Abstract
Background
Extracellular vesicles (EVs) are a group of nanoscale cell-derived membranous structures secreted by all cell types, containing molecular cargoes involved in intercellular communication. EVs can be used to mimic “nature’s delivery system” to transport nucleic acids, peptides, lipids, and metabolites to target recipient cells. EVs offer a range of advantages over traditional synthetic carriers, thus paving the way for innovative drug delivery approaches that can be used in different diseases, including cancer. Here, by using breast cancer (BC) cells treated with the multi-kinase inhibitor sorafenib, we generated EVs enriched in specific non-coding RNAs (miR-23b-3p, miR-126-3p, and the long ncRNA GAS5) and investigated their potential impact on the aggressive properties of the BC in vitro and in vivo using zebrafish.
Methods
EVs were collected from 4 different BC cell lines (HCC1937, MDA-MB-231, MCF-7, and MDA-MB-453) and characterized by western blotting, transmission electron microscopy and nanoparticle tracking analysis. Levels of encapsulated miR-23b-3p, miR-126-3p, and GAS5 were quantified by ddPCR. The role of the EVs as carriers of ncRNAs in vivo was established by injecting MDA-MB-231 and MDA-MB-453 cells into zebrafish embryos followed by EV-based treatment of the xenografts with EVs rich in miR-23b-3p, miR-126-3p and GAS5.
Results
ddPCR analysis revealed elevated levels of miR-23b-3p, miR-126-3p, and GAS5, encapsulated in the EVs released by the aforementioned cell lines, following sorafenib treatment. The use of EVs as carriers of these specific ncRNAs in the treatment of BC cells resulted in a significant increase in the expression levels of the three ncRNAs along with the inhibition of cellular proliferation in vitro. In vivo experiments demonstrated a remarkable reduction of xenograft tumor area, suppression of angiogenesis, and decreased number of micrometastasis in the tails after administration of EVs enriched with these ncRNAs.
Conclusions
Our study demonstrated that sorafenib-induced EVs, enriched with specific tumor-suppressor ncRNAs, can effectively inhibit the aggressive BC characteristics in vitro and in vivo. Our findings indicate an alternative way to enrich EVs with specific tumor-suppressor ncRNAs by treating the cells with an anticancer drug and support the development of new potential experimental molecular approaches to target the aggressive properties of cancer cells.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01936-9.
Keywords: EVs, ddPCR, Breast cancer, ncRNAs, Zebrafish xenograft, Angiogenesis
Background
Breast cancer (BC) is the second most commonly diagnosed cancer and the leading cause of cancer-related deaths among females, with an incidence of 2.29 million new cases in 2022, according to GLOBOCAN [1]. The curative options for BC depend on the activation of human epidermal growth factor receptor 2 (HER2), hormonal receptors (estrogen receptor, ER, and progesterone receptor, PR), and gene mutations (e.g., mutations of BRCA1/2 genes) [2]. The localized treatment of BC consists of surgery and/or radiation therapy; systemic treatment includes chemotherapy, antihormone therapy such as tamoxifen, cyclin-dependent kinase 4/6 (CDK4/6) inhibitors, immunotherapy based on PD-1/PD-L1 immune checkpoint inhibitors and monoclonal antibodies against HER2 such as trastuzumab [3]. In the era of precision medicine, novel approaches such as efficient and safe molecular therapies and targeted drug delivery systems are urgently required. In this context, extracellular vesicles (EVs) represent a very promising tool as therapeutic carriers due to their biocompatibility and stability. EVs are a group of nano-size membrane-bound vesicles ranging from 50 to 1000 nm in diameter carrying cell-derived cargo and released from all types of cells under normal physiological and pathological conditions [4]. EVs can transfer nucleic acids, proteins, lipids, and metabolites to neighboring cells [5]. In cancer, EVs cover every step of BC carcinogenesis up to metastatic dissemination and are a potential source of novel BC diagnosis, prognosis, and chemoresistance biomarkers [6]. The engineering approaches employed to modify their cargos highlighted their capability as vectors to convey conventional or innovative therapies to targeted cancer cells [5, 7]. Among these, EVs engineered to incorporate non-coding RNAs, including miRNAs and long ncRNAs, have shown very promising anti-cancer effects in vitro and in vivo [8]. Indeed, ncRNAs are widely appreciated as pervasive regulators of multiple cancer hallmarks such as proliferation, apoptosis, invasion, migration, and metastasis [9, 10]. ncRNAs have been widely researched for their potential as therapeutic targets and molecular biomarkers in BC [11]. Aberrant expression of miRNAs and lncRNAs in BC can contribute to its development, progression, recurrence, and therapeutic resistance [12]. The aim of this study was to explore how EVs containing certain ncRNAs released by BC cells influence the aggressive behavior of BC in vitro and in vivo. To accomplish this, we generated and isolated EVs from BC cells treated with a multikinase inhibitor, sorafenib that increases the levels of three selected ncRNAs: miR-23b-3p, miR-126-3p, and GAS5. These ncRNAs were specifically selected due to their established roles as tumor suppressors and their documented dysregulation across various cancer types in response to sorafenib treatment [13]. The lncRNA GAS5 is expressed at low levels in BC tissues and its downmodulation was associated with a bad clinical outcome for BC patients [14]. It has been demonstrated that GAS5 acts as a competitive endogenous RNA for various miRNAs with oncogenic functions (onco-miRs) [15] and plays a role in inhibiting proliferation and promoting the apoptosis of BC by sensitizing them to different types of treatment [16]. Several data demonstrated that miR-23b-3p targeted different genes involved in cancer cell aggressiveness such as cell migration, invasion, and metastasis. miR-23b-3p plays a central role in cytoskeletal remodelling by enhancing cell–cell interactions, reducing cell motility and invasion during BC progression [17, 18]. In vivo experiments have shown that miR-23b-3p expression is inversely correlated with BC metastases and tumour growth. OncoDB (https://oncodb.org/index.html /) database (accessed on 20-10-2024) reported a significant downregulation of miR-23b-3p in BC tissues (n = 1135) respect to normal tissues (n = 114) [19]. miR-23b-3p may be considered a promising diagnostic biomarker [20] and it may be able to modulate the response/resistance to some anticancer therapies for BC [21, 22]. Similarly, miR-126-3p was found to be decreased in triple-negative breast cancer (TNBC) cells [23]. Restoring its expression led to the inhibition of cancer cell proliferation, migration, and invasion, and it enhanced the activity of specific BC drugs in vitro [24]. Recently, it was reported that the levels of miR-126-3p encapsulated in the EVs isolated from the serum of overweight BC patients were down-regulated compared to normal-weight patients [25]. In summary, we tested the efficacy of using sorafenib-induced EVs as efficient nanovectors to modulate the expression levels of the selected ncRNAs in target BC cells. We aimed to investigate whether such modulation could also impact the aggressive properties of BC cells in vitro and in vivo. In the last few years, zebrafish (Danio rerio) has become an important vertebrate animal model for cancer, immune, and stem cell research [26]. Embryos lack an efficient immune system, which enables the induction of xenografts through injected cancer cells without rejection. This unique biological characteristic leads to the rapid formation of primary tumors and micrometastases, facilitating in vivo analysis of tumor progression as well as the interactions between tumor cells, the host microenvironment, and the angiogenesis process [27]. Here, our main focus was the treatment of BC xenografts induced in zebrafish with their cognate EVs. We aimed to verify the effect of these EVs on tumor growth and metastasis formation, with the goal of identifying innovative experimental targeted therapies.
Methods
Cell cultures and treatment with sorafenib
The following BC cell lines were used in this study: HCC1937 (ATCC CRL-2336), MDA-MB-453 (ATCC HTB-131), MCF-7 (ATCC HTB-22), and MDA-MB-231 (ATCC HTB-26). Cells were maintained in RPMI-1640 (Thermo Fisher Scientific); all culture media were supplemented with 20% (HCC1937 cells) or 10% (MDA-MB-453, MCF-7, and MDA-MB-231 cells) of Fetal Bovine Serum (Thermo Fisher Scientific) and 10,000 U/ml penicillin/streptomycin (ThermoFisher Scientific); cells were grown at 37 ℃ with 5% CO2. Sorafenib was synthesized and provided by Bayer Corporation (West Haven, CT, USA). This compound was dissolved in 100% dimethyl sulfoxide (DMSO; Sigma-Aldrich) and diluted with RPMI-1640 to achieve the required concentration. 0.1% DMSO was added to the cell cultures as a solvent-only negative control for in vitro studies.
EVs isolation
All cell lines were seeded in number of 300,000/ 10 cm Ø dish, in order to isolate EVs via three methods. The first method employed the use of Total Exosome Isolation Reagent (from cell culture media) (Thermo Fisher Scientific) as follows: cells were seeded in a 10 cm Ø dish until they reached 90% confluence. The cells monolayer was washed 3 times with 1x PBS and 15µM sorafenib or 0.1% DMSO, was added as treatment for 24 h, in the absence of FBS; cell culture media represented by RPMI-1640 (- FBS) was harvested, centrifuged at 2,000 x g for 30 min to remove the remaining cells and debris, and the supernatant was transferred into a new tube without disturbing the pellet; afterward, the appropriate volume of Total Exosome Isolation reagent was added (500 µL of reagent per 1 mL of cell media), vortexed until obtaining a homogenous solution and incubated overnight at 4 ℃; following incubation, tubes were centrifuged at 10,000 x g for 1 h at 4 ℃, the supernatant was discarded without disturbing the pellet (not visible), where EVs were found, and resuspended in 1X PBS (for 10 mL of cell media 200 µL of 1X PBS were required). For the second used method, EVs were purified by ultracentrifugation as indicated by the International Society of Extracellular Vesicles (ISEV) [28]. Briefly, the conditioned medium was centrifuged at 300 g for 10 min at 4 °C to pellet the cells. The supernatant was collected and centrifuged at 16,500 x g for 20 min at 4 °C to eliminate apoptotic bodies and cell debris. The supernatant was then filtered through 0.22 μm filters and ultracentrifuged at 110,000 x g for 70 min at 4 °C. Pellets were resuspended in 1x PBS and ultracentrifuged at 110,000 x g for 70 min at 4 °C. Finally, the pellet containing EVs was resuspended in 1x PBS [29]. As a third method, EVs were purified by immunoprecipitation using the Exosomes Isolation Kit Pan (Miltenyi Biotec, Germany) following the manufacturer’s instructions. EVs obtained with the first and second methods were quantified by BCA Protein Assay Kit (Thermo Fisher Scientific). In contrast, immunoprecipitated EVs were not quantified because magnetic beads interfere with quantification methods.
Western blotting
EVs were lysed in Laemmli Buffer (100 mM Tris–HCl pH 6.8, 4% SDS, 20% glycerol, and 0.2% blue bromophenol) and then quantified using the BCA assay (Thermo Fisher Scientific). Thus, the proteins of EVs were separated by SDS-PAGE, and precisely 2 µg of EV lysate obtained with the precipitation method was loaded. For immunoprecipitated EV, the number of cells was used as the normalization parameter. Subsequently, electroblotting onto polyvinylidenefluoride (PVDF) Immobilon-P membranes was performed (Millipore, Billerica, MA, USA), and the membrane was blocked in 5% milk in PBS for 30 min at room temperature [32]. Primary antibodies were incubated overnight and anti-mouse and anti-rabbit peroxidase-conjugated secondary antibodies were incubated for 1 h at room temperature. The presence of the cytosolic protein, Tumor Susceptibility gene 101 (TSG101), and three tetraspanins CD63, CD9, and CD81 (positive controls), and the absence of the non-EV component Ribosomal Protein S6 (RPS6) (negative control) were verified.
