Abstract
Restriction endonucleases and other nucleic acid cleaving enzymes form a large and extremely diverse superfamily that display little sequence similarity despite retaining a common core fold responsible for cleavage. The lack of significant sequence similarity between protein families makes homology inference a challenging task and hinders new family identification with traditional sequence-based approaches. Using the consensus fold recognition method Meta-BASIC that combines sequence profiles with predicted protein secondary structure, we identify nine new restriction endonuclease-like fold families among previously uncharacterized proteins and predict these proteins to cleave nucleic acid substrates. Application of transitive searches combined with gene neighborhood analysis allow us to confidently link these unknown families to a number of known restriction endonuclease-like structures and thus assign folds to the uncharacterized proteins. Finally, our method identifies a novel restriction endonuclease-like domain in the C-terminus of RecC that is not detected with structure-based searches of the existing PDB database.
INTRODUCTION
Restriction endonucleases and their nuclease relatives function to cleave a variety of nucleic acid substrates in various cellular processes. The SCOP (1) database currently assigns 20 different families to the restriction endonuclease-like superfamily, including 13 restriction endonucleases (2) and various other nucleic acid cleaving enzymes such as lambda exonuclease (3,4), DNA mismatch repair protein (MutH) (5), very short patch repair (Vsr) endonuclease (6), N-terminal domain of TnsA endonuclease (7), endonuclease I (8), archaeal Holliday junction resolvase (Hjc) (9) and XPF/Rad1/Mus81 nuclease (10). The cleavage reactions performed by these highly diverse proteins contribute to important biological functions, such as protecting host organisms against foreign DNA invasion (restriction endonucleases), repairing damaged DNA (MutH and Vsr), resolving Holliday junctions (endonuclease I, Hjc and XPF/Rad1/Mus81-dependent nuclease) and performing additional cleavage events in DNA recombination (lambda exonuclease and TnsA).
The restriction endonuclease-like superfamily is defined by a common core fold that includes a four-stranded, mixed β-sheet flanked on either side by an α-helix (αβββαβ topology, Figure 1). Residues within a relatively conserved PD-(D/E)XK motif (Motifs II and III, Figure 2) mark the active site and contribute to cleaving the nucleic acid phosphodiester bond (4,11,12). In addition to this named motif, a conserved acidic residue often resides at the N-terminus of the first core α-helix (Motif I, Figure 2), while a conserved residue from the second helix points toward the active site (Motif IV, Figure 2) in a subset of families. These residues play various catalytic roles, which include coordination of up to three divalent metal ion cofactors, depending on the family. The shared structural and functional features of restriction endonuclease-like families have been interpreted as evidence for a common evolutionary origin and have been exploited by various groups to identify and group endonuclease sequences (2,12–14).
Figure 1.
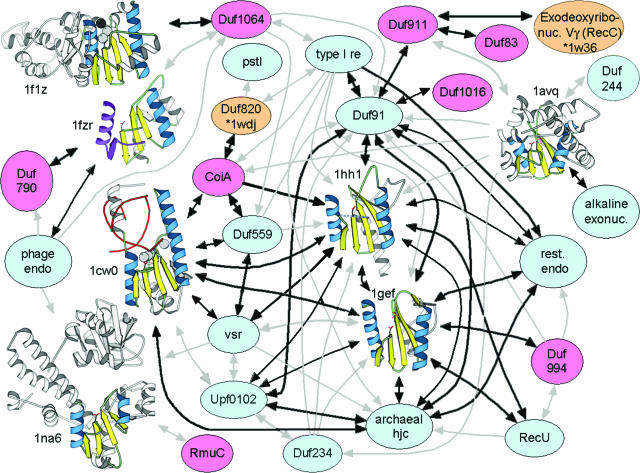
Structural connectivity network. Arrows indicate Meta-BASIC connections between PfamA families (colored circles) and PDB90 families (structural models). Arrows stem from the input query and are colored black or gray according to the Meta-BASIC score (above or below the confidence threshold of 12, respectively). PfamA families colored pink represent newly predicted restriction endonuclease-like domains, while those colored blue represent previously known predictions. Two PfamA families with structures released to the PDB after Meta-BASIC analysis are colored orange. Structural elements of the PDB90 models are colored blue (core α-helices), yellow (core β-strands), white (insertions) and purple (domain swaps). Invariant aspartic acid, phosphate ligand and DNA substrate (where present) are shown in stick representation, while bound ions are shown in CPK.
Figure 2.
