Abstract
Lipoproteins are produced by both Gram-positive and Gram-negative bacteria. Once secreted, lipoproteins are quickly acylated, anchoring them into the plasma membrane. Recent work has shown that Gram-positive bacteria are able to generate considerable diversity in the acylation of their lipoproteins, though the mechanisms involved are only just beginning to emerge. In Gram-negative organisms, most lipoproteins are subsequently trafficked to the outer membrane (OM). Lipoprotein trafficking is an essential pathway in these bacteria. At least one OM lipoprotein component is required by each of the essential machines that assemble the OM (such as the Bam and Lpt machines) and build the peptidoglycan cell wall (Lpo-penicillin-binding protein complexes). The Lol pathway has been the paradigm for OM lipoprotein trafficking: a complex of LolCDE extracts lipoproteins from the plasma membrane, LolA shuttles them through the periplasmic space, and LolB anchors them into the OM. The peptide signals responsible for OM-targeting via LolCDE have long been known for Escherichia coli. Remarkably, production of novel lipoprotein acyl forms in E. coli has reinforced the idea that lipid signals also contribute to OM targeting via LolCDE. Moreover, recent work has shown that lipoprotein trafficking can occur in E. coli without either LolA or LolB. Therefore, current evidence suggests that at least one additional, LolAB-independent route for OM lipoprotein trafficking exists. This chapter reviews the posttranslocation modifications of all lipoproteins, with a focus on the trafficking of lipoproteins to the OM of Gram-negative bacteria.
Lipoproteins are a family of secreted proteins that are acylated after their translocation across the plasma membrane (1–3). Acylation spatially confines lipoproteins by anchoring them into membranes. Lipoproteins are bioinformatically identifiable by the highly conserved lipobox motif in their short signal peptides (4). Within the lipobox is a cleavage site for signal peptidase II (SPII; Lsp). Immediately adjacent is an invariant Cys residue which is the target of acylation reactions. Most lipoproteins are secreted from the cytosol via the SecYEG translocon (5–7), though secretion via the twin-arginine transport (Tat) system has also been identified (8–11). Following translocation, the inner membrane (IM) enzyme Lgt attaches a diacyl moiety to the lipobox Cys of prolipoproteins via a thioester linkage (Fig. 1) (12–14). The diacylated product is a substrate for Lsp, which releases the apolipoprotein from its signal peptide (Fig. 1) (15–17). The diacylated Cys residue then becomes the first amino acid of the lipoprotein (Cys+1). In Gram-negative bacteria, a third acyl group is attached by the enzyme Lnt to the Cys+1 amino group (which was made available following Lsp cleavage) (Fig. 1) (18–21). The acyl chain donors in Lgt and Lnt reactions are plasma membrane phospholipids (Fig. 1). Gram-negative bacteria produce triacylated lipoproteins; lnt, lsp, and lgt are therefore conserved and essential in the majority of these organisms. Low-GC Gram-positive bacteria lack lnt homologs and generate considerable diversity in lipoprotein acylation; in addition to the triacyl form, these bacteria can variously generate diacyl, lyso, peptidyl, and N-acetyl lipoprotein forms (22, 23) (Fig. 1). How such diversity is generated largely awaits discovery, although recent progress has identified the enzyme, Lit, that is responsible for producing lyso-form lipoproteins in Enterococcus faecalis and Bacillus cereus (24).
Figure 1.
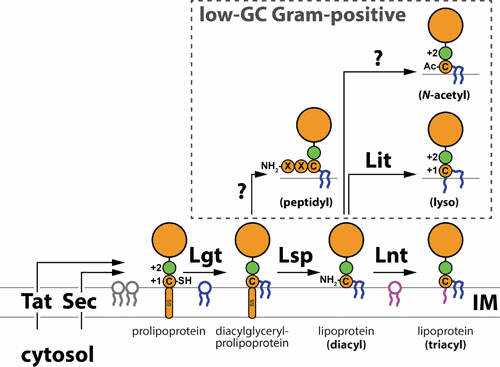
Posttranslocation lipoprotein maturation. Secreted lipoproteins are first diacylated at an invariant Cys residue by Lgt using resident phospholipids as acyl donors. The signal sequence is then cleaved by the peptidase Lsp to yield diacyl-form lipoproteins. In almost all Gram-negative bacteria, Lnt attaches another acyl chain to the amino group of Cys+1 to yield triacyl-form lipoproteins. Low-GC Gram-negative bacteria can also produce peptidyl forms (likely due to an Lsp-type enzyme that yields Cys+3), as well as N-acetyl and lyso forms that are derived from diacyl lipoproteins. Triacyl- and lyso-lipoproteins can efficiently interact with LolCDE for trafficking to the OM in Gram-negative organisms. Diacyl-form lipoproteins can be trafficked to the OM via LolDF.