Transmission electron microscopy (TEM)
TEM analysis of isolated EVs was performed with a JEOL JEM-1011 transmission electron microscope at 100 kV operating voltage, equipped with a 7.1 megapixel CCD camera (Orius SC1000, Gatan, Pleasanton, CA). TEM image analysis was achieved with Gatan Digital Micrograph™ (DM) software. For sample preparation, a 5 µL drop of a concentrated vesicle suspension was dropped on a Formvar-coated copper grid (placed on parafilm); after 20 min the grid was upside down infiltrated onto a 50 µL drop of cold carboxymethyl dextran solution (1 mg/mL in UP water) for 5 min. The excess liquid was slowly adsorbed tangentially by using Grade 1 Whatman paper. The resulting ultrathin polysaccharide layer prevents the vesicle collapse on the dried grids. The above-described protocol was slightly modified from the one used by Kreger B.T. et al. [30]. The grids were finally stained by UranyLess EM Stain (Electron Microscopy Sciences), by following the standard protocol provided by the manufacturer.
Nanoparticle tracking analysis
Nanoparticle tracking analysis (NTA) was performed using the Nanosight NS300 instrument (Malvern). A standard operating procedure was applied to acquire three independent videos of 60 s of each sample under a syringe pump speed 30. Samples were systematically diluted in 1x PBS to reach the recommended number of particles per frame and were recorded at Camera level 11. Particle analysis was performed with a detection threshold 5.0, always using NanoSight NS300 software NTA 3.4 Build 3.4.003 (Malvern).
RNA isolation and reverse transcription (RT)
Total RNA was isolated from 200 µL of EVs resuspended in 1x PBS (Thermo Fisher Scientific) using miRNeasy Mini Kit (Qiagen), according to the manufacturer’s instructions. TRIzol RNA Isolation Reagent (Thermo Fisher Scientific) was used for the total RNA isolated from cell cultures, according to the manufacturer’s instructions.
For lncRNA analysis, cDNA was synthesized from 5 µL of total RNA from EVs and 1 µg of intracellular RNA, in a 20 µL reaction volume, using M-MLV Reverse Transcriptase (Sigma-Aldrich) according to the manufacturer’s instruction. The RT reaction was performed at 70 ℃ for 10 min, followed by incubation at room temperature for 10 min, a second incubation at 37 ℃ for 50 min, followed by inactivation at 94 ℃ for 10 min in a T100 Thermal Cycler (Bio-Rad Laboratories).
For miRNAs analysis, cDNA was synthesized from 5 µl of RNA from EVs and 50 ng of cellular RNA, in a 15 µL reaction volume, using the TaqMan microRNA Reverse Transcription Kit components (Thermo Fisher Scientific) and the stem-loop primer for miR-23b-3p (Thermo Fisher Scientific; Assay ID 000400) and miR-126-3p (Thermo Fisher Scientific; Assay ID 002228). RT reaction was performed at 16 ℃ for 30 min, 42 ℃ for another 30 min, followed by inactivation at 85 ℃ for 5 min in a T100 Thermal Cycler (Bio-Rad Laboratories).
Droplet digital PCR workflow
The synthesized cDNA was used as a template for the ddPCR experiments using the QX200 Droplet Digital PCR (ddPCR) System (Bio-Rad Laboratories), and ddPCR was performed according to the ddPCR Supermix for Probes (Bio-Rad Laboratories) protocol [31]. Briefly, 1.33 µl of the cDNA obtained using TaqMan microRNA Reverse Transcription Kit, and 3.96 µl of the cDNA obtained using M-MLV Reverse Transcriptase, were prepared for amplification in a 20 µL reaction volume containing 2x ddPCR Supermix for Probes (Bio-Rad Laboratories), 20x TaqMan assay (Thermo Fisher Scientific) specific for miR-23b-3p, miR-126-3p, and PrimeTime qPCR Assay specific for the lncRNA GAS5 (Integrated DNA Technologies) and water. Each ddPCR assay mixture (20 µL) was loaded into a disposable droplet generator cartridge (Bio-Rad). Then, 70 µL of droplet generation oil for probes (Bio-Rad) were loaded into the wells dedicated to oil. The cartridge was then placed inside the QX200 droplet generator (Bio-Rad). When droplet generation was completed, the droplets were transferred to a 96-well PCR plate using a multichannel pipette. The plate was heat-sealed with foil and placed in a T100 Thermal Cycler (Bio-Rad Laboratories). A negative control (NC) and a positive control (PC) were included. Concentration data for miR-23b-3p, miR-126-3p and GAS5 levels were obtained using QuantaSoft Software (Bio-Rad Laboratories) as copies/µL.
EVs labeling and uptake by target cells
MDA-MB-453 and MCF-7 BC cells were incubated with CellTracker™ CM-DiI Dye (Thermo Fisher Scientific) diluted 1:1,500 in RPMI (-FBS) for 24 h at 37 ℃ with 5% CO2 and labeled EVs were obtained. Coverslips were placed into 3 cm diameter plates and 3 × 105 cells were seeded and incubated at 37 ℃ with 5% CO2. When indicated, labeled EVs were added to the cells in a final volume of 3 mL of RPMI (-FBS) and incubated for 24 h at 37 °C. Samples were washed with 1x PBS, fixed with methanol 100% and the nuclei stained with DAPI (1:3,000, Merck, Inc., Darmstadt, Germany) for 15 min. After washing with 1x PBS, the coverslips were mounted with Vectashield mounting medium (Vector Labs, Newark, CA, USA) and visualized using a Leitz fluorescence microscope.
Cell proliferation assay
For cell proliferation, MTT assay was performed using CellTiter reagent (Promega) according to the manufacturer’s instructions. Briefly, cells were seeded in a 96-wells plate (5 replicates for each experimental condition) having a density of 8 × 103 cells/well in RPMI (-FBS) and treated with 4 µL of EVs, 0.1% DMSO or 15 µM sorafenib, all of which were in the range of 10^8 according to the NTA analysis. After 24 h viability was assessed with the addition of 10 µL/well of sterile CellTiter reagent (Promega). The plates were incubated at 37 °C for 2 h in a humidified, 5% CO2 atmosphere and the absorbance at 490 nm was recorded using the microplate reader EnSight (PerkinElmer, Waltham, MA). For assessing the viability, a 1:2 dilution of cell suspension was made in 0.4% trypan blue stain in a 0.2 ml tube (10 µl of cell suspension added to 10 µl of trypan blue). Ten microliters of the mixture were loaded into the opening of the TC20 counting slide (Bio-Rad). The slide was then inserted into the TC20 instrument (Bio-Rad) and cells counted within 10 min of trypan blue addition.
Zebrafish maintenance and eggs collection
Danio rerio (zebrafish) was maintained and used according to EU Directive 2010/63/EU for animal use following protocols approved by the local committee (OPBA) and authorized by the Ministry of Health (Authorization Number 287/2018). Fish were maintained in 3 L of water at a controlled- temperature (28.5 ℃) with 14 h light and 10 h dark cycle and fed 3 times per day, 2 times with dry food and 1 time with artemia [32]. Mating was set up in organized tanks, upon fertilization the eggs were collected and placed in a Petri dish containing fish water and incubated at 28 °C. Tricaine (MS222; E10521, Sigma–Aldrich St. Louis, MO, USA) was added to the fish water for zebrafish embryos and larvae anesthesia at 0.02% fnal concentration. The wild-type line used in this work included an AB strain (KIT Institute -Karlsruhe-Germany) and a transgenic line Tg(kdrl-EGFP) [33].
EVs uptake by immersion method
Stock solutions of EVs were prepared for embryo exposure at 5, 10, and 15 µl in 2 mL fish water. Embryos were collected, dead embryos were discarded, and alive embryos were transferred in new Petri-dish. At the gastrula stage (5 hpf) embryos were exposed to EVDMSO or EVSorafenib at the selected doses by the classic immersion method [34] up to 96 hpf. As a negative control, embryos were exposed to fish water plus 0.1% DSO (expected mortality rate < 15%). As a positive control for survival rate experiments, we used 3,4-dichloroaniline (DCA) (Sigma-Aldrich) dissolved in fish water at a concentration of 3.74 mg/L (expected mortality rate > 85–90%) [35]. For each treatment condition (EVDMSO and EVsorafenib), 30 embryos were used, and experiments were repeated three times. For all experiments only positive embryos (embryos labeled with marked EVs with DiI) were taken into consideration. The survival rate was recorded at 24, 48, 72 and 96 hpf respectively. The calculated percentages of dead embryos were below 15%, which is the expected mortality according to the OECD 2019 guidelines (Test Guideline No. 203, Fish Acute Toxicity Testing) [36] and a dose-response graph was plotted.
Induction of tumor xenografts
Tumor xenograft experiments were performed according to the protocol established by Ren J. et al. [37]. To evaluate the effect of EVs on tumor growth, 48 hpf Tg (kdrl:EGFP) zebrafish embryos were dechorionated, anesthetized with tricaine at 0.02% fnal concentration, followed by microinjection of the labeled MDA-MB-231 and MDA-MB-453 cells into the perivitelline space (PVS) [38]. Microinjections were performed with a FemtoJet electronic microinjector coupled with an InjectMan N12 manipulator (Eppendorf Italia, Milan, Italy). Approximately 250 cells/4 nL were injected into each embryo (about 25 embryos/group); embryos were maintained in PTU/fish water in a 32 °C incubator to allow tumor cell growth. Pictures of injected embryos were acquired using Zeiss Axiozoom V13 (Zeiss, Jena, Germany) fluorescence microscope, equipped with Zen pro software, 2 h after cell injection (T0). Using the same concentration range of 108 EVs, 15 µl of EVsorafenib or EVDMSO was added directly to the injected embryos in PTU/fish water. The calculated percentages of dead embryos were below 15% [36]. At 1-day post-injection (T1) and at 3-days post-injection (T3), the effects of the EVs-based treatment on tumor xenografts growth were scored by representative pictures, to measure the tumor areas of each group at T0, T1 and T3 using Zen Blue software from ZEISS. Embryos with micrometastases (indicated by the presence of at least one fluorescence dot outside the site of injection) and metastatic cells in the tails were counted and some representative xenografted embryos were fixed and embedded in low-melting agarose for image analysis.
Image acquisition and analysis
Bright-field and fluorescence images of embryos at different development stages (anesthetized with tricaine 0.16 mg/mL embedded in 0.8% low melting agarose and mounted on a depression slide) were captured using a Zeiss Axio Zoom V16 equipped with Zeiss Axiocam 506 color digital camera and processed using Zen 3.5 (Blue Version) software from Zeiss (Oberkochen, Germany), magnification 32x and 40x. Embryos were also observed using AxioObserver.Z1/7 with Apotome 3, Objective EC Plan-Neofluar 20x/0.50 (Carl Zeiss S.p.A., Milan Italy). For Light Sheet image acquisition embryos were first anesthetized using tricaine (0.02% in fish water) and subsequently included in a low melting agarose matrix (Top Vision Low Melting Point Agarose, Thermo FisherScientific) (0.5% in fish water). Images were acquired using Zeiss LightSheet microscope V1 supported by ZenPro software using a 488–30 nm laser and 505–545 nm filter. Images from the same experiment were taken with the same laser intensity and exposure time to generate comparable images. After the acquisition, 3D images were generated and manipulated using Arivis Vision 4D (Zeiss Oberkochen, Germany) 3D reconstructions of EGFP-positive cells were manipulated to obtain pictures comparable to each other in terms of fluorescence intensity. 3D reconstructions were exported as a single snap with the same compression setting.
