Abstract
Phosphorus is required for many biological molecules and essential functions, including DNA replication, transcription of RNA, protein translation, posttranslational modifications, and numerous facets of metabolism. In order to maintain the proper level of phosphate for these processes, many bacteria adapt to changes in environmental phosphate levels. The mechanisms for sensing phosphate levels and adapting to changes have been extensively studied for multiple organisms. The phosphate response of Escherichia coli alters the expression of numerous genes, many of which are involved in the acquisition and scavenging of phosphate more efficiently. This review shares findings on the mechanisms by which E. coli cells sense and respond to changes in environmental inorganic phosphate concentrations by reviewing the genes and proteins that regulate this response. The PhoR/PhoB two-component signal transduction system is central to this process and works in association with the high-affinity phosphate transporter encoded by the pstSCAB genes and the PhoU protein. Multiple models to explain how this process is regulated are discussed.
INTRODUCTION
Phosphorus is essential for life. In living matter, it is mostly present in its +5 oxidation state, as inorganic phosphate (Pi), or as phosphate esters (1, 2). In some organisms it is also found in its +3 oxidation state as phosphonates, which contain a direct C-P bond (3–5). Pi derivatives are the key hydrophilic components of the amphipathic phospholipids that define the boundaries of all cellular life. Together with the sugars ribose and deoxyribose, Pi derivatives make up the structural backbone of DNA and RNA through their phosphodiester bonds, thus contributing to information storage and processing. Moreover, the cell’s energy currency is based upon the energy released from the hydrolysis of the phosphoanhydride bonds between the phosphates of ATP or of other nucleoside triphosphates. In addition, the biochemical activities of many proteins are regulated by the phosphorylation of specific amino acids, for example, histidine, aspartate, serine, threonine, and tyrosine. As an abundant component of the cytoplasm, phosphate is also a biological buffer that contributes to pH homeostasis.
Because of its indispensable functions, cells must maintain intracellular Pi pools at optimal levels. In bacteria such as Escherichia coli or Salmonella, this is believed to be somewhere between 1 and 10 mM (6–8). In the cell, Pi is assimilated into biological molecules through the synthesis of ATP, which can then be converted to other molecules. Because of its negative charge, Pi is mostly coordinated with metal ions, such as Mg2+. Because most E. coli strains live a biphasic life cycle, alternating between an intestinal phase and an extraintestinal, environmental phase, they must be able to adapt to the alternating environments of a warm, anaerobic, crowded existence inside a host to the widely variable and ever-changing environmental conditions of soil, water, or plants (9). The Pi content within each of these environments varies greatly both spatially and temporally. Fluctuations in the intestinal tract depend upon diet, position within the intestine, and the time since food ingestion (10, 11). In the environment, Pi levels vary based on geography, geology, and competition from other organisms. Because Pi is essential for growth, cells have developed sophisticated systems to acquire Pi when it is limiting in the environment and to cope with excess Pi when it accumulates to high levels within the cell.
This article aims to address the mechanisms by which E. coli and Salmonella cells sense and respond to changes in environmental Pi by reviewing the known genes and proteins that regulate this response. The Pi response is characterized by its specificity, its speed, and its scale so as to contribute to maximal cell fitness. This review also aims to address how this response is integrated with other aspects of cellular physiology.
A BRIEF HISTORY OF THE PHO REGULON
The story of how environmental Pi influences gene expression in E. coli begins in the early days of molecular biology (12). Annamaria Torriani-Gorini was studying the regulation of carbon metabolism with Jacques Monod at the Pasteur Institute (12, 13). In the course of her studies on the proteolytic turnover of the inducible enzyme amylomaltase, she included a control with a noninducible, constitutive acid phosphatase. She observed that when cells were starved of Pi, a new phosphatase was produced that had a pH optimum of 8.2. This observation suggested that the synthesis of alkaline phosphatase was repressed by Pi, an idea that did not entirely fit the model of gene regulation being pursued by Monod at that time (14). Further work on the regulation of alkaline phosphatase by Horiuchi et al. confirmed that the synthesis of this enzyme was not induced by Pi but rather was repressed by it (15). Additional genetic studies by Garen, Echols, and others showed that there were two unlinked genetic regions within the circular E. coli chromosome called R1 and R2 that controlled expression of alkaline phosphatase (16–18). Further research by several groups revealed that the R1 region contained two genes, phoB and phoR, and the R2 region contained five genes, pstS, pstC, pstA, pstB, and phoU (19–23). We now know that the phoBR operon encodes a two-component system (TCS) that regulates the expression of genes involved in the acquisition of Pi and its derivatives and the utilization of alternate sources of phosphorus, such as phosphonates. The PhoB response regulator (RR) and PhoR histidine kinase were some of the earliest known members of the TCS family of proteins, and studies on these proteins have helped establish the fundamental biochemistry and structural analysis of this large class of proteins (24–30).
Following the discovery that the synthesis of alkaline phosphatase is stimulated when environmental Pi is exhausted, other proteins were also identified as having similar expression patterns. The periplasmic phosphate-binding protein (PhoS, later named PstS) was found to be coregulated with alkaline phosphatase (31). Subsequently added to this list were an outer membrane porin specific for Pi called PhoE (32), and the genes for the transport and utilization of sn-glycerol-3-phosphate (33). Because these genes mapped at various locations on the circular E. coli chromosome and were all coregulated with alkaline phosphatase, they were called the Pho regulon (23), a term that had been originally coined to describe genes coregulated for arginine biosynthesis (34). In a comprehensive genetic approach to identify genes of the Pho regulon, Wanner and McSharry used a Mu transposon carrying a lacZ gene to generate lacZ transcriptional fusions whose expression was dependent upon Pi depletion (35). At least 22 promoters were identified through this approach that were regulated by Pi starvation (36). It turned out that several of these genes were also regulated by other environmental factors, including the general stress response that is induced upon entry into stationary phase. Additional researchers from the Neidhardt lab used two-dimensional polyacrylamide gels to identify proteins that were differentially expressed upon Pi starvation (37, 38). They observed >400 proteins whose expression levels changed when cells were subjected to Pi limitation. It was assumed that many of these genes were regulated by multiple systems, in addition to regulation by the Pho regulon. At the time of these experiments it was known that the genes for the utilization of phosphonates were also part of the Pho regulon (39, 40). Looking at the intersection of proteins regulated by Pi starvation and those regulated during growth on 2-aminoethylphosphonate, it was suggested that 137 gene products comprise the Pho regulon (38). This set of proteins probably also included genes from other stress response pathways. More recently, in the omics age, additional genes of the Pho regulon have been identified through global transcriptional analysis using microarrays (41–43), through chromatin immunoprecipitation experiments (44), and also through mass spectrometric approaches (43, 45).
MEMBERS OF THE Pho REGULON
For the purpose of this review article, Pho regulon gene members are defined as those genes directly regulated by PhoB and PhoR and do not include other genes whose expression is changed due to Pi limitation. Upon Pi depletion, expression patterns of numerous genes are altered in a PhoB-independent manner, underscoring the cell’s complex regulatory schemes and the importance of Pi to cell metabolism. Because it is known that ppGpp levels increase upon Pi starvation (46, 47), at least some of the PhoB-independent gene expression may be due to the production of this alarmone and its effects on transcription. For example, the expression of IraP, which is an anti-adapter protein involved in activating the general stress response following Pi starvation, is ppGpp dependent (48, 49). Many other PhoB-independent genes that are induced upon Pi starvation are also controlled by the general stress sigma factor σS, which is induced upon exhaustion of environmental Pi (48–50).
The genes of the Pho regulon from E. coli strain MG1655 for which the strongest evidence has been provided are shown in Table 1. Their descending order in the table reflects the level of expression of these genes upon Pi starvation as determined by transcriptome sequencing (RNA-seq) analysis. These genes include both those of known function and those of unknown function. The Pho regulon genes with known functions are those that are mostly involved in Pi import, the scavenging of Pi from other sources of phosphorus, or the regulation of these processes.
Table 1.
Members of the Pho regulon in E. coli K-12 MG1655 identified by RNA-seq
| Gene/operon | Function | Pho box sequencea | RNA read count in LoPib/log2 fold changec |
|---|---|---|---|
| pstSCAB-phoU | Phosphate ABC transporter + regulator | CTGTCATAAAACTGTCATATTCCTTACATATAACTGTCACCTGT | 73,000/6.5 |
| phoA | Alkaline phosphatase | CTGTCATAAAGTTGTCACGGCC | 67,000/7.5 |
| waaH (yibD) | LPS glycosyltransferase | CTGTAAAAATAATATCTCACAGGCTTAATAGTCTCTTAATACAA | 46,000/10 |
| phoH | Unknown function – predicted ATP-binding protein | CTGTCATCACTCTGTCATCTTT | 46,000/5.1 |
| phnCDEFGHIJKLMNOP | Phosphonate transport + catabolism | AATTAACCAAATCGTCACAATA | 27,000/12 |
| phoBR | Two-component regulatory proteins | TTTTCATAAATCTGTCATAAAT | 11,000/5.4 |
| phoE | Outer membrane porin | CTGTAATAAAAGCGTAAACAACCTGTAATATATCTTTAACAATC | 10,000/7.4 |
| ytfK | H2O2 tolerance | TTGTAACCTTTAGGTAAAAAAAGTTATACGCGGTGGAAACATTG | 8,700/2.4 |
| ugpBAECQ | sn-Glycerol 3-P transporter + phosphodiesterase | TTGTCATCTTTCTGACACCTTACTATCTTACAAATGTAACAAAAAAGTTATTTTTCTGTAATTCGA | 8,600/2.9 |
| psiE | Unknown function | AATATAGATCTCCGTCACATTT | 6,700/5.7 |
Pho box sequences of Pho regulon genes as determined by footprinting assay or EMSA. The most highly conserved residues are shown in bold.
The normalized read counts that were generated using the program DESeq (170) from cells grown in morpholinepropanesulfonic acid (MOPS) minimal medium containing 0.1 mM K2HPO4 (LoPi).
Log2 ratio of gene expression from an RNA-seq experiment from the McCleary lab comparing normalized RNA read counts from E. coli strain MG1655 grown in MOPS minimal media containing 0.2% glucose and either 0.1 or 3.3 mM K2HPO4. The cells were harvested at an optical density at 600 nm of 0.6 and treated with RNAprotect reagent as directed by the manufacturer (Qiagen). RNA was purified using an RNeasy Plus kit (Qiagen), tested for quality, and sent to Novogene (Chula Vista, CA) for rRNA depletion, library construction, sequencing, and analysis.
The genes whose products are directly involved with Pi transport include pstSCAB and phoE. Those involved with Pi scavenging include phoA, encoding bacterial alkaline phosphatase, which hydrolyzes Pi from a wide variety of organophosphates, thus making Pi available for uptake by multiple Pi transporters. The ugpBAECQ operon is also in this group, as it encodes the sn-glycerol-3-phosphate ABC transporter (UgpBAEC) and includes ugpQ, which encodes a phosphodiesterase that releases Pi from this substrate (51, 52). The use of alternate phosphorus sources is made possible by the phnCDEFGHIJKLMNOP operon, which encodes proteins involved in the uptake and metabolism of phosphonates (53, 54).
In addition to those genes involved in Pi metabolism, the Pho regulon also includes genes involved in cell envelope modifications and other stress responses and in virulence. Two genes that encode cell envelope modification enzymes have been identified as being regulated by PhoB. They are waaH and mipA (44, 55). waaH (also known as yibD) encodes an enzyme for the modification of lipopolysaccharide (LPS) in which a phosphate on the inner core region of LPS is replaced by glucuronic acid (56), allowing maintenance of a negative charge at the site. The function of this modification is not known, but it would decrease the demand for Pi under Pi-limiting growth conditions. MipA is a scaffolding protein for penicillin-binding protein function in peptidoglycan synthesis and degradation. Other regulon members YtfK and CusC appear to participate in H2O2 tolerance (57) and copper export (44), respectively.
Those genes whose functions are not yet known include phoH, psiE, and psiF. The purified PhoH protein binds ATP and shows sequence similarity to a family of helicases (58, 59). psiE encodes a membrane protein of unknown function that is predicted to span the membrane 4 times, with the C terminus being located in the cytoplasm (60). The only thing known about psiF is that it is expressed in an operon with phoA (61).