In Gram-positive bacteria, lipoprotein maturation completes their biogenesis. In Gram-negative organisms, many lipoproteins await a new journey. These diderm bacteria traffic many of their lipoproteins from the plasma IM to the outer membrane (OM) (3, 25). The model organism Escherichia coli targets almost 90% of the lipoprotein species it produces to the OM (26). In fact, the very first lipoprotein identified was Lpp, a highly abundant E. coli OM lipoprotein (27). Lpp forms a covalent C-terminal attachment to the cell wall peptidoglycan (PG) and functions as an architectural element in the cell envelope that ensures accurate spacing between the OM and cell wall (28–30). Lpp that is mislocalized to the IM also forms PG cross-links, but these are lethally toxic for E. coli (31). Hence, in order to avoid such toxicity, trafficking of lipoproteins to the OM must be highly efficient. Lipoproteins face daunting hurdles to reaching the OM: the highly hydrophobic acyl moieties must leave a favorable IM lipid bilayer, cross an adverse aqueous periplasmic environment, and then be inserted into the OM bilayer. Some lipoproteins are subsequently translocated from the periplasm and across the OM to become surface exposed.
THE Lol PATHWAY
The major trafficking route that brings lipoproteins from the IM to the OM was discovered entirely by the lab of Hajime Tokuda, who named this pathway Lol (localization of lipoproteins) (Fig. 2) (1, 32). The Lol pathway has components in each compartment of the cell envelope: LolCDE, an ATP-binding cassette (ABC) transporter in the IM; LolA, a soluble chaperone protein in the periplasm; and LolB, itself a lipoprotein, at the OM. In model organisms with a full LolABCDE pathway, mature lipoproteins are extracted from the IM by LolCDE, transferred to LolA, and shuttled to the OM, where LolB receives and then anchors them into the bilayer. Each of the Lol proteins was found to be essential for viability of wild-type E. coli (33–35). However, recent work has unexpectedly discovered conditions under which both lolA and lolB can be deleted in E. coli, revealing that at least one other unknown trafficking route can deliver lipoproteins to the OM (discussed below) (36). Additionally, phylogenetic analysis suggests that several Gram-negative genera do not encode a lolB homolog. The emergence of LolAB-independent trafficking has prompted a reassessment of our understanding of how lipoproteins reach the OM.
Figure 2.
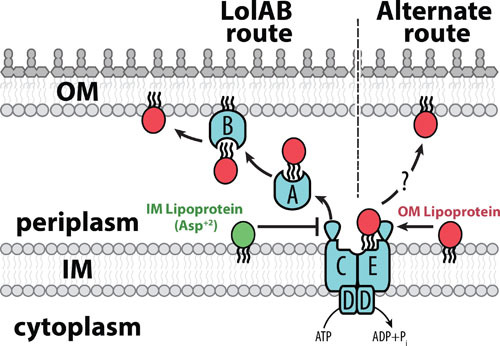
Trafficking routes for OM lipoproteins. The OM lipoprotein trafficking routes of E. coli are shown. Once mature, OM-targeted lipoproteins engage with the LolCDE transporter in the IM. LolE interacts with lipoproteins and LolC recruits the periplasmic chaperone protein LolA. At the expense of ATP hydrolysis by LolD, the LolCDE complex extracts lipoproteins from the IM bilayer and transfers them to LolA. Lipoproteins are shuttled through the periplasm in a LolA-bound complex. At the OM, the lipoprotein LolB receives LolA-bound client lipoproteins and anchors them into the OM bilayer. Since ΔlolAB mutants are viable, an alternate trafficking route must exist that can traffic essential OM lipoproteins to support cell viability. LolCDE remains essential in such ΔlolAB mutants, suggesting that lipoproteins originate from this complex and are then trafficked to the OM via an unknown mechanism.
