Abstract
We have developed a new universal influenza B vaccination strategy based on inactivated influenza B viruses displaying mosaic hemagglutinins (mHAs). Recombinant mHA viruses were constructed by replacing the four major antigenic sites of influenza B virus HAs, with those from exotic avian influenza A virus HAs. Sequential vaccination of naïve mice with mHA-based vaccines elicited higher immune responses towards the immuno-subdominant conserved epitopes of the HA than vaccination with wildtype viruses. Among the different preparations tested, mHA split vaccines were less immunogenic than their whole inactivated virus counterparts. This lower immunogenicity was overcome by the combination with adjuvants. mHA split vaccines adjuvanted with a Toll-like receptor-9 agonist (CpG 1018) increased Th1 immunity and in vivo cross-protection, whereas adjuvanting with an MF59-like oil-in-water nano-emulsion (AddaVax) enhanced and broadened humoral immune responses and antibody-mediated cross-protection. The mHA vaccines with or without adjuvant were subsequently evaluated in mice that were previously immunized to closely mimic human pre-existing immunity to influenza B viruses and the contribution of innate and cellular immunity was evaluated in this model. We believe these preclinical studies using the mHA strategy represent a major step toward the evaluation of a universal influenza B virus vaccine in clinical trials.
Subject terms: Influenza virus, Inactivated vaccines, Vaccines, Adjuvants
Introduction
Influenza is a contagious respiratory disease caused by influenza viruses that infect the upper and sometimes the lower respiratory tract of humans. This respiratory infection can cause mild to severe disease and, at times, can lead to death1,2. Influenza viruses undergo frequent changes in their surface glycoproteins that evade host pre-existing immunity acquired by a previous infection or vaccination. Mutations are mainly accumulated in the hemagglutinin (HA) and neuraminidase (NA) proteins through a process known as antigenic drift3. Due to this immune evasion, influenza outbreaks occur every year and seasonal influenza vaccines need to be updated annually to match the circulating viruses4. Among the four influenza types, influenza A and B are the most epidemiologically relevant for humans2. Influenza A viruses undergo frequent antigenic changes and can be found in many animal reservoirs, which pose a risk for the emergence of new viruses with pandemic potential1. Historically, influenza B viruses have attracted less attention since they are antigenically not as diverse as influenza A viruses5 and lack a known animal reservoir4,6. Despite their low pandemic risk, influenza B viruses account for 20 to 30% of all influenza cases (Fig. 1A)6,7. In some years, they have been the dominant circulating strains, causing high mortality in infants and children8–11. The first influenza B virus was isolated in 1940 (B/Lee/1940) and two antigenically distinct lineages of influenza B viruses have been co-circulating in the past decades: the B/Yamagata/16/1988-like (Yam) strains and the B/Victoria/2/1987-like (Vic) strains (Fig. 1B, C)4,12. Interestingly, since the coronavirus disease 2019 (COVID-19) pandemic, circulation of Yam-like viruses has not been confirmed in the human population (as of March 18th 2024 from the Global Influenza Programme13). Nonetheless, the development of a universal influenza B virus vaccine that protects against the ever-mutating influenza B viruses is key to overcome the present limitations of seasonal influenza vaccines since a mismatched virus in the formulation has been shown to reduce vaccine efficacy remarkably4. Here, we present the preclinical evaluation of a novel universal influenza B virus vaccine based on recombinant mosaic hemagglutinin-based (mHA) viruses.
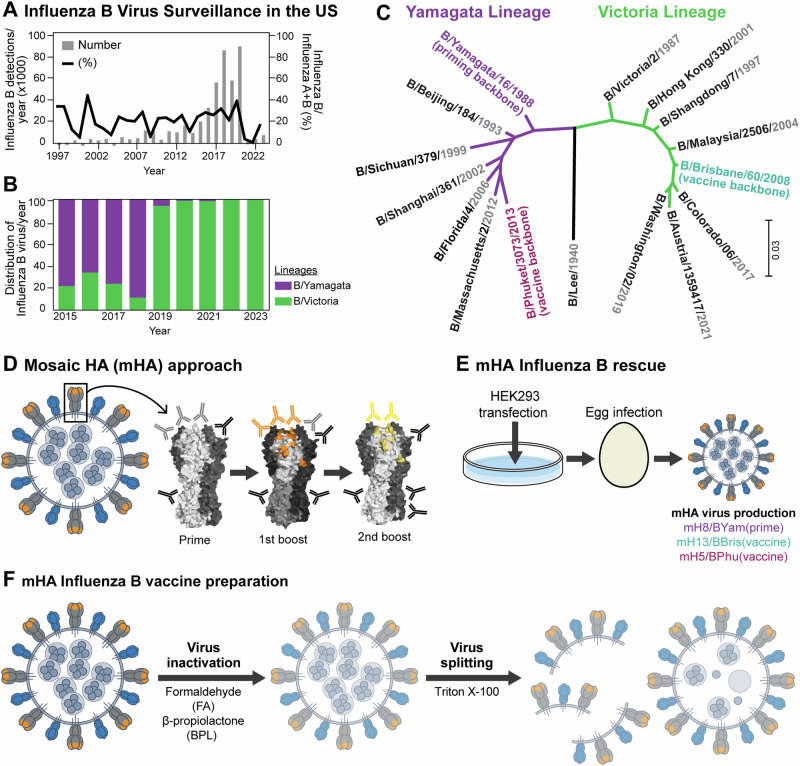
Fig. 1. Influenza B virus surveillance and universal influenza virus vaccine design.
A Surveillance of influenza B viruses in the US from 1997 to 2023 and (B) percentage of influenza B/Yamagata/16/1988-like and B/Victoria/2/1987-like lineage viruses identified from 2015 to 2023 (data obtained from FluNet, (who.int) as of 1st Aug 2023). C Phylogenetic tree of HA sequences of historical annual formulations for IBV vaccine strains from 1999 to 2023 (data obtained from the Global Influenza Programme (who.int) as of 1st Aug 2023 and reported in Supplementary Fig. 113). Influenza B virus HA sequences used in this study as prime vaccination (B/Yamagata/16/1988) and as universal influenza virus vaccines (B/Brisbane/60/2008 and B/Phuket/3073/2013) are highlighted in green and dark red, respectively. The phylogenetic tree was constructed using the maximum likelihood method and Tamura-Nei model and visualized with Mega1163,64. D Mosaic HA (mHA) universal influenza vaccine approach. Sequential vaccination with mHA vaccines, where the major immunodominant epitopes are replaced in each vaccination to refocus the immune response to subdominant head and stalk epitopes of the HA glycoprotein (representation of an HA glycoprotein based on PDB accession no. 4M44). E mHA influenza B virus rescue scheme. B mHA and wildtype (WT) viruses were rescued following the reverse genetics method as previously described17. Thereafter, viruses were propagated in embryonated chicken eggs and harvested in the allantoic fluid. The mosaic viruses were based on B/Yamagata/16/1988 (Yam), B/Brisbane/60/2008 (Bris) and B/Phuket/3073/2013 (Phu) resulting in the mH8/BYam, mH13/BBris and mH5/BPhu viruses, respectively18. F mHA influenza B virus vaccine preparation. Whole inactivated viruses (WIV) were generated by inactivating the harvested viruses with either formaldehyde (FA) or beta-propiolactone (BPL) and purified by sucrose cushion ultracentrifugation. To produce split versions of these vaccines, BPL inactivated and purified virus preparations were treated with Triton X-100 and the remaining detergent was removed using hydrophobic beads in batch mode chromatography19.
Results
Inactivated mHA-based vaccines induce higher levels of antibodies targeting the conserved HA epitopes than their wild type virus counterparts
Three different B mHA viruses were constructed by replacing the four major antigenic regions in the head domain of the influenza B virus HA by those of exotic avian HA sequences (Fig. 1D)14–16. Immunodominant regions in the B/Yamagata/16/1988 (BYam), B/Brisbane/60/2008 (BBris) and B/Phuket/3073/2013 (BPhu) HAs were replaced by those of H8, H13 and H5 to generate mH8/BYam, mH13/BBris and mH5/BPhu viruses (Fig. 1E)17–19. These immunodominant regions are subjected to antigenic drift, which allows the virus to escape pre-existing immunity. We hypothesize that sequential vaccination with different mHA viruses will enhance the immune responses toward the conserved regions of the HA given that the immunodominant antigenic sites presented in each vaccine strain are antigenically different (i.e. epitopes from H8 versus H13 versus H5 HAs) (Fig. 1D). Since most seasonal influenza virus vaccines are manufactured using the inactivated virus vaccine platform, we developed inactivated B mHA whole inactivated virus (WIV) and inactivated split vaccines (Fig. 1F)20. WIV was generated by inactivation with beta-propiolactone (BPL), and split vaccines were obtained by further splitting with Triton X-10021,22.
