Abstract
Background
Silver nanomaterials have been widely proven to have antifungal effects against Trichosporon asahii. However, the antifungal mechanism of silver nanomaterials with different morphologies still needs to be explored.
Methods
Herein, the antifungal effect of silver nanomaterials against fungus was comparative investigated via silver nanowires and silver nanoparticles with a similar size (30 nm).
Results
The optimal antifungal concentration of silver nanowires is 6.24 μg/mL, meanwhile the antifungal concentration of silver nanoparticles is 100 μg/mL. The silver nanowires are significantly superior to the silver nanoparticles. SEM and TEM results indicated that both silver nanoparticles and silver nanowires showed significant morphological changes in the mycelium of the strain, compared with the control. The lower MFC value of silver nanowires indicates good sterilization effect and suitability for eradication treatment, which is slower than that of silver nanoparticles. Moreover, we also investigated the toxicological effects of silver nanoparticles and silver nanowires.
Conclusion
We comparative studied and transcriptomic analyzed the antifungal mechanism of Ag nanoparticles and nanowires against Trichosporon asahii. The antifungal effects of silver nanowires were better than the silver nanoparticles, especially in the metabolic processes and oxidative phosphorylation. RNA sequencing results indicated that 15 key targets were selected for experimental verification to interpret the potential antifungal mechanism of Ag nanomaterials against fungus. This work proves that silver nanomaterials with different morphologies have potential applications in fungus therapy such as T. asahii.
Keywords: Ag, nanoparticles, nanowires, fungus, antifungal mechanism, Trichosporon asahii, transcriptomics
Introduction
Trichosporon asahii is a yeast-like basidiomycete, a conditionally pathogenic fungus is widely present in humans and nature.1–3 In the past 20 years, there has been an increasing number of patients infected with Trichosporon asahii, especially those with immunodeficiency, malignancy and hematological diseases.3–5 Invasive fungal infections caused by T. asahii can result in a mortality rate of up to 70%, posing an increasing threat to humans.6 At present, there are only a few antifungal agents for invasive fungal infections, including azoles, echinocandins, and polyenes.7,8 Moreover, each type of antifungal drug has only one antibacterial mechanism, with azoles disrupting ergosterol synthesis, echinocandins causing fungal cell wall abnormalities, and polyenes disrupting ergosterol structure.9,10 Moreover, fungi were prone to producing variants targeting each antibacterial mechanism, leading to drug resistance.11 The emergence of fungal drug resistance is a complex process involving multiple factors, such as genetic variations in the fungus itself, drug usage methods, and the patient’s immune status. Therefore, it is important to explore the potential antifungal mechanism against fungus.
In the literature, T. asahii was naturally resistant to echinocandins and had poor sensitivity to amphotericin B.12,13 Therefore, azole antifungal drugs have wide applications in the treatment for invasive T. asahii.14–16 However, the development speed of antifungal drugs is far from keeping up with the emergence of drug resistance. Therefore, there is an urgent need to develop novel antifungal drugs to address the increasing threat of antifungal resistance.17,18 Silver nanomaterials have been widely proven to have antifungal effects.19–21 The antifungal effect of silver nanomaterials was achieved through various mechanisms, such as mechanical damage mechanism, oxidative stress mechanism, ion release mechanism, etc.22–24 This is also the reason why silver nanomaterials are less prone to drug resistance. However, there are many reports on the impact of the size of silver nanomaterials on their antifungal effect,25,26 but few reports on the impact of morphologies of silver nanomaterials.27,28 In the literature, El-Zahry reported that hexagonal Ag nanoparticles displayed the highest antibacterial effect when compared to other nanoparticle shapes, with triangular Ag nanoparticles exhibiting no antibacterial effect under the adopted conditions.29
Gene sequencing technology, proteomics, cell biology technology, molecular biology technology, and RNA sequencing technology are widely used to explore the molecular mechanisms of organisms in developmental, physiological, or pathological states.30 Compared with other methods, RNA sequencing has the advantages of high throughput, comprehensiveness, accurate quantification, dynamic monitoring, and discovery of new genes and transcripts, which has become one of the important means of modern biological research.31 For example, Kafantaris et al32 investigated the antibacterial effects of pine honey against P. aeruginosa PA14 at the molecular level using a global transcriptome approach via RNA-sequencing, indicating that multiple mechanisms of action were implicated in antibacterial activity exerted by pine honey against P. aeruginosa. Shaaban et al33 reported antibacterial activities of hexadecanoic acid methyl ester and green-synthesized silver nanoparticles against multidrug-resistant bacteria after 2, 4, and 6 days in light and dark conditions. The silver nanoparticles possessed good inhibition activity against the tested pathogenic bacteria. In the literature, it reported that the antibacterial effect of silver nanomaterials mainly depends on its nanoscale size effect and the release of silver ions and the bioaccumulation of silver nanomaterials in living organisms is mainly influenced by factors, such as particle size, concentration, and dispersibility.34–36 However, there is no report about the antifungal effects of silver nanomaterials with different morphologies against fungus (T. asahii) via RNA sequencing.
Silver nanomaterials are reported to exhibit excellent antibacterial properties, but its toxicological effects cannot be ignored.37–39 In the literature, Wang et al reported the first demonstration of the bioaccumulation and speciation of Ag nanoparticles in a marine organism (N. virens).40 Yang et al found that ionic strength significantly enhanced the reproductive toxicity and neurotoxicity of silver nanoparticles in Caenorhabditis elegans.41 However, Lehmann et al reported that crumpling of silver nanowires by endolysosomes strongly reduces toxicity and nanowire-bending stiffness controls the cytotoxicity of silver nanowires to nonimmune cells from humans, mice, and fish.42 Therefore, when studying and applying silver nanomaterials, it is necessary to fully consider its antibacterial mechanism and toxicological properties, as well as its bioaccumulation and exposure pathways in different cell models and organisms.
In this work, we select silver nanowires and silver nanoparticles with a similar size (30 nm) and elucidate their antifungal mechanism of silver nanomaterials with different morphologies at the molecular levels via RNA sequencing. The optimal antifungal concentration of silver nanowires are 6.24 μg/mL, meanwhile the silver nanoparticles is 100 μg/mL. The silver nanowires is significantly superior to the silver nanoparticles. SEM and TEM results indicated that both silver nanoparticles and silver nanowires showed significant morphological changes in the mycelium of the strain, compared with the control. The antifungal effects of silver nanowires were better than the silver nanoparticles, especially in the metabolic processes and oxidative phosphorylation. Fifteen key targets were selected for experimental verification to interpret the potential antifungal mechanism of Ag nanomaterials against fungus. This work proves that silver nanomaterials with different morphologies have potential applications in the treatment of T. asahii.
Materials and Methods
Characterizations
The morphology of samples was observed by environmental scanning electron microscopy (ESEM, Quanta 200 FEI, Netherlands) and scanning electron microscopy (SEM, Tecnai F20 FEI, Netherlands). The morphology and size of samples were characterized by transmission electron microscope (TEM, H-7650B Hitachi, Japan).
Materials
Silver nanoparticles (1 mg/mL, 30 nm) and silver nanowires (20 mg/mL, 30 nm) are customized by Nanjing Xianfeng Nanotechnology Company. The silver nanoparticles were synthesized employed AgNO3 as Ag sources, sodium citrate as reducing agent by seed growth method. The silver nanowires were obtained employed polyhydric alcohol as reducing agent to control the growth direction of silver nanocrystals. Fluconazole was purchased from Beijing Solarbio Technology Co., Ltd. Three strains of Trichosporon asahii were preserved in the dermatology laboratory of the Seventh Medical Center of the People’s Liberation Army General Hospital. One is a sensitive strain of Trichosporon asahii (705), one is a clinical drug-resistant strain of Trichosporon asahii (705R), and one is an in vitro induced drug-resistant strain of Trichosporon asahii (705I). The drug resistance of the strains was confirmed by in vitro drug sensitivity tests,43,44 and these strains were stored in the laboratory at −80°C. Before the experiment, the strain was inoculated onto potato glucose agar (PDA) and subcultured at 35°C to maintain its optimal vitality.