GETTING TO THE OM
Amino Acid Targeting Signals
Given that all lipoproteins mature in the IM, the Gram-negative cell must first decide which of these will be targeted for trafficking to the OM and which will remain in the IM. Gene fusion experiments proved that targeting signals were encoded within lipoproteins themselves; heterologous proteins could be targeted to the OM by fusing them to N-terminal sequences of OM lipoproteins (37). Studies of E. coli soon revealed an elegantly simple OM targeting signal. In these bacteria, the identity of the second amino acid (adjacent to the lipidated Cys+1) determines localization. The presence of Asp+2 retains lipoproteins in the IM (37–39). Most other residues result in OM localization (39). This targeting mechanism became known as the “+2 rule.” There is compelling biochemical evidence that IM retention is caused by strong electrostatic interactions between Asp+2 and the headgroups of anionic IM phospholipids (40). These interactions likely prevent Asp+2 lipoproteins from interacting with the LolCDE transporter (41). Hence, the Asp+2 targeting mechanism has been coined “Lol avoidance.” Alternate +2 residues can also cause IM retention of E. coli lipoproteins, including Trp, Phe, Pro, Gly, and Tyr (42, 43), though none of these alternate retention signals are found in native E. coli lipoproteins. Given their chemistry, it is clear that the alternate retention signals do not cause IM retention via the same electrostatic mechanism as Asp+2. Most likely, alternate signals cause aberrant or inefficient interactions between lipoproteins and the LolCDE complex (44). Pseudomonas aeruginosa employs more complex retention signals that also involve the +3 and +4 residues (45–47). How these signals function is unclear; Asp+2 does function in this organism as a potent retention signal, though it is found infrequently among pseudomonal lipoproteins (45–47). More distantly related organisms employ altogether different targeting signals. For example, Borrelia burgdorferi relies on an acidic N-terminal linker region to retain lipoproteins in the IM (48, 49). It is remarkable that highly disparate lipoprotein targeting strategies appear to have evolved. Presumably, each strategy reflects the different cell envelope compositions and structures of diverse organisms. It is possible that the common goal of all these strategies is “Lol avoidance”—preventing lipoproteins that should be retained in the IM from interacting with the Lol ABC transporter—although this hypothesis awaits clear confirmation in many organisms.
Lipoprotein Acylation
The acylation state of lipoproteins is important for trafficking to the OM. For example, in Yersinia pestis, lack of modification by Lnt is proposed to act as a retention signal that prevents lipoprotein release from the IM (50). This proposal is based on the finding that the LolCDE complex in E. coli has very low affinity for diacyl-form lipoproteins. In wild-type E. coli, Lnt is an essential protein, but lnt can be deleted if LolCDE is highly overexpressed (51). Recent work suggests that Lnt N-acylation of Cys+1 is perhaps the key determinant for lipoprotein interaction with LolCDE. Armbruster and Meredith were able to complement the lethal loss of Lnt in E. coli by expressing a transacylase, Lit, from the low-GC Gram-positive organisms Enterococcus faecalis and Bacillus cereus (24). Lit removes one of the two thioester-linked acyl chains generated by the Lgt modification and attaches it to the Cys+1 amino group (Fig. 1) (24). The complementation of Lnt with Lit was successful without requiring LolCDE overproduction, and OM lipoprotein trafficking remained efficient enough to support viability (24). The requirement of LolCDE for lipoprotein N-acylation—the final maturation step—seems to serve as a secretion checkpoint mechanism that avoids premature trafficking of earlier maturation intermediates. Yet some Gram-negative species do not produce a LolCDE complex; rather than having a heterodimer of the IM LolC and LolE proteins, these organisms produce a homodimer of LolF proteins (52). LolF appears to be a hybrid of LolC and LolE proteins, containing key motifs from both (52). Bacteria that produce an IM complex of LolDF are suggested to not require Lnt for viability (52). Indeed, this was directly demonstrated for Francisella tularensis, Neisseria meningitidis, Acinetobacter baylyi, and Acinetobacter baumannii, from which lnt has successfully been deleted (52, 53). However, it should be noted that each of these organisms natively produces triacyl-form lipoproteins, at least when grown under laboratory conditions (52, 53). An intriguing possibility is that in some bacteria N-acylation may be a regulatable process that is linked to pathogenesis (52). In any case, it appears that LolDF complexes recognize triacylated lipoproteins but do not require N-acylation for lipoprotein trafficking.
DEPARTING THE IM VIA LolCDE
In the E. coli LolCDE complex, lipoprotein clients seem to interact primarily with LolE (54). This conclusion is based on site-specific photocross-linking at LolE, which can capture abundant clients in vivo (54). Meanwhile, LolC does not seem to interact with clients, despite having a hydrophobic cavity similar to that of LolE (54). The key role for LolC appears to be in recruiting LolA to the IM complex (54–57). LolC and LolE each contain one large periplasmic domain that is homologous between the proteins. LolA can be captured only at the LolC loop (54). Indeed, the specific LolA interaction with the LolC periplasmic domain was recently confirmed in a cocrystal structure and by biochemical methods (57). So LolC and LolE contribute to different functions in the early trafficking step: LolE recruits incoming lipoprotein clients, and LolC recruits the chaperone. In LolDF complexes, each of the LolF monomers in the homodimeric complexes must perform both of these functions. Arguably, segregating these functions between LolC and LolE may increase trafficking efficiency by generating a unidirectional flow of clients through the complex.
