Abstract
Sugar mimics are valuable tools in medicinal chemistry, offering the potential to overcome the limitations of carbohydrate inhibitors, such as poor pharmacokinetics and non-selectivity. In our continued efforts to develop heterocyclic galectin-1 inhibitors, we report the synthesis and characterization of thiazole-linked coumarin piperazine hybrids (10a–10i) as Gal-1 inhibitors. The compounds were characterized using 1H NMR, 13C NMR and HRMS. Among the synthesized molecules, four compounds demonstrated significant inhibitory activity, with more than 50% inhibition observed at a concentration of 20 μM in a Gal-1 enzyme assay. Fluorescence spectroscopy was further utilized to elucidate the binding constant for the synthesized compounds. 10g exhibited the highest affinity for Gal-1, with a binding constant (Ka) of 9.8 × 104 M−1. To elucidate the mode of binding, we performed extensive computational analyses with 10g, including 1.2 μs all-atom molecular dynamics simulations coupled with a robust machine learning tool. Our findings indicate that 10g binds to the carbohydrate binding site of Gal-1, with the coumarin moiety playing a key role in binding interactions. Additionally, our study underscores the limitations of relying solely on docking scores for conformational selection and highlights the critical importance of performing multiple MD replicas to gain accurate insights.
Synthesis of Gal-1 inhibitors with binding site analysis using 1.2 μs MD replicas and machine learning.
1. Introduction
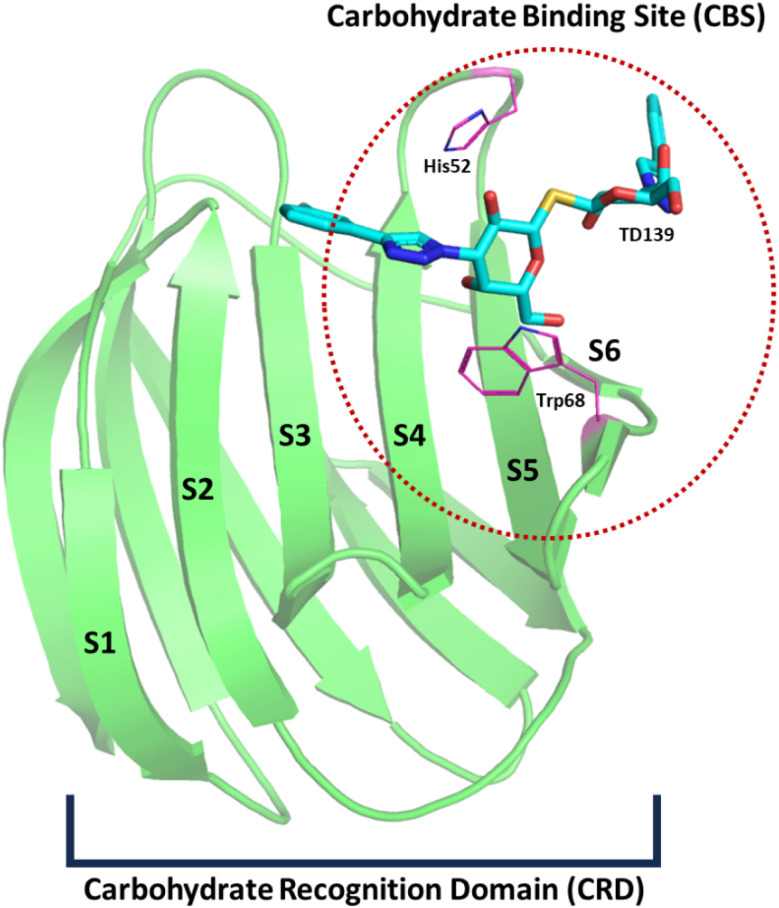
Galectins are a family of animal lectins characterized by their affinity for β-galactosides and notable sequence similarity in their carbohydrate-binding sites.1 Each galectin contains one or two highly conserved carbohydrate recognition domains (CRDs), typically composed of approximately 135 amino acids.2 These proteins are implicated in various pathological processes, including cancer and infections,3,4 among others. The first identified member of the galectin family, galectin-1 (Gal-1), has been demonstrated to play a significant role in several biological processes. Its involvement in tumour progression, inflammatory diseases, neurodegenerative disorders, immune escape and muscular dystrophies, underscores the importance of targeted therapeutic strategies. Specifically, enhancing the targeted overexpression or delivery of Gal-1 may be beneficial for treating certain inflammation-related conditions, while the development of selective Gal-1 inhibitors is essential for combating cancer progression.5 Gal-1 is a homodimer characterized by a β-sandwich fold, consisting of two antiparallel β-sheets – one with five (F1–F5) and the other with six (S1–S6) β-strands. The carbohydrate-binding site (CBS) of Gal-1 is located on the surface of the β-sheet formed by the S4–S6 strands (Fig. 1).6 The highly conserved amino acid residues involved in Gal-1 interactions include His44, Asn46, Arg48, Val59, Asn61, Trp68, Glu71, and Arg73.6 The specificity and affinity of galectin-1 for various carbohydrates are partially governed by a complex cooperative hydrogen bond network in addition to the CH–π interactions between the carbohydrate and residues with aromatic sidechains.7,8 Notably, computational studies have shown that the conserved tryptophan residue present within the CBS when mutated to alanine results in the complete rotation of the galactose ligand within the CBS due to the loss of stabilization.9 This has been further corroborated by another study which demonstrated that His52 and Trp68 are essential for sugar recognition prior to binding.7
Fig. 1. Depiction of galectin-1 (Gal-1) PDB ID: 4Y24 (green) bound to the co-crystal TD-139 (cyan). The structure represents a single chain of the Gal-1 dimer, with strands S1–S6 labelled. The carbohydrate-binding site (CBS) is circled with a red dotted line, located among strands S4–S6. The carbohydrate recognition domain (CRD) is highlighted by a black bracket. Key residues responsible for sugar recognition, His52 and Trp68, are shown in magenta. The loops have been smoothened for representational purposes.
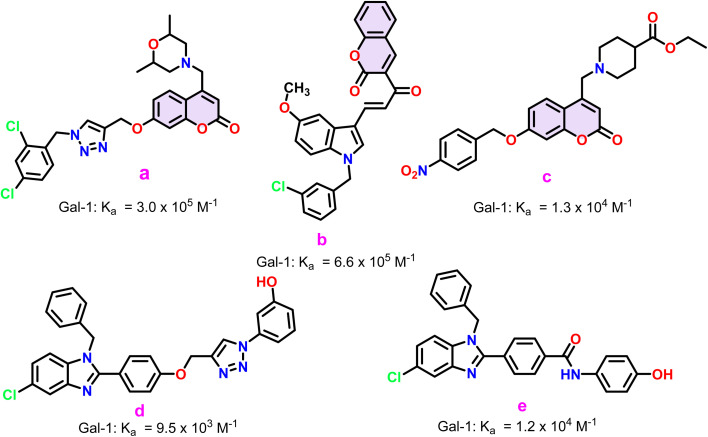
Most Gal-1 inhibitors reported so far are carbohydrates or their derivatives.8,10,11 However, these inhibitors present several challenges. The synthesis and characterization of carbohydrate-based inhibitors are complex, costly and often require extensive modifications of protecting groups.12 Additionally, they lack cell membrane permeability and selectivity.13 Recently, however, there has been a growing interest in heterocyclic compounds as Gal-1 inhibitors.13,14 These compounds have been extensively discussed in recent reviews on the topic.13,14 They offer a promising alternative due to their rich background in medicinal chemistry, allowing for easier development and optimization. Selected heterocyclic Gal-1 inhibitors along with their determined binding affinity values have been succinctly discussed in Fig. 2. Encouraged by these prior developments of heterocyclic Gal-1 inhibitors, coupled with the fact that hydrophobic/aromatic substitutions aid in enhancing binding to galectins,13,15 and reports of cyclic amino analogues acting as biologically active substitutions for the noviose (carbohydrate) moiety in novobiocin;16,17 we set out to develop thiazole linked coumarin-piperazine hybrids as potential Gal-1 inhibitors. Moreover, while numerous crystallographic structures have been resolved for carbohydrate-based Gal-1 inhibitors, which gives insight into their binding mode; studies pertaining to determination of binding site and the binding mode for heterocyclic inhibitors remains elusive. Thus, we also wanted to investigate and throw some light on the probable binding site of these compounds. This was explored by a comprehensive study utilizing all-atom molecular dynamic simulations.
