Abstract
To survive in a dynamic and unpredictable environment, cells must correctly interpret and integrate extracellular signals with internal factors. In particular, internal stores of nutrients must be managed for use during periods of nutrient limitation. To gain insight into this complex process, we combined biochemical and spectroscopic techniques to follow the dynamics of the phosphate responsive signaling pathway in both single yeast cells and populations. We demonstrate that the phosphate-responsive genes PHO5 and PHO84 exhibit different kinetics of transcriptional induction in response to phosphate starvation, and that transient phosphate limitation causes induction of PHO84 but not PHO5. This differential kinetic behavior is largely eliminated in cells that lack the ability to store phosphate internally in the form of polyphosphate, but the threshold of external phosphate required for induction of PHO5 and PHO84 is unaffected. Our observations indicate that polyphosphate acts as a buffer that can be mobilized during periods of phosphate limitation and enables the phosphate-responsive signaling pathway to filter transient fluctuations in extracellular phosphate levels.
Keywords: homeostasis, nutrients, polyphosphate, signaling
Cellular responses to environmental cues involve a complex interplay between intracellular conditions and extracellular factors, coordinated via elaborate signal transduction cascades that initiate changes in metabolism and gene regulation. Insight into how cellular responses facilitate survival in dynamic environments can be gained by following the response to deprivation of the essential nutrient, phosphate, in the model organism, Saccharomyces cerevisiae.
In response to phosphate starvation, budding yeast cells have three major responses: they mobilize internal stores of phosphate (1, 2), up-regulate production of a plasma membrane transporter involved in phosphate uptake (Pho84), and increase production of phosphatases (e.g., Pho5) that are secreted into the extracellular environment to liberate inorganic phosphate (Pi) by hydrolysis of organic phosphates (3, 4). These responses are mediated primarily at the level of gene regulation and are controlled by a signaling pathway known as the phosphate-responsive signaling (PHO) pathway (5–7). Key components in the PHO signaling pathway include a cyclin/cyclin-dependent kinase complex, Pho80/Pho85, whose activity is regulated in response to phosphate conditions, and its transcription factor substrate, Pho4, which regulates phosphate-responsive gene transcription (5, 8, 9). Phosphorylation of Pho4 on four serine residues by Pho80/Pho85 controls Pho4 subcellular localization and its interaction with another transcription factor, Pho2 (10).
When cells are grown in medium containing a high concentration of Pi, Pho80/Pho85 is active, Pho4 is fully phosphorylated and localized to the cytoplasm, and transcription of phosphate-responsive genes such as PHO5 and PHO84 is turned off (8, 9). In response to phosphate limitation, Pho80/Pho85 is inactivated, and unphosphorylated Pho4 accumulates in the nucleus where it activates transcription of phosphate-responsive genes such as PHO84 and PHO5 (8, 9). Interestingly, when cells are grown in intermediate concentrations of phosphate, Pho4 accumulates in a form that is partially phosphorylated, localized to the nucleus, not able to efficiently interact with Pho2, and capable of activating transcription of PHO84 but not PHO5 (11). It is thought that Pho80/Pho85 is partially active in intermediate phosphate conditions, leading to accumulation of Pho4 in a partially phosphorylated form. Under these conditions, Pho4 that is primarily phosphorylated on serine 223 accumulates, presumably because Pho80/Pho85 preferentially phosphorylates this site, which is involved in regulating the interaction of Pho4 with Pho2 (12).
Our working model is that changes in extracellular phosphate availability lead to alterations in internal levels of phosphate, which in turn lead to modulation of Pho80/Pho85 kinase activity and Pho4 phosphorylation state. Internal levels of phosphate are influenced by cellular usage, uptake into the cell through transporters, and mobilization of internal stores. It is known that cells store large amounts of phosphate internally in the form of polyphosphate (polyP), a linear polymer of phosphate, within the yeast vacuole, and that these stores are mobilized during phosphate starvation (1, 2, 13–16). If our model is correct, mobilization of internal stores of phosphate during phosphate limitation might temporarily increase intracellular levels of phosphate and keep the Pho80/Pho85 kinase partially active. Although previous studies have demonstrated that the PHO pathway is not grossly affected by mutations that impair usage or storage of polyP (17, 18), we speculated that the kinetics of the PHO response might be altered in cells that exhibit impaired storage or use of polyP.
In this study, we demonstrate that PHO5 and PHO84, genes that are regulated by the same transcription factor (Pho4), exhibit different kinetics of transcriptional induction in response to phosphate starvation. We examine the role of intracellular polyP stores in differential gene expression and in modulating the cellular response to fluctuations in the availability of extracellular phosphate.
