Abstract
Purpose
The aim of the present study was to identify more effective laboratory markers to assess the severity of corona virus disease 2019 and predict the progression of the disease by collecting more laboratory markers and variables.
Patients and Methods
In this study, most risk factors, including epidemiological characteristics, blood cell counts, cytokines, and infection markers, were collected from 126 patients with COVID-19 to assess their predictive value.
Results
The area under curve (AUC) of Albumin (Alb) to fibrinogen (Fib) ratio (AFR) (0.791), Lactate dehydrogenase (LDH) (0.792), myoglobin (MYO) (0.795), C-reactive protein (CRP) (0.801) and lymphocyte count (0.859) were higher than other markers to distinguish severe from non-severe patients in receiver operating characteristic (ROC) analysis. In the univariate logistic regression analysis, thirty-six out of 46 risk factors, including 34 laboratory markers, were significantly associated with increased odds of severe patients. Multivariate logistic regression analysis showed that the CD19+ lymphocyte count, MYO, LDH, and AFR were associated with increased odds of severe disease. Moreover, Lymphocyte count and AFR levels increased, LDH and CRP levels decreased during hospitalization in recovered severe patients, whereas severe lymphocytopenia and continuously increasing LDH levels were observed in deteriorated patients. AFR level increased and CRP level decreased before the disease worsened in the deteriorated patients; however, when the patients deteriorated, AFR decreased and CRP increased significantly.
Conclusion
CD19+ lymphocyte count, MYO, LDH, and AFR are independent biomarkers for early identification of severe COVID-19. Lymphocyte count, AFR, LDH, and CRP levels were helpful in predicting the clinical progression of the disease.\.
Keywords: Albumin to fibrinogen ratio, C-reactive protein, Lymphocyte, Coronavirus Disease 2019, Inflammation
Graphical Abstract
Introduction
Since December 2019, an acute respiratory disease caused by the novel coronavirus (SARS-CoV-2) has occurred in Wuhan, China and has received worldwide attention. This coronavirus was previously known as 2019-novel coronavirus (2019-nCoV), soon after being named corona virus disease 2019 (COVID-19). Although the origin of the SARS-CoV-2 outbreak has not yet been clarified, it has been reported that SARS-CoV-2 may be transmitted from person-to-person.1
With the cooperation of departments, including government administrations, scientific research workers, and medical workers, the outbreak was brought under control in China in 2020. However, the number of new infections and deaths subsequently increased in many countries.By July 17, 2021, COVID-19 had caused over 190 million confirmed cases and more than 4 million deaths globally. SARS-CoV-2 is transmitted through respiratory droplets, contact, and feces, and aerosol transmission is highly possible.2 The clinical symptoms of patients with COVID-19 mainly include fever, dry cough, fatigue, diarrhea, and severe manifestations including ARDS, acute cardiac injury, and multiple organ failure.3 COVID-19 is classified into four levels according to the severity of its symptoms: mild, moderate, severe, and critical. The laboratory features of COVID-19 include reduced lymphocyte count, prolonged prothrombin time, elevated lactate dehydrogenase (LDH), elevated creatine kinase (CK), elevated myoglobin (MYO), elevated D-dimer levels, and elevated C-reactive protein levels. The constant emergence of mutating strains of the virus renders newly developed drugs ineffective. Therefore, the group of patients with COVID-19 and the assessment of prognosis have become more important.
Although some studies have been published, the albumin-to-fibrinogen ratio (AFR) of COVID-19 in severe and non-severe cases has not yet been fully reported. Many studies did not collect as much laboratory data and as many variables as possible. In this study, we present the details of patients admitted to The First Affiliated Hospital of Nanchang University as of April 1, 2020. We aimed to investigate the differences between severe and non-severe cases, which may have prognostic value and help to identify important therapeutic targets.
Materials and Methods
Participants
One hundred twenty-six eligible COVID-19 patients confirmed in the First Affiliated Hospital of Nanchang University from January 2020 to February 2020 were included according to the following inclusion and exclusion criteria: 1) All patients were over 17 years old; 2) All patients had positive SARS-CoV-2 real-time RT-PCR results; 3) the eligible cases were not suffered from bacterium and other virus infection, malignancies, hematonosis, mmunodeficiency and autoimmune as well as cardiovascular and cerebrovascular disease before admission; 4) all included patients did not undergo hormone and antiviral treatment before hospitalization; All patients were infected with the wild-type strain and had not been vaccinated. This study was approved by the ethical committee of the local hospital.
Grouping Criteria and Data Collection
The illness severity of COVID-19 was defined according to the management guidelines for COVID-19 established by the Chinese National Health Commission. 1) Mild: mild clinical symptoms, no pneumonia on imaging; 2) moderate: fever, respiratory tract symptoms, and pneumonia on imaging; 3) severe: the patient met any of the following criteria: respiratory distress (respiratory rate ≥30 breaths/min), blood oxygen saturation (SpO2) ≤93% at rest, partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) ≤ 300 mmHg; 4) critical: the patient met one of the following criteria: respiratory failure requiring mechanical ventilation, septic shock, and/or multiple organ dysfunction or failure requiring ICU treatment. The mild and moderate groups were merged into a non-severe group in our study.
