Abstract
Bacteriophages, the natural predators of bacteria, are incredibly potent candidates to counteract antimicrobial resistance (AMR). However, the rapid development of phage-resistant mutants challenges the potential of phage therapy. Understanding the mechanisms of bacterial adaptations to phage predation is crucial for phage-based prognostic applications. Phage cocktails and combinatorial therapy, using optimized dosage patterns of antibiotics, can negate the development of phage-resistant mutations and prolong therapeutic efficacy. In this study, we describe the characterization of a novel bacteriophage and the physiology of phage-resistant mutant developed during infection. M12PA is a P. aeruginosa-infecting bacteriophage with Myoviridae morphology. We observed that prolonged exposure of P. aeruginosa to M12PA resulted in the selection of phage-resistant mutants. Among the resistant mutants, pyomelanin-producing mutants, named PA-M, were developed at a frequency of 1 in 16. Compared to the wild-type, we show that PA-M mutant is severely defective in virulence properties, with altered motility, biofilm formation, growth rate, and antibiotic resistance profile. The PA-M mutant exhibited reduced pathogenesis in an allantoic-infected chick embryo model system compared to the wild-type. Finally, we provide evidence that combinatory therapy, combining M12PA with antibiotics or other phages, significantly delayed the emergence of resistant mutants. In conclusion, our study highlights the potential of combinatory phage therapy to delay the development of phage-resistant mutants and enhance the efficacy of phage-based treatments against P. aeruginosa.
Keywords: Pyomelanin, Antimicrobial resistance, Anti-phage mutation, Phage cocktails, Antibiotics, Pigments
Graphical abstract
Highlights
-
•
Phage predatory stress allows the selection of mutants with distinct fitness abilities to adapt to a niche.
-
•
Inconsistent in-situ phage amplification and resistance development invoked the need for phage dosing/phage priming.
-
•
Pyomelanogenic phage-resistant mutant had altered biofilm-forming ability, lesser growth rate and changes in motility.
-
•
Production of pyomelanin confers protection against phage adsorption and increased antibiotic resistance profiles.
-
•
Gentamicin & streptomycin (single dosage of MIC) combinations with phage (MOI 1) delayed the resistance formation by 12 h.
1. Introduction
The Global Antimicrobial Resistance and Use Surveillance System (GLASS) report developed by the World Health Organization (WHO) has described the rapid emergence of life-threatening bacterial pathogens that are resistant to even last-resort drugs like carbapenem and colistin [1,2]. The Global Burden of Disease (GBD) study emphasize P. aeruginosa among the top five bacteria causing resistant infections and deaths worldwide [3]. Resistant infections have forced clinicians to use second-line antibiotics, which can cause serious side effects, prolonged recovery periods, and even organ failure. Many first-line antibiotics have become clinically obsolete due to widespread resistance. The World Health Organization (WHO) has reported that by 2050, the death toll from antibiotic-resistant bacterial infections could rise to 10 million per year, with a yearly increase of 10 %. This could potentially surpass the death toll from cancer. The rapid evolution of antibiotic-resistant bacteria, fuelled by the extensive use of antibiotics in animal husbandry, agriculture, and improper clinical practices, has exacerbated resistance profiles. This situation necessitates the development of innovative strategies to combat antibiotic resistance [4].
Bacteriophage are natural predators of bacteria and highly potent candidates to combat drug resistance. Bacteriophages have been used as an alternative therapeutical approach in several countries, especially regions of the Soviet Union, Poland and Georgia [5]. Phage therapy has been used in clinical practice. Although the success rate varies according to the case complication, bacterial infection, and phages, the success rate has occasionally been as high as 85–90 % [5,6]. Despite the high global success rate of phage therapy, resistant variants have been reported in 80 % of studies involving the intestinal milieu, 50 % of sepsis models, and in 3 out of 4 clinical trials [7]. In spite of the fact that phage resistance was frequently observed in majority of the in vitro studies, in vivo animal models, and also in human phage treatments, phage therapy has still been successful in clearing the infection [8]. This positive outcome can be attributed to the effective bacterial clearance by phages and the poor survival of phage-resistant variants that develop during treatment.
Several studies have reported the occurrence of spontaneous mutations and chromosomal DNA alterations in the phage-resistant variants [9]. Even when the fitness of the resistant and parent strain was analysed on a treatment basis, not many studies focus on the environment or niche fitness of phage-induced selection of variants. Bacteria can attain phage resistance through random spontaneous mutations that can alter the surface receptor to hinder phage adsorption or can produce competitive inhibitors that can bind to the receptors. Enhanced extracellular matrix production can also inhibit the adsorption of phage to the bacterial surface [10,11]. Bacteria can also exhibit immunity against phage through restriction enzymes and CRISPR-Cas systems [12]. During phage exposure, bacteria are under predatory selection pressure which allows the enrichment of variants that can withstand phage stress. Bacteria-phage co-evolution can lead to the emergence of phage with a wider host range [13]. When bacteria evolve against phages, the trade-off costs can end up detrimentally affecting the bacteria in most cases. These fitness or trade-off costs are nullified by compensatory mutations after multiple generations which allow the bacteria to survive. The trade-off costs in the case of phage resistance may result in loss of virulence due to changes in lipopolysaccharide (LPS), outer membrane proteins like OmpC, capsules, flagella (motility), receptors etc [14].
During a bacterial infection, the bacteria attain a certain threshold of bacterial density after which the host immune defence is incapable to fighting the infection resulting in the patient's death, most commonly seen in bacterial sepsis [15]. The success rate of phage therapy can be attributed to the synergism between phages and the host immune system in the clearance of bacterial load. This can be one of the reasons for a high success rate of phage therapy even with the emergence of resistant mutants. Induced bacterial infection in immune-deficient and immune-competent mice challenged with phages resulted in the survival of only immune-competent mice, more specifically, the synergy of phage and neutrophils was necessary for infection reduction and mice survival [16]. The initial drop in the bacterial load due to phage therapy can provide a window for host immune activation, as well as the use of other additional therapeutics. In the later stages, when phage resistance emerges the host defence itself can fight off the infections, which were successfully demonstrated in calves infected with E. coli. Bacteria were isolated even after days of therapy but no clinical disease manifestations were reported in the calves, denoting the inefficiency of the resistant strains to establish pathogenesis [17].
Even when multiple pieces of evidence report the advantages of phage therapy, the rapid resistance development has limited its translational and clinical potential. In this work, we have analysed the phage susceptible PA-WT (P. aeruginosa-parent/wild type strain) and phage resistant variant PA-M (P. aeruginosa-mutant) for changes in their virulence factors, adaptation to phage stress, and fitness in nutrient-deprived niche. Moreover, we have evaluated the synergistic efficacy of the phage and three antibiotics, categorized as highly susceptible, moderately susceptible, or resistant to the host P. aeruginosa, for prolonged reduction of P. aeruginosa infection and screening re-sensitization.
2. Materials and methods
2.1. Maintenance of bacteria and phage stocks
Pseudomonas aeruginosa cultures ATCC 15442, ATCC 27853 and isolates from various soil samples were cultured and maintained in Cetrimide agar, Luria Bertani Broth (LB broth), and Luria Bertani Agar (LB agar). Lysates of M12PA phage, or phage cocktail were maintained in SM buffer (3 % 5 M NaCl, 4 % 1 M TrisCl, 1 % 1 M MgSO4, 1 % 0.1 M CaCl2 in 100 mL sterile milliQ H2O, pH 7.5) at 4° C. Phages were propagated by standard double-layer agar method, as described previously [18]. Bacterial cultures S. aureus, K. pneumoniae, E. coli, Micrococcus spp., Proteus vulgaris, Bacillus subtilis, Bacillus megaterium and Alkaligens was grown in LB agar and LB broth. Bacterial cultures of L. plantarum, Lactobacillus spp., L. salivarius, L. brevis and fungi Candida albicans were cultured in MRS media (agar/broth) and SDA agar respectively. All the bacterial growth media (LB agar, MRS media, SDA and cetrimide agar) and all chemicals were procured from HiMedia, India.
2.2. Phage isolation and purification
Soil samples were collected from sewage and degradable waste-contaminated soil, which are potential sites of host and phage persistence, using standard phage isolation techniques [19]. 25 g of soil sample was weighed, and approximately 40 mL of SM buffer was added. The mixture was vortexed until no soil clumps were present and then kept undisturbed for around 24 h. After 24 h of incubation, the buffer containing phage was collected, and the soil sediment was discarded. The collected buffer was centrifuged at 10,000 rpm for 10 min at 22 °C to remove solid contaminants or particles. The supernatant was collected and filter-sterilized using 0.22 μm syringe-driven filters. LB broth was prepared using the obtained filtrate and filter-sterilized. PA-WT was inoculated into the broth and grown for 18–24 h for the enrichment of phages specific to P. aeruginosa (host). Following incubation, the culture was centrifuged at 10,000 rpm for 10 min, and the supernatant containing host-specific phage was collected while the pellet containing bacterial cells was discarded. The supernatant was filter-sterilized. This obtained phage lysate was serially diluted (10-fold), and 50 μL of each dilution was allowed to infect 400 μL of overnight-grown host culture for 30 min, mixed with top agar (approximately 5 mL at <40 °C), and plated by layering on base agar (LB with 2 % agar), as described in the standard double-layer agar method. Following multiple plating's, individual plaques were picked and dispensed in 90 μL buffer, serially diluted, and plated. This picking and plating were carried out at least three times to obtain a purified single isolate of the phage.
2.3. Propagation and titration of phage
The enumeration of phage in the purified phage lysate was carried out by serially diluting 10 μL of the lysate in 90 μL of phage buffer up to 10−15 and spotting on top agar containing host culture, as done for the standard double-layer agar method [20]. The phage titre was increased by both broth and plate propagation methods. For broth propagation, phage was allowed to amplify in LB broth for 18–24 h and the phage lysate was collected. The collected lysates were again subjected to titration by serial dilution and spotting. The plates were incubated at 37°C overnight and the number of plaque-forming units (PFU) was determined by manual counting.
2.4. Host range of M12PA
The host range of M12PA was evaluated by spotting 3 μL of high titre phage lysate (109 PFU/mL) on top agar containing bacterial cultures of S. aureus, K. pneumonia, E. coli, P. aeruginosa ATCC 27853 and P. aeruginosa (wild types) cultured on individual agar plates [21]. The spots were left undisturbed to avoid smudging and once the spots were dry, the plates were incubated at 37° C overnight. Following the incubation period, the plates were observed for lysis zones. The host range were also evaluated by performing a double agar overlay assay, where 10 μL of phage and was added to 90 μL (1.5 × 108 CFU/mL) of the host in 400 μL of LB broth and incubated for a period of 30 min. Following incubation 4.5 mL od top agar was added, mixed and poured to base agar plates and was allowed to solidify. The plates were then incubated at 37 °C overnight. Experiments were carried out in triplicates.
