Abstract
To gain insight into the expression pattern and functional importance of the forkhead transcription factor Foxs1, we constructed a Foxs1-β-galactosidase reporter gene “knock-in” (Foxs1β-gal/β-gal) mouse, in which the wild-type (wt) Foxs1 allele has been inactivated and replaced by a β-galactosidase reporter gene. Staining for β-galactosidase activity reveals an expression pattern encompassing neural crest-derived cells, e.g., cranial and dorsal root ganglia as well as several other cell populations in the central nervous system (CNS), most prominently the internal granule layer of cerebellum. Other sites of expression include the lachrymal gland, outer nuclear layer of retina, enteric ganglion neurons, and a subset of thalamic and hypothalamic nuclei. In the CNS, blood vessel-associated smooth muscle cells and pericytes stain positive for Foxs1. Foxs1β-gal/β-gal mice perform significantly better (P < 0.01) on a rotating rod than do wt littermates. We have also noted a lower body weight gain (P < 0.05) in Foxs1β-gal/lβ-gal males on a high-fat diet, and we speculate that dorsomedial hypothalamic neurons, expressing Foxs1, could play a role in regulating body weight via regulation of sympathetic outflow. In support of this, we observed increased levels of uncoupling protein 1 mRNA in Foxs1β-gal/β-gal mice. This points toward a role for Foxs1 in the integration and processing of neuronal signals of importance for energy turnover and motor function.
When the gene responsible for the Drosophila homeotic mutant fork head was isolated (55) and compared to HNF3α, β, and γ, a common highly conserved DNA-binding domain of some 100 amino acids was identified (54), hence the name forkhead. According to a widely adopted nomenclature, “Fox” (Forkhead box) is used as the unifying root symbol for all forkhead genes (31). Today, we know of some forty genes encoding this highly conserved DNA-binding region (8, 33). In many instances, forkhead genes such as Foxa2 and Foxc2 play different roles during embryogenesis, e.g., guiding morphogenetic events (2, 28, 56), than they do in the adult organism, e.g., regulating metabolic pathways (11, 57). A number of forkhead genes have been inactivated by homologous recombination using embryonic stem (ES) cells, providing evidence of important roles in development (2, 26, 27, 38, 56). Furthermore, mutations in forkhead genes have been linked to human disease (15, 18, 42, 58). Fox genes have been shown to be directly involved in cellular differentiation (6, 21) and in regulation of gene expression in fully differentiated cells/tissues, e.g., the liver and pancreas (reviewed by Kaestner in reference 30). There is also extensive evidence that these proteins are components of different signal transduction pathways, including those downstream of insulin, activin, and other transforming growth factor β-related ligands (1, 13, 14, 44, 47).
In a screen for forkhead genes expressed in human adipose tissue, we identified a gene, which we called FKHL18 (10), with many similarities to what had been reported for the mouse gene Fkh3 (32). Despite extensive efforts, we were not able to confirm its expression in either isolated adipocytes or adipose tissue. Hybridization to a panel of RNA from 50 different tissues identified FKHL18 transcripts only in the aorta and, to a lesser extent, also in the kidney (10). This is different from what had been published for Fkh3, with a reportedly strong expression in the lung, ovary, and testis and weaker expression in the heart, thymus, and spleen (32). With these contradictory data at hand, we decided to make an Fkh3 β-galactosidase reporter gene knock-in mouse targeting the Fkh3 locus. In this way, β-galactosidase activity can be used as a marker for Fkh3 expression. This approach also allows for assessment of any particular phenotype associated with a lack of Fkh3. A more extensive sequence analysis showed that the mouse orthologue of the human gene FKHL18 is indeed Fkh3. Recently, the nomenclature committee has approved FOXS1 and Foxs1 as the new names for the human and mouse genes, respectively.
Using Foxs1β-gal/β-gal mice, we have been able to identify expression of Foxs1 in neural crest-derived cells, particularly cranial sensory, dorsal root, and enteric ganglia. Furthermore, we have identified Foxs1 expression in regions of the central nervous system (CNS) of importance for integration and processing of balance, hearing, and motor functions. Foxs1 is also expressed in hypothalamic nuclei known to be involved in regulating energy balance. Functional tests show that Foxs1 appears to be implicated in regulating events important for motor function as well as body weight regulation.
MATERIALS AND METHODS
Targeting construct.
Foxs1 clones were isolated from a mouse 129/SvJ genomic library (Stratagene) using a Foxs1-specific probe, and correct identity of the clones was confirmed by sequencing. To construct the targeting vector, two fragments of the genomic DNA flanking the DNA-binding region were subcloned at convenient restriction sites into the pPNT vector (52). The targeting vector contained 1.5 and 8.1 kb of homologous genomic DNA on either side of a neomycin resistance cassette. The 1.5-kb short arm was fused in frame with an NLS (nuclear localization signal)-LacZ cassette obtained from the pCH110-nls plasmid, leaving the first 16 amino acids of the Foxs1 open reading frame intact.
ES cells and Foxs1-NLS-β-galactosidase (Foxs1β-gal/β-gal) mutant mice.
R1 ES cells (2 × 107) were electroporated in Dulbecco's modified Eagle medium containing linearized targeting construct (30 μg). Selection was performed on neomycin-resistant mouse embryonic fibroblasts with G418 (300 μg/ml) and ganciclovir (2 μM). Double-resistant colonies were screened by Southern blot analysis. DNA samples were either digested with EcoRV and probed with a 5′ external probe (RsaI-RsaI, 169 bp, genomic fragment) or digested with SmaI and probed with a 3′ external probe (SacI, 444 bp, genomic fragment). Homologous recombination was detected in 1% (13/1,128) of the ES cell clones screened. Four correctly targeted ES cell clones were injected into C57BL/6 host blastocysts to generate chimeric animals. Male chimeras were bred with C57BL/6 females, and two of the cell lines were found to generate germ line transmission, determined by Southern blot analysis after EcoRV digestion. Heterozygous mice were bred for six generations onto a C57BL/6 background. Subsequent progeny and embryos were genotyped by PCR. The PCR primers were designed to use a common 5′ primer located upstream of the ATG (5′-CAGATCGCCAGCTCTGAATA-3′) and specific primers for the lacZ gene (5′-CGGGGTCTTCTACCTTTCTCTT-3′) and the Foxs1 open reading frame (5′-CCCATGATGTAGCGGTAGATG-3′). These primers generate products of 148 bp specific for the mutant allele and 206 bp for the wild type (wt). Mice were given either standard chow with 4% fat content or a high-fat diet containing 36% fat.
Functional testing of Foxs1β-gal/β-gal mice. (i) Open-field activity.
Locomotor activity of mice in the open field (40 by 40 cm) was detected by 5 photocells by 5 photocells at two different levels from the floor. Beam breaks were quantified by a computer connected to the system. Activity was measured during 1 h, and the number of beam breaks was summed into 12 5-minute blocks.
(ii) Rotarod.
Rotarod testing was done essentially as previously described (3). In brief, mice were initially put through a 3-day training program and then given four 10-min acceleration trials over 2 days. Mean results from four trials were ranked and subjected to the Mann-Whitney nonparametric test.
(iii) Balance beam.
Balance skills were tested by letting the mice traverse a graded series of narrow beams essentially as described previously (9).
(iv) Reflex responses.
The Preyer reflex and reaching response were assessed as described previously (27).
(v) Mechanosensitivity.
Mechanosensitivity was assessed by application of calibrated von Frey hairs to the plantar surface of the hindpaws of mice. The 50% withdrawal threshold was determined by gradual increases or decreases of the stimulus strength as described previously (12).
(vi) Tail flick.
