Abstract
The study of cerebral microvessels is becoming increasingly important in a wide variety of conditions, such as stroke, sepsis, traumatic brain injury and neurodegenerative diseases. However, the molecular mechanisms underlying cerebral microvascular dysfunction in these conditions are largely unknown. The molecular characterization of cerebral microvessels in experimental disease models has been hindered by the lack of a standardized method to reproducibly isolate intact cerebral microvessels with consistent cellular compositions and without the use of enzymatic digestion, which causes undesirable molecular and metabolic changes. Herein, we describe an optimized protocol for microvessel isolation from mouse brain cortex that yields microvessel fragments with consistent populations of discrete blood–brain barrier (BBB) components (endothelial cells, pericytes and astrocyte end feet) while retaining high RNA integrity and protein post-translational modifications (e.g., phosphorylation). We demonstrate that this protocol allows the quantification of changes in gene expression in a disease model (stroke) and the activation of signaling pathways in mice subjected to drug administration in vivo. We also describe the isolation of genomic DNA (gDNA) and bisulfite treatment for the assessment of DNA methylation, as well as the optimization of chromatin extraction and shearing from cortical microvessels. This optimized protocol and the described applications should improve the understanding of the molecular mechanisms governing cerebral microvascular dysfunction, which may help in the development of novel therapies for stroke and other neurologic conditions.
Introduction
The cerebrovascular endothelium is a highly specialized vascular bed, which—in coordination with pericytes, vascular smooth muscle cells and glial cells—forms a physical, transport and metabolic barrier (the BBB), maintains an anticoagulant and anti-inflammatory state and regulates microcirculatory flow in order to meet neuronal demands and maintain homeostasis. Increasing preclinical and clinical evidence indicate that these fundamental functions of the endothelium are severely compromised in cerebrovascular and neurodegenerative diseases and substantially contribute to the exacerbation of neuronal injury in these conditions1–6. In addition, the therapeutic potential of the cerebrovascular endothelium is also well established, because it is positioned at the interface between the blood and brain parenchyma. However, despite extensive research, our understanding of the molecular mechanisms underlying cerebral microvascular dysfunction is still very limited.
Working toward this goal, several studies have attempted to characterize the molecular signature of the cerebrovascular endothelium by transcriptome and proteome profiling of isolated cerebral endothelial cells or mural cells7–10. These approaches have allowed the identification of molecules enriched in the cerebrovascular endothelium as compared to the endothelium of other organs or to other components of the neurovascular unit. To investigate the activation of signaling pathways in the cerebrovascular endothelium upon pharmacological treatments or upon ischemic or inflammatory challenge, numerous in vitro studies with cultured brain endothelial cells have been conducted11,12. However, both the unique phenotype and molecular signature of the endothelia from specific organs are lost when cells are removed from their tissue microenvironment, deprived of blood flow and cultured in vitro13,14. This treatment of cells also prohibits the accurate study of junctional protein expression, post-translational modifications and subcellular localization in health and disease states within a living organism.
The ability to study the intact microvasculature—isolated from the brain parenchyma while retaining the integrity of intercellular connections between the endothelium and mural cells—is important to fully understanding in vivo function. Various methods have been used to isolate microvessel fragments while preserving the interactions of the endothelium with mural cells. Many of these methods use enzymatic digestion to achieve purity and consistency between preparations15,16, causing undesirable molecular and metabolic changes, compromising RNA and protein integrity and impairing the ability to detect low-abundance transcripts and quantify changes in gene expression or the activation of signaling pathways. To overcome these challenges, we have developed and extensively validated a method for the isolation, processing and molecular characterization of mouse cortical microvessels by optimizing and supplementing various protocols12,17,18. Importantly, the optimized protocol described in this article (Fig. 1) yields intact microvessel fragments with consistent populations of discrete BBB components (endothelial cells, pericytes and astrocyte end feet; Figs. 2–4) without a requirement for enzymatic digestion, thereby retaining high RNA integrity (Fig. 5). We also demonstrate that the described protocol can be applied to quantify changes in gene expression in a disease model (stroke)19 (Fig. 6) and the activation of signaling pathways in mice subjected to drug administration (Fig. 7). In addition, we describe in detail other successful and novel applications of this protocol for molecular characterization of cerebral microvessels, such as the determination of DNA methylation status by isolation of gDNA, bisulfite treatment and methylation-specific PCR (Fig. 8), as well as the extraction of chromatin and shearing to the appropriate size for chromatin immunoprecipitation (ChIP) assays (Fig. 9). Finally, we recently published a study using the described protocol in which quantitative changes in subcellular localization of tight junction (TJ) proteins in cerebral microvessels were quantified from endothelial-specific genetically engineered mice and were correlated with in vivo BBB and neuronal function20.
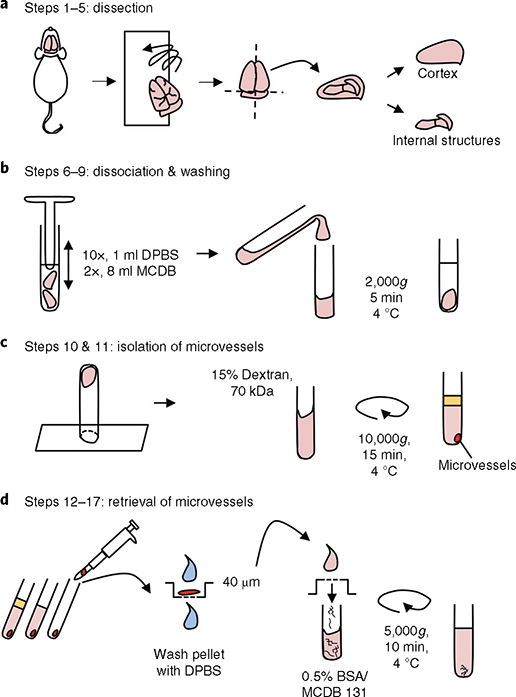
Fig. 1 |. Cortical microvessel isolation protocol overview.
a, Dissection. After mouse euthanasia, all steps are conducted in a cold room. Meninges are removed by rolling the brains on blotting paper, and cortices are dissected in cold Dulbecco’s phosphate-buffered saline (DPBS). b, Dissociation of tissue and washing. Cortices are homogenized using a loose-fit Dounce grinder and centrifuged (4 °C) for 5 min at 2,000g. c, Separation of the microvessel fraction by gradient centrifugation. Microvessel pellet is resuspended in a 15% (wt/vol) 70-kDa dextran solution and centrifuged (4 °C) for 15 min at 10,000g. d, Retrieval of microvessels. The top layer, containing myelin and brain parenchymal cells, is removed. Microvessels are retrieved by pipetting and transferred to a 40-μm cell strainer. After being washed with cold DPBS, the microvessels are collected in a tube by inverting the filter and adding 10 ml of MCDB 131 medium containing 0.5% (wt/vol) endotoxin-, fatty-acid- and protease-free BSA. The suspension can be centrifuged (4 °C) for 10 min at 5,000g to pellet the microvessels for some further applications.
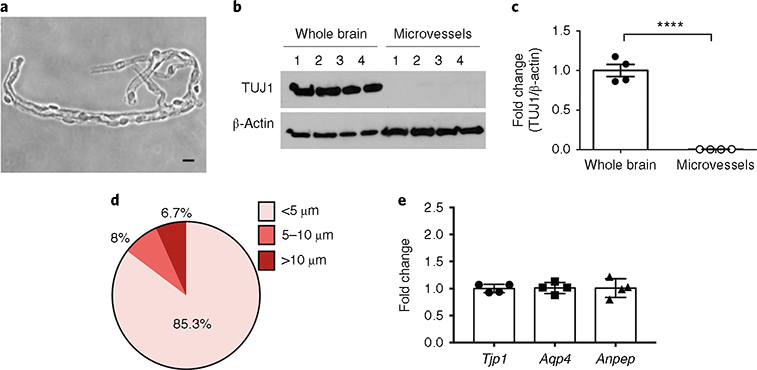
Fig. 2 |. Characterization of microvessel preparations: purity, structural features (size) and consistency in cellular composition.
a, Bright-field (phase-contrast) image of microvessels plated on microscope slides (scale bar, 10 μm). b,c, Purity of the microvessel preparations was assessed by the absence of the neuronal marker TUJ1. Western blot analysis of the neuronal marker TUJ1 in whole-brain and microvessel samples (b) and quantification after normalization with β-actin (c) are shown. ****P < .0001 (t-test). d, The diameters of the microvessel fragments were measured under an inverted microscope (Olympus CKX41) using the built-in tools of the CellSens Entry software. ~1,000 microvessel fragments were measured. e, The cellular compositions of the microvessel preparations were tested. The content of endothelial cells, astrocytes and pericytes in four independent microvessel preparations was assessed by RT-qPCR quantification of Tjp1 (ZO-1, endothelial marker), Aqp4 (astrocyte end-foot marker) and Anpep (CD13, pericyte marker) mRNA levels. Notice the consistent cell composition of the different preparations, as assessed by similar levels of cell-specific markers. RIN is ≥9.8. The individual values and the average ± s.e.m. are shown. Full blots for b are shown in Supplementary Fig. 1.
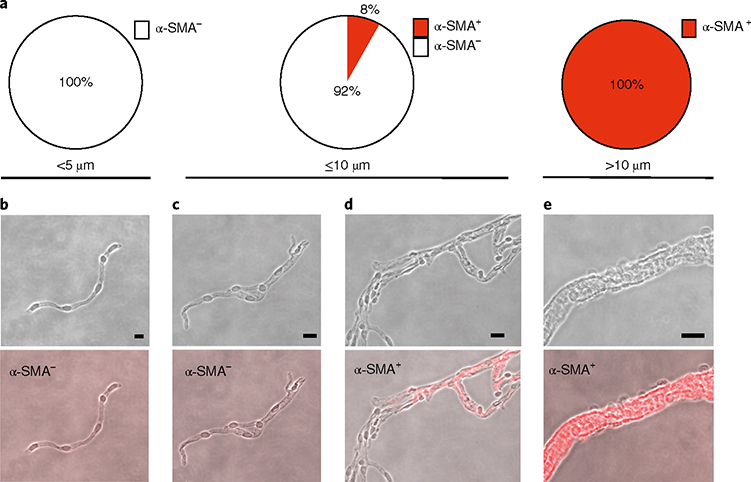
Fig. 4 |. Characterization of microvessel fragments: size and α-SMA content.
a, Percentage of α-SMA+ and α-SMA− microvessel fragments in microvessels <5 μm (85.3% of all microvessels), in microvessels ≤10 μm (93.3% of all microvessels) and microvessels >10 μm (6.7% of all microvessels). All vessels <5 μm were α-SMA−, 8% of vessels ≤10 μm were α-SMA+ and all vessels >10 μm were α-SMA+. In total, 85.2% of all microvessels were α-SMA−. b–e, Representative phase-contrast and immunofluorescence (α-SMA, red channel) pictures of microvessels from the <5 μm group (b), ≤10 μm group (c,d) and >10 μm group (e). Scale bars, 5 μm (b); 10 μm (c,d); 20 μm (e).