Materials and Methods
Strain Construction. Strains of S. cerevisiae used in this study were constructed by standard genetic methods and are listed in Table 1, which is published as supporting information on the PNAS web site (19). To generate pho5::GFP (EY0831) and pho5::YFP (YFP, yellow fluorescent protein) (EY1640) strains, the entire PHO5 ORF was replaced with the coding sequence of either GFP or yeast codon-optimized YFP by using a standard homologous recombination technique (20–22). The PHO84pr:GFP (EY1109) and PHO84pr:YFP (EY1624) strains were generated by inserting a cassette containing the PHO84 promoter followed by either the GFP or yeast codon-optimized YFP coding sequence at the LEU2 locus (23). In this strain, it was necessary to maintain a functional copy of PHO84 to prevent constitutive expression of the PHO genes characteristic of PHO84 deletion strains (24). PHM deletion strains were made as described (17). Additional strains listed in Table 1 were derived from the above parent strains by standard methods of mating, sporulation, and tetrad dissection (19). All strains were screened by using selection medium and confirmed by phenotypic testing.
Media and Growth Conditions. For all experiments, yeast were grown at 30°C in complete synthetic medium with dextrose (SD), which is high-phosphate medium containing ≈7.3 mM KH2PO4, to mid-log phase (OD600 0.4–0.7) (25). Before use, the pH of all media was titrated from ≈4.8 to 4.0 by addition of 1 M HCl. For phosphate starvation experiments, phosphate-free SD medium (no Pi) was prepared by substituting KCl for KH2PO4 (26). Media containing various phosphate concentrations were prepared from phosphate-free SD by adding 1 M KH2PO4 to give final concentrations ranging from 10 μM to 10 mM phosphate. For phosphate starvation, mid-log phase cells grown in SD were pelleted, washed, and resuspended in SD medium lacking phosphate (no Pi). Aliquots of cells were harvested for analysis at various time points from cultures grown at 30°C for up to 3 h after inoculation into medium lacking phosphate. Similarly, phosphate threshold experiments were conducted by washing and resuspending mid-log phase cultures in SD medium containing various concentrations of phosphate. Additional experiments required following gene expression over the course of 48 h. During this time course, cultures were periodically diluted to maintain appropriate cell densities. To avoid depletion of phosphate during the phosphate threshold experiments, cultures were diluted to a cell density of 4 × 105 cells per ml for growth in low phosphate conditions (<250 μM phosphate). Results obtained for experiments conducted at low cell density did not differ from those conducted with cells at mid-log phase densities (0.5–1.0 × 107 cells per ml). For transient phosphate deprivation experiments, cells grown to mid-log phase in SD medium were washed and resuspended in no Pi medium. Cells were grown at 30°C for varying lengths of phosphate starvation (10 min, 30 min, 1 h, 2 h). At designated times, aliquots of 1 M KH2PO4 were added to the culture medium (10 mM final concentration), allowing cells to continue growth in phosphate containing media until 2.5 h after the initial removal of phosphate.
Quantitation of Extracellular Phosphate. The concentration of phosphate in the medium was determined by using an acidified ammonium molybdate/malachite green G solution (Sigma–Aldrich) (27, 28). The phosphate concentration was depleted by no more than 10% throughout experiments conducted with low-phosphate medium (10–250 μM phosphate).
Northern Blot Analysis. RNA was extracted by standard phenolchloroform techniques from cells harvested at various times after inoculation into medium lacking phosphate (29). Probes to PHO5, PHO84, and ACT1 (the gene encoding actin) were made by random priming from a PCR product of each full-length gene (11). PHO5 and PHO84 signals were normalized to the signal from ACT1 to control for loading differences. Data are shown normalized to the maximum intensity observed for PHO5 and PHO84 after phosphate starvation for 2 h.
Fluorescence Microscopy. Images were captured with a Zeiss Axiovert 200M inverted microscope by using metamorph software (Version 6.2, Universal Imaging, Downington, PA) and a Photometrics Cascade 512F charge-coupled device camera. For phosphate starvation, separate aliquots of cells were collected and quickly visualized at various times after inoculation into SD medium lacking phosphate. Images were analyzed by using metamorph software to quantify the percentage of cells containing Pho4 localized to the nucleus. Using false color images representing pixel intensity, we established nuclear localization of Pho4 by comparing average pixel densities of nuclear and cytoplasmic signals relative to background. Cells containing nuclear signals of greater than twice that of the cytoplasmic signals were scored as containing nuclear-localized Pho4. Data were tabulated according to the percentage of cells containing a significant nuclear Pho4-GFP signal. For reference, cells exhibiting nuclear Pho4-GFP signals were compared with cells grown in high phosphate medium for which Pho4-GFP localization is cytoplasmic.
Flow Cytometry. Cells expressing pho5::GFP or PHO84pr:GFP were grown for 3 h at 30°C in SD medium containing the indicated concentration of Pi. Cells were collected, chilled on ice and sonicated for 2 s before FACS analysis using a BD FACS-Calibur system (BD Biosciences, San Jose, CA). Alternatively, expression of YFP under the control of the native PHO5 promoter was monitored periodically over the course of 48 h by using a BD LSRII system (BD Biosciences). For transient phosphate starvation experiments, cells expressing YFP under the control of either the PHO5 or the PHO84 promoter were grown as described above, collected, chilled on ice, and sonicated for 2 s before analysis with a BD LSRII system. All flow cytometry was conducted by using excitation at 488 nm and emission filters centered around 530 nm (515–545 nm).
