Abstract
In mammalian cells, the replication of genetic and epigenetic information is directly coupled; however, little is known about the maintenance of epigenetic information in DNA repair. Using a laser microirradiation system to introduce DNA lesions at defined subnuclear sites, we tested whether the major DNA methyltransferase (Dnmt1) or one of the two de novo methyltransferases (Dnmt3a, Dnmt3b) are recruited to sites of DNA repair in vivo. Time lapse microscopy of microirradiated mammalian cells expressing GFP-tagged Dnmt1, Dnmt3a, or Dnmt3b1 together with red fluorescent protein-tagged proliferating cell nuclear antigen (PCNA) revealed that Dnmt1 and PCNA accumulate at DNA damage sites as early as 1 min after irradiation in S and non-S phase cells, whereas recruitment of Dnmt3a and Dnmt3b was not observed. Deletion analysis showed that Dnmt1 recruitment was mediated by the PCNA-binding domain. These data point to a direct role of Dnmt1 in the restoration of epigenetic information during DNA repair.
Keywords: DNA methylation, Dnmt1, microirradiation, proliferating cell nuclear antigen
In higher eukaryotes, maintenance and propagation of genetic and epigenetic information is essential for cellular identity and survival. By-products of normal cellular metabolism, spontaneous mutations, and environmental agents can lead to various types of DNA damage. Numerous DNA repair pathways reestablishing the genetic information are known and have been intensively described (1, 2). However, very little is known about enzymes and mechanisms involved in the restoration of the epigenetic information. There are two main epigenetic marks, DNA methylation and histone modifications, which are essential for cell type-specific gene expression and maintained over multiple cell divisions (3–7). Recently, chromatin assembly and remodeling have been linked to DNA repair (8–10).
DNA methylation is a postreplicative modification occurring mostly at cytosine residues of CpG dinucleotides and is essential for mammalian development (11), parental imprinting (12), X inactivation (13), and genome stability (14, 15). In mammalian cells, DNA methylation is catalyzed by two types of enzymes, maintenance (Dnmt1) and de novo methyltransferases (Dnmt3a, Dnmt3b) (16). The maintenance methyltransferase Dnmt1 has a preference for hemimethylated CpG sites generated during DNA replication and is ubiquitously expressed (16). Dnmt1 associates with replication sites by directly binding to proliferating cell nuclear antigen (PCNA) and thus maintains DNA methylation patterns in the newly synthesized strand after DNA replication (17, 18). In contrast to the maintenance methyltransferase Dnmt1, the de novo methyltransferases Dnmt3a and Dnmt3b are responsible for establishing new DNA methylation patterns during development and show a low and tissue-specific expression (19–21).
The importance of maintaining the epigenetic information was recently underscored by knockdown experiments. Lowering Dnmt1 to rate-limiting amounts in transgenic mice lead to a loss of DNA methylation, affecting gene expression and development, and even caused cancer (14, 15, 22).
These results clearly demonstrate the importance of DNA methylation, and raise the question whether and how this epigenetic information is maintained during DNA repair. Therefore, we investigated whether and which DNA methyltransferases are present at DNA repair sites. As an experimental system, we choose local DNA damage induction at preselected subnuclear sites by UVA laser microirradiation (23), which allows the study of protein dynamics involved in the repair process in living cells.
We showed that Dnmt1 and PCNA are recruited to microirradiated sites in S and non-S phase cells immediately after irradiation, whereas Dnmt3a and Dnmt3b are not accumulated. Recruitment of Dnmt1 to DNA repair sites is mediated by the PCNA-binding domain (PBD) of Dnmt1. These results suggest that PCNA recruits enzymes involved in DNA synthesis, DNA methylation, and chromatin assembly and that Dnmt1 contributes to the restoration of epigenetic information in DNA repair.