2.5. Phage thermal and pH stability
The thermal and pH stability of the phage was determined for a range of temperatures and pH. 1.5 × 108 PFU of phage was made up to a final volume of 1 mL using SM buffer to a final titre of 107 PFU/mL. The vials containing the phage was incubated at 30° C, 40° C, 50° C, 60° C, 70° C, 80° C and 90° C for 1 h to assess their thermal stability. The concentration of phage was enumerated at every 10 min interval by sampling 10 μL of phage and serially diluting and spotting on top agar containing the host culture [20]. The spots were allowed to dry and the plates were then incubated at 37° C overnight. The PFU was manually counted.
The pH stability of M12PA was analysed for pH ranging from 1 to 11. SM buffer was prepared and the final pH of 1, 3, 5, 7, 9 and 11 was achieved by adjusting the pH using an acid (0.1 N HCl) and base (10 N NaOH) solution. Following this, 1.5 × 108 PFU of phage was made up to a final volume of 1 mL using phage buffer of various pH to a final titre of 107 PFU/mL. The vials were incubated for 24 h and the readings were taken at every 2 h interval by sampling 10 μL of phage. This was then serially diluted and spotted on top agar containing the host culture followed by incubation at 37° C overnight [20]. The PFU was manually counted on the next day. The experiments were carried out at least three times.
2.6. TEM analysis
For TEM imaging, 10 μL of high-titer purified phage lysate was spotted onto a 200-mesh carbon film grid and allowed to set at room temperature for 5 min. Excess phage lysate was then removed using Kimwipes. The adsorbed phages were negatively stained with Uranyless (Electron Microscopy Sciences) stain. Subsequently, the phage-coated carbon grids were imaged using a Joel JEM 2100 plus electron microscope at 200 kV voltage. Head length and widths, tail lengths were measured as previously described [22].
2.7. Assessing bacterial growth during phage infection
Bacterial growth kinetics of PA-WT (ATCC 15442) was analysed by growing the culture in LB broth and intermittent samples every 2 h [23]. The absorbance was read at 600 nm with technical replicates at each sampling. Bacterial culture and LB broth were maintained as negative and positive controls, respectively. Phage at various multiplicity of infection (MOI) levels were added to the host bacteria to determine the growth inhibition pattern at different phage-to-bacterial ratios. Phage MOI of 100, 10, 1, 0.1, 0.01, 0.001, 0.0001, 0.00001, 0.000001 was used to infect bacteria. The bacterial growth was recorded by measuring the absorbance of cultures for up to 18 h. The 18th hour culture was used to isolate resistant bacteria (PA-M). Experiments were carried out in triplicate.
2.8. Phage priming
To determine the effect of phage dosing in therapeutic applications as well as the inhibition of bacteria during priming, phage at MOI 1 was added to cultures grown up to 6 h, 7 h, 8 h, 9 h, 10 h, 11 h and 12 h. The cultures were infected prior to priming at 0th hour using M12PA at an MOI of 1. Priming was also carried out using MOI of 10 (higher titre M12PA), phage lysate obtained at 18th hour after infection (probability of co-evolved phage), and a phage cocktail. The absorbance of cultures was read at 600 nm at every 2-h interval for up to 24 h. Experiments were carried out in triplicate.
To understand the probability of occurrence of pyomelanogenic strains, P. aeruginosa (1.5 × 106 CFU/mL) was grown in 180 μL of media in multiple 96 well-plates and infected with phage at an MOI of 1. The plates were incubated at 37° C in a shaking incubator and observed at 12 h intervals for pyomelanogenic positive wells. Experiments were carried out in triplicate.
2.9. Pigment isolation
Pyomelanin was isolated and purified as previously described [24], with certain modifications. Pyomelanin was isolated by collecting 48 h grown PA-M cultures and subsequent removal of bacterial cells (biomass) by centrifuging at 8500 rpm for 30 min at 22° C. The supernatant was collected and filter-sterilized. Pyomelanin is present extracellularly, and can be precipitated in the presence of acid. The obtained filtrate was then acidified using 0.1 N HCl to a final pH of 2 and was incubated in dark for more than 24 h. Following the incubation period, the filtrate was centrifuged at 8500 rpm for 30 min. The pellet is washed in water at least thrice and finally washed with absolute ethanol. The pellet is collected and the ethanol is evaporated by placing the pellet at 70° C (dry heat).
2.10. Pigment characterization
The obtained pigment was subjected to FT-IR and NMR spectroscopic characterization to confirm its structure [25,26]. The KBr pressed disc/pellet technique was used to prepare samples for IR analysis. About 10 mg of the sample was mixed with oven-dried KBr crystals to obtain pellets, which were then characterized using a Spectrum Two FT-IR Spectrometer with LiTaO3 Detector (PerkinElmer), with 40 scans averaging at a resolution of 2 cm−1. The 1H NMR and 13C –NMR Spectra were recorded in Bruker 400 MHz instrument.
2.11. Phage adsorption analysis
Phage adsorption assay was performed as previously described with certain modifications [27]. Phage and host bacterium were added at a MOI of 1 and a cell density of 0.5 McFarland respectively, followed by incubation for 5 min in static conditions. 1 mL of the incubated samples was collected in 3 vials, and 2 vials were centrifuged at 10,000 rpm for 3 min at 22° C 10 μL from the supernatant of one of the centrifuged vials was sampled and plated to determine the free phage. The supernatant from the second vial is discarded and the pellet is re-suspended in phage buffer and 10 μL was sampled and plated to determine the number of adsorbed phages. A 10 μL sample was taken from the third vial without centrifugation to determine the total phage count. All three vials were sampled and plated at the same time without delay or time variation to avoid time-dependent adsorption and errors in the experiment. The adsorption efficiency of the phage in presence of pigment was also evaluated by adding 50 μL of isolated pigment during the 5 min incubation period and plated. Experiments were carried out in triplicate.
2.12. Toxicity of the pigment
The purified pyomelanin was spotted on bacterial lawn of pathogenic strains and soil bacteria including S. aureus, K. pneumoniae, E. coli, Micrococcus spp., Proteus vulgaris, Bacillus subtilis, Bacillus megaterium, Alkaligens and also on probiotic organisms like L. plantarum, Lactobacillus spp., L. salivarius, L. brevis and on fungi Candida albicans. The plates were incubated for 18–24 h and was observed for zone of inhibition.
2.13. Fitness survival – competition
2.13.1. Competition in broth
500 μL of each PA-WT and PA-M containing 1.5 × 108 CFU were inoculated in 19 mL LB broth and 18 mL LB broth containing additional 1 mL of phage at an MOI of 1 (1.5 × 108 PFU). PA-WT and PA-M were grown separately in 19.5 mL LB broth and 18.5 mL of LB broth with phage (MOI 1) by inoculating 500 μL of 1.5 × 108 CFU and were maintained as controls. The cultures were grown up to 10 h, with intermittent sampling at every 2 h. The collected samples were serially diluted, and all the dilutions were plated on LB agar plates. PA-WT and PA-M were also grown in combination in nutrient-deficient condition (water) to evaluate their survival ability and viability ratios.
2.13.2. Competition in plate
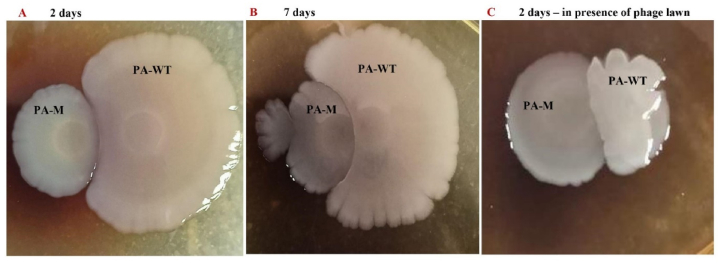
PA-WT and PA-M were grown up to an OD of 0.5, and 5 μL of each culture was spotted on LB plates at a distance of 4 mm. Spots were made in different combinations, including PA-WT/PA-WT, PA-WT/PA-M and PA-M/PA-M. Two sets of LB plates were prepared: one containing only LB agar and the other with a phage lawn. The phage lawn was prepared by swabbing 100 μL of 1.5 × 108 PFU of phages onto the agar. The plates were incubated for a total of 7 days and the diameter of the spots was measured every 2 days. Experiments were carried out in triplicate.
2.14. Bacterial motility
PA-WT and PA-M were grown up to a 0.5 of OD, and 10 μL these grown cultures were spotted on 0.7 % LB top agar medium. The plates were incubated at 37° C for 4 days and observed every 24 h.
2.15. Antibiotic profiling and MIC determination
The antibiotic sensitivity profiles of the bacterial strains (PA-WT and PA-M) were determined using the disc diffusion method [28]. A total of 23 antibiotics (tobramycin 10 μg, gentamicin 10 μg, norfloxacin 10 μg, ofloxacin 5 μg, streptomycin 10 μg, cefixime 5 μg, amoxycillin 10 μg, doxycycline 30 μg, levofloxacin 5 μg, penicillin 10 μg, amikacin 30 μg, cefpodoxime 10 μg, nafcillin 1 μg, kanamycin 30 μg, erythromycin 15 μg, rifamycin 5 μg, vancomycin 30 μg, methicillin 5 μg, clindamycin 2 μg, tetracycline 30 μg, ciprofloxacin 5 μg, chloramphenicol 30 μg, ampicillin 10 μg) obtained from HiMedia were screened and categorized as susceptible, moderate/intermediate susceptible, and resistant according to the Clinical & Laboratory Standards Institute (CLSI) guidelines [29]. PA-M was also screened to determine its antibiotic sensitivity profiles. Three antibiotics from the profile was chosen for combination therapy, highly susceptible (gentamicin), moderately susceptible (streptomycin) and resistant (ampicillin). The minimum inhibitory concentration of the selected antibiotics was determined by preparing a range of antibiotic concentrations by micro-dilution method in a 96 well plate (HiMedia). Concentrations ranging from 200 μg/mL to 0.8 μg/mL for gentamicin and streptomycin and from 0.8 μg/mL to 1 mg/mL for ampicillin were used according to the trend observed in trials. McFarland standard solutions of host bacteria were prepared and 106 CFU/mL of bacteria was used as the inoculum. The plates were incubated at 37° C for overnight with shaking at 80 rpm. Experiments were carried out in triplicate.