LolD is the ATPase that powers the LolCDE transporter (34, 58, 59). Recruitment of LolA does not require ATP binding or hydrolysis. Likely, the energy released by ATP hydrolysis is needed for the unfavorable step of extracting the acyl chains from the IM bilayer. However, current in vitro evidence suggests that the initial step of ATP binding alters the LolCDE-client complex in a way that makes the lipoprotein removable with detergent (58). This finding implies that lipoproteins are extracted upon ATP binding by LolD. In this case, the hydrolysis step should be important for the subsequent release reaction that transfers lipoproteins to LolA, for resetting the LolCDE complex, or for both activities.
TRAFFICKING TO THE OM VIA LolAB
LolA adopts an incomplete β-barrel structure with an enclosed hydrophobic cavity (60). LolA is recruited to LolC via a recently identified hook-and-pad interaction (57). The hook is a solvent exposed β-hairpin loop extending from the LolC periplasmic domain (57). The pad consists of three residues in the LolC periplasmic domain to which LolA binds (57). Interactions with both regions of LolC are involved in recruiting LolA (57). The periplasmic domains of LolC and LolA share sequence homology, and lipoprotein transfer is suggested to occur by the hydrophobic cavities lining up in a mouth-to-mouth orientation (56). The function of LolA must be to shield the acyl chains of its client from the aqueous periplasm. Yet E. coli LolA structures suggest that its hydrophobic cavity might not accommodate all three acyl chains of client lipoproteins (60). An alternate proposal for lipoprotein binding suggests that some acyl chains might bind hydrophobic patches on the surface of LolA (61).
At the OM, LolB receives lipoproteins from LolA and completes the trafficking route in E. coli by anchoring the lipoprotein into the OM bilayer (62). LolB is structurally similar to LolA (60). Surprisingly, LolB acylation is not required for its anchoring activity. A freely soluble, periplasmic LolB (termed mLolB; generated by replacing the native lipobox-containing signal sequence) remains able to receive lipoproteins from LolA and anchor them to membranes (63). However, mLolB perceives the periplasmic phospholipid headgroups of the IM and OM as equivalent, and it inserts lipoproteins into both the IM and the OM (63). Misinserted lipoproteins reenter the LolCDE transporter and try once more to reach the OM. Such a trafficking pathway is clearly inefficient; however, mLolB can complement inactivation of the native lolB gene if Lpp is either deleted or prevented from forming lethal PG cross-links from the IM (63). By anchoring LolB into the OM as a lipoprotein, the Lol pathway ensures accurate and unidirectional trafficking. A loop of LolB is important for the anchoring reaction, though the mechanism of anchoring remains unknown (64). Curiously, many Gram-negative organisms natively lack any lolB homolog (3). How such bacteria complete the trafficking pathway is an outstanding question. However, evidence from the artificial mLolB system suggests that an OM-localized lipoprotein membrane transferase might not be a strict requirement for trafficking.
TESTING THE ESSENTIALITY OF LolA AND LolB
Soon after they were discovered, LolA and LolB were identified as essential proteins for E. coli. Neither lolA nor lolB could be deleted, and depleting levels of either protein caused a decrease in cell viability (35, 62). LolCDE were discovered later and likewise determined to be essential (34). The finding that the Lol pathway is essential may have been initially puzzling—at the time, the only known essential OM lipoprotein was LolB itself. LolB depletion studies with E. coli revealed that lipoproteins mislocalize to the IM and also accumulate in the periplasm, complexed with LolA (33). The lethality of mislocalized Lpp was already known, but this did not explain Lol pathway essentiality since lolA, lolB, and lolCDE remained essential even when lpp was deleted (33, 35, 65).Hence, the reasonable conclusion was made that Lol proteins were essential because mislocalization of some lipoproteins may be toxic or may severely perturb the cell envelope (33). Essential OM lipoprotein clients were discovered in subsequent years and seemed to rationalize Lol protein essentiality. BamD and LptE are essential components of the OM assembly machinery that fold β-barrel OM proteins (the Bam machine) and transport lipopolysaccharide (the Lpt system), respectively (66–69). Accessory BamBCE OM lipoproteins and cell wall synthesis-regulating LpoAB OM lipoproteins are also collectively essential in E. coli to build a robust cell envelope, and combination mutants are lethal (70–74). It seemed that the Lol proteins were essential because they needed to deliver critical Bam, Lpt, and Lpo lipoproteins to the OM.
