Abstract
Glycosylation, a key mode of protein modification in living organisms, is critical in regulating various biological functions by influencing protein folding, transportation, and localization. Changes in glycosylation patterns are a significant feature of cancer, are associated with a range of pathological activities in cancer‐related processes, and serve as critical biomarkers providing new targets for cancer diagnosis and treatment. Glycoproteins like human epidermal growth factor receptor 2 (HER2) for breast cancer, alpha‐fetoprotein (AFP) for liver cancer, carcinoembryonic antigen (CEA) for colon cancer, and prostate‐specific antigen (PSA) for prostate cancer are all tumor biomarkers approved for clinical use. Here, we introduce the diversity of glycosylation structures and newly discovered glycosylation substrate—glycosylated RNA (glycoRNA). This article focuses primarily on tumor metastasis, immune evasion, metabolic reprogramming, aberrant ferroptosis responses, and cellular senescence to illustrate the role of glycosylation in cancer. Additionally, we summarize the clinical applications of protein glycosylation in cancer diagnostics, treatment, and multidrug resistance. We envision a promising future for the clinical applications of protein glycosylation.
Keywords: Glycosylation, immunity, cellular senescence, tumor biomarkers, cancer therapy
List of abbreviations
- ARPLA
aptamer and RNA in situ hybridization‐mediated proximity ligation assay
- Asn
asparagine
- B3GNT5
beta 1,3‐N‐acetylglucosaminyltransferase 5
- B4GALT1
beta 1,4‐galactosyltransferase 1
- B7H3
B7 homolog 3 protein
- BLBC
basal‐like breast cancer
- CAR‐T
chimeric antigen receptor T
- CSC
cancer stem cell
- EGFR
epidermal growth factor receptor
- EMT
epithelial‐mesenchymal transition
- ER
endoplasmic reticulum
- FADS2
fatty acid desaturase 2
- FASN
fatty acid synthase
- Fuc
fucose
- FUT2
fucosyltransferase 2
- FUT8
fucosyltransferase 8
- Gal
galactose
- GalNAc
N‐acetylgalactosamine
- GALNT14
N‐acetylgalactosaminyltransferase 14
- GALNT6
N‐acetylgalactosaminyltransferase‐6
- GALNT7
N‐acetylgalactosaminyltransferase 7
- GCNT2
β‐1,6‐N‐acetylglucosaminyltransferase 2
- Glc
glucose
- GlcA
glucuronic acid
- GlcNAc
N‐acetylglucosamine
- GLT8D1
glycosyltransferase 8 domain‐containing protein 1
- GLUT1
glucose transporter type 1
- GPI
glycosylphosphatidylinositol
- GPI‐AP
GPI‐anchored protein
- GPX4
glutathione peroxidase 4
- GRP78
glucose regulatory protein 78
- HCC
hepatocellular carcinoma
- IDOA
iduronic acid
- IGF1R
insulin‐like growth factor 1 receptor
- LRP1
lipoprotein receptor‐related protein 1
- Man
mannose
- MDH1
malate dehydrogenase 1
- MDR
multidrug‐resistant
- NCOA4
nuclear receptor coactivator 4
- NGI‐1
N‐linked glycosylation inhibitor‐1
- NSCLC
non‐small cell lung cancer
- OGA
O‐GlcNAcase
- OGT
O‐GlcNAc transferase
- OIS
oncogene‐induced senescence
- OST
oligosaccharyltransferase
- PDAC
pancreatic ductal adenocarcinoma
- PDGF‐C
platelet‐derived growth factor C
- PGK1
phosphoglycerate kinase 1
- PHB2
prohibitin 2
- PIGT
phosphatidylinositol glycan biosynthesis class T
- POFUT1
protein O‐fucosyltransferase 1
- ROS
reactive oxygen species
- RTK
tyrosine kinase
- SA
sialic acid
- SCAP
SREBP cleavage‐activating protein
- Ser
serine
- SLC35A2
solute carrier family 35 member A2
- SLC3A2
solute carrier family 3 member 2
- SLC7A11
solute carrier family 7a member 11
- SREBP
sterol regulatory element‐binding protein
- ST3GAL1
ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 1
- STT3A
STT3 oligosaccharyltransferase complex catalytic subunit A
- TCA
trichloroacetic acid
- Thr
threonine
- TMUB1
transmembrane and ubiquitin‐like domain‐containing protein 1
- TNBC
triple‐negative breast cancer
- Trp
tryptophan
- Tu
tunicamycin
- Xyl
xylose
- ZEB1
zinc finger E‐box binding homeobox 1
1. INTRODUCTION
Post‐translational changes in proteins add variety to the proteome by affecting its molecular makeup, impacting nearly every aspect of cellular function and disease, and establishing the basis for the intricacy of living beings [1, 2, 3, 4]. So far, there are over 650 protein modifications described, the most types of which are phosphorylation, glycosylation, ubiquitination, lactylation, methylation, N‐acetylation, and S‐palmitoylation [5]. Glycosylation is one of the most common and variable forms of post‐translational protein modification. Glycosylation is a process by which sugar chains are attached to proteins and lipids under the control of enzymes. Glycosyltransferases transfer sugar chains to proteins, creating glycosidic bonds with the amino acid residues on the protein, leading to the production of glycoproteins [6]. The majority of nuclear and cytoplasmic proteins undergo active O‐linked β‐N‐acetylglucosamine (O‐GlcNAc) glycosylation, while the majority of secreted proteins are glycosylated [7]. Proteins undergo glycosylation through a combination of metabolism and a complex network of various glycosylation pathways. Different glycoproteins participate in different functions of proteins in vivo, and affect various cell biological processes, such as allergic reactions and immune responses [8, 9]. For example, the cell surface glycoprotein human leukocyte antigen (HLA) serves as a recognition marker for the immune system [10]. Abnormal glycosylation has been implicated in several diseases, including rheumatic [11], cardiovascular [12], neurological [13], respiratory [14] and other diseases. The regulation of glycosylation and related enzymes is associated with abnormal activity in neuronal vasculopathy [13]. In numerous rheumatic disorders and chronic respiratory diseases, the protein glycosylation process is altered, which is associated with inflammatory processes and disease progression [11, 14]. The role of glycosylation in tumor cells is not negligible and is involved in all aspects of tumor development, from the initial stages to metastasis [15, 16, 17].
Tumor transformation is regulated by multiple factors. Among various post‐translational modifications associated with tumors, abnormal glycosylation is recognized as an important factor in tumor. Abnormal glycosylations are involved in many tumor malignancy progression processes, including proliferation, apoptosis, invasion, metastasis and immune escape [18, 19]. CD24, a heavily glycosylated protein, functions as a controller of cancer cell movement, infiltration, and growth [20]. Numerous tumor‐related polysaccharides and glycoproteins such as alpha‐fetoprotein (AFP), carcinoembryonic antigen (CEA), and human epidermal growth factor receptor 2 (HER2), are considered key markers of tumors, and are closely linked to the progression and prognosis of tumors. These molecules provide new targets for diagnosing, predicting and treating cancer [21]. Recently, research on the role of glycosylation in tumorigenesis has focused on cancer cell invasion and metastasis and immune evasion [22, 23]. Interestingly, the emerging roles of glycosylation in metabolic reprogramming, senescence, and ferroptosis have attracted increased interest [24, 25, 26]. Here, we outline the variety of structures involved in glycosylation and present a recently identified glycosylation target, glycosylated RNA (glycoRNA), a class of RNAs that are glycosylated. We focus on tumor metastasis, immune evasion, metabolic reprogramming, ferroptosis, and cellular senescence to illustrate the role of glycosylations in cancer. In addition, this review highlights the clinical applications of protein glycosylation modifications in cancer diagnostic markers, cancer therapy and multidrug resistance and provides future perspectives.
2. STRUCTURAL AND BIOLOGICAL FUNCTIONS OF GLYCOSYLATION
Glycans are polymers with carbohydrates as their main component. Glycans are essential in organisms and are involved in cell attachment, communication between cells, immune system control as well as energy processing [27, 28]. Sugar derivatives like glycoproteins, proteoglycans, glycosaminoglycans, and glycolipids are formed when glycans interact with proteins and lipids. These compounds are crucial for the structure of cells and organisms, and are involved in many physiological and pathological processes [29, 30].
Protein glycosylation is the attachment of sugar chains to certain amino acid residues on proteins through glycosyltransferases and nucleoside diphosphate sugars. Primarily happening in the endoplasmic reticulum (ER) and Golgi apparatus [6]. The glycan chain is primarily composed of monosaccharides such as glucose, galactose, mannose, xylose, fucose, N‐acetyl glucosamine, N‐acetyl galactosamine, glucuronic acid, iduronic acid, and sialic acids. These monosaccharides form complex carbohydrate molecules. Glycoconjugate molecules are then linked to protein structures to form glycoproteins, which regulate protein structure and cellular function. Glycosylation helps proteins to fold properly, facilitates interactions between the protein surface of folded glycoproteins and classical chaperones and other proteins that contribute to folding, and inhibits protein degradation by protecting proteins from proteases [31]. Glycosylation promotes cell‐cell adhesion through lectin‐polysaccharide interactions and contributes to cell‐cell transport [32]. Aside from protein secretion and translocation, glycosylation plays a role in various biological processes, including receptor interactions and the facilitation of intercellular signaling [33].
Variation in glycan chains and the diversity of glycosylation pathways can be influenced by the type and quantity of oligosaccharide residues present, as well as variations in the locations of glycosidic bonds [34]. Various processes can lead to changes in glycosylation, such as variations in the levels of glycosyltransferases and glycosidases, their distribution within the secretory pathway, alterations in chaperone function, and shifts in metabolism [35, 36]. Different types of glycosylation modifications include N‐glycosylation, O‐glycosylation, C‐glycosylation, and glycosyl phosphatidylinositol (GPI) anchoring, each corresponding to specific glycosylation sites [34] (Figure 1). N‐glycosylation and O‐glycosylation are the most common types include. N‐linked glycoproteins are formed when the protein backbone is connected covalently to the nitrogen element in asparagine (Asn), whereas O‐linked glycoproteins are formed when it is connected to the oxygen atom in the serine/threonine (Ser/Thr) side chain.
FIGURE 1.
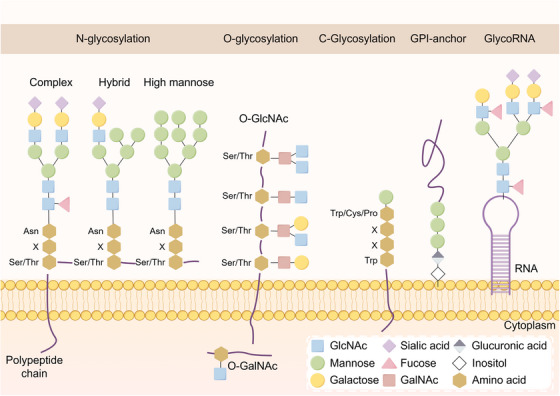
Structural diversity of glycosylation. The types and numbers of oligosaccharide residues in the glycan chain, as well as the differences in the sites of glycosidic bonds, lead to the diversity of the glycan chains and glycosylations pathways. Depending on the glycosylation site, glycosylations are mainly classified as N‐glycosylation, O‐glycosylation, C‐glycosylation, or glycosylphosphatidylinositol anchoring. In addition to proteins and lipids, RNA is the third major carrier of glycosylation. Abbreviations: Asn, asparagine; Cys, cysteine; GalNAc, N‐acetylgalactosamine; GlcNAc, N‐acetylglucosamine; GlycoRNA, glycosylated RNA; GPI, glycosyl phosphatidylinositol; Ser, serine; Thr, threonine; Trp, tryptophan; X, any amino acid.