Alkaline phosphatase (AP) assay
AP assay was performed according to Serbedzija et al. [39]. Briefly, embryos (n = 25) at 1-day post-injection (72 hpf) were fixed in 4% paraformaldehyde (PFA) and then put in 100% (v/v) methanol. The embryos were then equilibrated in Tris buffer (100 mM Tris HCl pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween-20) and stained with nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3’-indolyphosphate p-toluidine salt (BCIP) solution. The images were taken in a lateral and dorsal position at 32× magnification with a Zeiss AxiozoomV13 (Zeiss, Jena, Germany) microscope, equipped with a PlanNeoFluar Z 1×/0.25 FWD 56 mm lens and Zen Pro software.
Statistics and reproducibility
Statistical analysis was carried out using GraphPad Prism v8.0 (GraphPad Software, Inc., San Diego, CA, USA) software. Unpaired Student’s t-test was used to determine the differences of ncRNAs levels in EVs between the sorafenib treated cells and their control (0.1% DMSO) and for cell proliferation after the EV-based treatment. Analysis of variance (ANOVA) followed by post-hoc Tukey’s test was used to determine the significant differences in the ncRNAs levels after EV-based treatment as well as in the measurement of the tumor area, the assessment of micrometastases and angiogenesis in xenograft. All experiments conducted in vitro were performed two times, while in vivo experiments conducted in zebrafish, were performed three times and the number of embryos for each experimental point was n = 30. Data was considered statistically significant when the P-value ≤ 0.05.
Results
EVs derived from sorafenib-treated BC cells were enriched in miR-23b-3p, miR-126-3p, and GAS5
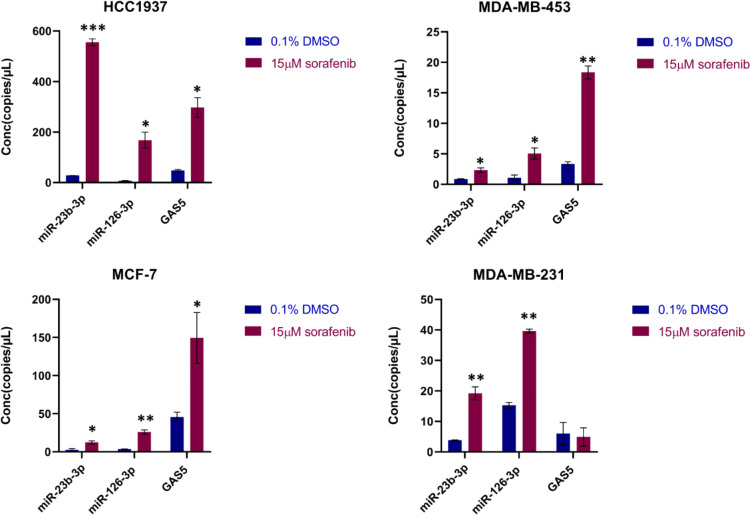
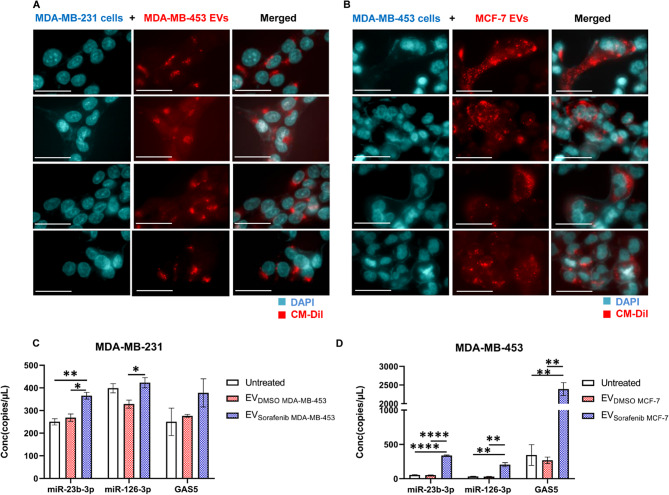
According to our previously reported data, the levels of cellular and circulating miR-23b-3p, miR-126-3p, and GAS5 were dysregulated in both hepatocellular carcinoma and BC, following treatment with sorafenib [13, 40]. Here, we investigated whether the sorafenib treatment could influence the levels of miR-23b-3p, miR-126-3p, and GAS5 encapsulated in EVs released by BC cells. Therefore, HCC1937, MDA-MB-453, MCF-7, and MDA-MB-231 cells were treated for 24 h with 15 µM sorafenib or 0.1% DMSO as negative control (DMSO is used as dissolving compound). The conditioned medium from serum-starved BC cells was collected and EVs were isolated as described in the material and methods section. Using ddPCR technology, we quantified the levels of miR-23b-3p, miR-126-3p and GAS5, measured as copies/µL, within EVs derived from BC cells subjected to sorafenib treatment (EVsorafenib) or those treated with DMSO (EVDMSO). We observed higher levels of the selected ncRNAs in EVSorafenib compared to EVDMSO (Fig. 1 and Additional file 1: Fig. 1). Notably, the EVs obtained using precipitation method exhibited the highest levels of the ncRNAs in comparison to those obtained through ultracentrifugation (Additional file 1: Figures 2 and 3) and immunoprecipitation (Additional file 1: Figures 4 and 5). Therefore, we opted to proceed with subsequent experiments by utilizing this specific vesicle type. The highest fold increases (F.I.) calculated as a ratio between (copies/µl) EVsorafenib/ (copies/µl) EVDMSO were detected in EVsorafenib produced by HCC1937 cells (F.I. miR-23b-3p = 19.8; F.I. miR-126-3p = 24.4; F.I. GAS5 = 6.2). Similar trend was also shown for all ncRNAs in EVsorafenib secreted by MDA-MB-453, MCF-7 and MDA-MB-231, where F.I. was between 2.7 and 7.4 with the exception of GAS5 in EVsorafenib from MDA-MB-231 showing an opposite trend with a fold decrease (F.D.) of -1.2, calculated as a ratio between (copies/µl) EVDMSO/ (copies/µl) EVsorafenib (Additional file 1: Table 1). The fold increase/ decrease determined in EVs obtained by ultracentrifugation and immunoprecipitation are reported in Additional file 1: Tables 2 and 3.
Fig. 1.
Levels of miR-23b-3p, miR-126-3p, and GAS5 in EVs from breast cancer cells after sorafenib treatment. Levels of miR-23b-3p, miR-126-3p, and GAS5 encapsulated in the EVs were determined using ddPCR technology. Concentration of each target is expressed as copies/µL. Treatment with sorafenib caused dysregulation of the level of the 3 selected ncRNAs. The graphics represent mean value; bars, SD. Unpaired t-test was used; *p < 0.05, **p < 0.01, ***p < 0.001. Results are representative of two independent experiments
Characterization of the EVs
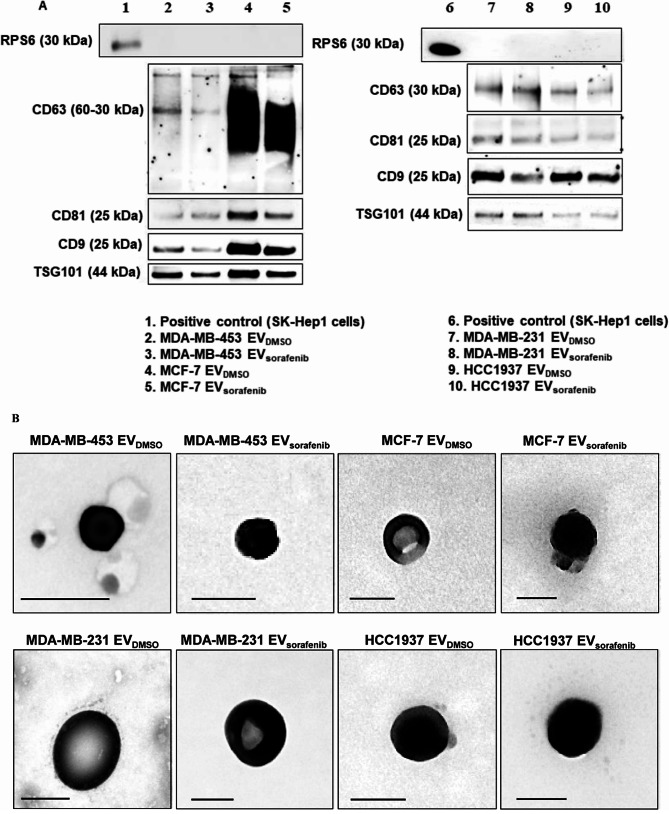
To characterize the EVs released by the selected BC cells, EVDMSO and EVSorafenib isolated through immunoprecipitation were tested via WB for specific tetraspanins, including CD63, CD81, and CD9 that are regarded as specific markers of EVs. The presence of Tumor Susceptibility gene 101 (TSG101), a protein commonly utilized to differentiate small EVs named exosomes from other vesicles of similar size, was also examined [41]. The occurrence of these proteins, in particular CD63 and TSG101, may indicate the endosomal origin of these vesicles, but since we cannot demonstrate a specific biogenesis, we named this particle EVs, as suggested by Minimum Information for Studies of Extracellular Extracellular Vesicles 2023 (MISEV2023) [28]. Moreover, WB analysis revealed specific bands, indicating that all four proteins were expressed on the EVs’ membrane of interest. CD63 appeared as a smear in the range of 30–60 kDa in EVs derived from MCF-7 cells and this is due to its heavy glycosylation (Fig. 2A, lanes 3–4) [42]. Since immunoprecipitation using magnetic beads that remain attached to EVs interfere with electron microscopy, EVs were purified using an alternative approach that involved a series of ultra-centrifugation and filtration steps of the conditioned media prior analysis by TEM [43]. Images showed nanovesicles with a typical round shape and an average size between 50 and 150 nm in diameter (Fig. 2B).
Fig. 2.
