Abstract
Background and purpose
Tumor bed (TB) is the residual cavity of resected tumor after surgery. Delineating TB from CT is crucial in generating clinical target volume for radiotherapy. Due to multiple surgical effects and low image contrast, segmenting TB from soft tissue is challenging. In clinical practice, titanium clips were used as marks to guide the searching of TB. However, this information is limited and may cause large error. To provide more prior location information, the tumor regions on both pre-operative and post-operative CTs are both used by the deep learning model in segmenting TB from surrounding tissues.
Materials and methods
For breast cancer patient after surgery and going to be treated by radiotherapy, it is important to delineate the target volume for treatment planning. In clinical practice, the target volume is usually generated from TB by adding a certain margin. Therefore, it is crucial to identify TB from soft tissue. To facilitate this process, a deep learning model is developed to segment TB from CT with the guidance of prior tumor location. Initially, the tumor contour on the pre-operative CT is delineated by physician for surgical planning purpose. Then this contour is transformed to the post-operative CT via the deformable image registration between paired pre-operative and post-operative CTs. The original and transformed tumor regions are both used as inputs for predicting the possible region of TB by the deep-learning model.
Results
Compared to the one without prior tumor contour information, the dice similarity coefficient of the deep-learning model with the prior tumor contour information is improved significantly (0.812 vs. 0.520, P = 0.001). Compared to the traditional gray-level thresholding method, the dice similarity coefficient of the deep-learning model with the prior tumor contour information is improved significantly (0.812 vs.0.633, P = 0.0005).
Conclusions
The prior tumor contours on both pre-operative and post-operative CTs provide valuable information in searching for the precise location of TB on post-operative CT. The proposed method provided a feasible way to assist auto-segmentation of TB in treatment planning of radiotherapy after breast-conserving surgery.
Keywords: Tumor bed, Radiotherapy, Deep learning, Segmentation
Introduction
For breast cancer patient after surgery and proceeding to radiotherapy, it is important to accurately delineate tumor bed (TB) for radiation treatment planning [1, 2]. However, contouring TB from post-operative CT is affected by many factors including artifacts of titanium clips, size and clarity of seroma, inter-observer variability, surgical specimen volume, etc [3, 4]. In practice TB contour is manually delineated by physicians in referencing tumor location before surgery and the surgical marks on post-operative image [5]. Given the intrinsic characteristics, there are several difficulties in delineating TB from post-operative CT. First, the contrast of soft tissue on CT is relatively low and the clarity of seroma is poor. Also, the high-density marker (lead wires and titanium clips) cause metal artifacts. Second, the contrast between TB and surrounding normal tissue is lower. Third, the size, shape and location of TB varied considerably between patients. In brief, delineating TB from CT is difficult and challenging.
As deep-learning methods popularly used in automatic segmentation of medical image, it also draws attention by researchers in TB segmentation [6, 7]. Certain efforts were made in this field and the results are encouraging. Dai employed a 3D U-Net to segment TB and several organs at risk (OAR) on planning CT and CBCT-generated synthetic CT [8]. The results showed that the tumor bed on synthetic CT is obviously larger than the one manually delineated by physicians. The dice similarity coefficient (DSC) is higher compared with those achieved in traditional medical image segmentation methods. Kazemimoghadam proposed a saliency-based deep-learning method for segmenting TB from CT [9]. It incorporated the salient information provided by titanium clip into the deep-learning model. The DSC is improved comparing with that of Dai’s method.
Differing from Kazemimoghadam’s method which encoded titanium clips’ location information into the model learning, we propose a method to incorporate prior tumor contour information in the deep-learning model for segmenting TB on post-operative CT. Instead of titanium clip related location information, the original tumor contour on pre-operative CT and its transformed contour on post-operative CT are both used in model learning. The rest of the paper is organized as follows. In Sect. 2, the delineation of regions of interests (ROIs), deformable image registration (DIR), and deep-learning model are introduced. In Sect. 3, the performances of the proposed model and the other existing models are reported. In Sect. 4, the strengths and weaknesses of the proposed method are discussed, and future work is prospected.
