Abstract
Background
Autophagy is a crucial mechanism for maintaining cellular homeostasis and responding to environmental stress, and it is closely linked to tumor drug resistance. Through multi-omics analysis, this study explores the expression patterns, functions, and potential role of the autophagy-related gene Angiotensinogen (AGT) in colorectal cancer (CRC), particularly in relation to chemotherapy resistance.
Methods
This study first compared AGT expression between CRC and normal tissues using the GTEx and TCGA databases. Differences in expression were assessed using Wilcoxon Rank Sum Tests, and the prognostic impact of AGT was evaluated through univariate Cox survival analysis and meta-analysis. Functional enrichment was performed using the limma and fgsea packages. Drug sensitivity analysis was conducted based on the CTRP database, while immune infiltration was assessed using the CIBERSORT and ESTIMATE methods. Spatial transcriptomic characteristics were explored through 10x Visium technology and deconvolution analysis to investigate the correlation between AGT expression levels and tumor cell content.scRNA-seq data from CRC tissues were sourced from Tumor Immune Single Cell Hub (TISCH).Functional annotation was performed with Single-sample gene set enrichment analysis (SSGSEA), and pseudotime analysis using Monocle 2 mapped their developmental trajectories. The potential of AGT inhibitors in the treatment of CRC was analyzed using drug-target Mendelian randomization.Finally, Phenome-Wide Association Study (PheWAS) was conducted to evaluate genetic associations and potential side effects of AGT inhibitors.
Results
AGT expression was significantly higher in CRC tissues compared to normal tissues and was associated with shorter recurrence-free survival (RFS). Autophagy signaling pathways were markedly enriched in the high AGT expression group. AGT expression was positively correlated with resistance to chemotherapeutic agents such as gemcitabine, cisplatin, paclitaxel, and 5-fluorouracil. Spatial transcriptomic analysis revealed that AGT was predominantly expressed in malignant tumor regions. Single-cell analysis identified 21 distinct cell subpopulations across 13 major types. AGT expression was significantly higher in tumor samples, especially in the fibroblast C6 subpopulation. Tumor-related pathways were enriched in C1, C5, C6, and C8 subpopulations. Pseudotime analysis revealed that these subpopulations, particularly C6, were in terminal developmental stages.Drug-target Mendelian randomization analysis indicated a negative causal relationship between AGT inhibitors and the risk of both heart failure(ORdrug = 0.950, 95% CI, 0.912–0.990; P = 0.014) and CRC(ORdrug = 0.874, 95% CI: 0.792–0.964; P = 0.007).PheWAS analysis showed no genetic associations between AGT inhibitors and other traits, indicating its specificity and low risk of side effects.
Conclusion
Elevated AGT expression in CRC is associated with resistance to chemotherapy, and its inhibition may offer a therapeutic avenue for the treatment of colorectal cancer.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-024-01545-5.
Keywords: AGT, Colorectal cancer, Autophagy, Chemotherapy resistance, Drug-target mendelian randomization
Introduction
CRC is a common malignant tumor of the digestive tract. According to literature reports, as of 2020, the incidence of CRC ranked third globally, while its mortality rate ranked second [1]. In Asia, CRC accounts for nearly half of new global cases, and over half of the global mortality rate [2, 3]. Currently, the primary clinical treatment for CRC remains surgical removal of the tumor, supplemented by chemotherapy and radiotherapy [4, 5]. Although various treatment options are currently available for colorectal cancer, including surgery, chemotherapy, targeted therapies, and immunotherapies, the five-year survival rate for advanced-stage cases remains low, at approximately 14% [6–8].Given the progressive trend of CRC incidence, it has become urgent to explore new therapeutic approaches.
AGT is the precursor of angiotensin and a critical component of the renin-angiotensin-aldosterone system (RAAS), which regulates blood pressure and renal sodium retention [9]. Through a series of physiological transformations, the AGT gene produces angiotensin II (AGTII), a process that promotes the cell cycle and induces pronounced autophagy [10]. Autophagy, as a distinctive mechanism of programmed cell death, has emerged as a promising new strategy in cancer treatment [11, 12]. Recent studies have identified AGT as a potential biomarker driving colorectal cancer progression, facilitating CRC cell proliferation, migration, and invasion [13]. However, no research has yet explored the relationship between AGT gene expression and resistance to chemotherapy or immunotherapy in CRC.
RNA interference (RNAi) is a promising gene-silencing technology, and it has already been demonstrated in various animal models that targeted interference with hepatic AGT gene expression can significantly reduce blood pressure [14]. The ALN-AGT Phase I study also confirmed its significant antihypertensive effect, suggesting that this small interfering RNA could become a novel RAS inhibitor, potentially revolutionizing the treatment of hypertension and heart failure [15]. Hence, whether AGT-targeted RNAi therapies can be used in the treatment of colorectal cancer is the primary focus of this study. We employed Mendelian randomization (MR) analysis for drug targeting, using genetic variants that simulate pharmacological inhibition of drug gene targets as instrumental variables. This approach elucidates the long-term effects of medications and strengthens causal inference regarding the impact of these drug gene targets on diseases. Evidence shows that drug-target MR is nearly 70% effective in target exploration [16]. We collected recently published genome-wide association study (GWAS) summary statistics and, through MR analysis targeting drug genes, investigated the causal relationship between genetically predicted AGT inhibition and colorectal cancer.
Materials and methods
Data collection and processing
Transcriptomic data from 607 CRC samples were downloaded from the TCGA database, along with the corresponding clinical data and follow-up prognostic information. Additionally, datasets were retrieved from the GEO database using the following selection criteria: human colon cancer whole-genome expression profiling data, with samples containing both cancerous and adjacent non-cancerous tissues. This study selected the following datasets for further analysis: GSE17537, GSE103479, GSE106584, GSE12945, GSE28722, GSE29621, GSE39084, GSE87211, GSE31595, and GSE41258. The GSE17537 dataset includes 55 CRC samples; GSE103479 contains 60 samples; GSE106584 comprises 156 samples; GSE12945 includes 62 samples; GSE28722 contains 125 samples; GSE29621 has 65 samples; GSE39084 includes 70 samples; GSE87211 comprises 363 samples; GSE87211 contains 37 samples, and GSE41258 includes 182 samples. The transcriptomic datasets used in the study were derived from the “Human Colorectal Cancer: Whole Transcriptome Analysis” dataset available on the 10x Genomics website(https://www.10xgenomics.com/).
Expression and prognostic analysis of AGT in CRC
The TPM expression levels of normal samples from GTEx were paired with tumor TPM expression levels from TCGA. The Wilcoxon Rank Sum Test was employed to compare the statistical differences in expression levels between tumor tissues in the CRC dataset and normal tissues from TCGA and GTEx. The Human Protein Atlas (HPA) is an open-access database, providing free access to researchers from academia and industry to explore the human proteome. In this study, the HPA database (http://www.proteinatlas.org) was utilized to validate AGT protein expression in normal and tumor cells through immunohistochemistry. Meta-analysis was conducted on the results of univariate Cox survival analysis using the inverse variance method, with the logarithmic HR value as the primary measure. HR values were classified into two categories: less than 1 (indicating tumor suppressive activity) and greater than 1 (suggesting oncogenic potential). Statistical analysis and visualization were performed using the “Meta” package in R (version 4.3.2).