Measurement of PolyP by 31P-NMR Spectroscopy. For in vivo NMR spectroscopy, cells were grown according to the phosphate starvation procedure described above. Cells were harvested by centrifugation at 10°C (400 ml of OD600 = 0.5). Cells were resuspended in medium containing 20% D2O (750 μl final volume) and transferred directly to a 5-mm NMR tube for immediate analysis. 31P-NMR spectra were obtained at 161.76 MHz by using a Varian Inova 400 spectrometer. Spectra were acquired with 90° pulses at a repetition rate of 2.5 s and 256 or 512 acquisitions. Chemical shifts were referenced to 50 mM methylene diphosphonate (δ = 17.98 ppm) as an external standard (1-mm capillary) calibrated against 85% phosphoric acid (δ = 0.0 ppm). Identical external reference samples were used for determining integrals for all spectra, including time-dependent phosphate starvation experiments. Peaks were assigned by reference to published chemical shifts (30, 31) and verified by addition of reference chemicals to samples. Integral values reported for polyP are normalized to the cell mass of each sample. An additional method for 31P-NMR of cellular extracts is described in Supporting Text, which is published as supporting information on the PNAS web site.
Results
PHO5 and PHO84 Exhibit Different Kinetics of Transcriptional Induction in Response to Phosphate Starvation. Previous studies have demonstrated that PHO5 and PHO84 exhibit different thresholds for induction in response to phosphate limitation (11). Specifically, PHO84 is significantly induced when cells are grown in intermediate phosphate conditions for 1–6 h, whereas PHO5 is not. Although both genes are regulated by the same transcription factor, Pho4, partially phosphorylated Pho4 can efficiently activate transcription of PHO84, but not PHO5 (11). The results of that study suggest that partial inhibition of Pho80/Pho85 in intermediate phosphate conditions leads to accumulation of partially phosphorylated Pho4 and activation of PHO84, but activation of PHO5 requires further kinase inhibition.
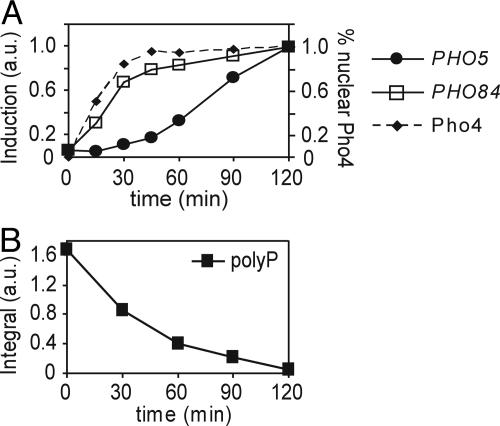
To gain a better understanding of how the PHO pathway responds to changing external phosphate levels, we examined the kinetics of PHO5 and PHO84 induction and Pho4 localization in response to phosphate starvation. We monitored induction of PHO5 and PHO84 mRNA levels by Northern blotting and Pho4 localization by fluorescence microscopic analysis of a strain expressing a functional Pho4-GFP fusion protein. Pho4 becomes concentrated in the nucleus within 15–30 min after transfer of cells to medium lacking phosphate (Fig. 1A), reflecting changes in its phosphorylation state due to modulation of the PHO pathway. Although PHO84 is induced on a timescale similar to that observed for Pho4 nuclear accumulation, we observe a significant delay (≈30 min) in induction of PHO5. As phosphate limitation continues, PHO5 is eventually induced.
Fig. 1.
Kinetics of induction of the phosphate-responsive signaling pathway and polyP utilization in WT cells. (A) Induction of PHO5 mRNA (solid circles), PHO84 mRNA (squares) monitored by Northern blotting, and percent of cells with nuclear Pho4-GFP (dashed line) at time points after inoculation into medium lacking phosphate. Expression data are normalized to maximal induction at t = 2 h. (B) Quantitation for in vivo 31P-NMR spectra of polyP in WT cells after inoculation into medium lacking phosphate. Integral values are measured relative to an external standard of 50 mM methylene diphosphonic acid.
We wished to understand how PHO5 repression was maintained immediately after phosphate deprivation. We presume that PHO5 repression arises from partially phosphorylated Pho4 that is sustained as long as internal phosphate levels remain above a certain threshold. It has been suggested that internal phosphate concentrations are temporarily maintained during phosphate starvation through mobilization of internal stores of phosphate from the vacuole (1, 2, 18). If intracellular phosphate stores act as a reserve that can be mobilized during phosphate limitation to help prevent rapid PHO5 induction, we expect that: (i) intracellular stores will be depleted on a timescale similar to that of PHO5 induction, and (ii) cells lacking intracellular stores will induce PHO5 on a timescale comparable to that observed for PHO84.