2.1. N‐glycosylation
N‐glycosylation is the most prevalent form and has been studied the most extensively [34]. N‐glycosylation involves a covalent connection between N‐acetylglucosamine (GlcNAc) found in N‐glycans and the nitrogen atom of the asparagine (Asn) amino group via a β1‐glycosidic bond. The change usually occurs at a specific Asn site in the conserved glycosylation pattern Asn‐X‐Ser/Thr, with “X” standing for any amino acid other than proline. The core glycan of an N‐glycan contains three mannose (Man) residues and two GlcNAc residues. N‐glycans are formed into glycan chains in the ER, then transported to the Golgi apparatus for further processing and modification. Within the Golgi apparatus, glycosidases remove mannose residues, and various glycosyltransferases add other monosaccharides, resulting in the formation of monosaccharides with intricate structures [37]. N‐glycosylated proteins are primarily categorized as high‐mannose, hybrid, and complex proteins. N‐linked glycosylation is a tightly regulated process involving a series of coordinated post‐translational events that result in the modification and alteration of cell surface, secretory, and cycling proteins. N‐linked polysaccharide synthesis is closely linked to protein folding, ER homeostasis, and many roles in lysosomes and autophagy [38, 39, 40]. Proteins lacking N‐glycosylation are unable to be properly identified by calnexin (CNX) and calreticulin (CRT) in the ER, causing misfolding of the hypoglycosylated proteins and their degraded [41].
2.2. O‐glycosylation
O‐glycosylation occurs when polysaccharides are bonded to the hydroxyl group of the oxygen atom of a polypeptide's serine or threonine residue. The main sugars linked to serine and threonine amino acids are N‐acetylglucosamine (GlcNAc) and N‐acetylgalactosamine (GalNAc). In contrast to N‐polysaccharides, O‐linked polysaccharides are synthesized in a more direct and polyphasic manner, since they are not dependent on common sequences or polyol precursors. Metabolized glucose is broken down by the hexosamine biosynthetic pathway and then combined with amino acids, fatty acids, and nucleotides to create uridine diphosphate‐N‐acetylglucosamine (UDP‐GlcNAc). UDP‐GlcNAc serves as a donor substrate for intracellular GlcNAc modification. It facilitates the O‐GlcNAc modification of proteins through the action of O‐GlcNAc transferase. O‐GlcNAc glycosylation affects the conformation of protein structure and stability, mediates protein‐protein interactions and molecular recognition of its bioreceptors [42]. O‐GlcNAc in the IXI domain of certain small heat shock proteins (sHSPs) prevents sHSPs from binding to substrates, thereby activating their anti‐amyloid activity [43].
Galactosyltransferase transferase adds GalNAc to proteins and catalyzes mucin‐type O‐glycosylation [44]. More than 20 different peptide GalNAc transferase enzymes initiate O‐glycosylation, an extremely complex mechanism [45]. O‐GalNAc glycosylation allows proteins to perform significant structural, protective, and signaling functions. Mucin‐like O‐glycans are widely present on numerous glycoproteins that are found outside the cell or are secreted, including mucins. Mucins are identified by an inconsistent number of consecutive repeat sequences containing elevated levels of proline (Pro), serine (Ser), and threonine (Thr), resulting in multiple locations for O‐glycosylation [46]. The process of O‐GalNAc glycosylation is crucial for shielding glycoproteins and cell surfaces from external stress, as well as for facilitating self‐recognition by the immune system. Abnormal glycosylation, such as that found in oncogenic MUC1, has been demonstrated to increase the ability of proteins to bind to antigens and bypass tolerance to cytotoxic lymphocytes [47].
2.3. C‐glycosylation
C‐glycosylation is a relatively rare glycosylation modification of α‐D‐mannopyranosyl attached to a tryptophan (Trp) residue via a C‐C glycosidic bond. Trp‐X‐X‐Trp/Cys/Pro is the amino acid sequence for C‐glycosylation sites, with X representing any amino acid. C‐glycoproteins account for around approximately 20% of proteins that are either secreted or embedded in cell membranes [48]. The process of C‐mannosylation is crucial for the proper folding, secretion, and stability of proteins [49, 50]. C‐mannose plays a role in ER folding by assisting in the creation of a tryptophan‐arginine ladder, which impacts the positioning of cysteine residues and the development of disulfide bonds [50]. Additionally, the signaling functions of specific important proteins, such as the PDGF‐C protein, rely on C‐glycosylation [51]. C‐glycosylation has also been found on certain pathogen antigens. Research on Toxoplasma gondii and Plasmodium falciparum revealed that C‐glycosylation is associated with the firmness and longevity of the cyst wall, as well as oxygen detection and the durability of proteins crucial for host invasion [52]. The transmission of Plasmodium falciparum is associated with C‐mannosylation [53].
2.4. Glycosyl phosphatidylinositol
Glycolipids are lipid compounds containing a glycosyl ligand. Glycosyl phosphatidylinositol (GPI) is a structurally complex glycolipid compound that is widely expressed in eukaryotes [54]. GPI anchoring is the process of targeting proteins to the cell membrane by attaching protein amide bonds to phosphoacetamide, which is attached to 3 mannose and 1 glucosamine, and finally to phosphatidylinositol [55]. The GPIT complex, a transmembrane enzyme, facilitates the binding of GPI to proteins within the ER, aiding in the maturation of GPI‐anchored proteins (GPI‐APs) [56]. At least 150 human proteins that are widely distributed throughout various cell types have been identified as GPI‐APs [55]. These GPI‐APs are involved in other biological processes such as biosynthesis, transport, cell membrane distribution and signal transduction, cell adhesion, and antigen presentation. [57, 58, 59]. CD48 is a member of the CD2 molecular family and is a cell surface protein anchored by GPI. It acts as a co‐stimulatory molecule, inducing various effects during the process of immunization [58]. Abnormalities in the biosynthesis of GPI‐ APs often lead to a variety of disorders, such as paroxysmal nocturnal hemoglobinuria, mental retardation, and epileptic seizures [60, 61].
2.5. GlycoRNAs: a new substrate for glycosylation
Previously, glycosylation was believed to be a post‐translational modification process in which one glycoconjugate is enzymatically catalyzed and added to another glycoconjugate, protein or lipid. In addition to proteins and lipids, which are the primary vehicles for glycosylation changes, there have been reports of glycosylation changes occurring on RNA at the cellular membrane [62]. Carolyn et al. [62] reported for the first time that RNA is the third major carrier of glycosylation in addition to proteins and lipids and revealed that it may have important physiological functions at the surface of cell membranes. This newly discovered RNA that can be glycosylated is called “glycoRNA”. This discovery created a new area of research by linking glycobiology to RNA for the first time.
The presence of glycoRNAs on the cell membrane indicates their potential role as ligands controlling cell‐to‐cell signaling. Carolyn et al. [62] also revealed that glycoRNAs can act as direct ligands for cell membrane Siglec receptors. Given the significant role of Siglec receptors in immune modulation, glycoRNAs play crucial roles in various physiological and pathological processes, such as the host immune response, tumor evasion of the immune system, and autoimmune conditions, thereby paving the way for further exploration in this area of study [62, 63]. Until recently, the biological function of glycoRNAs was largely unknown because of the absence of visualization techniques. However, a recent investigation revealed that sialic acid aptamer and RNA in situ hybridization‐mediated proximity linkage assay (ARPLA) can effectively visualize glycoRNAs in individual cells with exceptional sensitivity and specificity [64]. Research on the correlation between glycoRNAs and interactions between monocytes and endothelial cells has indicated that glycoRNAs can facilitate cell‐cell communication in the immune system [64]. GlycoRNAs on the cell surface are crucial for neutrophil recruitment. P‐selectin (SELP) facilitates communication between neutrophils and endothelial cells, allowing their identification [65]. These findings may lead to new ways of thinking about the relationships between tumors and immunity and inflammation, leading to new avenues for tumor immunotherapy.
3. ROLE OF GLYCOSYLATION IN CANCER CELLS
Glycosylation‐modified proteins are involved in a variety of biological processes within the cell. Abnormal glycosylation is strongly linked to numerous disease processes, and when glycosylation is not properly regulated in cancer, it can result in disrupted signaling, the spread of cancer cells, and the avoidance of immune system detection [66, 67]. Glycosylation has become a topic of interest for researchers to explore the pathophysiological processes of tumors. Abnormally glycosylated proteins play a role in controlling the aggressive nature of cancer cells, facilitating their transformation and contributing to various aspects of cancer progression, including tissue invasion, spread to other parts of the body, resistance to the immune response, promotion of inflammation within tumors, and development of senescent cells [68]. Changes in glycosylation are involved in the process of carcinogenesis and therefore have considerable promise as targets for preventing and treating cancer. Here, we discuss aberrant glycosylation‐mediated tumor promotion, including tumor cell metastasis, tumor immune evasion, and tumor metabolic reprogramming, as well as the relationships between glycosylation and tumor cell ferroptosis and between glycosylation and cellular senescence.
3.1. Glycosylation and tumor metastasis
Healthy cells rely on cell surface molecules, transmembrane proteins, and growth factors to facilitate proper communication between cells. Changes in glycosylation patterns can trigger tumor cell growth, invasion, and spread by activating signaling pathways and their subsequent targets [69]. Cancer is characterized by tissue invasion and metastasis, with adhesion proteins and proteases playing crucial roles in this process [23]. One example is the involvement of MUC1 in the progression of cancer to a malignant state at different points [70]. Glycosylation, which involves glycan formation, proteins with attached glycans, and enzymes with attached glycans, contributes to the spread of cancer cells. It affects events associated with the spread of tumors, such as the characteristics of cancer stem cells (CSCs), epithelial mesenchymal transition (EMT), and migration and invasion (Figure 2).
FIGURE 2.
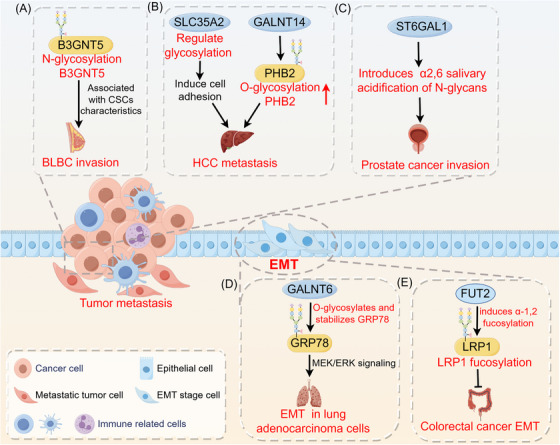
Role of glycosylation in tumor metastasis. Glycosylation has an impact on events related to tumor metastasis, including the stem cell properties of tumor cells, EMT, migration, and invasion. (A) B3GNT5 glycosylation is associated with cancer stem cell properties in basal‐like breast cancer. (B) SLC35A2 promotes HCC metastasis by regulating glycosylations to increase cell adhesion capacity. GALNT14‐mediated PHB2 O‐glycosylation promotes hepatocellular carcinoma cell growth and migration. (C) ST6GAL1 induces α2,6 salivary acidification of N‐glycans, promoting prostate cancer growth and invasion. (D) GALNT6 interacts with the O‐glycosylated chaperone protein GRP78, enhancing the MEK1/2/ERK1/2 signaling pathway in lung adenocarcinoma cells to promote EMT and invasion. (E) FUT2 induces α‐1,2 fucosylation and inhibits colorectal cancer EMT and metastasis via LRP1 fucosylation. Abbreviations: B3GNT5, β1,3‐N‐acetylglucosaminyltransferase 5; BLBC, basal‐like breast cancer; CSCs, cancer stem cells; EMT, epithelial‐mesenchymal transition; FUT2, fucosyltransferase 2; GALNT14, polypeptide N‐acetylgalactosaminyltransferase 14; GALNT6, N‐acetylgalactosaminyltransferase‐6; GRP78, glucose regulatory protein 78; HCC, hepatocellular carcinoma; LRP1, lipoprotein receptor‐related protein 1; PHB2, prohibitin 2; SLC35A2, solute carrier family 35 member A2.
