Abstract
In Shark Bay, wild bottlenose dolphins (Tursiops sp.) apparently use marine sponges as foraging tools. We demonstrate that genetic and ecological explanations for this behavior are inadequate; thus, “sponging” classifies as the first case of an existing material culture in a marine mammal species. Using mitochondrial DNA analyses, we show that sponging shows an almost exclusive vertical social transmission within a single matriline from mother to female offspring. Moreover, significant genetic relatedness among all adult spongers at the nuclear level indicates very recent coancestry, suggesting that all spongers are descendents of one recent “Sponging Eve.” Unlike in apes, tool use in this population is almost exclusively limited to a single matriline that is part of a large albeit open social network of frequently interacting individuals, adding a new dimension to charting cultural phenomena among animals.
Keywords: culture, Tursiops sp., social learning, tradition
The presence of culture within animal societies is a matter of considerable debate (e.g., 1–3). Among biologists and an increasing number of anthropologists, there is general consensus that an inclusive definition of culture should reflect a continuity between animals and humans (4). Here, we argue that the by far most parsimonious explanation for the first case of tool use in bottlenose dolphins (5) is that it is socially learned and transmitted. It is known that bottlenose dolphins are highly imitative and capable of social learning, both in the wild and in captivity. We show that ecological explanations for the observed behavioral pattern are inadequate and present genetic and long-term behavioral data showing that genetic explanations are extremely unlikely.
A behavioral trait is considered to vary culturally if it is acquired through social learning from conspecifics (6) and transmitted repeatedly within or between generations (7). Strong evidence for the existence of culture in animals comes from comparative long-term field studies of primate communities (1, 2), suggesting that there is significant cultural variation between populations for different behaviors. A study of six chimpanzee (Pan troglodytes) communities with no geographic overlap identified >39 behavioral patterns that were found in only some of the communities studied. Fifteen of these patterns were directly related to foraging and involved tool use (sensu 8), such as probing of vegetation to gather ants (9) or the use of stone hammers and anvils to crack nuts (10). Material culture also has been described in orangutans (Pongo pygmaeus) from six different sites on Sumatra and Borneo, where there was significant geographic variation among 19 “very likely cultural variants,” four of which involved tool use (2). In both studies, ecological explanations for the variation found between sites could be discounted. No conclusive evidence of social learning in the wild was presented, but claims for cultural transmission were further corroborated by additional evidence favoring the inference of social learning such as intense visual attention by offspring to the expert actions of their mothers and experimental evidence showing that captive apes will learn tool use observationally.
In wild populations, it is difficult to identify the transmission of a behavior based on social or observational learning. In cetaceans, this problem is exacerbated because of the habitat they live in and the limited observational opportunities. Therefore, we chose to adopt the approach suggested by Whiten et al. (1), which hinges on dismissing alternative explanations for the observed behavior, in particular ecological and genetic ones (3, 11).
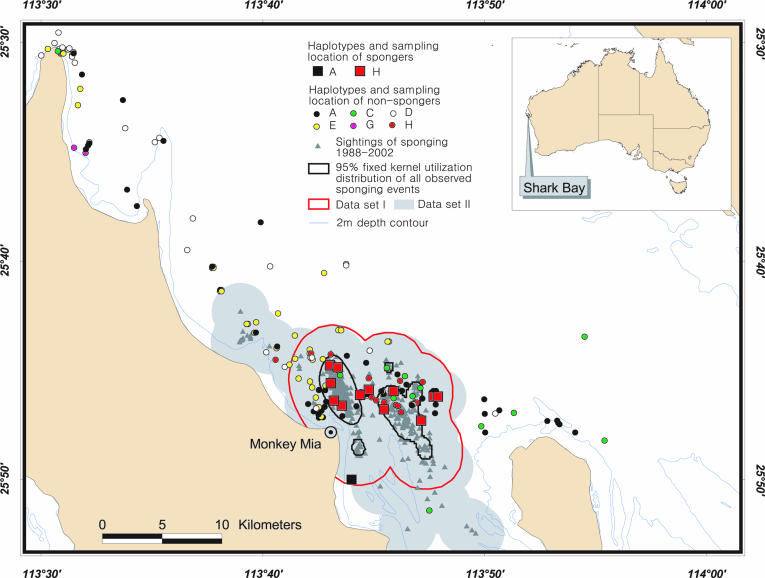
In Shark Bay, Western Australia (113°45′E, 25°48′S), a longitudinal study of bottlenose dolphins has been conducted since 1984. Previous genetic studies using both nuclear and mitochondrial markers have shown that all animals in our study area, comprising the Eastern Gulf of Shark Bay, are part of the same population (12) and interbreed (13). Within this population, as many as 11 different tactics related to foraging have been identified (14), exhibiting a diversity comparable with that of chimpanzees and orangutans. The only tactic involving tool use is sponging (Fig. 1), which has been observed in 15 of 141 known mothers in the Shark Bay population (14) and at least 7 offspring. In sponging, a dolphin breaks a marine sponge off the seafloor and wears it over its closed rostrum to apparently probe into the substrate for fish (5, 14). Sponging is significantly sex-biased to females (14), making it comparable with sex differences in learning tool use in chimpanzees (15). In dolphins' early development, which is critical for acquiring foraging skills, both males and females spend the same time with their mother (16), but all but one of the adult spongers observed to date are female, and male offspring of spongers have not been shown to take up sponging (14). It was previously suggested that sponging is a tradition (3, 14) but that more detailed genetic data are needed, which this paper provides.