Materials and methods
Patient dataset
The dataset consisted of 110 Paris of CTs that were obtained when patients were positioned in supine. These patients were diagnosed with early-staged breast cancer and underwent breast-conserving surgery and post-operative radiotherapy in our hospital. The pre-operative CT were acquired about one week before surgery and used for diagnostic purpose. The parameters of pre-operative CT are: pixel size 0.68–0.94 mm, matrix 512 × 512, and slice thickness 5 mm. The post-operative CT were acquired about ten weeks after surgery and used for treatment planning purpose in radiotherapy. The parameters of post-operative CT are: pixel size 1.18–1.37 mm, matrix 512 × 512, and slice thickness 5 mm. All CTs were pre-processed using 3D Slicer [10, 11]. They were first resampled as isotropic resolution of 1 × 1 × 5 mm and then identically cropped to dimensions of 256 × 256 × 32 around the breast’s centroid [12].
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The need for informed consent was waived by the ethics committee/Institutional Review Board of [Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College], because of the retrospective nature of the study. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The patient identification information on these CT data is anonymized before they are processed by the subsequent processing in this study. The patient data is stored and processed in workstation located in our institute. We don’t share and distribute the patient data with the other institutes and organizations.
Delineation of ROIs
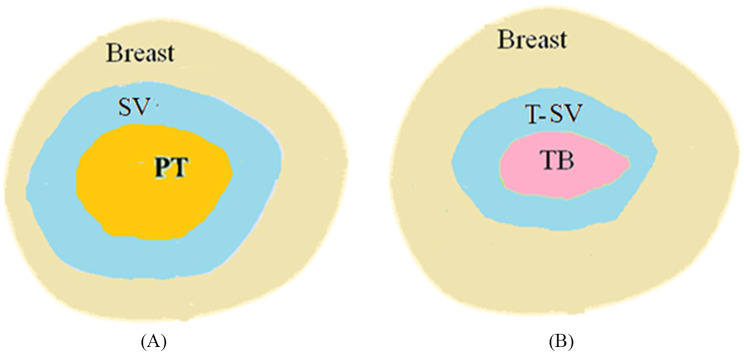
Before surgery, patient is scanned and the contour of primary tumor (PT) is delineated manually by physicians for surgical planning purpose. After surgery, the major tumor volume is resected and its pathological volume (PV) is measured as shown in Fig. 1. Accordingly, the surgical volume (SV) of tumor on pre-operative CT is created based on the PV as illustrated in Fig. 2A. In practice, certain margins (1 –3 cm) are added to PT to form SV in approximating the measured sizes of PV. 2 cm margin is mostly used which result in the size of SV on CT closed to the measured size of PV.
Fig. 1.
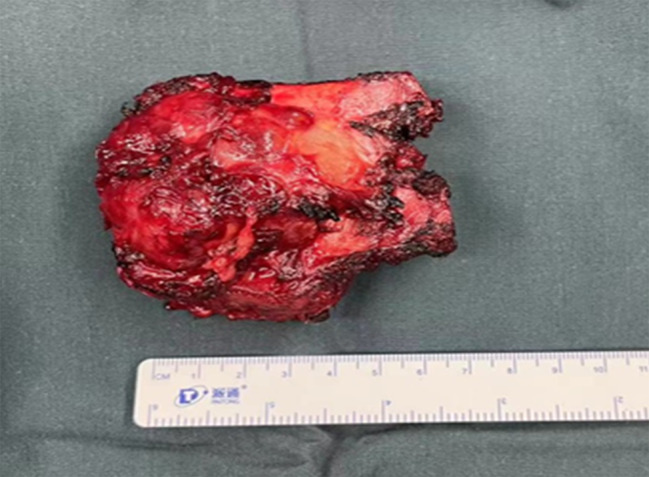
The pathological volume (PV) of the primary tumor (PT) after surgery
Fig. 2.
Illustration of target volume on pre- and post-operative CTs. (A) The primary tumor and surgical volume on pre-operative CT; (B) The tumor bed and transformed surgical volume on post-operative CT
For treatment planning purpose another CT, post-operative CT, is obtained after 3–6 months of surgery. The important ROIs are illustrated in Fig. 2B. Due to complex surgical effects during this period, such as seroma and fibrosis, the volume of TB varies rapidly and changes considerably between patients. Also, the contrast of TB is nearly identical to the surrounding soft tissue which is hardly identified visually. In clinical practice, TB is usually contoured based on the marks such as surgical clips, seroma, and fibrosis. Also, the contour of PT and SV on pre-operative CT provides an important guidance in searching for the possible contour of TB on post-operative CT.