Multiple family sequence alignment. Representative sequences (labeled according to gi number) from each newly identified restriction endonuclease-like fold family (labeled in bold above representative sequence gi numbers) are aligned with sequences corresponding to PDB90 entries (labeled in bold according to PDB ID) within the conserved structural core. Sequence gi numbers are colored according to taxonomy: with bacterial in black, viral in green and archaeal in red. Secondary structure predictions are indicated above each PfamA family and observed secondary structure elements of 1hh1_A are indicated above the PDB90 sequences. First residue numbers are indicated before each sequence, while omitted residues are enclosed in parenthesis. Residues are highlighted according to property conservations: hydrophobic (yellow), small (gray), and polar/charged in typical restriction endonuclease-like active site positions (black) and alternative active site positions (blue). Restriction endonuclease-like motifs are labeled above the corresponding residue columns. Italicized sequence corresponds to domain-swapped region of 1fzr.
In addition to sequence- and structure-based methods, analysis of genomic context and domain fusions have led to identifying new restriction endonuclease-like domains (15,16). Restriction endonuclease-like proteins frequently cooperate with their genome neighbors to perform specific biological functions. For example, restriction-modification systems include a restriction endonuclease and a methyltransferase that function together to protect cells against foreign DNA. The two genes encoding these enzymes often reside adjacent to each other in genomes or can be found fused in a single gene. Similarly, domain fusions exist between restriction endonuclease-like proteins and superfamily I/II helicases, suggesting a close functional association of nuclease activity with ATP-dependent DNA helix unwinding. Other functional restriction endonuclease-like fold fusions include several different types of Zn-binding and DNA-binding domains. Analysis of these conserved gene neighborhoods/gene fusions has allowed the prediction of a novel prokaryotic DNA repair system (16) that includes previously identified members of the restriction endonuclease-like fold group [RecB nuclease domain (12)]. In addition to implying functional associations, similar conservations of domain fusions and genomic organizations help justify new restriction endonuclease-like fold predictions of increasingly divergent families.
To further expand the realm of the restriction endonuclease-like superfamily, we combine the concept of transitivity (‘If A is B and B is C, then A is C’) with a fold recognition approach Meta-BASIC (http://basic.bioinfo.pl) to identify nine new uncharacterized protein families as endonucleases. Meta-BASIC combines the use of sequence profiles and secondary structure predictions (meta profiles) for given protein families [currently PfamA (17) or PDB90 (18)] with various scoring systems and meta profile alignment algorithms to quickly establish links between families of both known and unknown structure. Using a query endonuclease of known structure (1gef), numerous hits to potential and known restriction endonuclease-like fold families arise, including a domain from an existing structure that was not identified by structure-based methods. Although many of the scores assigned to these hits fall below a confident threshold of 12 [predictions with Z-score above 12 have <5% probability of being incorrect (19)], we can further extend the limits of Meta-BASIC detection by applying the concept of transitivity to identified sequence groups. Results for the restriction endonuclease-like fold superfamily suggest that our sequence-based fold recognition approach can provide additional information about structural similarities in the realm of the ‘midnight zone (2)’ and can identify divergent folds in new and existing structures that structure-based identification methods may fail to find.
METHODS
Identification of novel restriction endonuclease-like families
Identification and linkage of restriction endonuclease-like families was carried out using GRDB system (http://basic.bioinfo.pl), which includes precalculated results of Meta-BASIC mappings between all PfamA families (represented as consensus sequences) and PDB entries (representatives at 90% of sequence identity) ranked according to a confidence score. Initially, an endonuclease of known structure (1gef) was used as a starting point to detect remotely homologous families and structures. We manually inspect all hits in ranked order, including those hits assigned with below threshold scores, for conservation of the fold-specific secondary structure pattern [predicted with PSIPRED (20)] and critical active site residues. We include hits with below threshold scores if (i) fold conservations and active site residues hold and (ii) no incorrect or unconfident hit exists with a higher confidence score. Interestingly, some structurally characterized proteins with restriction endonuclease-like folds (e.g. 1gef finds restriction enzyme MspI PDB ID 1sa3 at rank 179) show up in the ranked lists below incorrect or uncertain hits; however, we do not include these hits in our analysis. All confident hits were then used as queries in transitive Meta-BASIC searches until no new families were detected. To confirm correctness of difficult assignments and to generate reliable sequence-to-structure alignments, representative sequences for all newly identified restriction endonuclease-like fold families were submitted to the Meta Server (21) (http://bioinfo.pl/meta) that assembles various secondary structure prediction and top-of-the-line fold recognition methods. Predictions collected from diverse structure prediction services were screened with the consensus 3D-Jury system (21). These newly identified families were also subjected to transitive PSI-BLAST (22) searches performed against the NCBI non-redundant protein sequence database (filtered nr, posted September 3, 2004; 1 986 685 sequences) to detect other distantly related sequences and to neighborhood analysis by the STRING database (23) or visual inspection of the MBGD database (24) to detect possible functional associations.