Cell Cytotoxicity
The 3-(4.5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was employed to examine the cytotoxicity of different shapes of silver nanomaterials on the growth of non-malignant fibroblast cells. Concisely, various concentrations of silver nanoparticles and nanowires (0, 1.56, 3.12, 6.24, 12.5, 25, 50, 100 μg) were added to the fibroblast cell lines. After 24 h of incubation in Dulbecco’s modified eagle medium (DMEM) media, the cells were analyzed. The 10 μL of MTT reagent was mixed into each well for 2 h and then the formed formazan depositions were cleared by the addition of dimethyl sulfoxide (DMSO), and absorbance was taken at 570 nm.
Antifungal Susceptibility
Measurement of Minimum Inhibitory Concentration (MICs)
Measurement of MIC refers to the implementation of the M27-A3 standardization plan released by Clinical and Laboratory Standards Institute (CLSI). The silver nanoparticle (1 mg/mL, 30 nm) and silver nanowires colloid (20 mg/mL, 30 nm) were dispersed into distilled water, and the original solution of fluconazole (2.56 mg/mL) was also dissolved in sterile double distilled water. Then, the working solution was prepared by diluting the backup solution with Roswell Park Memorial Institute (RPMI) 1640 medium (pH 7.0 with 0.165 M morpholinepropanesulfonic acid [MOPS]). In the 96-well plate, the concentrations of silver nanoparticles and silver nanowires were 100 μg/mL, 50 μg/mL, 25 μg/mL, 12.5 μg/mL, 6.24 μg/mL, 3.12 μg/mL, 1.56 μg/mL, 0.78 μg/mL, 0.39 μg/mL, and 0.195 μg/mL, respectively. The highest concentration of fluconazole was 256 μg/mL, and the lowest concentration was 0.5 μg/mL. When drug sensitivity test results are obtained using yeast micro dilution method, the interpretation standard of MIC value for azole drugs is the minimum drug concentration that can be suppressed by 50% using the micro method. For comparison, the MIC values of silver nanomaterials and fluconazole are uniformly determined using the minimum drug concentration that can be suppressed by 50% using the micro method, with an interpretation time of 48 h. SPSS27.0 statistical software was used to process the collected experimental data. An unpaired test with Welch’s correction was used to analyze the difference between the two groups.
Determination of Minimum Fungicidal Concentration (MFCs)
In vitro drug susceptibility testing (MIC and MFC) is conducted using a 96 well plate. The preparation of MIC and MFC fungicidal suspensions is the same. According to the fungal in vitro drug sensitivity test, the fungicidal suspension after MIC determination was further cultured for 24 h. 20 µL of fungicidal suspension was taken from each concentration test well and the drug-free control well, and poured into the Sabouraud glucose agar plate. After cultivation at 35°C for 48 h, viable fungal were counted. The minimum drug concentration less than 3 CFU was MFC. More research suggests that compared with MIC, MFC can more accurately reflect the efficacy of antifungal drugs.
Statistical Methods for MIC and MFC Results
SPSS27.0 statistical software is used to process the collected experimental data. The three sets of data for MIC were analyzed using one-way ANOVA and Dunnett T3 post hoc test, while MFC was analyzed using two sample t-test. An unpaired t-test with Welch’s correction was used to analyze the difference between the two groups.
Transcriptome Sequencing Analysis
RNA Sequencing
The samples were divided into control, Treat 1 (silver nanoparticles treatment group), and Treat 2 (silver nanowires treatment group), with three replicates for each group. The clinical drug-resistant strain (705R) of T. asahii was adjusted to a bacterial suspension with a Mach turbidity of 0.5. About 150 μL was taken and placed in a 150 mL yeast peptone dextrose (YPD) shaker at 35°C and 120 rpm for 12 h. An appropriate amount of T. asahii was taken, and silver nanoparticles and silver nanowires were added to achieve a final concentration of 100 μg/mL for silver nanoparticles and 6.24 μg/mL for silver nanowires. After counting shaking the bed at 30°C and 120 rpm for 8 h, cells are collected. After cleaning with PBS solution, centrifuging three times, freezing with liquid nitrogen, storing in a refrigerator at −80°C, and finally Treat 1 and Treat 2 are obtained.
Total RNA was extracted from the control and treated T. asahii named control, Treat 1, and Treat 2 tissues using TRIzol® Reagent according the manufacturer’s instruction. RNA quality was determined by 5300 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Then the RNA Purification, reversed transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd (Shanghai, China). The raw data of transcriptomics sequencing was uploaded into the National Genomics Data Center (NGDC: https://ngdc.cncb.ac.cn/) and can be accessible through GSA accession number CRA024183.
Differential Expression Analysis
Differential expression analysis between two groups (Treat 1 vs Control, Treat 2 vs Control, Treat 2 vs Treat 1) was performed by the “limma” package in R. Differential expressed genes (DEGs) met the criteria (|log2(Fold Change)| > 0.58 and p value < 0.05) were screened out between groups.
Enrichment Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the DEGs between the Treat 1 vs Control groups, Treat 2 vs Control groups, and Treat 1 vs Treat 2 groups were carried out with the DAVID database, and visualized with “ggplot2” package in R.
Network Construction and Key Genes Screening
The protein–protein network (PPI) of DEGs between the different groups was construction by STRING database (https://cn.string-db.org/) with the criteria of combined score >0.7, and visualized in Cytoscape 3.7.2 platform. Then, topological parameter of nodes and interaction edges in the networks were calculated by cytoHubba plug-in, and nodes with top degree of networks between groups were considered as key genes. Then the enrichment of nodes in PPI networks was performed by Metascape (https://metascape.org/gp/index.html).
Results and Discussion
Morphological Characterization of Silver Nanoparticles and Silver Nanowires
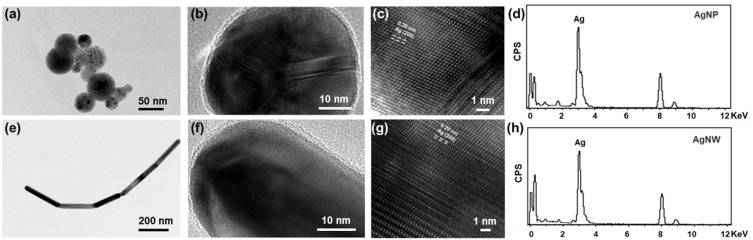
The morphological characterization of silver nanoparticles and silver nanowires was explored, as shown in Figure 1. The silver nanoparticles display a sphere-like shape (Figure 1a), which had diameters about 30 nm with uneven size distribution (Figure S1). Figure 1b shows HR-TEM image of typical silver nanoparticles, demonstrating a structure of high crystallization. Correspondingly, as exhibited in Figure 1c, the lattice fringe with a spacing of 0.20 nm was assigned to the (200) crystal face of highly crystallization silver.45 In addition, it can be clearly seen Ag element from the EDS spectrum in silver nanoparticles (Figure 1d). This result further proves the as-synthesized silver nanoparticles. Meanwhile, silver nanowires had lengths ranging from 100 μm to 200 μm (Figure 1e). Silver nanowires obtained similar results, compared with those of silver nanoparticles, demonstrating the structure of high crystallization, (200) crystal face, and Ag element from the EDS spectrum (Figure 1f-h).
Figure 1.
(a, e) TEM images, (b, c, f, g) HR-TEM images, and (d, h) EDS of silver nanoparticles (a-d) and silver nanowires (e-h).
In vitro Drug Sensitivity Test of Silver Nanoparticles and Silver Nanowires
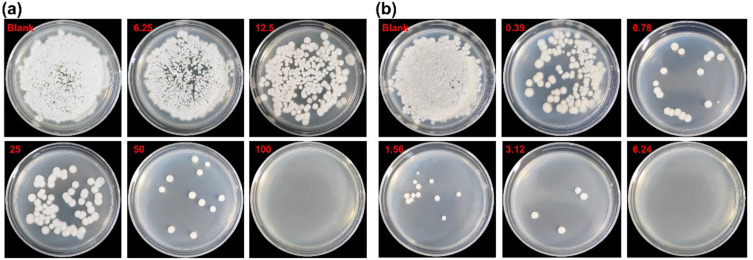
Figure 2 shows the colony growth of 705R strain on SDA culture plates after the action of different concentrations of silver nanoparticles and silver nanowires. The results show that T. asahii is displayed on the SDA tablet. The colony growth of T. asahii clinical fluconazole-resistant strains is concentration dependent on different forms of silver nanomaterials, and the higher the concentration, the fewer colonies compared to the control group. When the concentration of silver nanowires was 6.24 μg/mL, the colony was completely inhibited, while when the concentration of silver nanoparticles was 100 μg/mL, the colony was completely inhibited. It can be seen that the concentration gradient has a greater impact on the silver nanowires, displaying better antifungal effect.