ALTERNATE TRAFFICKING ROUTE(S) FOR OM LIPOPROTEINS
Recently, the underlying reasons for LolA and LolB essentiality were directly tested (36). While conditions permitting deletion of both lolA and lolB were identified, lolCDE could not be deleted (36). Thus, the LolCDE complex is fundamentally required for all routes of lipoprotein trafficking. These findings revealed that LolAB are not truly essential for trafficking (36). Rather, their essential function in wild-type cells is to provide an efficient trafficking route that mitigates toxicities caused by OM lipoproteins mislocalizing to the IM. When the LolAB route was depleted, two OM-targeted lipoproteins were found kill the cell, most likely by accumulating in the IM: Lpp, which (as discussed above) forms toxic PG cross-links from the IM, and OsmB, which may form pores across the IM that dissipate the proton motive force, killing the cell (36). There must be at least one other alternate trafficking route that can perform the essential task of bringing lipoproteins from LolCDE, through the periplasm, and into the OM in ΔlolAB mutants. Indeed, the Bam lipoproteins were directly shown to reach the OM even when LolAB were absent (36). Remarkably, Helicobacter pylori, which lacks a lolB homolog, also appears to tolerate inactivation of lolA (75). Therefore, it is tempting to speculate that the same alternate trafficking route that functions in E. coli ΔlolAB cells is required to support essential OM lipoprotein trafficking in H. pylori ΔlolA mutants. If this is true, the alternate trafficking route may even be ancestral to the LolAB route. LolAB may have emerged to provide increased trafficking efficiency and capacity, a requirement for evolving OM lipoproteins (such as Lpp and OsmB) whose activity at the OM is beneficial but whose accumulation in the IM is potently toxic. Indeed, Lpp and OsmB are narrowly conserved to a subset of Gram-negative bacteria that possess both lolA and lolB.
SUMMARY
Efforts in recent years have yielded considerable insights into the maturation of lipoproteins and, in Gram-negative bacteria, their trafficking towards the OM. New questions have emerged: how is acylation diversity achieved in Gram-positive bacteria, and why is the triacyl form the apparent default among Gram-negative organisms? How are lipoproteins trafficked when the known LolAB route is inactivated? How does lipoprotein trafficking occur in organisms lacking lolB? Moreover, what are the molecular mechanisms that underlie the highly efficient trafficking via LolAB? As answers to these questions are found, we will be rewarded with an increasingly sophisticated and comprehensive understanding of lipoprotein biogenesis and trafficking. Given that the OM is a major barrier against antibiotics and that OM lipoproteins are essential for OM assembly, insights into trafficking may prove invaluable to the goal of developing new drugs to treat increasingly antibiotic-resistant Gram-negative infections.
ACKNOWLEDGMENTS
I thank Kerrie May and the anonymous reviewers for comments that have improved this manuscript.
This work was supported by institutional startup funding from Emory University.
Contributor Information
Marcin Grabowicz, Emory Antibiotic Resistance Center, Emory University School of Medicine, Atlanta, GA 30322; Department of Microbiology & Immunology, Emory University School of Medicine, Atlanta, GA 30322; Division of Infectious Diseases, Department of Medicine, Emory University School of Medicine, Atlanta, GA 30322.
Maria Sandkvist, Department of Microbiology and Immunology, University of Michigan, Ann Arbor, Michigan.
Eric Cascales, CNRS Aix-Marseille Université, Mediterranean Institute of Microbiology, Marseille, France.
Peter J. Christie, Department of Microbiology and Molecular Genetics, McGovern Medical School, Houston, Texas
REFERENCES
- 1.Narita S-I, Tokuda H. 2017. Bacterial lipoproteins; biogenesis, sorting and quality control. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1414–1423. 10.1016/j.bbalip.2016.11.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Buddelmeijer N. 2015. The molecular mechanism of bacterial lipoprotein modification—how, when and why? FEMS Microbiol Rev 39:246–261. 10.1093/femsre/fuu006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Grabowicz M. 2018. Lipoprotein transport: greasing the machines of outer membrane biogenesis. Bioessays 40:e1700187. 10.1002/bies.201700187. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188:2761–2773. 10.1128/JB.188.8.2761-2773.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayashi S, Wu HC. 1985. Accumulation of prolipoprotein in Escherichia coli mutants defective in protein secretion. J Bacteriol 161:949–954. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugai M, Wu HC. 1992. Export of the outer membrane lipoprotein is defective in secD , secE , and secF mutants of Escherichia coli. J Bacteriol 174:2511–2516. 10.1128/jb.174.8.2511-2516.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fröderberg L, Houben ENG, Baars L, Luirink J, de Gier J-W. 2004. Targeting and translocation of two lipoproteins in Escherichia coli via the SRP/Sec/YidC pathway. J Biol Chem 279:31026–31032. 10.1074/jbc.M403229200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Thompson BJ, Widdick DA, Hicks MG, Chandra G, Sutcliffe IC, Palmer T, Hutchings MI. 2010. Investigating lipoprotein biogenesis and function in the model Gram-positive bacterium Streptomyces coelicolor. Mol Microbiol 77:943–957. [DOI] [PubMed] [Google Scholar]