Characterization of the EVs. A Western blot for tetraspanins (CD63, CD81, CD9), TSG101 and RPS6 was performed on EVs purified by immunoprecipitation and derived from MDA-MB-453, MCF-7, MDA-MB-231, and HCC1937 cells treated with sorafenib. B Transmission electron microscopy (TEM) on EVs obtained by ultracentrifugation showed vesicles with characteristic morphology and size, between 50 and 150 nm in diameter. Scale bar, 100 nm
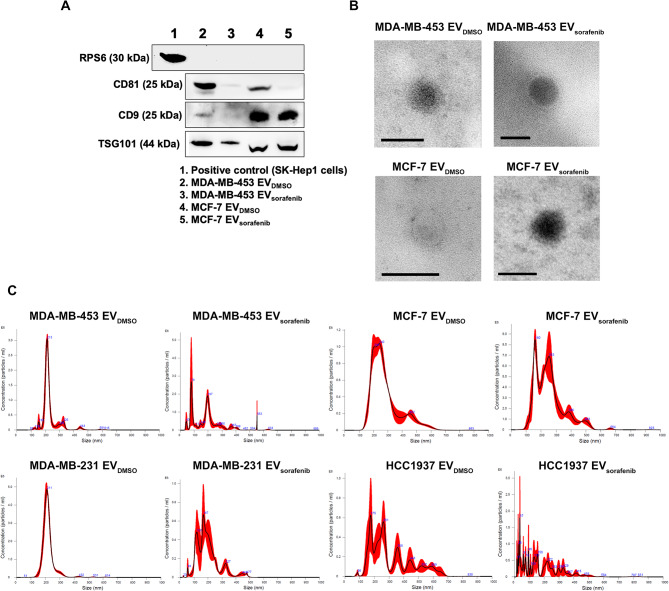
Western blot and TEM analyses were also performed for EVs purified using the Total Exosome Isolation Reagent (from cell culture media) from MDA-MB-453 and MCF-7 treated with DMSO and sorafenib. As shown in Fig. 3A, CD9 and CD81 tetraspanins and TSG101 were present, while the non-EV component RPS6, used as the negative control, was absent, as expected (Fig. 3A). Furthermore, TEM revealed that these EVs were also characterized by a typical round shape and an average size between 50 and 150 nm in diameter (Fig. 3B). Altogether, these results indicate that by using any of the three methods (precipitation, immunoprecipitation and ultracentrifugation), it is possible to isolate EVs from culture media of cell lines with typical markers, shapes, and dimensions. EVs isolated using the Total Exosome Isolation Reagent were characterized by nanoparticle tracking analysis (NTA) (Fig. 3C). Particles recovered from the cell lines treated with DMSO showed heterogeneous size-distribution profiles. To provide a numerical indication of the heterogeneity of particle sizes (Additional file 1: Fig. 6A-B), we quantified the relative abundance of particles below 300 nm in diameter, which includes the most represented peaks, thereby encompassing small EVs and the exosome size range. Sorafenib treatment caused an approximately 20% increase in small particles derived from HCC1937 cells. We also quantified the fraction of particles larger than 300 nm in diameter, including larger EVs within the range analyzed by NTA. Larger particles derived from MCF-7 and HCC1937 cell lines represented around 30% of the total particles detected, with the latter showing an opposite fluctuation in smaller particles. In contrast, larger particles composed about 10% of the total in MDA-MB-231 and MDA-MB-453 cell lines (Additional file 1: Table 4). EVs from MDA-MB-231 cells were characterized by a relevant subpopulation peak around 210 nm and similar mean and mode diameters (~ 220 nm vs. ~ 210 nm, respectively) (Additional file 1: Fig. 6C-D). In comparison, the ones from HCC1937 cells were characterized by high polydispersity with multiple subpopulation peaks and, as expected, the highest difference between mean and mode diameters (312 nm vs. ~ 175 nm, respectively). EVs from MCF7 and MDA-MB-453 cells showed intermediate profiles compared to the other two cell lines (Fig. 3C and Additional file 1: Table 4). In our culture conditions, the calculated EVs concentration was in the range of 108/ml and MDA-MB-231 cells appeared as the most efficient in particle release. Treatment with sorafenib induced size shifts in favor of both subpopulation - smaller and larger EVs- as can be appreciated for all the cell lines tested notwithstanding the underestimation/exclusion of particles > 600 nm in dynamic light scattering (particularly relevant in the case of HCC1937 cells) and smaller particles, as demonstrated by the general increase in the mean/mode diameter ratio. Treatment resulted in a small reduction in the concentration of EVs released by MCF-7 cells and a larger reduction by MDA-MB-231 cells, while an opposite trend of relative EVs abundance in the case of HCC1937 and MDA-MB-453 cells (Additional file 1: Table 4).
Fig. 3.
Characterization of EVs obtained with precipitation method. A Western blot for tetraspanins (CD81, CD9), TSG101 and RPS6 was performed on EVs purified with precipitation method and derived from MDA-MB-453 and MCF-7 cells treated with DMSO and Sorafenib. Lysate from SK-Hep1 cell was used as a positive control. B Transmission electron microscopy (TEM) precipitated EVs showed vesicles with characteristic morphology and size of EVs. Scale bar, 100 nm. C Nanoparticle tracking analysis showed heterogeneous size-distribution profiles of the EVs derived from DMSO or sorafenib treated breast cancer cells
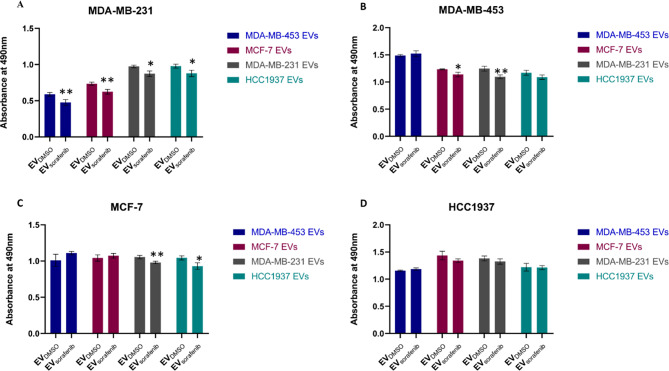
Enriched EVs-based treatment impaired the proliferation ability of BC cells
To assess both the biological effects and treatment efficacy of EVs on the proliferation ability of BC cells, HCC1937, MDA-MB-453, MCF7, and MDA-MB-231 cells were treated with their cognate EVDMSO and EVsorafenib (Fig. 4A-D). Intriguingly, the MTT assay results demonstrated a significant inhibition in the proliferation ability of MDA-MB-231 cells when treated with EVsorafenib derived from any BC cells, in comparison to the corresponding EVDMSO. The inhibition percentages were as follows: 19% (p < 0.01, MDA-MB-453 EVsorafenib), 15% (p < 0.01, MCF-7 EVsorafenib), 10% (p < 0.05, MDA-MB-231 EVsorafenib) and 10% (p < 0.05, HCC1937 EVsorafenib) (Fig. 4A). Therefore, the highest percentage of cellular proliferation inhibition was obtained when treating MDA-MB-231 cells with EVsorafenib derived from MDA-MB-453 cells. Furthermore, EVsorafenib from MDA-MB-231 significantly reduced proliferation in 3 out of 4 BC cell lines while the ones from MDA-MB-453 significantly affected only one cell line. MDA-MB-231 cells treated with EVs derived from MDA-MB-453 cells produced the highest percentage of cell proliferation inhibition.
Fig. 4.
EV-based treatment affected BC cell proliferation. A MDA-MB-231, B MDA-MB-453, C MCF-7, and D HCC1937 cells were treated with different types of enriched EVs for 24 h, and the effects on cell proliferation were assessed by MTT assay. Results are representative of one of two experiments. The graphics represent the average value of five replicates for each condition; the bars represent SD. Unpaired t-test was used; *p < 0.05, **p < 0.01 (n = 53)
EVs as ncRNAs delivery vehicles to target BC cells
To investigate the potential role of EVs as carriers of miR-23b-3p, miR-126-3p, and GAS5, which were found to be over-represented in EVsorafenib, we initially generated membrane-labeled EVs by introducing the fluorescent dye CM-DiI into the culture media of the parental BC cells. MDA-MB-453 and MCF-7 BC cells were incubated with CM-DiI Dye, diluted 1:1,500 in RPMI (-FBS) for 24 h. Afterward, cell culture media was harvested, and EVs were isolated. diI Dye was removed through first centrifugation at 2,000 x g for 30 min, followed by a second one at 10,000 x g for 1 h. Among the cell lines used in this study, we first focused on MDA-MB-231 cells which showed the highest significant reduction of proliferation ability after treatment with EVs. In fact, MDA-MB-231 cells treated with EVs derived from MDA-MB-453 cells were the combination that produced the highest percentage of cell proliferation inhibition. To confirm the results, we used a second cell line, MDA-MB-453, treated with EVs released by MCF-7 cells, which showed an intermediate response in terms of proliferation reduction. We used these sets of cells and EVs to assess the uptake of the labeled EVs by the recipient cells using fluorescent microscopy analysis. When treating MDA-MB-231 cells with MDA-MB-453-derived EVs (Fig. 5A) or MDA-MB-453 cells with MCF-7-derived EVs (Fig. 5B), we detected the presence of a distinct red fluorescent signal corresponding to the labeled EVs. In both cases, the EVs were detected within the cytoplasmic compartment of the cells, surrounding the nuclei that were labeled with DAPI. This observation confirms the successful uptake of EVs by the targeted cancer cells. To further evaluate whether EVs rich in miR-23b-3p, miR-126-3p, and GAS5, were able to release their cargo and to modulate the cellular expression levels, we used ddPCR technology to determine the intracellular amount of the 3 selected ncRNAs. Therefore, MDA-MB-231 cells were treated with EVDMSO and EVsorafenib produced by MDA-MB-453 cells (Fig. 5C) and MDA-MB-453 cells were treated with EVDMSO and EVsorafenib released by MCF-7 cells (Fig. 5D). miR-23b-3p increased by 1.36 times in MDA-MB-231 cells and by 6.51 times in MDA-MB-453 cells. miR-126-3p increased by 1.29 times in MDA-MB-231 cells and by 6.85 times in MDA-MB-453 cells. GAS5 increased by 1.37 times in MDA-MB-231 cells and by 8.89 times in MDA-MB-453. Taken together, these results collectively indicated that the enriched EVs were efficiently taken up by the recipient cells, allowing for the release of their cargo. Furthermore, the uptake of these EVs resulted in increased expression levels of miR-23b-3p, miR-126-3p, and GAS5 in the targeted BC cells.
Fig. 5.
EVs-based ncRNAs delivery to target breast cancer cells. Representative fluorescent microscopy images showing the uptake of MDA-MB-453 and MCF-7 derived extracellular EVs labeled in red by A MDA-MB-231 and B MDA-MB-453 recipient BC cells at 24 h post-treatment. Scale bars correspond to 30 μm for 63x magnification, and the fluorescent dyes used were DAPI (blue) for nuclei and CM-DiI (red) for EVs. The expression levels of cellular miR-23b-3p, miR-126-3p, and GAS5 were determined in terms of copies/µL using ddPCR technology. C Treatment with EVsorafenib derived from MDA-MB-453 resulted in an increased expression level of the three selected ncRNAs in MDA-MB-231 target cells. D Similarly, MDA-MB-453 target cells treated with EVsorafenib released by MCF-7 showed the same outcome. The histograms represent mean value of two replicates for each condition; bars, SD. *p < 0.05, **p < 0.01, ****p < 0.0001 in one-way ANOVA followed by Tukey’s test. Results are representative of two independent experiments
Treatment and uptake of the EVs in the zebrafish model
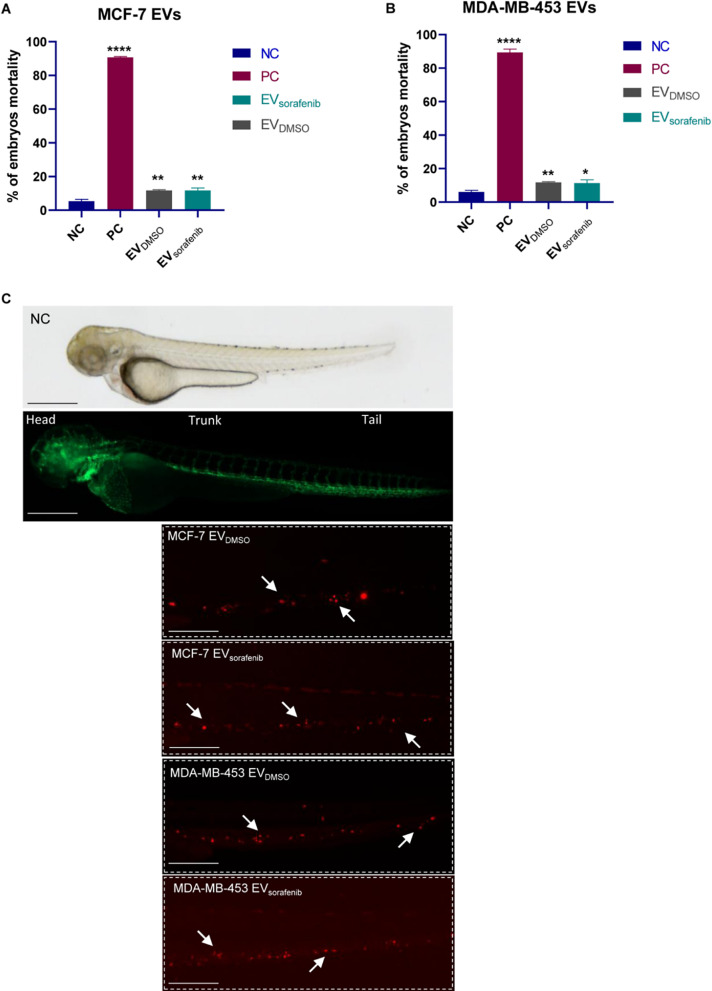
Preliminary experiments were conducted to assess the toxicity of EVs and determine the optimal dose. We carefully monitored the embryos for phenotypic changes, focusing on the most important features described in the OECD 2019 guidelines (Test Guideline No. 203, Fish Acute Toxicity Testing), educed head size and/or delays in development, underdeveloped body, and/or undeveloped eyes [36]. The percentage of zebrafish presenting abnormal phenotype was below 10% for both control EV (EVDMSO) and the enriched EV (EVsorafenib). A dose-response curve was generated at 96 h post-fertilization (hpf) to evaluate the effects. Briefly, zebrafish embryos were treated with 5, 10, and 15 µL of either EVDMSO or EVsorafenib, to determine the appropriate volume to be used (Additional file 1: Fig. 7A-B). Since the percentage of mortality was lower than 15% in all volumes tested, we decided to use the highest dose (15µL) thus ensuring the highest amount of EVs to be administrated [35]. To investigate the uptake of EVs in the zebrafish model, the presence of labeled EVs in zebrafish embryos was examined using fluorescence microscopy [44]. After 24 h, stained EVs were isolated from the media. Based on the results obtained from the dose-response curve, 15 µL of labeled EVs (either EVDMSO or EVsorafenib) were added to the fish-water at 48 hpf. For this study, the transgenic line Tg (kdrl:EGFP) was utilized, which expresses green fluorescent protein (EGFP) specifically in endothelial cells, driven by the kdrl promoter [33]. At 72 hpf, the embryo mortality was determined (Fig. 6A-B) and images of CM-DiI labeled EVs were acquired. EVs migrated towards the caudal region of the zebrafish embryo and the fluorescent signals are indicated by arrows; EVs detected at the tail level (Fig. 6C) demonstrated the successful uptake of the EVs. The absence of the fluorescent signal in untreated fish was used as a negative control (Fig. 6C).