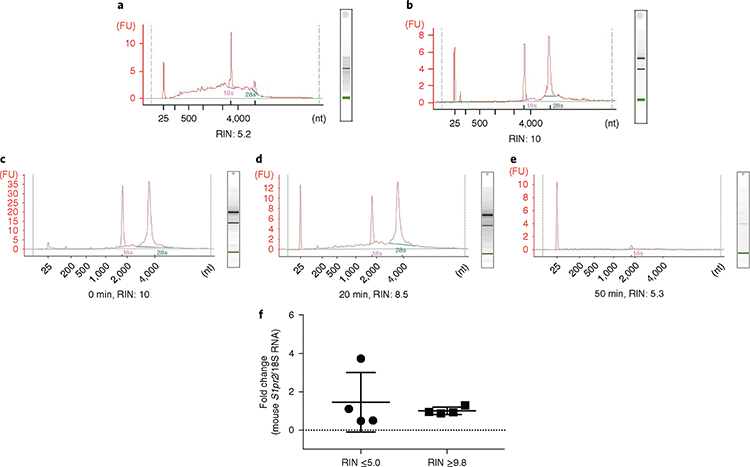
Fig. 5 |. Impact of the microvessel isolation method on RNA integrity and sample-to-sample variation.
a,b, Isolation of microvessels was performed in two different environmental conditions. RNA was isolated from microvessels and RNA integrity was determined using the Bioanalyzer 2100. Bioanalyzer profiles of RNA isolated from microvessels when the brain dissection and preparation of microvessels were conducted at room temperature, but all reagents, tools and equipment were kept on ice or cold (a) and when the same procedures were conducted at 4 °C in a cold room (b). c–e, Impact of the time of enzymatic digestion on RNA integrity. Microvessel preparations from one brain were incubated in 1.3 ml of Liberase TH at a concentration of 0.625 mg/ml at 37 °C on a rotor for 0 (c), 20 (d) or 50 min (e). DPBS (20 ml) was added to stop the digestion, and the cell suspensions were spun at 276g for 10 min at 4 °C. f, Impact of RNA quality on mRNA quantification. Sphingosine-1-phosphate receptor 2 (S1pr2) mRNA levels were quantified by reverse transcription followed by quantitative PCR (RT-qPCR) analysis and normalized by 18S ribosomal RNA. The individual values and the average ± s.e.m. are shown, n = 4. The coefficient of variation between preparations was 106.2% when RIN ≤5 and 18.8% when RIN ≥9.8. FU, fluorescence units; nt, nucleotides.
Fig. 6 |. Quantification of changes in gene expression in cerebral microvessels after stroke.
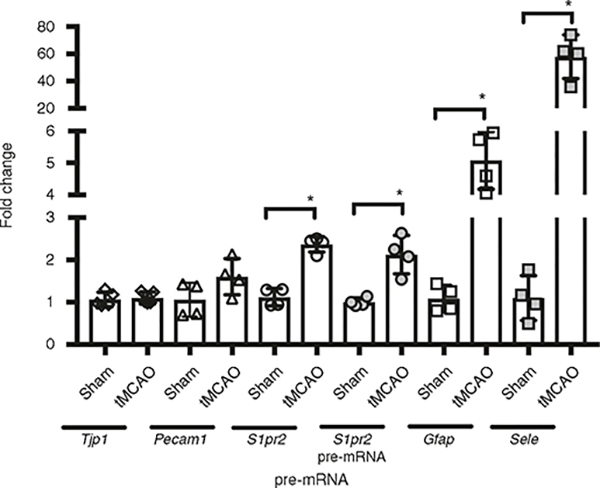
Mice were subjected to sham surgery or transient middle cerebral artery occlusion (tMCAO, 45-min occlusion followed by reperfusion). 24 h later, microvessels were isolated from the ischemic (ipsilateral) cortex (tMCAO) or sham. Changes in Tjp1 (ZO-1), Pecam1 (CD31), S1pr2, glial fibrillary acidic protein (Gfap), Sele (E-selectin) mRNA and S1pr2 pre-mRNA levels (were quantified by RT-qPCR and normalized by 18S ribosomal RNA. RIN ≥9.8. Fold induction of tMCAO versus sham is shown. The individual values and the average ±s.e.m. are shown. *P < 0.05 (t-test); n = 4. The use of laboratory animals was approved by the Weill Cornell Medicine Institutional Animal Care and Use Committee.
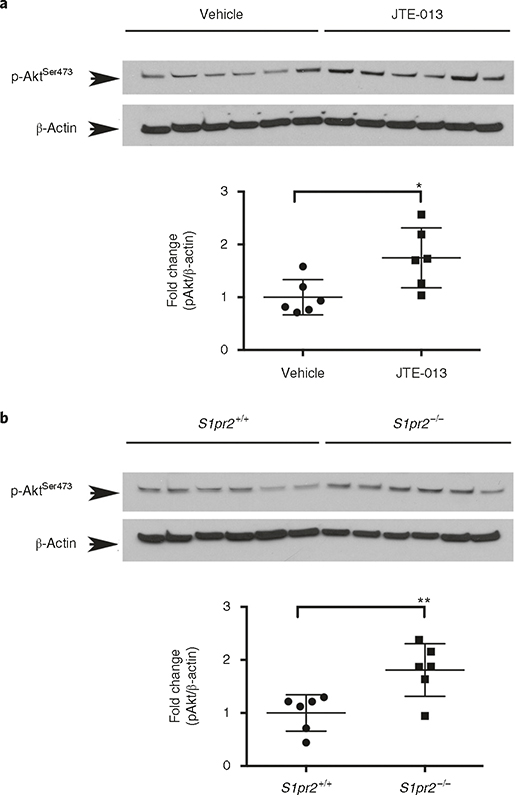
Fig. 7 |. Quantification of phospho-Ser473 Akt levels in cerebral microvessels in wild-type mice after administration of the S1PR2 antagonist JTE-013.
a, Mice were treated with vehicle or JTE013 (30 mg/kg) by gavage. Six hours later, microvessels were isolated and proteins were subjected to western blot analysis. b, Microvessels were isolated from 8-week-old adult S1pr2 knockout (S1pr2−/−) mice or wild-type littermates (S1pr2+/+), and western blot analysis for phospho-Ser473 Akt (pAkt) was conducted; n = 6. Optical density of each lane was measured using Image J. Fold induction of p-Akt (normalized by β-actin) in JTE013-treated versus vehicle-treated mice (a) and S1pr2−/− versus wild-type mice (b) are shown. The individual values and the average ± s.e.m. are shown. *P < 0.05, **P < 0.005 (t-test). The use of laboratory animals was approved by the Weill Cornell Medicine Institutional Animal Care and Use Committee. Full western blots for a and b are shown in Supplementary Fig. 1.
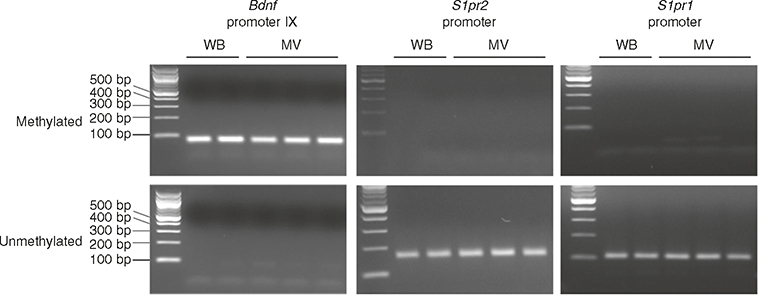
Fig. 8 |. Detection of DNA methylation in Bdnf, S1pr2 and S1pr1 promoter regions in microvessels and whole brain.
Methylation status was examined using methylation-specific PCR with specific primers located in Bdnf promoter IX and S1pr1 and S1pr2 core promoter regions. Genomic DNA was isolated from whole-brain (WB) and the microvessel fraction of adult brain (MV; 2 months old, C57BL/6J) and subjected to bisulfite treatment. The PCR products were analyzed on a 2% (wt/vol) agarose gel. Left lane:100-bp DNA ladder.
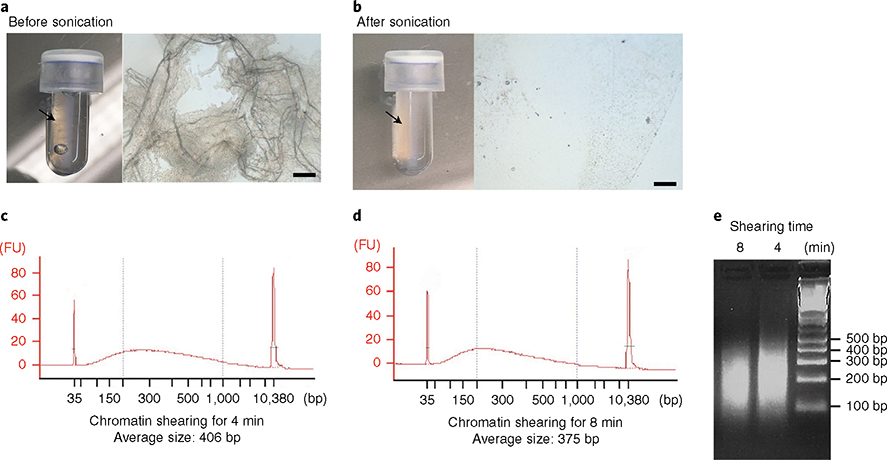
Fig. 9 |. Optimization of chromatin extraction and shearing from cortical microvessels.
Isolated microvessels were cross-linked with 1% (wt/vol) methanol-free paraformaldehyde and subjected to lysis and chromatin extraction protocol. a,b, Optimization of microvessel disruption and lysis. a, Images of lysate after vortexing for 15 min in lysis buffer B, per the manufacturer’s instructions. (Left) Arrow indicates visible microvessels in the microTUBE. (Right) Microscopic image of the lysate taken under inverted microscope (Olympus CKX41; scale bar, 100 μm). b), Images of lysate after optimization of the conditions to efficiently disrupt microvessels. (Left) Microvessels are no longer visible. (Right) Microscopic image of the lysate. c–e, Optimization of chromatin shearing. Impact of the time of sonication on the size of chromatin fragments. After microvessel disruption, extracted chromatin was sonicated for 4 min (c,e) or 8 min (d,e). The size of the sheared chromatin was determined with a Bioanalyzer (c,d) and electrophoresis on a 2% (wt/vol) agarose gel (e). Right lane: 100-bp DNA ladder. FU, fluorescence units.
This protocol will be of great interest to researchers intending to investigate the molecular mechanisms governing cerebrovascular dysfunction in disease models or to study the activation of signaling pathways and intercellular interactions in the cerebral microvasculature in vivo. Ultimately, these studies will aid the development of novel therapies targeting the endothelium to mitigate the progression and/or exacerbation of neuronal injury in cerebrovascular and neurodegenerative diseases.