Fig. 2. Select heterocyclic Gal-1 inhibitors (a–e) alongside their binding constants. For a comprehensive and up-to-date list of all known heterocyclic Gal-1 inhibitors, refer to recent review articles on the topic.13,14.
2. Methods
2.1. Chemistry
All starting materials, reagents and solvents used in this study were sourced from commercial suppliers. Analytical thin-layer chromatography (TLC) was conducted on pre-coated silica gel 60-F254 aluminium plates from MERCK. Visualization of the TLC plates was performed using UV light or in some instances, an iodine chamber. Melting points were measured with a Stuart® SMP30 melting point apparatus. Column chromatography was performed using silica gel (60–120 mesh) and compounds were eluted with a mixture of ethyl acetate and hexane. NMR spectra were recorded on Bruker 500 (500 MHz for 1H NMR and 125 MHz for 13C NMR) using DMSO or CDCl3 as solvent. Chemical shift was reported in parts per million (ppm) with respect to internal standard Tetra Methyl Silane (TMS). Coupling constants were quoted in Hertz (Hz). High-resolution mass spectra (HRMS) was obtained on Agilent Q-TOF-Mass Spectrometer 6540-UHD LC/HRMS operating at 70 eV using direct inlet.
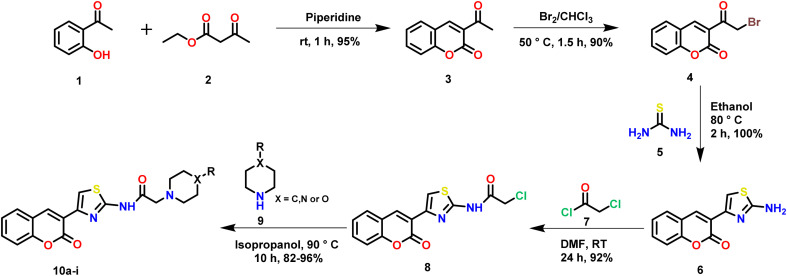
The target thiazole linked coumarin-piperazine hybrids (10a–i) were synthesized by a 5-step method (Scheme 1). One standout feature of the depicted scheme was that “no column chromatography purification” was required until the last step of the reaction. The products were obtained by simple purification. Initially, Knoevenagel condensation between aldehydic functionality of salicylaldehyde (1) and active methylene of ethyl acetoacetate (2) in presence of piperidine followed by cyclization gave us 3-acetyl coumarin (3) in high yields. Next, we performed electrophilic substitution on bromine to obtain bromoacetyl coumarin (4). Further, 4 was heated in ethanol in presence of thiourea (5) to afford cyclized product 2-amino thiazole coumarin (6). Subsequently, a nucleophilic substitution was performed by amino group of 6 on chloroacetyl chloride (7) to obtain 8. Finally, another nucleophilic substitution reaction was performed on 8 by the amine present in the cyclic amino ring (9) to give the final product (10a–i) in excellent yields. The final step of the reaction was optimized based on a previous report.18 All synthesized compounds (10a–i) were characterized by 1H-NMR, 13C-NMR and High-Resolution Mass Spectrometry (HRMS) spectral techniques [ESI†]. The characteristic thiazole proton peak was observed at > δ 8.5 ppm for all the compounds. The amide NH proton was highly acidic and it rapidly underwent deuterium exchange. Thus, it was not found for all the compounds. The protons associated with the saturated heterocyclic rings were observed in the range of δ 1–4 ppm. The characteristic CH2 protons next to the amide functionality were observed in the range of δ 3–4 ppm as a sharp singlet. The 13C NMR spectrum displayed the characteristic peak of carbonyl carbon of chalcone at > δ 160. The HRMS (ESI) of compounds showed corresponding [M + H]+ peaks based on their molecular weights.
Scheme 1. Synthesis of thiazole linked coumarin-piperazine hybrids (10a–i).
2.2. Biology
2.2.1. Enzymatic assay
All the synthesized derivatives were screened for their reduction in Gal-1 levels with the aid of Gal-1 ELISA enzymatic study. HCT-116 cells were grown in 12- well plate and treated with 20 μM concentration of compounds for 24 h. Then the supernatant was collected as Gal-1 is an extracellular secreted protein. Equal amounts of supernatant were subjected to quantikine enzyme-linked immunosorbent assay (ELISA) as per manufacturer's protocol (DGAL 10, R&D systems). The supernatant was diluted 2-fold with calibrator diluent and incubated with human Gal-1 coated plates for 2 h at room temperature on a horizontal orbital microplate shaker and washed to remove any unbound protein. Gal-1 specific polyclonal antibody conjugated to horseradish peroxidase was added and incubated for 2 h on shaker. It was then washed and incubated with substrate solution for 30 minutes before terminating the reaction with 50 microlitres of stop solution. Amount of protein expression was detected at 450 nm using UV spectrophotometer.
2.2.2. Fluorescence studies
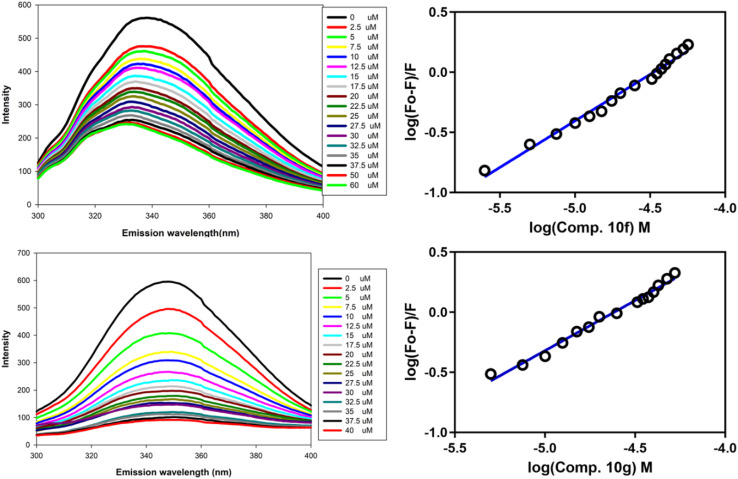
The selected compounds were further subjected to fluorescence spectroscopy to determine their binding constants. For fluorescence binding analysis, human Gal-1 was cloned into the pET-28a(+) expression vector and expressed into E. coli BL21(DE3) cells, followed by its purification through Ni2+-NTA affinity chromatography according to the previous report,19 with few modifications. Similarly, human Gal-3 and Leishmania donovani phosphatase (LdPase) were cloned, expressed, and purified. The buffer used for human Gal-1 and Gal-3 was 25 mM Tris-HCl pH 8, while LdPase was analysed in sodium phosphate buffer pH 7. The intrinsic fluorescence emission spectra of Gal-1, Gal-3, and LdPase were recorded from 300 to 400 nm by Jasco Spectrofluorometer (FP-8500). The excitation wavelength of all the proteins was 280 nm and the scan speed was kept to 100 nm min−1. The binding constant (Ka) was deduced for compounds 10f and 10g employing a modified Stern–Volmer plot as described earlier,20 with the log of ligand concentration on the X-axis and log of F0 − F/F on the Y-axis, where F0 is the fluorescence intensity without any ligand and F is fluorescence intensity with different ligand concentrations.
