Summary
The long-term complications of COVID-19, known as the post-acute sequelae of SARS-CoV-2 infection (PASC), significantly burden healthcare resources. Quantifying the demand for post-acute healthcare is essential for understanding patients’ needs and optimizing the allocation of valuable medical resources for disease management. Driven by this need, we developed a heterogeneous latent transfer learning framework (Latent-TL) to generate critical insights for individual health systems in a distributed research network. Latent-TL enhances learning in a specific health system by borrowing information from all other health systems in the network in a data-driven fashion. By identifying subpopulations with varying healthcare needs, our Latent-TL framework can provide more effective guidance for decision-making. Applying Latent-TL to electronic health record (EHR) data from eight health systems in PEDSnet, a national learning health system in the US, revealed four distinct patient subpopulations with heterogeneous post-acute healthcare demands following COVID-19 infections, varying across subpopulations and hospitals.
Keywords: COVID-19, electronic health records, healthcare utilization, learning health system, Long COVID, real-world data, transfer learning
Highlights
-
•
Assess the healthcare demands of COVID-19 to inform strategic decision-making
-
•
Utilize diverse hospital data to generate tailored, actionable insights for each hospital
-
•
Identify patient subpopulations with varying healthcare needs
-
•
Enhance personalized care through data-driven approaches
The bigger picture
A challenge in understanding the long-term health burden of COVID-19, or any widespread disease, is that clinical effects are observed through the experiences and data of individual hospitals, and yet those hospitals vary in both the distribution of patients they see and the care they provide. Existing studies pool data from multiple hospitals but fail to consider this heterogeneity, limiting the applicability of their findings to local decision-making. Latent transfer learning (Latent-TL) leverages electronic health record data from multiple heterogeneous hospitals to provide actionable insights tailored to each individual institution. As health systems seek to offer more adaptive, personalized care, our work highlights the power of transfer learning to enhance evidence-based clinical decision-making, advocating for broader data sharing to improve healthcare responses to future public health challenges.
Wu et al. introduce latent transfer learning, a framework designed to address hospital-specific healthcare demands related to post-acute sequelae of SARS-CoV-2 infection (PASC). Using electronic health records from eight health systems in the PEDSnet network, the approach identifies distinct patient subpopulations with varied post-acute healthcare needs. By leveraging data across hospitals, it enhances accuracy at individual hospitals and supports stratified, evidence-based decision making tailored to specific patient groups and health systems.
Introduction
Electronic health record (EHR) systems have allowed data capture and use within and across health systems to an extent that was impossible when records consisted largely of handwritten ink on individual charts. During the COVID-19 pandemic, the usage of EHR data played a critical role in knowledge generation to better inform decision-making and public health policies.1,2,3,4,5 The global surge of COVID-19, accounting for over 600 million cases, prompted governments to implement a range of preventive measures, from quarantine protocols to social distancing initiatives.6 To understand the dynamic nature of the pandemic and subsequently inform public health policies, collaborative networks and research initiatives such as RECOVER,7 N3C,8 and 4CE9 were established as critical resources for clinical evidence generation. These initiatives facilitate the collaboration of diverse organizations and stakeholders, fostering a collective learning environment that leverages insights from EHRs across multiple health systems for enhanced learning and response.
The COVID-19 pandemic strained healthcare infrastructures globally, pushing capacities and workforces to their limits, especially in nations like the US. Quantifying the demand for healthcare resources following a COVID-19 diagnosis is crucial for health systems and providers to more effectively assess patient needs and optimize the allocation of health resources.10,11,12 As the pandemic progressed, the focus shifted from acute management of the disease to the long-term impacts on patients suffering from post-acute sequelae of SARS-CoV-2 infection (PASC).13 Emerging evidence shows that a significant subset of patients in both adult and pediatric demographics, after recovering from COVID-19, report a diverse spectrum of PASC symptoms,14,15,16 which necessitate additional medical care and contribute to a prolonged healthcare demand. A thorough understanding of the healthcare burden attributed to PASC is critical for anticipating future healthcare needs and enhancing care delivery systems. While research has focused on the adult population,10,11 there is limited knowledge regarding the utilization pattern during the post-acute phase of COVID-19 among the pediatric population.
PASC in the pediatric population is having a significant impact on communities, highlighting an urgent need for its assessment and management.17,18 Driven by the goal of generating insights for local decision-making, our research aims to understand hospital-specific utilization patterns in children and adolescents during the post-acute phase of COVID-19, leveraging data from multiple health systems through a multi-site transfer learning pipeline. Our study offers several appealing features. First, our analysis is targeted at the level of individual hospitals rather than estimating a global effect by “averaging” across multiple hospitals. Given the heterogeneity of hospitals in patient demographics, severity of illness, healthcare resources and facilities, and patient and physician socio-economic factors, analyses that offer hospital-specific estimates are essential to provide insights that are immediately applicable to local decision-making. Second, our study employs a transfer learning pipeline that leverages data from multiple hospitals to enhance learning within the target hospital of interest, which yields more precise results when the outcome of interest is rare. Third, our study goes beyond assessing the overall healthcare burden to pinpoint patient subgroups that may have additional medical needs. We implement data-driven and interpretable identification of subpopulations with potentially heterogeneous healthcare needs, which may lead to more actionable guidance in practice. Fourth, we focus on a pediatric population with pre-existing chronic conditions, who may require considerable attention within health systems due to the long-term and complex nature of their healthcare needs.
To address the need for reliable inference, we introduce an end-to-end heterogeneous latent transfer learning (Latent-TL) pipeline. Our approach has two components. First, recognizing that pediatric patients with chronic conditions form a heterogeneous population, Latent-TL leverages collaborative learning across multiple participating health systems within a distributed research network15 to identify latent patient subpopulations. Second, Latent-TL estimates causal effects tailored to each subpopulation within each health system while borrowing information from the remaining health systems. Together, these capabilities provide a robust analytical tool that has the potential to significantly enhance evidence-based policy decision-making.
Our approach starts with multi-site latent class analysis (MLCA),19 a robust tool for discerning multimorbidity patterns and characterizing clinically significant subpopulations among patients with chronic conditions. Then, to estimate the subpopulation-specific effects of COVID-19 on healthcare utilization within a specific hospital, we implement a transfer causal learning framework. This method demonstrates increased precision over independently learning from individual health systems by leveraging data from other institutions within a distributed research network.
We applied our Latent-TL method to analyze EHR data from eight institutions affiliated with PEDSnet,20 a pediatric learning health system in the US. Our application identified four clinically meaningful patient subpopulations characterized by distinct comorbidity patterns: mental health conditions, atopic/allergic conditions, non-complex chronic conditions, and complex chronic conditions. Using Latent-TL, for each participating institution, we identified varying impacts of COVID-19 on post-acute healthcare utilization across these distinct groups. To account for possible unmeasured confounding and other systematic biases, we performed calibration using negative control outcomes.21,22 This analysis serves as an example of a principled approach to the adaptive integration of diverse data sources to perform causal inference while accounting for heterogeneity both within hospital-level patient populations and between different health systems. Our approach facilitates an informed, stratified decision-making process tailored to distinct patient subgroups and individual health systems, which has general applications in informing patient care strategies and improving healthcare outcomes.
Results
Overview of the proposed Latent-TL pipeline
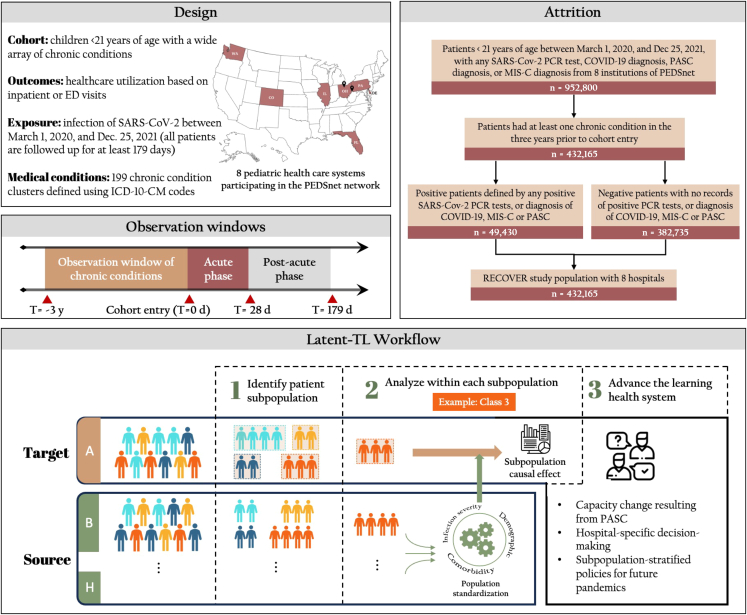
Figure 1 illustrates the workflow of the proposed Latent-TL pipeline. The proposed Latent-TL pipeline aims to determine the varied impacts of COVID-19 infections on healthcare use during the post-acute phase within a designated target hospital, borrowing relevant data from additional source hospitals (i.e., additional hospitals that may provide useful information in learning at the target hospital) within a distributed research network. We developed the Latent-TL pipeline based on the study cohort from the EHR data collected from eight hospitals participating in the PEDSnet network, focusing on the pediatric population with underlying chronic conditions.