Fig. 1.
Wild bottlenose dolphin foraging with marine sponge (photograph by M.R.H.).
Ecological data show that sponging is mainly, but not exclusively (5, 14), confined to deep-water channels (≥8 m) (Fig. 2). At least four nonsponging females regularly forage in at least one of these channels without using a sponge (14), strongly indicating that both spongers and nonspongers use the same habitat for foraging. Males also are regularly seen foraging in all sponging channels (R.C.C., unpublished data), ruling out the possibility that the occurrence of sponging is due to ecological differences alone.
Fig. 2.
Distribution of mtDNA haplotypes and sampling locations of spongers and nonspongers.
Given the recent findings in New Caledonian crows (Corvus moneduloides) (17), we examine the possibility that the behavior is transmitted genetically as a single-locus trait, such as the “green-beard” locus in red fire ants (Solenopsis invicta) (18). Possible genetic influences on such behavior also include polygenic inheritance. If genetic inheritance of sponging could be discounted, then it seems highly likely that sponging is culturally transmitted mainly within a matriline, i.e., daughters learn this behavior from their mothers. This social transmission would add an interesting new dimension to the mapping of cultural phenomena among animals by showing that unlike in apes, tool use in this population of bottlenose dolphins is limited almost exclusively to the social transmission within a matriline that is part of a larger population.
Members of a matriline should share mitochondrial DNA haplotypes, but what is expected of bi-parentally inherited nuclear DNA variants? Under random mating, the nuclear coancestry coefficient among noninbred individuals of the same matriline is expected to rapidly approach zero, because the number of alleles that are identical by descent shared by two individuals is expected to halve every generation. Thus, if the shared mitochondrial haplotypes derived from very ancient coancestry, nuclear relatedness of spongers should not be above population average. In contrast, if there is recent coancestry, nuclear genes also should show high relatedness. Moreover, there are two possible reasons for recent coancestry: (i) that the behavior is genetically inherited or (ii) that it is culturally transmitted between relatives. Previous genetic analyses showed that random mating can be assumed for the Shark Bay population (12). Hence, if the relatedness levels among all spongers were significantly above the population average, then it would be likely that sponging was a fairly recent invention.
Materials and Methods
Behavioral Observations. Both spongers and nonspongers were identified by using one-zero sampling methods (19) during 9,029 boat surveys with 14,447 different sightings, which were opportunistically conducted off Monkey Mia in the Eastern Gulf of Shark Bay (Fig. 2) between 1988 and 2002. Unusual behaviors, such as sponging, were recorded separately during the surveys, and the individual performing the behavior was identified by using a photographic database containing >850 different individuals.
Genetic Analysis. Genetic data were obtained from biopsy samples (20) of 185 individually identified dolphins (Fig. 2), 13 of which were adult spongers (Table 2). Each individual was genotyped for 12 hypervariable microsatellite loci (12), a genetic marker that is bi-parentally inherited. Additionally, a 355-bp segment of the maternally inherited control region of mtDNA was sequenced to obtain mtDNA haplotypes for each dolphin (12).