Fig. 6.
EVs uptake in zebrafish. Fluorescent microscopy images of CM-DiI labeled EVs uptake by the recipient transgenic zebrafish line Tg(kdrl:EGFP). Mortality analysis of embryos treated with EVs derived from A MCF-7 and B MDA-MB-453 at 72 hpf; data are representative of three replicates (n = 30 for each group) and are shown as the mean ± standard deviation; negative control embryos (NC) were exposed to 0.1% DMSO in fish water, while the positive control embryos (PC) were exposed to 3,4-DCA dissolved in fish water at a concentration of 3.74 mg/L; unpaired t-test was used; *p < 0.05, **p < 0.01, ****p < 0.0001. C EVDMSO and EVsorafenib derived from MCF-7 and MDA-MB-453 cells can be observed at the tail level. Green signal indicates fluorescent vasculature, while red signal indicates the cancer cells. The absence of the luminescent signal marks the untreated fish used as negative control (NC). Magnification 20x and 32x. Scale bars correspond to 500 μm
Enriched EV-based treatment inhibited the growth of tumor xenografts and micrometastasis formation in zebrafish
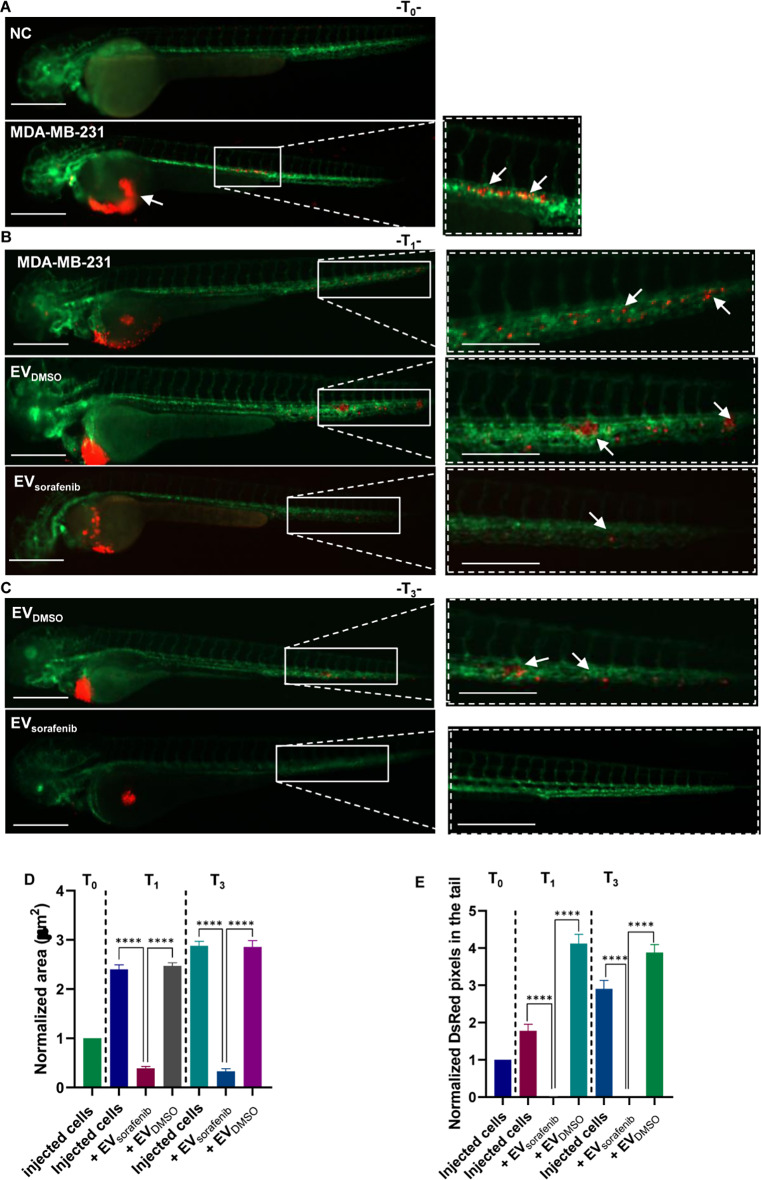
To assess the potential of enriched EV-based treatment to inhibit the aggressive properties of BC cells in vivo, we generated a xenograft model by injecting BC cells in zebrafish, followed by treatment with EVDMSO and EVsorafenib. At 48 hpf, zebrafish embryos were dechorionated, and CM-DiI stained MDA-MB-231 BC cells were microinjected into the PVS. Subsequently, EVDMSO or EVsorafenib collected from MDA-MB-453 cells were added to the fish-water. Fluorescent microscopy pictures revealed that at T0 (2 h post injection- hpi) MDA-MB-231 cells were primarily localized at the site of microinjection. However, fluorescent cells were also localized in the first part of the trunk into the circulation. Not injected fish served as negative control (NC) for comparison (Fig. 7A). Pictures acquired at T1 (24 hpi) exposed the growth of tumor xenografts and the formation of micrometastases in the tails of both untreated and EVDMSO-treated fish. On the contrary, the treatment with EVsorafenib in zebrafish resulted in a notable reduction in tumor xenograft size and a decrease in the number of micrometastases in the tail region (Fig. 7B). We found an inhibition of 84% (p < 0.0001) of the tumor xenografts area (Fig. 7D) and a decrease of about 99% (p < 0.0001) of the tail micrometastases (Fig. 7E). At T3 (72 hpi) the presence of micrometastases and clusters of cancer cells was evident in the tail region of fish that were treated with EVDMSO. The decrease of tumor xenografts area of about 88% (p < 0.0001; Fig. 7D) in EVsorafenib -treated fish was marked by the lessening of CM-DiI signal in the trunk (Fig. 7C). Furthermore, at T3 the micrometastases formation was quite completely abolished with a percentage of inhibition of about 100% (p < 0.0001; Fig. 7E).
Fig. 7.
Effects of enriched EVs-based treatment on MDA-MB-231 xenografts at T0, T1, and T3. A Representative lateral view pictures of not injected Tg (kdrl:EGFP) fish were used as negative control (NC); CM-Dil labeled MDA-MB-231 breast cancer (BC) cells were injected into the perivitelline space of the 48 hpf zebrafish. Representative images of the zebrafish were acquired 2 h post-injection (hpi) (T0); B at 24 hpi (T1) trunk and tail pictures were obtained, migrated cell number is regarded as an indicator of the aggressivity of MDA-MB-231 BC cells. C At 72 hpi (T3) illustrative pictures were acquired, and arrows indicate the cancer cell cluster formation at the tail level. Green signal indicates fluorescent vasculature, while red signal indicates the cancer cells. Scale bars correspond to 500 μm for magnification of 20x and 32x. D The quantification of the tumor area in the xenografts and E the assessment of the tumor cell number in the tail were conducted at T0, T1, and T3. Data are representative of three replicates (n = 30 for each group) and are shown as the mean ± standard deviation. Tumor area and the number of pixels in the tail were normalized to the same parameters obtained in fish injected with MDA-MB-231 cells at T0. ****p < 0.0001 in one-way ANOVA followed by Tukey’s test
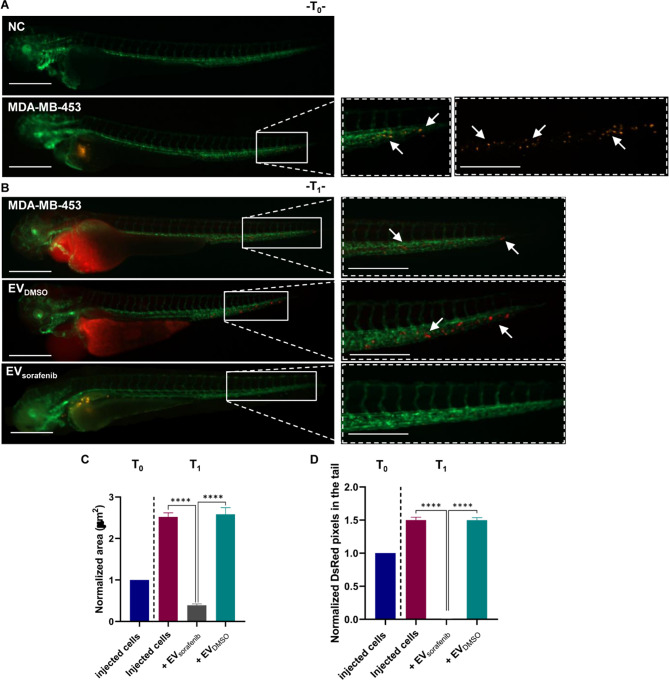
To further validate the efficacy of the treatment in vivo, we injected a second BC cell line, MDA-MB-453, to establish a xenograft model. Subsequently, the xenograft model was treated with EVDMSO or EVsorafenib released by MCF-7 cells. Images obtained by fluorescence microscopy images revealed the presence of MDA-MB-453 cells at the site of microinjection and in the first part of the trunk into the circulation at T0. Fish not injected with MDA-MB-453 were used as negative control (NC) (Fig. 8A).
Fig. 8.