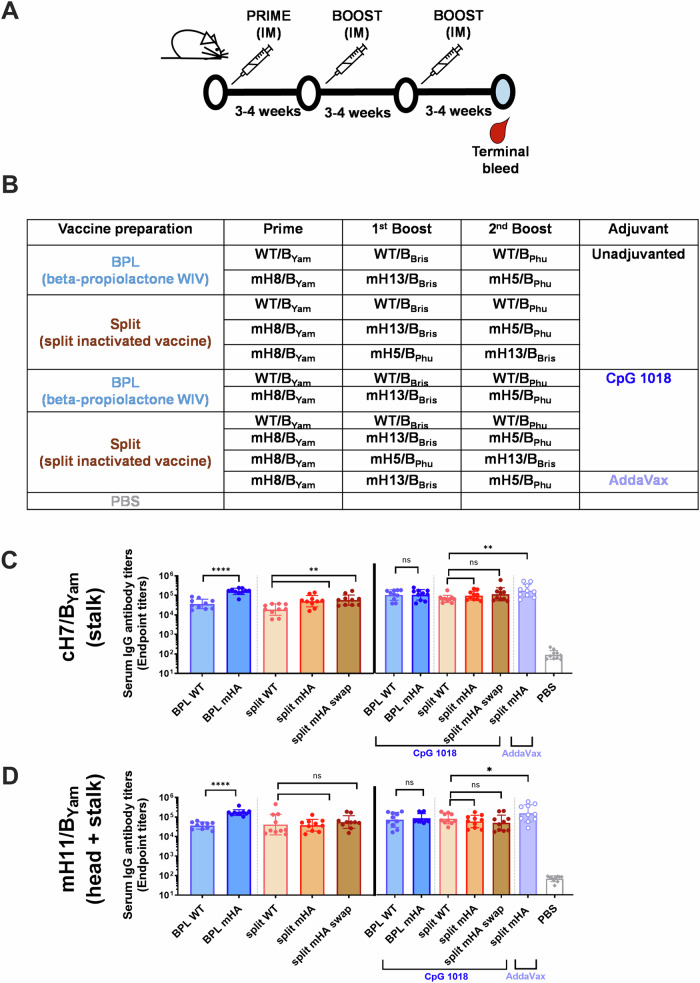
Immunogenicity of inactivated virus vaccines was assessed in BALB/c mice following a three-dose sequential vaccination regimen17,18 (Fig. 2A, B). The mH8/BYam vaccine or wild type (WT) BYam vaccine was given as prime, followed by two boosters of mH13/BBris and mH5/BPhu, or WT BBris and WT BPhu, respectively. An additional group was included in which the order of split B mHA vaccine boosters were swapped (mH5/BPhu and mH13/BBris, swap). We explored the adjuvanting potential of CpG 1018, a Toll-like receptor-9 agonist. CpG 1018 is present in the HEPLISAV-B hepatitis B virus vaccine and has been recently tested with group 2 cHA constructs in preclinical studies23. As an adjuvanted control group, split B mHA vaccines were tested in combination with AddaVax, a squalene-based oil-in-water nano-emulsion with a formulation similar to that of MF59 that has been licensed for adjuvanted influenza virus vaccines and used in previous studies18,24. Inactivation of whole virus vaccines with formaldehyde (FA) was also included for comparison with previous studies (Supplementary Figs. 2 and 3)18. Vaccination was performed in a 3- to 4-week interval at a dose of 1 µg of HA per mouse in each vaccination. Then, mice were bled for serological analyses and serum passive transfer/viral challenge studies were performed (Figs. 2, 3 and Supplementary Fig. 4).
Fig. 2. Antibody responses to the conserved HA stalk and immuno-subdominant head domains elicited by wildtype and mHA vaccines prepared by two different methods.
Whole inactivated virus (WIV) with beta-propiolactone (BPL) and inactivated split vaccine combining BPL inactivation and Triton X-100 splitting were compared for two different vaccination strategies with WT viruses or mHA viruses. Vaccines were tested without adjuvant or with the combination with CpG 1018 (30 µg per mouse). A, B Vaccination regimen and groups. BALB/c mice (n = 10) were vaccinated in a three-dose vaccination experiment with a dose of 1 µg of HA in a 3–4-week interval. mHA inactivated split vaccines adjuvanted with AddaVax (1:1 v:v) and an unvaccinated group (PBS) were included as controls. C, D Binding of serum antibodies towards the immuno-subdominant epitopes. A cH7/BYam protein with a group 2 avian H7 head and the B/Yamagata/16/1988 HA stalk was used to measure stalk-specific antibodies in the unadjuvanted and adjuvanted groups (C). A mH11/BYam protein displaying the H11 major antigenic sites in the B/Yamagata/16/1988 HA (mH11/BYam) was used to measure antibody binding to conserved epitopes in the HA head and stalk domains in the unadjuvanted and adjuvanted groups (D). The geometric mean endpoint titer was calculated as the readout. The statistics were calculated using an unpaired one-tailed t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
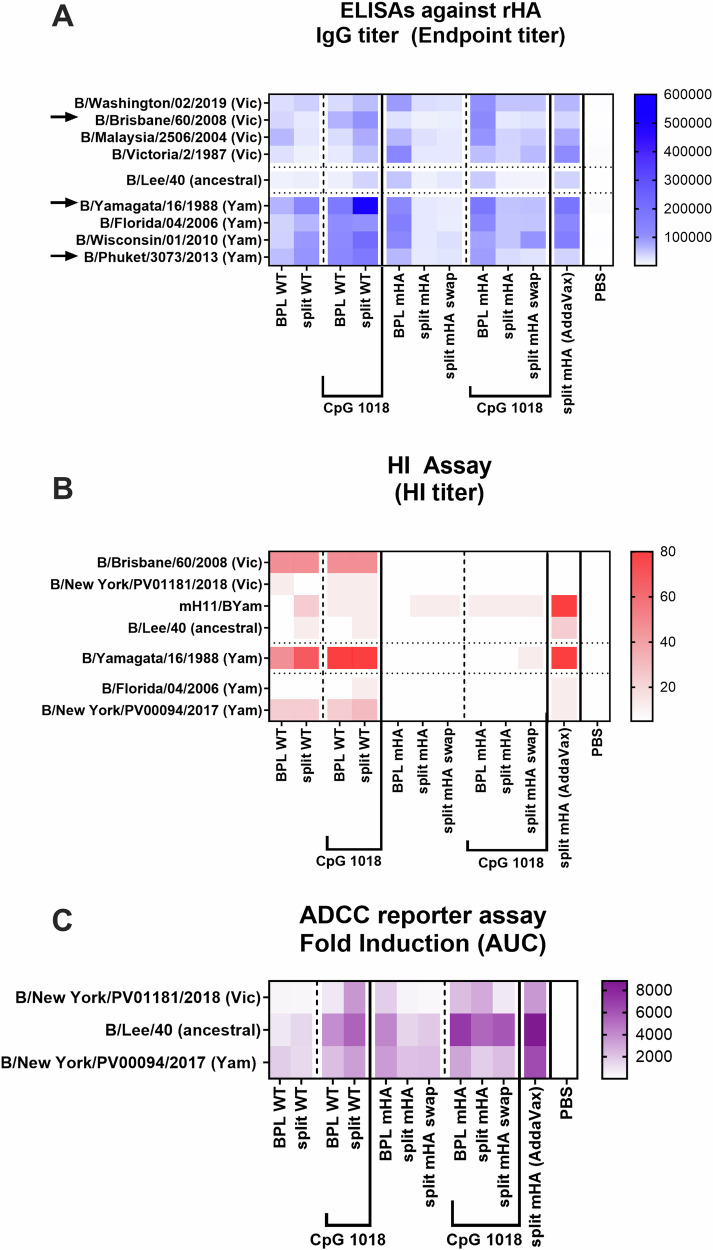
Fig. 3. Cross-reactivity, HI-activity, and Fc effector function antibody responses elicited by wildtype and mHA vaccines prepared by two different methods.
Whole inactivated virus (WIV) with beta-propiolactone (BPL) and inactivated split vaccine combining BPL inactivation and Triton X-100 splitting were compared for two different vaccinations with WT viruses or mHA viruses. Vaccines were tested without adjuvant or with the addition of CpG 1018 (30 µg) as explained in Fig. 2. A Heatmap of IgG binding serum antibodies against a panel of recombinant influenza B HA proteins. B Heatmap of hemagglutination inhibition (HI) titer against a panel of influenza B viruses. C Heatmap of ADCC activity against three influenza B viruses. To perform the ADCC reporter assay, MDCK cells were infected with each virus at an MOI of 5 (single-cycle replication). One day after infection, mouse sera were added to the cells and incubated for 30 min and genetically modified Jurkat cells expressing the mouse FcγRIV with a luciferase reporter gene under transcriptional control of the nuclear factor-activated T (NFAT) cell promoter were then added and incubated for 6 h. Later luciferase activity was measured. The geometric mean AUC of fold induction of each group (pooled sera) was measured in technical duplicate.
First, we determined the antibody responses to the conserved HA stalk domains, and to the immuno-subdominant HA head domains by ELISA using cH7/BYam and mH11/BYam recombinant proteins, respectively (Fig. 2C, D). cH7/BYam has an H7 head domain on top of the BYam HA stalk domain and the mosaic mH11/BYam protein displays the H11 sequences at the major antigenic sites of the BYam HA. Unadjuvanted mHA vaccines induced higher levels of antibodies targeting the HA conserved regions than their WT counterparts (Fig. 2C, D). Among unadjuvanted conditions, the BPL mHA WIV group elicited the highest antibody titers. Interestingly, no differences were observed between the mHA and WT vaccine groups adjuvanted with CpG 1018, independently of the vaccine type (Fig. 2C, D). On the other hand, the combination of the mHA split vaccine with AddaVax elicited the highest levels of binding antibodies to the conserved HA epitopes. We did not observe a significant impact of swapping the order of mHA split vaccine boosters.
Mosaic HA-based vaccines elicit a cross-reactive and HI-inactive humoral immune response with Fc-mediated effector functions
Antibody titers against a panel of 9 phylogenetically distinct influenza B virus HA recombinant proteins were analyzed by ELISA (Fig. 3A). In WT vaccinated groups, a higher level of antibody titers was detected for all three HAs in the Yam-lineage compared to the Vic-lineage, especially in the adjuvanted groups (Fig. 3A, left). This was likely due to the two vaccinations with the Yam-lineages viruses21. On the other hand, mHA vaccinated groups developed a more balanced immune response against all the HAs tested, although overall titers may be lower than their WT counterparts in the Yam-lineage. This is particularly evident in the BPL mHA group, which showed the highest antibody titers among unadjuvanted mHA groups (Fig. 3A, right). Of note, mHA split vaccines combined with AddaVax elicited a higher booster effect than CpG 1018 (Fig. 3A, last column).
