Abstract
Background
Peritoneal metastasis, a major complication of colorectal cancer (CRC), often leads to poor quality of life and unfavorable outcomes. Despite numerous studies characterizing its biological features in CRC, intratumor heterogeneity and interactions between cancer cells and tumor microenvironment cells remain poorly understood.
Methods
To explore these aspects, we performed single-cell transcriptome analysis of matched primary tumor and peritoneal metastasis samples from a treatment-naïve patient.
Results
Our analysis revealed enrichment of “tip” endothelial cells in the primary tumor, driving angiogenic sprouting, whereas these cells were absent in peritoneal metastases. Moreover, cancer cells in peritoneal metastasis displayed a distinct expression signature associated with epithelial–mesenchymal transition and tumor invasiveness. Analysis of cell–cell communication between endothelial and tumor cells revealed decreased VEGF signaling and increased CXCL–ACKR1 interactions in peritoneal metastasis.
Conclusions
Although limited by its N-of-1 design and requiring further validation, our study provides preliminary observations suggesting that alterations in cancer-endothelial cell interactions could reduce dependence on VEGF signaling and influence immune cell infiltration in CRC peritoneal metastasis.
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related mortality worldwide [1]. Metastases, particularly to the liver and lungs, are a major contributor to this mortality. Systemic chemotherapy has shown efficacy in treating these metastases, leading to improved overall survival. However, patients with peritoneal metastasis (PM) typically have poor outcomes due to the lack of effective treatment options [2]. Although synchronous PM is diagnosed in only around 5% of CRC cases, prior studies suggest that its prevalence may be higher than previously recognized, underscoring the persistent gaps in our understanding of its biological characteristics [3].
The tumor microenvironment (TME) comprises various nonmalignant cells, including immune, fibroblast, and endothelial cells (ECs), playing critical roles in cancer development and progression [4, 5]. Tumor-associated ECs, generated through tumor angiogenesis, are crucial for supplying sufficient nutrients and oxygen to cancer cells and facilitating metastasis [6]. The vascular endothelial growth factor (VEGF) signaling pathway is a major driver of tumor angiogenesis, and tumor cells often produce VEGFA to promote EC proliferation, migration, and vascular formation [7]. Although VEGF inhibitors, such as bevacizumab, have improved outcomes in metastatic and recurrent CRC [8], resistance to these drugs has been observed. Additionally, despite the development of many VEGF or VEGF receptor inhibitors, some clinical trials have failed to demonstrate their ability to prolong survival in patients with metastatic CRC [9]. Therefore, a better understanding of EC features in CRC is imperative for developing effective strategies targeting tumor angiogenesis.
Single-cell analysis has emerged as a potent tool for studying human tumors at the individual cell level [10]. Traditional methods, such as fluorescence-activated cell sorting, allow analysis of tumor and TME cells via their physical separation; however, they show limitations in capturing the complexity of tumor ecosystems and cell interactions. Single-cell RNA sequencing (scRNA-seq) profiling has revealed intratumor heterogeneity and cell–cell interactions among immune cells, fibroblasts, and cancer cells [11, 12], highlighting its advantages in improving our understanding of tumor ecosystems.
In the present study, we conducted single-cell transcriptome profiling of a primary tumor (PT) and a PM from a patient with untreated stage IV CRC. Our analysis successfully classified each cell type, and we focused our investigation on endothelial and tumor cells. Subclustering analysis of ECs showed that “tip” ECs, leading to angiogenic sprouting, were enriched in the PM but absent in PT tissue. Additionally, the PM exhibited an increased number of epithelial–mesenchymal transition (EMT)/stem-like and invasive tumor cells, whereas differentiated cells were enriched in the PT. Cell–cell interaction analysis between endothelial and tumor cells revealed decreased VEGF signaling intensity in PM tissue. Overall, our study highlights the distinction in cell properties and intercellular communication between the PT and PM in CRC.