Figure 2.
Colony growth of 705R strain in SDA culture dishes of silver nanoparticles (a) and silver nanowires (b).
According to CLSI M27-A3, it was confirmed that 705 is a fluconazole sensitive strain, 705R and 705I are fluconazole-resistant strains. As shown in Table 1, we compare the MICs and MFCs of silver nanoparticles, silver nanowires, and fluconazole against T. asahii. As for 705, MIC50 is 0.20, 1.56, and 8 μg/mL for silver nanoparticles, silver nanowires, and fluconazole against T. asahii, respectively. MFCs of silver nanoparticles and silver nanowires are 50 and 3.12 μg/mL against T. asahii. We separately measured the effects of silver nanoparticles and silver nanowires on three T. asahii’s in vitro drug sensitivity (Tables S1–S4). The results showed that the MIC value of silver nanoparticles (0.327 ± 0.098 μg/mL) was significantly lower than that of silver nanowires (2.6 ± 0.806 μg/mL, P < 0.001, Welch’s t test). The MIC of silver nanoparticles and silver nanowires was significantly higher than that of FLC (p < 0.05, Dunnett T3 multiple comparison). However, the MFC value of silver nanoparticles (83.333 ± 25.820 μg/mL) was significantly higher than that of silver nanowires (5.72 ± 1.274 μg/mL, P < 0.001, Welch’s t test). Obviously, silver nanoparticles had low MIC50 value and high MFC value compared with those of silver nanowires, indicating that the shape of silver had an effect on the against T. asahii. The low MIC50 value of silver nanoparticles indicated their relatively good and fast antibacterial effect. A lower MFC value indicated the good antibacterial effect of silver nanowires, but the effect is slower than that of silver nanoparticles. As for both 705R and 705I, it observes the similar results. The results showed that in vitro T. asahii planktonic cells on antifungal effect of silver nanowires and silver nanoparticles are superior to that of fluconazole-resistant strains in sensitive strains, clinical isolates, and in vitro induction of fluconazole-resistant strains. The antifungal effect of silver nanoparticles on T. asahii in vitro planktonic cells is good, while the bactericidal effect of silver nanowires is better. At the same concentration, silver nanoparticles had diameters about 30 nm and silver nanowires had diameters about 30 nm and lengths ranging from 100 μm to 200 μm. Therefore, silver nanoparticles had larger surface area compared with silver nanowires, which can be more in contact with the surface of fungi, thus inhibiting the activity of fungi. Therefore, the MIC50 value of silver nanoparticles is lower than that of silver nanowires (Table 1). Different from the spherical morphology of silver nanoparticles, silver nanowires are more likely to puncture the cell wall and membrane of fungi and cause cell apoptosis. In the literature, it reported that silver nanowires punctured the enclosing membrane and released silver ions and lysosomal contents to the cytoplasm, thereby initiating oxidative stress, leading to cellular dysfunction, cell death, inflammation, and disease.42 The optimal bactericidal concentration of silver nanowires is 6.24 μg/mL, and the silver nanoparticles are 100 μg/mL in Fluconazole resistant strain. The silver nanowires are significantly superior to the silver nanoparticles. In addition, silver nanowires are easier to produce and cheaper than silver nanoparticles. From an economic perspective, silver nanowires also have higher advantages compared with silver nanoparticles.
Table 1.
MICs and MFCs of Silver Nanoparticles, Silver Nanowires, and Fluconazole Against T. Asahii
| Strains | Concentration (μg/mL) | Fluconazole MIC50 |
|||
|---|---|---|---|---|---|
| Silver Nanowires | Silver Nanoparticles | ||||
| MIC50 | MFC | MIC50 | MFC | ||
| 705 | 1.56 | 3.12 | 0.20 | 50 | 8 |
| 705R | 3.12 | 6.24 | 0.39 | 100 | 64 |
| 705I | 3.12 | 6.24 | 0.39 | 100 | 64 |
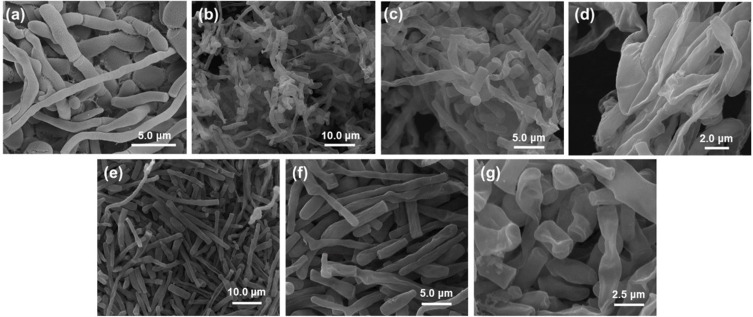
Morphology of selected 705R colonies that had a significant inhibitory effect on the colony but were not completely inhibited, by scanning electron microscopy (SEM) and transmission electron microscopy (TEM), as shown in Figures 3 and 4. The 705R colonies that have not been affected by silver nanomaterials are selected as the control group. We chose the colony treated with 50 μg/mL silver nanoparticles and the colony treated with 3.12 μg/mL silver nanowires. The results showed that the control group had orderly, long, plump, smooth, and mycelium growth (Figure 3a). Both silver nanoparticles and silver nanowires induced significant morphological changes in the mycelium of the strain. The mycelium was disordered, fractured, shrunk, deformed, and even dried and dissolved due to the leakage of cell contents (Figure 3b-g). When silver nanomaterials act on fungi, their cell walls and membranes are first damaged. This destruction leads to the loss of cell integrity, which in turn triggers changes in the morphology of the mycelium. For example, mycelium may become twisted, broken, or atrophied.
Figure 3.
SEM images of (a) control and mycelial growth of 705R strains using (b-d) silver nanowires and (e-g) silver nanoparticles.
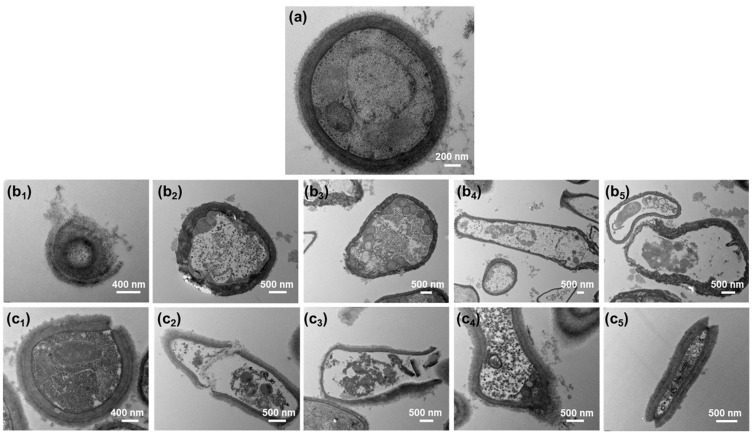
Figure 4.
TEM images of (a) control, (b, c) mycelial growth of 705R strains treated with (b) silver nanoparticles and (c) silver nanowires. (b1) Lysis, (b2) Wrinkling, (b3) Thinning, (b4 and b5) Vacuolization and thinning of cell of silver nanoparticles; (c1) Rupture, (c2 and c3) Vacuolization, thinning, and rupture, (c4) Lysis, (c5) Rupture and disintegration of cell of silver nanowires.
