Abstract
The dynamic modulation of the aggregation process of small molecules represents an important research objective for scientists. However, the complex and dynamic nature of internal environments in vivo impedes controllable aggregation processes of single molecules. In this study, we successfully achieve tumor-targeted aggregation of an aggregation-induced emission photosensitizer (AIE-PS), TBmA, with the catalysis of a tumor-overexpressed enzyme, γ-Glutamyl Transferase (GGT). Mechanistic investigations reveal that TBmA-Glu can be activated by GGT through cleavage of the γ-glutamyl bond and releasing TBmA. The poor water solubility of TBmA induces its aggregation, leading to aggregation-enhanced emission and photodynamic activities. The TBmA-Glu not only induces glutathione (GSH) depletion through GGT photo-degradation but also triggers lipid peroxidation accumulation and ferroptosis in cancer cells through photodynamic therapy. Finally, the in vivo studies conducted on female mice using both tumor xenograft and orthotopic liver cancer models have also demonstrated the significant anti-cancer effects of TBmA-Glu. The exceptional cancer-targeting ability and therapeutic efficiency demonstrated by this GGT activatable AIE-PS highlights enzymatic-mediated modulation as an effective approach for regulating small molecule aggregation intracellularly, thereby advancing innovative therapeutic strategies for various diseases.
Subject terms: Mechanism of action, Biocatalysis
Intracellular regulation of small molecule aggregation is challenging. Here, the authors achieved tumor-targeted aggregation of an aggregation-induced emission photosensitizer (AIE-PS), TBmA, using γ-glutamyl transferase, an enzyme overexpressed in tumors, and showed this causes ferroptosis in cancer cells through photodynamic therapy.
Introduction
Molecular science is the foundation of modern scientific investigation, offering invaluable insights into the comprehension and exploration of our world1,2. As an implementation of the research philosophy of reductionism in the physical science area, molecular science underlines that the properties and performance of a substance across different length scales are determined mainly by its molecular structure. This fundamental principle holds true in numerous instances, which, however, also exhibit its limitations. In intricate and extensive systems that are beyond mere replicas of individual molecules, it is impossible for researchers to derive all system properties from their molecular structure alone. More and more research demonstrated that the aggregation of molecules could also generate functions beyond molecular properties. For instance, the crystalline and amorphous states of the same molecules could exhibit totally different properties just because the aggregation forms are different. More interestingly, some aggregates could even generate properties that the molecular species lacked or could not present in the isolated state. One of the prototype examples is the phenomenon of aggregation-induced emission (AIE), whose luminescence is undetectable in the single molecular state due to molecular motion but is much enhanced at aggregate state due to the suppression of excited state molecular motion upon aggregation3–5. Since most of the materials utilized by humans exist in the form of aggregates, understanding the aggregation process and realizing the precise tuning of the aggregation are highly valued in materials science.
Living organisms possess their unique regulatory mechanisms for the aggregation of internal biomolecules6,7, including the formation of hierarchical protein structures8 and the assembly and disassembly of chromatin, etc9. However, current methods for tuning the aggregation of exogenous molecules in the organism’s environment are inadequate, let alone achieving targeted aggregation at specific sites within the organism. Despite significant progress made thus far, the aggregation of exogenous molecular species mainly depends on their inherent properties like the hydrophilic-hydrophobic interaction and electrostatic interaction within the physiological microenvironment10–14. It is significant to achieve precise and controlled molecular aggregation within living systems and harness novel properties exhibited by the aggregates distinct for diagnosing and treating diseases. Nevertheless, due to the complex and dynamic nature of living cells and in vivo environments, implementing controlled processes from single molecules to aggregates still presents substantial challenges.
Enzymes are indispensable constituents of living organisms, capable of catalyzing diverse biochemical reactions. Aberrant enzyme activity or expression is intricately associated with the occurrence and progression of cancers and other diseases15–18. By identifying a molecule that selectively targets overexpressed enzymes in tumors, the aggregation of the molecules can be modulated by utilizing the catalytic reaction characteristics of the enzyme. Therefore, rational utilization of enzymatic reactions potentially enables precise molecular aggregation. Furthermore, the resulting properties of molecular aggregates, such as fluorescence and photodynamic activity, can be used in cancer imaging and therapeutic applications19–22. Although enzymatic catalysis has been extensively explored in developing “turn-on” fluorescent probes, investigations into utilizing enzymatic reactions to regulate molecular aggregation states to realize more sophisticated and multifunctional biomedical applications remain rare23–25.
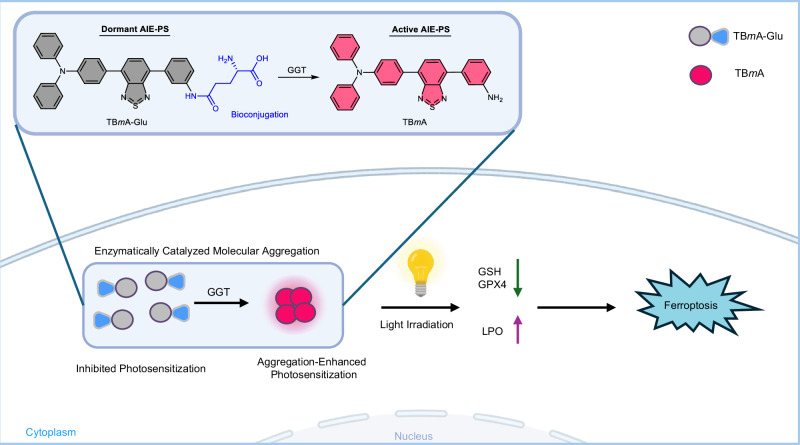
In this study, we developed a γ-glutamyltransferase (GGT) activated aggregation-induced emission photosensitizer (AIE-PS), TBmA-Glu, that can target and aggregate in cancer cells with the help of GGT. GGT is an enzyme highly expressed in various tumor cells, which has been extensively employed as a marker for tumor tracking and therapy23,26–29. By incorporating a GGT-targeting, water-soluble glutamate group (-Glu)30–32 into the TBmA molecule (Fig. 1), we obtained a water-soluble TBmA-Glu and successfully silenced its AIE property and photodynamic activity (dormant AIE-PS) in aqueous media. GGT can catalyze the cleavage of the water-soluble Glu moiety, inducing the aggregation of poorly soluble TBmA and reactivating its AIE luminescence and photodynamic activity (active AIE-PS). In contrast to conventional photosensitizers, which typically exhibit aggregation-caused quenching properties and poor tumor selectivity33, TBmA-Glu displays aggregation-enhanced luminescence and photodynamic activity in philological conditions through GGT-catalyzed aggregation in cancer cells. Through this strategy, targeted molecular aggregation of AIE-PS is achieved at tumor sites. Meanwhile, tumor tracking, and targeted photodynamic therapy (PDT) are realized by taking advantage of the luminescence properties and photodynamic activity of the aggregate.
