed] [Google Scholar]
- 13.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-513. [DOI] [PubMed] [Google Scholar]
- 14.Han, J., B. Das, W. Wei, L. Van Aelst, R. D. Mosteller, R. Khosravi-Far, J. K. Westwick, C. J. Der, and D. Broek. 1997. Lck regulates Vav activation of members of the Rho family of GTPases. Mol. Cell. Biol. 17:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harnois, T., B. Constantin, A. Rioux, E. Grenioux, A. Kitzis, and N. Bourmeyster. 2003. Differential interaction and activation of Rho family GTPases by p210bcr-abl and p190bcr-abl. Oncogene 22:6445-6454. [DOI] [PubMed] [Google Scholar]
- 17.Heisterkamp, N., G. Jenster, J. ten Hoeve, D. Zovich, P. Pattengale, and J. Groffen. 1990. Acute leukemia in BCR/ABL transgenic mice. Nature 344:251-253. [DOI] [PubMed] [Google Scholar]
- 18.Heisterkamp, N., and J. Groffen. 2002. Philadelphia-positive leukemia: a personal perspective. Oncogene 21:8536-8540. [DOI] [PubMed] [Google Scholar]
- 19.Hemmeryckx, B., A. van Wijk, A. Reichert, V. Kaartinen, R. de Jong, P. K. Pattengale, I. Gonzalez-Gomez, J. Groffen, and N. Heisterkamp. 2001. Crkl enhances leukemogenesis in BCR/ABL P190 transgenic mice. Cancer Res. 61:1398-1405. [PubMed] [Google Scholar]
- 20.Hu, Y., Y. Liu, S. Pelletier, E. Buchdunger, M. Warmuth, D. Fabbro, M. Hallek, R.A. van Etten, and S. Li. 2004. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat. Genet. 36:453-461. [DOI] [PubMed] [Google Scholar]
- 21.Joneson, T., M. McDonough, D. Bar-Sagi, and L. van Aelst. 1996. Rac regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science 274:1374-1376. [DOI] [PubMed] [Google Scholar]
- 22.Joyce, P. L., and A. D. Cox. 2003. Rac1 and Rac3 are targets for geranyl-geranyltransferase I inhibitor-mediated inhibition of signaling, transformation, and membrane ruffling. Cancer Res. 63:7959-7967. [PubMed] [Google Scholar]
- 23.Kaartinen, V., I. Gonzalez-Gomez, J. W. Voncken, E. Faure, L. Haataja, J. Groffen, and N. Heisterkamp. 2001. Abnormal function of astroglia lacking abr and bcr RacGAPs. Development 128:4217-4227. [DOI] [PubMed] [Google Scholar]
- 24.Katzav, S., D. Martin-Zanca, and M. Barbacid. 1989. vav, a novel human oncogene derived from a locus ubiquitously expressed in hematopoietic cells. EMBO J. 8:2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khosravi-Far, R., P. A. Solski, G. J. Clark, M. S. Kinch, and C. J. Der. 1995. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol. Cell. Biol. 15:6443-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamarche, N., N. Tapon, L. Stowers, P. D. Burbelo, P. Aspenstrom, T. Bridges, J. Chant, and A. Hall. 1996. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65Pak and the JNK/SAPK MAP kinase cascade. Cell 87:519-529. [DOI] [PubMed] [Google Scholar]
- 27.Leung, K., A. Nagy, I. Gonzalez-Gomez, J. Groffen, N. Heisterkamp, and V. Kaartinen. 2003. Targeted expression of activated Rac3 in mammary epithelium leads to defective postlactational involution and benign mammary gland lesions. Cells Tissues Organs 175:72-83. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y. S., K. Hayakawa, and R. R. Hardy. 1993. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 178:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malosio, M. L., D. Gilardelli, S. Paris, C. Albertinazzi, and I. de Curtis. 1997. Differential expression of distinct members of Rho family GTP-binding proteins during neuronal development: identification of Rac1B, a new neural-specific member of the family. J. Neurosci. 17:6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mareel, M., and A. Leroy. 2003. Clinical, cellular, and molecular aspects of cancer invasion. Physiol. Rev. 83:337-376. [DOI] [PubMed] [Google Scholar]