The control colony cells without silver nanomaterials treatment have a complete structure, and complete cell walls, cell membranes, vacuoles, nuclei, and cytoplasm can be seen with uniform color and distribution (Figure 4a). After the action of silver nanoparticles, the fungal colony undergoes deformation, wrinkling, thinning, and detachment of the cell wall and membrane, thinning and condensation of the cytoplasm, rupture of vacuoles and nuclei, and vacuolization and dissolution due to the exposure of cell contents (Figure 4b). For example, one can see the lysis of cell wall (Figure 4b1), wrinkling of cell wall (Figure 4b2), thinning of cell wall (Figure 4b3), and Vacuolization and thinning of cell (Figure 4b4 and b5) of silver nanoparticles. After the action of silver nanowires, some cell walls and membranes break, and some become thinner (Figure 4c). For example, one can see rupture of cell wall (Figure 4c1), vacuolization, thinning, and rupture of cell wall (Figure 4c2 and c3), lysis of cell wall (Figure 4c4), and rupture of cell wall and disintegration of nucleus and vacuole (Figure 4c5) of silver nanowires. Organelles such as nuclei, vacuoles, ribosomes, and mitochondria are severely disrupted, and some may experience vacuolization and dissolution due to the leakage of cellular contents. The cytoplasm also appears lighter in color and condensed. After silver ions enter fungal cells, they interfere with the synthesis of the cell wall, inhibit the cross-linking between polysaccharide chains, and cause the cell wall to lose its integrity. This reduces the protective effect of the cell wall on osmotic pressure, leading to leakage of cell contents. At the same time, silver ions also react with the phospholipid bilayer on the cell membrane, enhancing its permeability and allowing small molecules such as H ⁺ inside the cell to leak out, further damaging the integrity of the cell membrane.
Cell Cytotoxicity of the Silver Nanoparticles and Silver Nanowires
In the literature, silver nanomaterials are reported to have toxicological effects. Therefore, the cell cytotoxicity is important for the biomedical applications of silver nanomaterials. Hasan et al reported that Ag nanoparticles showed less toxicity, only when higher concentrations >100 mg/L were applied.38 When the concentration of silver nanoparticles and silver nanowires is less than 25 μg/mL, the decrease in cell activity is not significant with the increase of silver nanoparticles and silver nanowires concentration, especially the cell activity of silver nanowires remains basically unchanged (Figure S2). However, the cell activity decreases more significantly after the concentration of silver nanoparticles and silver nanowires increases to 50 and 100 μg/mL, but the cell activity decreases more significantly after the action of silver nanoparticles than that of silver nanowires. When the concentration of silver nanomaterials is less than 25 μg/mL, the toxicity of silver nanomaterials to experimental cells is relatively small. As the concentration increases to 50 and 100 μg/mL, toxicity of silver nanomaterials is produced to cells, and the toxicity of silver nanoparticles is more pronounced. This result is consistent with previous literature reports.38,42
Antifungal Effect of Silver Nanoparticles and Silver Nanowires
Differential Expression Analysis
In this article, a total of 592,824,680 raw reads with adapters and low-quality reads were identified. Following quality control, 6,410,040 reads were filtered out, leaving 586,414,640 high-quality clean reads for further analysis. The RNA mapped reads were removed before read alignment, and approximately 90.41% of the paired-end clean reads were mapped to the reference genome, containing approximately 87.07% uniquely mapped reads. In this section, the samples treated with silver nanoparticles and silver nanowires are marked as Treat 1 and Treat 2, respectively.
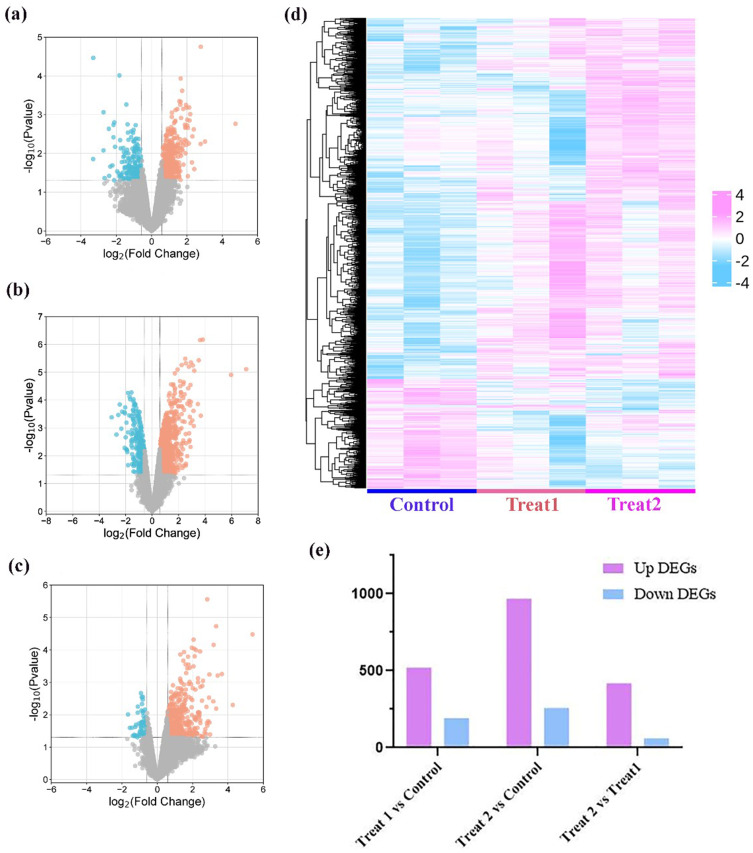
RNA-sequencing technology was applied to systematically explore the potential antifungal mechanism of Ag nanomaterials against Fungus. The gene expression level was estimated using the gene expression abundance corresponding to the FPKM value, and differentially analysis was performed with |log2(Fold Change)| >0.58 and p value <0.05 as the screening criteria. As shown in the Figure 5a and b, 708 DEGs including 119 downregulated and 517 upregulated genes were identified in the Treat 1 group compared with that of the control group, 1213 DEGs with 255 downregulated and 958 upregulated genes in Treat 2, compared with that of control groups. Among these, 473 DEGs with 61 downregulated genes and 412 upregulated genes were also differentially regulated between Treat 2 and Treat 1 groups (Figure 5c). As shown in Figure 5d, the results of hierarchical heatmap illustrated that the gene expression after Treat 1 and Treat 2 treatment were significant reversed, especially in Treat 2 vs control. Figure 5e further indicated that the number of differential gene expressions in Treat 2 vs control group is more than in Treat 1 vs control group. Above results suggested that Ag nanomaterials resulted in notable changes in gene expression by transcriptomics sequencing.
Figure 5.
Differential analysis of transcriptome sequencing. (a-c) Differential expression gene volcano map of (a) Treat 1 vs control, (b) Treat 2 vs control, (c) Treat 2 vs Treat 1; (d) Hierarchical heatmap analysis of differentially expressed genes of control, Treat 1, and Treat 2. (e) Up DEGs and down DEGs of Treat 1 vs control, Treat 2 vs control, Treat 2 vs Treat 1.
Enrichment Analysis
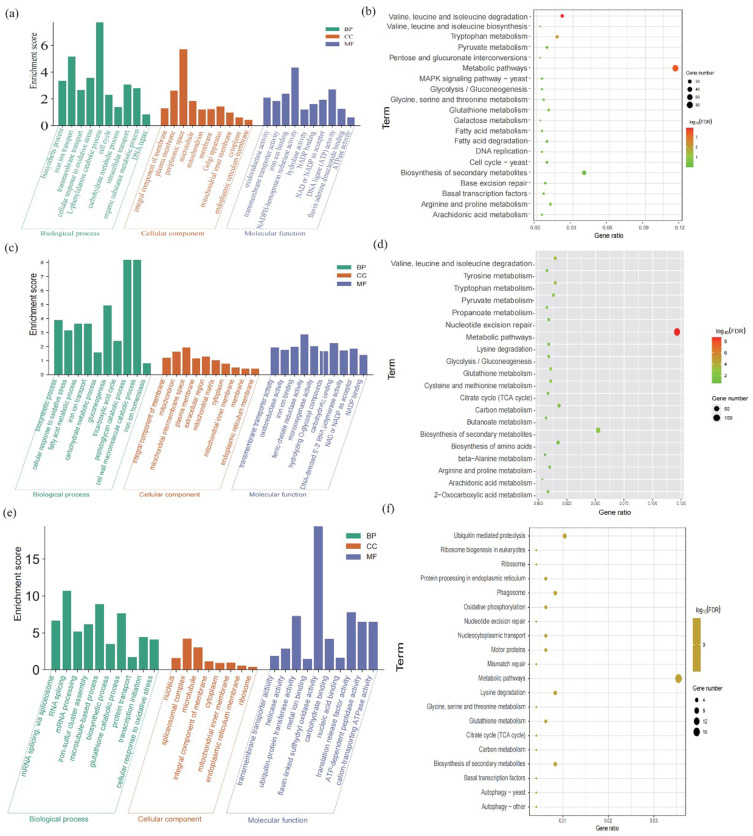
We further mapped the DEGs in Treat 1 vs control groups, Treat 2 vs control groups to DAVID database to interpret the underlying mechanism of Ag nanomaterials against Fungus based on GO and KEGG enrichment analysis. GO analysis revealed that DEGs were classified into three ontologies, including biological processes (BPs), molecular functions (MFs), and cellular components (CCs). As shown in Figure 6a, biosynthetic process, iron ion transport, transmembrane transport, cellular response to oxidative stress, L-phenylalanine catabolic process, cell cycle, carbohydrate metabolic process, intracellular transport, organic substance metabolic process, DNA repair in BP terms, integral component of membrane, plasma membrane, periplasmic space, microtubule, mitochondrion, membrane, golgi apparatus, mitochondrial inner membrane, cytoplasm, endoplasmic reticulum membrane in CC terms, and oxidoreductase activity, transmembrane transporter activity, iron ion binding, NADPH-hemoprotein reductase activity, hydrolase activity, NADP binding, NAD or NADP as acceptor, DNA ligase (ATP) activity, Flavin adenine dinucleotide binding, and ATPase activity in MF terms are observed among the Treatment 1 vs Control groups. We also found that as in the Treatment 2 vs control groups, the biosynthetic process, cellular response to oxidative stress, fatty acid metabolic process, iron ion transport, carbohydrate metabolic process, gluconeogenesis, tricarboxylic acid cycle, peptidoglycan catabolic process, cell wall macromolecule catabolic process, iron ion homeostasis enriched in the BP terms, integral component of membrane, mitochondrion, mitochondrial intermembrane space, plasma membrane, extracellular region, mitochondrial matrix, cytoplasm, mitochondrial inner membrane, membrane, endoplasmic reticulum membrane enriched in the CC terms.
