Abstract
Osteosarcoma (OSA) is a primary bone malignancy characterized by its aggressive nature and high propensity for metastasis. Despite advancements in multimodal therapies, the clinical outcomes for OSA patients remain suboptimal, necessitating deeper molecular insights for improved therapeutic strategies. Here, we employed single-cell RNA sequencing (scRNA-seq) to elucidate the cellular heterogeneity and transcriptional dynamics of OSA tumors. Our study identified eleven distinct tumor cell subpopulations, including osteoblastic, chondroblastic, and myeloid lineages, each exhibiting unique transcriptional profiles associated with disease progression and metastasis. Epithelial-mesenchymal transition (EMT) emerged as a critical process driving aggressive phenotypes, supported by gene set enrichment analyses (GSVA) and transcription factor regulatory network analyses. Integration of copy number variation (CNV) data highlighted genomic alterations in osteoblastic and chondroblastic cells, implicating potential therapeutic targets. Furthermore, immune cell infiltration analyses revealed distinct immune profiles across OSA subtypes, correlating with tumor mutational burden (TMB) and clinical outcomes. Our findings underscore the complexity of OSA biology and provide a foundation for developing personalized treatment strategies targeting tumor heterogeneity and immune interactions.
Keywords: Osteosarcoma, Single-cell analysis, Heterogeneity, Epithelial–mesenchymal transition, Personalized medicine
Introduction
Osteosarcoma, the most common primary bone malignancy, predominantly affects adolescents and young adults, presenting a unique challenge in oncology due to its aggressive nature and high potential for metastasis [1–3]. Despite advancements in multimodal therapies, including surgery and chemotherapy, the prognosis for osteosarcoma patients, especially those with metastatic or recurrent disease, remains dismal [4, 5]. The five-year survival rate for non-metastatic osteosarcoma patients has plateaued at approximately 60–70%, with significantly poorer outcomes for those with metastatic disease. This stagnation in therapeutic advancements highlights the pressing need for innovative treatment strategies and deeper molecular insights into the pathogenesis of osteosarcoma [6, 7].
The pathogenesis of OSA involves a complex interplay of genetic mutations, epigenetic alterations, and interactions within the tumor microenvironment (TME) [7, 8]. Originating from primitive bone-forming mesenchymal cells, OSA disrupts normal bone architecture and function, leading to debilitating consequences for patients [9, 10]. Current diagnostic and prognostic strategies, largely reliant on histopathological features and clinical staging systems, often fall short in accurately predicting individual patient outcomes [11–13]. Thus, there is a critical demand for molecular profiling approaches that can discern the heterogeneous nature of OSA tumors and tailor treatment strategies accordingly.
The rationale for undertaking this study is rooted in several pivotal gaps in current OSA research and clinical management. Firstly, while significant strides have been made in unraveling the genomic and transcriptomic landscapes of OSA, comprehensive studies integrating single-cell resolution analyses and multi-omics data remain sparse [14–16]. Such integrative approaches are essential for deciphering the heterogeneity of OSA, identifying rare cell populations, and elucidating the genomic drivers of treatment resistance. Secondly, the phenomenon of epithelial–mesenchymal transition (EMT), implicated in promoting invasion, metastasis, and therapeutic resistance in various cancers, including OSA, remains incompletely understood [17–19]. Exploring the role of EMT and its functional implications across different OSA subtypes could provide critical insights into disease progression and inform targeted therapies.
Furthermore, the advent of single-cell technologies offers unprecedented opportunities to dissect cellular heterogeneity and clonal evolution within OSA tumors. By employing advanced computational tools and bioinformatics analyses, this study aims to characterize distinct tumor cell populations, unravel their transcriptional dynamics, and identify novel therapeutic vulnerabilities [20–22]. Integration of single-cell RNA sequencing (scRNA-seq) with copy number variation (CNV) analyses will allow mapping of genomic alterations associated with aggressive OSA phenotypes and metastatic potential [23, 24].
The overarching objectives of this study are to comprehensively delineate the cellular and molecular landscapes of OSA and to translate these insights into improved clinical outcomes. By characterizing tumor cell heterogeneity, investigating EMT dynamics, elucidating transcriptional regulatory networks, developing prognostic models, and exploring interactions within the immune microenvironment, this research aims to pave the way for precision medicine approaches in OSA. Ultimately, the findings are anticipated to contribute to the development of novel biomarkers for patient stratification, the discovery of targeted therapies, and the optimization of treatment strategies to enhance survival rates and quality of life for individuals affected by OSA.
Methods
Acquisition and processing of transcriptome data
RNA expression profiles and corresponding clinical data of osteosarcoma were obtained from the TARGET database (n = 85). Data were transformed to TPM format and log2 transformed for subsequent analyses.
Acquisition and processing of scRNA-seq data
Single-cell dataset was retrieved from the GEO database (GSE152048), comprising 9 primary tumor samples (including 2 metastatic and 7 non-metastatic samples). R software (version 4.1.3) was used for analysis with Seurat package. Cell quality control criteria included <20% mitochondrial content, <3% erythrocyte content, and UMI and gene count thresholds of 200–50000 and 200–7000, respectively. Data normalization, selection of highly variable genes (2000), and cell cycle effect removal (parameters: vars.to.regress = c("S.Score", "G2M.Score")) were performed using NormalizeData, FindVariableFeatures, and ScaleData functions in Seurat. Batch effects were adjusted using harmony. Dimensionality reduction (UMAP, tSNE) and clustering (Louvain) were conducted using Seurat's methods. Differential gene expression analysis between clusters or cell types was performed using FindAllMarkers function with parameters: pvalue < 0.05, log2FC > 0.25, and expression in >10% of cells.
Acquisition of EMT-related genes
EMT-related genes were obtained from the Msigdb database under the HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION category (gsea-msigdb.org).