The process of glycosylation is associated with the characteristics of CSCs and has multiple functions in the biology, such as facilitating cell attachment and spread and preventing cell death [71, 72]. N‐glycosylation of the B3GNT5 protein promotes protein stabilization. Elevated B3GNT5 expression is associated with high breast cancer grade and poor survival, suggesting a poor prognosis for breast cancer patients. B3GNT5 glycosylation is linked to the traits of CSCs in basal‐like breast cancer (BLBC) and enhances its invasiveness [73]. Glycosylation affects the transformation of cancer cells between epithelial and mesenchymal phenotypes through several mechanisms. EMT is characterized by changes in cell shape and alterations in the expression or function of adhesion molecules. Research has demonstrated that the progression of EMT relies on the presence of glycosphingolipids mediated by 1,4‐galactosyltransferase [74]. Glycosylation plays a role in breast cancer progression and metastasis [75]. Irregular changes in N‐glycosylation in the Golgi apparatus are associated with EMT and cancer metastasis in breast cancer patients. Fluvastatin, an approved medication, slows breast cancer metastasis by inhibiting the mevalonate pathway, which results in decreased N‐glycosylation levels and branching [76]. Elevated levels of GALNT6 expression in lung adenocarcinoma are linked to lymph node spread and an unfavorable prognosis. In lung adenocarcinoma cells, GALNT6 directly interacts with the O‐glycosylated chaperone protein GRP78, leading to an increase in the MEK1/2/ERK1/2 signaling pathway and subsequently promoting EMT, migration, and invasion of lung cancer cells [77]. Cancer metastasis is also linked to fucosyltransferase 2 (FUT2) and the related process of α‐1,2 fucosylation. FUT2 triggers α‐1,2 fucosylation and hinders EMT and the spread of colorectal cancer through LRP1 fucosylation [78]. Moreover, in liver cancer, the O‐glycosylation of PHB2 was positively associated with the GALNT14 expression. O‐glycosylation of PHB2 at serine‐161, which is mediated by GALNT14, has been shown to promote hepatocellular carcinoma (HCC) cell growth and migration by activating the IGF1R signaling cascade [79]. SLC35A2 is crucial in increasing HCC metastasis through the regulation of cellular glycosylations that increase cell adhesion [80]. Increased α2,6 sialylation of N‐glycans is a common type of glycosylation in tumor cells, that is driven by the sialyltransferase ST6GAL1. For example, ST6GAL1 is overexpressed in prostate cancer tissue and enhances the growth and invasion of prostate tumors [81]. In conclusion, disruptions in glycosylation processes, especially in glycan chain structures, glycosyltransferase activities, and glycosylation substrate expression, influence a range of oncogenic signaling pathways to trigger tumor invasion and metastasis.
3.2. Glycosylation and tumor immune evasion
Cells and tissues are perpetually monitored by a hypervigilant immune system that recognizes, destroys, and swiftly expels mutated cells from the body, thereby eradicating most newly transformed tumor cells. However, as cancer progresses, tumor cells adjust to the immune system's selective pressures, slowly developing ways to avoid detection.
In the context of cancer, the glycosylation of tumor cells plays a pivotal role in circumventing an effective immune response (Figure 3). In prostate cancer tissues, glycosyltransferase GALNT7 expression is upregulated, which leads to alterations in O‐glycosylation within prostate cancer cells. This alteration accelerates the development of prostate cancer and is closely connected to the cell cycle and immune signaling pathways [82]. O‐linked salivary modification of CD55 by ST3GAL1 contributes to tumor immune evasion [83]. In breast cancer cells, the overexpression of ST3GAL1 is a key factor driving tumorigenesis and correlated with increased tumor grades. In breast cancer cells, the inhibition of ST3GAL1 causes O‐linked desialylation of CD55, leading to elevated C3 deposition and complement‐induced lysis. This change also heightened their vulnerability to antibody‐triggered cell destruction. In triple‐negative breast cancer (TNBC) patients, improving anti‐tumor immune responses by focusing on B7H3 glycosylation could be a potential treatment approach. B7H3, which highly glycosylated, becomes physiologically and clinically important in individuals with TNBC as a result of abnormal glycosylation patterns induced by the upregulation of fucosyltransferase 8 (FUT8). The knockdown of FUT8 mitigates B7H3 glycosylation‐mediated immunosuppression in TNBC cells [84]. Hypersialylation, a key feature of glycosylation in cancer, increases disease progression and helps tumors avoid detection by interacting with Siglec receptors on immune cells that have infiltrated the tumor. Desialylation therapy is a new approach to immunotherapy that focuses on changing macrophage characteristics to improve the body's ability to combat against tumors [85].
FIGURE 3.
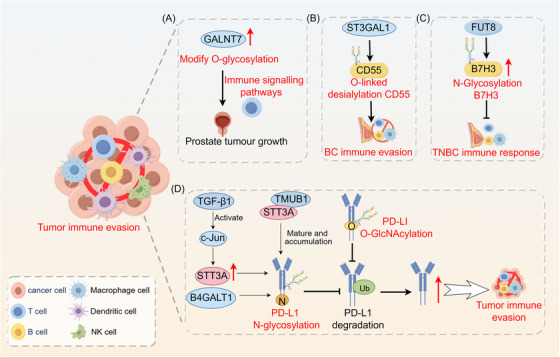
Role of glycosylation in tumor immune evasion. (A) GALNT7 modifies O‐glycosylation, which is associated with the immune signaling pathways, in prostate cancer cells. (B) O‐linked salivary modification of CD55 by ST3GAL1 contributes to breast cancer cell immune evasion. (C) Abnormal B7H3 glycosylation mediated by FUT8 suppresses the immune response in TNBC cells. B4GALT1 mediates the N‐linked glycosylation of PD‐L1 protein, thereby preventing PD‐L1 degradation at the post‐transcriptional level. (D) TGF‐β1‐mediated glycosylation of PD‐L1 promotes immune evasion through the c‐Jun/STT3A signaling pathway. TMUB1 enhances PD‐L1 N‐glycosylation and stability by recruiting STT3A, thereby promoting PD‐L1 maturation and tumor immune evasion. O‐GlcNAcylation hinders the lysosomal degradation of PD‐L1 to promote tumor immune evasion. Abbreviations: B4GALT1, beta1,4‐galactosyltransferase 1; B7H3, B7 homolog 3 protein; BC, breast cancer; FUT8, fucosyltransferase 8; GALNT7, N‐acetylgalactosaminyltransferase 7; NK cell, natural killer cell; PD‐L1, programmed death‐ligand 1; ST3GAL1, ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 1; STT3A, STT3 oligosaccharyl transferase complex catalytic subunit A; TGF‐β1, transforming growth factor‐beta1; TMUB1, transmembrane and ubiquitin‐like domain‐containing protein 1; TNBC, triple negative breast cancer; Ub, ubiquitin.
Therapies that block immune checkpoints, especially those that focus on the PD‐L1/PD‐1 pathway, have shown important advantages in clinical settings. Effectively targeting cancer glycosylation constitutes a pivotal approach for blocking this immune checkpoint. O‐GlcNAcylation may aid in tumor immune evasion by inhibiting the lysosomal breakdown of PD‐L1 [86]. Transmembrane and ubiquitin‐like structural domain protein 1 (TMUB1), a protein with transmembrane and ubiquitin‐like structural domains, has been recognized as a crucial controller of post‐translational changes in PD‐L1 in cancerous cells. TMUB1 protein levels are correlated with PD‐L1 expression in human tumor tissues, and higher TMUB1 expression levels are linked to lower patient survival rates. TMUB1 enhances PD‐L1 maturation and facilitates tumor immune evasion by increasing PD‐L1 N‐glycosylation and stability through the recruitment of STT3A [87]. B4GALT1 plays a direct role in N‐linked glycosylation of PD‐L1, leading to the prevention of PD‐L1 degradation after transcription. Furthermore, B4GALT1 stabilizes the TAZ protein through glycosylation, consequently activating CD274 at the transcriptional level. These mechanisms contribute to immune escape in lung cancer. Blocking B4GALT1 increases the abundance and function of CD8+ T cells, boosting the body's ability to fight tumors and improving the effectiveness of PD‐1 treatment in living organisms [88]. PD‐L1 glycosylation mediated by TGF‐β1 in nasopharyngeal cancer is essential for avoidance of the immune system through the c‐Jun/STT3A pathway [89].
3.3. Glycosylation and tumor metabolic reprogramming
Metabolic reprogramming is a hallmark of cancer. Tumor cells meet increasing bioenergetic and biosynthetic demands by altering various metabolic pathways [24]. The glycosylation of proteins is crucial in the metabolic reprogramming of cancer cells, coordinating metabolic pathways to produce necessary metabolites that promote tumor development (Figure 4).
FIGURE 4.
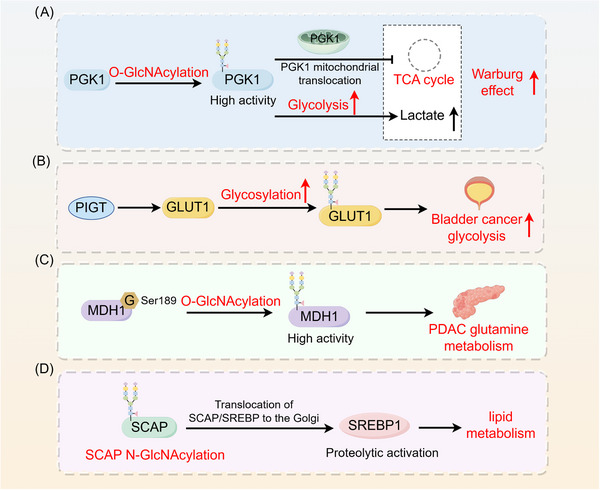
Role of glycosylation in tumor metabolic reprogramming. (A) Glycosylation increases the metabolic enzyme activity of PGK1 and induces PGK1 translocation to mitochondria to inhibit the TCA cycle, thereby enhancing the Warburg effect in cancer cells. (B) PIGT enhances glycolysis in bladder cancer cells through the regulation of GLUT1 glycosylation. (C) O‐GlcNAcylation regulates the metabolic activity of MDH1, promoting glutamine metabolism in pancreatic cancer. (D) SCAP N‐glycosylation promotes SCAP/SREBP translocation to the Golgi apparatus, which in turn activates SREBP1 to regulate lipid metabolism in tumors through SREBP‐dependent lipids. Abbreviations: G, glycosylation; GLUT1, glucose transporter type 1; MDH1, malate dehydrogenase 1; PDAC, pancreatic ductal adenocarcinoma; PGK1, phosphoglycerate kinase 1; PIGT, phosphatidylinositol glycan biosynthesis class T; SCAP, SREBP cleavage‐activating protein; SREBP, sterol regulatory element‐binding protein.
O‐GlcNAcylation functions as a vital sensor of nutrients, playing a complex role in the breakdown of sugars, proteins, fats, and genetic material. The hexosamine biosynthesis pathway controls the production of O‐GlcNAcylation through the exclusive donor uridine diphosphate‐N‐acetylglucosamine, which is influenced by glucose, glutamine, acetyl coenzyme A, and uridine triphosphate. Through its function as a vital sensor of molecular changes, O‐GlcNAcylation is established as a central component in the processing of carbohydrates, proteins, fats, and nucleic acids [90]. Anaerobic glycolysis serves as the primary energy source for tumor cells, with glycosylations playing a crucial role in regulating the Warburg effect in tumors. Glycosylation not only upregulates the metabolic enzyme activity of phosphoglycerate kinase 1 (PGK1), thereby increasing glycolysis, but also induces the mitochondrial translocation of PGK1, subsequently inhibiting the TCA cycle. The cumulative effect of these two functions exacerbates the Warburg effect in cancer cells, consequently promoting tumor growth [91]. PIGT is a regulator of GPI‐anchor biosynthesis that enhances glycolysis in bladder cancer cells through the regulation of GLUT1 glycosylation and membrane translocation, consequently fostering tumor growth and metastasis [92]. O‐GlcNAc glycosylation plays a pivotal role in regulating glutamine metabolism. Malate dehydrogenase 1 (MDH1), a crucial enzyme in this breakdown process, is enhanced by O‐GlcNAcylation at serine 189 [93]. Notably, increased levels of MDH1 O‐GlcNAcylation have been observed in clinical samples of pancreatic ductal adenocarcinoma (PDAC). Research has demonstrated that O‐GlcNAcylation increases the growth of pancreatic cancer by controlling the metabolic function of MDH1. Reducing O‐GlcNAcylation depletes MDH1 function, hinders glutamine processing, increases PDAC cell vulnerability to oxidative stress, and decreased cell proliferation, ultimately hindering tumor growth [93]. Glycosylation may play a role in controlling lipid metabolism in tumors by affecting SREBP‐dependent pathways. The N‐glycosylation of SCAP activates SREBP‐1, an ER‐binding transcription factor crucial for lipid metabolism, in this process [94]. The addition of glycans strengthens the stability of SCAP, reducing its binding with Insig‐1, which helps move SCAP/SREBP to the Golgi apparatus, ultimately activating SREBP protein breakdown. Notably, inhibiting SCAP N‐glycosylation suppresses the growth of EGFRvIII‐driven glioblastoma [94]. Furthermore, cells exhibiting elevated O‐GlcNAc levels exhibited elongated mitochondria, increased mitochondrial membrane potential, and energy metabolism reprogramming. In SH‐SY5Y neuroblastoma cells, increased O‐GlcNAc levels led to an upregulation of O‐GlcNAcase (OGA) expression, coupled with a reduction in cellular respiration and reactive oxygen species (ROS) generation [95].