As TB is the region surrounds the SV, the contour of SV on pre-operative CT should overlap with the contour of TB on post-operative CT. Therefore, in clinical practice the contour of SV is usually mapped onto the post-operative CT. With the mapped region of SV in mind, physicians can derive TB contour based on the few visible surgical marks. Thus the transformed SV (T-SV) contour on post-operative CT is a good indication of the potential TB contour. For this purpose, the pre-operative and post-operative CTs are aligned via DIR. The resulted deformation vector field (DVF) is then applied to the SV contour on pre-operative CT to create the T-SV contour on post-operative CT as shown in Fig. 2B.
Deformable image registration
The intensity-based B-Splines registration algorithm is used to align pre-operative CT with post-operative CT. The resulted DVF is applied to transform the SV contour on pre-operative CT to T-SV contour on post-operative CT. The two contours then act as prior information in the deep-learning model. To achieve this goal, the similarity metric between both CTs is used to evaluate the quality of image alignment. In our work mutual information is employed as it is suitable for the situations in which intensities of the corresponding structures are inherently different. The registration code was developed based on the functions provided by Elastix registration software (https://elastix.lumc.nl) [13, 14].
Initially, the images are registered by rigid and affine transformation. Then, multi-resolution strategy for B-Splines transformation is performed. Gaussian pyramid (3 scales) is used to smooth and down-sample the image at different scales. The control points’ grid size is set to 12. Each scale’s grid space is set to [4 2 1] times a physical unit (mm). The larger grid size is used to match larger structures and skip smaller structures, while the smaller grid size is used to match detailed structures. An iterative stochastic gradient descent method is used for optimization in each scale. The desktop computer equipped with Inter® dual core processor (3.0 GHz) is used to perform all the tasks of image registrations.
Deep-learning model
V-Net is a popular model used to segment objects from background in computer vision and medical imaging [15–18]. It is specially designed for volumetric images instead of planar images in segmentation application, such as U-Net [19–21]. It composed of a contractive and an expanding path which aims to build a bottleneck in its centermost part through a combination of convolution and down-sampling. After the bottleneck, the image is reconstructed through a combination of convolution and up-sampling. To improve the training, skip connections are added to assist the backward flow of gradients.
As both SV and T-SV provide prior tumor contour information, their effects on CT images are enhanced. In practice, the values of pixels of SV and T-SV are multiplied by an integer number such as 25, while the values of pixels outside SV and T-SV are multiplied by a fraction number such as 0.1. After the preprocessing of image enhancement, the regions of SV and T-SV are highlighted on CT images and become more visible by the deep-learning model. In this study, there were two 3D input channels (pre-operative and post-operative CTs) and one 3D output channel (for predicted label image) in the deep-learning model. To improve the capability of the model, a five-fold cross-validation strategy is applied to the 110 patient data. In the process of cross-validation, three folds (66 patients) are used for training and one fold (22 patients) is used for validation to tune hyper-parameters, and the remaining one fold (22 patients) is used for testing.
The weights of convolution layers are initialized by a normal distribution according to the published studies [19]. The loss function used for model training is DSC [20]. The Adaptive moment estimation (Adam) optimizer with batch size of 4 and weight decay of 3e-5 is used [22]. The main parameters are described as follows: initial learning rate 0.0005, learning rate drop factor 0.95, and validation frequency 20. The V-Net model is implemented on Matlab (MathWorks, Natick, MA 01760) and trained with maximal 500 epochs. The experiments are performed on a workstation equipped with one NVIDIA Geforce GTX 1080 TI GPU and 64GB DRR RAM. It would be more favorable if the advanced GPU, such as NVIDIA Geforce GTX 4090, is used in the test and the learning time would be greatly reduced. Actually, the training of V-Net is time-consuming but can be performed at background nightly. When the model is trained and applied to the new case, the predication task of tumor bed can be completed within few seconds. Therefore, it would be more convenient to install the trained V-Net model on a personal computer, such as laptop and desktop, and use it to perform the segmentation task on the new cases in clinical setting.