2.16. Combination therapy
The synergistic effect of antibiotics and phage was analysed by infecting host bacteria with phage at an MOI of 1 along with antibiotics [30]. Different sets of antibiotic concentrations, including the MIC, ½ MIC, 1/3 MIC and ¼ MIC, were used along with phage. PA-WT infected with the antibiotic concentrations and MOI 1 separately was maintained as controls to compare the growth pattern of the host. PA-WT alone was used as a negative control. LB broth was maintained as a positive control. The experiment was carried out for the three chosen antibiotics (gentamicin, streptomycin and ampicillin). Experiments were carried out in triplicate.
2.17. Biofilm assay-MTT
PA-WT and PA-M were inoculated in LB media with additional glucose (2 g in 100 mL media), to promote biofilm formation [31]. To understand the effect of phage, antibiotics, and phage-antibiotic combinations, both cultures were allowed to grow in 8 wells along with control groups in 96 well plates. The plates were incubated for 24–48 h to ensure biofilm formation. Following the incubation period, the samples phage, gentamicin, streptomycin, ampicillin, phage + gentamicin, phage + streptomycin and phage + ampicillin were added to the assigned wells and were incubated for 4 h. After 4 h of treatment period, the planktonic cells were pipetted out, and the wells were washed twice with PBS. 50 μL of MTT dye and 150 μL of PBS were added to all wells and incubated for 3–4 h, after which DMSO was added to dissolve the formazan product, and the plate was read at 600 nm using a plate reader. PA-WT and PA-M were also grown in catheters and the dry weight of biomass was quantified by weighing.
2.18. Cell aggregation assay
To evaluate the auto aggregation of PA-WT and PA-M and the changes in aggregation rate in presence of M12PA and antibiotics, cell aggregation was performed [32]. PA-WT and PA-M were grown in LB broth up to an OD of 0.5. The bacterial cells were collected by centrifugation, and the supernatant was discarded. The cells were re-suspended in sterile PBS and then aliquot into 12 test tubes. To this, phage, antibiotics and antibiotic-phage combinations were added. The cells were incubated at statically for 10 h after sampling at 0th hour. Following incubation, the clear supernatant was sampled at the 10th hour, and the absorbance was read at 600 nm. The cell aggregation rate was determined using equation (1);
| (1) |
where, At is the absorbance at time ‘t’ and A0 is the absorbance at time 0.
2.19. In-ovo virulence assay
Fertilized chick embryos were procured from local farmers around Thiruvarur, India. All the eggs were cleaned and incubated at 37°C for up to 7 days. The eggs were rotated manually at an interval of 6 h. The eggs were candled on day 4,5, and 6. Eggs that showed a blood ring or no visible embryo at the time of candling were removed prior to the experiments. On day 7, the eggs were candled, and the air sac was marked on the blunt end. An incision was made in the air sac carefully to inject the cultures. To evaluate the lethality of bacterial cultures, two sets of eggs with three groups each containing 6 eggs were labelled for inoculation of PA-WT, PA-M and sterile PBS (control) [33]. The eggs were candled and the eye of the embryo was marked and the inoculations were done on the opposite side of the marked eye. The cultures were inoculated at a cell density of 1.5 × 106 CFU using a 2 mL syringe through the incision at the air sac. The needles were allowed to penetrate to at least 1.5 cm deep into the eggs to reach and infect the allantoic fluid. Experiments were carried out in triplicate.
2.20. Statistical analysis
All experiments were done in triplicates, and the mean values were plotted with error bars representing the standard deviation from the mean values. The linear graphs were compared based on the slope and the area under the curve. The exponential and logistic growth curves of PA-WT and PA-M were plotted using the GeoGebra classic graphing tool. The carrying capacity and growth rate were obtained from the absorbance values using R software by loading growth curver package. Other data sets were analysed using one-way and two-way ANOVA with Tukey's post-hoc test. The significance values were calculated with a 95 % confidence interval (CI). A P value < 0.05 was considered statistically significant. The in-ovo analysis and percentage survival of eggs were plotted by using the Kaplan-Meier survival curve.
3. Results
3.1. M12PA isolation and TEM analysis
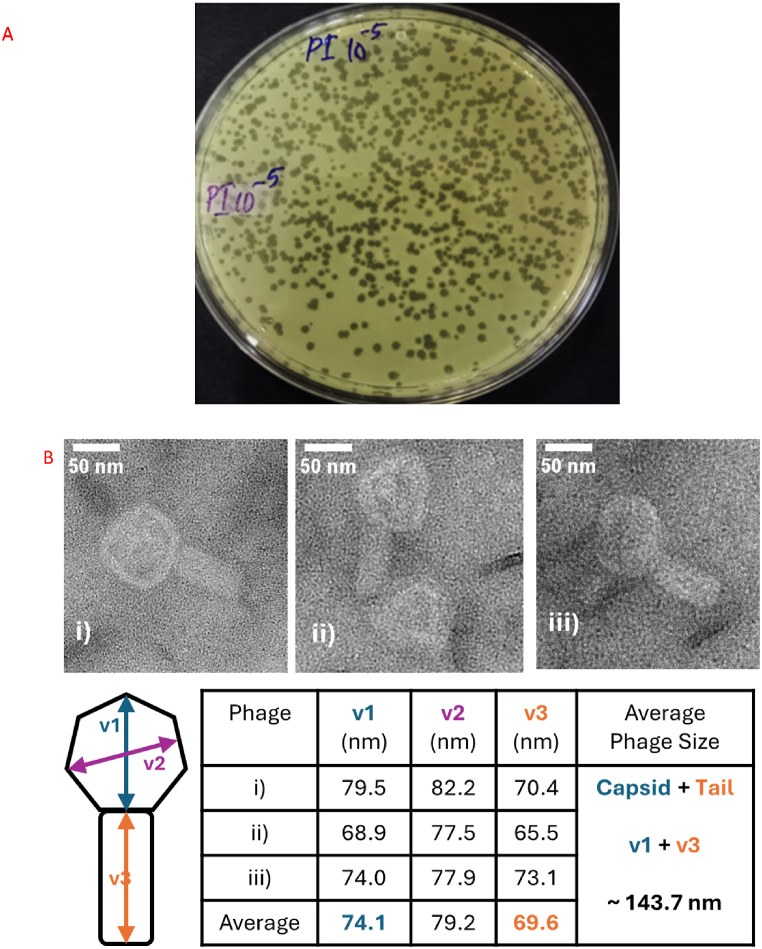
Phages infecting wild-type P. aeruginosa, PA-WT (ATCC 15442), were isolated from soil samples (Table S1). Plaques with inconsistent lysis and turbid plaques were not taken up. One phage producing a clear lytic zone and larger plaque size was chosen for the study (Fig. 1A). The purified phage was labelled M12PA. TEM analysis revealed that M12PA formed Myovirus morphologically, with a size corresponding to 143.7 nm (Fig. 1B (i-ii)). Genome sequencing revealed that M12PA has a genome size of 66,388 bp (GenBank accession number - PP668090) [34]. M12PA exhibited high specificity to PA-WT and did not show any lytic activity towards other strains of P. aeruginosa as well as other gram-negative organisms (Table S2).
Fig. 1.
A. Lytic plaques produced by M12PA B. TEM image of M12PA phage belonging to the Myoviridae family.
3.2. Temperature and pH stability of M12PA
M12PA was highly stable at a range of temperatures and pH. M12PA was stable at temperatures ranging from 30 to 70°C (Figs. S2 and S3). Above 70°C, the phage titre steeply dropped to zero. The phage was stable at pH ranging from 5 to 11. The phage was viable at pH 3 for 2 h and at pH 11 for 10 h, after which no viable phage could be recovered. M12PA showed greater stability towards alkaline pH, with no significant reduction observed at pH 7 and 9 for up to 24 h.
3.3. Multiplicity of infection – M12PA
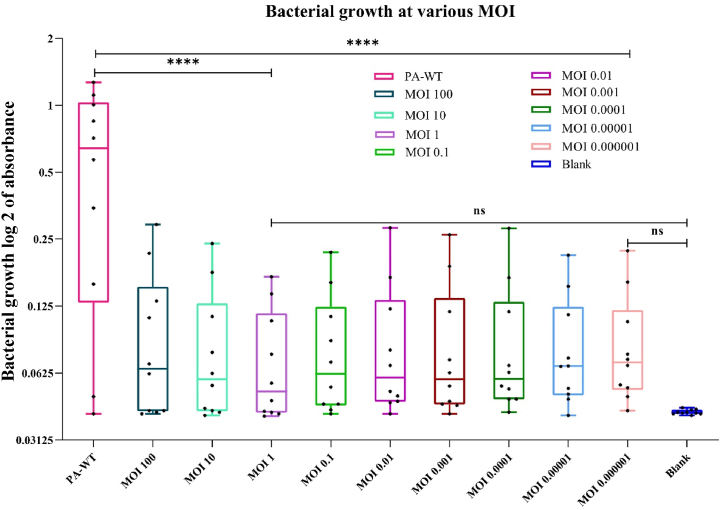
M12PA was allowed to infect PA-WT at various MOIs, all of which showed complete inhibition of PA-WT for 10 h (Fig. 2). MOI 1 was chosen for further studies based on the results of MTT assay at the end of 18 h, showing the least percentage of cell viability, even though there was no significant difference among all the MOIs used. Notably, we observed emergence of phage resistant variants after 12 h.
Fig. 2.
MOI optimization for M12PA against P. aeruginosa (PA-WT). MOI ranging from 100 to 0.000001 was used to infect the host. The box and whiskers graph represent the least to the highest obtained values for each MOI and the statistical significance is represented (ns-non significant, ∗ p value < 0.05, ∗∗ <0.01, ∗∗∗ <0.005, ∗∗∗∗ <0.0001).