Results
Colon cancer cell and tumor heterogeneity
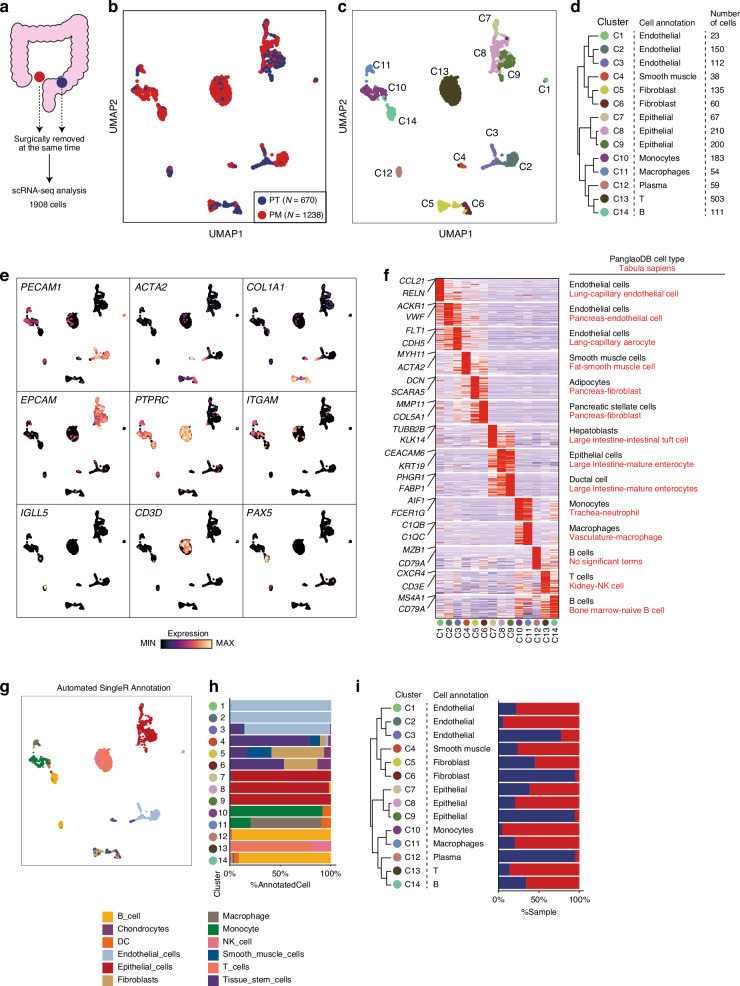
To compare the heterogeneity of gene expression between the PT and PM, we performed single-cell transcriptome analysis of matched PT and PM samples simultaneously removed via surgery from a 61-year-old woman (age at the time of surgery) with no history of cancer (Fig. 1a). The PT originated from the sigmoid colon, whereas the PM was located at the Douglas pouch. Histologically, both were adenocarcinomas, and the TNM stage was IVC (T4aN1bM1).
Fig. 1. Single-cell transcriptome profiling of human CRC with PM from a single patient.
a Study design schema. Uniform manifold approximation and projection (UMAP) colored according to samples (b) or clusters (c). d Consensus cell type annotation table for each cluster. Hierarchical clustering was performed using gene expression values of the top 50 cluster-specific genes. e UMAP overlay of selected cell type markers. f Heatmap showing the top 50 cluster-specific genes. Annotations (right) indicate the most significant cell types predicted via gene enrichment analysis using cell type gene databases: PanglaoDB (black) and Tabula Sapiens (red). g UMAP colored according to SingleR automated cell type annotation. h Bar plot showing the proportion of each SingleR-predicted cell type in each cluster. i Bar plot showing the proportion of each sample in each cluster.
Following quality filtering, we obtained 1,908 high-quality transcriptome profiles from PT and PM samples (refer to Methods). Dimensionality reduction, graph-based clustering, and uniform manifold approximation and projection (UMAP) performed via Seurat were used to classify cell types based on gene expression patterns, with 14 distinct cell clusters identified (C1–C14; Fig. 1b–d). For cell type identification, consensus annotation was conducted using three approaches: (1) examining cell type marker gene expression levels (Figs. 1e), (2) identifying differentially expressed genes (DEGs) in each cluster (Fig. 1f), and (3) performing automated cell type annotation (Fig. 1g, h).
Based on a consensus of the three approaches, clusters were annotated as ECs (C1–3), smooth muscle cells (C4), fibroblasts (C5 and C6), epithelial cells (C7–9), monocytes (C10), macrophages (C11), plasma cells (C12), T cells (C13), and B cells (C14) (Fig. 1d; Supplementary Table 1). Notably, immune cells, including monocytes, macrophages, T cells, and B cells, were enriched in the PM sample compared with the PT sample (Fig. 1l). The number of DEGs identified in these immune cells between the PT and the PM was low (Supplementary Table 2), implying that the difference in the expression program of immune cells between the PT and the PM may be small in this patient. Taken together, we were able to identify cancer and TME cells, highlighting the proportional differences between PT and PM samples.