Fig. 1. Schematic elucidation of the aggregation-enhanced photodynamic therapy mechanism mediated by TBmA-Glu.
TBmA-Glu can be activated by GGT to form the aggregates of TBmA, which will induce ferroposis of cancer cells.
Results
Synthesis and characterization
The synthetic route to TBmA-Glu and TBpA-Glu is depicted in Supplementary Fig. 1. TBmA and TBpA were synthesized via two successive Suzuki reactions of 4,7-dibromobenzo[c][1,2,5]thiadiazole with 4-(diphenylamino)phenylboronic acid and the corresponding aminophenyl boric acid, respectively. Subsequently, TBmA-Glu and TBpA-Glu were obtained by coupling TBmA or TBpA with Boc-L-glutamic acid 1-tert-butyl ester followed by a TFA-catalyzed deprotection reaction. All intermediates and final products were characterized using 1H NMR spectroscopy, 13C NMR spectroscopy, and high-resolution mass spectrometry (Supplementary Figs. 2–19, Supporting Information).
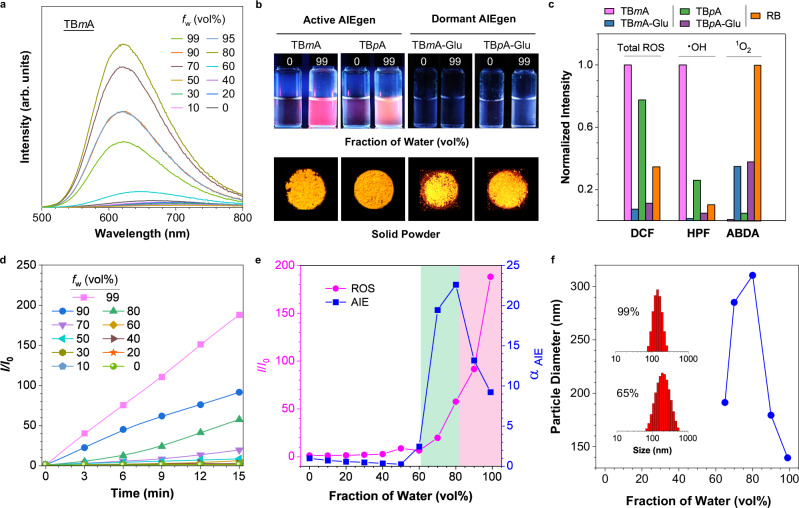
The photophysical properties of TBmA-Glu, TBmA, TBpA-Glu, and TBpA were first explored in MeOH and phosphate buffer solution (PBS). All compounds exhibited a maximum absorption peak at around 450 nm in methanol and showed a slight red shift in PBS (Supplementary Fig. 20). Notably, the emission intensity of all compounds was much stronger in water than in methanol, suggesting their potential AIE activity (Supplementary Fig. 21). The AIE properties of the four compounds were then investigated further. The results demonstrated that TBmA functioned as a typical AIE luminogen (AIEgen), exhibiting enhanced emission intensity upon increasing the water fraction (fw) in the N,N-dimethylformamide (DMF)/water solution while simultaneously displaying strong solid powder emission (Fig. 2a, b). In contrast, TBmA-Glu solely exhibited solid-state emission, adding water into its DMF solution could not cause the aggregation and induce the emission enhancement (Fig. 2b), indicating the Glu-moiety modification endows the TBmA superior water solubility, which is favorable to removing the nonspecific aggregation resulted in background luminescence in physiological conditions (Supplementary Figs. 22, 23). Similarly, both TBpA and TBpA-Glu exhibited identical AIE characteristics as observed for TBmA and TBmA-Glu (Fig. 2b and Supplementary Figs. 22–24).
Fig. 2. Construction and characterization of AIE photosensitizers.
a Emission spectra of TBmA (10 μM) in DMF/water mixtures with different fractions of water (fw). b Photographs of TBmA, TBpA, TBmA-Glu, and TBpA-Glu powders and in DMF solutions (fw = 0%) and DMF/water mixture (fw = 99%) taken at room temperature under 365 nm UV illumination. c The generation of total ROS generation (2’,7’-dichlorofluorescein, DCF), hydroxyl radical (hydroxyphenyl fluorescein, HPF) and singlet oxygen (9,10-anthracenediyl-bis(methylene)dimalonic Acid, ABDA) by photosensitizers (5 μM) after white LED light (predominant emission peaks at 450 and 570 nm, Supplementary Fig. 28) irradiation (20 mW·cm−2) for 15 min using the corresponding ROS indicator in PBS/DMSO (v/v = 99:1). DCF, λex = 488 nm. d The plot of the relative emission intensity (I/I0) of DCF (10 μM) in solutions with different fw containing TBmA (5 μM) versus the irradiation (20 mW·cm−2) time, where I0 = PL intensity of DCFH in solutions with different fw without light irradiation. λex = 488 nm. e plot of αAIE (I/I0, where I0 = PL intensity in pure DMF) of TBmA and I/I0 (where I0 = PL intensity of DCF in pure DMF after irradiation with a white LED array (20 mW·cm−2) for 15 min) of DCF in the presence of TBmA (5 μM) versus fw. f The size of the aggregates of TBmA versus fw. Insertion: the size distribution of the TBmA aggregates in 99% (up) and 65% (down) water fraction.
The aggregation-enhanced photosensitivity
Subsequently, the reactive oxygen species (ROS) generation capabilities of the four compounds were investigated in PBS (Fig. 2c and Supplementary Fig. 25). It was found that TBmA and TBpA produced significantly higher ROS compared to TBmA-Glu and TBpA-Glu, even surpassing the commercial photosensitizer, Rose Bengal (RB). Moreover, TBmA was identified as the most potent photosensitizer among the four compounds. Further analysis revealed that TBmA and TBpA functioned as strong type I photosensitizers (Fig. 2c and Supplementary Fig. 26), while TBmA-Glu and TBpA-Glu acted as very weak type II photosensitizers (Fig. 2c and Supplementary Fig. 27). Although the reason for the different photodynamic processes of the AIE-PSs with/without Glu-modification is still unknown, the findings highlight the significant impact of PS structure on photodynamic activity. Although TBmA and TBpA exhibited aggregation-enhanced ROS generation abilities in the aggregate state (Fig. 2d), they failed to generate any detectable levels of ROS in high DMF concentrations (fw < 40%) solutions (Fig. 2d, e). These results suggest that TBmA and TBpA are not effective photosensitizers in their molecular states but gain ROS generation capabilities when present as aggregates.