- 9.Widdick DA, Hicks MG, Thompson BJ, Tschumi A, Chandra G, Sutcliffe IC, Brülle JK, Sander P, Palmer T, Hutchings MI. 2011. Dissecting the complete lipoprotein biogenesis pathway in Streptomyces scabies. Mol Microbiol 80:1395–1412. 10.1111/j.1365-2958.2011.07656.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Shruthi H, Anand P, Murugan V, Sankaran K. 2010. Twin arginine translocase pathway and fast-folding lipoprotein biosynthesis in E. coli: interesting implications and applications. Mol Biosyst 6:999–1007. 10.1039/b916510j. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Randall LB, Dobos K, Papp-Wallace KM, Bonomo RA, Schweizer HP. 2015. Membrane-bound PenA β-lactamase of Burkholderia pseudomallei. Antimicrob Agents Chemother 60:1509–1514. 10.1128/AAC.02444-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokunaga M, Tokunaga H, Wu HC. 1982. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci U S A 79:2255–2259. 10.1073/pnas.79.7.2255. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankaran K, Wu HC. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem 269:19701–19706. [PubMed] [PubMed] [Google Scholar]
- 14.Mao G, Zhao Y, Kang X, Li Z, Zhang Y, Wang X, Sun F, Sankaran K, Zhang XC. 2016. Crystal structure of E. coli lipoprotein diacylglyceryl transferase. Nat Commun 7:10198. 10.1038/ncomms10198. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogeley L, El Arnaout T, Bailey J, Stansfeld PJ, Boland C, Caffrey M. 2016. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 351:876–880. 10.1126/science.aad3747. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Tokunaga M, Loranger JM, Wu HC. 1984. Prolipoprotein modification and processing enzymes in Escherichia coli. J Biol Chem 259:3825–3830. [PubMed] [PubMed] [Google Scholar]
- 17.Inouye S, Franceschini T, Sato M, Itakura K, Inouye M. 1983. Prolipoprotein signal peptidase of Escherichia coli requires a cysteine residue at the cleavage site. EMBO J 2:87–91. 10.1002/j.1460-2075.1983.tb01386.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta SD, Gan K, Schmid MB, Wu HC. 1993. Characterization of a temperature-sensitive mutant of Salmonella typhimurium defective in apolipoprotein N-acyltransferase. J Biol Chem 268:16551–16556. [PubMed] [PubMed] [Google Scholar]
- 19.Noland CL, Kattke MD, Diao J, Gloor SL, Pantua H, Reichelt M, Katakam AK, Yan D, Kang J, Zilberleyb I, Xu M, Kapadia SB, Murray JM. 2017. Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase. Proc Natl Acad Sci U S A 114:E6044–E6053. 10.1073/pnas.1707813114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiktor M, Weichert D, Howe N, Huang C-Y, Olieric V, Boland C, Bailey J, Vogeley L, Stansfeld PJ, Buddelmeijer N, Wang M, Caffrey M. 2017. Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis. Nat Commun 8:15952. 10.1038/ncomms15952. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal-Ingigliardi D, Lewenza S, Buddelmeijer N. 2007. Identification of essential residues in apolipoprotein N-acyl transferase, a member of the CN hydrolase family. J Bacteriol 189:4456–4464. 10.1128/JB.00099-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakayama H, Kurokawa K, Lee BL. 2012. Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J 279:4247–4268. 10.1111/febs.12041. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa K, Ryu K-H, Ichikawa R, Masuda A, Kim M-S, Lee H, Chae J-H, Shimizu T, Saitoh T, Kuwano K, Akira S, Dohmae N, Nakayama H, Lee BL. 2012. Novel bacterial lipoprotein structures conserved in low-GC content gram-positive bacteria are recognized by Toll-like receptor 2. J Biol Chem 287:13170–13181. 10.1074/jbc.M111.292235. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armbruster KM, Meredith TC. 2017. Identification of the lyso-form N-acyl intramolecular transferase in low-GC Firmicutes. J Bacteriol 199:e00099-17. 10.1128/JB.00099-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konovalova A, Silhavy TJ. 2015. Outer membrane lipoprotein biogenesis: Lol is not the end. Philos Trans R Soc Lond B Biol Sci 370:20150030. 10.1098/rstb.2015.0030. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horler RSP, Butcher A, Papangelopoulos N, Ashton PD, Thomas GH. 2009. EchoLOCATION: an in silico analysis of the subcellular locations of Escherichia coli proteins and comparison with experimentally derived locations. Bioinformatics 25:163–166. 10.1093/bioinformatics/btn596. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Hantke K, Braun V. 1973. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur J Biochem 34:284–296. 10.1111/j.1432-1033.1973.tb02757.x. [DOI] [PubMed] [Google Scholar]
