Abstract
Once considered unconventional cellular structures, membraneless organelles (MLOs), cellular substructures involved in biological processes or pathways under physiological conditions, have emerged as central players in cellular dynamics and function. MLOs can be formed through liquid-liquid phase separation (LLPS), resulting in the creation of condensates. From neurodegenerative disorders, cardiovascular diseases, aging, and metabolism to cancer, the influence of MLOs on human health and disease extends widely. This review discusses the underlying mechanisms of LLPS, the biophysical properties that drive MLO formation, and their implications for cellular function. We highlight recent advances in understanding how the physicochemical environment, molecular interactions, and post-translational modifications regulate LLPS and MLO dynamics. This review offers an overview of the discovery and current understanding of MLOs and biomolecular condensate in physiological conditions and diseases. This article aims to deliver the latest insights on MLOs and LLPS by analyzing current research, highlighting their critical role in cellular organization. The discussion also covers the role of membrane-associated condensates in cell signaling, including those involving T-cell receptors, stress granules linked to lysosomes, and biomolecular condensates within the Golgi apparatus. Additionally, the potential of targeting LLPS in clinical settings is explored, highlighting promising avenues for future research and therapeutic interventions.
Subject terms: Cell biology, Molecular biology
Introduction
Eukaryotic cells are equipped with two distinct types of organelles: membrane-bound organelles, which encompass the nucleus, endoplasmic reticulum, synaptic vesicles, mitochondria, lysosomes, etc., and MLOs like stress granules (SGs), processing bodies (P-bodies), nucleolus, and Cajal bodies (Fig. 1).1,2
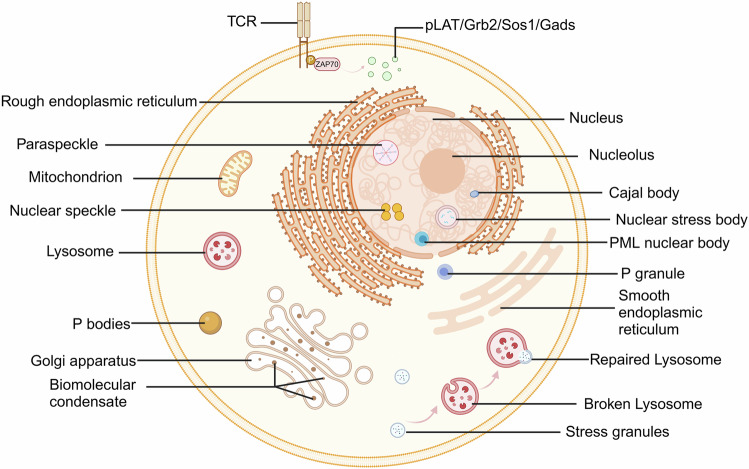
Fig. 1.
The localization of membraneless organelles. The cells contain membrane-bound organelles as well as MLOs such as Nucleolus, P granules, Paraspeckles, Stress Granules, Processing Bodies, Cajal Bodies, and Nuclear Speckles. Condensates associated with membranes include T-cell receptor, stress granules associated with lysosomes, and biomolecular condensates within the Golgi apparatus
Membrane-bound organelles facilitate organized biochemical reactions and regulatory processes and protect the cells by acting as barriers against harmful substances. For example, apoptosis can be triggered by releasing cytochrome c into the cytoplasm, while the release of nucleic acids activates innate immune pathways. In contrast, MLOs are cellular compartments lacking a surrounding lipid membrane yet exhibit distinctive organization within the cell. These structures are characterized by the dynamic assembly and disassembly of proteins and nucleic acids, creating specialized microenvironments. Unlike traditional membrane-bound organelles, most MLOs are characterized by their ability to undergo liquid-liquid phase separation (LLPS).3–6 MLOs engage in the exchange of various molecular substances with the surrounding environment, selectively concentrating specific substrates or enzymes to expedite specific biochemical reactions.7 Through phase separation, membraneless particles can temporarily store surplus biological macromolecules and even organelles, facilitating quick mobilization without the need for synthesis. An example of an MLO is the nucleolus, a distinct structure found in the nucleus. The nucleolus participates in ribosome biogenesis, with its assembly driven by the LLPS of nucleolar proteins and RNA molecules.8 Studies have shown that specific protein-protein and protein-RNA interactions govern the formation of nucleoli, highlighting the role of non-covalent interactions in the organization of MLOs.9,10 Another well-known MLO is the SGs, which form in response to cellular stress.11 Composed of RNA, proteins, and other biomolecules, SGs aid in the temporary storage of mRNA during stress conditions. This blocks the translation of unnecessary proteins, enabling the cell to prioritize its response to stress.11,12
The groundbreaking study conducted by Brangwynne et al. uncovered that cell function can be influenced through LLPS.13 Their research illustrated that the specification of germ cells involves the movement and condensation of P granules, which contain RNA and RNA-binding proteins (RBPs) and are evenly distributed in the unpolarized one-cell embryo. The relocation of P granules to the posterior half of the cell was linked to cell division. Notably, P granules exhibit liquid-like behavior, experiencing rapid dissolution/condensation and possessing the ability to fuse.13 Three years later, Rosen’s team established that small multivalent proteins can associate and form large gel-like complexes through LLPS.14 Within the same timeframe, McKnight’s group demonstrated that RNA granules are formed by ribonucleoproteins (RNPs) with LCR, facilitating a reversible process of phase transition.15
LLPS emerges as a possible mechanism involved in the formation of MLOs.7 This process entails the aggregation of molecules in a solution due to intermolecular interactions. Within living cells, only specific sets of proteins can undergo LLPS. Banani et al.16 proposed the subdivision of the membraneless particle component into scaffold and client. In the cellular context, a distinct group of biomolecules, known as scaffolds, can form condensates through multivalent interactions, typically involving repeating domains. Once the scaffold is established, it attracts other molecules, referred to as clients, which become bound to it. Certain proteins achieve LLPS through intrinsically disordered regions (IDRs),17–21 characterized by the presence of low-complexity sequence regions (LCRs) containing particular amino acids in high frequency (Table 1).22–31 One example is the prion-like LCRs,32 which exhibit chaperone-like functions that protect proteins from proteotoxic damage by regulating protein phase behavior.33 These LCRs are frequently found in proteins linked to neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS).34,35 The distribution of LCR sequences is not random but enriched in RNA- and DNA-binding proteins associated with transcription and translation.14 The low-complexity domains (LCDs) are typically not found in isolation and are usually connected to folded domains, either as tails or internal linkers between folded domains.36 Multi-domain proteins that contain one or more folded domains along with linker and disordered regions are ideal candidates for modulating LLPS.37 Martin et al. studied the interaction between the folded and disordered domains of the RNA-binding protein hnRNPA1.38 They discovered that the folded RNA recognition motifs (RRMs) have a higher fraction of charged residues compared to the LCD. Electrostatic interactions between the folded domains and the LCD of hnRNPA1 contribute to phase separation at low ionic strengths.38 These interactions are disrupted at high ionic strengths, where the folded domains help to solubilize the LCD.38 Phase separation can also occur when the proline-rich motif (PRM) ligand binds to its Src homology 3 (SH3) domain.39 Both IDRs and SH3/PRM interactions are multivalent, meaning phase separation relies on interactions between multiple domains.7 Biogenesis of MLOs often begins with the nucleation of proteins or nucleic acids, which act as seeds for condensate formation.40,41 Nucleation can be facilitated by various factors,42 which may occur spontaneously or be triggered by specific cellular signals or stimuli.43 Once nucleation occurs, biomolecules undergo LLPS, resulting in the formation of a dense, coalesced phase within the cytoplasm or nucleoplasm.44–46 However, condensates do not need to be fluid, and not all condensates are MLOs.
Table 1.
Database to predict LLPS formation
| Database | Function | Website | Reference |
|---|---|---|---|
| D2P2 | Prediction of disorder proteins | http://d2p2.pro | 22 |
| DrLLPS | Provide annotations of known and computationally detected LLPS-associated proteins, including IDR, post-translational modification, disease-associated information, etc. | http://llps.biocuckoo.cn/ | 23 |
| LLPSDB | Provide protein sequence, modifications on specific amino acids, ability of coalescing with nucleic acid, phase behavior, experimental conditions | http://bio-comp.org.cn/llpsdb | 24 |
| MloDisDB | Provide information on LLPS and related diseases | http://mlodis.phasep.pro/ | 25 |
| PhaSepDB | Provide annotation for phase separation (PS) entries, including the material states of the PS droplet, verification experiments, PS partners, mutations, modification, etc. | http://db.phasep.pro/ | 26 |
| PhaSePred | Provides self-assembling and partner-dependent phase-separating protein prediction, and integrates scores from several PS-related predicting tools. | http://predict.phasep.pro | 27 |
| PhaSePro | Provides information on the biophysical driving force, biological function and regulation of LLPS. | https://phasepro.elte.hu | 28 |
| Pi-Pi predictor | Predicts LLPS formation in a given sequence based on π-π interaction. | 10.7554/eLife.31486.021 | 30 |
| PLAAC | Search protein sequences to identify the prion-like domains | http://plaac.wi.mit.edu/ | 29 |
| PSPredictor | A sequence-based tool for the prediction of proteins with LLPS potential | http://www.pkumdl.cn/PSPredictor | 31 |
In this review, we discuss the regulation of MLOs as a dynamic and multifaceted process that involves a combination of molecular interactions, signaling pathways, and cellular responses. We also discuss how dysregulation of these regulatory mechanisms can lead to aberrant condensate formation and contribute to various cellular dysfunctions and disease states.
The mechanism and validation of LLPS
The mechanism of LLPS
Phase separation is driven by weak, multivalent interactions between molecules, such as protein-protein interactions, protein-RNA interactions, or RNA-RNA interactions (Fig. 2).47,48 Following phase separation, the condensates undergo maturation and growth through the recruitment of additional biomolecules and the coalescence of small condensates into larger complexes.49,50 This process often involves specific interactions among proteins and RNAs that stabilize the condensate and contribute to its structural integrity. Maturation may also involve the incorporation of additional components to regulate condensate dynamics and function. MLOs exhibit dynamic properties, including fusion, fission, and exchange of components with the surrounding environment. Cellular signals or changes in the cellular environment can regulate the assembly, disassembly, or remodeling of MLOs in response to specific physiological cues or stress conditions. The dissolution of MLOs may occur through the reversal of phase separation, where the weak interactions holding the condensate together are disrupted.51 Dissolution can also be actively regulated by cellular processes, such as protein degradation, or by changes in cellular conditions that alter the stability or composition of the condensate.52–54 Overall, the biogenesis of MLOs is a highly dynamic and precisely regulated process that involves the self-assembly, phase separation, and maturation of biomolecules to form functional and dynamic structures within cells.
Fig. 2.
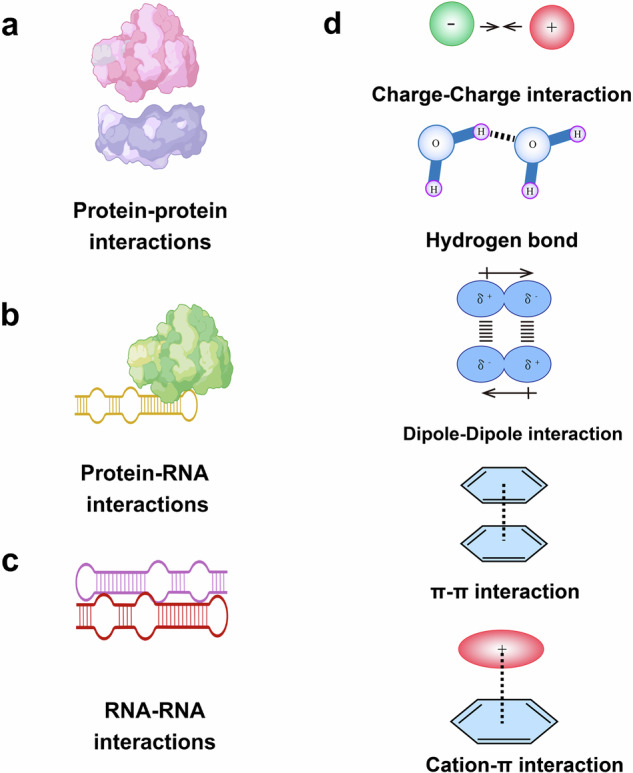
Multivalent interactions involved in phase separation. a Protein-protein interactions. b Protein-RNA interactions. c RNA-RNA interactions. d Multivalent interactions between IDRs include charge-charge interaction, hydrogen bond, Dipole-Dipole interaction, π–π stacking, and cation–π interaction
The stickers-and-spacers polymer framework describes the multivalent homotypic protein-protein and heterotypic protein-RNA interactions driving biomolecular condensation.55–59 In IDRs, residues that enable inter-chain attractive interactions include arginine (R) in R/G-rich IDRs and tyrosine (Y) in prion-like IDRs,60 commonly referred to as stickers. The linker residues connecting these stickers are known as spacers. The patterning of stickers and spacers can influence the physical properties of condensates and their phase behavior.60,61
The stickers are regions in the disordered protein that drive its compaction, making them crucial for the interactions leading to phase separation.60 Using nuclear magnetic resonance spectroscopy and small-angle X-ray scattering, aromatic residues were identified as the stickers in the protein hnRNAP1. These stickers alone are sufficient to explain phase behavior. Additionally, the arrangement of stickers (their patterning and the spacers between them) is necessary for functional LLPS. Clustering of stickers in the sequence led to the formation of solid aggregates rather than liquid droplets.60
The primary components of MLOs are proteins and nucleic acids, although other molecules such as lipids and metabolites may also contribute.62–64 These molecules interact via various non-covalent interactions, including electrostatic interactions, hydrogen bond, π–π, and dipole-dipole interactions (Fig. 2).65 RNA molecules, particularly long noncoding RNAs (lncRNAs) and ribosomal RNAs (rRNAs), are often found within MLOs (Fig. 3). RNA molecules can act as scaffolds or contribute to the material properties of the organelles.66 DNA can also be present, especially in the context of transcriptional condensates. MLOs serve diverse functions within cells, including the spatial and temporal organization of biological processes such as responses to stress, transcription and translation.7,67,68 By concentrating reactants and enzymes, MLOs can accelerate reaction rates and regulate reaction specificity. MLOs help spatially organize cellular components and play crucial roles in oxidative stress, heat shock, and nutrient deficiency.69 They can dynamically assemble to sequester and protect essential molecules, prevent the aggregation of misfolded proteins or facilitate the degradation of damaged components. Certain MLOs, such as transcriptional condensates, facilitate gene expression.70 They concentrate transcription factors, RNA polymerases, and regulatory RNAs to control the transcription of specific genes in response to developmental cues or environmental signals (Fig. 3).51,71,72 By bringing together signaling molecules and effectors, they facilitate the efficient propagation and regulation of cellular signals.73
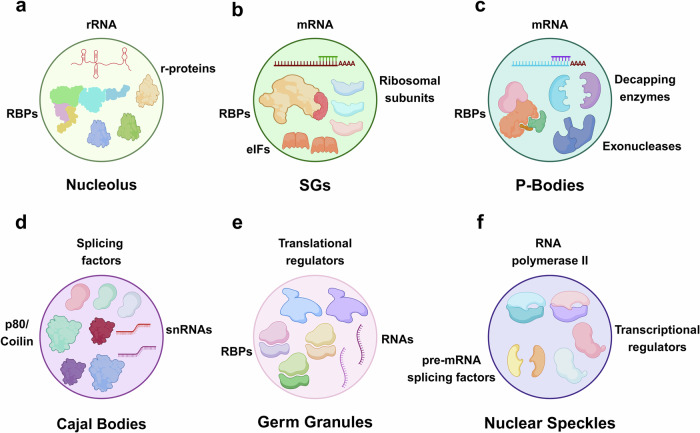
Fig. 3.
Molecular composition of various MLOs. a Nucleolus contains RBP: RNA-binding proteins (RBP), ribosomal RNA and proteins (rRNA and r-proteins). b SGs contain RBP, mRNA, ribosomal subunits, eIFs. c P-Bodies contain mRNA, decapping enzymes, RBP, exonucleases. d Cajal Bodies contain small nuclear RNAs (snRNAs), splicing factors and p80/coilin. e Germ Granules RNAs, RBPs, and translational regulators. f Nuclear Speckles contain pre-mRNA splicing factors, RNA polymerase II, and transcriptional regulators
RNA molecules, including lncRNAs and specific mRNAs, can regulate the formation and properties of MLOs.74 RNA molecules can act as scaffolds, regulators, or structural components within condensates, influencing their assembly and function.75 RNA regulation modulates the composition, stability, and activity of MLOs, impacting processes such as RNA metabolism, translation, and RNA-based regulation of gene expression.
Conceptualizing the dynamic assembly of mRNAs into RNA granules has been challenging. To address this issue, Han et al. simulated the formation of these granules by treating mouse brain extracts and human cell lysates with biotinylated isoxazole (b-isox).76 Deep sequencing of the associated RNAs revealed an enrichment of mRNAs known to be recruited to neuronal granules, which are used for dendritic transport and localized translation at synapses. The precipitated mRNAs contained extended 3′UTR sequences and were enriched in binding sites for known granule-associated proteins. The mRNAs enriched by b-isox precipitation had an average 3′UTR length roughly five times longer than those excluded from the precipitate. They demonstrated that hydrogels composed of the low-complexity (LC) sequence domain of FUS recruited and retained the same mRNAs selectively precipitated by the b-isox chemical. Interestingly, phosphorylation of the LC domain of FUS prevented hydrogel retention.
Protein-protein interactions and the composition of MLOs are tightly regulated by specific factors, including chaperones, binding partners, and regulatory proteins. Competitive binding and allosteric regulation influence the recruitment and retention of proteins within condensates.77,78 Regulation of protein interactions and composition modulates the structural integrity, function, and adaptability of MLOs, influencing cellular processes such as metabolism, signaling, and stress responses.
Cellular signals and stress conditions regulate the formation, dissolution, or remodeling of MLOs in response to specific physiological cues. Signaling pathways, such as those mediated by kinases, phosphatases, and transcription factors, can directly modulate the properties of condensates.79,80 Regulation by cellular signaling and stress responses allows MLOs to dynamically adapt to changing cellular environments, modulating processes such as gene expression, SG formation, and cellular homeostasis.
Molecular chaperones and quality control mechanisms regulate the maturation, stability, and turnover of proteins within MLOs. Chaperones facilitate protein folding, prevent protein aggregation, and promote the degradation of misfolded or damaged proteins within condensates.81,82 Chaperone-mediated regulation ensures the proper assembly, function, and maintenance of MLOs, preventing aberrant condensate formation and maintaining cellular proteostasis.
Using passive microrheology with optical tweezers (pMOT), Alshareedah et al. quantified the viscoelastic properties of a series of artificial condensates formed by disordered sticker-spacer polypeptides and RNA.83 They demonstrated that at shorter timescales, peptide-RNA condensates exhibit an elastically dominant rheological response, whereas at longer timescales, these condensates behave as predominantly viscous liquids.84 The network relaxation time, or the timescale at which the condensate transitions from an elastically dominant to a viscous behavior, is determined by the chemical identities of the sticker and spacer residues in the polypeptide chain.84 Additionally, they showed that the viscous and elastic regimes of these condensates can be tuned by the sequences of the polypeptides and RNA, as well as their mixture compositions.84
It is theoretically conceivable that the fluidity and stiffness of a network can be encoded by the sequence composition and sticker identity in a polypeptide chain.85 Recent studies have shown that biomolecular condensates are network fluids with variable viscoelastic properties.10,45,86,87 This viscoelasticity is likely due to transient network-like structures formed through physical crosslinking among protein and/or RNA chains with finite bond lifetimes.45 Consequently, there is growing interest in using experimental methods to probe the material properties of condensates across different timescales.
RNA-binding proteins such as FUS, TDP43, and hnRNPA1 form dynamic liquid-like condensates that can transition to a solid state over time, a process known as maturation or aging of condensates. This transition can result in pathological aggregates.34,35,88,89 It is now widely accepted that this transition is associated with the viscoelastic properties of these proteins.90–92
Traffic into and out of the nucleus occurs through nuclear pore complexes that span the two membrane bilayers of the nuclear envelope. Small molecules and ions passively diffuse through these pores, while larger molecules are restricted from entry. The transport of large macromolecules requires binding to nuclear transport receptors (NTRs). The nuclear pore complex is mainly composed of proteins known as nucleoporins (Nups), some of which contain IDRs. These disordered proteins, called FG nucleoporins (FG-Nups), have multiple phenylalanine–glycine repeats (FG repeats) in their amino acid sequences.93 It is now understood that mixtures of FG-domains and NTRs undergo phase separation to facilitate passage of NTR carried cargos through the nuclear pore. This process involves a competitive disruption of adjacent inter-repeat contacts, transiently opening adjoining meshes.94 NTRs may act as crosslinkers between FG-domains, converting them into elastic and reversible hydrogels.