Materials and Methods
Cell Culture and Transfection. Mouse C2C12 myoblasts and human HeLa cells were cultured in DMEM containing 50 μg/ml gentamicin supplemented with 20% and 10% FCS, respectively. For transfection, cells grown on gridded coverslips or on Lab-Tek chamber slides (Nunc) were either microinjected with plasmid DNA by using an automated microinjection system (Eppendorf) or cotransfected with TransFectin transfection reagent (Bio-Rad) according to the manufacturer's instructions. Cells were subsequently incubated overnight before performing microirradiation and live cell analyses or immunostainings.
Expression Plasmids. Mammalian expression constructs encoding translational fusions of mouse Dnmt1, Dnmt1ΔPBD, and the PBD alone with enhanced GFP were described (24). Red variants of the previously described GFP-PCNA (25) and GFP-Dnmt1 were generated by replacing GFP with monomeric red fluorescent protein (mRFP1) (26) and termed RFP-PCNA and RFP-Dnmt1. GFP-Dnmt3a and GFP-Dnmt3b1 expression constructs were as described (27). The GFP-Ligase 3 construct was generated by cloning the human DNA Ligase 3 cDNA (28) into the peGFP-C1 (Clontech) vector. DNA Ligase 3 was amplified by using the following oligonucleotides as primers for the PCR: forward, 5′-GGGG GTCGAC GCT GAG CAA CGG TTC-3′; reverse, 5′-CCCC GGATCC GCA GGG AGC TAC CAG-3′. The residues in bold indicate a SalI and a BamHI site encoded in the forward and reverse primer, respectively, for subcloning the PCR fragment into the SalI and BamHI sites of the peGFP-C1 vector downstream of GFP.
In all cases, expression was under the control of the CMV promoter. We tested all fusion proteins by expression in COS7 or 293T cells followed by Western blot analysis (29).
Immunofluorescence. Cells were fixed with 3.7% formaldehyde in PBS and permeabilized with ice-cold methanol for 5 min or with 0.2% Triton-X-100 for 3 min. The following primary antibodies (diluted 1:200 in PBS containing 2% BSA) were used: anti-γ H2AX (Ser-139) rabbit antibodies (Upstate Biotechnology), anti-Dnmt1 rabbit antibodies raised against the N-terminal domain (18), and anti-PCNA (clone PC10) mouse monoclonal antibodies (Santa Cruz Biotechnology). Primary antibodies were detected by using secondary antibodies (diluted 1:400 in PBS containing 2% BSA) conjugated to Alexa Fluor 488, 635 (Molecular Probes) and Cy3 (Amersham Pharmacia), respectively. Cells were counterstained with DAPI and mounted in Vectashield (Vector Laboratories).
Microscopy. Stained cells were analyzed by using a Zeiss Axiovert 135 TV epifluorescence microscope equipped with a ×63/1.4 numerical aperture Plan-Apochromat oil immersion objective. Images were recorded with a cooled charge-coupled device camera using metamorph software and appropriate filter sets.
For time lapse analysis, light optical sections were acquired with a Zeiss LSM410 confocal laser scanning microscope using the 488-nm Ar laser line and the 543-nm HeNe laser line, respectively. Six mid z sections at 0.5-μm intervals were taken every 3–10 min, and cells were followed up to several hours. Focus drift over time was compensated with a macro that uses the reflection at the coverslip to medium interface as reference.
After image acquisition, a projection of all six z sections was performed from each time point by using imagej 1.34.
Alternatively, time series were taken with a Leica TCS SP2/AOBS confocal laser scanning microscope using the 488-nm Ar laser line and the 561-nm DPSS laser line. Before and after microirradiation, confocal image series of one mid z section were recorded at 2-s time interval (typically 1 preirradiation and 60–120 postirradiation frames) followed by an image series with 5-min time intervals.