Figure 6.
Functional enrichment of DEGs. Biological processes (BPs), molecular functions (MFs), and cellular components (CCs) of (a) Treat 1 vs control, (c) Treat 2 vs control, (e) Treat 2 vs Treat 1. KEGG enrichment analysis of (b) Treat 1 vs control, (d) Treat 2 vs control, (f) Treat 2 vs Treat 1.
As shown in Figure 6b, the results of KEGG enrichment analysis suggested that DEGs between the Treatment 1 and control groups were mapped to valine, leucine and isoleucine degradation, metabolic pathways, tryptophan metabolism, arginine and proline metabolism, glutathione metabolism, arachidonic acid metabolism, base excision repair, fatty acid degradation, pyruvate metabolism, basal transcription factors, biosynthesis of secondary metabolites, glycine, serine and threonine metabolism, valine, leucine and isoleucine biosynthesis, fatty acid metabolism, cell cycle, DNA replication, galactose metabolism, glycolysis/gluconeogenesis, pentose and glucuronate interconversions, MAPK signaling pathway. And in the Treatment 1 vs Treatment 2, transmembrane transporter activity, oxidoreductase activity, iron ion binding, ferric-chelate reductase activity, monooxygenase activity, hydrolyzing o-glycosyl compounds, carbohydrate binding, DNA-directed 5’-3’ RNA polymerase activity, NAD or NADP as acceptor, NADP binding was enriched in the MF terms (Figure 6c). Also, the DEGs between Treatment 2 vs control groups (Figure 6d) were enriched in the signaling pathway, including metabolic pathways, tryptophan metabolism, valine, leucine and isoleucine degradation, glutathione metabolism, biosynthesis of secondary metabolites, pyruvate metabolism, citrate cycle (TCA cycle), cysteine and methionine metabolism, propanoate metabolism, carbon metabolism, arginine and proline metabolism, tyrosine metabolism, Lysine degradation, 2-oxocarboxylic acid metabolism, butanoate metabolism, arachidonic acid metabolism, biosynthesis of amino acids, glycolysis/gluconeogenesis, nucleotide excision repair, beta-alanine metabolism.
Furthermore, to obtain the further insight into the mechanism, the difference between the Treatment 2 vs Treatment 1, the DEGs between Treatment 2 vs Treatment 1 groups were also performed enriched analysis, as shown in Figure 6e and f. The BP analysis indicated that the enriched DEGs were closely associated with the protein transportation, and cellular response to oxidative stress. The CC terms were mainly related to the integral component of membrane and extracellular matrix, and the MF terms were related to the transmembrane transporter activity, ubiquitin-protein transferase activity, and carbohydrate binding. Pathway analysis has suggested that the DEGs were enriched mainly in oxidative phosphorylation, and metabolism processes, such as glutathione metabolism, citrate cycle (TCA cycle), serine and threonine metabolism, carbon metabolism, biosynthesis of secondary metabolites, metabolic pathways. Given these above results, we preliminary speculated that the Treat 1 and Treat 2 were also played antifungal effects, and also found that the antifungal effects of Treat 2 were better than the Treat 1, especially in the oxidative phosphorylation and metabolic processes.
Network Analysis and Key Target Identified
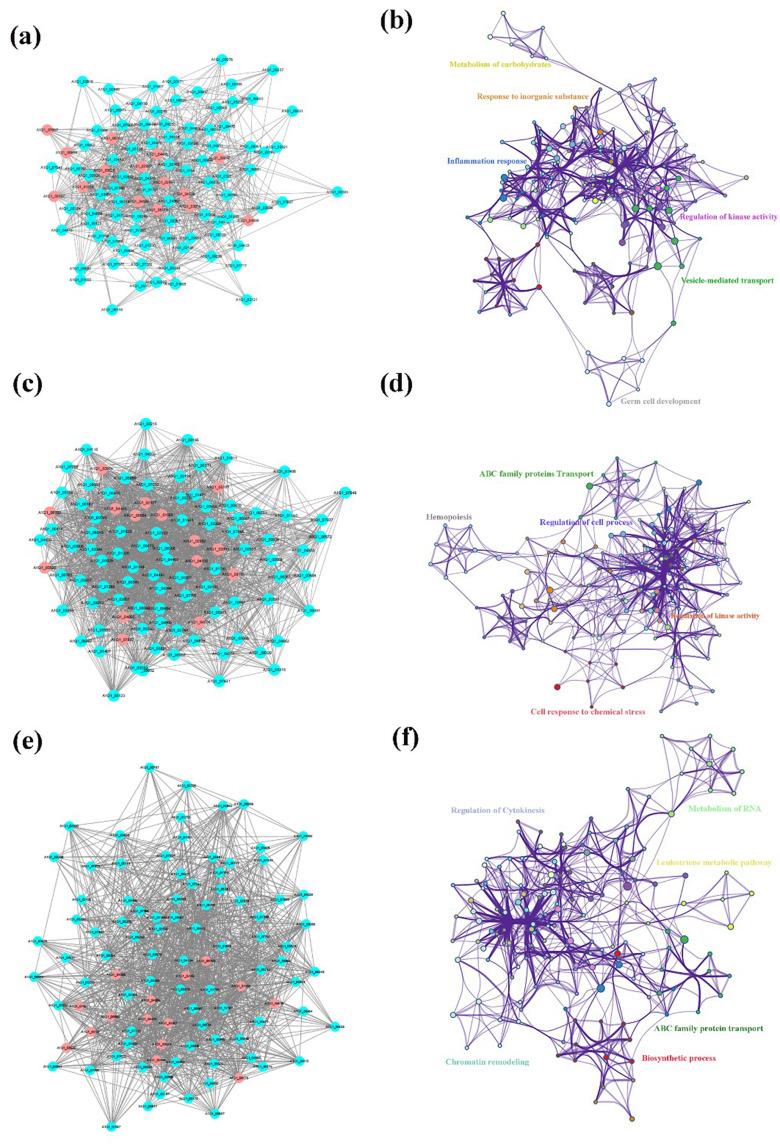
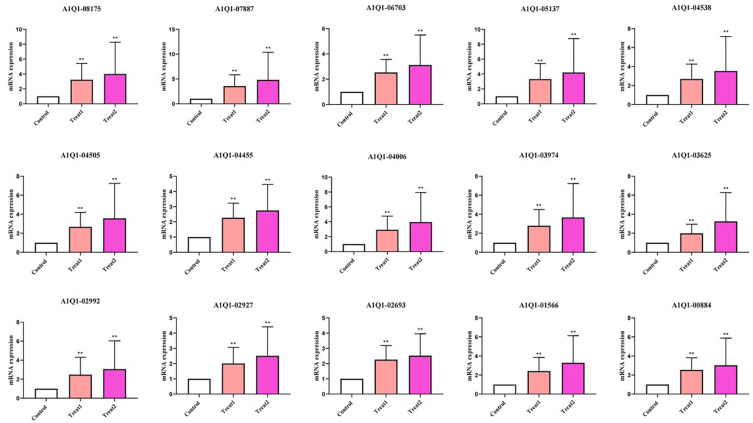
To further explore the potential antifungal mechanism of Ag nanomaterials against fungus, the identified DEGs in the Treat 1 vs control, Treat 2 vs control, and Treat 2 vs Treat 1 groups were uploaded into the STRING online database (combined score > 0.7). PPI networks of DEGs was constructed, and the Cytoscape 3.7.2 platform was used to visualize the PPI network with 139 nodes and 918 interaction edges in Treat 1 vs control group, with 269 nodes and 3600 interaction edges in Treat 2 vs control, and with 196 nodes and 1804 interaction edges in Treat 2 vs Treat 1 groups (Figure S3). Based on the network’s topological properties calculated using the cytoHubba plug-in, the PPI network of top 100 degree nodes was shown in Figure 7a and b with 100 nodes and 802 interaction edges. Furthermore, we found that the targets in PPI network were enriched in GO terms including metabolism of carbohydrates, response to inorganic substance, inflammation response, regulation of kinase activity, vesicle-mediated transport, and germ cell development. Also, the PPI network of top 100 degree nodes between the Treat 2 vs control, and Treat 2 vs Treat 1 was constructed (Figure 7c-f), and visualized in the Cytoscape 3.7.2 platform. In Figure 7c and d, the PPI network of Top 100 degree in Treat 2 vs control groups with 100 nodes and 1813 interaction edges, and the targets in the PPI network were mainly enriched in the GO terms associated with ABC family proteins transport, hemopoiesis, regulation of cell process, cell response to chemical stress, and regulation of kinase activity. Additionally, the PPI network with 100 nodes and 1175 interaction edges in the Treat 2 vs Treat 1 groups (Figure 7e and f), the targets in the PPI networks were enriched mainly in GO terms associated with regulation of cytokinesis, metabolism of RNA, leukotriene metabolism pathway, ABC family protein transport, biosynthetic process, chromatin remodeling. Among the above results, we further found 15 common targets with high degree in the top 100 degree PPI network among Treat 1 vs control, Treat 2 vs control, and Treat 1 vs Treat 2 groups, including the A1Q1-00884, A1Q1-01566, A1Q1-02693, A1Q1-02927, A1Q1-02992, A1Q1-03625, A1Q1-03974, A1Q1-04006, A1Q1-04455, A1Q1-04505, A1Q1-04538, A1Q1-05137, A1Q1-06703, A1Q1-07887, A1Q1-08715, were regarded as the key targets in antifungal mechanism of Ag nanomaterials against Fungus based on the topological analysis (Figure 8). Therefore, 15 key targets were then selected for experimental verification to interpret the potential antifungal mechanism of Ag nanomaterials against fungus.
Figure 7.
Network analysis and Go analysis of (a and b) Treat 1 vs control, (c and d) Treat 2 vs control, (e and f) Treat 2 vs Treat 1.