3.4. Glycosylation and ferroptosis
In 2012, Dixon et al. [96] from Columbia University described ferroptosis, a unique form of cell death that relies on iron and is different from apoptosis, necrosis, and autophagy. Ferroptosis is characterized primarily by a reduction in glutathione levels. Decreased function of glutathione peroxidase (GPX4) results in the inability to process lipid oxides through the GPX4‐mediated glutathione reductase reaction. Subsequently, divalent iron ions oxidize lipids and produce ROS, thereby contributing to the process of ferroptosis [97]. Owing to its distinct mechanisms and morphology compared with other types of cell death, ferroptosis has garnered considerable interest among cancer researchers. In recent years, many studies have elucidated the role and mechanism of ferroptosis in tumor suppression and tumor immunity [98, 99], underscoring its significant potential in cancer therapy.
Glycosylations and ferroptosis exhibit crosstalk in cancer, thereby modulating tumor progression through the regulation of iron metabolism (Figure 5). Dynamic modifications in O‐GlcNAc glycosylation drive ferroptosis by orchestrating ferritin phagocytosis and mitosis. Exposure to ferroptosis triggers such as RSL3, causes a two‐phase change in protein O‐GlcNAcylation, which in turn affects the ferroptosis process. Removing O‐GlcNAc from the ferritin heavy chain at serine 179 increases its binding with the ferritin receptor NCOA4.This modification leads to the accumulation of labile iron within mitochondria, contributing to iron‐induced instability and cell death [100]. N‐glycosylation of asparagine on 4F2hc (SLC3A2) is crucial for the induction of ferroptosis in PDAC. SLC3A2 knockdown or inhibition of 4F2hc N‐glycosylation increases the susceptibility of PDAC cells to ferroptosis by suppressing the activity of system Xc‐ in the glutamate‐cystine retrotranslocator protein system. Furthermore, the use of tunicamycin (TM), an N‐glycosylation inhibitor, significantly increases the sensitivity of PDAC cells to ferroptosis, particularly when combined with ferroptosis inducers [101]. In HCC, promoting ferroptosis can be achieved by targeting USP8, leading to decreased stability of OGT and subsequent inhibition of SLC7A11 O‐GlcNAcylation [102]. Glucose‐induced O‐GlcNAcylation of the Ser555 site of ZEB1 stabilizes the protein and translocates it to the nucleus, leading to increased transcriptional activity of genes associated with lipogenesis, including FASN and FADS2. This series of events eventually results in ferroptosis in mesenchymal pancreatic cancer cells due to lipid peroxidation [103].
FIGURE 5.
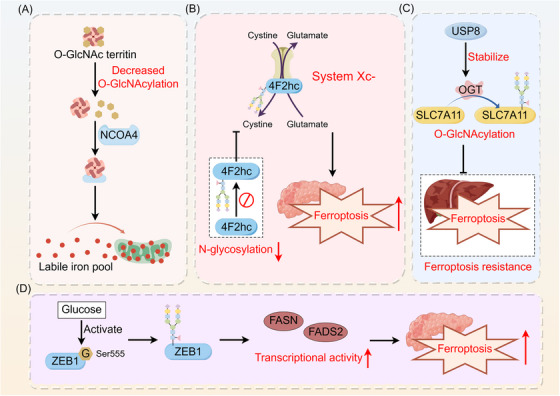
Role of glycosylation in tumor ferroptosis. Glycosylations interact with ferroptosis in cancer, modulating tumor progression. (A) Deglycosylation of the ferritin heavy chain enhances its interaction with the ferritin phagocytic receptor NCOA4, leading to the accumulation of unstable iron in mitochondria, which contributes to ferroptosis. (B) Inhibiting the N‐glycosylation of 4F2hc enhances the ferroptosis sensitivity of PDAC cells by suppressing the activity of the glutamate‐cystine reverse transport system Xc‐. (C) USP8 inhibits ferroptosis sensitivity in hepatocellular carcinoma by stabilizing OGT, which promotes the O‐GlcNAcylation of SLC7A11. (D) Glucose‐induced ZEB1 O‐GlcNAcylation activates the transcriptional activity of the adipogenesis‐related genes FASN and FADS2, leading to lipid peroxidation‐dependent ferroptosis in mesenchymal pancreatic cancer cells. Abbreviations: FADS2, fatty acid desaturase 2; FASN, fatty acid synthase; G, glycosylation; NCOA4, nuclear receptor coactivator 4; OGT, O‐GlcNAc transferase; PDAC, pancreatic ductal adenocarcinoma; SLC7A11, solute carrier family 7a member 11; USP18, ubiquitin‐specific peptidase 18; ZEB1, zinc finger E‐box binding homeobox 1.
3.5. Glycosylation and cellular senescence
Cellular senescence acts as a protective mechanism to maintain tissue homeostasis. There is increasing evidence that, under certain conditions, senescent cells may contribute to tumorigenesis and progression through various mechanisms, including DNA damage, oxidative stress, tumor microenvironment modulation, and immune responses [104, 105, 106, 107]. Senescent cells are increasingly recognized as hallmarks of cancer [68]. Senescence and cancer exhibit numerous overlapping characteristics. Several aspects of aging, such as genomic instability, epigenetic alterations, chronic inflammation, and ecological imbalance, share similarities with specific characteristics of cancer, leading to their classification as “meta‐hallmarks” [25].
Recent research has revealed a link between glycosylation mechanisms and the interaction between cellular senescence and the progression of cancer (Figure 6). The inhibition of oligosaccharyl transferase (OST) triggers senescence in receptor tyrosine kinase (RTK)‐driven tumor cells. Scientists have conducted high‐throughput screening using cells, followed by the optimization of a lead compound, resulting in the creation of a cell‐permeable inhibitor called N‐linked glycosylation inhibitor‐1 (NGI‐1), which specifically targets OST. NGI‐1 efficiently inhibits the cell‐surface positioning and communication of the epidermal growth factor receptor (EGFR) glycoprotein in non‐small‐cell lung cancer (NSCLC) cells. Specifically, it inhibits the growth of specific cell lines that rely on EGFR or fibroblast growth factor receptor (FGFR) for their survival. Within these specific cell lines, OST inhibition leads to cell‐cycle arrest, characterized by the induction of p21, increased autofluorescence, and notable changes in cell morphology, all of which are indicative of senescence. These results highlight the possibility of using OST inhibition as a treatment approach for tumors that rely on receptor tyrosine kinases [108]. Additionally, another study revealed that mutated KRAS promotes glycolytic activity in lung cancer, which may result in abnormal protein glycosylation associated with aging. In lung cancer cells with mutant KRAS, the levels of O‐linked β‐N‐acetylglucosamine (O‐GlcNAcylation) post‐translational modifications, which play a crucial role in inhibiting senescence, are increased during this process. Notably, O‐GlcNAcylation effectively suppresses senescence induced by the KrasG12D oncogene, thereby accelerating the process of lung tumor development [109].
FIGURE 6.
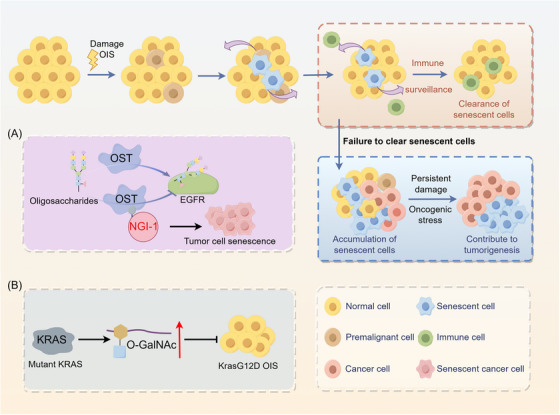
Role of glycosylation in cellular senescence. Under certain conditions, the escape of senescent cells from immune surveillance may promote tumorigenesis and progression through mechanisms such as DNA damage and oxidative stress. (A) The cell‐permeable inhibitor NGI‐1 targets oligosaccharyl transferase and blocks cell surface localization and signaling of EGFR glycoproteins, promoting cellular senescence. (B) Increased levels of O‐GlcNAcylation in KRAS‐mutant lung cancer cells inhibit KrasG12D OIS and accelerate lung tumorigenesis. Abbreviations: EGFR, epidermal growth factor receptor; NGI‐1, N‐linked glycosylation inhibitor‐1; O‐GalNAc, O‐N acetylgalactosamine; OIS, oncogene‐induced senescence; OST, oligosaccharyl transferase.
4. THERAPEUTIC POTENTIAL OF GLYCOSYLATION IN CANCER
4.1. Glycosylation‐based tumor biomarkers
Despite the substantial investment of time and resources in cancer research, the effects of the developed therapies are not satisfactory. Early detection and treatment are key to cancer therapy, so there is an urgent clinical need for new methods for the early diagnosis and treatment of cancer. Changes in the glycosylation patterns of cancer cells often appear in the initial phases of tumor growth. Specific tumor‐associated glycosylation patterns have been identified in precursor lesions across various cancer types, establishing them as potent early diagnostic biomarkers [22, 110] (Table 1).
TABLE 1.
Clinical application of glycosylation alterations in tumor markers.
| Type | Biomarker | Clinical application | Reference |
|---|---|---|---|
| Glycoproteins | AFP | Prediction of tumorigenesis and recurrence in HCC | [111] |
| CEA | A well‐known marker for colorectal cancer staging and surveillance | [112] | |
| PSA | Prediction of aggressive prostate cancer | [113] | |
| HE4 | An ideal biomarker for ovarian and endometrial cancers | [114, 115] | |
| HER2 | Highly expressed in breast cancer tissues and elevated in gastric and lung cancers | [116] | |
| HCG | Diagnosis, stage and monitor choriocarcinoma and testicular cancer | [117] | |
| CA125 | An important marker for the prognosis and diagnosis of ovarian cancer | [118] | |
| CA19‐9 | A biomarker and predictor in pancreatic cancer | [119] | |
| CA724 | Adjuvant diagnosis, dynamic progression monitoring and prognostic assessment of gastric cancer | [120] | |
| CA153 | Overexpressed in many types of cancer, including lung, breast and cervical cancer. | [121, 122, 123] | |
| Glycosyltransferases | GLT8D1, GLT8D2 | Potential prognostic biomarkers with immunological implications in gastric cancer | [124] |
| B4GALNT2 | Predictor of good prognosis in colon cancer | [125] | |
| POFUT1 | Potential biomarker for gastric cancer | [126] | |
| GCNT2 | Emerging biomarker and therapeutic target for melanoma | [127] | |
| GALNT6 | Novel prognostic biomarker for colorectal cancer | [128] |
Abbreviations: AFP, alpha‐fetoprotein; B4GALNT2, β‐1,4‐N‐acetyl‐galactosaminyltransferase 2; CA125, carbohydrate antigen 125; CA153, carbohydrate antigen 153; CA19‐9, cancer antigen 19‐9; CA724, carbohydrate antigen 724; CEA, carcinoembryonic antigen; GALNT6, N‐acetylgalactosaminyl transferase 6; GLT8D1, glycosyltransferase 8 domain‐containing protein 1; GLT8D2, glycosyltransferase 8 domain containing 2; HCG, human chorionic gonadotropin; HE4, human epididymis protein 4; HER2, human epidermal growth factor receptor 2; POFUT1, protein O‐fucosyltransferase 1;GCNT2, β‐1,6‐N‐acetylglucosaminyltransferase 2; PSA, prostate‐specific antigen.