Laboratory findings and demographic, clinical, and medical histories were extracted from electronic medical records. Albumin to fibrinogen ratio (AFR), neutrophil-to-lymphocyte ratio (NLR), platelet-to-lymphocyte ratio (PLR), and lymphocyte-to-monocyte ratio (LMR) were calculated. SARS-CoV-2 PCR tests of throat swab specimens were repeated daily or on other days. The standard for discharge was no fever for at least 3 days, significant improvement in the lungs on CT, clinical relief of respiratory symptoms, and two throat swab samples SARS-CoV-2 RNA negative at least 24 h apart.
Statistics
Continuous variables are expressed as median and interquartile range (IQR) and compared between different groups using the Mann–Whitney U-test. The optimal cutoff points of laboratory markers and the calculated ratios for distinguishing the severe group from the non-severe group were determined by analyzing the ROC curve. Significant variables in the univariate logistic regression analysis were chosen as candidates for the multivariate logistic regression analyses. Highly collinear variables are not included in the model. To avoid overfitting, the number of variables in the model was less than four. All statistical analyses were performed using the SPSS software. 22.0 (IBM Corp, Armonk, NY, USA). All analyses were two sided.
Results
The 126 enrolled patients, 53% of whom were men, were divided into three groups according to illness severity: non-severe (n=90), severe (n =28), and critical (n=8). The median age of the patients was 45 years. The severe (52.5, IQR 44–66) and critical (66, IQR 51.75–75) groups were significantly older than the non-severe group (42, IQR 30–50). The most common comorbidities were hypertension (7.9%) and diabetes mellitus (8.7%). The proportion of hypertension in critical patients was higher than that in non-severe patients (p=0.021). The most common symptoms are fever, cough, sputum production, and fatigue. Other symptoms observed included a runny nose, myalgia, nausea or vomiting, diarrhea, chills, headache, dizziness and chest distress. Patients with severe pneumonia had acute respiratory distress syndrome (ARDS) and refractory hypoxemia, causing symptoms such as chill (critical vs non-severe, p<0.001; critical vs severe, p=0.005), fatigue (critical vs non-severe, p=0.011; critical vs severe, p=0.015) and dyspnea (severe vs non-severe: p<0.001). The median duration of viral shedding after illness onset was 15 days (IQR 10–21). The severe (16, IQR 14–23.5) and critical groups (23, IQR 16.25–28.5) last longer than the non-severe group (13, IQR 9–19.25) (severe vs non-severe: p=0.042, critical vs non-severe: p=0.004). The median hospital stay was 16 days. The median hospital stay of the non-severe, severe, and critical groups was 13, 20.5, 33.5 days, respectively. Hospital stay was related to illness severity; the more severe the illness, the longer the hospital stay (severe vs non-severe, p<0.001; critical vs non-severe, p<0.001; critical vs severe, p=0.015) (Table 1).
Table 1.
Clinical Characteristics of Patients with COVID-19
| Total (n=126) | Non severe (n=90) | Severe (n=28) | Critical (n=8) | P value | |||
|---|---|---|---|---|---|---|---|
| N vs S | N vs C | S vs C | |||||
| Age(years) | 45.00(31.00–54.00) | 42.00(30.00–50.00) | 52.50(44.00–66.00) | 66.00(51.75–75.0) | <0.001 | 0.001 | 0.118 |