We conducted a study to understand the relationship between ROS generation abilities and the aggregation state of the AIE-PSs. To do this, we investigated the correlation between ROS generation, AIE emission, and aggregate size using TBmA (Fig. 2e, f). We found that increased water content did not induce aggregation of TBmA at a low water content (fw < 60%) and we could not detect any emission or ROS production. However, when the fw reached approximately 60%, TBmA started to aggregate, resulting in enhanced emission and ROS generation. The emission intensity peaked at a fw of 80% and decreased afterward as the fw increased, while ROS generation continued to increase with no decrease. The sizes of aggregates began to decrease after reaching their maximum at a fw of 80%, which had a similar change tendency to the emission intensity. The reduction in nanoparticle size resulted in an increased surface area, promoting interactions between PS molecules on the nanoparticle surface and surrounding molecules, thereby promoting efficient photodynamic processes. Considering the competition between luminescence and ROS generation for energy absorbed from excitation light, an increase in ROS generation will result in lower emission intensity. Moreover, the TBmA aggregates exhibited excellent long-term stability (Supplementary Fig. 29a,c) and photodynamic stability (Supplementary Fig. 29a, b), no significant aggregation or degradation was found after dispersed in FBS (fetal bovine serum) solution for 72 h or light irradiated for 30 min. In summary, TBmA exhibits not only AIE property but also an aggregation-enhanced ROS generation, the excellent properties make it a suitable material for therapeutic application. If the targeting aggregation of TBmA in cancer cells can be achieved, the resulting aggregation-induced emission and photodynamic activity of TBmA would not only facilitate tumor tracing but also enable targeted treatment.
Activation of the dormant AIE-PS by GGT
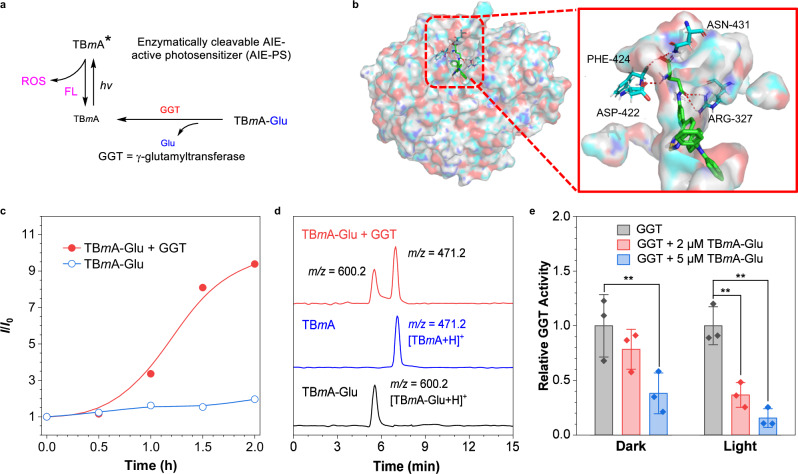
In our proposal, the well-designed dormant AIE-PS TBmA-Glu can be converted into active AIE-PS TBmA in the presence of GGT, thereby inducing the aggregation of TBmA (Fig. 3a). The aggregates of TBmA have been proven to possess potent ROS generation activity under light irradiation. Therefore, we subsequently explored the potential of GGT in inducing the aggregation of TBmA-Glu. The docking results revealed that TBmA-Glu could interact with the amino acid residues (ARG-327, ASP-422, PHE-424, and ASN-431) in the active pocket of GGT (Fig. 3b). The inhibitor constant (Ki value) for TBmA-Glu was determined to be 0.17 μM, which is lower than that of TBpA-Glu and glutamic acid (Supplementary Fig. 30 and Supplementary Table 1), indicating a strong affinity between TBmA-Glu and GGT. Subsequently, the luminescence changes of TBmA-Glu in the presence of GGT were studied. A marked increase in luminescence intensity was observed after co-incubation of TBmA-Glu and GGT (Fig. 3c and Supplementary Fig. 31a), indicating successful cleavage of the -Glu moiety of TBmA-Glu. To validate if the luminescence enhancement was induced by TBmA, a liquid chromatograph mass spectrometer (LC-MS) analysis was performed on the product obtained after a 2-h co-incubation of TBmA-Glu and GGT. A product (m/z = 471.2) with an identical retention time and m/z value of pure TBmA was found (Fig. 3d). These findings provide evidence supporting that GGT can convert dormant AIE-PS (TBmA-Glu) into its active state (TBmA).
Fig. 3. Characterization of the GGT-dependent PDT properties of AIE photosensitizers.
a The schematic diagram of the activation of the AIEgen photosensitizer by GGT. b The molecular docking images of TBmA-Glu with GGT (PDB: 4GG2). The TBmA-Glu is colored green, and the molecular surface of GGT is shown as a colorful surface with transparency. The enlarged images show the hydrogen bonds formed between TBmA-Glu and GGT. c Fluorescence intensity changes of TBmA-Glu incubated in the HEPES buffer at 37 °C in the presence and absence of GGT without light irradiation. d LC-MS curves of TBmA-Glu, TBmA-Glu + GGT (incubated in the buffer for 2 h), and TBmA. e The GGT activity changes in the presence of different concentrations of TBmA-Glu under light irradiation (12 J·cm−2). All assays (n = 3) were biologically independent samples, data expressed as average ± standard error; statistical significance: P values, ns: **P < 0.01, calculated with the one-sided Student’s t-test.
Furthermore, we investigated the changes in TBmA aggregates formed during the GGT catalytic reaction (12 h) by dynamic light scattering (DLS, Supplementary Fig. 32a) and transmission electron microscopy (TEM). The DLS results revealed a gradual increase in the size of aggregates to ~158.0 nm over the initial 4-h period (Supplementary Fig. 32b–e). Subsequently, a steady state size was attained after 4 h of incubation, with no further changes observed even when extending the incubation time to 12 h (Supplementary Fig. 32f–g). The TEM analysis revealed that GGT facilitated the formation of spherical TBmA aggregates, exhibiting an average diameter of ~100 nm (Supplementary Fig. 32h). Additionally, evaluation of the total ROS generation properties of stable TBmA aggregates showed that it induced an ~164-fold increase in the intensity of DCFH after 15 min light irradiation (Supplementary Fig. 33). While this enhancement was slightly lower than the aggregates formed in 99% PBS (188-fold), it was significantly higher than Rose Bengal (67.0-fold). These findings are consistent with previous observations that smaller aggregate size tends to enhance photodynamic efficiency. Our results demonstrate that TBmA aggregates generated by GGT possess potent photodynamic activity.