There are also condensates that are membrane-associated, including T-cell receptor, stress granules associated with lysosomes, and biomolecular condensates within the Golgi apparatus (Fig. 1). Activation of various cell surface receptors leads to the reorganization of downstream signaling molecules into micrometer- or submicrometer-sized clusters. The functional implications of such clustering have remained unclear. Su et al. biochemically reconstituted a 12-component signaling pathway on model membranes, starting with T-cell receptor (TCR) activation and culminating in actin assembly. Upon TCR phosphorylation, downstream signaling proteins spontaneously formed liquid-like clusters that enhanced signaling outputs both in vitro and in human Jurkat T cells. These reconstituted clusters were rich in kinases but excluded phosphatases, thereby promoting actin filament assembly by recruiting and organizing actin regulators. These findings illustrate that protein phase separation can create distinct physical and biochemical compartments that facilitate signaling.95
Bussi et al. explored mechanisms of lysosomal membrane repair. Using super-resolution microscopy on human stem cell-derived macrophages, they observed that chemically induced lysosomal rupture led to the formation of G3BP-positive granules in a plug-like pattern at the sites of membrane damage. Their findings revealed that stress granules nucleate near damaged endolysosomes, acting as protective plugs that stabilize the ruptured membrane and facilitate efficient repair.96
Extensive studies have revealed a wide variety of nuclear and cytosolic MLOs. However, there is a growing interest in protein condensates associated with membranes of the secretory pathway, such as the endoplasmic reticulum and the Golgi apparatus. The Golgi apparatus is essential for protein sorting and lipid metabolism, characterized by its stacked, flattened cisternal structure and distinct polarity with cis- and trans-faces that coordinate various protein maturation and transport processes. Central to its structural integrity and organization are the Golgi Matrix Proteins (GMPs), mainly composed of Golgins and GRASPs. These proteins contribute to the unique stacked and polarized structure of the Golgi, ensuring the precise localization of Golgi-resident enzymes crucial for accurate protein processing. Research has shown that GMPs across different eukaryotic lineages have a significant tendency to form biomolecular condensates. Rebane et al. demonstrated that GM130, a member of the Golgin family, can form droplets with internal component mobility in vitro.97 Furthermore, when overexpressed in cells, GM130 exhibited dynamic condensates in the nucleus.15 Using optical and fluorescence microscopy, Mendes et al. observed the formation of protein-rich, round-shaped condensates of GRASP55.98
Neuronal transmission depends on the regulated release of neurotransmitters, which are stored in synaptic vesicles (SVs). These SVs are highly mobile, allowing them to be quickly recruited to the plasma membrane for rapid release during neuronal activity. At synapses, SVs form tight clusters, acting as a reservoir from which they are drawn for exocytosis during sustained activity. Synapsin, a family of proteins, is a major component of the matrix connecting SVs and has long been implicated in regulating neurotransmitter release at synapses. Milovanovic et al. discovered that synapsin can form a distinct liquid phase in an aqueous environment.99 Hoffmann et al., using two-color SMT and super-resolution imaging in living axons, demonstrated that synapsin 1 drives the accumulation of SVs in boutons. They found that synapsin 1 condensation is sufficient to ensure the reliable confinement and motility of SVs in vivo.100
Golgins are a plentiful class of peripheral membrane proteins found in the Golgi apparatus. Rebane et al. demonstrated that overexpression of GM130, the most abundant Golgin at the cis Golgi, leads to the formation of liquid droplets in cells. This behavior is similar to that observed in many intrinsically disordered proteins with low-complexity sequences, even though GM130 itself is neither low in complexity nor intrinsically disordered.97 A subsequent study by the same group revealed that other members of the Golgin family, including golgin160, GMAP210, golgin97, golgin245, GCC88, and GCC185, also form condensates when overexpressed.101
A prominent feature in the oocytes of many diverse organisms is the Balbiani body (Bb).102 The Bb is a non-membrane-bound compartment that, in addition to localized RNAs and proteins, contains a high number of membrane-bound organelles such as mitochondria and endoplasmic reticulum. Velo1 has been identified as the most enriched protein in the Xenopus Bb that is not part of the membranous organelles. When Velo1-GFP is injected into oocytes, it localizes to Balbiani bodies and fills the gaps between mitochondria. Velo1 forms a stable matrix in the Bb, as evidenced by its very slow turnover after photobleaching.86 These observations suggest that Velo1 acts as a structural glue, holding the organelles together in the Balbiani body.103 How can a protein act as a glue to bring organelles together in a stable yet reversible matrix in the cytoplasm? Velo1 is a highly disordered protein with a prion-like domain (PLD) in its N-terminus and a positively charged C-terminus that binds to RNA. In vivo and in vitro experiments have demonstrated that Velo1 is a physiological amyloid that forms cages around organelles.103
MLOs have a distinct organization of proteins at their interfaces, which regulate their interactions with membranes.104 Using graphene-based sensors, Hoffmann et al. discovered that synapsin condensates generate strong electrical responses, which are absent when synapsin is present in a single phase.105 These experiments suggest that synapsin/synaptic vesicle condensates could act as charge centers at synaptic boutons, adding a new layer of regulation to neurotransmission.105 Additionally, Dai et al. demonstrated that the interface of condensates can drive spontaneous redox reactions both in vitro and in living cells.106
Although LLPS is believed to be the main mechanism behind the formation of many MLOs, some MLOs, like glycogen granules, do not rely on LLPS because they are not liquid, which prevents them from readily exchanging their contents with the environment.83 Glycogen granules serve as energy reservoirs that can be mobilized when needed.83
Validation methods
Disordered regions can be identified using predictive algorithms such as PLAAC.29 The prediction can then be tested experimentally by reconstitution of LLPS with minimal components and verified using mutants.107 The formation of LLPS is characterized by the appearance of spherical droplets, which can be visualized under a microscope. LLPS can also be detected via turbidity measurement through either optical density or direct static light scattering. The material state of condensates can be monitored by measuring the ratio of the viscosity to surface tension using fluorescence or transmitted light microscopy.1
An optogenetic platform was developed to track the formation of the droplet in vivo using blue light-activated IDR-Cry2 fusion protein.108 To achieve this, the researchers combined the “sticky” IDR from different proteins to the photolyase homology region (PHR) of Arabidopsis thaliana Cry2, which interacts autonomously when exposed to blue light.108 The researchers demonstrated that by activating light-sensitive proteins, they could induce phase transitions and form MLOs.109 These transitions could be reversed simply by turning off the light. Through enhancing light intensity and protein levels, the investigators achieved greater control over the transitions, allowing them to dictate the formation of condensed liquid protein droplets and solid-like protein aggregates, which may be associated with diseases.108
The liquidity of droplets can be measured quantitatively in living cells using fluorescence recovery after photobleaching (FRAP).1,110 Considering the difficulties in validating phase separation in vivo, the LLPS phenomenon may have been over-interpreted in the literature. As pointed out by McSweiggen et al., many reported LLPS studies were descriptive rather than quantitative. According to their criterion, a study reporting the roundness of the droplet is considered qualitative. In contrast, a study that measures the degree of roundness is considered quantitative.110 It is important to note that not all proposed MLOs adhere to these criteria.
MLOs and biomolecule condensates formed by LLPS
MLOs, also known as biomolecular condensates, encompass a diverse array of structures found within cells. These condensates form through LLPS, driven by the self-assembly of specific proteins and nucleic acids. MLOs encompass a diverse array of structures found within cells, containing specific components.
Ribosomes
Ribosomes are essential macromolecular machines found in all cells, responsible for translating messenger RNA (mRNA) into proteins. They link amino acids together in the order specified by mRNA codons to form polypeptide chains. Ribosomes consist of two primary subunits: the small and large ribosomal subunits, each containing one or more rRNA molecules and numerous ribosomal proteins (r-proteins). Collectively, ribosomes and their associated molecules are known as the translational apparatus. Unlike many other organelles, ribosomes are not membrane-bound; they are MLOs that float freely in the cytoplasm. The lack of a membrane is crucial as it facilitates the efficient transport of newly synthesized proteins, reducing the energy required for this process.
Nucleolus
Nucleolus is located within the nucleus, enriched in rRNA, ribosomal proteins, and RNA-binding proteins (RBPs) such as RBM28 (RNA-binding motif protein 28).54,111 Its function is involved in ribosome biogenesis, where it serves as the site of rRNA transcription, processing, and assembly of ribosomal subunits.112,113 The nucleolus is the largest MLO driven by LLPS. Experiments using FRAP technique and nucleolar proteins have demonstrated a significant level of exchange between these proteins and the surrounding nucleoplasm.114 Many nucleolar proteins contain IDRs, often with charged domains, which are crucial for driving phase separation. It is known that nucleolar proteins such as fibrillarin and nucleolin possess Gly–Arg-rich (GAR) domains.115 As nascent transcripts come out from the fibrillar center (FC)–dense fibrillar component (DFC) interface, they bind to the RNA-binding domain of fibrillarin (FBL). FBL interacts with itself via its GAR domain, which is composed of IDRs. This self-association facilitates the formation of the DFC phase and initiates the processing of pre-rRNA.116
P granules
P granules are RNA granules that can be found in the perinuclear area of the germ cells of C. elegans. During most of germline development, P granules are perinuclear, but they become cytoplasmic during the transition from oocyte to embryo.117 The defining constituents of P granules include two classes of RBPs: the RGG-domain proteins PGL-1 and PGL-3, and the DEAD-box proteins GLH1-4.118 P granules were the first cytoplasmic RNA granules identified to exhibit liquid-like property.95 They are roughly spherical, can fuse with one another, and exchange constituents with the cytoplasm. The spontaneous LLPS of PGL-3 with RNA drives P granule formation. However, the PGL-3 phase is unstable and needs a second phase for stabilization in embryos. This second phase is created by congregations of the disordered protein MEG-3, which is associated with PGL-3 liquid droplets in the embryo’s posterior. The congregations of these gel and liquid phases provide regional stability and dynamic properties essential for localized P granule assembly.119
Paraspeckles
Paraspeckles are nuclear bodies involved in gene expression regulation in mammalian cells, though their exact function is not yet fully understood. They are believed to control gene expression by segregating proteins or mRNAs with inverted repeats in their 3′ UTRs.109,120 Paraspeckles are protein-rich nuclear organelles constructed around a specific long noncoding RNA (lncRNA) scaffold. Initially identified in 2002 as nuclear foci containing paraspeckle component 1 (PSPC1),121 paraspeckles are now recognized to contain at least three RBPs such as PSP1/2 (paraspeckle protein 1/2), p54/nrb and nuclear paraspeckle assembly transcript 1 (NEAT1), which is an architectural lncRNA that attracts proteins containing LCDs to initiate the LLPS process to form paraspeckles.
Promyelocytic leukemia protein nuclear bodies (PML NBs)
PML NBs are macromolecular multi-protein complexes exhibiting properties of phase-separated liquid-like droplets, undergoing LLPS through heterotypic multivalent interactions between proteins and RNA molecules.122,123 The participation of LLPS in the formation of PML NBs has been confirmed by FRAP.124 PML NBs were first observed under electron microscopy, and their importance was realized due to their reversible disruption by treatment in a rare form of leukemia.125 These bodies recruit multiple partner proteins and have emerged as sumoylation factories responsive to interferon and oxidative stress. The composition of PML NBs is controlled by PML sumoylation and mRNA concentration.122 PML NBs mediate interferon-induced viral constraint and implement stress-induced senescence.126 The PML protein (TRIM19) is the crucial component of PML NBs. It contains the Ring finger, B-box, and coiled-coil domains that function as a scaffold protein necessary for the assembly of these bodies.127 Other proteins that reside in PML NBs are termed client proteins. One such client protein is TRIM33, which mediates nodal signaling in mouse embryonic stem cells.128 Malfunction of PML NBs often leads to acute leukemia and other severe diseases. The molecular basis of arsenic’s success in treating acute promyelocytic leukemia (APL) lies in rescuing PML NBs. Compared to wild-type PML NBs, the PML A216V variant from arsenic-resistant leukemia patients significantly impairs LLPS.124
Nuclear stress bodies (nSBs)
nSBs were initially recognized as the primary site of heat-shock factor-1 (HSF1) buildup in stressed cells.129 These structures are typically located close to the nucleoli. Like nuclear speckles,130 nSBs are rapidly evolving, as evidenced by the swift and easily reversible movement of HSF1.131 This dynamic nature challenges the notion that nSBs are merely clusters of misfolded proteins and suggests an involvement in cell recovery from stress. The formation of nSBs necessitates the engagement of HSF1, which modulates the transcription factor’s transactivating capacity.132 Originally believed to be accumulations of denatured proteins, this perspective shifted when the Morimoto’s group demonstrated that the formation of nSB can be triggered by various stressors, not all of which cause the denaturation of proteins.133 The amount of stress bodies correlates with cell ploidy;134 for instance, up to ten nSBs can be observed in cells that acquired infinite growth, while only two can be observed in primary cells. Notably, nSBs can be found in all primate cells but not in rodents.135 This observation suggests that the noncoding regions of the genome poorly conserved among different species might be involved in the formation of nSB.135 Under stress conditions. nSBs regulate gene expression by altering chromatin structure and attracting transcription and splicing factors. The nSB proteome includes at least 133 proteins, with over 90% being highly disordered and 66% having a high probability of promoting LLPS.136
Amyloid bodies (A-bodies)
The aggregation of amyloid can occur under physiological conditions.137 The A-bodies share the same biophysical characteristics as plaques associated with various amyloid-related diseases. The formation of A-bodies under stress conditions allows cells to gather enough proteins before entering an inactive state. It is proposed that cells can convert native proteins into an amyloid-like solid phase in a reversible process involving post-translational pathway. Different subtypes of A-bodies with discrete protein compositions can be formed in response to different stimuli. The formation of A-bodies may have evolved as a highly specialized mechanism for enduring various stressors.138
Stress granules (SGs)
SGs are located in the cytoplasm and are formed in response to various stresses, such as heat, over-production of free radicals, oxygen or nutrient deprivation. It contains mRNAs, RBPs (e.g., TIA-1, G3BP), translation initiation factors, and ribosomal subunits.96,139 SGs sequester and store untranslated mRNAs during stress conditions, allowing cells to prioritize essential processes and protect mRNAs from degradation.78 SGs contain non-translating messenger ribonucleoproteins (mRNPs) that are formed when mRNAs stall during the initiation phase of translation, either due to drug treatments or stress responses.140 SGs are fluid architectures, exhibiting quick turnover of components, dissociation into translating mRNPs, and removal through autophagy. The current paradigm suggests that various RNP granules are formed through LLPS and directed by the interplay between IDRs.16,17,33,141,142
SG formation can impact cellular reactions in two main ways. First, the increased amount of components within stress granules shifts the equilibrium of interacting molecules toward linked states. For instance, amidst a viral infection, SGs enhance the innate immune response by engaging and activating antiviral proteins such as OAS, RIG-1, RnaseL and PKR. Second, SGs can regulate signaling pathways by trapping components from the bulk cytosol, thereby limiting their interactions. This sequestration can affect pathways involving TOR, RACK1, or TRAF2.143 Mutations that influence the establishment or maintenance of SGs are linked to myopathies, ALS, and frontotemporal lobar degeneration (FTLD).144,145 Additionally, stress granules are implicated in both cancer progression and management, with many chemotherapeutic agents promoting their formation.146,147
Processing bodies (P-Bodies)
P-bodies are distinct cytoplasmic foci in eukaryotic cells, formed by phase separation and comprising decapping factors Dcp1 and Dcp2 and other enzymes that break down mRNAs in the 5′ to 3′ direction.148 These highly conserved structures are found in somatic cells of vertebrates, invertebrates, plants, and yeast. P-bodies play essential roles in several RNA-related processes, including general mRNA degradation and microRNA (miRNA)-induced mRNA suppression.149 They serve as sites for mRNA storage, degradation, and surveillance, playing a role in regulating gene expression and mRNA quality control.150
Cajal bodies (CBs)
CBs are unique sub-nuclear structures found in eukaryotic cells, typically located in the nucleus near the nucleolus. They contain components involved in RNA processing and modification, including splicing factors and small nuclear ribonucleoproteins (snRNPs). CBs participate in snRNP biogenesis and the assembly of the spliceosome, which is involved in the splicing of pre-mRNA.54 They are crucial for RNA metabolism and the assembly of RNPs involved in processes such as telomere maintenance, splicing, transcription, and ribosome biogenesis.
Germ granules
Germ granules are specialized structures enriched in RNAs found exclusively in the cytoplasm of germ cells (e.g., oocytes, spermatocytes). These granules house essential factors for germ cell development and likely serve as central hubs for the posttranscriptional regulation of gene expression. It contains germ cell-specific RNAs, RBPs, and translational regulators. Germ granules are involved in germ cell development and germline specification. They play roles in RNA regulation, mRNA localization, and translational control within germ cells.151
Nuclear speckles
Nuclear speckles are found in the nucleus. They are abundant in pre-mRNA splicing factors, RNA polymerase II, and transcriptional regulators. Nuclear speckles are evolving complexes involved in the storage and assembly of pre-mRNA splicing factors.152 They also regulate gene expression and mRNA processing.153
These are just a few examples of MLOs found in cells. Each organelle has unique compositions and functions, contributing to different aspects of cellular physiology, including gene expression regulation, RNA metabolism, and stress responses.
MLOs and biomolecule condensates in physiological conditions
MLOs have garnered significant attention in cell and molecular biology owing to their roles across different normal physiological states, such as gene expression, mRNA processing, translation, stress response and signal transduction (Fig. 4). For example, the nucleolus orchestrates the intricate process of ribosome assembly. The nucleolus and P-body are involved in the regulation of stem cell fate decision. Additionally, following cellular stress or viral infections, cells form SGs, which are assemblies of RNA-binding proteins, ribosome subunits, and stalled mRNAs following the general arrest of protein translation.
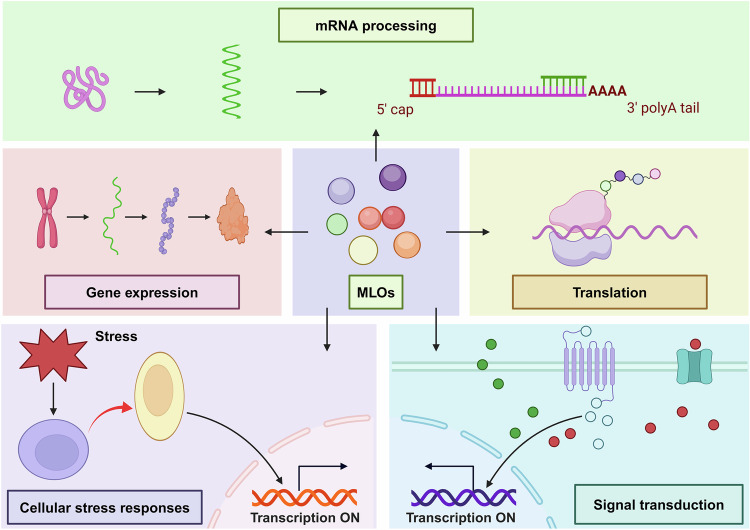
Fig. 4.