Tip ECs exist in the PT but not in the PM
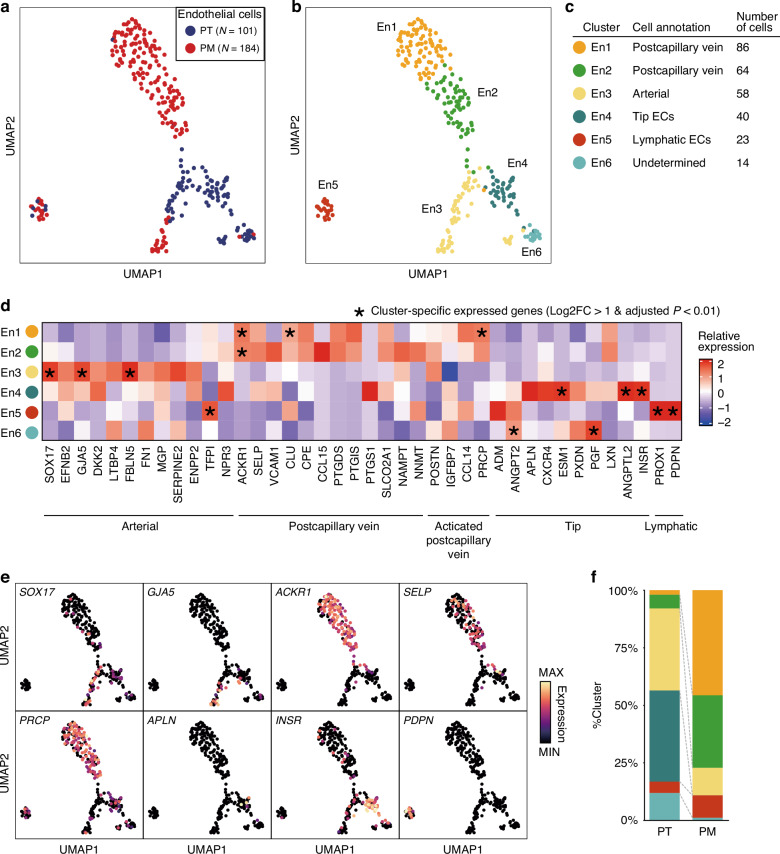
To further investigate EC differences between PT and PM tissues, we performed subclustering of 295 ECs, identifying 6 clusters (En1–En6; Fig. 2a–c). To classify these EC clusters, we examined the expression levels of typical EC subtype markers (Fig. 2d, e; Supplementary Table 3). En1 and En2 showed relatively heightened expression levels of postcapillary vein markers, including ACKR1 and SELP. En1 and En2 also expressed certain marker genes, including CCL14 and PRCP, typically associated with high endothelial venules, which are involved in immune cell recruitment. En3 displayed significantly high expression levels of arterial EC markers, including SOX17 and GJA5. En4 was classified as “tip” ECs, leading to angiogenic sprouting, expressing APLN, INSR, and several tip endothelial markers [13]. En5 exhibited a distinct expression profile compared with the other clusters (Fig. 2b), with high expression levels of lymphatic EC genes. En6, the smallest cluster (N = 14), remained unclassified.
Fig. 2. Subclustering of endothelial cells in the PT and the PM.
UMAP visualization colored according to samples (a) and clusters (b). c Endothelial subtype annotation table. d Heatmap illustrating the relative expression of the endothelial subtype markers in each cluster. Asterisks (*) mean cluster-specific expressed genes (Log2FC > 1 and adjusted P-value < 0.01) compared to other clusters. e UMAP overlay representing expression levels of selected subtype markers. f Bar plot showing the cluster population in each sample.
Comparison of PT and PM EC components revealed abundant tip ECs (En4) in the PT but not in the PM (Fig. 2f). Most ECs in the PM were postcapillary vein cells, whereas arterial ECs were observed less frequently in the PM than in the PT. Our findings illustrate differences in EC types between the PT and the PM, notably the enrichment of tip ECs in the PT, implying heightened and diminished angiogenesis in the PT and the PM, respectively.