Subsequently, we evaluated the hemagglutination inhibition (HI) activity of serum antibodies in Fig. 3B. A panel of 7 different influenza B viruses of both lineages and the ancestral strain was evaluated. In addition, the recombinant mH11/BYam virus was included to present a strain antigenically distinct from all the other influenza B viruses to which these sera should not have HI activity. Overall, HI activity was only detected in WT vaccinated groups17,25,26. Higher HI titers were observed against the matched strains compared with titers against the mismatched ones. The combination with CpG 1018 did not improve HI antibody titers of the WT virus vaccines nor the mHA vaccines. Interestingly, AddaVax substantially improved HI activity of the mHA split vaccine (Fig. 3B, last column), suggesting the possible generation of HI active antibodies targeting the head epitopes outside of the immunodominant sites (i.e., receptor binding site or unknown HI reactive sites) of the HA.
Finally, we evaluated Fc-mediated effector functions of serum antibodies in a reporter assay (Fig. 3C) and the antibody-mediated protection in a passive transfer/challenge experiment (Supplementary Fig. 4) against three antigenically different viruses: the ancestral B/Lee/1940 strain and the two recent influenza B isolates B/New York/PV01181/2018 (Vic-like) and B/New York/PV00094/2017 (Yam-like) strains18. Fc-mediated effector functions have been associated with protection provided by antibodies targeting the conserved domains, such as the HA stalk domain27–32. An antibody dependent cellular cytotoxicity (ADCC) reporter assay was performed to examine the Fc-mediated effector activity (Fig. 3C). To do so, genetically modified Jurkat cells expressing the mouse FcγRIV with a luciferase reporter gene under transcriptional control of the nuclear factor-activated T (NFAT) cell promoter were used. An increased ADCC reporter activity was observed in all adjuvanted groups compared to the unadjuvanted ones, suggesting the boosting of the antibody responses towards the HA conserved regions (Fig. 3C). In the passive transfer/challenge experiment, 100 µL of serum was administered intraperitoneally to each mouse two hours before virus infection, and the weight loss and survival was followed for 14 days. For a simplified comparison, the minimum weight observed in each challenge is represented in Supplementary Fig. 4E. For B/Lee/1940 and Vic-like viruses, a similar antibody-mediated protection profile was found for all groups. Among the unadjuvanted preparations, the BPL mHA vaccines provided the highest protection and resulted in the least weight loss. When vaccines were combined with CpG 1018, a lower morbidity and higher survival was observed for all conditions compared to their unadjuvanted counterparts. As for the Yam-like virus challenge, an advantage of all WT groups versus the mHA ones was clearly observed (Supplementary Fig. 4E). This could be associated with the antibody titers and HI activity against the Yam-like viruses described in Fig. 3A, B. The mHA split group adjuvanted with AddaVax also provided a high protection and minimal weight loss against all three challenge viruses.
Given the higher cross-protection of mHA candidates in the absence of adjuvant, and the different humoral immune profiles observed with CpG 1018 versus the AddaVax control group, we decided to further test these constructs and both adjuvants in animals with pre-existing immunity to influenza B. The combination of WT viruses with CpG 1018, which also presented increased cross-protection, will be explored in more detail in future studies.
Adjuvanting mHA WIV and mHA split vaccines with CpG 1018 and AddaVax induces different Th1/Th2 immune profiles in mice with low pre-existing immunity
In a first study, a low pre-existing immunity model was generated by intramuscular vaccination with mH8/BYam recombinant protein at 1 µg of HA per mouse (Supplementary Figs. 5 and 6) adapted from17. It was decided to prime with the mHA protein since priming with WT protein may interfere with the WT virus challenge. Immunity acquired with prime-only vaccination are presented in Supplementary Fig. 5C. Primed mice that were vaccinated with a commercial seasonal quadrivalent inactivated vaccine (QIV) and an unvaccinated group were included as the standard of care and negative controls, respectively (Supplementary Fig. 5A).
Unadjuvanted mHA WIV developed a Th1-polarized immune response, whereas QIV and unadjuvanted split mHA vaccine biased toward Th2 responses33,34. Adjuvanting with AddaVax increased the levels of cH7/BYam and mH11/BYam-specific antibodies (Supplementary Fig. 5D, E), and germinal center (GC) reaction in the draining lymph nodes (Supplementary Fig. 5J)18,35,36. Mosaic HA split vaccine + AddaVax boosted B/Lee/1940-specific antibodies to a higher degree than its WIV counterpart (Supplementary Fig. 5F), which correlated with highest protection in a passive transfer/challenge experiment with the same virus (Supplementary Fig. 6D–F). On the other hand, adjuvanting with CpG 1018 increased IgG2a/IgG1ratio (Supplementary Fig. 5G) and reduced morbidity after direct virus challenge (Supplementary Fig. 6C).
Mosaic HA split vaccines are highly immunogenic in the context of high pre-existing immunity in mice
The same vaccine groups were tested in the context of high pre-existing immunity induced by viral infection (Fig. 4). We decided to more thoroughly investigate other arms of the immune response in this model, including acute responses generated by the vaccine (Fig. 4C), and T cell immunity (Supplementary Figure 7). The cytokine profile was measured 4 h post vaccination in serum samples to monitor the vaccine response (Fig. 4C). CpG 1018 adjuvanted groups showed higher levels of IFN-γ and TNF-α in sera which is associated with a Th1-skewed immune response. In the case of AddaVax adjuvanted groups, AddaVax did not boost IFN-γ and TNF-α levels as CpG 1018 did. In contrast, a boosting of IL-5 was observed when combining it with WIV or split vaccines, which indicates a Th2-skewed immune response and correlates with the IgG2a/IgG1 ratio previously shown (Supplementary Figure 5). Antibodies targeting the conserved HA stalk and head domains were tested after the two vaccine doses. The “high-priming” induced similar levels of antibodies in all vaccinated groups, including the QIV (Fig. 4D, E). Interestingly, mHA split vaccines induced higher frequency of GC B cell formation than the mHA WIV group (Fig. 4I). This contrasts with what was observed in animals primed with recombinant protein (Supplementary Fig. 5J). Consistent with previous observations, AddaVax but not CpG 1018, significantly increased GC B cell formation for mHA WIV (Fig. 4I). Since unadjuvanted split vaccine already induced high levels of GC reaction, the combination with any of the adjuvants did not further improve this response.
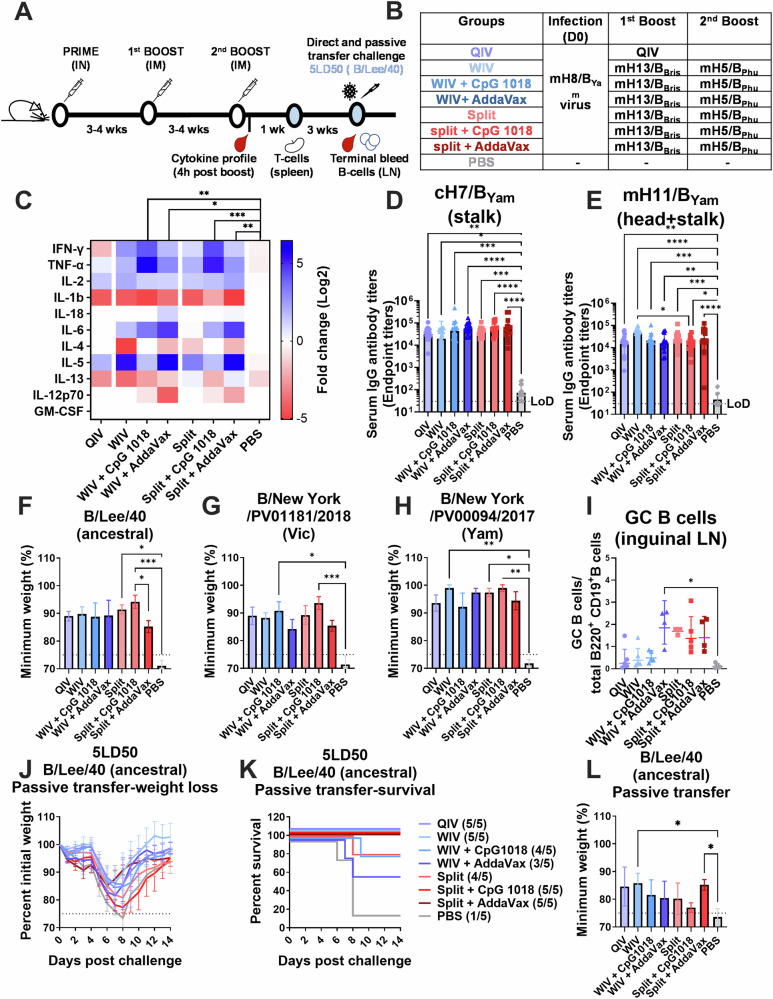
Fig. 4. Cross-reactivity, Th1/Th2 immunogenicity, germinal center reaction and protection of mHA vaccines in a high pre-existing immunity model in mice.