We measured the kinetics of changes in intracellular phosphate stores using 31P-NMR spectroscopy, taking advantage of the relatively simple spectra and distinct chemical shift values characteristic of phosphorus-containing biomolecules (14, 16, 31, 32). In WT cells, the dominant spectral feature, polyP, accumulates during growth in high-phosphate medium (Fig. 6A, which is published as supporting information on the PNAS web site) (1, 18). During phosphate starvation, polyP levels decrease on a timescale that correlates with PHO5 induction (Fig. 1B). Phosphate is liberated from polyP, presumably enabling cells to transiently mitigate decreasing internal phosphate levels (1), as well as sustain partial phosphorylation of Pho4 and PHO5 repression during the onset of phosphate limitation.
Disrupting PolyP Accumulation and Mobilization Eliminates Differences in the Kinetics of Induction of PHO5 and PHO84. To investigate whether loss of intracellular phosphate stores affects the kinetics of PHO5 induction, we monitored the PHO pathway response in strains unable to generate or mobilize polyP. Disruption of genes linked to vacuolar storage of polyP, PHM1-PHM4 (VTC1-VTC4), results in the inability of cells to accumulate polyP in yeast vacuoles (17, 33, 34). Additionally, disruption of PHM5 (PPN1) results in accumulation of long chains of polyP in the vacuole, suggesting that this gene encodes an activity required for mobilization of phosphate from polyP (17, 35). Subsequent studies have demonstrated that the gene product of PHM5 encodes an endopolyphosphatase (Ppn1) that cleaves long polyP chains into shorter chains and tripolyphosphate molecules (polyP3) (36).
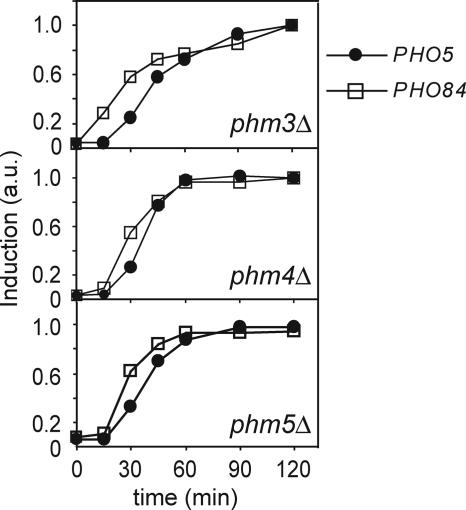
In contrast to the kinetic differences observed in WT cells, we observed similar kinetics of induction for PHO5 and PHO84 in the PHM deletion strains (Fig. 2). In cells deficient in storage of vacuolar polyP (i.e., phm3Δ or phm4Δ strains, as confirmed by 31P NMR, Fig. 6A), PHO5 is induced with similar kinetics as observed for PHO84. A phm5Δ strain, which exhibits impaired catabolism of polyP due to a lack of the endopolyphosphatase Phm5 (Ppn1) (31P NMR, Fig. 6), also induces PHO5 more rapidly than WT cells (Fig. 2). In the phm5Δ strain, we presume that long polyP chains are not cleaved into shorter chains, thereby decreasing the overall rate of phosphate liberation that occurs only at the polyP chain termini (36–38). The resulting decreased rate of polyP catabolism relative to WT renders phm5Δ cells temporarily unable to maintain internal phosphate concentrations high enough to repress PHO5 transcription upon phosphate deprivation.
Fig. 2.
Kinetics of induction of phosphate-responsive genes in phm3Δ, phm4Δ, and phm5Δ strains. Induction of PHO5 (solid circles) and PHO84 (squares) mRNA monitored by Northern blotting in phm3Δ (Top), phm4Δ (Middle) and phm5Δ (Bottom) cells at time points after inoculation into medium lacking phosphate. Data are normalized to maximal induction at t = 2 h.
These results are consistent with the model that polyP serves as a buffer that can be mobilized during phosphate limitation to temporarily maintain internal phosphate levels. For a period after phosphate starvation, the efficient metabolism of polyP sustains internal phosphate levels such that partially phosphorylated Pho4 accumulates and activates transcription of PHO84, but not PHO5.
PolyP Does Not Alter the Phosphate Threshold for PHO5 Induction. Our working model is that the intracellular level of phosphate is maintained within an appropriate range by balancing cellular usage of Pi with uptake from the environment and mobilization from internal stores. Cells mobilize internal stores when usage exceeds uptake from the environment, and such mobilization will temporarily maintain intracellular phosphate levels. However, eventually the contents of the buffer will be depleted, the internal phosphate level will decrease further, and PHO5 will be fully induced. In addition to the observed kinetic delay in PHO5 expression, we wished to determine whether polyP affects the threshold of external phosphate at which induction of phosphate-responsive genes occurs.