Focused light of two different intensities was directed at the tail 3 cm from the tip. Time was automatically measured until the tail was twitched away from the heat stimulus, and 15 s was used as a cutoff to prevent tissue injury.
(vii) Capsaicin injection.
Capsaicin (5 μg/20 μl, dissolved in 5% ethanol, 5% Tween 80, and 90% saline) was injected into the dorsal surface of the hindpaw, and the time spent attending the paw, biting or licking, during the following 15 min was measured using a stop watch.
(viii) Water maze.
Swimming abilities, spatial learning, and memory were tested in a water task as described previously (53). Swimming paths and times were recorded and processed by use of the Water 2020 system (HVS Image).
Histology.
For detection of β-galactosidase expression, whole embryos or free-floating sections were incubated in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining solution [5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 5 mM EGTA, 0.01% deoxycholate, 0.02% NP-40, 2 mM MgCl2, and 1 mg/ml of X-Gal in phosphate-buffered saline (PBS)] at 37°C. Whole embryos were incubated overnight, and sections were incubated for 5 to 6 h. β-Galactosidase-expressing tissues were located and photographed under a dissecting microscope. To aid in identifying β-galactosidase-expressing nuclei in the postnatal day 14 (P14) brain, a number of X-Gal-stained 20-μm sections were attached to slides, counterstained for 5 min in 0.1% cresyl violet acetate in distilled H2O, and washed for 15 min in 96% ethanol and for 5 min in PBS. Double-stained sections were examined and photographed using an Eclipse E800 microscope (Nikon). For dorsal root ganglia (DRG) immunohistochemistry, P7 mice were transcardially perfused with 4% paraformaldehyde. DRG were postfixed in the same fixative, embedded in Tissue Tek, and cryosectioned at 14 μm. Embryos and P14 brains for immunohistochemistry or histology were fixed by immersion in 4% paraformaldehyde. For adequate penetration of fixative in embryonic day 15.5 (E15.5) and E18.5 embryos, skin was partly removed and the peritoneum and skulls were opened up. Embryos and brains for immunostaining were embedded in acrylamide and vibratome sectioned at 100 μm. Tissues that were to be used for X-Gal and cresyl violet (Nissl) staining were embedded in a mixture of 25% bovine serum albumin (BSA) and 0.4% gelatin in PBS. The solution was hardened by addition of glutaraldehyde to a final concentration of 2.5%, and the resulting tissue blocks were serial sectioned on a vibratome at 20, 100, or 200 μm. Whole-mount RNA in situ hybridization was performed using an InsituPro robot (Inatavis AG) as described previously (39). The probe consisted of a HindIII/ApaI fragment constituting nucleotides 936 to 1268 of the Foxs1 coding sequence (GenBank accession no. NM010226). In an effort to reduce nonspecific background due to substrate entrapment, the head of the embryo was carefully perforated with a fine cannula.
Immunohistochemistry.
Cryosections were permeabilized and blocked in 1% BSA and 0.2% Triton X-100 in PBS and then incubated for 1 h with primary antibodies. After washes in PBS, sections were incubated for another hour with secondary antibodies. All incubations were at room temperature. Vibratome floating sections were blocked and permeabilized in either 1% BSA or 5% donkey serum together with 0.5% Triton X-100 in PBS. Incubation with primary antibody was overnight followed by washes in PBS and incubation with secondary antibody for 6 h. Sections for isolectin B4 staining were, after blocking but before antibody staining, incubated for 1 h in PBLEC (1% Triton X-100, 0.1 mM CaCl2, 0.1 mM MgCl2, 0.1 mM MnCl2 in PBS, pH 6.8) and then incubated overnight in isolectin B4-biotin (200 ng/μl stock solution; Sigma) diluted 1:10 in PBLEC. All incubations with floating sections were at 4°C. For nuclear counterstaining, Topro3 (1:1,000; Molecular Probes) was either added to secondary antibody solution or diluted in PBS and added separately after antibody incubations. Sections were mounted in ProLong antifade (Molecular Probes). Primary antibodies used were rabbit anti-β-galactosidase (1:3,000; Cappel), guinea pig anti-calcitonin gene-related peptide (anti-CGRP, 1:1,000; Peninsula Laboratories), guinea pig anti-substance P (1:1,000; Peninsula Laboratories), rabbit anti-parvalbumin (1:1,000; Swant), mouse anti-β-tubulin III (1:500; Covance), mouse anti-neurofilament 160 (1:100; Developmental Studies Hybridoma Bank), mouse anti-calbindin D28k (1:2,000; Sigma), goat anti-GABAA receptor α6 (1:100; Santa Cruz), and fluorescein isothiocyanate-conjugated anti-smooth muscle actin (1:100; Sigma). Species-specific secondary antibodies were from Jackson Immunoresearch and Molecular Probes. For double staining with two rabbit antibodies, the Zenon labeling kit (Molecular Probes) was used according to the manufacturer's protocol. Sections were visualized using an LSM 510 Meta laser confocal microscope (Carl Zeiss).
DNA sequences and real-time PCR.
To establish that the mouse gene Foxs1/Fkh3/Fkhl18 is a true orthologue of the human gene FKHL18, we compared the following FKHL18 (cDNA GenBank accession no. NM004118; genomic GenBank accession no. AL160175) and Foxs1/Fkhl18 (cDNA GenBank accession no. NM0102266; genomic GenBank accession no. AL833801) sequences with other reported forkhead sequences in GenBank (http://www.ncbi.nlm.nih.gov). For quantitative real-time PCR, total RNA was extracted from brown adipose tissue using Trizol (Invitrogen) according to the manufacturer's protocol and first-strand cDNA was generated with the random hexamer primer in the first-strand cDNA synthesis kit for reverse transcription-PCR (Roche). The mRNA levels of uncoupling protein 1 (ucp1) were analyzed by real-time PCR in a ABI Prism 7900HT detection system with 36B4 as an internal control. Samples were run in triplicate using the SYBR Green PCR master mix (Applied Biosystems). The primers used were as follows: Ucp1 forward, 5′-CACCTTCCCGCTGGACACT-3′; Ucp1 reverse, 5′-CCCTAGGACACCTTTATACCTAA-3′; 36B4 forward, 5′-GAGGAATCAGATGAGGATATGGGA-3; 36B4 reverse, 5′-AAGCAGGCTGACTTGGTTGC-3′.
RESULTS
Construction of a Foxs1 knock-in β-galactosidase reporter (Foxs1β-gal/β-gal) mouse.
The coding sequence of Foxs1, contained within a single exon, is well suited for an experimental strategy in which a β-galactosidase reporter gene is fused into the open reading frame of Foxs1 to report of its expression. We constructed a targeting vector that recombined with the wild-type allele in such a way that the β-galactosidase reporter was fused downstream of the 16 most N-terminal amino acids of Foxs1 (Fig. 1). An NLS was ligated to the N terminus of β-galactosidase to ensure a distinct nuclear staining of β-galactosidase-positive cells. Localization of β-galactosidase to the low-volume nuclear compartment will, as opposed to a cytoplasmic stain, increase the sensitivity of detection. In this way a Foxs1-NLS-β-galactosidase fusion protein was created with the potential to faithfully report of Foxs1 expression levels, since transcription start, cis elements, and exon/intron structure will be very similar to that of the wt Foxs1 allele. To ensure this, we compared Foxs1 staining patterns of wt and Foxs1+/β-gal embryos (Fig. 2A and D) using a Foxs1 cRNA probe for the wt (Fig. 2A) and X-Gal (a β-galactosidase substrate) for Foxs1+/β-gal embryos (Fig. 2D) (see Materials and Methods). As can be deduced from Fig. 2D, the staining patterns are essentially the same in E11.5 Foxs1+/β-gal X-Gal-stained embryos compared with wt E11.5 embryos stained using cRNA in situ whole-mount hybridization (Fig. 2A). In both embryos, staining is seen in trunk and cranial nerve ganglia, e.g., the trigeminal ganglion. This finding supports the idea that the staining pattern in Foxs1+/β-gal mice reflects the Foxs1 mRNA expression pattern of wt mice. There is some unspecific background activity in embryos subjected to whole-mount cRNA experiments (Fig. 2A), derived from cephalic vesicles and limb buds, also found in the sense control (not shown). In spite of careful comparisons between Foxs1+/β-gal and Foxs1β-gal/β-gal, we have not been able to detect any difference in the pattern of X-Gal staining or immunoreactivity in sections or whole-mount analysis.