2.3. Computational studies
2.3.1. Molecular docking and machine learning
Molecular docking studies were conducted using the crystal structure of Gal-1 (PDB ID: 4Y24). The protein was prepared by removing all water molecules and heteroatoms, followed by the addition of hydrogen atoms and determination of protonation states of the protein residues at physiological pH (7.4) using the H++ server.21,22 For compound 10g, the protonation state was determined using the MolGpka.23 The smiles string for the compound was converted to the PDBQT format using Open Babel.24 Docking simulations were performed with AutoDock Vina,25 using the following grid box parameters: center_x = 10.867, center_y = −6.230, center_z = −16.693, size_x = 110, size_y = 110, size_z = 110 and an exhaustiveness value of 15. The docking pose generated were analyzed using PyMOL.26
The prediction of binding site using machine learning was carried using RoseTTAFold All-Atom,27 on neurosnap platform.28 Briefly, the sdf file for the TD-139 (co-crystal in PDB ID: 4Y24) and compound 10g were uploaded along with the Gal-1 sequence and the run was subsequently executed. The output pdb file generated was downloaded and analyzed using PyMOL.26
2.3.2. Molecular dynamics
The protein was modeled using the AMBER ff19SB force field,29 while the ligand, compound 10g, was parameterized using the General Amber Force Field (GAFF). The protein–ligand complex was prepared with AmberTools23.30 Solvation of the complex was carried out using the Placevent algorithm,31 based on the 3D reference interaction site model theory.32 The solvated complex was then placed in an octahedral box filled with OPC water molecules, maintaining a 10 Å buffer around the complex. The system was neutralized with counterions (Na+ and Cl−) to achieve an ionic strength of 0.1 M. The hydrogen mass repartitioning (HMR) method was applied to generate topologies that enabled a 4 fs time step during simulations.33 Finally, the resulting trajectories were analyzed using PyContact.34
2.3.3. ADMET analysis
The ADMET analysis was carried out using two tools: pkCSM,35 and ADMETlab 3.0.36 Simply, the smiles string of compound 10g was added in the dedicated field on the online server and subsequently the run was executed.
3. Results
3.1. Chemistry
3.1.1. Synthesized analogues
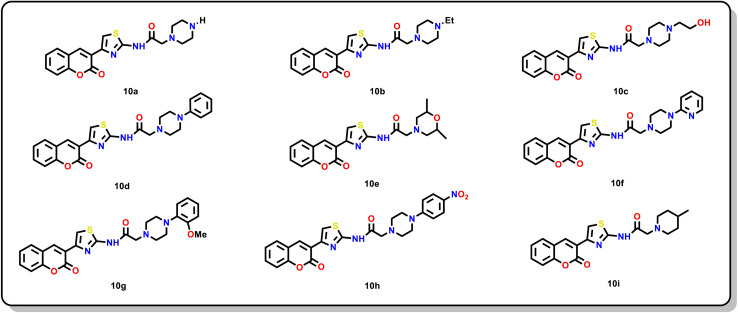
Based on resources at our disposal, we synthesized a series of compounds 10a–i, which include cyclic amino analogues such as piperazine, piperidine and morpholine (Fig. 3). The selection of piperazine-containing compounds was strategically designed to encompass both aliphatic and aromatic groups. The rationale behind this was to explore the potential for aromatic residues to engage in additional π–π or hydrophobic interactions within the Gal-1 CBS, a hypothesis supported by prior studies suggesting that aromatic groups could enhance affinity towards galectins.13,15
Fig. 3. Structure of synthesized compounds (10a–i).
3.1.2. Spectral data (1H NMR, 13C NMR and HRMS)
3.1.2 1. N-(5-(2-Oxo-2H-chromen-3-yl)thiazol-2-yl)-2-(piperazin-1-yl)acetamide (10a)
White solid; 95% yield; mp: 170–172 °C; 1H NMR (500 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.03 (s, 1H), 7.87 (dd, J = 7.7, 1.2 Hz, 1H), 7.69–7.61 (m, 1H), 7.48 (d, J = 8.2 Hz, 1H), 7.41 (t, J = 7.5 Hz, 1H), 3.29 (s, 2H), 2.81–2.68 (m, 4H), 2.49–2.46 (m, 4H); 13C NMR (125 MHz, DMSO-d6) δ 169.23, 159.32, 157.50, 152.66, 142.98, 139.28, 132.79, 129.92, 125.56, 121.15, 119.75, 116.74, 115.08, 67.57, 61.13, 54.11. HRMS (ESI): m/z calc. for C18H18N4O3S, 371.1173 found (M + H)+ 371.1157.
3.1.2 2. 2-(4-Ethylpiperazin-1-yl)-N-(5-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)acetamide (10b)
White solid; 91% yield; mp: 161–163 °C; 1H NMR (500 MHz, CDCl3) δ 8.63 (s, 1H), 8.03 (s, 1H), 7.86 (dd, J = 7.7, 1.4 Hz, 1H), 7.65 (ddd, J = 8.6, 7.5, 1.5 Hz, 1H), 7.47 (d, J = 8.3 Hz, 1H), 7.41 (td, J = 7.6, 0.9 Hz, 1H), 2.56 (s, 4H), 2.43 (dd, J = 17.4, 13.2 Hz, 4H), 2.34 (dd, J = 14.3, 7.1 Hz, 2H), 1.00 (t, J = 7.2 Hz, 3H); 13C NMR (125 MHz, DMSO-d6) δ 169.38, 159.25, 157.63, 152.95, 142.54, 139.09, 132.40, 129.37, 125.26, 120.83, 119.55, 116.43, 114.80, 60.68, 53.12, 52.75, 52.06, 12.47; HRMS (ESI): m/z calc. for C20H22N4O3S, 398.1486 found (M + H)+ 399.1511.