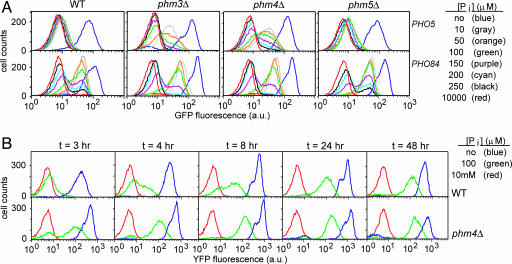
To more thoroughly characterize this threshold, we constructed WT and mutant strains in which the gene encoding GFP was placed under the control of the PHO5 or PHO84 promoter. We transferred these cells from high-phosphate medium to different phosphate concentrations, grew them for 3 h, and characterized GFP expression in single cells using flow cytometry (Fig. 3A). As expected, the GFP reporter strains exhibit maximal PHO5 and PHO84 expression in medium lacking phosphate, whereas basal levels of expression observed in high-phosphate medium were similar to autofluorescence levels observed in control cells lacking GFP reporter constructs. As observed in previous population studies, PHO84 is induced at a higher concentration of external phosphate than is PHO5; in WT cells, a significant fraction of the population expressed PHO84 when grown in phosphate concentrations ranging from 10 to 200 μM, whereas PHO5 is only minimally induced in this concentration range.
Fig. 3.
Effect of polyP on the threshold for PHO pathway induction. (A) FACS analysis of pho5::GFP and PHO84pr:GFP expression for WT (Left), phm3Δ (Center Left), phm4Δ (Center Right), and phm5Δ (Right) cells at 3 h after inoculation into medium containing various phosphate concentrations: no phosphate (blue), 10 μM (gray), 50 μM (orange), 100 μM (green), 150 μM (purple), 200 μM (cyan), 250 μM (black), and 10 mM (red). (B) FACS analysis of PHO5 expression during extended phosphate limitation. Shown is pho5::YFP expression for WT (Upper) and phm4Δ (Lower) cells maintained for 48 h in medium containing a range of phosphate concentrations: 10 mM (red), 100 μM (green), and no phosphate (blue). Samples of the cultures were collected and analyzed at various times (as indicated) throughout the experiment.
The phm5Δ strain exhibits PHO5 and PHO84 induction profiles similar to WT cells. We speculated that the phm5Δ strain is able to liberate phosphate from polyP, but at a slower rate than the WT strain. In the phm5Δ strain, intracellular phosphate obtained from mobilization of some polyP in addition to uptake from the environment is apparently sufficient to repress PHO5 transcription in intermediate phosphate conditions (Fig. 3A). However, in medium lacking phosphate, the slower rate of polyP metabolism in phm5Δ cells does not offset usage, resulting in rapid induction of PHO5 (Fig. 2). In contrast, phm3Δ and phm4Δ strains lacking vacuolar polyP stores had striking alterations in the response to various external phosphate concentrations; we observed significant expression of PHO5 in phm3Δ and phm4Δ cells grown in intermediate phosphate concentrations (50–200 μM). However, PHO84 induction is not altered by the absence of vacuolar polyP stores. The effect of phm3Δ and phm4Δ on the response to varying external phosphate suggests that upon elimination of polyP stores, PHO5 may be partially induced even in intermediate phosphate concentrations. These observations led us to examine more carefully the threshold for induction of PHO5 and PHO84 in WT and mutant cells, particularly once cells reach steady state.
To test whether polyP alters the threshold for PHO5 induction in cells under steady-state conditions, we monitored PHO5 expression for WT and phm4Δ cells grown in medium containing no, 100 μM, or 10 mM phosphate over the course of 48 h. We hypothesized that if polyP were acting as a phosphate reserve capable of delaying PHO5 induction in WT cells, then perhaps PHO5 would be induced upon depletion of polyP stores, even in intermediate phosphate conditions. In cells lacking polyP, PHO5 expression increases and stabilizes within 4–8 h of growth in 100 μM phosphate medium (Fig. 3B). Although PHO5 induction in cells containing polyP initially lags behind that of cells without polyP, prolonged growth in intermediate phosphate leads to PHO5 expression at levels comparable to those of the phm4Δ strain (Fig. 3B). The data indicate that both WT and phm4Δ strains exhibit a stable, intermediate level of PHO5 expression in cells grown in intermediate levels of extracellular phosphate. These observations underscore the idea that the experiments conducted at 3 h after phosphate limitation do not represent steady-state conditions but rather reflect differences in the kinetics of transcriptional induction of PHO5 and PHO84.