Biological functions of MLOs. The biological functions of MLOs include gene expression, mRNA processing, translation, cellular stress responses, and signal transduction
Stem cell fate determination and embryonic development
Stem cells have the potential to differentiate into different types of cells. Recent studies suggest that LLPS participates in asymmetric cell division,154 where two daughter cells with distinct fates are produced. Stem cells use asymmetric division for self-renew and generate specialized daughter cells. The capacity of stem cells to undergo asymmetric division is essential for the diversity of cell types and tissue maintenance. LLPS facilitates the polarized distribution of proteins during the asymmetric division of Drosophila neuroblasts.155–157 In Drosophila neuroblasts, the Par complex undergoes condensation dependent on the cell cycle, facilitated by LLPS. Disruptions in the phase separation of Par3/Par6 hinder the formation of apical-basal polarity during asymmetric divisions of neuroblasts, resulting in faulty lineage development.155
Recent discoveries propose that nucleoli actively regulate pluripotency and differentiation by influencing the expression of key regulatory genes.158 Stem cell self-renewal entails the division of stem cells to generate additional stem cells, sustaining the stem cell reservoir over time. Although the nucleolus is implicated in this crucial process, the precise molecular mechanisms governing it remain elusive. TMF1-regulated nuclear protein 1 (Trnp1) is a low-complexity protein with the ability to regulate the self-renewal of neural stem cells through phase separation.159 It was shown that Trnp1 maintains neural stem cells in a self-renewal proliferation state by interacting with factors present in various nuclear MLOs, including the nuclear speckles, nucleolus, and condensed chromatin.160 Reducing Trnp1 levels in mice have been demonstrated to decrease glia cell proliferation while concurrently enhancing their differentiation.159 This is the first nuclear protein that regulates stem cell fate by organizing the size, structure, and function of various MLOs.
Nucleolin, the principal nucleolar protein in actively dividing eukaryotic cells, is primarily known for its role in ribosome biogenesis. The precise localization of nucleolin proves to be crucial for myogenic differentiation. An anti-nucleolin aptamer, iSN04, was able to induce the differentiation of induced pluripotent stem cells (iPSCs) into Nkx2.5+ beating cardiomyocytes.160 iSN04 forms a guanine quadruplex-like structure, which was recognized by the RNA-binding domains of nucleolin. This unique conformation of iSN04 is pivotal for facilitating cardiomyogenesis.160 These findings are significant as iPSCs derived from patients offer immune-compatible cell sources for transplantation. Nucleolin can also be induced by β-crystallin B2 (CRYβB2) to promote the proliferation of cancer stem cells. In breast cancer cells, the increased expression of CRYβB2 correlates with enhanced stemness, growth, and metastasis.161 Within tumors, CRYβB2 fosters de-differentiation, amplifies mesenchymal markers, and promotes the presence of cancer-associated fibroblasts, along with an enlargement of nucleoli. CRYβB2 initiates nucleolin expression, subsequently activating AKT and EGFR signaling pathways.161
The nucleolus is also involved in embryonic development. Embryonic cells at the two-cell (2C) stage have totipotent potential with the capability to differentiate into the full range of cell types. Within the mouse embryonic stem (mES) cell cultures, there is a subset of cells that can spontaneously transition into a 2C stage embryo.162 However, the precise molecular mechanisms driving this transition remain elusive. Recent research has demonstrated that CX-5461, an agent that can induce nucleolar stress by inhibiting RNA polymerase I (Pol I), can promote the expansion of 2C-like cell population.163 A recent study confirmed the significance of nucleolar phase separation in determining stem cell fate.132 It demonstrated that the nucleolus-localized RBP LIN28A undergoes LLPS in both mES cells and in vitro conditions, and the ability of pluripotent cells to transition between states relies on this capacity for phase separation.164
The fate of cells is regulated by the modification known as N6-methyladenosine (m6A). It was shown that the LLPS phenomenon involving YTH N6-methyladenosine RNA-binding protein 1 (YTHDF1), a crucial “reader” protein for m6A, plays a significant role in driving spermatogonial stem cells to undergo transdifferentiation into cells resembling neural stem cells.165 This process is facilitated by activating the IκB-nuclear factor κB (NF-κB)-CCND1 pathway.165 Cell fate is also influenced by topologically associating domain (TAD), which is a region of the genome that interacts with itself. Changes in TAD organization might influence cell fate changes by controlling important genes that determine cell identity.166 However, the precise relationship between the reorganization of TAD and cell fate decision remains unclear. Recent research has discovered that TADs undergo reorganization during cellular reprogramming, which is linked to phase separation of the pluripotent protein OCT4.167
P-bodies contribute to stem cell decision-making by controlling the stability and translation of mRNAs, mediating miRNA activity, responding to cellular stress, interacting with key signaling pathways, and potentially influencing epigenetic regulation.168 Kedia et al. demonstrate that, during the development of murine cerebral cortex, the assembly of P-body promotes neural stem cell self-renewal. They further showed that the ubiquitination of 4E-T leads to the assembly of P-body in neural progenitor cells.169 Notably, 4E-T inhibits translation to ensure the stem cell pool is not depleted during a period of rapid cell genesis.170 However, in mES cells, increased levels of P-body primes mES cells for differentiation.171 Mechanistically, the O-GlcNAc modification of proteasome activator subunit 3 (Psme3) enhances the breakdown of DEAD box polypeptide 6 (Ddx6), an essential component for P-body assembly, thereby maintaining mES cells pluripotency. Conversely, Ddx6 is stabilized in the absence of Psme3 O-GlcNAcylation, leading to the spontaneous transition of mES cells out of the pluripotent state. These findings indicate that P-bodies play different roles in cell fate decisions depending on cell types.171
Regulation of gene expression and RNA metabolism
MLOs play a critical role in regulating gene expression and RNA metabolism at various levels, including transcription, RNA processing, and translation. Studying these dynamic structures provides valuable insights into their functions in health and disease, potentially leading to new therapeutic interventions.
Transcription regulation
The nucleolus is essential for rRNA synthesis and ribosome assembly, influencing rRNA gene transcription by concentrating the necessary machinery and substrates. The transcriptional activity within the nucleolus is linked to its internal pH, which regulates the recruitment and condensation of the DEAD-box RNA helicase DDX21.111 Nuclear speckles, which contain pre-mRNA splicing factors, play an important role in mRNA processing. The dynamic three-dimensional spatial organization of genomic DNA drives the high concentrations of splicing factors within these nuclear speckles.172
RNA processing and modification
CBs participate in regulating the maturation and assembly of RNPs, which are crucial for splicing pre-mRNA. These bodies form at specific locations within the genome due to high transcriptional activity. Depletion of CBs disrupts splicing dynamics by inhibiting the transcription of small nuclear RNA (snRNA) and small nuclear RNPs (snRNPs).173 P-bodies play a significant role in mRNA decapping and degradation, thereby affecting mRNA turnover and gene expression levels. These dynamic cytoplasmic MLOs contain components for mRNA storage and degradation, such as deadenylase and decapping factors. Additionally, various mRNA metabolic regulators, including m6A readers and those involved in miRNA-mediated gene silencing, are linked to P-bodies.174
Signal transduction
MLOs are essential for the spatial and temporal regulation of signal transduction pathways. Their ability to compartmentalize signaling molecules, dynamically assembles and disassembles, and enhances reaction specificity and efficiency makes them integral to cellular signaling. Understanding their roles in health and disease can open new avenues for therapeutic interventions targeting dysregulated signaling pathways.
By concentrating specific signaling molecules, MLOs create microenvironments where signaling reactions can occur more efficiently and with higher specificity. For example, PML-NBs sequester tumor suppressor proteins like p53, regulating their stability and activity. They also manage reactive oxygen species (ROS) homeostasis by linking ROS to p53 signaling, enforcing basal ROS protection, and mediating their acute toxicity.175 PML-NBs are also implicated in regulating interferon signaling pathways. Upon infection with IE1-deficient HCMV, PML-NBs rearrange into enlarged PML cages. This process requires interferon signaling and DNA damage response, causing the invading HCMV genomes to become trapped within PML-NBs in a transcriptionally repressed state. This functions as a defensive approach to combat viral infections by combining interferon and DNA damage signaling to capture both nucleic acids and protein components.126
SGs modulate signaling pathways implicated in the cellular stress reaction by sequestering key signaling molecules and mRNAs, thereby regulating their translation and activity. The stress reaction induced by nucleic acids is vital for antiviral defense and innate immunity. SARS-CoV-2 evades the immune response by attenuating antiviral SG formation.176 SG assembly inhibits apoptosis and enhances cell survival under stress. Using a proximity-labeling technique, Fujikawa et al. demonstrated that the buildup of caspase-3/7 in SGs is necessary to inhibit caspase activation and prevent apoptosis.177
MLOs and biomolecule condensates in diseases
Dysfunctional MLOs and condensates have the capacity to interfere with protein localization, signal transduction, and gene expression, ultimately contributing to the development of various diseases.
Cardiovascular diseases
The condensates formed via LLPS are vital in organizing signaling molecules and transcription factors that participate in cardiac differentiation and remodeling. An imbalance in phase separation has been linked to cardiomyopathies and heart failure.178,179 Grasping the fundamentals of LLPS within cardiac biology presents a transformative perspective for understanding the molecular complexities underlying heart health and disease.
Myofibroblasts play a pivotal role in causing cardiac fibrosis by producing collagens. Collagen production can be induced by TGF-β and matrix stiffness. Vestigial-like family member 3 (VGLL3) is a protein sensitive to mechanical stimulation180 implicated in myogenesis,181 cell proliferation182 and autoimmune disorders.183 Recent studies have revealed that substrate stiffness can cause VGLL3 to translocate to the nucleus to induce the production of collagen.184 Within the nucleus, VGLL3 undergoes LLPS facilitated by its LCD (Table 2), forming condensates alongside the non-paraspeckle NONO condensates possessing the EWS RNA-binding protein 1 (EWSR1).184 Upon binding to EWSR1, VGLL3 effectively suppressing miR-29b, a molecule that inhibits collagen production.184 Consistent with these findings, cardiac fibrosis is notably diminished in VGLL3-knockout mice, accompanied by increased expression of miR-29b after infarction (Table 2).184 These reports suggest that VGLL3 phase separation is implicated in the development of cardiac remodeling and heart failure (Fig. 5).
Table 2.
Proteins form LLPS in the cardiovascular system
| Name | Disease | Mechanism | Reference |
|---|---|---|---|
| HIP55 | Heart failure | Phase separation of HIP55 relies on Akt-mediated phosphorylation; prolonged sympathetic stimulation and stress inhibit the phosphorylation, leading to dysregulated phase separation and aggregate formation. | 179 |
| RBM20 | Dilated cardiomyopathy | The pathogenic R636S variant of RBM20 induces abnormal accumulation of RNP granules in the sarcoplasm. | 187 |
| VGLL3 | Cardiac fibrosis | VGLL3 (LCD, aa63-78) is translocated to the nucleus triggered by substrate stiffness and undergoes LLPS, which promotes collagen production by suppressing miR29b. | 184 |
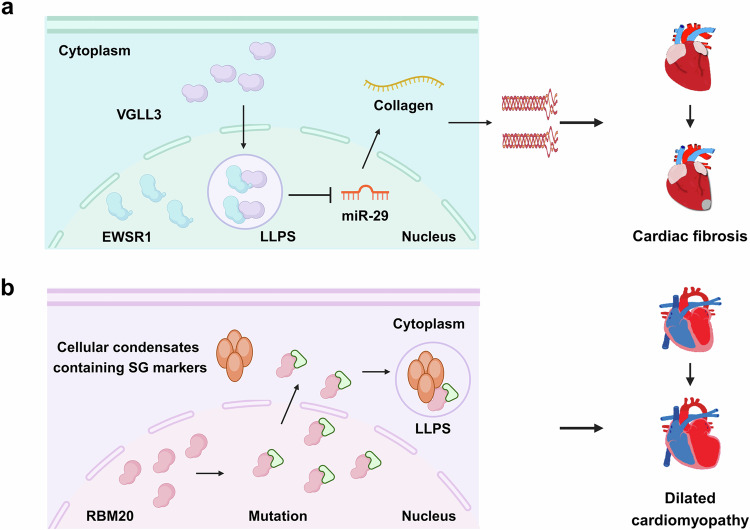
Fig. 5.
The role of MLOs in cardiovascular diseases. a Within the nucleus, VGLL3 undergoes LLPS facilitated by its LCD, forming condensates alongside the EWSR1, which inhibits miR-29, leading to increased collagen production and cardiac fibrosis. b The mutated RBM20 from nuclear splicing speckles relocates to SG, leading to dilated cardiomyopathy
RNA-binding motif protein-20 (RBM20) is a splicing factor highly expressed in the heart.185 Linkage analysis has revealed that RBM20 mutations are linked to dilated cardiomyopathy (DCM), highlighting a mutation hotspot in the arginine/serine (RS)-rich region.186 This mutation is characterized by a defect in RNP condensates, leading to abnormal heart development and function.154 The mutations cause RBM20 to relocate from the nucleus to cytoplasm and merged with other components within the SGs.187 This condensatopathy results in the restriction and isolation of polysomes, mRNA, and cytoskeleton proteins. Therefore, DCM caused by RBM20 has been considered a RNP granule disease (Fig. 5),187 which may be cured by either antisense oligonucleotides or adeno-associated virus-mediated gene therapy.185
Neurodegenerative diseases
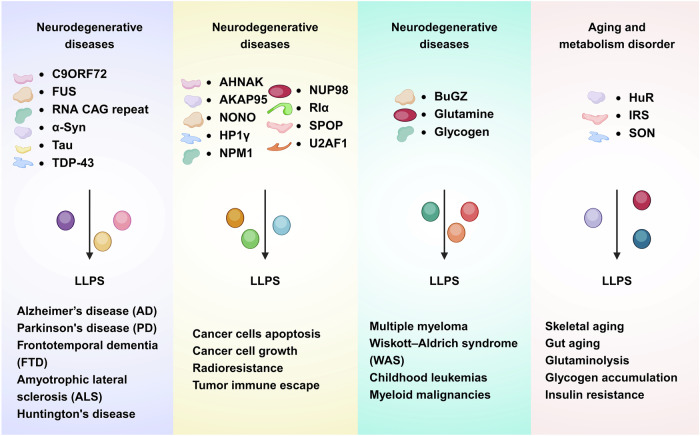
Neurodegenerative diseases present a significant global health challenge, imposing a growing burden on individuals and societies. Recent progress in cell biology has underscored the importance of MLOs in neurodegeneration. These dynamic structures, shaped through LLPS, have become pivotal components in the complex network of cellular dysfunction linked to disorders such as Amyotrophic lateral sclerosis (ALS), Frontotemporal dementia (FTD), Parkinson’s disease (PD), Alzheimer’s disease (AD), and Huntington’s disease (Fig. 6).188–190
Fig. 6.
Diseases linked to dysregulation of MLOs. Abnormal phase separation or dysfunction of specific biomolecular condensates can disrupt cellular homeostasis and contribute to a range of diseases, including neurodegenerative diseases, cancer, hematological diseases, aging and metabolism disorder
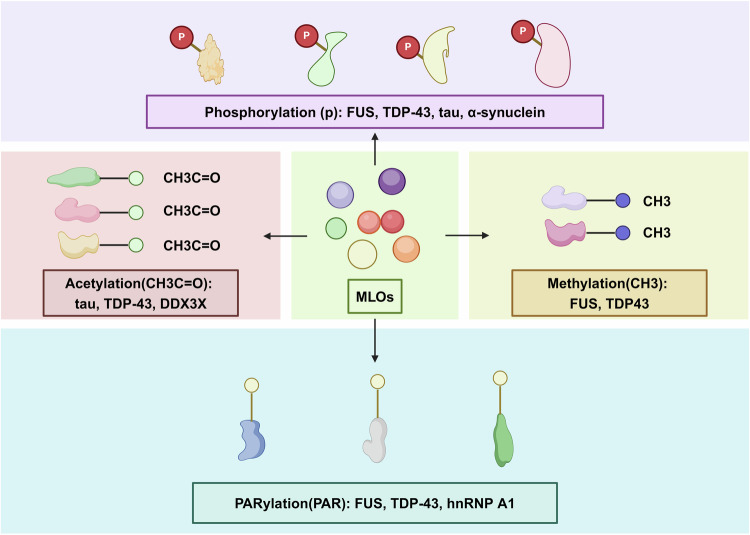
Tau, a crucial neuronal protein implicated in AD, typically resides in axons under normal physiological conditions, participating in microtubule assembly.191–195 In tauopathies, it becomes hyperphosphorylated and detaches from the microtubule, exhibiting amyloid fibril characteristics.191–194,196,197 Recent studies highlight that purified full-length tau (tau441) readily undergoes LLPS in vitro, especially when crowding agents are present to mimic cellular macromolecule concentrations (Table 3). This LLPS phenomenon occurs regardless of tau phosphorylation status, driven mainly by interactions between N-terminal and C-terminal regions of tau through electrostatic attraction.198–202 Additionally, tau condensation into droplets is facilitated by polyanions such as heparin or RNA.203,204 These in vitro observations find partial support in cellular studies.205–209 Contrary to previous suggestions, a recent study showed that point mutations in the pseudorepeat region do not significantly alter tau’s tendency for LLPS.210 However, these mutations notably accelerate liquid-to solid phase transition and promotes the formation of fibrillar aggregate.210 These studies suggest that various forms of interactions are involved in phase separation and subsequent fibrillar aggregate formation.211 Studies using electron cryo-microscopy showed that tau filaments form distinct structures in different neurodegenerative diseases.212 Further studies are needed to find out whether different tau isoforms or mutants are involved in forming these distinct structures and whether other co-factors are involved in assembling the filaments.
Table 3.
Proteins form LLPS in neurodegenerative diseases
| Name | Disease | Mechanism | Reference |
|---|---|---|---|
| C9ORF72 | ALS/FTD | Expansion of GGGGCC repeats in the gene C9ORF72 alters its LLPS dynamics | 247 |
| FUS | ALS | Wild-type FUS forms reversible fibrils | 256 |
| FUS with glycine mutations undergo rapid loss of fluidity | 240 | ||
| RNA CAG repeat | Huntington’s disease | RNA molecules with expanded CAG repeat (eCAGr) form cytoplasmic gel-like foci that significantly reduce the global protein synthesis rate | 248 |
| αS (α-synuclein) | Parkinson | Phase-separated droplet forms a precursor for the pathogenic αS fibrils. | 190 |
| Tau | Alzheimer | The phosphorylated or mutant tau can initiate aggregation through LLPS | 198 |
| The microtubule-binding repeats of Tau form liquid droplets in a phosphorylation-specific manner. | 197 | ||
| TDP-43 | ALS | Two familial variants within the312 NFGAFS317 segment of TDP-43(A315T and A315E), together with phosphorylation, create pathogenic aggregation. | 224 |
Alpha-synuclein (α-syn) is a synuclein protein mainly found in neurons and is involved in the pathogenesis of PD, which manifests as a multisystem disorder with a spectrum of motor and non-motor symptoms. Pathologically, these manifestations are linked to extensive aggregated proteins known as Lewy bodies (LBs) within neurons.213 A key constituent of LBs is α-syn encoded by the SNCA gene.214 Amplification of the wild-type SNCA gene leads to early-onset PD and dementia.215 Various investigations have suggested that aggregates formed by misfolded α-Syn are implicated in cell death during PD progression.216 It was shown that α-Syn has the ability to undergo LLPS, typically resulting in the formation of amyloid fibrils (Table 3).217 Recent findings indicate that LLPS of α-Syn takes place during the nucleation phase of aggregation. The α-Syn droplets with liquid-like properties undergo an irreversible shift into amyloid-like hydrogels, encapsulating oligomers and fibrils.218 Research also demonstrated that α-Syn directly influences P-bodies, which are MLOs responsible for mRNA turnover and storage. The α-Syn binds with various decapping proteins closely positioned on the Edc4 scaffold. Elevated levels of α-Syn, as observed in pathological conditions, lead to increased association with Edc4, thereby interfering with interactions with other decapping-module proteins. Consequently, certain mRNAs, such as those involved in protein trafficking and RNA metabolism, are stabilized.219 Questions remain as to why only a selected group of mRNAs are stabilized and how they contribute to the formation of LBs. Although highly expressed in neurons, α-Syn is also found in other tissues such as blood220 and kidney.221 So the question is why only neuron is affected.
Neurons efficiently transport all essential components for translation, such as ribosomes, mRNA, and translation factors, to synthesize proteins at distant locations. The localization pattern of mRNAs is tightly regulated by several RBPs, such as FUS, TDP-43, and hnRNPA1, which are associated with ALS. TDP-43 and FUS can form condensates with various material conditions involved in both normal cellular process and disease. These RBPs play pivotal roles in regulating pre-mRNA splicing and transcription. Moreover, they constitute constituents of RNP granules triggered by stress, comprising RBPs and RNA, and are prominently present in cytoplasmic aggregates within neurons in degeneration, serving as essential disease markers of ALS and FTD.222
The presence of pathological aggregation of phosphorylated TDP-43 (p-TDP-43) is a key feature of ALS and FTD. ALS stands as the predominant clinical manifestation of upper and lower motor neuron disease.223,224 A mounting body of evidence substantiates the concept of overlapping genetic and pathological features between ALS and FTD.225 The pathology of both ALS and FTD has been associated with environmental factors and an array of genetic changes, encompassing multiple point mutations in the LCR of proteins localized within RNP granules, as well as repeat expansions.