3.1.2 3. 2-(4-(2-Hydroxyethyl)piperazin-1-yl)-N-(5-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)acetamide (10c)
White solid; 96% yield; mp: 184–186 °C; 1H NMR (500 MHz, DMSO-d6) δ 8.63 (s, 1H), 8.03 (s, 1H), 7.87 (dd, J = 7.8, 1.6 Hz, 1H), 7.66 (td, J = 8.3, 2.0 Hz, 1H), 7.48 (d, J = 8.2 Hz, 1H), 7.41 (t, J = 7.5 Hz, 1H), 4.39 (brs, 1H), 3.50 (t, J = 6.3 Hz, 2H), 3.31 (s, 2H), 2.55 (s, 4H), 2.47 (s, 4H), 2.44–2.28 (m, 2H); 13C NMR (125 MHz, DMSO-d6) δ 169.37, 159.26, 157.62, 152.95, 142.54, 139.10, 132.40, 129.38, 125.26, 120.83, 119.55, 116.43, 114.81, 60.69, 58.97, 53.56, 53.15, 53.09; HRMS (ESI): m/z calc. for C20H22N4O4S, 415.1435 found (M + H)+ 415.1466.
3.1.2 4. N-(5-(2-Oxo-2H-chromen-3-yl)thiazol-2-yl)-2-(4-phenylpiperazin-1-yl)acetamide (10d)
White solid; 90% yield; mp: 199–201 °C; 1H NMR (500 MHz, DMSO-d6) δ 8.63 (s, 1H), 8.03 (s, 1H), 7.85 (dd, J = 7.8, 1.8 Hz, 2H), 7.65 (dd, J = 4.4, 3.2 Hz, 1H), 7.47 (d, J = 8.2 Hz, 1H), 7.41 (dd, J = 7.6, 3.8 Hz, 1H), 7.21 (dd, J = 6.7, 1.8 Hz, 2H), 6.95 (d, J = 8.5 Hz, 1H), 6.79 (t, J = 7.2 Hz, 1H), 3.41 (s, 2H), 3.25–3.04 (m, 4H), 2.81–2.56 (m, 4H); 13C NMR (125 MHz, DMSO-d6) δ 170.03, 159.64, 158.24, 153.09, 152.41, 142.55, 141.68, 139.65, 133.01, 129.95, 125.41, 122.93, 121.07, 120.62, 119.55, 118.38, 116.08, 60.94, 54.15, 51.24. HRMS (ESI): m/z calc. for C24H22N4O3S, 447.1486 found (M + H)+ 447.1514.
3.1.2 5. 2-(2,6-Dimethylmorpholino)-N-(5-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)acetamide (10e)
White solid; 95% yield; mp: 173–175 °C; 1H NMR (500 MHz, DMSO-d6) δ 12.02 (s, 1H), 8.63 (s, 1H), 8.03 (s, 1H), 7.86 (dd, J = 7.7, 1.4 Hz, 1H), 7.69–7.62 (m, 1H), 7.48 (d, J = 8.3 Hz, 1H), 7.44–7.39 (m, 1H), 3.63 (dtd, J = 8.3, 6.2, 2.0 Hz, 2H), 3.33 (s, 2H), 2.79 (d, J = 10.1 Hz, 2H), 1.91 (t, J = 10.7 Hz, 2H), 1.06 (d, J = 6.3 Hz, 6H); 13C NMR (125 MHz, DMSO-d6) δ 169.25, 159.24, 157.63, 152.94, 142.54, 139.06, 132.38, 129.34, 125.25, 120.84, 119.53, 116.42, 114.78, 71.42, 60.47, 59.06, 19.34; HRMS (ESI): m/z calc. for C20H21N3O4S, 400.1326 found (M + H)+ 400.1327.
3.1.2 6. N-(5-(2-Oxo-2H-chromen-3-yl)thiazol-2-yl)-2-(4-(pyridin-2-yl)piperazin-1-yl)acetamide (10f)
White solid; 85% yield; mp: 235–238 °C; 1H NMR (500 MHz, DMSO-d6) δ 8.64 (s, 1H), 8.12 (dd, J = 4.9, 2.3 Hz, 1H), 8.04 (s, 1H), 7.86 (dd, J = 7.8, 1.8 Hz, 1H), 7.65 (td, J = 7.1, 3.6 Hz, 1H), 7.54 (ddd, J = 8.8, 7.0, 1.9 Hz, 1H), 7.48 (d, J = 8.1 Hz, 1H), 7.41 (dd, J = 9.6, 5.4 Hz, 1H), 6.84 (d, J = 8.5 Hz, 1H), 6.64 (dd, J = 7.1, 4.9 Hz, 1H), 3.63–3.43 (m, 4H), 3.40 (s, 2H), 2.77–2.55 (m, 4H); 13C NMR (125 MHz, DMSO-d6) δ 169.32, 159.47, 159.25, 157.66, 152.94, 148.03, 142.55, 139.07, 137.97, 132.39, 129.36, 125.26, 120.84, 119.54, 116.43, 114.82, 113.46, 107.58, 60.64, 52.80, 45.11; HRMS (ESI): m/z calc. for C23H21N5O3S, 448.1438 found (M + H)+ 448.1472.
3.1.2 7. 2-(4-(2-Methoxyphenyl)piperazin-1-yl)-N-(5-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)acetamide (10g)
White solid; 94% yield; mp: 210–212 °C; 1H NMR (500 MHz, DMSO-d6) δ1H NMR (500 MHz, DMSO) δ 8.64 (s, 1H), 8.04 (s, 1H), 7.86 (dd, J = 7.7, 1.2 Hz, 1H), 7.68–7.63 (m, 1H), 7.48 (d, J = 8.2 Hz, 1H), 7.43–7.38 (m, 1H), 6.98–6.93 (m, 2H), 6.92–6.86 (m, 2H), 3.78 (s, 3H), 3.41 (s, 2H), 3.02 (s, 4H), 2.72 (s, 4H); 13C NMR (125 MHz, DMSO-d6) δ 169.38, 159.26, 157.65, 152.94, 152.49, 142.55, 141.68, 139.09, 132.39, 129.37, 125.26, 122.93, 121.31, 120.83, 119.55, 118.49, 116.43, 114.82, 112.41, 60.70, 55.81, 53.27, 50.53; HRMS (ESI): m/z calc. for C25H24N4O4S, 476.1592 found (M + H)+ 477.1591.
3.1.2 8. 2-(4-(4-Nitrophenyl)piperazin-1-yl)-N-(5-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)acetamide (10h)
Bright yellow solid; 82% yield; mp: 249–251 °C; 1H NMR (500 MHz, DMSO-d6) δ 8.62 (s, 1H), 8.11 (dd, J = 7.3, 2.2 Hz, 2H), 8.08–8.05 (m, 2H), 8.04 (s, 1H), 7.88–7.82 (m, 1H), 7.69–7.62 (m, 1H), 7.48 (d, J = 8.2 Hz, 1H), 7.43–7.38 (m, 1H), 3.71 (dd, J = 6.4, 4.3 Hz, 4H), 3.22 (dd, J = 6.2, 4.5 Hz, 4H); 13C NMR (125 MHz, DMSO-d6) δ 170.32, 160.37, 158.24, 153.55, 148.43, 143.27, 139.76, 137.89, 132.39, 129.79, 125.36, 121.89, 120.22, 116.95, 116.03, 114.19, 108.99, 61.01, 53.34, 47.24. HRMS (ESI): m/z calc. for C24H21N5O5S, 491.1337 found (M + H)+ 492.1364.