Functional enrichment analysis of AGT-related differentially expressed genes in CRC
Samples with the top 30% highest gene expression were categorized as the high-expression group, while those in the lowest 30% were designated as the low-expression group. Differential analysis was conducted using the limma package [17, 18] to obtain the log2FC for each gene. All genes were ranked according to their log2FC, and gene set enrichment analysis was performed using the fgsea package [19, 20], based on the KEGG gene sets. The enrichment score (ES) for each gene set was calculated, followed by significance testing and multiple hypothesis testing of the ES values. A p-value less than 0.05 and an adjusted p-value below 0.25 were considered significant and visualized accordingly.
The role of AGT gene in tumor immune infiltration and somatic mutations
Based on gene expression profile data, the samples were divided into high and low expression groups using the median expression value as a threshold. The CIBERSORT package [21] was employed to calculate immune cell content in the samples, and the non-parametric Wilcoxon rank-sum test was used to compare the differences in immune cell content between the two groups as evaluated by various software. Subsequently, the ESTIMATE algorithm [22, 23] was used to estimate the immune and stromal cell content in CRC individuals, and to assess immune scores, stromal scores, and tumor purity. The “maftools” package [24, 25] was utilized to analyze and visualize the top 20 most frequently mutated genes between the high and low expression groups.
Correlation between AGT and drug sensitivity in CRC
Spearman correlation analysis was used to compute the correlation between gene expression and the half-maximal inhibitory concentration (IC50) of antagonists measured in the CTRP database [26]. A negative correlation indicates that as gene expression increases, the cell line becomes more sensitive to the drug. Conversely, a positive correlation suggests that as gene expression increases, the cell line becomes more resistant to the drug. The pRRophetic package is primarily used to predict clinical chemotherapy response based on gene expression levels [27]. This R package, developed from the research of Paul Geeleher, Nancy Cox, and R. Stephanie Huang at the University of Minnesota, establishes ridge regression models based on the relationship between baseline gene expression levels and drug sensitivity in vitro cell lines. The Cancer Genome Project (CGP) cell line data was utilized to predict the IC50 of TCGA patients, aiming to forecast chemotherapy response. Spearman correlation analysis was employed to calculate the relationship between IC50 and gene expression levels.
Single-cell transcriptomic landscape of CRC
Single-cell RNA sequencing data from CRC tissues were extracted from the EMTAB8107 single-cell RNA sequencing dataset, available in the Tumor Immune Single-cell Hub (TISCH) database (http://tisch.compgenomics.org/home/). The values in the single-cell expression matrix were normalized using the “NormalizeData” method in Seurat, adjusting the raw counts (UMI) in each cell to 10,000. A unified analytical algorithm (MAESTRO) was employed for quality control, clustering, and cell type annotation for each dataset. The EMTAB8107 dataset comprises 23,176 cells from seven CRC tissues, and the single-cell RNA sequencing was performed using the 10× Genomics platform.
Single-cell transcriptomic landscape and enrichment analysis of fibroblasts
Fibroblast subpopulations were isolated, and principal component analysis was performed. The FindNeighbors and FindClusters functions were utilized to group cells into distinct subclusters based on their similarities, selecting the top 20 principal components with a resolution set at 0.2 for clustering. Finally, the SSGSEA algorithm was employed to evaluate the activity of each fibroblast subcluster across various hallmark gene sets, thereby assigning functional annotations to each subpopulation.
Pseudotime analysis of fibroblasts in scRNA-seq
Pseudotime analysis was conducted across all fibroblast subpopulations, using previously identified subpopulation markers to order the cells and assess the relationships between overall gene expression changes. The Monocle 2 algorithm was employed to construct the single-cell pseudotime trajectory of the fibroblast subpopulations, revealing distinct fates and functions exhibited by cells along each branch.
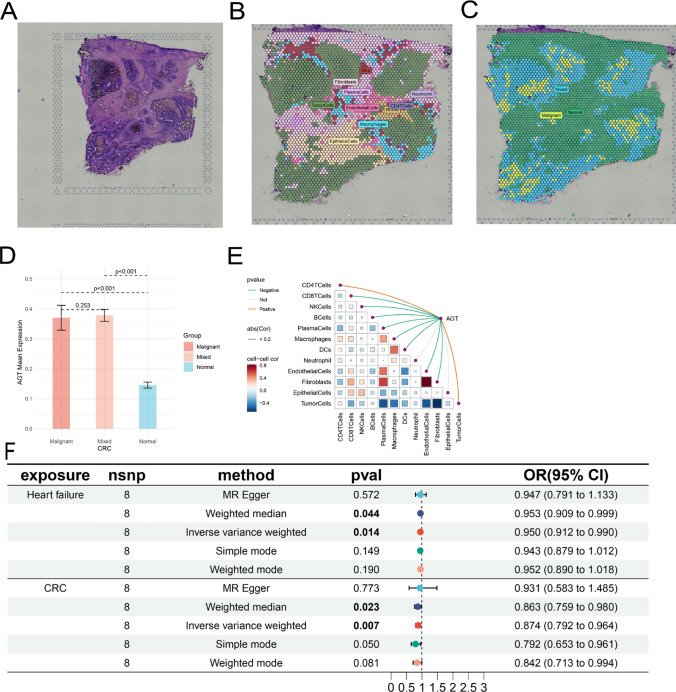
Molecular characteristics of AGT at the spatial transcriptomic level
To accurately assess the cellular composition at each spot on the 10x Visium slide, we employed deconvolution analysis. This technique is based on spatial transcriptomics (ST) and single-cell transcriptomics data, with particular consideration given to the specific cancer type. Initially, we gathered scRNA-seq data from multiple samples sharing the same cancer type, thereby constructing a comprehensive single-cell RNA reference library. To ensure the reliability of the analysis, we applied stringent quality control measures to the scRNA-seq data, based on the number of expressed genes, unique molecular identifier (UMI) counts, and the percentage of mitochondrial RNA per cell. The screening parameters were meticulously selected in reference to studies on the source of the single-cell transcriptomics data, ensuring both scientific rigor and accuracy in the filtering criteria. Next, we constructed a signature score matrix by calculating the average expression of the top 25 cell-type-specific genes in the scRNA-seq reference for each spot. Using the get_enrichment_matrix and enrichment_analysis functions from the Cottrazm package, we generated an enrichment score matrix, which provided robust support for the subsequent analysis of cellular composition. The SpatialFeaturePlot function from the Seurat package was then employed to visualize the enrichment scores for each cell type; the darker the color, the higher the enrichment score, indicating a greater presence of that cell type in the spot. Spots with a malignant cell score of 1 were classified as the Malignant group, those with a score of 0 as the Normal group, and others as the Mixed group. The Wilcoxon Rank Sum Test was used to evaluate the statistical significance of gene expression differences between the three groups. Spearman correlation analysis was employed to calculate the correlations between cell contents across all spots and between cell content and gene expression. The linkET package was used to visualize these correlations.