Table 2. Haplotype distribution within study area and for both data sets.
| Study area (12)
|
Data set 1
|
Data set 2
|
||||
|---|---|---|---|---|---|---|
| Haplotype | Males (%) | Females (%) | Males (%) | Females (%) | Males (%) | Females (%) |
| A | 30 (16.2) | 37 (20.0) | 19 (17.0) | 24 (21.3) | 21 (16.2) | 25 (19.2) |
| C | 9 (4.9) | 5 (2.7) | 5 (4.5) | 4 (3.6) | 6 (4.6) | 4 (3.1) |
| D | 8 (4.3) | 19 (10.3) | 3 (2.7) | 3 (2.7) | 3 (2.3) | 4 (3.1) |
| E | 27 (14.6) | 17 (9.2) | 14 (12.5) | 11 (9.8) | 21 (16.1) | 16 (12.3) |
| G | 0 | 2 (1.1) | 0 | 0 | 0 | 0 |
| H | 10 (5.4) | 21 (11.4) | 10 (8.9) | 19 (17.0) | 10 (7.7) | 20 (15.4) |
| Total | 84 | 101 | 51 | 61 | 61 | 69 |
Statistical Analyses. The analysis was limited to the most conservative possible delineation of a randomly breeding group. Such conservatism reduces the chance of obtaining a high relatedness of spongers simply because the analysis includes many animals that would have little chance of interbreeding with them. Two different ways of limiting the data were tried, creating data sets 1 and 2, both of which included not only dolphins from the sponging area, but also close neighbors. Dolphins sampled in proximity to the sponging area were included because they have a high likelihood of having been exposed to a sponger and/or sponging behavior in their lifetime (i.e., they are familiar with sponging), and they also were potential mating partners to spongers. Data set 1 was produced by calculating a fixed kernel utilization distribution (21) from all location points where sponging had been observed (Fig. 2), by using the Animal Movement Analysis add-in (22) in arcview 3.3 (Environmental Systems Research Institute, Redlands, CA). A buffer was then created around the 95% kernel at a distance of 3.17 km (Fig. 2), which represents the radius of the average circular home range of a female bottlenose dolphin in Shark Bay (23). Data set 1 included animals sampled within the kernel and the buffer zone. Data set 2 was produced by creating a buffer at a distance of 3.17 km around the location points where sponging had been observed. This method is slightly less conservative because it does not take into account the probability distribution of the location points. The hypothesis that there is a nonrandom association between haplotype and sponging behavior was tested by using a resampling simulation. Twelve females and 1 male were resampled with replacement 10,000 times for data sets 1 and 2, and the number of occasions on which the resampled average proportion of spongers with the same haplotype exceeded the observed was counted. Pairwise relatedness among individuals that were sampled within the area of data sets 1 and 2, excluding known offspring of spongers (Table 1) and those of other females, was calculated from the 12 microsatellite loci by using relatedness 5.08 software (www.gsoftnet.us/GSoft.html) (24). Sponger status was randomly assigned 10,000 times to 12 females and 1 male among all 112 and 130 individuals for data sets 1 and 2, respectively. Significance was applied by counting the number of occasions on which the observed mean relatedness among all spongers was exceeded by the randomized mean.
Table 1. Individual characteristics of spongers.
| Individual, ID code | Sex | MtDNA haplotype | No. of sightings with sponge (%) | No. of years observed sponging | Known offspring, sex, sponger |
|---|---|---|---|---|---|
| CKY | F | H | 14 (35.7) | 2 | SPZ, U, U |
| ANT | M | H | 49 (30.6) | 6 | |
| RUF | F | H | 9 (33.3) | 4 | |
| RSH | F | H | 9 (33.3) | 2 | RAW, U, U |
| MOO | F | H | 28 (21.4) | 2 | IND, F, Y; KOK, U, Y |
| SPE | F | H | 36 (41.7) | 4 | |
| BUS | F | A | 21 (42.9) | 6 | |
| WOB | F | H | 16 (31.3) | 1 | |
| DEM | F | H | 154 (52.0) | 9 | DOD, U, Y |
| KIT | F | H | 87 (71.3) | 7 | |
| BYT | F | H | 137 (56.2) | 12 | PIE, F, Y; BIN, M, once |
| SPO | F | H | 34 (35.3) | 5 | GRG, F, Y |
| GUM | F | H | 64 (20.3) | 8 | MUM, M, N; GOO, F, once; GMP, F, U |
M, male; F, female; U, unknown; Y, yes; N, no.