UVA Laser Microirradiation. Cells were seeded on 40-mm-i.d. round coverslips and sensitized for microirradiation by incubation in medium containing BrdUrd (10 μg/ml) for 20 h. For live cell microscopy and irradiation, coverslips were mounted in a FCS2 live-cell chamber (Bioptechs) and maintained at 37°C. Microirradiation was carried out with a laser microdissection system (PALM) coupled into a Zeiss LSM410 confocal laser scanning microscope. A pulsed N2 laser (337 nm) coupled into the epifluorescence path of the microscope was focused through a UV transmitting Plan-Neofluar ×63/1.25 numerical aperture objective to locally irradiate preselected spots of ≈1 μm i.d. within the nucleus. The pulse energy could be tuned with a rotatable absorption filter and was measured before passing through the objective with a power meter. Taking into account the 10% transmission of the objective at 337 nm, the energy delivered to the target was estimated to be 8 nJ per pulse. BrdUrd-sensitized cells were usually irradiated with 30 pulses that corresponded to an estimated energy of 240 nJ per irradiated site. For the acquisition of time series, the objective was changed in some cases to a Plan-Apochromat ×63/1.4 numerical aperture objective after irradiation.
Alternatively, BrdUrd-sensitized cells grown on Lab-Tek chamber slides (Nunc) were microirradiated with a 405-nm diode laser coupled into a Leica TCS SP2/AOBS confocal laser scanning microscope. The 405-nm laser was focused through a UV transmitting Leica HCX PL APO ×63/1.40 numerical aperture oil objective to locally irradiate preselected spots of ≈1 μm i.d. within the nucleus. For microirradiation, a region of interest was selected and irradiated with an intense 405-nm diode laser beam (laser set to maximum power, 50 mW, at 100% transmission) for 1 s. Under these conditions, thymine dimers and double strand breaks are generated as demonstrated by staining with specific antibodies (data not shown).
Results
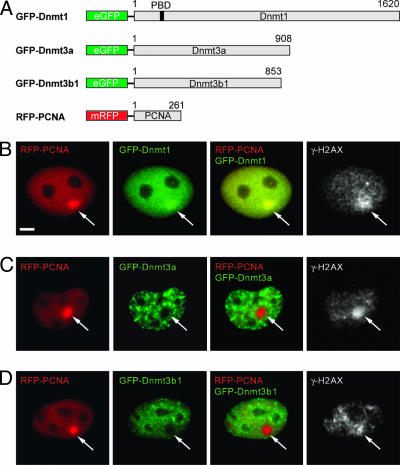
Dnmt1 and PCNA Localize at DNA Repair Sites. To study whether and which DNA methyltransferases are recruited to sites of DNA repair, we used a UVA laser microirradiation system with a pulsed 337-nm N2 laser to introduce DNA lesions. Local irradiation with this system causes a variety of different types of DNA damage at defined nuclear sites. Replicative incorporation of BrdUrd into DNA sensitizes cells and enhances the double strand break (DSB) formation upon irradiation (23, 30, 31). We microirradiated BrdUrd-sensitized mouse C2C12 myoblasts or human HeLa cells and determined DNA damage induction by immunostaining for γ-H2AX. The histone variant H2AX becomes phosphorylated (γ-H2AX) upon induction of DSBs within a few minutes and therefore serves as a DSB marker (31). Cells coexpressing GFP-Dnmt and RFP-PCNA fusion constructs were microirradiated and incubated for 25–30 min before fixation and immunodetection of γ-H2AX. At irradiated sites, an accumulation of RFP-PCNA and GFP-Dnmt1 colocalizing with γ-H2AX was observed (Fig. 1). In contrast, GFP-tagged Dnmt3a and Dnmt3b1 were found bleached at the irradiated region, indicating that de novo methyltransferases were not recruited to sites of induced DNA damage. Recruitment of endogenous Dnmt1 and PCNA at irradiated sites was confirmed by immunostaining of cells fixed ≈20 min after irradiation (Fig. 6, which is published as supporting information on the PNAS web site). Accumulation of PCNA and Dnmt1 depended on BrdUrd treatment and the energy of the UVA laser beam (data not shown). These results clearly show that PCNA and Dnmt1 are recruited to UVA-induced nuclear DNA repair sites.
Fig. 1.
