Abstract
X-ray structures of aspartate transcarbamoylase in the absence and presence of the first substrate carbamoyl phosphate are reported. These two structures in conjunction with in silico docking experiments provide snapshots of critical events in the function of the enzyme. The ordered substrate binding, observed experimentally, can now be structurally explained by a conformational change induced upon the binding of carbamoyl phosphate. This induced fit dramatically alters the electrostatics of the active site, creating a binding pocket for aspartate. Upon aspartate binding, a further change in electrostatics causes a second induced fit, the domain closure. This domain closure acts as a clamp that both facilitates catalysis by approximation and also initiates the global conformational change that manifests homotropic cooperativity.
Keywords: allosteric transition, induced fit, homotropic cooperativity
Enzymes that exhibit positive cooperativity play a vital role in the regulation of the rates of key metabolic pathways by amplifying the response of a pathway to an effector molecule. In the case of aspartate transcarbamoylase (ATCase), substrate-induced domain closure triggers a quaternary conformational change that results in the observed homotropic cooperativity, one mechanism by which this enzyme controls the rate of de novo pyrimidine biosynthesis. Understanding the molecular features of domain closure not only provides insights into the mechanism of homotropic cooperativity but also demonstrates how ligand-induced domain closure can be used as part of catalytic mechanism of many enzymes (1, 2).
The Escherichia coli ATCase is composed of six chains (Mr 34,000 each) grouped into two trimeric catalytic subunits and six chains (Mr 17,000 each) grouped into three dimeric regulatory subunits. The three active sites in the catalytic subunit are shared across the interface between adjacent chains (3, 4), whereas the regulatory subunits contain the binding sites for the heterotropic activator, ATP, as well as the heterotropic inhibitors, CTP and UTP. Each catalytic chain is composed of two structural domains, the carbamoyl phosphate (CP) domain (residues 1–135 and 292–310) and the l-aspartate (Asp) domain (residues 136–291), which contain the binding sites for CP and Asp, respectively. Each regulatory chain is also composed of two structural domains, the AL domain (residues 1–100) and the Zn domain (residues 101–153), which contain the binding sites for the allosteric effectors and the structural Zn, respectively. In mammals ATCase exists as a component of the multienzyme complex CAD (carbamoyl phosphate synthetase, aspartate transcarbamoylase, and dihydroorotase) (5), and in humans, CAD has become a target for the development of antiproliferation drugs (6). The E. coli enzyme and the ATCase portion of CAD are 44% conserved, and all residues known to be involved in substrate binding and catalysis are conserved.
Domain closure is triggered by the ordered binding of the substrates (7), with CP binding before Asp, and N-carbamoyl-l-aspartate leaving before phosphate (Pi). In the E. coli enzyme, domain closure induces a dramatic quaternary structural change, from the tense (T) to the relaxed (R) structure, involving an elongation of the molecule by 11 Å as well as rotations of the catalytic and regulatory subunits (8). This domain closure promotes catalysis by correctly orientating many of the side chains of the active site that are thought to be necessary for high affinity substrate binding and catalysis. Because of structural constraints, the closure of the domains of the catalytic chains cannot occur without the global quaternary conformational change, which allows domain closure without steric interference. The quaternary conformational change can be triggered by the binding of a single molecule of a bisubstrate analog to just one of the six active sites (9).
The binding of substrates to ATCase is ordered with CP binding before Asp (10, 11). Small-angle x-ray scattering (12) and sedimentation velocity (13) studies have shown that CP does not cause significant global quaternary changes, whereas local changes are observed by circular dichroism in the aromatic region (14). These data suggest that upon CP binding, local conformational changes occur that create a viable Asp-binding site. Here, we report the high-resolution x-ray structures of E. coli ATCase in the absence and presence of the first substrate, CP. A comparison of these two structures in conjunction with in silico docking experiments not only establishes a molecular mechanism for ordered substrate binding but also describes the manner by which the binding of Asp induces the domain closure that initiates the concerted quaternary conformational change.