| Body mass index | 23.9(17.6–39.0) | 23.7(17.6–39.0) | 25.8(21.0–29.4) | NA | 0.234 | NA | NA |
| Sex | |||||||
| Female | 53.0(42.1%) | 42.0(46.7%) | 9.0(32.1%) | 2.0(25.0%) | 0.177 | 0.240 | 0.703 |
| Male | 73.0(57.9%) | 48.0(53.3%) | 19.0(67.9%) | 6.0(75.0%) | |||
| Smoking | 7(5.6) | 7(7.8%) | 0 | 0 | 0.130 | 0.415 | NA |
| Alcohol | 10(7.9%) | 5(5.6%) | 3(10.7%) | 2(25%) | 0.345 | 0.042 | 0.310 |
| Chronic medical illness | |||||||
| Hypertension(yes) | 10.0(7.9%) | 4.0(4.4%) | 4.0(14.3%) | 2.0(25.0%) | 0.072 | 0.021 | 0.480 |
| Diabetes mellitus(yes) | 11.0(8.7%) | 6.0(6.7%) | 3.0(10.7%) | 2.0(25.0%) | 0.483 | 0.071 | 0.310 |
| Symptom | |||||||
| Fever | 111.0(88.1%) | 76.0(84.4%) | 27.0(96.4%) | 8.0(100.0%) | 0.098 | 0.231 | 0.593 |
| Cough | 75.0(59.5%) | 50.0(55.6%) | 19.0(67.9%) | 6.0(75.0%) | 0.251 | 0.289 | 0.703 |
| Expectoration | 30.0(23.8%) | 22.0(24.4%) | 6.0(21.4%) | 2.0(25.0%) | 0.744 | 0.972 | 0.833 |
| Fatigue | 20.0(15.9%) | 13.0(14.4%) | 3.0(10.7%) | 4.0(50.0%) | 0.616 | 0.011 | 0.015 |
| Throat discomfort | 26.0(20.6%) | 20.0(22.2%) | 4.0(14.3%) | 2.0(25.0%) | 0.364 | 0.858 | 0.480 |
| Dyspnea | 13.0(10.3%) | 2.0(2.2%) | 10.0(35.7%) | 1.0(12.5%) | <0.001 | 0.108 | 0.215 |
| Polypnea | 5.0(4.0%) | 2.0(2.2%) | 3.0(10.7%) | 0 | 0.052 | 0.672 | 0.340 |
| Runny nose | 6.0(4.8%) | 5.0(5.6%) | 1.0(3.6%) | 0 | 0.678 | 0.496 | 0.593 |
| Myalgia | 12.0(9.5%) | 8.0(8.9%) | 3.0(10.7%) | 1.0(12.5%) | 0.773 | 0.736 | 0.889 |
| Nausea or vomiting | 5.0(4%) | 4.0(4.4%) | 1.0(3.6%) | 0 | 0.842 | 0.545 | 0.593 |
| Diarrhea | 9.0(7.1%) | 6.0(6.7%) | 3.0(10.7%) | 0 | 0.483 | 0.453 | 0.340 |
| Chill | 23.0(18.3%) | 11.0(12.2%) | 6.0(21.4%) | 6.0(75%) | 0.228 | <0.001 | 0.005 |
| Headache or dizziness | 10.0(7.9%) | 9.0(10.0%) | 1.0(3.6%) | 0 | 0.288 | 0.350 | 0.593 |
| Chest distress | 14.0(11.1%) | 7.0(7.8%) | 7.0(25.0%) | 0 | 0.014 | 0.415 | 0.120 |
| Time from illness onset to first negative result of pharyngeal swab(days) | 15.00(10.00–21.00) | 13.00(9.00–19.25) | 16.00(14.00–23.50) | 23.00(16.25–28.5) | 0.042 | 0.004 | 0.105 |
| Length of stay(days) | 16.00(10.75–21.25) | 13.00(8.75–19.00) | 20.5(18.00–25.75) | 33.5(20.75–44.75) | <0.001 | <0.001 | 0.015 |
Abbreviations: N, non-severe; S, Severe; C, Critical; NA, not available.
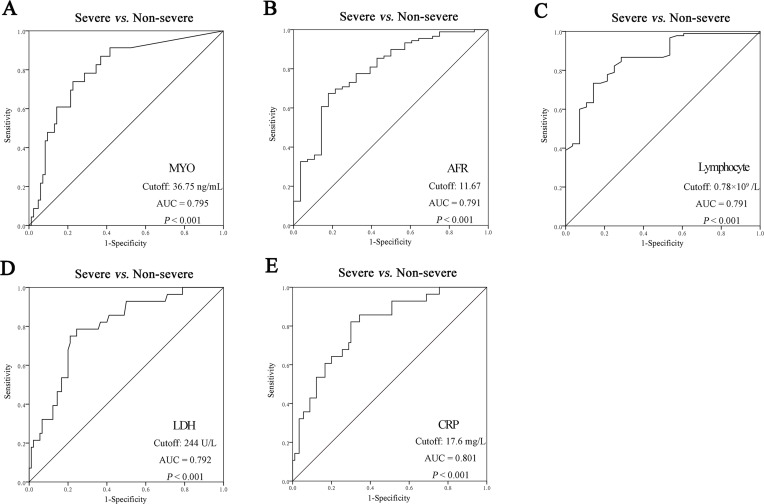
Fibrinogen, CRP, D-dimer, high-sensitivity cardiac troponin T(hs-cTNT), erythrocyte sedimentation rate (ESR), Urea, IL-6, IL-10, NLR were significantly increased, while lymphocyte count, Albumin, LMR and AFR were significantly decreased in the severe and critical groups compared with those in the non-severe group. The fibrinogen, LDH, D-dimer, PLR, CRP, and procalcitonin (PCT) levels significantly increased with disease severity. Lymphocyte subsets, including CD3+CD4+, CD3+CD8+, CD56+CD16+ (NK cells), CD19+ (B cells), and CD3+CD4+CD25+CD127+ lymphocytes, were significantly lower in the severe and critical groups than in the non-severe group. IL-2R and MYO levels were remarkably increased in severe patients compared to those in the non-severe group (Table 2). ROC analysis showed that the AUCs of AFR (0.791), LDH (0.792), MYO (0.795), CRP (0.801), and lymphocyte count (0.859) were higher than those of the other markers for differentiating between severe and non-severe patients (Figure 1).
Table 2.