- 28.Braun V, Rehn K. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10:426–438. 10.1111/j.1432-1033.1969.tb00707.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Asmar AT, Ferreira JL, Cohen EJ, Cho S-H, Beeby M, Hughes KT, Collet J-F. 2017. Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol 15:e2004303. 10.1371/journal.pbio.2004303. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen EJ, Ferreira JL, Ladinsky MS, Beeby M, Hughes KT. 2017. Nanoscale-length control of the flagellar driveshaft requires hitting the tethered outer membrane. Science 356:197–200. 10.1126/science.aam6512. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yakushi T, Tajima T, Matsuyama S, Tokuda H. 1997. Lethality of the covalent linkage between mislocalized major outer membrane lipoprotein and the peptidoglycan of Escherichia coli. J Bacteriol 179:2857–2862. 10.1128/jb.179.9.2857-2862.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda S, Tokuda H. 2011. Lipoprotein sorting in bacteria. Annu Rev Microbiol 65:239–259. 10.1146/annurev-micro-090110-102859. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Tanaka K, Matsuyama S-I, Tokuda H. 2001. Deletion of lolB , encoding an outer membrane lipoprotein, is lethal for Escherichia coli and causes accumulation of lipoprotein localization intermediates in the periplasm. J Bacteriol 183:6538–6542. 10.1128/JB.183.22.6538-6542.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakushi T, Masuda K, Narita S, Matsuyama S, Tokuda H. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat Cell Biol 2:212–218. 10.1038/35008635. [DOI] [PubMed] [Google Scholar]
- 35.Tajima T, Yokota N, Matsuyama S, Tokuda H. 1998. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins. FEBS Lett 439:51–54. 10.1016/S0014-5793(98)01334-9. [DOI] [PubMed] [Google Scholar]
- 36.Grabowicz M, Silhavy TJ. 2017. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci U S A 114:4769–4774. 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi K, Yu F, Inouye M. 1988. A single amino acid determinant of the membrane localization of lipoproteins in E. coli. Cell 53:423–432. 10.1016/0092-8674(88)90162-6. [DOI] [PubMed] [Google Scholar]
- 38.Gennity JM, Inouye M. 1991. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J Biol Chem 266:16458–16464. [PubMed] [Google Scholar]
- 39.Terada M, Kuroda T, Matsuyama S-I, Tokuda H. 2001. Lipoprotein sorting signals evaluated as the LolA-dependent release of lipoproteins from the cytoplasmic membrane of Escherichia coli. J Biol Chem 276:47690–47694. 10.1074/jbc.M109307200. [DOI] [PubMed] [Google Scholar]
- 40.Hara T, Matsuyama S, Tokuda H. 2003. Mechanism underlying the inner membrane retention of Escherichia coli lipoproteins caused by Lol avoidance signals. J Biol Chem 278:40408–40414. 10.1074/jbc.M307836200. [DOI] [PubMed] [Google Scholar]
- 41.Masuda K, Matsuyama S, Tokuda H. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc Natl Acad Sci U S A 99:7390–7395. 10.1073/pnas.112085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seydel A, Gounon P, Pugsley AP. 1999. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol 34:810–821. 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 43.Lewenza S, Vidal-Ingigliardi D, Pugsley AP. 2006. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J Bacteriol 188:3516–3524. 10.1128/JB.188.10.3516-3524.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto C, Satou R, Tokuda H, Narita S. 2010. Novel mutations of the LolCDE complex causing outer membrane localization of lipoproteins despite their inner membrane-retention signals. Biochem Biophys Res Commun 401:586–591. 10.1016/j.bbrc.2010.09.106. [DOI] [PubMed] [Google Scholar]
- 45.Lewenza S, Mhlanga MM, Pugsley AP. 2008. Novel inner membrane retention signals in Pseudomonas aeruginosa lipoproteins. J Bacteriol 190:6119–6125. 10.1128/JB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narita S, Tokuda H. 2007. Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J Biol Chem 282:13372–13378. 10.1074/jbc.M611839200. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka S-Y, Narita S, Tokuda H. 2007. Characterization of the Pseudomonas aeruginosa Lol system as a lipoprotein sorting mechanism. J Biol Chem 282:13379–13384. 10.1074/jbc.M611840200. [DOI] [PubMed] [Google Scholar]
- 48.Schulze RJ, Zückert WR. 2006. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol 59:1473–1484. 10.1111/j.1365-2958.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- 49.Kumru OS, Schulze RJ, Rodnin MV, Ladokhin AS, Zückert WR. 2011. Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J Bacteriol 193:2814–2825. 10.1128/JB.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva-Herzog E, Ferracci F, Jackson MW, Joseph SS, Plano GV. 2008. Membrane localization and topology of the Yersinia pestis YscJ lipoprotein. Microbiology 154:593–607. 10.1099/mic.0.2007/013045-0. [DOI] [PubMed] [Google Scholar]