Additional Proposed Pho Regulon Genes
Many other genes have been proposed as members of the Pho regulon (41, 44, 62), such as eda, which encodes 2-keto-3-deoxygluconate 6-phosphate aldolase (62). This Entner-Doudoroff pathway enzyme catalyzes the cleavage of 2-keto-3-deoxygluconate 6-phosphate to pyruvate and glyceraldehyde-3-phosphate. Recent microarray data (44) and unpublished RNA-seq experiments from our lab have not confirmed Pho regulon dependence; moreover, the putative Pho box sequence upstream of eda has very low sequence similarity to confirmed Pho box sequences, and electrophoretic mobility shift assays (EMSAs) to examine PhoB binding required high levels of PhoB (62). Nevertheless, it is possible that eda is a member of the Pho regulon but requires other signals for stimulation in addition to Pi limitation.
In addition to core genes that are regulated by the PhoBR TCS, there are multiple examples of accessory virulence genes that are also under PhoBR control in E. coli O157:H7, a pathogenic strain of E. coli. Yoshida et al. used lacZ operon fusions to a genomic library to identify novel genes of the Pho regulon (63, 64). Chekabab et al. employed microarray analysis of wild-type and ΔphoB cells in both Pi-replete and Pi-deficient media to uncover many genes that are differentially regulated by PhoB in response to Pi starvation in this strain (65). In that report, Chekabab et al. then extended their transcriptome study by showing by EMSAs that PhoB bound to sequences within the LEE1, LEE2, and stx2 promoter regions. These genes and operons are major players involved in the virulence of E. coli O157:H7. Thus, it now appears that PhoB is part of the exceedingly complex regulatory cascade (66) that governs expression of critical O157:H7 virulence determinants. Finding PhoB-mediated regulation of virulence factors leads to questions which may drive future research. Where and when in the gastrointestinal tract is the PhoBR system induced? Does activation of the Pho regulon increase or decrease virulence? How does E. coli integrate Pi signals with other signaling pathways to promote virulence?
A recent examination of changes to the proteome of Salmonella enterica serovar Typhimurium upon phosphate starvation revealed more than 380 gene products whose levels changed upon Pi depletion (45). The response to Pi starvation is important because the intracellular membrane-bound compartment in which these organisms survive and replicate, called SCVs for Salmonella-containing vacuoles, is a Pi-deficient environment. The authors demonstrated that the expression of most of these 380 proteins was independent of PhoB, suggesting activation through the stringent or stationary-phase responses under these conditions. However, the authors did note that the expression of the NagA and NagB proteins was partially controlled by PhoB. These proteins are part of an operon that allows for the catabolism of N-acetylglucosamine to fructose-6-phosphate and the release of Pi from fructose-6-phosphate via the phosphatase NagC.
Moreover, it has been well documented that the virulence of pathogenic strains of E. coli is attenuated when the Pho regulon is constitutively activated by disabling the PstSCAB transporter (67–72). Because the PstSCAB transporter is a negative regulator of the Pho regulon, mutations that affect its synthesis lead to high-level expression of the entire regulon, even when Pi is abundant. An example of a mechanism by which virulence attenuation occurs was recently reported in a paper by Crépin et al. (71). They showed that the transcription of yaiC, a gene that encodes a diguanylate cyclase, is increased in a pst mutant. The yaiC gene is immediately downstream of the phoA operon, and increased expression of yaiC led to increased production of c-di-GMP, which, in turn, repressed the expression of the fim operon, encoding type 1 fimbriae. Reduced production of type 1 fimbriae then led to diminished virulence in the uropathogenic E. coli strain. These types of studies underscore the importance of the correct regulation of the Pho regulon in maximizing fitness during pathogenesis.
CONTROL OF THE Pho REGULON
The Pho Box
Genes and operons of the Pho regulon are directly controlled by PhoB binding to a DNA sequence in their promoters called a Pho box. This sequence was first identified by Makino et al. as 18 bp in length (26). These researchers used DNase I footprinting and DNA methylation protection experiments with purified phospho-PhoB bound to the pstS and phoB promoter regions to visualize the DNA sequences that were protected by PhoB. By comparing these sequences to the promoter regions from phoA and phoE, they identified a sequence common to each promoter. The consensus sequence from these four promoters was described as a direct repeat of CTGTCAT separated by a 4-bp AT-rich linker. The elucidation of the crystal structure of PhoB bound to DNA showed that it binds as a dimer, protecting 22 bp with contacts within both the major and minor grooves. The current Pho box sequence has therefore been extended by four bases to include 22 bp (73).
Gao and Stock have recently shown using reporter expression data and mathematical modeling that there is a correlation between the information content of a Pho box (its closeness to a consensus sequence) to the affinity of phospho-PhoB for that DNA sequence (73). They showed for a few examples that this differential affinity for Pho box sequences led to a temporal pattern of gene expression in which cells first scavenge for Pi from the environment by expressing pstSCAB-phoU genes and then utilize alternate sources of phosphorus, such as phosphate esters or phosphonates (73). As noted in Table 1, some of the Pho regulon promoters contain multiple Pho boxes. This arrangement may lead to higher-ordered PhoB structures beyond the dimeric form.
Control of the Pho regulon is fully dependent upon the PhoBR TCS, the PstSCAB transporter, and PhoU. PhoR is the transmembrane sensor histidine kinase (SHK), and PhoB is the soluble RR. Pi starvation is the stimulus that triggers PhoR autophosphorylation and the subsequent transfer of a phosphoryl group to an aspartate residue of PhoB. Phosphorylated PhoB then binds to Pho box DNA sequences upstream of genes involved in the acquisition of Pi or in the utilization of alternate sources of phosphorus. Once bound to DNA, it recruits RNA polymerase to regulate gene expression. When environmental Pi levels are abundant, PhoR functions as a phospho-PhoB phosphatase to remove the phosphoryl group from PhoB. The phosphatase function prevents further transcription of Pho regulon genes, allowing their products to diminish by degradation and dilution by cellular growth to return cells to a pre-Pi-starvation state (74). The phosphatase function of PhoR also insulates the Pho regulon from activation by other noncognate SHKs and small-molecule phosphodonors to PhoB (74–78). Pi sensing, which controls the alternate kinase and phosphatase activities of PhoR, is mediated by the PstSCAB transporter and an auxiliary protein called PhoU.
REGULATORS OF THE Pho REGULON
The following sections provide structural and biochemical details about each of the seven signaling proteins that are involved in sensing environmental Pi and controlling the expression of Pho regulon genes. These proteins are PhoR, PhoB, PstS, PstC, PstA, PstB, and PhoU.
PhoR
PhoR is a homodimeric, bifunctional histidine autokinase/phospho-PhoB phosphatase. When environmental Pi is limiting, it autophosphorylates on a conserved histidine residue and subsequently donates this phosphoryl group to PhoB, but when Pi is plentiful, it removes the phosphoryl group from phospho-PhoB (79, 80). Thus, PhoR like many other SHKs, displays autokinase, phosphotransferase, and phosphatase functions (81). PhoR is 431 amino acids in length and has a predicted molecular weight of 49,498. It is an integral membrane protein that, unlike many other membrane-bound SHKs, is not predicted to contain a significant periplasmic domain. It does include a membrane-spanning region, a cytoplasmic charged region (CR), a Per-ARNT-Sim (PAS) domain (Pfam: PF00989, PAS) (82), and prototypical dimerization/histidine phosphorylation (DHp; Pfam: PF06580, His_kinase) and catalytic ATP-binding (CA; Pfam: PF02518, HATPase_c) domains at its C terminus (Fig. 1). There are no three-dimensional structures of PhoR or its isolated domains in the databases, so the following structural descriptions are based upon homologies with structures from other SHKs.
Figure 1.
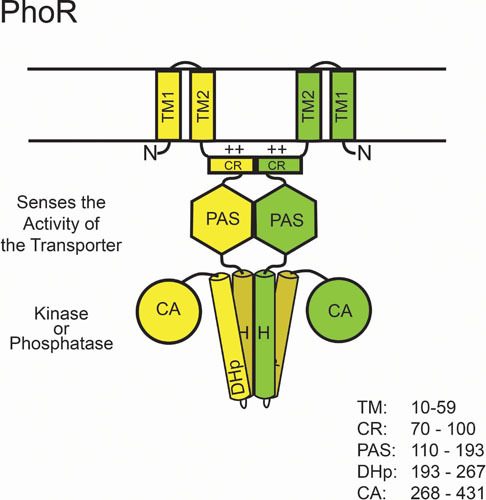
Predicted domain structure of PhoR. PhoR, like other SHKs, is a dimer that is composed of multiple domains. It autophosphorylates on His-213. TM, transmembrane domain; CR, charged region; PAS domain; DHp, dimerization and histidine phosphorylation domain; CA, catalytic and ATP-binding domain.
The membrane-spanning region is predicted to span residues 10 to 59, and it may comprise two transmembrane α helices. The cytoplasmic CR extends from about residue 70 to residue 100. It is predicted to be helical in structure and includes a central core with the sequence RNKKRRR. The function of this region is unknown, but it is completely conserved between E. coli and Salmonella. The PAS domain extends from residue 110 to residue 193 and was originally identified by sequence similarity to other PAS domains (83). PAS domains are generally 90 to 100 amino acids in length and function in signal perception activities (84). The structural fold for PAS domains, based upon the X-ray structures of many sensory proteins, is composed of a single antiparallel, five-stranded β-sheet with a 2-1-5-4-3 strand order with several α helices flanking the sheet. In some proteins, PAS domains bind cofactors that recognize signals; for example, the FixL PAS domain binds a heme group that binds O2 (85). Alternatively, it has been proposed that some other PAS domains may modulate binding to protein partners, either an identical or a nonidentical partner (84). Because PhoR does not contain a significant periplasmic sensory domain, it is assumed that its PAS domain senses a cytoplasmic signal that reflects extracellular Pi concentrations, but the nature of the signal is not completely understood.
The catalytic core of SHKs contains both DHp and CA domains. In PhoR, the DHp domain includes residues 194 to 266 and the CA domain includes residues 267 to 431. Structural analysis of SHKs in the same class as PhoR show that they function as homodimers. In the DHp domain, each monomer within the dimer contributes a helical hairpin to form a four-helix bundle with the conserved phospho-accepting histidine residues being positioned midway on one face of the first helix (His-213) (86). It has been shown that phosphorylation of PhoR occurs in cis, where the CA domain of one of the monomers phosphorylates the His residue of the same polypeptide chain (87). Many SHKs autophosphoryate in trans, the difference being the handedness of the loops connecting the two helices in the hairpin structure. A left-handed loop directs cis autophosphorylation, while a right-handed loop directs trans autophosphorylation. The DHp domain also contains all of the residues necessary for its phospho-PhoB phosphatase activity (80). If PhoR is similar to other SHKs of the HisKA subfamily, then Thr-217 would be required for this phospho-PhoB phosphatase activity (88). Thr-217 may guide a water molecule to the phosphoryl group on Asp-53 to facilitate the nucleophilic attack that removes the phosphate (89). The CA domain of PhoR harbors the enzymatic activity for transferring a phosphoryl group from ATP to the conserved histidine residue of the DHp domain. It has an α/β sandwich fold that harbors a very conserved ATP-binding pocket. This structure directs a nucleophilic substitution reaction in which His-213 functions as the nucleophile attacking the γ-phosphate of ATP, leading to autophosphorylation.
It is predicted that there is structural flexibility between the DHp and CA domains to enable the interdomain phosphorylation reaction (90, 91). Therefore, the control of the opposing kinase and phosphatase activities of PhoR may involve the constraint of the CA domains to prevent their access to the DHp domains while simultaneously exposing the residues of the DHp domain that are required for phosphatase function (Fig. 1). The nature of these constraints is suggested from several structures of other SHKs in kinase and phosphatase conformations. In other SHK structures, the kinase conformation involves an asymmetry along the DHp domain permitting the CA domain access to the phosphorylation site, whereas the phosphatase conformation involves a symmetric DHp conformation that reduces access (92).
PhoB
PhoB is the RR protein of this TCS. It is composed of an N-terminal receiver domain and a C-terminal DNA-binding domain (DBD). This protein family is defined by the N-terminal receiver domain of approximately 120 amino acids (Pfam: PF00072, response_reg). Structural studies involving PhoB have used the isolated receiver or DBDs and have generated a wealth of information about how its structure dictates its function. The receiver domain has a β5α5 fold with a central five-stranded β-sheet (29). Two of the helices are located on one side of the β-sheet (α1 and α5), with the other three helices being positioned on the other side (α2, α3, and α4). This domain contains the site of aspartyl phosphorylation, which in PhoB is Asp53. This completely conserved residue is found in the loop connecting the end of β3 and the beginning of α3. The receiver domain of PhoB contains the necessary catalytic residues to transfer a phosphoryl group from the phosphohistidine residue of phospho-PhoR (93). The conserved residues surrounding Asp-53 provide an environment where a metal ion can be bound, which facilitates a nucleophilic attack on the phosphorus atom from the phosphohistidine residue of PhoR. PhoB can also be phosphorylated by small-molecule phosphodonors such as acetyl phosphate and phosphoramidate (93, 94).