Cell annotation analysis
Cell annotation was based on markers: osteoblastic cells (COL1A1, CDH11, RUNX2), proliferating osteoblastic cells (TOP2, PCNA, MKI67), chondroblastic cells (ACAN, COL2A1, SOX9), osteoclastic cells (CTSK, MMP9), TILs (IL7R, CD3D, NKG7), myeloid cells (CD74, CD14, FCGR3A), fibroblasts (COL1A1, LUM, DCN), pericytes (ACTA2, RGS5), MSCs (CXCL12, SFRP2, MME), myoblasts (MYLPF, MYL1), endothelial cells (PECAM1, VWF). Visualization included tSNE plots and violin plots of cell markers.
Single-cell CNV analysis
InferCNV software was used with endothelial cells as reference to analyze CNV profiles across cell subgroups, focusing on identification of malignant cells and CNV score assessment.
Single-cell pseudotime analysis
Monocle2 software was employed for pseudotime analysis of tumor cell subgroups, using DDRTree for dimensionality reduction and default parameters to infer cellular differentiation trajectories.
Single-cell transcription factor analysis
SCENIC software was used to analyze transcription factors in tumor cell subgroups, employing RcisTarget and GRNBoost databases for motif discovery and analysis of regulons' activity across cell types using AUCell.
Cell communication analysis
CellChat package was utilized to assess potential cell-cell communication networks. Normalized gene expression matrices were processed using identifyOverExpressedGenes, identifyOverExpressedInteraction, and ProjectData functions with default parameters, followed by computeCommunProb, filterCommunication, and computeCommunProbPathway functions to determine ligand-receptor interactions and generate communication networks with aggregateNet function.
Calculation of EMT signature score
EMT-related genes were used to compute signature scores across single-cell data using gsva and ssGSEA algorithms from the GSVA package.
Immune infiltration analysis
IOBR package was used to evaluate immune infiltration levels in high-risk groups of patients using ESTIMATE, CIBERSORT, and xCell algorithms.
Gene enrichment analysis
clusterProfiler package was employed for enrichment analysis using KEGG and GO databases. Enriched functions were visualized using ggplot2 package after BH correction with p < 0.05.
Comparison of genomic variability landscapes between two groups
Maftools package was used to compare mutation profiles between two groups, assessing differences in TMB and its correlation with risk scores, and integrating TMB into survival analysis.
LASSO-Cox prognostic model establishment based on C0 tumor cells' markers
Marker genes from C0 tumor cells (highest EMT score) were used for univariate Cox analysis to identify survival-associated genes (p < 0.05). LASSO+Cox regression analysis was then performed using glmnet package. Model accuracy was evaluated with timeROC package for 1, 3, and 5-year AUC values.
Statistical analysis
All data processing, statistical analyses, and plotting were conducted in R 4.1.3 software. Pearson correlation coefficient was used to assess correlation between continuous variables. Chi-square test was employed for categorical variables, while Wilcoxon rank-sum test or t-test was used for continuous variables. survival package was used for Cox regression and Kaplan-Meier analysis.
Results
Single-cell expression atlas of OSA
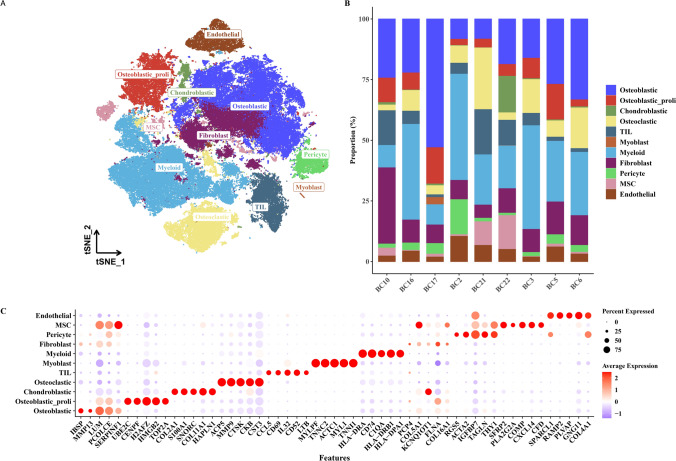
We conducted a series of analyses including quality control and dimensionality reduction on single-cell data, resulting in a total of 99,732 cells. Using classical cell type markers, we classified the cells into 11 major categories: Osteoblastic, Osteoblastic_proli, Chondroblastic, Osteoclastic, TIL (Tumor-Infiltrating Lymphocytes), Myoblast, Myeloid, Fibroblast, Pericyte, MSC (Mesenchymal Stem Cell), and Endothelial, as shown in Fig. 1A. Subsequently, we illustrated the distribution of cell types among patients (Fig. 1B) and presented the expression profiles of marker genes for each cell type to verify classification accuracy (Fig. 1C).
Fig. 1.
Single-cell classification of osteosarcoma (OSA). A t-SNE plot showing the clustering of single cells based on their transcriptomic profiles into 11 major cell types. B Bar chart depicting the composition of cell types across different patients. C Bubble plot illustrating the expression levels of marker genes for each identified cell type to confirm classification accuracy
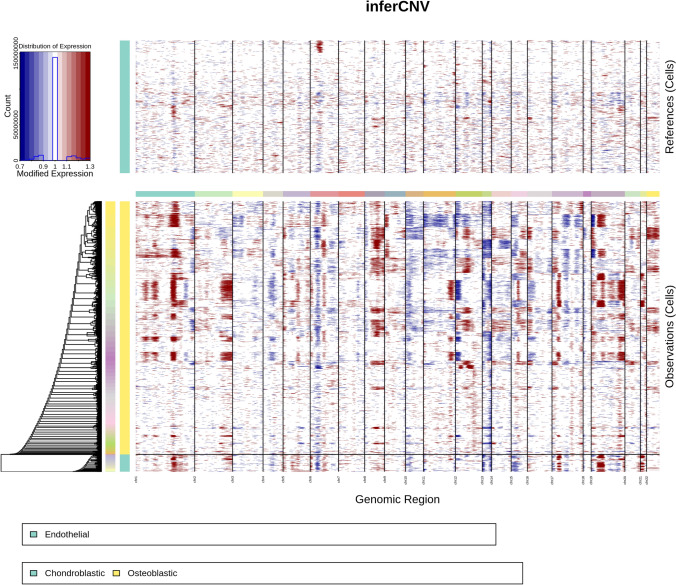
CNV analysis of single cells
Using inferCNV software with endothelial cells as a reference, we assessed the CNV profiles of various cells (Fig. 2). Significant CNV variations were observed particularly in Osteoblastic and Chondroblastic cells.