TDP-43 is involved in many cellular activities, such as regulating mRNA splicing, RNA transportation, and forming cytoplasmic SGs that halt translation.226 These functions often occur within RNP granules, which are formed through LLPS.227 In typical physiological circumstances, TDP-43 is mainly found in the nucleus, forming oligomers and residing within biomolecular condensates assembled through LLPS. However, in disease states, TDP-43 forms inclusions either in the cytoplasm or intranuclearly. Studies indicate that the carboxy-terminal domain of TDP-43 alone can induce phase separation. Mutations linked to ALS interfere with interactions and hinder this phase separation process, potentially explaining the functional impairment observed.228 The significance of TDP-43 in pathological condition is emphasized by the fact that dominant missense mutations alone are adequate to induce disease. Studies have revealed that cytoplasmic RNP granules formed by TDP-43 facilitate the delivery of target mRNA to distant neuronal regions through microtubule-dependent transport system. The ability to transport mRNA is impaired by TDP-43 mutations that cause ALS. Thus, TDP43 mutations associated with ALS result in a partial loss of the physiological function of TDP-43 (Table 3).229
By conducting whole genome linkage analysis and exome sequencing, Ervilha Pereira et al. identified a frameshift mutation in TDP-43 that was conclusively linked, resulting in a C-terminally altered PrLD (TDP-43p.Trp385IlefsTer10). Muscle biopsies obtained from patients showcased TDP-43-positive sarcoplasmic inclusions. In vitro phase separation assays revealed that TDP-43Trp385IlefsTer10 formed solid-like fibrils instead of liquid-like condensates, indicating an increased tendency for aggregation compared to wild-type TDP-43. Collectively, these findings affirm that TDP-43p.Trp385IlefsTer10 is a partial loss-of-function and susceptible to aggregation variant responsible for autosomal dominant vacuolar myopathy.230
A lingering question has revolved around the consequences of the processes involved in TDP-43 mutations and how their impacts correspond to the events accompanying the pathological re-localization of TDP-43 in patients. TDP-43 WT RNP granules display discrete biophysical characteristics according to where they are situated in the axon, while granules generated by ALS-associated mutant TDP-43 exhibit increased viscosity and impaired axonal transport function.231 Through the utilization of various cellular systems expressing variants of TDP-43 based on structure, Perez-Berlanga et al. demonstrated that oligomerization and RNA binding play pivotal roles in governing the stability of TDP-43, its splicing activity, LLPS, and cellular distribution.232 As an RNA-binding protein, mislocalization of TDP-43 could potentially alter RNA metabolism. However, TDP-43-regulated RNAs in motor neurons are yet to be discovered. In this context, Klim et al. illustrated that the expression of STMN2, a microtubule-modulating protein crucial for normal axonal expansion and regrowth, decreased following TDP-43 silencing, aberrant TDP-43 distribution, and spinal cords from deceased patients.233 Neuropathological investigations substantiate the notion that TDP-43 aggregates might spread from one cell to another, leading to the dissemination of pathological inclusions in the brain. The transmission of TDP43 from cell to cell has been observed in cell cultures, potentially contributing to the pathological propagation of TDP43 in FTLD-TDP.234 A recent study confirmed these findings in vivo, demonstrating that a single intracerebral injection of pathological TDP43 derived from human brains affected by FTLD-TDP initiates the onset and propagation of TDP43 inclusions in the brain in a spatiotemporal-dependent manner.235
Using the optogenetic platform, Mann et al. showed that the generation of pathologically relevant TDP-43 phase transitions can be prevented by adding a “bait” RNA oligonucleotide.236 Compared to the antisense oligonucleotides, which suppress protein expression, the “bait RNA” regulates protein solubility via reversible interaction; therefore, it is a useful approach for treating either gain- or loss-of-function protein aggregates.
Although it is generally believed that the generation of insoluble protein aggregates is responsible for the development of neurodegenerative diseases, a recent study showed that some mutations that increase TDP-43 aggregation actually decrease toxicity in yeast cells by titrating proteins away from toxic interactions.237 Therefore, this yeast cell toxicity model may be useful to evaluate antisense or “bait RNAs” before initiating clinical trials.
Uncommon mutations affecting the LCD of the RBP T-cell-restricted intracellular antigen-1 (TIA1) have been detected in individuals with ALS and FTD. TIA1 holds an important position as an SG constituent, and its LCD is essential for SG congregation. The mutations associated with the disease affect the biophysical behavior of TIA1, enhancing its tendency for phase separation, causing a delay in SG disassembly, and fostering the buildup of static SGs. These SGs house TDP-43, which experiences reduced mobility and increased insolubility.238 Therefore, it was suggested that delayed disassembly of SG might increase the accumulation of insoluble TDP-43.238 Along this line, it was shown that TIA1 facilitates the phase separation and formation of toxic tau.239
Fused in sarcoma (FUS), a nuclear RBP, manifests its pathogenic signature in ALS and FTD through cytoplasmic aggregation. FUS is involved in various RNA metabolism and the assembly of RNP bodies, including SGs. These RNP bodies are thought to arise through LLPS, driven by temporary RNA and RBP interactions that possess IDRs and RNA recognition motifs (RRMs). FUS forms fluidic structures at DNA breakpoints in cells and in the cytoplasm during stress, and FUS liquid droplets transition over time from a fluid to an aggregated state, a process accelerated by mutations observed in patients (Table 3).34 The mechanisms by which FUS-RNA interactions drive phase separation and whether ALS-associated mutations affect this behavior are not fully understood. A recent study showed that wild-type FUS binds single-stranded RNA in a manner dependent on length, forming small, fluid condensates through dynamic interactions with RNA multimers.240 In contrast, glycine mutations in FUS result in a rapid decrease in fluidity, underscoring the pivotal role of glycine in promoting fluidity.240 Interestingly, although both FUS and TDP-43 can form aggregates in SGs, they do so in a mutually exclusive manner.241,242 The mechanisms responsible for this phenomenon remain incompletely understood but may be due to different “molecular grammars” involved in their phase separation.
Another RBP that is recruited to SGs is hnRNPA1,243 a part of the heterogeneous nuclear ribonucleoproteins (hnRNPs) family, which predominantly functions as nuclear RBPs, forming complexes with RNA polymerase II transcripts. These proteins engage in diverse cellular activities, including transcription, pre-mRNA processing, and translation. Recent investigations propose that numerous intrinsic features of hnRNPs contribute to their participation in various regulatory pathways.244 Genetic evidence establishes a connection between persistent SG and the buildup of abnormal inclusions.33 The disease-associated hnRNPA1 undergoes LLPS, forming protein-rich droplets mediated by LCD.33 When SGs consist of RBPs containing LCD mutations that promote fibrillization or when SGs persist as a consequence of interruptions in the disassembly process, pathogenic fibrils can form and evade the surveillance of quality control.43 Similar to disease-causing mutations observed in hnRNPA1, the variants D290V and P298L promote aggregation in hnRNPA2.245 Interestingly, deficiency of nuclear hnRNPA1 in motor neurons alongside concurrent cytoplasmic aggregation of TDP-43 is a key feature in progressive neuronal death in ALS.246 These findings suggest that the disappearance of nuclear hnRNPA1 might be related to TDP-43 in the SGs.
The occurrence of repeat expansions of small nucleotide segment represents another unique type of genetic modification linked to diseases like Huntington’s disease, myotonic dystrophy, ALS/FTD, and spinocerebellar ataxias. The extent of these illnesses is proportional to the repeat’s length. The resultant abnormal polypeptides or RNAs form condensates and recruit other crucial molecules. One example is the GGGGCC repeat, a common cause of ALS/FTD. The buildup of the repeat that contains RNAs within nuclear regions might drive the disease by sequestering RBPs.247 Another example is the expanded CAG repeats (eCAGr). RNA molecules carrying eCAGr have the potential to undergo sol-gel phase transitions and form cytoplasmic gel-like foci, which may substantially decrease the rate of protein production, possibly through the sequestration of the elongation factor eEF2 for translation. In brain tissue sections from a knock-in mouse model and from patients affected by Huntington’s disease, eEF2 puncta were notably enhanced. Furthermore, the injection of adeno-associated virus containing eCAGr RNA resulted in significant behavioral impairment in mice.248
In summary, there is substantial evidence indicating a strong association between the LLPS of specific proteins and the progression of neurodegenerative diseases.211,249–255 However, the precise molecular basis for this connection remains inadequately understood. A significant argument supporting a clear mechanistic link between LLPS and the development of disease is the observation that mutations in proteins such as TDP43,228 FUS,34,256 hnRNPA1,33 hnRNPA2,245 or TIA1238 induce abnormality of MLOs within cells or liquid droplets formed in laboratory settings. Nevertheless, the nature of these abnormalities appears to vary depending on the specific protein and mutation involved. For instance, some mutations, like those in TIA1, promote LLPS,238 while others, notably most mutations in TDP43, have the opposite effect.228 Moreover, the implications of LLPS in terms of protein aggregation also differ depending on the protein in question.253
Cancer
Recent advancements have unveiled a connection between abnormal phase separation and various types of cancer. The abnormal phase separation leads to genomic instability and disruption of transcription and signal transduction.257–259
p53 is a tumor suppressor protein involved in many signaling pathways in cells under stress. The p53-binding protein 1 (53BP1) is an interactor of p53.260 Ghodke et al. identified AHNAK as a scaffolding protein that binds to 53BP1. AHNAK prevents overactive interaction between 53BP1 and p53. This regulatory mechanism protects cancer cells from apoptotic stimuli. The loss of AHNAK results in enhanced p53-mediated apoptosis due to excessive buildup of 53BP1 on chromatin and enhanced phase separation (Table 4).260 Thus, AHNAK functions as a rheostat of p53 by restraining 53BP1 phase separation.
Table 4.
Proteins form LLPS in cancer
| Name | Disease | Mechanism | Reference |
|---|---|---|---|
| AHNAK | Cancer cell survival | AHNAK is a G1-enriched interactor of 53BP1 that ensures optimal partitioning of 53BP1 into phase-separated condensates and limits excessive interaction with p53, which leads to apoptosis in cancer cells | 260 |
| AKAP95 | Cancer | AKAP95 is a nuclear protein that regulates transcription and RNA splicing by forming liquid-like condensates in nucleus. | 261 |
| HP1γ | Myeloma | The deacetylation of HP1γ promotes nuclear condensation, and this condensed form of HP1γ plays a crucial role in drug resistance by facilitating DNA repair in multiple myeloma cell. | 286 |
| NONO | Tumor radioresistance | LLPS of NONO recruits nuclear EGFR and DNA-PK and promotes DNA repair, leading to radioresistance. | 262 |
| NPM1 (Nucleophosmin) | Triple-negative breast cancer (TNBC) | NPM1 undergoes LLPS through interactions with nucleolar components, including rRNA and proteins featuring multivalent arginine-rich linear motifs (R-motifs). NPM1 binds to the PD-L1 promoter in TNBC cells, activating PD-L1 transcription. | 263,264 |
|
NUP98 (Nucleoporin 98) |
Leukemia | The biomolecular condensation is embedded within the N-terminus of NUP98 and possesses the ability to induce leukemia-specific gene expression. | 290 |
|
RIα (Type I regulatory subunit of PKA) |
Cell transformation | Loss of RIα LLPS in normal cells induces cell transformation. | 44 |
| SPOP | Prostate, breast cancer | Cancer-associated mutations in tumor suppressor SPOP disrupt LLPS and correlate with a loss of function. | 275 |
| U2AF1 | Myeloid malignancies | U2AF1 splicing factor mutations, lead to an increased SG response, indicating a new function for biomolecular condensates in adaptive oncogenic mechanisms. | 292 |
AKAP95, a nuclear protein, is overexpressed in clinical samples of cancer tissues. AKAP95 contributes to tumor growth by facilitating the splicing of cyclin A2, an important regulator of the cell cycle.261 The regulatory functions of AKAP95 in gene expression and tumorigenesis are contingent on its capacity to establish condensates with appropriate liquidity and dynamicity (Table 4).261 The data suggest that AKAP95 is involved in tumorigenesis by promoting the proliferation of cancer cells. Radioresistance stands out as a primary contributor to the failure of cancer treatment, resulting in relapse and diminished survival outcomes for cancer patients. NONO, an RNA/DNA-binding protein with LLPS capability, has become an essential regulator of tumor radioresistance. The LLPS of NONO facilitates the recruitment of nuclear EGFR and DNA-PK, intensifying their interaction. This cascade leads to an augmented initiation of DNA damage-induced pT2609-DNA-PK and fosters non-homologous end joining (NHEJ)-mediated DNA repair, ultimately culminating in tumor radioresistance. The phase separation-mediated radioresistance mediated by NONO presents a potential novel molecular target for sensitizing tumor cells to radiotherapy (Table 4).262
The interaction between programmed cell death protein-1 (PD-1) and its ligand (PD-L1) is pivotal in tumor immune escape mechanisms. Triple-negative breast cancer (TNBC) exhibits elevated expression of PD-L1 compared to other subtypes. Nucleophosmin (NPM1) activates the transcription of PD-L1 in TNBC cells by specifically binds to its promoter. Consequently, this activation hinders T-cell activity both in vitro and in vivo.263 NPM1, a highly prevalent oligomeric protein located in the nucleolus, is actively involved in ribosome biogenesis by interacting with nucleolar components through self-interaction mediated phase separation (Table 4).264
Oncogenic receptor tyrosine kinase fusion proteins exhibit elevated assembly, forming membraneless cytoplasmic protein granules. These granules play a crucial role in coordinating local RAS activation and organizing RAS/MAPK signaling within lung cancer cells.70 Additionally, RIα, the type I subunit of cAMP-dependent protein kinase A (PKA), undergoes LLPS in response to cAMP signaling, leading to the generation of cAMP-enriched biomolecular condensates (Table 4). Normal cells use this LLPS process to sequester and constrain cAMP. However, in the presence of PKA fusion oncoprotein, the phase separation of Riα is blocked, leading to enhanced cell proliferation and the induction of cell transformation.44
RNA-binding protein 14 (RBM14) is a coactivator of nuclear receptors, and its expression is increased in castration-resistant prostate cancer (CRPC). Despite androgen deficiency, the androgen receptor signaling pathway remains an important driving force in CRPC. Tsuji et al. demonstrated that RBM14 promotes phase separation, sustaining prostate-specific antigen expression during androgen suppression in human prostate cancer.265
SPOP (speckle-type POZ protein) is a cancer inhibitor, and its mutations cause solid tumors.266–270 SPOP functions as a binding scaffold of the cullin3-RING ubiquitin ligase and attracts substrate through LLPS.271–274 Cancer-associated mutations in SPOP impair LLPS and are associated with a loss of function (Table 4).275
UTX/KDM6A, a gene encoding a histone H3K27 demethylase, acts as a crucial cancer suppressor commonly mutated in human cancers.276 However, subsequent studies revealed that the demethylase activity of UTX is frequently independent of its role as a tumor suppressor.277–282 A recent study revealed that the chromatin regulatory activity of UTX in tumor suppression is rooted in its phase separation.283 The core intrinsically disordered region (cIDR) of UTX gives rise to condensates through phase separation, the loss of cIDR due to mutations is a key factor in nullifying tumor suppression.283
Hematological disorders
In hematological disorders, which encompass a diverse range of conditions affecting blood cells and their precursors, recent progress in cell biology has brought attention to the crucial role of MLOs in orchestrating cellular processes. This provides novel insights into the pathophysiology of blood diseases.
Heterochromatin protein 1γ (HP1γ) is a reader of H3K9me2/3284 that is involved in efficient transcriptional elongation.285 A significant challenge in managing multiple myeloma is the resistance to proteasome inhibitors. Elevated HP1γ levels are linked to poorer clinical outcomes.286 Mechanistically, heightened HDAC1 activity in bortezomib-resistant myeloma cells leads to the deacetylation of HP1γ at lysine 5, promoting nuclear condensation. This condensed form of HP1γ is essential for chemotherapy resistance in multiple myeloma cells (Table 4). These findings suggest that targeting HP1γ could effectively overcome drug resistance in patients with relapsed multiple myeloma.286
Wiskott–Aldrich syndrome protein (WASP) is an actin nucleation factor.287 Mutations in WASP cause Wiskott–Aldrich syndrome (WAS), which is an X chromosome-linked disease characterized by thrombocytopenia, eczema, and immune deficiency.288 WASP controls the transcription of splicing factors via phase separation. Mutations in the WASP gene result in abnormal RNA splicing.289
The nucleoporin 98 (NUP98) protein is a member of the nuclear pore complexes that regulate the movement of macromolecules between the nucleus and cytoplasm. NUP98 gene is recurrently translocated, particularly in pediatric leukemia cases. Terlecki-Zaniewicz et al. explored the protein interactomes of several NUP98 proteins in human leukemia cells using confocal imaging. They demonstrated that various NUP98 fusion proteins with distinct structures are concentrated within biomolecular condensates by LLPS. Additionally, they showed that the structural characteristics directing the condensation are embedded within the N-terminus of NUP98 and possess the ability to trigger leukemia-specific gene expression when incorporated into oncogenic fusion proteins (Table 4).290 These observations were confirmed by a subsequent study showing that NUP98 fusion oncoproteins contain an LCR that is prone to LLPS, driving the transformation of hematopoietic cells.291
Mutations in splicing factors, particularly U2AF1, have become prevalent key factors in myeloid malignancies. However, the precise influence of these mutations on splicing and their role in promoting cancer has remained ambiguous. Biancon et al. utilized high-resolution interactome to reveal that U2AF1 splicing factor mutations lead to an intensified stress response in myeloid malignancies.292 Cell lines carrying U2AF1 mutations and blasts derived from MDS/AML patients exhibited an increased SG response, indicating a new function for biomolecular condensates in adaptive oncogenic mechanisms (Table 4).
Aging and metabolism disorder
Some studies suggest that changes in the composition, dynamics, or functionality of MLOs could contribute to aging-related cellular dysfunction by altering the content or the formation of SGs.293
Human antigen R (HuR) is a bone-associated RBP that forms SGs. Research has demonstrated that both the expression of HuR and the generation of HuR-positive SGs decrease in primary osteoblasts obtained from aging mice.294 Inhibiting the HuR-positive SGs led to a reduction in osteoblastic differentiation, indicating the vital involvement of HuR and SGs in osteogenesis (Table 5).294 These findings underscore the importance of HuR and the associated SGs in enhancing bone formation in skeletal aging and provide a basis for developing innovative therapeutic approaches for age-related skeletal diseases.
Table 5.
Proteins form LLPS in metabolism and aging
| Name | Disease | Mechanism | Reference |
|---|---|---|---|
| BuGZ (a coacervating mitotic effector) | Aging | Inhibiting the phase transition of BuGZ extends the lifespan of Drosophila | 295 |
| Glutamine | Glutamine deprivation stress | When glutamine level is decreased, lncRNA GIRGL inhibits glutaminase activity through phase separation to enable cancer cell survival | 297 |
| Glycogen | Tumor initiation | Cancer-initiating cells proliferate by increasing glycogen storage and block Hippo signaling through glycogen phase separation. | 298 |
| HuR (Human antigen R) | Age-related bone loss | HuR is a bone-associated RBP that forms SGs and facilitates osteogenesis during aging | 294 |
| IRS (Insulin receptor substrates) | Type 2 diabetes | LLPS of IRS functions as intracellular signal hubs to transmit insulin signals | 299 |
| SON (Nuclear speckle scaffolding protein) | Dysregulated proteostasis | The XBP1s-SON axis dictates a nuclear speckle LLPS dynamics that protects cells from proteome stress | 300 |
Research has demonstrated that BuGZ, a coacervating mitotic effector, undergoes condensation in Drosophila intestinal stem cells (ISCs) nuclei during interphase, particularly in association with aging and injury. This condensation of BuGZ promotes the proliferation of ISCs, impacting gut repair and longevity in Drosophila (Table 5).295 Inhibiting the phase transition of BuGZ enhances functions of the intestine and extends the duration of life of these flies. These findings suggest that manipulating protein phase transitions could potentially delay or counteract aging associated with ISCs.295
The theory of antagonistic pleiotropy in aging suggests that genes beneficial for early-life fitness may have detrimental effects on lifespan, yet molecular evidence supporting this notion remains largely unexplored. Through a study mapping translatome alterations in Caenorhabditis elegans during the restoration phase after starvation, Wu et al. discovered that trl-1 becomes significantly active after refeeding. Deficiency of trl-1 leads to aberrant upregulation of vitellogenin translation, which enhances reproduction but at the expense of lifespan. They demonstrated that trl-1 undergoes LLPS, forming granules that recruit vitellogenin mRNA and suppress the translation.296 These data demonstrate that trl-1 influences the balance between reproduction and longevity by enhancing nutrient provision for the next generation.