A Vaccination regimen and experimental workflow. B Vaccination groups. BALB/c mice were vaccinated in a two-dose vaccination scheme with 1 µg of HA of the different WIV or split mHA vaccines in a 3-4 week interval after priming with 105 PFU of mH8/BYam virus per mouse. WIV and split mHA vaccines were tested without adjuvant or with the addition of CpG 1018 (30 µg) or AddaVax (1:1 v:v). A QIV (Flulaval Quadrivalent) vaccinated group and an unvaccinated group (PBS) were included as controls. mH8/BYam virus infection was given intranasally in a total volume of 30 µL. C Heatmap of Th1/Th2 cytokine panel measured in sera taken 4 h after second boost vaccination (n = 4) using a 11-plex Luminex panel. Log2 fold change over PBS group and 2-way ANOVA test corrected using Dunnet’s test for multiple comparisons is shown (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). D, E Binding of serum antibody titers (n = 13–15) against subdominant epitopes analyzed as previously described in Fig. 3. Minimum weight loss (n = 5) of direct virus challenge with the ancestral B/Lee/40 virus (F), B/New York/PV01181/2018 virus (Victoria-lineage) (G) and B/New York/PV00094/2017 virus (Yamagata-lineage) (H). I Frequency of germinal center B cell formation. Inguinal lymph nodes were collected 4 weeks after second boost and frequency of germinal center B cells (live CD3-B220+CD19+IgD-GL7+CD38low) was measured by FACS (n = 3–5). Serum passive transfer challenge with the B/Lee/40 virus (J–L) weight loss (J), survival of vaccination groups (K) and minimum weight loss (L) is shown. For direct challenge, BALB/c mice (n = 4–5) were challenged 8 weeks after second boost intranasally in a total volume of 30 µL. In the serum passive transfer experiment, BALB/c mice (n = 5) received 100 µL of pooled sera intraperitoneally and 2 hours later were challenged intranasally as previously described. In all cases a dose of 5× mLD50 was used. Weight loss and survival of mice were monitored for 2 weeks with a humane endpoint of ≥25% loss of the initial weight. Statistical significance was calculated using Kruskal-Wallis test corrected using Dunn’s test for multiple comparisons (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Eight weeks after the second boost, mice were challenged with three different influenza B viruses (Fig. 4F–H). All vaccinated mice survived independently of the vaccination strategy (survival data not shown). When the minimum weight was compared between groups, mice vaccinated with mHA split vaccines adjuvanted with CpG 1018 presented the lowest morbidity (Fig. 4F–H), in line with previous results (Supplementary Fig. 6). A serum passive transfer experiment was performed with B/Lee/1940 as challenge virus (Fig. 4J–L). A higher survival rate was observed for all groups compared with the low pre-existing immunity experiment (Supplementary Fig. 6). Unadjuvanted mHA WIV and mHA split vaccine + AddaVax showed the lowest morbidity, also in line with previous findings (Supplementary Figs. 4 and 6).
The elicitation of antigen-specific effector memory T cells after vaccination was evaluated using an intracellular cytokine staining assay (Supplementary Fig. 7). T cell activity was measured against NP and M1 internal proteins, which have been described as the main target for CD4+ and CD8+ T cell immunity37–39. mHA split vaccines induced a higher level of antigen-specific CD4+ and CD8+ T cells than QIV and mHA WIV. The combination with CpG 1018 increased the percentage of stimulated monofunctional and polyfunctional CD8+ T cells (Supplementary Fig. 7B, D, F, H), whereas AddaVax stimulated more monofunctional and polyfunctional CD4+ T cells Supplementary Fig. 7C, E, G). These results highlight the different immune responses induced by these adjuvants when combined with different types of mHA vaccines.
Mosaic HA-based vaccines induce stronger in vivo protection than QIV in a dose de-escalation experiment in mice
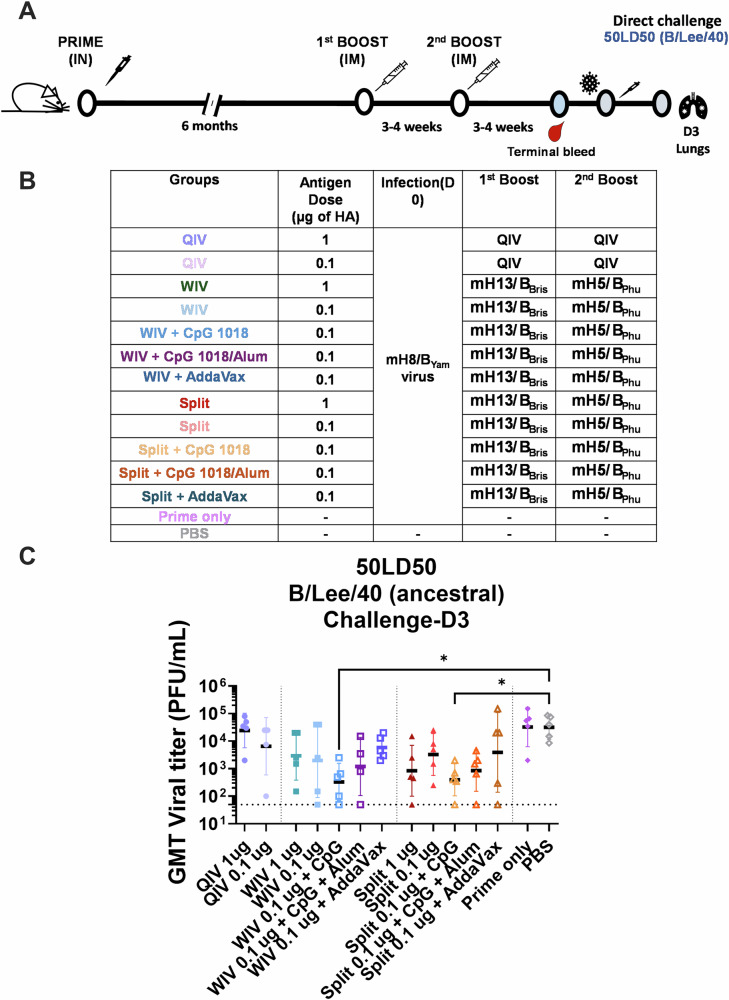
To better understand the immunogenicity of mHA vaccines relative to the standard of care (QIV) in the “high-priming” model, we tested mHA vaccines in a dose de-escalation experiment in mice (Fig. 5A, B). A similar immunization schedule was performed as in previous experiments, except that a longer interval between the high-prime and the mHA vaccination was implemented allowing the immune responses to plateau. In addition to CpG 1018 and AddaVax, we included a new adjuvant formulation combining CpG 1018 (10 µg) with Alum (50 µg). The combination of CpG 1018 with Alum has been tested in Phase II/III clinical trials with a recombinant severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) trimeric spike protein subunit vaccine (NCT04672395), showing an improvement on spike immunogenicity40–42.
Fig. 5. In vivo cross-protection of mice vaccinated with a low dose of mHA vaccines in a direct virus challenge and serum passive transfer/challenge experiment.
A, B Vaccination regimen and experimental workflow. BALB/c mice were vaccinated in a two-dose vaccination scheme with 1 or 0.1 µg of HA of the different vaccines in a 3–4-week interval after priming with 105 PFU of mH8/BYam virus. WIV or split mHA vaccines were tested without adjuvant or with the addition of CpG 1018 (10 µg), CpG 1018 (10 µg) + Alum (50 µg) or AddaVax (1:1 v:v). A QIV (FluLaval Quadrivalent) vaccinated group and an unvaccinated group (PBS) were included as controls. mH8/BYam virus prime infection was given intranasally in a total volume of 30 µL 6 months prior to the two-dose immunization. C Four weeks after the second dose, mice (n = 5) were challenged with 50× mLD50 dose and viral titers were measured from harvested lung homogenate tissues 3 days post infection. Kruskal–Wallis test corrected using Dunn’s test for multiple comparisons against PBS is shown (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
The cross-protection conferred by these different vaccines was again tested in a direct challenge experiment with the B/Lee/1940 virus (Fig. 5) and passive transfer/challenge (Supplementary Figure 8). To improve the resolution of protection, mice were challenged with 50× LD50 of the B/Lee/1940 virus in the direct challenge (Fig. 5C). Peak viral titers in the lungs three days post challenge were used as the readout. Vaccination with unadjuvanted mHA WIV and mHA split vaccines reduced viral titers to a higher extent than QIV at the two doses tested (Fig. 5C, 1st group vs 3rd group and 8th group). Vaccine combination with CpG 1018 allowed to better control viral replication, showing the lowest viral titers for those groups in which the adjuvant was used (Fig. 5C, 5th group and 10th group). AddaVax did not improve virus clearance in comparison with the unadjuvanted groups. The combination of Alum with CpG 1018-adjuvanted vaccines showed an intermediate protection between the CpG 1018 only-adjuvanted vaccines and AddaVax.
Discussion
We designed a universal vaccine against influenza B virus disease based on sequential vaccination with recombinant mHA B viruses in which the immunodominant regions of the HA had been replaced by those of exotic avian influenza viruses17,18,25,26,43. Compared to vaccination with WT viruses, mHA-based vaccines provided protection based on cross-reactive HI-inactive antibodies with Fc-mediated functions17,18. Here, we assessed the immunogenicity of inactivated mHA virus vaccines in the mouse model. Inactivated split viruses are the main platform for seasonal influenza virus vaccine development4,44,45 due to their low reactogenicity46,47. In this work we compared WIV versus inactivated split vaccines in the context of mHA constructs. mHA BPL WIV was more immunogenic than its mHA split vaccine counterpart in the absence of adjuvant (Table 1, Unadjuvanted column), in line with previous observations46,47.
Table 1.
Summary of the effects of CpG 1018 and AddaVax on the immunogenicity of different mHA vaccine platforms
| Unadjuvanted | CpG 1018-adjuvanted | AddaVax-adjuvanted | |
|---|---|---|---|
| mHA WIV Vaccine | High immunogenicity with Th1-biased immune response | Limited increase in in vivo cross-protection | Decrease in serum passive transfer cross-protection |
| mHA split vaccine | Moderate immunogenicity with Th2-biased immune response | Increase in in vivo cross-protection | Increase in serum passive transfer cross-protection |
When mHA WIV and mHA split vaccines were combined with CpG 1018, an enhanced Th1 immune response was elicited in which viral replication in the lungs and weight loss were reduced in the context of direct virus challenge in mice with pre-existing immunity. These results are consistent with recent data published on universal group 2 cHA split vaccines in mice23. Although CpG 1018 also stimulated Th1 immunity when combined with WIV, its final contribution to vaccine protection was marginal due to the high immunogenicity of the mHA WIV vaccine itself (Table 1, CpG 1018 adjuvanted column). The presence of viral RNA in the virion and the repetitiveness of viral surface may, in part, explain the higher immunogenicity of WIV vaccines48,49. It is noteworthy that naïve mice that received three vaccinations of different WT viruses in combination with CpG 1018 also developed good cross-protective immune responses, especially in the split vaccine format. Due to complex exposures histories of humans50, it remains unclear if broader protection can be achieved by vaccination with WT viruses. We hope to continue investigating this approach in future studies.