GFP-Dnmt1 and RFP-PCNA but not GFP-Dnmt3a and GFP-Dnmt3b1 colocalize with γ-H2AX at microirradiated sites. Wide-field fluorescence images of cotransfected C2C12 cells formaldehyde fixed 25–30 min after UVA laser microirradiation. Double strand breaks were detected with rabbit polyclonal antibodies against γ-H2AX. Arrows mark sites of irradiation. (A) Schematic representation of the fusion proteins. (B) GFP-Dnmt1 and RFP-PCNA accumulate at sites of DNA damage and colocalize with γ-H2AX. (C and D) GFP-Dnmt3a and GFP-Dnmt3b1 are bleached at irradiated sites and do not redistribute to sites of DNA damage after microirradiation. (Scale bar, 5 μm.)
Kinetics of Dnmt1 and PCNA Recruitment. To study the kinetics of DNA methyltransferase and PCNA recruitment in vivo, we performed time lapse microscopy of microirradiated C2C12 cells expressing various combinations of fusion constructs for GFP- or RFP-tagged Dnmt1, Dnmt3a, Dnmt3b1, and PCNA. Short-term confocal live cell series of irradiated cells were recorded with time intervals of 3 min. Recruitment of RFP-PCNA and GFP-Dnmt1 to microirradiation sites could be observed as early as 1 min after irradiation, reached a maximum ≈5–10 min after irradiation, and persisted throughout the observation period of ≈30 min. As was observed before, neither GFP-Dnmt3a nor GFP-Dnmt3b1 accumulated at sites of DNA repair after microirradiation (Fig. 2). Instead, GFP fluorescence at the irradiated spot was bleached and did not recover over the total observation period, indicating that both Dnmt3a and Dnmt3b1 were rather immobile. These results were confirmed with further FRAP analyses of cells expressing RFP-Dnmt1 and GFP-Dnmt3a or GFP-Dnmt3b1 using a 488-nm Ar laser (data not shown).
Fig. 2.
Dynamics of DNA methyltransferase recruitment to repair sites in living cells. Live cell imaging of microirradiated C2C12 cells in S phase coexpressing fluorescent fusion constructs of DNA methyltransferases and PCNA is shown. The constructs used were the same as depicted in Fig. 1 A except for the RFP-Dnmt1 construct where the GFP was replaced by monomeric RFP. Maximum projections of confocal midsections are shown and times after microirradiation are indicated. (A) A C2C12 cell in early to mid S phase coexpressing GFP-Dnmt1 and RFP-PCNA shows accumulation of RFP-PCNA and GFP-Dnmt1 at sites of DNA damage (arrows) as early as 1 min after irradiation. (B) An early S phase cell coexpressing GFP-Dnmt3a and RFP-Dnmt1 shows accumulation of RFP-Dnmt1 at the irradiated site (arrow), whereas GFP-Dnmt3a is bleached (arrow) and does not recover during the entire observation period. (C) An early S phase cell coexpressing GFP-Dnmt3b1 and RFP-PCNA shows bleaching of GFP-Dnmt3b1 (arrow) at the irradiation spot, whereas RFP-PCNA accumulates at this site (arrow). (Scale bars, 5 μm.)
Next we tested whether the recruitment of RFP-PCNA and GFP-Dnmt1 occurs in S phase and non-S phase cells. The characteristic focal distribution of RFP-PCNA allowed identification of S phase in living cells (25, 32). After irradiation, we followed cotransfected S and non-S phase cells over several hours, recording confocal z stacks every 5–10 min. Accumulations of GFP-Dnmt1 and RFP-PCNA at DNA damage sites could be observed in S and non-S phase cells and both proteins could still be detected at the irradiated sites as late as several hours after irradiation (Fig. 3). Relative fluorescence intensities at the irradiated sites decreased with a half time of ≈50 min.
Fig. 3.
Recruitment of GFP-Dnmt1 and RFP-PCNA to repair sites in S and non-S phase C2C12 cells. The structure of the fusion proteins is depicted in Fig. 1 A. Live cell imaging of microirradiated C2C12 cells in G1 (A) and late S phase (B) coexpressing GFP-Dnmt1 and RFP-PCNA shows accumulation of RFP-PCNA and GFP-Dnmt1 at sites of DNA damage (arrows). Maximum projections of confocal midsections are shown and times after microirradiation are indicated. (Scale bars, 5 μm.)