Figure 1.
Overview of study design, cohort attrition, and the heterogeneous Latent-TL pipeline
The figure delineates the three core stages to implement the latent transfer learning (Latent-TL) pipeline: (1) identification of latent patient subpopulations characterized by specific multimorbidity patterns based on data from multiple health systems, (2) causal estimation tailored to the patient population in the target hospital, and (3) adaptive integration across hospitals for enhanced estimation.
The Latent-TL pipeline was designed with a three-step procedure. The initial phase of the Latent-TL uses a MLCA19 to discern four unique patient subpopulations characterized by specific multimorbidity patterns. Utilizing data aggregated from multiple institutions, these subpopulations are collaboratively identified, even though their distribution (i.e., prevalence of four subpopulations) might vary across individual hospitals. Following this, the transfer learning pipeline assesses the effects of COVID-19 infection on healthcare utilization during the post-acute phase within each subpopulation in the target hospital. Recognizing the differences in patient characteristics between the target and source hospitals, in the second step, patient populations from the source hospitals are “standardized” through a weighting process, which results in comparable covariate profiles across hospitals. Then, in the third step, the pipeline ascertains the influence of COVID-19 on healthcare utilization within each subpopulation in the target hospital by collaboratively analyzing data from both the target and source hospitals. Importantly, this approach selectively down-weights the source hospitals that exhibit significantly different COVID-19 impacts compared to the target hospital.
A comprehensive explanation of the techniques embedded within the Latent-TL pipeline and the analytical models are given in the experimental procedures section.
Characteristics of the study population
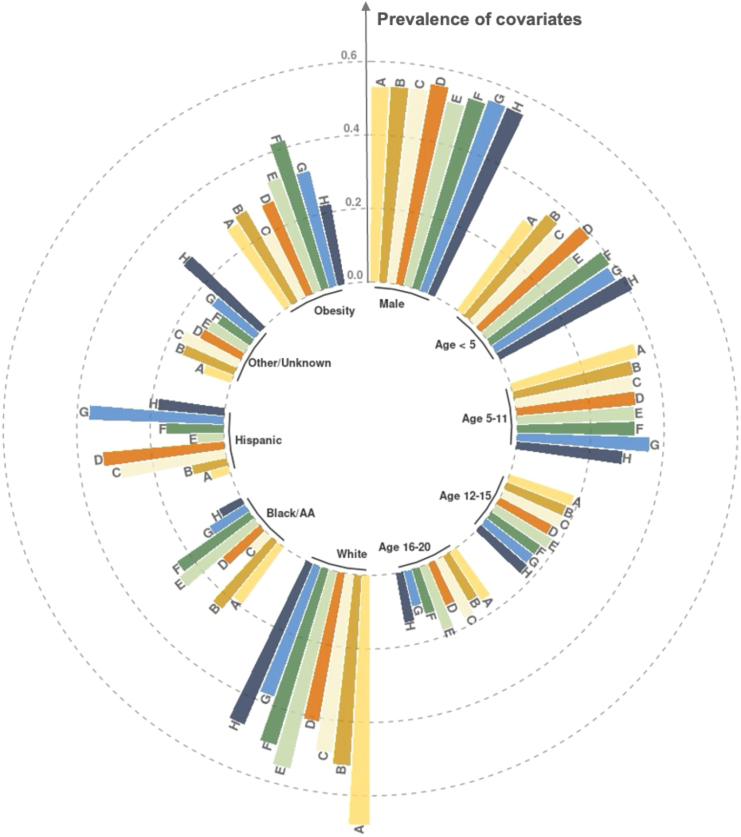
The study cohort includes 432,165 individuals with 49,430 from the COVID-19 infection cohort and 382,735 from the comparator cohort. We summarize the baseline patient characteristics of the study cohort in Table 1. A comprehensive breakdown of all chronic conditions is detailed in Table S2. Overall, 34.47% of patients were younger than 5 years old, and slightly more than half were male (53.32%). Over half were non-Hispanic White (52.73%), while 18.20% were non-Hispanic Black. Additionally, 28.42% of the patients were categorized as having obesity. Most patients entered the cohorts with an outpatient test (74.41%) as opposed to an inpatient or emergency department (ED) test. In the COVID-19 infection cohort, 98.4% of the patients were confirmed to have COVID-19 through a positive PCR test, while the remainder were confirmed via diagnosis codes. An analysis of patient characteristics summarized at the hospital level, displayed in Figure 2, reveals similar age and gender distributions across health systems. However, there is evidence of substantial variation in the ethnicity and obesity metrics across different health systems, underlining the heterogeneity of patient demographics between hospitals. This underscores the point that a one-size-fits-all approach would be inadequate in measuring the impact of COVID-19 on diverse healthcare systems.
Table 1.
Baseline characteristics of patients aged <21 with chronic conditions in the study analyzing the post-acute healthcare utilization outcomes following COVID-19 diagnosis in the pediatric population utilizing the Latent-TL pipeline
| COVID-19 infection (N = 49,430) | No COVID-19 infection (N = 382,735) | Overall (N = 432,165) | |
|---|---|---|---|
| Age, years (%) | |||
| <5 | 13,497 (27.31) | 135,466 (35.39) | 148,963 (34.47) |
| 5–11 | 16,172 (32.72) | 126,709 (33.11) | 142,881 (33.06) |
| 12–15 | 10,120 (20.47) | 64,936 (16.97) | 75,056 (17.37) |
| 16–20 | 9,641 (19.50) | 55,624 (14.53) | 65,265 (15.10) |
| Gender (%) | |||
| Female | 23,852 (48.25) | 177,879 (46.48) | 201,731 (46.68) |
| Male | 25,578 (51.75) | 204,856 (53.52) | 230,434 (53.32) |
| Ethnicity (%) | |||
| Non-Hispanic White | 22,489 (45.50) | 205,405 (53.67) | 227,894 (52.73) |
| Non-Hispanic Black | 12,238 (24.76) | 66,434 (17.36) | 78,672 (18.20) |
| Hispanic | 8,923 (18.05) | 57,619 (15.05) | 66,542 (15.40) |
| Other/unknown | 5,780 (11.69) | 53,277 (13.92) | 59,057 (13.67) |
| Obesity | 16,217 (32.81) | 106,589 (27.85) | 122,806 (28.42) |
| Test location (%) | |||
| ED | 11,645 (23.56) | 69,517 (18.16) | 81,162 (18.78) |
| Inpatient | 2,227 (4.51) | 27,192 (7.10) | 29,419 (6.81) |
| Outpatient | 35,558 (71.94) | 286,026 (74.73) | 321,584 (74.41) |
| Hospital (%) | |||
| A | 9,926 (20.08) | 64,213 (16.78) | 74,139 (17.16) |
| B | 12,335 (24.95) | 92,020 (24.04) | 104,355 (24.15) |
| C | 6,970 (14.10) | 56,241 (14.69) | 63,211 (14.63) |
| D | 1,790 (3.62) | 22,463 (5.87) | 24,253 (5.61) |
| E | 10,843 (21.94) | 68,153 (17.81) | 78,996 (18.28) |
| F | 3,126 (6.32) | 28,642 (7.48) | 31,768 (7.35) |
| G | 3,374 (6.83) | 27,944 (7.30) | 31,318 (7.25) |
| H | 1,066 (2.16) | 23,059 (6.02) | 24,125 (5.58) |
| Entry time (%) | |||
| 2020Q1Q2 | 1,713 (3.47) | 24,202 (6.32) | 25,915 (6.00) |
| 2020Q3 | 2,824 (5.71) | 48,504 (12.67) | 51,328 (11.88) |
| 2020Q4 | 11,288 (22.84) | 59,940 (15.66) | 71,228 (16.48) |
| 2021Q1 | 8,383 (16.96) | 52,815 (13.80) | 61,198 (14.16) |
| 2021Q2 | 4,747 (9.60) | 55,311 (14.45) | 60,058 (13.90) |
| 2021Q3 | 9,786 (19.80) | 70,337 (18.38) | 80,123 (18.54) |
| 2021Q4 | 10,689 (21.62) | 71,626 (18.71) | 82,315 (19.05) |
| Acute-phase illness severitya(%) | |||
| Asymptomatic | 31,701 (64.13) | 199,907 (52.23) | 231,608 (53.59) |
| Mild | 13,338 (26.98) | 88,453 (23.11) | 101,791 (23.55) |
| Moderate | 2,695 (5.45) | 78,463 (20.50) | 81,158 (18.78) |
| Severe | 1,696 (3.43) | 15,912 (4.16) | 17,608 (4.07) |
| Baseline visitsb (averaged per year) | 3.3 (5.8) | 3.1 (5.3) | 3.1 (5.4) |
| Underlying chronic conditionsc(%) | |||
| Allergies | 13,912 (0.281) | 89,489 (0.234) | 103,401 (0.239) |
| Asthma | 9,158 (0.185) | 56,659 (0.148) | 65,817 (0.152) |
| Implant device or graft-related encounter | 5,625 (0.114) | 48,012 (0.125) | 53,637 (0.124) |
| Sleep-wake disorders | 5,722 (0.116) | 47,911 (0.125) | 53,633 (0.124) |
| Esophageal disorders | 4,929 (0.100) | 38,172 (0.100) | 43,101 (0.100) |
This table summarizes the characteristics of patients within the cohort of COVID-19 infection, no infection, and the overall study cohort.