Workflow of auto-segmentation
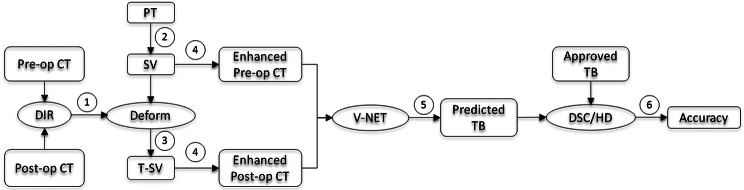
The workflow of the prior information guided auto-segmentation process is shown in Fig. 3:(1) DIR is performed on both pre-operative and post-operative CTs and the DVF is achieved; (2) PT is obtained from pre-operative CT by physician for surgical planning purpose, and SV contour is generated from PT by adding a certain margin; (3) SV contour on pre-operative CT is transformed onto post-operative CT via DVF to create T-SV; (4)The SV and T-SV contours are enhanced on pre-operative and post-operative CTs, respectively; (5) These CTs with enhanced contours are processed by the deep-learning model and the TB contour is predicted on post-operative CT; (6) The similarity coefficients (DSC and HD) between the predicted and clinically approved TB contours is computed and the segmentation accuracy is evaluated.
Fig. 3.
The workflow of prior information guided auto-segmentation of TB. Pre-op: Pre-operative; Post-op: Post-operative
Experiments
The segmentation accuracy was quantified by the averaging errors of five-fold cross-validation results. The similarity between the predicted and clinically approved TB contours is assessed with dice similarity coefficient (DSC) [23] and 95% percentile Hausdorff distance (HD95) [24]. In order to validate the effectiveness of prior contour information on the high accuracy of the segmentation model, the ablation study is performed by four V-Net models with different input CTs. The input of V-Net Model 1 consists of both sets of the original pre-op CT and post-op CT. The input of V-Net Model 2 consists of pre-op CT with the enhanced SV contour and the original post-op CT. The input of V-Net Model 3 consists of the original pre-op CT and the post-op CT with the enhanced T-SV contour. The input of V-Net Model 4 consists of the pre-op CT with the enhanced SV contour and the post-op CT with the enhanced T-SV contour. Note that Model 4 is the primary model used in the following testing.
In addition, the deep-learning model is also compared with the traditional gray-level thresholding method [25] and the other three existing deep-learning methods [8, 9, 20]. Gray-level thresholding method generates a binary image from a given gray-scale image by separating it into two regions based on a threshold value. For statistical comparison, if the data are in normal distribution, the paired t-test is performed. Otherwise, if the data are in abnormal distribution, the Wilcoxon signed-rank test for paired samples (non-parametric test) is performed. P < 0.05 is regarded as statistically significant. R Project for Statistical Computing (version 3.6.3) is used for statistics analysis.
Results
Ablation test
The results of V-Net Models with and without prior contour information are compared in Table 1. Among the four models with different input CTs, V-Net model 4 has the best performance. The average values of DSC (mean ± standard deviation) are 0.812 ± 0.053 and 0.520 ± 0.095 for V-Net Model 4 (with prior contour information) and Model 1 (without prior contour information). The average values of HD95 (mean ± standard deviation) are 18.243 ± 5.135 for V-Net Model 4 and 59.570 ± 26.012 for V-Net Model 1. The Shapiro-Wilk normality test confirmed that the results are in normal distribution, thus the paired t-test is employed for statistical comparison. Statistically significant differences between V-Net Model 4 and V-Net Model 1, respectively are found in DSC (0.812 vs. 0.520, P = 0.001) and HD95 (18.243 vs. 59.570, P = 0.002).
Table 1.
The result of ablation tests with different inputs
| Types of prior contour information | DSC (%) | HD95 (mm) |
|---|---|---|
| V-Net Model 1: [Pre-op CT & Post-op CT] | 0.520 ± 0.095 | 59.570 ± 26.012 |
| V-Net Model 2: [Pre-op CT with SV & Post-op CT] | 0.751 ± 0.081 | 46.876 ± 21.265 |
| V-Net Model 3: [Pre-op CT & Post-op CT with T-SV] | 0.765 ± 0.078 | 39.240 ± 15.790 |
| V-Net Model 4:[Pre-op CT with SV & Post-op CT with T-SV] | 0.812 ± 0.053 | 18.243 ± 5.135 |
The average values of DSC (mean ± standard deviation) are 0.751 ± 0.081 for V-Net Model 2 (with SV contour on pre-operative CT) and 0.765 ± 0.078 for V-Net Model 3 (without T-SV contour on post-operative CT). While The average values of HD95 (mean ± standard deviation) are 46.876 ± 21.265 for V-Net Model 2 (with SV contour on pre-operative CT) and 39.240 ± 15.790 for V-Net Model 3 (without T-SV contour on post-operative CT). The DSCs and HD95s of V-Net Model 2 and V-Net Model 3 are both higher than that of V-Net Model 1 and lower than that of V-Net Model 4. The result shows that the inclusion of prior tumor contour information in V-Net model can improve the prediction accuracy of TB contour significantly.