The effector domain of PhoB is involved in DNA binding and RNA polymerase interactions. This ∼99-amino-acid domain has a winged-helix structure (Pfam: PF00486, trans_reg_c) (30), which represents the largest group within the RR family (95, 96). This structure consists of a helix-turn-helix motif that contains the DNA recognition helix. This central core is flanked by an N-terminal four-stranded β-sheet and a C-terminal β-hairpin that is considered the wing. When PhoB becomes phosphorylated, it forms a stable dimer that binds cooperatively to Pho box sequences in the DNA (26, 30, 93, 97). Each PhoB protomer within the dimer binds to an 11-bp direct repeat within the 22-bp Pho box. Recognition of the TGTCA tract within the DNA major groove is mediated by the α3 recognition helix (30). The β-hairpin wing mediates several minor groove interactions, mostly with less conserved A/T residues. These short Pho box sequences are located upstream of Pho regulon genes to recruit RNA polymerase and initiate transcription by remodeling the RNA polymerase holoenzyme-DNA complex (26, 30, 98).
Activation of PhoB by phosphorylation
How does phosphorylation of the receiver domain of PhoB control the activity of its effector domain? As is the case for other RRs, phosphorylation influences the oligomeric state of the protein by favoring a conformational change mediated by the reorientation of conserved Thr-83 and Tyr-102 “switch” residues within this receiver domain (28). Crystal structures of PhoB’s receiver domain in inactive and active conformations showed two different dimeric structures (28, 29); one is a symmetric dimer whose interface is along its α1/α5 surface and the other through its α4-β5-α5 surface. The α4-β5-α5 dimer was formed in the presence of BeF3−, which is a phosphoryl analog that is predicted to stabilize an active conformation. It has been suggested, based upon recent nuclear magnetic resonance (NMR) studies, that unphosphorylated PhoB is in equilibrium between monomer and multiple dimeric forms, not just the α1/α5 dimer (99). Phosphorylation may stabilize the α4-β5-α5 dimeric conformation that enhances DNA binding and transcriptional activation. Research by Creager-Allen et al. suggests that PhoB activation is complex (100). The kinetics of PhoB autophosphorylation with the small-molecule phosphodonor phosphoramidate clearly demonstrate that PhoB/phospho-PhoB heterodimers exist and that phosphorylation is cooperative.
It is interesting that the structure of the isolated receiver domain shows a dimeric structure with a head-to-head symmetry but the structure of the DNA-bound dimeric DBD shows a head-to-tail arrangement. This arrangement requires flexibility of the loop connecting the two domains (101).
The stable PhoB dimer binds to and bends DNA containing a Pho box sequence. Bound PhoB interacts with the σ4 domain of the σ70 subunit of RNA polymerase to recruit the enzyme to the chromosome. A crystal structure of a PhoB/σ4/Pho box complex suggests that σ4 makes contacts with the upstream PhoB protomer within a PhoB dimer and residues within the major groove of DNA and that PhoB may help to remodel the ternary complex to facilitate open complex formation and transcription initiation (98, 102).
PstSCAB
The PstSCAB proteins form a high-affinity Pi transporter that has a Km of 0.4 μM Pi and a maximum velocity of 16 nmol of Pi mg (dry weight)−1 min−1 (103). It is a member of the ATP-binding cassette (ABC) superfamily from the Transporter Classification and Pfam databases (95, 104). This protein superfamily employs the hydrolysis of ATP to bring a variety of substrates across biological membranes, both as importers and as exporters (105). Members of this protein superfamily are found among the bacteria, archaea, and eukaryotes. Prokaryotic importers, such as the PstSCAB proteins, utilize an extracytoplasmic substrate-binding protein that binds substrates and presents them to their membrane-spanning proteins (106). PstS is the periplasmic substrate-binding protein of the Pst system, with a molecular weight of 34 kDa. Its structure contains two globular domains separated by a linker with the Pi binding site located in a crevice between the two domains (107). PstS binds Pi under physiological conditions with a Kd (dissociation constant) of ∼10 μM (108). Molecular dynamics analysis suggest that PstS binds the 1H form of Pi, not the 2H form (109), although this issue is not entirely settled (110). PstC and PstA compose the membrane-spanning components of the transporter (111, 112). The most highly conserved feature within the superfamily is the nucleotide-binding domain, also called the ATP-binding cassette, which binds ATP, hydrolyzes it, and then releases it in order to provide the energy for transport (113). PstB contains the nucleotide-binding domain for this transporter (114). The crystal structures of several ABC importers have been solved, which has shed some light on the mechanisms of transport (115).
To understand how the Pst transporter is involved in the regulation of the PhoBR TCS, it is important to understand its structure and mechanisms of action (Fig. 2). Apart from the structure of the PstS protein (107), no structures are available for the PstS, PstC, or PstB protein. Therefore, a comparison of PstSCAB to transporters of known structure has been helpful. Of particular note is the structure of the putative molybdate transporter, called ModABC, from the archaeon Archaeoglobus fulgidus (116). Like the PstSCAB transporter, this protein also imports an oxyanion. A clue to understanding the molecular mechanisms of Pi transport through the PstCAB proteins comes based upon sequence similarities between the molybdate, sulfate, and Pi transporters. The most highly conserved sequences within this group are found in a region of the protein that creates a cavity and a gate within the membrane-spanning region that most likely represent the pathway through which the anion must pass. The published ModABC structure is of the proteins in a nucleotide-free conformation and shows 12 transmembrane helices situated in an inward-facing conformation with the gate at the periplasmic surface of the membrane. It has been proposed that PstSCAB, like other transporters in this superfamily, utilize an alternating access mechanism to transport their substrates, in which they alternate between inward- and outward-facing states that are driven by substrate binding, ATP hydrolysis, ADP release, and subsequent ATP binding (Fig. 2) (115). ATP binding across the PstB dimer interface would be predicted to close the cavity and lead to an outward-facing structure that can receive Pi from the substrate-loaded, periplasmic PstS protein. This event would trigger ATP hydrolysis, which would flip the outward-facing transmembrane components to an inward-facing conformation, thereby opening the gate and allowing Pi to gain access to the cytoplasm. The cycle would be continued as ADP was released and ATP was rebound.
Figure 2.
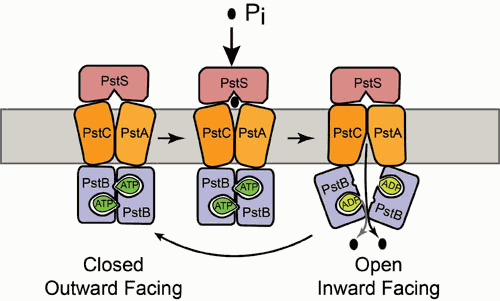
Transport cycle of PstSCAB transporter. The PstS protein is the periplasmic phosphate-binding protein that binds and presents Pi to the transmembrane components PstC and PstA. PstB binds ATP, which stabilizes a closed nucleotide-binding domain with PstC and PstA adopting an outward-facing conformation. Pi-loaded PstS triggers ATP hydrolysis, which causes a conformational change between the PstB protomers that switch the PstC/PstA proteins into an inward-facing structure. ATP binding resets the system to the outward-facing structure.
The Pst transporter is most highly expressed when environmental Pi levels are low. For this reason, it was assumed that it played its most important role in Pi transport under those conditions. More recently, it has been proposed that it plays the primary role in Pi transport under all conditions (117). The expression of the pstSCAB genes is controlled by the PhoBR TCS. The primary promoter for this operon, and the one which is regulated by Pi levels, is found upstream of the pstS gene (118). Other promoters that are internal to the operon have been identified upstream of the pstC, pstB, and phoU genes and are rather weak, but they may play a role in maintaining basal levels of the PstSCAB proteins under Pi-replete conditions (119).
PhoU
In addition to the PstSCAB protein, PhoU is also required for Pi signal transduction, but it is not required for transport through the complex (120). PhoU is a peripheral membrane protein that modulates Pi transport through the PstSCAB complex (121–123). When Pi is plentiful, PhoU acts like a brake to prevent too much Pi import, with its accompanying ATP hydrolysis (123). Several ABC transporters have domains within their nucleotide-binding domain that participate in a type of negative feedback, a process called transinhibition (124–128). PhoU most likely performs this function for the PstSCAB transporter. Multiple crystal structures have been reported for PhoU proteins from various organisms (129–131). PhoU consists of two symmetric, three-α-helix bundles, and metal ions are associated with two of these structures. The metals are coordinated by highly conserved amino acid residues that are found in each three-helix bundle. PhoU from Thermotoga maritima coordinates iron clusters (129), while PhoU from Streptococcus pneumoniae shows zinc ions bound (130). Gardner et al. have recently shown that the soluble form of PhoU from E. coli is a dimer that binds manganese or magnesium (132). Mutagenesis experiments were consistent with the hypothesis that these divalent metals are bound by the same conserved amino acid residues as bind the iron and zinc ions in the two crystal structures. It was also suggested in this study that metal binding may be important for PhoU interactions with the membrane. Alternatively, PhoU may bind Pi through its interactions with these metals.
When phoU is mutated or deleted, PhoR is constitutively active as an autokinase, leading to high-level expression of Pho regulon genes. phoU deletion mutants show poor growth and frequently accumulate compensatory mutations in phoR, phoB, or the pstSCAB genes (120, 121, 123). This suggests that cell growth is impaired when intracellular Pi levels are too high.
Two general classes of models have been previously suggested for how PhoU participates in the signaling pathway. It may mediate the formation of a signaling complex between the PstSCAB transporter and PhoR (36, 130), or it may produce a soluble messenger that is recognized by the cytoplasmic domains of PhoR (consistent with observations reported by Hoffer and Tommassen [133] and by Rao et al. [134]).
PI SENSING
Signal transduction pathways sense stimuli from the environment and respond by altering intracellular processes. For the Pho system, the response is an altered transcriptome that is mediated by the action of the phosphorylated PhoB protein interacting with RNA polymerase. How, then, is environmental Pi sensed? It has generally been accepted that the Pho regulon is turned on when environmental Pi levels dip below 0.4 μM (36). Because the PhoR protein does not have a significant periplasmic domain, it was thought that this SHK did not sense environmental Pi directly but rather something else. Pi sensing requires the PstSCAB transporter and PhoU, because mutations in their genes led to constitutive expression of the Pho regulon regardless of environmental Pi levels (36). Put differently, these mutants are Pi blind. Thus, the default biochemical activity of PhoR is as an autokinase and the signaling roles of the Pst transporter and PhoU are to negatively regulate this activity and to stimulate its phospho-PhoB phosphatase activity in response to environmental Pi.
Two possibilities for how the Pst transporter may function to control the activity of PhoR are as follows. The first is by controlling the intracellular levels of Pi, which would be sensed by PhoR, most likely through its PAS domain. According to this model, intracellular Pi levels should show fluctuations that reflect environmental Pi levels. A problem with this model is that intracellular Pi levels have been measured by 31P NMR and have been shown to be constant under conditions in which the Pho regulon is both repressed and derepressed (7). Another problem with this model is that there are several mutations in pstC and pstA that lead to defective transporters that would be unable to maintain intracellular Pi levels but that retain their signaling capacity; i.e., they can still stimulate the phospho-PhoB phosphatase activity of PhoR even though they are not able to transport Pi (111, 112).
A second model for how the Pst transporter senses Pi proposes that it may physically interact with PhoR to signal its transport activity within a signaling complex (135). That is to say, it is not necessarily the intracellular level of Pi that is sensed but rather how active the transporter is. It has been postulated for many years that the Pi signaling proteins operate within a stable complex that involves PhoR, PhoU, and the PstSCAB transporter (36, 117, 136). Circumstantial evidence for protein-protein interactions had been provided by experiments that suggest that PhoU modulates the transport activity of the PstSCAB transporter, which would require physical interactions between the proteins (123). Further work establishing a signaling complex came from Gardner et al., who used bacterial two-hybrid and coelution experiments to demonstrate that the PhoU protein physically interacts with both the PAS domain of PhoR and the PstB protein (132). PhoU is a central part of this signaling complex, and insight into its function comes from the analysis of several phoU alleles (phoU was originally known as phoT) (19, 137). The original allele of phoU was called phoU35 and was identified as having normal Pi transport but lost proper Pho regulon control (138), and it was later determined that it contained an A147E substitution (139). This mutant is unique because while phoU35 has normal Pi transport, a ΔphoU allele shows a severe growth defect resulting from uncontrolled Pi import (120, 121). It was postulated that the phoU35 version of PhoU lost its ability to communicate with PhoR, hence its derepression, but retained its ability to interact with and control the PstSCAB proteins to prevent hyperimport of Pi with its accompanying growth defect. These observations suggested that PhoU had at least two genetically separable functions and that structural changes resulting from a substitution at Ala-147 disrupted the PhoU/PhoR interaction (140). A scanning-mutagenesis study of the PAS domain of PhoR pointed to several regions of PhoR that were also important for maintaining interactions between PAS and PhoU (140). From these data, a structural PhoR/PhoU docking model was created using the ClusPro web-based server (141). Direct coupling analysis, which is a bioinformatic technique to identify covarying residues between interacting proteins, supported and extended this structural model, which shows PhoU interacting not only with the PAS domain of PhoR but also with its CA domain. Taken together, the findings show that the interaction between PhoU and the PstB protein may be responsible for environmental input, whereas the interactions between PhoU and the PAS and CA domains of PhoR may control output. This may occur as PhoU constrains the interdomain motions of the CA domain of PhoR that could modulate its two biochemical activities.