Multiple sequence-structure alignment
Initially, optimal superposition of all identified structures generated with Swiss-PdbViewer program (25) was used to derive structure-based alignment of their sequences in the structurally conserved regions encompassing the core four β-strands and two α-helices. Multiple sequence alignments for new restriction endonuclease-like families were prepared using PCMA (26) followed by manual adjustments. Sequence-to-structure alignments between considered protein families and their distantly related templates within the conserved core elements were built using the consensus alignment approach and 3D assessment procedure (21) based on 3D-Jury and Meta-BASIC results. These alignments were subsequently merged to final multiple family alignment. In many cases, sequence-structure mapping was derived manually guided by the results of secondary structure predictions and the preservation of functionally critical residues as well as the hydrophobic core of the fold.
Comparison of identified restriction endonuclease-like structures
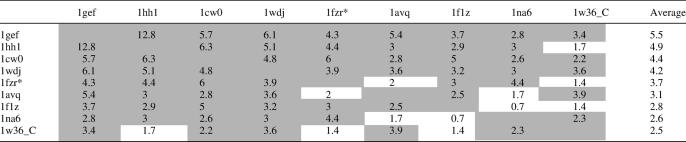
Comparison of restriction endonuclease-like structures identified in the transitive Meta-BASIC approach was carried out using DaliLite (27). Pairwise Dali Z-scores were generated for each chain within the PDB structure files. The domain-swapped structure 1fzr (asterisk in Table 1) was modified prior to DaliLite comparisons so that a single chain encompassed residues 17–44 from chain B and residues 47–145 from chain A to form a complete restriction endonuclease-like fold domain. The reported Z-scores for each structure pair correspond to the highest score computed between pairwise comparisons of all complete chains. The average Z-scores reported for each structure were calculated as the mean of pairwise Z-scores between that structure and each other identified structure.
Table 1.
Pairwise structure comparisons (Dali Z-scores)
Asterisk indicates a modified domain-swapped PDB (see Methods); DaliLite Z-scores above the threshold for structurally similar proteins are highlighted in gray.
RESULTS AND DISCUSSION
Detecting novel restriction endonuclease-like families and establishing a linkage network
Figure 1 illustrates the connectivity of Meta-BASIC hits between restriction endonuclease-like families contained within the PfamA and the PDB databases. Two PDB structures (1gef and 1hh1) and one PfamA family (DUF91) display the largest number of connections (represented by arrows) and form the center of the linkage network. Each of the structures functions to resolve Holliday junctions and is contained within the PfamA archaeal Hjc family (Figure 1, green highlights). Common to all of the highly connected families is a lack of inserted elements within the restriction endonuclease-like core (αβββαβ). Pairwise structure comparisons suggest that the Hjc proteins (1gef and 1hh1) display the highest average structural similarity (as measured by Dali Z-score) to all other identified proteins, while those structures with less connectivity (1f1z, 1na6 and 1w36_C) exhibit lower average Z-scores (Table 1). Less connected structures contain insertions of elements within the core (insertions bolded: 1f1z topology αβαββααβ, 1na6 topology αββαβαβ and 1w36_C topology αααβββαβ) and distortions of core element lengths and packing. Thus, the network built from Meta-BASIC-detected connections reflects the structural relatedness of identified restriction endonuclease-like families. Unfortunately, their high degree of sequence diversity does not allow us to extend the network of connectivity to evolutionary relatedness, although a tempting speculation follows that the Hjc structures would most closely resemble a common ancestor.
Strikingly, consensus sequences of all identified restriction endonuclease-like families extend across the entire structural core and include several identified motifs associated with cleavage (Figure 2). Consistent with motif conservations of existing restriction endonuclease-like families, an acidic residue (D/E from Motif II) is almost invariant in the newly identified families. Together with two other relatively conserved amino acids (H/E from Motif I and D/E/Q from Motif III), this residue coordinates the essential divalent metal ion cofactor(s) seen in several structures [T7 endonuclease I (28), TnsA (7), lambda exonuclease (3) and recB (29)]. Alternatively, the Vsr endonuclease (1cw0), missing one of the acidic residues from Motif III, binds metal with a family conserved residue substituted from a different position (uses His69 located in the loop preceding the second helix) (6). The relatively conserved basic residue of Motif III is located near the metal ion coordinating residues in position to stabilize the transition state or the product of the cleavage reaction. Consistent with the essential role of these residues in catalyzing the nuclease reaction, their mutations lead to a loss of activity (7,30–33).