Combination of mHA WIV and mHA split vaccines with AddaVax induced a Th2-skewed immune response51, stimulating IL-5 secretion after vaccination that was associated with a strong germinal center B cell formation. The combination of mHA WIV with AddaVax negatively impacted vaccine protection in serum passive transfer studies, which was associated with the elicitation of higher levels of IgG1 antibodies51. On the other hand, the combination of the mHA split vaccine with AddaVax provided the highest antibody-mediated protection in serum passive transfer/challenge experiments (Table 1, AddaVax-adjuvanted column). Human clinical trials using MF59 adjuvant with split influenza virus vaccine have reported a higher percentage of adverse reactions in MF59 vaccinees compared to the unadjuvanted groups52, which may be caused, in part, by this Th2-skewed immunity which is also associated with cell-mediated inflammation53.
Furthermore, we tested these vaccines in two models representing low and high pre-existing immunity, by priming with recombinant protein (low prime), or by priming with an influenza virus infection (high prime)17. A higher level of protection was observed for all groups when mice were primed with a virus infection. In the case of the split vaccine, higher germinal center B cell formation and antigen-specific T cell immune responses were observed in the high pre-existing immunity model. As most human adults have been previously exposed to influenza B viruses54, mHA split vaccines are an interesting vaccine platform in the context of high pre-existing immunity.
Ferrets are the gold standard to study virus pathogenesis, transmission and immunogenicity of influenza A viruses, but there are few influenza B viruses that can effectively infect this animal model45. In this work, we have used the most phylogenetically distant influenza B virus described so far (B/Lee/1940 virus) as a surrogate of cross-protection in the mouse model. Broad cross-protection remains a priority given the continuous drift of influenza B viruses and the limited breadth of current seasonal vaccines. Several vaccine updates have been needed in the last decade for the Victoria-lineage component (Fig. 1). Furthermore, since the beginning of the COVID-19 pandemic, the B/Yamagata-lineage has not been detected. This fact poses a new epidemiological scenario where pre-existing immunity against this lineage will disappear and will bring a new risk of re-emergence of similar viruses against which we will not be protected. Despite of the limitations of the mouse model, we believe the in-depth evaluation reported in this study will guide clinical development of mHA split vaccines in humans. We characterized the innate immunity, humoral immune response, germinal center B cell formation in draining lymph nodes and antigen-specific T cell immunity. Our data shows mHA split vaccines provide better immunogenicity against phylogenetically distant influenza B viruses than QIV. The combination of these vaccines with CpG 1018 is shown to reduce morbidity and control viral infection in the lungs, which is a critical mechanism to prevent severe disease. On the other side, the combination with Addavax enhances germinal center B cell reactions and antibody production. Future clinical trials investigating these different mechanisms of protection will help to guide which type of immune response is more optimal to achieve broad and long-lasting immunity to influenza B viruses. Ultimately, the influenza B mHA vaccine will be combined with group 155,56 and group 2 cHA constructs23 into a trivalent vaccine for universal protection against influenza A and B viruses19.
Methods
Study design
This study was designed to evaluate the immunogenicity and in vivo protection of recombinant mHA constructs as universal influenza B inactivated virus vaccines in the mouse model. Different inactivated vaccine preparations and adjuvants were tested. Humoral immune responses measuring the level of antibodies targeting the conserved regions of the HA, cross-reactivity against a panel of HAs, HI antibodies, Fc mediated effector functions and antibody subclasses were evaluated in mice sera post-vaccination. Germinal center activation and T cell immunity was evaluated post-vaccination in inguinal lymph nodes and spleens, respectively. In vivo protection was tested in direct and serum passive transfer challenge experiments using three different influenza B viruses. Randomization was achieved by randomly distributing mice into different cages upon arrival. No animal subjects were excluded from the sample collection or analysis unless the sample was exhausted. This study was performed in preparation for phase I clinical trials.
Cell culture
Madin-Darby canine kidney (MDCK) cells were maintained in minimum essential medium (MEM; Gibco) supplemented with 10% (vol/vol) of heat inactivated fetal bovine serum (FBS), 100 units/mL of penicillin, 100 µg/mL of streptomycin (P/S; Gibco), 2 mM of L-glutamine (Gibco), 0.15% (w/vol) of sodium bicarbonate (Corning) and 20 mM of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, HEPES; (Gibco). Cell lines were maintained at 37 °C with 5% CO2.
Rescue of recombinant wildtype and mosaic HA influenza B viruses
Wildtype (WT) recombinant viruses contained the HA gene sequences of the B/Phuket/3073/2013, B/Yamagata/16/1988 or B/Brisbane/60/2008 strains. The mosaic HA (mHA) gene segments were designed by replacing the major antigenic sites of the HA gene sequences of B/Phuket/3073/2013, B/Yamagata/16/1988 and B/Brisbane/60/2008 with the corresponding sequences of H5 (A/Vietnam/1203/2004 H5N1-PR8-IBCDC-RG/GLP), H8 (A/mallard/Sweden/24/2002 H8N4) and H13 (A/black-headed gull/Sweden/1/1999 H13N6) to generate mH5/BPhu, mH8/BYam, and mH13/BBris, respectively. Sequences were reported in previous works17,18. The HA gene fragments were ordered as synthetic DNA gBlocks from Integrated DNA Technologies (IDT) and cloned into the pDZ plasmid17,18, and mHA and WT viruses were rescued using a reverse genetics protocol, as previously described5,17,18.
Hemagglutination inhibition (HI) assay
Mouse sera were treated with receptor destroying enzyme (RDE; Denka Seiken) to eliminate non-specific inhibitors in the sera. Briefly, serum was mixed with RDE in a 1:3 ratio (vol/vol). RDE-treated samples were incubated at 37 °C for 18–20 h and the reaction was stopped by the addition of 2.5% sodium citrate solution in a 1:3 ratio (vol/vol). The samples were then heat-treated at 56 °C for 30 min. Serum was finally diluted with sterile phosphate-buffered saline (PBS) to reach a final dilution of 1:10. To perform HI assays, virus stocks were diluted in PBS to a final HA titer of 8 HA units (4 wells of HA) per 50 μL sample. Two-fold dilutions (25 μL) of RDE-treated serum in PBS prepared in 96-well V-bottom microtiter plates (Thermo Fisher Scientific) were then combined with 25 μL of the diluted virus. The plates were incubated for 30 min at room temperature (RT) to allow HA-specific antibodies to bind to virus. Then 50 μL of a 0.5% suspension of turkey red blood cells (Lampire) were added to each well. HI titers were defined as the reciprocal of the highest dilution of serum that inhibited hemagglutination of red blood cells.
Preparation of inactivated viruses for vaccination
Production of mHA and WT influenza B virus vaccine preparations was performed in specific pathogen free embryonated chicken eggs (Charles River Laboratories) incubated at 33 °C. After a 3-day incubation, eggs were cooled to 4 °C overnight, and allantoic fluid was harvested and clarified by low-speed centrifugation using a Sorvall Legend RT Plus Refrigerated Benchtop Centrifuge (Thermo Fisher Scientific). The presence of virus in the allantoic fluid was measured by hemagglutination (HA) assay and the HA sequence was confirmed by Sanger sequencing (Genewiz). Clarified allantoic fluids were treated with formaldehyde (FA) or with beta-propiolactone (BPL). FA inactivation was performed with 0.03% (vol/vol) FA at 4 °C under continuous shaking for 72 h18. BPL inactivation was performed with the addition of 13 mM disodium phosphate (DSP) to stabilize the pH followed by 0.05% (vol/vol) BPL and rocked at 4 °C under continuous shaking for 30 min57. The BPL mixture was then placed in a 37 °C water bath for 2 h shaken every 15 min. The inactivated allantoic fluid was clarified by centrifugation at 4000 rpm for 30 min using a Sorvall Legend RT Plus Refrigerated Benchtop Centrifuge (Thermo Fisher Scientific). Clarified allantoic fluids were laid on top of a 30% sucrose cushion in NTE buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM Ethylenediamine tetraacetic acid (EDTA), pH 7.4). Ultracentrifugation in a Beckman L7-65 ultracentrifuge at 25,000 rpm for 2 h at 4 °C using a Beckman SW28 rotor (Beckman Coulter, Brea, CA, USA) was performed to pellet the viruses through the sucrose cushion while soluble egg proteins were removed. The virus pellets were re-suspended in PBS (pH 7.4). The splitting process was performed with 1% (vol/vol) Triton X-100 (Fisher Bioreagents) at room temperature (18–20 °C) under continuous shaking for 1 h. Triton X-100 was removed by mixing the split virus with Bio-beads SM-2 (BioRad) at 4 °C under continuous shaking overnight. The total protein content was determined using the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) according to the manufacturer’s protocol. HA content was measured using an in-house ELISA and densitometry in protein gels of the different vaccine preparations19,58,59.