Fig. 2.
CNV analysis of single cells. CNV heatmap derived using inferCNV software, with endothelial cells as a reference, displaying significant CNV alterations in osteoblastic and chondroblastic cells
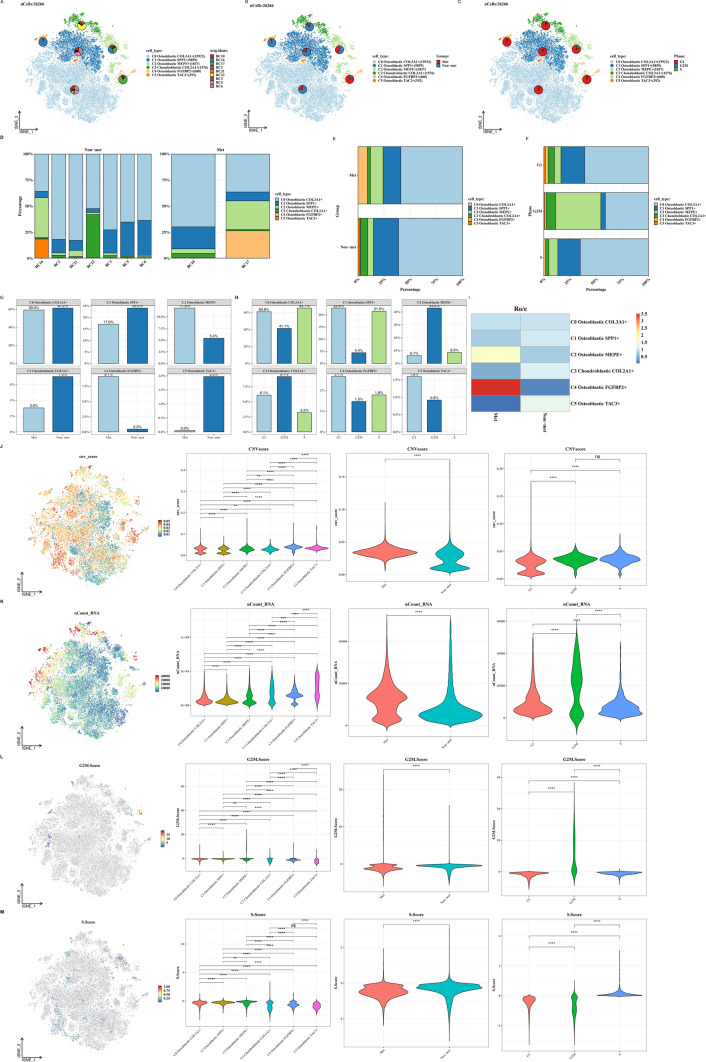
Subclassification analysis of tumor cells
We isolated tumor cells (mainly comprising Osteoblastic and Chondroblastic cells) for re-clustering analysis, resulting in 6 clusters. Each cluster was characterized by specific marker genes: C0 Osteoblastic COL3A1+, C1 Osteoblastic SPP1+, C2 Osteoblastic MEPE+, C3 Chondroblastic COL2A1+, C4 Osteoblastic FGFBP2+, and C5 Osteoblastic TAC3+. We analyzed the distribution of these clusters among patients, metastatic status, and cell cycle phases (Fig. 3A–F). Subsequently, we evaluated the proportions of clusters in relation to metastasis and cell cycle phases (Fig. 3G–I), followed by comparing CNV scores, UMI counts, G2M scores, and S scores across clusters, metastatic status, and cell cycle phases (Fig. 3J–M).
Fig. 3.
Subtyping analysis of tumor cells. A–C t-SNE plots of tumor cells showing the distribution of each clustering cluster across patients, metastatic status, and cell cycle phases. D–H Bar graphs illustrating the proportions of each clustering cluster across patients, metastatic status, and cell cycle phases. I Ratio of observed to expected cell numbers (Ro/e) for each tumor cell subtype to assess tissue preference. J–M Violin plots and t-SNE overlays showing differences in CNV scores, nCount_RNA, G2M.Score, and S.Score across clustering clusters, metastatic status, and cell cycle phases
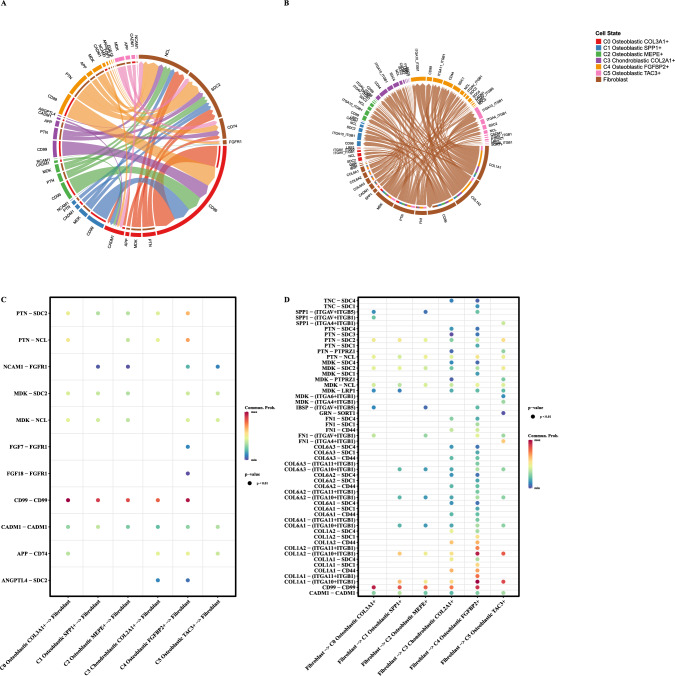
Cell communication analysis
Figure 4A illustrates the communication from tumor cells to fibroblasts, while Fig. 4B shows communication from fibroblasts to tumor cells. Corresponding bubble plots depict specific receptor interactions (Fig. 4C, D).