Diversity of detected restriction endonuclease-like structures
The architecture of the restriction endonuclease-like fold allows different modes of substrate recognition. The recognized patterns vary from specific palindrome DNA sequences (type II restriction endonuclease and Vsr) to unique DNA backbone structures (Hjc). In many cases, specificity arises from various insertions to the core fold, including the addition of secondary structural elements [i.e. Vsr N-terminal helix inserts into the minor DNA groove (34)] or entire domains [i.e. EcoRII N-terminal DNA effector-binding domain (32)], while in other cases the quaternary structure of the fold plays a role in substrate recognition [i.e. lambda exonuclease I forms a toroidal structure thought to enclose the DNA substrate (3)].
Such a broad range of specificities recognized by various restriction endonuclease-like proteins have undoubtedly led to the observed sequence and structural diversities that hinder identification of new families. Nevertheless, application of Meta-BASIC successfully establishes an extensive network of connections between many PfamA families (even between two families of unknown structure) and restriction endonuclease-like structures. The detected structures display significant diversity, with some pairwise Dali Z-scores falling below the suggested cutoff for structurally similar proteins (Table 1, non-gray highlights). Importantly, the Meta-BASIC connections between diverse structures are only established transitively. Among the remaining connections, nine new endonuclease families that are mostly annotated in the PfamA database as domains of unknown function (DUFs) were detected. Many of these assignments are not possible with standard sequence similarity search tools, such as PSI-BLAST or RPS-BLAST, or even with other fold recognition methods that require a known reference protein structure. In the following section, we outline these newly identified restriction endonuclease-like fold families and use various sequence-based and context-based criteria to help justify our predictions.
RecC
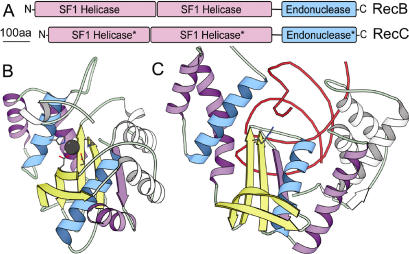
RecC functions together with RecB and RecD in a multifunctional enzyme complex to process DNA ends resulting from a double-strand break, leading to repair by homologous recombination. In this process, helicase and nuclease activities lead to ATP-dependent unwinding and digestion of DNA until encountering a specific site, where the complex pauses and subsequently initiates recombination through RecA. In the recently solved crystal structure of RecBCD, each subunit contains ATP-dependent helicase domains (29). The RecB exonuclease subunit comprises two N-terminal helicases (with domains 1A and 2A forming canonical helicase motors) and a C-terminal restriction endonuclease-like domain that includes identified active site motif residues (H in diverged Motif I, D in Motif II and DXK in motif III). Previous predictions have identified this C-terminal domain as having a restriction endonuclease-like active site (35) and have classified the RecB-like family in a superfamily with the Holliday junction resolvase and lambda exonuclease structures shown to be connected here (12). Our Meta-BASIC strategy does not identify the RecB exonuclease subunit of RecBCD due to its exclusion from PfamA. Alternatively, Meta-BASIC connects the C-terminal domain of the RecC subunit (Figure 1) to the other restriction endonuclease-like families through a link to DUF911 (see below). Interestingly, the RecC C-terminal domain displays such divergence from the other restriction endonuclease-like structures that it is not easily identified by structure comparison searches (DALI server finds closest existing nuclease structure at rank 48) and was therefore suggested to form a new fold in the crystal structure of the RecBCD complex (29).
Similar to RecB of the RecBCD complex, the RecC subunit contains two N-terminal domains that resemble helicases (lacking characteristic active site residues) (29). Both subunits possess long linker sequences leading to the C-terminal endonuclease-like domain (Figure 3A), which has also lost the characteristic active site residues in RecC (Figure 2). Structural similarities between the two restriction endonuclease-like domains (Dali Z-score = 9.9) extend beyond the nuclease core (Figure 3, yellow and blue). These similarities consist of an N-terminal helical insertion with a diverged N-terminal helix and a C-terminal topological extension (βα) to the core (Figure 3B, purple). In the RecBCD complex structure, the RecC C-terminal domain mediates binding to the DNA substrate and forms a channel for the 5′ tail to pass through to the RecD subunit. RecC binds DNA with residues contributed from the N-terminus of the first α-helix, the loops following the third and the fourth β-strands, and a C-terminal α-helical sub-domain unique to RecC. This mode of DNA binding generally resembles that of other restriction endonuclease structures. The structural and DNA binding similarity, along with a retained domain organization and preserved genome proximity, suggests that the RecB and RecC subunits of the RecBCD multienzyme complex arose as a gene duplication from a single ancestor with subsequent loss of catalytic residues in RecC.