3.4. Phage priming/dosing
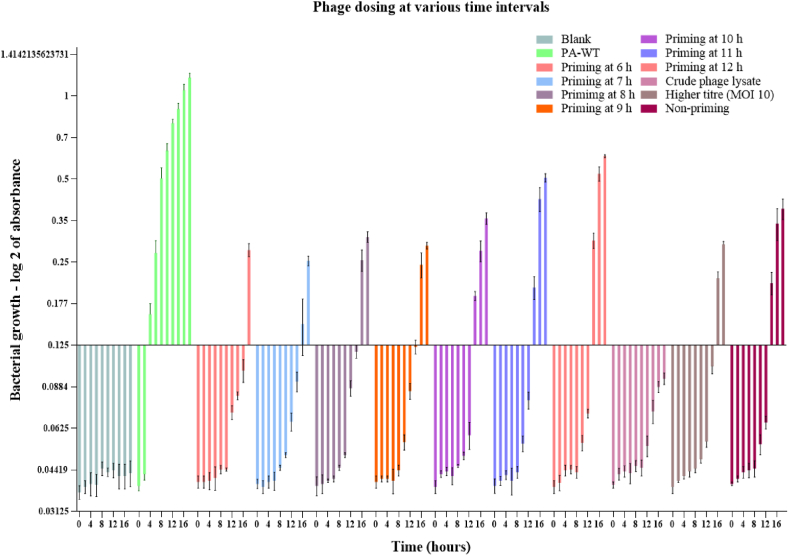
To combat the formation of phage resistance after 12 h, we performed phage priming experiment in which purified M12PA phage at MOI 1, enriched phage lysate sample, or higher titer M12PA phage (MOI 10) were added at different time points and resistance formation was analysed (Fig. 3). Priming at a very early stage showed a decrease in bacterial density at the time of resistance development but did not overcome the resistance formation. Addition of phage-enriched cocktail was effective for an additional 6 h, after which there was emergence of resistant mutants. Interestingly, we observed that, after 36- hour time period, all the treatments resulted in the development of dark brown to black pigmented cultures. The resistant mutants were isolated from the mixed culture in which multiple mutants showed white to cream-coloured colonies whereas few colonies produced brown pigmentation. We focused on the brown mutant, which was subcultured multiple times to obtain pure colonies, and one of the mutants was labelled as PA-M studied further. The probability of occurrence of the brown mutant was analysed by plating PA-WT followed by infection using phage. Without any priming, the probability of development of brown mutant was 6.25 % (Fig. S4), which was significantly increased to over 90 % with priming, denoting the selection of rare mutants with high predatory stress using phage.
Fig. 3.
Phage priming – A second dose of phage (MOI 1) was provided at hour 6 h, 7 h, 8 h, 9 h, 10 h, 11 h and 12 h from the initial dosage time. The growth curve depicts the least cellular abundance by 18th hour with early dosage. Priming with cocktail was effective for an extended period of 6 h after which resistant mutants emerged.
3.5. Pigment characterization
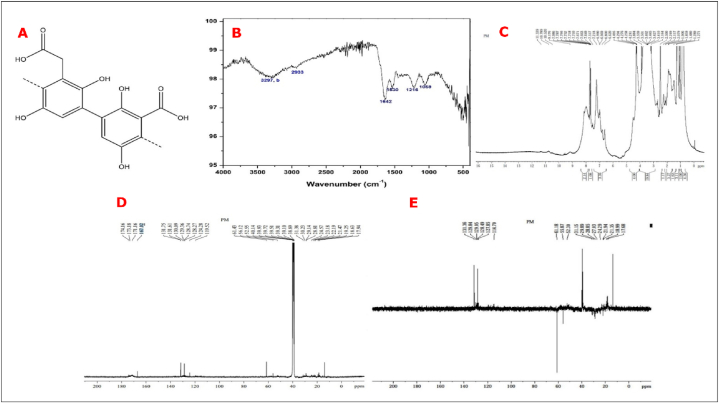
To characterize the pigment produced by the PA-M mutant, we isolated the pigment and analysed it through FT-IR and NMR spectroscopic analysis. The IR spectrum (Fig. 4B) shows a characteristic broad hydroxyl peak spanning from 3600 to 2500 cm−1 and a sharp absorption at 1642 cm−1, corresponding to carbonyl stretching, confirming the presence of carboxyl group. A weak signal at about 2933 cm−1 indicates C-H stretching vibrations. The IR signals at 1530 and 1059 cm−1, indicate the presence of C=C in a conjugated ring and = C-H = bending, respectively, confirming the presence of an aromatic ring. The broad absorption above 3300 cm−1 indicates a hydroxyl group, and the sharp signal at 1216 cm−1 corresponds to C-O stretching of an aromatic hydroxyl group, featuring the presence of aromatic (phenolic) hydroxyl group. The 1H NMR (Fig. 4C) shows characteristic peaks at δ = 3.35, 6.7 to 7.5, 8.1 and 11.4 ppm. Likewise, 13C NMR (Fig. 4D) displays characteristic chemical shift values at δ = 61.43, 119–129, 132, 171–174 and 167 ppm. A broad signal in the off-scale region at δ = 11.4 ppm corresponded to the exchangeable proton of the carboxylic group. The peak at 8.1 is a clear indicator of a phenolic hydroxyl group in the unprotonated carbon of the ring bearing the -OH group, with shoulders, and at 6.7–7.5 ppm, protons in the aromatic ring (-CH = C-) together in broad region. The peak at δ 3.35 corresponds to the methylenic H in the deshielded phenyl ring. Prominent signals in slightly varied positions, at δ 171–174 ppm that corresponded to the unprotonated carbon of the carboxylic group, then at 132 of the unprotonated carbon of the ring bearing the -OH group, with shoulders, and at 119–129 ppm, protonated and unprotonated carbons of the aromatic ring (-CH = C-) together in broad band with a positive DEPT signal. The peak at δ = 61.43 corresponds to the methylenic C bonded to the phenyl ring, which is confirmed by the negative peak in the DEPT-135 spectra (Fig. 4E).
Fig. 4.
A) Structure of pyomelanin molecule, C3-C6 (β-bindings) B) IR spectrum of the sample recorded in Spectrum Two FT-IR Spectrometer (PerkinElmer) C) 1H NMR Spectra of the sample recorded in Bruker 400 MHz instrument -Avance NEO {Solvent d6-DMSO, number of scans −32}D) 13C NMR Spectra of the sample recorded in Bruker 400 MHz instrument -Avance NEO {Solvent d6-DMSO, number of scans −15360 E) DEPT-135 13C NMR Spectra of the sample recorded in Bruker 400 MHz instrument -Avance NEO {Solvent d6-DMSO, number of scans −4625}.
The spectral analysis confirms the presence of pyomelanin pigment as shown in Fig. 4A.
3.6. M12PA adsorption
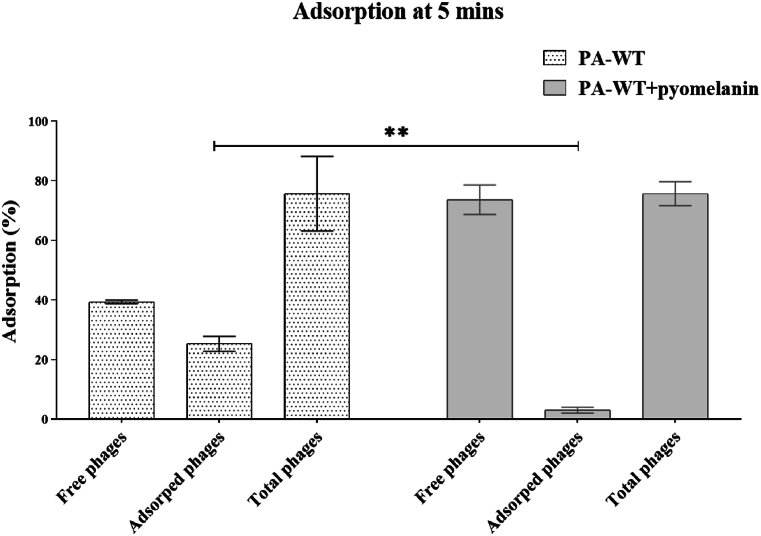
The adsorption of M12PA in the presence and absence of pyomelanin was evaluated to be 28 % and 5 %, respectively. The PFU of total, free, and adsorbed phages are shown in Fig. 5.
Fig. 5.
Phage adsorption percentage in 5 min against PA-WT in presence and absence of pyomelanin.
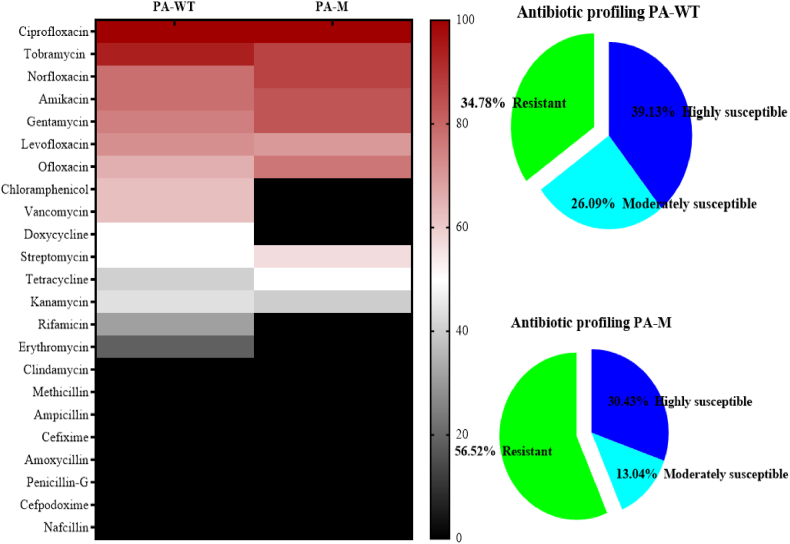
3.7. Antibiotic resistance profiling of PA-WT and PA-M
The resistance profiles of PA-WT and PA-M against 23 antibiotics were analysed using Kirby-Bauer disc diffusion assay. In general, PA-M mutant exhibited increased resistance towards antibiotics compred to the PA-WT. The obtained zones were normalized and categorized into highly susceptible, moderately susceptible, and resistant antibiotics. Antibiotics chloramphenicol, vancomycin, rifampicin, erythromycin, and doxycycline produced zones of inhibition (not necessarily indicating susceptible) for PA-WT but not in case of PA-M. Vancomycin produced a turbid and hollow double ring zone for PA-WT. The resistance profile of PA-WT increased from 34.78 % to 56.52 % for PA-M (Fig. 6). Even though the heat map shows a slight increase in susceptibility for gentamicin and streptomicin against PA-WT, the MIC values were found to be same for both strains.
Fig. 6.
Resistant profiles of PA-WT and PAM.