Exclusion of Genetic Explanations. There are several possible modes of genetic inheritance that might explain the presence or absence of a behavioral trait in certain individuals. We used a combination of long-term behavioral and genetic data to test whether sponging might be transmitted genetically by evaluating 10 different modes of genetic inheritance and expression patterns for their agreement with the data both on a family and population level. We also assessed whether the nonrandom (i.e., assortative) mating based on the phenotype between spongers and nonspongers might explain the observed transmission patterns of sponging; although it appears unlikely, assortative mating could possibly reduce recombination between multiple loci that determine a trait, and thus enhance the trait's transmissibility and variation between lineages. The number of occasions a sponging female was consorted by either nonsponging or the one sponging male between 2001 and 2004 was determined using focal data from an ongoing study of alliance affiliation of >100 males. Consortships are conducted by pairs or trios of males and are the only recognized mating strategy in Shark Bay (25, 26). A consortship was recorded if a pair or trio with a single female was observed for 1 h or on two consecutive surveys separated by more than 1 h, or if there was evidence of aggression toward the female (25, 26). Because assortative mating is expected to lead to a decrease of heterozygote genotype frequencies among the progeny (27), we investigated this possibility by calculating f (28), which is an estimator of the inbreeding coefficient FIS (29) among all spongers, using fstat 2.9.3 software (www.unil.ch/izea/softwares/fstat.html) (30).
Results
We sampled 13 of the 15 documented adult spongers (Table 1). Among all sampled individuals, six of the eight different haplotypes common in Shark Bay occur in our study area (Fig. 2 and Table 2). Only one of the regular spongers is male, and all but one sponger share the same haplotype H (Table 1), which in the broader sample is found in 11.35% of all females, and 5.41% of all males (Fig. 2 and Table 2). We found a significant nonrandom association between haplotype and sponging (P < 0.001 for each data set), indicating that sponging is mainly passed on within a single matriline. Moreover, the observed mean pairwise relatedness using nuclear markers among all 13 adult spongers is significantly above random expectations, regardless of the two methods we used to limit the data to the most conservative delineation of a randomly breeding group [data set 1: mean relatedness population, r(p), = –0.0041, mean pairwise relatedness of 13 adult spongers, r(s), = 0.0784, n = 112 individuals, P = 0.0002; data set 2: r(p) = –0.0042, r(s) = 0.0797, n = 130, P = 0.0001]. This result strongly indicates that spongers are closely related to each other and share a recent common ancestor.
None of the 10 different modes of single-locus inheritance that we considered agrees with the data (Table 3). In particular for all autosomal non-sex-limited modes, the observed highly significant correlation between haplotype and sponging would not be expected. If sponging was polygenic, then nongenetic effects would blur inheritance; additionally the loss of sponging genotypes due to recombination between generations could be avoided only if these sponging loci were closely linked, in which case they were expected to be inherited like a single locus, i.e., the mechanism we just discounted (Table 3).
Table 3. Inheritance and expression patterns in the first generation if sponging was coded by a single locus or by several very closely linked loci.