Figure 3.
RecC degenerated nuclease domain. The RecB and RecC subunits of the multienzyme complex RecBCD probably arose from a duplication event. (A) Multi-domain organization of each subunit includes two helicase domains (colored pink) followed by a nuclease domain (blue). Domains are scaled according to indicated residue number. Asterisks designate a lack of conservation within active site motifs, which suggests a loss of catalytic function. Structural diagrams depict (B) the RecB nuclease domain and (C) the RecC nuclease domain. Secondary structural elements of the core are colored blue (α-helix) and yellow (β-strand), with similar elements extending beyond the core colored purple and unique insertions colored white. RecB active site residues (stick representation) corresponding to typical restriction endonuclease-like motifs (Figure 2) coordinate a calcium ion (black sphere). Alternatively, a less conserved residue (stick representation) from Motif III (Figure 2) mediates DNA (red coil) binding to the RecC subunit.
DUF911 and DUF83
DUF911 (COG4343) includes several sequences of unknown function found in one or two copies in select archaeal genomes. Although this family remains uncharacterized, both the genome organization and the connectivity of DUF911 sequences suggest a functional similarity to the RecB exonuclease family. Inspection of DUF911 neighborhoods (24) reveals a connection to DUF83 (COG1468), annotated in COG as a RecB family exonuclease. Both DUF911 and DUF83 sequences include relatively conserved restriction endonuclease-like active site motifs (Figure 2) that resemble those of RecB. PSI-BLAST connects the three families (DUF911 query gi|15920200 finds DUF83 sequence gi|23124021 in iteration 2 with E-value 8 × 10−4 and finds RecB sequence gi|19703859 in iteration 4 with E-value 8 × 10−5). The two uncharacterized families (DUF911 and DUF83) find each other as first Meta-BASIC hits and establish the only link to RecC (Figure 1). DUF911 and DUF83 possess similar additional elements at the N-terminus (α) and the C-terminus (βα) that probably pack onto the restriction endonuclease-like core to form a single domain, unlike the multi-domain helicase/nuclease composition of RecB and RecC. Inspection of genome neighborhoods reveals a predicted helicase (COG1203) in close proximity to both DUF911 and DUF83 sequences (confident DUF911/DUF83 neighborhood STRING score 0.627 and highly confident DUF83/helicase neighborhood STRING score 0.939). Together with sequence and structure similarities, this predicted helicase association suggests that DUF91 and DUF83 proteins function as nucleases in DNA repair similar to RecB.
DUF1016
Sequences from bacteria, archaea and viruses form DUF1016 (COG4804), which includes a predicted α/β N-terminal extension to the restriction endonuclease-like domain. This N-terminal extension does not display any significant sequence similarity to any known protein, although its secondary structure prediction is similar to that of the DUF790 N-terminus. DUF1016 sequences include typical active site motifs (Figure 2), and inspection of their genomic neighborhoods reveals several potential functional associations. DUF1016 sequences reside near type I/III restriction-modification system enzymes and ATP-dependent DNA helicases. For example, in the Nitrosomonas europa genome, a DUF1016 sequence (NE2308) belongs to a potential operon that includes a type III restriction enzyme (NE2306), an adenine-specific DNA methylase (NE2309), and a superfamily II DNA helicase (NE2311). Interestingly, biochemical evidence suggests a functional association between a type III restriction enzyme (EcoP15I) and an exonuclease activity, which allows EcoP15I to perform multiple enzymatic turnovers (36). Perhaps DUF1016 nuclease activity plays a similar role in enhancing bacterial restriction reactions.