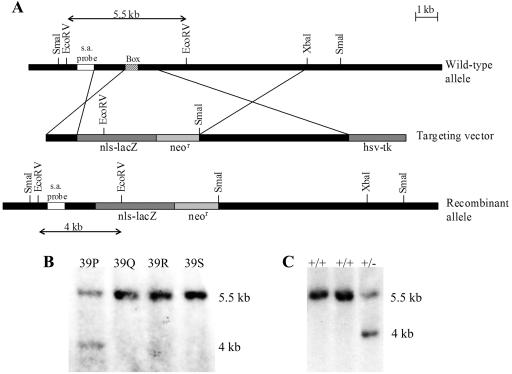
FIG. 1.
Strategy for disruption of the Foxs1 gene. (A) Schematic representation of the Foxs1 locus, the targeting vector, and the recombinant allele. The targeting vector carried a neomycin resistance gene (neor) and an Escherichia coli lacZ gene fitted with a nuclear localization signal (nls-lacZ) that had been fused in frame with the Foxs1 coding sequence 16 amino acids downstream of its initiation codon. Homologous recombination resulted in a mutant allele where most of the Foxs1 gene, including the DNA binding region (Box), had been deleted. Arrows indicate the sizes of restriction fragments detected by the short arm (s.a.) probe in the wt and recombinant alleles. Southern blot analysis of EcoRV-digested DNA from wild-type and targeted ES cells (B) and mice (C). The short arm probe detected a 5.5-kb band for the wild-type allele and a 4-kb fragment for the targeted allele. The ES cell clone denoted 39P was used for production of mice.
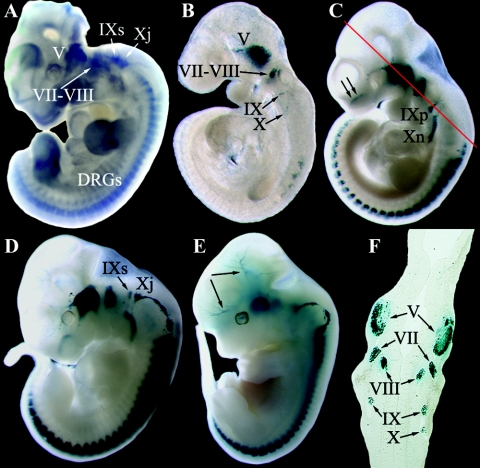
FIG. 2.
Early embryonic expression pattern of Foxs1. (A) Whole-mount Foxs1 in situ hybridization using an Foxs1 cRNA probe. Transcripts were detected in the trigeminal (V), facio-acoustic (VII-VIII), superior (IXs), and jugular (Xj) ganglia and also in DRG. (B to E) Whole-mount X-Gal staining of Foxs1+/β-gal heterozygotic embryos of ages ranging from E9.5 to E12.5. Expression of Foxs1 is initiated after E8.5 and by E9.5 (B), X-Gal staining can be detected in the trigeminal ganglion and the facio-acoustic ganglia and in a few DRG. By E10.5 (C), expression has extended to DRG along the rostro-caudal axis and to the petrosal (IXp) and nodose (Xn) ganglia as well as in the nasal region of the embryo (arrows in panel C). In the E11.5 embryo (D), Foxs1 expression can be detected in the superior and jugular ganglia, and by E12.5 (E), Foxs1 expression can be detected along major cephalic blood vessels (arrow in panel E). (F) Staining in all cranial sensory ganglia evident in transverse section of E10.5 embryo. The level of the transverse section is indicated by the red line in panel C.
Expression of Foxs1 during embryogenesis.
The pattern of staining of Foxs1+/β-gal embryos (Fig. 2B to F) is consistent with the observation that most neural crest cells acquire Foxs1 expression after migration, when they have reached the ganglia. Foxs1-positive cells in the trigeminal and facio-acoustic ganglia of cranial nerves V and VII-VIII, respectively, were evident already at E9.5 (Fig. 2B). We have not been able to detect any β-galactosidase activity at stages earlier than E9.5, i.e., at E8.5 (not shown). At E10.5 (Fig. 2C and F), expression is seen in the nasal region and along the anterior posterior axis including petrosal and nodose ganglia of the glossopahryngeal (IX) and vagal (X) nerves, respectively. Staining of the superior and jugular ganglia of cranial nerves IX and X is observed at E11.5 (Fig. 2D). This expression pattern is compatible with recruitment of Foxs1-positive cells from both neural crest and placodal cell populations (20, 46). In the E12.5 embryo, Foxs1 reporter gene activity is also found along major cranial blood vessels (Fig. 2E). At a later stage, E15.5, Foxs1-specific staining is found in the external granule layer (EGL) of the developing cerebellum and in the deep fastigial nucleus (Fig. 3A). Staining remains strong in the sensory divisions of cranial ganglia (V, VII, VIII, and IX) and DRG (Fig. 3B and C). Interestingly, cells along the ventral ramus of the spinal nerve, most likely glia cells, stain positive for Foxs1 (Fig. 3C). Whereas at E18.5, cells of the cartilage primordium of nasal septum cells stain for Foxs1, there are, at this stage, no β-galactosidase-expressing cells to be found in the retina (Fig. 3D), as opposed to the situation in the adult retina (see Fig. 7B). Furthermore, expression is found in the cochlear nucleus and in cells lining the cochlear and vestibular nerves, extending from the vestibulocochlear ganglion (VIII) (Fig. 3E). Foxs1-positive cells, most likely glia cells, are located along a trigeminal branch that extends toward the follicles of vibrissae (Fig. 3F). It also appears that, in contrast to the situation at E15.5 (Fig. 3C), β-galactosidase activity is found in the gray matter of the spinal cord (Fig. 3G).
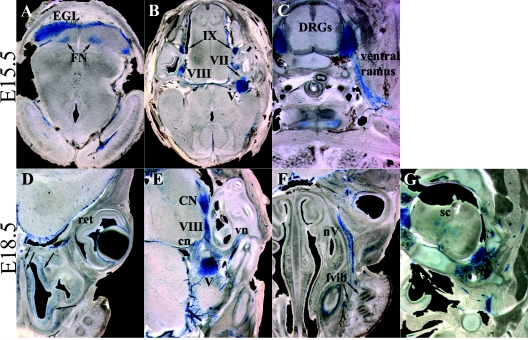
FIG. 3.
Late embryonic expression of Foxs1. X-Gal staining of 100-μm vibratome sections of E15.5 (A to C) and E18.5 (D to G) embryos is shown. (A) Foxs1 expression is present in the EGL of the forming cerebellum and in the deep fastigial nucleus (FN). Staining remains strong in sensory division of cranial ganglia (B) as well as in DRG, but at this stage, Foxs1 is also expressed in peripherally located Schwann cells along the ventral ramus of the spinal nerve (C). In the E18.5 embryo, cells just beneath the nasal epithelium (arrows in panel D), in the cartilage primordium of the nasal septum, stain positive for β-galactosidase. At this stage, the retina (ret) is negative for β-galactosidase (D). Expression is evident in the cochlear nucleus (CN) and in Schwann cells of the cochlear (cn) and vestibular (vn) nerves extending from the vestibulocochlear ganglion (VIII) (E) and also in Schwann cells of a branch of the trigeminal nerve (nV) extending toward follicles of vibrissae (fvib) (F). (G) Foxs1 expression has been initiated in the gray matter of the spinal cord (sc).