Figure 8.
The key targets of control, Treat 1, and Treat 2. ** indicate P < 0.01.
As reported in the literature, nanomaterials have excellent antifungal activity because of their high specific surface area and good physicochemical properties.46 It is confirmed that silver nanomaterial is a broad spectrum antifungal agent, which is more of a fungicide than a bacteriostatic agent.47 In the literature, the antibacterial effect of silver nanomaterials mainly depends on its nanoscale size effect and the release of silver ions, including destruction of bacterial structure, blocking the respiratory chain, DNA damage, and strong permeability.48,49 The bioaccumulation of silver nanomaterials in living organisms is mainly influenced by factors, such as particle size, concentration, and dispersibility.40,50 There are various exposure pathways for silver nanomaterials, mainly including respiratory exposure, digestive tract exposure, skin exposure, and direct injection.51,52 As mentioned above, toxicological effects of silver nanomaterials cannot be ignored. The long-term cost-effectiveness of using silver nanomaterials in eukaryotes is a complex issue that involves multiple considerations, including the performance, preparation cost, biocompatibility, environmental impact, and potential health risks of silver nanomaterials. The long-term cost-effectiveness of using silver nanomaterials in eukaryotes is significant, but attention should also be paid to their potential challenges and risks. In the future, with the continuous advancement of technology and in-depth research, the application prospects of silver nanomaterials in the biomedical field will be even broader. In this study, in vitro drug sensitivity tests confirmed that both silver nanoparticles and nanowires had better antifungal effects against Trichosporon asahii compared to fluconazole (Table 1). Moreover, silver nanoparticles have better antibacterial effects, while silver nanowires have better antifungal effects. The reason is that under the same concentration, silver nanoparticles are more dispersed than silver nanowires, allowing for more contact with fungal surfaces and better inhibition of fungal activity. Unlike the spherical morphology of silver nanoparticles, silver nanowires are more likely to puncture fungal cell walls and membranes, leading to cell apoptosis, resulting in better antifungal effects. It has been confirmed that silver nanowires are more likely to cause damage to cell walls and membranes Figure 4c1–c5, demonstrating the antifungal advantage of silver nanowires. The analysis of GO enrichment results showed that the silver nanoparticles and silver nanowires all exert antifungal mechanisms from three aspects of biological processes (BPs), cellular components (CCs), and molecular functions (MFs), thereby confirming that the antifungal effect of silver nanomaterials is multi-faceted and less prone to drug resistance (Figure 6a,c,e). Furthermore, the roles of silver nanoparticles and silver nanowires in the membrane structure are prominent in cellular components (CCs), while in biological processes (BPs), silver nanowires have a more prominent effect on the decomposition and metabolism of cell walls. This also confirms that silver nanowires can exert better antifungal effects by puncturing fungal cell walls. Based on the analysis of KEGG enrichment results, both silver nanoparticles and silver nanowires mainly participate in the metabolic pathways of pyruvate metabolism, citrate cycle (TCA cycle), cysteine and metal metabolism, propanoate metabolism, carbon metabolism, which are upregulated for the synthesis, decomposition, and transformation of substances, as well as energy metabolism related genes.
Therefore, we further screened 15 common genes based on PPI network analysis (Treat 2 vs Control, Treat 1 vs Control, Treat 2 vs Control), including A1Q1-00884, A1Q1-01566, A1Q1-02693, A1Q1-02927, A1Q1-02992, A1Q1-03625, A1Q1-03974, A1Q1-04006, A1Q1-04455, A1Q1-04505, A1Q1-04538, A1Q1-05137, A1Q1-06703, A1Q1-07887, A1Q1-08715 (Figure 8), using the expression levels of 18S rRNA as the reference gene and calculated by the 2−ΔΔCt method. After the interaction of silver nanoparticles and silver nanowires, these genes showed an upregulation trend, and the upregulation effect of silver nanowires was more significant, confirming that silver nanowires have better antifungal effects. By querying the UniProt database, the components and functions of 9 genes involved were obtained (Table S5). The cell composition and functions of the other 6 genes involved have not been reported yet, which needs to be further explored in the near future. Through these 9 genes, we can determine the antifungal mechanism of silver nanowires in fluconazole resistant Trichosporon asahii. Among them, 5 genes, such as A1Q1-01566, A1Q1-05137, A1Q1-06703, A1Q1-08175, and A1Q1-02992, are involved in the composition and function of the membrane, while A1Q1-00884, A1Q1-02693, A1Q1-04455, and A1Q1-04505 are involved in the composition and function of the nucleus, ribosome, ribonucleoprotein complex, etc. As is well known, the cell membrane controls the exchange of substances between the cell and the outside world, which has a selective permeation function, making the chemical composition inside and outside the cell different. According to the gene functions of A1Q1-01566, A1Q1-05137, A1Q1-06703, A1Q1-08175, and A1Q1-02992, it can be inferred that silver nanowires increased the activity of transmembrane transporters and potassium ion transporters, sphingolipid metabolism, oxidoreductase activity, and pyruvate metabolism, alters the structure of fungal cell membranes and intracellular signaling pathways, and regulates energy metabolism. The remaining four genes play an antifungal role in cellular genetic information storage, transcription, replication, and protein synthesis due to their involvement in the composition of the nucleus, ribonucleoproteins, and ribosomes.