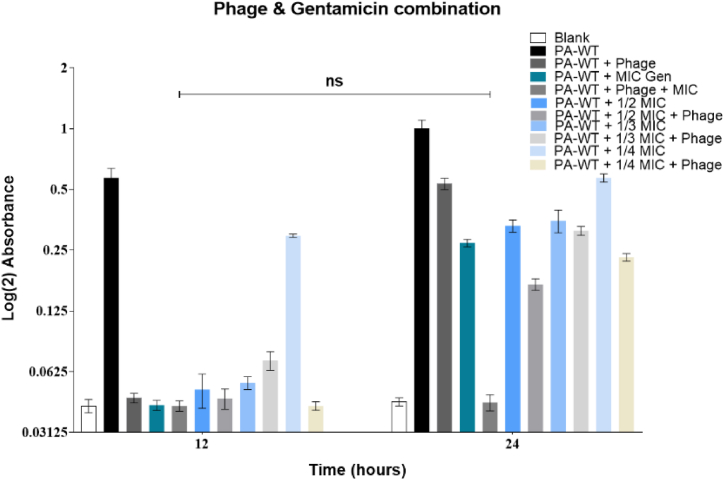
3.8. Combination therapy
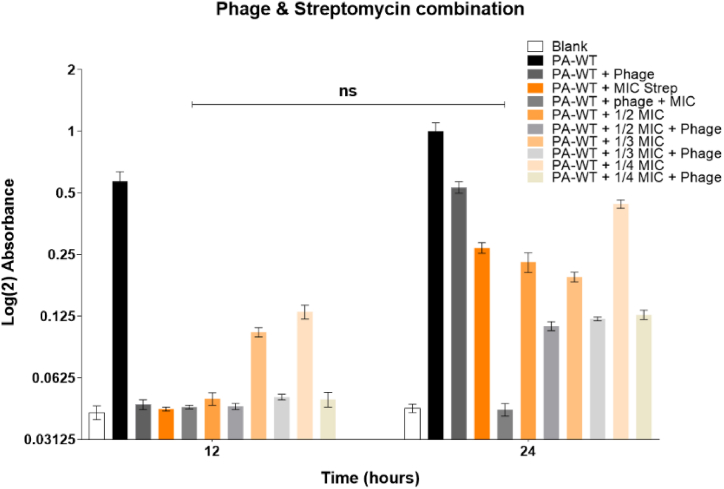
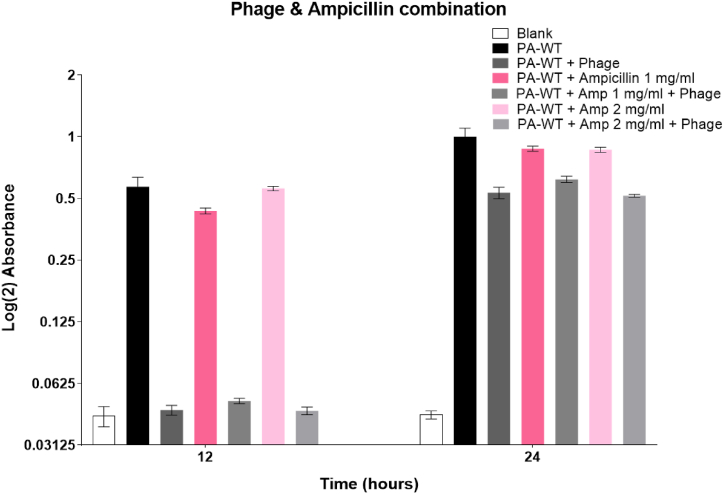
The MIC values of gentamicin and streptomycin were evaluated to be 0.75 μg/mL and 2.5 μg/mL, respectively. A combinatory therapy approach with MIC, ½ MIC, 1/3 MIC, and ¼ MIC values of both gentamicin and streptomycin was used along with M12PA at a MOI of 1. Cultures treated with combination therapy showed lower bacterial density compared to either antibiotic or phage alone. All fractions of antibiotics from MIC to ¼ MIC had a decreasing effect on inhibiting bacterial growth but showed a significant difference from the growth of PA-WT alone. The hypersusceptibility of PA-WT with the combinatorial approach was evident for MIC and ½ MIC values of gentamicin and streptomycin with an MOI of 1 (Fig. 7A and B). Combination therapy with MIC and phage (MOI of 1) inhibited bacterial growth up to 24 h for both gentamicin and streptomicin, with only a singe dose of antibiotic and phage. Ampicillin did not show any hypersusceptibility, unlike gentamicin and streptomycin (Fig. 7C).
Fig. 7A.
Combination therapy using gentamicin and phage Fig. 7B: Combination therapy using streptomycin and phage Fig. 7C: Combination therapy using ampicillin and phage.
3.9. Fitness costs of PA-M
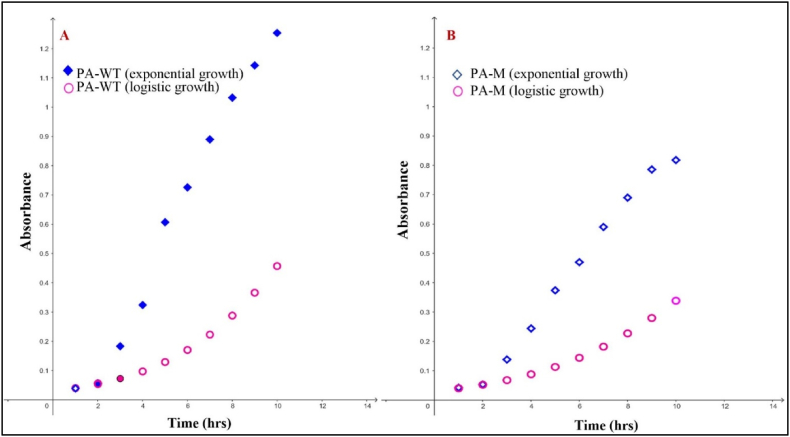
PA-M was analysed in comparison with PA-WT to determine possible changes in growth, colony morphology, resistance profile, biofilm-forming ability, motility (Fig. S5), and virulence. PA-M showed a significantly lower growth rate compared to PA-WT. The differences in the growth rate were evident with significant differences between the slopes and area under the growth curves. The growth rates were estimated to be 0.359 and 0.324 for PA-WT and PA-M, respectively. Exponential growth of PA-WT (Fig. 8A) and PA-M (Fig. 8B) depicts the growth of bacterium while utilizing infinite resources and unrestricted population expansion, whereas logistic growth curve is depictive of bacterial growth in limiting resources, thus entitiled to a carrying capacity rendered by the medium. The carrying capacity at 20 h of growth was determined to be 99 % and 66 %, respectively, for PA-WT and PA-M.
Fig. 8.
(A) and (B) Exponential and logistic growth curve of PA-WT and PAM with growth rate and carrying capacity derived using growthcurver package in R software and plotted using Geogebra graphing tool. The growth rate of PA-WT and PA-M was 0.359 and 0.324 with carrying capacity at 20 h being 99 % and 66 % respectively.
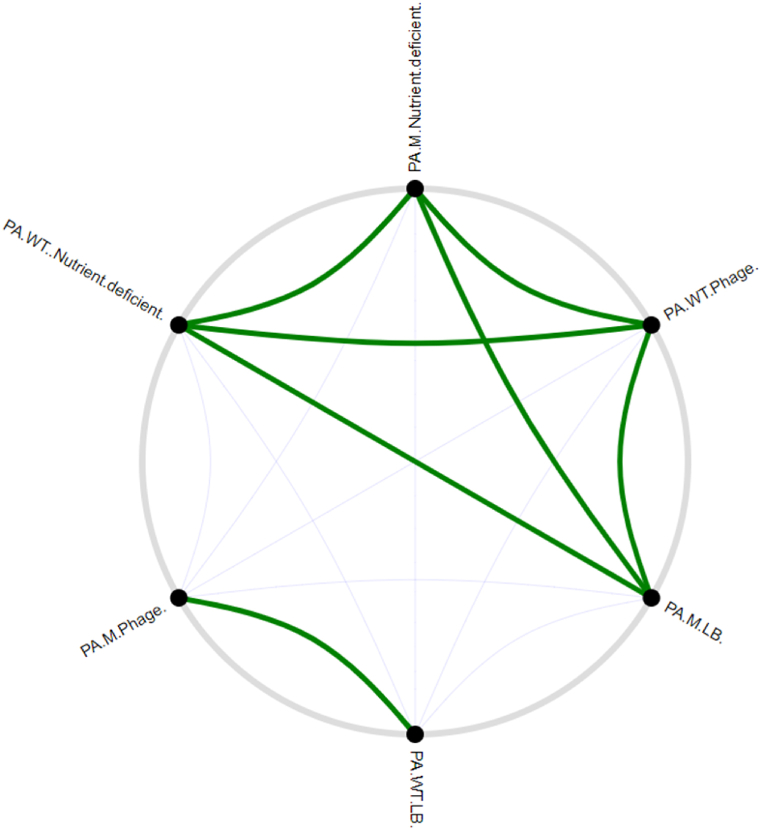
The relative fitness of PA-WT and PA-M while co-cultured was analysed under normal growth conditions, phage stress, and under nutrient-deprived niches. PA-WT showed dominance over PA-M under normal physiological and nutrient conditions after 10 h, whereas PA-M showed complete dominance under phage stress with no viable PA-WT at the end of 10 h. Experiments were not prolonged beyond 12 h to avoid the development of other resistant variants. In nutrient-deficient niches without any phage stress, both PA-WT and PA-M exhibited loss in viability, but PA-M had half the viable colonies of normally grown cultures and significantly more than PA-WT (Fig. 9, Fig. 10) & (Figs. S6 and S7). These changes in viability clearly indicate the relative fitness of PA-M over PA-WT under stress and nutrient-deprived conditions. Bacterial competition on solid media was visualized by growing PA-WT and PA-M on agar media at 0.4 cm distance. In the absence of phage PA-WT fluorished and displayed dominance over PA-M and vice-versa in presence of phage (Fig. 10A–C).
Fig. 9.
Positive and negative correlations among the bacterial populations, PA-WT in LB broth and PA-M grown in presence of phage show a higher degree of positive correlation (represented by thicker lines) whereas no other conditions produced positive correlation of growth model with PA-WT in LB and PAM in phage stress denoting the extent of variation of growth pattern for both strains in other niche.
Fig. 10.
PA-WT and PA-M competition in solid media indicates no production of antagonistic metabolites.
3.10. Biofilm formation
Bacterial cell aggregation is a defensive mechanism of bacterial cells to overcome immediate changes in their niche. The normal cell aggregations of PA-WT and PA-R were analysed. The changes in the pattern of cell aggregation with the addition of phage, antibiotic and phage-antibiotic combination are shown in Fig. S8. The aggregation is high for ampicillin and M12PA-ampicillin combination in PA-WT, which could be one of the mechanisms conferring resistance. The difference in aggregation due to phage has decreased in PA-M and is unaffected by its presence.