Effects of enriched EVs-based treatment on MDA-MB-453 xenografts at T0 and T1. A Lateral view images of not injected Tg (kdrl: EGFP) zebrafish were selected as the negative control (NC). CM-Dil labeled MDA-MB-453 breast cancer (BC) cells were injected into the perivitelline space of zebrafish at 48 hpf. Representative images of the zebrafish were captured at 2 hpi (T0). B At 24 hpi (T1), illustrative pictures of the trunk and tail were taken, with arrows indicating the presence of numerous cancer cells in the tail, reflecting the progression and aggressiveness of MDA-MB-453 BC cells. Green signal indicates fluorescent vasculature, while red signal indicates the cancer cells. Scale bars correspond to 500 μm for magnification of 20x and 32x. C Quantification of the tumor area of the xenografts as well as D quantification of tumor cells in the tail were determined at T0 and T1. Data are representative of three replicates (n = 30 for each group) and are shown as the mean ± standard deviation. Tumor area and the number of pixels in the tail were normalized to the same parameters obtained in fish injected with MDA-MB-453 cells at T0. ****p < 0.0001 in one-way ANOVA followed by Tukey’s test
Images captured at T1 (24 hpi) demonstrated similar outcomes in both untreated and EVDMSO-treated fish, with observable tumor xenograft growth and a significant presence of migrating cancer cells towards the tail region. However, following treatment with EVsorafenib, a notable reduction in tumor xenograft size and a significant decrease in the number of cancer cells in the tail region were detected in zebrafish (Fig. 8B). We found an inhibition of 85% (p < 0.0001) of the tumor xenografts area (Fig. 8C) and a decrease of about 99% (p < 0.0001) of the tail micrometastases (Fig. 8D). Due to the severity of the phenotype and the presence of pericardial edema at 1 dpi, it was not possible to acquire pictures at T3 (Additional file 1: Fig. 8).
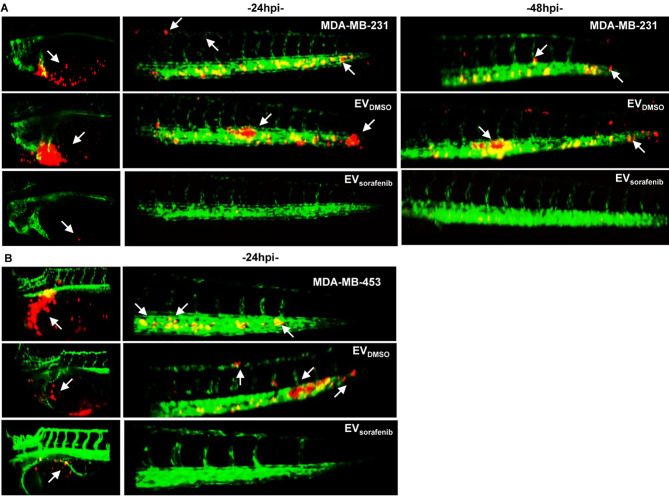
Light Sheet microscopy analysis was conducted to acquire more detailed images that could elucidate the efficacy of the EV-based treatment. We developed the tumor xenografts by injecting CM-DiI labeled BC cells into the PVS of 48 hpf zebrafish embryos followed by treatment with EVDMSO or EVsorafenib. Light Sheet microscopy pictures of the yolk and tail were acquired at 24 hpi and 48 hpi for the fish injected with MDA-MB-231 cells and treated with EVs released by MDA-MB-453 cells (Fig. 9A). A considerable number of BC cells were observed migrating toward the tail, enabling the visualization of cell extravasation and the formation of micrometastases at both time points. When comparing the treatments with EVs, a perceptible decrease in tumor mass within the yolk sac and reduction in tail micrometastases were observed in the EVsorafenib-treated zebrafish in comparison to those treated with EVDMSO. Pictures acquired at 24hpi for the fish injected with MDA-MB-453 cells and treated with EVs derived from MCF-7 cells displayed similar outcomes. A notable reduction in tumor mass and a decrease in the presence of cancer cells in the tail were observed in the fish treated with EVsorafenib (Fig. 9B). Taken together these results highlighted the effectiveness of the EVs rich in miR-23b-3p, miR-126-3p, and GAS5 in reducing the tumorigenicity and the experimental metastasis in the zebrafish model.
Fig. 9.
Imaging of cancer xenografts and micrometastases in zebrafish acquired by digital light sheet microscopy. A Representative image of the yolk sac and tail regions were captured at 24 h and 48 h after microinjection of MDA-MB-231 breast cancer (BC) cells labeled in red. B Additionally, images were taken at 24 h after microinjection of MDA-MB-453 BC (red). Arrows were used to indicate the presence of tumor masses, micrometastases in the tail, as well as the formation of clusters of cancer cells (n = 30 for each group)
The treatment with enriched EVs affected angiogenesis in vivo
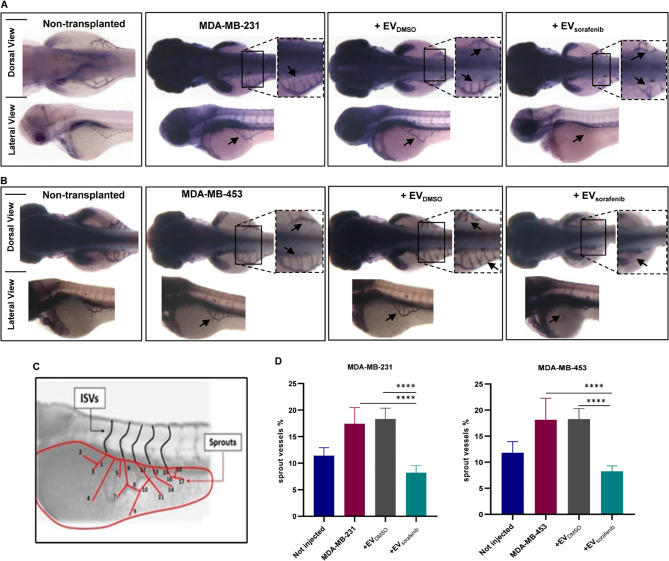
Among the invasiveness and metastatic behavior, the induction of angiogenesis is frequently assessed to evaluate the response to anticancer therapy, particularly in the successful engraftment of human malignant cells. In the zebrafish model, the nascent vessels of the sub-intestinal venous plexus (SIVP) in the anterior region of the yolk were used as a tool to analyze the related mechanisms of the angiogenesis process. To further verify the effects of miR-23b-3p, miR-126-3p, and GAS5 over-represented in enriched EVs on the angiogenesis process, we micro-injected MDA-MB-231 and MDA-MB-453 BC cells in proximity of the developing SIVP at 48 hpf. After the micro-injection, embryos were raised in fish water or treated with EVs until 72 hpf, then fixed in 4% (v/v) paraformaldehyde (PFA) and stained with alkaline phosphatase (AP) assay to visualize the ectopic sprouts [45]. In the case of MDA-MB-231 xenografts, there was a notable stimulation of ectopic sprout formation originating from the SIVP basket. When EVsorafenib were administered, a reduction in sprout formation was observed to EVDMSO (Fig. 10A). Similarly, inducing MDA-MB-453 xenografts also resulted in evident stimulation of abnormal sprout formation from the SIVP basket that was decreased by the administration of EVsorafenib (Fig. 10B). A schematic representation was devised to facilitate a more accurate assessment and measurement of sprout formation and the percentage of angiogenesis inhibition was determined based on it. Treatment with EVsorafenib for either MDA-MB-453 or MDA-MB-231 led to a 55% angiogenesis inhibition compared to treatment with EVDMSO supporting a significant reduction in sprout formation (p < 0.0001; Fig. 10D).
Fig. 10.
Xenotransplantation-induced angiogenesis in the zebrafish embryos. The alkaline phosphatase assay was performed to assess the angiogenic potential of two different cancer cell lines, A MDA-MB-231 and B MDA-MB-453, in embryos that were either untreated or treated, with enlargement of the sub-intestinal venous plexus (SIVP) region at 72 hpf (dorsal and lateral view). Magnification 32x. C The scheme depicts the counting of ectopic sprouts. D To present the results, a graph was generated showing the average number of SIVP branches at 72 hpf. Data are representative of three replicates (n = 30 for each group) and are shown as the mean ± standard deviation. ****p < 0.0001 in one-way ANOVA followed by Tukey’s test
Discussion
The objective of this research was to investigate the potential impact of specific ncRNAs enriched EVs released by BC cells on the aggressive characteristics of the BC in vitro and in vivo. To achieve this, we generated EVs that were particularly abundant in 3 ncRNAs with tumor-suppressor functions: miR-23b-3p, miR-126-3p, and GAS5. The selection of these specific ncRNAs was guided by prior observations regarding their dysregulation in different types of cancer subsequent to treatment with a multikinase inhibitor, sorafenib [13, 40]. Sorafenib, a compact compound, was reported to inhibit various serine/threonine and tyrosine kinases, such as CRAF, BRAF, VEGFR-2 and − 3, PDGFR-ß, FGFR-1, c-kit, and Fms-like tyrosine kinase 3 (Flt-3), involved in numerous cancer-causing signaling pathways [13]. We treated the BC cells with sorafenib for 24 h followed by EVs isolation from culture media, a more efficient approach than the laborious and time-consuming conventional transfection technique commonly used to enhance the loading of specific ncRNA into the vesicles [46, 47]. ddPCR analysis revealed an increased level of all 3 ncRNAs encapsulated in EVs released by sorafenib-treated cells, confirming that the vesicles were successfully enriched. To date, many techniques have been developed for EVs isolation but still, an optimal one has yet to be identified [48]. For this reason, we assessed the levels of these ncRNAs in EVs obtained by 3 methods described in MISEV2023 [28] with different recovery and specificity characteristics: precipitation (commercial isolation reagent), ultracentrifugation, and immunoprecipitation. Precipitation method has the highest recovery and the lowest specificity, meaning that it generates a mixture of various extracellular particles (EPs). Immunoprecipitation shows usually the highest specificity but the lowest recovery, while differential ultracentrifugation shows intermediate specificity and recovery. We characterized EVs purified by the precipitation method using WB analysis. The results demonstrate that these EVs present typical markers of small EVs, suggesting they are most likely exosomes. Furthermore, through TEM and NTA analyses, we identified EVs with a typical round shape, ranging from 50 to 150 nm, which aligns with the size range of exosomes. This may indicate that our EV preparation predominantly contains exosome subtypes that may enhance ncRNA delivery. Moreover, our NTA data indicate that the majority of EVs are below 300 nm in diameter, with exosomes being the most abundant subtype. This suggests that exosomes, given their size and abundance in our preparation, likely play a significant role in ncRNA delivery efficiency. Importantly, we used all three methods to prepare EVs and found similar variation trends of the 3 ncRNAs with very few exceptions. These differences may be due to the specificity and recovery associated with each method. The results confirmed that the vesicles enrichment occurred regardless of the isolation method used, with the highest yield achieved when utilizing the precipitation method. While numerous studies have focused on finding the functional role of EV-mediated ncRNA transfer in various biological processes [49, 50], only a few of them have determined the molecular effect. Given the high interest in utilizing EVs as natural carriers for ncRNAs [51], in our study we subjected two distinct BC cell lines to two different types of enriched EVs. To monitor the uptake of EVs by target BC cells, various tracking methods have been employed, including PKHs and DiD staining [52, 53]. We used CM-DiI to stain the vesicle membranes and tracked the fate of two distinct types of EVs within two different types of recipient cells. In both cases, our results demonstrated that the EVs successfully entered the recipient cells within 24 h of incubation. ddPCR analysis revealed that the treatment caused the increased expression levels of miR-23b-3p, miR-126-3p, and GAS5 in the recipient cells at 24 h-post treatment in both cases. These findings support the idea that enriched EVs can serve as effective in vitro delivery systems for specific ncRNAs, inducing clear molecular changes in target cells but also biological changes. In fact, the proliferation capacity of BC cells treated with enriched EVs was noticeably suppressed. This