Laboratory Indexes of Patients with COVID-19
| Total (n=126) | Non severe (n=90) | Severe (n=28) | Critical (n=8) | P value | |||
|---|---|---|---|---|---|---|---|
| N vs S | N vs C | S vs C | |||||
| Cell count | |||||||
| White blood cell count(× 109 cells per L) | 5.17(3.77–7.12) | 5.06(3.79–6.67) | 4.83(3.63–8.52) | 7.64(4.54–14.50) | 0.793 | 0.049 | 0.202 |
| Platelet count (×109 per L) | 170.5(133.3–220.3) | 174.5(147.3–223.3) | 147.0(121.0–189.8) | 162(117.8–245.5) | 0.012 | 0.707 | 0.493 |
| Neutrophil count (× 109 cells per L) | 3.72(2.39–5.30) | 3.47(2.29–4.85) | 3.98(2.64–7.40) | 5.98(3.88–9.20) | 0.109 | 0.007 | 0.209 |
| Lymphocyte count (× 109 cells per L) | 1.03(0.68–1.48) | 1.20(0.91–1.65) | 0.69(0.41–0.87) | 0.36(0.30–0.67) | <0.001 | <0.001 | 0.052 |
| Monocyte count (× 109 cells per L) | 0.33(0.24–0.45) | 0.34(0.26–0.46) | 0.30(0.18–0.40) | 0.27(0.20–0.59) | 0.029 | 0.240 | 0.924 |
| NLR | 3.40(1.91–5.99) | 2.58(1.72–4.19) | 6.47(3.28–13.37) | 12.07(8.30–21.69) | <0.001 | <0.001 | 0.062 |
| PLR | 170.8(122.8–247.7) | 153(109.5–210.5) | 210.4(167.6–336.9) | 450(346.5–540.2) | <0.001 | <0.001 | 0.011 |
| LMR | 3.20(2.36–4.27) | 3.60(2.55–4.64) | 2.69(1.48–3.35) | 1.36(1.07–3.03) | <0.001 | 0.006 | 0.305 |
| Nutrition and Coagulation index | |||||||
| Albumin (g/L) | 44.40(40.60–47.53) | 45.80(42.65–48.15) | 39.65(36.38–44.38) | 30.95(29.45–34.4) | <0.001 | <0.001 | <0.001 |
| Fibrinogen (g/L) | 3.62(2.78–4.91) | 3.40(2.67–4.25) | 4.71(3.72–5.74) | 5.14(2.92–6.07) | <0.001 | 0.051 | 0.775 |
| AFR | 11.93(8.21–15.93) | 13.09(10.20–17.62) | 8.14(6.84–11.29) | 7.16(5.12–10.04) | <0.001 | <0.001 | 0.171 |
| D-dimer (µg/mL) | 0.33(0.20–0.60) | 0.27(0.18–0.46) | 0.55(0.29–1.12) | 2.47(1.26–3.56) | <0.001 | <0.001 | 0.004 |
| Biochemical test | |||||||
| Lactate dehydrogenase(U/L) | 216.0(183.5–282.0) | 204.0(176.5–230.5) | 273.0(236.3–356.0) | 377(336.0–691.0) | <0.001 | <0.001 | 0.017 |
| Urea (mmol/L) | 4.10(3.20–5.20) | 3.90(3.00–4.80) | 4.90(4.05–6.13) | 7.20(2.90–15.80) | 0.002 | 0.022 | 0.132 |
| Creatine kinase (U/L) | 85.0(60.0–130.0) | 81.0(58.5–122.0) | 111.0(70.0–194.0) | 61.00(23.0–160.0) | 0.025 | 0.477 | 0.136 |
| High-sensitive cardiac troponin I(ng/mL) | 7.96(5.81–10.11) | 6.89(4.99–9.19) | 10.20(8.12–13.41) | 10.59(7.03–38.30) | <0.001 | 0.009 | 0.518 |
| Myoglobin (ng/mL) | 29.30(21.00–43.55) | 22.70(21.00–36.30) | 52.70(34.80–76.80) | 27.1(21.00–33.20) | <0.001 | 0.832 | 0.088 |
| ESR (mm/h) | 33.00(21.00–63.00) | 27.50(14.00–51.25) | 63.00(28.50–99.25) | 59.5(41.75–101.5) | <0.001 | 0.012 | 0.864 |
| C-reactive protein (mg/L) | 13.03(2.81–49.73) | 7.77(1.56–23.56) | 46.8(18.1–105.5) | 111.3(87.1–155.3) | <0.001 | <0.001 | 0.026 |
| Procalcitonin (ng/mL) | 0.04(0.02–0.08) | 0.03(0.02–0.05) | 0.07(0.05–011) | 0.17(0.13–0.21) | <0.001 | <0.001 | 0.004 |
| Lymphocyte subgroup | |||||||
| CD3 Lymphocyte (per µL) | 560.0(363.0–898.3) | 696.5(497–1022.8) | 333.5(155.0–505.3) | 189(104.8–433.8) | <0.001 | <0.001 | 0.183 |
| CD3+ CD4+ Lymphocyte (per µL) | 364.5(208.8–539.0) | 413.5(283.3–617.5) | 207.0(102.5–301.0) | 133.0(69.0–204.3) | <0.001 | <0.001 | 0.209 |
| CD3+ CD8+ Lymphocyte (per µL) | 184.5(104.0–291.3) | 230.0(136.5–344.3) | 101.0(53.5–169.25) | 43.0(34.0–118.8) | <0.001 | 0.001 | 0.133 |
| CD56+CD16+ Lymphocyte (per µL) | 109.5(72.8–176.25) | 142.0(87.8–192.5) | 76.0(53.0–103.25) | 81.0(51.75–95.25) | <0.001 | 0.004 | 0.970 |
| CD19+ Lymphocyte(per µL) | 106.0(69.50–173.0) | 127.0(83.0–210.25) | 69.0(40.5–108.5) | 71.5(32.9–107.75) | <0.001 | 0.016 | 0.985 |
| CD3+CD4+CD25+CD127+ Lymphocyte (per µL) | 32.37(17.10–58.35) | 44.18(29.16–72.08) | 21.60(10.54–33.75) | 10.61(4.50–13.07) | <0.001 | 0.003 | 0.047 |
| Cytokine | |||||||
| IL-2r | 508.0(408.3–611.0) | 471.5(400.8–577.8) | 689.0(518.5–780.5) | NA | 0.006 | NA | NA |
| IL-6 | 5.67(2.55–12.05) | 4.57(2.46–10.50) | 10.13(2.41–26.74) | 33.0(12.56–94.41) | 0.077 | 0.005 | 0.053 |
| IL-10 | 2.36(1.50–4.26) | 1.78(1.26–2.47) | 3.41(1.72–4.77) | 5.80(4.21–10.84) | 0.036 | 0.005 | 0.066 |
Abbreviations: AFR, albumin/fibrinogen ratio; PLR, Platelet/lymphocyte ratio; NLR, neutrophil-to-lymphocyte ratio; LMR, lymphocyte/monocyte ratio; ESR, erythrocyte sedimentation rate; IL-2r, interleukin-2 receptor; IL-6, interleukin-6; IL-10, interleukin-10; N, non-severe; S, Severe; C, Critical; NA, not available.