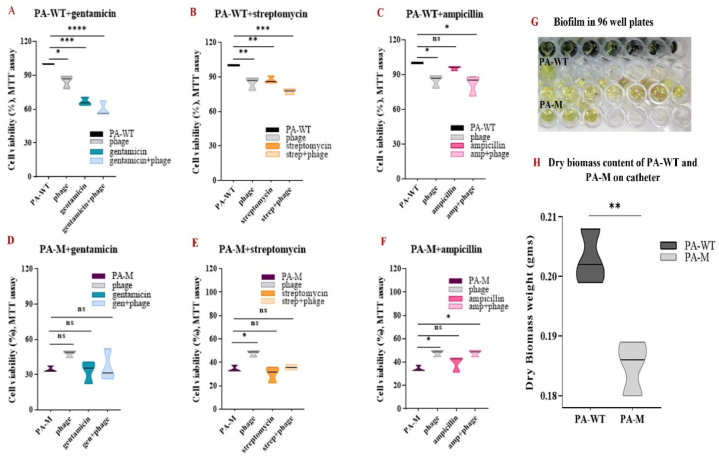
The differences in the biofilm formation of PA-WT and PA-M were confirmed using MTT assay, where the viabilty of the sessile cells was evaluated (Fig. 11A–F). The resistant strain PA-M showed significant variation in the viable cells (little to no viable cells) compared to PA-WT. In comparison to PA-WT, which was considered as 100 % biofilm-forming, the relative absorbance observed for PA-M was <40 %. There was no deveopment of purple formazan product in any of the wells with PA-M, and the absorance was similar to that of the dye alone (Fig. 11G). In case of PA-WT, combination therapy using gentamicin with phage and streptomycin with phage resulted in a significant decrease in the biofilm formation and was more effective than antibiotic or phage therapy alone (Fig. 11A). Gentamycin and streptomycin are aminoglycosides that inhibit protein synthesis and are clinically used againt gram-negative organisms. In contrast, ampicillin is an aminopenicillin that inhibits gram-positive and a few gram-negative organisms. The use of ampicillin in an attempt to resensitize PA-WT was not successful, as the presence of ampicillin allowed more cell aggregation and biofilm formation (Fig. 11C–F). The dry mass (mg) of biofilm formed by PA-WT and PA-M on the catheter surface is shown in Fig. 11H.
Fig. 11.
A, B, C, D, E, F) MTT assay on biofilm formed by PA-WT and PA-M treated using gentamicin, streptomycin and ampicillin G) The MTT wells for PA-WT and PA-M, where PA-M did not develop any purple formazan product denoting the absence of viable cells. H) Difference in the dry weight of biomass obtained in a catheter represented using violin plots. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.11. In-ovo virulence assay
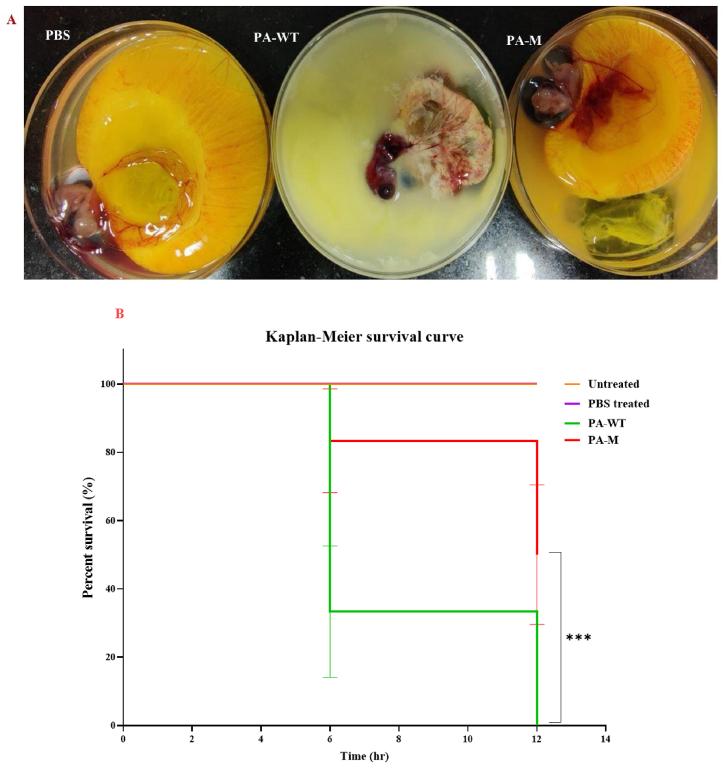
A few 7 day-old chicken eggs were infected with PA-WT, PA-M, and PBS (control). Fig. 12A shows the extent of pathogenesis of each sample. PA-WT exhibited a higher degree of pathogenesis. The bacteria were inoculated into the allantoic fluid that surrounds the egg yolk and embryo. PA-WT infected the egg yolk, disrupted the amniotic membrane, and infected the embryo with characteristic pyoverdin pigmentation. By the 6th and 12th hours, all eggs infected with PA-WT endured a high bacterial load, and the embryos were dead (confirmed ex vivo by lack of heart beat). In contrast, PA-M did not cause structural deformation in any infected eggs with intact egg yolk, amniotic membrane, and embryo. The brown pigmentation was visible in the allantoic fluid, while the embryos remained viable up to 15 h for 50 % of the subjects (eggs) by 12 h. The in-ovo lethality and percentage of survival of PA-WT and PA-M are plotted using the Kaplan-Meier survival curve (Fig. 12B).
Fig. 12.
(A) Degree of pathogenesis and structural deformity in eggs infected with PBS, PA-WT and PA-M. PA-WT shows extensive damage of egg yolk, allantoic cavity and the embryo. (B) Kaplan-Meier survival curve depicts the percentage of eggs survived with respect to time of infection. The p-value indicated the significant difference between the survival percentage of infected eggs. Log-Rank (Mantel-Cox) test was used to determine the significance.
4. Discussion
The present study aims to understand the role of phage-based fitness in P. aeruginosa mutants emerged during phage infection/therapy. The model organism, P. aeruginosa ATCC 15442, is a phenotypically and genotypically characterized strain sourced from a water bottle in an animal room and is used as a test organism for studying novel disinfectants and potential antibacterial compounds [35]. Sewage and hospital garbage-contaminated soil are among the natural habitats that harbour P. aeruginosa, making them a reliable source for phage isolation [36]. Host-specific enrichment and isolation of M12PA phage were successfully carried out from sewage contaminated soil. A study comparing the phage source and its corresponding efficiency of plating (EoP) demonstrated higher retrieval and efficacy of phage isolated from sewage as well as sewage-contaminated soil [36]. The initial enrichment of host-specific phages yielded phage lysates (cocktail 1 & 2 – Table S1), which produced halo/turbid/clear zones of lysis. Subsequent phage selection and isolation were performed by analysing the plaque size and morphology (M12PA showed transparent plaques without any halo appearance). Following the successful selection of P. aeruginosa specific phage – ‘M12PA’, its properties including host tropism, MOI, and phage stability were investigated to assess the phage applicability. Phage storage, shelf-life, viability, and application rely on the phage's stability at distinct temperature and pH [37]. M12PA exhibited remarkable stability across a range of temperatures (30–70 °C & storage at 8 °C) and pH (5–9), allowing phage storage in phosphate buffer (pH 7) at 4 °C. We were able to retrieve phages for more than 6 months from the lysates though loss of titre was observed with time. The stability of the phage across various temperatures and pH levels enhances its utility and has enabled the incorporation of P. aeruginosa-specific phages into formulations like PEG-based ointment for in vivo applications [38].
The purified phage was morphologically analysed by TEM, which revealed a large head and tail, consistent with the morphological characteristics of Myoviridae. Myoviridae are highly evolved double-stranded DNA containing bacteriophages that primarily infect gram-negative organisms like P. aeruginosa and E. coli. M12PA has a narrow host range and high host specificity (host tropism), as detailed in Table S2. Phage adsorption and infectivity are two distinct factors that are closely correlated with MOI, which subsequently affects phage efficacy. Phage adsorption is never 100 % and this discrepancy between MOIinput and MOIactual underscores the need for optimization of phage titres and dosage patterns [39].
The growth curve of P. aeruginosa in presence of phage demonstrated a revival from inhibition at 12 h post-infection. During phage-based biocontrol of bacteria, the bacterial population is prone to a loss of viability, which is an uncontrollable variable. We anticipated that the decrease in bacterial numbers after the initial phage adsorption and lysis (due to the lack of host cells) could be one among the reasons for the fall in phage titre, following the predator-prey model of population dynamics. Low bacterial density may adversely affect phage adsorption, phage penetration, infectivity and potentially leading to an in-situ loss of phage titre.
Pharmacologically, the efficacy of any drug is high at low bacterial populations, resulting in larger clearance. Conversely, phage infectivity is affected at low bacterial populations since the population dynamics do not follow the normal absorption-distribution-metabolism-elimination peak model of drugs but rather predatory-prey models where the number of prey affects the populations of predator and vice-versa [40]. The predictable loss in phage titres was mitigated by priming multiple dosages at distinct time periods using M12PA, which allowed the emergence of variants producing pyomelanin (PA-M), also reported in a few antibiotic-resistant clinical isolates of P. aeruginosa [41,42]. In some cases, it has been observed that phage-resistant pyomelanogenic strains exhibit a large genomic deletion of approximately 300 kb, resulting in the loss of several genes, including hmgA, which encodes pyomelanin [43].Priming at an early stage effectively reduced the bacterial load but did not prevent the formation of resistant variants, whereas the cocktail was effective in inhibiting bacterial growth for up to 18 h. Phage dosing/priming also resulted in the emergence of resistant strains against the cocktail, indicating the presence of phages possessing the same receptors or bacterial mutation that confers broad protection over similar phages. Studies have shown that synthetic cocktails with two or more families of phages binding different receptors can significantly enhance the efficiency of cocktails [44,45].
At 48–72 h after phage infection, cultures under phage priming (multiple phage doses) exhibited deep brown to black pigmentation. The population of P. aeruginosa that emerged after 12 h of phage inhibition is likely to contain subtypes of persister and resistant cells. Persister cells withstand environmental stress through epigenetic regulation and can revert back once the stress is eliminated; in contrast, resistant cells undergo irreversible gene mutations that are inherited by the progeny [46]. To rule out the presence of persister cells, we observed the efficiency of PA-M (isolated after 48 h) to produce pyomelanin in absence of phage over multiple generations, ensuring that pyomelanin production was indeed due to mutational adaptations, confirming PA-M as a resistant variant.