DUF1064
DUF1064 (NOG09405) contains phage and bacterial proteins of unknown function. The predicted restriction endonuclease-like core includes all conserved Motif residues (Figure 2). Neighborhood analysis suggests functional associations with several other phage protein families (all with medium confidence STRING scores 0.399), including phage terminase large and small subunits involved in packaging viral DNA, a DNA repair protein that binds single-stranded DNA and promotes complementary DNA renaturation, and a phage-related protein predicted as another endonuclease. PSI-BLAST links DUF1064 sequences with other restriction endonuclease-like fold families. For example, one DUF1064 sequence (gi|29028685) finds a bacteriophage T7 endonuclease with known structure (1fzr with E-value 2 × 10−5, iteration 5) and a DUF790 sequence also identified in this study (gi|23126860 with E-value 0.003, iteration 4). This family most likely plays a role in phage genome segregation (37), or in repairing double-stranded breaks introduced during this process or during DNA replication.
DUF790
DUF790 (COG3372) includes hypothetical sequences from archaea and cyanobacteria (including one sequence from Actinobacteria). The identified restriction endonuclease-like core is located C-terminal to a region predicted as α/β, and includes a family conserved residue (Figure 2, K in Motif IV) that may substitute for the residue normally found in Motif III. Every member of this family resides next to a DEXH-box helicase (COG1061) in their respective genomes, suggesting a functional association (STRING score 0.656). DEXH-box helicases function in the ATP-dependent unwinding of nucleic acids required for a number of cellular processes that include some types of bacterial restriction-modification and nucleic acid excision repair. Together with a transitive PSI-BLAST connection to T7 endonuclease through DUF1064, this context-based association with a typical endonuclease functional partner supports our prediction for DUF790.
DUF994
DUF994 (NOG13964) includes several bacterial and phage proteins of unknown function. DUF994 sequences include a conservative substitution in Motif I (Q for E) that can presumably retain metal binding. In a genomic context, DUF994 associates with a family (NOG12860, STRING score 0.95) similar to ATP-dependent recombinase, which binds DNA and facilitates strand exchange during homologous recombination. Using a DUF994 sequence query (gi|28895436), PSI-BLAST detects several close eukaryotic homologs (gi|39595012, e.g. in iteration 1 with E-value 0.002), while PSI-BLAST using one of the DUF994-detected sequences (gi|48860544) finds an archaeal Holliday junction resolvase (gi|15897494 in iteration 6 with E-value 1 × 10−6). The predicted nuclease domain present in eukaryotic DUF994 sequence homologs maps to the extreme C-terminus of hypothetical human protein (gi|28839601), with the N-terminal sequence extension including a Rad18-like zinc finger domain. This zinc finger domain is found in a number of other proteins involved in DNA repair [Rad18 (38) and polkappa (39)] and maintenance of genome stability [WHIP/MGS1 (40)]. Such a domain fusion supports a role for the human DUF994 nuclease homolog (KIAA1018) in DNA repair or maintenance of genome stability.
DUF820
DUF820 (COG4636) sequences form a family of hypothetical proteins from bacteria that have greatly expanded in a number of cyanobacterial species. Recently, a structure (1wdj) was solved for a hypothetical protein Tt1808 from Thermus thermophilus with unknown function belonging to this family. Accordingly, this structure retains the restriction endonuclease core with a C-terminal extension (ββββ) and a small N-terminal sub-domain (βαββ) that appears to mediate oligomerization. DUF820 sequences include the conserved motifs (Figure 2) that make up the nuclease active site. However, the position of the N-terminal sub-domain with respect to this site may block nucleic acid binding, therefore requiring a structural rearrangement for activity. The absence of this domain in one of the three chains making up the crystallographic asymmetric unit could support such a hypothesis. Some DUF820 sequences are fused to a ClpA N-terminal domain, an α-helical structure (1khy) that targets binding of ClpA ATPase to unfolded protein substrates destined for cleavage by the ClpP protease. Perhaps the Clp pathway facilitates DUF820 ability to bind nucleic acid.
Competence protein CoiA
The CoiA family (COG4469) includes proteins from a diverse set of bacteria, including naturally competent species such as Streptococcus pneumoniae and Bacillus subtilis. Interestingly, the active site residues are switched in this family, with the K typically found in Motif III occupying the Motif I position, and the metal ion coordinating residue (Q) typically found in Motif I occupying the Motif III position (Figure 2). CoiA sequences include an N-terminal extension (predicted as all β) with two conserved motifs (CXXC and HXXH) resembling a zinc finger ribbon (41). CoiA is an uncharacterized protein that belongs to a competence-specific operon in S.pneumoniae (42). Natural competence, or the ability to take up and incorporate foreign DNA into the chromosome, proceeds in several distinguishable stages: DNA binding, DNA processing and uptake, and integration of foreign DNA into the host chromosome. The STRING database assigns a confident (score 0.789) functional association between the CoiA family (COG4469) and a late competence protein superfamily II DNA/RNA helicase (COG4098) required for DNA uptake, suggesting that the CoiA protein mediates DNA cleavage in one of the stages of genetic transformation (DNA processing or integration).