To extend this signaling complex model, recent research explored how the PstSCAB transporter may communicate with PhoR. Vuppada et al. asked whether the alternating conformations of the PstSCAB protein, which are adopted as part of the normal Pi transport cycle, are the signal that is sensed by PhoR (142). Two variants of PstB that were predicted to lock the protein in either an open or a closed state were tested for Pi signaling by assaying expression of the phoA gene. Similar mutations had been shown to lock the maltose transporter into either an open or a closed conformation (143). It was observed that the mutant predicted to reside in an inward-facing, open conformation (PstBQ160K) signaled Pi sufficiency, whereas the mutant predicted to reside in an outward-facing, closed conformation (PstBE179Q) signaled Pi starvation. Neither mutant was able to transport Pi. The inward-facing conformation of the Pst transporter would normally be formed only following Pi transport, consistent with a Pi-replete environment. These results were consistent with the hypothesis that the signaling activity of the PstSCAB transporter is the signal sensed by PhoR.
A recent study from the Groisman lab has prompted a reexamination of these competing hypotheses (144). They observed through chromatin immunoprecipitation-sequencing experiments that when Salmonella enterica serovar Typhimurium cells were grown for 4 h in medium with 10 μM Mg2+, RNA polymerase occupancy increased for mgtA, mgtB, and mgtC, as well as for phoBR, pstSCAB, and phoU (144). mRNA levels for these genes were also increased. Increased transcription of the mgt genes was expected because they are part of the phoPQ regulon that responds to limiting Mg2+, whereas the increased expression of the Pho regulon genes was unexpected. Using green fluorescent protein reporter assays, they also demonstrated that the activation of the phoB promoter was transient and that it occurred after several hours of incubation in low-Mg2+ medium. The proposed mechanism for these observations, supported by many experiments, is that low Mg2+ levels inhibit ribosome assembly, which leads to decreased protein synthesis. Because translation is the greatest consumer of cellular ATP, decreased protein synthesis would lead to increases in cellular ATP levels with a concomitant decrease in free cytoplasmic Pi. This response is transient because the activation of the PhoPQ regulon leads to normalized Mg2+ levels and the activation of the PhoBR regulon leads to increased Pi uptake. This work underscores the complex regulatory schemes connecting protein synthesis and Pi homeostasis. The implication of this work is that the Pho system senses intracellular Pi, which is contradictory to earlier work showing Pi levels remaining constant during induction of the Pho regulon.
Additional evidence also favors a signaling model that incorporates sensing intracellular Pi levels. PitA and PitB are two Pi transporters of the major facilitator superfamily. They are synporters that utilize the proton motive force to transport Pi-metal ion complexes. Hoffer and Tommassen (133) showed that overexpression of either pitA or pitB in a pst mutant background could lead to downregulation of the Pho regulon. Normally, the Pho regulon is fully induced in a pstS mutant. The researchers also showed that the pstC, pstA, pstB, and phoU genes were required for this downregulation. Although not measured directly, these results implied that high intracellular levels of Pi could lead to downregulation of the Pho regulon, but only when the transmembrane and cytoplasmic parts of the transporter and PhoU were present.
It is possible that aspects of both competing models are correct (Fig. 3). The PhoR protein may interact with the Pst transporter to detect its signaling activity by recognizing alternate conformations of the transporter, and it may also sense cytoplasmic Pi levels. Under normal growth conditions, intracellular Pi levels would be buffered to remain nearly constant, but when Pi transport through the Pst complex is reduced because the extracellular Pi concentrations are too low, then a signal represented by the conformational state of the transporter would be sent to PhoR informing it to adopt its autokinase conformation. Alternatively, when the Pst transporter is actively importing Pi, the PstB protein would hydrolyze ATP, causing the transporter to adopt a conformation that would signal PhoR to adopt its phospho-PhoB phosphatase conformation. The conformations adopted by the signaling complex may also be sensitive to intracellular Pi levels, particularly if there exists an equilibrium between the two conformational states. For example, if intracellular Pi concentrations are elevated, such as when PitA is overexpressed, Pi may bind to the signaling complex and shift its equilibrium towards the phosphatase conformation. Alternatively, when intracellular Pi levels are depleted, such as when protein synthesis is inhibited and ATP levels are elevated, the conformations of the signaling complex may be shifted towards the autokinase conformation.
Figure 3.
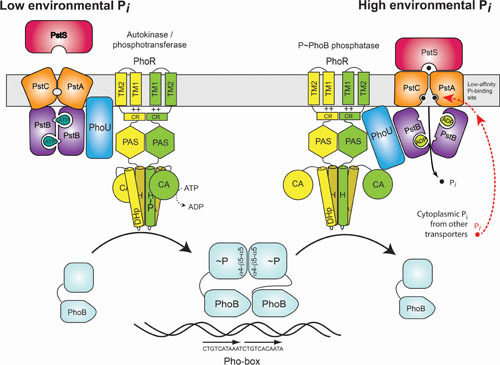
Model for transmembrane signal transduction to regulate the Pho regulon in E. coli. The output of this signal transduction system is based upon the amount and phosphorylation state of the response regulator PhoB. Phospho-PhoB forms a dimer and binds to DNA sequences containing a Pho box. The phosphorylation state of PhoB is, in turn, controlled by the opposing autokinase/phosphotransferase and phosphatase activities of PhoR, the SHK of the system. The activities of PhoR are ultimately controlled by the PstSCAB transporter and PhoU. When environmental Pi levels are high, the transporter signals through PhoU to favor PhoR in its phosphatase conformation. When environmental Pi levels are low, the transporter and PhoU signal PhoR to favor its autokinase conformation. In an alternate (or supplementary) mechanism, the PstSCAB transporter may also signal Pi sufficiency to PhoR through a process in which its conformation is sensitive to intracellular Pi levels by binding Pi at a low-affinity, cytoplasm-accessible site.
Other systems may also contribute to regulation of the Pho regulon by interacting with PhoR to control its activities. For example, Lüttmann et al. published a report linking the nitrogen phosphotransferase system (PTSNtr) to Pho gene regulation (145). The PTSNtr is paralogous to the phosphoenolpyruvate:carbohydrate PTS system that transports various sugars and controls multiple aspects of metabolism based upon carbohydrate availability (146). The PTSNtr consists of three proteins (EINtr, NPr, and EIIANtr) that also participate in a phosphorylation cascade. It is thought that this PTSNtr system plays regulatory roles in potassium (K+) transport (146), σ factor selectivity (147), the stringent response (148), and virulence (149–152). These researchers demonstrated that EIIANtr directly binds to PhoR to stimulate its kinase activity (145). Moreover, they showed that ptsN mutants (lacking EIIANtr) show a muted response to Pi limitation and a growth delay upon shift to Pi-deficient growth media.
LOGIC OF THE Pho REGULON
The Pho regulatory circuitry displays specificity and appropriate temporal responsiveness to produce maximal cellular fitness. To understand and model the Pho regulatory circuit, it is important to determine the amounts and characteristics of the individual proteins. These parameters can help answer some of the following questions. For example, under native cellular conditions, how do the kinase and phosphatase activities of PhoR partition? How does phospho-PhoB binding to target DNA control the temporal pattern of gene expression? How is the Pho TCS insulated from other TCS proteins and other activating signals within the cell to maintain signal integrity?
The answers to some of these questions have been provided through careful biochemical experiments, mathematical modeling of the signaling reactions, and bioinformatic analysis (73, 74, 77, 87, 153–156). For example, it has been shown through quantitative Western blotting that the concentrations of PhoB and PhoR under Pi-limited growth conditions are ∼9.3 μM and 1 μM, respectively (77). These amounts of PhoB and PhoR are about 20-fold higher than under Pi-replete conditions. If a protein concentration of 1 μM roughly corresponds to ∼600 molecules/cell, then PhoB increases from ∼300 copies per cell to ∼6,000 upon Pi starvation and PhoR increases from ∼30 molecules per cell to ∼600. By employing Phos-tag acrylamide electrophoresis to quantify the amount of phospho-PhoB produced under activating conditions and analyzing these quantities using a kinetic model that encompasses PhoR autophosphorylation, phosphotransfer to PhoB, and phospho-PhoB dephosphorylation, it was demonstrated that in the steady state the amount of phospho-PhoB depended upon the total amount of PhoB and that this amount saturated under high expression levels, near 4 μM. This means that significant fractions of PhoB are unphosphorylated, even when the system is fully induced. The PhoB/phospho-PhoB heterodimers reported by Creager-Allen et al. may account for a sizeable fraction of PhoB in the fully induced state (100). In another study using similar experimental strategies, Gao and Stock demonstrated that the PhoR phosphatase activity was quite robust. This activity was strong enough to attenuate the low-Pi response following Pi feeding and also strong enough to suppress nonspecific activation of PhoB by acetyl phosphate or noncognate SHKs (74). These observations underscore the importance of the phosphatase activity of PhoR in controlling the output of the Pho regulon, especially in a cell with multiple potential phosphodonors, such as noncognate SHKs and small metabolites, like acetyl phosphate.
Signal insulation has also been studied by examining the interactions between SHKs and RRs. The analysis of coevolving residues has been used to identify sites within the DHp domains of SHKs and the receiver domains of RRs that provide specificity to the interaction between these two classes of proteins (156). In an examination of divergent evolution in TCS pathways, the Laub group studied the PhoB/PhoR signaling system. They showed that the residues directing the specificity between PhoR and PhoB are subject to strong purifying selection. As expected, the specificity residues are solvent exposed within the DHp domain of PhoR and are located below His-213 on both α helices of its helical hairpin. In PhoR, these specificity residues correspond to Thr-220, Val-221, Gly-224, Tyr-225, Glu-227, and Met-228. In PhoB, the specificity residues are spatially near the site of aspartyl phosphorylation (Asp-53) and are Glu-11, Ala-12, Ile-14, Met-17, Phe-20, Val-21, and Ser-108. The importance of these coevolving specificity residues is that they constitute an important mechanism to insulate signaling pathways from one another and prevent cross talk, even when they utilize identical chemistry with similar overall structures.
As has been stated above, the amounts of PhoB and PhoR are autoregulated through a positive-feedback loop with phospho-PhoB binding to the phoB promoter to activate its own transcription when environmental Pi levels are low. A fitness advantage of an autoregulated signaling pathway is that output genes are not expressed when an activating signal is absent, thereby conserving biosynthetic capacity. It has been elegantly demonstrated through competition assays that the PhoB/PhoR TCS is tuned for optimal expression of gene products under both nonactivating and activating conditions (153). Maximal fitness was observed when PhoB was expressed at a concentration near where phospho-PhoB starts to saturate (4 μM). This type of autoregulation leads to high amplification of a response, i.e., production of enzymes for the acquisition of Pi only when needed and low production costs when Pi is abundant.
However, one downside to an autoregulatory circuit, like that found in the PhoB/PhoR TCS, is that such circuits are predicted to slow the timing of expression due to the time that it takes to build up the levels of the transcription factor in the cell. To study this phenomenon in the Pho system, Gao and Stock used transcriptional reporters and sophisticated modeling to show that the Pho system is both fast and optimized for maximal fitness (155). They demonstrated that the positive-feedback loop for PhoB expression has a coupled negative autoregulation. The mechanism for this feature is the presence of a previously unidentified second Pho box in the phoB promoter whose binding affinity is weaker than the activating Pho box and that faces in the opposite direction. Binding of phospho-PhoB late in a low-Pi response to this site represses transcription of phoB by blocking access to the −10 region of the promoter.