Development of the protocol and comparison with other methods
To isolate cortical microvessels, we initially used the protocol described by Wu et al.15. While the protocol produced consistent samples in terms of microvessel size and cellular composition, it included an enzymatic digestion step. Although samples would potentially be suitable for immunostaining and some biochemical analyses, application of exogenous enzymes and incubation at 37 °C caused undesirable molecular and metabolic changes, induced cell activation21 and compromised the integrity of the RNA (Fig. 5). On exclusion of the digestion step, samples contained unwanted debris, as assessed by low-power bright-field images, whereby cellular debris and detached cells could easily be observed. To increase the purity of the preparation without the use of enzymatic digestion, we optimized the dissection (Steps 2–5) and homogenization processes (Steps 6 and 7) and used an additional wash step (Step 9). To speed up and optimize the dissection process, we found that gently rolling the brains on blotting paper (Step 3) enabled swift and very efficient removal of the meninges as compared to the use of forceps to pry them away. This technique, which was described in an endothelial cell isolation protocol by Ruck et al.12, considerably shortened the dissection process. In addition, we found that a consistent tissue homogenization technique is essential for producing samples of similar composition and containing minimal debris. Although existing protocols recommended mincing brain tissue into small pieces with scissors, followed by three or 20 strokes using a Dounce homogenizer15,17, we found that not mincing the tissue and using ten-stroke homogenization is enough to homogenize cortices and obtain pure microvessel preparations of consistent cellular composition (Fig. 2). This modification substantially shortened the time of preparation, allowing the preservation of microvessel structure and RNA integrity.
After optimization of our protocol to obtain pure microvessel preparations of consistent cellular composition, the next critical factor for the molecular characterization and quantification of changes in gene expression was the processing of the samples to preserve the quality of the RNA. Therefore, we used RNA quality as the ultimate benchmark in developing our optimized protocol and next examined the impact of the microvessel isolation method on RNA integrity. We compared the RNA integrity of microvessel preparations isolated using two different conditions. In one condition, all reagents, tools and equipment were kept on ice, but the isolation was performed at room temperature (20–25 °C), as previously described17. In the other condition, all steps (from the dissection of the brain to the final step) were conducted in a cold room (4 °C). Interestingly, we found marked differences between the RNA integrities of the microvessel preparations obtained by following these two experimental protocols. As shown in Fig. 5a, RNA from microvessels isolated at room temperature was partially degraded as assessed by the RNA integrity number (RIN = 5.2). By contrast, the RNA quality was markedly increased and RNA showed no degradation (RIN = 10) when microvessels were isolated in a cold room (Fig. 5b). Next, we tested the effect of enzymatic digestion of microvessel fragments on RNA integrity. We incubated each microvessel preparation isolated from one brain in 1.3 ml of Liberase TH (0.625 mg/ml) at 37 °C on a rotor for 0, 20 and 50 min. Complete dissociation of the microvessels, as assessed by microscopy, was observed upon incubation for 50 min with Liberase TH. DPBS (20 ml) was added to stop the digestion, and cell suspensions were spun at 276g for 10 min at 4 °C. Then RNA was isolated to determine RNA integrity. We found that RNA integrity decreased upon incubation at 37 °C with Liberase TH in a time-dependent manner (Fig. 5c–e). RNA degradation may be caused in part by the presence of dead cells. Although non-viable cells could be removed by FACS sorting or by plating cells in tissue culture, our results indicate that the enzymatic digestion step negatively impacts RNA quality in the microvessel preparations.
Finally, we tested how RNA integrity can affect the reliability of molecular characterization results. We chose the sphingosine-1-phosphate receptor 2 (S1pr2) gene, because we have previously shown that S1PR2 is a low-abundance transcript in the endothelium (1–5 mRNA copies/cell)19 and, therefore, is particularly difficult to study in terms of generating consistent results if RNA integrity is compromised. cDNAs were synthesized using RNA isolated from microvessel preparations with RIN ≥9.8 or RIN ≤5.0, and the change in cycle threshold (ΔCt) value of each sample in relation to the average value of the group was calculated (Fig. 5f). We found that the coefficient of variation between the preparations was 18.8% when RIN was ≥9.8 and was as high as 106.2% when RIN was ≤5.0. Together, these data indicate that the method of microvessel fraction preparation has a great impact on the integrity of the RNA preparation, the accurate quantification of transcript levels and, thus, the ability to detect changes in gene expression.
In summary, we found that, compared to previously published protocols, the method described here expedited the preparation and optimized sample processing (i.e., dissection and homogenization techniques, the absence of an enzymatic digestion step and the fact that the entire protocol is conducted at 4 °C in a cold room), which had a great impact on the purity, consistency of the cellular composition, integrity of the preparation, and quality of the RNA. This greatly reduced sample-to-sample variation and thereby improved the ability to detect changes in gene expression in disease models.
Applications of the protocol
Our validation studies have focused on quantification of changes in gene expression in disease models or phosphorylated signaling protein levels after drug administration. In Fig. 6, we show that this protocol can successfully be used to quantify changes in gene expression of low-abundance and high-abundance transcripts in cerebral microvessels in a mouse model of ischemic stroke. We also demonstrate that the protocol is adequate to determine changes in protein phosphorylation upon drug administration in vivo (Fig. 7), which allows the investigation of the activation of signaling pathways in the cerebral microvasculature in which blood-flow conditions and the cellular interactions with mural and parenchymal cells are preserved during the treatments.
DNA methylation is an epigenetic modification that occurs at position 5 of cytosine to generate 5-methylcytosine and is commonly observed to occur in the context of a cytosine followed by a guanine (CpG). DNA methylation at gene promoters leads to transcriptional repression and is an important regulatory mechanism governing cell-specific regulation of gene expression22. In addition, in preclinical and clinical studies, aberrant DNA methylation has been linked to an inflammatory phenotype of the endothelium in atherosclerosis23–25. We describe in detail how to isolate gDNA from microvessels and treat it with bisulfite to determine the DNA methylation status of gene promoters with methylation-specific PCR (Fig. 8). Bisulfite-treated DNA can be used for further methylation analysis, such as pyrosequencing or large-scale bisulfite sequencing. ChIP assay is a very valuable technique for determining histone modifications, as well as binding of proteins (e.g., RNA polymerases, transcription factors) to specific regions of the chromatin, both in vitro and in vivo. Herein, we provide a detailed protocol to properly disrupt microvessels to enable the efficient extraction and shearing of chromatin to the adequate size (Fig. 9) to be used for ChIP assays and further analysis such as ChIP-seq. These protocol applications have the potential to reveal important insights into the role of epigenetics in transcriptional regulation of gene expression in cerebral microvessels and to advance our understanding of the molecular mechanisms governing cerebrovascular dysfunction.
In summary, we describe a protocol to isolate microvessel fragments and extract RNA, protein, DNA and chromatin, which are suitable for many different applications in molecular biology and for biochemical assays in which the quality of the material used is of vital importance in producing reliable, quantitative and reproducible data.
Experimental design and quality controls
Each microvessel isolation procedure should include control and experimental groups conducted simultaneously to minimize any variation between procedures. The protocol described in this article usually takes ~90–120 min when conducted using 4–5 mice. The overall timing of the procedure may differ depending on the level of expertise of the researcher. Therefore, we encourage planning the number of samples in accordance with what can feasibly be completed within 2 h, in order to preserve microvessel structural integrity and prevent degradation of RNA or signaling molecules such as phosphorylated proteins. The expected yields from a microvessel preparation from one mouse are ~3 μg of RNA, 3 μg of gDNA and 160 μg of protein.
Microvessel preparations should be assessed for (i) purity and structural features such as size distribution; (ii) consistency in the cellular composition of endothelial cells, pericytes and astrocytes; (iii) structural integrity and morphological characterization; and (iv) RNA integrity for gene expression analysis.
The purity and structural features of the microvessel preparations should be first examined using a bright-field microscope. When the procedure is performed successfully, microvessels should retain their structure, as shown in Fig. 2a, and brain debris, myelin, white matter and single cells should not be observed in the final preparation. Pure microvessel preparations will also exhibit an absence of neuronal markers (e.g., TUJ1) when assessed by western blot analysis (Fig. 2b,c). In addition, we encourage evaluating the size distribution of the isolated microvessels. We found that the majority (~85%) of microvessel fragments isolated as described in this protocol are <5 μm in diameter and 8% of them are between 5 and 10 μm (overall, ~93% ≤10 μm). A small percentage of the microvessels isolated following the described protocol had a larger caliber (6.7% of the microvessels had a diameter between 10 and 30 μm; Fig. 2d). In our protocol we include a step to remove the pial vessels, so these bigger vessels are expected to be mainly penetrating parenchymal arterioles and venules.
The consistency in the cellular composition between microvessel preparations should be quantified by western blotting or reverse transcription followed by quantitative PCR (RT-qPCR) (Fig. 2e). When microvessel fragments are isolated to obtain quantitative data (e.g., changes in gene expression or activation of signaling pathway analysis), it is critical to confirm the consistency of the cellular composition between preparations by measuring the expression levels of endothelial (e.g., zonula occludens-1, ZO-1, Tjp1), pericyte (e.g., cluster of differentiation 13, CD13, Anpep) and astrocyte markers (e.g., aquaporin-4, Aqp4). We have found that microvessels isolated using this protocol retain consistent cell composition. As shown in Fig. 2e, we found that four different preparations contained similar levels of these three cellular components of the BBB.