Many members of this family are annotated as transcription factors, stemming from a noted weak similarity (43) of a single sequence (gi|1771202) to vsf-1 transcription factor. Although simple BLAST with the query (gi|1771202) detects this proposed similarity (to gi|1076603 with an insignificant E-value 0.14), the same sequence after a single iteration of PSI-BLAST disappears from the list of hits (E-value > 56). Such a result brings into the question the transcription factor annotation for members of the CoiA family.
RmuC
The RmuC family (COG1322) includes a number of widely distributed bacterial sequences with a central restriction endonuclease-like fold domain surrounded by predicted coiled-coil regions. Although the molecular basis of RmuC function remains unknown, the Escherichia coli rmuC genetic locus influences the rate of DNA inversions at short inverted repeats (44), suggesting a general role of the gene product in recombination events. The rmuC gene is reported to belong to the LexA regulon (45), which controls bacterial cellular response to metabolic stresses that damage DNA. Finally, STRING analysis of RmuC (COG1322) suggests a functional association with ATP-dependent double-strand break repair endonuclease (COG0419, score 0.584). In fact, mutation of this ATP-dependent endonuclease in E.coli is required for the phenotype associated with the RmuC genetic locus (44). The presence of active site residues (Figure 2), together with the described genetic profiles for rmuC, is consistent with a role in DNA cleavage that suppresses the rate of inversions at short inverted repeats.
CONCLUSIONS
By applying the concept of transitivity to fold recognition, we identify restriction endonuclease-like domains in nine families of previously unknown function (Figure 1). These hypothetical proteins possess conserved active site residues typically responsible for nucleic acid cleavage and display similar hydrophobicity profiles in multiple sequence alignments (Figure 2). Furthermore, the families help to establish a network of connectivity between known restriction endonuclease-like structures (Figure 1). The connectivity network reflects their structural relatedness, with highly connected structures closely resembling (higher average Z-scores, Table 1) the defined restriction endonuclease-like fold core (αβββαβ). Those structures linked by fewer connections tend to include insertions to and/or deviations in length or packing of core secondary structure elements (Figure 1). In fact, some of the transitively linked structures display such significant diversity that they would be classified as dissimilar (Dali Z-score < 2 for some pairs, Table 1) in the absence of intermediate links. One such structure (the C-terminal domain of RecC, Figure 3) detected by our method is not identified with a structure-based search (Dali) of the existing PDB database.
Acknowledgments
This work was supported in part by the NIH grant GM67165 to N.V.G. and by MNI and the European Commission with grants LSHG-CT-2003-503265 and LSHG-CT-2004-503567 to L.R. Funding to pay the Open Access publication charges for this article was provided by Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Bujnicki J.M. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the ‘midnight zone’ of homology. Curr. Protein Pept. Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- 3.Kovall R., Matthews B.W. Toroidal structure of lambda-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 4.Kovall R.A., Matthews B.W. Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol. 1999;3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 5.Ban C., Yang W. Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J. 1998;17:1526–1534. doi: 10.1093/emboj/17.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsutakawa S.E., Jingami H., Morikawa K. Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell. 1999;99:615–623. doi: 10.1016/s0092-8674(00)81550-0. [DOI] [PubMed] [Google Scholar]
- 7.Hickman A.B., Li Y., Mathew S.V., May E.W., Craig N.L., Dyda F. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 8.Hadden J.M., Convery M.A., Declais A.C., Lilley D.M., Phillips S.E. Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I. Nature Struct. Biol. 2001;8:62–67. doi: 10.1038/83067. [DOI] [PubMed] [Google Scholar]
- 9.Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K. Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition. Structure (Camb.) 2001;9:197–204. doi: 10.1016/s0969-2126(01)00576-7. [DOI] [PubMed] [Google Scholar]
- 10.Nishino T., Komori K., Ishino Y., Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure (Camb.) 2003;11:445–457. doi: 10.1016/s0969-2126(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 11.Galburt E.A., Stoddard B.L. Catalytic mechanisms of restriction and homing endonucleases. Biochemistry. 2002;41:13851–13860. doi: 10.1021/bi020467h. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L., Makarova K.S., Koonin E.V. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujnicki J.M., Rychlewski L. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 2001;3:69–72. [PubMed] [Google Scholar]