The blackish-brown pigment pyomelanin, is a bacterial extracellular pigment suspected to provide protection against the antibiotic penetration in clinical setting [25]. It also possesses iron-scavenging ability, promotes bacterial electron transfer, and provides protection against UV and ROS damage [24]. The extracted pyomelanin pigment from PA-M shows a very broad, featureless UV–visible absorption spectrum spanning from 700 to 200 nm. The molar extinction coefficient is as low as 7400 ± 100 M−1cm−1 at 350 nm, which is comparable to the literature value and is spectroscopically pure (>99 %) [47]. Unlike pyoverdin (produced by PA-WT), pyomelanin (produced by PA-M) is proven to be non-toxic to prokaryotic and eukaryotic cells [48]. Extracellular pigment production and accumulation allows the bacteria to overcome phage absorptivity; we observed a 5-fold reduction in the number of adsorbed phages. This could also be a factor that enhances the resistance profile of PA-M by inhibiting antibiotic penetration [49]. It has been proven that phage therapy alters the antibiotic resistance and sensitivity pattern. A study evaluating the E. coli mutants during phage therapy reported that only 1 in 4 mutants resulted in trade-offs relating to antibiotic resistance [50].
The niche competition and spatial orientation of multi species consortia are specifically crucial during predatory stress [51]. Bacterial strains with low growth rate are evolutionarily supported to be more adaptable and capable of surviving harsh environment [52]. When PA-WT and PA-M were grown in competition with one another in nutrient-deprived conditions, PA-M demonstrated a the higher survival rate compared to PA-WT by more than 40 %. PA-M's slow growth rate potentially ensured fewer nutrient requirements, resulting in high survival rates. Another aspect of pyomelanin overproduction is its negative regulation of hundreds of genes, which simultaneously impairs the virulence, quorum sensing, and fitness of bacterial strains [53]. Diard et al., 2010 emphasized that bacterial evolution does not necessarily select for high virulence strains, but may occur while adapting to other parameters [54]. Similarly, bacterial adaptations result in mutants that produce defective LPS (one of a common phage binding receptor), thereby inhibiting phage adsorption. In our study, the MTT assay confirmed that PA-M, the phage resistant mutant, was defective for biofilm formation. Gafoor et al., in 2011, demonstrated the inefficiency of Pseudomonas mutants lacking exopolysaccharides to produce biofilms following phage therapy [55]. Similarly, the loss of bacterial receptors like porins, T4P, teichoic acids, and OmpC can also inhibit phage adsorption, thereby conferring phage resistance [56]. The dry biomass formed by PA-WT and PA-M on the catheter surface supported by the in-vitro viability percentage of biofilm-forming cells, revealed the inefficiency of PA-M to produce biofilms.
To analyse the changes in virulence of PA-WT and PA-M, the allantoic cavity of fertilized chicken eggs was infected using PA-WT and PA-M. Blanco et al., in 2017, evaluated the virulence of E. faecalis in chick embryos and discussed the importance of the administering the pathogen via allantoic cavity over yolk sac and chorion-allantoic administration [57]. The survival rates of PA-M infected chick embryos indicate a weakened degree of pathogenesis. Li et al., in 2023, evaluated the virulence of K. pneumoniae phage-resistant mutants isolated from an inpatient who received phage therapy and observed that the mutants were non-virulent [58]. Moreover, the combinatorial approach using antibiotic (gentamicin 0.75 μg/mL and streptomycin 2.5 μg/mL) and phage (MOI 1) delayed resistance formation by 12 h and the bacterial load was significantly reduced at sub-inhibitory concentrations. The profound inhibition pattern observed with a single dosage suggests that the success of a combinatorial therapy is predictably higher than either treatment alone. Altamirano et al., 2022, in a pre-clinical study, used phage along with ceftazidime antibiotics to treat Acinetobacter baumannii bacteraemia, isolating capsule-deficient phage-resistant mutants from organs after 10 and 16 h, which were re-sensitized to the antibiotic [59]. With the idea that the loss of LPS could re-sensitize resistant bacteria to drugs, ampicillin, an aminopenicillin drug, was chosen to evaluate re-sensitization. Phage therapy did not alter the MIC antibiotics gentamycin and streptomycin. Phage and ampicillin combination therapy did not re-sensitize P. aeruginosa, whereas the cell-aggregation of bacterial cells was high in presence of this combination. Apart from these selected antibiotics, PA-M exhibited an altered antibiogram with no clearance for vancomycin, erythromycin, doxycycline, rifamycin and chloramphenicol; all showing negligible zones of inhibition towards PA-WT and is generally not used for P. aeruginosa infections.
According to Gause's law of competition, ‘no two organisms will be able to grow at the same pace while utilizing the same resources’. This principle holds true for the bacterial mutants evolved due to the predatory stress, and this study provides further evidence for the enhanced fitness of evolutionary mutants [60]. Prolonged phage stress undeniably allows more fitted strains with loss-of-function genes to grow and multiply. While the theory supports these findings, the bacterial interactions and niche associations during prolonged treatment in vivo are yet to be fully understood.
5. Conclusion
Resistance is one of the most significant challenges in expanding phage therapy. This study emphasizes the development of phage-resistant mutants with loss of functions and virulence. Phage steering, or evolutionary adaptions leading to the development of less virulent and antibiotic-sensitive strains, can alter the course of treatment regimen. PA-M, a phage resistant bacterial mutant that emerged during M12PA phage exposure, was analysed in our study. We observed that besides resistance development, certain associated trade-off or fitness costs were inflicted upon the wild-type strain (PA-WT). The interplay of such fitness costs along with a phage-antibiotic combinatorial approach will not only render enhanced efficacy and reduces the chances of resistant formation but will also increase the reliability of therapeutic approaches, since the popularity of antibiotics is quite fixated. The key factor for developing successful combinations lies in understanding the synergistic mechanisms of both phage and antibiotic entities. This work also substantiates the use of antibiotics in combination with phage to prolong the inhibition period and to avoid resistance formation.
CRediT authorship contribution statement
Aarcha Shanmugha Mary: Writing – original draft, Methodology, Investigation. Nashath Kalangadan: Methodology, Investigation, Formal analysis. John Prakash: Writing – review & editing, Validation, Formal analysis, Data curation. Srivignesh Sundaresan: Writing – review & editing, Validation, Data curation. Sutharsan Govindarajan: Investigation, Data curation. Kaushik Rajaram: Writing – review & editing, Validation, Supervision, Methodology, Investigation.
Ethical approval
Not required.
Data availability
Data that are not directly available in the manuscript will be made available upon request. M12PA genome sequence is available in NCBI with the accession number PP668090.
Funding
No financial support was obtained during the course of the work.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank Dr Jayalakshmi Krishnan, Assistant Professor, Central University of Tamil Nadu (CUTN), for providing technical support. We also extend our gratitude to Ms. Aiswarya Mohan, Senior Research Fellow, Department of Epidemiology and Public Health, CUTN, Ms. Gouthami P, Technical staff, Department of Microbiology, CUTN and to Mr. Barath Sivaraj, SRM University for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e40076.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Diseases T.L.I. Antimicrobial resistance through the looking-GLASS. Lancet Infect. Dis. Feb. 2023;23(2):131. doi: 10.1016/S1473-3099(23)00012-9. [DOI] [PubMed] [Google Scholar]
- 2.Ghasemian A., Mobarez A.M., Peerayeh S.N., Abadi A.T.B., Khodaparast S., Nojoomi F. Report of plasmid-mediated colistin resistance in Klebsiella oxytoca from Iran. Reviews and Research in Medical Microbiology. Apr. 2018;29(2):59. doi: 10.1097/MRM.0000000000000134. [DOI] [Google Scholar]
- 3.Ikuta K.S., et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2022;400(10369):2221–2248. doi: 10.1016/S0140-6736(22)02185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikhimiukor O.O., Odih E.E., Donado-Godoy P., Okeke I.N. A bottom-up view of antimicrobial resistance transmission in developing countries. Nat Microbiol. Jun. 2022;7(6) doi: 10.1038/s41564-022-01124-w. Art. no. 6. [DOI] [PubMed] [Google Scholar]
- 5.Karn S.L., Gangwar M., Kumar R., Bhartiya S.K., Nath G. Phage therapy: a revolutionary shift in the management of bacterial infections, pioneering new horizons in clinical practice, and reimagining the arsenal against microbial pathogens. Front. Med. Oct. 2023;10 doi: 10.3389/fmed.2023.1209782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abedon S.T., Danis-Wlodarczyk K.M., Alves D.R. Phage therapy in the 21st century: is there modern, clinical evidence of phage-mediated efficacy? Pharmaceuticals. Nov. 2021;14(11):1157. doi: 10.3390/ph14111157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oechslin F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses. Jun. 2018;10(7):351. doi: 10.3390/v10070351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egido J.E., Costa A.R., Aparicio-Maldonado C., Haas P.-J., Brouns S.J.J. Mechanisms and clinical importance of bacteriophage resistance. FEMS Microbiol. Rev. Sep. 2021;46(1):fuab048. doi: 10.1093/femsre/fuab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le S., et al. Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci. Rep. Apr. 2014;4(1):4738. doi: 10.1038/srep04738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abedon S.T. Phage ‘delay’ towards enhancing bacterial escape from biofilms: a more comprehensive way of viewing resistance to bacteriophages. AIMS Microbiol. Mar. 2017;3(2):186–226. doi: 10.3934/microbiol.2017.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alkhudhairy M.K., Alshadeedi S.M.J., Mahmood S.S., Al-Bustan S.A., Ghasemian A. Comparison of adhesin genes expression among Klebsiella oxytoca ESBL-non-producers in planktonic and biofilm mode of growth, and imipenem sublethal exposure. Microb. Pathog. Sep. 2019;134 doi: 10.1016/j.micpath.2019.103558. [DOI] [PubMed] [Google Scholar]
- 12.Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. May 2010;8(5):317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 13.Blazanin M., Turner P.E. Community context matters for bacteria-phage ecology and evolution. ISME J. Nov. 2021;15(11):3119–3128. doi: 10.1038/s41396-021-01012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangalea M.R., Duerkop B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. Jun. 2020;88(7) doi: 10.1128/IAI.00926-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma D., Misba L., Khan A.U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. May 2019;8(1):76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roach D.R., et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe. Jul. 2017;22(1):38–47.e4. doi: 10.1016/j.chom.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Smith H.W., Huggins M.B. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. Aug. 1983;129(8):2659–2675. doi: 10.1099/00221287-129-8-2659. [DOI] [PubMed] [Google Scholar]