According to the logic of the Pho regulon, the genes of this system are not all expressed at the same time, nor at the same levels. Gao and Stock carefully analyzed the temporal patterns of gene expression by examining the timing of fluorescent reporters that were under the control of various Pho regulon promoters (73). They observed that the early genes include those for the phoBR autofeedback loop that amplifies the signal, as well as for the genes encoding phosphate import proteins (pstSCAB-phoU). Later genes include those for the acquisition and utilization of alternate phosphorus sources, such as through the expression of alkaline phosphatase, the glycerol-3-phosphate transporter, or the phosphonate operon. The researchers demonstrated that this temporal expression pattern reflected the binding affinity of phospho-PhoB to the various Pho boxes throughout the chromosome by measuring apparent dissociation rates of phospho-PhoB from various promoter fragments using Bio-layer interferometry (73). They noted that the binding affinities correlated well with the information content of each promoter based upon the position weight matrix from conserved promoters. However, the binding affinities did not correlate with the levels of gene expression, and it is assumed that promoter architecture, the positioning of Pho boxes, and the presence of other regulatory factors may influence the levels of expression output.
The kinetics of the phosphate starvation response can also be influenced by previous activation of the Pho regulon. Hoffer et al. demonstrated that E. coli cells induced alkaline phosphatase faster if the cultures had recently been starved for phosphate (157). This type of memory may represent the carryover of PhoB/PhoR proteins from a previously induced state. This memory model suggests that cells should accumulate higher levels of Pho regulon proteins if they have previously been starved of phosphate. However, this is not observed (154). To explain this robust behavior for the Pho system, Gao and Stock again employed sophisticated modeling of response kinetics, with the assessment of promoter activities to show that there exists a counterbalancing negative regulation to bring about output homeostasis (154). This phenotypic memory seemed to be dependent upon the induction of σs, although the exact mechanism remains unknown.
Pi HOMEOSTASIS
The phenotypes of a ΔphoU mutation are instructive in regard to the importance of phosphate homeostasis. Steed and Wanner showed that ΔphoU mutants have a severe growth defect and accumulate compensatory mutations in genes that control the Pho regulon (120). It has been interpreted that these mutants may accumulate too much intracellular phosphate and that this is toxic to the cells, potentially due to magnesium starvation caused by excess polyphosphate accumulation in the cell (158). Therefore, it seems likely that E. coli has mechanisms not only to acquire and maintain intracellular Pi when it is limiting in the environment but also to ensure that Pi does not accumulate to toxic levels when Pi is in abundance. The target of excess intracellular Pi is not known, but too much intracellular Pi may feedback inhibit various enzymatic reactions, and this could affect nucleic acid biochemistry or central metabolism. Mechanisms to maintain intracellular Pi may include Pi export and Pi sequestration through the production of polyphosphate (polyP). Pi export may involve the PitA or PitB transporters, which are secondary transporters that translocate metal-phosphate substrates (159). Other proteins may also be involved in Pi export. For example, one interesting observation was that yjbB overexpression led to decreased polyP accumulation in a phoU mutant strain (160). This suggests that YjbB may function as a Pi exporter. The N-terminal half of yjbB consists of two repeats of an Na+/Pi cotransporter domain, and the C-terminal half consists of two PhoU-like domains.
PolyP may also influence expression of the Pho regulon during the stationary phase (161). Grillo-Puertas et al. showed that during the stationary phase PhoB could be activated when cells were grown in a medium containing high levels of phosphate (∼40 mM Pi) (161). It was demonstrated that this activation depended upon the production of acetyl phosphate and the production of polyP.
Pi Sensing and Stationary-Phase Response
Many of the genes that are upregulated in response to extended Pi limitation belong to the RpoS stimulon, or members of the general stress response, controlled by the alternate sigma factor σS. The Gottesman laboratory has beautifully demonstrated a mechanism for the regulation of entry into stationary phase when cells are starved of Pi (48, 49). When Pi becomes limiting, cells accumulate ppGpp in a SpoT-dependent manner (49). ppGpp is a second messenger that mediates the stringent response (162). ppGpp, with DksA, reprograms RNA polymerase to decrease transcription of many genes involved in amino acid biosynthetic pathways and for the synthesis of ribosomal proteins. This reprogrammed RNA polymerase also increases the synthesis of a variety of genes, one of which is iraP (163). IraP is an anti-adapter protein that interacts with RssB to prevent it from binding to σS to direct it to the ClpXP protease for degradation. The synthesis of IraP therefore leads to increased stability of σS, which leads to an induction of the general stress response.
It has also been suggested that a small regulatory RNA may also contribute to the induction of the general stress response following Pi limitation (164, 165). Ruiz and Silhavy showed that a Tn cam minitransposon mutation in the pstS gene, oriented so that transcription of its cam cassette was in the same direction as that of pstS, led to constitutive expression of the Pho regulon and resulted in increased levels of σS during exponential growth (165). They demonstrated that Hfq was required for this increase in σS and concluded that an uncharacterized small RNA controls translation of the rpoS message during Pi limitation. They also showed that this effect was dependent upon a functional PhoB protein. In a follow-up to these studies, Schurdell et al. provided genetic evidence that the processed 3′ end of the pstA message stimulates translation of rpoS (50). An unusual feature of the pstSCAB-phoU operon is an extended intergenic region between the pstA and pstB genes. Schurdell et al. created a series of deletions of the intergenic region between pstA and pstB and showed that mutants missing this sequence displayed a deficiency in the accumulation of σS and the induction of σS-dependent genes. It had previously been demonstrated that several small RNAs bind to the long untranslated leader of the rpoS mRNA in an Hfq-dependent manner to unmask the ribosome-binding site for rpoS and stimulate translation (166). It was proposed that the processed pstA message played the same role as the several small RNAs do to promote translation of the rpoS mRNA.
CONCLUSION
Control of the Pho regulon provides E. coli and Salmonella strains the ability to adapt to an ever-changing environment and integrate signals from a variety of inputs. While feasible models exist to describe the signaling pathway that monitors environmental Pi, additional work is still needed to more fully understand the molecular mechanisms of this pathway. It is anticipated that much progress will be made as signaling complexes are purified and studied in vitro. Perhaps more importantly, further research is needed to better understand how the phosphate signaling pathway is integrated into other aspects of cellular physiology.
Different bacteria use different mechanisms for adapting to changes in environmental phosphate (167). For example, while PhoU is essential for proper signaling in E. coli, there are many bacteria that do not have an obvious phoU gene in their genomes and there are other bacteria with multiple phoU genes in theirs (168, 169). Given a wide variety of available genes and gene arrangements, it is important to remember that not all bacteria use the same mechanisms for phosphate signaling as does E. coli. However, research about the E. coli Pho system provides a benchmark that can be used to help understand other systems. In addition to contributing to a better understanding of how cells adapt to changes in the availability of this essential nutrient, continued research of this system may also identify future targets for antimicrobial strategies against specific bacteria.
ACKNOWLEDGMENTS
This work was supported by The College of Life Sciences at Brigham Young University.
We thank Joel Griffitts, David Erickson, and Steve Fields for helpful suggestions in the preparation of the manuscript.
We offer our apologies to authors of related research who were not included in this review; the omission was unintentional.
Contributor Information
Stewart G. Gardner, Department of Biological Sciences, Emporia State University, Emporia, KS 66801
William R. McCleary, Microbiology and Molecular Biology Department, Brigham Young University, Provo, UT 84602
James M. Slauch, The School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL
REFERENCES
- 1.Westheimer FH. 1987. Why nature chose phosphates. Science 235:1173–1178. 10.1126/science.2434996. [PubMed] 10.1126/science.2434996 [DOI] [PubMed] [Google Scholar]
- 2.Kamerlin SC, Sharma PK, Prasad RB, Warshel A. 2013. Why nature really chose phosphate. Q Rev Biophys 46:1–132. 10.1017/S0033583512000157. [PubMed] 10.1017/S0033583512000157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin JP, McGrath JW, Quinn JP. 2016. Microbial transformations in phosphonate biosynthesis and catabolism, and their importance in nutrient cycling. Curr Opin Chem Biol 31:50–57. 10.1016/j.cbpa.2016.01.010. [PubMed] 10.1016/j.cbpa.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 4.Manav MC, Sofos N, Hove-Jensen B, Brodersen DE. 2018. The Abc of phosphonate breakdown: a mechanism for bacterial survival. Bioessays 40:e1800091. 10.1002/bies.201800091. [PubMed] 10.1002/bies.201800091 [DOI] [PubMed] [Google Scholar]
- 5.Metcalf WW, van der Donk WA. 2009. Biosynthesis of phosphonic and phosphinic acid natural products. Annu Rev Biochem 78:65–94. 10.1146/annurev.biochem.78.091707.100215. [PubMed] 10.1146/annurev.biochem.78.091707.100215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shulman RG, Brown TR, Ugurbil K, Ogawa S, Cohen SM, den Hollander JA. 1979. Cellular applications of 31P and 13C nuclear magnetic resonance. Science 205:160–166. 10.1126/science.36664. [PubMed] 10.1126/science.36664 [DOI] [PubMed] [Google Scholar]
- 7.Rao NN, Roberts MF, Torriani A, Yashphe J. 1993. Effect of glpT and glpD mutations on expression of the phoA gene in Escherichia coli. J Bacteriol 175:74–79. 10.1128/jb.175.1.74-79.1993. [PubMed] 10.1128/jb.175.1.74-79.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xavier KB, Kossmann M, Santos H, Boos W. 1995. Kinetic analysis by in vivo 31P nuclear magnetic resonance of internal Pi during the uptake of sn-glycerol-3-phosphate by the pho regulon-dependent Ugp system and the glp regulon-dependent GlpT system. J Bacteriol 177:699–704. 10.1128/jb.177.3.699-704.1995. [PubMed] 10.1128/jb.177.3.699-704.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blount ZD. 2015. The unexhausted potential of E. coli. eLife 4:e05826. 10.7554/eLife.05826. [PubMed] 10.7554/eLife.05826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko I, Tatsumi S, Segawa H, Miyamoto KI. 2017. Control of phosphate balance by the kidney and intestine. Clin Exp Nephrol 21(Suppl 1):21–26. 10.1007/s10157-016-1359-4. [PubMed] 10.1007/s10157-016-1359-4 [DOI] [PubMed] [Google Scholar]
- 11.Marks J, Debnam ES, Unwin RJ. 2013. The role of the gastrointestinal tract in phosphate homeostasis in health and chronic kidney disease. Curr Opin Nephrol Hypertens 22:481–487. 10.1097/MNH.0b013e3283621310. [PubMed] 10.1097/MNH.0b013e3283621310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torriani-Gorini A. 1996. History of the Pho system, p 291–294. In Lin ECC, Lynch AS (ed), Regulation of Gene Expression in Escherichia coli. RG Landes Company, Austin, TX. 10.1007/978-1-4684-8601-8_14. 10.1007/978-1-4684-8601-8_14 [DOI] [Google Scholar]
- 13.Torriani-Gorini A. 1987. The birth and growth of the Pho regulon, p 3–11. In Torriani-Gorini A, Rothman FG, Silver S, Wright A, Yagil E (ed), Phosphate Metabolism and Cellular Regulation in Microorganisms. ASM Press, Washington, DC. [Google Scholar]