Given that GGT is a key enzyme in glutathione (GSH) homeostasis34, the high level of GGT could alleviate the oxidative stress of cancer cells35–37. We subsequently investigated the effects of TBmA-Glu on GGT activity. The results suggest that a high concentration of TBmA-Glu (5 μM) can significantly inhibit the catalytic activity of GGT independent of light irradiation (Fig. 3e). This could be ascribed to the potent affinity of TBmA-Glu towards GGT and the aggregation of TBmA, which can block the entry of substrate into the active pocket of the enzyme. Moreover, when light is irradiated, the catalytic activity of GGT decreases sharply with a low concentration of TBmA-Glu (2 μM, Fig. 3e and Supplementary Fig. 31a, b). This suggests that the cleaved product, TBmA, can accumulate near GGT and induce in situ damage to GGT function by generating ROS through photodynamic activity.
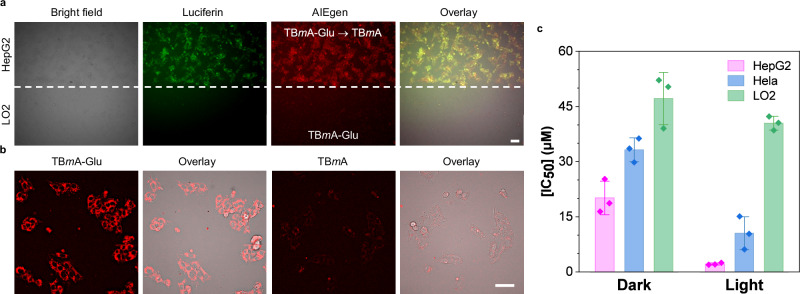
The upregulated expression of GGT in various cancer cell lines has identified it as an appealing target for cancer detection38. The strong GGT binding ability and GGT activable property of TBmA-Glu aroused our interest in investigating its potential in antitumor application. To investigate the selectivity of TBmA-Glu towards cancer cells, co-incubation assays were conducted using a GGT upregulated liver cancer cell line (HepG2) and a normal liver cell line (LO2). The results demonstrated that TBmA-Glu has long-term retention ability in HepG2 cells (Supplementary Fig. 34a). TBmA-Glu was activated in luciferase-expressed HepG2 cells and displayed co-localization with luciferin emission (Fig. 4a), while no significant emission was detected in LO2 cells after 12 h incubation with TBmA-Glu (Supplementary Fig. 34b). Additionally, TBmA-Glu showed remarkable specific accumulation in HepG2 cells compared to TBmA (Fig. 4b and Supplementary Fig. 34c), along with exceptional photostability within living cells (Supplementary Fig. 35a), as evidenced by the absence of significant bleaching even after continuous light irradiation (Supplementary Fig. 35b). These findings highlight the exceptional cancer selectivity of TBmA-Glu as a probe for cancer imaging.
Fig. 4. Characterization of GGT-dependent PDT properties of AIE photosensitizers in cells.
a Confocal laser scanning microscope (CLSM) images of co-incubated cancer (HepG2; luciferase-transfected) and normal (LO2) cells after treatment with TBmA-Glu (5 μM, 12 h). TBmA-Glu, λex = 465 nm, λem = 700 ± 20 nm. Luciferin, λem = 520 ± 20 nm, 120 ms. Three independent experiments were performed. Scale bar: 30 μm. b CLSM images of HepG2 cells after being treated by TBmA-Glu (5 μM) or TBmA (5 μM) for 12 h. Three independent experiments were performed. Scale bar: 30 μm. c The cytotoxicity (IC50, μM) of TBmA-Glu against the cell lines with different GGT expression levels (HepG2 > HeLa > LO2). All assays (n = 3) were biologically independent samples, data expressed as average ± standard error.
As TBmA-Glu functioned as a GGT-activated AIE-PS, we next studied its anticancer activity against cells with different GGT-expressing levels using cytotoxicity tests. The cytotoxicity of TBmA-Glu at both the dark state and photoirradiated state is positively related to the GGT expression level in cells (Fig. 4c). Among all tested compounds, TBmA-Glu showed the most potent photocytotoxicity against the HepG2 cell line, affording IC50 of 20.12 μM (dark condition) and 2.15 μM (light condition), respectively, resulting in the highest phototoxicity index (PI) values of 9.36 (Supplementary Table 2). TBmA-Glu demonstrated minimal toxicity in both dark and light conditions (IC50 > 40 μM) towards the normal cell line (LO2), confirming its excellent cancer selectivity. The broad-spectrum anticancer potential of TBmA-Glu against GGT-overexpressing cancer cells was further demonstrated through subsequent investigations, including OVCAR-5 and murine 4T1 cancer cells (Supplementary Fig. 36). The IC50 values for photodynamic therapy were determined to be 5.13 ± 0.69 μM (OVCAR-5) and 5.28 ± 1.56 μM (4T1), respectively. Moreover, no significant photocytotoxicity was observed in HLF-1 cells with regular GGT expression, nor any significant dark cytotoxicity. Therefore, the potential effectiveness of TBmA-Glu could extend to other cancers that exhibit elevated GGT levels.