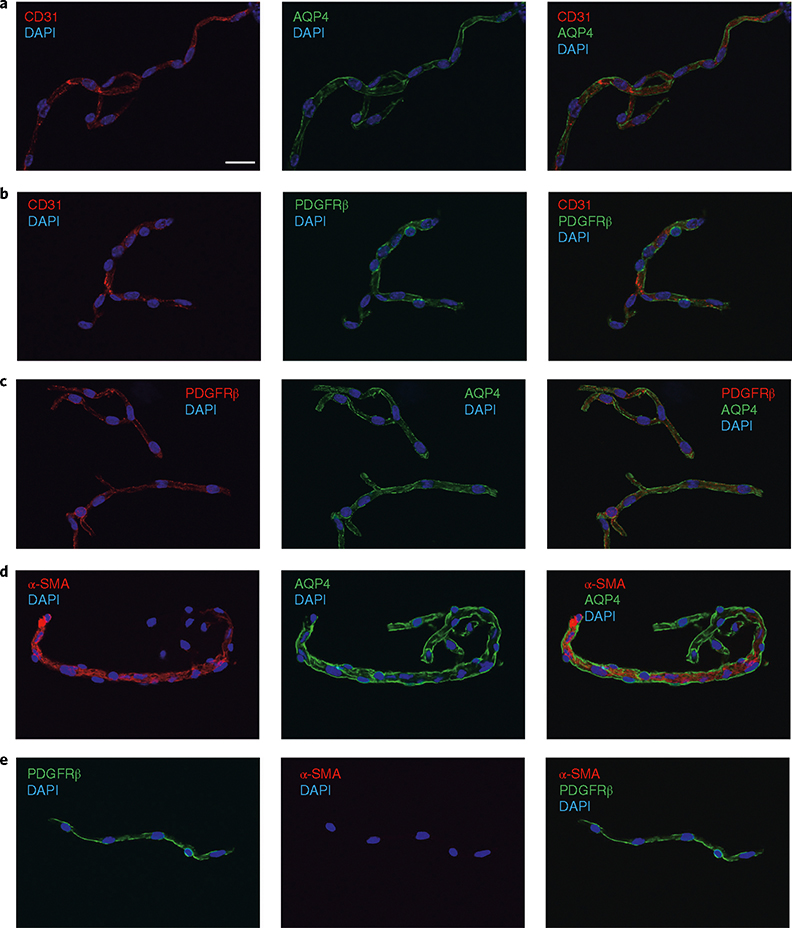
The structural integrity of microvessel fragments and their morphological characterization should be assessed by immunofluorescence staining using antibodies for endothelial (e.g., CD31), pericyte (platelet-derived growth factor receptor-beta, PDGFRβ), vascular smooth muscle cell (α-smooth muscle actin, α-SMA) and astrocytic end foot (AQP4) markers (Figs. 3 and 4). In Fig. 3, representative pictures of the microvessel fragments (diameter ≤10 μm, which are ~93% of the microvessels) are shown. When the protocol is performed successfully, microvessel fragments should contain endothelial cells surrounded by mural cells (pericytes or vascular smooth muscle cells) and the end feet of the astrocytes, as assessed by the presence of CD31, PDGFRβ and AQP4, respectively (Fig. 3). In Fig. 3d, the capillary branches (α-SMA−) of a precapillary arteriole (α-SMA+) are shown. Figure 3e is a representative picture of an α-SMA− microvessel fragment (capillary). We further characterized the microvessel fragment preparations according to size and α-SMA content (assessed by immunodetection). As shown in Fig. 4a, we found that all vessels <5 μm in diameter (85.3% of all microvessel fragments, Fig. 2d) were negative for α-SMA, indicating that they are capillaries and post-capillary venules26,27. Only 8% of the vessel fragments ≤10 μm in diameter (93.3% of all microvessel fragments) were α-SMA+ (probably precapillary arterioles26,27), whereas we found that all microvessels >10 μm (6.7% of all microvessel fragments) were positive for α-SMA (penetrating arterioles and venules). Thus, when performed successfully, the described protocol yields intact microvessel fragments (comprising endothelial cells surrounded by pericytes/smooth muscle cells and the end feet of astrocytes), the vast majority of which are capillaries on the basis of size and cell marker analysis (85.3% of all microvessel fragments were α-SMA−).
Fig. 3 |. Morphological characterization of microvessel preparations: structural integrity and cell composition.
a–e, Representative pictures of immunofluorescence analysis of isolated microvessels (≤10 μm, ~93% of all microvessels) stained with antibodies for each cell component. Endothelial markers (CD31, red channel; a,b), astrocyte end-foot markers (AQP4, green channel; a,c) and pericyte/smooth muscle cell markers (PDGFRβ, green or red channel; b,c) were detected in all microvessel fragments. d, Notice the transition of a precapillary arteriole (α-SMA+) to capillaries (α-SMA−) in the microvessel fragment. e, Example of an α-SMA− microvessel. DAPI (nuclear stain, blue channel). Images were taken using confocal laser scanning microscope (Olympus FV10i) and processed using Olympus Fluoview v.3.0. Scale bar, 20 μm.
Last, when microvessels are to be used to determine changes in RNA expression between experimental groups, each RNA sample isolated should be tested for RNA integrity. We have demonstrated that RNAs from cortical microvessels can be easily degraded during prolonged procedures and incubations (Fig. 5); microvessel isolation is a long process relative to routine whole-tissue harvesting and pulverization for RNA extraction, which prolongs the time that microvessels are exposed to endogenous RNases. Although we found that performing the entire procedure at 4 °C protects against RNA degradation very effectively (Fig. 5a,b), we strongly recommend examining RNA integrity for each sample before proceeding with further analysis. We used a Bioanalyzer to test RNA integrity because it gives quantitative values (RIN 1–10) and requires only a small amount of RNA for each assay, but alternative methods such as gel electrophoresis can be used if a Bioanalyzer is unavailable.
In summary, when performed successfully, the described method yields pure microvessel preparations of consistent cell composition and very high structural, as well as RNA and phosphoprotein, integrity. If the quality controls described in this section are met, the microvessel preparations can be confidently used for quantitative applications such as those described in this protocol.
Level of expertise needed to implement the protocol
In regard to the level of expertise needed to implement this protocol, it is important to consider that the isolation of microvessels requires dissection of the mouse brain. Although a microscope is not necessary for the dissection, competent handling of forceps and other appropriate dissecting tools is required to cleanly and swiftly separate the brain from the skull and to remove the cortex from the deeper structures of the brain. Beyond dissection skills, a person who has both some general rodent-handling experience and familiarity with basic biological techniques (such as preparation of solutions) and equipment (such as the centrifuge) should be able to conduct the procedure competently with 5–10 practice experiments. In order to obtain intact preparations, the entire protocol needs to be completed in 90–120 min.
Advantages and limitations of this approach
The key advantages of the proposed method for the isolation of cerebral microvessels are the following:
This method permits the prompt isolation of high-purity cortical microvessel fragments of consistent cell composition without the need for enzymatic digestion, thereby preserving RNA and phosphoprotein integrity and minimizing cell activation. Therefore, it enables quantification of changes in gene expression (even in low-abundance transcripts or pre-mRNA) and cell signaling pathway analysis in disease models upon in vivo pharmacological treatments or genetic deletion in mice.
Owing to the high integrity and cellular consistency of the preparations obtained using this method, additional molecular studies can be conducted to gain mechanistic insights into the molecular mechanisms governing these changes in gene expression (e.g., to determine epigenetic changes). We provide detailed protocols for the determination of the methylation status of specific gene promoters as well as for chromatin isolation and shearing for ChIP analysis.
Because protein post-translational modifications are also retained (e.g., phosphorylation), the method can be used for the quantification of the activation of signaling pathways or changes in the subcellular localization of TJ proteins upon ligand or drug administration in vivo. The data obtained can be correlated with in vivo functional assays, such as the determination of BBB function via injection of specific vascular markers20. Compared to alternative approaches, such as brain endothelial cell isolation and in vitro culture, the proposed method preserves the physiological blood-flow conditions and the cellular interactions with mural cells during the treatments.
When mastered, the technique is easy and fast, and it yields intact cerebral microvessel fragments with consistent cell composition.
As with any technique, this protocol has limitations, which must be kept in mind during the experimental design and the interpretation of the results. The main limitations of this technique are the following:
The preservation of the cellular interactions between the endothelium and mural cells is critical to maintaining the phenotype and the molecular signature of the cerebrovascular endothelium13,14. The microvessel fragments consist of three main cellular components (i.e., endothelial cells, pericytes and end feet of astrocytes). For this reason, acquisition of quantitative data is possible only when cell composition is consistent between control and experimental groups. Therefore, we recommend assessing the quality controls described above (‘Experimental design and quality controls’ section) in order to detect any differences in cell composition due to experimental treatments. If the disease model alters the cellular composition, we recommend choosing an earlier time point or modulating the severity of the disease to prevent changes in the cellular components. If this is not feasible, alternative methods, such enzymatic digestion to achieve single-cell isolation, followed by endothelial cell sorting, need to be used. However, it is important to keep in mind that this further processing of the samples will compromise the RNA quality and will lead to cell activation, which can cause changes in gene expression21 (e.g., induction of immediate–early genes), hindering the ability to detect molecular changes between treatment groups.
There is enrichment of cerebral capillaries in the preparation (~85% of microvessels). Therefore, for the study of larger-caliber cerebral vessels, other protocols should be followed28.
As in other organs, the brain vasculature is heterogeneous; with the proposed method, it is possible to quantify only average changes across the microvessel preparation. This approach can be combined with semi-quantitative techniques such as immunofluorescence analysis or in situ hybridization to detect changes in discrete regions or specific cell populations. If information regarding cell-specific changes in gene expression or the global distribution of transcript levels across cell populations needs to be obtained, other approaches, such as enzymatic digestion followed by cell sorting or single-cell RNA sequencing, need to be used. Nevertheless, the data obtained with the protocol described herein can provide very useful information that can complement the data obtained by these other approaches.
Materials
Biological materials
Adult C57BL/6J mice (6 weeks old to 4 months old; Jackson Laboratory, cat. no. 000644) and/or a transgenic mouse line. In our protocol, we have used S1pr2 null mice29 to confirm the data obtained with the S1PR2 antagonist JTE-013
 All animal experiments should be conducted in accordance with the relevant institutional guidelines and regulations. Our protocol for the use of laboratory animals was approved by the Weill Cornell Medicine Institutional Animal Care and Use Committee.
All animal experiments should be conducted in accordance with the relevant institutional guidelines and regulations. Our protocol for the use of laboratory animals was approved by the Weill Cornell Medicine Institutional Animal Care and Use Committee.
Reagents
JTE-013 (Cayman Chemical, cat. no. 10009458)
(2-Hydroxypropyl)-β-cyclodextrin (Sigma-Aldrich, cat. no. C0926)
Dimethyl sulfoxide (DMSO; Thermo Fisher Scientific, cat. no. D1391)
Ultrapure water
Microvessel isolation
DPBS (10×; Thermo Fisher Scientific, cat. no. 14190250)
Dextran (molecular weight (MW) ~70,000 Da; Sigma-Aldrich, cat. no. 31390)
MCDB 131 medium (no glutamine; Thermo Fisher Scientific, cat. no. 10372019)
Fatty-acid-free BSA (protease–nuclease free; Millipore Sigma, cat. no. 126609)
Liberase TH (Roche, cat. no. 5401135001)
NP-40 (Thermo Fisher Scientific, cat. no. 28324)
Immunofluorescence staining
Paraformaldehyde (PFA; Sigma-Aldrich, cat. no. P6148)
 PFA is classified as a known cancer hazard. It should be handled only in a working chemical fume hood with personal protective equipment (PPE) such as lab coat, gloves and face shield.
PFA is classified as a known cancer hazard. It should be handled only in a working chemical fume hood with personal protective equipment (PPE) such as lab coat, gloves and face shield.NaOH (1 N; Fisher Scientific, cat. no. SS266–1)
HCl (1 N; Fisher Scientific, cat. no. SA48–1)
BSA (Fisher Scientific, cat. no. BP-1605)
Anti-CD31 (Dianova, cat. no. DIA-310, RRID: AB_2631039)
Anti-Aquaporin 4 (Sigma-Aldrich, cat. no. A5871, RRID: AB_258270)
Anti-PDGFRβ (R&D Systems, cat. no. AF1042-SP, RRID: AB_2162633)
Anti-α-SMA-Cy3 (Sigma-Aldrich, cat. no. C6198, RRID: AB_476856)
DAPI (Sigma-Aldrich, cat. no. D9542)
Vectashield antifade mounting medium (Vector Labs, cat. no. H1000)
RNA isolation
Diethyl pyrocarbonate (DEPC; Sigma-Aldrich, cat. no. D5758)
β-Mercaptoethanol (Bio-Rad, cat. no. 1610710)
 β-Mercaptoethanol is toxic upon inhalation and contact with skin. It should be handled only in a working chemical fume hood with PPE such as lab coat, gloves and face shield.