Fig. 4.
Cell communication analysis. A Chord diagrams depicting ligand-receptor interactions between tumor cell subtypes and fibroblasts. B Chord diagrams illustrating ligand-receptor interactions between fibroblasts and tumor cell subtypes. C, D Bubble plots displaying specific ligand-receptor pairs between tumor cell subtypes and fibroblasts
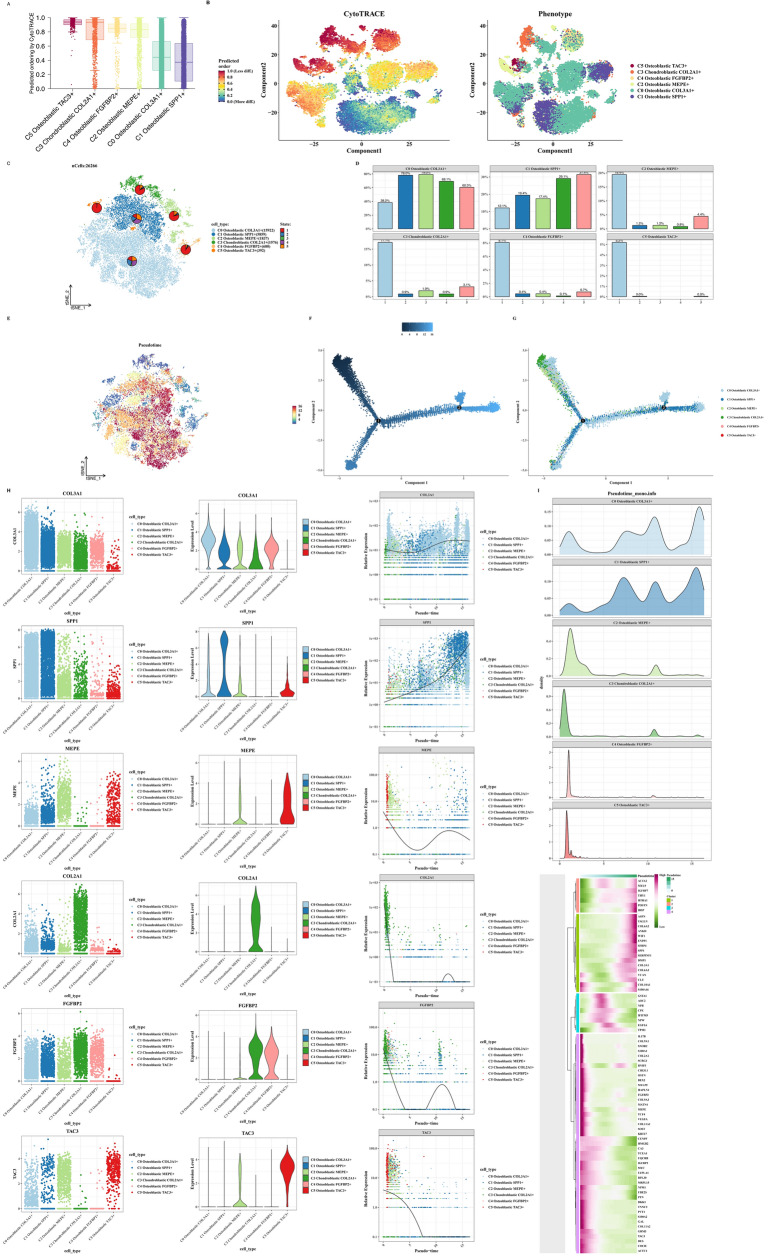
CytoTRACE and cell trajectory analysis
Using CytoTRACE, we assessed the differentiation degree of tumor cell subtypes, with C5 showing the highest and C0, C1 the lowest differentiation levels (Fig. 5A, B). Subsequently, using monocle software, we performed cell trajectory analysis, revealing C5 at the pseudotime origin and C0, C1 at the endpoint, indicating higher differentiation levels for C0 and C1 (Fig. 5C–G). We presented the marker gene expression profiles of each tumor cell cluster and their expression patterns over pseudotime (Fig. 5H, I).
Fig. 5.