MLOs also play important roles in various metabolic processes within the cell by organizing and regulating the dynamics of metabolic reactions across space and time, RNA metabolism, and protein synthesis. Glutamine is a vital supplier of carbon and nitrogen for various biological processes. The initial and crucial step in glutaminolysis is the conversion of glutamine to glutamate, facilitated by the enzyme glutaminase-1 (GLS1). When there is a shortage of glutamine, levels of GLS1 decrease, although the exact mechanisms remain unclear. Research has revealed the existence of a long noncoding RNA called glutamine insufficiency regulator of glutaminase lncRNA (GIRGL), which is upregulated during glutamine deficiency. In the absence of glutamine, elevated levels of GIRGL lead to the formation of a complex between dimers of CAPRIN1 and GLS1 mRNA. CAPRIN1 is a cytosolic phosphoprotein expressed ubiquitously and implicated in various stress responses. This complex formation induces the LLPS of CAPRIN1 and triggers the formation of SG. By inhibiting the translation of GLS1 mRNA, cancer cells can adapt and survive prolonged periods of glutamine deprivation stress.297
Cancer cells adapt their metabolic processes to enhance survival and facilitate invasion. Research indicates that glycogen accumulation is crucial in promoting oncogenesis and is likely a prevalent metabolic trait among initially transformed liver cells. The gathered glycogen undergoes LLPS, interfering with Hippo signaling and promoting tumor initiation (Table 5).298
Insulin is a metabolic hormone that orchestrates various cellular processes to uphold cell functionality and overall metabolic well-being. The emergence of insulin resistance, characterized by compromised insulin function within cells, is a driving factor behind the escalating global incidence of type 2 diabetes (T2D) in recent decades. Given the widespread prevalence of T2D, an urgent need exists for a comprehensive understanding of the mechanisms governing insulin action and resistance. Upon binding with insulin, the insulin receptor (IR)’s tyrosine kinase activity is activated through autophosphorylation on multiple tyrosine residues, subsequently leading to the phosphorylation of insulin receptor substrates (IRS). Research has revealed that IRS condensates formed via LLPS may serve as crucial intracellular signaling hubs, facilitating the transmission of insulin signals deep into the cellular milieu (Table 5).299 However, the formation of IRS condensates is impaired in insulin-resistant cells.299
Significant strides have been taken in comprehending the properties and functions of biomolecular condensates and LLPS. However, it remains uncertain whether LLPS dynamics are governed by “autonomous clocks.” It was shown that the XBP1s-SON axis incorporates a 12-hour rhythm with LLPS in nuclear speckles to regulate proteostasis across space and time.300 These findings indicate that by influencing the timing of proteostasis dynamics, nuclear speckle LLPS could serve as a target for conditions linked to disrupted proteostasis.
Regulation of LLPS phenomenon
Regulation by post-translational modifications (PTMs)
PTMs such as phosphorylation,71 acetylation,49 methylation,80 and ubiquitination301 can influence gene expression, signaling, and stress responses by modulating the properties of proteins through phase separation, and the recruitment of specific components to the condensates (Fig. 7). PTMs play an essential role in the assembly and disassembly of condensates by introducing covalent changes to protein components.302 These modifications significantly influence the ability of IDRs to form condensates.303
Fig. 7.
Regulation of MLOs through modifications occurring after translation. The formation and function of MLOs are regulated by modifications occurring after translation, including phosphorylation, acetylation, methylation, and PARylation
Phosphorylation
Protein phosphorylation involves the attachment of a phosphoryl group, typically to the hydroxyl group of serine, threonine, or tyrosine. At physiological pH, each phosphate group introduces two negative charges to the protein. The heightened negative charge can be harnessed to modify the phase separation of proteins. Notably, serine/threonine phosphorylation of FUS disrupts phase separation and prevents its aggregation.304 A phosphomimetic substitution at S48 impairs the polymeric assembly of TDP-43.305 These findings reveal the potential for LLPS and aggregation modulation through phosphorylation, presenting intriguing prospects for therapeutics. However, further research is essential to pinpoint the active kinases, delineate the timing and pattern of protein phosphorylation, and elucidate their effects on LLPS-mediated granule formation.
The excessive phosphorylation of the neuronal tau protein stands out as a characteristic feature of Alzheimer’s disease. The phosphorylation of tau by MARK2 was demonstrated to promote K18 LLPS.197 An unanswered query revolves around the mechanisms driving tau’s progressive hyperphosphorylation. Recent findings suggest an interdependence mechanism, establishing a connection between initial site-specific and subsequent multi-site phosphorylation. It was shown that a primary phosphorylation site determines the spread of phosphorylation across multiple sites.306 It would be interesting to find out whether pathological tau levels can be reduced by inhibiting the master site.
The advancement of Parkinson’s disease hinges significantly on the propagation of pathological α-synuclein (α-Syn). This aggregation of α-Syn propagates in a manner akin to prions, gradually advancing in the brain and from peripheral organs to the brain throughout the disease’s course. Certain proteins on the cell surface, including lymphocyte activation gene 3 (LAG3)307,308 and amyloid precursor-like protein 1 (APLP1),309 have been identified as receptors facilitating the uptake and spread of α-Syn preformed fibrils (PFFs). However, the specific molecular basis for this preferential binding remains unclear. Recent research suggests that phosphorylation on serine 129 (pS129) enhances the interaction between α-Syn fibrils and these specific receptors.310 This discovery is corroborated by the heightened efficiency of pS129 fibrils in cellular uptake, initiation, and induction of PD-like α-Syn pathology in transgenic mice.310
Acetylation
Lysine acetylation, a reversible PTM, is orchestrated by lysine acetyltransferases and lysine deacetylases, contributing to both histone and non-histone targets. Lysine’s significance in cellular function is underscored by lysine-rich forms of the Alzheimer’s disease-associated protein tau. These variants exhibit coacervation with RNA and are associated with SGs in cellular environments. Acetylation of lysine is demonstrated to reverse LLPS, diminishing the association of tau with SGs.205
In the case of TDP-43, acetylation disrupts RNA binding, fostering the buildup of insoluble and hyperphosphorylated TDP-43 forms closely resembling abnormal inclusions in ALS and FTLD-TDP. The presence of acetylated TDP-43 lesions in the spinal cord of ALS patients further suggests a link between abnormal TDP-43 acetylation and the loss of RNA binding in TDP-43-mediated diseases.309
Traumatic brain injury (TBI), a prominent risk factor for Alzheimer’s disease (AD), induces tau acetylation at positions similarly acetylated in human AD brain, suggesting a potential molecular connection between TBI and AD.311 Targeting post-TBI-induced tau acetylation using acetylation inhibitors, such as salsalate, correlates with diminished neurodegeneration in humans. This approach prevents tau abnormal localization, loss of solubility, and cognitive deficits in animal models, offering potential therapeutic avenues.311
Precise control over IDR action is paramount to ensuring that LLPS occurs only when needed. Recent findings indicate that acetylation/deacetylation of IDRs regulates LLPS and MLOs formation in response to stress conditions. Acetylome analysis uncovered the RNA helicase DDX3X, an essential SG constituent, as a new target for the deacetylase HDAC6. The acetylation of DDX3X impedes the generation of liquid droplets.49
Methylation
Methylation, which involves adding methyl (CH3) groups to arginine and lysine, stands out as a modification that alters biomolecular interactions. Unlike phosphorylation, methylation doesn’t add charge but increases the size and hydrophobicity of amino acids and changes the distribution of charge. Arginine methylation is prevalent among RNPs, influencing SG assembly through interactions with LCR regions.
In a subset of individuals with FTD, the presence of cytoplasmic FUS aggregates serves as a disease hallmark. Phase separation is inhibited by arginine methylation, which is missing in FTD-FUS patients.312 Loss of arginine methylation results in accumulation and aggregation of FUS in SG.312 In vitro studies demonstrate that FUS experiences concentration-dependent, reversible LLPS. These liquid FUS structures eventually transition into solid, fibrous aggregates over time, known as liquid-to-solid phase transition. Dimethylation of arginine residues on FUS diminishes LLPS and its association with SG, suggesting that the absence of arginine methylation, observed in FTD-FUS patients, contributes directly to FUS aggregation and pathology.312
The widely prevalent RNA modification, adenosine N6 methylation (m6A), affects RNA stability, transport, and translation. It has been demonstrated that the composition of the phase-separated transcriptome can be influenced by the number and distribution of m6A sites in mRNAs. This highlights the governance of cellular characteristics of m6A-modified mRNAs by LLPS principles.313 TDP43 has been identified as a recognizer of m6A RNA, with extensive RNA hypermethylation playing a crucial role in both TDP43 binding and self-regulation. Extensive hypermethylation of RNA observed in ALS spinal cord aligns with methylated TDP43 substrates, unveiling RNA modification targeting as an emerging avenue to regulate pathological protein phase transitions.314 Importantly, the investigator showed that TDP43-related neurotoxicity can be alleviated by knocking out the m6A reader YTHDF2,314 opening a potential new avenue for treating ALS.
PARylation (poly-ADP-ribosylation)
PARylation is a PTM where ADP-ribose molecules are incorporated into proteins by poly(ADP-ribose) (PAR) polymerases (PARPs). Recognizing PARylated proteins in MLOs is crucial for comprehending the impact of PARylation on MLOs and exploring its therapeutic potential.
FUS has been identified as a novel participant in the DNA damage response. FUS swiftly localizes DNA double-strand breaks, relying on PARPs activity. The interaction between FUS and PAR is mediated by arginine/glycine-rich domains, highlighting their direct connection.315 Notably, elevated PAR levels at DNA damage sites induce phase separation, leading to disease-associated protein aggregation.316 These findings might explain the mechanisms behind the therapeutic effects of PARP inhibitor in treating cancer317 and PD.318
Increased nuclear PARP-1/2 activity was found within ALS spinal cord motor neurons. Veliparib inhibited the generation of cytoplasmic TDP-43 aggregates by reducing nuclear PARP-1/2 activity. These findings suggest that PARP-1/2 inhibitors may have therapeutic potential for treating ALS.319 In the context of Drosophila, PARP inhibitors were shown to prevent TDP-43 accumulation and alleviate neurodegeneration.320 Furthermore, PARP silencing reduces neurotoxicity by preventing the condensate formation of hnRNPA1 and TDP-43 in a Drosophila model of ALS.321
Regulation by small molecules
Amidst the quest for ALS therapeutics, the concept of ‘druggable’ condensates emerges as a tempting prospect.258,322–329 Approved drugs like Cisplatin and Mitoxantrone exhibit a tendency for partitioning into transcriptional condensates in tumor cells, opening avenues for exploration (Table 6).330 Compounds such as daunorubicin, pyrvinium, and pararosaniline, identified through high-content cellular screening, exhibit the potential to modulate condensate behaviors selectively. These compounds were able to prevent the accumulation of TDP-43, FUS, and HNRNPA2B1 in SGs and ameliorate neurotoxicity in ALS patients.331 Additionally, specific compounds have been pinpointed for their role in reducing stress-induced TDP-43 aggregation in PC12 cells, marking significant progress in the pursuit of effective ALS treatments.332
Table 6.
Drugs modulate LLPS in disease
| Drug | Function | Mechanism | Reference |
|---|---|---|---|
| Adriamycin (doxorubicin) | Anticancer drug | Adriamycin induces conformational change of chromatin by triggering the phase transition of H1 and forming fibrous aggregates | 326 |
| AR (Androgen Receptor) | AR antagonist | ET516 inhibits the transcriptional activity of AR by disrupting AR condensates, thereby inhibiting the proliferation and tumor growth of prostate cancer cells expressing AR-resistant mutant. | 328 |
| Cisplatin and Mitoxantrone | Antineoplastic drugs | The antineoplastic drugs are concentrated in specific protein condensates in tumor cells. The therapeutic efficacy of the drugs is related to their ability to partition into the condensate that harbors their target. | 330 |
| HIV | Anti-HIV drug | LLPS of SRC-1 is required for activating YAP oncogenic transcription. EVG inhibits YAP transcription by disrupting the LLPS of SRC-1. | 327 |
| SHP099 | Protein tyrosine phosphatase (PTP)SHP2 inhibitor | LLPS serves as a gain-of-function mechanism involved in the pathogenesis of SHP2-associated diseases. SHP2 allosteric inhibitors attenuate LLPS of SHP2 mutants. | 329 |
The interactions between TDP-43 and RNA drive the formation of abnormal inclusions, culminating in neurotoxicity. However, a promising therapeutic avenue unfolds as oligonucleotides, comprising TDP-43 target sequences, demonstrate their ability to prevent inclusions and neurotoxicity. This approach unveils the potential of oligonucleotides as a ray of hope in mitigating the neurotoxic impact of TDP-43 aberrations, providing a novel and targeted strategy in ALS treatment.
Progress of targeting LLPS in clinic
Neurodegenerative diseases
The aggregation of α-Syn is associated with the development of Parkinson’s disease. Research using in vitro reconstitution and cellular models has demonstrated that the LLPS of α-Syn occurs before its aggregation.216 Therefore, direct targeting of α-Syn has become a potential therapeutic strategy for Parkinson’s disease. Minzasolmin (UCB0599) is a small molecule targeting α-Syn misfolding and is currently being investigated as a potential treatment for Parkinson’s disease.333 Using the Line 61 transgenic mouse model of Parkinson’s disease, Price et al. found that minzasolmin-induced reductions in α-Syn pathology were associated with improvements in gait and decreased inflammatory markers.333 These findings provide evidence for determining appropriate human doses (ClinicalTrials.gov ID: NCT04658186; EudraCT Number 2020-003265).
The accumulation of over-phosphorylated, tangled microtubule-associated protein tau (MAPT) is a defining feature of AD.334–341 An exciting recent discovery is that tau has a strong tendency to undergo LLPS.211 A randomized phase 1b clinical trial investigated the effect of BIIB080, an antisense oligonucleotide targeting MAPT, on tau synthesis in patients with mild AD. The trial reported that BIIB080 was well tolerated and resulted in a dose-dependent decrease in total tau (t-tau) and phosphorylated tau (p-tau181) levels within the cerebrospinal fluid. Tau PET imaging showed reduced tau accumulation compared to placebo at week 25.342
The misfolding and mutation of Cu/Zn superoxide dismutase (SOD1) are frequently linked to ALS. SOD1 can accumulate within stress granules via LLPS.343 In a clinical trial involving 50 ALS patients with SOD1 mutations, CSF SOD1 concentrations decreased following 12 weeks of intrathecal administration of the antisense oligonucleotide tofersen at the highest dose.344
Huntington’s disease is caused by the expansion of a CAG trinucleotide repeat in the Huntingtin (HTT) gene.345–347 The HTT protein can assemble into liquid-like structures, which can transition into solid-like fibrillar assemblies within cells.348 This transition was further enhanced when the R200/205 methylation sites were modified.349 Therapeutic strategies aimed at lowering HTT levels are being developed to slow or halt the progression of Huntington’s disease.350–355 HTTRx, an antisense oligonucleotide developed to block HTT mRNA, reduces levels of mutant huntingtin protein. In patients with early Huntington’s disease, intrathecal administration of HTTRx did not result in serious adverse events and led to dose-dependent reductions in mutant huntingtin levels.356
The expansions of GGGGCC hexanucleotide repeat in C9ORF72 are the most common genetic cause of ALS and FTD.223,357–362 The hexanucleotide repeat causes a gain of toxicity to the neurons through phase separation.363,364 A one-time injection of antisense oligonucleotides targeting RNAs containing the repeat resulted in prolonged reductions in RNA foci and dipeptide-repeat proteins with improvements in behavioral deficits.365
Cancer
The impact of genes related to LLPS on melanoma prognosis was investigated using the DrLLPS database, identifying TROAP as a gene marker associated with poor prognosis.366 Additionally, using The Cancer Genome Atlas and PhaSepDB datasets, Lai et al. identified five LLPS-related genes (LRGs) (BMX, FYN, KPNA2, PFKFB4, and SPP1) linked to the overall survival of hepatocellular carcinoma patients.367
LRGs extracted from PhaSepDB have proven valuable in predicting anticancer drug sensitivity in prostate cancer patients.368 FUS showed efficacy in predicting sensitivity to Nelarabine, Hydroxyurea, and Pemetrexed. USH1C emerged as a reliable predictor for Fluorouracil, Arsenic trioxide, and Everolimus sensitivity. Additionally, TAZ demonstrated predictive capabilities for Cladribine, Nelarabine, and Fludarabine sensitivity, while TPX2 was also effective in predicting Nelarabine sensitivity.368
Cardiovascular diseases
RBM20 is a splicing factor involved in cardiovascular development.185 RBM20 mutations cause the relocation of RBM20 from nuclear splicing speckles to cytoplasmic condensates, resulting in the sequestration of mRNA, polysomes, and cardiac cytoskeleton proteins.187 Wyles et al. investigated the impact of β-adrenergic stress on familial DCM using human-induced pluripotent stem cell (hiPSC)-derived cardiomyocytes (CMs) from a patient with RBM20-related DCM. Their findings revealed that RBM20-deficient familial DCM hiPSC-CMs were more susceptible to stress, a vulnerability that could be therapeutically reduced by the β-blocker carvedilol and the Ca2+ channel blocker verapamil.369 This study paves the way for the development of therapeutic strategies to modify the progression of DCM.
Hematological disorders
WASP mutations cause Wiskott–Aldrich syndrome (WAS), characterized by eczema, thrombocytopenia, immune deficiency, and thrombocytopenia.288 WASP regulates RNA splicing through a phase-separation process, and mutations in the WASP gene result in abnormal RNA splicing.289 Gene therapy using modified autologous CD34+ cells is an emerging treatment for WAS. Ferrua et al. presented safety and efficacy data from an interim analysis of a phase 1/2 clinical study on patients with severe WAS who underwent lentiviral vector-based gene therapy.370 The treatment involved a single intravenous infusion of autologous CD34+ cells genetically modified with a lentiviral vector encoding human WAS cDNA. The overall survival rate was 100%, with successful and sustained engraftment of genetically corrected HSPCs in all patients. The study also demonstrated improved immune function, as indicated by normalized in vitro T-cell function and the discontinuation of immunoglobulin supplementation in seven patients with follow-up longer than one year. Additionally, severe infections decreased, and platelet counts significantly improved. These findings were corroborated by a recent phase 1/2 clinical trial, which reported outcomes for five patients with severe WAS who received gene therapy using a self-inactivating lentiviral vector expressing human WAS cDNA.371 All patients were alive and in good health, with sustained multilineage vector gene marking. Universal clinical improvements were observed in eczema, infections, and bleeding diathesis. Notably, the most significant enhancements in platelet count and cytoskeletal function in myeloid cells occurred in patients with higher vector copy numbers in the transduced product.
Conclusions and perspective
The exploration into MLOs has revealed a dynamic aspect of cellular organization, challenging traditional compartmentalization and adding depth to our comprehension of cellular dynamics. This extensive analysis delves into the varied functions of MLOs, encompassing SGs, P-bodies, the nucleolus, and the principles of LLPS across diverse biological scenarios. As we wrap up this investigation, we contemplate the current state of knowledge and offer insights into the future directions of this rapidly evolving field.
The identification and characterization of MLOs have unveiled the intricate nature of cellular organization. These formations, facilitated by LLPS, exemplify the adaptable and responsive characteristics of cellular constituents. The complex interaction between membrane-bound and MLOs highlights the precise coordination governing cellular activities. MLOs assume diverse roles in cellular mechanisms like mRNA regulation, protein equilibrium, and stress response, underscoring their significance in preserving cellular equilibrium.