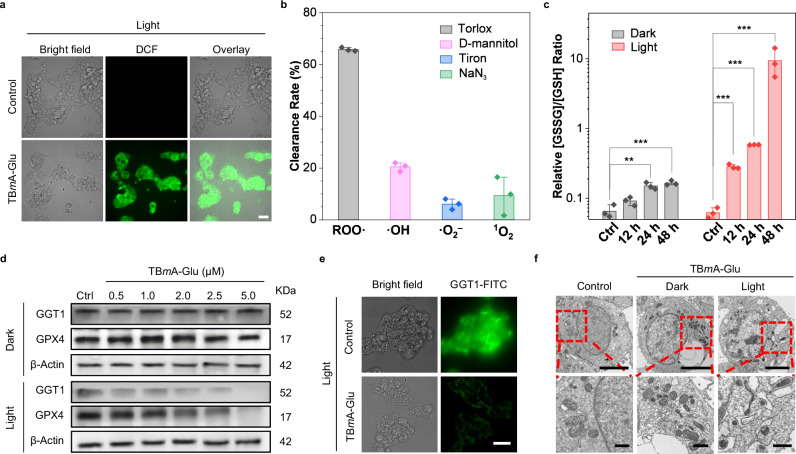
To elucidate the antitumor mechanism of TBmA-Glu, we characterized the ROS generated in HepG2 cells during the PDT process. Firstly, the total ROS level was detected using DCFH-DA (Dichlorodihydrofluorescein diacetate) as a ROS indicator. As shown in Fig. 5a, HepG2 cells exhibited bright green fluorescence in the TBmA-Glu + light treatment group, which indicated that the photosensitizer could promote cells to produce ROS under light irradiation. Interestingly, a slight green fluorescence was observed in the TBmA-Glu dark treatment group (Supplementary Fig. 37), which might be attributed to the inhibition of GGT activity. Since GGT plays a significant role in cancer cells to produce GSH to relieve oxidative stress, inhibiting the GGT activity would lead to elevated ROS levels39. Subsequently, the ROS generated during PDT were identified by the ROS scavenger. Flow cytometry analysis (Supplementary Fig. 38) revealed that using HepG2 cells treated with TBmA-Glu (2 μM) plus irradiation as the control, pre-incubating cells with NaN3 and Tiron (1O2 and O2− specific scavengers, respectively) only caused a slight decrease in the fluorescence intensity of DCF (Fig. 5b and Supplementary Fig. 39). In contrast, pre-incubation with Trolox (ROO· specific scavenger) and D-mannitol (·OH specific scavenger) effectively quenched the generation of DCF fluorescence, indicating that lipid peroxidation and ·OH accounted for the vast majority of TBmA-Glu PDT induced ROS. Lipid oxidation in vitro and in living cells by TBmA was further confirmed using an unsaturated lipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and the lipid peroxidation assay kit. The peroxidation products of DOPE and the lipid peroxidation products, malondialdehyde (MDA), were also detected in the DOPE/TBmA mixture (Supplementary Fig. 40) and the cells after light irradiation (Supplementary Fig. 41).
Fig. 5. Investigation of anticancer mechanism for TBmA-Glu.
a Intracellular ROS generation in HepG2 cells PDT treatment groups, DCFH-DA (10 μM, 10 min) used as the indicator. DCF, λex = 488 nm, λem = 500 ± 20 nm. Scale bar, 20 μm. b The Clearance rates of different ROS scavengers (Trolox: 50 μM (ROO· scavenger); d-mannitol: 50 mM (·OH scavenger); Tiron: 10 mM (·O2– scavenger); NaN3: 5 mM (1O2 scavenger)) on the ROS induced by PDT of TBmA-Glu were evaluated. All assays (n = 3) were biologically independent samples, data expressed as average ± standard error. c Impact of TBmA-Glu (2 μM) on cellular GSSG/GSH ratios. All assays (n = 3) were biologically independent samples, data expressed as average ± standard error. Statistical significance: P values, ***P < 0.001, calculated with the one-sided Student’s t-test. d The expression levels of GGT and GPX4 in HepG2 cells of TBmA-Glu dark and PDT groups. Three independent experiments were performed. e The expression level of GGT1 in HepG2 cells in TBmA-Glu (2 μM) or 1% DMSO (Ctrl) for PDT groups. Three independent experiments were performed. Scale bar, 20 μm. f Transmission electron microscopy images revealed morphological changes in HepG2 cells treated with TBmA-Glu (2 μΜ, 24 h) without and under light irradiation conditions. Enlarged regions are indicated by red rectangles. Three independent experiments were performed. Scale bars represent lengths of 5 μm and 500 nm (for the enlarged images), respectively.
High-level GSH not only facilitates the elimination of ROS generated during cancer cell metabolism but also helps cancer cells overcome drug-induced ROS stress, conferring drug resistance38,40. GSH levels are closely related to GGT function. Therefore, we checked the ratio change of oxidative glutathione (GSSH) to reductive glutathione (GSH) in TBmA-Glu (2 μM) treated HepG2 cells. A time-dependent increase in the ratio of GSSG/GSH was observed upon TBmA-Glu + light treatment (Fig. 5c), attributable to ROS and GGT damage induced by TBmA-Glu mediated PDT. Furthermore, we confirmed the detrimental effects of TBmA-Glu on GGT activity through western blotting and immunofluorescence analysis, which revealed a decreased level of GGT in HepG2 cells after PDT treatment (Fig. 5d, e and Supplementary Fig. 42). This implied that the GGT catalysis resulted in the accumulation of TBmA, which conversely caused damage to GGT by producing ROS. Additionally, prolonged incubation in dark conditions resulted in an increased GSSG/GSH ratio, attributed once again to GGT inhibition by aggregated TBmA (Fig. 5c). These findings indicated that TBmA-Glu can induce lipid peroxidation accumulation and depletion of GSH in HepG2 cells through photodynamic effect.
Since GPX4 plays a crucial role in the clearance of lipid peroxidation by utilizing GSH as its substrate, serving as the primary inhibitor of cellular ferroptosis41,42, we examined the impact of TBmA-Glu on the expression level of GPX4 in HepG2 cells. Compared to the control group, GPX4 levels significantly decreased upon TBmA-Glu + light treatment (Fig. 5d). Moreover, TEM revealed alternations in cell morphology, including shrunken mitochondria with increased bilayer density43 in TBmA-Glu + light-treated cells, which were the symbols of ferroptosis (Fig. 5f). However, no discernible alterations were observed in nuclear morphology. These findings suggested that TBmA-Glu inhibited and impaired GGT through photodynamic activity, leading to oxidative stress, lipid peroxidation accumulation, and GPX4 level decrease, ultimately inducing ferroptosis in HepG2 cells.