CytoTRACE and cell trajectory analysis. A, B CytoTRACE scores evaluating stemness across tumor cell subtypes and exploring aspects of tumor cell subtype differentiation. C–G Monocle results displaying differentiation trajectories of tumor cell subtypes (C0, C1) compared to others. H Along the pseudotime trajectory, changes in the expression of tumor cell subtype markers (COL3A1, SPP1, MEPE, COL2A1, FGFBP2, TAC3) are observed. I A heatmap based on pseudotime trajectory analysis provides the key indicators for each tumor cell subtype
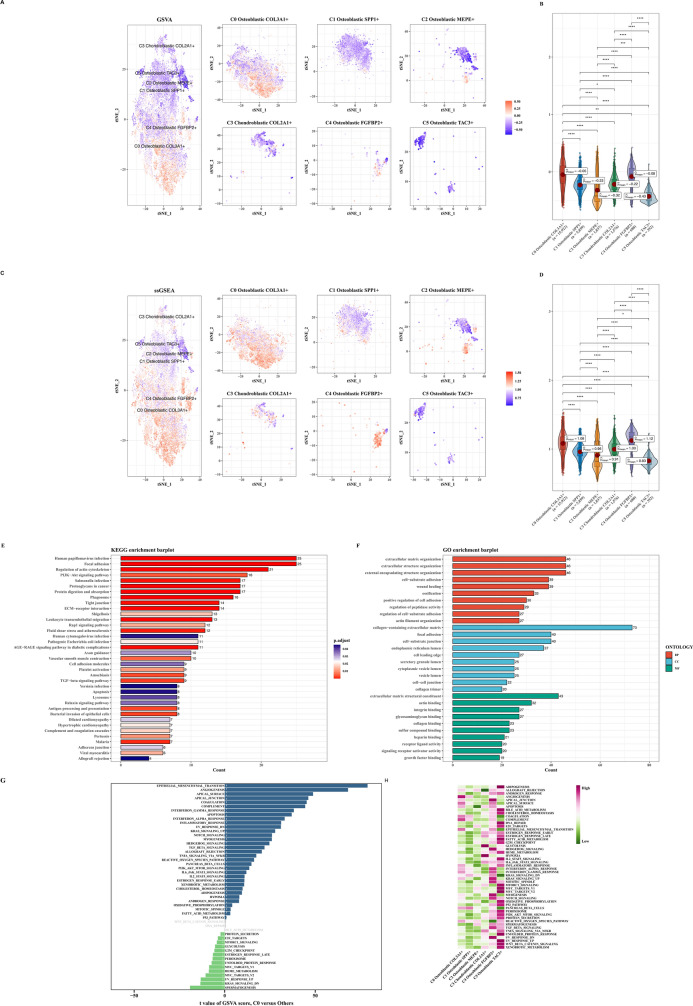
Exploration of EMT functional scores in various tumor cell subtypes
We employed GSVA and ssGSEA algorithms to calculate EMT scores across tumor cell data. Results were consistent, showing C0 with the highest score and significant differences compared to other subtypes (Fig. 6A–D). We identified differential genes between C0 and other subtypes and performed enrichment analyses (KEGG, GO) revealing associations with functions such as extracellular matrix organization (Fig. 6E, F). Subsequently, we explored differential patterns in the Hallmark gene set (Fig. 6G, H).
Fig. 6.
Functional assessment of EMT in tumor cell subtypes. A, B Differential activity of EMT-related genes across tumor cell subtypes assessed using GSVA. C, D Differential activity of EMT-related genes across tumor cell subtypes assessed using ssGSEA. E KEGG pathway enrichment analysis highlighting pathways enriched in C0 tumor cells compared to other tumor cell subtypes. F GO enrichment analysis demonstrating functional differences between C0 tumor cells and other tumor cell subtypes. G GSVA enrichment analysis revealing pathway differences between C0 tumor cells and other tumor cell subtypes. H GSEA enrichment analysis depicting pathway differences between C0 tumor cells and other tumor cell subtypes
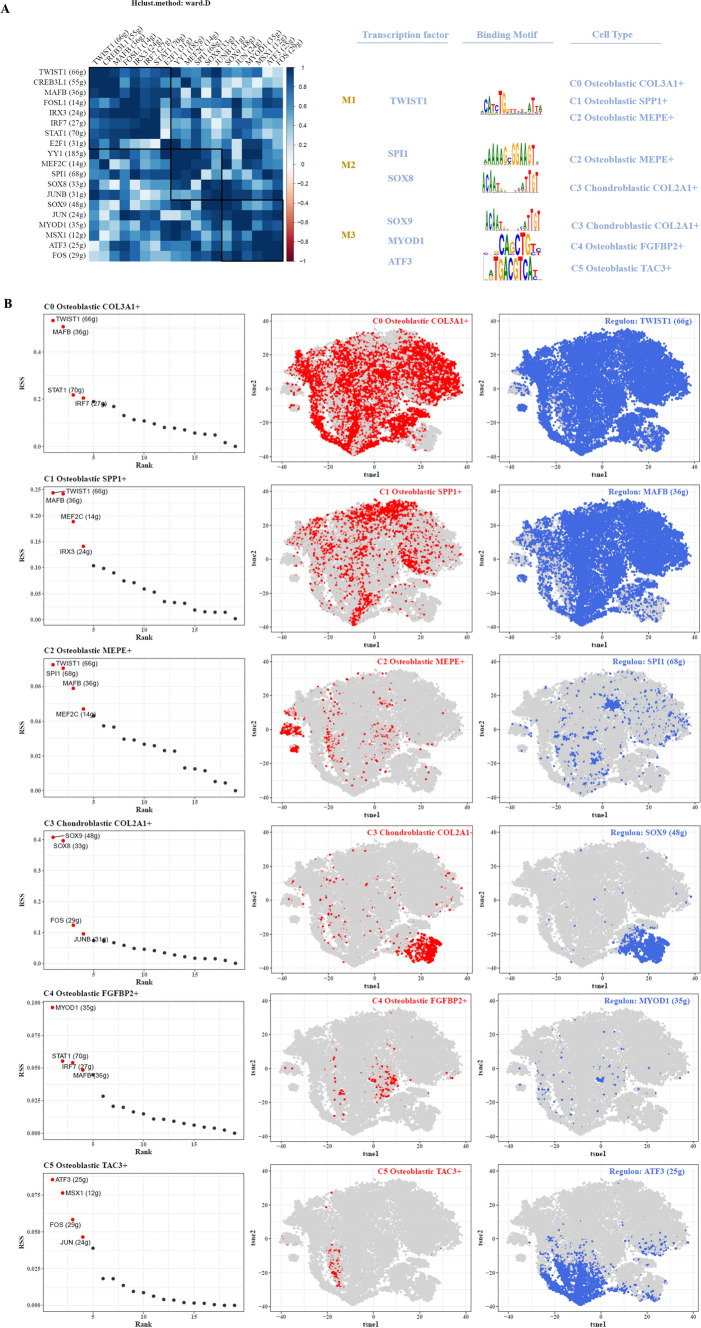
Transcription factor analysis
By computing CSI matrices for various transcription factors and applying clustering algorithms, we categorized them into three classes (M1, M2, M3). We illustrated representative transcription factors, their associated motifs, and relevant cell types (Fig. 7A). Next, we calculated RSS scores for each transcription factor across tumor cell subtypes and visualized these scores (Fig. 7B).