Figure 1.
The ROC analysis of laboratory markers to differentiate severe from non-severe patients. (A) The ROC analysis of MYO; (B) The ROC analysis of AFR; (C) The ROC analysis of Lymphocyte; (D) The ROC analysis of LDH; (E) The ROC analysis of CRP.
Abbreviations: MYO, Myoglobin; AFR, albumin/fibrinogen ratio; LDH, Lactate dehydrogenase; CRP, C-reactive protein.
In the univariate logistic regression analysis, 36 of 46 risk factors, including 34 laboratory markers, were significantly associated with increased odds of severe disease. Older age, longer time from onset to admission, elevated white blood cell count, neutrophil granulocyte count, CRP, D-dimer, ESR, PCT, hs-TNT, creatine kinase(CK), CKMB, MYO, LDH, creatinine, urea, IL-2r IL-6, IL4, IL5, IL10, IL17, IFN-γ, fibrinogen, PLR, NLR, decreased platelet count, albumin, lymphocytes, AFR, and LMR were significantly associated with severe disease. In multivariate logistic regression analysis to predict patients with severe COVID-19, we found that B cell count, MYO, LDH, and AFR are associated with increased odds of severe disease (Table 3). The prediction model had a sensitivity of 73.9% and a specificity of 94.9% for distinguishing between severe and non-severe patients, with the AUC of 0.916. Moreover, the AFR was positively correlated with lymphocyte count (p<0.001) and negatively correlated with age (p<0.001), CRP (p<0.001), LDH (p<0.001), ESR (p<0.001), PCT (p<0.001), hs-cTNT (p=0.018), MYO (p<0.001), IL10 (p=0.002) and IL6 (p<0.002).
Table 3.
Univariate and Multivariate Logistic Analyses of Risk Factors for Severe Group
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p-value | OR (95% CI) | p-value | |
| Sex(male) | 1.155 (0.711–4.308) | 0.223 | - | - |
| Age(years)* | 1.065(1.029–1.102) | <0.001 | - | - |
| Body mass index(kg/m2)* | 1.051(0.924–1.195) | 0.452 | - | - |
| Alcohol(yes) | 2.040(0.456–9.135) | 0.351 | - | - |
| Hypertension(yes) | 3.583(0.834–15.397) | 0.086 | - | - |
| Diabetes(yes) | 1.680(0.392–7.206) | 0.485 | - | - |
| Time from onset to admission(days)* | 1.155(1.017–1.312) | 0.026 | - | - |
| White blood cell count(×109 per L)* | 1.164(1.003–1.352) | 0.046 | - | - |
| Neutrophil granulocyte(×109 per L)* | 1.302(1.096–1.547) | 0.003 | - | - |
| Lymphocyte count(×109 per L)* | 0.016(0.003–0.097) | <0.001 | - | - |
| CD3 Lymphocyte(>473 per µL) | 0.101(0.037–0.272) | <0.001 | - | - |
| CD3+ CD4+ Lymphocyte(>268 per µL) | 0.091(0.034–0.244) | <0.001 | - | - |
| CD3+ CD8+ Lymphocyte(>174 per µL) | 0.132(0.048–0.361) | <0.001 | - | - |
| CD56+CD16+ Lymphocyte(>83 per µL) | 0.125(0.049–0.324) | <0.000 | - | - |
| CD19+ Lymphocyte(>116 per µL) | 0.120(0.038–0.376) | <0.001 | 0.190(0.040–0.911) | 0.038 |
| CD3+CD4+CD25+CD127+Lymphocyte(>24 per µL) | 0.077(0.022–0.276) | <0.001 | - | - |
| Platelet count(>150×109 per L) | 0.273(0.113–0.659) | 0.004 | - | - |
| C-reactive protein(>17.6 mg/L) | 10.733(3.693–31.199) | <0.001 | - | - |
| D-Dimer(>0.48 mg/L) | 7.100(2.801–17.997) | <0.001 | - | - |
| ESR(>62.5 mm/h) | 7.897(3.049–20.454) | <0.001 | - | - |
| Procalcitonin(>0.04 ng/mL) | 8.653(2.980–25.124) | <0.001 | - | - |