As research progresses, the influence of MLOs expands beyond traditional cellular mechanisms. Their significance is apparent in diverse biological contexts, from regulating gene expression in cancer cells to coordinating metabolism and cardiac function and impacting the dynamics of aging and neurodegenerative diseases. This broadening biological scope underscores the ubiquity and versatility of MLOs, positioning them as central actors in cellular physiology and pathology.
The therapeutic potential of MLOs is an emerging area of exploration. Targeting these dynamic structures provides a novel avenue to intervene in cellular processes associated with various ailments. Small molecules that modulate LLPS or influence the dynamics of specific MLOs hold promise as prospective therapeutics. Ongoing endeavors to translate knowledge into therapeutic tactics underscore the potential impact of this field on future medical interventions.
As therapeutic strategies targeting MLOs progress, ethical considerations must remain paramount. Understanding potential off-target effects and unintended consequences of modulating LLPS or MLOs dynamics is critical. Ethical frameworks should guide the development and implementation of therapeutic interventions to ensure safety and efficacy.
Despite significant progress in understanding MLOs, challenges persist in comprehending their assembly, dynamics, and functionalities. The ephemeral and dynamic nature of these structures presents experimental hurdles, necessitating innovative approaches to observe and manipulate MLOs within living cells. Further inquiry is essential to decipher the principles dictating LLPS behavior and the factors influencing condensate formation.
To fully elucidate the complexities of MLOs, a multidisciplinary approach and reagents (Table 7) are imperative.372 Integration of techniques from cell biology, biophysics, biochemistry, and computational biology is essential to attain a comprehensive understanding of MLOs formation, regulation, and functionalities. Collaborative endeavors across disciplines will drive innovation, enabling researchers to effectively address the intricacies of these dynamic structures.
Table 7.
Reagents modulate LLPS
| Reagent | Function | Mechanism | Reference |
|---|---|---|---|
| Dextran | Crowding reagent | Crowding reagents to promote the formation of MLO | 3 |
| Ficoll | Crowding reagent | Crowding reagents to promote the formation of MLO | 3 |
| 1,6-Hexanediol (1,6-HD) | 1,6-HD is an inhibitor and indicator of phase separation | 1,6-HD disrupts LLPS by interfering the weak hydrophobic protein-protein or protein-RNA interactions. | 3 |
| Lipoamide and lipoic acid | Inhibit stress granule formation | Dissolve stress granule by affecting the kinetics of phase separation | 374 |
| Poly(ethylene glycol) | Crowding reagent | Crowding reagents to promote MLO | 3 |
The field of MLOs stands on the brink of ongoing expansion and exploration. Future investigations may focus on elucidating the functions of specific proteins, RNA molecules, and PTMs in governing MLOs. Given the evolutionary connection between PTM and LLPS, it is likely that additional types of PTMs remain to be discovered. For instance, the strong electronic neutralization effect of O-glycosylation may inhibit potential LLPS. Investigating how PTMs regulate LLPS will undoubtedly become a key focus of future research.373 The potential of lysosome-associated condensates in therapeutic applications should be further explored. For instance, Aloperine (ALO), a natural alkaloid, has demonstrated the ability to inhibit tumor growth by directly targeting lysosomes in glioma cells.374 Progressing technologies, including advanced imaging methodologies and CRISPR-based techniques, are expected to bolster our capacity to manipulate and scrutinize these structures in real time.
In summary, MLOs serve as connectors between cellular compartments, challenging traditional perceptions of cellular organization. Their dynamic essence, illuminated through LLPS, underscores the inherent flexibility within cellular processes. The convergence of technological advancement, interdisciplinary cooperation, and ethical considerations will steer us toward unlocking the complete potential of these dynamic structures in both health and disease. The pursuit of comprehending MLOs persists, promising a future where their contributions to cellular dynamics are fully understood and leveraged for therapeutic progress.
Unanswered questions
While significant advancements have been achieved in understanding MLOs and their roles in cellular function, several unanswered questions persist, particularly regarding their involvement in human diseases. Here are some of the key unanswered questions in this field:
Disease-specific mechanisms
What are the specific molecular mechanisms underlying the dysregulation of MLOs in different human diseases? While it is known that disruptions in MLOs play a role in disease development, the precise molecular events and interactions involved in this process remain incompletely understood.
Contributions to disease progression
How do alterations in MLOs contribute to the progression of various human diseases? While some studies have linked MLOs dysfunction to disease initiation, it is unclear how these alterations evolve over time and exacerbate disease severity.
Cell-type specificity
Are MLOs present in all cell types, or are they restricted to particular cell types? Do MLOs dysfunctions exhibit cell-type specificity in the context of human diseases? Different cell types may have distinct MLOs compositions and functions, and understanding how MLOs dysregulation affects specific cell types could provide insights into disease heterogeneity.
Interplay with genetic and environmental factors
How do genetic and environmental factors interact with MLOs dysregulation to influence disease susceptibility and progression? When might the reprogramming of MLOs occur? Can MLOs be passed down to daughter cells during cell division? Genetic variants and environmental stressors may modulate MLOs function and contribute to disease risk, but the underlying mechanisms are not fully elucidated.
Diagnostic and prognostic markers
Can MLOs dysregulation serve as diagnostic or prognostic indicators for human diseases? Identifying specific MLOs alterations associated with disease states could facilitate the advancement of biomarkers for early disease detection and prognostication.
Concept issue
The concept of LLPS is somewhat inaccurate, as there is also a gel state and solid state. Therefore, whether it should be changed to “phase separation” or “phase transition” is debatable. The intrinsically disordered region (IDR) is important for LLPS, but protein phase transition doesn’t necessarily require an IDR. Proteins with disordered regions don’t necessarily undergo phase transition. Therefore, the mechanisms underlying the formation of LLPS remain to be explored.
The interaction and location
The interactions between MLOs and membrane-bound organelles under normal and abnormal conditions require further research. The half-life and number of MLOs within a particular type of cells need further clarification. Previous studies focus on individual molecule, their upstream and downstream regulation. MLOs concentrate key molecules together, acting in specific times and spaces (nucleus, cytoplasm, potential mitochondria). However, the dynamic network mediated by MLOs remains largely unexplored.
Addressing these unanswered questions will require interdisciplinary approaches combining genetics, molecular biology, cell biology, biochemistry, and clinical research. By elucidating the complex roles of MLOs in human diseases, researchers will uncover novel therapeutic targets and diagnostic tools to improve patient outcomes.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82370264 to Y.Li, 82370501 to Y.H.S., 81870194 and 91849122 to Y.Li, 92168203 to Z.S.). Project of State Key Laboratory of Radiation Medicine and Protection, Soochow University (No. GZK12023023 to Y.Li). Jiangsu Cardiovascular Medicine Innovation Center (CXZX202210). China Postdoctoral Science Foundation (No. 2021M702394 to Y.W.), Jiangsu Funding Program for Excellent Postdoctoral Talent (No. 2022ZB578 to Y.W.). The project for the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The National Key Research and Development Program of China (2022YFE0209700) to X.Y. Translational Research Grant of NCRCH (2020WSB07). We thank Dr. Ali J Marian for insightful comments and advice in structuring the manuscript. BioRender (https://www.biorender.com) was used to create the figures. We thank Shiyuan Han, Yu Zhang, Weiliang Wu, Tonggan Lu, and Sha Geng for editing the tables and figures.
Author contributions
There are five first authors in this manuscript and they have equally contributed to this project. Y.X.L., Y.Z.L., X.Y.Y., Y.X., X.B.P., Y.S., and Y.L. drafted the manuscript, prepared the tables and figures, and participated in the discussion. We have three corresponding authors in this manuscript. Y.X.L. and Z.Y.S. conceptualized this work. Y.H.S. revised the manuscript and provided intellectual input. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yangxin Li, Yuzhe Liu, Xi-Yong Yu, Yan Xu, Xiangbin Pan
Contributor Information
Yangxin Li, Email: yangxin_li@yahoo.com.
Yao-Hua Song, Email: yaohua_song1@yahoo.com.
Zhenya Shen, Email: uuzyshen@aliyun.com.
References
- 1.Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren, J., Zhang, Z., Zong, Z., Zhang, L. & Zhou, F. Emerging implications of phase separation in cancer. Adv. Sci.9, e2202855 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao, Y., Li, X., Li, P. & Lin, Y. A brief guideline for studies of phase-separated biomolecular condensates. Nat. Chem. Biol.18, 1307–1318 (2022). [DOI] [PubMed] [Google Scholar]
- 4.Volkov, V. A. & Akhmanova, A. Phase separation on microtubules: from droplet formation to cellular function? Trends Cell Biol.34, 18–30 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Sahin, C., Leppert, A. & Landreh, M. Advances in mass spectrometry to unravel the structure and function of protein condensates. Nat. Protoc.18, 3653–3661 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Nesterov, S. V., Ilyinsky, N. S. & Uversky, V. N. Liquid-liquid phase separation as a common organizing principle of intracellular space and biomembranes providing dynamic adaptive responses. Biochim. Biophys. Acta Mol. Cell Res.1868, 119102 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long, Q., Zhou, Y., Guo, J., Wu, H. & Liu, X. Multi-phase separation in mitochondrial nucleoids and eukaryotic nuclei. Biophys. Rep.9, 113–119 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisvert, F. M., van Koningsbruggen, S., Navascues, J. & Lamond, A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol.8, 574–585 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, Q. et al. Diverse CMT2 neuropathies are linked to aberrant G3BP interactions in stress granules. Cell186, 803–820.e825 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Kedersha, N., Ivanov, P. & Anderson, P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci.38, 494–506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science324, 1729–1732 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Michelitsch, M. D. & Weissman, J. S. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl Acad. Sci. USA97, 11910–11915 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banani, S. F. et al. Compositional control of phase-separated cellular bodies. Cell166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, J. et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell174, 688–699.e616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holehouse, A. S. & Kragelund, B. B. The molecular basis for cellular function of intrinsically disordered protein regions. Nat. Rev. Mol. Cell Biol.25, 187–211 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moses, D., Ginell, G. M., Holehouse, A. S. & Sukenik, S. Intrinsically disordered regions are poised to act as sensors of cellular chemistry. Trends Biochem. Sci.48, 1019–1034 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oates, M. E. et al. D2P2: database of disordered protein predictions. Nucleic Acids Res.41, D508–D516 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning, W. et al. DrLLPS: a data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res.48, D288–D295 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Q. et al. LLPSDB: a database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res.48, D320–D327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou, C., Xie, H., Fu, Y., Ma, Y. & Li, T. MloDisDB: a manually curated database of the relations between membraneless organelles and diseases. Brief Bioinform22, bbaa271 (2021). [DOI] [PubMed]
- 26.You, K. et al. PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res.48, D354–D359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, Z. et al. Screening membraneless organelle participants with machine-learning models that integrate multimodal features. Proc. Natl Acad. Sci. USA119, e2115369119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meszaros, B. et al. PhaSePro: the database of proteins driving liquid-liquid phase separation. Nucleic Acids Res.48, D360–D367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lancaster, A. K., Nutter-Upham, A., Lindquist, S. & King, O. D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics30, 2501–2502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullick, P. & Trovato, A. Sequence-based prediction of protein phase separation: the role of beta-pairing propensity. Biomolecules12, 1771 (2022). [DOI] [PMC free article] [PubMed]
- 31.Chu, X. et al. Prediction of liquid-liquid phase separating proteins using machine learning. BMC Bioinformatics23, 72 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberti, S., Halfmann, R., King, O., Kapila, A. & Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell137, 146–158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franzmann, T. M. & Alberti, S. Prion-like low-complexity sequences: key regulators of protein solubility and phase behavior. J. Biol. Chem.294, 7128–7136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Ramirez, D. A., Hough, L. E. & Shirts, M. R. Coiled-coil domains are sufficient to drive liquid-liquid phase separation of proteins in molecular models. Biophys. J.123, 703–717 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohanty, P. et al. Principles governing the phase separation of multidomain proteins. Biochemistry61, 2443–2455 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin, E. W. et al. Interplay of folded domains and the disordered low-complexity domain in mediating hnRNPA1 phase separation. Nucleic Acids Res.49, 2931–2945 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature483, 336–340 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin, Y. et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell175, 1481–1491.e1413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markaki, Y. et al. Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell184, 6212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, H. Y. et al. Coupling of protein condensates to ordered lipid domains determines functional membrane organization. Sci. Adv.9, eadf6205 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimobayashi, S. F., Ronceray, P., Sanders, D. W., Haataja, M. P. & Brangwynne, C. P. Nucleation landscape of biomolecular condensates. Nature599, 503–506 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Mehta, S. & Zhang, J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer22, 239–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, J. Z. et al. Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell182, 1531–1544.e1515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsang, B., Pritisanac, I., Scherer, S. W., Moses, A. M. & Forman-Kay, J. D. Phase separation as a missing mechanism for interpretation of disease mutations. Cell183, 1742–1756 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Mittag, T. & Pappu, R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell82, 2201–2214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong, X. et al. Liquid-liquid phase separation in tumor biology. Signal Transduct. Target. Ther.7, 221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ugolini, I. et al. Chromatin localization of nucleophosmin organizes ribosome biogenesis. Mol. Cell82, 4443–4457.e4449 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito, M. et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol.15, 51–61 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Wang, L. et al. Multiphase coalescence mediates Hippo pathway activation. Cell185, 4376–4393.e4318 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson, J. L. et al. Macromolecular condensation buffers intracellular water potential. Nature623, 842–852 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gui, T. et al. Targeted perturbation of signaling-driven condensates. Mol. Cell83, 4141–4157.e4111 (2023). [DOI] [PubMed] [Google Scholar]
- 54.Fritsch, A. W. et al. Local thermodynamics govern formation and dissolution of Caenorhabditis elegans P granule condensates. Proc. Natl Acad. Sci. USA118, e2102772118 (2021). [DOI] [PMC free article] [PubMed]
- 55.Choi, J. M., Holehouse, A. S. & Pappu, R. V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys.49, 107–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peran, I. & Mittag, T. Molecular structure in biomolecular condensates. Curr. Opin. Struct. Biol.60, 17–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gouveia, B. et al. Capillary forces generated by biomolecular condensates. Nature609, 255–264 (2022). [DOI] [PubMed] [Google Scholar]
- 58.Choi, J. M., Dar, F. & Pappu, R. V. LASSI: a lattice model for simulating phase transitions of multivalent proteins. PLoS Comput. Biol.15, e1007028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harmon, T. S., Holehouse, A. S., Rosen, M. K. & Pappu, R. V. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife6, e30294 (2017). [DOI] [PMC free article] [PubMed]
- 60.Martin, E. W. et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science367, 694–699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bremer, A. et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem.14, 196–207 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature581, 209–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol.22, 183–195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Glauninger, H., Wong Hickernell, C. J., Bard, J. A. M. & Drummond, D. A. Stressful steps: progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell82, 2544–2556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaur, T. et al. Sequence-encoded and composition-dependent protein-RNA interactions control multiphasic condensate morphologies. Nat. Commun.12, 872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo, H. et al. Spatial engineering of E. coli with addressable phase-separated RNAs. Cell185, 3823–3837.e3823 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Galvanetto, N. et al. Extreme dynamics in a biomolecular condensate. Nature619, 876–883 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol.22, 196–213 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Ma, W. & Mayr, C. A membraneless organelle associated with the endoplasmic reticulum enables 3’UTR-mediated protein-protein interactions. Cell175, 1492–1506.e1419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lyons, H. et al. Functional partitioning of transcriptional regulators by patterned charge blocks. Cell186, 327–345.e328 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tulpule, A. et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell184, 2649–2664.e2618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, H. et al. Phosphorylation-dependent membraneless organelle fusion and fission illustrated by postsynaptic density assemblies. Mol. Cell84, 309–326.e307 (2024). [DOI] [PubMed] [Google Scholar]
- 73.Qiu, H. et al. Short-distance vesicle transport via phase separation. Cell187, 2175–2193.e2121 (2024). [DOI] [PubMed] [Google Scholar]
- 74.Cardona, A. H. et al. Self-demixing of mRNA copies buffers mRNA:mRNA and mRNA:regulator stoichiometries. Cell186, 4310–4324.e4323 (2023). [DOI] [PubMed] [Google Scholar]
- 75.Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature573, 144–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell149, 768–779 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Patil, A. et al. A disordered region controls cBAF activity via condensation and partner recruitment. Cell186, 4936–4955.e4926 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu, Y. et al. Disrupting the phase separation of KAT8-IRF1 diminishes PD-L1 expression and promotes antitumor immunity. Nat. Cancer4, 382–400 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rawat, P. et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell81, 1013–1026.e1011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boyd-Shiwarski, C. R. et al. WNK kinases sense molecular crowding and rescue cell volume via phase separation. Cell185, 4488–4506.e4420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell173, 720–734.e715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoo, H., Bard, J. A. M., Pilipenko, E. V. & Drummond, D. A. Chaperones directly and efficiently disperse stress-triggered biomolecular condensates. Mol. Cell82, 741–755.e711 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bratek-Skicki, A., Van Nerom, M., Maes, D. & Tompa, P. Biological colloids: unique properties of membraneless organelles in the cell. Adv. Colloid Interface Sci.310, 102777 (2022). [DOI] [PubMed] [Google Scholar]
- 84.Alshareedah, I., Moosa, M. M., Pham, M., Potoyan, D. A. & Banerjee, P. R. Programmable viscoelasticity in protein-RNA condensates with disordered sticker-spacer polypeptides. Nat. Commun.12, 6620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dias, C. S., Araujo, N. A. M. & Telo da Gama, M. M. Dynamics of network fluids. Adv. Colloid Interface Sci.247, 258–263 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Woodruff, J. B., Hyman, A. A. & Boke, E. Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci.43, 81–94 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Espinosa, J. R. et al. Liquid network connectivity regulates the stability and composition of biomolecular condensates with many components. Proc. Natl Acad. Sci. USA117, 13238–13247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.St George-Hyslop, P. et al. The physiological and pathological biophysics of phase separation and gelation of RNA binding proteins in amyotrophic lateral sclerosis and fronto-temporal lobar degeneration. Brain Res.1693, 11–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murakami, T. et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron88, 678–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nair, S. J. et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol.26, 193–203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Risso-Ballester, J. et al. A condensate-hardening drug blocks RSV replication in vivo. Nature595, 596–599 (2021). [DOI] [PubMed] [Google Scholar]
- 92.Lyon, A. S., Peeples, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol.22, 215–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nag, N., Sasidharan, S., Uversky, V. N., Saudagar, P. & Tripathi, T. Phase separation of FG-nucleoporins in nuclear pore complexes. Biochim. Biophys. Acta Mol. Cell Res.1869, 119205 (2022). [DOI] [PubMed] [Google Scholar]
- 94.Frey, S., Richter, R. P. & Gorlich, D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science314, 815–817 (2006). [DOI] [PubMed] [Google Scholar]