Methods
Enzyme Preparation. The wild-type ATCase was overexpressed by using E. coli strain EK1104 (15) containing plasmid pEK152 (16). The isolation and purification were as described in ref. 15. The concentration of purified enzyme was determined by absorption at 280 nm with an extinction coefficient of 0.59 (mg/ml)–1·cm–1. The purity of the enzyme was checked by SDS/PAGE (17) and nondenaturing PAGE (18, 19).
Crystallization and Freezing of Crystals. ATCase was crystallized by microdialysis, using 50-μl wells. The enzyme solution, at ≈18 mg/ml, was dialyzed against a solution of 40 mM citric acid/3 mM sodium azide/1 mM 2-mercaptoethanol/1 mM cytidine 5′-triphosphate/0.2 mM EDTA (pH 5.7). Crystals grew to average dimensions of 0.5 × 0.5 × 0.1 mm within 2 weeks. For the structure determination in the presence of CP, the crystals in the microdialysis chambers were dialyzed over 2 h against crystallization buffer to which 10 mM CP had been added. This procedure was repeated a second time before the crystals were used. Crystals were transferred into a freezing solution containing 30% (vol/vol) 2-methyl-2,4-pentanediol in crystallization buffer for ≈1 min before freezing in liquid nitrogen.
X-Ray Data Collection and Processing. The data for the structure in the absence of CP were collected by using a Rigaku/MSC (The Woodlands, TX) R-axis IV++ detector, whereas x-rays were generated by using a Rigaku RU-200 rotating anode generator operating at 50 kV and 100 mA. All data were collected at the Crystallographic Facility in the Chemistry Department of Boston College. The diffraction data were integrated, scaled, and averaged by using d*trek (Rigaku/MSC) (20).
The data for the structure in the presence of CP were collected at the National Synchrotron Light Source (Beamline X29) at Brookhaven National Laboratory (Upton, New York). The diffraction data were integrated, scaled, and averaged with hkl2000 (21).
Structural Refinement. The initial model for the structure in the absence of CP was derived from the coordinates of the published T-state structure in ref. 22 (PDB ID code 1NBE). Before refinement, all of the waters and ligands were removed. The refinement was carried out by using Crystallography & NMR System (cns) (23). After initial rigid body, simulated annealing, minimization, and B-factor refinement, initial maps were inspected. The N terminus of the regulatory chain was disordered; for this reason, the first 15 aa were removed from the model, then replaced through several rounds of manual rebuilding using xtalview (24). Corrections were also made to the catalytic chains. After rebuilding of the N terminus of the regulatory chains, the CTP was fit into the difference density in the allosteric binding pocket.
The initial model for the structure in the presence of CP was the T-state structure of the E50A catalytic chain mutant with phosphonoacetamide bound (PDB ID code 1TU0) (25). After initial rigid body, simulated annealing, minimization, and B-factor refinement, inspection of initial maps and the value of the R factor indicated that model building was necessary. The regulatory chains of the 1NBE structure (22) were overlaid and incorporated into the model. Another round of refinement was performed before CTP and CP were introduced into the structure. Several rounds of manual rebuilding of both catalytic chains and regulatory chains were performed to improve the quality of the electron density maps, particularly for residues 77–85 of the catalytic chains and 50–54 of regulatory chains.
Waters were added to both structures by using cns and xtalview on the basis of Fo – Fc electron density maps at or above the 2.5 σ level and were checked and retained only when they could be justified by hydrogen bonds. Both models were checked for correctness by using procheck (26). The details of data processing and refinement statistics for both structures are given in Table 1.
Table 1. Data collection and refinement statistics.