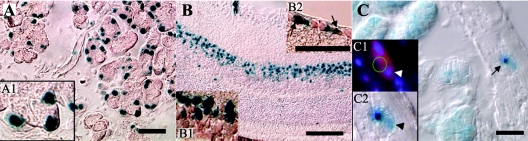
FIG. 7.
Sites of Foxs1 expression in adult tissues. (A, A1) In adult mouse tissues, β-galactosidase staining reveals Foxs1 expression in lachrymal glands. In the retina, neurons in the external part of the outer nuclear layer (B, B1) and some ganglion neurons (B2) express Foxs1. (C) Staining can also be found in enteric ganglia (arrow). Arrowheads in inset panels C1 and C2 point to Foxs1-expressing peripherin-positive (red in panel C1) cells. Bars, 50 μm (A and C), 200 μm (B).
Postnatal Foxs1 expression in CNS.
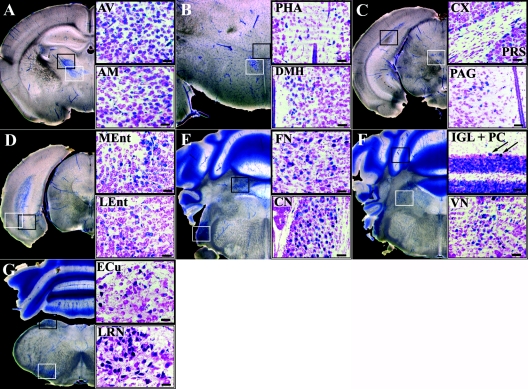
To investigate the expression pattern of Foxs1 in postnatal brains (P14), we used vibratome sections that were double stained: X-Gal was used to identify Foxs1-positive cells and cresyl violet (Nissl stain) was then applied to visualize CNS nuclei (see Materials and Methods). Foxs1-positive neurons were seen in the anteroventral and anteromedial thalamic nuclei (Fig. 4A) as well as in the posterior regions of the hypothalamus, the posterior hypothalamic area, and dorsomedial hypothalamic nucleus (Fig. 4B). Cortical and presubicular layer VI neurons and periaqueductal gray matter also display neurons positive for Foxs1 (Fig. 4C). A layer of neurons bordering the white matter in the lateral and medial entorhinal cortices (Fig. 4D) and the fastigial and cochlear nuclei (Fig. 4E) are all positive for Foxs1. In the cerebellum, the internal granule layer, a subpopulation of Purkinje cells, and the vestibular nuclei (Fig. 4F) display reporter gene activity. We can also identify Foxs1-expressing neurons in the external cuneate and lateral reticular nuclei (Fig. 4G). Taken together, Foxs1 appears to be expressed in a multitude of CNS neurons, forming a complex pattern of expression.
FIG. 4.
Sites of prominent Foxs1 expression in the P14 brain. To the left are low-magnification pictures of X-Gal-stained 200-μm coronal sections indicating the approximate level of the higher-magnification pictures, to the right, of 20-μm sections stained with both X-Gal and cresyl violet. Close-ups are depicted in black and white frames referring to sections marked in low-magnification pictures. (A) Anteroventral (AV) and anteromedial (AM) nuclei of thalamus. (B) Posterior hypothalamic area (PHA) and dorsomedial nucleus (DMH) of hypothalamus. (C) Cortical (Cx) and presubicular (PRS) layer VI neurons and periaqueductal gray matter (PAG). (D) Layer bordering the white matter in medial (MEnt) and lateral (LEnt) entorhinal cortices. (E) Fastigial (FN) and cochlear (CN) nuclei. (F) Cerebellar IGL and a subpopulation of Purkinje cells (PC, arrows) and vestibular nuclei (VN). (G) External cuneate (ECu) and lateral reticular (LRN) nuclei. Bars, 50 μm.
Cell type-specific expression of Foxs1.
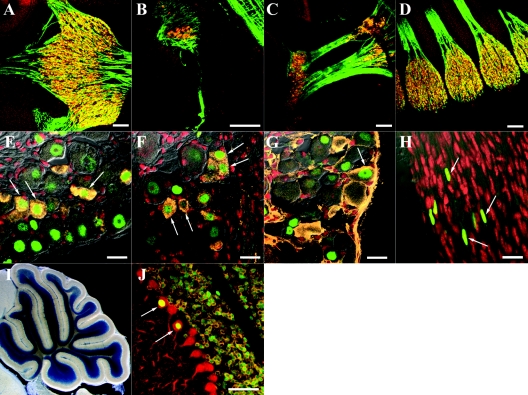
To examine particular cell types that express Foxs1, as indicated by β-galactosidase activity, we used a set of markers for various cell types of interest and compared the staining pattern of these markers with that of β-galactosidase. To study the cranial ganglia (Fig. 2, 3) in greater detail, we used antibodies against neurofilament 160 and β-galactosidase together with a nuclear counterstain (see Materials and Methods). This immunostaining procedure showed that most if not all of the neurons at E11.5 express Foxs1 at the following locations: the trigeminal ganglion (Fig. 5A), vestibulocochlear ganglion (Fig. 5B), proximal and distal ganglia of glossopharyngeal and vagal nerves (Fig. 5C), and DRG (Fig. 5D). The fact that Foxs1-negative cells can hardly be found in these ganglia indicate that glia cells might also express Foxs1. We have not been able to identify Foxs1 expression in neural crest cell populations prior to delamination nor in migrating neural crest cells (not shown). Thus, Foxs1 seems to mark neural crest cells that have reached their final migratory destination, e.g., cranial sensory and dorsal root ganglia. Expression appears to persist throughout embryonic and early postnatal development and is localized to DRG and other neural crest- and placode-derived cranial sensory ganglia, suggesting that Foxs1-positive cells are recruited from the ventral route of neural crest migration, cells that eventually will give rise to neurons and glia and form DRG and cranial sensory ganglia (41). At this stage, cells of neural crest and placode origin form sensory cranial ganglia. Most likely, placodal cells start to express Foxs1 when the ganglion is formed, since we have not seen any placodal expression of β-galactosidase. This is also supported by the fact that we have not been able to detect β-galactosidase-positive cells during migration nor have we seen any Foxs1-positive cells in the skin of Foxs1+/β-gal or Foxs1β-gal/β-gal mice. This is in agreement with a ventral route of migration for neural crest cells that at a later stage will express Foxs1. Within the DRG at P7, Foxs1 expression colocalizes with several markers for different sensory modalities, such as CGRP expressed by trkA-positive neurons, predominately unmyelinated and small myelinated neurons, involved in nociception (Fig. 5E) (19, 50). Foxs1 expression is also found in trkA-positive neurons expressing substance P, nonmyelinated nociceptive neurons (Fig. 5F) (19, 50). Furthermore, Foxs1 is expressed in parvalbumin-positive trkC, large-diameter neurons that convey proprioceptive information (Fig. 5G) (22, 24, 34). Thus, several different types of DRG neurons express Foxs1. In the outer perimeters of DRG and connecting rami, the glia cell population (Schwann and satellite cells) was examined, and a subpopulation of such cells was shown to express Foxs1 (Fig. 5H) (20). Cerebellar expression of Foxs1, at P14, is restricted to the internal granule layer (Fig. 5I); here, virtually all cells analyzed express both Foxs1 and GABAA receptor α6, a marker for granule cells (40), with the exception of a subpopulation of Purkinje cells, defined as positive for calbindin D28K, that appear to express Foxs1 (Fig. 5J). We have examined cerebellar sections from earlier time points (E15.5) (Fig. 3A), and here we can demonstrate β-galactosidase activity in the external granule layer. To assess a potential loss of neurons, serial sections of dorsal root ganglia were probed with antibodies directed against markers for DRG neuron subfamilies, e.g., CGRP, parvalbumin, and substance P, together with anti-β-tubulin III, a general neuronal marker (29). Cell counts revealed no differences between wt and Foxs1β-gal/β-gal mice in DRG total neuron count nor for any of the specific markers (not shown).