β-Mercaptoethanol is toxic upon inhalation and contact with skin. It should be handled only in a working chemical fume hood with PPE such as lab coat, gloves and face shield.RNase-free DNase set (Qiagen, cat. no. 79254)
RNeasy Mini Kit (Qiagen, cat. no. 74106)
Tris–HCl (1 M, pH 8.0; Boston BioProducts, cat. no. BM-320)
EDTA (0.5 M, pH 8.0; Boston BioProducts, cat. no. BM-150)
Liquid nitrogen (TechAir, cat. no. NI230L22)
Protein isolation
BCA Protein Assay Kit (Pierce, cat. no. 23227)
Protease Inhibitor Cocktail Set I (Calbiochem, cat. no. 539131)
HEPES buffer (1 M, pH 7.3; Fisher Scientific, cat. no. BP299100)
Triton X-100 detergent (Bio-Rad, cat. no. 1610407)
Sodium deoxycholate (Sigma-Aldrich, cat. no. D6750)
Sodium dodecyl sulfate (SDS; Fisher Scientific, cat. no. BP166–100)
Sodium chloride (NaCl; 5 M; Promega, cat. no. V4221)
Magnesium chloride solution (MgCl2; 1 M; Sigma-Aldrich, cat. no. M1028)
β-Glycerophosphate disodium salt hydrate (Sigma-Aldrich, cat. no. G6251)
Sodium orthovanadate (Na3VO4; Sigma-Aldrich, cat. no. S6508)
Sodium fluoride (NaF; Sigma-Aldrich, cat. no. 215309)
Tris-buffered saline (TBS; Bio-Rad, cat. no.170–6435)
Monoclonal anti-β-actin antibody (Sigma-Aldrich, cat. no. A5316, RRID: AB_476743)
Phospho-Akt (Ser473) (193H12) rabbit mAb (Cell Signaling Technology, cat. no. 4058, RRID: AB_331168)
gDNA isolation and bisulfite conversion
DNeasy Blood & Tissue Kit (Qiagen, cat. no. 69504)
Sodium acetate (3 M; Sigma-Aldrich, cat. no. S7899)
100% (vol/vol) Ethanol (Thermo Fisher Scientific, cat. no. 07–678-004)
EpiTect Bisulfite Kit (Qiagen, cat. no. 59104)
EpiTect MSP Kit (Qiagen, cat. no. 59305)
Agarose (Denville Scientific, cat. no. GR140)
Ethidium bromide solution (Bio-Rad, cat. no. 1610433)
DNA ladder (100 bp; New England BioLabs, cat. no. N3231)
Crosslinking and chromatin shearing
RNase A (Sigma-Aldrich, cat. no. R4642)
Proteinase K (Roche, cat. no. 50-100-3393)
truChIP Chromatin Shearing Kit with Formaldehyde (Covaris, cat. no. 520155)
PCR purification kit (Qiagen, cat. no. 28104)
Equipment
Dounce tissue grinder set (Sigma-Aldrich, cat. no. D9063)
Round-bottom tubes (14 ml; Nunc, cat. no. 150268)
Ultracentrifuge (Sorvall, model no. RC-5C Plus with model no. 05-SS-34 rotor)
Microcentrifuge tube (USA Scientific, cat. no. 1615–5500)
Surgical fine forceps (Fine Science Tools, cat. no. 11271–30)
Petri dishes (Thermo Fisher Scientific, cat. no. FB0875712)
Inverted light microscope (Olympus, model no. CKX41)
Freezer (Panasonic, model no. MDF-U700VXC)
Shaker (Bellco Glass, model no. 7744–02020)
Rotator (Boekel Scientific, model no. 260200)
Superfrost microscope slides (Fisher Scientific, cat. no. 12–550-123)
Cover glasses (Fisher Scientific, cat. no. 12-544-12)
Confocal laser scanning microscope (Olympus, model no. FV10i)
RT-PCR system (Applied Biosystems, model no. ABI 7500 Fast)
Pulsing vortex mixer (Fisher Scientific, model no. 02-215-375)
Electrophoresis instrument (Agilent, model no. Bioanalyzer 2100)
Sonicator (Fisherbrand Q55 Sonicator; Fisher Scientific, model no.FB50)
Gel documentation system (Bio-Rad, model no. 1708195EDU)
Focused ultrasonicator (Covaris, model no. M220)
microTUBEs with snap-cap, AFA fiber (Covaris, cat. no. 520045)
Falcon cell strainer (Corning, cat. no. 352340)
Shredder (Qiagen, cat. no. 79654)
Blotting paper (Bio Rad, cat. no. 1703966)
Bottle-top filter (0.2 μm; Corning, cat. no. 430769)
CO2 chamber (VetEquip, model no. 941443)
Thermal cycler (Bio Rad, model no. 1851148)
Kimwipes (Kimtech, cat. no. 34133)
Reagent setup
S1pr2 null mice
Mice carrying targeted deletion of the S1pr2 gene are maintained on a mixed C57BL/6;129Sv genetic background (five times back-crossed to C57BL/6)29. Wild-type littermates should be used as controls for experiments on S1pr2 knockout (KO) mice. In our protocol, we have used S1pr2 null mice29 to confirm the data obtained with the S1PR2 antagonist JTE-013. When using other antagonists or inhibitors, a relevant transgenic mouse line can be used, if available.
JTE-013 stock solution
Dissolve JTE-013 in DMSO at a final concentration of 100 mM. Make sure that it completely goes into solution. Aliquots can be stored for up to 6 months at −20 °C.
JTE-013 gavage administration
Prepare a solution of 2% (wt/vol) (2-hydroxypropyl)-β-cyclodextrin in DPBS, saline or H2O. Filter to sterilize and prepare aliquots of the solution to keep it sterile. Aliquots can be stored at −20 °C for 3 months. Just before gavage, prepare a suspension of 3 mg/ml JTE-013 in 2% (wt/vol) (2-hydroxypropyl)-β-cyclodextrin solution. Vortex right before gavage to homogenize the suspension.
15% (wt/vol) dextran–DPBS
Weigh 1.5 g of dextran into a 50-ml conical tube and add 8 ml of DPBS. Rotate the tube at 4 °C until the dextran completely dissolves. This takes about 30 min. Adjust the final volume to 10 ml with DPBS. The solution can be stored for 1 week at 4 °C.
0.5% (wt/vol) BSA in MCDB 131 medium
Dissolve 0.5 g of fatty acid–, protease-, endotoxin-free BSA in 10 ml of 1× DPBS in a 15-ml conical tube to make a 5% (wt/vol) BSA–DPBS solution. Once the solution looks clear, add 1 ml of 5% (wt/vol) BSA–DPBS to 9 ml of MCDB 131 medium. The 0.5% (wt/vol) BSA in MCDB 131 medium can be stored for up to 6 months at −20 °C.
4% (wt/vol) PFA–DPBS
Weigh 20 g of PFA and add it to pre-heated ultrapure water at 60 °C in a glass beaker. Cover and maintain at 60 °C. Add 1 N NaOH until the solution becomes clear, remove the beaker from the heat and add 50 ml of 10× DPBS. Adjust the pH to 7.2 using HCl and adjust the final volume to 500 ml. Sterile-filter with a 0.2 μm bottle-top filter, prepare aliquots of small volume and store for up to 3 months at −20 °C  PFA is classified as a known cancer hazard. It should be handled only in a working chemical fume hood with PPE such as lab coat, gloves and face shield.
PFA is classified as a known cancer hazard. It should be handled only in a working chemical fume hood with PPE such as lab coat, gloves and face shield.
0.1% (vol/vol) NP-40–DPBS
Add 1 ml of NP-40 to 999 ml of 1× DPBS in a plastic beaker and mix well using a magnetic stirrer. Sterile-filter using 0.2 μm bottle-top filter and store at room temperature protected from light for up to 6 months.
Blocking solution for immunofluorescence staining
Dissolve 5 g of BSA in 100 ml of 1× DPBS and mix well. Once the solution looks clear, sterile-filter and make 10-ml aliquots. This solution can be stored for up to 6 months at −20 °C.
DEPC water
Add 1 ml of DEPC to 1 liter of ultrapure water in a clean glass bottle, mix well and incubate for 3 h at room temperature with occasional swirling. Autoclave for 15 min and store at room temperature for up to 12 months.
1× TE buffer
Add 5 ml of 1 M Tris–HCl, pH 8.0, and 1 ml of 0.5 M EDTA, pH 8.0, to 500 ml of ultrapure water and mix well. Autoclave for 15 min and store at room temperature for up to 12 months.
1 M Na3VO4
Weigh 1.839 g of Na3VO4 and dissolve it in 10 ml of ultrapure water. Make 500-μl aliquots and store for up to 12 months at −20 °C. Avoid free–thaw cycles.
1 M NaF
Weigh 0.42 g of NaF and dissolve it in 10 ml of ultrapure water. Make 500-μl aliquots and store for up to 12 months at −20 °C. Avoid free–thaw cycles.
Equipment setup
Ultracentrifuge
Pre-chill the ultracentrifuge to 4 °C before use.
Focused ultrasonicator
Fill the chamber with water according to the manufacturer’s instructions and pre-chill before using.
Procedure
Microvessel isolation  1.5–2 h
1.5–2 h
 The entire procedure should be carried out in a cold room.
The entire procedure should be carried out in a cold room.
-
1
Sacrifice mice by placing them into a CO2 chamber for 2–3 min and performing cervical dislocation; then bring the mice immediately to a cold room. Use at least one control and one treated mouse (or equivalent) per procedure.
-
2
Carefully remove the brain and transfer it immediately to a Petri dish containing MCDB 131 medium; rinse the brain in the medium. Ensure that the entire brain remains submerged in the medium.
 RNAs start to degrade as soon as the mouse is euthanized. Perform Steps 1 and 2 on one mouse at one time and keep the brain in the cold MCDB 131 medium on ice until the entire group is processed. We recommend having 4–5 mice per isolation because a larger number of mice can slow down the entire process.
RNAs start to degrade as soon as the mouse is euthanized. Perform Steps 1 and 2 on one mouse at one time and keep the brain in the cold MCDB 131 medium on ice until the entire group is processed. We recommend having 4–5 mice per isolation because a larger number of mice can slow down the entire process. -
3
Using closed forceps, gently and evenly roll the brain on blotting paper to remove the meninges and meningeal vessels. Place the brain in DPBS (which provides greater visibility than MCDB 131 medium for optimal dissection).
-
4
Using a razor blade, slice the brain sagittally and remove the cerebellum.
-
5
Isolate the cortices by removing all deep-brain structures, including the hippocampus and residual white matter, being careful to avoid damaging the cortices. Although as much of the above should be removed as possible, a certain amount of white matter is likely to remain attached. Because this will be removed in future steps, do not attempt to remove all white matter tissue at the cost of damaging the cortices.
? TROUBLESHOOTING
 If you cannot proceed immediately to the next step, place the isolated cortices in MCDB 131 medium on ice (4 °C), but do not leave the dissected cortices for more than 15 min.
If you cannot proceed immediately to the next step, place the isolated cortices in MCDB 131 medium on ice (4 °C), but do not leave the dissected cortices for more than 15 min. -
6
Homogenize the cortical tissue using a loose-fit, 7-ml Dounce tissue grinder. Perform an initial ten strokes in 1 ml of MCDB 131 medium at a steady pace, without twisting the pestle.