Model comparison
The average values of DSC (mean ± standard deviation) is 0.633 ± 0.086 for the traditional gray-level thresholding method. The average value of HD95 (mean ± standard deviation) is 58.275 ± 27.659 for the traditional gray-level thresholding method. The Shapiro-Wilk normality test confirmed that the results are in normal distribution, thus the paired t-test is employed for statistical comparison. Statistically, significant differences between V-Net Model 4 (with prior contour information) and the traditional gray-level thresholding method are also found in DSC (0.812 vs. 0.633, P = 0.0005) and HD95 (18.243 vs. 58.275, P = 0.001).
The result of V-Net Model 4 is also compared with the other three existing deep-learning models, Res-SE-U-Net [8], SDL-Seg-U-Net [9] and U-Net [20], recently published for TB segmentation. As the data and source codes for them are not publically available, we can only assess them based on the published results. As shown in Table 2, the two similarity metrics, DSC and HD95, and the sizes of training and test sets for each model are listed. V-Net Model 4 has the highest DSC, while SDL-Seg-U-Net [9] has the lowest HD95 among four models. Res-SE-U-Net [8] has the largest training and test sets but from two institutions. The CT and contours from two different institutions may vary considerably which cause lower DSC and higher HD95 of this model.
Table 2.
The comparison of the proposed and existing deep-learning models for TB segmentation task
Discussion
In this study, a deep-learning model was developed for auto-segmenting TB from post-operative CT. The results show that the prior tumor contour information provides important information in searching for the precise location of TB from low-contrast soft tissue on post-operative CT. This improvement of DSC would be attributed to the introduction of SV and T-SV contours in the deep-learning model, which brings the high-probability region of TB on post-operative CT. The improvement of segmentation accuracy also means that planning target volume can be more close to the real volume and then the radiation dose can be more precisely delivered to it. This would be favorable for promoting tumor control probability and lowering normal tissue complication probability in clinical radiotherapy. In addition to the introduction of prior tumor contour information, the image enhancement technique also brings a simple way to burn footprint on CT for more efficient network training. Without it, another two channels for SV and T-SV masks are needed in addition to the two existing CT channels, which would cause increased cost in network training.
The deep-learning model was initially used in auto-segmentation of TB on planning/synthetic CT and the result was not desired.8 Later, Kazemimoghadam incorporated the salient information provided by titanium clip in deep-learning model and the result was improved.9 In our study DSC is further improved compared with the previous reports. Compared with the DSC (0.63 ± 0.08) of Dai’s method, the DSC (0.764 ± 0.027) of Kazemimoghadam’s method shows a 21% improvement and the DSC (0.812 ± 0.053) of our method shows a 28% improvement. It should be noted that both Kazemimoghadam and our methods incorporated prior information in the deep-learning model to assist the refinement of more precise contour of TB. Our prior contour information covers more region than that of Kazemimoghadam’s method. The proposed segmentation method not only provides an accurate tool for TB contouring, also improves the accuracy of treatment planning and delivery of radiotherapy. As suggested by clinical trials, the high contour quality improves patient survival [26].
To implement the deep-learning model in clinic, both pre-operative and post-operative CTs are extracted from clinical radiotherapy treatment planning system, such as Eclipse™ (Varian Medical System). These CT images are handled by an in-house developed program for subsequent image processing. First, DVF is obtained by performing DIR on both sets of CTs. Then, the SV contour is automatically created based on the PT contour with a given margin and mapped onto post-operative CT via DVF to form T-SV contour. Next, the SV and T-SV contours are enhanced on both sets of CTs for model learning. Finally, the TB contour on post-operative CT is predicted by the deep-learning model. The predicted TB contour is saved in DICOM RT structure set by the in-house developed program and imported to the treatment planning system for clinical use.