FIG. 5.
Cellular localization of Foxs1 expression. (A to D) Immunostaining of sagittal sections of E11.5 Foxs1β-gal/β-gal embryos with antibodies against β-galactosidase (orange), neurofilament 160 (green), and a nuclear counterstain (red). Foxs1 is expressed in most if not all of the neurons located in the trigeminal (A), vestibulocochlear (B), proximal and distal ganglia of glossopharyngeal and vagal nerves (C), and DRG (D). (E to H) Foxs1 expression in P7 dorsal root ganglia. Immunofluorescence with anti-β-galactosidase antibody (green) and a nuclear counterstain (red) reveals that Foxs1 is not restricted to neurons of a specific sensory modality but colocalizes with several different markers in P7 Foxs1β-gal/β-gal DRG. Colocalization is seen with CGRP (orange) (E), substance P (orange) (F), and parvalbumin (orange) (G). (H) β-Galactosidase-positive cells are also seen along the nerve sheet extending from the DRG. (I and J) Cerebellar expression of Foxs1 in P14 brain. (I) Low-magnification view of X-Gal-stained sagittal section identifying Foxs1 in the internal granule layer. (J) Immunofluorescence analysis with antibodies against β-galactosidase (green), calbindin D28K (red, a marker of Purkinje cells), and GABAA receptor α6 (orange, a marker of granule cells) confirms Foxs1 expression in most or all granule cells as well as in a subpopulation of Purkinje cells (arrows). Bars, 100 μm (A to D), 20 μm (E to H), 50 μm (J).
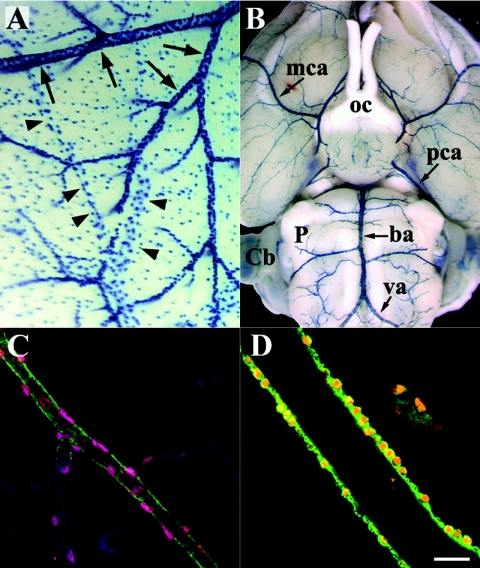
Blood vessels located in the cephalic region of vertebrates have been shown to be of neural crest origin (16, 17). In the Foxs1β-gal/β-gal and Foxs1+/β-gal mice β-galactosidase-positive cells are found in cephalic vessels, such as the brain surface vessels (Fig. 6A and B). Using a confocal technique together with markers for anti-smooth muscle actin and endothelium, we were able to localize β-galactosidase expression to pericytes and vascular smooth muscle cells (vSMCs) (Fig. 6C and D). Other sites of Foxs1 expression include lachrymal glands (Fig. 7A) and the outer nuclear layer of the retina as well as some of its ganglion cells (Fig. 7B). β-Galactosidase activity can also be found in enteric ganglion neurons, based on colocalization with peripherin, a marker for autonomic and peripheral sensory neurons (Fig. 7C) (48).
FIG. 6.
Foxs1 expression in pericytes and vSMCs of cephalic blood vessels. (A) Brain surface blood vessels of X-Gal-stained P12 mouse brain. Arteries and arterioles (arrows) appear densely stained due to extensive coverage of Foxs1-expressing vSMCs, whereas veins (arrowheads) exhibit a less-dense staining pattern. (B) Ventral view of X-Gal-stained adult brain. β-Galactosidase-positive vSMCs line the blood vessels of the entire circle of Willis surrounding the optic chiasm (oc) and major vessels extending from it, such as middle cerebral arteries (mca), posterior cerebral arteries (pca), the basilar artery (ba), and vertebral arteries (va). Also marked are the cerebellum (Cb) and pons (P). (C) Immunofluorescence with antibodies against β-galactosidase (pink), anti-smooth muscle actin (ASMA; green), and isoloectin B4 (blue), an endothelial marker. An arteriole with ASMA immunoreactivity has smaller branches that appear to be devoid of vSMCs, while pericytes stain for β-galactosidase (pink) and the endothelium stains for isoloectin B4 (blue). (D) β-Galactosidase seen in the nuclei of ASMA-positive (green) vSMCs. Bar, 20 μm.
Foxs1β-gal/β-gal mice outperform wt mice on the rotarod.
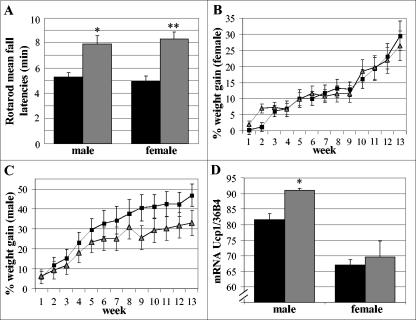
In terms of fertility, lifespan, and general behavior, we have not noted any difference between wt, Foxs1+/β-gal, and Foxs1β-gal/β-gal mice. To further investigate possible phenotypes associated with DRG, cranial sensory ganglia, and cerebellar function, wt and Foxs1β-gal/β-gal mice were compared in a series of functional tests designed to reveal altered sensory afference and/or motor efference. In the following tests, open-field activity (general behavior), balance beam performance (motor function/balance), Preyer reflex (inner ear, hearing), reaching response (inner ear, vestibular function), mechanosensitivity, tail flick (nociception), capsaicin injections (nociception), and water maze (balance, spatial learning) (see Materials and Methods), there were no differences between wt and Foxs1β-gal/β-gal mice. These findings rule out any major cerebellar or DRG/sensory cranial ganglia-related phenotype. However, Foxs1β-gal/β-gal mice perform significantly better (P < 0.01) than wt littermates on a rotating rod, i.e., a rotarod test, in which the time mice can stay on an accelerating rotating rod is measured (Fig. 8A). The rotarod is a widely employed test of a rodent's ability to sustain complex coordinated movements over time. There are other examples of gene-targeting experiments in which null mutants outperform wt mice on a rotarod test, e.g., mice lacking the dopamine D4 receptor (49) or the growth factor neuregulin1 (25).
FIG. 8.
(A) Rotarod performance. Bars represent mean fall latencies for wt (black bars) and Foxs1β-gal/β-gal (gray bars) male (n = 7 per genotype) and female mice (n = 8 per genotype). Minutes on the rotating rod differed significantly: *, P < 0.05; **, P < 0.01 (Mann-Whitney U test). Weight curves for female (B) and male (C) wt (squares) and Foxs1β-gal/β-gal (triangles) mice. Mice were put on a high-fat diet for 13 weeks. Repeated measures of analysis of variance indicate a significantly (P < 0.05) lower percent weight gain in Foxs1β-gal/β-gal (n = 7) male mice than in wt (n = 5) male mice. No difference was seen for female mice (n = 8 per genotype). (D) Relative mRNA levels of ucp1 were measured using quantitative real-time PCR. A small but significant difference was seen between wt (n = 4) (black bars) and Foxs1β-gal/β-gal (n = 3) (gray bars) male mice, whereas there was no difference in female mice (n = 4 per genotype). *, P < 0.05 (Student's t test). Error bars (all panels) indicate standard errors of the means.