-
7
Add 7 ml of MCDB 131 medium to the homogenate from Step 6 and perform an additional two strokes in a total of 8 ml.
-
8
Pour the homogenized tissue into a 14-ml round-bottom centrifuge tube. Take care to adjust the volumes of the tubes with additional MCDB 131 medium as needed, so that all samples contain the same amount of mixture.
-
9
Centrifuge at 2,000g for 5 min at 4 °C. Pour out the supernatant and rest the inverted tube on a paper towel to absorb excess medium/debris clinging to the sides.
 This is a wash step; it helps with the removal of small pieces of white-matter debris that have remained attached to the sample.
This is a wash step; it helps with the removal of small pieces of white-matter debris that have remained attached to the sample. -
10
Resuspend the pellet in 15% (wt/vol) dextran–DPBS. To dissociate the pellet evenly, add an initial 1 ml of 15% (wt/vol) dextran solution to the pellet and use it to sluice the pellet gently from the sides of the tube. Resuspend the floating pellet with 5–6 steady uses of the pipette, taking up the entire pellet with each use. Then add a further 7 ml of 15% (wt/vol) dextran solution and balance with additional 15% (wt/vol) dextran solution.
-
11
Centrifuge at 10,000g for 15 min at 4 °C. A red, microvessel-containing pellet will appear.
 If the pellet does not appear to be a clean red color, it indicates contamination with white matter, myelin or debris.
If the pellet does not appear to be a clean red color, it indicates contamination with white matter, myelin or debris.? TROUBLESHOOTING
-
12
Remove the supernatant carefully to avoid contaminating the pellet with debris. Begin by pipetting off the top, brain tissue–containing layer completely and then aspirate the underlying solution, which will contain a small amount of debris.
? TROUBLESHOOTING
-
13
Remove the pellet with 1 ml of DPBS. Sluice the pellet from the sides of the tube and take it up into the pipette tip with the absolute minimum of DPBS that has passed into the tube.
 Using more than 1 ml of DPBS is discouraged. You will see some debris on the wall of the tube that can be easily taken up by pipetting. Use of a small volume is beneficial in minimizing the contamination.
Using more than 1 ml of DPBS is discouraged. You will see some debris on the wall of the tube that can be easily taken up by pipetting. Use of a small volume is beneficial in minimizing the contamination. -
14
Transfer the pellet to a 40-μm cell strainer and wash it through with <10 ml of DPBS.
 This step is intended to remove debris that has remained slightly adhered to the microvessels, as well as to remove loose debris.
This step is intended to remove debris that has remained slightly adhered to the microvessels, as well as to remove loose debris. Depending on the downstream application, you may need to proceed directly to Step 18 immediately after this step.
Depending on the downstream application, you may need to proceed directly to Step 18 immediately after this step.? TROUBLESHOOTING
-
15
Reverse the filter and retrieve the microvessels using 10–20 ml of MCDB 131 medium containing 0.5% (wt/vol) fatty-acid–, protease- and endotoxin-free BSA.
Bright-field and phase-contrast imaging
-
16
To quickly evaluate the purity of the microvessels by a microscopic method, take 50 μl of the microvessel suspension from Step 15, plate it onto a 35-mm culture dish and leave it for 5 min at room temperature. Once microvessels attach to the surface of the dish, observe them under an inverted microscope.
-
17
Centrifuge at 5,000g for 10 min at 4 °C. This will yield the final microvessel pellet.
? TROUBLESHOOTING
Analysis of microvessels
-
18For characterization by immunofluorescence, follow option A. For RNA isolation, follow option B. For protein extraction, follow option C. For gDNA isolation and bisulfite conversion, follow option D. For crosslinking and chromatin shearing, follow option E.
- Immunofluorescence characterization
 24 h
24 h
- Add 8 ml of 4% (wt/vol) PFA–DPBS to a 35-mm Petri dish.
- Submerge the cell strainer containing the microvessels from Step 14 in 4% (wt/vol) PFA–DPBS and fix the microvessels for 15 min with gentle shaking.
- Place the cell strainer on a 50-ml tube and wash the microvessels through with 10 ml of DPBS.
- Reverse-filter on another 50-ml tube and collect the microvessels by thoroughly applying 15–20 ml of 1% (wt/vol) BSA–DPBS to the surface of the filter.
- Centrifuge the suspension at 2,000g for 10 min at 4 °C.
- Aspirate and discard the 1% (wt/vol) BSA–DPBS and resuspend the pellet in 400 μl of DPBS.
-
Drop 50 μl of the suspension onto each Superfrost microscope slide and air-dry the slides. It takes about 30 min.
 The slides can be stored at −80 °C for up to 12 months before processing.
The slides can be stored at −80 °C for up to 12 months before processing. - Wet the microvessels with 1× DPBS for 10 min.
- Permeabilize the microvessels with 0.1% (vol/vol) NP-40–DPBS for 15 min.
- Block the microvessels with 5% (wt/vol) BSA–DPBS for 1 h.
- Incubate the microvessels overnight at 4 °C with the primary antibodies CD31 (1:200), PGDFRβ (1:200), AQP4 (1:500) or SMAα-Cy3 (1:200) diluted in 5% (wt/vol) BSA–DPBS.
- After three washes with 0.1% (vol/vol) NP-40–DPBS, incubate the microvessels for 1 h at room temperature in the dark with the appropriate secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 568.
- Wash three times with 0.1% (vol/vol) NP-40–DPBS.
- Counterstain the microvessels with DAPI for 1 min.
- Wash once with 0.1% (vol/vol) NP-40–DPBS.
- Air-dry the slides and mount with Vectashield mounting medium. Protect the slides from the light and store at 4 °C for up to 2 weeks
- Observe the staining under a confocal microscope.
- RNA isolation
 2–3 h
2–3 h
- Centrifuge at 5,000g for 10 min at 4 °C to yield the final microvessel pellet.
- Discard the supernatant and quickly remove the residual medium by inverting the tube onto a paper towel.
- Add 700 μl of RLT buffer from the RNeasy Mini Kit.
- Resuspend the microvessel pellet in RLT buffer with 20–25 times of pipetting and vigorous vortexing for ~30 s to completely disrupt the microvessels.
-
Snap-freeze in liquid nitrogen.
 The lysate can be stored at −80 °C for up to 6 months before processing.
The lysate can be stored at −80 °C for up to 6 months before processing. - Isolate the RNA according to the instructions in the RNeasy Mini Kit and include the DNaseI treatment step as described there.
-
Measure the RNA integrity with a Bioanalyzer instrument.
 RNA integrity should be tested before downstream analysis of the mRNA.
RNA integrity should be tested before downstream analysis of the mRNA.
- Protein extraction
 2 h
2 h
- Centrifuge at 5,000g for 10 min at 4 °C to yield the final microvessel pellet.
- Discard the supernatant by inverting the tube containing the pelleted microvessels and standing it on a paper towel for a few seconds.
-
Add 80 μl of ice-cold protein lysis buffer (50 mM HEPES, pH 7.5; 1% (vol/vol) Triton X-100; 0.5% (wt/vol) sodium deoxycholate; 0.1% (wt/vol) SDS; 500 mM NaCl; 10 mM MgCl2; 50 mM β-glycerophosphate; 1× protease inhibitor cocktail; 1 mM Na3VO4; 1 mM NaF).
 Add protease inhibitor cocktail and phosphatase inhibitors (Na3VO4 and NaF) to fresh protein lysis buffer immediately before use.
Add protease inhibitor cocktail and phosphatase inhibitors (Na3VO4 and NaF) to fresh protein lysis buffer immediately before use. - Sonicate the pelleted microvessels for 3 × 5 s at 50 amplitude with a probe-type sonicator on ice.
-
Vortex the lysate at maximum speed for 15 min at 4 °C. This step will help lyse the microvessels adequately.
 Keep the tip of the probe in the lysate to avoid foaming during sonication.
Keep the tip of the probe in the lysate to avoid foaming during sonication. -
Centrifuge the lysate at 10,000g for 10 min at 4 °C and transfer the supernatant to a new tube.
 Proteins can be stored at −80 °C for up to 6 months before processing.
Proteins can be stored at −80 °C for up to 6 months before processing. -
Measure the protein concentration using a bicinchoninic acid (BCA) assay kit according to the manufacturer’s instructions.
 We normally obtain 150–160 μg of protein from one brain.
We normally obtain 150–160 μg of protein from one brain.
- gDNA isolation and bisulfite conversion
 12–24 h
12–24 h
- Centrifuge at 5,000g for 10 min at 4 °C to yield the final microvessel pellet.
- Discard the supernatant by inverting the tube containing the pelleted microvessels and standing it on a paper towel for a few seconds.
- Add 180 μl of ATL buffer from the DNeasy Blood & Tissue Kit.
- Add 20 μl of proteinase K and mix vigorously by vortexing for ~10 s.
-
Incubate the tube at 56 °C until the microvessel pellet is completely dissolved. This takes 1.5–2 h.
 Make sure the pellet is completely dissolved.
Make sure the pellet is completely dissolved. -
Isolate gDNA according to the instructions from Qiagen.
 gDNA can be stored at −20 °C up to 6 months before processing.
gDNA can be stored at −20 °C up to 6 months before processing. - Add 2 μl of 25 mg/ml RNase A to 180 μl of eluate.
- Incubate the tube at 37 °C for 1 h.
- Precipitate the gDNA by adding a 1/10 volume of 3 M sodium acetate (NaOAc) and 2 volumes of 100% (vol/vol) ethanol.
-
Determine the concentration of the RNase-treated gDNA.
 RNase-treated gDNA can be stored at −20 °C for up to 6 months before processing.
RNase-treated gDNA can be stored at −20 °C for up to 6 months before processing. - Use 2 μg of gDNA to perform the bisulfite conversion. Add 85 μl of Bisulfite Mix, 35 μl of DNA Protect Buffer, and RNase-free water from the EpiTect Bisulfite Kit to the PCR tube containing 2 μg of DNA and perform the reaction using a thermal cycler according to the instructions from Qiagen.
- Proceed to the cleanup of bisulfite-converted DNA as instructed by the kit.
- In the PCR reaction, use 3–5 μl of eluted DNA and primers designed to detect DNA methylation.
-
Crosslinking and chromatin shearing
 24 h
24 h We used a commercially available kit, the truChIP Chromatin Shearing Kit with Formaldehyde from Covaris. Here, we describe only steps optimized from the manufacturer’s instructions for usage of microvessels.
We used a commercially available kit, the truChIP Chromatin Shearing Kit with Formaldehyde from Covaris. Here, we describe only steps optimized from the manufacturer’s instructions for usage of microvessels.
- Add 8 ml of 1% (wt/vol) methanol-free PFA solution from the kit to a 35-mm Petri dish.
 PFA is classified as a known cancer hazard. It should be handled only in a working chemical fume hood with PPE such as lab coat, gloves and face shield.