The acute-phase severity was evaluated within 27 days after the index date based on criteria developed in Forrest et al.23
The baseline visits were reported as the average number of visits within 3 years to 7 days prior to the index date.
The period for underlying chronic conditions is from 3 years to 7 days prior to the index date.
Figure 2.
Distribution of demographic attributes including age, gender, race or ethnicity, and obesity prevalence among patients from each of the eight participating health systems in the PEDSnet network
The eight hospitals are indexed from A to H. The height of the bars indicates the prevalence of each demographic variable within each hospital. The outermost circle corresponds to a prevalence of 0.6, with inner circles indicating lower prevalence levels in increments of 0.2. This visualization reveals variations in ethnicity and obesity metrics across different health systems, highlighting the heterogeneity of patient demographics between hospitals.
Subpopulations identified via MLCA
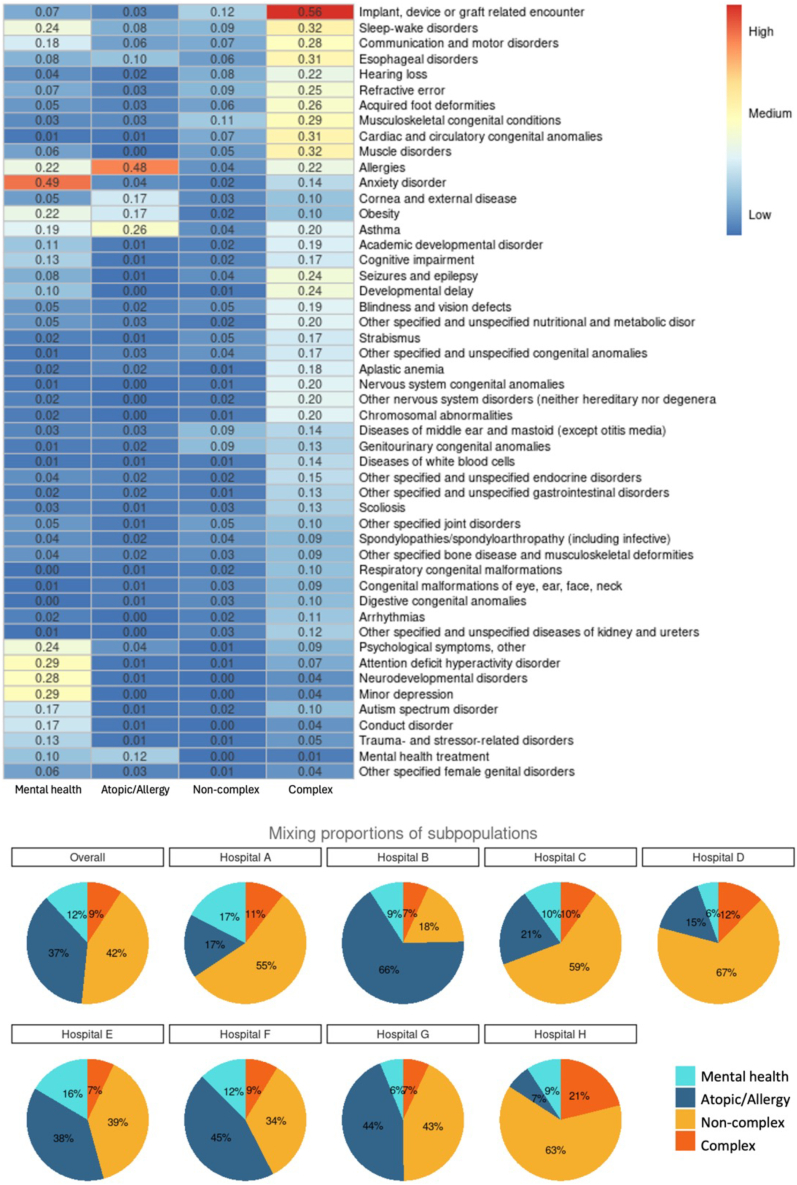
Through the collaborative MLCA approach, we identified four distinct subpopulations, each characterized by a unique mix of chronic conditions. Figure 3 shows the prevalence of different chronic conditions across subpopulations. Here, each column denotes a subpopulation, and each row represents a specific chronic condition cluster. We show only the 50 most prevalent chronic conditions. (A heatmap showing all conditions is available in the supplemental information.) We summarize the four identified subpopulations as follows.
-
•
Class 1 (mental health conditions) includes anxiety disorder, psychological symptoms, attention-deficit hyperactivity disorder, neurodevelopmental disorder, major depression, minor depression, autism spectrum disorder, etc.
-
•
Class 2 (atopic/allergic chronic conditions) encompasses patients predominantly affected by atopic conditions, like allergies and asthma.
-
•
Class 3 (non-complex chronic conditions) consists of patients with non-complex chronic ailments.
-
•
Class 4 (complex chronic conditions) identifies patients with complex conditions, evidenced by a significant reliance on medical technology and severe multi-system disorders.
Figure 3.
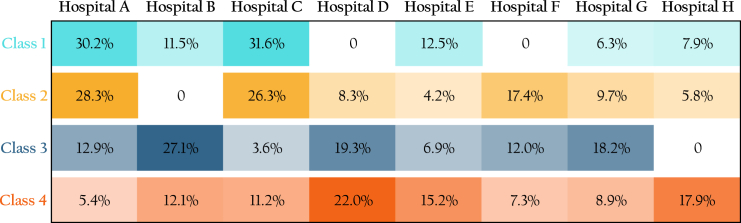
Collaborative identification of subpopulations based on underlying chronic conditions through MLCA approach
Top: a heatmap detailing the four discerned subpopulations (or latent classes). Columns symbolize individual subpopulations, while rows indicate the prevalence of specific chronic condition clusters. The heatmap emphasizes the 50 most prevalent chronic conditions. Each subpopulation is characterized by its unique distribution of chronic condition cluster incidences. Bottom: pie charts illustrating the overall prevalence of each subpopulation and its respective prevalence within individual hospitals.
There is substantial heterogeneity in the prevalence of these subpopulations across hospitals. For instance, only 6.7% of patients in hospital H belonged to class 2, while in hospital B, this percentage was nearly ten times as large, at 66.3%. Such disparities could be attributed to a range of factors, including, for example, the geographical location of the hospital, the types of facilities available, the range of services offered, and the organization of clinics. The R code to compute class membership probabilities for each individual is available in Data S1. By inputting the diagnosis information of chronic conditions for an individual, the R function will output the estimated probabilities of that individual belonging to each subpopulation, along with the subpopulation membership determined by the maximum posterior probability assignment.