So far, we implemented a V-NET model with prior tumor contour information in auto-segmentation of the TB contour. There are certain aspects to be improved for this method in regard to the input data and model configuration. First, the SV contour can be generated more precisely from the PV with additional imaging. In previous study the SV contour is simply estimated from the PT contour by adding a given margin. This is not the true volume of SV. If feasible the on-site imaging (CT/CBCT/MRI) of PV can be performed in surgery room and then fuse with the pre-operative CT, this will make the SV more close to the real one. Second, the attention mechanism should be introduced to calculate different weight parameters for the input feature map, so that the model pays more attention to key information and ignores irrelevant information such as background. For this purpose, the model architecture could be changed by adding attention modules, including channel self-Attention module, spatial self-Attention module, and hybrid/dual attention module [27].
There are several limitations in this study. First, the dataset is limited, which requires massive cross-validation to ensure the stability of deep-learning model. More data will be collected in the future to improve the generalization and robustness of the model. Second, the traditional intensity-based DIR is limited for certain clinical scenarios, especially when organs suffering from not only deformation but also mass changes. This mass change could cause difficulty in mapping voxel-to-voxel, such as in the case of tumor removal. The mechanical model based method, such as finite element method (FEM), computes a deformation field to satisfy both the elasticity constraint and the similarity metric constraint, which could be a better solution for this case. Third, MRI scan is not available in this study. Since soft tissue contrast on MRI is better than CT, the tumor bed tissue would be more visible on MRI than that on CT. Thus it is beneficial to include MRI in the deep-learning model in the future. Fourth, this method is only tested on the site of breast. Tumor bed after surgery on the other sites, such as pelvic and oral tumor, would be tested in the future. As the tumor bed volumes are larger and image contrast of these sites is better than the one in the site of breast, we are optimistic about the future of applying this method in these clinical scenarios.
Conclusions
Incorporating the prior tumor contour information in the deep-learning model can effectively improve the segmentation accuracy of TB. Comparing with Dai’s and Kazemimoghadam’s methods, the DSC of the proposed method increases with 28% and 6%, respectively. Instead of point-related location information such as titanium clips, the region-related information such as tumor contour is utilized in training deep-learning model in this study. The proposed method demonstrated a feasible way to auto-segment TB from CT in assisting the physician to delineate target volume for treatment planning in breast cancer radiotherapy.
Acknowledgements
Not applicable.
Author contributions
PH, JS and XX performed the experiments and have made substantial contributions to the conception. PH and XX provided study materials or patients. JS and XX collected the data. PH and JS analyzed and interpreted the data. XX and HY supervised the whole study. All authors wrote and have approved the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (No. 11975312), Beijing Municipal Natural Science Foundation (No.7202170), Joint Funds for the Innovation of Science and Technology, Fujian province (No.2023Y9442), Fujian Provincial Health Technology Project (No. 2023GGA052) and National High Level Hospital Clinical Research Funding (No. 2022-CICAMS-80102022203). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The need for informed consent was waived by the ethics committee/Institutional Review Board of [Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College], because of the retrospective nature of the study. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The institutional Ethics Committee of the Cancer Hospital, Chinese Academy of Medical Sciences, and Peking Union Medical College approved this study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peng Huang, Hui Yan and Jiawen Shang contributed equally to this work.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Litiere S, Werutsky G, Fentiman IS, Rutgers E, Christiaens MR, Van Limbergen E, Baaijens MH, Bogaerts J, Bartelink H. Breast conserving therapy versus mastectomy for stage I-II breast cancer: 20 year follow-up of the EORTC 10801 phase 3 randomised trial. Lancet Oncol. 2012;13(4):412–9. [DOI] [PubMed] [Google Scholar]
- 3.van Mourik AM, Elkhuizen PH, Minkema D, Duppen JC, Dutch Young Boost Study G, van Vliet-Vroegindeweij C. Multiinstitutional study on target volume delineation variation in breast radiotherapy in the presence of guidelines. Radiother Oncol. 2010;94(3):286–91. [DOI] [PubMed] [Google Scholar]