Differentiated and VEGF-expressing cancer cell numbers are decreased while EMT and invasive cancer cell numbers are increased in the PM
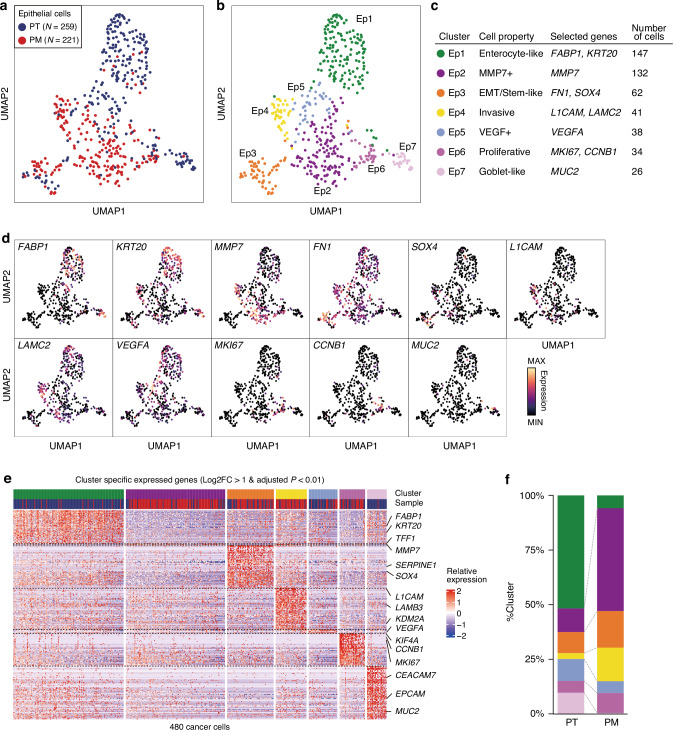
To delineate cancer cell heterogeneity between the PT and PM, we subclustered 480 epithelial cells, identifying 7 clusters (Ep1–Ep7; Fig. 3a–c). To describe the properties of each epithelial cell cluster, we identified each cluster’s DEGs (Log2FC > 1 and adjusted P-value < 0.01; Fig. 3d, e; Supplementary Table 4). Ep1 expressed enterocyte differentiation markers, including FABP1 and KRT20. Ep2 showed few significantly expressed genes, one of which was MMP7, associated with poor prognosis in CRC and potentially involved in invasion and metastasis [14, 15]. Ep3 exhibited significantly high expression of EMT markers, including FN1 and SERPINE1 [16, 17], as well as SOX4, linked to stemness and tumor metastasis [18]. Ep4 expressed L1CAM, involved in metastatic ability [19], as well as laminins, associated with cancer dissemination [20]. Ep5 showed high VEGFA expression. Ep6, a proliferating cell cluster, expressed cell cycle markers, including MKI67 and CCNB1. Ep7, a goblet-like cell cluster, expressed MUC2, typically synthesized by secretory intestinal goblet cells. In summary, the clusters were annotated as follows: Ep1, enterocyte-like; Ep2, MMP7+; Ep3, EMT/stem-like; Ep4, invasive; Ep5, VEGF+; Ep6, proliferative; and Ep7, goblet-like (Fig. 3c).
Fig. 3. Subclustering of epithelial cells in the PT and the PM.
UMAP visualization colored according to samples (a) and clusters (b). c Epithelial cell property annotation table. d UMAP overlay representing expression levels of distinct markers. e Heatmap depicting the relative expression of cluster-specific expressed genes (Log2FC > 1 and adjusted P-value < 0.01) in each cell. f Bar plot showing the cluster population in each sample.
PT and PM tissues showed different proportions of each cluster. Compared with the PT, differentiated cell types, including enterocyte-like (Ep1) and goblet-like (Ep7) clusters, were markedly decreased in the PM (Fig. 3f). Conversely, MMP7+ (Ep2), EMT/stem-like (Ep3), and invasive (Ep4) clusters were more abundant in the PM. Importantly, VEGF+ cell (Ep5) counts were higher in the PT than in the PM, consistent with observed tip EC enrichment in PT tissue. Taken together, these findings suggest that PT epithelial cells were differentiated and angiogenesis-related, whereas PM epithelial cells were dedifferentiated and invasive.