Induction of PHO84, but Not PHO5, Is Sensitive to Transient Starvation. PolyP acts as a buffer whose mobilization during periods of phosphate limitation can temporarily keep PHO5 repressed. The polyP buffer might also have a role in filtering transient fluctuations in external phosphate levels, preventing PHO5 from being inappropriately induced in response to transient phosphate deprivation. It may be advantageous for cells to induce PHO84 in response to even minor phosphate limitation, because it encodes a transporter that allows cells to bring in more phosphate from the environment. By contrast, PHO5 encodes a secreted phosphatase whose production demands considerable cellular resources and whose expression is restricted to conditions of extreme phosphate deprivation (39, 40).
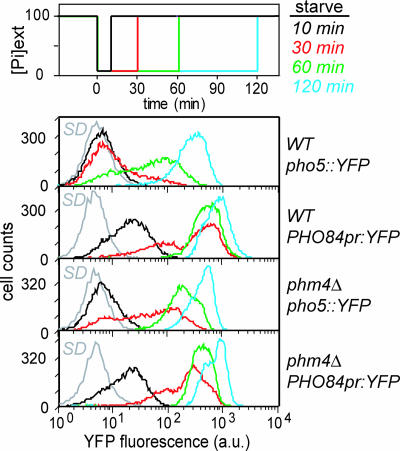
To test this idea, we subjected cells to phosphate deprivation for different lengths of time and monitored PHO5 and PHO84 induction by analysis of fluorescent reporter strains by flow cytometry. Cells grown in high phosphate medium to mid-log phase were inoculated into medium lacking phosphate for varying lengths of time (10, 30, 60, or 120 min) before readdition of phosphate to high phosphate levels. We observed that cells induce PHO84 during phosphate limitation as short as 10 min, whereas PHO5 induced only in response to significantly longer periods of phosphate starvation in WT cells (Fig. 4). In contrast, in the absence of polyP, both PHO84 and PHO5 are induced in response to phosphate deprivation as short as 30 min (Fig. 4). We conclude that polyP is rapidly mobilized in response to phosphate limitation to temporarily maintain intracellular phosphate concentrations. The presence of a phosphate buffer allows the cell to filter out transient fluctuations in external phosphate concentrations so that PHO5 is induced only in response to prolonged periods of starvation.
Fig. 4.
PHO pathway induction in response to transient phosphate starvation in WT and phm4Δ cells. Cells were subjected to varying durations of phosphate deprivation, before readdition of phosphate, as illustrated (Top). FACS profiles depict induction of pho5::YFP and PHO84pr:YFP for WT (Middle) and phm4Δ (Bottom) strains in response to: 10 (black), 30 (red), 60 (green), and 120 (cyan) min of phosphate deprivation. FACS profiles of cells grown in high-phosphate (SD) medium are depicted in gray.
Discussion
We have demonstrated that phosphate-responsive genes are induced with different kinetics when cells are deprived of phosphate. In WT cells, PHO5 induction is delayed relative to PHO84, but in cells lacking polyP, PHO5 is induced with kinetics similar to those of PHO84. We have shown that polyP consumption occurs on a time scale similar to that of PHO5 induction during initial phosphate starvation, consistent with the model that polyP is mobilized during phosphate limitation to generate Pi, which temporarily keeps cytoplasmic phosphate levels high enough to allow induction of PHO84 but repression of PHO5 transcription. As expected for a buffer or reserve, although polyP affects the kinetics of PHO5 induction, it does not alter the threshold of intracellular phosphate that triggers PHO5 expression. Finally, we demonstrate that transient phosphate deprivation activates transcription of PHO84, but not PHO5, in WT cells, whereas cells lacking polyP induce both PHO5 and PHO84 in response to transient starvation. Hence, we deduce that polyP functions as an internal nutrient buffer that is mobilized during periods of transient external phosphate deprivation, as well as during the initial phase of phosphate starvation, while an appropriate response ensues.
Our results agree with recent work showing that the phm3Δ strain induces PHO5 faster than WT cells (2). In that work, the need for a phosphate reservoir was related to nutrient demand for nucleic acid synthesis preceding mitosis. Additionally, 31P-NMR spectra of WT cells have suggested that a decrease in polyP levels coincides with a stabilizing of cytoplasmic phosphate concentration during phosphate starvation (1). Other work has shown that polyP levels are inversely correlated with phosphatase activity induced in response to external phosphate limitation (18). We focused on whether polyP can buffer internal phosphate concentrations, thereby affecting transcriptional regulation during the initial response to phosphate deprivation and filtering fluctuations in external phosphate availability. Implicit in this hypothesis is the observation that intracellular phosphate is the target of the unidentified upstream sensor for the PHO pathway (18).
We previously observed that PHO84 was significantly induced when cells were grown in intermediate phosphate conditions, but that PHO5 was not, suggesting that there are differences in the threshold of external phosphate that triggers induction of these genes (11). This study reveals that, although there are significant differences in the kinetics of induction, both PHO84 and PHO5 are eventually induced after culturing cells for prolonged periods of time in intermediate phosphate conditions.