Immunization studies
For animal immunizations, 8- to 10-week-old female BALB/c mice (Jackson Laboratories) were used for all experiments. Experiments were performed in accordance with protocols approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee (IACUC). Vaccination with QIV (Flulaval Quadrivalent, GSK) vaccinated group and an unvaccinated group (PBS) were included as controls. Vaccines were administered intramuscularly (IM) at a dose of 0.1 or 1 μg HA per mouse diluted in a total volume of 100 μL with or without 10 or 30 μg of CpG 1018 adjuvant (DynaVax), 10 μg of CpG 1018 + 50 μg of aluminum hydroxide gel (Invivogen), or 50 μL of AddaVax adjuvant (1:1 v:v, Invivogen), respectively. Vaccines were diluted in sterile PBS (GIBCO) except for the combination of the CpG 1018 and aluminum hydroxide gel 2% (alum) adjuvants, when vaccines were prepared in a saline solution composed of 20 mM Tris and 100 mM NaCl (pH = 7.5). Intranasal administration of mH8/BYam virus was performed in a total volume of 30 μL in a drop-by-drop manner distributing equal volumes between each nostril of the mice after anesthesia with a ketamine/xylazine cocktail administered intraperitoneally. PBS-vaccinated mice were included as negative controls. Vaccinations were given in 3- or 4-week intervals, unless otherwise indicated. After the final immunization, mice were euthanized at defined time points. Terminal bleed was collected using cardiac puncture method, in which mice were administered a terminal anesthesia cocktail of ketamine/xylazine at a dose of 80–100 mg/kg of ketamine and 5–10 mg/kg of xylazine intraperitoneally. Euthanasia was performed following approved IACUC protocols with CO2 followed by cervical dislocation. After euthanasia, spleen, lymph nodes and blood were collected. Spleen and lymph nodes were stored in PBS (Gibco) on ice until further processing. Blood was obtained by cardiac puncture or cheek bleeding and sera were isolated by low-speed centrifugation and were stored at −20 °C until use.
Passive transfer and challenge studies
The challenge viruses B/New York/PV01181/2018 and B/New York/PV00094/2017 were isolated by the Personalized Virology Initiative at the Icahn School of Medicine at Mount Sinai (ISMMS) and were mouse adapted in a previous work18. Mice were infected intranasally with 5 murine 50% lethal doses (5× LD50) of the mouse-adapted B/New York/PV01181/2018 virus, mouse-adapted B/New York/PV00094/2017 virus or B/Lee/1940 virus in a volume of 30 μL of sterile PBS after anesthesia with a ketamine/xylazine cocktail administered intraperitoneally. In direct challenge, mice were challenged 3-4-weeks after the second boost. For passive transfer experiments, 8- to 10-week-old female BALB/c mice (Jackson Laboratories) received 100 μL of pooled sera via intraperitoneal injection 2 h before the challenge. Animals were monitored for survival and weight loss for 14 days post-challenge and were scored dead and humanely euthanized if they lost more than 25% of their initial body weight.
Enzyme-linked immunosorbent assay (ELISA)
Recombinant HA proteins were produced using the baculovirus expression system as described previously59,60. Proteins were coated onto Immulon® 4 HBX 96-well microtiter plates (Thermo Fisher Scientific) at 2 μg/mL in 1× coating buffer (SeraCare Life Sciences Inc.) at 50 μL/well overnight at 4 °C. All plates were washed 3 times with 225 μL PBS containing 0.1% (vol/vol) Tween-20 (PBST) and 220 μL blocking solution (3% v/v goat serum, 0.5% w/v non-fat dried milk powder in PBST) was added to each well and incubated for 1 h at RT. Individual serum samples or pooled sera were serially diluted 3-fold in blocking solution followed by a 2-h incubation at RT. ELISA plates were washed 3 times with PBST and 50 μL of secondary antibody conjugated with horseradish peroxidase (HRP) was added. For total IgG quantification, a 1:3000 dilution of anti-mouse IgG (H&L) peroxidase conjugated (Cytiva) in blocking solution was added. For IgG1 and IgG2a quantification, a 1:2000 dilution in blocking solution of anti-mouse IgG1 or anti-mouse IgG2a (Abcam) was added, respectively. After 1 h, plates were washed 3 times with PBST and developed using SigmaFast o-Phenylenediamine dihydrochloride (Sigma-Aldrich) for 10 min. Reactions were stopped by adding 50 μL 3 M hydrochloric acid (HCl) and absorbance at 492 nm was determined on a plate reader. For each ELISA plate, the average plus 3 standard deviations of absorbance values of blank wells was used as a cutoff to determine endpoint titers and the area under the curve (AUC) using GraphPad Prism version 10.0.0 for Windows, (GraphPad Software, Boston, Massachusetts USA, www.graphpad.com).
Antibody dependent cell-mediated cytotoxicity (ADCC) reporter assay
White flat-bottom 96-well plates (Corning) were seeded with 2× 104 MDCK cells per well. After 24 h at 37 °C, the MDCK cells were washed once with PBS and then infected with influenza B viruses at a multiplicity of infection (MOI) of 5 for a single cycle of virus replication for one hour, then the infection media was removed and infected MDCK cells were incubated overnight at 33 °C. The next day, the culture medium was removed and 25 μL of assay buffer (Roswell Park Memorial Institute (RPMI) 1640 supplemented with 4% low-IgG FBS [Gibco]) was added to each well. Pooled mouse sera were diluted 1:2 (from a starting dilution of 1:30) in RPMI 1640 medium (Gibco) and added (25 μL per well) to the virus infected MDCK cells in duplicates for 30 min. Genetically modified Jurkat cells expressing the mouse FcγRIV with a luciferase reporter gene under transcriptional control of the nuclear factor-activated T (NFAT) cell promoter were then added to the plate at 7.5× 104 cells in 25 μL/well (Promega) and incubated for 6 hours at 37 °C. FcγRIV receptor will bind to anti-influenza antibodies and the genetically modified Jurkat cells will express luciferase through activation of NFAT promoter, At the end of the incubation time, a volume of 75 μL of Bio-Glo Luciferase assay reagent (Promega) was added to each well and luminescence was measured using a Synergy 4 microplate reader (BioTek) using Gen5 2.09 software. Fold induction was calculated as follows: (RLUinduced−RLUbackground)/(RLUuninduced−RLUbackground) and the geometric mean of AUC was calculated using GraphPad Prism version 10.0.0 for Windows, (GraphPad Software, Boston, Massachusetts USA, www.graphpad.com).
Multiplex ELISA
A Th1/Th2 cytokine panel was used with simultaneous measurements of different cytokines in serum samples taken 4 h after vaccine administration adapted from61,62 using the Th1/Th2 Cytokine 11-Plex Mouse ProcartaPlex™ Panel (Cat#EP100-20820-901, Thermo Fisher Scientic). The following cytokines were quantified: GM-CSF, IFN-γ, IL-1 beta, IL-2, IL-4, IL-5, IL-6, IL-12p70, IL-13, IL-18 and TNF-α. Serum samples were serially diluted 1:2. Cytokine levels were measured using a MAGPIX device (Luminex xMAP detection system, TX, USA) following the manufacturer’s instructions. Fold change (log 2) values over the PBS vaccination group were reported.
Spleen processing and intracellular cytokine staining
Splenocytes were resuspended in complete RPMI 1640 media (cRPMI, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% w/v FBS (Gibco), 100 U/mL of P/S (Gibco), and 2 mM L-Glutamine (Gibco). Cells were seeded in V-bottom 96-well plates (CELLSTAR, Greiner Bio-One North America Inc., Monroe, NC, USA) at an average of 2 ×106 cells/well in cRPMI media containing anti-mouse CD28 (1:500, BD), brefeldin A (1:1000, GolgiPlug™, BD Biosciences), and monensin (1:1,000, GolgiStop™, BD Biosciences). Splenocytes were stimulated with B/Florida/4/2006 Nucleoprotein and Matrix Protein 1 (NP, NR-36045 and M1, NR-36046, BEI Resources) pooled peptides at a final individual peptide concentration of 5 µg/mL at 37 °C with 5% CO2 10 h. Negative control cells were stimulated with an equivalent volume of dimethyl sulfoxide (DMSO). Positive control cells were stimulated with a cocktail containing phorbol 12-myristate 13-acetate (0.5 µg/mL, Sigma-Aldrich) and ionomycin (1 µg/mL, Sigma-Aldrich). The unstimulated control cells were only treated with the cRPMI media. After stimulation, cells were washed with PBS containing 2% FBS and centrifuged at 500 × g for 5 min and then stained with Zombie Red™ diluted in PBS (1:500, BioLegend) for 15 min at RT in the dark. Cells were washed in PBS containing 2% FBS (500 × g for 5 min) and incubated with surface staining cocktail containing Fc Block CD16/CD32 1:50 (BD) and the anti-mouse antibodies BV 711 CD3 (1:300, clone 17A2 BioLegend Cat#100241, AB_2563945), Pacific Blue CD4 (1:400, clone GK1.5 BioLegend Cat#100428 RRID: AB_493647), PerCP/Cy5.5 CD8 (1:200, clone 53-6.7 BioLegend Cat#100734 RRID: AB_2075238) for 30 min at 4 °C in FACS buffer. Cells were washed in FACS buffer and then incubated in fixation/permeabilization buffer (CytoFix, BD Biosciences) for 5 min at 4 °C. After fixation, cells were washed in 1× permeabilization buffer (CytoPerm, BD Biosciences), then incubated with the intracellular staining cocktail containing anti-mouse antibodies Alexa Fluor 647 IFN-γ (1:400, clone XMG1.2 BioLegend Cat#505814, RRID:AB_493314), Alexa Fluor 488 TNF-α (1:300, clone MP6-XT22 BioLegend Cat#506313 RRID:AB_493328), PE/Cy7 IL-2 (1:300, clone JES6-5H4, BioLegend Cat#503832 RRID:AB_2561750) in 1× permeabilization buffer for 1 h at 4 °C. Samples were then washed in 1× permeabilization buffer and resuspended in PBS buffer for acquisition. Samples were acquired on an Aurora spectral cytometer (Cytek, Fremont, CA, USA) using SpectroFlo® software (Cytek), with the relevant single fluorochrome compensation controls set by the daily acquisition of Cytometer Setup and Compensation beads (Ultracomp beads, Invitrogen). Analysis was performed with FCS Express 7 (DeNovo Software) and analyzed using the GraphPad Prism version 10.0.0 for Windows, (GraphPad Software, Boston, Massachusetts USA, www.graphpad.com). Briefly, the percentage of cytokine CD4+T or CD8+T cells was calculated, and the value of stimulated groups was subtracted from that of the unstimulated condition.