| Proposed mode of inheritance | Phenotypic expression of sponging in first generation | Result on population scale | Possible? | Reason |
|---|---|---|---|---|
| mtDNA (through mother) | ♂ and ♀ | No sex bias | No | Sponging significantly sex-biased toward females in population (ref. 14 and Table 1) |
| One female sponger has nonsponging haplotype A, but other females with haplotype A in sponge area do not sponge (Fig. 2) | ||||
| Not all animals with haplotype H in sponge area use sponge | ||||
| Y-linked | All ♂ | No ♀ | No | Most spongers are female |
| No ♀ | ||||
| X-linked recessive | ss × s-: ♂ and ♀ | Bias to ♂ | No | Sponging is significantly sex-biased toward females (ref. 14 and Table 1) |
| sn × s-: 1/2 of ♂ and 1/2 of ♀ | ||||
| ss × n-: ♂ | ||||
| sn × n-: 1/2 of ♂ | ||||
| X-linked recessive but only expressed in ♀ | ss × s-: only ♀ | No ♂ | No | At least one male sponger in population |
| sn × s-: 1/2 of ♀ | ||||
| X-linked dominant | ss × s-: ♂ and ♀ | Bias to ♀; strength of bias would depend on frequency of s | Very low likelihood | No significant correlation between sponging and haplotype expected |
| sn × s-: 1/2 of ♂ and all ♀ | ||||
| nn × s-: ♀ | ||||
| ss × n-: ♂ and ♀ | Mother offspring similarity approaches significance (ref. 14) | |||
| sn × n-: 1/2 of ♂ and 1/2 of ♀ | ||||
| X-linked dominant but only expressed in ♀ | Only ♀ | No ♂ | No | At least one male sponger in population |
| Autosomal recessive | ♂ and ♀ if homozygote for s | No sex bias | No | Sponging is significantly sex-biased toward females (ref. 14 and Table 1) |
| Autosomal recessive but only expressed in ♀ | Only ♀ if homozygote for s | No ♂ | No | At least one male sponger in population |
| Autosomal dominant | ♂ and ♀ if carrying allele s | No sex bias | No | Sponging is significantly sex-biased toward females (ref. 14 and Table 1) |
| Autosomal dominant but only expressed in ♀ | All ♀ if carrying allele s | No ♂ | No | At least one male sponger in population |
Matings are only considered if they lead to a sponging phenotype in the first generation. s, sponging allele; n, nonsponging allele. Crosses are ♀ genotype × ♂ genotype.
Between 2001 and 2004, 45 of the 65 adult males whose home ranges overlap with the sponging area consorted 11 sponge carriers in 34 documented consortships. Males engaging in this behavior account for at least 88.2% of all paternities that were assigned in a previous study (13). Only 2 (2.2%) of all 90 consortship places were occupied by the single known adult male sponge carrier, suggesting that almost all offspring of spongers are sired by nonsponging males. Furthermore, there was no observed heterozygote deficit among all 13 spongers (FIS = –0.045, P = 0.85 for 12 microsatellite loci), providing further evidence that sponging is not based on assortative mating.
Discussion
Our study is a significant step toward broadening the growing body of evidence for culture in cetaceans (3, 31), because it involves tool use. We showed that sponge-carrying qualifies as material culture in a marine mammal species, first by demonstrating that it is extremely unlikely that a genetic propensity or habitat differences alone account for a sponging phenotype; the latter was first suggested by Rendell and Whitehead (3). The observed patterns of occurrence and transmission of tool use are most unlikely to be explained by any single-locus mode of inheritance, with any sex-limitation or other special expression pattern. Furthermore, multilocus inheritance also is unlikely to produce the observed pattern unless the multiple loci are so tightly genetically linked that they essentially behave as a single locus or if there is very strong assortative mating. In the first case, the arguments of Table 3 would necessarily apply, whereas assortative mating is an extremely unlikely explanation for the transmission of tool use in dolphins for the following reasons. First, because adult males virtually never sponge, any assortment would have to be based on some other (unknown) correlated trait. Second, sponging females have been shown to conceive from nonsponging males (13). Third, our focal data show that almost all offspring of spongers are sired by nonsponging males. Fourth, we did not observe the predicted heterozygosity deficit among all 13 spongers. Given the small sample size, this test is not statistically very powerful, but certainly offers no support for the existence of assortative mating.
The only other possible transmission mechanism of sponging is cultural transmission within a matriline, and bottlenose dolphins are certainly capable of such transmission. Previous studies have revealed complex cognitive and imitative skills that have rarely been documented in other taxa (32), as well as the ability of vocal learning both in captivity (33) and in the wild (34). Most importantly, experimental evidence from captive dolphins demonstrates that bottlenose dolphins are capable of social learning (35, 36), have been successfully trained to imitate behaviors of their conspecifics and even human models (36–38), and are even thought to be superior in this ability compared with primates (11). Hence, among all marine mammals, bottlenose dolphins are prime candidates for a social transmission of a behavioral trait in the wild.
Cultural diversity within a single population has rarely been documented. In non-human primates, the only comparable case of a subculture within a population appears to be the grooming hand-clasp of chimpanzees in Mahale, Tanzania (39). In contrast with non-human primates, where culture has been demonstrated by using regional comparisons between populations (1, 2), the observed vertical transmission of culture with a matriline of a single population creates a different spatial pattern. Thus, the operational definition of culture used in primatology (1) does not capture cases as described in our study. We propose, therefore, to extend this definition to “include traditions that are habitual in some but not other individuals with overlapping home ranges, within the same population, where genetic and behavioral data are inconsistent with those individuals having acquired the behavior genetically.”