Male Foxs1β-gal/β-gal mice on a high-fat diet gain less weight than wt counterparts.
Based on the fact that the dorsomedial hypothalamic nuclei have been shown to be involved in body weight regulation (4), particularly in response to high-fat diets (5), we set out to study weight curves for Foxs1β-gal/β-gal and wt mice on a high-fat diet. A significantly lower weight gain was seen in male Foxs1β-gal/β-gal mice, while no difference was observed when female Foxs1β-gal/β-gal and wt were compared (Fig. 8B and C). Since differences in sympathetic outflow to brown adipose tissue (BAT) have been postulated as a mechanism responsible for this (7), we measured ucp1 mRNA levels in BAT from wt and Foxs1β-gal/β-gal mice (Fig. 8D). Whereas ucp1 mRNA levels were significantly up-regulated in Foxs1β-gal/β-gal male mice compared to male wt mice (P < 0.05) (Fig. 8D), no difference was observed in females (Fig. 8D), a fact that agrees well with their weight curves (Fig. 8B and C). There was no difference in the amount of food eaten between Foxs1β-gal/β-gal and wt mice (not shown).
DISCUSSION
Granule cells, the most numerous neuron in CNS, are thought to originate from the rostral portion of the rhombomeric lip. Granule cell precursors migrate over the cerebellar surface and then accumulate to form the EGL. From this site, a second wave of mitosis within the EGL initiates a new migratory movement into the cerebellar cortex forming the internal granule layer (IGL). Foxs1 appears to be expressed during the formation of the EGL (Fig. 3A) as well as that of the IGL (Fig. 5I and J). β-Galactosidase activity was also found in more ventrally located regions, such as the fastigial, cochlear, vestibular, external cuneate, and lateral reticular nuclei (Fig. 4). At present, it is unclear whether these nuclei share a common origin with granule cells. Even though there is overwhelming evidence for a rostral rhombomeric lip origin of granule cell precursors, some authors claim that this by no means excludes contribution to more ventrally located precerebellar nuclei, outside the cerebellum proper (43).
In the truncal neural crest, Foxs1 is first present at E9.5, and at the same time, in the cephalic neural crest, Foxs1 is expressed in the trigeminal ganglion (Fig. 2B). The Foxs1 expression seen at E9.5 along the neural tube at the position of condensing DRG and the persistent expression at E10.5 and E11.5 is consistent with a coincident expression in postmigratory cells (Fig. 2B to D). We have not been able to identify any Foxs1-positive cells at E8.5. This is compatible with a view in which most Foxs1-positive cells follow a ventral route of migration and that, in most cells, Foxs1 is turned on in neural crest cells that have reached their destination in the dorsal root or cranial sensory ganglia. This is further supported by the fact that we have not been able to identify Foxs1-positive cells in melanocytes of epidermis (not shown). Differences in gene expression between various neural crest cell populations have been described, e.g., TRP2/DT, a melanoblast marker expressed by neural crest cells of the dorsolateral route (51). In this context, Foxs1 could be regarded as a marker for neural crest cells that have used the ventral migratory route. At P7, Foxs1 expression is found in the mature DRG (Fig. 5E to G). Foxs1 expression is not restricted to any specific neuronal subtype (Fig. 5E to G). Furthermore, it appears that some neural crest cells migrating along the ventral route and eventually ending up in the enteric ganglia also turn on Foxs1 expression (Fig. 7C). Foxs1 is also expressed in nonneural crest-derived cell types/organs, e.g., the outer nuclear layer of retina and lachrymal glands (Fig. 7A and B). In the cephalic neural crest, the situation is analogous, Foxs1 is expressed from the time point when the cranial nerve ganglia are formed and appear to persist in these locations at least to P7 (Fig. 2, 3, 5). In sensory portions of cranial ganglia, neurons of both placodal and neural crest origin appear to express Foxs1 (Fig. 2, 3, 5). Neural crest-derived glia cells express Foxs1 in the nerves extending from the ventral rami (Fig. 3C), trigeminal branches (Fig. 3F), and DRG (Fig. 5H). Thus, both neural crest and rostral rhombomeric lip cells start to express Foxs1 at their destination, e.g., DRG and EGL, respectively. Future efforts will be directed toward exploring whether these two migratory cell populations have more in common than temporal similarity in Foxs1 expression.
While essentially all nuclei of pericytes and vSMCs of cephalic blood vessels stain distinctly for β-galactosidase (Fig. 6), vessels from other locations, e.g., limbs and abdomen, lack β-galactosidase activity (not shown). Based on this, we suggest that Foxs1 should be regarded as a marker for cephalic neural crest-derived pericytes and vSMCs in contrast to vasculature of mesodermal origin. No phenotypic difference has been observed between Foxs1+/β-gal, Foxs1β-gal/β-gal, and wt mice with regard to macro- and microscopic morphology of cephalic vessels. Based on experiments using quail chick chimeras, it has been proposed that two distinct cell populations give rise to pericytes and vSMCs in the vasculature of CNS. Neural crest-derived pericytes/vSMCs are found in the forebrain (telencephalon and diencephalon), and pericytes/vSMCs of mesodermal origin are localized to the veins and arteries that irrigate dorsal/posterior parts of the head and neck. The boundary is located in the circle of Willis; here, the anterior part (e.g., internal carotid artery) has been shown to be of neural crest origin, whereas the posterior part (anastomotic posterior communicante arteries and the basilar artery) has blood vessels lined with pericytes/vSMCs derived from the mesoderm (23). As depicted in Fig. 6B, it is evident that, in mice, Foxs1 is expressed in pericytes populating both anterior parts as well as posterior parts of the circle of Willis. This would argue for a difference in pericyte/vSMC recruitment between birds and mammals, assuming that, in mice, Foxs1-positive pericytes/vSMCs are derived from a common progenitor of neural crest origin. Alternatively, a Foxs1-positive, non-neural crest-derived cell population might supply pericytes and vSMCs to dorsal/posterior parts of head and neck. Other investigators, using the same methodology, argue that neuroectoderm is able to supply all the blood vessels of the brain with pericytes/vSMCs (36).
Foxs1β-gal/β-gal mice perform significantly better (P < 0.01) than wt littermates in a rotarod test (Fig. 8A). This test measures the ability to learn, sustain, and execute complex coordinate movements, processes in which the cerebellum and its processing of incoming and outgoing signals plays a crucial role. In this context, it is interesting that other Foxs1-positive regions, such as the fastigial nucleus, vestibular nuclei, and external cuneate nucleus, have been implicated as targets for cerebellar projections (37, 45). Thus, it is possible that alterations in processing and integration of cerebellar afference/efference, at several levels, might be altered in response to lack of Foxs1, rendering null mutants more capable in the rotarod test.
In rodents, lesions in the dorsomedial hypothalamic nucleus (DMH) has been shown to specifically offer protection against excessive fat accumulation in response to a high-fat diet (5). The reason for this remains obscure. However, it was recently demonstrated that neurons in the DMH control sympathetic cardiovascular and thermogenic efference via sympathetic premotor neurons. Chemical disinhibition of such neurons, using bicuculline, caused a significant increase in BAT sympathetic nerve activity (7). The same authors conclude that blockade or inhibition of tonically active inhibitory GABAergic terminals, in close apposition to DMH neurons in sympathetic pathways, increases sympathetic outflow to thermogenic targets such as BAT. We would like to speculate that Foxs1 could act as a positive regulator, direct or indirect, of GABAergic inhibition in DMH and the release of such inhibition in Foxs1β-gal/β-gal mice might explain the observed reduction of weight gain on the basis of a more active ucp1-dependent BAT thermogenesis (35).