Crosstalk switch from VEGF to CXCL signaling between cancer cells and ECs in the PT and PM
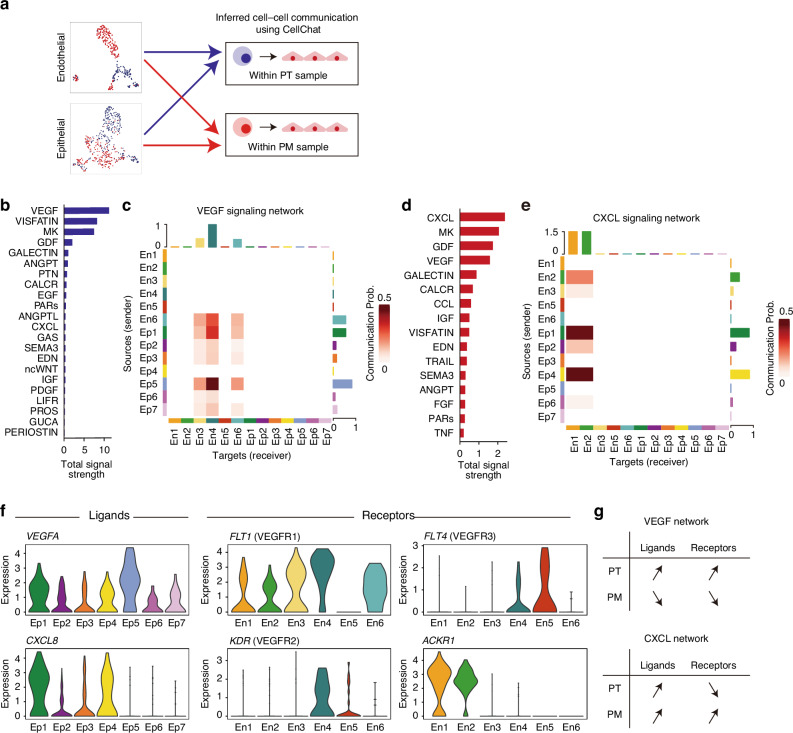
To investigate the interaction between TME cells and cancer cells, we next performed cell-cell communication analysis between TME cell clusters and epithelial cells. Interestingly, the endothelial clusters (Cluster 3 in the PT; Cluster 2 in the PM) are the strongest receiver of the interaction signals in both PT and PM (Supplementary Fig. 1). Subset analysis of endothelial and epithelial cells highlighted substantial differences in cell characteristics between the PT and PM (Figs. 2 and 3). Then, we conducted the interaction analysis between only ECs and epithelial cells in each sample (Fig. 4a). In PT tissue, VEGF signaling was the strongest pathway between each cluster (Fig. 4b). Communication analysis revealed that the VEGF+ cluster (Ep5) was the primary sender of VEGF signals, whereas the tip ECs (En4) were the most prominent receiver of VEGF signals (Fig. 4c). Conversely, in PM tissues, the CXCL pathway exhibited the strongest signal between clusters (Fig. 4d). The invasive cell cluster (Ep4) was the principal sender of CXCL signals, whereas postcapillary vein cells (En1 and En2) were the most prominent receivers of CXCL signals (Fig. 4e). These results indicate a shift from VEGF signaling in the PT to CXCR signaling in the PM occurring in the crosstalk pathway between endothelial and epithelial cells.
Fig. 4. Cell–cell communication across endothelial and epithelial cells.
a Cell–cell interaction analysis schema. b Bar plot showing total signal strength in endothelial and epithelial cells in the PT. c Heatmap illustrating the communication probability of the VEGF signaling pathway across each endothelial or epithelial cluster. d Bar plot showing total signal strength in endothelial and epithelial cells in the PM. e Heatmap depicting the communication probability of the CXCL signaling pathway across each endothelial or epithelial cluster. f Violin plot showing expression levels of selected ligands and receptors in each cluster. g Expression patterns of the ligands and receptors in the VEGF and CXCL signaling pathways.
In the VEGF signaling pathway, the VEGFA ligand was significantly highly expressed in the VEGF+ cluster (Ep5; log2FC = 2.3, adjusted P = 0.005, compared to other clusters), as previously shown (Figs. 3e and 4f). The receptor FLT1 (VEGFR1) was significantly highly expressed in tip ECs (En4; log2FC = 2.1, adjusted P = 1.1e-9), enriched in the PT (Fig. 4f). KDR (VEGFR2) was highly expressed in tip endothelial (En4; log2FC = 0.81, adjusted P = 0.056), whereas FLT4 (VEGFR3), a lymphatic EC-specific VEGF receptor, was relatively expressed in lymphatic ECs (En5; log2FC = 1.6, adjusted P = 0.65). In the CXCL pathway, the ligand CXCL8 was significantly expressed in both enterocyte-like cell clusters (Ep1; log2FC = 1.7, adjusted P = 3.0e-19), enriched in the PT, and invasive cell clusters (Ep4), enriched in the PM (Fig. 4f), whereas the receptor ACKR1 was expressed only in postcapillary veins (En1; log2FC = 1.7, adjusted P = 9.6e-9 and En2; log2FC = 1.0, adjusted P = 1.2e-8), enriched in the PM. Collectively, these results highlight the differences between the two signaling pathways: both ligands and receptors in the VEGF pathway were highly expressed in the PT but not in the PM, whereas ligands in the CXCL pathway were expressed in both the PT and PM while receptors were specifically expressed in the PM (Fig. 4g).