- 14.Jacob F, Monod J. 1959. Genes of structure and genes of regulation in the biosynthesis of proteins. C R Hebd Seances Acad Sci 249:1282–1284. (In French.) [PubMed] [Google Scholar]
- 15.Horiuchi T, Horiuchi S, Mizuno D. 1959. A possible negative feedback phenomenon controlling formation of alkaline phosphomonoesterase in Escherichia coli. Nature 183:1529–1530. 10.1038/1831529b0. [PubMed] 10.1038/1831529b0 [DOI] [PubMed] [Google Scholar]
- 16.Echols H, Garen A, Garen S, Torriani A. 1961. Genetic control of repression of alkaline phosphatase in E. coli. J Mol Biol 3:425–438. 10.1016/S0022-2836(61)80055-7. [PubMed] 10.1016/S0022-2836(61)80055-7 [DOI] [PubMed] [Google Scholar]
- 17.Garen A, Echols H. 1962. Genetic control of induction of alkaline phosphatase synthesis in E. coli. Proc Natl Acad Sci U S A 48:1398–1402. 10.1073/pnas.48.8.1398. [PubMed] 10.1073/pnas.48.8.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garen A, Echols H. 1962. Properties of two regulating genes for alkaline phosphatase. J Bacteriol 83:297–300. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willsky GR, Bennett RL, Malamy MH. 1973. Inorganic phosphate transport in Escherichia coli: involvement of two genes which play a role in alkaline phosphatase regulation. J Bacteriol 113:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracha M, Yagil E. 1969. Genetic mapping of the phoR regulator gene of alkaline phosphatase in Escherichia coli. J Gen Microbiol 59:77–81. 10.1099/00221287-59-1-77. [PubMed] 10.1099/00221287-59-1-77 [DOI] [PubMed] [Google Scholar]
- 21.Shinagawa H, Makino K, Nakata A, Brenner S. 1983. Regulation of the pho regulon in Escherichia coli K-12. Genetic and physiological regulation of the positive regulatory gene phoB. J Mol Biol 168:477–488. 10.1016/S0022-2836(83)80297-6. 10.1016/S0022-2836(83)80297-6 [DOI] [PubMed] [Google Scholar]
- 22.Amemura M, Makino K, Shinagawa H, Kobayashi A, Nakata A. 1985. Nucleotide sequence of the genes involved in phosphate transport and regulation of the phosphate regulon in Escherichia coli. J Mol Biol 184:241–250. 10.1016/0022-2836(85)90377-8. 10.1016/0022-2836(85)90377-8 [DOI] [PubMed] [Google Scholar]
- 23.Wanner BL, Latterell P. 1980. Mutants affected in alkaline phosphatase, expression: evidence for multiple positive regulators of the phosphate regulon in Escherichia coli. Genetics 96:353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stock JB, Ninfa AJ, Stock AM. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev 53:450–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol 210:551–559. 10.1016/0022-2836(89)90131-9. 10.1016/0022-2836(89)90131-9 [DOI] [PubMed] [Google Scholar]
- 26.Makino K, Shinagawa H, Amemura M, Kimura S, Nakata A, Ishihama A. 1988. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J Mol Biol 203:85–95. 10.1016/0022-2836(88)90093-9. [PubMed] 10.1016/0022-2836(88)90093-9 [DOI] [PubMed] [Google Scholar]
- 27.Wanner BL. 1995. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by PhoR, CreC, and acetyl phosphate, p 203–221. In Hoch JA, Silhavy TJ (ed), Two-Component Signal Transduction. ASM Press, Washington, DC. [Google Scholar]
- 28.Bachhawat P, Swapna GV, Montelione GT, Stock AM. 2005. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure 13:1353–1363. 10.1016/j.str.2005.06.006. [PubMed] 10.1016/j.str.2005.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solá M, Gomis-Rüth FX, Serrano L, González A, Coll M. 1999. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol 285:675–687. 10.1006/jmbi.1998.2326. [PubMed] 10.1006/jmbi.1998.2326 [DOI] [PubMed] [Google Scholar]
- 30.Blanco AG, Sola M, Gomis-Rüth FX, Coll M. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701–713. 10.1016/S0969-2126(02)00761-X. 10.1016/S0969-2126(02)00761-X [DOI] [PubMed] [Google Scholar]
- 31.Medveczky N, Rosenberg H. 1970. The phosphate-binding protein of Escherichia coli. Biochim Biophys Acta 211:158–168. 10.1016/0005-2736(70)90090-8. 10.1016/0005-2736(70)90090-8 [DOI] [Google Scholar]
- 32.Tommassen J, Lugtenberg B. 1980. Outer membrane protein e of Escherichia coli K-12 is co-regulated with alkaline phosphatase. J Bacteriol 143:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H, Grussenmeyer T, Boos W. 1982. Mapping of two ugp genes coding for the pho regulon-dependent sn-glycerol-3-phosphate transport system of Escherichia coli. J Bacteriol 150:1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maas WK, Clark AJ. 1964. Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli. II. Dominance of repressibility in diploids. J Mol Biol 8:365–370. 10.1016/S0022-2836(64)80200-X. [PubMed] 10.1016/S0022-2836(64)80200-X [DOI] [PubMed] [Google Scholar]
- 35.Wanner BL, McSharry R. 1982. Phosphate-controlled gene expression in Escherichia coli K12 using Mudl-directed lacZ fusions. J Mol Biol 158:347–363. 10.1016/0022-2836(82)90202-9. [PubMed] 10.1016/0022-2836(82)90202-9 [DOI] [PubMed] [Google Scholar]
- 36.Wanner BL. 1996. Phosphorous assimilation and control of the phosphate regulon, p 1357–1381. In Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 37.VanBogelen RA, Hutton ME, Neidhardt FC. 1990. Gene-protein database of Escherichia coli K-12: edition 3. Electrophoresis 11:1131–1166. [PubMed] 10.1002/elps.1150111205 [DOI] [PubMed] [Google Scholar]
- 38.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J Bacteriol 178:4344–4366. 10.1128/jb.178.15.4344-4366.1996. [PubMed] 10.1128/jb.178.15.4344-4366.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C-M, Ye QZ, Zhu ZM, Wanner BL, Walsh CT. 1990. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn ( psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem 265:4461–4471. [PubMed] [PubMed] [Google Scholar]
- 40.Wanner BL, Boline JA. 1990. Mapping and molecular cloning of the phn ( psiD) locus for phosphonate utilization in Escherichia coli. J Bacteriol 172:1186–1196. 10.1128/jb.172.3.1186-1196.1990. [PubMed] 10.1128/jb.172.3.1186-1196.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baek JH, Lee SY. 2006. Novel gene members in the Pho regulon of Escherichia coli. FEMS Microbiol Lett 264:104–109. 10.1111/j.1574-6968.2006.00440.x. [PubMed] 10.1111/j.1574-6968.2006.00440.x [DOI] [PubMed] [Google Scholar]
- 42.Baek JH, Lee SY. 2007. Transcriptome analysis of phosphate starvation response in Escherichia coli. J Microbiol Biotechnol 17:244–252. [PubMed] [Google Scholar]
- 43.Crépin S, Chekabab SM, Le Bihan G, Bertrand N, Dozois CM, Harel J. 2011. The Pho regulon and the pathogenesis of Escherichia coli. Vet Microbiol 153:82–88. 10.1016/j.vetmic.2011.05.043. [PubMed] 10.1016/j.vetmic.2011.05.043 [DOI] [PubMed] [Google Scholar]
- 44.Yang C, Huang TW, Wen SY, Chang CY, Tsai SF, Wu WF, Chang CH. 2012. Genome-wide PhoB binding and gene expression profiles reveal the hierarchical gene regulatory network of phosphate starvation in Escherichia coli. PLoS One 7:e47314. 10.1371/journal.pone.0047314. [PubMed] 10.1371/journal.pone.0047314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang J, Yu K, Qi L, Liu Y, Cheng S, Wu M, Wang Z, Fu J, Liu X. 2018. A proteomic view of Salmonella Typhimurium in response to phosphate limitation. Proteomes 6:E19. 10.3390/proteomes6020019. [PubMed] 10.3390/proteomes6020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spira B, Yagil E. 1998. The relation between ppGpp and the PHO regulon in Escherichia coli. Mol Gen Genet 257:469–477. 10.1007/s004380050671. [PubMed] 10.1007/s004380050671 [DOI] [PubMed] [Google Scholar]
- 47.Spira B, Silberstein N, Yagil E. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol 177:4053–4058. 10.1128/jb.177.14.4053-4058.1995. [PubMed] 10.1128/jb.177.14.4053-4058.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bougdour A, Wickner S, Gottesman S. 2006. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev 20:884–897. 10.1101/gad.1400306. [PubMed] 10.1101/gad.1400306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bougdour A, Gottesman S. 2007. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci U S A 104:12896–12901. 10.1073/pnas.0705561104. [PubMed] 10.1073/pnas.0705561104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schurdell MS, Woodbury GM, McCleary WR. 2007. Genetic evidence suggests that the intergenic region between pstA and pstB plays a role in the regulation of rpoS translation during phosphate limitation. J Bacteriol 189:1150–1153. 10.1128/JB.01482-06. [PubMed] 10.1128/JB.01482-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer H, Boos W. 1985. Regulation of ugp, the sn-glycerol-3-phosphate transport system of Escherichia coli K-12 that is part of the pho regulon. J Bacteriol 163:392–394. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wuttge S, Bommer M, Jäger F, Martins BM, Jacob S, Licht A, Scheffel F, Dobbek H, Schneider E. 2012. Determinants of substrate specificity and biochemical properties of the sn-glycerol-3-phosphate ATP binding cassette transporter (UgpB-AEC2) of Escherichia coli. Mol Microbiol 86:908–920. 10.1111/mmi.12025. [PubMed] 10.1111/mmi.12025 [DOI] [PubMed] [Google Scholar]
- 53.Metcalf WW, Wanner BL. 1991. Involvement of the Escherichia coli phn ( psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J Bacteriol 173:587–600. 10.1128/jb.173.2.587-600.1991. [PubMed] 10.1128/jb.173.2.587-600.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metcalf WW, Wanner BL. 1993. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129:27–32. 10.1016/0378-1119(93)90692-V. [PubMed] 10.1016/0378-1119(93)90692-V [DOI] [PubMed] [Google Scholar]
- 55.Yoshida Y, Sugiyama S, Oyamada T, Yokoyama K, Kim SK, Makino K. 2011. Identification of PhoB binding sites of the yibD and ytfK promoter regions in Escherichia coli. J Microbiol 49:285–289. 10.1007/s12275-011-0360-6. [PubMed] 10.1007/s12275-011-0360-6 [DOI] [PubMed] [Google Scholar]
- 56.Klein G, Müller-Loennies S, Lindner B, Kobylak N, Brade H, Raina S. 2013. Molecular and structural basis of inner core lipopolysaccharide alterations in Escherichia coli: incorporation of glucuronic acid and phosphoethanolamine in the heptose region. J Biol Chem 288:8111–8127. 10.1074/jbc.M112.445981. [PubMed] 10.1074/jbc.M112.445981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iwadate Y, Kato JI. 2017. Involvement of the ytfK gene from the PhoB regulon in stationary-phase H2O2 stress tolerance in Escherichia coli. Microbiology 163:1912–1923. 10.1099/mic.0.000534. [PubMed] 10.1099/mic.0.000534 [DOI] [PubMed] [Google Scholar]
- 58.Kim S-K, Makino K, Amemura M, Shinagawa H, Nakata A. 1993. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J Bacteriol 175:1316–1324. 10.1128/jb.175.5.1316-1324.1993. [PubMed] 10.1128/jb.175.5.1316-1324.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koonin EV, Rudd KE. 1996. Two domains of superfamily I helicases may exist as separate proteins. Protein Sci 5:178–180. 10.1002/pro.5560050124. [PubMed] 10.1002/pro.5560050124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rapp M, Drew D, Daley DO, Nilsson J, Carvalho T, Melén K, De Gier JW, Von Heijne G. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci 13:937–945. 10.1110/ps.03553804. [PubMed] 10.1110/ps.03553804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metcalf WW, Steed PM, Wanner BL. 1990. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi:lacZ(Mu d1) transcriptional fusions. J Bacteriol 172:3191–3200. 10.1128/jb.172.6.3191-3200.1990. [PubMed] 10.1128/jb.172.6.3191-3200.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray EL, Conway T. 2005. Multiple regulators control expression of the Entner-Doudoroff aldolase (Eda) of Escherichia coli. J Bacteriol 187:991–1000. 10.1128/JB.187.3.991-1000.2005. [PubMed] 10.1128/JB.187.3.991-1000.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida Y, Sugiyama S, Oyamada T, Yokoyama K, Makino K. 2010. Identification and characterization of novel phosphate regulon genes, ecs0540-ecs0544, in Escherichia coli O157:H7. Mol Genet Genomics 284:197–205. 10.1007/s00438-010-0559-y. [PubMed] 10.1007/s00438-010-0559-y [DOI] [PubMed] [Google Scholar]
- 64.Yoshida Y, Sugiyama S, Oyamada T, Yokoyama K, Makino K. 2012. Novel members of the phosphate regulon in Escherichia coli O157:H7 identified using a whole-genome shotgun approach. Gene 502:27–35. 10.1016/j.gene.2012.03.064. [PubMed] 10.1016/j.gene.2012.03.064 [DOI] [PubMed] [Google Scholar]