3.1.2 9. 2-(4-Methylpiperidin-1-yl)-N-(5-(2-oxo-2H-chromen-3-yl)thiazol-2-yl)acetamide (10i)
White solid; 88% yield; mp: 168–170 °C; 1H NMR (500 MHz, DMSO-d6) δ 11.71 (s, 1H), 8.64 (s, 1H), 8.03 (s, 1H), 7.87 (dd, J = 7.7, 1.2 Hz, 1H), 7.66 (dd, J = 15.6, 1.5 Hz, 1H), 7.48 (d, J = 8.3 Hz, 1H), 7.41 (dd, J = 11.6, 4.2 Hz, 1H), 3.33 (s, 2H), 2.85 (d, J = 11.4 Hz, 2H), 2.17 (td, J = 11.5, 2.0 Hz, 2H), 1.59 (d, J = 11.2 Hz, 2H), 1.34 (ddd, J = 14.4, 12.4, 7.1 Hz, 1H), 1.21 (ddd, J = 15.3, 12.1, 3.6 Hz, 2H), 0.91 (d, J = 6.4 Hz, 3H); 13C NMR (125 MHz, DMSO-d6) δ 169.71, 159.26, 157.66, 152.94, 142.54, 139.10, 132.39, 129.37, 125.26, 120.82, 119.56, 116.43, 114.78, 61.17, 53.75, 34.37, 30.28, 22.28; HRMS (ESI): m/z calc. for C20H21N3O3S, 384.1377 found (M + H)+ 384.1406.
3.2. Biology
All synthesized compounds were initially screened using a Gal-1 enzymatic assay to assess their inhibitory potential (Table 1). Compounds that demonstrated >20% inhibition at a 20 μM concentration were further analysed for their affinity to Gal-1 using fluorescence spectroscopy. Among the tested compounds, 10f exhibited the highest inhibition in the enzymatic assay, achieving >80% inhibition. Compounds 10a, 10d, and 10g also showed significant inhibitory activity, with >50% inhibition. In contrast, compounds 10h and 10i exhibited <10% inhibition and thus were not pursued in subsequent analyses. To enumerate the binding constants of selected compounds (10f and 10g) through fluorescence spectroscopy, recombinant human Gal-1, Gal-3 and LdPase were purified to homogeneity using affinity chromatography. The fluorescence spectra for compounds 10f and 10g are displayed in Fig. 4, which revealed that compounds 10f and 10g had the highest affinity for Gal-1 (Table 1). Importantly, both compounds displayed significantly lower binding to Gal-3 compared to Gal-1, and no detectable binding to LdPase (Table 1). These studies indicate that compound 10g holds significant promise as a hit candidate suitable for further development.
Enzymatic Assay (EA) and binding constant (Ka) values of thiazole-linked coumarin piperazine hybrids (10a–i).
| Compound | % Inhibition (EA) | Gal-1 Ka (104 M−1) | Gal-3 Ka (10−6 M−1) | LdPasea |
|---|---|---|---|---|
| 10a | 58.19 ± 1.12 | 8.8 | NE | NE |
| 10b | 25.93 ± 0.81 | 2.1 | NE | NE |
| 10c | 22.46 ± 1.67 | 4.1 | NE | NE |
| 10d | 56.73 ± 2.13 | 6.6 | NE | NE |
| 10e | 27.80 ± 0.76 | 6.5 | NE | NE |
| 10f | 80.57 ± 1.77 | 9.3 | 1.1 | NB |
| 10g | 58.19 ± 3.28 | 9.8 | 1.0 | NB |
| 10h | <10 | NE | NE | NE |
| 10i | <10 | NE | NE | NE |
NE – not evaluated, NB – no binding.
Fig. 4. Fluorescence spectra of compound 10f (top panel) and 10g (bottom panel).
3.3. Computational studies
3.3.1. Molecular docking and machine learning
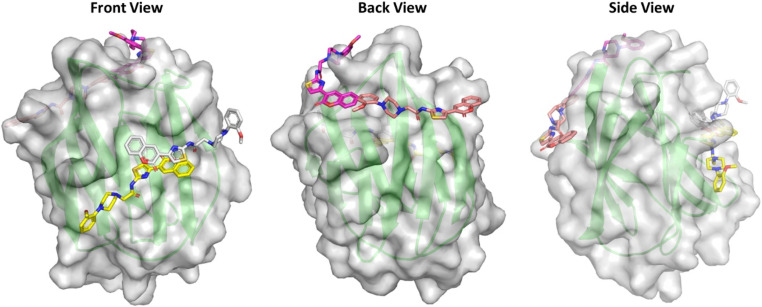
To validate our docking protocol, we attempted to reproduce the cocrystal pose in a blind docking experiment. Initially, we were unable to satisfactorily reproduce a RMSD of less than 2.0 Å, a widely accepted benchmark for validation,37 in the first two runs. To improve accuracy, we increased the exhaustiveness parameter in AutoDock Vina, which controls the computational effort applied during docking. This adjustment allowed us to successfully reproduce the cocrystal pose, with a docking score of −8.3 kcal mol−1. Following this validation, we proceeded to dock compound 10g, which had shown the highest affinity for galectin-1 in our experimental studies. Using the same parameters from the successful validation run, we generated 20 docking poses, which were manually inspected. Among these, two poses, pose 3 and pose 15, were located within the carbohydrate-binding site (CBS) of Gal-1, with docking scores of −5.7 kcal mol−1 and −4.7 kcal mol−1, respectively. Additionally, we identified two other significant poses: pose 1, the top-scoring pose with a docking score of −6.4 kcal mol−1, positioned just above the CBS (Fig. 5), and pose 5, located at an allosteric site where a galectin-1 inhibitor had previously been shown to bind,38 with a docking score of −5.4 kcal mol−1.
Fig. 5. Binding site for the top four poses of compound 10g identified via molecular docking. The front, back and side views of Gal-1 are depicted. The four poses are represented as follows: pose 1 in magenta, pose 3 in yellow, pose 5 in brownish-red and pose 15 in grey.
To further validate our findings, we utilized the RoseTTAFold All-Atom,27 a robust machine learning model, to predict the binding site of compound 10g. Such machine learning models are now gained prominence are now at the forefront of drug discovery.39 Before applying RoseTTAFold All-Atom to 10g, we first validated the model by predicting the co-crystal pose, which achieved a remarkable RMSD of 1.037 Å compared to its crystallographic pose. For compound 10g, the model indicated a binding location within the CBS, consistent with the location predicted by pose 3 in our docking analysis. This alignment between the docking and machine learning predictions reinforced the potential accuracy of our identified binding sites.