PFA is classified as a known cancer hazard. It should be handled only in a working chemical fume hood with PPE such as lab coat, gloves and face shield. -
Transfer the cell strainer containing the washed microvessels from Step 14 to a Petri dish and incubate for 5 min at room temperature with gentle shaking.
 Make sure the strainer is submerged completely in the 1% (wt/vol) methanol-free PFA solution during the crosslinking.
Make sure the strainer is submerged completely in the 1% (wt/vol) methanol-free PFA solution during the crosslinking. - Add quenching buffer provided the kit as described in the manufacturer’s instructions to stop crosslinking.
- After a brief wash with 2 ml of DPBS, retrieve the fixed microvessels from the strainer with 10 ml of 1% (wt/vol) BSA–DPBS, and centrifuge at 2,000g for 10 min at 4°C.
-
Discard the supernatant and resuspend the pellet in lysis buffer B from the kit. We combine microvessels from two brains in 300 μl of lysis buffer B.
 Resuspended microvessel pellet can be stored at −80 °C before processing for up to 6 months.
Resuspended microvessel pellet can be stored at −80 °C before processing for up to 6 months. - Incubate the lysate for 15 min at 4 °C on a rotator and transfer the lysate to the microTUBEs.
-
Set the Covaris ultrasonicator to 5% duty factor, 50 peak incident power and 200 cycles per burst and sonicate the lysates for 3 min.
 We incorporated a mild sonication step because of difficulties with pulverizing microvessels. To maximize the yield of chromatin extraction, it is critical to disrupt and lyse the microvessels properly and completely. After Step 18E(vii), the structure of the microvessels should not be visible.
We incorporated a mild sonication step because of difficulties with pulverizing microvessels. To maximize the yield of chromatin extraction, it is critical to disrupt and lyse the microvessels properly and completely. After Step 18E(vii), the structure of the microvessels should not be visible. - Centrifuge the lysate from Step 18E(vii) at 1,700g for 5 min at 4 °C.
- Add 600 μl of wash buffer from the kit and incubate for 5 min on a rotator at 4 °C, followed by centrifugation at 1,700g for 5 min at 4°C to collect the nuclear extract.
- Resuspend the pellet in 550 μl of shearing buffer from the kit and vortex until it is mixed.
- Transfer the chromatin to the microTUBE and sonicate with the default setting from the ‘Low Cell Chromatin Shearing Protocol for M-Series’, as described in the manufacturer’s instructions, for 4 or 8 min.
- Process the sheared chromatin according to the instructions in the truChIP Chromatin Shearing kit with formaldehyde, to check the size of the sheared chromatin by electrophoresis with a 2% (wt/vol) agarose gel or with a Bioanalyzer instrument.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
|
| |||
| 5 | Delayed or improper dissection of cortices | Usage of inappropriate tools can cause difficulties | Use curved-end forceps |
| 11 | The pellet does not seem red or is covered with a white layer | The pellet was not properly resuspended | Mix the pellet from Step 9 completely with 15% (wt/vol) dextran–DPBS by pipetting |
| The pellet does not appear at the bottom of tube | The concentration of dextran solution was incorrect | Confirm that the dextran completely dissolves. Do not exceed the correct final volume | |
| 12 | Debris is too close to the pellet. It is difficult to get a clean pellet | Dextran was not mixed thoroughly with brain lysate or a small volume of dextran solution was used | Increase the volume of dextran and mix the pellet thoroughly by pipetting. If necessary, wipe out debris from the walls of the tube with a Kimwipe |
| 14 | The pellet becomes trapped inside the pipette tip | The microvessels can easily adhere to the surface of plastic | Pre-wet the pipette tip by pipetting 1× DPBS in and out repeatedly |
| 17 | The pellet is very small or invisible | The microvessels remain in the cell strainer or BSA was not added to the medium | Use enough volume to retrieve the microvessels from the strainer. Add BSA (0.5% (wt/vol)) to pellet the microvessels |
Timing
Microvessel isolation (90–120 min)
Steps 1–5, euthanasia and dissection of the brain: 5–7 min per mouse
Steps 6–8, homogenization of cortices and centrifugation: 20 min
Steps 9 and 10, removal of the supernatant and resuspension of the pellet in dextran: 10–15 min
Steps 11–15, centrifugation and retrieval of microvessels pellet: 30–40 min
Step 16, microscopic evaluation: 5 min
Step 17, final centrifugation and resuspension in proper lysis buffer: 15–20 min
Analysis of microvessels
Step 18A, immunofluorescence characterization: 24 h
Step 18B, RNA isolation: 2–3 h
Step 18C, protein extraction: 2 h
Step 18D, gDNA isolation and bisulfite conversion: 12–24 h
Step 18E, crosslinking and chromatin shearing: 24 h
Anticipated results
Quantification of changes in gene expression
Isolated microvessels retained the expected molecular changes in the brain vasculature in a disease model, namely, the transient middle cerebral artery occlusion (tMCAO) mouse model of ischemic stroke. Two-month-old C57/Bl6 mice were subjected to sham surgery or transient focal ischemia (tMCAO) for 45 min as we previously described19. Twenty-four hours after tMCAO or sham surgery, microvessels were isolated from the whole ipsilateral cortex (i.e., penumbral region). Ipsilateral cortices from two mice were pooled.
RNA was isolated from the microvessels of each group, and levels of gene expression were quantified by reverse transcription and quantitative RT-PCR. The primer sequences used for analysis are indicated in Table 2. Figure 6 shows typical results seen when the protocol is conducted successfully. The endothelial pro-inflammatory gene, Sele (E-selectin), and the astrocyte activation marker Gfap were significantly upregulated in cerebral microvessels in the ipsilateral cortex as compared to sham (57.9 ± 14.8-fold and 5.1 ± 0.8-fold induction, respectively, N = 4/group, mean ± s.e.m.). These findings are in line with the induction of post-ischemic neurovascular inflammation30–33. The mRNA levels of S1pr2, one of sphingosine-1-phosphate (S1P) receptors34 upregulated in endothelial cells upon in vitro ischemia–reperfusion (I-R) and inflammatory injury19,35, were increased 2.4 ± 0.2-fold (N = 4/group, mean ± s.e.m.) in mouse cortical microvessels upon tMCAO (Fig. 6). In addition, the levels of S1pr2 pre-mRNA, which is normally less abundant than mature mRNA, were also increased 2.1 ± 0.4-fold (N = 4/group, mean ± s.e.m.) in microvessels after tMCAO, suggesting that S1PR2 is upregulated at the transcriptional level upon I-R injury. The mRNA levels of the endothelial markers Pecam1 (CD31) and the TJ protein Tjp1 (ZO-1) were not significantly changed (Fig. 6), nor were the mRNA levels of astrocyte (Aqp4) and pericyte markers (Anpep, CD13) (data not shown), indicating that the cellular composition was similar between preparations. Together with claudin-5 and occludin, ZO-1 is one of the most abundant TJ proteins in mouse cerebral microvessels20,26,36. ZO-1 is an intracellular component of the TJ, which links the transmembrane components (e.g., claudin-5, occludin) with the actin cytoskeleton37,38. It is well established that, upon ischemia, TJ proteins undergo redistribution from the cytoskeletal insoluble fraction to the cytosolic fraction, leading to BBB leakage39,40. Although several studies have reported a modest decrease in mRNA levels of ZO-1, or a transient decrease in protein levels41, these changes have been determined in the total brain, and they do not necessarily indicate changes in cerebral microvessels. Indeed, we did not find significant changes in the mRNA levels of TJ proteins such as ZO-1 in cortical microvessels upon stroke. These data are consistent with recently published transcriptome analysis of isolated endothelial cells after stroke36 and emphasize the need for separating microvessels from brain parenchyma in order to study changes in gene expression at the BBB.
Table 2 |.
Primer sequences
| Gene | Sequence (5′–3′) |
|---|---|
|
| |
| Primers used for gene expression analysis by qPCR | |
| Tjp1 (ZO1) F | CCA AGG TCA CAC TGG TGA AGT C |
| Tjp1 (ZO1) R | CTT GAA TGT TAC CAT CTC TTG CTG |
| Aqp4 F | TGG TTG GAG GAT TGG GAG TCA C |
| Aqp4 R | CAG TTC GTT TGG AAT CAC AGC TG |
| Anpep (CD13) F | CAA GGG AGC CTC AGT CAT CAG GAT GC |
| Anpep (CD13) R | GAC AGC TGT CTG TTG GTT CAC G |
| S1pr2 F | ATGGGCGGCTTATACTCAGAG |
| S1pr2 R | GCGCAGCACAAGATGATGAT |
| S1pr2 pre-mRNA F | GCC ATA ACT GTG ACC TGC TGT C |
| S1pr2 pre-mRNA R | TCT CAG GAT TGA GGT ACT CTG AG |
| Gfap F | GCTGGAGGGCGAAGAAAAC |
| Gfap R | TGGCCTTCTGACACGGATTT |
| Sele (E-selectin) F | CCGTCCCTTGGTAGTTGCA |
| Sele (E-selectin) R | CAAGTAGAGCAATGAGGACGATGT |
| Pecam1 (CD31) F | GCG GTG TTC AGC GAG ATC CTG |
| Pecam1 (CD31) R | ACT GTA CCT CGT TAC TCG ACA G |
| Primers used for DNA methylation analysis by PCR | |
| BDNF promoter IX, Chr2: 109722792–109722873 (GeneBank: GRCm38.p4 C57BL/6J) | |
| Methylation-specific primer F | TCGTAAAATATGGTGGTTTATATCGT |
| Methylation-specific primer R | AATATAATAAAACAACGATCGATCGA |
| Unmethylation-specific primer F | TTGTAAAATATGGTGGTTTATATTGT |
| Unmethylation-specific primer R | CAATATAATAAAACAACAATCAATCAA |
| S1PR2 promoter, Chr9: 20976033–20976175 (GeneBank: GRCm38.p4 C57BL/6J) | |
| Methylation--specific primer F | TGTTACGGACGAGCGAGCGG |
| Methylation-specific primer R | CTT CGA TAC AAA ATAA TTCTTCCGA |
| Unmethylation-specific primer F | AGGTGTTATGGATGAGTGAGTGG |
| Unmethylation-specific primer R | TCC TTC AAT ACAAAATAATTCTTCCAA |
| S1PR1 promoter, Chr3: 115714705–115714785 (GenBank: GRCm38.p4 C57BL/6J) | |
| Methylation-specific primer F | GTGAGTTGGTCGTCGTTAGTCG |
| Methylation-specific primer R | CTACCTCGCTCAAACCGAACG |
| Unmethylation-specific primer F | GAGTGAGTTGGTTGTTGTTAGTTG |
| Unmethylation-specific primer R | CCTACCTCACTCAAACCAAACAA |
F, forward; R, reverse.