Many glycoproteins, such as AFP for liver cancer, CEA for colon cancer, and prostate‐specific antigen (PSA) for prostate cancer, have been clinically approved as tumor biomarkers. AFP, an N‐linked glycoprotein, is found in fetal serum, but the level decreases quickly after birth. AFP is elevated in HCC and has been used to predict tumorigenesis and recurrence [111]. CEA is an acidic glycoprotein with a molecular weight of 180 kDa that has human antigenic properties and is usually expressed during fetal development. CEA is a commonly used indicator for monitoring and assessing colorectal cancer progression and observation [112]. PSA is an N‐linked glycoprotein that is secreted by the epithelium of the prostate and glands in the urethra. Combining serum and urine PSA ratios with urine and serum tests enhances the accuracy of predicting aggressive prostate cancer [113]. Human epididymis protein 4 (HE4) has been identified as a valuable biomarker for ovarian and endometrial cancers [114, 115]. HER2, a 185 kDa glycoprotein produced by the ERBB2 gene, is expressed at high levels in breast cancer tissues and is increased in gastric and lung cancers. HER2 is commonly targeted by immunosuppressant therapy in cancer treatment [116]. Human chorionic gonadotropin (HCG), a glycoprotein composed of two different subunits, is frequently used to detect pregnancy. The beta subunit of HCG is elevated in choriocarcinoma and testicular cancer, making it a valuable tool for diagnosing, staging, and monitoring the disease [117]. Highly glycosylated HCG is a marker for the early invasion of human trophoblasts [129]. The glycan antigen CA125, a peptide epitope of the 3000‐5000 kDa mucin MUC16, is an important marker for the prognosis and diagnosis of ovarian cancer [118]. CA19‐9 accelerate pancreatic cancer progression by modifying proteins with sugar molecules, attaching to E‐selectin, increasing the formation of new blood vessels, influencing immune reactions, and serving as a predictor and facilitator in pancreatic cancer [119]. The role of CA724 in the adjuvant diagnosis, dynamic progression monitoring and prognostic assessment of gastric cancer should not be neglected [120]. CA153 is upregulated in various cancer types, such as lung, breast, and cervical cancer. Approval has been granted for its use as an indicator in tracking breast cancer therapy response [121, 122, 123]. Recent advancements in glycobiology have led to the development of detection technologies that utilize abnormal glycoproteins on the cell surface as indicators for cancer prediction. This advancement has created opportunities for improved cancer detection and evaluation of the success of treatments. A variety of glycoproteins and glycolipids are increasingly being identified as potential novel tumor markers [21].
The aberrant expression of glycosyltransferases and glycosidases plays a pivotal role in the irregularities of glycan chains and glycoproteins. Consequently, these enzymes can be considered as potential and actionable biomarkers for cancer [130]. Potential prognostic biomarkers with immunological implications in gastric cancer tumors, GLT8D1 and GLT8D2 glycosyltransferases have been identified [124]. The glycosyltransferase B4GALNT2 can be used to predict a positive outcome in colon cancer patients [125]. POFUT1 expression increased in gastric cancer patients and could potentially function as a biomarker for this disease [126]. The glycosyltransferase β‐1,6‐N‐acetylglucosaminyltransferase 2 (GCNT2) has the potential to serve as both a biomarker and a treatment target in melanoma [127]. The levels of GALNT6 mRNA and protein can be used as new prognostic indicators for colorectal cancer, emphasizing the impact of glycogen imbalance on glycan production in cancer and predicting a negative outcome [128].
Compared with total exosomal PD‐L1, glycosylated exosomal PD‐L1 is a more reliable biomarker for cancer diagnosis and predicting immunotherapy effectiveness. Nevertheless, using exosomal PD‐L1 levels in cancer diagnosis faces challenges from high incidence of false positive and negative results [131]. Exosomal PD‐L1 proteins display important variations in glycosylation, playing a critical role in the interactions between PD‐L1 and PD‐1, leading to the suppression of CD8+ T‐cell proliferation [132]. Innovatively, a method involving an aptamer and lectin dual recognition‐induced neighbor‐joining reaction, coupled with fluorescence quantitative PCR, was developed for the precise quantification of glycosylated exosomal PD‐L1. This method has confirmed that glycosylated exosomal PD‐L1 is a reliable indicator for tumors detection, with the ability to accurately differentiate between individuals with and without cancer. In contrast, total exosomal PD‐L1 lacks this precise discriminative ability [133].
Consequently, focusing on tumor‐associated glycosylation abnormalities could increase the diagnostic and prognostic accuracy in clinical settings, offering improved sensitivity and specificity.
4.2. Glycosylation and tumor therapy
The effective treatment of patients with multidrug‐resistant cancer remains a major challenge. New research has shown that glycosylation is crucial in the emergence of acquired resistance to multiple drugs. Changes in N‐glycosylation linked to tumors disrupt interferon γRα, leading to interferon γ resistance in colorectal cancer [134]. Abnormal N‐glycosylation leads to reduced expression of interferon γRα and impairs interferon γ R signaling by diminishing protein stability via proteasome‐dependent degradation [134]. In HCC, the O‐glycosylation of PHB2 by GALNT14 plays a major role in promoting tumor growth and migration, as well as enhancing the resistance of cancer cells to chemotherapy [79]. Research on pancreatic cancer has recognized beta‐1,4‐galactosyltransferase 1 (B4GALT1) as a significant clinical biomarker and a crucial factor in resistance to chemotherapy. Elevated B4GALT1 expression correlates with decreased survival rates, increased tumor size, increased frequency of lymph node metastasis, accelerated tumor progression, and higher recurrence rates in PDAC patients [135]. B4GALT1 expression is notably upregulated in patient‐derived organoids and in gemcitabine‐resistant cancer cell lines. B4GALT1 interacts with and stabilizes the CDK11p110 protein, a component of the cytosolic protein‐dependent protein 11 heterodimer, through N‐linked glycosylation. This interaction contributes to promoting cancer progression and chemoresistance [135]. Tunicamycin (Tu), an effective glycosylation inhibitor, has demonstrated remarkable antitumor efficacy across various cancer types. Tu markedly increases chemotherapy‐induced apoptosis in gastric cancer cells, particularly in MDR cells, by inducing ER stress, an effect largely attributed to glycosylation inhibition. Targeted inhibition of tumor glycosylation represents a viable approach to overcome chemoresistance in gastric cancer patients [136]. Targeting the glycosylation of MUC1 is a promising strategy for improving the sensitivity and effectiveness of chemotherapeutic agents in the treatment of breast cancer [137]. Increased secretory fucosylation serves as a universal indicator of both the response and resistance to multitargeted therapy across various types of cancer. Large‐scale pharmacogenomic studies indicate a broad association between fucosylation genes and drug resistance [138].
Furthermore, glycosylation has been associated with cancer immunotherapy, and resistance to radiotherapy. Clinical studies have revealed that abnormal glycosylation patterns in patients are associated with decreased survival rates. Mechanistic studies have revealed that branching N‐glycans can protect tumor cells from being killed by CAR‐T cells, are less likely to form immune synapses, and block the release of cytokines. Interfering with the N‐glycosylation of tumor cells can significantly increase the efficacy of CAR‐T cell therapy for solid tumors [139]. ALG3 has been demonstrated to increase resistance to radiation in breast cancer patients and promote the growth of tumor stem cells by modifying the glycosylation of TGFBR2 [140].
The process of glycosylation is essential for tumor progression, and focusing on glycosylation presents promising opportunities for the creation of drugs to combat tumors. Notably, considerable advancements have been made in the development of selectin inhibitors, glycoconjugated antibodies, vaccines with modified glycosylation of antigens, and innovative glycovaccines [69, 141]. For example, the specific E‐selectin antagonist uproleselan (GMI‐1271) is used in combination with chemotherapy for treating relapsed/refractory acute myeloid leukemia (AML) [142]. Advances in immunotherapy research have significantly increased the potential for cancer vaccine development [143]. Elevated glycan levels found on cancer cells surfaces suggest that vaccines utilizing tumor‐associated glycan antigens could be a viable option for cancer immunotherapy [144]. Recent studies have shown that MUC1 glycopeptide vaccines with GalNAc glycocluster modifications targeting macrophage galactose c‐type lectins on dendritic cells induce higher anti‐Tn‐MUC1 antibody titers [145]. Currently, many drugs and vaccines that target glycosylation are being tested in clinical trials for the treatment of cancer (Table 2).
TABLE 2.
Clinical trials related to cancer therapeutic agents and vaccines that target glycosylation.
| Drug | Mechanism of action | Indications | Clinical phase | Clinical Trial# |
|---|---|---|---|---|
| GR‐MD‐02 | Galectin inhibitor | Melanoma, non‐small cell lung cancer, squamous cell carcinoma of the head and neck | I | NCT02575404 |
| GR‐MD‐02 | Galectin inhibitor | Metastatic melanoma | I | NCT02117362 |
| GM‐CT‐01 | Galectin inhibitor | Colorectal cancer, lung cancer, breast cancer | I | NCT00054977 |
| GM‐CT‐01 | Galectin inhibitor | Advanced metastatic melanoma | I/II | NCT01723813 |
| GM‐CT‐01 | Galectin inhibitors | Metastatic colorectal cancer | II | NCT00110721 |
| GMI‐1271 | E‐selectin inhibitor | Acute myeloid leukemia | I/II | NCT02306291 |
| GMI‐1271 | E‐selectin inhibitor | Acute myeloid leukemia | III | NCT03616470 |
| MUC‐1 CAR‐T | CAR‐T cells against aberrant glycosylation of MUC‐1 | Intrahepatic cholangiocarcinoma | I/II | NCT03633773 |
| 2‐fluorofucose | Fucosylation inhibitor | Advanced solid tumors | I/II | NCT02952989 |
| Globo‐H‐GM2‐sTn‐TF‐Tn | Cancer vaccination with truncated O‐glycans | Epithelial ovarian, fallopian tube, or peritoneal cancer | 1 | NCT01248273 |
| MUC‐2‐KLH vaccine | Cancer vaccination with truncated O‐glycans | Prostate cancer | I | NCT00003819 |
| MUC1 peptide‐Poly‐ICLC vaccine | MUC1 peptide vaccine | Lung carcinoma | I | NCT03300817 |
Abbreviations: CAR‐T, cimeric antigen receptor T; KLH, keyhole limpet hemocyanin; MUC1, mucin 1; MUC‐2, mucin‐2; Poly‐ICLC, polyinosinic‐polycytidylic acid‐poly‐l‐lysine carboxymethylcellulose.
In conclusion, significant advances in elucidating the role of glycosylation in the clinical treatment of cancer have been made in recent years. These insights have substantially contributed to the further discovery of novel cancer biomarkers and potential therapeutic targets.