- 51.Narita S, Tokuda H. 2011. Overexpression of LolCDE allows deletion of the Escherichia coli gene encoding apolipoprotein N-acyltransferase. J Bacteriol 193:4832–4840. 10.1128/JB.05013-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LoVullo ED, Wright LF, Isabella V, Huntley JF, Pavelka MS, Jr. 2015. Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol 197:1705–1715. 10.1128/JB.02414-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gwin CM, Prakash N, Christian Belisario J, Haider L, Rosen ML, Martinez LR, Rigel NW. 2018. The apolipoprotein N-acyl transferase Lnt is dispensable for growth in Acinetobacter species. Microbiology 164:1547–1556. 10.1099/mic.0.000726. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizutani M, Mukaiyama K, Xiao J, Mori M, Satou R, Narita S, Okuda S, Tokuda H. 2013. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex. FEBS Lett 587:23–29. 10.1016/j.febslet.2012.11.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Okuda S, Watanabe S, Tokuda H. 2008. A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins. FEBS Lett 582:2247–2251. 10.1016/j.febslet.2008.05.022. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Okuda S, Tokuda H. 2009. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc Natl Acad Sci U S A 106:5877–5882. 10.1073/pnas.0900896106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaplan E, Greene NP, Crow A, Koronakis V. 2018. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain. Proc Natl Acad Sci U S A 115:E7389–E7397. 10.1073/pnas.1806822115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito Y, Kanamaru K, Taniguchi N, Miyamoto S, Tokuda H. 2006. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins. Mol Microbiol 62:1064–1075. 10.1111/j.1365-2958.2006.05378.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.Taniguchi N, Tokuda H. 2008. Molecular events involved in a single cycle of ligand transfer from an ATP binding cassette transporter, LolCDE, to a molecular chaperone, LolA. J Biol Chem 283:8538–8544. 10.1074/jbc.M800026200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takeda K, Miyatake H, Yokota N, Matsuyama S, Tokuda H, Miki K. 2003. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J 22:3199–3209. 10.1093/emboj/cdg324. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Remans K, Pauwels K, van Ulsen P, Buts L, Cornelis P, Tommassen J, Savvides SN, Decanniere K, Van Gelder P. 2010. Hydrophobic surface patches on LolA of Pseudomonas aeruginosa are essential for lipoprotein binding. J Mol Biol 401:921–930. 10.1016/j.jmb.2010.06.067. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Matsuyama S, Yokota N, Tokuda H. 1997. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J 16:6947–6955. 10.1093/emboj/16.23.6947. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukahara J, Mukaiyama K, Okuda S, Narita S, Tokuda H. 2009. Dissection of LolB function—lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J 276:4496–4504. 10.1111/j.1742-4658.2009.07156.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Hayashi Y, Tsurumizu R, Tsukahara J, Takeda K, Narita S, Mori M, Miki K, Tokuda H. 2014. Roles of the protruding loop of factor B essential for the localization of lipoproteins (LolB) in the anchoring of bacterial triacylated proteins to the outer membrane. J Biol Chem 289:10530–10539. 10.1074/jbc.M113.539270. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narita S, Tanaka K, Matsuyama S, Tokuda H. 2002. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane. J Bacteriol 184:1417–1422. 10.1128/JB.184.5.1417-1422.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164. 10.1111/j.1365-2958.2006.05211.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. 10.1073/pnas.0604744103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konovalova A, Kahne DE, Silhavy TJ. 2017. Outer membrane biogenesis. Annu Rev Microbiol 71:539–556. 10.1146/annurev-micro-090816-093754. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. 10.1038/nrmicro.2016.25. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lupoli TJ, Lebar MD, Markovski M, Bernhardt T, Kahne D, Walker S. 2014. Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J Am Chem Soc 136:52–55. 10.1021/ja410813j. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. 10.1016/j.cell.2010.11.037. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. 10.1016/j.cell.2010.11.038. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Misra R, Stikeleather R, Gabriele R. 2015. In vivo roles of BamA, BamB and BamD in the biogenesis of BamA, a core protein of the β-barrel assembly machine of Escherichia coli. J Mol Biol 427:1061–1074. 10.1016/j.jmb.2014.04.021. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. 2012. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol 194:1002–1008. 10.1128/JB.06426-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, Brinkman KK, Warren PV, Lupas A, Stanhope MJ, Brown JR, Hoffman PS. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J Bacteriol 183:1259–1268. 10.1128/JB.183.4.1259-1268.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