3.3.2. Molecular dynamic simulations
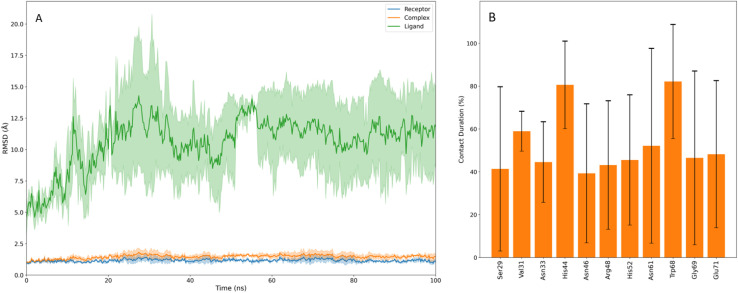
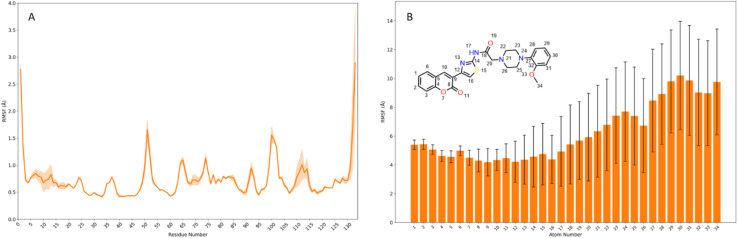
The four poses identified through docking were subjected to molecular dynamics (MD) simulations. For each pose, 100 ns (n = 3) run in triplicate was conducted. Running multiple simulations provides more reliable and significant conclusions than relying on a single extended simulation.40 Across all simulations, the protein/receptor remained stable; however, the stability of compound 10g (referred to as the ligand) varied depending on its initial pose. For poses 1 and 3, 10g remained stable within the binding site only during the initial few nanoseconds before completely dissociating into the bulk solvent (Fig. S1†). In the case of pose 5, 10g dissociated almost immediately upon the start of the simulation. In contrast, for pose 15, compound 10g remained bound within the CBS and reached equilibration in all three simulations, although it exhibited a significantly higher RMSD than considered desirable, with a mean value of 10.654 Å across the runs (Fig. 6). The high RMSD can primarily be attributed to the fluctuations of unbound units of 10g throughout the simulation (Fig. 7). At no point during the simulation were all core units of compound 10g simultaneously bound to the protein. Instead, while some subunits-maintained interaction within the CBS, others were observed to fluctuate freely, contributing to the increased RMSD. Finally, we assessed the contact duration between Gal-1 residues and compound 10g across the three simulations (Fig. 6). Those residues were chosen for analysis which maintained >50% contact in atleast one simulation.
Fig. 6. (A) shows the RMSD of ligand i.e. compound 10g (green), complex (orange) and the receptor (blue) for pose 15 over the course of the simulation. The shaded regions represent the standard deviation of the RMSD values. (B) illustrates the contact duration of key residues of Gal-1 interacting with 10g, with error bars indicating the standard deviation of each interaction.
Fig. 7. (A) displays the root mean square fluctuation (RMSF) of Gal-1 across the three simulations for pose 15, with the shaded region representing the standard deviation. (B) Shows the atom-wise RMSF for compound 10g, with standard deviation represented by error bars.
3.3.3. ADMET analysis
ADMET analysis of compound 10g was conducted using two different computational tools to ensure robustness of the findings. The analysis predicted that 10g lacks substructures associated with Pan Assay Interference Compounds (PAINS), reducing the likelihood of false positives in bioassays. Both tools consistently predicted 10g to have high Caco-2 cell permeability and a very low probability of penetrating the blood–brain barrier. Additionally, 10g was predicted to have high human intestinal absorption and demonstrated stability in human liver microsomes. Importantly, both tools classified 10g as non-mutagenic, though it was flagged as potentially hepatotoxic and a hERG channel blocker. Complete details with all predicted values from the tools can be found in ESI.† These findings indicate that while 10g shows promise, careful consideration of its potential hepatic and cardiac effects will be important in its further development.
4. Discussion
In this study, we synthesized and characterized a series of novel thiazole-linked coumarin piperazine derivatives, compounds 10a–10i, utilizing 1H NMR, 13C NMR, and HRMS. These compounds were subsequently evaluated for their inhibitory activity against Gal-1 (Table 1). Among the synthesized compounds, 10a, 10d, 10f, and 10g demonstrated significant inhibitory activity, with >50% inhibition at a concentration of 20 μM. Further analysis was carried out to determine the binding constants using fluorescence spectroscopy. It revealed that compound 10g exhibited the highest affinity for Gal-1, with a binding constant (Ka) of 9.8 × 104 M−1. Further, it also suggested that both aromatic and aliphatic groups attached to the piperazine moiety are well-tolerated, though aromatic groups appear to have a higher affinity, as predicted in prior studies.13,15 Interestingly, compound 10h, which contains a para-nitro substituent, exhibited <10% inhibition of Gal-1. We hypothesize that the para-nitro group may introduce steric hindrance that disrupts the optimal fit of the coumarin moiety within the binding site, thereby reducing its inhibitory potential. This result highlights the importance of steric considerations in the design of effective Gal-1 inhibitors. Even more surprising was the finding that compound 10i, which shares significant structural similarity with compounds 10a and 10b, also showed <10% inhibition. This observation suggests that the presence of a hydrogen bond donor is critical in the absence of an aromatic moiety. This is further supported by the moderate inhibition observed for compounds 10b, 10c and the morpholine-containing 10e, which lack a hydrogen bond donor.
Despite the development of numerous heterocyclic Gal-1 inhibitors, the precise mode of binding has remained largely unexplored. To address this, we employed a combination of molecular docking and RoseTTAFold All-Atom simulations to predict the potential binding modes of compound 10g. Four plausible binding poses were identified and subjected to MD simulations, with each pose being run for 100 ns in replicates of three to ensure the robustness of the findings. It was observed that in three of the four poses, compound 10g dissociated from the Gal-1 binding site and diffused into the bulk solvent, suggesting a lack of stable interactions. However, in pose 15, located within the CBS of Gal-1, compound 10g remained bound throughout the simulation, albeit with a high RMSD value. This elevated RMSD can primarily be attributed to the significant fluctuations of the piperazine moiety (Fig. 7), which failed to maintain consistent interactions with the protein. Nevertheless, the core coumarin-thiazole structure remained bound, interacting with key residues Trp68 and His52, which are crucial for sugar recognition within the binding site.7 These findings also underscore the importance of the hydrogen bond network that natural sugars typically exploit within the CBS of Gal-1. Our data suggest that the piperazine moiety, in its current form, does not adequately mimic these interactions, leading to the observed instability. Future inhibitor designs should focus on incorporating additional hydrogen bond donors or acceptors into the piperazine ring or similar scaffolds to better replicate the hydrogen bonding capabilities of sugars, thereby enhancing their affinity. Additionally, this study highlights the limitations of relying solely on docking scores to select conformations for further investigation. While pose 1 had the highest docking score, it was pose 15, with a least docking score among the four poses that was found to be stable during MD simulations. This discrepancy reinforces the need for comprehensive post-docking analyses, such as MD simulations, to validate and refine docking predictions. Moreover, the importance of conducting MD simulations in replicates was underscored by the observation that, even among the discarded poses, there was at least one run in which the compound remained stable. This variability suggests that relying on a single MD simulation could lead to misleading conclusions.
In summary, the synthesized thiazole-linked coumarin-piperazine hybrids were found to effectively inhibit Gal-1. The study provides significant insights into the binding mode of synthesized compounds within the Gal-1 binding site, particularly highlighting the significance of the coumarin moiety. The findings suggest that future development of heterocyclic Gal-1 inhibitors should prioritize the incorporation of functional groups capable of enhancing the hydrogen bonding network, akin to the hydroxy groups in natural sugars, while leveraging the stabilizing effect of the coumarin scaffold.