Recruitment of RFP-PCNA and GFP-Dnmt1 to sites of DNA damage could also be observed in human HeLa cells in S and non-S phase (Fig. 7, which is published as supporting information on the PNAS web site). These results show that PCNA and Dnmt1 are recruited with similar kinetics in both human and mouse cells.
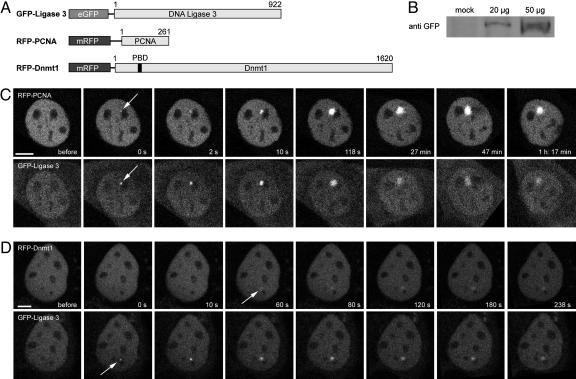
To compare the recruitment kinetics of PCNA and Dnmt1 with a known repair protein, we microirradiated C2C12 cells cotransfected with human DNA Ligase 3 fused to GFP (GFP-Ligase 3) and RFP-PCNA or RFP-Dnmt1 expression vectors. DNA Ligase 3 is known to be involved in base excision repair, single strand break repair, and error-prone nonhomologous end joining (33–35). Immediately after laser microirradiation, recruitment of GFP-Ligase 3 and RFP-PCNA could be observed, whereas Dnmt1 became visible with a delay of ≈1 min (Fig. 4). The distinct recruitment kinetics at repair sites probably reflects their different functions in repair. Thus, the slightly delayed accumulation of Dnmt1 at repair sites fits well with a role in post synthetic maintenance of DNA methylation.
Fig. 4.
Recruitment of human Ligase 3 fused to GFP (GFP-Ligase 3) and RFP-PCNA or RFP-Dnmt1 to DNA repair sites after microirradiation. (A) Schematic representation of the fusion proteins. (B) Correct expression of the GFP-Ligase 3 construct was determined by Western blot analysis. (C and D) Live cell imaging of C2C12 cells coexpressing GFP-Ligase 3 and either RFP-PCNA (C) or RFP-Dnmt1 (D). After microirradiation with a 405-nm diode laser, GFP-Ligase 3, RFP-PCNA, and RFP-Dnmt1 accumulate at sites of DNA damage (arrows). One confocal midsection is shown and times after microirradiation are indicated. (Scale bars, 5 μm.)
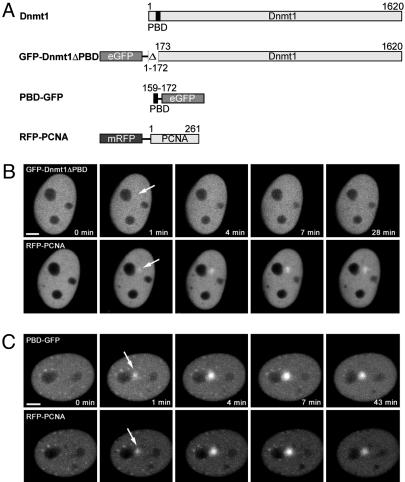
Dnmt1-Recruitment to Sites of DNA Damage via PCNA. During S phase, Dnmt1 is targeted to sites of DNA replication via its PBD (17, 24). To test whether PCNA is also responsible for the recruitment of Dnmt1 during DNA repair, we deleted the PBD of Dnmt1 (GFP-Dnmt1ΔPBD) and expressed this deletion construct in C2C12 cells together with RFP-PCNA. After UVA laser microirradiation, GFP-Dnmt1ΔPBD, unlike RFP-PCNA, remained diffuse and showed no accumulation at irradiated sites (Fig. 5). This finding suggests that the PBD plays a critical role in the recruitment of Dnmt1. To directly study the function of the PBD, we fused the PBD alone with GFP (PBD-GFP). Besides association to replication sites, the PBD fusion protein was recruited to sites of DNA damage with kinetics similar to the full-length Dnmt1 fusion construct (Fig. 5). These results demonstrate that the PBD of Dnmt1 is necessary and sufficient for localization at repair sites.