| ATCase without CP | ATCase with CP | |
|---|---|---|
| Data collection | ||
| Space group | P321 | P321 |
| Cell dimensions | ||
| a, b, c, Å | 120.56, 120.56, 142.42 | 121.44, 121.44, 142.55 |
| α, β, γ, ° | 90, 90, 120 | 90, 90, 120 |
| Resolution, Å | 30-2.0 (2.07-2.00) | 50-2.50 (2.59-2.50) |
| Rmerge,* % | 6.3 (44.4) | 6.8 (44.7) |
| Average, l/σ | 14.7 (3.8) | 36.7 (3.8) |
| Completeness, % | 99.5 (99.0) | 99.9 (99.0) |
| Total reflections | 435,580 | 381,778 |
| Unique reflections | 80,778 | 42,642 |
| Redundancy | 5.35 (5.23) | 9.0 (8.1) |
| Refinement | ||
| Resolution, Å | 30-2.15 | 50-2.50 |
| σ cutoff | 0 | 0 |
| Reflections | 74,750 | 40,371 |
| R factor/Rfree | 0.205/0.252 | 0.197/0.251 |
| rms deviations | ||
| Bonds, Å | 0.012 | 0.006 |
| Angles, ° | 1.58 | 1.40 |
| Dihedrals, ° | 23.8 | 23.8 |
| Total number waters | 590 | 381 |
| Average temperature factors | ||
| Catalytic chain | ||
| CP domain, Å2 | 35.3 | 39.8 |
| Asp domain, Å2 | 38.6 | 49.6 |
| Regulatory chain | ||
| AL domain, Å2† | 71.4 | 79.3 |
| Zn domain, Å2 | 46.4 | 52.1 |
Values in parentheses are for the highest-resolution shell.
Rmerge= Σhkl Σi|Ii(hkl) — 〈I(hkl)〉|/Σkhl ΣiIhkl.
First 10 residues omitted from calculation.
Automated Docking Procedure. The program autodock (27) was used for the automated docking of ligands to the active site of the structures. To have one complete active site, two adjacent catalytic chains were used for the docking procedure. The charges on CP and Asp were set at –2. The grid box was set at 30 Å3 centered at the middle of the active site, with a grid spacing of 0.275 Å between grid points. In each case, 25 docking runs were performed by using the Genetic Algorithm with a maximum of 500,000 energy evaluations.
Graphics. The figures were prepared with povscript+ (28) and chimera (29). The electrostatic potential maps were calculated with grasp (30).
Results and Discussion
Structure of ATCase in the Absence and Presence of CP. CP is a relatively unstable metabolic intermediate with a t1/2 of 5 min for decomposition in solution at 37°C and 2 sec at 100°C. We have found that when CP is bound to ATCase, the CP is protected from thermal decomposition. The stabilization of CP by ATCase has allowed us to prepare crystals of the ATCase·CP complex suitable for x-ray diffraction. Crystals without CP diffracted to 2.0 Å, significantly better than determined in ref. 31. However, the crystals soaked with CP diffracted to lower resolution. Therefore, data for these crystals were collected on Beamline X29 at the National Synchrotron Light Source at Brookhaven National Laboratory, yielding data to 2.5-Å resolution. The structures of the enzyme in the absence and presence of CP were refined to an R factor/Rfree of 20.5%/25.5% and 19.7%/25.1% respectively, by using cns (23).
To evaluate whether the binding of CP altered the quaternary structure of the enzyme, the vertical separation between the upper and lower catalytic subunits was compared. The vertical separation for the wild-type enzyme in the T and R states is 45.6 Å and 56.6 Å, respectively. The difference between the R- and T-state vertical separations corresponds to the vertical expansion of the enzyme observed during the T-to-R transition (56.6 – 45.6 = 11 Å). For the wild-type enzyme in the presence of CP, the vertical separation was 45.9 Å, indicating that the binding of CP does not significantly alter the quaternary structure of the enzyme.
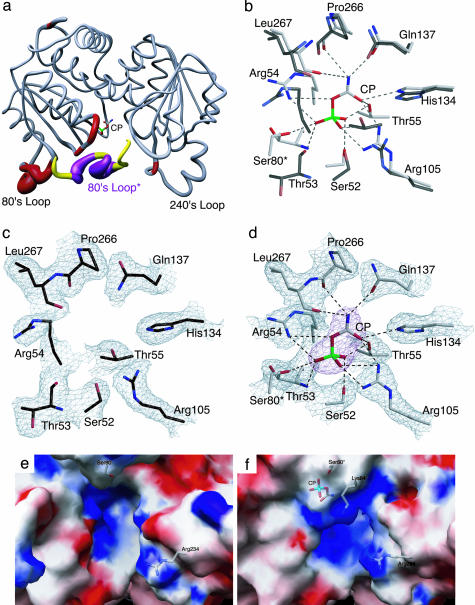
Fig. 1a shows the backbone changes that occur when CP binds to the active site of ATCase. The portion of the active site contributed by the 80's loop, a loop in the catalytic chain of ATCase composed of residues 73–88, of the adjacent chain is also shown. This comparison shows considerable changes in the tertiary structure of the catalytic chains induced upon CP binding in the 50's loop, a loop of residues in the catalytic chain from 50–56; the 80's loop; and smaller changes in the 240's loop, a loop of residues in the catalytic chain from 230–245. The largest differences between the two structures occur in the 80's loop from residues 76–84.