We interpret our observations in the context of the following model (Fig. 5). When cells are transferred from high-phosphate to phosphate-starvation conditions, the internal concentration of phosphate decreases, triggering mobilization of polyP, which will mitigate decreasing internal phosphate levels. Intracellular phosphate levels are temporarily sustained, resulting in partially active Pho80/Pho85 kinase and accumulation of partially phosphorylated Pho4 that can activate transcription of PHO84, but not PHO5. The observed timing and magnitude of expression of these genes is governed by the balance between uptake of external phosphate, the size of the polyP buffer, and the rate of its mobilization. Even in the absence of external phosphate, increasing the size of the polyP reserve further extends the duration of kinetic delay of PHO5 induction by providing additional intracellular phosphate (data not shown). However, once polyP is depleted, internal phosphate levels decrease, permitting accumulation of nuclear Pho4 sufficient to activate PHO5 transcription.
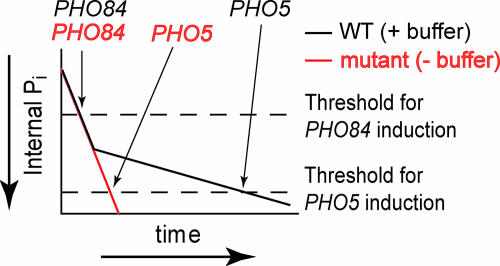
Fig. 5.
Model of gene induction in response to phosphate starvation. Schematic diagram of decreasing internal phosphate levels (in arbitrary units) for cells with (WT, black line) and without polyP (mutant, red line) after phosphate starvation (time, in arbitrary units). Once intracellular phosphate decreases below the threshold for gene induction (dashed lines), induction occurs. Arrows indicate relative timing for the onset of PHO84 and PHO5 expression in each strain. In cells containing polyP, buffer utilization sustains phosphate levels longer, delaying PHO5 induction.
Buffering or sequestering of essential nutrients is an important feature that may enhance cell fitness in the presence of environmental stress. Our work demonstrates that fluctuating environmental conditions are filtered by the mobilization of a buffer capable of overriding the effect of such changes on a signaling pathway. In effect, a buffer can dampen spikes or oscillations in external nutrient availability, minimizing unnecessary synthesis of genes that encode energetically expensive molecules, such as secreted enzymes that cannot be recovered or recycled. Additionally, buffering the response to fluctuating environmental sources of nutrients such as transition metals that are toxic in excess prevents cytotoxicity during transient spikes in extracellular concentrations. Cells manage nutrient demand by balancing uptake and mobilization of metal ion stores, while preventing toxicity due to overaccumulation of metals by sequestration in the form of vacuolar stores (41, 42). As demonstrated in this work, cells use buffering systems as a source of nutrients during deprivation as well as a means to integrate intracellular nutrient concentrations into signal transduction pathways. Nutrient buffers filter fluctuations in the extracellular environment, providing cells with fine-tuning and robust control to optimize their response to changing environmental conditions.
Supplementary Material
Acknowledgments
We thank members of the O'Shea laboratory for discussion and critical commentary. This work was supported by the National Institutes of Health (GM51377), the Howard Hughes Medical Institute, and the David and Lucile Packard Foundation (to E.K.O.). M.R.T. is supported by Postdoctoral Fellowship PF-03-083-01-TBE from the American Cancer Society.
Author contributions: M.R.T. and E.K.O. designed research; M.R.T. performed research; M.R.T. contributed new reagents/analytic tools; M.R.T. and E.K.O. analyzed data; and M.R.T. and E.K.O. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Pi, inorganic phosphate; polyP, polyphosphate; SD, synthetic medium with dextrose; YFP, yellow fluorescent protein; PHO, phosphate responsive signaling.