Fig. 5.
Recruitment of Dnmt1 to DNA damage sites is mediated by PCNA. (A) Schematic representation of the fusion proteins. (B) Live cell imaging of a microirradiated C2C12 cell coexpressing GFP-Dnmt1ΔPBD and RFP-PCNA. Deletion of the PBD in GFP-Dnmt1ΔPBD abolishes recruitment to sites of DNA damage, whereas RFP-PCNA accumulates at these sites as seen before (arrow). (C) A late S phase cell coexpressing PBD-GFP and RFP-PCNA shows accumulation of both RFP-PCNA and PBD-GFP at sites of microirradiation (arrows). Maximum projections of confocal midsections are shown and times after microirradiation are indicated. (Scale bars, 5 μm.)
Discussion
Higher eukaryotes have established a number of DNA repair pathways to deal with various types of DNA damage occurring during normal cellular metabolism. Whereas repair of the genetic information has been intensively studied, the mechanism by which the epigenetic information is reestablished during DNA repair is poorly understood.
The low and tissue-specific expression of Dnmt3a and -3b (19), as well as their binding to pericentromeric heterochromatin (36), makes these de novo methyltransferases rather unlikely candidates for an involvement in the genome-wide restoration of DNA methylation in DNA repair. In contrast, Dnmt1 is ubiquitously expressed at high levels and has a highly mobile fraction in the nucleus (unpublished data). Using a laser microirradiation system to induce DNA lesions at defined nuclear sites, we observed recruitment of Dnmt1 but did not detect any accumulation of Dnmt3a or Dnmt3b at repair sites in living mammalian cells. These results fit well with the recent identification of Dnmt1 as a potential component of the mismatch repair (MMR) pathway in a genetic screen for MMR mutants using Bloom′s syndrome protein (Blm)-deficient embryonic mouse stem cells (37). The accumulation of Dnmt1 at repair sites suggests that, like in DNA replication, Dnmt1 maintains the DNA methylation pattern in the DNA newly synthesized during the repair process. Thus, Dnmt1 likely prevents a loss of DNA methylation in repair, which otherwise could cause epigenetic deregulation (38) and genomic instability (14, 15). In addition, Dnmt1 has been reported to interact with histone deacetylases (39, 40) and could thus, together with chromatin assembly factor 1 (CAF-1) (8), contribute to the reestablishment of chromatin structures and respective histone modifications. Finally, Dnmt1 may also participate in the identification of the template strand in various repair pathways as was suggested for MMR (41, 42). Scope and details of Dnmt1 function(s) at DNA repair sites remain to be elucidated.
Key steps in DNA repair are recognition of the DNA damage and recruitment of the repair machinery. We could follow the accumulation of PCNA at DNA damage sites in living cells, which fits well with earlier reports identifying PCNA at DNA lesions (8, 43–45). Here, we could demonstrate that Dnmt1 is recruited to DNA damage sites via PCNA and that the PBD of Dnmt1 is necessary and sufficient for this recruitment. Our results show that PCNA mediates recruitment of the maintenance methyltransferase Dnmt1 not only to replication sites but also to DNA repair sites. Interestingly, PCNA is controlled by ubiquitination and sumoylation leading to a switch between alternative repair pathways (45, 46). In summary, PCNA plays a central role in DNA replication and repair, thus serving as a versatile loading platform for enzymes involved in DNA synthesis, chromatin assembly, and maintenance of DNA methylation.