- 31.Matsuguchi, T., R. C. Inhorn, N. Carlesso, G. Xu, B. Druker, and J. D. Griffin. 1995. Tyrosine phosphorylation of p95Vav in myeloid cells is regulated by GM-CSF, IL-3 and Steel factor and is constitutively increased by p210BCR/ABL. EMBO J. 14:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mira, J. P., V. Benard, J. Groffen, L. C. Sanders, and U. G. Knaus. 2000. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc. Natl. Acad. Sci. USA 97:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra, S., A. Reichert, J. Cunnick, D. Senadheera, B. Hemmeryckx, N. Heisterkamp, and J. Groffen. 2003. Protein kinase CKIIalpha interacts with the Bcr moiety of Bcr/Abl and mediates proliferation of Bcr/Abl-expressing cells. Oncogene 22:8255-8262. [DOI] [PubMed] [Google Scholar]
- 34.Nimnual, A. S., B. A. Yatsula, and D. Bar-Sagi. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279:560-563. [DOI] [PubMed] [Google Scholar]
- 35.Olson, M. F., A. Ashworth, and A. Hall. 1995. An essential role for Rho, Rac and Cdc42 GTPases in cell cycle progression through G1. Science 269:1270-1272. [DOI] [PubMed] [Google Scholar]
- 36.Ptasznik, A., Y. Nakata, A. Kalota, S. G. Emerson, and A. M. Gewirtz. 2004. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug-resistant, BCR-ABL1+ leukemia cells. Nat. Med. 10:1187-1189. [DOI] [PubMed] [Google Scholar]
- 37.Qiu, R-G., J. Chen, D. Kirn, F. McCormick, and M. Symons. 1995. An essential role for Rac in Ras transformation. Nature 374:457-459. [DOI] [PubMed] [Google Scholar]
- 38.Roberts, A. W., C. Kim, L. Zhen, J. B. Lowe, R. Kapur, B. Petryniak, A. Spaetti, J. D. Pollock, J. B. Borneo, G. B. Bradford, S. J. Atkinson, M. C. Dinauer, and D. A. Williams. 1999. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity 10:183-196. [DOI] [PubMed] [Google Scholar]
- 39.Scita, G., J. Nordstrom, R. Carbone, P. Tenca, G. Giardina, S. Gutkind, M. Bjarnegard, C. Betsholtz, and P. P. Di Fiore. 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401:290-293. [DOI] [PubMed] [Google Scholar]
- 40.Skorski, T., P. Wlodarski, L. Daheron, P. Salomoni, M. Nieborowska-Skorska, M. Majewski, M. Wasik, and B. Calabretta. 1998. BCR/ABL-mediated leukemogenesis requires the activity of the small GTP-binding protein Rac. Proc. Natl. Acad. Sci. USA 95:11858-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanley, E. R. 1997. Murine bone marrow-derived macrophages. Methods Mol. Biol. 75:301-304. [DOI] [PubMed]
- 42.Sugihara, K., N. Nakatsuji, K. Nakamura, K. Nakao, R. Hashimoto, H. Otani, H. Sakagami, H. Kondo, S. Nozawa, A. Aiba, and M. Katsuki. 1998. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 17:3427-3433. [DOI] [PubMed] [Google Scholar]
- 43.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 44.Voncken, J. W., S. Griffiths, M. Greaves, P. K. Pattengale, N. Heisterkamp, and J. Groffen. 1992. J. Restricted oncogenicity of BCR/ABL P190 transgenic mice. Cancer Res. 52:4534-4539. [PubMed] [Google Scholar]
- 45.Voncken, J. W., V. Kaartinen, J. Groffen, and N. Heisterkamp. 1998. Bcr/Abl associated leukemogenesis in bcr null mutant mice. Oncogene 16:2029-2032. [DOI] [PubMed] [Google Scholar]
- 46.Walmsley, M. J., S. K. Ooi, L. F. Reynolds, S. H. Smith, S. Ruf, A. Mathiot, L. Vanes, D. A. Williams, M. P. Cancro, and V. L. Tybulewicz. 2003. Critical roles for Rac1 and Rac2 GTPases in B cell development and signaling. Science 302:459-462. [DOI] [PubMed] [Google Scholar]
- 47.Westwick, J. K., Q. T. Lambert, G. J. Clark, M. Symons, L. Van Aelst, R. G. Pestell, and C. J. Der. 1997. Rac regulation of transformation, gene expression and actin organization by multiple, Pak-independent pathways. Mol. Cell. Biol. 17:1324-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, X., K. A. Carnevale, and M. K. Cathcart. 2003. Human monocytes use Rac1, not Rac2, in the NADPH oxidase complex. J. Biol. Chem. 278:40788-40792. [DOI] [PubMed] [Google Scholar]