- 65.Chekabab SM, Jubelin G, Dozois CM, Harel J. 2014. PhoB activates Escherichia coli O157:H7 virulence factors in response to inorganic phosphate limitation. PLoS One 9:e94285. 10.1371/journal.pone.0094285. [PubMed] 10.1371/journal.pone.0094285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tree JJ, Wolfson EB, Wang D, Roe AJ, Gally DL. 2009. Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli. Trends Microbiol 17:361–370. 10.1016/j.tim.2009.06.001. [PubMed] 10.1016/j.tim.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 67.Lamarche MG, Dozois CM, Daigle F, Caza M, Curtiss R, III, Dubreuil JD, Harel J. 2005. Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect Immun 73:4138–4145. 10.1128/IAI.73.7.4138-4145.2005. [PubMed] 10.1128/IAI.73.7.4138-4145.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crépin S, Lamarche MG, Garneau P, Séguin J, Proulx J, Dozois CM, Harel J. 2008. Genome-wide transcriptional response of an avian pathogenic Escherichia coli (APEC) pst mutant. BMC Genomics 9:568. 10.1186/1471-2164-9-568. [PubMed] 10.1186/1471-2164-9-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamarche MG, Kim SH, Crépin S, Mourez M, Bertrand N, Bishop RE, Dubreuil JD, Harel J. 2008. Modulation of hexa-acyl pyrophosphate lipid A population under Escherichia coli phosphate (Pho) regulon activation. J Bacteriol 190:5256–5264. 10.1128/JB.01536-07. [PubMed] 10.1128/JB.01536-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lamarche MG, Wanner BL, Crépin S, Harel J. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev 32:461–473. 10.1111/j.1574-6976.2008.00101.x. [PubMed] 10.1111/j.1574-6976.2008.00101.x [DOI] [PubMed] [Google Scholar]
- 71.Crépin S, Porcheron G, Houle S, Harel J, Dozois CM. 2017. Altered regulation of the diguanylate cyclase YaiC reduces production of type 1 fimbriae in a pst mutant of uropathogenic Escherichia coli CFT073. J Bacteriol 199:e00168-17. 10.1128/JB.00168-17. [PubMed] 10.1128/JB.00168-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crépin S, Houle S, Charbonneau ME, Mourez M, Harel J, Dozois CM. 2012. Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infection. Infect Immun 80:2802–2815. 10.1128/IAI.00162-12. [PubMed] 10.1128/IAI.00162-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao R, Stock AM. 2015. Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics. mBio 6:e00686-15. 10.1128/mBio.00686-15. [PubMed] 10.1128/mBio.00686-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao R, Stock AM. 2017. Quantitative kinetic analyses of shutting off a two-component system. mBio 8:e00412-17. 10.1128/mBio.00412-17. [PubMed] 10.1128/mBio.00412-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siryaporn A, Goulian M. 2010. Characterizing cross-talk in vivo avoiding pitfalls and overinterpretation. Methods Enzymol 471:1–16. 10.1016/S0076-6879(10)71001-6. [PubMed] 10.1016/S0076-6879(10)71001-6 [DOI] [PubMed] [Google Scholar]
- 76.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70:494–506. 10.1111/j.1365-2958.2008.06426.x. [PubMed] 10.1111/j.1365-2958.2008.06426.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao R, Stock AM. 2013. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc Natl Acad Sci U S A 110:672–677. 10.1073/pnas.1214587110. [PubMed] 10.1073/pnas.1214587110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao R, Stock AM. 2018. Quantitative analysis of intracellular response regulator phosphatase activity of histidine kinases. Methods Enzymol 607:301–319. 10.1016/bs.mie.2018.04.004. [PubMed] 10.1016/bs.mie.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shinagawa H, Makino K, Yamada M, Amemura M, Sato T, Nakata A. 1994. Signal transduction in the phosphate regulon of Escherichia coli: dual functions of PhoR as a protein kinase and a protein phosphatase, p 285–289. In Torriani-Gorini A, Yagil E, Silver S (ed), Phosphate in Microorganisms: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC. [Google Scholar]
- 80.Carmany DO, Hollingsworth K, McCleary WR. 2003. Genetic and biochemical studies of phosphatase activity of PhoR. J Bacteriol 185:1112–1115. 10.1128/JB.185.3.1112-1115.2003. [PubMed] 10.1128/JB.185.3.1112-1115.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. 10.1146/annurev.biochem.69.1.183. [PubMed] 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- 82.Etzkorn M, Kneuper H, Dünnwald P, Vijayan V, Krämer J, Griesinger C, Becker S, Unden G, Baldus M. 2008. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat Struct Mol Biol 15:1031–1039. 10.1038/nsmb.1493. [PubMed] 10.1038/nsmb.1493 [DOI] [PubMed] [Google Scholar]
- 83.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Möglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294. 10.1016/j.str.2009.08.011. [PubMed] 10.1016/j.str.2009.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci U S A 95:15177–15182. 10.1073/pnas.95.26.15177. [PubMed] 10.1073/pnas.95.26.15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. 10.1146/annurev.micro.091208.073214. [PubMed] 10.1146/annurev.micro.091208.073214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ashenberg O, Keating AE, Laub MT. 2013. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J Mol Biol 425:1198–1209. 10.1016/j.jmb.2013.01.011. [PubMed] 10.1016/j.jmb.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Willett JW, Kirby JR. 2012. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet 8:e1003084. 10.1371/journal.pgen.1003084. [PubMed] 10.1371/journal.pgen.1003084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zschiedrich CP, Keidel V, Szurmant H. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. 10.1016/j.jmb.2016.08.003. [PubMed] 10.1016/j.jmb.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang C, Sang J, Wang J, Su M, Downey JS, Wu Q, Wang S, Cai Y, Xu X, Wu J, Senadheera DB, Cvitkovitch DG, Chen L, Goodman SD, Han A. 2013. Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. PLoS Biol 11:e1001493. 10.1371/journal.pbio.1001493. [PubMed] 10.1371/journal.pbio.1001493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marina A, Waldburger CD, Hendrickson WA. 2005. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J 24:4247–4259. 10.1038/sj.emboj.7600886. [PubMed] 10.1038/sj.emboj.7600886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhate MP, Molnar KS, Goulian M, DeGrado WF. 2015. Signal transduction in histidine kinases: insights from new structures. Structure 23:981–994. 10.1016/j.str.2015.04.002. [PubMed] 10.1016/j.str.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCleary WR. 1996. The activation of PhoB by acetylphosphate. Mol Microbiol 20:1155–1163. 10.1111/j.1365-2958.1996.tb02636.x. [PubMed] 10.1111/j.1365-2958.1996.tb02636.x [DOI] [PubMed] [Google Scholar]
- 94.McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem 269:31567–31572. [PubMed] [Google Scholar]
- 95.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44(D1):D279–D285. 10.1093/nar/gkv1344. [PubMed] 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13:150–159. 10.1016/j.mib.2010.01.005. [PubMed] 10.1016/j.mib.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blanco AG, Canals A, Coll M. 2012. PhoB transcriptional activator binds hierarchically to pho box promoters. Biol Chem 393:1165–1171. 10.1515/hsz-2012-0230. [PubMed] 10.1515/hsz-2012-0230 [DOI] [PubMed] [Google Scholar]
- 98.Canals A, Blanco AG, Coll M. 2012. σ70 and PhoB activator: getting a better grip. Transcription 3:160–164. 10.4161/trns.20444. [PubMed] 10.4161/trns.20444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kou X, Liu Y, Li C, Liu M, Jiang L. 2018. Dimerization and conformational exchanges of the receiver domain of response regulator PhoB from Escherichia coli. J Phys Chem B 122:5749–5757. 10.1021/acs.jpcb.8b01034. [PubMed] 10.1021/acs.jpcb.8b01034 [DOI] [PubMed] [Google Scholar]
- 100.Creager-Allen RL, Silversmith RE, Bourret RB. 2013. A link between dimerization and autophosphorylation of the response regulator PhoB. J Biol Chem 288:21755–21769. 10.1074/jbc.M113.471763. [PubMed] 10.1074/jbc.M113.471763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buckler DR, Zhou Y, Stock AM. 2002. Evidence of intradomain and interdomain flexibility in an OmpR/PhoB homolog from Thermotoga maritima. Structure 10:153–164. 10.1016/S0969-2126(01)00706-7. 10.1016/S0969-2126(01)00706-7 [DOI] [PubMed] [Google Scholar]
- 102.Blanco AG, Canals A, Bernués J, Solà M, Coll M. 2011. The structure of a transcription activation subcomplex reveals how σ(70) is recruited to PhoB promoters. EMBO J 30:3776–3785. 10.1038/emboj.2011.271. [PubMed] 10.1038/emboj.2011.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Willsky GR, Malamy MH. 1980. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol 144:356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saier MH, Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The transporter classification database (TCDB): recent advances. Nucleic Acids Res 44(D1):D372–D379. 10.1093/nar/gkv1103. [PubMed] 10.1093/nar/gkv1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. 10.1128/MMBR.00031-07. [PubMed] 10.1128/MMBR.00031-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Quiocho FA, Ledvina PS. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol 20:17–25. 10.1111/j.1365-2958.1996.tb02484.x. [PubMed] 10.1111/j.1365-2958.1996.tb02484.x [DOI] [PubMed] [Google Scholar]
- 107.Luecke H, Quiocho FA. 1990. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature 347:402–406. 10.1038/347402a0. [PubMed] 10.1038/347402a0 [DOI] [PubMed] [Google Scholar]
- 108.Wang Z, Choudhary A, Ledvina PS, Quiocho FA. 1994. Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies. J Biol Chem 269:25091–25094. 10.2210/pdb1pbp/pdb [DOI] [PubMed] [Google Scholar]
- 109.Qi R, Jing Z, Liu C, Piquemal JP, Dalby KN, Ren P. 2018. Elucidating the phosphate binding mode of phosphate-binding protein: the critical effect of buffer solution. J Phys Chem B 122:6371–6376. 10.1021/acs.jpcb.8b03194. [PubMed] 10.1021/acs.jpcb.8b03194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sippel KH, Bacik J, Quiocho FA, Fisher SZ. 2014. Preliminary time-of-flight neutron diffraction studies of Escherichia coli ABC transport receptor phosphate-binding protein at the Protein Crystallography Station. Acta Crystallogr F Struct Biol Commun 70:819–822. 10.1107/S2053230X14009704. [PubMed] 10.1107/S2053230X14009704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cox GB, Webb D, Godovac-Zimmermann J, Rosenberg H. 1988. Arg-220 of the PstA protein is required for phosphate transport through the phosphate-specific transport system in Escherichia coli but not for alkaline phosphatase repression. J Bacteriol 170:2283–2286. 10.1128/jb.170.5.2283-2286.1988. [PubMed] 10.1128/jb.170.5.2283-2286.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cox GB, Webb D, Rosenberg H. 1989. Specific amino acid residues in both the PstB and PstC proteins are required for phosphate transport by the Escherichia coli Pst system. J Bacteriol 171:1531–1534. 10.1128/jb.171.3.1531-1534.1989. [PubMed] 10.1128/jb.171.3.1531-1534.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rees DC, Johnson E, Lewinson O. 2009. ABC transporters: the power to change. Nat Rev Mol Cell Biol 10:218–227. 10.1038/nrm2646. [PubMed] 10.1038/nrm2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chan FY, Torriani A. 1996. PstB protein of the phosphate-specific transport system of Escherichia coli is an ATPase. J Bacteriol 178:3974–3977. 10.1128/jb.178.13.3974-3977.1996. [PubMed] 10.1128/jb.178.13.3974-3977.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beis K. 2015. Structural basis for the mechanism of ABC transporters. Biochem Soc Trans 43:889–893. 10.1042/BST20150047. [PubMed] 10.1042/BST20150047 [DOI] [PubMed] [Google Scholar]
- 116.Hollenstein K, Frei DC, Locher KP. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446:213–216. 10.1038/nature05626. [PubMed] 10.1038/nature05626 [DOI] [PubMed] [Google Scholar]
- 117.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. 10.1016/j.mib.2010.01.014. [PubMed] 10.1016/j.mib.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kimura S, Makino K, Shinagawa H, Amemura M, Nakata A. 1989. Regulation of the phosphate regulon of Escherichia coli: characterization of the promoter of the pstS gene. Mol Gen Genet 215:374–380. 10.1007/BF00427032. [PubMed] 10.1007/BF00427032 [DOI] [PubMed] [Google Scholar]
- 119.Spira B, Aguena M, de Castro Oliveira JV, Yagil E. 2010. Alternative promoters in the pst operon of Escherichia coli. Mol Genet Genomics 284:489–498. 10.1007/s00438-010-0584-x. [PubMed] 10.1007/s00438-010-0584-x [DOI] [PubMed] [Google Scholar]