References
- 1.Shirahama, K., Yazaki, Y., Sakano, K., Wada, Y. & Ohsumi, Y. (1996) Plant Cell Physiol. 37, 1090-1093. [DOI] [PubMed] [Google Scholar]
- 2.Neef, D. W. & Kladde, M. P. (2003) Mol. Cell. Biol. 23, 3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bun-ya, M., Nishimura, M., Harashima, S. & Oshima, Y. (1991) Mol. Cell. Biol. 11, 3229-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshida, K., Ogawa, N. & Oshima, Y. (1989) Mol. Gen. Genet. 217, 40-46. [DOI] [PubMed] [Google Scholar]
- 5.Lenburg, M. E. & O'Shea, E. K. (1996) Trends Biochem. Sci. 10, 383-387. [PubMed] [Google Scholar]
- 6.Oshima, Y. (1997) Genes Genet. Syst. 72, 323-334. [DOI] [PubMed] [Google Scholar]
- 7.Lau, W.-T. W., Schneider, K. R. & O'Shea, E. K. (1998) Genetics 150, 1349-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaffman, A., Herskowitz, I., Tjian, R. & O'Shea, E. K. (1994) Science 263, 1153-1156. [DOI] [PubMed] [Google Scholar]
- 9.O'Neill, E. M., Kaffman, A., Jolly, E. R. & O'Shea, E. K. (1996) Science 271, 209-212. [DOI] [PubMed] [Google Scholar]
- 10.Komeili, A. & O'Shea, E. K. (1999) Science 284, 977-980. [DOI] [PubMed] [Google Scholar]
- 11.Springer, M., Wykoff, D. D., Miller, N. & O'Shea, E. K. (2003) PLOS Biol. 1, 261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffery, D. A., Springer, M., King, D. S. & O'Shea, E. K. (2001) J. Mol. Biol. 306, 997-1010. [DOI] [PubMed] [Google Scholar]
- 13.Urech, K., Duerr, M., Boller, T., Wiemken, A. & Schwencke, J. (1978) Arch. Microbiol. 116, 275-278. [DOI] [PubMed] [Google Scholar]
- 14.Castro, C. D., Koretsky, A. P. & Domach, M. M. (1999) Biotechnol. Prog. 15, 65-73. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg, A., Rao, N. N. & Ault-Riche, D. (1999) Annu. Rev. Biochem. 68, 89-125. [DOI] [PubMed] [Google Scholar]
- 16.Vagabov, V. M., Trilisenko, L. V. & Kulaev, I. S. (2000) Biochemistry (Moscow) 65, 349-354. [PubMed] [Google Scholar]
- 17.Ogawa, N., DeRisi, J. L. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auesukaree, C., Homma, T., Tochio, H., Shirakawa, M., Kaneko, Y. & Harashima, S. (2004) J. Biol. Chem. 279, 17289-17294. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie, C. & Fink, G. R., eds. (2002) Methods in Enzymology (Academic, Boston), Vol. 350–351.
- 20.Longtine, M. S., McKenzie, A., Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P. & Pringle, J. R. (1998) Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- 21.Griesbeck, O., Baird, G. S., Campbell, R. E., Zacharias, D. A. & Tsien, R. Y. (2001) J. Biol. Chem. 276, 29188-29194. [DOI] [PubMed] [Google Scholar]
- 22.Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K. & Miyawaki, A. (2002) Nat. Biotechnol. 20, 87-90. [DOI] [PubMed] [Google Scholar]
- 23.Raser, J. M. & O'Shea, E. K. (2004) Science 304, 1811-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wykoff, D. D. & O'Shea, E. K. (2001) Genetics 159, 1491-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman, F. (1991) Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- 26.Huang, S., Jeffery, D. A., Anthony, M. D. & O'Shea, E. K. (2001) Mol. Cell. Biol. 21, 6695-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter, S. G. & Karl, D. W. (1982) J. Biochem. Biophys. Methods 7, 7-13. [DOI] [PubMed] [Google Scholar]
- 28.Van Veldhoven, P. P. & Mannaerts, G. P. (1987) Anal. Biochem. 161, 45-48. [DOI] [PubMed] [Google Scholar]
- 29.Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R. & O'Shea, E. K. (2003) Science 299, 114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.den Hollander, J. A., Ugurbil, K. & Shulman, R. G. (1986) Biochemistry 25, 212-219. [DOI] [PubMed] [Google Scholar]
- 31.Teleman, A., Richard, P., Toivari, M. & Penttila, M. (1999) Anal. Biochem. 272, 71-79. [DOI] [PubMed] [Google Scholar]
- 32.Gillies, R. J., Ugurbil, K., den Hollander, J. A. & Shulman, R. G. (1981) Proc. Natl. Acad. Sci. USA 78, 2125-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen, A., Perzov, N., Nelson, H. & Nelson, N. (1999) J. Biol. Chem. 274, 26885-26893. [DOI] [PubMed] [Google Scholar]
- 34.Müller, O., Bayer, M. J., Peters, C., Andersen, J. S., Mann, M. & Mayer, A. (2002) EMBO J. 21, 259-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumble, K. D. & Kornberg, A. (1996) J. Biol. Chem. 271, 27146-27151. [DOI] [PubMed] [Google Scholar]
- 36.Sethuraman, A., Rao, N. N. & Kornberg, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulakovskaya, T. V., Andreeva, N. A., Karpov, A. V., Sidorov, I. A. & Kulaev, I. S. (1999) Biochemistry (Moscow) 64, 990-993. [PubMed] [Google Scholar]
- 38.Lichko, L., Kulakovskaya, T. & Kulaev, I. (2002) Biochim. Biophys. Acta 1599, 102-105. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, G., Bartsch, G., Laumont, M. C., Herman, T. & Liss, M. (1963) Biochemistry 2, 126-131. [DOI] [PubMed] [Google Scholar]
- 40.Bostian, K. A., Lemire, J. M. & Halvorson, H. O. (1983) Mol. Cell. Biol. 3, 839-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eide, D. J. (2003) J. Nutr. 133, 1532S-1535S. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford, J. C. & Bird, A. J. (2004) Eukaryot. Cell 3, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.