Conclusion
In summary, we comparative explored the bactericidal/fungicidal mechanism of silver nanowires and silver nanoparticles against T. asahii. The optimal bactericidal concentration of silver nanowires is 6.24 μg/mL, and the silver nanoparticles is 100 μg/mL. The lower MFC value of silver nanowires indicates good sterilization effect and suitability for eradication treatment, which is slower than that of silver nanoparticles. The silver nanowires is significantly superior to the silver nanoparticles. SEM and TEM results indicated that both silver nanoparticles and silver nanowires showed significant morphological changes in the mycelium of the strain, compared with the control. The in vitro antifungal effects of silver nanowires were better than the silver nanoparticles, especially in the metabolic processes and oxidative phosphorylation. Transcriptome analysis and in vitro PCR experiments further demonstrated that 15 key targets of silver nanowires participated in regulating the structure and energy metabolism of the membrane to exert antifungal effects. This work proves that silver nanomaterials with different morphologies have potential applications in the treatment for T. asahii. Future research should further explore the antibacterial mechanism, biological toxicity, and influencing factors of silver nanomaterials, the in vivo studies in a more complex biological system to correlate in vitro findings to clinical applications, providing scientific basis for its widespread applications.
Acknowledgments
The financial support in part from the Beijing Natural Science Foundation (7212105) and National Natural Science Foundation of China (82073466) is gratefully acknowledged.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zhang MW, Zhu ZH, Xia ZK, et al. Comprehensive circRNA-microRNA-mRNA network analysis revealed the novel regulatory mechanism of Trichosporon asahii infection. Military Med Res. 2021;8(1):19. doi: 10.1186/s40779-021-00311-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurakado S, Miyashita T, Chiba R, Sato C, Matsumoto Y, Sugita T. Role of arthroconidia in biofilm formation by Trichosporon asahii. Mycoses. 2021;64(1):42–47. doi: 10.1111/myc.13181 [DOI] [PubMed] [Google Scholar]
- 3.Ozkaya-Parlakay A, Karadag-Oncel E, Cengiz AB, et al. Trichosporon asahii sepsis in a patient with pediatric malignancy. J Microbiol Immunol Infect. 2016;49(1):146–149. doi: 10.1016/j.jmii.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 4.Almeida JND, Hennequin C. Invasive trichosporon infection: a systematic review on a re-emerging fungal pathogen. Front Microbiol. 2016;7:1629. doi: 10.3389/fmicb.2016.01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan SY, Chien JY, Hsueh PR. Invasive trichosporonosis caused by Trichosporon asahii and other unusual Trichosporon species at a medical center in Taiwan. Clin Infectious Dis. 2009;49(1):e11–e17. doi: 10.1086/599614 [DOI] [PubMed] [Google Scholar]
- 6.Lee EH, Choi MH, Lee KH, Song YG, Han SH. Differences of clinical characteristics and outcome in proven invasive Trichosporon infections caused by asahii and non-asahii species. Mycoses. 2023;66(11):992–1002. doi: 10.1111/myc.13635 [DOI] [PubMed] [Google Scholar]
- 7.Yang X, Bai S, Wu JM, et al. Antifungal activity and potential action mechanism of Allicin against Trichosporon asahii. Microbiol Spectrum. 2023;11(3):e0090723. doi: 10.1128/spectrum.00907-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins N, Wright GD, Cowen LE, Heitman J. Antifungal drugs: the current armamentarium and development of new agents. Microbiol Spectrum. 2016;4(5). doi: 10.1128/microbiolspec.FUNK-0002-2016 [DOI] [PubMed] [Google Scholar]
- 9.Weiss ZF, Little J, Hammond S. Evolution of antifungals for invasive mold infections in immunocompromised hosts, then and now. Exp Rev Anti-Infective Ther. 2023;21(5):535–549. doi: 10.1080/14787210.2023.2207821 [DOI] [PubMed] [Google Scholar]
- 10.Ma X, Liu H, Liu Z, et al. Trichosporon asahii PLA2 gene enhances drug resistance to azoles by improving drug efflux and biofilm formation. Int J Mol Sci. 2023;24(10):8855. doi: 10.3390/ijms24108855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arastehfar A, De Almeida Júnior JN, Perlin DS, Ilkit M, Boekhout T, Colombo AL. Multidrug-resistant Trichosporon species: underestimated fungal pathogens posing imminent threats in clinical settings. Crit Rev Microbiol. 2021;47(6):679–698. doi: 10.1080/1040841X.2021.1921695 [DOI] [PubMed] [Google Scholar]
- 12.Itoh K, Iwasaki H, Negoro E, et al. Successful treatment of breakthrough trichosporon asahii fungemia by the combination therapy of fluconazole and liposomal amphotericin B in a patient with follicular lymphoma. Mycopathologia. 2021;186(1):113–117. doi: 10.1007/s11046-020-00525-x [DOI] [PubMed] [Google Scholar]
- 13.Francisco EC, De Almeida Junior JN, De Queiroz Telles FD, et al. Species distribution and antifungal susceptibility of 358 Trichosporon clinical isolates collected in 24 medical centres. Clin Microbiol Infect. 2019;25(7):909.e1–e5. doi: 10.1016/j.cmi.2019.03.026 [DOI] [PubMed] [Google Scholar]
- 14.de Almeida JN, Jimenez-Ortigosa C, Francisco EC, Colombo AL, Perlin DS. ERG11 analysis among clinical isolates of trichosporon asahii with different azole susceptibility profiles. Antimicrob Agents Chemother. 2022;66(12). doi: 10.1128/aac.01101-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kushima H, Tokimatsu I, Ishii H, et al. Cloning of the lanosterol 14-α-demethylase (ERG11) gene in Trichosporon asahii: a possible association between G453R amino acid substitution and azole resistance in T. asahii. Fems Yeast Res. 2012;12(6):662–667. doi: 10.1111/j.1567-1364.2012.00816.x [DOI] [PubMed] [Google Scholar]
- 16.Kushima H, Tokimatsu I, Ishii H, Kawano R, Watanabe K, Kadota JI. A new amino acid substitution at G150S in Lanosterol 14-alpha Demethylase (Erg11 protein) in Multi-azole-resistant Trichosporon asahii. Med Mycology J. 2017;58(1):E23–E28. doi: 10.3314/mmj.16-00027 [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Yue ZG, Wang W, Cao ZQ. Strategies on designing multifunctional surfaces to prevent biofilm formation. Front Chem Sci Eng. 2015;9(3):324–335. doi: 10.1007/s11705-015-1529-z [DOI] [Google Scholar]
- 18.Shao XY, Wang J, Liu ZT, et al. Nano-copper ions assembled cellulose-based composite with antibacterial activity for biodegradable personal protective mask. Front Chem Sci Eng. 2023;17(10):1544–1554. doi: 10.1007/s11705-022-2288-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia ZK, Ma QH, Li SY, et al. The antifungal effect of silver nanoparticles on Trichosporon asahii. J Microbiol Immunol Infect. 2016;49(2):182–188. doi: 10.1016/j.jmii.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 20.Krishnan PD, Banas D, Durai RD, et al. Silver nanomaterials for wound dressing applications. Pharmaceutics. 2020;12(9):821. doi: 10.3390/pharmaceutics12090821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aziz AT, Alshehri MA, Alanazi NA, et al. Phytochemical analysis of Rhazya stricta extract and its use in fabrication of silver nanoparticles effective against mosquito vectors and microbial pathogens. Sci Total Environm. 2020;700:134443. doi: 10.1016/j.scitotenv.2019.134443 [DOI] [PubMed] [Google Scholar]