Data availability
The 1H NMR, 13C NMR spectra and HRMS data alongside RMSD plots for pose 1, 3 and 5 are provided in ESI.† MD trajectories, along with all necessary parameters, are publicly accessible via the Zenodo repository (10.5281/zenodo.13774410). Any additional data can be obtained from the authors upon through direct correspondence.
Conflicts of interest
The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgments
The authors would like to thank Prof. Jan Brezovsky for granting access to the internal cluster of Laboratory of Biomolecular Interactions and Transport for performing the MD simulations. A part of the computations was also performed at the Poznan Supercomputing and Networking Center.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06715k
References
- Leffler H. Carlsson S. Hedlund M. Qian Y. Poirier F. Introduction to Galectins. Glycoconjugate J. 2002;19(7):433–440. doi: 10.1023/B:GLYC.0000014072.34840.04. doi: 10.1023/B:GLYC.0000014072.34840.04. [DOI] [PubMed] [Google Scholar]
- Hughes R. C. Secretion of the Galectin Family of Mammalian Carbohydrate-Binding Proteins. Biochim. Biophys. Acta, Gen. Subj. 1999;1473(1):172–185. doi: 10.1016/S0304-4165(99)00177-4. doi: 10.1016/S0304-4165(99)00177-4. [DOI] [PubMed] [Google Scholar]
- Mariño K. V. Cagnoni A. J. Croci D. O. Rabinovich G. A. Targeting Galectin-Driven Regulatory Circuits in Cancer and Fibrosis. Nat. Rev. Drug Discovery. 2023;22(4):295–316. doi: 10.1038/s41573-023-00636-2. doi: 10.1038/s41573-023-00636-2. [DOI] [PubMed] [Google Scholar]
- Liu F.-T. Stowell S. R. The Role of Galectins in Immunity and Infection. Nat. Rev. Immunol. 2023;23(8):479–494. doi: 10.1038/s41577-022-00829-7. doi: 10.1038/s41577-022-00829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camby I. Le Mercier M. Lefranc F. Kiss R. Galectin-1: A Small Protein with Major Functions. Glycobiology. 2006;16(11):137R–157R. doi: 10.1093/glycob/cwl025. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- López-Lucendo M. F. Solís D. André S. Hirabayashi J. Kasai K. Kaltner H. Gabius H.-J. Romero A. Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding. J. Mol. Biol. 2004;343(4):957–970. doi: 10.1016/j.jmb.2004.08.078. doi: 10.1016/j.jmb.2004.08.078. [DOI] [PubMed] [Google Scholar]
- Echeverria I. Amzel L. M. Disaccharide Binding to Galectin-1: Free Energy Calculations and Molecular Recognition Mechanism. Biophys. J. 2011;100(9):2283–2292. doi: 10.1016/j.bpj.2011.03.032. doi: 10.1016/j.bpj.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modenutti C. P. Capurro J. I. B. Di Lella S. Martí M. A. The Structural Biology of Galectin-Ligand Recognition: Current Advances in Modeling Tools, Protein Engineering, and Inhibitor Design. Front. Chem. 2019;7:823. doi: 10.3389/fchem.2019.00823. doi: 10.3389/fchem.2019.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynier C. Guerlesquin F. Roche P. Computational Studies of Human Galectin-1: Role of Conserved Tryptophan Residue in Stacking Interaction with Carbohydrate Ligands. J. Biomol. Struct. Dyn. 2009;27(1):49–57. doi: 10.1080/07391102.2009.10507295. doi: 10.1080/07391102.2009.10507295. [DOI] [PubMed] [Google Scholar]
- Blanchard H. Bum-Erdene K. Bohari M. H. Yu X. Galectin-1 Inhibitors and Their Potential Therapeutic Applications: A Patent Review. Expert Opin. Ther. Pat. 2016;26(5):537–554. doi: 10.1517/13543776.2016.1163338. doi: 10.1517/13543776.2016.1163338. [DOI] [PubMed] [Google Scholar]
- Sethi A. Sanam S. Alvala R. Alvala M. An Updated Patent Review of Galectin-1 and Galectin-3 Inhibitors and Their Potential Therapeutic Applications (2016–Present) Expert Opin. Ther. Pat. 2021;31(8):709–721. doi: 10.1080/13543776.2021.1903430. doi: 10.1080/13543776.2021.1903430. [DOI] [PubMed] [Google Scholar]
- Sethi A. Sasikala K. Jakkula P. Gadde D. Sanam S. Qureshi I. A. Talla V. Alvala M. Design, Synthesis and Computational Studies Involving Indole-Coumarin Hybrids as Galectin-1 Inhibitors. Chem. Pap. 2021;75(6):2791–2805. doi: 10.1007/s11696-021-01534-w. doi: 10.1007/s11696-021-01534-w. [DOI] [Google Scholar]
- Sethi A. Sanam S. Alvala M. Non-Carbohydrate Strategies to Inhibit Lectin Proteins with Special Emphasis on Galectins. Eur. J. Med. Chem. 2021;222:113561. doi: 10.1016/j.ejmech.2021.113561. doi: 10.1016/j.ejmech.2021.113561. [DOI] [PubMed] [Google Scholar]
- Nerella S. G. Non-Carbohydrate Galectin-1 Inhibitors as Promising Anticancer Agents: Design Strategies, Structure Activity Relationship and Mechanistic Insights. Eur. J. Med. Chem. Rep. 2024;11:100170. doi: 10.1016/j.ejmcr.2024.100170. [DOI] [Google Scholar]
- Sethi A. Sanam S. Alvala M. New Clues Arising from Hunt of Saccharides Binding to Galectin 3 via 3D QSAR and Docking Studies. Inform. Med. Unlocked. 2020;21:100411. doi: 10.1016/j.imu.2020.100411. doi: 10.1016/j.imu.2020.100411. [DOI] [Google Scholar]
- Zhao H. Reddy Kusuma B. Blagg B. S. J. Synthesis and Evaluation of Noviose Replacements on Novobiocin That Manifest Antiproliferative Activity. ACS Med. Chem. Lett. 2010;1(7):311–315. doi: 10.1021/ml100070r. doi: 10.1021/ml100070r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G. Zhao H. Blagg B. S. J. Design, Synthesis, and Biological Evaluation of Ring-Constrained Novobiocin Analogues as Hsp90 C-Terminal Inhibitors. ACS Med. Chem. Lett. 2015;6(2):204–209. doi: 10.1021/ml5004475. doi: 10.1021/ml5004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A. Alvala M. β-CD/Lewis Acid Dual Catalytic System for Efficient Synthesis of Pyranopyrazoles in Aqueous Environment. Lett. Org. Chem. 2023;20(7):585–595. doi: 10.2174/1570178620666221122121021. doi: 10.2174/1570178620666221122121021. [DOI] [Google Scholar]
- Prato C. A. Carabelli J. Cattaneo V. Campetella O. Tribulatti M. V. Purification of Recombinant Galectins Expressed in Bacteria. STAR Protoc. 2020;1(3):100204. doi: 10.1016/j.xpro.2020.100204. doi: 10.1016/j.xpro.2020.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J. Jyotisha Qureshi R. Jagruthi P. Arifuddin M. Qureshi I. A. Discovery of 8-Hydroxy-2-Quinoline Carbaldehyde Derivatives as Inhibitors for M1 Aminopeptidase of Leishmania Donovani. Int. J. Biol. Macromol. 2024;279:135105. doi: 10.1016/j.ijbiomac.2024.135105. doi: 10.1016/j.ijbiomac.2024.135105. [DOI] [PubMed] [Google Scholar]