Lymph node processing and B cell studies
Inguinal lymph nodes were dispersed into single-cell suspensions by mechanical disruption through 40 μm cell strainer (Fisher Scientific) using syringe plungers into cold PBS. Cells were pelleted by centrifugation (500 × g, 5 min) and re-suspended in FACS buffer (PBS containing 1% bovine serum albumin [BSA] and 2 mM EDTA) containing Fc block anti-mouse CD16/CD32 (1:50, eBioscience) in 50 μL for 10 min at 4 °C and then stained with primary antibodies cocktail (30 min, 4 °C). The primary antibody cocktail contained (50 μL): Zombie Red (1:400, BioLegend, Cat# 423109) as the viability dye, Pacific Blue anti-mouse CD3 (1:200, clone 17A2, BioLegend, Cat# 100214, RRID: AB_493645), Alexa Fluor 700 anti-mouse/human CD45R/B220 (1:200, clone RA3-6B2, BioLegend, Cat# 103232, RRID: AB_493717), Brilliant Violet 785 anti-mouse CD19 (1:100, clone 6D5, BioLegend, Cat# 115543, RRID: AB_11218994), Brilliant Violet 605 anti-mouse IgD (1:200, clone 11–26 c.2a, BioLegend, Cat# 405727, RRID: AB_2562887), APC/Cyanie7 anti-mouse IgM (1:400, clone RMM-1, BioLegend, Cat# 406516, RRID: AB_10660305), PerCP/Cyanine 5.5 anti-MU/HU GL7 antigen (1:50, clone GL7, BioLegend, Cat# 144610, RRID: AB_2562979), and PE/Cyanine 7 anti-mouse CD38 (1:400, clone 90, BioLegend, Cat# 102718, RRID: AB_2275531). Cells were washed in FACS buffer (500 × g, 5 minutes) and then incubated in 4% methanol-free paraformaldehyde (PFA) fixation buffer for 30 min at 4 °C. Cells were then washed and resuspended in FACS buffer and kept in the dark at 4 °C before acquisition. Samples were measured on an Aurora spectral cytometer (Cytek, Fremont, CA, USA) using SpectroFlo® software (Cytek), with the relevant single fluorochrome unmixing reference controls set by the daily acquisition of Cytometer Setup and Tracking beads. Analysis was performed with FCS Express 7 (DeNovo Software) and GraphPad Prism version 10.0.0 for Windows, (GraphPad Software, Boston, Massachusetts USA, www.graphpad.com). Germinal center B cells were gated as live CD3-B220+CD19+IgD-IgM-GL7+CD38low population. The frequency of germinal center B cells out of the B220+CD19+ was calculated and graphed.
Viral lung titers
Virus titers in the lungs of mice challenged with 50× LD50 of B/Lee/1940 harvested at day 3 after challenge were analyzed by the plaque assay method in 12-well plates. Harvested lungs were homogenized in 2 disruption cycles (10 s/cycle) using tubes that contained high impact zirconium beads (Andwin Scientific) and 1 mL of PBS. MDCK cells were seeded onto 12-well plates in growth media at 4× 105 cells per well and cultured for one day. Tissue homogenates were 10-fold serially diluted in infection medium (PBS with 0.21% (w/v) BSA (MP Biomedicals), 100 unit/mL of penicillin, 100 µg/mL of streptomycin (P/S; Gibco) and 0.83 mM CaCl2 and 0.1 mM MgCl2). Two hundred microliters of each dilution were inoculated onto each well. The plates were incubated at 33 °C for 1 h with occasional rocking every 10 min. The inoculum in each well was then removed and 1 mL of agar overlay containing 0.7% of agar in 2× MEM was placed onto each well. Once the agar was solidified, the plates were incubated at 33 °C with 5% CO2. Three days later, the plates were fixed with 4% (v/v) formaldehyde in PBS for 2 h and cells were later permeabilized with 0.5% Triton X-100 (v/v) for 15 min. The plaques were immuno-stained with a mice polyclonal serum vaccinated with a Yamagata-like virus at a 1:1000 dilution in 5% (w/v) dried milk in PBS. An HRP-conjugated goat anti-mouse secondary antibody was used at 1:2000 dilution in 5% (w/v) dried milk in PBS and the plaques were visualized using TrueBlueTM Peroxidase Substrate (SeraCare Life Sciences Inc.). Virus titers are presented as the log10 plaque forming units (PFU) per mL.
Phylogenetic tree
Phylogenetic tree of HA sequences was performed from the historical annual formulation sequences for IBV vaccine strains from 1999 to 2023 (Data taken from Global Influenza Programme (who.int) as of 1st Aug 2023 and reported in Supplementary Fig. S1). The phylogenetic tree was constructed using the Maximum Likelihood method and Tamura-Nei model and was visualized through Mega1163,64. Sequences were searched in the Influenza Research Database. The phylogenetic tree was constructed using raxML treeAlgorithm (https://www.fludb.org/)65 and was visualized through FigTree66.
Statistics
Normality/lognormality test, unpaired one-tailed t test, Kruskal–Wallis test corrected using Dunn’s test and two-way ANOVA test corrected using Dunnet’s test for group comparison were performed using GraphPad Prism version 10.0.0 for Windows (GraphPad Software, Boston, Massachusetts USA, www.graphpad.com). T-test data passed a normality/lognormality.
Supplementary information
Acknowledgements
We thank Dynavax Technologies for providing the CpG 1018 adjuvant. We would like to thank Jill Gregory for her help with the data visualization. We thank the Flow Cytometry Core at ISMMS for their assistance in flow cytometry analyses. The assistance of Dr. Shirin Strohmeier and previous members of F.K. laboratory in the recombinant HA production is greatly appreciated. We also thank Viviana Simon and Harm van Bakel and the Mount Sinai Pathogens Surveillance Program for the initial isolation of the two New York influenza B virus strains. This work was partially funded by NIH (Centers of Excellence for influenza Research and Response, CEIRR,75N93021C00014, P.P., F.K.), (NIAID grant P01 AI097092-07, P.P.), (NIAID grant R01 AI145870-03, PP) and by the Collaborative Influenza Vaccine Innovation centers (CIVICs) contract 75N93019C00051 (P.P., F.K.), Collaborative Influenza Vaccine Innovation centers (CIVICs) contract 75N93019C00051 Option 17E (W.S.). E.P.M. was supported by a postdoctoral fellowship from Fundación Ramón Areces.
Author contributions
W.S. and P.P. initiated this study. W.S., P.P. and I.G.D. designed the experiments. I.G.D., E.P.M., A.A., T.Y.L., Y.L., N.L., M.B., V.D., S.S. performed the experiments. I.G.D. and W.S. analyzed the data. M.L., B.F., E.P.M., J.L.M.G. and F.K. provided protein and virus reagents. I.G.D., W.S. and P.P. wrote the manuscript. All authors have read and approved the manuscript.
Data availability
Raw data can be found in ImmPort under the identifiers: SDY2873.
Competing interests
The Icahn School of Medicine at Mount Sinai and Dynavax have filed a patent application in which P.P., W.S., I.G.D. and F.K. are listed as co-inventors. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays, NDV-based SARS-CoV-2 vaccines, influenza virus vaccines and influenza virus therapeutics which list F.K. as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2 and another company, Castlevax, to develop SARS-CoV-2 vaccines. F.K., P.P. and W.S. are co-founder of Castlevax. F.K. has consulted for Merck, Curevac, Seqirus and Pfizer and is currently consulting for 3rd Rock Ventures, GSK, Gritstone and Avimex. The Krammer laboratory is also collaborating with Dynavax on influenza A virus vaccine development. All other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Irene González-Domínguez, Email: irene.gonzalez@mssm.edu.
Peter Palese, Email: peter.palese@mssm.edu.
Weina Sun, Email: weina.sun@mssm.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-01014-8.
References
- 1.Krammer, F. et al. Influenza. Nat. Rev. Dis. Prim.4, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. https://www.cdc.gov/flu/index.htm (2023).
- 3.Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol Rev.56, 152–179 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannoun, C. The evolving history of influenza viruses and influenza vaccines. Expert Rev. Vaccines12, 1085–1094 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Fulton, B. O., Sun, W., Heaton, N. S. & Palese, P. The Influenza B Virus Hemagglutinin Head Domain Is Less Tolerant to Transposon Mutagenesis than That of the Influenza A Virus. J. Virol. 92, 10.1128/JVI.00754-18 (2018). [DOI] [PMC free article] [PubMed]
- 6.Koutsakos, M. & Kent, S. J. Influenza B viruses: underestimated and overlooked. Microbiol. Aust.42, 110–115 (2021). [Google Scholar]
- 7.Koutsakos, M., Nguyen, T. H., Barclay, W. S. & Kedzierska, K. Knowns and unknowns of influenza B viruses. Future Microbiol.11, 119–135 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Molinari, N. A. et al. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine25, 5086–5096 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Dijkstra, F., Donker, G. A., Wilbrink, B., Van Gageldonk-Lafeber, A. B. & Van Der Sande, M. A. Long time trends in influenza-like illness and associated determinants in The Netherlands. Epidemiol. Infect.137, 473–479 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Terho, H., Niina, I. & Ziegler, T. Impact of Influenza B Lineage-Level Mismatch Between Trivalent Seasonal Influenza Vaccines and Circulating Viruses, 1999–2012. Clin. Infect. Dis.59, 1519–1524 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Herman, K. et al. Surveillance of Influenza in Indonesia, 2003–2007. Influenza Other Resp.7, 313–320 (2012). [Google Scholar]
- 12.Paul, A. R. et al. Cocirculation of Two Distinct Evolutionary Lineages of Influenza Type B Virus since 1983. Virology175, 59–68 (1990). [DOI] [PubMed] [Google Scholar]
- 13.WHO. Global Influenza Programme, https://www.who.int/tools/flunet (2024).