Selection of instrumental variables
The GWAS data for heart failure were derived from a study encompassing 208,187 individuals of European descent(GWAS ID: finn-b-I9_HEARTFAIL). Systolic blood pressure was chosen as the biomarker due to its established reduction through AGT inhibition, as demonstrated in clinical trials. Instrumental variables that could target AGT to lower systolic blood pressure were selected to simulate the effects of AGT inhibitors. The instrumental variables comprised single nucleotide polymorphisms (SNPs) located within ± 100 kb of the AGT locus, and these SNPs were associated with systolic blood pressure levels. The threshold for genome-wide significance was set at p < 5 × 10^−8. To minimize the influence of strong linkage disequilibrium (LD) on the results, an LD threshold of r^2 < 0.3 was applied [28]. Ultimately, eight significant AGT SNPs were retained (Supplementary File 1: Table S2). There was no overlap between the samples from the exposure and outcome GWAS studies, and all participants were of European ancestry.
Outcome data
Heart failure was used as a positive control dataset, with all datasets drawn from European populations. Additionally, we collected a colorectal cancer GWAS summary dataset, including 1,803 cases and 174,006 controls(GWAS ID: finn-b-C3_COLON_EXALLC), as the primary outcome. These data were obtained from the Finnish database (https://www.finngen.fi/fi).
Drug target mendelian randomization analysis
AGT inhibitors have been employed in the treatment of refractory hypertension and heart failure. Therefore, we utilized the GWAS summary data for heart failure as a positive control outcome to validate the effectiveness of the instrumental variables. First, we harmonized the drug-targeted instrumental variables associated with the exposure and the outcome dataset. Five MR methods were applied to assess the causal relationship between the drug-targeted instrumental variables and the outcome dataset. These methods included: MR-Egger regression(MR-Egger), Weighted Median Estimator (WME), Inverse Variance Weighted (IVW), Simple Mode(SM), and Weighted Mode(WM).The IVW method serving as the primary MR analysis. A p-value < 0.05 was set as the significance threshold for the MR results. Cochran’s Q statistic was used to evaluate the heterogeneity of the genetic instruments, with p > 0.05 indicating no significant heterogeneity. MR-Egger regression and MR-PRESSO were employed to assess the horizontal pleiotropy of the genetic instruments, where p > 0.05 suggested no evidence of pleiotropy. To verify whether weak instrument bias was present in the selected instrumental variables, we calculated the F-statistic, with F > 10 indicating no evidence of weak instrument bias, further reinforcing the validity of the causal inference. The F-statistic was calculated using the formula F = [(N - K − 1) / K] × [R^2 / (1 - R^2)], where N represents the sample size of the exposure, K denotes the number of instrumental variables, and R^2 refers to the proportion of variance in the exposure explained by the instrumental variables. These steps ensure the reliability of the instrumental variables in evaluating causal relationships.
Phenome-wide association study
To thoroughly evaluate the broad effects of potential drug targets and their possible side effects, we employed AstraZeneca’s Phenome-Wide Association Study (PheWAS) platform (https://azphewas.com/) for phenotype association analysis. This analysis was based on exome sequencing data publicly available from the UK Biobank, covering a subset of approximately 450,000 participants, specifically including around 15,500 binary phenotypes and approximately 1,500 continuous phenotype data points.
Results
Expression and prognostic analysis of AGT in CRC
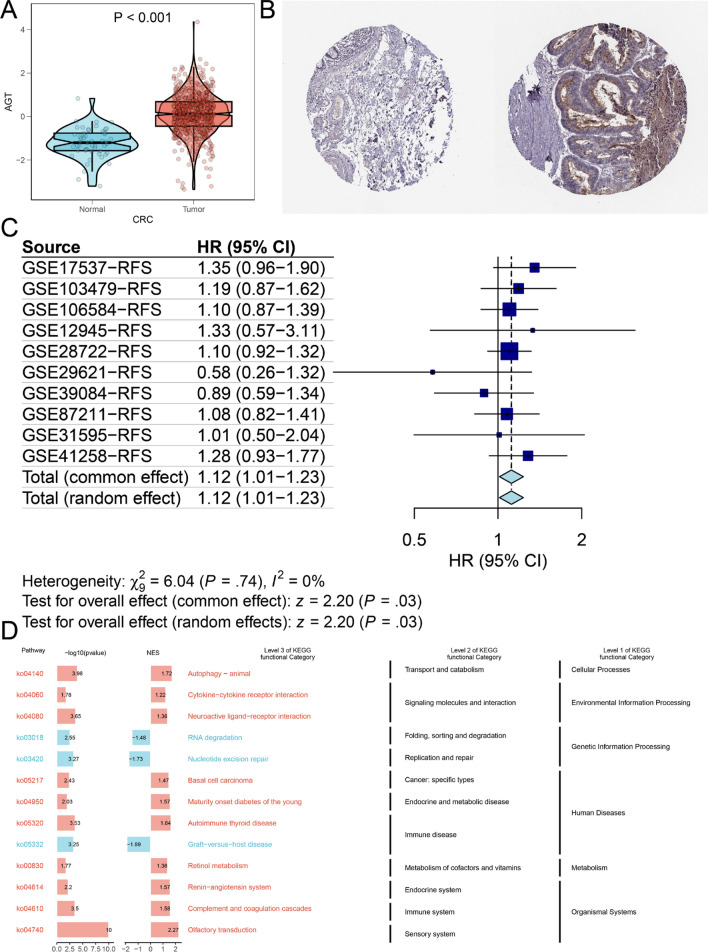
According to the analysis of the TCGA database, AGT expression is elevated in CRC tissues compared to normal tissues (Fig. 1A). Immunohistochemical validation of AGT expression in normal and CRC tissues, using data from the HPA database, revealed that AGT protein levels were significantly higher in CRC (Fig. 1B). A meta-analysis of univariate Cox survival results from 10 datasets reporting RFS showed no significant heterogeneity among the studies (I² = 0.00%, p = 0.740), thus a fixed-effects model was applied. The combined effect result yielded an HR of 1.12 (95% CI: 1.01–1.23), indicating a significant correlation between AGT expression levels and RFS (z = 2.20, p < 0.05), suggesting that patients with higher AGT expression have shorter RFS(Fig. 1C).
Fig. 1.