Fig. 1.
Comparison of the x-ray structures of ATCase in the absence and presence of CP. (a) One catalytic chain (gray, red) of ATCase along with the 80's loop of the adjacent catalytic chain (yellow, purple, 80's loop*). The width of the tube is proportional to the rms deviation between the α-carbon positions of the two structures. The CP domain is on the left (with CP shown) and the Asp domain is on the right. (b) Overlay of the CP binding site in the absence and presence of CP. The two structures were first superimposed by using sequoia (37). ATCase in the presence of CP is shown with gray side-chain carbons, whereas the structure without CP is shown with black side-chain carbons. (c) Active site of ATCase in the absence of CP. Orientation is the same as in b. The refined coordinates of the side chains are overlayed on the 2Fo – Fc electron density map shown contoured at 0.8 σ.(d) Active site of ATCase in the presence of CP. Orientation is the same as in b. The refined coordinates of the side chains are overlayed on the 2Fo – Fc electron density map shown contoured at 1.2σ. The CP is overlayed on the Fo – Fc electron density map (magenta) shown contoured at 1.6 σ.(e) Electrostatic potentials mapped onto the surface of ATCase in the absence of CP. (f) Electrostatic potentials mapped onto the surface of ATCase in the presence of CP. In e and f, the positions of Ser-80, Lys-84, Arg-234, and CP are overlaid onto the electrostatic map.
A comparison of the active site region of the structures in the absence and presence of CP is shown in Fig. 1 c and d. When CP binds, a number of residues, such as Ser-52, Thr-53, Arg-54, Thr-55, Arg-105, and Leu-267, along with Ser-80 from the adjacent chain, reorient to create the CP-binding pocket. A superposition of the active site with and without CP is shown in Fig. 1b. There are ≈15 hydrogen bonding interactions between CP and side chain or backbone atoms in the active site along with a smaller number of hydrophobic interactions. In total, these interactions explain the micromolar affinity of CP (32). The significance of this structural change is that the 80's loop from the adjacent chain provides critical residues involved in the stabilization of the tetrahedral intermediate (33). The binding of CP causes local alterations in the 50's loop, which draws side chains such as Ser-80 and Lys-84 of the adjacent 80's loop toward the active site. This movement induced upon CP binding is the first critical step in the preparation of the active site for domain closure and the allosteric transition.
Alterations in the Shape and Electrostatics of the Active Site upon CP Binding. The number of positively charged residues that are involved in the interactions with CP strongly suggests that electrostatic neutralization is important for binding. Fig. 1 e and f show the shape and electrostatic environment of the active site structure of the enzyme in the absence and presence of CP. A comparison of these two electrostatic potential maps reveals that the binding of CP has two critical effects on the active site. First, the volume of the active site is reduced because of both the binding of CP and conformational changes induced by its binding. Second, there is a dramatic increase in the electropositive nature of the active site. These changes taken together suggest that CP has a profound influence on preparing the active site for Asp binding. Electrostatic differences are caused mainly by the alterations in the 50's and 80's loops as well as smaller scale side-chain repositioning in the Asp domain, and a shift in interactions at the interface between the CP and Asp domains. For example, in the CP ligated structure, the T-state stabilizing interaction between Arg-54 and Leu-267 (backbone interaction) is lost (34). The loss of this interaction has the effect of repositioning the Arg-54 side chain toward the active site, contributing to the increased electropositive nature of the active site (compare Fig. 1 e and f).