Supplementary Material
Acknowledgments
We thank Dr. E. Li (Novartis Institutes for Biomedical Research, Cambridge, MA) (GFP-Dnmt3a and GFP-Dnmt3b1), Dr. R. Tsien (Howard Hughes Medical Institute, University of California, San Francisco) (mRFP1), and Dr. T. Lindahl (Imperial Cancer Research Center, Clare Hall Laboratories, London) (hLigase 3) for providing cDNAs and expression vectors. We are grateful to I. Grunewald, A. Gahl, Dr. U. Rothbauer, and K. Zolghadr for help and advice. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Volkswagenstiftung (to H.L. and M.C.C.).
Author contributions: L.S., M.C.C., and H.L. designed research; O.M. performed research; J.W. and M.C.C. contributed new reagents/analytic tools; O.M., L.S., and J.W. analyzed data; and O.M., L.S., M.C.C., and H.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PCNA, proliferating cell nuclear antigen; PBD, PCNA-binding domain; RFP, red fluorescent protein.
References
- 1.Friedberg, E. C. (2003) Nature 421, 436–440. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers, J. H. (2001) Nature 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch, R. & Bird, A. (2003) Nat. Genet. 33, Suppl., 245–254. [DOI] [PubMed] [Google Scholar]
- 4.Robertson, K. D. (2002) Oncogene 21, 5361–5379. [DOI] [PubMed] [Google Scholar]
- 5.Leonhardt, H. & Cardoso, M. C. (2000) J. Cell Biochem., Suppl. 35, 78–83. [DOI] [PubMed]
- 6.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 7.Felsenfeld, G. & Groudine, M. (2003) Nature 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 8.Green, C. M. & Almouzni, G. (2003) EMBO J. 22, 5163–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okano, S., Lan, L., Caldecott, K. W., Mori, T. & Yasui, A. (2003) Mol. Cell Biol. 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Attikum, H., Fritsch, O., Hohn, B. & Gasser, S. M. (2004) Cell 119, 777–788. [DOI] [PubMed] [Google Scholar]
- 11.Li, E., Bestor, T. H. & Jaenisch, R. (1992) Cell 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 12.Li, E., Beard, C. & Jaenisch, R. (1993) Nature 366, 362–365. [DOI] [PubMed] [Google Scholar]
- 13.Panning, B. & Jaenisch, R. (1996) Genes Dev. 10, 1991–2002. [DOI] [PubMed] [Google Scholar]
- 14.Eden, A., Gaudet, F., Waghmare, A. & Jaenisch, R. (2003) Science 300, 455. [DOI] [PubMed] [Google Scholar]
- 15.Gaudet, F., Hodgson, J. G., Eden, A., Jackson-Grusby, L., Dausman, J., Gray, J. W., Leonhardt, H. & Jaenisch, R. (2003) Science 300, 489–492. [DOI] [PubMed] [Google Scholar]
- 16.Bestor, T. H. (2000) Hum. Mol. Genet. 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 17.Chuang, L. S., Ian, H. I., Koh, T. W., Ng, H. H., Xu, G. & Li, B. F. (1997) Science 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- 18.Leonhardt, H., Page, A. W., Weier, H. U. & Bestor, T. H. (1992) Cell 71, 865–873. [DOI] [PubMed] [Google Scholar]
- 19.Okano, M., Xie, S. & Li, E. (1998) Nat. Genet. 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 20.Okano, M., Bell, D. W., Haber, D. A. & Li, E. (1999) Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 21.Xu, G. L., Bestor, T. H., Bourc'his, D., Hsieh, C. L., Tommerup, N., Bugge, M., Hulten, M., Qu, X., Russo, J. J. & Viegas-Pequignot, E. (1999) Nature 402, 187–191. [DOI] [PubMed] [Google Scholar]