Discussion
In recent years, studies have emphasized the heterogeneity of tumor-associated ECs. For instance, a study using scRNA-seq to investigate lung tumor ECs resulted in a molecular atlas of various EC types, highlighting the importance of tip cells for patient survival and sensitivity to VEGF signaling blockade [13]. Additionally, this study showed that treatment with a VEGFR tyrosine kinase inhibitor, vatalanib (PTK787), reduced the proportion of tip cells, whereas postcapillary veins exhibited lower sensitivity to this drug. Despite complete vatalanib-mediated blockade of the VEGFR pathway, two clinical trials, namely CONFIRM-1 (for first-line treatment) and CONFIRM-2 (for second-line treatment), evaluating the drug’s efficacy against metastatic CRC failed to show a significant improvement in overall survival [21, 22].
Our study, although limited by its N-of-1 design and the relatively small number of cells analyzed, offers preliminary observations that may guide future research. Specifically, we found distinct cellular characteristics and interactions between PT and PM in CRC, suggesting potential differences in endothelial-cancer cell communication. Below, we explore key findings from our analysis and discuss their implications.
In the present study, we observed the presence of tip cells specifically in PT tissue and an increased number of postcapillary veins in the PM, suggesting that ECs in metastatic tumors may lack expression of VEGFRs, particularly VEGFR2, a critical factor in tumor angiogenesis [23]. This finding implies that VEGFR blockade may be less effective for treating CRC with PM. We also observed a significant increase in postcapillary vein ECs expressing ACKR1, also known as Duffy antigen receptor chemokines, in PM tissue. ACKR1 is a chemokine receptor classified as a “silent” heptahelical receptor. These receptors do not bind to G proteins; therefore, they do not produce detectable intracellular signals [24]. A previous study revealed that ACKR1 promotes inflammation by binding to CXCL8 and other chemokines, facilitating their internalization and transport from the basolateral side to the apical side of ECs [25]. This process enhances the immobilization and presentation of these chemokines to leukocytes, ultimately promoting their extravasation and contributing to the inflammatory response. In the present study, the increased infiltration of immune cells observed in PM tissue compared with PT tissue (Fig. 1i) aligns with the function of ACKR1. These findings suggest that the reprogramming of interactions between ECs and cancer cells may have transformed the PM into a “hot tumor.”
In conclusion, while our study provides insights into endothelial-cancer cell interactions, its N-of-1 design and limited cell numbers restrict the generalizability of the findings. Further investigations with large sample sizes and mechanistic studies will be essential to validate these observations and better understand how these interactions influence metastatic processes.
Methods
Clinical specimens
Colon cancer samples were obtained from surgically removed tumors. Tissues were dissociated into single cells using the MACS Tumor Dissociation Kit and a gentle MACS dissociator (Miltenyi Biotec) following the manufacturer’s instructions. Written informed consent was obtained from the participant prior to sample collection. The protocol received approval from the institutional ethical committee of Cancer Institute Hospital, Japanese Foundation for Cancer Research (No. 2013-1003). The study was performed in accordance with the Declaration of Helsinki.
Single-cell RNA-seq experiment
scRNA-seq libraries were generated using the BD Rhapsody Single-Cell Analysis System (BD, New Jersey, USA) according to the manufacturer’s instructions. Briefly, cells were labeled using the BD Single-Cell Multiplexing Kit after thawing. The labeled cells were then washed, pooled, and loaded onto the BD Rhapsody microwell cartridge, and cDNA was synthesized using the BD Rhapsody Whole Transcriptome Analysis Amplification Kit. The resulting gene expression and cellular label libraries were sequenced on an Illumina NextSeq 550 platform (Illumina, California, USA) with paired-end reads (read1, 75 bp; index1, 8 bp; read2, 75 bp). Sequencing data were processed using the BD Rhapsody Analysis pipeline on the Seven Bridges Genomics platform and subsequently converted into a gene expression count matrix.