Phospho-protein changes
Given the relative difficulties in measuring changes in protein phosphorylation in vivo, we tested whether our microvessel isolation protocol is suitable to capture these changes. We and others have previously shown that S1P, which is very abundant in plasma and is produced and released mainly by endothelial cells, erythrocytes and platelets42, activates S1PR2 in the endothelium, leading to inhibition of Akt phosphorylation (Ser473) via the Rho-Rho dependent kinase (ROCK)–phosphatase and tensin homolog (PTEN) pathway43–45. Therefore, we aimed to compare phospho-Ser473-Akt levels in the cortical microvessels of mice upon acute inhibition of S1PR2 signaling by administration of the S1PR2 antagonist JTE-013. As shown Fig. 7a, phospho-Akt levels were significantly increased (1.75 ± 0.23-fold) in cerebral microvessels 6 h after gavage administration of JTE-013 (30 mg/kg) as compared to those of vehicle-treated mice. Then we compared Akt phosphorylation levels in cortical microvessels isolated from S1pr2−/− mice with those of wild-type mice. As shown in Fig. 7b, we found that phospho-Akt levels were significantly higher in S1pr2 null mice as compared to their wild-type littermates (1.8 ± 0.2-fold). These data are consistent with our previous in vitro signaling studies in endothelial cells43,46 and indicate that the described method allows the quantification of changes in protein phosphorylation, and thus, it can be used to conduct signaling studies in the brain microvasculature in vivo.
Assessment of DNA methylation
Given the critical role of DNA methylation in the control of gene expression and its implications in cardiovascular disease, we also tested whether gDNA isolated from microvessels can be used to detect epigenetic modifications such as methylation of gene promoters. Primers to detect specific methylated or unmethylated DNA were designed using MethPrimer (http://www.urogene.org/methprimer/) and are shown in Table 2. gDNA isolation, bisulfite treatment and methylation-specific PCR were performed. We first tested promoter IX of the brain-derived neurotrophic factor (Bdnf) gene because it has been shown to be methylated in adult mouse brain47. We isolated gDNA from microvessels, as well as whole brains, of adult mice and treated them with bisulfite. Then we conducted PCR with primers to detect methylated or unmethylated DNA (Table 2). As shown in Fig. 8, we were able to detect methylation of Bdnf promoter IX, but no bands were amplified with the primers for detection of unmethylated DNA. These data indicate that Bdnf promoter IX is methylated in brain as well as in cerebral microvessels, consistent with previous reports47. We then tested the methylation status of S1PR1 and S1PR2 CpG islands in their core promoters. The information on genomic features, including core promoter regions and CpG islands, was obtained from Ensemble (http://www.ensembl.org) and the UCSC Genome Browser (https://genome.ucsc.edu). Interestingly, we found that both S1PR1 and S1PR2 CpG islands were unmethylated, which is consistent with the expression of S1PR1 and S1PR2 in the cerebrovascular endothelium. These data indicate that microvessel preparations can serve as useful materials for the detection of gDNA methylation. As described earlier, the bisulfite-treated gDNA can be subjected to pyrosequencing to quantify the degree of methylation in CpG islands of the specific gene promoters or it can be subjected to bisulfite whole-genome sequencing48 to discover a broad range of methylation statuses, which should generate valuable information to improve the understanding of mechanisms regulating gene expression in cerebral microvessels.
Optimization of chromatin extraction and shearing from cortical microvessels
When conducting ChIP assays in tissues, chromatin extraction and shearing to the adequate size is one of the major challenges. The type and concentration of fixative, crosslinking and sonication conditions need to be optimized for each specific tissue. We used a commercially available kit to extract chromatin from cortical microvessels and described the optimized method for isolation and shearing of chromatin fragments for ChIP assays. In particular, it is critical to properly disrupt the tissue to extract chromatin completely, which generally can be done by pulverizing frozen tissue in liquid nitrogen using a mortar and pestle. Owing to the challenges of pulverizing a small pellet of microvessels, we first incubated the microvessel preparation in lysis buffer alone, as recommended by the kit manufacturer’s instructions. However, this procedure was insufficient to completely disrupt them, as assessed by the presence of visible structure of intact microvessels in the lysate (Fig. 9a). In addition, incomplete disruption yielded very low amount of chromatin. However, when we incorporated a mild sonication step after incubation in lysis buffer, the lysate turned cloudy without any visible intact microvessels (Fig. 9b), resulting in a marked increase in the yield.
In addition, we have tested various conditions for crosslinking and sonicating the extracted chromatin and found that crosslinking with 1% (wt/vol) methanol-free formaldehyde for 5 min and sonication for 4–8 min gives rise to 200- to 400-bp chromatin fragments (Fig. 9c–e), which is the optimal shearing size for most applications.
The protocol we provide here may need to be further optimized, depending on the levels of expression and the location of the specific DNA-binding protein to be pulled down and/or the genomic region/s involved49. However, our data indicate that the sheared chromatin obtained with the described protocol would be suitable for ChIP assay and whole-genome sequencing.
Supplementary Material
Acknowledgements
This work was supported by funds from the American Heart Association (Grant-in-Aid, 12GRNT12050110), the NIH (HL094465) and the Leducq Foundation (14CVD02) to T.S. A.I. was supported by a grant from the Roche Foundation.
Related link
Key reference using this protocol
Yanagida, K. et al. Proc. Natl. Acad. Sci. USA 114, 4531–4536 (2017): https://doi.org/10.1073/pnas.1618659114
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information Nature Protocols thanks Xavier Declèves, Sven Meuth and other anonymous reviewer(s) for their contribution to the peer review of this work.
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-019-0212-0.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. No datasets were generated or analyzed during the current study.
References
- 1.Zlokovic BV The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 57, 178–201 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Shi Y et al. Rapid endothelial cytoskeletal reorganization enables early blood–brain barrier disruption and long-term ischaemic reperfusion brain injury. Nat. Commun. 7, 10523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackman K et al. Progranulin deficiency promotes post-ischemic blood-brain barrier disruption. J. Neurosci. 33, 19579–19589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell RD et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montagne A et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 85, 296–302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nation DA et al. Blood-brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 25, 270–276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo S et al. The vasculome of the mouse brain. PLoS ONE 7, e52665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daneman R et al. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS ONE 5, e13741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L et al. Analysis of the brain mural cell transcriptome. Sci. Rep. 6, 35108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paolinelli R et al. Wnt activation of immortalized brain endothelial cells as a tool for generating a standardized model of the blood brain barrier in vitro. PLoS ONE 8, e70233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruck T, Bittner S, Epping L, Herrmann AM & Meuth SG Isolation of primary murine brain microvascular endothelial cells. J. Vis. Exp. 2014, e52204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aird WC Endothelial cell heterogeneity. Cold Spring Harb. Perspect. Med. 2, a006429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regan ER & Aird WC Dynamical systems approach to endothelial heterogeneity. Circ. Res. 111, 110–130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Z, Hofman FM & Zlokovic BV A simple method for isolation and characterization of mouse brain microvascular endothelial cells. J. Neurosci. Methods 130, 53–63 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Munikoti VV, Hoang-Minh LB & Ormerod BK Enzymatic digestion improves the purity of harvested cerebral microvessels. J. Neurosci. Methods 207, 80–85 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Boulay AC, Saubamea B, Decleves X & Cohen-Salmon M Purification of mouse brain vessels. J. Vis. Exp. 2015, e53208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousif S, Marie-Claire C, Roux F, Scherrmann JM & Decleves X Expression of drug transporters at the blood-brain barrier using an optimized isolated rat brain microvessel strategy. Brain Res. 1134, 1–11 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Kim GS et al. Critical role of sphingosine-1-phosphate receptor-2 in the disruption of cerebrovascular integrity in experimental stroke. Nat. Commun. 6, 7893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagida K et al. Size-selective opening of the blood-brain barrier by targeting endothelial sphingosine 1-phosphate receptor 1. Proc. Natl. Acad. Sci. USA 114, 4531–4536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacar B et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7, 11022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan Y et al. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J. Biol. Chem. 279, 35087–35100 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Zaina S et al. DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 7, 692–700 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Jiang YZ, Manduchi E, Stoeckert CJ Jr. & Davies PF Arterial endothelial methylome: differential DNA methylation in athero-susceptible disturbed flow regions in vivo. BMC Genomics 16, 506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn J et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J. Clin. Invest. 124, 3187–3199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanlandewijck M et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Hill RA et al. Regional blood flow in the normal and ischemic brain is controlled by arteriolar smooth muscle cell contractility and not by capillary pericytes. Neuron 87, 95–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowyer JF et al. A visual description of the dissection of the cerebral surface vasculature and associated meninges and the choroid plexus from rat brain. J. Vis. Exp. 2012, e4285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kono M et al. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J. Biol. Chem. 279, 29367–29373 (2004). [DOI] [PubMed] [Google Scholar]
- 30.del Zoppo GJ & Mabuchi T Cerebral microvessel responses to focal ischemia. J. Cereb. Blood Flow. Metab. 23, 879–894 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Faraci FM Vascular protection. Stroke 34, 327–329 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Iadecola C & Anrather J The immunology of stroke: from mechanisms to translation. Nat. Med. 17, 796–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terasaki Y et al. Mechanisms of neurovascular dysfunction in acute ischemic brain. Curr. Med. Chem. 21, 2035–2042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez T Sphingosine-1-phosphate signaling in endothelial disorders. Curr. Atheroscler. Rep. 18, 31 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Zhang G et al. Critical role of sphingosine-1-phosphate receptor 2 (S1PR2) in acute vascular inflammation. Blood 122, 443–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez-Lopez D et al. Blood-brain barrier permeability is increased after acute adult stroke but not neonatal stroke in the rat. J. Neurosci. 32, 9588–9600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bazzoni G & Dejana E Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol. Rev. 84, 869–901 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Luissint AC, Artus C, Glacial F, Ganeshamoorthy K & Couraud PO Tight junctions at the blood brain barrier: physiological architecture and disease-associated dysregulation. Fluids Barriers CNS 9, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mark KS & Davis TP Cerebral microvascular changes in permeability and tight junctions induced by hypoxia-reoxygenation. Am. J. Physiol. Heart Circ. Physiol. 282, H1485–1494 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Jin X, Liu KJ & Liu W Matrix metalloproteinase-2-mediated occludin degradation and caveolin-1-mediated claudin-5 redistribution contribute to blood-brain barrier damage in early ischemic stroke stage. J. Neurosci. 32, 3044–3057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McColl BW, Rothwell NJ & Allan SM Systemic inflammation alters the kinetics of cerebrovascular tight junction disruption after experimental stroke in mice. J. Neurosci. 28, 9451–9462 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venkataraman K et al. Vascular endothelium as a contributor of plasma sphingosine 1-phosphate. Circ. Res. 102, 669–676 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez T et al. PTEN as an effector in the signaling of antimigratory G protein-coupled receptor. Proc. Natl. Acad. Sci. USA 102, 4312–4317 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 7, 399–404 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Wolfrum S et al. Inhibition of Rho-kinase leads to rapid activation of phosphatidylinositol 3-kinase/protein kinase Akt and cardiovascular protection. Arterioscler. Thromb. Vasc. Biol. 24, 1842–1847 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez T et al. Induction of vascular permeability by the sphingosine-1-phosphate receptor-2 (S1P2R) and its downstream effectors ROCK and PTEN. Arterioscler. Thromb. Vasc. Biol. 27, 1312–1318 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Ma DK et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lister R et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson JD, Denisenko O & Bomsztyk K Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. No datasets were generated or analyzed during the current study.