- 120.Steed PM, Wanner BL. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol 175:6797–6809. 10.1128/jb.175.21.6797-6809.1993. [PubMed] 10.1128/jb.175.21.6797-6809.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haldimann A, Daniels LL, Wanner BL. 1998. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol 180:1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Surin BP, Rosenberg H, Cox GB. 1985. Phosphate-specific transport system of Escherichia coli: nucleotide sequence and gene-polypeptide relationships. J Bacteriol 161:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rice CD, Pollard JE, Lewis ZT, McCleary WR. 2009. Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl Environ Microbiol 75:573–582. 10.1128/AEM.01046-08. [PubMed] 10.1128/AEM.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. 2008. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321:250–253. 10.1126/science.1157987. [PubMed] 10.1126/science.1157987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson E, Nguyen PT, Yeates TO, Rees DC. 2012. Inward facing conformations of the MetNI methionine ABC transporter: implications for the mechanism of transinhibition. Protein Sci 21:84–96. 10.1002/pro.765. [PubMed] 10.1002/pro.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. 2008. Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 321:246–250. 10.1126/science.1156213. [PubMed] 10.1126/science.1156213 [DOI] [PubMed] [Google Scholar]
- 127.Chen S, Oldham ML, Davidson AL, Chen J. 2013. Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature 499:364–368. 10.1038/nature12232. [PubMed] 10.1038/nature12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang JG, Rees DC. 2015. The allosteric regulatory mechanism of the Escherichia coli MetNI methionine ATP binding cassette (ABC) transporter. J Biol Chem 290:9135–9140. 10.1074/jbc.M114.603365. [PubMed] 10.1074/jbc.M114.603365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu J, Lou Y, Yokota H, Adams PD, Kim R, Kim SH. 2005. Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J Biol Chem 280:15960–15966. 10.1074/jbc.M414117200. [PubMed] 10.1074/jbc.M414117200 [DOI] [PubMed] [Google Scholar]
- 130.Oganesyan V, Oganesyan N, Adams PD, Jancarik J, Yokota HA, Kim R, Kim SH. 2005. Crystal structure of the “PhoU-like” phosphate uptake regulator from Aquifex aeolicus. J Bacteriol 187:4238–4244. 10.1128/JB.187.12.4238-4244.2005. [PubMed] 10.1128/JB.187.12.4238-4244.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee SJ, Park YS, Kim SJ, Lee BJ, Suh SW. 2014. Crystal structure of PhoU from Pseudomonas aeruginosa, a negative regulator of the Pho regulon. J Struct Biol 188:22–29. 10.1016/j.jsb.2014.08.010. [PubMed] 10.1016/j.jsb.2014.08.010 [DOI] [PubMed] [Google Scholar]
- 132.Gardner SG, Johns KD, Tanner R, McCleary WR. 2014. The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J Bacteriol 196:1741–1752. 10.1128/JB.00029-14. [PubMed] 10.1128/JB.00029-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoffer SM, Tommassen J. 2001. The phosphate-binding protein of Escherichia coli is not essential for P(i)-regulated expression of the pho regulon. J Bacteriol 183:5768–5771. 10.1128/JB.183.19.5768-5771.2001. [PubMed] 10.1128/JB.183.19.5768-5771.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rao NN, Wang E, Yashphe J, Torriani A. 1986. Nucleotide pool in pho regulon mutants and alkaline phosphatase synthesis in Escherichia coli. J Bacteriol 166:205–211. 10.1128/jb.166.1.205-211.1986. [PubMed] 10.1128/jb.166.1.205-211.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Peterson CN, Mandel MJ, Silhavy TJ. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J Bacteriol 187:7549–7553. 10.1128/JB.187.22.7549-7553.2005. [PubMed] 10.1128/JB.187.22.7549-7553.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wanner BL. 1996. Signal transduction in the control of phosphate-regulated genes of Escherichia coli. Kidney Int 49:964–967. 10.1038/ki.1996.136. [PubMed] 10.1038/ki.1996.136 [DOI] [PubMed] [Google Scholar]
- 137.Rosenberg H, Gerdes RG, Chegwidden K. 1977. Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol 131:505–511. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zuckier G, Torriani A. 1981. Genetic and physiological tests of three phosphate-specific transport mutants of Escherichia coli. J Bacteriol 145:1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Muda M, Rao NN, Torriani A. 1992. Role of PhoU in phosphate transport and alkaline phosphatase regulation. J Bacteriol 174:8057–8064. 10.1128/jb.174.24.8057-8064.1992. [PubMed] 10.1128/jb.174.24.8057-8064.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gardner SG, Miller JB, Dean T, Robinson T, Erickson M, Ridge PG, McCleary WR. 2015. Genetic analysis, structural modeling, and direct coupling analysis suggest a mechanism for phosphate signaling in Escherichia coli. BMC Genet 16(Suppl 2):S2. 10.1186/1471-2156-16-S2-S2. [PubMed] 10.1186/1471-2156-16-S2-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. 2017. The ClusPro web server for protein-protein docking. Nat Protoc 12:255–278. 10.1038/nprot.2016.169. [PubMed] 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vuppada RK, Hansen CR, Strickland KAP, Kelly KM, McCleary WR. 2018. Phosphate signaling through alternate conformations of the PstSCAB phosphate transporter. BMC Microbiol 18:8. 10.1186/s12866-017-1126-z. [PubMed] 10.1186/s12866-017-1126-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Daus ML, Grote M, Müller P, Doebber M, Herrmann A, Steinhoff HJ, Dassa E, Schneider E. 2007. ATP-driven MalK dimer closure and reopening and conformational changes of the “EAA” motifs are crucial for function of the maltose ATP-binding cassette transporter (MalFGK2). J Biol Chem 282:22387–22396. 10.1074/jbc.M701979200. [PubMed] 10.1074/jbc.M701979200 [DOI] [PubMed] [Google Scholar]
- 144.Pontes MH, Groisman EA. 2018. Protein synthesis controls phosphate homeostasis. Genes Dev 32:79–92. 10.1101/gad.309245.117. [PubMed] 10.1101/gad.309245.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lüttmann D, Göpel Y, Görke B. 2012. The phosphotransferase protein EIIA(Ntr) modulates the phosphate starvation response through interaction with histidine kinase PhoR in Escherichia coli. Mol Microbiol 86:96–110. 10.1111/j.1365-2958.2012.08176.x. [PubMed] 10.1111/j.1365-2958.2012.08176.x [DOI] [PubMed] [Google Scholar]
- 146.Lee CR, Cho SH, Yoon MJ, Peterkofsky A, Seok YJ. 2007. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc Natl Acad Sci U S A 104:4124–4129. 10.1073/pnas.0609897104. [PubMed] 10.1073/pnas.0609897104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lee CR, Cho SH, Kim HJ, Kim M, Peterkofsky A, Seok YJ. 2010. Potassium mediates Escherichia coli enzyme IIA(Ntr)-dependent regulation of sigma factor selectivity. Mol Microbiol 78:1468–1483. 10.1111/j.1365-2958.2010.07419.x. [PubMed] 10.1111/j.1365-2958.2010.07419.x [DOI] [PubMed] [Google Scholar]
- 148.Ronneau S, Petit K, De Bolle X, Hallez R. 2016. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun 7:11423. 10.1038/ncomms11423. [PubMed] 10.1038/ncomms11423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoo W, Kim D, Yoon H, Ryu S. 2017. Enzyme IIA(Ntr) regulates Salmonella invasion via 1,2-propanediol and propionate catabolism. Sci Rep 7:44827. 10.1038/srep44827. [PubMed] 10.1038/srep44827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Muriel-Millán LF, Moreno S, Gallegos-Monterrosa R, Espín G. 2017. Unphosphorylated EIIANtr induces ClpAP-mediated degradation of RpoS in Azotobacter vinelandii. Mol Microbiol 104:197–211. 10.1111/mmi.13621. [PubMed] 10.1111/mmi.13621 [DOI] [PubMed] [Google Scholar]
- 151.Choi J, Shin D, Yoon H, Kim J, Lee CR, Kim M, Seok YJ, Ryu S. 2010. Salmonella pathogenicity island 2 expression negatively controlled by EIIANtr-SsrB interaction is required for Salmonella virulence. Proc Natl Acad Sci U S A 107:20506–20511. 10.1073/pnas.1000759107. [PubMed] 10.1073/pnas.1000759107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J Bacteriol 192:3055–3067. 10.1128/JB.00213-10. [PubMed] 10.1128/JB.00213-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gao R, Stock AM. 2013. Evolutionary tuning of protein expression levels of a positively autoregulated two-component system. PLoS Genet 9:e1003927. 10.1371/journal.pgen.1003927. [PubMed] 10.1371/journal.pgen.1003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gao R, Godfrey KA, Sufian MA, Stock AM. 2017. Counterbalancing regulation in response memory of a positively autoregulated two-component system. J Bacteriol 199:e00390-17. 10.1128/JB.00390-17. [PubMed] 10.1128/JB.00390-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gao R, Stock AM. 2018. Overcoming the cost of positive autoregulation by accelerating the response with a coupled negative feedback. Cell Rep 24:3061–3071.e6. 10.1016/j.celrep.2018.08.023. [PubMed] 10.1016/j.celrep.2018.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. 10.1146/annurev-micro-092611-150039. [PubMed] 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hoffer SM, Westerhoff HV, Hellingwerf KJ, Postma PW, Tommassen J. 2001. Autoamplification of a two-component regulatory system results in “learning” behavior. J Bacteriol 183:4914–4917. 10.1128/JB.183.16.4914-4917.2001. [PubMed] 10.1128/JB.183.16.4914-4917.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rudat AK, Pokhrel A, Green TJ, Gray MJ. 2018. Mutations in Escherichia coli polyphosphate kinase that lead to dramatically increased in vivo polyphosphate levels. J Bacteriol 200:e00697-17. 10.1128/JB.00697-17. [PubMed] 10.1128/JB.00697-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.van Veen HW, Abee T, Kortstee GJ, Konings WN, Zehnder AJ. 1994. Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry 33:1766–1770. 10.1021/bi00173a020. [PubMed] 10.1021/bi00173a020 [DOI] [PubMed] [Google Scholar]
- 160.Motomura K, Hirota R, Ohnaka N, Okada M, Ikeda T, Morohoshi T, Ohtake H, Kuroda A. 2011. Overproduction of YjbB reduces the level of polyphosphate in Escherichia coli: a hypothetical role of YjbB in phosphate export and polyphosphate accumulation. FEMS Microbiol Lett 320:25–32. 10.1111/j.1574-6968.2011.02285.x. [PubMed] 10.1111/j.1574-6968.2011.02285.x [DOI] [PubMed] [Google Scholar]
- 161.Grillo-Puertas M, Rintoul MR, Rapisarda VA. 2016. PhoB activation in non-limiting phosphate condition by the maintenance of high polyphosphate levels in the stationary phase inhibits biofilm formation in Escherichia coli. Microbiology 162:1000–1008. 10.1099/mic.0.000281. [PubMed] 10.1099/mic.0.000281 [DOI] [PubMed] [Google Scholar]
- 162.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. 10.1146/annurev.micro.62.081307.162903. [PubMed] 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- 163.Girard ME, Gopalkrishnan S, Grace ED, Halliday JA, Gourse RL, Herman C. 2017. DksA and ppGpp regulate the sigma(S) stress response by activating promoters for the small RNA DsrA and the anti-adapter protein IraP. J Bacteriol 200:e00463-17. 10.1128/JB.00463-17. [PubMed] 10.1128/JB.00463-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Peterson CN, Ruiz N, Silhavy TJ. 2004. RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J Bacteriol 186:7403–7410. 10.1128/JB.186.21.7403-7410.2004. [PubMed] 10.1128/JB.186.21.7403-7410.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ruiz N, Silhavy TJ. 2003. Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J Bacteriol 185:5984–5992. 10.1128/JB.185.20.5984-5992.2003. [PubMed] 10.1128/JB.185.20.5984-5992.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11:494–507. 10.4161/rna.28867. [PubMed] 10.4161/rna.28867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Santos-Beneit F. 2015. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol 6:402. 10.3389/fmicb.2015.00402. [PubMed] 10.3389/fmicb.2015.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brokaw AM, Eide BJ, Muradian M, Boster JM, Tischler AD. 2017. Mycobacterium smegmatis PhoU proteins have overlapping functions in phosphate signaling and are essential. Front Microbiol 8:2523. 10.3389/fmicb.2017.02523. [PubMed] 10.3389/fmicb.2017.02523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang X, Han H, Lv Z, Lin Z, Shang Y, Xu T, Wu Y, Zhang Y, Qu D. 2017. PhoU2 but not PhoU1 as an important regulator of biofilm formation and tolerance to multiple stresses by participating in various fundamental metabolic processes in Staphylococcus epidermidis. J Bacteriol 199:e00219-17. 10.1128/JB.00219-17. [PubMed] 10.1128/JB.00219-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. 10.1186/gb-2010-11-10-r106. [PubMed] 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]