In conclusion, mice lacking Foxs1 seem to develop normally and tissues expressing Foxs1 also appear normal, as judged by macro- and microscopic examination. This could be attributed to redundancy with regard to other forkhead genes with overlapping expression patterns, e.g., Foxj3 (39). Many forkhead genes display a high degree of similarity in their DNA binding domains, and as a consequence hereof, they are likely to interact with identical cis elements. The phenotype observed might be related to cell types in which Foxs1 has a unique nonexchangeable role to play.
Acknowledgments
pCH110-nls plasmid was a kind gift from Jean-Jacques Panthier. ES cell culture work and blastocyst injections were performed by the Transgenic Core Facility at Göteborg University.
This work was supported by the Swedish Research Council (grants K2002-04X-03522-31D and K2002-31X-12186-06A to S.E. and K2004-32X-11283-10A to P.E.), EU grants (QLK3-CT-2002-02149 and LSHM-CT-2003-503041 to S.E.), The Arne and IngaBritt Foundation and The Söderberg Foundation (to S.E.), and Petrus and Augusta Hedlunds Foundation (to P.E.).
REFERENCES
- 1.Accili, D., and K. C. Arden. 2004. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117:421-426. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S. L., and J. Rossant. 1994. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78:561-574. [DOI] [PubMed] [Google Scholar]
- 3.Bergquist, F., H. N. Shahabi, and H. Nissbrandt. 2003. Somatodendritic dopamine release in rat substantia nigra influences motor performance on the accelerating rod. Brain Res. 973:81-91. [DOI] [PubMed] [Google Scholar]
- 4.Bernardis, L. L. 1975. The dorsomedial hypothalamic nucleus in autonomic and neuroendocrine homeostasis. Can. J. Neurol. Sci. 2:45-60. [DOI] [PubMed] [Google Scholar]
- 5.Bernardis, L. L., and L. L. Bellinger. 1991. Brown (BAT) and white (WAT) adipose tissue in high-fat junk food (HFJF) and chow-fed rats with dorsomedial hypothalamic lesions (DMNL rats). Behav. Brain Res. 43:191-195. [DOI] [PubMed] [Google Scholar]
- 6.Blomqvist, S. R., H. Vidarsson, S. Fitzgerald, B. R. Johansson, A. Ollerstam, R. Brown, A. E. Persson, G. G. Bergstrom, and S. Enerback. 2004. Distal renal tubular acidosis in mice that lack the forkhead transcription factor Foxi1. J. Clin. Investig. 113:1560-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, W. H., W. Fan, and S. F. Morrison. 2004. Medullary pathways mediating specific sympathetic responses to activation of dorsomedial hypothalamus. Neuroscience 126:229-240. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson, P., and M. Mahlapuu. 2002. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250:1-23. [DOI] [PubMed] [Google Scholar]
- 9.Carter, R. J., L. A. Lione, T. Humby, L. Mangiarini, A. Mahal, G. P. Bates, S. B. Dunnett, and A. J. Morton. 1999. Characterization of progressive motor deficits in mice transgenic for the human Huntington's disease mutation. J. Neurosci. 19:3248-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cederberg, A., R. Betz, S. Lagercrantz, C. Larsson, M. Hulander, P. Carlsson, and S. Enerback. 1997. Chromosome localization, sequence analysis, and expression pattern identify FKHL 18 as a novel human forkhead gene. Genomics 44:344-346. [DOI] [PubMed] [Google Scholar]
- 11.Cederberg, A., L. M. Gronning, B. Ahren, K. Tasken, P. Carlsson, and S. Enerback. 2001. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell 106:563-573. [DOI] [PubMed] [Google Scholar]
- 12.Chaplan, S. R., F. W. Bach, J. W. Pogrel, J. M. Chung, and T. L. Yaksh. 1994. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53:55-63. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-beta signalling. Nature 383:691-696. (Erratum, 384:648.) [DOI] [PubMed] [Google Scholar]
- 14.Chen, X., E. Weisberg, V. Fridmacher, M. Watanabe, G. Naco, and M. Whitman. 1997. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389:85-89. [DOI] [PubMed] [Google Scholar]
- 15.Clifton-Bligh, R. J., J. M. Wentworth, P. Heinz, M. S. Crisp, R. John, J. H. Lazarus, M. Ludgate, and V. K. Chatterjee. 1998. Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat. Genet. 19:399-401. [DOI] [PubMed] [Google Scholar]
- 16.Couly, G., P. Coltey, A. Eichmann, and N. M. Le Douarin. 1995. The angiogenic potentials of the cephalic mesoderm and the origin of brain and head blood vessels. Mech. Dev. 53:97-112. [DOI] [PubMed] [Google Scholar]
- 17.Couly, G. F., P. M. Coltey, and N. M. Le Douarin. 1992. The developmental fate of the cephalic mesoderm in quail-chick chimeras. Development 114:1-15. [DOI] [PubMed] [Google Scholar]
- 18.Crisponi, L., M. Deiana, A. Loi, F. Chiappe, M. Uda, P. Amati, L. Bisceglia, L. Zelante, R. Nagaraja, S. Porcu, M. S. Ristaldi, R. Marzella, M. Rocchi, M. Nicolino, A. Lienhardt-Roussie, A. Nivelon, A. Verloes, D. Schlessinger, P. Gasparini, D. Bonneau, A. Cao, and G. Pilia. 2001. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet. 27:159-166. [DOI] [PubMed] [Google Scholar]
- 19.Crowley, C., S. D. Spencer, M. C. Nishimura, K. S. Chen, S. Pitts-Meek, M. P. Armanini, L. H. Ling, S. B. MacMahon, D. L. Shelton, A. D. Levinson, et al. 1994. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell 76:1001-1011. [DOI] [PubMed] [Google Scholar]
- 20.D'Amico-Martel, A., and D. M. Noden. 1983. Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. Am. J. Anat. 166:445-468. [DOI] [PubMed] [Google Scholar]
- 21.Enerback, S., B. G. Ohlsson, L. Samuelsson, and G. Bjursell. 1992. Characterization of the human lipoprotein lipase (LPL) promoter: evidence of two cis-regulatory regions, LP-alpha and LP-beta, of importance for the differentiation-linked induction of the LPL gene during adipogenesis. Mol. Cell. Biol. 12:4622-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernfors, P., K. F. Lee, J. Kucera, and R. Jaenisch. 1994. Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents. Cell 77:503-512. [DOI] [PubMed] [Google Scholar]
- 23.Etchevers, H. C., C. Vincent, N. M. Le Douarin, and G. F. Couly. 2001. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development 128:1059-1068. [DOI] [PubMed] [Google Scholar]
- 24.Farinas, I., K. R. Jones, C. Backus, X. Y. Wang, and L. F. Reichardt. 1994. Severe sensory and sympathetic deficits in mice lacking neurotrophin-3. Nature 369:658-661. [DOI] [PubMed] [Google Scholar]
- 25.Gerlai, R., P. Pisacane, and S. Erickson. 2000. Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav. Brain Res. 109:219-227. [DOI] [PubMed] [Google Scholar]
- 26.Hulander, M., A. E. Kiernan, S. R. Blomqvist, P. Carlsson, E. J. Samuelsson, B. R. Johansson, K. P. Steel, and S. Enerback. 2003. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development 130:2013-2025. [DOI] [PubMed] [Google Scholar]