- 95.Su, X. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bussi, C. et al. Stress granules plug and stabilize damaged endolysosomal membranes. Nature623, 1062–1069 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rebane, A. A. et al. Liquid-liquid phase separation of the Golgi matrix protein GM130. FEBS Lett.594, 1132–1144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mendes, L. F. S., Oliveira, C. G., Simoes, K. F., Kava, E. & Costa-Filho, A. J. Exploring liquid-liquid phase separation in the organisation of Golgi matrix proteins. Biochim. Biophys. Acta Proteins Proteom.1872, 141029 (2024). [DOI] [PubMed] [Google Scholar]
- 99.Milovanovic, D., Wu, Y., Bian, X. & De Camilli, P. A liquid phase of synapsin and lipid vesicles. Science361, 604–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoffmann, C. et al. Synapsin condensation controls synaptic vesicle sequestering and dynamics. Nat. Commun.14, 6730 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ziltener, P., Rebane, A. A., Graham, M., Ernst, A. M. & Rothman, J. E. The golgin family exhibits a propensity to form condensates in living cells. FEBS Lett.594, 3086–3094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.So, C., Cheng, S. & Schuh, M. Phase separation during germline development. Trends Cell Biol.31, 254–268 (2021). [DOI] [PubMed] [Google Scholar]
- 103.Boke, E. et al. Amyloid-like self-assembly of a cellular compartment. Cell166, 637–650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Farag, M. et al. Condensates formed by prion-like low-complexity domains have small-world network structures and interfaces defined by expanded conformations. Nat. Commun.13, 7722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoffmann, C. et al. Electric potential at the interface of membraneless organelles gauged by graphene. Nano Lett.23, 10796–10801 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dai, Y. et al. Interface of biomolecular condensates modulates redox reactions. Chem9, 1594–1609 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mitrea, D. M. et al. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J. Mol. Biol.430, 4773–4805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell168, 159–171.e114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hirose, T. et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell25, 169–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McSwiggen, D. T., Mir, M., Darzacq, X. & Tjian, R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev.33, 1619–1634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aryan, F. et al. Nucleolus activity-dependent recruitment and biomolecular condensation by pH sensing. Mol. Cell83, 4413–4423.e4410 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hirose, T., Ninomiya, K., Nakagawa, S. & Yamazaki, T. A guide to membraneless organelles and their various roles in gene regulation. Nat. Rev. Mol. Cell Biol.24, 288–304 (2023). [DOI] [PubMed] [Google Scholar]
- 113.Mensah, M. A. et al. Aberrant phase separation and nucleolar dysfunction in rare genetic diseases. Nature614, 564–571 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Phair, R. D. & Misteli, T. High mobility of proteins in the mammalian cell nucleus. Nature404, 604–609 (2000). [DOI] [PubMed] [Google Scholar]
- 115.Girard, J. P. et al. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J.11, 673–682 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yao, R. W. et al. Nascent pre-rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell76, 767–783.e711 (2019). [DOI] [PubMed] [Google Scholar]
- 117.Strome, S. Specification of the germ line. WormBook 1–10, 10.1895/wormbook.1.9.1 (2005). [DOI] [PMC free article] [PubMed]
- 118.Wang, J. T. & Seydoux, G. P granules. Curr. Biol.24, R637–R638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Putnam, A., Cassani, M., Smith, J. & Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol.26, 220–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Imamura, K. et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell53, 393–406 (2014). [DOI] [PubMed] [Google Scholar]
- 121.Fox, A. H. et al. Paraspeckles: a novel nuclear domain. Curr. Biol.12, 13–25 (2002). [DOI] [PubMed] [Google Scholar]
- 122.Corpet, A. et al. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res.48, 11890–11912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fonin, A. V. et al. New evidence of the importance of weak interactions in the formation of PML-bodies. Int. J. Mol. Sci.23, 1613 (2022). [DOI] [PMC free article] [PubMed]
- 124.Wu, W. et al. Phase separation is required for PML nuclear body biogenesis and function. FASEB J.37, e22986 (2023). [DOI] [PubMed] [Google Scholar]
- 125.Niu, D. et al. Hemolytic reactions in the hemolymph of bivalve Sinonovacula constricta show complement-like activity. Fish. Shellfish Immunol.79, 11–17 (2018). [DOI] [PubMed] [Google Scholar]
- 126.Scherer, M. et al. Dual signaling via interferon and DNA damage response elicits entrapment by giant PML nuclear bodies. Elife11, e73006 (2022). [DOI] [PMC free article] [PubMed]
- 127.Ishov, A. M. et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol.147, 221–234 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun, H. et al. Recruitment of TRIM33 to cell-context specific PML nuclear bodies regulates nodal signaling in mESCs. EMBO J.42, e112058 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sarge, K. D., Murphy, S. P. & Morimoto, R. I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell Biol.13, 1392–1407 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Misteli, T. Protein dynamics: implications for nuclear architecture and gene expression. Science291, 843–847 (2001). [DOI] [PubMed] [Google Scholar]
- 131.Jolly, C., Usson, Y. & Morimoto, R. I. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl Acad. Sci. USA96, 6769–6774 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Holmberg, C. I., Tran, S. E., Eriksson, J. E. & Sistonen, L. Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends Biochem. Sci.27, 619–627 (2002). [DOI] [PubMed] [Google Scholar]
- 133.Cotto, J., Fox, S. & Morimoto, R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci.110, 2925–2934 (1997). [DOI] [PubMed] [Google Scholar]
- 134.Jolly, C., Morimoto, R., Robert-Nicoud, M. & Vourc’h, C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell Sci.110, 2935–2941 (1997). [DOI] [PubMed] [Google Scholar]
- 135.Denegri, M. et al. Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol. Biol. Cell13, 2069–2079 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mokin, Y. I. et al. Nucleolar- and nuclear-stress-induced membrane-less organelles: a proteome analysis through the prism of liquid-liquid phase separation. Int. J. Mol. Sci.24, 11007 (2023). [DOI] [PMC free article] [PubMed]
- 137.Audas, T. E. et al. Adaptation to stressors by systemic protein amyloidogenesis. Dev. Cell39, 155–168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marijan, D. et al. Stress-specific aggregation of proteins in the amyloid bodies. FEBS Lett.593, 3162–3172 (2019). [DOI] [PubMed] [Google Scholar]
- 139.Yang, P. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell181, 325–345.e328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Anderson, P. & Kedersha, N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol.10, 430–436 (2009). [DOI] [PubMed] [Google Scholar]
- 141.Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kroschwald, S. et al. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife4, e06807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reineke, L. C., Kedersha, N., Langereis, M. A., van Kuppeveld, F. J. & Lloyd, R. E. Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. mBio6, e02486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol.201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ramaswami, M., Taylor, J. P. & Parker, R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell154, 727–736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kaer, P., Nielsen, T. R., Lodahl, P., Jauho, A. P. & Mork, J. Non-markovian model of photon-assisted dephasing by electron-phonon interactions in a coupled quantum-dot-cavity system. Phys. Rev. Lett.104, 157401 (2010). [DOI] [PubMed] [Google Scholar]
- 147.Kaehler, C., Isensee, J., Hucho, T., Lehrach, H. & Krobitsch, S. 5-Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Res.42, 6436–6447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Luo, Y., Na, Z. & Slavoff, S. A. P-bodies: composition, properties, and functions. Biochemistry57, 2424–2431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kulkarni, M., Ozgur, S. & Stoecklin, G. On track with P-bodies. Biochem. Soc. Trans.38, 242–251 (2010). [DOI] [PubMed] [Google Scholar]
- 150.Youn, J. Y. et al. Properties of stress granule and P-body proteomes. Mol. Cell76, 286–294 (2019). [DOI] [PubMed] [Google Scholar]
- 151.Du, Z. et al. Condensate cooperativity underlies transgenerational gene silencing. Cell Rep.42, 112859 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ilik, I. A. et al. SON and SRRM2 are essential for nuclear speckle formation. Elife9, e60579 (2020). [DOI] [PMC free article] [PubMed]
- 153.Takakuwa, H. et al. Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles. Nat. Cell Biol.25, 1664–1675 (2023). [DOI] [PubMed] [Google Scholar]
- 154.Deng, Q. & Wang, H. Re-visiting the principles of apicobasal polarity in Drosophila neural stem cells. Dev. Biol.484, 57–62 (2022). [DOI] [PubMed] [Google Scholar]
- 155.Liu, Z. et al. Par complex cluster formation mediated by phase separation. Nat. Commun.11, 2266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu, X. et al. Mitotic implantation of the transcription factor Prospero via phase separation drives terminal neuronal differentiation. Dev. Cell52, 277–293.e278 (2020). [DOI] [PubMed] [Google Scholar]
- 157.Shan, Z. et al. Basal condensation of Numb and Pon complex via phase transition during Drosophila neuroblast asymmetric division. Nat. Commun.9, 737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Long, Q. et al. Phase separation drives the self-assembly of mitochondrial nucleoids for transcriptional modulation. Nat. Struct. Mol. Biol.28, 900–908 (2021). [DOI] [PubMed] [Google Scholar]
- 159.Esgleas, M. et al. Trnp1 organizes diverse nuclear membrane-less compartments in neural stem cells. EMBO J.39, e103373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ishioka, M. et al. Myogenetic oligodeoxynucleotide induces myocardial differentiation of murine pluripotent stem cells. Int. J. Mol. Sci.24, 14380 (2023). [DOI] [PMC free article] [PubMed]
- 161.Yan, Y. et al. CRYbetaB2 enhances tumorigenesis through upregulation of nucleolin in triple negative breast cancer. Oncogene40, 5752–5763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Choi, Y. J. et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science355, eaag1927 (2017). [DOI] [PMC free article] [PubMed]
- 163.Yu, H. et al. rRNA biogenesis regulates mouse 2C-like state by 3D structure reorganization of peri-nucleolar heterochromatin. Nat. Commun.12, 6365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tan, T. et al. Dynamic nucleolar phase separation influenced by non-canonical function of LIN28A instructs pluripotent stem cell fate decisions. Nat. Commun.15, 1256 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fang, Q. et al. YTHDF1 phase separation triggers the fate transition of spermatogonial stem cells by activating the IkappaB-NF-kappaB-CCND1 axis. Cell Rep.42, 112403 (2023). [DOI] [PubMed] [Google Scholar]
- 166.Zhang, Y. et al. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat. Genet.51, 1380–1388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang, J. et al. Phase separation of OCT4 controls TAD reorganization to promote cell fate transitions. Cell Stem Cell28, 1868–1883.e1811 (2021). [DOI] [PubMed] [Google Scholar]
- 168.Buddika, K. et al. Coordinated repression of pro-differentiation genes via P-bodies and transcription maintains Drosophila intestinal stem cell identity. Curr. Biol.32, 386–397.e386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kedia, S. et al. Ubiquitination and deubiquitination of 4E-T regulate neural progenitor cell maintenance and neurogenesis by controlling P-body formation. Cell Rep.40, 111070 (2022). [DOI] [PubMed] [Google Scholar]
- 170.Kolaj, A. et al. The P-body protein 4E-T represses translation to regulate the balance between cell genesis and establishment of the postnatal NSC pool. Cell Rep.42, 112242 (2023). [DOI] [PubMed] [Google Scholar]
- 171.Pecori, F. et al. Site-specific O-GlcNAcylation of Psme3 maintains mouse stem cell pluripotency by impairing P-body homeostasis. Cell Rep.36, 109361 (2021). [DOI] [PubMed] [Google Scholar]
- 172.Bhat, P. et al. Genome organization around nuclear speckles drives mRNA splicing efficiency. Nature629, 1165–1173 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sawyer, I. A., Sturgill, D., Sung, M. H., Hager, G. L. & Dundr, M. Cajal body function in genome organization and transcriptome diversity. Bioessays38, 1197–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nsengimana, B. et al. Processing body (P-body) and its mediators in cancer. Mol. Cell Biochem.477, 1217–1238 (2022). [DOI] [PubMed] [Google Scholar]
- 175.Niwa-Kawakita, M. et al. PML is a ROS sensor activating p53 upon oxidative stress. J. Exp. Med.214, 3197–3206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zheng, Y. et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther.7, 22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fujikawa, D. et al. Stress granule formation inhibits stress-induced apoptosis by selectively sequestering executioner caspases. Curr. Biol.33, 1967–1981.e1968 (2023). [DOI] [PubMed] [Google Scholar]
- 178.Gregorich, Z. R., Zhang, Y., Kamp, T. J., Granzier, H. L. & Guo, W. Mechanisms of RBM20 cardiomyopathy: insights from model systems. Circ. Genom. Precis. Med.17, e004355 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jiang, Y. et al. Phosphorylation-regulated dynamic phase separation of HIP-55 protects against heart failure. Circulation150, 938–951 (2024). [DOI] [PMC free article] [PubMed]
- 180.Simon, E., Faucheux, C., Zider, A., Theze, N. & Thiebaud, P. From vestigial to vestigial-like: the Drosophila gene that has taken wing. Dev. Genes Evol.226, 297–315 (2016). [DOI] [PubMed] [Google Scholar]
- 181.Figeac, N. et al. VGLL3 operates via TEAD1, TEAD3 and TEAD4 to influence myogenesis in skeletal muscle. J. Cell Sci.132, jcs225946 (2019). [DOI] [PMC free article] [PubMed]
- 182.Hori, N. et al. Vestigial-like family member 3 (VGLL3), a cofactor for TEAD transcription factors, promotes cancer cell proliferation by activating the Hippo pathway. J. Biol. Chem.295, 8798–8807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liang, Y. et al. A gene network regulated by the transcription factor VGLL3 as a promoter of sex-biased autoimmune diseases. Nat. Immunol.18, 152–160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Horii, Y. et al. VGLL3 is a mechanosensitive protein that promotes cardiac fibrosis through liquid-liquid phase separation. Nat. Commun.14, 550 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lennermann, D., Backs, J. & van den Hoogenhof, M. M. G. New insights in RBM20 cardiomyopathy. Curr. Heart Fail. Rep.17, 234–246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brauch, K. M. et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol.54, 930–941 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Schneider, J. W. et al. Dysregulated ribonucleoprotein granules promote cardiomyopathy in RBM20 gene-edited pigs. Nat. Med.26, 1788–1800 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang, S., Pei, G., Li, B., Li, P. & Lin, Y. Abnormal phase separation of biomacromolecules in human diseases. Acta Biochim. Biophys. Sin.55, 1133–1152 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chakraborty, P. & Zweckstetter, M. Role of aberrant phase separation in pathological protein aggregation. Curr. Opin. Struct. Biol.82, 102678 (2023). [DOI] [PubMed] [Google Scholar]
- 190.Takamuku, M. et al. Evolution of alpha-synuclein conformation ensemble toward amyloid fibril via liquid-liquid phase separation (LLPS) as investigated by dynamic nuclear polarization-enhanced solid-state MAS NMR. Neurochem. Int.157, 105345 (2022). [DOI] [PubMed] [Google Scholar]
- 191.Iqbal, K., Liu, F. & Gong, C. X. Tau and neurodegenerative disease: the story so far. Nat. Rev. Neurol.12, 15–27 (2016). [DOI] [PubMed] [Google Scholar]
- 192.Goedert, M., Eisenberg, D. S. & Crowther, R. A. Propagation of Tau aggregates and neurodegeneration. Annu. Rev. Neurosci.40, 189–210 (2017). [DOI] [PubMed] [Google Scholar]
- 193.Guo, T., Noble, W. & Hanger, D. P. Roles of tau protein in health and disease. Acta Neuropathol.133, 665–704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang, Y. & Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci.17, 5–21 (2016). [DOI] [PubMed] [Google Scholar]
- 195.Praschberger, R. et al. Neuronal identity defines alpha-synuclein and tau toxicity. Neuron111, 1577–1590.e1511 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Congdon, E. E., Ji, C., Tetlow, A. M., Jiang, Y. & Sigurdsson, E. M. Tau-targeting therapies for Alzheimer disease: current status and future directions. Nat. Rev. Neurol.19, 715–736 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ambadipudi, S., Biernat, J., Riedel, D., Mandelkow, E. & Zweckstetter, M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun.8, 275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wegmann, S. et al. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J.37, e98049 (2018). [DOI] [PMC free article] [PubMed]
- 199.Ferreon, J. C. et al. Acetylation disfavors tau phase separation. Int. J. Mol. Sci.19, 1360 (2018). [DOI] [PMC free article] [PubMed]
- 200.Boyko, S., Qi, X., Chen, T. H., Surewicz, K. & Surewicz, W. K. Liquid-liquid phase separation of tau protein: the crucial role of electrostatic interactions. J. Biol. Chem.294, 11054–11059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kanaan, N. M., Hamel, C., Grabinski, T. & Combs, B. Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat. Commun.11, 2809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Boyko, S. & Surewicz, W. K. Domain-specific modulatory effects of phosphomimetic substitutions on liquid-liquid phase separation of tau protein. J. Biol. Chem.299, 104722 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang, X. et al. RNA stores tau reversibly in complex coacervates. PLoS Biol.15, e2002183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lin, Y., Fichou, Y., Zeng, Z., Hu, N. Y. & Han, S. Electrostatically driven complex coacervation and amyloid aggregation of Tau are independent processes with overlapping conditions. ACS Chem. Neurosci.11, 615–627 (2020). [DOI] [PubMed] [Google Scholar]
- 205.Ukmar-Godec, T. et al. Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat. Commun.10, 2909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wolozin, B. & Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci.20, 649–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tan, R. et al. Microtubules gate tau condensation to spatially regulate microtubule functions. Nat. Cell Biol.21, 1078–1085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang, X. et al. The proline-rich domain promotes Tau liquid-liquid phase separation in cells. J. Cell Biol.219, e202006054 (2020). [DOI] [PMC free article] [PubMed]
- 209.Ukmar-Godec, T., Wegmann, S. & Zweckstetter, M. Biomolecular condensation of the microtubule-associated protein tau. Semin. Cell Dev. Biol.99, 202–214 (2020). [DOI] [PubMed] [Google Scholar]
- 210.Boyko, S., Surewicz, K. & Surewicz, W. K. Regulatory mechanisms of tau protein fibrillation under the conditions of liquid-liquid phase separation. Proc. Natl Acad. Sci. USA117, 31882–31890 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Boyko, S. & Surewicz, W. K. Tau liquid-liquid phase separation in neurodegenerative diseases. Trends Cell Biol.32, 611–623 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Shi, Y. et al. Structure-based classification of tauopathies. Nature598, 359–363 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Morris, H. R., Spillantini, M. G., Sue, C. M. & Williams-Gray, C. H. The pathogenesis of Parkinson’s disease. Lancet403, 293–304 (2024). [DOI] [PubMed] [Google Scholar]
- 214.Simuni, T. et al. A biological definition of neuronal alpha-synuclein disease: towards an integrated staging system for research. Lancet Neurol.23, 178–190 (2024). [DOI] [PubMed] [Google Scholar]
- 215.Soto, C. α-Synuclein seed amplification technology for Parkinson’s disease and related synucleinopathies. Trends Biotechnol.42, 829–841 (2024). [DOI] [PMC free article] [PubMed]
- 216.Ray, S. et al. α-Synuclein aggregation nucleates through liquid-liquid phase separation. Nat. Chem.12, 705–716 (2020). [DOI] [PubMed] [Google Scholar]
- 217.Mahapatra, A. & Newberry, R. W. Liquid-liquid phase separation of α-synuclein is highly sensitive to sequence complexity. Protein Sci.33, e4951 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mukherjee, S. et al. Liquid-liquid phase separation of alpha-synuclein: a new mechanistic insight for alpha-synuclein aggregation associated with Parkinson’s disease pathogenesis. J. Mol. Biol.435, 167713 (2023). [DOI] [PubMed] [Google Scholar]
- 219.Hallacli, E. et al. The Parkinson’s disease protein alpha-synuclein is a modulator of processing bodies and mRNA stability. Cell185, 2035–2056.e2033 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Locascio, J. J. et al. Association between alpha-synuclein blood transcripts and early, neuroimaging-supported Parkinson’s disease. Brain138, 2659–2671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bozic, M. et al. Protective role of renal proximal tubular alpha-synuclein in the pathogenesis of kidney fibrosis. Nat. Commun.11, 1943 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Harrison, A. F. & Shorter, J. RNA-binding proteins with prion-like domains in health and disease. Biochem. J.474, 1417–1438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Akcimen, F. et al. Amyotrophic lateral sclerosis: translating genetic discoveries into therapies. Nat. Rev. Genet.24, 642–658 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guenther, E. L. et al. Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol.25, 463–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Burrell, J. R. et al. The frontotemporal dementia-motor neuron disease continuum. Lancet388, 919–931 (2016). [DOI] [PubMed] [Google Scholar]
- 226.Piol, D., Robberechts, T. & Da Cruz, S. Lost in local translation: TDP-43 and FUS in axonal/neuromuscular junction maintenance and dysregulation in amyotrophic lateral sclerosis. Neuron111, 1355–1380 (2023). [DOI] [PubMed] [Google Scholar]
- 227.Portz, B., Lee, B. L. & Shorter, J. FUS and TDP-43 phases in health and disease. Trends Biochem. Sci.46, 550–563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Conicella, A. E., Zerze, G. H., Mittal, J. & Fawzi, N. L. ALS mutations disrupt phase separation mediated by alpha-helical structure in the TDP-43 low-complexity C-terminal domain. Structure24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Alami, N. H. et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron81, 536–543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ervilha Pereira, P. et al. C-terminal frameshift variant of TDP-43 with pronounced aggregation-propensity causes rimmed vacuole myopathy but not ALS/FTD. Acta Neuropathol.145, 793–814 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gopal, P. P., Nirschl, J. J., Klinman, E. & Holzbaur, E. L. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc. Natl Acad. Sci. USA114, E2466–E2475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Perez-Berlanga, M. et al. Loss of TDP-43 oligomerization or RNA binding elicits distinct aggregation patterns. EMBO J.42, e111719 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Klim, J. R. et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci.22, 167–179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Feiler, M. S. et al. TDP-43 is intercellularly transmitted across axon terminals. J. Cell Biol.211, 897–911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Porta, S. et al. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat. Commun.9, 4220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Mann, J. R. et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron102, 321–338.e328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bolognesi, B. et al. The mutational landscape of a prion-like domain. Nat. Commun.10, 4162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mackenzie, I. R. et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron95, 808–816.e809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ash, P. E. A. et al. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc. Natl Acad. Sci. USA118, e2014188118 (2021). [DOI] [PMC free article] [PubMed]