A Differential expression of AGT between tumor and normal groups (TCGA combined with GTEx analysis); B Immunohistochemical validation of AGT expression in normal and CRC tissues from the HPA database (Left: normal tissue, Right: tumor tissue); C Meta-analysis of univariate Cox survival analysis across multiple datasets; D KEGG gene set enrichment analysis
Functional enrichment analysis of AGT-related differentially expressed genes in CRC
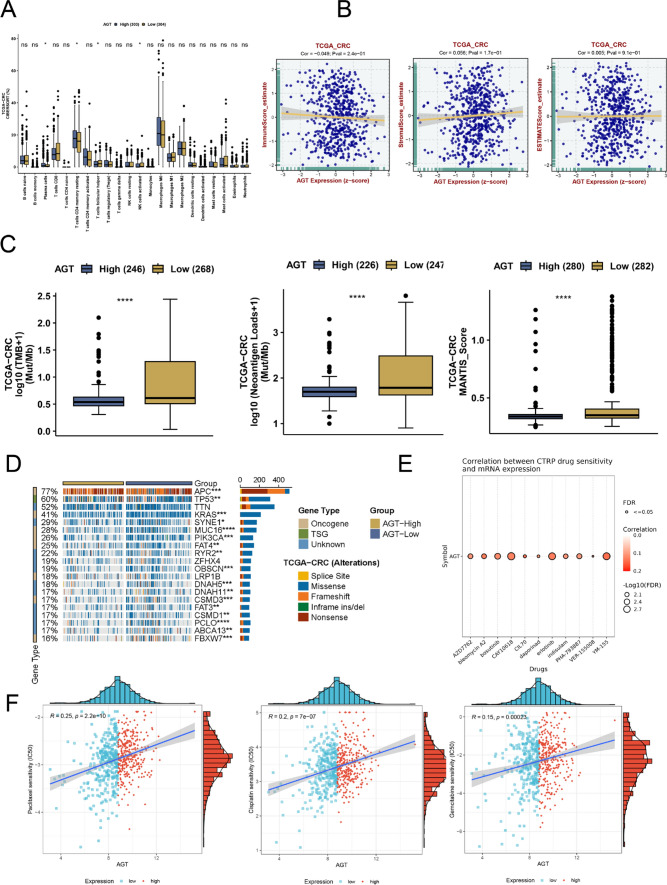
Further pathway enrichment analysis of AGT using fgsea revealed that autophagy-related pathways were significantly enriched in the AGT high-expression group (Fig. 1D). Given the close relationship between autophagy and gene mutations, we examined the association between AGT and mutated genes in CRC. The findings suggested that varying AGT expression levels were linked to distinct genetic alterations. Figure 2C demonstrated higher tumor mutational burden, neoantigen load, and mantis scores in the AGT low-expression group. Additionally, Fig. 2D highlighted significant differences in mutation statuses between the low and high AGT expression groups. In the high-expression group, a higher frequency of TP53 and APC mutations was observed, with mutated APC and TP53 potentially playing a role in the progression of CRC under the influence of AGT, particularly TP53, whose importance in chemotherapy resistance has been documented across various cancers.
Fig. 2.
A Analysis of immune cell infiltration proportions between high and low AGT expression groups (CIBERSORT); B Correlation analysis of AGT expression with ESTIMATE scores, immune scores, and stromal scores in the TCGA colorectal cancer cohort; C Comparison of tumor mutational burden, neoantigen load, and MANTIS scores between high and low AGT expression groups; D Comparison of somatic mutation frequencies between high and low AGT expression groups; E Spearman correlation between AGT expression and IC50 values from the CTRP database; F Spearman correlation between AGT expression and drug sensitivity IC50 values
Correlation between AGT and immune infiltration
Using the CIBERSORT method, we predicted the scores of 22 types of immune cells for each colorectal cancer patient. The Wilcoxon test was employed to compare the differences in immune cell scores between high and low AGT expression groups. As shown in Fig. 2A, we observed significant differences in certain NK cell scores between the two groups. Following this, we used the ESTIMATE package to predict tumor purity. The results revealed no significant correlation between immune and stromal scores and tumor purity in the AGT high and low expression groups (Fig. 2B).
Correlation between AGT and drug sensitivity in CRC
AGT expression showed a positive correlation with sensitivity scores for common chemotherapeutic agents, such as gemcitabine, cisplatin, paclitaxel, and 5-fluorouracil (p < 0.05, Fig. 2E-F and Supplementary File 1: Table S1). These findings suggest that high AGT expression is associated with chemotherapy resistance in CRC patients.
Single-cell transcriptomic landscape of CRC
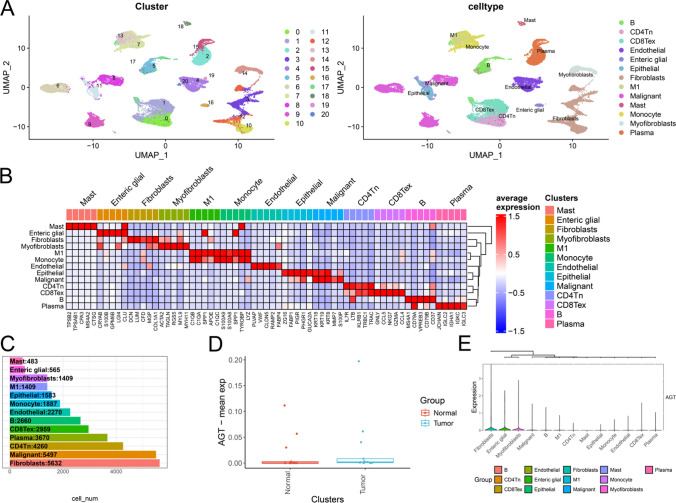
All cells were divided into 21 subclusters, which were further classified into 13 major cell types: B cells, CD4 + naive T cells (CD4Tn), exhausted CD8 + T cells (CD8Tex), endothelial cells, enteric glial cells, epithelial cells, fibroblasts, M1 macrophages, malignant cells, mast cells, monocytes, myofibroblasts, and plasma cells (Fig. 3A). Figure 3C illustrates the cell counts and their proportions across different groups. Subsequently, the FindAllMarkers function was employed with logfc set to 0.5 (fold change) and Minpct set to 0.35 (minimum expression ratio of differential genes) to identify marker genes for each subcluster, and the top 5 genes for each subcluster were visualized in a heatmap (Fig. 3B). Using the single-cell dataset, we analyzed AGT expression between normal and tumor samples as well as among various cell types. As shown in Fig. 3D, AGT expression in normal samples is slightly lower than in tumor samples, while Fig. 3E demonstrates that AGT expression in fibroblasts is significantly higher than in other cell types.
Fig. 3.
A UMAP plot showing 21 distinct cell subpopulations categorized into 13 major cell types; B Heatmap displaying the top five marker genes for each cell subpopulation; C Bar plot showing the number and proportion of each cell type across normal and tumor groups; D AGT expression levels in normal vs. tumor samples, indicating higher expression in tumor tissues; E AGT expression across different cell types
Single-cell transcriptomic landscape and enrichment analysis of fibroblasts
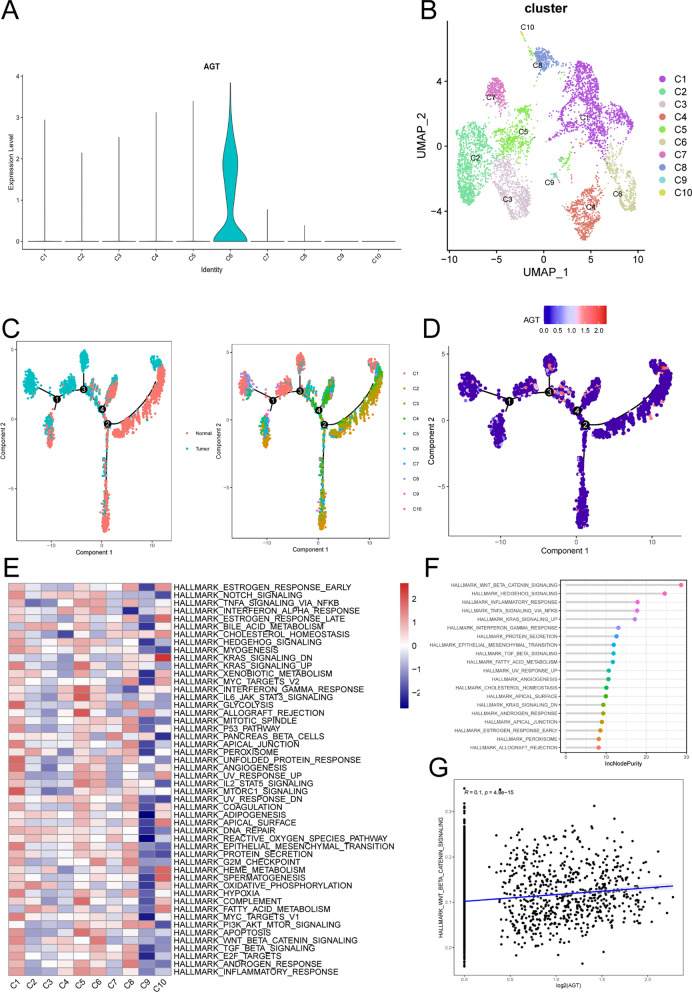
Re-clustering analysis of fibroblasts revealed that they remain divided into 10 distinct subpopulations (Fig. 4B). Among these, the C6 subpopulation exhibits significantly higher AGT expression compared to others (Fig. 4A). To uncover the association between fibroblast subpopulations and tumor progression, we investigated the characteristics of various tumor-related pathways within the 10 fibroblast subpopulations. Figure 4E illustrates the SSGSEA scores of multiple tumor-associated pathways across different fibroblast subpopulations. The results indicate that tumor-associated pathways are predominantly enriched in the C1, C5, C6, and C8 subpopulations. Using the random forest algorithm, we identified the WNT signaling pathway as the most significant cancer hallmark in the C6 subpopulation (Fig. 4F) and discovered a positive correlation between AGT expression and WNT signaling pathway activation (Fig. 4G). This suggests that AGT may play a pivotal role in activating the WNT signaling pathway in the C6 subpopulation, further supporting its potential role in cancer.