- 18.Mendoza S.D., et al. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature. Jan. 2020;577(7789):244–248. doi: 10.1038/s41586-019-1786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman P. Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals. Mar. 2019;12(1):35. doi: 10.3390/ph12010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luong T., Salabarria A.-C., Edwards R.A., Roach D.R. Standardized bacteriophage purification for personalized phage therapy. Nat. Protoc. Sep. 2020;15(9):2867–2890. doi: 10.1038/s41596-020-0346-0. [DOI] [PubMed] [Google Scholar]
- 21.Letarov A.V., Kulikov E.E. In: Bacteriophage Therapy: from Lab to Clinical Practice. Azeredo J., Sillankorva S., editors. Springer; New York, NY: 2018. Determination of the bacteriophage host range: culture-based approach; pp. 75–84. [DOI] [Google Scholar]
- 22.Attai H., Boon M., Phillips K., Noben J.-P., Lavigne R., Brown P.J.B. Larger than life: isolation and genomic characterization of a jumbo phage that infects the bacterial plant pathogen, agrobacterium tumefaciens. Front. Microbiol. 2018;9(Aug) doi: 10.3389/fmicb.2018.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maffei E., Fino C., Harms A. In: Bacterial Persistence: Methods and Protocols. Verstraeten N., Michiels J., editors. Springer US; New York, NY: 2021. Antibiotic tolerance and persistence studied throughout bacterial growth phases; pp. 23–40. [DOI] [PubMed] [Google Scholar]
- 24.Mahmood H., Mohammed A.K., Flayyih M. Purification and physiochemical characterization of pyomelanin pigment produced from local PSEUDOMONAS aeruginosa isolates. 2015. https://www.semanticscholar.org/paper/PURIFICATION-AND-PHYSIOCHEMICAL-CHARACTERIZATION-OF-Mahmood-Mohammed/56244f581dbf4cbbcb655c30795e4724fb1912e6 [Online]. Available:
- 25.Kurian N.K., Bhat S.G. Data on the characterization of non-cytotoxic pyomelanin produced by marine Pseudomonas stutzeri BTCZ10 with cosmetological importance. Data Brief. Jun. 2018;18:1889–1894. doi: 10.1016/j.dib.2018.04.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorquin F., et al. Production and properties of non-cytotoxic pyomelanin by laccase and comparison to bacterial and synthetic pigments. Sci. Rep. Apr. 2021;11(1):8538. doi: 10.1038/s41598-021-87328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S., et al. Isolation and characterization of a lytic bacteriophage against Pseudomonas aeruginosa. Sci. Rep. Sep. 2021;11(1) doi: 10.1038/s41598-021-98457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayot M.L., Bragg B.N. StatPearls, Treasure Island (FL) StatPearls Publishing; 2024. Antimicrobial susceptibility testing.http://www.ncbi.nlm.nih.gov/books/NBK539714/ [Online]. Available: [Google Scholar]
- 29.R. Humphries, A. M. Bobenchik, J. A. Hindler, and A. N. Schuetz, “Overview of changes to the clinical and laboratory standards Institute performance standards for antimicrobial susceptibility testing, M100, 31st edition,” J. Clin. Microbiol., vol. 59, no. 12, pp. e00213-e00221, doi: 10.1128/JCM.00213-21. [DOI] [PMC free article] [PubMed]
- 30.Ryan E.M., Alkawareek M.Y., Donnelly R.F., Gilmore B.F. Synergistic phage-antibiotic combinations for the control of Escherichia coli biofilms in vitro. FEMS Immunol. Med. Microbiol. Jul. 2012;65(2):395–398. doi: 10.1111/j.1574-695X.2012.00977.x. [DOI] [PubMed] [Google Scholar]
- 31.Mary A.S., et al. Enhanced in vitro wound healing using pva/B-pei nanofiber mats: a promising wound therapeutic agent against eskape and opportunistic pathogens. ACS Appl. Bio Mater. Dec. 2021;4(12):8466–8476. doi: 10.1021/acsabm.1c00985. [DOI] [PubMed] [Google Scholar]
- 32.Trunk T., Khalil H.S., Leo J.C. Bacterial autoaggregation. AIMS Microbiol. Mar. 2018;4(1):140–164. doi: 10.3934/microbiol.2018.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manders T.T.M., van Eck J.H.H., Buter G.J., Landman W.J.M. Assessment of the best inoculation route for virulotyping Enterococcus cecorum strains in a chicken embryo lethality assay. Avian Pathol. Nov. 2022;51(6):613–625. doi: 10.1080/03079457.2022.2130174. [DOI] [PubMed] [Google Scholar]
- 34.Mary A.S., et al. Formulation of dual-functional nonionic cetomacrogol creams incorporated with bacteriophage and human platelet lysate for effective targeting of MDR P. aeruginosa and enhanced wound healing. ACS Appl. Bio Mater. Sep. 2024 doi: 10.1021/acsabm.4c00747. [DOI] [PubMed] [Google Scholar]
- 35.Araújo G. de M.S., et al. Toward a platform for the treatment of burns: an assessment of nanoemulsions vs. Nanostructured lipid carriers loaded with curcumin. Biomedicines. Dec. 2023;11(12):3348. doi: 10.3390/biomedicines11123348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aghaee B.L., Mirzaei M.K., Alikhani M.Y., Mojtahedi A. Sewage and sewage-contaminated environments are the most prominent sources to isolate phages against Pseudomonas aeruginosa. BMC Microbiol. May 2021;21:132. doi: 10.1186/s12866-021-02197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shahdadi M., et al. A systematic review and modeling of the effect of bacteriophages on Salmonella spp. Reduction in chicken meat. Heliyon. Apr. 2023;9(4) doi: 10.1016/j.heliyon.2023.e14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piranaghl H., et al. The potential therapeutic impact of a topical bacteriophage preparation in treating Pseudomonas aeruginosa-infected burn wounds in mice. Heliyon. Jul. 2023;9(7) doi: 10.1016/j.heliyon.2023.e18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abedon S.T. Phage therapy dosing: the problem(s) with multiplicity of infection (MOI) Bacteriophage. Jul. 2016;6(3) doi: 10.1080/21597081.2016.1220348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dąbrowska K., Abedon S.T. Pharmacologically aware phage therapy: pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol. Mol. Biol. Rev. Oct. 2019;83(4) doi: 10.1128/MMBR.00012-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ketelboeter L.M., Potharla V.Y., Bardy S.L. NTBC treatment of the pyomelanogenic Pseudomonas aeruginosa clinical isolate PA1111 inhibits pigment production and increases sensitivity to oxidative stress. Curr. Microbiol. 2014;69(3):343–348. doi: 10.1007/s00284-014-0593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez-Rojas A., Mena A., Martín S., Borrell N., Oliver A., Blázquez J. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology. Apr. 2009;155(4):1050–1057. doi: 10.1099/mic.0.024745-0. [DOI] [PubMed] [Google Scholar]
- 43.Menon N.D., et al. Increased innate immune susceptibility in hyperpigmented bacteriophage-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. Jul. 2022;66(8) doi: 10.1128/aac.00239-22. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C., Shi T., Sun Y., Zhang Y. A novel method to create efficient phage cocktails via use of phage-resistant bacteria. Appl. Environ. Microbiol. Mar. 2022;88(6) doi: 10.1128/aem.02323-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo S., Lee K.-M., Kim N., Vu T.N., Abadie R., Yong D. Designing phage cocktails to combat the emergence of bacteriophage-resistant mutants in multidrug-resistant Klebsiella pneumoniae. Microbiol. Spectr. Jan. 2024;12(1) doi: 10.1128/spectrum.01258-23. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brauner A., Fridman O., Gefen O., Balaban N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. May 2016;14(5) doi: 10.1038/nrmicro.2016.34. , Art. no. 5. [DOI] [PubMed] [Google Scholar]
- 47.Sarna T., Swartz H.A. The Pigmentary System. John Wiley & Sons, Ltd; 2006. The physical properties of melanins; pp. 311–341. [DOI] [Google Scholar]
- 48.Ben Tahar I., Kus-Liśkiewicz M., Lara Y., Javaux E., Fickers P. Characterization of a nontoxic pyomelanin pigment produced by the yeast Yarrowia lipolytica. Biotechnol. Prog. 2020;36(2):e2912. doi: 10.1002/btpr.2912. [DOI] [PubMed] [Google Scholar]
- 49.Hernando-Amado S., Sanz-García F., Martínez J.L. Rapid and robust evolution of collateral sensitivity in Pseudomonas aeruginosa antibiotic-resistant mutants. Sci. Adv. Aug. 2020;6(32):eaba5493. doi: 10.1126/sciadv.aba5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGee L.W., Barhoush Y., Shima R., Hennessy M. Phage-resistant mutations impact bacteria susceptibility to future phage infections and antibiotic response. Ecol. Evol. Jan. 2023;13(1):e9712. doi: 10.1002/ece3.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W., Jacquiod S., Brejnrod A., Russel J., Burmølle M., Sørensen S.J. Deciphering links between bacterial interactions and spatial organization in multispecies biofilms. ISME J. Dec. 2019;13(12) doi: 10.1038/s41396-019-0494-9. Art. no. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray D.A., Dugar G., Gamba P., Strahl H., Jonker M.J., Hamoen L.W. Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat. Commun. Feb. 2019;10(1):890. doi: 10.1038/s41467-019-08719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., et al. Vibrio campbellii hmgA-mediated pyomelanization impairs quorum sensing, virulence, and cellular fitness. Front. Microbiol. 2013;4 doi: 10.3389/fmicb.2013.00379. https://www.frontiersin.org/articles/10.3389/fmicb.2013.00379 [Online]. Available: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diard M., Garry L., Selva M., Mosser T., Denamur E., Matic I. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J. Bacteriol. Oct. 2010;192(19):4885–4893. doi: 10.1128/JB.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghafoor A., Hay I.D., Rehm B.H.A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. Aug. 2011;77(15):5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nobrega F.L., et al. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. Dec. 2018;16(12):760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- 57.Blanco A.E., et al. Chicken embryo lethality assay for determining the lethal dose and virulence of Enterococcus faecalis. Avian Pathol. Sep. 2017;46(5):548–555. doi: 10.1080/03079457.2017.1324942. [DOI] [PubMed] [Google Scholar]
- 58.Li J., et al. Development of phage resistance in multidrug-resistant Klebsiella pneumoniae is associated with reduced virulence: a case report of a personalised phage therapy. Clin. Microbiol. Infect. Dec. 2023;29(12):1601.e1–1601.e7. doi: 10.1016/j.cmi.2023.08.022. [DOI] [PubMed] [Google Scholar]
- 59.Altamirano F.L.G., Kostoulias X., Subedi D., Korneev D., Peleg A.Y., Barr J.J. Phage-antibiotic combination is a superior treatment against Acinetobacter baumannii in a preclinical study. EBioMedicine. 2022;80(Jun) doi: 10.1016/j.ebiom.2022.104045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holt R.D. Reference Module in Life Sciences. Elsevier; 2017. Species coexistence. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that are not directly available in the manuscript will be made available upon request. M12PA genome sequence is available in NCBI with the accession number PP668090.