Transfer learning approach produces more adequate and effective estimates
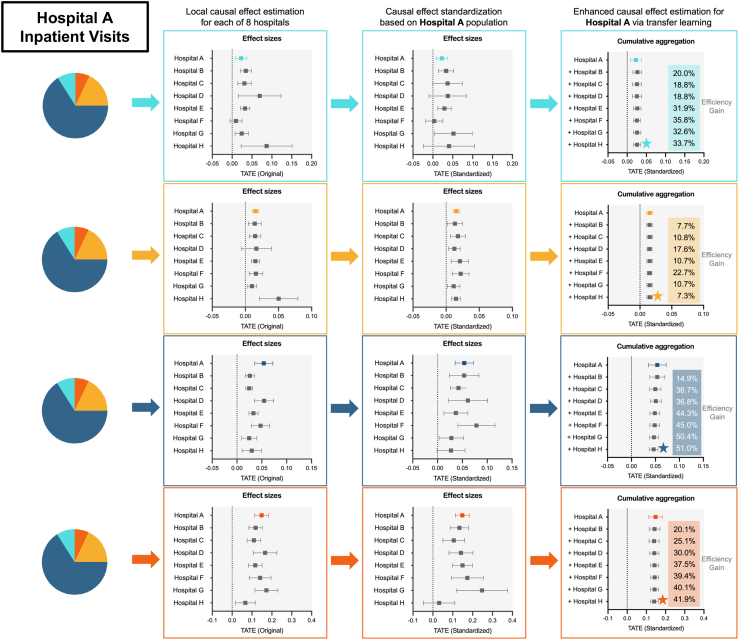
In this section, we demonstrates the effectiveness of the transfer learning approach in estimating the subpopulation- and hospital-specific effects of COVID-19 on post-acute healthcare utilization. This approach is designed to use data from multiple health systems to improve estimation precision. Figure 4 illustrates how this method estimates the effect of COVID-19 on inpatient visits at hospital A.
Figure 4.
Estimation of subpopulation-specific impacts of COVID-19 infection on post-acute inpatient visits at hospital A (target hospital) using Latent-TL pipeline
The first column illustrates effect sizes estimated by applying standard causal inference within each hospital. The second column shows effect sizes after standardizing the source samples via a weighting mechanism in alignment with the patient characteristics in the target hospital. The final column showcases the causal effects by incorporating the source hospital in a cumulative manner, highlighting the efficiency gains by involving data from source hospitals cumulatively using the Latent-TL pipeline. The error bars represent 95% confidence intervals of estimates. The estimates, marked with the star, have the highest estimation efficiency and are showcased in Figure 6.
The forest plot in the first column of the figure displays effect sizes when a standard causal inference approach was applied individually to each subpopulation in each hospital (see supplemental information for more details). These estimates exhibit variability across hospitals due to the distinct impact of COVID-19 on each hospital, as well as the diverse patient populations served by these institutions. The variation in the impacts of COVID-19 across various hospitals suggests that utilizing global estimates for a specific hospital setting may not yield accurate insights. The Latent-TL approach aims to derive causal effects specifically tailored to an individual hospital through two key adjustments.
First, focusing on a specific target hospital of interest (for instance, hospital A in this illustration), a weighting algorithm is used to harmonize baseline patient characteristics from source hospitals, as depicted in the second column of the forest plots. This "standardization" of samples through weighting ensures that the data sourced from the additional hospitals more accurately mirror the distribution of potential confounding variables at the target hospital, thereby producing estimates in each source hospital that are tailored to the patient population in the target hospital.
Next, to achieve more precise estimation for the target hospital, the Latent-TL pipeline adaptively integrates estimates from source hospitals. Specifically, estimates from all hospitals are aggregated, and source hospitals with a higher degree of similarity to the target hospital are given greater weight in the aggregation process. Figure 5 illustrates the role of each hospital (both target and source) in estimating the subpopulation-specific impact of COVID-19 on inpatient visits in hospital A. The magnitude of each hospital’s contribution is determined based on the estimated degree of similarity in the effects of COVID-19, which can vary across subpopulations and hospitals.
Figure 5.
Hospital contributions in estimating the subpopulation-specific effect of COVID-19 infection on post-acute inpatient visits in hospital A
Contribution magnitudes are determined based on the similarity of COVID-19 effects across different hospitals.
Finally, Latent-TL synthesizes all of these elements to estimate the impact COVID-19 has on inpatient visits at hospital A, as shown in the last column of forest plots in Figure 4. To underscore the precision gained by incorporating data from source hospitals, as opposed to relying solely on data from the target hospital, we sequentially incorporated the source hospitals in a cumulative manner. We observe that including source hospitals results in a reduction in estimated standard errors. This efficiency gain shows how the Latent-TL pipeline can improve our estimates by effectively sharing knowledge across hospitals.
Latent-TL pipeline quantifies hospital- and subpopulation-specific post-acute healthcare demands
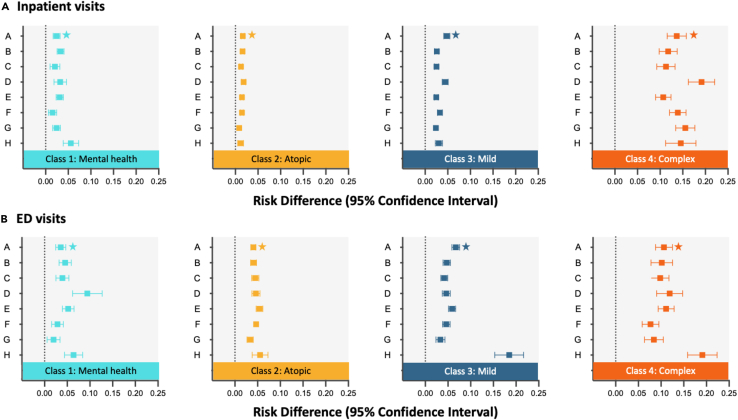
After identifying patient subpopulations, we used the transfer learning approach described above to investigate the effects of COVID-19 infection on both inpatient and ED visits during the post-acute phase across both hospitals and subpopulations. Figure 6 presents the estimated causal effects, along with 95% confidence intervals, as determined by the Latent-TL method. The figure is divided into two rows, with Figure 6A focusing on inpatient admissions and Figure 6B on ED visits.
Figure 6.
Hospital- and subpopulation-specific COVID-19 effects on post-acute inpatient and ED visits in eight health systems from PEDSnet
Hospital- and subpopulation-specific COVID-19 effects on post-acute inpatient (A) and ED (B) visits in eight health systems from PEDSnet. The error bars represent 95% confidence intervals of estimates. The estimates concerning inpatient visits at hospital A, marked with a star, serve as illustrative examples in Figure 4.
From Figure 6, it is evident that COVID-19 has variable impacts on inpatient and ED visits, both across patient subpopulations and across hospitals. For example, at hospital D, patients with mental health and complex chronic conditions had an increased probability of an ED visit after COVID-19 infection. In contrast, at hospital H, those with non-complex and complex chronic conditions saw a significant rise in ED visits. The rate of inpatient visits by patients with complex chronic conditions at hospital D was significantly affected by COVID-19, more so than at other hospitals. Similarly, hospital H observed the most pronounced impact among patients with mental health conditions.
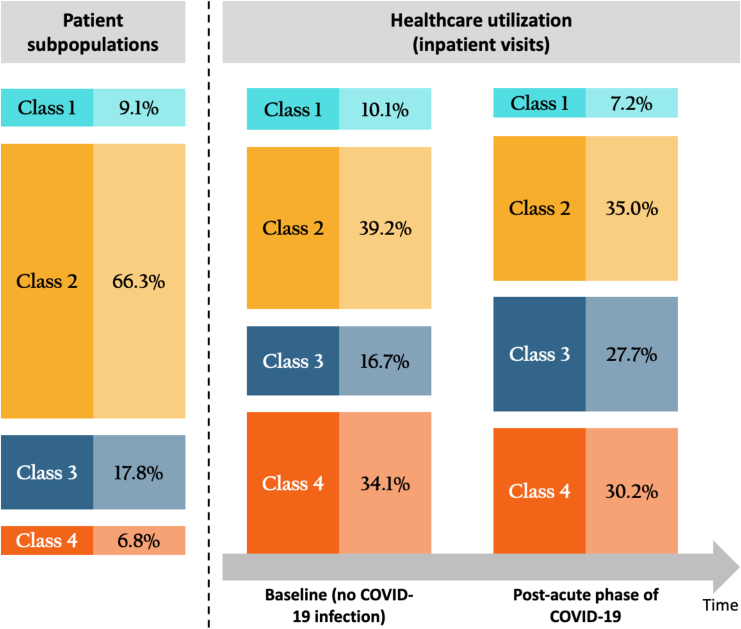
Figure 7 highlights the distribution of patient subpopulations in hospital A along with estimates of healthcare utilization (in terms of inpatient visits) in two counterfactual scenarios: one in which no patients were infected with COVID-19 and one in which all patients were infected. The former provides an estimate of baseline utilization during the study period had the patients remained uninfected. It is notable that while patients with complex chronic conditions only constituted 6.8% of the overall population, they accounted for a significant portion of healthcare utilization—34.1% without COVID-19 infection and 30.2% during the post-acute phase of COVID-19. Meanwhile, the subpopulation with non-complex chronic conditions had the most significant increase in healthcare utilization after COVID-19 infection. Estimates of contemporaneous utilization for other hospitals can be found in Figure S3.