Fig. 4.
A AGT expression across 10 fibroblast subpopulations; B UMAP plot showing 10 fibroblast subpopulations (C1–C10) identified in CRC tissues; C Pseudotime trajectory analysis of fibroblast subpopulations; E Heatmap displaying the activity of hallmark pathways across the fibroblast subpopulations; F Random forest analysis indicating the importance of WNT signaling in the C6 subpopulation; G Correlation plot showing the relationship between AGT expression and WNT pathway activation
Pseudotime analysis of fibroblasts in scRNA-seq
To further validate the developmental stages of fibroblast subpopulations, we performed pseudotime analysis using Monocle2 software. The results indicate that cells within the C1, C5, C6, and C8 subpopulations are at the terminal stages of development (Fig. 4C). Figure 4D illustrates the trend of AGT gene expression along the pseudotime trajectory, revealing that AGT is predominantly expressed during the terminal stages of tumor-associated fibroblast development.
Molecular characteristics of AGT at the spatial transcriptomic level
AGT expression appears to colocalize with tumor cells, suggesting that in colorectal cancer, AGT is predominantly expressed within the tumor. AGT expression is highest in malignant regions and lowest in normal areas, indicating that AGT is primarily expressed by tumor cells (Fig. 5A–D). Consistent with previous localization results, AGT expression is significantly positively correlated with the proportion of tumor cells in each spot (Fig. 5E).
Fig. 5.
A Tissue imaging; (B) Post-deconvolution cell composition for each spot, showing the maximum value; C Spatial resolution of malignant (Malignant), mixed malignant (Mixed), and normal (Normal) regions; D Differential AGT expression across malignant, mixed, and normal regions; E Spearman correlation between gene expression and microenvironment components at spatial resolution; F Results and forest plot of Mendelian randomization analysis of the causal relationship between AGT inhibition and CRC
Positive control analysis
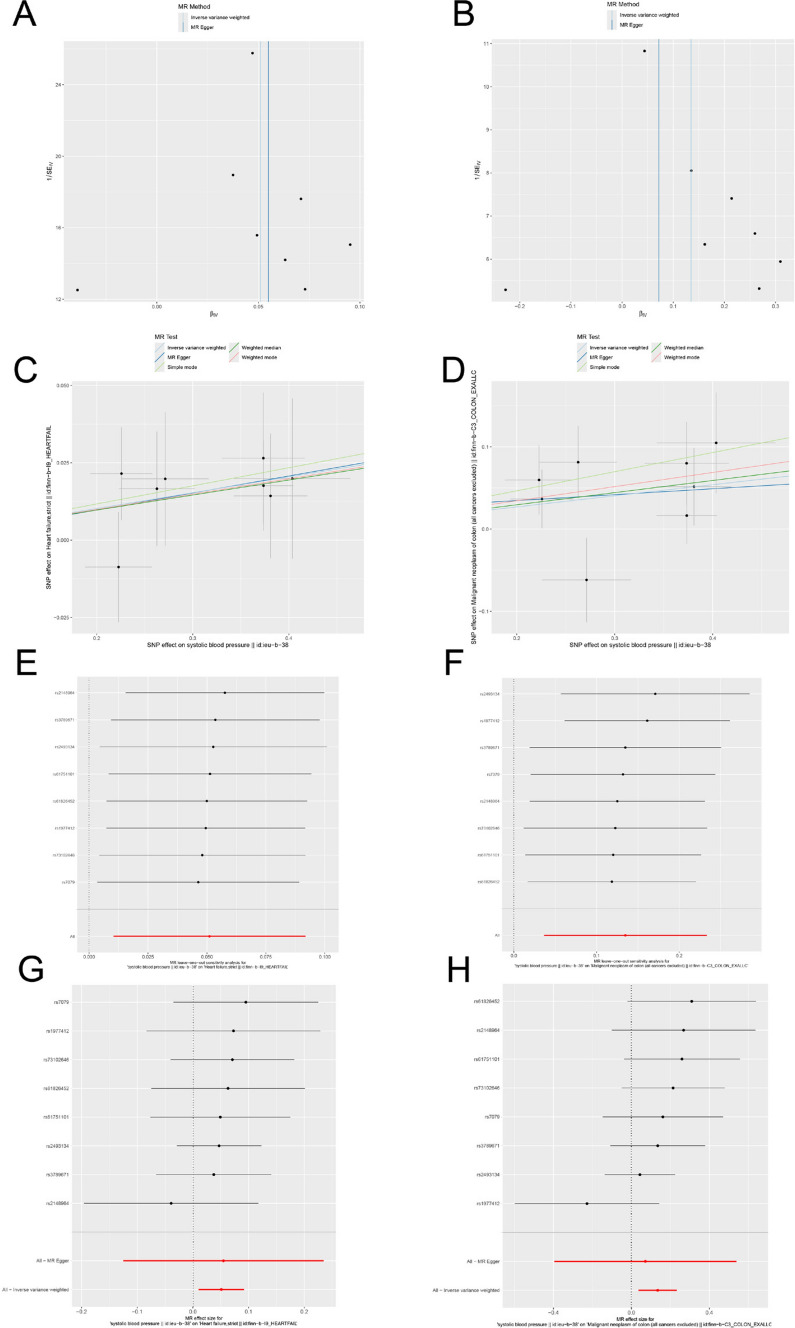
The MR analysis was conducted with a stringent p-value threshold of 5 × 10^−8. Using these criteria (r^2 < 0.3, kb = 100 KB), eight significant AGT SNPs were identified. The F-statistics for the instrumental variables (IVs) were all greater than 10, indicating the absence of weak instrument bias and confirming the reliability of the results (Supplementary File 1: Table S2). Genetic variation at the AGT inhibitor target was associated with a reduced risk of heart failure, specifically showing a negative causal relationship between AGT inhibitors and heart failure (ORdrug = 0.950, 95% CI, 0.912–0.990; P = 0.014). Cochran’s Q test revealed no significant heterogeneity between genetic variants at the AGT inhibitor target and gout (P > 0.05). Based on the MR-Egger intercept and MR-PRESSO tests, there was no evidence to support horizontal pleiotropy in the genetic variants at the AGT inhibitor target (Fig. 5F and Supplementary File 1: Table S3). The effect of each SNP on these diseases is shown in Fig. 6C and G, and the funnel plot similarly demonstrated the absence of bias in the study (Fig. 6A). Additionally, leave-one-out sensitivity analysis was employed to assess the impact of each SNP on the overall causal relationship, and the results showed no significant differences in the observed causal effect when systematically removing individual SNPs and repeating the MR analysis (Fig. 6E), proving that the estimated effect cannot be attributed to any single genetic instrument. The positive control analysis confirmed that the genetic instruments selected for AGT inhibition were appropriate.