| High-sensitive troponin T(>8.8 pg/mL) | 6.400(2.497–16.404) | <0.001 | - | - |
| Creatine kinase(>97.5 U/L) | 3.343(1.364–8.193) | 0.008 | - | - |
| Creatine kinase-MB(>15.5 U/L) | 4.286(1.658–11.078) | 0.003 | - | - |
| Myoglobin(>36.8 ng/mL) | 9.693(3.352–28.029) | <0.001 | 5.645(1.373–23.206) | 0.016 |
| Lactate dehydrogenase(>244 U/L) | 11.211(4.149–30.290) | <0.001 | 4.829(1.233–18.911) | 0.024 |
| Creatinine(>52.3 µmol/L) | 5.319(1.149–24.623) | 0.033 | - | - |
| Urea(>4.4 mmol/L) | 4.472(1.604–12.472) | 0.004 | - | - |
| IL-2r(>615 U/mL) | 18.667(3.390–102.772) | 0.001 | - | - |
| IL-6(>12.25 pg/mL) | 4.431(1.505–13.048) | 0.007 | - | - |
| IL-1β(>11.8 pg/mL) | 1.894(0.713–5.034) | 0.200 | ||
| IL-2(>1.99pg/mL) | 2.000(0.430–9.293 | 0.376 | ||
| IL-4(>1.31pg/mL) | 6.000(1.052–34.212) | 0.044 | ||
| IL-5(>1.86 pg/mL) | 7.467(1.385–40.245) | 0.019 | ||
| IL-8(>20.5 pg/mL) | 0.390(0.079–1.931) | 0.248 | ||
| IL-10(>2.82 pg/mL) | 9.000(1.550–52.266) | 0.014 | ||
| IL-17(>1.48 pg/mL) | 9.917(1.747–56.304) | 0.010 | ||
| IL-12p70(>0.52 pg/mL) | 4.667(0.916–23.785) | 0.064 | ||
| IFN-α(>1.98 pg/mL) | 4.400(0.749–25.842) | 0.101 | ||
| IFN-γ(>1.41 pg/mL) | 0.167(0.029–0.950) | 0.044 | ||
| TNF(>2.58 pg/mL) | 6.750(0.719–63.372) | 0.095 | ||
| Albumin(>40.2 g/L) | 0.062(0.020–0.188) | <0.001 | - | - |
| Fibrinogen(>4.3 g/L) | 7.283(2.855–18.577) | <0.001 | - | - |
| PLR(>165.9) | 5.252(1.941–14.214) | 0.001 | - | - |
| NLR(>6.31) | 11.857(4.069–34.551) | <0.001 | - | - |
| LMR(>3.41) | 0.190(0.066–0.547) | 0.002 | - | - |
| AFR (>11.67) | 0.105(0.036–0.304) | <0.001 | 0.116(0.026–0.512) | 0.005 |
Note: *Per 1 unit increase.
Abbreviations: OR, Odd ratio; CI, confidence interval; AFR, albumin/fibrinogen ratio; PLR, Platelet/lymphocyte ratio; NLR, Neutrophil granulocyte/lymphocyte ratio; LMR, lymphocyte/monocyte ratio; ESR, erythrocyte sedimentation rate; IL-2r, interleukin-2 receptor; IL-6, interleukin-6; IL-1β, interleukin-1β; IL-2, interleukin-2; IL4, interleukin-4; IL-5, interleukin-5; IL-8, interleukin-8; IL-10, interleukin-10; IL-17, interleukin-17; IL-12p70, interleukin-12p70; IFN-α, interferon-α; IFN-γ, interferon-γ; TNF, Tumor Necrosis Factor.
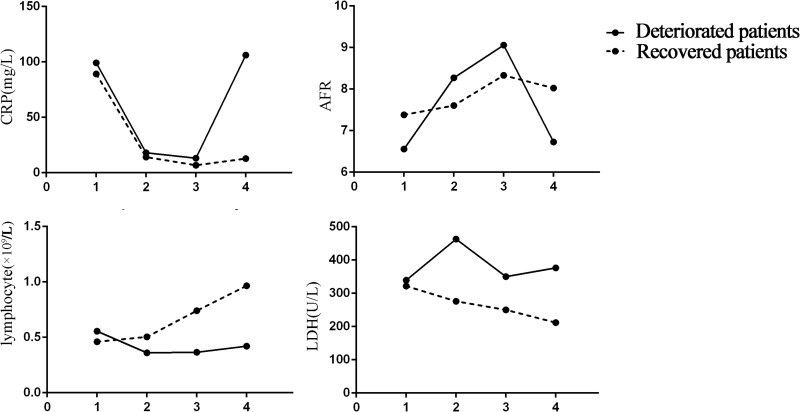
Furthermore, we tracked changes in lymphocyte count, LDH, AFR, and CRP levels from illness onset in patients with severe disease (Figure 2). In recovered patients, the lymphocyte count increased and LDH levels decreased during hospitalization, whereas severe lymphopenia and continuously increased LDH levels were observed in deteriorated patients. AFR level increased and CRP level decreased before the disease worsened in deteriorated patients and throughout the clinical course in recovered patients; However, AFR decreased and CRP level increased significantly when the disease deteriorated.
Figure 2.
Dynamic changes in CRP, AFR, Lymphocyte, LDH levels in patients with severe disease. 1: Time on admission; 2:2–5 days after admission; 3: The time of previous test before time point 4; 4: The time when disease deteriorated to a critical condition or the last data point before discharge if patients recovered.