Figure 7.
Distribution of patient subpopulation and corresponding healthcare utilization with no COVID-19 infection and healthcare utilization during post-acute phase of COVID-19 for four identified subpopulations
This analysis involves estimating healthcare utilization in two hypothetical scenarios where all patients were presumed to be either non-infected or infected with COVID-19.
In summary, this application evaluates the post-acute healthcare demand within the pediatric population, focusing on each individual hospital participating in the PEDSnet network. Our findings reveal a heterogeneous pattern of healthcare demands across various hospitals, indicating that a global- or national-level estimate may not be adequately informative for localized decision-making. The data-driven subpopulations exhibit diverse healthcare needs, particularly noting that patients with complex chronic conditions demanded more healthcare services after COVID-19 in comparison to other subpopulations. These findings suggest stratified healthcare strategies for patient care and disease management of PASC.
Addressing residual bias in EHR data through negative control experiments
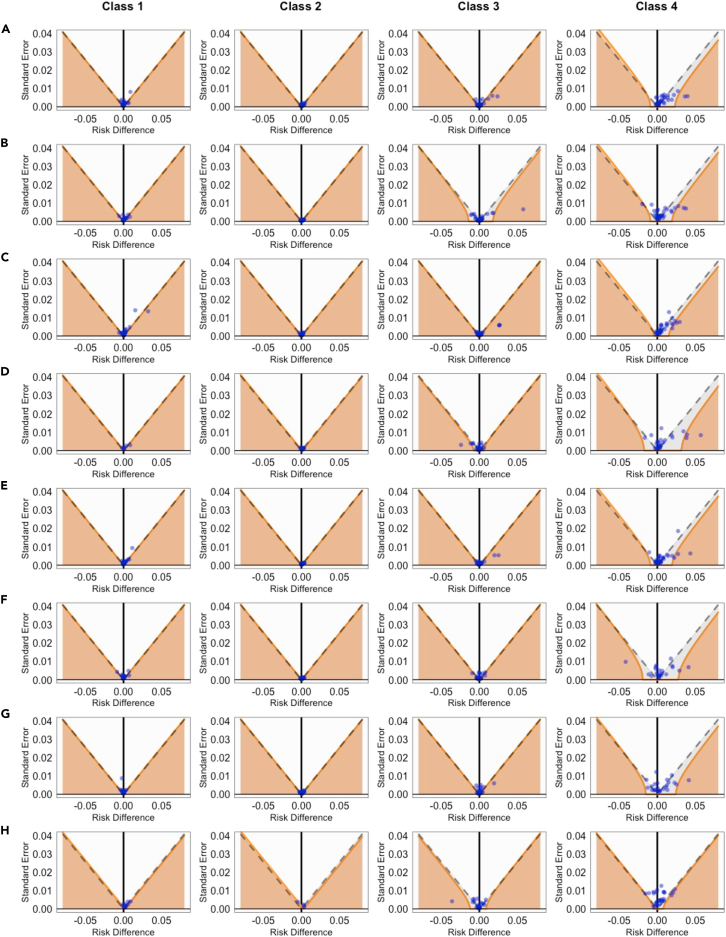
While our analysis adjusted for a large number of measured potential confounding variables, hidden biases due to unmeasured confounders could still influence results. To mitigate this, we employed a negative control experiment, a technique popularized by Schuemie and colleagues21,22 found to be effective in pharmacoepidemiologic studies.24,25 We identified 33 outcome variables not thought to be influenced by COVID-19 infection as negative controls (see supplemental information for details). By applying the same causal transfer learning pipeline to the set of negative control outcomes, we established a baseline or "null" distribution, which helps to measure and adjust for the bias in estimating the effects of interest. Figure 8 presents the estimated impacts of COVID-19 on these control outcomes using our method, denoted by blue dots. While traditional significance testing rendered 70.9% of these effects as non-significant (indicated by dots below the dashed line), calibration using the null distribution increased this rate to 91.1%, as shown by solid orange lines. Figure S4 shows the results from the Latent-TL analysis of healthcare utilization outcomes after calibrating using the empirical null distribution. There was a negligible shift in point estimates, accompanied by wider confidence intervals, suggesting a minimal degree of systematic bias.
Figure 8.
Funnel plot of traditional and calibrated significance testing
(A)–(H) corresponds to the results from hospital A through H, respectively. Areas below the dashed line indicate p < 0.05 based on traditional p value calculations. Estimates in orange areas have p < 0.05 after calibrating the empirical null distribution. Blue dots indicate estimates corresponding to negative control variables. The overall coverage of the null hypothesis changed from 70.9% to 91.1% after calibration.
Discussion
In this study, we introduced Latent-TL, a multi-site transfer learning framework designed to identify patient subpopulations and estimate hospital- and subpopulation-specific causal effects. Our approach combined MLCA, a method that collaboratively identifies latent subpopulations of patients from multi-site data, and causal transfer learning, which adaptively incorporates data from source hospitals to enhance learning within a target hospital. To further ensure the integrity of our results, we employed a set of negative control outcomes, which served as a mechanism to detect and correct any residual biases in our analysis. We used the Latent-TL pipeline to evaluate the effects of COVID-19 on healthcare utilization during the post-acute phase in each of the eight pediatric hospitals participating in PEDSnet. Our analysis identified four clinically significant patient subpopulations that experienced heterogeneous effects of COVID-19 on healthcare utilization.
Numerous studies have been conducted to support decision-making in the context of COVID-19. While a significant portion of them focus on mitigating the overall burden of the virus or projecting future infection rates, hospitalizations, and fatalities at a population level,3,26,27,28,29 our research delves into the care-seeking behaviors of patients, offering insights critical for patient management within hospital settings. We have segmented the patient population into distinct categories based on types of chronic conditions. By doing so, we aim to generate knowledge on the varied healthcare utilization patterns across these subgroups, which ideally can be translated into clinical care. For instance, in our illustration example from hospital A, we observed that patients with non-complex chronic conditions experienced the most significant increase in inpatient visits following COVID-19 infection. This insight highlights the potential for increased demand in this patient subgroup, prompting healthcare providers to allocate resources accordingly, prioritize follow-up care, and personalize treatment plans for these patients. In addition, the focused analysis by Latent-TL provides a more tailored approach for guiding hospital decisions, moving beyond broad national-level estimates. Appropriate situational awareness in COVID-19, at the provider and administrative levels, required balancing broad, national-level data with the patterns in care seen locally. An ability to confidently apply national-level data to a local system was not readily available, and so the impact of demographic and subpopulation differences between centers, such as those demonstrated in Figures 2 and 3, were challenging to interpret. In Figure 6, hospital H serves as an illustrative example of the impact that application of the hetero-Latent-TL approach could have at an individual center. A multi-center estimate of ED visit risk for complex patients would likely underestimate what providers and administrators at site H were observing at their care center. However, applying this model could inform hospital H that, in fact, their complex population was at higher risk of ED utilization, with estimation improved by the inclusion of the multi-site data without losing insight into the site-level differences. Armed with these data, one could imagine that hospital H could more appropriately plan for ED care at the time of COVID-19 surge or evaluate why their local practice and ED referral patterns might drive higher ED utilization without a concurrent increase in inpatient utilization for the complex subpopulation. Lastly, we can expect greater statistical power due to leveraging information across health systems. The effective deployment of Latent-TL in COVID-19 research underscores the invaluable role of data sharing in enhancing hospital-specific decision-making processes.
Several contemporary studies have also emphasized the importance of borrowing information across multiple sites for causal inference. For instance, Dahabreh et al.30 introduced a causally interpretable meta-analysis. This method is applicable when individual-level data are only available for the target population but summary-level data from multiple studies with potential population heterogeneity are also available. This approach first estimates the treatment effect from the external studies and subsequently translates it to the designated target population, utilizing covariates that could modify the effect. The effectiveness of Dahabreh et al.'s technique, in comparison to conventional meta-analysis, was evaluated by Rott et al.31 Meanwhile, Han et al.32,33 proposed federated algorithms to assimilate data from numerous source sites to deduce the causal effect in target sites. Their strategy modifies the covariate distribution in each source site to match the target site. It then aggregates the source and target site estimates by optimizing the site-level weights. Clark et al.34 proposed a method that bridges causally interpretable meta-analysis with random-effect meta-analysis, allowing for site-level heterogeneity beyond differences in the covariate distribution. Our Latent-TL framework is unique in its accommodation of latent subpopulation structures spanning multiple hospitals. It addresses site-level heterogeneity in several dimensions: by estimating the variation in subpopulation composition via the MLCA, adjusting source samples to align with the target hospital, and considering the variability in subpopulation-level causal effects and adaptively weighting source estimates in accordance with resemblance to the target population.