In vivo PDT efficacy
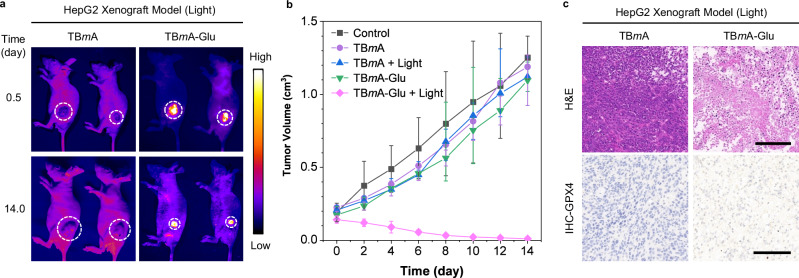
Motivated by the excellent anticancer effect of TBmA-Glu, the in vivo phototherapeutic efficacy of TBmA-Glu was further evaluated on the HepG2 xenograft nude mice model. The results demonstrated that TBmA-Glu (5 mg/kg) was selectively activated in the tumor region 12 h after intravenous injection compared with TBmA (Fig. 6a). Additionally, the tumor volume in the TBmA-Glu treated group under light irradiation significantly decreased compared to other groups at the end of the therapeutic process (Fig. 6a, b and Supplementary Fig. 43). Hematoxylin and eosin (H&E) staining results also confirmed the pronounced anticancer PDT efficacy of TBmA-Glu than other groups (Fig. 6c and Supplementary Fig. 44). Meanwhile, the immunohistochemistry (IHC) results validated the photodynamic effect of TBmA-Glu induced ferroptosis in cancer cells (Fig. 6c and Supplementary Fig. 45). Notably, no noticeable weight loss (Supplementary Fig. 46) or organ damage (Supplementary Fig. 47) occurred during the therapeutic process in mice treated with TBmA-Glu, indicating its excellent biocompatibility and tumor selectivity. In contrast, significant weight loss and liver damage were observed in mice in the TBmA-treated group (Supplementary Figs. 46, 47), which is attributed to the potent photodynamic activity but low tumor selectivity of TBmA.
Fig. 6. Investigation of the in vivo therapeutic effect of TBmA-Glu mediated phototherapy.
a The fluorescence images represent mice in TBmA-Glu and TBmA PDT treatment groups at the start and end of the treatment process. The white circles indicate the region of tumors. TBmA-Glu and TBmA, λex = 500 nm, λem = 700 nm (Long-pass filter). b Tumor growth curves of tumor-bearing mice (n = 5), data expressed as average ± standard error. The red arrows indicate the time points for drug administration. c The H&E (upper panel) and immunohistochemistry of GPX4 (lower panel) in tumor sections from mice in TBmA-Glu treatment groups. Three independent experiments were performed. Scale bars: 50 μm.
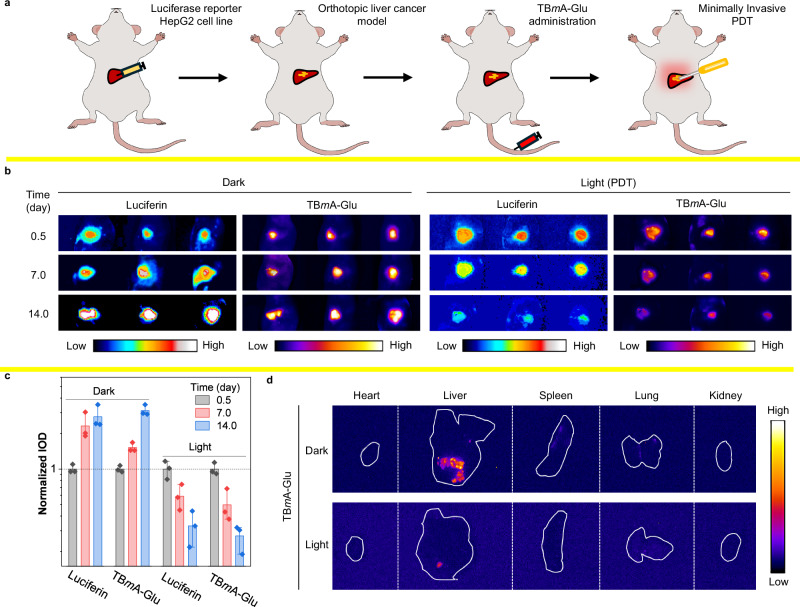
Although TBmA-Glu exhibited excellent PDT efficiency in the subcutaneous tumor model, it is considered that the subcutaneous tumor model could not well simulate the actual characteristics of the tumor microenvironment44,45. Hence, the anticancer efficiency of TBmA-Glu was re-evaluated in an orthotopic liver cancer mouse model. Luciferase reporter HepG2 (HepG2-Luc) cells were injected into the livers of nude mice to establish the orthotopic liver cancer model. TBmA-Glu was given intravenously at a dosage of 5 mg/kg, followed by minimally invasive PDT and in vivo imaging 12 h later (Fig. 7a). The well co-localization of TBmA-Glu luminescence and luciferin fluorescence in HepG2 cells provided compelling evidence to support the advantages of TBmA-Glu as a promising tool for tumor tracking. (Fig. 7b and Supplementary Fig. 48). Furthermore, the decrease of the fluorescence intensity in the PDT groups signified that TBmA-Glu inhibited orthotopic liver cancer (Inhibitory rate = 99.1%, Fig. 7c, d and Supplementary Fig. 49), which is further supported by the H&E stain (Supplementary Fig. 50a), TUNEL stain (Supplementary Fig. 50b) and IHC-Ki67 stain (Supplementary Fig. 51) results of the cancer tissues. These findings highlighted the remarkable PDT efficacy of TBmA-Glu in orthotopic liver cancer models.
Fig. 7. Investigation of the therapeutic effect of TBmA-Glu mediated phototherapy in orthotopic liver cancer mouse models.
a Schematic diagram of the modeling process of a mouse model of liver tumor in situ. b The fluorescence images of the in situ hepatoma mice at the start, middle, and end of the treatment process by TBmA-Glu (5 mg/kg). Luminescence, λem = 550 ± 50 nm (Band-pass filter), 1000 ms. TBmA-Glu, λex = 500 nm, λem = 650 nm (Long-pass filter). c The normalized integrated optical density of cancer in the mice of in situ hepatoma models (n = 3) after treated TBmA-Glu (5 mg/kg) with or without light irradiation, data expressed as average ± standard error. d The fluorescence images of the separated organs of the orthotopic liver cancer mice at the end of the therapeutic process. TBmA-Glu, λex = 500 nm, λem = 650 nm (long-pass filter).