Fig. 6.
A–B Funnel plots demonstrating the symmetry of the causal effect of AGT inhibitor on heart failure and colorectal cancer; The black dots in the figure represent single nucleotide polymorphisms, with the horizontal axis displaying the β values of these polymorphisms, and the vertical axis showing their standard errors. C–D Scatter plots of the five MR analysis methods; The vertical axis denotes the influence of single nucleotide polymorphisms on exposure variables, the horizontal axis delineates their impact on outcome variables. E–F Leave-one-out analysis of statistically significant variations in the causal effects of AGT inhibitor on heart failure and colorectal cancer; The Y-axis corresponds to each excluded rsid number and the ‘all’ condition not excluded by the IVW method. The X-axis corresponds to specific IVW values. G–H Forest plot of each SNP’s causal relationship with heart failure and colorectal cancer
Causal relationship between simulated AGT inhibitors and CRC
The MR analysis revealed a significant association between CRC and AGT inhibitors (Fig. 5F and Supplementary File 1: Table S4). Specifically, AGT inhibition showed a negative causal relationship with CRC (OR = 0.874, 95% CI: 0.792–0.964; P = 0.007). Cochran’s Q test indicated no significant heterogeneity between genetic variants at the AGT inhibitor target and CRC (P > 0.05). The MR-Egger intercept and MR-PRESSO tests provided no evidence of horizontal pleiotropy in the genetic variants associated with AGT inhibition (Supplementary File 1: Table S4), and the funnel plot similarly confirmed the absence of bias in the study (Fig. 6B). The impact of each SNP on the disease is illustrated in Fig. 6D and H. Additionally, leave-one-out sensitivity analysis was performed to assess the influence of each SNP on the overall causal relationship. The results demonstrated no significant differences in the causal effect when systematically removing individual SNPs and repeating the MR analysis (Fig. 6F), confirming that the estimated effect cannot be attributed to any single genetic instrument.
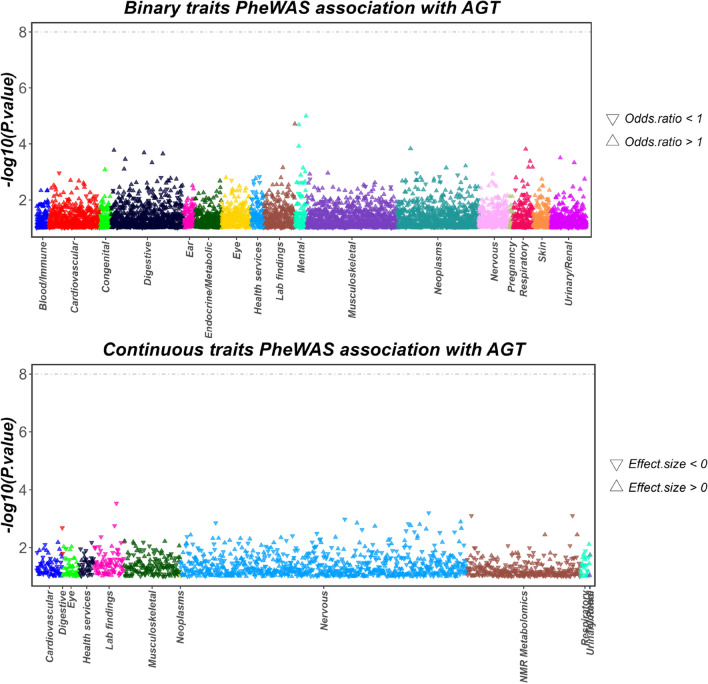
Phenome-wide association study
According to the PheWAS results, no significant genetic associations were found between the AGT gene and other traits (P < 5E − 8)(Fig. 7). This/ outcome suggests that drugs targeting AGT are unlikely to trigger pleiotropic effects at the genetic level, meaning they are less likely to influence multiple traits or produce unintended side effects. The lack of a significant association further strengthens the reliability of the study’s findings, indicating that AGT, as a drug target, possesses high specificity and potentially carries a lower risk of adverse effects.
Fig. 7.
PheWAS analysis showing associations of AGT with both binary and continuous traits
Discussion
In this study, we systematically explored the role of elevated expression of the autophagy-related gene AGT in tumorigenesis and its contribution to drug resistance in cancer, leading us to hypothesize that targeting AGT inhibition may serve as an effective therapeutic approach for colorectal cancer. AGT, an autophagy-related gene, is also the sole known source of Angiotensin downstream of the RAAS, primarily produced in the liver, and is a key regulator of RAAS [29]. Inhibiting liver-derived AGT not only circumvents the body’s compensatory mechanisms but also protects renal RAS activity, making it an ideal target for hypertension treatment [30]. With advances in RNAi and gene-editing technologies, targeting liver-derived AGT for hypertension therapy has already progressed to clinical trials [15, 31]. Consequently, we employed drug-target Mendelian randomization analysis to simulate the effects of AGT inhibitors on colorectal cancer treatment. To our knowledge, this is the first large-scale genetic consortium-based MR analysis establishing a causal link between AGT inhibitors and colorectal cancer. Through drug-target Mendelian randomization analysis, we conclude that AGT inhibitors can significantly reduce the risk of developing colorectal cancer.
Through the analysis of the CTRP database and the pRRophetic software package, we investigated the impact of AGT gene expression on the sensitivity of common chemotherapeutic agents used in colorectal cancer treatment. The results revealed a significant positive correlation between AGT expression and the IC50 values of fluorouracil, paclitaxel, cisplatin, and gemcitabine, indicating that patients with high AGT expression exhibited resistance to these four chemotherapeutic drugs.
Given the critical influence of the tumor immune microenvironment on treatment outcomes and prognosis, we further examined the relationship between AGT expression and the immune microenvironment. Although the CIBERSORT algorithm showed higher NK cell infiltration in the low AGT expression group, potentially associated with an immunoinflammatory phenotype, the ESTIMATE score indicated no significant correlation between immune and stromal scores and tumor purity across high and low AGT expression groups [12, 32]. This suggests that the immune microenvironment has a limited impact on the chemotherapy and immunotherapy resistance observed in patients with high AGT expression.