We acknowledge several limitations of this study. First, to ensure an adequate follow-up period, our study excluded patients infected during the Omicron surge, which means recent infections predominantly from the Omicron variant of SARS-CoV-2 (from December 25, 2021) were not considered. Future studies updating the analysis to include these variants would be beneficial for a more current understanding of the findings. Second, our primary data sources were diagnoses, symptoms, and indications from EHRs, which might lack pertinent laboratory, imaging, and procedural findings. Third, our study aimed to infer the causal effect of COVID-19 infection on healthcare utilization from observational data. While a list of potential confounders was carefully identified by domain experts in the field, there is no assurance that all influential confounding variables were included in the analysis. To mitigate this, we conducted negative control experiments to assess and calibrate the potential residual bias from unmeasured confounders. These experiments revealed negligible systematic error in point estimates. Fourth, the comparator cohort was assembled from patients without any recorded evidence of COVID-19 infection but with at least one negative COVID-19 test. This approach might potentially target a cohort with a higher frequency of healthcare visits, subsequently introducing bias to the estimates. The use of negative control experiments serves as a vital tool in assessing and mitigating these possible biases. Lastly, evidence suggests that long-COVID can persist for longer than 6 months.35 We acknowledge that the interpretation of the findings in this study is limited to the first 6 months following acute COVID-19 infection. Future studies that extend the follow-up period and analyze trends over time would be beneficial for a more comprehensive understanding of the condition.
In conclusion, our proposed Latent-TL method underscores the transformative potential of adopting transfer learning to distributed research networks to generate evidence for local decision-making. By discerning latent subpopulations and harnessing transfer learning, our methodology elevates the applicability and accuracy of healthcare utility analyses. Our findings highlight the profound implications of this approach in delivering more nuanced, data-informed insights for patient care and management. As health systems strive for more adaptive and personalized care, the integration of such advanced methodologies may be pivotal in navigating future health challenges.
Experimental procedures
Database
The cohorts for this study were extracted from the large-scale EHR database of PEDSnet,20 which is a national patient-centered clinical research network consisting of eight participating systems: Children’s Hospital of Philadelphia (CHOP), Cincinnati Children’s Hospital Medical Center, Children’s Hospital of Colorado, Ann & Robert H. Lurie Children’s Hospital of Chicago, Nationwide Children’s Hospital, Nemours Children’s Health System (sites in Delaware and Florida), Seattle Children’s Hospital, and Stanford Children’s Health. Routinely collected EHR data on more than 7 million children and adolescents were available in PEDSnet. Data from different institutions in PEDSnet are harmonized based on the common data model,36 an extension of the Observational Medical Outcomes Partnership common data model.37 The PEDSnet COVID-19 Database v.2022-03-17 was used for this study.
Cohort construction
We constructed the COVID-19 infection cohort and comparator cohort by selecting individuals <21 years of age between March 1, 2020, and December 25, 2021, who have a history of chronic conditions.
-
•
The COVID-19 infection cohort was determined by a positive SARS-CoV-2 PCR test during any type of healthcare visit, a COVID-19 diagnosis associated with an inpatient or ED encounter, or a diagnosis of multi-system inflammatory syndrome in children (MIS-C) or PASC during any healthcare encounter. The index date for patients in this infected cohort was defined as the date of either the first positive PCR test or the earliest COVID-19 diagnosis or the date of the earliest PASC/MIS-C diagnosis minus 28 days, whichever occurred first.
-
•
The comparator cohort consisted of patients who had at least one negative PCR test, with no recorded positive test results or COVID-19, MIS-C, or PASC diagnoses. The index date for patients in this non-infected cohort was determined as the date of a randomly selected negative SARS-CoV-2 PCR test from among multiple tests.
Patients having a minimum of 179 days of follow-up time after the index date were included, enabling us to examine healthcare utilization outcomes during the post-acute phase of COVID-19. All patients were required to have a history of chronic conditions, identified by 199 chronic condition clusters based on ICD-10-CM codes (drawing from the Agency for Healthcare Research and Quality [AHRQ] Clinical Classification Software Refined16) documented at any time up within 3 years up to 7 days before the index date. A population selection workflow is available in Figure S1. For a more detailed list of diagnosis codes used to define the study cohort, please refer to the supplemental information.
Outcome and confounding variables
The outcome of interest in this study was a binary variable indicating a specific type of medical visit between 28 and 179 days after the index date. Specifically, we defined two outcomes as (1) at least one inpatient visit 28–179 days after the index date and (2) at least one ED visit 28–179 days after the index date. We focused on evaluating general patterns of healthcare demands in the post-acute period without focusing on any specific cause. Besides 199 pre-existing chronic conditions, we further collected patient baseline covariates including age groups (categorized into <5, 5–11, 12–15, or 16–20 years) at index date, gender (female, male), race/ethnicity (non-Hispanic White, Non-Hispanic Black, Hispanic, other, or unknown), test location (ED, inpatient, outpatient), obesity (defined as age-sex-standardized BMI Z score ≥ 95th percentile based on weight measurement at the index date and height within 60 days of index date), cohort entry period (March 2020–June 2020, July 2020–September 2020, October 2020–December 2020, January 2021–March 2021, April 2021–June 2021, July 2021–September 2021, October 2021–December 2021), and the number of visits per year associated with existing chronic conditions during the 3-year time period prior to the index date. See supplemental information for a detailed list of study variables.
Development of the Latent-TL pipeline
The Latent-TL pipeline aims to assess the impact of COVID-19 on post-acute healthcare demands using data from multiple health systems, i.e., hospitals. The study is focused on gaining a deeper understanding of healthcare patterns within an individual health system (target hospital) while utilizing data collected from additional relevant hospitals (source hospitals).
Let be the hospital index for the -th subject such that , indicating that the subject was collected from the target hospital, and if the subject belongs to a source hospital. Let be the sample size of each hospital and be the total sample size. In the application of the Latent-TL pipeline on EHR data from PEDSnet, each hospital was in turn designated as the target hospital, while knowledge was leveraged from the other relevant source hospitals. Given that the variability in underlying chronic comorbidities significantly influences the clinical manifestation of COVID-19 and PASC and the corresponding healthcare-seeking behavior, we hypothesize that the overall patient population across different hospitals is divided into subpopulations or classes based on chronic conditions. This allows for potential heterogeneity in the healthcare utilization pattern across different subpopulations.
Identification of patient subpopulation via MLCA
To better understand the heterogeneity of baseline conditions in the cohort, we characterized patient subpopulations based on their distinct patterns of chronic comorbidities. We employed a MLCA19 to collaboratively identify patient subpopulations with distinct patterns of pre-existing chronic conditions using data from multiple hospitals.
Let be the binary manifest variables, which are pre-existing chronic conditions, observed for -th subject. Assume that the manifest variables are conditionally independent given the class membership , that is,
, , is denoted as the probability of a subject from the -th hospital belonging to class . Through the formulation of MLCA, these subpopulations are characterized by distinct patterns in the prevalence of chronic conditions, establishing a unified definition for subpopulations across different healthcare systems. The MLCA model accommodates variations in the distribution or prevalence of these subpopulations among different hospitals, recognizing the inherent between-site heterogeneity. Our model aligns with the family of latent class regression (LCR)38 models, wherein the covariate for the subject is a categorical variable indicating the health system to which the subject belongs. The expectation maximization (EM) algorithm,39 an effective method for estimating LCR models, is utilized to estimate the parameters, specifically the prevalence of chronic conditions within subpopulations and the distribution/prevalence of these subpopulations across individual hospitals. In this study, we assume that the number of latent subpopulations is known a priori. However, in practical applications, one can employ data-driven approaches like the Akaike information criterion (AIC) or Bayesian information criterion (BIC) to ascertain the optimal number of subgroups.
Causal estimation with population standardization
We focus on estimating the causal effects of COVID-19 on post-acute healthcare utilization specifically tailored to each identified patient subpopulation, adapting to the unique characteristics of a target hospital (). Let be a -dimensional vector of covariates for the -th subject (i.e., all confounding variables in the study), where . denotes the binary exposure in the study (i.e., infection or non-infection of SARS-CoV-2), and denotes the observed outcomes (e.g., inpatient visits 28–179 days after cohort entry). The membership of each subject toward each class is indicated by . Following the potential outcome framework, we let and denote the potential outcomes when and . Then, the Latent-TL pipeline aims to estimate the average treatment effect for the target population (TATE) of each subpopulation of the target hospital:
It is crucial to acknowledge that the patient populations across source hospitals might markedly vary from the target population, primarily owing to differences in geographical locations and the range of hospital services offered. To account for this, Latent-TL facilitates the accommodation of heterogeneous covariate distributions across different hospitals. It is achieved by modeling the sampling probability for each hospital, conditional on covariates and subpopulation membership, and subsequently re-weighting the source patients to more accurately represent the target population.