5. CONCLUSION AND PERSPECTIVES
Cancer research represents a complex challenge in the field of medical science and remains a focal point of intensive study. To improve our comprehension of the fundamental processes of cancer, we must explore new therapeutic strategies to increase treatment effectiveness. Previous studies have shown that alterations in the glycosylation process and glycan chain configuration are strongly associated with the progression of cancer. However, although numerous studies have confirmed the critical biological role of glycosylation, current research is still constrained by the complexity and diversity of glycan chain structures, the dynamics of the glycan chain synthesis process, and the absence of a direct one‐to‐one linear relationship between structure and function. The inherent heterogeneity of tumor tissues further complicates glycosylation studies, as various cells and tissues can display distinct glycosylation patterns. Therefore, thoroughly understanding and studying this complex biochemical process remain challenging. Moreover, considering the clinical application of these studies is equally important, and the clinical translation of glycosylation research faces many challenges. Currently, clinical glycoprotein‐based assays, exemplified by tumor markers, are predominantly targeted to the protein fraction. The lack of glycoform information affects the application value of glycoprotein‐based markers. Furthermore, the non‐tumor specificity of these tumor biomarkers affects their clinical diagnostic specificity. The development of glycoprotein markers with well‐defined glycoforms or tumor‐specific glycosylation patterns may provide a way forward for precision proteins. Targeting the glycosylation pathway in tumor treatment using small molecules or glycan‐modifying enzymes offers new possibilities for enhancing the effectiveness of antitumor medications or combination cancer therapy. In the future, advancements in methodologies such as proteomics, genomics, and glycomics will surely improve our investigation of glycosylation regulation and its structure‐function interactions. This task is essential for deciphering the complex processes of glycosylation and comprehending its crucial biological function during tumorigenesis and development. Advancements in this area will make it easier to create drugs such as inhibitors of glycosylation‐related enzymes, antagonists of glycan chains, and modulators of glycan function, ultimately improving the use of clinical translation results.
In summary, this review of aberrant glycosylation has provided new insights into the progression of cancer. Investigating the mechanisms of glycosylation and identifying glycoprotein biomarkers will contribute to the advancement of cancer treatment, offering scientific guidance for early detection and therapy. In the future, the evolution of glycomics and glycosylation research may significantly improve the current landscape of cancer treatment.
AUTHOR CONTRIBUTIONS
Conceptualization: Qianjin Liao, Yujuan Zhou and Hui Wang. Data accumulation, writing, and original draft preparation: Xuemeng Xu, Qiu Peng, Xianjie Jiang, Shiming Tan and Wenjuan Yang. Editing: Yaqian Han, Linda Oyang, Jinguan Lin, Mengzhou Shen, Jiewen Wang, Haofan Li, Longzheng Xia, Mingjing Peng, Nayiyuan Wu and Yanyan Tang. Figure preparation: Xuemeng Xu and Qiu Peng. Revision: Qianjin Liao and Yujuan Zhou. Supervision: Qianjin Liao, Yujuan Zhou and Hui Wang. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
ACKNOWLEDGEMENTS
We thank Figdraw(www.figdraw.com) for the assistance in creating figures. This work was supported in part by grants from the following sources: the National Natural Science Foundation of China (82472882, 82302987, 82303534, 82203233, 82202966, 82173142), the Natural Science Foundation of Hunan Province (2024JJ4025, 2023ZJ1122, Z2023086, 2023JJ60469, 2023JJ40413, 2023JJ30372, 2023JJ30375, 2022JJ80078, 2020JJ5336), Key Research and Development Program of Hunan Province (2022SK2051), Science and Technology Innovation Program of Hunan Province (2023RC3199, 2023SK4034, 2023RC1073), the Research Project of Health Commission of Hunan Province (R2023040, R2023093, 202203034978, 202202055318, 202109031837, 202109032010, 20201020), the Changsha Science and Technology Board (kh2201054), Ascend Foundation of National cancer center (NCC201909B06), and by Hunan Cancer Hospital Climb Plan (ZX2020001‐3, YF2020002, 2023NSFC‐A001, 2023NSFC‐A002, 2023NSFC‐A004).
Xu X, Peng Q, Jiang X, Tan S, Yang W, Han Y, et al. Altered glycosylation in cancer: molecular functions and therapeutic potential. Cancer Commun. 2024;44:1316–1336. 10.1002/cac2.12610
Xuemeng Xu and Qiu Peng contribute equally to this work.
Contributor Information
Hui Wang, Email: wanghui710327@163.com.
Qianjin Liao, Email: march-on@126.com.
Yujuan Zhou, Email: yujany_zhou@163.com.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Wang WH, Yuan T, Qian MJ, Yan FJ, Yang L, He QJ, et al. Post‐translational modification of KRAS: potential targets for cancer therapy. Acta Pharmacol Sin. 2021;42(8):1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen L, Liu S, Tao Y. Regulating tumor suppressor genes: post‐translational modifications. Signal Transduct Target Ther. 2020;5(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gupta R, Sahu M, Srivastava D, Tiwari S, Ambasta RK, Kumar P. Post‐translational modifications: Regulators of neurodegenerative proteinopathies. Ageing Res Rev. 2021;68:101336. [DOI] [PubMed] [Google Scholar]
- 4. Millan‐Zambrano G, Burton A, Bannister AJ, Schneider R. Histone post‐translational modifications ‐ cause and consequence of genome function. Nat Rev Genet. 2022;23(9):563–580. [DOI] [PubMed] [Google Scholar]
- 5. Zhong Q, Xiao X, Qiu Y, Xu Z, Chen C, Chong B, et al. Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications. Med Comm (2020). 2023;4(3):e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eichler J. Protein glycosylation. Curr Biol. 2019;29(7):R229–R231. [DOI] [PubMed] [Google Scholar]
- 7. Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol. 2020;21(12):729–749. [DOI] [PubMed] [Google Scholar]
- 8. Newby ML, Allen JD, Crispin M. Influence of glycosylation on the immunogenicity and antigenicity of viral immunogens. Biotechnol Adv. 2024;70:108283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hale RC, Morais D, Chou J, Stowell SR. The role of glycosylation in clinical allergy and immunology. J Allergy Clin Immunol. 2024;153(1):55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoek M, Demmers LC, Wu W, Heck AJR. Allotype‐Specific Glycosylation and Cellular Localization of Human Leukocyte Antigen Class I Proteins. J Proteome Res. 2021;20(9):4518–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kissel T, Toes REM, Huizinga TWJ, Wuhrer M. Glycobiology of rheumatic diseases. Nat Rev Rheumatol. 2023;19(1):28–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wojcik I, Wuhrer M, Heeringa P, Stegeman CA, Rutgers A, Falck D. Specific IgG glycosylation differences precede relapse in PR3‐ANCA associated vasculitis patients with and without ANCA rise. Front Immunol. 2023;14:1214945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaur D, Khan H, Grewal AK, Singh TG. Glycosylation: A new signaling paradigm for the neurovascular diseases. Life Sci. 2024;336:122303. [DOI] [PubMed] [Google Scholar]
- 14. Xie X, Kong S, Cao W. Targeting protein glycosylation to regulate inflammation in the respiratory tract: novel diagnostic and therapeutic candidates for chronic respiratory diseases. Front Immunol. 2023;14:1168023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10:473–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bangarh R, Khatana C, Kaur S, Sharma A, Kaushal A, Siwal SS, et al. Aberrant protein glycosylation: Implications on diagnosis and Immunotherapy. Biotechnol Adv. 2023;66:108149. [DOI] [PubMed] [Google Scholar]
- 17. Grzesik K, Janik M, Hoja‐Lukowicz D. The hidden potential of glycomarkers: Glycosylation studies in the service of cancer diagnosis and treatment. Biochim Biophys Acta Rev Cancer. 2023;1878(3):188889. [DOI] [PubMed] [Google Scholar]
- 18. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. [DOI] [PubMed] [Google Scholar]
- 19. Mereiter S, Balmana M, Campos D, Gomes J, Reis CA. Glycosylation in the Era of Cancer‐Targeted Therapy: Where Are We Heading? Cancer Cell. 2019;36(1):6–16. [DOI] [PubMed] [Google Scholar]
- 20. Altevogt P, Sammar M, Huser L, Kristiansen G. Novel insights into the function of CD24: A driving force in cancer. Int J Cancer. 2021;148(3):546–559. [DOI] [PubMed] [Google Scholar]
- 21. Silsirivanit A. Glycosylation markers in cancer. Adv Clin Chem. 2019;89:189–213. [DOI] [PubMed] [Google Scholar]
- 22. RodrIguez E, Schetters STT, van Kooyk Y. The tumour glyco‐code as a novel immune checkpoint for immunotherapy. Nat Rev Immunol. 2018;18(3):204–211. [DOI] [PubMed] [Google Scholar]
- 23. Oliveira‐Ferrer L, Legler K, Milde‐Langosch K. Role of protein glycosylation in cancer metastasis. Semin Cancer Biol. 2017;44:141–152. [DOI] [PubMed] [Google Scholar]
- 24. Xu X, Peng Q, Jiang X, Tan S, Yang Y, Yang W, et al. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Exp Mol Med. 2023;55(7):1357–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lopez‐Otin C, Pietrocola F, Roiz‐Valle D, Galluzzi L, Kroemer G. Meta‐hallmarks of aging and cancer. Cell Metab. 2023;35(1):12–35. [DOI] [PubMed] [Google Scholar]
- 26. Kang N, Son S, Min S, Hong H, Kim C, An J, et al. Stimuli‐responsive ferroptosis for cancer therapy. Chem Soc Rev. 2023;52(12):3955–3972. [DOI] [PubMed] [Google Scholar]
- 27. Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5(7):526–542. [DOI] [PubMed] [Google Scholar]
- 28. Zhou JY, Cobb BA. Glycans in Immunologic Health and Disease. Annu Rev Immunol. 2021;39:511–536. [DOI] [PubMed] [Google Scholar]
- 29. Song Y, Zhang F, Glycosaminoglycans Linhardt RJ.. Adv Exp Med Biol. 2021;1325:103–116. [DOI] [PubMed] [Google Scholar]
- 30. Varki A. Biological roles of glycans. Glycobiology. 2017;27(1):3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. [DOI] [PubMed] [Google Scholar]
- 32. Gu J, Isaji T, Xu Q, Kariya Y, Gu W, Fukuda T, et al. Potential roles of N‐glycosylation in cell adhesion. Glycoconj J. 2012;29(8‐9):599–607. [DOI] [PubMed] [Google Scholar]
- 33. Gedaj A, Gregorczyk P, Zukowska D, Chorazewska A, Ciura K, Kalka M, et al. Glycosylation of FGF/FGFR: An underrated sweet code regulating cellular signaling programs. Cytokine Growth Factor Rev. 2024;77:39–55. [DOI] [PubMed] [Google Scholar]
- 34. Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, et al. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol Cell. 2019;75(2):394–407 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma J, Wu C, Hart GW. Analytical and Biochemical Perspectives of Protein O‐GlcNAcylation. Chem Rev. 2021;121(3):1513–1581. [DOI] [PubMed] [Google Scholar]
- 36. Yang W, Tian E, Chernish A, McCluggage P, Dalal K, Lara A, et al. Quantitative mapping of the in vivo O‐GalNAc glycoproteome in mouse tissues identifies GalNAc‐T2 O‐glycosites in metabolic disorder. Proc Natl Acad Sci U S A. 2023;120(43):e2303703120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suzuki T. Catabolism of N‐glycoproteins in mammalian cells: Molecular mechanisms and genetic disorders related to the processes. Mol Aspects Med. 2016;51:89–103. [DOI] [PubMed] [Google Scholar]