Discussion
In summary, we have developed an efficient AIE-PS named TBmA-Glu, which can be activated by GGT. Unlike traditional AIE-PSs, TBmA-Glu is soluble in water and exists in molecular form in an aqueous solution. When the water-soluble Glu moiety in TBmA-Glu is cleaved by GGT, it triggers the aggregation of insoluble TBmA. By harnessing the “Glu” moiety recognition property of cancer-overexpressed GGT and GGT’s “Glu” cleavage catalytic activities, we realized tumor-targeting aggregation of AIE-PS. The aggregation of TBmA resulted in enhanced emission and boosted photodynamic activity. The PDT process mediated by TBmA-Glu not only damaged GGT, leading to depletion of GSH, but also induced ferroptosis in cancer cells through lipid peroxidation induction and GPX4 downregulation. Consequently, both the HepG2 xenograft and the orthotopic liver cancer models demonstrated excellent targeting efficiency and PDT efficacy of TBmA-Glu. Overall, the results underscore the significant promise of TBmA-Glu as a clinical photosensitizer for tumor theranostics. The enzymatic-mediated modulation strategy also proves to be an effective approach for realizing the regulation of the aggregation process of small molecules within cells.
Methods
Cell lines and culture conditions
The HepG2, HeLa, and LO2 cell lines were sourced from the Cell Bank of the Shanghai Institute of Biochemistry and Cell Biology. HepG2 and HeLa cells were cultured using the Dulbecco’s modified Eagle’s medium (DMEM, Gibco BRL), and LO2 cells were cultured using the Roswell Park Memorial Institute 1640 medium. (RPMI 1640, Gibco BRL) medium. 10% FBS (fetal bovine serum; Gibco BRL), 100 μg/mL streptomycin (Gibco BRL), and 100 U/mL penicillin (Gibco BRL) were added to the culture medium as the supplements. All cells were cultivated in an incubator under conditions of 95% air and 5% CO2, with a temperature set at 37 °C. The cells in the control groups of all cell assays were treated with culture media containing 1% DMSO.
Cell viability assay
The cytotoxicity of TBmA, TBmA-Glu, TBpA, and TBpA-Glu against human cancer cell lines (HepG2 and HeLa) and normal liver cell line (LO2) was measured by MTT assays as previously described46. Briefly, cells (HepG2, 1 × 104 cells per well, HeLa and LO2, 5 × 103 cells per well) were seeded in 96-well plates and cultured for 24 h before treating with the tested compounds. For the PDT groups, light irradiation was performed 12 h after the test compound treatment. 20 μL of MTT solution (5 mg/mL) was added to each well 20 h after compound administration. Then, the cells were cultured for another 4 h before replacing the culture media in each well with 150 μL DMSO. Finally, the absorbance at 595 nm was recorded by a microplate reader after shaking for 5 min in the dark.
Molecular docking
The structure optimization of TBmA-Glu and TBpA-Glu was applied by Gaussian 1647 with B3LYP function and 6-31G** basis set48. All the docking calculations were performed with AutoDock 4.249 using the Genetic Algorithm method. The PDB structure of the cocrystallized human γ-Glutamyltransferase 1 (GGT1, PDB code: 4GG250), and the binding site of the l-glutamine ligand were included and used as the grid box for docking assays. The best-ranked pose was selected as the final binding mode.
GGT activity assays
The GGT activity was assayed spectrophotometrically with slight modifications of the method reported previously51. 20 μL of diluted enzyme solution was added to 1 mL work solution with various concentrations of TBmA-Glu. The mixtures were incubated at 37 °C for 2 h, followed by light irradiation (12 J·cm−2). Subsequently, 5 mM γ-glutamyl-p-nitroanilide, 80 mM glycylglycine, and 0.1 M Tris-HCl (pH 10.0) were added to the mixtures of dark and light treatment groups. The enzymatic reaction was performed at 37 °C for 30 min and terminated by adding 0.4 mL of 4 M acetic acid. The transferase activity was calculated from the absorbance difference at 410 nm with a microplate reader between the reaction mixture with and without glycylglycine. The GGT activity in dark treatment groups were detected using the mentioned method without light irradiation.
Confocal microscopy
Cancer discrimination assays: The luciferase reporter HepG2 (HepG2-Luc) cells (5 × 105 cells per dish) were cultured in a confocal dish for 24 h, after which half of the HepG2-Luc cells were removed using a cell scraper. Following this step, LO2 cells were introduced into the culture and incubated for 24 h. A concentration of 5 μM TBmA-Glu was then added and further incubated for another 12 h. Before imaging with confocal microscopy, the cells were treated with a PBS solution containing d-luciferin potassium salt at a concentration of 300 μg/mL). TBmA-Glu, λex = 465 nm, λem = 700 ± 20 nm. Luciferin, λem = 520 ± 20 nm, 200 ms.
Cellular uptake assays
HepG2 cells (5 × 105 cells per dish) were mounted in confocal dishes and incubated for 24 h. The cells were treated with 5 μM TBmA-Glu or TBmA. Imaging was conducted after a further 12-h incubation period. TBmA-Glu and TBmA, λex = 465 nm, λem = 700 ± 20 nm.
Total ROS generation
HepG2 cells (5 × 105 cells per dish) were seeded in the confocal dishes for 24 h. The cells were treated with 2 μM TBmA-Glu for 12 h. Then, the light treatment groups were irradiated with a white led array (12 J·cm−2). DCFH-DA (10 μM) was added and the cells were incubated in dark conditions for another 15 min, followed by imaging with confocal microscopy. DCF, λex = 488 nm, λem = 500 ± 20 nm.
Reactive oxygen species (ROS) assays
HepG2 cells (8 × 105 cells per well) were seeded in six-well plates and incubated for 24 h, followed by treatment with TBmA-Glu at the indicated concentrations for 12 h. Subsequently, the PDT groups underwent light irradiation. Both the dark and light treatment groups were incubated with basal DMEM medium (without FBS) containing 10 μM DCFH-DA at 37 °C in the dark for 20 min and harvested. For the scavenging experiment, cells were pre-incubated with media having TBmA-Glu supplemented with ROS scavengers (Trolox: 100 μM, mannitol: 50 mM, Tiron: 10 mM, NaN3:5 mM) for 12 h before light irradiation (12 J·cm−2). Afterward, the cells were incubated with basal DMEM medium (without FBS) containing 10 μM DCFH-DA for an additional duration of 20 min. The cells were then harvested, centrifuged, washed twice with PBS, resuspended in a volume of PBS (500 μL), and analyzed using flow cytometry. DCF: λex = 488 nm, λem = 525 nm. A total of 104 cells were collected per sample, and data analysis was performed using FlowJo 10.8.1 software.
Measurement of GSSH/GSH ratios
HepG2 cells (8 × 105 cells per well) were cultured in 6 cm dishes and allowed to reach a density of ~80% confluency before treatment with TBmA-Glu (2 μM) for various durations (12, 24, and 48 h). For the PDT groups, light irradiation was conducted 12 h later of TBmA-Glu administration. The GSSH/GSH ratios were determined using the GSH and GSSG Assay Kit (Beyotime, China) according to the manufacturer’s instructions.