Specifically, in each source hospital, we implement a doubly robust estimator for the causal effect of -th subpopulation in the target hospital30,31:
where and represent the conditional inclusion probabilities for the target and source hospitals, respectively, which can be determined through the application of a classification model; is a regression model of the outcome on covariates and COVID-19 infection in subpopulation of the target hospital; and is a propensity score model for subpopulation of source hospital . The proposed estimator is doubly robust in the sense that if either the outcome model or both the sampling probabilities and propensity scores are consistently estimated, then the overall estimator is consistent for the TATE.
Adaptive integration of source hospitals
To effectively incorporate insights from source hospitals, we constructed the final estimator as a linear combination of estimates derived from both the target and source hospitals. The weights used in the linear combination were estimated based on the degree of similarity between the estimates from the target and source hospitals. Specifically, we define the final estimator as . Intuitively, the final estimator offers an enhanced precision compared to the estimator that relies solely on data from the target hospital without introducing substantial bias from integrating dissimilar source hospitals. Besides, a source estimator that presents a minimal discrepancy with the target should exert a greater influence on the final estimator through a larger weight, while the weights associated with dissimilar sources should be reduced, converging toward zero. As a result, our strategy anchors to the point estimate that relies solely on data from the target hospital. The weights of hospitals were determined by minimizing the mean-squared error (MSE) of the final estimator, all while penalizing the discrepancy between source and target estimates30,31:
where the MSE is defined as
represents the influence function of the doubly robust estimator in -th hospital. defines a penalty term on the discrepancy between target and source estimators. The asymptotic variance is calculated by applying appropriate weights to these influence functions, which are subsequently utilized to construct confidence intervals in the results section.
Resource availability
Lead contact
For further details, questions, or requests, please send direct correspondence to Yong Chen (ychen123@pennmedicine.upenn.edu).
Materials availability
No physical materials were used in the conduction of this research.
Data and code availability
-
•
The data are not publicly available due to privacy concerns. The individual de-identified participant data will not be shared. The individual de-identified participant data that support our findings can only be shared with qualified researchers after a successful application for PEDSnet Study Approval (https://pedsnet.org/research/accessing-pedsnet/request-pedsnet-study-approval/). These requests will be evaluated by the steering committee and processed by the coordinating center.
-
•
The source code and an illustration based on synthetic data are publicly available on GitHub at https://github.com/Penncil/Latent-TL and have been archived at Zenodo.40
Acknowledgments
This work was supported in part by the National Institutes of Health (U01TR003709, U24MH136069, RF1AG077820, 1R01LM014344, 1R01AG077820, R01LM012607, R01AI130460, R01AG073435, R56AG074604, R01LM013519, R56AG069880, R21AI167418, and R21EY034179). This work was supported partially through Patient-Centered Outcomes Research Institute (PCORI) Project Program Awards (ME-2019C3-18315 and ME-2018C3-14899). All statements in this report, including its findings and conclusions, are solely those of the authors and do not necessarily represent the views of the PCORI, its board of governors, or its methodology committee.
Author contributions
Q.W. and Y.C. designed the model and the Latent-TL pipeline. Q.W., N.M.P., C.B.F., D.A.A., and Y.C. devised the project. V.L. and C.B.F. coordinated the data harmonization. Q.W., N.M.P., D.A.A., and Y.C. designed the real-data analysis, and Q.W. performed the real-data analysis. Q.W. and Y.C. drafted the manuscript. N.M.P., C.B.F., and D.A.A. provided clinical interpretations of the clinical findings. All co-authors provided critical edits of the early draft and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2024.101079.
Contributor Information
David A. Asch, Email: asch@upenn.edu.
Yong Chen, Email: ychen123@pennmedicine.upenn.edu.
Supplemental information
References
- 1.Dickerman B.A., Gerlovin H., Madenci A.L., Kurgansky K.E., Ferolito B.R., Figueroa Muñiz M.J., Gagnon D.R., Gaziano J.M., Cho K., Casas J.P., Hernán M.A. Comparative Effectiveness of BNT162b2 and mRNA-1273 Vaccines in U.S. Veterans. N. Engl. J. Med. 2022;386:105–115. doi: 10.1056/NEJMoa2115463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Q., Tong J., Zhang B., Zhang D., Chen J., Lei Y., Lu Y., Wang Y., Li L., Shen Y., et al. Real-World Effectiveness of BNT162b2 Against Infection and Severe Diseases in Children and Adolescents. Ann. Intern. Med. 2024;177:165–176. doi: 10.7326/M23-1754. [DOI] [PubMed] [Google Scholar]
- 3.Lauring A.S., Tenforde M.W., Chappell J.D., Gaglani M., Ginde A.A., McNeal T., Ghamande S., Douin D.J., Talbot H.K., Casey J.D., et al. Clinical severity of, and effectiveness of mRNA vaccines against, covid-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: prospective observational study. BMJ. 2022;376 doi: 10.1136/bmj-2021-069761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith J.C., Williamson B.D., Cronkite D.J., Park D., Whitaker J.M., McLemore M.F., Osmanski J.T., Winter R., Ramaprasan A., Kelley A., et al. Data-driven automated classification algorithms for acute health conditions: applying PheNorm to COVID-19 disease. J. Am. Med. Inf. Assoc. 2024;31:574–582. doi: 10.1093/jamia/ocad241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J., Grabowska M.E., Kerchberger V.E., Smith J.C., Eken H.N., Feng Q., Peterson J.F., Trent Rosenbloom S., Johnson K.B., Wei W.-Q. ConceptWAS: A high-throughput method for early identification of COVID-19 presenting symptoms and characteristics from clinical notes. J. Biomed. Inf. 2021;117 doi: 10.1016/j.jbi.2021.103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellenius G.A., Vispute S., Espinosa V., Fabrikant A., Tsai T.C., Hennessy J., Dai A., Williams B., Gadepalli K., Boulanger A., et al. Impacts of social distancing policies on mobility and COVID-19 case growth in the US. Nat. Commun. 2021;12:3118. doi: 10.1038/s41467-021-23404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz L.I., Thaweethai T., Brosnahan S.B., Cicek M.S., Fitzgerald M.L., Goldman J.D., Hess R., Hodder S.L., Jacoby V.L., Jordan M.R., et al. Researching COVID to Enhance Recovery (RECOVER) adult study protocol: Rationale, objectives, and design. PLoS One. 2023;18 doi: 10.1371/journal.pone.0286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haendel M.A., Chute C.G., Bennett T.D., Eichmann D.A., Guinney J., Kibbe W.A., Payne P.R.O., Pfaff E.R., Robinson P.N., Saltz J.H., et al. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. J. Am. Med. Inf. Assoc. 2021;28:427–443. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brat G.A., Weber G.M., Gehlenborg N., Avillach P., Palmer N.P., Chiovato L., Cimino J., Waitman L.R., Omenn G.S., Malovini A., et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. npj Digit. Med. 2020;3:109. doi: 10.1038/s41746-020-00308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salerno S., Sun Y., Morris E.L., He X., Li Y., Pan Z., Han P., Kang J., Sjoding M.W., Li Y. Comprehensive evaluation of COVID-19 patient short- and long-term outcomes: Disparities in healthcare utilization and post-hospitalization outcomes. PLoS One. 2021;16 doi: 10.1371/journal.pone.0258278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koumpias A.M., Schwartzman D., Fleming O. Long-haul COVID: healthcare utilization and medical expenditures 6 months post-diagnosis. BMC Health Serv. Res. 2022;22:1010. doi: 10.1186/s12913-022-08387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B.Z., Creekmur B., Yoo M.S., Broder B., Subject C., Sharp A.L. Healthcare Utilization Among Patients Diagnosed with COVID-19 in a Large Integrated Health System. J. Gen. Intern. Med. 2022;37:830–837. doi: 10.1007/s11606-021-07139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thaweethai T., Jolley S.E., Karlson E.W., Levitan E.B., Levy B., McComsey G.A., McCorkell L., Nadkarni G.N., Parthasarathy S., Singh U., et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA. 2023;329:1934. doi: 10.1001/jama.2023.8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proal A.D., VanElzakker M.B., Aleman S., Bach K., Boribong B.P., Buggert M., Cherry S., Chertow D.S., Davies H.E., Dupont C.L., et al. SARS-CoV-2 reservoir in post-acute sequelae of COVID-19 (PASC) Nat. Immunol. 2023;24:1616–1627. doi: 10.1038/s41590-023-01601-2. [DOI] [PubMed] [Google Scholar]