In the Absence of Ligands, Asp and CP Bind to the Same Site. Because substrate binding to ATCase is ordered, with CP binding before Asp, it has been hypothesized that the binding of CP induces conformational changes that allow Asp to bind. To further investigate the precise mechanism of ordered-substrate binding to ATCase we carried out in silico docking experiments with autodock (27). The use of autodock was validated by testing the ability of the program to dock the bisubstrate analog N-phosphonacetyl-l-aspartate (PALA) into the x-ray structure of PALA bound to ATCase (PDB ID code 1D09) (33) and by docking CP into the x-ray structure of the ATCase·CP complex as reported here. The ligand (PALA or CP) was removed from the active site of the respective structure. The coordinates of the ligand were translated away from the protein, and its conformation was energy minimized before autodock (27) was used to redock the ligand. The docked positions of PALA and CP into their respective active sites were excellent, with a rms deviation of 0.95 Å for PALA and 0.73 Å for CP.
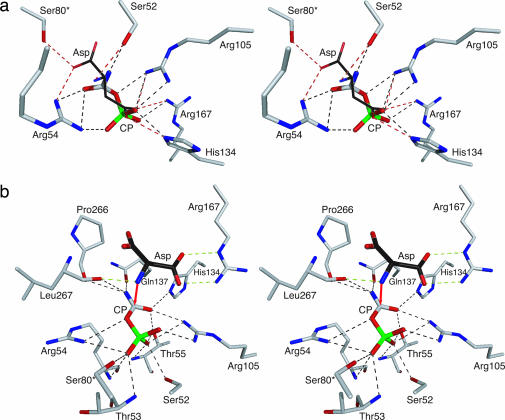
In separate experiments, CP and Asp were docked into the active site of the unligated structure. As seen in Fig. 2a, the docked positions of CP and Asp overlap in the active site, suggesting that, in this form of the enzyme, there is only one primary binding site, which can bind either CP or Asp.
Fig. 2.
Stereoview of a portion of the active site of ATCase. (a) The side chains are from the structure of the enzyme in the absence of CP. The positions of CP and Asp shown are from autodock (27). Interactions with CP and Asp are shown with black and red dashed lines, respectively. (b) The side chains are from the structure of the enzyme in the presence of CP. The position of CP is from the x-ray structure and the position of Asp is from the docking of Asp to the ATCase·CP complex with autodock (27). Interactions with CP and Asp are shown with black and green dashed lines, respectively. Red arrow shows the proposed attack of the nitrogen of the α-amino of Asp onto the carbonyl carbon of CP.
The results of these autodock experiments in conjunction with the electrostatic calculations provide an explanation for the experimental observation that the binding of the substrates to ATCase is ordered, with CP binding before Asp. When the active site of the enzyme is devoid of substrates, the position of the side chains is such that there is essentially one positively charged binding pocket (see Fig. 2a) that can bind either CP or Asp. However, the binding of CP alone not only induces local conformational changes in the active site that enhance its own binding but also brings about a significant alteration in the electrostatic environment of the active site (compare Fig. 1 e and f), creating the Asp-binding site. The lack of a functional Asp-binding site in the absence of CP binding provides a direct structural explanation for the required ordered binding of substrates to ATCase.
Asp Bound to the ATCase·CP Complex Is Positioned Correctly for the Nucleophilic Attack. To evaluate the interaction of Asp with the ATCase·CP complex autodock (27) was also used. The results show that the docked Asp was oriented in the exact position expected immediately before the nucleophilic attack. As seen in Fig. 2b, the nitrogen of the α-amino group of Asp is only 2.8 Å from the carbonyl carbon of CP. Thus, the formation of the ATCase·CP complex both creates a binding site for Asp and makes the electrostatic environment of the active site more electropositive (compare Fig. 1 e and f). This increase in the electropositive nature of the active site would effectively lower the pKa of the α-amino group of Asp, thereby promoting catalysis.
The Binding of CP Weakens Interactions That Are Required for Stabilization of the Enzyme in the T State. Previous site-specific mutagenesis experiments identified interactions that are necessary to stabilize the T and R states of ATCase, such as the T-state stabilizing interactions of Glu-239 found in the upper catalytic subunit toward both Lys-164 and Tyr-165 in the lower catalytic subunit (31). When the structures with and without CP are compared and averaged over the two catalytic chains in the asymmetric unit, the Glu-239–Lys-164 and Glu-239–Tyr-165 interactions increase in length by 0.4 and 0.7 Å, respectively. Other T-state stabilizing interactions also increase in length, but to a smaller extent. The lengthening of these interactions suggests that the binding of CP alone has a destabilizing influence on the T-state of the enzyme. These results also explain the ability of CP, in the absence of aspartate or an aspartate analog, to convert T-state-destabilized mutants (35, 36) completely into the R-state quaternary structure, whereas CP alone has little influence on the quaternary structure of the wild-type enzyme.