Discussion
Accumulating evidence has confirmed the transmission of SARS-COV-2 through asymptomatic patients via person-to-person contact; this patient showed no symptoms during the entire period of monitoring and isolation.4,5 Individuals showing symptoms of the disease, COVID-19 is now classified into four levels based on the severity of their symptoms: mild, moderate, severe, and critical. Mild patients presented with low fever, mild fatigue, and no pneumonia. In severe cases, the symptoms usually include dyspnea/hypoxemia. Critically ill patients can rapidly develop ARDS, shock, metabolic acidosis, coagulation dysfunction, and multiple organ failure. Many potential approaches have been developed to fight COVID-19, including small-molecule drugs, interferon therapies, vaccines, oligonucleotides, peptides and monoclonal antibodies.6 Unfortunately, no approach has shown significant efficacy in patients with COVID-19. Our findings help to estimate the development trend and prognosis of the disease at an early stage.
In our study, we enrolled 126 patients infected with SARS-COV-2, of which 53% were men, 71.4% were considered mild/common (non-severe) pneumonia, 22.2% were severe cases, and 6.34% were critical cases. The confirmed patients were aged between 17 and 88 years old. Patients in the severe and critical groups were older than those in the non-severe group. The most common comorbidities were hypertension (7.9%) and diabetes mellitus (8.7%). The proportion of hypertension in critical patients was higher than that in non-severe patients, which may be related to the different ages of the two groups of patients. Studies have already stated that blood pressure control remains an important approach to reduce the burden of COVID-19 infection, even if it has no effect on susceptibility to viral infection.7 In the early phase of the disease, patients can only exhibit chills and respiratory symptoms including fever, cough, and sore throat. Patients with severe pneumonia suffer from acute respiratory distress syndrome (ARDS) and refractory hypoxemia, causing symptoms such as fatigue, which are similar to previous reports.8 The table show that median duration of viral shedding after illness onset was 15 days (IQR 13–16), and the duration of viral shedding in the severe and critical groups was longer than that in the non-severe group. The median hospital stay of the non-severe, severe, and critical groups was 13, 20.5, 33.5 days, respectively. This result indicates that hospitalization time is strongly related to disease severity.
The laboratory evaluation of the patients in our study mostly consisted of complete blood count, coagulation testing, nutrition index, and serum biochemical tests. Many basic laboratory tests, such as white blood cell count, neutrophil count, monocyte count, Urea, CK, LDH, hs-cTNT, MYO, PCT, CRP, ESR, and D-dimer levels, were elevated in severe and critical patients. A significant decrease in the total number of lymphocytes in severe and critical groups compared with non-severe cases and absolute numbers of CD4+T cells (Th cells), CD8+T cells (Ts cells), NK cells, and B cells showed similar trends, suggesting that lymphocytes are likely targets of SARS-CoV-2. IL-2r, IL-6, and IL-10 levels were higher in patients with severe disease than in those with non-severe disease. A cytokine storm caused by virus infection was present in patients with COVID-19, and the decreased number of circulating lymphocyte counts and subgroups could be considered as a diagnostic marker for SARS-CoV-2 infection and provide insights into distinct severities of the disease.9,10 Cytokines such as IL-2r, IL-6, and IL-10 were increased in more serious cases. These results suggest that the host immunity of COVID-19 patients differs depending on the severity of the disease.11 A decreased platelet count was observed in the severe cases compared with the non-severe group. A lower platelet count usually implies serious organ malfunction, and is associated with an increased risk of severe COVID-19.12 NLR and LMR were elevated, whereas PLR was decreased in severe and critical cases, similar to those reported in previous studies.13,14
In the univariate logistic regression analysis, many factors were associated with severe disease. In the multivariate logistic regression analysis, B cell count, MYO, LDH, and AFR were independent predictors for severe patients, and the prediction model had high sensitivity and specificity for distinguishing severe patients from non-severe patients. The AUC of the prediction model is higher than that of many previous reports.15–17 To the best of our knowledge, our results proved our hypothesis and indicated that AFR was prognostic biomarkers for predicting the severity of pneumonia in COVID-19 patients. Albumin is synthesized exclusively in the liver and plays an important role in a number of physiological mechanisms, including the regulation of osmotic pressure and the transport of poorly water-soluble molecules. Albumin level is usually regarded as an indicator of nutritional status. A previous study showed that low serum albumin levels are associated with poor outcomes in patients with COVID-19.18–20 Recent studies have shown that increased fibrinogen levels are associated with severe COVID 19.15,21 Fibrinogen is a predictor of thromboelastography maximum amplitude, which has been demonstrated to correlate with the degree of hypercoagulability in multiple clinical conditions.22 Emerging evidence shows that patients with severe COVID-19 have prothrombotic characteristics and a high risk of venous thromboembolism.23 Fibrinogen plays an important role in many diseases as a key regulator of inflammation. Our previous work reported that AFR could be a promising biomarker for predicting the clinical outcomes of patients with non-small cell lung cancer individuals.24,25 In this study, we found that circulating AFR gradually increased to its highest value from the time of admission to the following days, and decreased before the disease deteriorated in severe patients, indicating that the indicator could effectively monitor disease progression in COVID-19 patients. The AFR at admission was associated with inflammatory markers; low albumin and high fibrinogen levels can predict disease progression and may be early biomarkers for admission to the intensive care unit.