- 39. Fahie K, Zachara NE. Molecular Functions of Glycoconjugates in Autophagy. J Mol Biol. 2016;428(16):3305–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cherepanova N, Shrimal S, Gilmore R. N‐linked glycosylation and homeostasis of the endoplasmic reticulum. Curr Opin Cell Biol. 2016;41:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tannous A, Pisoni GB, Hebert DN, Molinari M. N‐linked sugar‐regulated protein folding and quality control in the ER. Semin Cell Dev Biol. 2015;41:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saha A, Bello D, Fernandez‐Tejada A. Advances in chemical probing of protein O‐GlcNAc glycosylation: structural role and molecular mechanisms. Chem Soc Rev. 2021;50(18):10451–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Balana AT, Levine PM, Craven TW, Mukherjee S, Pedowitz NJ, Moon SP, et al. O‐GlcNAc modification of small heat shock proteins enhances their anti‐amyloid chaperone activity. Nat Chem. 2021;13(5):441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chatham JC, Zhang J, Wende AR. Role of O‐Linked N‐Acetylglucosamine Protein Modification in Cellular (Patho)Physiology. Physiol Rev. 2021;101(2):427–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gill DJ, Clausen H, Bard F. Location, location, location: new insights into O‐GalNAc protein glycosylation. Trends Cell Biol. 2011;21(3):149–158. [DOI] [PubMed] [Google Scholar]
- 46. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol. 2019;15(6):346–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pathangey LB, Lakshminarayanan V, Suman VJ, Pockaj BA, Mukherjee P, Gendler SJ. Aberrant Glycosylation of Anchor‐Optimized MUC1 Peptides Can Enhance Antigen Binding Affinity and Reverse Tolerance to Cytotoxic T Lymphocytes. Biomolecules. 2016;6(3):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Julenius K. NetCGlyc 1.0: prediction of mammalian C‐mannosylation sites. Glycobiology. 2007;17(8):868–876. [DOI] [PubMed] [Google Scholar]
- 49. Crine SL, Acharya KR. Molecular basis of C‐mannosylation ‐ a structural perspective. FEBS J. 2022;289(24):7670–7687. [DOI] [PubMed] [Google Scholar]
- 50. Shcherbakova A, Preller M, Taft MH, Pujols J, Ventura S, Tiemann B, et al. C‐mannosylation supports folding and enhances stability of thrombospondin repeats. Elife. 2019;8:e52978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu W, Zhang R, Chen W, Lin D, Wei K, Li J, et al. Glycosylation at Asn254 Is Required for the Activation of the PDGF‐C Protein. Front Mol Biosci. 2021;8:665552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bandini G, Albuquerque‐Wendt A, Hegermann J, Samuelson J, Routier FH. Protein O‐ and C‐Glycosylation pathways in Toxoplasma gondii and Plasmodium falciparum. Parasitology. 2019;146(14):1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lopaticki S, McConville R, John A, Geoghegan N, Mohamed SD, Verzier L, et al. Tryptophan C‐mannosylation is critical for Plasmodium falciparum transmission. Nat Commun. 2022;13(1):4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kinoshita T, Fujita M. Biosynthesis of GPI‐anchored proteins: special emphasis on GPI lipid remodeling. J Lipid Res. 2016;57(1):6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kinoshita T. Biosynthesis and biology of mammalian GPI‐anchored proteins. Open Biol. 2020;10(3):190290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang H, Su J, Li B, Gao Y, Liu M, He L, et al. Structure of human glycosylphosphatidylinositol transamidase. Nat Struct Mol Biol. 2022;29(3):203–209. [DOI] [PubMed] [Google Scholar]
- 57. Guo Z. Glycosphingolipid and Glycosylphosphatidylinositol Affect Each Other in and on the Cell. Chembiochem. 2023;24(13):e202200761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Elishmereni M, Levi‐Schaffer F. CD48: A co‐stimulatory receptor of immunity. Int J Biochem Cell Biol. 2011;43(1):25–28. [DOI] [PubMed] [Google Scholar]
- 59. White D, Cote‐Martin A, Bleck M, Garaffa N, Shaaban A, Wu H, et al. Programmed Cell Death‐1 (PD‐1) anchoring to the GPI‐linked co‐receptor CD48 reveals a novel mechanism to modulate PD‐1‐dependent inhibition of human T cells. Mol Immunol. 2023;156:31–38. [DOI] [PubMed] [Google Scholar]
- 60. Bravo‐Perez C, Guarnera L, Williams ND, Visconte V. Paroxysmal Nocturnal Hemoglobinuria: Biology and Treatment. Medicina (Kaunas). 2023;59(9):1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Paprocka J, Hutny M, Hofman J, Tokarska A, Klaniewska M, Szczaluba K, et al. Spectrum of Neurological Symptoms in Glycosylphosphatidylinositol Biosynthesis Defects: Systematic Review. Front Neurol. 2021;12:758899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, et al. Small RNAs are modified with N‐glycans and displayed on the surface of living cells. Cell. 2021;184(12):3109–3124 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Disney MD. A glimpse at the glycoRNA world. Cell. 2021;184(12):3080–3081. [DOI] [PubMed] [Google Scholar]
- 64. Ma Y, Guo W, Mou Q, Shao X, Lyu M, Garcia V, et al. Spatial imaging of glycoRNA in single cells with ARPLA. Nat Biotechnol.2024;42(4):608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang N, Tang W, Torres L, Wang X, Ajaj Y, Zhu L, et al. Cell surface RNAs control neutrophil recruitment. Cell. 2024;187(4):846–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Caval T, Alisson‐Silva F, Schwarz F. Roles of glycosylation at the cancer cell surface: opportunities for large scale glycoproteomics. Theranostics. 2023;13(8):2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lumibao JC, Tremblay JR, Hsu J, Engle DD. Altered glycosylation in pancreatic cancer and beyond. J Exp Med. 2022;219(6):e20211505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12(1):31–46. [DOI] [PubMed] [Google Scholar]
- 69. Thomas D, Rathinavel AK, Radhakrishnan P. Altered glycosylation in cancer: A promising target for biomarkers and therapeutics. Biochim Biophys Acta Rev Cancer. 2021;1875(1):188464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Supruniuk K, Radziejewska I. MUC1 is an oncoprotein with a significant role in apoptosis (Review). Int J Oncol. 2021;59(3):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mallard BW, Tiralongo J. Cancer stem cell marker glycosylation: Nature, function and significance. Glycoconj J. 2017;34(4):441–452. [DOI] [PubMed] [Google Scholar]
- 72. Le Minh G, Reginato MJ. Role of O‐GlcNAcylation on cancer stem cells: Connecting nutrient sensing to cell plasticity. Adv Cancer Res. 2023;157:195–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miao Z, Cao Q, Liao R, Chen X, Li X, Bai L, et al. Elevated transcription and glycosylation of B3GNT5 promotes breast cancer aggressiveness. J Exp Clin Cancer Res. 2022;41(1):169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jacob F, Alam S, Konantz M, Liang CY, Kohler RS, Everest‐Dass AV, et al. Transition of Mesenchymal and Epithelial Cancer Cells Depends on alpha1‐4 Galactosyltransferase‐Mediated Glycosphingolipids. Cancer Res. 2018;78(11):2952–2965. [DOI] [PubMed] [Google Scholar]
- 75. Gupta R, Ponangi R, Indresh KG. Role of glycosylation in breast cancer progression and metastasis: implications for miRNA, EMT and multidrug resistance. Glycobiology. 2023;33(7):545–555. [DOI] [PubMed] [Google Scholar]
- 76. Yu R, Longo J, van Leeuwen JE, Zhang C, Branchard E, Elbaz M, et al. Mevalonate Pathway Inhibition Slows Breast Cancer Metastasis via Reduced N‐glycosylation Abundance and Branching. Cancer Res. 2021;81(10):2625–2635. [DOI] [PubMed] [Google Scholar]
- 77. Song J, Liu W, Wang J, Hao J, Wang Y, You X, et al. GALNT6 promotes invasion and metastasis of human lung adenocarcinoma cells through O‐glycosylating chaperone protein GRP78. Cell Death Dis. 2020;11(5):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. He L, Guo Z, Wang W, Tian S, Lin R. FUT2 inhibits the EMT and metastasis of colorectal cancer by increasing LRP1 fucosylation. Cell Commun Signal. 2023;21(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chu YD, Fan TC, Lai MW, Yeh CT. GALNT14‐mediated O‐glycosylation on PHB2 serine‐161 enhances cell growth, migration and drug resistance by activating IGF1R cascade in hepatoma cells. Cell Death Dis. 2022;13(11):956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cheng H, Wang S, Gao D, Yu K, Chen H, Huang Y, et al. Nucleotide sugar transporter SLC35A2 is involved in promoting hepatocellular carcinoma metastasis by regulating cellular glycosylation. Cell Oncol (Dordr). 2023;46(2):283–297. [DOI] [PubMed] [Google Scholar]
- 81. Scott E, Archer Goode E, Garnham R, Hodgson K, Orozco‐Moreno M, Turner H, et al. ST6GAL1‐mediated aberrant sialylation promotes prostate cancer progression. J Pathol. 2023;261(1):71–84. [DOI] [PubMed] [Google Scholar]
- 82. Scott E, Hodgson K, Calle B, Turner H, Cheung K, Bermudez A, et al. Upregulation of GALNT7 in prostate cancer modifies O‐glycosylation and promotes tumour growth. Oncogene. 2023;42(12):926–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lin WD, Fan TC, Hung JT, Yeo HL, Wang SH, Kuo CW, et al. Sialylation of CD55 by ST3GAL1 Facilitates Immune Evasion in Cancer. Cancer Immunol Res. 2021;9(1):113–122. [DOI] [PubMed] [Google Scholar]
- 84. Huang Y, Zhang HL, Li ZL, Du T, Chen YH, Wang Y, et al. FUT8‐mediated aberrant N‐glycosylation of B7H3 suppresses the immune response in triple‐negative breast cancer. Nat Commun. 2021;12(1):2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stanczak MA, Rodrigues Mantuano N, Kirchhammer N, Sanin DE, Jacob F, Coelho R, et al. Targeting cancer glycosylation repolarizes tumor‐associated macrophages allowing effective immune checkpoint blockade. Sci Transl Med. 2022;14(669):eabj1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhu Q, Wang H, Chai S, Xu L, Lin B, Yi W, et al. O‐GlcNAcylation promotes tumor immune evasion by inhibiting PD‐L1 lysosomal degradation. Proc Natl Acad Sci U S A. 2023;120(13):e2216796120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shi C, Wang Y, Wu M, Chen Y, Liu F, Shen Z, et al. Promoting anti‐tumor immunity by targeting TMUB1 to modulate PD‐L1 polyubiquitination and glycosylation. Nat Commun. 2022;13(1):6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cui Y, Li J, Zhang P, Yin D, Wang Z, Dai J, et al. B4GALT1 promotes immune escape by regulating the expression of PD‐L1 at multiple levels in lung adenocarcinoma. J Exp Clin Cancer Res. 2023;42(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ma XM, Luo YF, Zeng FF, Su C, Liu X, Li XP, et al. TGF‐beta1‐Mediated PD‐L1 Glycosylation Contributes to Immune Escape via c‐Jun/STT3A Pathway in Nasopharyngeal Carcinoma. Front Oncol. 2022;12:815437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hu YJ, Zhang X, Lv HM, Liu Y, Li SZ. Protein O‐GlcNAcylation: The sweet hub in liver metabolic flexibility from a (patho)physiological perspective. Liver Int. 2024;44(2):293–315. [DOI] [PubMed] [Google Scholar]
- 91. Nie H, Ju H, Fan J, Shi X, Cheng Y, Cang X, et al. O‐GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nat Commun. 2020;11(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tan M, Pan Q, Yu C, Zhai X, Gu J, Tao L, et al. PIGT promotes cell growth, glycolysis, and metastasis in bladder cancer by modulating GLUT1 glycosylation and membrane trafficking. J Transl Med. 2024;22(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhu Q, Zhou H, Wu L, Lai Z, Geng D, Yang W, et al. O‐GlcNAcylation promotes pancreatic tumor growth by regulating malate dehydrogenase 1. Nat Chem Biol. 2022;18(10):1087–1095. [DOI] [PubMed] [Google Scholar]