The Binding of Aspartate Induces the Allosteric Conformational Change. Based on the docking experiments shown in Fig. 2b, the binding of Asp to the ATCase·CP complex not only positions Asp correctly for the nucleophilic attack but also makes the electrostatic environment of the active site more electronegative. This second change in the electrostatics of the active site would induce movements of the positively charged residues of the 80's and 240's loops, such as Lys-84 and Arg-234, to reposition closer to the substrates. Such backbone motions correlate with the domain closure required to complete the formation of the high-affinity high-activity active site that is characteristic of the R-quaternary structure (33).
The Link Between Catalysis and Homotropic Cooperativity. The conformational changes induced by the binding of Asp cause the Asp domain, the 240's loop, and the 80's loop of the adjacent chain to close in on the two substrates. This motion has two consequences: First, it forces the substrates closer together, thereby lowering the activation energy of the reaction, and, second, it further weakens the intersubunit interactions that specifically stabilize the T quaternary structure. The weakening of these interactions triggers the initiation of the global quaternary conformational change. In fact, the 240's loops cannot attain their final domain-closed conformation without an expansion of the enzyme along the threefold axis, which allows the 240's loops from the upper and lower catalytic subunits to slide past each other.
The Link Between Domain Closure and Homotropic Cooperativity. The structures of ATCase reported here provide two snapshots of critical events in the function of the enzyme. The ordered substrate binding observed experimentally is explained by an induced-fit conformational change upon the binding of CP. This change dramatically transforms the electrostatics of the active site, creating the binding site for Asp. The subsequent binding of Asp changes the electrostatics once more, causing a second induced fit, the domain closure, which both facilitates the catalytic reaction and induces the quaternary conformational change that manifests homotropic cooperativity in ATCase.
Acknowledgments
We thank Howard Robinson of Brookhaven National Laboratory (Upton, NY) for data collection and assistance with data processing.This work was supported in part by Grant GM26237 (to E.R.K.) from the National Institutes of Health. Data for the crystals with CP bound were measured at Beamline X29 of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and from the National Center for Research Resources of the National Institutes of Health.
Author contributions: J.W., K.A.S., J.P.C., and E.R.K. designed research, performed research, analyzed data, and wrote the paper.
Abbreviations: ATCase, aspartate transcarbamoylase (EC 2.1.3.2, aspartate carbamoyltransferase); CP, carbamoyl phosphate; Asp, l-aspartate; T structure, the tense structure of ATCase in which the vertical separation between catalytic subunits is approximately 46 Å; R structure, the relaxed structure of ATCase in which the vertical separation between catalytic subunits is approximately 57 Å; PALA, N-(phosphonoacetyl)-l-aspartate; 80's loop, a loop in the catalytic chain of ATCase composed of residues 73–88; 50's loop, a loop in the catalytic chain of ATCase composed of residues 50–56; 240's loop, a loop in the catalytic chain of ATCase composed of residues 230–245.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1ZA1 and 1ZA2).