The study that included almost all risk factors is the first time for us to evaluate the predictive and monitoring role of AFR in COVID-19 patients. Our study had some limitations. First, it was a single-center study used wide-type strain, and the accuracy of the data required more multicenter data support. Then for we just displayed the changes of laboratory index and explored possible explanations, more mechanism researches are needed to confirm our hypothesis in the future.
In conclusion, thirty-six risk factors, including 34 laboratory markers, were associated with COVID-19 severity. AFR, LDH, MYO, CRP and lymphocyte count had higher AUCs than other markers to differentiate severe from non-severe patients Multivariate logistic regression model including AFR, B cells count, MYO and LDH had a sensitivity of 73.9% and a specificity of 94.9% to distinguish severe patients from non-severe patients. Continuous monitoring of AFR, LDH, CRP, and lymphocyte counts may indicate changes in illness severity.
Acknowledgments
We are grateful to the Major Public Health Event Medical Center of Jiangxi for answering our questions regarding the clinical data.
Funding Statement
This study was supported by the Science and Technology Project of the Health Commission of Jiangxi Province (grant number:20175134) and the Natural Science Foundation of Jiangxi Province (grant number: 20242BAB20430).
Ethical Approval and Informed Consent
The study was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University. Informed consent was waived by the Ethics Committee due to the retrospective study. All patient data were collected anonymously and ensured the confidentiality of their information. The study completely followed the guiding principles in the Declaration of Helsinki.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai Y, Yao L, Wei T, et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong ZD, Tang A, Li KF, et al. Potential Presymptomatic Transmission of SARS-CoV-2, Zhejiang Province, China, 2020. Emerg Infect Dis. 2020;26(5):1052–1054. doi: 10.3201/eid2605.200198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domling A, Gao L. Chemistry and Biology of SARS-CoV-2. Chem. 2020;6(6):1283–1295. doi: 10.1016/j.chempr.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozkurt B, Kovacs R, Harrington B. Joint HFSA/ACC/AHA Statement Addresses Concerns Re: using RAAS Antagonists in COVID-19. J Card Fail. 2020;26(5):370. doi: 10.1016/j.cardfail.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meckiff BJ, Ramirez-Suastegui C, Fajardo V, et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4(+) T Cells in COVID-19. Cell. 2020;183(5):1340–1353e1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Hou H, Luo Y, et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5(10). doi: 10.1172/jci.insight.137799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin C, Zhou L, Hu Z, et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang AP, Liu JP, Tao WQ, et al. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharm. 2020;84:106504. doi: 10.1016/j.intimp.2020.106504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi X, Su Z, Yan H, et al. Prediction of severe illness due to COVID-19 based on an analysis of initial Fibrinogen to Albumin Ratio and Platelet count. Platelets. 2020;31(5):674–679. doi: 10.1080/09537104.2020.1760230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu J, Zhu HH, Huang XJ, et al. Abnormal Indexes of Liver and Kidney Injury Markers Predict Severity in COVID-19 Patients. Infect Drug Resist. 2021;14:3029–3040. doi: 10.2147/IDR.S321915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallo Marin B, Aghagoli G, Lavine K, et al. Predictors of COVID-19 severity: a literature review. Rev Med Virol. 2021;31(1):1–10. doi: 10.1002/rmv.2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Yang H, Wang J, et al. Serum Albumin Levels are a Predictor of COVID-19 Patient Prognosis: evidence from a Single Cohort in Chongqing, China. Int J Gene Med. 2021;14:2785–2797. doi: 10.2147/ijgm.s312521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popadic V, Klasnja S, Milic N, et al. Predictors of Mortality in Critically Ill COVID-19 Patients Demanding High Oxygen Flow: a Thin Line between Inflammation, Cytokine Storm, and Coagulopathy. Oxid Med Cell Longev. 2021;2021:6648199. doi: 10.1155/2021/6648199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin S, Li W, Shi X, et al. 3044 Cases reveal important prognosis signatures of COVID-19 patients. Comput Struct Biotechnol J. 2021;19:1163–1175. doi: 10.1016/j.csbj.2021.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui J, Noubouossie D, Gandotra S, et al. Elevated Plasma Fibrinogen Is Associated With Excessive Inflammation and Disease Severity in COVID-19 Patients. Front Cell Infect Microbiol. 2021;11:734005. doi: 10.3389/fcimb.2021.734005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandel A, Patolia S, Looby M, et al. Association of D-dimer and Fibrinogen With Hypercoagulability in COVID-19 Requiring Extracorporeal Membrane Oxygenation. J inT Care Med. 2021;36(6):689–695. doi: 10.1177/0885066621997039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kollias A, Kyriakoulis K, Dimakakos E, et al. Thromboembolic risk and anticoagulant therapy in COVID-19 patients: emerging evidence and call for action. Br J Haematol. 2020;189(5):846–847. doi: 10.1111/bjh.16727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davalos D, Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin Immunopathol. 2012;34(1):43–62. doi: 10.1007/s00281-011-0290-8 [DOI] [PubMed] [Google Scholar]
- 25.Li SQ, Jiang YH, Lin J, et al. Albumin-to-fibrinogen ratio as a promising biomarker to predict clinical outcome of non-small cell lung cancer individuals. Cancer Med. 2018;7(4):1221–1231. doi: 10.1002/cam4.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]