- 4.Major T, Gutierrez C, Guix B, van Limbergen E, Strnad V, Polgar C, Breast Cancer Working Group of G-E: Recommendations from GEC ESTRO Breast Cancer Working Group (II). Target definition and target delineation for accelerated or boost partial breast irradiation using multicatheter interstitial brachytherapy after breast conserving open cavity surgery. Radiother Oncol. 2016;118(1):199–204. [DOI] [PubMed] [Google Scholar]
- 5.Landis DM, Luo W, Song J, Bellon JR, Punglia RS, Wong JS, Killoran JH, Gelman R, Harris JR. Variability among breast Radiation oncologists in Delineation of the Postsurgical Lumpectomy Cavity. Int J Radiation Oncology*Biology*Physics. 2007;67(5):1299–308. [DOI] [PubMed] [Google Scholar]
- 6.Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):640–51. [DOI] [PubMed] [Google Scholar]
- 7.Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481–95. [DOI] [PubMed] [Google Scholar]
- 8.Dai Z, Zhang Y, Zhu L, Tan J, Yang G, Zhang B, et al. Geometric and dosimetric evaluation of Deep Learning-based automatic delineation on CBCT-Synthesized CT and planning CT for breast Cancer adaptive radiotherapy: a multi-institutional study. Front Oncol 2021;11:725507. [DOI] [PMC free article] [PubMed]
- 9.Kazemimoghadam M, Chi W, Rahimi A, Kim N, Alluri P, Nwachukwu C, Lu W, Gu X. Saliency-guided deep learning network for automatic tumor bed volume delineation in post-operative breast irradiation. Phys Med Biol. 2021;66(17):175019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, et al. 3D slicer as an image computing platform for the quantitative Imaging Network. Magn Reson Imaging. 2012;30(9):1323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinter C, Lasso A, Wang A, Jaffray D, Fichtinger G. SlicerRT: radiation therapy research toolkit for 3D slicer. Med Phys. 2012;39(10):6332–8. [DOI] [PubMed] [Google Scholar]
- 12.Xie X, Song Y, Ye F, Yan H, Wang S, Zhao X, Dai J. Prior information guided auto-contouring of breast gland for deformable image registration in postoperative breast cancer radiotherapy. Quant Imaging Med Surg. 2021;11(12):4721–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29(1):196–205. [DOI] [PubMed] [Google Scholar]
- 14.Shamonin DP, Bron EE, Lelieveldt BP, Smits M, Klein S, Staring M. Alzheimer’s Disease Neuroimaging I: fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front Neuroinform. 2013;7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milletari F, Navab N, Ahmadi S-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV): 2016 2016; 2016: 565–571.
- 16.Orlando N, Gillies DJ, Gyacskov I, Romagnoli C, D’Souza D, Fenster A. Automatic prostate segmentation using deep learning on clinically diverse 3D transrectal ultrasound images. Med Phys. 2020;47(6):2413–26. [DOI] [PubMed] [Google Scholar]
- 17.Hsu DG, Ballangrud A, Shamseddine A, Deasy JO, Veeraraghavan H, Cervino L, Beal K, Aristophanous M. Automatic segmentation of brain metastases using T1 magnetic resonance and computed tomography images. Phys Med Biol 2021, 66(17). [DOI] [PMC free article] [PubMed]
- 18.Wang T, Lei Y, Tian S, Jiang X, Zhou J, Liu T, Dresser S, Curran WJ, Shu HK, Yang X. Learning-based automatic segmentation of arteriovenous malformations on contrast CT images in brain stereotactic radiosurgery. Med Phys. 2019;46(7):3133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W, editors. Medical image computing and computer-assisted intervention – MICCAI 2016. Lecture notes in computer science, Springer, Cham, 2016. p. 424–32. 10.1007/978-3-319-46723-8_49.
- 20.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv:150504597 [cs] 2015.
- 21.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. arXiv:1512.00567. 2015.
- 22.Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014.
- 23.Dice LR. Measures of the Amount of Ecologic Association between Species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 24.Huttenlocher DP, Rucklidge WJ, Klanderman GA. Comparing images using the Hausdorff distance under translation. In: Proceedings 1992 IEEE Computer Society Conference on Computer Vision and Pattern Recognition: 15–18 June 1992 1992; 1992: 654–656.
- 25.Al-Bayati M, El-Zaart A. Mammogram images thresholding for breast Cancer detection using different thresholding methods. Adv Breast Cancer Res. 2013;2(3):72–7.
- 26.Chua DT, Sham JS, Kwong DL, Tai KS, Wu PM, Lo M, Yung A, Choy D, Leong L. Volumetric analysis of tumor extent in nasopharyngeal carcinoma and correlation with treatment outcome. Int J Radiat Oncol Biol Phys. 1997;39(3):711–9. [DOI] [PubMed] [Google Scholar]
- 27.Hu R, Yan H, Nian F, Mao R, Li T. Unsupervised computed tomography and cone-beam computed tomography image registration using a dual attention network. Quant Imaging Med Surg. 2022;12(7):3705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.