Fig. 7.
Transcription factor analysis. A Classification of tumor cell subtypes based on Connectivity Specificity Index (CSI) matrix, identifying regulatory modules, representative transcription factors, associated binding motifs, and relevant tumor cell subtypes. B Ranking of transcription factors based on Regulatory Specificity Score (RSS) across tumor cell subtypes highlighted on t-SNE
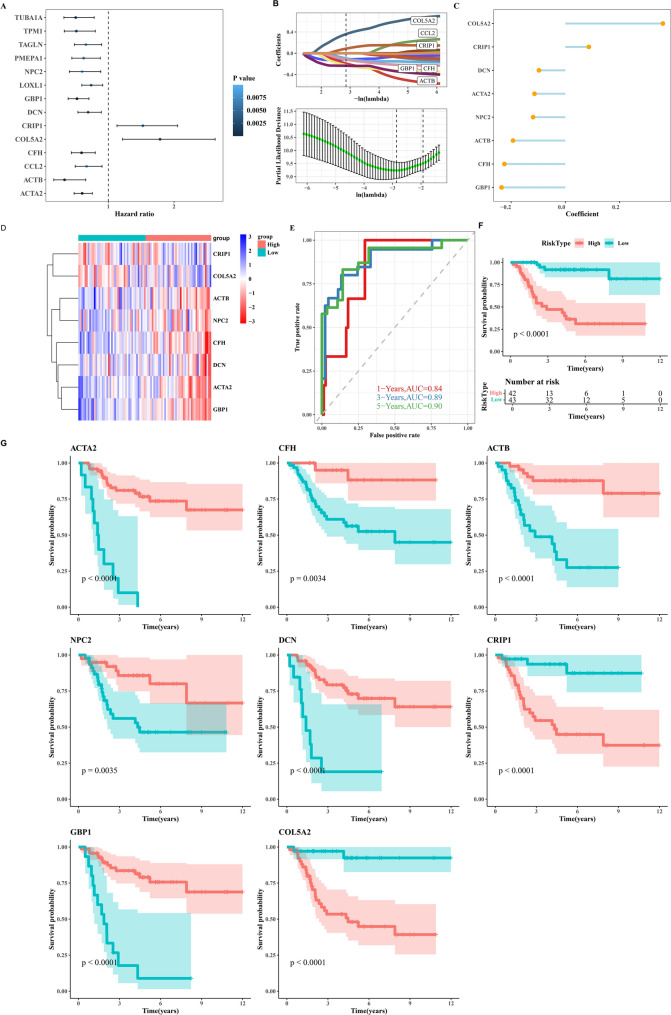
Construction of prognostic model using markers of C0 tumor cells
Using marker genes of C0 tumor cells (highest EMT score), we conducted single-factor Cox analysis to identify genes significantly associated with prognosis (p < 0.01, Fig. 8A). We established a prognostic model using LASSO+Cox algorithms (Fig. 8B) and displayed the coefficients of each gene in the model (Fig. 8C) along with their corresponding expression levels (Fig. 8D). Subsequently, survival analysis results and timeROC outcomes were presented (Fig. 8E, F). Figure 8G illustrates survival analysis results for each prognostic gene.
Fig. 8.
Prognostic model construction for C0 tumor cells. A Forest plot depicting results from univariate Cox analysis of marker genes in C0 tumor cells (highest EMT score). B Identification of 8 prognosis-related genes using LASSO (Least Absolute Shrinkage and Selection Operator) Cox regression analysis. C Dot plot displaying coefficients (coef) of genes comprising the risk score model. D Heatmap showing expression profiles of these genes across different groups. E ROC curves for risk scores at 1-year, 3-year, and 5-year time points. F Kaplan-Meier curves demonstrating survival differences between high-risk and low-risk groups based on the risk score. G Survival analysis results for the 8 prognosis-related genes
Immune infiltration and TMB analysis
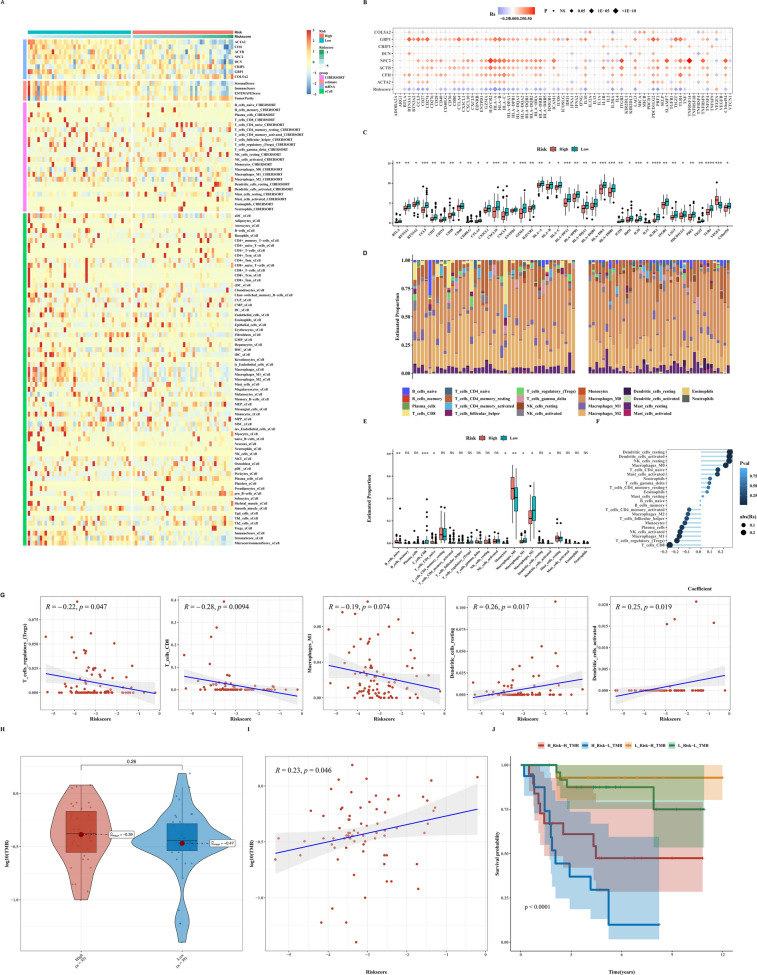
We depicted expression levels of prognostic genes in OSA and heatmap predictions of immune infiltration levels using ESTIMATE, CIBERSORT, and Xcell algorithms (Fig. 9A). We calculated risk scores, assessed correlations between prognostic genes and immune checkpoint genes, and examined differences in immune checkpoint genes between two groups (Fig. 9B, C). We then presented bar and box plots of immune cell proportions predicted by CIBERSORT and their correlations (Fig. 9D, E). Further analysis explored differences in TMB between groups, correlations between TMB and risk scores, and associations with prognosis (Fig. 9H–J).