- 27.Hulander, M., W. Wurst, P. Carlsson, and S. Enerback. 1998. The winged helix transcription factor Fkh10 is required for normal development of the inner ear. Nat. Genet. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 28.Iida, K., H. Koseki, H. Kakinuma, N. Kato, Y. Mizutani-Koseki, H. Ohuchi, H. Yoshioka, S. Noji, K. Kawamura, Y. Kataoka, F. Ueno, M. Taniguchi, N. Yoshida, T. Sugiyama, and N. Miura. 1997. Essential roles of the winged helix transcription factor MFH-1 in aortic arch patterning and skeletogenesis. Development 124:4627-4638. [DOI] [PubMed] [Google Scholar]
- 29.Ji, R. R., Q. Zhang, P. Y. Law, H. H. Low, R. Elde, and T. Hokfelt. 1995. Expression of mu-, delta-, and kappa-opioid receptor-like immunoreactivities in rat dorsal root ganglia after carrageenan-induced inflammation. J. Neurosci. 15:8156-8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaestner, K. H. 2000. The hepatocyte nuclear factor 3 (HNF3 or FOXA) family in metabolism. Trends Endocrinol. Metab. 11:281-285. [DOI] [PubMed] [Google Scholar]
- 31.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142-146. [PubMed] [Google Scholar]
- 32.Kaestner, K. H., K. H. Lee, J. Schlondorff, H. Hiemisch, A. P. Monaghan, and G. Schutz. 1993. Six members of the mouse forkhead gene family are developmentally regulated. Proc. Natl. Acad. Sci. USA 90:7628-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann, E., and W. Knochel. 1996. Five years on the wings of fork head. Mech. Dev. 57:3-20. [DOI] [PubMed] [Google Scholar]
- 34.Klein, R., I. Silos-Santiago, R. J. Smeyne, S. A. Lira, R. Brambilla, S. Bryant, L. Zhang, W. D. Snider, and M. Barbacid. 1994. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements. Nature 368:249-251. [DOI] [PubMed] [Google Scholar]
- 35.Kopecky, J., G. Clarke, S. Enerback, B. Spiegelman, and L. P. Kozak. 1995. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J. Clin. Investig. 96:2914-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korn, J., B. Christ, and H. Kurz. 2002. Neuroectodermal origin of brain pericytes and vascular smooth muscle cells. J. Comp. Neurol. 442:78-88. [DOI] [PubMed] [Google Scholar]
- 37.Kotchabhakdi, N., and F. Walberg. 1978. Cerebellar afferent projections from the vestibular nuclei in the cat: an experimental study with the method of retrograde axonal transport of horseradish peroxidase. Exp. Brain Res. 31:591-604. [DOI] [PubMed] [Google Scholar]
- 38.Labosky, P. A., G. E. Winnier, T. L. Jetton, L. Hargett, A. K. Ryan, M. G. Rosenfeld, A. F. Parlow, and B. L. Hogan. 1997. The winged helix gene, Mf3, is required for normal development of the diencephalon and midbrain, postnatal growth and the milk-ejection reflex. Development 124:1263-1274. [DOI] [PubMed] [Google Scholar]
- 39.Landgren, H., and P. Carlsson. 2004. FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Dev. Dyn. 231:396-401. [DOI] [PubMed] [Google Scholar]
- 40.Laurie, D. J., P. H. Seeburg, and W. Wisden. 1992. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. II. Olfactory bulb and cerebellum. J. Neurosci. 12:1063-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Douarin, N. M., and M. A. Teillet. 1974. Experimental analysis of the migration and differentiation of neuroblasts of the autonomic nervous system and of neurectodermal mesenchymal derivatives, using a biological cell marking technique. Dev. Biol. 41:162-184. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann, O. J., N. D. Ebenezer, T. Jordan, M. Fox, L. Ocaka, A. Payne, B. P. Leroy, B. J. Clark, R. A. Hitchings, S. Povey, P. T. Khaw, and S. S. Bhattacharya. 2000. Chromosomal duplication involving the forkhead transcription factor gene FOXC1 causes iris hypoplasia and glaucoma. Am. J. Hum. Genet. 67:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, J. C., L. Cai, and C. L. Cepko. 2001. The external granule layer of the developing chick cerebellum generates granule cells and cells of the isthmus and rostral hindbrain. J. Neurosci. 21:159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakae, J., W. H. Biggs III, T. Kitamura, W. K. Cavenee, C. V. Wright, K. C. Arden, and D. Accili. 2002. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 32:245-253. [DOI] [PubMed] [Google Scholar]
- 45.Newlands, S. D., J. T. Vrabec, I. M. Purcell, C. M. Stewart, B. E. Zimmerman, and A. A. Perachio. 2003. Central projections of the saccular and utricular nerves in macaques. J. Comp. Neurol. 466:31-47. [DOI] [PubMed] [Google Scholar]
- 46.Noden, D. M. 1993. Spatial integration among cells forming the cranial peripheral nervous system. J. Neurobiol. 24:248-261. [DOI] [PubMed] [Google Scholar]
- 47.Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee, H. A. Tissenbaum, and G. Ruvkun. 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389:994-999. [DOI] [PubMed] [Google Scholar]
- 48.Parysek, L. M., and R. D. Goldman. 1988. Distribution of a novel 57 kDa intermediate filament (IF) protein in the nervous system. J. Neurosci. 8:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubinstein, M., T. J. Phillips, J. R. Bunzow, T. L. Falzone, G. Dziewczapolski, G. Zhang, Y. Fang, J. L. Larson, J. A. McDougall, J. A. Chester, C. Saez, T. A. Pugsley, O. Gershanik, M. J. Low, and D. K. Grandy. 1997. Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine. Cell 90:991-1001. [DOI] [PubMed] [Google Scholar]
- 50.Smeyne, R. J., R. Klein, A. Schnapp, L. K. Long, S. Bryant, A. Lewin, S. A. Lira, and M. Barbacid. 1994. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature 368:246-249. [DOI] [PubMed] [Google Scholar]
- 51.Steel, K. P., D. R. Davidson, and I. J. Jackson. 1992. TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development 115:1111-1119. [DOI] [PubMed] [Google Scholar]
- 52.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 53.Weeber, E. J., M. Levy, M. J. Sampson, K. Anflous, D. L. Armstrong, S. E. Brown, J. D. Sweatt, and W. J. Craigen. 2002. The role of mitochondrial porins and the permeability transition pore in learning and synaptic plasticity. J. Biol. Chem. 277:18891-18897. [DOI] [PubMed] [Google Scholar]
- 54.Weigel, D., and H. Jackle. 1990. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell 63:455-456. [DOI] [PubMed] [Google Scholar]
- 55.Weigel, D., G. Jurgens, F. Kuttner, E. Seifert, and H. Jackle. 1989. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57:645-658. [DOI] [PubMed] [Google Scholar]
- 56.Winnier, G. E., L. Hargett, and B. L. Hogan. 1997. The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes Dev. 11:926-940. [DOI] [PubMed] [Google Scholar]
- 57.Wolfrum, C., D. Q. Shih, S. Kuwajima, A. W. Norris, C. R. Kahn, and M. Stoffel. 2003. Role of Foxa-2 in adipocyte metabolism and differentiation. J. Clin. Investig. 112:345-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yildirim-Toruner, C., K. Subramanian, L. El Manjra, E. Chen, S. Goldstein, and E. Vitale. 2004. A novel frameshift mutation of FOXC2 gene in a family with hereditary lymphedema-distichiasis syndrome associated with renal disease and diabetes mellitus. Am. J. Med. Genet. 131:281-285. [DOI] [PubMed] [Google Scholar]