- 14.Yang J., Malik H.S., Eickbush T.H. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl Acad. Sci. USA. 1999;96:7847–7852. doi: 10.1073/pnas.96.14.7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravind L., Walker D.R., Koonin E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova K.S., Aravind L., Grishin N.V., Rogozin I.B., Koonin E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L., Sander C. Protein folds and families: sequence and structure alignments. Nucleic Acids Res. 1999;27:244–247. doi: 10.1093/nar/27.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginalski K., von Grotthuss M., Grishin N.V., Rychlewski L. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004;32:W576–W581. doi: 10.1093/nar/gkh370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 21.Ginalski K., Elofsson A., Fischer D., Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchiyama I. MBGD: microbial genome database for comparative analysis. Nucleic Acids Res. 2003;31:58–62. doi: 10.1093/nar/gkg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 26.Pei J., Sadreyev R., Grishin N.V. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 27.Holm L., Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 28.Hadden J.M., Declais A.C., Phillips S.E., Lilley D.M. Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I. EMBO J. 2002;21:3505–3515. doi: 10.1093/emboj/cdf337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 30.Komori K., Sakae S., Daiyasu H., Toh H., Morikawa K., Shinagawa H., Ishino Y. Mutational analysis of the Pyrococcus furiosus holliday junction resolvase hjc revealed functionally important residues for dimer formation, junction DNA binding, and cleavage activities. J. Biol. Chem. 2000;275:40385–40391. doi: 10.1074/jbc.M006294200. [DOI] [PubMed] [Google Scholar]
- 31.Tsutakawa S.E., Muto T., Kawate T., Jingami H., Kunishima N., Ariyoshi M., Kohda D., Nakagawa M., Morikawa K. Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell. 1999;3:621–628. doi: 10.1016/s1097-2765(00)80355-x. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X.E., Wang Y., Reuter M., Mucke M., Kruger D.H., Meehan E.J., Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson M.J., Pohler J.R., Lilley D.M. Catalytic and binding mutants of the junction-resolving enzyme endonuclease I of bacteriophage t7: role of acidic residues. Nucleic Acids Res. 1999;27:682–689. doi: 10.1093/nar/27.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusov R.L., Koonin E.V. A simple tool to search for sequence motifs that are conserved in BLAST outputs. Comput. Appl. Biosci. 1994;10:457–459. doi: 10.1093/bioinformatics/10.4.457. [DOI] [PubMed] [Google Scholar]
- 35.Yu M., Souaya J., Julin D.A. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 1998;283:797–808. doi: 10.1006/jmbi.1998.2127. [DOI] [PubMed] [Google Scholar]
- 36.Raghavendra N.K., Rao D.N. Functional cooperation between exonucleases and endonucleases—basis for the evolution of restriction enzymes. Nucleic Acids Res. 2003;31:1888–1896. doi: 10.1093/nar/gkg275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer L.M., Makarova K.S., Koonin E.V., Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyase S., Tateishi S., Watanabe K., Tomita K., Suzuki K., Inoue H., Yamaizumi M. Differential regulation of Rad18 through Rad6-dependent mono-and polyubiquitination. J. Biol. Chem. 2005;280:515–524. doi: 10.1074/jbc.M409219200. [DOI] [PubMed] [Google Scholar]
- 39.Okada T., Sonoda E., Yamashita Y.M., Koyoshi S., Tateishi S., Yamaizumi M., Takata M., Ogawa O., Takeda S. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 2002;277:48690–48695. doi: 10.1074/jbc.M207957200. [DOI] [PubMed] [Google Scholar]
- 40.Hishida T., Iwasaki H., Ohno T., Morishita T., Shinagawa H. A yeast gene, MGS1, encoding a DNA-dependent AAA(+) ATPase is required to maintain genome stability. Proc. Natl Acad. Sci. USA. 2001;98:8283–8289. doi: 10.1073/pnas.121009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishna S.S., Majumdar I., Grishin N.V. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova E.V., Morrison D.A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nardi M., Renault P., Monnet V. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J. Bacteriol. 1997;179:4164–4171. doi: 10.1128/jb.179.13.4164-4171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slupska M.M., Chiang J.H., Luther W.M., Stewart J.L., Amii L., Conrad A., Miller J.H. Genes involved in the determination of the rate of inversions at short inverted repeats. Genes Cells. 2000;5:425–437. doi: 10.1046/j.1365-2443.2000.00341.x. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyk T.K., DeRose E.J., Gonye G.E. LuxArray, a high-density, genomewide transcription analysis of Escherichia coli using bioluminescent reporter strains. J. Bacteriol. 2001;183:5496–5505. doi: 10.1128/JB.183.19.5496-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]