effect could be attributed to the influence of the three ncRNAs, with tumor-suppressor functions, released into the recipient cells by the EVs. In light of this, the identification of the key molecular pathways affected by these specific ncRNAs is challenging. Future study, based on omics technologies (RNAseq and proteomics) will allow deeper insight into their anti-cancer role upon their release following EVs treatment. Establishment of models to investigate the in vivo uptake, fate, targets, and impacts of enriched EVs has faced challenges due to their small size and the limited techniques that are suitable for their labeling [44]. Therefore, we set out to test whether the labeled EVs would be absorbed by a vertebrate animal model, zebrafish. Our findings, including dose-curve results and fluorescent microscopy images, demonstrated a successful uptake of these vesicles by the zebrafish, followed by rapid dispersion throughout the fish’s body. To our knowledge, up to now, no previous reports have described the utilization of EVs as a therapeutic tool for inhibiting the progression of BC xenografts in zebrafish. However, a recent study successfully established a preclinical zebrafish xenograft model by injecting metastatic BC cells, which enabled researchers to evaluate cancer cell behavior. In that study, it was observed—similar to our findings—that the cancer cells primarily populated the peripheral regions of the larvae [54]. In contrast, our investigation takes a different approach by exploring the therapeutic potential of enriched EVs. Specifically, this study focuses on assessing the effectiveness of vesicles enriched with miR-23b-3p, miR-126-3p, and GAS5 in treating tumor xenografts. Until now, the focus regarding cancer-derived EVs has predominantly centered on their potential to promote tumor progression [55, 56]. However, one study evaluated the effect of bacterial EVs on oral squamous cell carcinoma and found that EVs isolated from a specific strain of bacteria were able to reduce the metastasis rate [57]. Notably, in our study, we have demonstrated, among other findings, that EVs derived from human BC and enriched with the three specific tumor suppressor ncRNAs can effectively inhibit tumor growth in vivo. To investigate this, we conducted microinjections of two distinct cell lines, MDA-MB-231 and MDA-MB-453, into zebrafish, followed by the administration of enriched EVs as a treatment. Analysis of images taken 24 h and 72 h post-treatment illustrates the major impact of the treatment, characterized by an approximately 80% decrease in tumor area. Furthermore, we analyzed the metastatic behavior of microinjected BC cells, which is indicative of their aggressive characteristics. Our pictures clearly show an almost complete eradication of micrometastases located in the tail of the xenograft-induced zebrafish following treatment with EVsorafenib. While the role of cancer-derived EVs in cell-to-cell communication to promote tumor metastasis is clear [58, 59], our study revealed that EVs can serve as promising therapeutic strategy to effectively reduce the formation of micrometastases probably through the targeted enrichment of their cargo by elevating the levels of miR-23b-3p, miR-126-3p and GAS5. The effect of EVs treatment was validated in both BC cell lines with which we conducted our experiments and further consolidated through the light-sheet microscopy pictures. We opted for this microscope to enhance the visualization of the impact of this enriched EV-based treatment on the two most aggressive properties involved in tumor progression, one of which is angiogenesis. This plays a crucial role in the development of tumors and the spread of metastases in various types of cancer, including BC [60]. In our study, our objective was to demonstrate that not only MDA-MB-231 and MDA-MB-453 BC cells microinjection induce angiogenesis in zebrafish but also to assess the effectiveness of enriched-EVs in inhibiting this process. Blocking the development of the angiogenesis led to a significant reduction of abnormal sprout formation from the SIVP basket, with approximately a 55% decrease observed in both cases. In recent years, the clinical use of EVs has made significant progress due to promising results obtained in preclinical models across various fields, such as cancer treatment, drug delivery systems, and immune diseases [48, 61]. In clinical studies where EVs derived from tumor cells were used to deliver the desired chemotherapeutic compound to target cancer cells, the results demonstrated that EVs exhibited low toxicity while showing promising effects, such as inducing the death of tumor-repopulating cells [62]. Here, for the first time to our knowledge, we have demonstrated the ability of sorafenib-induced EVs to significantly limit the aggressive properties of BC both in vitro and in vivo, establishing a proof-of-concept for this approach. EVs carry a variety of biomolecules that could be influenced by sorafenib in addition to the three selected ncRNAs, further characterization of the entire EV cargo may contribute to their potential as promising candidates for innovative treatments in cancer.
Conclusion
EVs represent an important field of interest for the development of innovative therapeutic tools, particularly as delivery systems for ncRNAs. Thanks to their physiological attributes, including biocompatibility, minimal toxicity, and heightened stability within organisms, EVs can serve as highly effective carriers for a diverse array of therapeutic compounds, including nucleic acids such as mRNA, siRNA, miRNA, and shRNA [63]. The sorafenib-induced EVs were particularly enriched in miR-23b-3p, miR-126-3p, and GAS5 and we have provided the proof-of-principal for their ability to limit the aggressive properties of breast cancer in vitro and in vivo. Given the fact that EVs carry a wide range of biomolecules (i.e. proteins, metabolites, mRNAs, ncRNAs) and that the cargo may determine specific effects on the recipient cells, we cannot exclude the possibility that other dysregulated molecules may have an impact on the outcomes. More research is required to elucidate the antitumoral function of EVs in BC. Although data using zebrafish appeared very promising, more in-depth investigations using other experimental animal models are needed to validate the use of EVs as delivery systems for tumor targeted treatments.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. M. Crosatti (University of Leicester; UK) for the linguistic revision of the manuscript.
Abbreviations
- BC
Breast cancer
- HER2
Human epidermal growth factor receptor 2
- ER
Estrogen receptor
- PR
Progesterone receptor
- ncRNA
Non-coding RNA
- TNBC
Triple-negative breast cancer
- EVs
Extracellular vesicles
- TEM
Transmission Electron Microscopy
- NTA
Nanoparticle tracking analysis
- WB
Western Blot
- RT
Reverse transcription
- ddPCR
Droplet digital PCR
- miRNA
microRNA
- lncRNAs
Long non-coding RNAs
- FI
Fold increase
- FD
Fold decrease
- TSG101
Tumor Susceptibility gene 101
- ISEV
International Society of Extracellular Vesicles
- MISEV
Minimum Information for Studies of Extracellular Extracellular Vesicles
- PVS
Perivitelline space
- hpf
Hours post-fertilization
- hpi
Hours post-injection
- SIVP
Sub-intestinal venous plexus
Author contributions
I.A.P. carried out most of the experiments and wrote the manuscript; D.Z. performed the experiments in zebrafish; F.G., G.G. and C.B. carried out the purification of EVs by ultracentrifugation, immunoprecipitation and the characterization of EVs by WB; L.M. acquired the images by LightSheet microscopy; R.D.C. characterized the EVs by TEM; V.G.D.A. performed the Nanoparticles Tracking Analysis (NTA); I.G. performed cell proliferation and viability assays; E.M. helped to collect raw data; AS and GDP supervised the experiments, AS conceived, designed the study and contributed in writing the draft. All the authors contributed to the drafting of the manuscript. All authors have read and approved the manuscript.
Funding
This research was supported by Lega Italiana Lotta ai Tumori (LILT) grant number 02/2023, Liberamente Association (Gambara, BS, Italy) grant number 03/2023, and the University of Brescia (local grants) grant numbers 60/2022 and 60/2023.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Iulia Andreea Pelisenco and Daniela Zizioli contributed equally to this work and co-first authors.
References
- 1.Bray Bsc F et al. Laversanne | Mathieu, Hyuna |, Phd S, Ferlay J, Siegel Mph RL,. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. 2024 [cited 2024 Jun 3]; https://acsjournals.onlinelibrary.wiley.com/doi/10.3322/caac.21834 [DOI] [PubMed]
- 2.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Primers. 2019;5(1). [DOI] [PubMed]
- 3.Waks AG, Winer EP. Breast Cancer Treatment: a review. JAMA. 2019;321(3):288–300. [DOI] [PubMed] [Google Scholar]
- 4.Dellar ER, Hill C, Melling GE, Carter DRF, Baena-Lopez LA. Unpacking extracellular vesicles: RNA cargo loading and function. J Extracell Biology. 2022;1(5). [DOI] [PMC free article] [PubMed]
- 5.Lee Y, Graham P, Li Y. Extracellular vesicles as a novel approach for breast cancer therapeutics. Cancer Letters. Volume 555. Elsevier Ireland Ltd; 2023. [DOI] [PubMed]
- 6.Lee Y, Ni J, Beretov J, Wasinger VC, Graham P, Li Y. Recent advances of small extracellular vesicle biomarkers in breast cancer diagnosis and prognosis. Mol Cancer [Internet]. 2023;22:33. http://creativecommons.org/licenses/by/4.0/.TheCreativeCommonsPublicDomainDedicationwaiver [DOI] [PMC free article] [PubMed]
- 7.Zhang F, Guo J, Zhang Z, Duan M, Wang G, Qian Y, et al. Application of engineered extracellular vesicles for targeted tumor therapy. Journal of Biomedical Science. Volume 29. BioMed Central Ltd; 2022. [DOI] [PMC free article] [PubMed]
- 8.Kalluri R, McAndrews KM. The Role of Extracellular Vesicles in Cancer. Cell [Internet]. 2023 Apr 13 [cited 2024 Oct 25];186(8):1610. https://pmc.ncbi.nlm.nih.gov/articles/PMC10484374/ [DOI] [PMC free article] [PubMed]
- 9.Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. [cited 2023 Jun 7]; 10.1038/s41392-022-00975-3 [DOI] [PMC free article] [PubMed]
- 10.Chen G, Qiu Z, Du L, Shi L, Luo Z, Qian Y. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Frontiers in Medicine | www.frontiersin.org [Internet]. 2020 [cited 2023 Oct 4];7:612393. Available from: www.frontiersin.org. [DOI] [PMC free article] [PubMed]
- 11.Schwarzenbach H, Gahan PB. Interplay between LncRNAs and microRNAs in Breast Cancer. 2023; 10.3390/ijms24098095 [DOI] [PMC free article] [PubMed]
- 12.Peinado P, Herrera A, Baliñas C, Martín-Padrón J, Boyero L, Cuadros M, et al. Long noncoding RNAs as cancer biomarkers. In: Chakrabarti DJ, Mitra DS, editors. Cancer and noncoding RNAs. 2018. p. 95–114.