- 14.Weina, S. et al. Antibody Responses toward the Major Antigenic Sites of Influenza B Virus Hemagglutinin in Mice, Ferrets, and Humans. J. Virol.93, e01673–01618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhirnov, O. P. & Klenk, H. D. Influenza A virus proteins NS1 and hemagglutinin along with M2 are involved in stimulation of autophagy in infected cells. J. Virol.87, 13107–13114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qinghua, W., Feng, C., Mingyang, L., Xia, T. & Ma, J. Crystal Structure of Unliganded Influenza B Virus Hemagglutinin. J. Virol.82, 3011–3020 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun, W. et al. Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol.93, 10.1128/JVI.00333-19 (2019). [DOI] [PMC free article] [PubMed]
- 18.Liu, Y. et al. Mosaic Hemagglutinin-Based Whole Inactivated Virus Vaccines Induce Broad Protection Against Influenza B Virus Challenge in Mice. Front. Immunol.12, 746447 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puente-Massaguer, E. et al. Bioprocess development for universal influenza vaccines based on inactivated split chimeric and mosaic hemagglutinin viruses. Front Bioeng. Biotechnol.11, 1097349 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sparrow, E. et al. Global production capacity of seasonal and pandemic influenza vaccines in 2019. Vaccine39, 512–520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vieira, M. C. et al. Lineage-specific protection and immune imprinting shape the age distributions of influenza B cases. Nat. Commun.12, 4313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edler, P. et al. Differential cross-reactivity to the influenza B virus haemagglutinin underpins lineage-specific susceptibility between birth cohorts, 10.1101/2023.08.25.554879 (2023).
- 23.Puente-Massaguer, E. et al. Chimeric hemagglutinin split vaccines elicit broadly cross-reactive antibodies and protection against group 2 influenza viruses in mice. Sci. Adv.9, eadi4753 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samuele, C. et al. The adjuvant effect of MF59 is due to the oil-in-water emulsion formulation,none of the individual components induce a comparable adjuvant effect. Vaccine31, 3363–3369 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Liu, S. T. H. et al. Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J. Clin. Invest128, 4992–4996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broecker, F. et al. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines4, 31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Vries, R. D. et al. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine35, 238–247 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ermler, M. E. et al. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 91, 10.1128/JVI.00286-17 (2017). [DOI] [PMC free article] [PubMed]
- 29.He, W. et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Natl Acad. Sci. USA113, 11931–11936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen, H. et al. Influenza Virus Hemagglutinin Stalk-Specific Antibodies in Human Serum are a Surrogate Marker for In Vivo Protection in a Serum Transfer Mouse Challenge Model. mBio8, 10.1128/mBio.01463-17 (2017). [DOI] [PMC free article] [PubMed]
- 31.Jegaskanda, S., Reading, P. C. & Kent, S. J. Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine. J. Immunol.193, 469–475 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Von Holle, T. A. & Moody, M. A. Influenza and Antibody-Dependent Cellular Cytotoxicity. Front Immunol.10, 1457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox, R. J., Brokstad, K. A. & Ogra, P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol.59, 1–15 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Hovden, A. O., Cox, R. J., Madhun, A. & Haaheim, L. R. Two doses of parenterally administered split influenza virus vaccine elicited high serum IgG concentrations which effectively limited viral shedding upon challenge in mice. Scand. J. Immunol.62, 342–352 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Lofano, G. et al. Oil-in-Water Emulsion MF59 Increases Germinal Center B Cell Differentiation and Persistence in Response to Vaccination. J. Immunol.195, 1617–1627 (2015). [DOI] [PubMed] [Google Scholar]
- 36.O’Hagan, D. T., van der Most, R., Lodaya, R. N., Coccia, M. & Lofano, G. “World in motion” - emulsion adjuvants rising to meet the pandemic challenges. NPJ Vaccines6, 158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sridhar, S. et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med19, 1305–1312 (2013). [DOI] [PubMed] [Google Scholar]
- 38.van de Sandt, C. E. et al. Influenza B virus-specific CD8 + T-lymphocytes strongly cross-react with viruses of the opposing influenza B lineage. J. Gen. Virol.96, 2061–2073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, L. et al. Immunodominant CD4 + T-cell responses to influenza A virus in healthy individuals focus on matrix 1 and nucleoprotein. J. Virol.88, 11760–11773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bravo, L. et al. Efficacy of the adjuvanted subunit protein COVID-19 vaccine, SCB-2019: a phase 2 and 3 multicentre, double-blind, randomised, placebo-controlled trial. Lancet399, 461–472 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richmond, P. et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. Lancet397, 682–694 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smolenov, I. et al. Impact of previous exposure to SARS-CoV-2 and of S-Trimer (SCB-2019) COVID-19 vaccination on the risk of reinfection: a randomised, double-blinded, placebo-controlled, phase 2 and 3 trial. Lancet Infect. Dis.22, 990–1001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, W. et al. Antibody Responses toward the Major Antigenic Sites of Influenza B Virus Hemagglutinin in Mice, Ferrets, and Humans. J. Virol.93, 10.1128/JVI.01673-18 (2019). [DOI] [PMC free article] [PubMed]
- 44.FDA. Vaccines Licensed for Use in the United States, https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states (2023).
- 45.Roubidoux, E. K. & Schultz-Cherry, S. Animal Models Utilized for the Development of Influenza Virus Vaccines. Vaccines (Basel)9, 10.3390/vaccines9070787 (2021). [DOI] [PMC free article] [PubMed]
- 46.Krammer, F. & Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov.14, 167–182 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Kon, T. C. et al. Influenza Vaccine Manufacturing: Effect of Inactivation, Splitting and Site of Manufacturing. Comparison of Influenza Vaccine Production Processes. PLoS One11, e0150700 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeisy-Scott, V. et al. TLR7 recognition is dispensable for influenza virus A infection but important for the induction of hemagglutinin-specific antibodies in response to the 2009 pandemic split vaccine in mice. J. Virol.86, 10988–10998 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, A. J. et al. Toll-like receptor 7 agonist imiquimod in combination with influenza vaccine expedites and augments humoral immune responses against influenza A(H1N1)pdm09 virus infection in BALB/c mice. Clin. Vaccin. Immunol.21, 570–579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edler, P. et al. Immune imprinting in early life shapes cross-reactivity to influenza B virus haemagglutinin. Nat. Microbiol.10.1038/s41564-024-01732-8 (2024). [DOI] [PubMed] [Google Scholar]
- 51.Jangra, S. et al. RIG-I and TLR-7/8 agonists as combination adjuvant shapes unique antibody and cellular vaccine responses to seasonal influenza vaccine. Front. Immunol.13, 974016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, Z., Zhao, Y., Li, Y. & Chen, X. Adjuvantation of Influenza Vaccines to Induce Cross-Protective Immunity. Vaccines (Basel)9, 10.3390/vaccines9020075 (2021). [DOI] [PMC free article] [PubMed]
- 53.Spellberg, B. & Edwards, J. E. Jr Type 1/Type 2 immunity in infectious diseases. Clin. Infect. Dis.32, 76–102 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Monto, A. S. & Sullivan, K. M. Acute respiratory illness in the community. Frequency of illness and the agents involved. Epidemiol. Infect.110, 145–160 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nachbagauer, R. et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med27, 106–114 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Nachbagauer, R. et al. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines1. 10.1038/npjvaccines.2016.15 (2016). [DOI] [PMC free article] [PubMed]
- 57.Sun, W. et al. A Newcastle Disease Virus (NDV) Expressing a Membrane-Anchored Spike as a Cost-Effective Inactivated SARS-CoV-2 Vaccine. Vaccines8, 10.3390/vaccines8040771 (2020). [DOI] [PMC free article] [PubMed]
- 58.Guha Asthagiri, A. et al. Broadly Cross-Reactive, Nonneutralizing Antibodies against Influenza B Virus Hemagglutinin Demonstrate Effector Function-Dependent Protection against Lethal Viral Challenge in Mice. J. Virol.93, e01696–01618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirkpatrick, E. et al. Characterization of Novel Cross-Reactive Influenza B Virus Hemagglutinin Head Specific Antibodies That Lack Hemagglutination Inhibition Activity. J. Virol.94, 10.1128/JVI.01185-20 (2020). [DOI] [PMC free article] [PubMed]
- 60.Margine, I., Palese, P. & Krammer, F. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp., e51112 10.3791/51112 (2013). [DOI] [PMC free article] [PubMed]
- 61.McKay, P. F. et al. Identification of potential biomarkers of vaccine inflammation in mice. Elife8, 10.7554/eLife.46149 (2019). [DOI] [PMC free article] [PubMed]
- 62.Jangra, S. et al. A Combination Adjuvant for the Induction of Potent Antiviral Immune Responses for a Recombinant SARS-CoV-2 Protein Vaccine. Front Immunol.12, 729189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol.38, 3022–3027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol.10, 512–526 (1993). [DOI] [PubMed] [Google Scholar]
- 65.Han, M. V. & Zmasek, C. M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinforma.10, 356, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stamatakis, A. Using RAxML to Infer Phylogenies. Curr. Protoc. Bioinforma.51, 6 14 11–16 14 14, (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data can be found in ImmPort under the identifiers: SDY2873.