References
- 1.Gerstein, M., Lesk, A. M. & Chothia, C. (1994) Biochemistry 33, 6739–6749. [DOI] [PubMed] [Google Scholar]
- 2.Hayward, S. (2004) J. Mol. Biol. 339, 1001–1021. [DOI] [PubMed] [Google Scholar]
- 3.Krause, K. L., Voltz, K. W. & Lipscomb, W. N. (1985) Proc. Natl. Acad. Sci. USA 82, 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robey, E. A. & Schachman, H. K. (1985) Proc. Natl. Acad. Sci. USA 82, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grayson, D. R. & Evans, D. R. (1983) J. Biol. Chem. 258, 4123–4129. [PubMed] [Google Scholar]
- 6.Swyryd, E. A., Seaver, S. S. & Stark, G. R. (1974) J. Biol. Chem. 249, 6945–6950. [PubMed] [Google Scholar]
- 7.Wedler, F. C. & Gasser, F. J. (1974) Arch. Biochem. Biophys. 163, 57–68. [DOI] [PubMed] [Google Scholar]
- 8.Ke, H.-M., Lipscomb, W. N., Cho, Y. & Honzatko, R. B. (1988) J. Mol. Biol. 204, 725–747. [DOI] [PubMed] [Google Scholar]
- 9.Macol, C. P., Tsuruta, H., Stec, B. & Kantrowitz, E. R. (2001) Nat. Struct. Biol. 8, 423–426. [DOI] [PubMed] [Google Scholar]
- 10.Porter, R. W., Modebe, M. O. & Stark, G. R. (1969) J. Biol. Chem. 244, 1846–1859. [PubMed] [Google Scholar]
- 11.Hsuanyu, Y. & Wedler, F. C. (1987) Arch. Biochem. Biophys. 259, 316–330. [DOI] [PubMed] [Google Scholar]
- 12.Fetler, L., Tauc, P. & Vachette, P. (1997) J. Appl. Crystallogr. 30, 781–786. [Google Scholar]
- 13.Kirschner, M. W. & Schachman, H. K. (1971) Biochemistry 10, 1900–1919. [DOI] [PubMed] [Google Scholar]
- 14.Griffin, J. H., Rosenbusch, J. P., Weber, K. K. & Blout, E. R. (1972) J. Biol. Chem. 247, 6482–6490. [PubMed] [Google Scholar]
- 15.Nowlan, S. F. & Kantrowitz, E. R. (1985) J. Biol. Chem. 260, 14712–14716. [PubMed] [Google Scholar]
- 16.Baker, D. P. & Kantrowitz, E. R. (1993) Biochemistry 32, 10150–10158. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 18.Davis, B. J. (1964) Ann. N.Y. Acad. Sci. 121, 680–685. [Google Scholar]
- 19.Ornstein, L. (1964) Ann. N.Y. Acad. Sci. 121, 321–349. [DOI] [PubMed] [Google Scholar]
- 20.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Williams, M. K., Stec, B. & Kantrowitz, E. R. (1998) J. Mol. Biol. 281, 121–134. [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 24.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 25.Stieglitz, K., Stec, B., Baker, D. P. & Kantrowitz, E. R. (2004) J. Mol. Biol. 341, 853–868. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 27.Goodsell, D. S., Morris, G. M. & Olson, A. J. (1996) J. Mol. Recognit. 9, 1–5. [DOI] [PubMed] [Google Scholar]
- 28.Fenn, T. D., Ringe, D. & Petsko, G. A. (2003) J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- 29.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004) J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 31.Kosman, R. P., Gouaux, J. E. & Lipscomb, W. N. (1993) Proteins Struct. Funct. Genet. 15, 147–176. [DOI] [PubMed] [Google Scholar]
- 32.Suter, P. & Rosenbusch, J. P. (1976) J. Biol. Chem. 251, 5986–5991. [PubMed] [Google Scholar]
- 33.Jin, L., Stec, B., Lipscomb, W. N. & Kantrowitz, E. R. (1999) Proteins Struct. Funct. Genet. 37, 729–742. [PubMed] [Google Scholar]
- 34.Jin, L., Stec, B. & Kantrowitz, E. R. (2000) Biochemistry 39, 8058–8066. [DOI] [PubMed] [Google Scholar]
- 35.Tauc, P., Vachette, P., Middleton, S. A. & Kantrowitz, E. R. (1990) J. Mol. Biol. 214, 327–335. [DOI] [PubMed] [Google Scholar]
- 36.Chan, R. S., Sakash, J. B., Macol, C. P., West, J. M., Tsuruta, H. & Kantrowitz, E. R. (2002) J. Biol. Chem. 277, 49755–49760. [DOI] [PubMed] [Google Scholar]
- 37.Bruns, C. M., Hubatsch, I., Ridderstrom, M., Mannervik, B. & Tainer, J. A. (1999) J. Mol. Biol. 288, 427–439. [DOI] [PubMed] [Google Scholar]