- 22.Hao Z, Wang M, Cheng L, Si MM, Feng ZZ, Feng ZY. Synergistic antibacterial mechanism of silver-copper bimetallic nanoparticles. Front Bioeng Biotechnol. 2023;11:1337543. doi: 10.3389/fbioe.2023.1337543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alavi M, Ashengroph M. Mycosynthesis of AgNPs: mechanisms of nanoparticle formation and antimicrobial activities. Exp Rev Anti-Infective Ther. 2023;21(4):355–363. doi: 10.1080/14787210.2023.2179988 [DOI] [PubMed] [Google Scholar]
- 24.Pachaiappan R, Rajendran S, Show PL, Manavalan K, Naushad M. Metal/metal oxide nanocomposites for bactericidal effect: a review. Chemosphere. 2021;272:128607. doi: 10.1016/j.chemosphere.2020.128607 [DOI] [PubMed] [Google Scholar]
- 25.Esfandiari N, Simchi A, Bagheri R. Size tuning of Ag-decorated TiO2 nanotube arrays for improved bactericidal capacity of orthopedic implants. J Biomed Mater Res Part A. 2014;102(8):2625–2635. doi: 10.1002/jbm.a.34934 [DOI] [PubMed] [Google Scholar]
- 26.Dogru E, Demirbas A, Altinsoy B, Duman F, Ocsoy I. Formation of Matricaria chamomilla extract-incorporated Ag nanoparticles and size-dependent enhanced antimicrobial property. J Photochem Photobiol B. 2017;174:78–83. doi: 10.1016/j.jphotobiol.2017.07.024 [DOI] [PubMed] [Google Scholar]
- 27.Stefan M, Hritcu L, Mihasan M, et al. Enhanced antibacterial effect of silver nanoparticles obtained by electrochemical synthesis in poly(amide-hydroxyurethane) media. J Mater Sci. 2011;22(4):789–796. doi: 10.1007/s10856-011-4281-z [DOI] [PubMed] [Google Scholar]
- 28.Wang L, He H, Yu YB, et al. Morphology-dependent bactericidal activities of Ag/CeO2 catalysts against Escherichia coli. J Inorganic Biochem. 2014;135:45–53. doi: 10.1016/j.jinorgbio.2014.02.016 [DOI] [PubMed] [Google Scholar]
- 29.El-Zahry MR, Mahmoud A, Refaat IH, Mohamed HA, Bohlmann H, Lendl B. Antibacterial effect of various shapes of silver nanoparticles monitored by SERS. Talanta. 2015;138:183–189. doi: 10.1016/j.talanta.2015.02.022 [DOI] [PubMed] [Google Scholar]
- 30.Mustafa G, Hasan M, Yamaguchi H, Hitachi K, Tsuchida K, Komatsu S. A comparative proteomic analysis of engineered and bio synthesized silver nanoparticles on soybean seedlings. J Proteomics. 2020;224:103833. doi: 10.1016/j.jprot.2020.103833 [DOI] [PubMed] [Google Scholar]
- 31.Yang YQ, Hao KY, Jiang MS, et al. Transcriptomic analysis of drug-resistance Acinetobacter baumannii under the stress condition caused by Litsea cubeba L. essential oil via RNA sequencing. Genes. 2021;12(7):1003. doi: 10.3390/genes12071003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafantaris I, Tsadila C, Nikolaidis M, et al. Transcriptomic analysis of pseudomonas aeruginosa response to pine honey via RNA sequencing indicates multiple mechanisms of antibacterial activity. Foods. 2021;10(5):936. doi: 10.3390/foods10050936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaaban MT, Ghaly MF, Fahmi SM. Antibacterial activities of hexadecanoic acid methyl ester and green-synthesized silver nanoparticles against multidrug-resistant bacteria. J Basic Microbiol. 2021;61(6):557–568. doi: 10.1002/jobm.202100061 [DOI] [PubMed] [Google Scholar]
- 34.Huang L, Chen R, Luo J, Hasan M, Shu X. Shu X.Synthesis of phytonic silver nanoparticles as bacterial and ATP energy silencer. J Inorganic Biochem. 2022;231:111802. doi: 10.1016/j.jinorgbio.2022.111802 [DOI] [PubMed] [Google Scholar]
- 35.Zulfiqar H, Zafar A, Rasheed MN, et al. Synthesis of silver nanoparticles using Fagonia cretica and their antimicrobial activities. Nanoscale Adv. 2019;1(5):1707–1713. doi: 10.1039/c8na00343b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussain R, Hasan M, Iqbal KJ, et al. Anjum S I.Nano-managing silver and zinc as bio-conservational approach against pathogens of the honey bee. J Biotechnol. 2023;365:1–10. doi: 10.1016/j.jbiotec.2023.01.009 [DOI] [PubMed] [Google Scholar]
- 37.Flores-López LZ, Espinoza-Gómez H, Somanathan R. Silver nanoparticles: electron transfer, reactive oxygen species, oxidative stress, beneficial and toxicological effects. J Appl Toxicol. 2019;39:16–26. doi: 10.1002/jat.3654 [DOI] [PubMed] [Google Scholar]
- 38.Hasan M, Mehmood K, Mustafa G, et al. Phytotoxic Evaluation of Phytosynthesized Silver Nanoparticles on Lettuce. Coatings. 2021;11(2):225. doi: 10.3390/coatings11020225 [DOI] [Google Scholar]
- 39.Hasan M, Sajjad M, Zafar A, et al. Blueprinting morpho-anatomical episodes via green silver nanoparticles foliation. Green Processing and Synthesis. 2022;11:697–708. doi: 10.1515/gps-2022-0050 [DOI] [Google Scholar]
- 40.Wang HH, Ho KT, Scheckel KG, et al. Toxicity, bioaccumulation, and biotransformation of silver nanoparticles in marine organisms. Environ Sci Technol. 2014;48:13711–13717. doi: 10.1021/es502976y [DOI] [PubMed] [Google Scholar]
- 41.Yang YN, Xu GM, Xu SM, Chen SP, Xu A, Wu LJ. Effect of ionic strength on bioaccumulation and toxicity of silver nanoparticles in Caenorhabditis elegans. Ecotoxicol Environ Saf. 2018;165:291–298. doi: 10.1016/j.ecoenv.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 42.Lehmann SG, Toybou D, Del Real AEP, et al. Crumpling of silver nanowires by endolysosomes strongly reduces toxicity. Proceedings of The National Academy of Sciences of The United States of America, 2019; 116: 14893–14898. DOI: 10.1073/pnas.1820041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortazar P, Johnson BE. Review of the efficacy of individualized chemotherapy selected by in vitro drug sensitivity testing for patients with cancer. J Clin Oncol. 1999;17:1625–1631. doi: 10.1200/JCO.1999.17.5.1625 [DOI] [PubMed] [Google Scholar]
- 44.Wayne PA; Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. In: CLSI Document M27-A3. 3rd ed. Clinical and Laboratory Standards; 2008. [Google Scholar]
- 45.Zhu JF, Zhu YJ. Microwave-assisted one-step synthesis of polyacrylamide - Metal (M) Ag, Pt, Cu) nanocomposites in ethylene glycol. J Phys Chem B. 2006;110:8593–8597. doi: 10.1021/jp060488b [DOI] [PubMed] [Google Scholar]
- 46.Mabrouk M, Das DB, Salem ZA, Beherei HH. Nanomaterials for biomedical applications: production, characterisations, recent trends and difficulties. Molecules. 2021;26:1077. doi: 10.3390/molecules26041077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luiz GR, Gabriella SCR, Rafael C, Ana ODS. Antifungal activity of mycogenic silver nanoparticles on clinical yeasts and phytopathogens. Antibiotics. 2023;12(1):91. doi: 10.3390/antibiotics12010091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000;52:662–668. doi: [DOI] [PubMed] [Google Scholar]
- 49.Marambio-Jones C, Hoek EMV. A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. J Nanopart Res. 2010;12:1531–1551. doi: 10.1007/s11051-010-9900-y [DOI] [Google Scholar]
- 50.Joo HS, Kalbassi MR, Yu IJ, Lee JH, Johari SA. Bioaccumulation of silver nanoparticles in rainbow trout (Oncorhynchus mykiss): influence of concentration and salinity. Aquat Toxicol. 2013;140:398–406. doi: 10.1016/j.aquatox.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 51.Ferdous Z, Nemmar A. Health impact of silver nanoparticles: a review of the biodistribution and toxicity following various routes of exposure. Int J Mol Sci. 2020;21:2375. doi: 10.3390/ijms21072375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scown TM, Santos EM, Johnston BD, et al. Effects of aqueous exposure to silver nanoparticles of different sizes in rainbow trout. Toxicol Sci. 2010;115:521–534. doi: 10.1093/toxsci/kfq076 [DOI] [PubMed] [Google Scholar]