Although bottlenose dolphins in Shark Bay live in a fission–fusion society and are not matrilineally organized, the analysis of the relationship between genetic variants, behavioral traits, and reproductive success in neighboring populations both within and outside Shark Bay could contribute to an examination of Whitehead's suggestion about the interaction between genes and culture in cetaceans (40, 41). Additionally, if the transmission is cultural, as we infer, one might wonder why males so rarely use this behavior. Sponging is a time-consuming solitary activity that may not be compatible with the requirement for males to associate at high levels with alliance partners, but this hypothesis awaits further investigation.
Acknowledgments
We thank A. Samuels, C. Flaherty, J. J. Watson-Capps, B. Sargeant, L. Barre, and R. Smolker for contributing data to this study. W. McGrew, C. van Schaik, H. Whitehead, A. Whiten, and R. Wrangham gave helpful comments on earlier drafts of this manuscript. The Monkey Mia Dolphin Resort and Monkey Mia Wildsights were very generous with logistical support. Biopsy sampling was conducted under Department of Conservation and Land Management Sampling Permit SF002958 (to M.K.), and ethics approval was obtained from the University of New South Wales (99/52). This work was supported by the Australian Research Council, the National Geographic Society, the W. V. Scott Foundation, and the Linnaean Society of New South Wales.
Author contributions: M.K. designed research; M.K., M.R.H., R.C.C., L.B., and W.B.S. performed research; M.K. contributed new reagents/analytic tools; M.K., J.M., and R.C.C. analyzed data; and M.K. and W.B.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Whiten, A., Goodall, J., McGrew, W. C., Nishida, T., Reynolds, V., Sugiyama, Y., Tutin, C. E. G., Wrangham, R. & Boesch, C. (1999) Nature 399, 682–685. [DOI] [PubMed] [Google Scholar]
- 2.van Schaik, C. P., Ancrenaz, M., Borgen, G., Galdikas, B., Knott, C. D., Singleton, I., Suzuki, A., Utami, S. S. & Merrill, M. (2003) Science 299, 102–105. [DOI] [PubMed] [Google Scholar]
- 3.Rendell, L. & Whitehead, H. (2001) Behav. Brain Sci. 24, 309–382. [DOI] [PubMed] [Google Scholar]
- 4.Laland, K. N. & Hoppitt, W. (2003) Evol. Anthropol. 12, 150–159. [Google Scholar]
- 5.Smolker, R. A., Richards, A., Connor, R., Mann, J. & Berggren, P. (1997) Ethology 103, 454–465. [Google Scholar]
- 6.Nishida, T. (1987) in Primate Societies, eds. Smuts, B. B., Cheney, D. L., Seyfarth, R. M., Wrangham, R. W. & Struhsaker, T. T. (Univ. of Chicago Press, Chicago), pp. 462–474.
- 7.Boyd, R. & Richerson, P. J. (1996) Proc. Br. Acad. 88, 77–93. [Google Scholar]
- 8.Beck, B. (1980) Animal Tool Behavior (Garland, New York).
- 9.Nishida, T. (1973) J. Hum. Evol. 2, 357–370. [Google Scholar]
- 10.Boesch, C. (1978) Rev. Ecol.-Terre Vie 32, 195–201. [Google Scholar]
- 11.Whiten, A. (2001) Behav. Brain Sci. 24, 359–360. [Google Scholar]
- 12.Krützen, M., Sherwin, W. B., Berggren, P. & Gales, N. J. (2004) Mar. Mammal Sci. 20, 28–47. [Google Scholar]
- 13.Krützen, M., Barre, L. M., Connor, R. C., Mann, J. & Sherwin, W. B. (2004) Mol. Ecol. 13, 1975–1990. [DOI] [PubMed] [Google Scholar]
- 14.Mann, J. & Sargeant, B. (2003) in The Biology of Traditions, eds. Fragaszy, D. & Perry, S. (Cambridge Univ. Press, Cambridge, U.K.), pp. 236–266.