Data analysis
Preprocess, clustering, visualization, and identification of DEGs
The web-based analysis pipeline for BD Rhapsody and BD Precise ASSAYS (https://www.sevenbridges.com/bdgenomics/) was employed to generate the count matrix, with RSEC counts used as the input count matrix. The Seurat package [26] was used for downstream analysis. Low-quality cells with >40% mitochondrial RNAs and <400 or >9000 features were filtered out. For filtered cells, the transcript count matrix was normalized to the total number of counts for the cell and multiplied by a scaling factor of 10,000. The normalized values were then natural-log transformed using Seurat’s “NormalizeData()” function, followed by a linear transformation applied via the “ScaleData()” function. Principal component analysis was performed using the “RunPCA()” function, with the top 2000 highest variable features identified using the “FindVariableFeatures()” function with the vst selection method. Subsequently, Seurat’s standard clustering procedures were performed using the “FindNeighbors()” and “FindClusters()” functions with the top 20 principal components (PCs) and a resolution of 0.5. Data visualization was achieved through the “RunUMAP()” function, with the same PCs employed to identify clusters. The “FindAllMarkers()” function with “only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25” was employed to identify DEGs for each cluster (Only test genes that are detected in a minimum of 25% of cells in one of the two groups, and that show at least a 0.25-fold difference between the two groups of cells). For subclustering of endothelial and epithelial cells, 10 and 20 PCs were used with resolutions of 0.8 and 0.6, respectively.
Cell type annotation
We performed three different analyses to identify cell types: (1) lineage marker gene estimation, such as PECAM1 for ECs, ACTA2 for smooth muscle cells, COL1A2 for fibroblasts, EPCAM for epithelial cells, ITGAM for myeloid cells, IGLL5 for plasma cells (C12: IGLL5+), CD3D for T cells, and PAX5 for B cells; (2) gene set enrichment analysis using the top 50 DEGs and cell type annotation gene sets, including PanglaoDB [27] and Tabula Sapiens [28]; and (3) automated annotation using SingleR [29].
Cell–cell interaction analysis
The CellChat package [30] was used for cell–cell communication analysis. The “secreted signaling” subset was employed as the communication database, obtained using “subsetDB(search = “Secreted Signaling”).” Each Seurat object of the PT and PM was employed as input to create a CellChat object using the “createCellChat()” function. Communication analysis was achieved using the standard CellChat flow.
Statistics and reproducibility
For identifying cluster-specific DEGs, we used Seurat’s “FindAllMarkers()” function and determined genes with log2FC > 1 and adjusted P-value < 0.01 as DEGs.
Supplementary information
Acknowledgements
We would like to thank Dr. Liying Yang for her experimental support. We also thank Enago (www.enago.jp) for their English language review.
Author contributions
YS, TY, TM, and TA recruited patients and obtained clinical specimens. KK performed data analysis. YS processed tumor samples and performed scRNA-seq experiments. SM, KO, SM, YK, TA, and RM supervised all the work. YS, KK, and RM wrote the manuscript. All the authors discussed the results and commented on the paper.
Funding
This work was supported in part by grants from JSPS KAKENHI (JP24K02312 to RM, JP24K18499 to KK), the Japan Agency for Medical Research and Development (AMED; JP24ama221606 to RM, JP23ama221129 to RM, JP23ama221228 to RM, and JP23ama221504 to RM), and the Project Mirai Cancer Research Grants to KK.
Data availability
The processed scRNA-seq data are available from the lead contact upon reasonable request.
Code availability
The R code used to conduct the analyses is available from the lead contact upon reasonable request.
Competing interests
All authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuri Sakimoto, Kohei Kumegawa.