We then evaluated tumor mutation profiles to explore potential links between AGT gene expression and treatment response. Our analysis of mutational burden and specific gene mutation frequencies revealed that the high AGT expression group had a lower mutational burden but a higher frequency of mutations in key genes, such as the chemotherapy resistance gene TP53. Chemotherapeutic drugs typically eliminate cancer cells by inducing DNA damage or inhibiting cell division. If TP53 functions normally, it triggers apoptosis upon detecting DNA damage, thereby preventing further proliferation of cancer cells. However, when TP53 is mutated, the apoptosis process fails to initiate properly, allowing cancer cells to survive chemotherapy, leading to resistance. Studies indicate that in colorectal cancer, TP53 mutations are not only associated with tumor aggressiveness and poor prognosis but also contribute to resistance against common chemotherapeutic agents, as these drugs rely on TP53’s intact function to be effective.Additionally, pathway analysis between high and low AGT expression groups indicated a significant enrichment of autophagy-related pathways in the high-expression group. We found that autophagy has been implicated in promoting tumor cell survival during various cancer treatments and contributes to the development of treatment resistance [33]. For instance, Samaddar et al. [34]demonstrated that estrogen receptor-positive breast cancer cell lines, which exhibit intrinsic resistance to the selective estrogen receptor modulator tamoxifen, regained sensitivity following autophagy inhibition. Similarly, in prostate cancer, autophagy inhibition overcame resistance to the androgen receptor inhibitor enzalutamide [35]. Furthermore, in gastrointestinal stromal tumors, imatinib induced autophagy in tumor cells, while autophagy inhibition by lysosome-promoting agents such as chloroquine enhanced tumor cell apoptosis [36]. Nevertheless, although autophagy inhibition demonstrates potential in overcoming drug resistance in certain cancer treatments, prolonged inhibition of autophagy can similarly lead to resistance, particularly in CRC. Research indicates that the Wnt/β-catenin signaling pathway plays a crucial role in suppressing tumor cell apoptosis and facilitating epithelial-mesenchymal transition. Cancer-associated fibroblasts activate the Wnt/β-catenin signaling pathway, inhibiting the expression of apoptosis regulator 1 and F-box/WD repeat-containing protein 7, thereby suppressing mitochondrial apoptosis and enhancing the resistance of CRC cells to the chemotherapeutic agent 5-FU [37]. Further studies corroborate this observation. For instance, CAF-secreted CXCL12 activates the CXCR4/Wnt/β-catenin signaling pathway in ovarian cancer cells, inducing EMT and increasing resistance to cisplatin [38].
These findings suggest that high AGT expression may be associated with a higher TP53 mutational burden and the activation of the Wnt pathway, resulting in the inhibition of autophagy and consequently influencing tumor sensitivity to chemotherapy.
The limitations of this study are as follows: (1) As the study is primarily based on the results from the TCGA database, it does not allow for an in-depth exploration of the underlying biological mechanisms [39, 40]. (2) Since the drug-target Mendelian randomization analysis was conducted exclusively on a European population, the generalizability of the conclusions may be restricted in other ethnic groups. Therefore, future research should not only include in vitro and in vivo experiments for further validation but also expand to different populations for more nuanced, stratified analysis.
Conclusion
In conclusion, this study assessed the correlation between AGT expression and CRC chemotherapy resistance, revealing that AGT inhibitors can reduce the risk of CRC development.
In future clinical practice, AGT expression levels could serve as a valuable biomarker to identify patients who are likely to benefit from AGT-targeted therapies, enabling more personalized and effective treatment strategies. Moreover, the study’s insights into the spatial distribution and cellular heterogeneity of AGT expression could facilitate the development of precision medicine approaches, integrating AGT inhibitors with other therapeutic modalities tailored to the tumor microenvironment. These findings provide promising preliminary evidence for broadening the therapeutic applications of AGT inhibitors.
Electronic supplementary material
Acknowledgements
We gratefully acknowledge the authors and participants of all data from which we used in the study.
Author contributions
T-X, XL-L and WJ-C designed this study. CF conducted analyses and drafted the manuscript. CF directed the analytical strategy and supervised the study from conception to completion. T-X, WJ-C, and CF performed the data analyses.CF and WJ-C revised the manuscript draft. All authors contributed to the interpretation of data and critically revised the manuscript.
Funding
This study is supported by the funding from the Anhui Provincial Health Commission (No.AHWJ2023BAa20028).
Data availability
Single-cell RNA sequencing data from CRC patients were retrieved from the TISCH database (http://tisch.compgenomics.org/home/). Batch RNA-seq data were obtained from TCGA database(https://portal.gdc.cancer.gov/) and GSE17537, GSE103479, GSE106584, GSE12945, GSE28722, GSE29621, GSE39084, GSE87211, GSE31595, and GSE41258. in the GEO database, with the raw datasets being annotated using the corresponding files. The transcriptomic datasets used in the study were derived from the “Human Colorectal Cancer: Whole Transcriptome Analysis” (https://www.10xgenomics.com/). GWAS data for CRC and heart failure were accessed from the Finngen database (https://www.finngen.fi/fi).
Declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenjiao Cai and Tao Xiang contributed equally.
Contributor Information
Xiaoli Liu, Email: Liuxli369@163.com.
Chong Fu, Email: fclucky777@gmail.com.
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Cancer J Clin 2023. 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 3.Gu Y, Jiang L, Shui M, Luo H, Zhou X, Zhang S, Jiang C, Huang J, Chen H, Tang J, et al. Revealing the association between east Asian oral microbiome and colorectal cancer through mendelian randomization and multi-omics analysis. Front Cell Infect Microbiol. 2024;14:1452392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin 2014, 64(1). [DOI] [PubMed] [Google Scholar]
- 5.Chi H, Huang J, Yan Y, Jiang C, Zhang S, Chen H, Jiang L, Zhang J, Zhang Q, Yang G, et al. Unraveling the role of disulfidptosis-related LncRNAs in colon cancer: a prognostic indicator for immunotherapy response, chemotherapy sensitivity, and insights into cell death mechanisms. Front Mol Biosci. 2023;10:1254232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakatos G, Köhne C-H, Bodoky G. Current therapy of advanced colorectal cancer according to RAS/RAF mutational status. Cancer Metastasis Rev. 2020;39(4):1143–57. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Zhang G, Wang M, Guo SB, Wang F, Li Y, Kadier K, Zhou Z, Zhang P, Chi H, et al. Artificial intelligence hybrid survival assessment system for robot-assisted proctectomy: a retrospective cohort study. JCO Precis Oncol. 2024;8:e2400089. [DOI] [PubMed] [Google Scholar]
- 8.He R, Huang S, Lu J, Su L, Gao X, Chi H. Unveiling the immune symphony: decoding colorectal cancer metastasis through immune interactions. Front Immunol. 2024;15:1362709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang L, Ren X, Yang J, Chen H, Zhang S, Zhou X, Huang J, Jiang C, Gu Y, Tang J, et al. Mitophagy and clear cell renal cell carcinoma: insights from single-cell and spatial transcriptomics analysis. Front Immunol. 2024;15:1400431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savoia C, Burger D, Nishigaki N, Montezano A, Touyz RM. Angiotensin II and the vascular phenotype in hypertension. Expert Rev Mol Med. 2011;13:e11. [DOI] [PubMed] [Google Scholar]
- 11.Huang X, Chi H, Gou S, Guo X, Li L, Peng G, Zhang J, Xu J, Nian S, Yuan Q. An aggrephagy-related LncRNA signature for the prognosis of pancreatic adenocarcinoma. Genes (Basel) 2023. 10.3390/genes14010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun H, Wang H, Li XIN, Hao Y, Ling JUN, Wang H, Wang F, Xu F. Increased MAD2L2 expression predicts poor clinical outcome in colon adenocarcinoma. Biocell. 2023;47(3):607–18. [Google Scholar]
- 13.Shimomoto T, Ohmori H, Luo Y, Chihara Y, Denda A, Sasahira T, Tatsumoto N, Fujii K, Kuniyasu H. Diabetes-associated angiotensin activation enhances liver metastasis of colon cancer. Clin Exp Metastasis. 2012;29:915–25. [DOI] [PubMed] [Google Scholar]
- 14.Daugherty A, Lu HS, Bakris GL. Angiotensinogen in sex and hypertension: new insights from the multi-ethnic study of atherosclerosis (MESA). American College of Cardiology Foundation: Washington DC. 2023;81:1260–2. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann TS, Karsten V, Chan A, Chiesa J, Boyce M, Bettencourt BR, Hutabarat R, Nochur S, Vaishnaw A, Gollob J. Clinical proof of concept for a novel hepatocyte-targeting GalNAc-siRNA conjugate. Mol Ther. 2017;25(1):71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu C, Wang L, Cai W. IL6 receptor inhibitors: exploring the therapeutic potential across multiple diseases through drug target mendelian randomization. Front Immunol. 2024;15:1452849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Jiang C, Jiang L, Chen H, Huang J, Gao X, Xia Z, Tran LJ, Zhang J, Chi H, et al. Construction of a diagnostic model for hepatitis B-related hepatocellular carcinoma using machine learning and artificial neural networks and revealing the correlation by immunoassay. Tumour Virus Res. 2023;16:200271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. Biorxiv 2016: 060012.