- 15.Lonsdorf, E. V., Everly, L. E. & Pusey, A. E. (2004) Nature 428, 715–716. [DOI] [PubMed] [Google Scholar]
- 16.Mann, J. & Watson-Capps, J. J. (2005) Anim. Behav. 69, 899–909. [Google Scholar]
- 17.Kenward, B., Weir, A. A. S., Rutz, C. & Kacelnik, A. (2005) Nature 433, 121. [DOI] [PubMed] [Google Scholar]
- 18.Keller, L. & Ross, K. G. (1998) Nature 394, 573–575. [Google Scholar]
- 19.Altmann, J. (1974) Behaviour 79, 227–267. [DOI] [PubMed] [Google Scholar]
- 20.Krützen, M., Barre, L. M., Möller, L. M., Heithaus, M. R., Simms, C. & Sherwin, W. B. (2002) Mar. Mammal Sci. 18, 863–878. [Google Scholar]
- 21.Worton, B. J. (1989) Ecology 70, 164–168. [Google Scholar]
- 22.Hooge, P. N. & Eichenlaub, B. (2000) Animal Movement Extension to arcview (Alaska Science Center–Biological Science Office, U.S. Geological Survey, Anchorage, AK), Version 2.0. (www.absc.usgs.gov/glba/gistools/animal_mvmt.htm)
- 23.Richards, A. F. (1996) Ph.D. thesis (Univ. of Michigan, Ann Arbor).
- 24.Queller, D. C. & Goodnight, K. F. (1989) Evolution (Lawrence, Kans.) 43, 258–275. [DOI] [PubMed] [Google Scholar]
- 25.Connor, R. C. & Smolker, R. A. (1996) Behaviour 133, 643–662. [Google Scholar]
- 26.Connor, R. C., Smolker, R. A. & Richards, A. F. (1992) Proc. Natl. Acad. Sci. USA 89, 987–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crow, J. F. & Kimura, M. (1970) An Introduction to Population Genetics Theory (Harper & Row, New York).
- 28.Weir, B. S. & Cockerham, C. C. (1984) Evolution (Lawrence, Kans.) 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- 29.Wright, S. (1969) Evolution and the Genetics of Populations: The Theory of Gene Frequencies (Univ. of Chicago Press, Chicago).
- 30.Goudet, J. (1995) J. Hered. 86, 485–486. [Google Scholar]
- 31.Noad, M. J., Cato, D. H., Bryden, M. M., Jenner, M.-N. & Jenner, K. C. S. (2000) Nature 408, 537. [DOI] [PubMed] [Google Scholar]
- 32.Herman, L. M., Pack, A. A. & Morrel-Samuels, P. (1993) in Language and Communication: Comparative Perspectives, eds. Roitblat, H. L., Herman, L. M. & Nachtigall, P. E. (Erlbaum, Hillsdale, NJ), pp. 403–442.
- 33.Caldwell, D. K., Caldwell, M. C. & Tyack, P. L. (1990) in The Bottlenose Dolphin, eds. Leatherwood, S. & Reeves, R. R. (Academic, New York), pp. 199–234.
- 34.Janik, V. M. & Slater, P. J. B. (1997) Adv. Study Behav. 26, 59–99. [Google Scholar]
- 35.Kuczaj, S. A., Gory, J. D. & Xitco, M. J. (1998) Behav. Brain Sci. 21, 695–696. [Google Scholar]
- 36.Xitco, M. J. & Roitblat, H. L. (1996) Anim. Learn. Behav. 24, 355–365. [Google Scholar]
- 37.Herman, L. M. (2002) in Imitation in Animals and Artifacts, eds. Dautenhahn, K. & Nehaniv, C. L. (MIT Press, Cambridge, MA), pp. 63–108.
- 38.Bauer, G. & Johnson, C. M. (1994) Percept. Motor Skills 79, 1307–1315. [DOI] [PubMed] [Google Scholar]
- 39.McGrew, W. C., Marchant, L. F., Scott, S. E. & Tutin, C. E. G. (2001) Cur. Anthropol. 42, 148–153. [Google Scholar]
- 40.Whitehead, H. (1998) Science 282, 1708–1711. [DOI] [PubMed] [Google Scholar]
- 41.Whitehead, H. (2005) Mar. Mammal Sci. 21, 58–79. [Google Scholar]