- 22.Gaudet, F., Rideout, W. M., III, Meissner, A., Dausman, J., Leonhardt, H. & Jaenisch, R. (2004) Mol. Cell Biol. 24, 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tashiro, S., Walter, J., Shinohara, A., Kamada, N. & Cremer, T. (2000) J. Cell Biol. 150, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Easwaran, H. P., Schermelleh, L., Leonhardt, H. & Cardoso, M. C. (2004) EMBO Rep. 5, 1181–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonhardt, H., Rahn, H. P., Weinzierl, P., Sporbert, A., Cremer, T., Zink, D. & Cardoso, M. C. (2000) J. Cell Biol. 149, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A. & Tsien, R. Y. (2002) Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hata, K., Okano, M., Lei, H. & Li, E. (2002) Development (Cambridge, U.K.) 129, 1983–1993. [DOI] [PubMed] [Google Scholar]
- 28.Wei, Y. F., Robins, P., Carter, K., Caldecott, K., Pappin, D. J., Yu, G. L., Wang, R. P., Shell, B. K., Nash, R. A., Schar, P., et al. (1995) Mol. Cell. Biol. 15, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Easwaran, H. P., Leonhardt, H. & Cardoso, M. C. (2005) Cell Cycle 4, e53–e55. [DOI] [PubMed] [Google Scholar]
- 30.Lukas, C., Falck, J., Bartkova, J., Bartek, J. & Lukas, J. (2003) Nat. Cell. Biol. 5, 255–260. [DOI] [PubMed] [Google Scholar]
- 31.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. (1999) J. Cell Biol. 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sporbert, A., Gahl, A., Ankerhold, R., Leonhardt, H. & Cardoso, M. C. (2002) Mol. Cell 10, 1355–1365. [DOI] [PubMed] [Google Scholar]
- 33.Caldecott, K. W., McKeown, C. K., Tucker, J. D., Ljungquist, S. & Thompson, L. H. (1994) Mol. Cell. Biol. 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackey, Z. B., Niedergang, C., Murcia, J. M., Leppard, J., Au, K., Chen, J., de Murcia, G. & Tomkinson, A. E. (1999) J. Biol. Chem. 274, 21679–21687. [DOI] [PubMed] [Google Scholar]
- 35.Gottlich, B., Reichenberger, S., Feldmann, E. & Pfeiffer, P. (1998) Eur. J. Biochem. 258, 387–395. [DOI] [PubMed] [Google Scholar]
- 36.Bachman, K. E., Rountree, M. R. & Baylin, S. B. (2001) J. Biol. Chem. 276, 32282–32287. [DOI] [PubMed] [Google Scholar]
- 37.Guo, G., Wang, W. & Bradley, A. (2004) Nature 429, 891–895. [DOI] [PubMed] [Google Scholar]
- 38.Jackson-Grusby, L., Beard, C., Possemato, R., Tudor, M., Fambrough, D., Csankovszki, G., Dausman, J., Lee, P., Wilson, C., Lander, E. & Jaenisch, R. (2001) Nat. Genet. 27, 31–39. [DOI] [PubMed] [Google Scholar]
- 39.Rountree, M. R., Bachman, K. E. & Baylin, S. B. (2000) Nat. Genet. 25, 269–277. [DOI] [PubMed] [Google Scholar]
- 40.Fuks, F., Burgers, W. A., Brehm, A., Hughes-Davies, L. & Kouzarides, T. (2000) Nat. Genet. 24, 88–91. [DOI] [PubMed] [Google Scholar]
- 41.Kim, M., Trinh, B. N., Long, T. I., Oghamian, S. & Laird, P. W. (2004) Nucleic Acids Res. 32, 5742–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, K. Y. & James Shen, C. K. (2004) Oncogene 23, 7898–7902. [DOI] [PubMed] [Google Scholar]
- 43.Jakob, B., Scholz, M. & Taucher-Scholz, G. (2002) Int. J. Radiat Biol. 78, 75–88. [DOI] [PubMed] [Google Scholar]
- 44.Balajee, A. S. & Geard, C. R. (2001) Nucleic Acids Res. 29, 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solomon, D. A., Cardoso, M. C. & Knudsen, E. S. (2004) J. Cell Biol. 166, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. (2002) Nature 419, 135–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.