- 20.Zhang S, Jiang C, Jiang L, Chen H, Huang J, Zhang J, Wang R, Chi H, Yang G, Tian G. Uncovering the immune microenvironment and molecular subtypes of hepatitis B-related liver cirrhosis and developing stable a diagnostic differential model by machine learning and artificial neural networks. Front Mol Biosci. 2023;10:1275897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steen CB, Liu CL, Alizadeh AA, Newman AM. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Stem Cell Transcriptional Networks: Methods Protocols 2020;2117:135–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang C, Zhang S, Jiang L, Chen Z, Chen H, Huang J, Tang J, Luo X, Yang G, Liu J, et al. Precision unveiled: synergistic genomic landscapes in breast cancer-integrating single-cell analysis and decoding drug toxicity for elite prognostication and tailored therapeutics. Environ Toxicol. 2024;39(6):3448–72. [DOI] [PubMed] [Google Scholar]
- 24.Mayakonda A, Lin D-C, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Chi H, Xu K, Li Y, Yin X, Chen S, Yang Q, He M, Zhu G, Li X. A novel classification model for Lower-Grade Glioma patients based on pyroptosis-related genes. Brain Sci 2022, 12(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12(2):109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolf B, Rajasundaram S, Cronjé HT, Yarmolinsky J, Burgess S, Gill D. A drug target for erectile dysfunction to help improve fertility, sexual activity, and wellbeing: mendelian randomisation study. BMJ. 2023. 10.1136/bmj-2023-076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soltysiak J, Skowronska B, Fichna P, Ostalska-Nowicka D, Stankiewicz W, Lewandowska-Stachowiak M, Lipkowska K, Zachwieja J. Urinary angiotensinogen and urinary sodium are associated with blood pressure in normoalbuminuric children with diabetes. Pediatr Nephrol. 2014;29:2373–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uijl E, Mirabito Colafella KM, Sun Y, Ren L, van Veghel R, Garrelds IM, de Vries R, Poglitsch M, Zlatev I, Kim JB. Strong and sustained antihypertensive effect of small interfering RNA targeting liver angiotensinogen. Hypertension. 2019;73(6):1249–57. [DOI] [PubMed] [Google Scholar]
- 31.Korin E, Bejerano T, Cohen S. GalNAc bio-functionalization of nanoparticles assembled by electrostatic interactions improves siRNA targeting to the liver. J Controll Releas. 2017;266:310–20. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Yu HAN, Liu Y, Wu W, Zeng H, Li EN. Hsa_circ_0036740 in familial adenomatous polyposis: immune regulation and neutrophil effects in CRC based on high-throughput assay. Biocell. 2023;47(11):2409–22. [Google Scholar]
- 33.Kudo Y, Sugimoto M, Arias E, Kasashima H, Cordes T, Linares JF, Duran A, Nakanishi Y, Nakanishi N, L’Hermitte A. PKCλ/ι loss induces autophagy, oxidative phosphorylation, and NRF2 to promote liver cancer progression. Cancer Cell. 2020;38(2):247–62 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samaddar JS, Gaddy VT, Duplantier J, Thandavan SP, Shah M, Smith MJ, Browning D, Rawson J, Smith SB, Barrett JT. A role for macroautophagy in protection against 4-hydroxytamoxifen–induced cell death and the development of antiestrogen resistance. Mol Cancer Ther. 2008;7(9):2977–87. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen H, Yang J, Kung H, Shi X, Tilki D, Lara P, DeVere White R, Gao A, Evans C. Targeting autophagy overcomes enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene. 2014;33(36):4521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta A, Roy S, Lazar AJ, Wang WL, McAuliffe JC, Reynoso D, McMahon J, Taguchi T, Floris G, Debiec-Rychter M. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST). Proceedings of the National Academy of Sciences 2010, 107(32):14333–14338. [DOI] [PMC free article] [PubMed]
- 37.Hu J, Wang W, Lan X, Zeng Z, Liang Y, Yan Y, Song F, Wang F, Zhu X, Liao W. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang F, Cui J-y, Gao H-f, Yu H, Gao F-f, Chen J-l, Chen L. Cancer-associated fibroblasts induce epithelial-mesenchymal transition and cisplatin resistance in ovarian cancer via CXCL12/CXCR4 axis. Future Oncol. 2020;16(32):2619–33. [DOI] [PubMed] [Google Scholar]
- 39.Shahrajabian MH, Sun W. Survey on multi-omics, and multi-omics data analysis, integration and application. Curr Pharm Anal. 2023;19(4):267–81. [Google Scholar]
- 40.Song Z, Yu J, Wang M, Shen W, Wang C, Lu T, Shan G, Dong G, Wang Y, Zhao J. CHDTEPDB: transcriptome expression profile database and interactive analysis platform for congenital heart disease. Congenit Heart Dis. 2023;18(6):693–701. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA sequencing data from CRC patients were retrieved from the TISCH database (http://tisch.compgenomics.org/home/). Batch RNA-seq data were obtained from TCGA database(https://portal.gdc.cancer.gov/) and GSE17537, GSE103479, GSE106584, GSE12945, GSE28722, GSE29621, GSE39084, GSE87211, GSE31595, and GSE41258. in the GEO database, with the raw datasets being annotated using the corresponding files. The transcriptomic datasets used in the study were derived from the “Human Colorectal Cancer: Whole Transcriptome Analysis” (https://www.10xgenomics.com/). GWAS data for CRC and heart failure were accessed from the Finngen database (https://www.finngen.fi/fi).