- 94. Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, et al. Glucose‐Mediated N‐glycosylation of SCAP Is Essential for SREBP‐1 Activation and Tumor Growth. Cancer Cell. 2015;28(5):569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tan EP, McGreal SR, Graw S, Tessman R, Koppel SJ, Dhakal P, et al. Sustained O‐GlcNAcylation reprograms mitochondrial function to regulate energy metabolism. J Biol Chem. 2017;292(36):14940–14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron‐dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22(7):381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang F, Xiao Y, Ding JH, Jin X, Ma D, Li DQ, et al. Ferroptosis heterogeneity in triple‐negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023;35(1):84–100 e8. [DOI] [PubMed] [Google Scholar]
- 99. Wang Y, Wu X, Ren Z, Li Y, Zou W, Chen J, et al. Overcoming cancer chemotherapy resistance by the induction of ferroptosis. Drug Resist Updat. 2023;66:100916. [DOI] [PubMed] [Google Scholar]
- 100. Yu F, Zhang Q, Liu H, Liu J, Yang S, Luo X, et al. Dynamic O‐GlcNAcylation coordinates ferritinophagy and mitophagy to activate ferroptosis. Cell Discov. 2022;8(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ma H, Chen X, Mo S, Zhang Y, Mao X, Chen J, et al. Targeting N‐glycosylation of 4F2hc mediated by glycosyltransferase B3GNT3 sensitizes ferroptosis of pancreatic ductal adenocarcinoma. Cell Death Differ. 2023;30(8):1988–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tang J, Long G, Hu K, Xiao D, Liu S, Xiao L, et al. Targeting USP8 Inhibits O‐GlcNAcylation of SLC7A11 to Promote Ferroptosis of Hepatocellular Carcinoma via Stabilization of OGT. Adv Sci (Weinh). 2023;10(33):e2302953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wang X, Liu M, Chu Y, Liu Y, Cao X, Zhang H, et al. O‐GlcNAcylation of ZEB1 facilitated mesenchymal pancreatic cancer cell ferroptosis. Int J Biol Sci. 2022;18(10):4135–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schmitt CA, Wang B, Demaria M. Senescence and cancer ‐ role and therapeutic opportunities. Nat Rev Clin Oncol. 2022;19(10):619–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Prieto LI, Sturmlechner I, Graves SI, Zhang C, Goplen NP, Yi ES, et al. Senescent alveolar macrophages promote early‐stage lung tumorigenesis. Cancer Cell. 2023;41(7):1261–1275 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhang JW, Zhang D, Yin HS, Zhang H, Hong KQ, Yuan JP, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression and chemoresistance by enhancing the secretion of chemotherapy‐induced senescence‐associated secretory phenotype via activation of DNA damage response pathway. Gut Microbes. 2023;15(1):2197836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kudryavtseva AV, Krasnov GS, Dmitriev AA, Alekseev BY, Kardymon OL, Sadritdinova AF, et al. Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget. 2016;7(29):44879–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lopez‐Sambrooks C, Shrimal S, Khodier C, Flaherty DP, Rinis N, Charest JC, et al. Oligosaccharyltransferase inhibition induces senescence in RTK‐driven tumor cells. Nat Chem Biol. 2016;12(12):1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Taparra K, Wang H, Malek R, Lafargue A, Barbhuiya MA, Wang X, et al. O‐GlcNAcylation is required for mutant KRAS‐induced lung tumorigenesis. J Clin Invest. 2018;128(11):4924–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chang X, Obianwuna UE, Wang J, Zhang H, Qi G, Qiu K, et al. Glycosylated proteins with abnormal glycosylation changes are potential biomarkers for early diagnosis of breast cancer. Int J Biol Macromol. 2023;236:123855. [DOI] [PubMed] [Google Scholar]
- 111. Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19(10):670–681. [DOI] [PubMed] [Google Scholar]
- 112. Lee TH, Kim JS, Baek SJ, Kwak JM, Kim J. Diagnostic Accuracy of Carcinoembryonic Antigen (CEA) in Detecting Colorectal Cancer Recurrence Depending on Its Preoperative Level. J Gastrointest Surg. 2023;27(8):1694–1701. [DOI] [PubMed] [Google Scholar]
- 113. Hoti N, Lih TS, Dong M, Zhang Z, Mangold L, Partin AW, et al. Urinary PSA and Serum PSA for Aggressive Prostate Cancer Detection. Cancers (Basel). 2023;15(3):960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Anastasi E, Farina A, Granato T, Colaiacovo F, Pucci B, Tartaglione S, et al. Recent Insight about HE4 Role in Ovarian Cancer Oncogenesis. Int J Mol Sci. 2023;24(13):10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Behrouzi R, Barr CE, Crosbie EJ. HE4 as a Biomarker for Endometrial Cancer. Cancers (Basel). 2021;13(19):4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cen S, Liu Z, Pan H, Han W. Clinicopathologic features and treatment advances in cancers with HER2 alterations. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188605. [DOI] [PubMed] [Google Scholar]
- 117. Pretorius CJ, Wilgen U, Klingberg S, Zournazi A, Sanders L, Ungerer JPJ. Comparison between free beta subunit of human chorionic gonadotropin (hCG) and total hCG assays in adults with testicular cancer. Clin Chem Lab Med. 2023;61(10):1841–1849. [DOI] [PubMed] [Google Scholar]
- 118. Felder M, Kapur A, Gonzalez‐Bosquet J, Horibata S, Heintz J, Albrecht R, et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer. 2014;13:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Luo G, Jin K, Deng S, Cheng H, Fan Z, Gong Y, et al. Roles of CA19‐9 in pancreatic cancer: Biomarker, predictor and promoter. Biochim Biophys Acta Rev Cancer. 2021;1875(2):188409. [DOI] [PubMed] [Google Scholar]
- 120. Xu Y, Zhang P, Zhang K, Huang C. The application of CA72‐4 in the diagnosis, prognosis, and treatment of gastric cancer. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188634. [DOI] [PubMed] [Google Scholar]
- 121. Li H, Li L, Sun J, Dong S, Li H. Value of TCT combined with serum CA153 and CA50 in early diagnosis of cervical cancer and precancerous lesions. Pak J Med Sci. 2022;38(6):1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tang S, Wei L, Sun Y, Zhou F, Zhu S, Yang R, et al. CA153 in Breast Secretions as a Potential Molecular Marker for Diagnosing Breast Cancer: A Meta Analysis. PLoS One. 2016;11(9):e0163030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Li G, Zhang H, Zhang L, Liu H, Li S, Wang Y, et al. Serum Markers CA125, CA153, and CEA along with Inflammatory Cytokines in the Early Detection of Lung Cancer in High‐Risk Populations. Biomed Res Int. 2022;2022:1394042. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124. Xu H, Huang K, Lin Y, Gong H, Ma X, Zhang D. Glycosyltransferase GLT8D1 and GLT8D2 serve as potential prognostic biomarkers correlated with Tumor Immunity in Gastric Cancer. BMC Med Genomics. 2023;16(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pucci M, Malagolini N, Dall'Olio F. Glycosyltransferase B4GALNT2 as a Predictor of Good Prognosis in Colon Cancer: Lessons from Databases. Int J Mol Sci. 2021;22(9):4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Dong S, Wang Z, Huang B, Zhang J, Ge Y, Fan Q, et al. Bioinformatics insight into glycosyltransferase gene expression in gastric cancer: POFUT1 is a potential biomarker. Biochem Biophys Res Commun. 2017;483(1):171–177. [DOI] [PubMed] [Google Scholar]
- 127. Perez M, Chakraborty A, Lau LS, Mohammed NBB, Dimitroff CJ. Melanoma‐associated glycosyltransferase GCNT2 as an emerging biomarker and therapeutic target. Br J Dermatol. 2021;185(2):294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Noda M, Okayama H, Tachibana K, Sakamoto W, Saito K, Thar Min AK, et al. Glycosyltransferase Gene Expression Identifies a Poor Prognostic Colorectal Cancer Subtype Associated with Mismatch Repair Deficiency and Incomplete Glycan Synthesis. Clin Cancer Res. 2018;24(18):4468–4481. [DOI] [PubMed] [Google Scholar]
- 129. Guibourdenche J, Handschuh K, Tsatsaris V, Gerbaud P, Leguy MC, Muller F, et al. Hyperglycosylated hCG is a marker of early human trophoblast invasion. J Clin Endocrinol Metab. 2010;95(10):E240–E244. [DOI] [PubMed] [Google Scholar]
- 130. Fernandez‐Ponce C, Geribaldi‐Doldan N, Sanchez‐Gomar I, Quiroz RN, Ibarra LA, Escorcia LG, et al. The Role of Glycosyltransferases in Colorectal Cancer. Int J Mol Sci. 2021;22(11):5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion. Nat Rev Immunol. 2020;20(4):209–215. [DOI] [PubMed] [Google Scholar]
- 132. Zhu L, Xu Y, Wei X, Lin H, Huang M, Lin B, et al. Coupling Aptamer‐based Protein Tagging with Metabolic Glycan Labeling for In Situ Visualization and Biological Function Study of Exosomal Protein‐Specific Glycosylation. Angew Chem Int Ed Engl. 2021;60(33):18111–18115. [DOI] [PubMed] [Google Scholar]
- 133. Zhu L, Xu Y, Kang S, Lin B, Zhang C, You Z, et al. Quantification‐Promoted Discovery of Glycosylated Exosomal PD‐L1 as a Potential Tumor Biomarker. Small Methods. 2022;6(9):e2200549. [DOI] [PubMed] [Google Scholar]
- 134. Krug J, Rodrian G, Petter K, Yang H, Khoziainova S, Guo W, et al. N‐glycosylation Regulates Intrinsic IFN‐gamma Resistance in Colorectal Cancer: Implications for Immunotherapy. Gastroenterology. 2023;164(3):392–406 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chen Y, Su L, Huang C, Wu S, Qiu X, Zhao X, et al. Galactosyltransferase B4GALT1 confers chemoresistance in pancreatic ductal adenocarcinomas by upregulating N‐linked glycosylation of CDK11(p110). Cancer Lett. 2021;500:228–243. [DOI] [PubMed] [Google Scholar]
- 136. Wu J, Chen S, Liu H, Zhang Z, Ni Z, Chen J, et al. Tunicamycin specifically aggravates ER stress and overcomes chemoresistance in multidrug‐resistant gastric cancer cells by inhibiting N‐glycosylation. J Exp Clin Cancer Res. 2018;37(1):272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Xi X, Wang J, Qin Y, Huang W, You Y, Zhan J. Glycosylated modification of MUC1 maybe a new target to promote drug sensitivity and efficacy for breast cancer chemotherapy. Cell Death Dis. 2022;13(8):708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Aldonza MBD, Cha J, Yong I, Ku J, Sinitcyn P, Lee D, et al. Multi‐targeted therapy resistance via drug‐induced secretome fucosylation. Elife. 2023;12:e75191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Greco B, Malacarne V, De Girardi F, Scotti GM, Manfredi F, Angelino E, et al. Disrupting N‐glycan expression on tumor cells boosts chimeric antigen receptor T cell efficacy against solid malignancies. Sci Transl Med. 2022;14(628):eabg3072. [DOI] [PubMed] [Google Scholar]
- 140. Sun X, He Z, Guo L, Wang C, Lin C, Ye L, et al. ALG3 contributes to stemness and radioresistance through regulating glycosylation of TGF‐beta receptor II in breast cancer. J Exp Clin Cancer Res. 2021;40(1):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Smith BAH, Bertozzi CR. The clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans. Nat Rev Drug Discov. 2021;20(3):217–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. DeAngelo DJ, Jonas BA, Liesveld JL, Bixby DL, Advani AS, Marlton P, et al. Phase 1/2 study of uproleselan added to chemotherapy in patients with relapsed or refractory acute myeloid leukemia. Blood. 2022;139(8):1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Liu J, Fu M, Wang M, Wan D, Wei Y, Wei X. Cancer vaccines as promising immuno‐therapeutics: platforms and current progress. J Hematol Oncol. 2022;15(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Padler‐Karavani V. Glycan Microarray Reveal the Sweet Side of Cancer Vaccines. Cell Chem Biol. 2016;23(12):1446–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gabba A, Attariya R, Behren S, Pett C, van der Horst JC, Yurugi H, et al. MUC1 Glycopeptide Vaccine Modified with a GalNAc Glycocluster Targets the Macrophage Galactose C‐type Lectin on Dendritic Cells to Elicit an Improved Humoral Response. J Am Chem Soc. 2023;145(24):13027–13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