Supplementary information
The online version contains supplementary material available at 10.1038/s44276-024-00112-3.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2.Franko J, Shi Q, Meyers JP, Maughan TS, Adams RA, Seymour MT, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol. 2016;17:1709–19. [DOI] [PubMed] [Google Scholar]
- 3.Bakkers C, Lurvink RJ, Rijken A, Nienhuijs SW, Kok NF, Creemers GJ, et al. Treatment strategies and prognosis of patients with synchronous or metachronous colorectal peritoneal metastases: a population-based study. Ann Surg Oncol. 2021;28:9073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41:374–403. [DOI] [PubMed] [Google Scholar]
- 6.Yao X, Zeng Y Tumor-associated endothelial cells: origin, characteristics and role in metastasis and anti-angiogenic resistance. Front Physiol 2023;14. 10.3389/fphys.2023.1199225. [DOI] [PMC free article] [PubMed]
- 7.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–27. [DOI] [PubMed] [Google Scholar]
- 8.Cervantes A, Adam R, Roselló S, Arnold D, Normanno N, Taïeb J, et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:10–32. [DOI] [PubMed] [Google Scholar]
- 9.Sobrero AF, Bruzzi P. Vatalanib in advanced colorectal cancer: two studies with identical results. J Clin Oncol. 2011;29:1938–40. [DOI] [PubMed] [Google Scholar]
- 10.Suvà ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell. 2019;75:7–12. [DOI] [PubMed] [Google Scholar]
- 11.Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M, et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12:670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goveia J, Rohlenova K, Taverna F, Treps L, Conradi L-C, Pircher A, et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020;37:21–36.e13. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Ke X. MMP7 as a potential biomarker of colon cancer and its prognostic value by bioinformatics analysis. Medicine. 2021;100:e24953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Nan X, Shi X, Mu X, Liu B, Zhu H, et al. SREBP1 promotes the invasion of colorectal cancer accompanied upregulation of MMP7 expression and NF-κB pathway activation. BMC Cancer. 2019;19:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshmukh AP, Vasaikar SV, Tomczak K, Tripathi S, den Hollander P, Arslan E, et al. Identification of EMT signaling cross-talk and gene regulatory networks by single-cell RNA sequencing. Proc. Natl. Acad. Sci. 2021;118. 10.1073/pnas.2102050118. [DOI] [PMC free article] [PubMed]
- 17.Hu B, Chen Z, Wang X, Chen F, Song Z, Cao C. MicroRNA-148a-3p directly targets SERPINE1 to suppress EMT-mediated colon adenocarcinoma progression. Cancer Manag Res. 2021;13:6349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno CS. SOX4: the unappreciated oncogene. Semin Cancer Biol. 2020;67:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganesh K, Basnet H, Kaygusuz Y, Laughney AM, He L, Sharma R, et al. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat Cancer. 2020;1:28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee S, Lo W-C, Majumder P, Roy D, Ghorai M, Shaikh NK, et al. Multiple roles for basement membrane proteins in cancer progression and EMT. Eur J Cell Biol. 2022;101:151220. [DOI] [PubMed] [Google Scholar]
- 21.Hecht JR, Trarbach T, Hainsworth JD, Major P, Jäger E, Wolff RA, et al. Randomized, placebo-controlled, phase III study of first-line oxaliplatin-based chemotherapy plus PTK787/ZK 222584, an oral vascular endothelial growth factor receptor inhibitor, in patients with metastatic colorectal adenocarcinoma. J Clin Oncol. 2011;29:1997–2003. [DOI] [PubMed] [Google Scholar]
- 22.Van Cutsem E, Bajetta E, Valle J, Köhne C-H, Randolph Hecht J, Moore M, et al. Randomized, placebo-controlled, phase III study of oxaliplatin, fluorouracil, and leucovorin with or without PTK787/ZK 222584 in patients with previously treated metastatic colorectal adenocarcinoma. J Clin Oncol. 2011;29:2004–10. [DOI] [PubMed] [Google Scholar]
- 23.Jiang X, Wang J, Deng X, Xiong F, Zhang S, Gong Z, et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res. 2020;39:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horuk R. The Duffy antigen receptor for chemokines DARC/ACKR1. Front Immunol. 2015;6. 10.3389/fimmu.2015.00279. [DOI] [PMC free article] [PubMed]
- 25.Crawford KS, Volkman BF. Prospects for targeting ACKR1 in cancer and other diseases. Front Immunol. 2023;14. 10.3389/fimmu.2023.1111960. [DOI] [PMC free article] [PubMed]
- 26.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franzén O, Gan L-M, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019; 2019. 10.1093/database/baz046. [DOI] [PMC free article] [PubMed]
- 28.Jones RC, Karkanias J, Krasnow MA, Pisco AO, Quake SR, Salzman J. et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 2022; 376. 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed]
- 29.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed scRNA-seq data are available from the lead contact upon reasonable request.
The R code used to conduct the analyses is available from the lead contact upon reasonable request.