- 13.Faranda T, Grossi I, Manganelli M, Marchina E, Baiocchi G, Portolani N et al. Differential expression profiling of long non-coding RNA GAS5 and mir-126-3p in human cancer cells in response to sorafenib. Sci Rep. 2019;9(1). [DOI] [PMC free article] [PubMed]
- 14.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. [DOI] [PubMed] [Google Scholar]
- 15.Filippova EA, Fridman MV, Burdennyy AM, Loginov VI, Pronina IV, Lukina SS, et al. Long noncoding rna gas5 in breast cancer: epigenetic mechanisms and biological functions. International Journal of Molecular Sciences. Volume 22. MDPI; 2021. [DOI] [PMC free article] [PubMed]
- 16.Grossi I, Marchina E, De Petro G, Salvi A. The Biological Role and Translational Implications of the Long Non-Coding RNA GAS5 in Breast Cancer. Cancers (Basel) [Internet]. 2023;15(13):3318. https://www.mdpi.com/2072-6694/15/13/3318 [DOI] [PMC free article] [PubMed]
- 17.Sciortino M, Del Camacho-Leal P, Orso M, Grassi F, Costamagna E, Provero A. P, Dysregulation of Blimp1 transcriptional repressor unleashes p130Cas/ErbB2 breast cancer invasion. Sci Rep. 2017;7(1). [DOI] [PMC free article] [PubMed]
- 18.Pellegrino L, Stebbing J, Braga VM, Frampton AE, Jacob J, Buluwela L, et al. miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013;41(10):5400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang G, Cho M, Wang X. OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. 2022 [cited 2023 Sep 27];50. 10.1093/nar/gkab970 [DOI] [PMC free article] [PubMed]
- 20.Sharifi Z, Talkhabi M, Taleahmad S. Identification of potential microRNA diagnostic panels and uncovering regulatory mechanisms in breast cancer pathogenesis. Scientific Reports | [Internet]. 123AD;12:20135. 10.1038/s41598-022-24347-7 [DOI] [PMC free article] [PubMed]
- 21.Zhang P, Yang Y, Qian K, Li L, Zhang C, Fu X, et al. A novel tumor suppressor ZBTB1 regulates tamoxifen resistance and aerobic glycolysis through suppressing HER2 expression in breast cancer. J Biol Chem. 2020;295(41):14140–52. [DOI] [PMC free article] [PubMed]
- 22.Rezaei Z, Sebzari A, Kordi-Tamandani DM, Dastjerdi K. Involvement of the dysregulation of miR-23b-3p, miR-195-5p, miR-656-5p, and mir-340-5p in Trastuzumab Resistance of HER2-Positive breast Cancer cells and System Biology Approach to Predict their targets involved in resistance. DNA Cell Biol. 2019;38(2):184–92. [DOI] [PubMed] [Google Scholar]
- 23.Ebrahimi F, Gopalan V, Smith RA, Lam AKY. MiR-126 in human cancers: clinical roles and current perspectives. Exp Mol Pathol. 2014;96(1):98–107. [DOI] [PubMed]
- 24.Hong Z, Hong C, Ma B, Wang Q, Zhang X, Li L, et al. MicroRNA-126-3p inhibits the proliferation, migration, invasion, and angiogenesis of triple-negative breast cancer cells by targeting RGS3. Oncol Rep. 2019;42(4):1569–79. [DOI] [PubMed] [Google Scholar]
- 25.Barone I, Gelsomino L, Accattatis FM, Giordano F, Gyorffy B, Panza S et al. RESEARCH Open Access Analysis of circulating extracellular vesicle derived microRNAs in breast cancer patients with obesity: a potential role for Let-7a. J Transl Med [Internet]. 2023 [cited 2023 Jun 30];21:232. http://creativecommons.org/licenses/by/4.0/.TheCreativeCommonsPublicDomainDedicationwaiver [DOI] [PMC free article] [PubMed]
- 26.Goessling W, North TE, Zon LI. New waves of discovery: modeling cancer in zebrafish. J Clin Oncol. 2007;25:2473–9. [DOI] [PubMed] [Google Scholar]
- 27.Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. 2002. [DOI] [PubMed]
- 28.Welsh JA, Goberdhan I, Buzas DC, Blenkiron EI, Bussolati C, Cai B. H, Minimal information for studies of extracellular vesicles (MISEV): From basic to advanced approaches. 2024 [cited 2024 May 29]; https://isevjournals.onlinelibrary.wiley.com/doi/10.1002/jev2.12404 [DOI] [PMC free article] [PubMed]
- 29.Guerra F, Paiano A, Migoni D, Girolimetti G, Perrone AM, De Iaco P et al. Modulation of RAB7A protein expression determines resistance to cisplatin through late endocytic pathway impairment and extracellular vesicular secretion. Cancers (Basel). 2019;11(1):52. [DOI] [PMC free article] [PubMed]
- 30.Kreger BT, Johansen ER, Cerione RA, Antonyak MA. The enrichment of survivin in exosomes from breast cancer cells treated with paclitaxel promotes cell survival and chemoresistance. Cancers (Basel). 2016;8(12). [DOI] [PMC free article] [PubMed]
- 31.Grossi I, Schiavone M, Cannone E, Grejdan OA, Tobia C, Bonomini F et al. Lasp1 expression is implicated in embryonic development of zebrafish. Genes (Basel). 2023;14(1). [DOI] [PMC free article] [PubMed]
- 32.Aleström P, D’Angelo L, Midtlyng PJ, Schorderet DF, Schulte-Merker S, Sohm F et al. Zebrafish: Housing and husbandry recommendations. Lab Anim [Internet]. 2020 Jun 1 [cited 2023 May 25];54(3):213. /pmc/articles/PMC7301644/ [DOI] [PMC free article] [PubMed]
- 33.Jin SW, Beis D, Mitchell T, Chen JN, Stainier DYR. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132(23):5199–209. [DOI] [PubMed] [Google Scholar]
- 34.Gao C, Huang Q, Lan Q, Feng Y, Tang F, Hoi MPM et al. A user-friendly herbicide derived from photo-responsive supramolecular vesicles. Nat Commun. 2018;9(1). [DOI] [PMC free article] [PubMed]
- 35.von Hellfeld R, Brotzmann K, Baumann L, Strecker R, Braunbeck T. Adverse effects in the fish embryo acute toxicity (FET) test: a catalogue of unspecific morphological changes versus more specific effects in zebrafish (Danio rerio) embryos. Environ Sci Eur [Internet]. 2020 Dec 1 [cited 2023 Jun 23];32(1):1–18. https://enveurope.springeropen.com/articles/10.1186/s12302-020-00398-3
- 36.Test No. 203: Fish, Acute Toxicity Test [Internet]. OECD. 2019 [cited 2024 Oct 24]. (OECD Guidelines for the Testing of Chemicals, Sect. 2). https://www.oecd-ilibrary.org/environment/test-no-203-fish-acute-toxicity-test_9789264069961-en
- 37.Ren J, Liu S, Cui C, Ten Dijke P. Invasive Behavior of Human Breast Cancer Cells in Embryonic Zebrafish. J Vis Exp [Internet]. 2017 [cited 2024 Jul 22];(122):55459. https://www.jove.com/video/55459 [DOI] [PMC free article] [PubMed]
- 38.Martinez-Lopez M, Póvoa V, Fior R. Generation of zebrafish larval xenografts and Tumor Behavior Analysis. J Visualized Experiments. 2021;2021:172. [DOI] [PubMed] [Google Scholar]
- 39.Serbedzija GN, Flynn E, Willett CE. Zebrafish angiogenesis: a new model for drug screening. Angiogenesis. 1999;3:353–9. [DOI] [PubMed] [Google Scholar]
- 40.Manganelli M, Grossi I, Ferracin M, Guerriero P, Negrini M, Ghidini M et al. Longitudinal circulating levels of mir-23b-3p, mir-126-3p and lncrna gas5 in hcc patients treated with sorafenib. Biomedicines. 2021;9(7). [DOI] [PMC free article] [PubMed]
- 41.Matejovič A, Wakao S, Kitada M, Kushida Y, Dezawa M. Comparison of separation methods for tissue-derived extracellular vesicles in the liver, heart, and skeletal muscle. FEBS Open Bio. 2021;11(2):482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tominaga N, Hagiwara K, Kosaka N, Honma K, Nakagama H, Ochiya T. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy [Internet]. 2014. http://www.molecular-cancer.com/content/13/1/134 [DOI] [PMC free article] [PubMed]
- 43.Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4(1). [DOI] [PMC free article] [PubMed]
- 44.Verweij FJ, Revenu C, Arras G, Dingli F, Loew D, Pegtel DM, et al. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev Cell. 2019;48(4):573–e5894. [DOI] [PubMed] [Google Scholar]
- 45.Basnet RM, Zizioli D, Muscò A, Finazzi D, Sigala S, Rossini E et al. Caffeine Inhibits Direct and Indirect Angiogenesis in Zebrafish Embryos. Int J Mol Sci [Internet]. 2021 [cited 2023 May 25];22:4856. 10.3390/ijms22094856 [DOI] [PMC free article] [PubMed]
- 46.Ohno SI, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microrna to breast cancer cells. Mol Ther. 2013;21(1):185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic potential of mesenchymal cell–derived miRNA-150-5p–Expressing exosomes in rheumatoid arthritis mediated by the modulation of MMP14 and VEGF. J Immunol. 2018;201(8):2472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanz-Ros J, Mas-Bargues C, Romero-García N, Huete-Acevedo J, Dromant M, Borrás C. Extracellular vesicles as Therapeutic resources in the clinical environment. International Journal of Molecular Sciences. Volume 24. MDPI; 2023. [DOI] [PMC free article] [PubMed]
- 49.Kenneweg F, Bang C, Xiao K, Boulanger CM, Loyer X, Mazlan S et al. Long Noncoding RNA-Enriched Vesicles Secreted by Hypoxic Cardiomyocytes Drive Cardiac Fibrosis. [cited 2023 Aug 29]; 10.1016/j.omtn.2019.09.003 [DOI] [PMC free article] [PubMed]
- 50.Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21(4):498–510. [DOI] [PubMed] [Google Scholar]
- 51.Born LJ, Harmon JW, Jay SM. Therapeutic potential of extracellular vesicle-associated long noncoding RNA. Bioengineering and Translational Medicine. Volume 5. Blackwell Publishing Ltd; 2020. [DOI] [PMC free article] [PubMed]
- 52.Reclusa P, Verstraelen P, Taverna S, Gunasekaran M, Pucci M, Pintelon I et al. improving extracellular vesicles visualization: from static to motion. [cited 2023 Aug 28]; 10.1038/s41598-020-62920-0 [DOI] [PMC free article] [PubMed]
- 53.Lerner N, Avissar S, Beit-Yannai E. Extracellular vesicles mediate signaling between the aqueous humor producing and draining cells in the ocular system. PLoS ONE. 2017;12(2). [DOI] [PMC free article] [PubMed]
- 54.Murali Shankar N, Ortiz-Montero P, Kurzyukova A, Rackwitz W, Künzel SR, Wels WS et al. Preclinical assessment of CAR-NK cell-mediated killing efficacy and pharmacokinetics in a rapid zebrafish xenograft model of metastatic breast cancer. Front Immunol. 2023;14. [DOI] [PMC free article] [PubMed]
- 55.Chang WH, Cerione RA, Antonyak MA. Extracellular vesicles and their roles in cancer progression. Methods Mol Biol. 2021;2174:143–70. [DOI] [PMC free article] [PubMed]
- 56.Bao Q, Huang Q, Chen Y, Wang Q, Sang R, Wang L, et al. Tumor-derived extracellular vesicles regulate Cancer Progression in the Tumor Microenvironment. Frontiers in Molecular Biosciences. Volume 8. Frontiers Media S.A.; 2022. [DOI] [PMC free article] [PubMed]
- 57.Metsäniitty M, Hasnat S, Öhman C, Salo T, Eklund KK, Oscarsson J, et al. Zebrafish larvae as a model for studying the impact of oral bacterial vesicles on tumor cell growth and metastasis. Human Cell. 2024;37:1696–1705. 10.1007/s13577-024-01114-6 [DOI] [PMC free article] [PubMed]
- 58.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383–90. [DOI] [PubMed] [Google Scholar]
- 59.Hyenne V, Ghoroghi S, Collot M, Bons J, Follain G, Harlepp S, et al. Studying the fate of Tumor Extracellular vesicles at high Spatiotemporal Resolution using the zebrafish embryo. Dev Cell. 2019;48(4):554–e5727. [DOI] [PubMed] [Google Scholar]
- 60.Madu CO, Wang S, Madu CO, Lu Y. Angiogenesis in Breast Cancer Progression, Diagnosis, and Treatment. J Cancer [Internet]. 2020 [cited 2023 Sep 13];11(15):4474–94. http://www.jcancer.org://creativecommons.org/licenses/by/4.0/ [DOI] [PMC free article] [PubMed]
- 61.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16(4):782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mengfei Guo F, Wu G, Chen HL, Xu J, Xu P, et al. Autologous tumor cell–derived microparticle-based targeted chemotherapy in lung cancer patients with malignant pleural effusion. Sci Transl Med. 2019;11(474):eaat5690. [DOI] [PubMed]
- 63.Androuin A, Verweij FJ, van Niel G. Zebrafish as a preclinical model for Extracellular Vesicle-based therapeutic development. Advanced Drug Delivery Reviews. Volume 176. Elsevier B.V.; 2021. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.