- 15.Groff D., Sun A., Ssentongo A.E., Ba D.M., Parsons N., Poudel G.R., Lekoubou A., Oh J.S., Ericson J.E., Ssentongo P., Chinchilli V.M. Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao S., Lee G.M., Razzaghi H., Lorman V., Mejias A., Pajor N.M., Thacker D., Webb R., Dickinson K., Bailey L.C., et al. Clinical Features and Burden of Postacute Sequelae of SARS-CoV-2 Infection in Children and Adolescents. JAMA Pediatr. 2022;176:1000. doi: 10.1001/jamapediatrics.2022.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao S., Gross R.S., Mohandas S., Stein C.R., Case A., Dreyer B., Pajor N.M., Bunnell H.T., Warburton D., Berg E., et al. Postacute Sequelae of SARS-CoV-2 in Children. Pediatrics. 2024;153 doi: 10.1542/peds.2023-062570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long COVID and kids: more research is urgently needed. Nature. 2022;602:183. doi: 10.1038/d41586-022-00334-w. [DOI] [PubMed] [Google Scholar]
- 19.Jing N., Liu X., Wu Q., Rao S., Mejias A., Maltenfort M., Schuchard J., Lorman V., Razzaghi H., Webb R., et al. Development and validation of a federated learning framework for detection of subphenotypes of multisystem inflammatory syndrome in children. medRxiv. 2024 doi: 10.1101/2024.01.26.24301827. Preprint at. [DOI] [Google Scholar]
- 20.Forrest C.B., Margolis P.A., Bailey L.C., Marsolo K., Del Beccaro M.A., Finkelstein J.A., Milov D.E., Vieland V.J., Wolf B.A., Yu F.B., Kahn M.G. PEDSnet: a National Pediatric Learning Health System. J. Am. Med. Inf. Assoc. 2014;21:602–606. doi: 10.1136/amiajnl-2014-002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuemie M.J., Hripcsak G., Ryan P.B., Madigan D., Suchard M.A. Empirical confidence interval calibration for population-level effect estimation studies in observational healthcare data. Proc. Natl. Acad. Sci. USA. 2018;115:2571–2577. doi: 10.1073/pnas.1708282114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuemie M.J., Ryan P.B., DuMouchel W., Suchard M.A., Madigan D. Interpreting observational studies: why empirical calibration is needed to correct p -values. Stat. Med. 2014;33:209–218. doi: 10.1002/sim.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forrest C.B., Burrows E.K., Mejias A., Razzaghi H., Christakis D., Jhaveri R., Lee G.M., Pajor N.M., Rao S., Thacker D., Bailey L.C. Severity of Acute COVID-19 in Children <18 Years Old March 2020 to December 2021. Pediatrics. 2022;149 doi: 10.1542/peds.2021-055765. [DOI] [PubMed] [Google Scholar]
- 24.Hripcsak G., Suchard M.A., Shea S., Chen R., You S.C., Pratt N., Madigan D., Krumholz H.M., Ryan P.B., Schuemie M.J. Comparison of Cardiovascular and Safety Outcomes of Chlorthalidone vs Hydrochlorothiazide to Treat Hypertension. JAMA Intern. Med. 2020;180:542–551. doi: 10.1001/jamainternmed.2019.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suchard M.A., Schuemie M.J., Krumholz H.M., You S.C., Chen R., Pratt N., Reich C.G., Duke J., Madigan D., Hripcsak G., Ryan P.B. Comprehensive comparative effectiveness and safety of first-line antihypertensive drug classes: a systematic, multinational, large-scale analysis. Lancet. 2019;394:1816–1826. doi: 10.1016/S0140-6736(19)32317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unwin H.J.T., Mishra S., Bradley V.C., Gandy A., Mellan T.A., Coupland H., Ish-Horowicz J., Vollmer M.A.C., Whittaker C., Filippi S.L., et al. State-level tracking of COVID-19 in the United States. Nat. Commun. 2020;11:6189. doi: 10.1038/s41467-020-19652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin J., Agarwala N., Kundu P., Harvey B., Zhang Y., Wallace E., Chatterjee N. Individual and community-level risk for COVID-19 mortality in the United States. Nat. Med. 2021;27:264–269. doi: 10.1038/s41591-020-01191-8. [DOI] [PubMed] [Google Scholar]
- 28.Pilishvili T., Gierke R., Fleming-Dutra K.E., Farrar J.L., Mohr N.M., Talan D.A., Krishnadasan A., Harland K.K., Smithline H.A., Hou P.C., et al. Effectiveness of mRNA Covid-19 Vaccine among U.S. Health Care Personnel. N. Engl. J. Med. 2021;385:e90. doi: 10.1056/NEJMoa2106599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei S., Yamana T.K., Kandula S., Galanti M., Shaman J. Burden and characteristics of COVID-19 in the United States during 2020. Nature. 2021;598:338–341. doi: 10.1038/s41586-021-03914-4. [DOI] [PubMed] [Google Scholar]
- 30.Dahabreh I.J., Robertson S.E., Petito L.C., Hernán M.A., Steingrimsson J.A. Efficient and Robust Methods for Causally Interpretable Meta-Analysis: Transporting Inferences from Multiple Randomized Trials to a Target Population. Biometrics. 2023;79:1057–1072. doi: 10.1111/biom.13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rott K.W., Bronfort G., Chu H., Huling J.D., Leininger B., Murad M.H., Wang Z., Hodges J.S. Causally interpretable meta-analysis: Clearly defined causal effects and two case studies. Res. Synth. Methods. 2024;15:61–72. doi: 10.1002/jrsm.1671. [DOI] [PubMed] [Google Scholar]
- 32.Han L., Hou J., Cho K., Duan R., Cai T. Federated Adaptive Causal Estimation (FACE) of Target Treatment Effects. arXiv. 2021;1234:1234. https://arxiv.org/abs/2112.09313 Preprint at. [Google Scholar]
- 33.Han L., Li Y., Niknam B.A., Zubizarreta J.R. Privacy-Preserving, Communication-Efficient, and Target-Flexible Hospital Quality Measurement. arXiv. 2022;1234:1234. https://arxiv.org/abs/2203.00768 Preprint at. [Google Scholar]
- 34.Clark J.M., Rott K.W., Hodges J.S., Huling J.D. Causally-Interpretable Random-Effects Meta-Analysis. arXiv. 2023;1234:1234. https://arxiv.org/abs/2302.03544 Preprint at. [Google Scholar]
- 35.Zheng Y.-B., Zeng N., Yuan K., Tian S.-S., Yang Y.-B., Gao N., Chen X., Zhang A.-Y., Kondratiuk A.L., Shi P.-P., et al. Prevalence and risk factor for long COVID in children and adolescents: A meta-analysis and systematic review. J. Infect. Public Health. 2023;16:660–672. doi: 10.1016/j.jiph.2023.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.PEDSnet Common Data Model https://pedsnet.org/data/common-data-model/.
- 37.OMOP Common Data Model https://ohdsi.github.io/CommonDataModel/.
- 38.Bandeen-roche K., Miglioretti D.L., Zeger S.L., Rathouz P.J. Latent Variable Regression for Multiple Discrete Outcomes. J. Am. Stat. Assoc. 1997;92:1375–1386. doi: 10.1080/01621459.1997.10473658. [DOI] [Google Scholar]
- 39.Dempster A.P., Laird N.M., Rubin D.B. Maximum Likelihood from Incomplete Data Via the EM Algorithm. J. Roy. Stat. Soc. B. 1977;39:1–22. doi: 10.1111/j.2517-6161.1977.tb01600.x. [DOI] [Google Scholar]
- 40.Wu Q. Zenodo; 2024. Penncil/Latent-TL: Source Code of “Post-Acute Healthcare Demands Following SARS-CoV-2 Infection: A Hospital-specific Investigation through Heterogeneous Latent Transfer Learning.”. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data are not publicly available due to privacy concerns. The individual de-identified participant data will not be shared. The individual de-identified participant data that support our findings can only be shared with qualified researchers after a successful application for PEDSnet Study Approval (https://pedsnet.org/research/accessing-pedsnet/request-pedsnet-study-approval/). These requests will be evaluated by the steering committee and processed by the coordinating center.
-
•
The source code and an illustration based on synthetic data are publicly available on GitHub at https://github.com/Penncil/Latent-TL and have been archived at Zenodo.40