- Anandakrishnan R. Aguilar B. Onufriev A. V. H++ 3.0: Automating pK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res. 2012;40(W1):W537–W541. doi: 10.1093/nar/gks375. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. C. Myers J. B. Folta T. Shoja V. Heath L. S. Onufriev A. H++: A Server for Estimating p Ka s and Adding Missing Hydrogens to Macromolecules. Nucleic Acids Res. 2005;33(suppl_2):W368–W371. doi: 10.1093/nar/gki464. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X. Wang H. Li C. Zhang J. Z. H. Ji C. MolGpka: A Web Server for Small Molecule pKa Prediction Using a Graph-Convolutional Neural Network. J. Chem. Inf. Model. 2021;61(7):3159–3165. doi: 10.1021/acs.jcim.1c00075. doi: 10.1021/acs.jcim.1c00075. [DOI] [PubMed] [Google Scholar]
- O'Boyle N. M. Banck M. James C. A. Morley C. Vandermeersch T. Hutchison G. R. Open Babel: An Open Chemical Toolbox. J. Cheminf. 2011;3(1):33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt J. Santos-Martins D. Tillack A. F. Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021;61(8):3891–3898. doi: 10.1021/acs.jcim.1c00203. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PyMOL | pymol.org, https://www.pymol.org/, accessed 2024-08-24
- Krishna R. Wang J. Ahern W. Sturmfels P. Venkatesh P. Kalvet I. Lee G. R. Morey-Burrows F. S. Anishchenko I. Humphreys I. R. McHugh R. Vafeados D. Li X. Sutherland G. A. Hitchcock A. Hunter C. N. Kang A. Brackenbrough E. Bera A. K. Baek M. DiMaio F. Baker D. Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom. Science. 2024;0(0):eadl2528. doi: 10.1126/science.adl2528. doi: 10.1126/science.adl2528. [DOI] [PubMed] [Google Scholar]
- Neurosnap – Computational Biology, Simplified, https://neurosnap.ai/?gad_source=1&gclid=CjwKCAjwiaa2BhAiEiwAQBgyHiATHtuQEi4rMTV-3JynNQGb1e1Y__o6I8Tecfzha5nCnizXLhoUBhoC2poQAvD_BwE, accessed 2024-08-24
- Tian C. Kasavajhala K. Belfon K. A. A. Raguette L. Huang H. Migues A. N. Bickel J. Wang Y. Pincay J. Wu Q. Simmerling C. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020;16(1):528–552. doi: 10.1021/acs.jctc.9b00591. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- Case D. A. Aktulga H. M. Belfon K. Cerutti D. S. Cisneros G. A. Vinicius W. D. Forouzesh N. Giese T. J. Gotz A. W. Gohlke H. Izadi S. Kasavajhala K. Kaymak M. C. King E. Kurtzman T. Lee T.-S. Li P. Liu J. Luchko T. Luo R. Manathunga M. Machado M. R. Nguyen H. M. O’Hearn K. A. Onufriev A. V. Pan F. Pantano S. Qi R. Rahnamoun A. Risheh A. Schott-Verdugo S. Shajan A. Swails J. Wang J. Wei H. Wu X. Wu Y. Zhang S. Zhao S. Zhu Q. Cheatham III T. E. Roe D. R. Roitberg A. Simmerling C. York D. M. Nagan M. C. Merz Jr K. M. J. Chem. Inf. Model. 2023;63(20):6183–6191. doi: 10.1021/acs.jcim.3c01153. doi: 10.1021/acs.jcim.3c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhikara D. J. Yoshida N. Hirata F. Placevent: An Algorithm for Prediction of Explicit Solvent Atom Distribution—Application to HIV-1 Protease and F-ATP Synthase. J. Comput. Chem. 2012;33(18):1536–1543. doi: 10.1002/jcc.22984. doi: 10.1002/jcc.22984. [DOI] [PubMed] [Google Scholar]
- Three-Dimensional Molecular Theory of Solvation Coupled with Molecular Dynamics in Amber | Journal of Chemical Theory and Computation, https://pubs.acs.org/doi/10.1021/ct900460m, accessed 2024-08-24 [DOI] [PMC free article] [PubMed]
- Hopkins C. W. Le Grand S. Walker R. C. Roitberg A. E. Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning. J. Chem. Theory Comput. 2015;11(4):1864–1874. doi: 10.1021/ct5010406. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- Scheurer M. Rodenkirch P. Siggel M. Bernardi R. C. Schulten K. Tajkhorshid E. Rudack T. PyC. Rapid, Customizable, and Visual Analysis of Noncovalent Interactions in MD Simulations. Biophys. J. 2018;114(3):577–583. doi: 10.1016/j.bpj.2017.12.003. doi: 10.1016/j.bpj.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E. V. Blundell T. L. Ascher D. B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. Shi S. Yi J. Wang N. He Y. Wu Z. Peng J. Deng Y. Wang W. Wu C. Lyu A. Zeng X. Zhao W. Hou T. Cao D. ADMETlab 3.0: An Updated Comprehensive Online ADMET Prediction Platform Enhanced with Broader Coverage, Improved Performance, API Functionality and Decision Support. Nucleic Acids Res. 2024;52(W1):W422–W431. doi: 10.1093/nar/gkae236. doi: 10.1093/nar/gkae236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. N. Head M. S. Kulkarni A. LaLonde J. M. Validation Studies of the Site-Directed Docking Program LibDock. J. Chem. Inf. Model. 2007;47(6):2159–2171. doi: 10.1021/ci6004299. doi: 10.1021/ci6004299. [DOI] [PubMed] [Google Scholar]
- Dings R. P. M. Miller M. C. Nesmelova I. Astorgues-Xerri L. Kumar N. Serova M. Chen X. Raymond E. Hoye T. R. Mayo K. H. Antitumor Agent Calixarene 0118 Targets Human Galectin-1 as an Allosteric Inhibitor of Carbohydrate Binding. J. Med. Chem. 2012;55(11):5121–5129. doi: 10.1021/jm300014q. doi: 10.1021/jm300014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A. Rathi B. Artificial Intelligence in Drug Discovery: A Mirage or an Oasis? Drug Discovery Today. 2024;29(6):103994. doi: 10.1016/j.drudis.2024.103994. doi: 10.1016/j.drudis.2024.103994. [DOI] [PubMed] [Google Scholar]
- Knapp B. Ospina L. Deane C. M. Avoiding False Positive Conclusions in Molecular Simulation: The Importance of Replicas. J. Chem. Theory Comput. 2018;14(12):6127–6138. doi: 10.1021/acs.jctc.8b00391. doi: 10.1021/acs.jctc.8b00391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 1H NMR, 13C NMR spectra and HRMS data alongside RMSD plots for pose 1, 3 and 5 are provided in ESI.† MD trajectories, along with all necessary parameters, are publicly accessible via the Zenodo repository (10.5281/zenodo.13774410). Any additional data can be obtained from the authors upon through direct correspondence.