Fig. 9.
Immune infiltration and TMB analysis. A Heatmap depicting ESTIMATE, CIBERSORT, and Xcell results in OSA, highlighting immune infiltration levels. B Bubble plot showing correlation analysis between immune checkpoint-related genes, risk scores, and prognosis-related genes. C Differential expression of immune checkpoint-related genes between high-risk and low-risk groups. D CIBERSORT analysis illustrating immune cell proportions in high-risk and low-risk groups. E Correlation analysis between immune infiltration cells and risk scores. F, G Correlation analysis between immune infiltration cells and risk scores. H–J TMB analysis results between low-risk and high-risk groups, and correlation analysis between TMB and risk scores, as well as with prognosis-related genes
Discussion
The comprehensive characterization of osteosarcoma (OSA) in this study provides valuable insights into the molecular and cellular mechanisms underlying tumor progression, metastasis, and therapeutic resistance. By integrating advanced genomic technologies and computational analyses, we aimed to address key gaps in current understanding and contribute to the development of targeted therapeutic strategies. This discussion explores the implications of our findings in relation to existing literature, highlights novel insights uncovered, and outlines future directions for research and clinical applications.
Our study utilized single-cell RNA sequencing (scRNA-seq) to dissect the heterogeneity of OSA tumors at unprecedented resolution. We identified distinct tumor cell subpopulations, characterized their transcriptional profiles, and delineated their roles in disease progression. This approach revealed previously unrecognized cellular states and rare cell populations within OSA, shedding light on potential drivers of treatment resistance and metastasis. These findings align with recent studies emphasizing the importance of tumor cell heterogeneity in influencing clinical outcomes and therapeutic responses [25].
One of the notable discoveries from our study is the role of epithelial-mesenchymal transition (EMT) in OSA pathogenesis. By employing gene set enrichment analyses (GSVA) and single-sample gene set enrichment analysis (ssGSEA), we demonstrated that high EMT scores correlate with aggressive tumor phenotypes and poor prognosis. This observation is consistent with findings in other cancer types, where EMT promotes invasion, metastasis, and resistance to therapy [26]. Our study extends this understanding to OSA, suggesting that targeting EMT pathways may represent a promising therapeutic strategy to mitigate disease progression and improve patient outcomes.
In addition to characterizing tumor cell heterogeneity, our study investigated the transcriptional regulatory networks governing OSA subtypes. Through computational analyses of transcription factor activity profiles, we identified key regulators associated with aggressive tumor phenotypes. This approach not only provides insights into the molecular underpinnings of OSA but also offers potential biomarkers for patient stratification and personalized treatment approaches. Similar studies have highlighted the significance of transcriptional dysregulation in cancer progression and its implications for therapeutic interventions [27].
Furthermore, our findings underscore the complex interplay between OSA tumors and the immune microenvironment. Analysis of immune cell infiltration patterns using algorithms such as ESTIMATE and CIBERSORT revealed distinct immune profiles associated with OSA subtypes. We observed correlations between immune cell signatures, tumor mutational burden (TMB), and patient outcomes, suggesting potential immunotherapeutic targets. These findings align with emerging evidence advocating for immune checkpoint inhibitors and adoptive cell therapies as promising avenues for treating refractory OSA [28].
Comparatively, our study contributes to the field by integrating multi-omics data to provide a holistic view of OSA biology. By correlating genomic alterations with clinical phenotypes, we identified candidate biomarkers predictive of treatment response and prognosis. These insights are crucial for advancing precision medicine approaches in OSA, where current treatment modalities often yield suboptimal outcomes due to tumor heterogeneity and acquired resistance mechanisms [29, 30].
Despite the advancements presented in this study, several limitations warrant consideration. First, while scRNA-seq offers unparalleled resolution in characterizing tumor cell populations, the interpretation of rare cell states and their functional significance requires further validation through experimental models and clinical cohorts. Secondly, the cross-sectional nature of our study limits temporal insights into tumor evolution and treatment responses over time. Longitudinal studies incorporating serial sampling could elucidate dynamic changes in tumor biology and therapeutic resistance mechanisms. Looking ahead, future research directions should focus on translating these molecular insights into clinical practice. Integration of multi-omics data with clinical outcomes data in large-scale prospective cohorts will be essential for validating biomarkers and therapeutic targets identified in our study. Additionally, leveraging artificial intelligence and machine learning algorithms could enhance predictive modeling and facilitate personalized treatment strategies for OSA patients.
In conclusion, our study contributes to advancing the understanding of OSA biology and highlights opportunities for precision medicine approaches. By unraveling the complexities of tumor heterogeneity, transcriptional regulation, and immune interactions, we aim to pave the way for novel therapeutic interventions that improve patient outcomes and ultimately achieve long-term disease control. Continued collaborative efforts between researchers, clinicians, and industry partners will be crucial in translating these scientific discoveries into clinical innovations for the benefit of patients with osteosarcoma worldwide.