- 240.Niaki, A. G. et al. Loss of dynamic RNA interaction and aberrant phase separation induced by two distinct types of ALS/FTD-linked FUS mutations. Mol. Cell77, 82–94.e84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ratti, A. & Buratti, E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem138, 95–111 (2016). [DOI] [PubMed] [Google Scholar]
- 242.Ling, S. C., Polymenidou, M. & Cleveland, D. W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Guil, S., Long, J. C. & Caceres, J. F. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell Biol.26, 5744–5758 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Geuens, T., Bouhy, D. & Timmerman, V. The hnRNP family: insights into their role in health and disease. Hum. Genet.135, 851–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ryan, V. H. et al. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell69, 465–479.e467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Honda, H. et al. Loss of hnRNPA1 in ALS spinal cord motor neurons with TDP-43-positive inclusions. Neuropathology35, 37–43 (2015). [DOI] [PubMed] [Google Scholar]
- 247.Freibaum, B. D. & Taylor, J. P. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci.10, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pan, Y. et al. Gelation of cytoplasmic expanded CAG RNA repeats suppresses global protein synthesis. Nat. Chem. Biol.19, 1372–1383 (2023). [DOI] [PubMed] [Google Scholar]
- 249.Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol.28, 420–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Elbaum-Garfinkle, S. Matter over mind: liquid phase separation and neurodegeneration. J. Biol. Chem.294, 7160–7168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ryan, V. H. & Fawzi, N. L. Physiological, pathological, and targetable membraneless organelles in neurons. Trends Neurosci.42, 693–708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nedelsky, N. B. & Taylor, J. P. Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol.15, 272–286 (2019). [DOI] [PubMed] [Google Scholar]
- 253.Babinchak, W. M. & Surewicz, W. K. Liquid-liquid phase separation and its mechanistic role in pathological protein aggregation. J. Mol. Biol.432, 1910–1925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Darling, A. L., Zaslavsky, B. Y. & Uversky, V. N. Intrinsic disorder-based emergence in cellular biology: physiological and pathological liquid-liquid phase transitions in cells. Polymers11, 990 (2019). [DOI] [PMC free article] [PubMed]
- 255.Mann, J. R. & Donnelly, C. J. RNA modulates physiological and neuropathological protein phase transitions. Neuron109, 2663–2681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Luo, F. et al. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol.25, 341–346 (2018). [DOI] [PubMed] [Google Scholar]
- 257.Jiang, S., Fagman, J. B., Chen, C., Alberti, S. & Liu, B. Protein phase separation and its role in tumorigenesis. Elife9, e60264 (2020). [DOI] [PMC free article] [PubMed]
- 258.Silva, J. L. et al. Targeting biomolecular condensation and protein aggregation against cancer. Chem. Rev.123, 9094–9138 (2023). [DOI] [PubMed] [Google Scholar]
- 259.Dall’Agnese, G. et al. Role of condensates in modulating DNA repair pathways and its implication for chemoresistance. J. Biol. Chem.299, 104800 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ghodke, I. et al. AHNAK controls 53BP1-mediated p53 response by restraining 53BP1 oligomerization and phase separation. Mol. Cell81, 2596–2610.e2597 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Li, W. et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol.22, 960–972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Fan, X. J. et al. NONO phase separation enhances DNA damage repair by accelerating nuclear EGFR-induced DNA-PK activation. Am. J. Cancer Res.11, 2838–2852 (2021). [PMC free article] [PubMed] [Google Scholar]
- 263.Qin, G. et al. NPM1 upregulates the transcription of PD-L1 and suppresses T cell activity in triple-negative breast cancer. Nat. Commun.11, 1669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mitrea, D. M. et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun.9, 842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Tsuji, K., Kawata, H., Kamiakito, T., Nakaya, T. & Tanaka, A. RNA-binding protein 14 promotes phase separation to sustain prostate specific antigen expression under androgen deprivation in human prostate cancer. J. Steroid Biochem. Mol. Biol.235, 106407 (2023). [DOI] [PubMed] [Google Scholar]
- 266.Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov.2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kim, B. et al. Breast cancer metastasis suppressor 1 (BRMS1) is destabilized by the Cul3-SPOP E3 ubiquitin ligase complex. Biochem. Biophys. Res. Commun.415, 720–726 (2011). [DOI] [PubMed] [Google Scholar]
- 268.Le Gallo, M. et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet.44, 1310–1315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kim, M. S., Je, E. M., Oh, J. E., Yoo, N. J. & Lee, S. H. Mutational and expressional analyses of SPOP, a candidate tumor suppressor gene, in prostate, gastric and colorectal cancers. APMIS121, 626–633 (2013). [DOI] [PubMed] [Google Scholar]
- 270.Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hernandez-Munoz, I. et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl Acad. Sci. USA102, 7635–7640 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kent, D., Bush, E. W. & Hooper, J. E. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development133, 2001–2010 (2006). [DOI] [PubMed] [Google Scholar]
- 273.Kwon, J. E. et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem.281, 12664–12672 (2006). [DOI] [PubMed] [Google Scholar]
- 274.Li, G. et al. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell25, 455–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Bouchard, J. J. et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell72, 19–36.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wang, L. & Shilatifard, A. UTX mutations in human cancer. Cancer Cell35, 168–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Gozdecka, M. et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet.50, 883–894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Andricovich, J. et al. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell33, 512–526.e518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wang, C. et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl Acad. Sci. USA109, 15324–15329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Shpargel, K. B., Sengoku, T., Yokoyama, S. & Magnuson, T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet.8, e1002964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Shpargel, K. B., Starmer, J., Wang, C., Ge, K. & Magnuson, T. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc. Natl Acad. Sci. USA114, E9046–E9055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Miller, S. A., Mohn, S. E. & Weinmann, A. S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell40, 594–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Shi, B. et al. UTX condensation underlies its tumour-suppressive activity. Nature597, 726–731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Casale, A. M., Cappucci, U. & Piacentini, L. Unravelling HP1 functions: post-transcriptional regulation of stem cell fate. Chromosoma130, 103–111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Zeng, W., Ball, A. R. Jr. & Yokomori, K. HP1: heterochromatin binding proteins working the genome. Epigenetics5, 287–292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Li, X. et al. Deacetylation induced nuclear condensation of HP1gamma promotes multiple myeloma drug resistance. Nat. Commun.14, 1290 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Thrasher, A. J. & Burns, S. O. WASP: a key immunological multitasker. Nat. Rev. Immunol.10, 182–192 (2010). [DOI] [PubMed] [Google Scholar]
- 288.Candotti, F. Clinical manifestations and pathophysiological mechanisms of the Wiskott-Aldrich syndrome. J. Clin. Immunol.38, 13–27 (2018). [DOI] [PubMed] [Google Scholar]
- 289.Yuan, B. et al. Wiskott-Aldrich syndrome protein forms nuclear condensates and regulates alternative splicing. Nat. Commun.13, 3646 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Terlecki-Zaniewicz, S. et al. Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat. Struct. Mol. Biol.28, 190–201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Chandra, B. et al. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov.12, 1152–1169 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Biancon, G. et al. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol. Cell82, 1107–1122.e1107 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Bauer, K. E., de Queiroz, B. R., Kiebler, M. A. & Besse, F. RNA granules in neuronal plasticity and disease. Trends Neurosci.46, 525–538 (2023). [DOI] [PubMed] [Google Scholar]
- 294.Huai, Y. et al. HuR-positive stress granules: potential targets for age-related osteoporosis. Aging Cell23, e14053 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhang, Q. et al. Phase separation of BuGZ regulates gut regeneration and aging through interaction with m6A regulators. Nat. Commun.14, 6700 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wu, D. et al. An antagonistic pleiotropic gene regulates the reproduction and longevity tradeoff. Proc. Natl Acad. Sci. USA119, e2120311119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wang, R. et al. LncRNA GIRGL drives CAPRIN1-mediated phase separation to suppress glutaminase-1 translation under glutamine deprivation. Sci. Adv.7, eabe5708 (2021). [DOI] [PMC free article] [PubMed]
- 298.Liu, Q. et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell184, 5559–5576.e5519 (2021). [DOI] [PubMed] [Google Scholar]
- 299.Zhou, K. et al. Spatiotemporal regulation of insulin signaling by liquid-liquid phase separation. Cell Discov.8, 64 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Dion, W. et al. Four-dimensional nuclear speckle phase separation dynamics regulate proteostasis. Sci. Adv.8, eabl4150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Yasuda, S. et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature578, 296–300 (2020). [DOI] [PubMed] [Google Scholar]
- 302.Hofweber, M. & Dormann, D. Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem.294, 7137–7150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Snead, W. T. & Gladfelter, A. S. The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell76, 295–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Monahan, Z. et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J.36, 2951–2967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Wang, A. et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J.37, e97452 (2018). [DOI] [PMC free article] [PubMed]
- 306.Stefanoska, K. et al. Alzheimer’s disease: ablating single master site abolishes tau hyperphosphorylation. Sci. Adv.8, eabl8809 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Mao, X. et al. Pathological alpha-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science353, aah3374 (2016). [DOI] [PMC free article] [PubMed]
- 308.Mao, X. et al. Aplp1 interacts with Lag3 to facilitate transmission of pathologic alpha-synuclein. Nat. Commun.15, 4663 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Cohen, T. J. et al. An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun.6, 5845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Zhang, S. et al. Mechanistic basis for receptor-mediated pathological α-synuclein fibril cell-to-cell transmission in Parkinson’s disease. Proc. Natl Acad. Sci. USA118, e2011196118 (2021). [DOI] [PMC free article] [PubMed]
- 311.Shin, M. K. et al. Reducing acetylated tau is neuroprotective in brain injury. Cell184, 2715–2732.e2723 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell173, 706–719.e713 (2018). [DOI] [PubMed] [Google Scholar]
- 313.Ries, R. J. et al. m6A enhances the phase separation potential of mRNA. Nature571, 424–428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.McMillan, M. et al. RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Mol. Cell83, 219–236.e217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Mastrocola, A. S., Kim, S. H., Trinh, A. T., Rodenkirch, L. A. & Tibbetts, R. S. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem.288, 24731–24741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Altmeyer, M. et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun.6, 8088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Garnett, M. J. et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature483, 570–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Mandir, A. S. et al. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc. Natl Acad. Sci. USA96, 5774–5779 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.McGurk, L. et al. Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol. Commun.6, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.McGurk, L. et al. Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell71, 703–717.e709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Duan, Y. et al. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res.29, 233–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Ladbury, J. E., Lin, C. C. & Suen, K. M. Phase separation enhances probability of receptor signalling and drug targeting. Trends Biochem. Sci.48, 428–436 (2023). [DOI] [PubMed] [Google Scholar]
- 323.Lin, C. C., Suen, K. M., Lidster, J. & Ladbury, J. E. The emerging role of receptor tyrosine kinase phase separation in cancer. Trends Cell Biol.34, 371–379 (2024). [DOI] [PubMed] [Google Scholar]
- 324.Li, T., Zeng, Z., Fan, C. & Xiong, W. Role of stress granules in tumorigenesis and cancer therapy. Biochim. Biophys. Acta Rev. Cancer1878, 189006 (2023). [DOI] [PubMed] [Google Scholar]
- 325.Hurtle, B. T., Xie, L. & Donnelly, C. J. Disrupting pathologic phase transitions in neurodegeneration. J. Clin. Invest.133, e168549 (2023). [DOI] [PMC free article] [PubMed]
- 326.Wang, T. et al. Chemical-induced phase transition and global conformational reorganization of chromatin. Nat. Commun.14, 5556 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Zhu, G. et al. Pharmacological inhibition of SRC-1 phase separation suppresses YAP oncogenic transcription activity. Cell Res.31, 1028–1031 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Xie, J. et al. Targeting androgen receptor phase separation to overcome antiandrogen resistance. Nat. Chem. Biol.18, 1341–1350 (2022). [DOI] [PubMed] [Google Scholar]
- 329.Zhu, G. et al. Phase separation of disease-associated SHP2 mutants underlies MAPK hyperactivation. Cell183, 490–502.e418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science368, 1386–1392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Fang, M. Y. et al. Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron103, 802–819.e811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Boyd, J. D. et al. A high-content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J. Biomol. Screen19, 44–56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Price, D. L. et al. In vivo effects of the alpha-synuclein misfolding inhibitor minzasolmin supports clinical development in Parkinson’s disease. NPJ Parkinsons Dis.9, 114 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ossenkoppele, R., van der Kant, R. & Hansson, O. Tau biomarkers in Alzheimer’s disease: towards implementation in clinical practice and trials. Lancet Neurol.21, 726–734 (2022). [DOI] [PubMed] [Google Scholar]
- 335.Scheltens, P. et al. Alzheimer’s disease. Lancet397, 1577–1590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Breijyeh, Z. & Karaman, R. Comprehensive review on Alzheimer’s disease: causes and treatment. Molecules25, 5789 (2020). [DOI] [PMC free article] [PubMed]
- 337.Litvinchuk, A. et al. Amelioration of Tau and ApoE4-linked glial lipid accumulation and neurodegeneration with an LXR agonist. Neuron112, 384–403.e388 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Jia, J. et al. Biomarker changes during 20 years preceding Alzheimer’s disease. N. Engl. J. Med.390, 712–722 (2024). [DOI] [PubMed] [Google Scholar]
- 339.Colom-Cadena, M. et al. Synaptic oligomeric tau in Alzheimer’s disease—a potential culprit in the spread of tau pathology through the brain. Neuron111, 2170–2183.e2176 (2023). [DOI] [PubMed] [Google Scholar]
- 340.Zhang, D. et al. P-tau217 correlates with neurodegeneration in Alzheimer’s disease, and targeting p-tau217 with immunotherapy ameliorates murine tauopathy. Neuron112, 1676–1693.e1612 (2024). [DOI] [PubMed] [Google Scholar]
- 341.Steward, A. et al. ApoE4 and connectivity-mediated spreading of Tau pathology at lower amyloid levels. JAMA Neurol.80, 1295–1306 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Edwards, A. L. et al. Exploratory Tau biomarker results from a multiple ascending-dose study of BIIB080 in Alzheimer disease: a randomized clinical trial. JAMA Neurol.80, 1344–1352 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Das, B. et al. A Zn-dependent structural transition of SOD1 modulates its ability to undergo phase separation. EMBO J.42, e111185 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Miller, T. et al. Phase 1-2 trial of antisense oligonucleotide tofersen for SOD1 ALS. N. Engl. J. Med.383, 109–119 (2020). [DOI] [PubMed] [Google Scholar]
- 345.Dhingra, H. & Gaidhane, S. A. Huntington’s disease: understanding its novel drugs and treatments. Cureus15, e47526 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Ayyildiz, D. et al. CAG repeat expansion in the Huntington’s disease gene shapes linear and circular RNAs biogenesis. PLoS Genet.19, e1010988 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Aviner, R. et al. Polyglutamine-mediated ribotoxicity disrupts proteostasis and stress responses in Huntington’s disease. Nat. Cell Biol.26, 892–902 (2024). [DOI] [PubMed] [Google Scholar]
- 348.Peskett, T. R. et al. A liquid to solid phase transition underlying pathological huntingtin exon1 aggregation. Mol. Cell70, 588–601.e586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Ratovitski, T. et al. Interaction of huntingtin with PRMTs and its subsequent arginine methylation affects HTT solubility, phase transition behavior and neuronal toxicity. Hum. Mol. Genet.31, 1651–1672 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Leavitt, B. R., Kordasiewicz, H. B. & Schobel, S. A. Huntingtin-lowering therapies for Huntington disease: a review of the evidence of potential benefits and risks. JAMA Neurol.77, 764–772 (2020). [DOI] [PubMed] [Google Scholar]
- 351.Saade, J. & Mestre, T. A. Huntington’s disease: latest frontiers in therapeutics. Curr. Neurol. Neurosci. Rep.24, 255–264 (2024). [DOI] [PubMed]
- 352.Kim, S. D. & Fung, V. S. An update on Huntington’s disease: from the gene to the clinic. Curr. Opin. Neurol.27, 477–483 (2014). [DOI] [PubMed] [Google Scholar]
- 353.Huntington Study Group Reach2HD Investigators. Safety, tolerability, and efficacy of PBT2 in Huntington’s disease: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol.14, 39–47 (2015). [DOI] [PubMed] [Google Scholar]
- 354.Reilmann, R. et al. Safety and efficacy of pridopidine in patients with Huntington’s disease (PRIDE-HD): a phase 2, randomised, placebo-controlled, multicentre, dose-ranging study. Lancet Neurol.18, 165–176 (2019). [DOI] [PubMed] [Google Scholar]
- 355.Bahat, A. et al. Lowering mutant huntingtin by small molecules relieves Huntington’s disease symptoms and progression. EMBO Mol. Med.16, 523–546 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Tabrizi, S. J. et al. Targeting huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med.380, 2307–2316 (2019). [DOI] [PubMed] [Google Scholar]
- 357.DeJesus-Hernandez, M. et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Ortega, J. A. et al. CLIP-Seq analysis enables the design of protective ribosomal RNA bait oligonucleotides against C9ORF72 ALS/FTD poly-GR pathophysiology. Sci. Adv.9, eadf7997 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Sachdev, A. et al. Reversal of C9orf72 mutation-induced transcriptional dysregulation and pathology in cultured human neurons by allele-specific excision. Proc. Natl Acad. Sci. USA121, e2307814121 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Sonobe, Y. et al. Translation of dipeptide repeat proteins in C9ORF72 ALS/FTD through unique and redundant AUG initiation codons. Elife12, e83189 (2023). [DOI] [PMC free article] [PubMed]
- 362.Gao, C. et al. Neuromuscular organoids model spinal neuromuscular pathologies in C9orf72 amyotrophic lateral sclerosis. Cell Rep.43, 113892 (2024). [DOI] [PubMed] [Google Scholar]
- 363.Geng, Y. & Cai, Q. Role of C9orf72 hexanucleotide repeat expansions in ALS/FTD pathogenesis. Front. Mol. Neurosci.17, 1322720 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Raguseo, F. et al. The ALS/FTD-related C9orf72 hexanucleotide repeat expansion forms RNA condensates through multimolecular G-quadruplexes. Nat. Commun.14, 8272 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Jiang, J. et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron90, 535–550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Liu, J. et al. Liquid-liquid phase separation throws novel insights into treatment strategies for skin cutaneous melanoma. BMC Cancer23, 388 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Lai, W. et al. A five-LLPS gene risk score prognostic signature predicts survival in hepatocellular carcinoma. Int. J. Genomics2023, 7299276 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.You, Q. et al. A liquid-liquid phase separation-related index associate with biochemical recurrence and tumor immune environment of prostate cancer patients. Int. J. Mol. Sci.24, 5515 (2023). [DOI] [PMC free article] [PubMed]
- 369.Wyles, S. P. et al. Pharmacological modulation of calcium homeostasis in familial dilated cardiomyopathy: an in vitro analysis from an RBM20 patient-derived iPSC model. Clin. Transl. Sci.9, 158–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Ferrua, F. et al. Lentiviral haemopoietic stem/progenitor cell gene therapy for treatment of Wiskott-Aldrich syndrome: interim results of a non-randomised, open-label, phase 1/2 clinical study. Lancet Haematol.6, e239–e253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Labrosse, R. et al. Outcomes of hematopoietic stem cell gene therapy for Wiskott-Aldrich syndrome. Blood142, 1281–1296 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Basu, S. et al. Unblending of transcriptional condensates in human repeat expansion disease. Cell181, 1062–1079.e1030 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Li, J. et al. Post-translational modifications in liquid-liquid phase separation: a comprehensive review. Mol. Biomed.3, 13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Tang, T. et al. Aloperine targets lysosomes to inhibit late autophagy and induce cell death through apoptosis and paraptosis in glioblastoma. Mol. Biomed.4, 42 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]