Author contributions
JZ, XJ, and JW designed the study. DH, XC, HZ, JG, LC, and JL performed data analysis. DH, XC, HZ, and JG drafted the manuscript. JZ, XJ, and JW revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by Zhejiang Province Traditional Chinese Medicine Science and Technology Plan Project (2023ZL128) and Zhejiang Medical and Health Science and Technology Project (2022504276).
Data availability
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dongan He, Xiaoqian Che, Haiming Zhang, and Jiandong Guo have contributed equally to the work.
Contributor Information
Jinxi Zhang, Email: ice1998@sina.com.
Xin Jin, Email: 541385033@163.com.
Jianfeng Wang, Email: lwzyyx123456789@163.com.
References
- 1.Belayneh R, Fourman MS, Bhogal S, Weiss KR. Update on osteosarcoma. Curr Oncol Reports. 2021;23(6):71. [DOI] [PubMed] [Google Scholar]
- 2.Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, et al. Molecular biology of osteosarcoma. Cancers (Basel). 2020;12(8):2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–5. [DOI] [PubMed] [Google Scholar]
- 4.Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18(10):609–24. [DOI] [PubMed] [Google Scholar]
- 5.Moukengue B, Lallier M, Marchandet L, Baud’huin M, Verrecchia F, Ory B, et al. Origin and therapies of osteosarcoma. Cancers (Basel). 2022;14(14):3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daw NC, Billups CA, Rodriguez-Galindo C, McCarville MB, Rao BN, Cain AM, et al. Metastatic osteosarcoma. Cancer. 2006;106(2):403–12. [DOI] [PubMed] [Google Scholar]
- 7.Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y, et al. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci. 2020;21(19):6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris MA, Hawkins CJ. Recent and ongoing research into metastatic osteosarcoma treatments. Int J Mol Sci. 2022;23(7):3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang W, Wei L, Weng J, Yu F, Qin H, Wang D, et al. Advances in the research of osteosarcoma stem cells and its related genes. Cell Biol Int. 2022;46(3):336–43. [DOI] [PubMed] [Google Scholar]
- 10.Zhu P, Li T, Li Q, Gu Y, Shu Y, Hu K, et al. Mechanism and role of endoplasmic reticulum stress in osteosarcoma. Biomolecules. 2022;12(12):1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamborsky R, Kokavec M, Harsanyi S, Danisovic L. Identification of prognostic and predictive osteosarcoma biomarkers. Med Sci (Basel, Switzerland). 2019;7(2):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng J, Peng Y, Wang D, Ayesha K, Chen S. The interaction between osteosarcoma and other cells in the bone microenvironment: From mechanism to clinical applications. Front Cell Dev Biol. 2023;11:1123065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun. 2020;11(1):6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynn E, Almonte-Loya A, Fragiadakis GK. Single-cell multiomics. Ann Rev Biomed Data Sci. 2023;6:313–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh WC, Budiarto BR, Wang YF, Lin CY, Gwo MC, So DK, et al. Spatial multi-omics analyses of the tumor immune microenvironment. J Biomed Sci. 2022;29(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang XJ, Song XY, Wu JL, Liu D, Lin BY, Zhou HS, et al. Advances in multi-omics study of prognostic biomarkers of diffuse large B-cell lymphoma. Int J Biol Sci. 2022;18(4):1313–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinton K, Kirk A, Paul P, Persad S. Regulation of the epithelial to mesenchymal transition in osteosarcoma. Biomolecules. 2023;13(2):398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra R, Nathani S, Varshney R, Sircar D, Roy P. Berberine reverses epithelial-mesenchymal transition and modulates histone methylation in osteosarcoma cells. Mol Biol Rep. 2020;47(11):8499–511. [DOI] [PubMed] [Google Scholar]
- 19.Tian H, Zhou T, Chen H, Li C, Jiang Z, Lao L, et al. Bone morphogenetic protein-2 promotes osteosarcoma growth by promoting epithelial-mesenchymal transition (EMT) through the Wnt/β-catenin signaling pathway. J Orthop Res. 2019;37(7):1638–48. [DOI] [PubMed] [Google Scholar]
- 20.Jovic D, Liang X, Zeng H, Lin L, Xu F, Luo Y. Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med. 2022;12(3): e694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamperman T, Karperien M, Le Gac S, Leijten J. Single-cell microgels: Technology, challenges, and applications. Trends Biotechnol. 2018;36(8):850–65. [DOI] [PubMed] [Google Scholar]
- 22.Rajan S, Franz EM, McAloney CA, Vetter TA, Cam M, Gross AC, et al. Osteosarcoma tumors maintain intra-tumoral transcriptional heterogeneity during bone and lung colonization. BMC Biol. 2023;21(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohlenova K, Goveia J, García-Caballero M, Subramanian A, Kalucka J, Treps L, et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metabolism. 2020;31(4):862-77.e14. [DOI] [PubMed] [Google Scholar]
- 24.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M, et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12(3):670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao S, Piao L, Wang J, Guo L, Wang J, Wang L, et al. Tspan9 induces EMT and promotes osteosarcoma metastasis via activating FAK-Ras-ERK1/2 pathway. Front Oncol. 2022;12: 774988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du X, Wei H, Zhang B, Wang B, Li Z, Pang LK, et al. Molecular mechanisms of osteosarcoma metastasis and possible treatment opportunities. Front Oncol. 2023;13:1117867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–35. [DOI] [PubMed] [Google Scholar]
- 29.Cheng D, Zhang Z, Mi Z, Tao W, Liu D, Fu J, et al. Deciphering the heterogeneity and immunosuppressive function of regulatory T cells in osteosarcoma using single-cell RNA transcriptome. Comput Biol Med. 2023;165: 107417. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida A. Osteosarcoma: Old and new challenges. Surg Pathol Clin. 2021;14(4):567–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.