Measurement of MDA levels
HepG2 cells (8 × 105 cells per dish) were cultured in 6 cm dishes until reached the density of ~80% confluency. The cells were treated with TBmA-Glu (2 μM) for 12 h before harvested, counted, and lysed with radioimmunoprecipitation assay (RAPI) lysis buffer at 4 °C for 15 min. Malondialdehyde (MDA) levels in each group were determined using the Lipid Peroxidation MDA Assay Kit (Beyotime, China), according to the manufacturer’s instructions.
Transmission electron microscopy (TEM) measurement
The HepG2 cells (2 × 106 cells per dish) were placed in 10 cm culture dishes and incubated for 24 h. Then the cells were treated with TBmA-Glu (2 μM) for 12 h. Then, light irradiation was conducted for the PDT groups. The collected cells in dark groups and PDT groups were washed three times with cold PBS and then fixed with glutaric dialdehyde.
Western blotting
The HepG2 cells (2 × 106 cells per dish) were seeded in 10 cm culture dishes and incubated for 24 h, followed by treatment with various concentrations of TBmA-Glu (0, 0.5, 1.0, 2.0, 2.5, and 5.0 μM) for 12 h. Subsequently, light irradiation was performed on PDT groups, followed by incubation for another 3 h. All cells were trypsinized and lysed, and the total proteins of each group were collected and determined using BCA (bicinchoninic acid) assays. Equal amounts of cellular total proteins (20 μg) were separated using SDS-PAGE and then transferred to polyvinylidene difluoride membranes (Millipore, MA, USA). Membranes were blocked and incubated with the primary antibodies at 4 °C for 12 h. Then, the membranes were washed with phosphate-buffered saline with Tween-20 three times before incubated with the appropriate horseradish peroxidase-conjugated secondary antibody. Images were captured using an imaging station (JP-K300, JiaPang, China) and analyzed manually with ImageJ software. The antibodies for GGT (1:1000, ab109427, rabbit mAb), GPX4 (1:1000, ab125066, rabbit mAb) and β-actin (1:1000, ab8226, mouse mAb), goat anti-rabbit IgG H&L (HRP) (1:5000, ab6721), and goat anti-mouse IgG H&L (HRP) (1:5000, ab205719) were used and obtained from Abcam.
Immunofluorescence assays
HepG2 cells (5 × 105 cells per dish) were seeded in the confocal dishes for 24 h. Then, the cells were treated with 2 μM TBmA-Glu for 12 h. The PDT treatment groups were irradiated with a white led array (12 J·cm−2) before all the cells were fixed, permeabilized, and blocked before incubated with GGT primary antibody (1:100, ab55138, rabbit mAb, Abcam) at 4 °C for 12 h and goat anti-rabbit IgG (H&L, Alexa Fluor® 488 labeled, 1:500, ab150077, Abcam) at 4 °C for 2 h.
In vivo antitumor assay
All animal operations followed the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the Animal Experiment Center of the Chinese University of Hong Kong (Shenzhen, China, protocol number, CUHKSZ-AE2022004). Housing conditions: temperature, 21–26 °C; humidity, 50–60%, light cycle, 10 h (on)/14 h (off).
HepG2 xenograft-tumor model
Twenty-five pathogen-free female BALB/c nude mice, 4–5 weeks of age, were purchased from the Guangdong Medical Laboratory Animal Center. The HepG2 xenograft-tumor model was established by subcutaneous injection of HepG2 cells (1 × 106) into the right axilla of each mouse. When the tumor volume reached ~100 mm3, the mice were randomly divided into five groups (5 mice per group): (a) Control group, (b) TBmA group, (c) TBmA + light group, (d) TBmA-Glu group, (c) TBmA-Glu + light group. Each group was subjected to the following treatments: the same volume of physiological saline solution (Control), TBmA solution (5 mg/kg in physiological saline), or TBmA-Glu (5 mg/kg in physiological saline) was injected through the tail vein of each mouse on 0, 5, and 10 days. For the light treatment groups, the light irradiation was performed after 12 h of the drug administration using a white LED light array (20 mW/cm2, 20 min). The tumor growth and body weight of each mouse were monitored every 2 days. The mice should be euthanized when the diameter of any single tumor exceeds 20 mm. No tumor diameter of xenograft-tumor mice was larger than 20 mm during the whole treatment. The tumor volume was calculated by measuring the perpendicular diameter of the tumor using calipers and calculated according to the formula:
Orthotopic liver cancer mouse model
Luciferase reporter HepG2 (HepG2-Luc) cells (1 × 105) were intraportal injected into the livers of nude mice. Luminescence was employed to assess the successful establishment of the orthotopic liver cancer mouse model. Mice exhibiting comparable luminescence intensity were selected and randomly divided into two groups: (a) TBmA-Glu group and (b) TBmA-Glu + light group. Each mouse received intravenous administration of TBmA-Glu (5 mg/kg) through the tail vein on days 0, 5, and 10 days, followed by minimally invasive photodynamic therapy after a 12-hour interval. The in vivo imaging was conducted at the startpoint (0 days), midpoint (7 days), and endpoint (14 days) of the therapeutic process.
For the minimally invasive photodynamic therapy, the mice were initially anesthetized using an isoflurane inhalant anesthesia apparatus. The TBmA fluorescence (activated TBmA-Glu) was utilized to precisely locate the tumor section in the liver and a small incision (2 mm) was meticulously made with a scalpel at the localized region. Subsequently, the laser probe was carefully inserted into the incision to execute the photodynamic therapy. Finally, the wound was sutured and disinfected.
For hematoxylin-eosin (H&E) and immunohistochemistry staining, tumor samples and organs were collected and fixed with 4% paraformaldehyde at 4 °C for 24 h. Then, all the samples were then transferred to a 10% formalin-neutral buffer solution and embedded in paraffin. Sections were stained by H&E (Beyotime, China) or corresponding antibodies (GPX4, 1:100, ab125066 and Ki67, 1:500, ab92742, rabbit mAb) and observed under a Zeiss inverted fluorescence microscope (Germany).
Statistical analyses
Origin 2023 was used to help finish the plotting and significance analyses. The statistical significance in this study is determined by a one-side Student’s t-test at *p < 0.05 **p < 0.01, and ***p < 0.001, with replicated three times. The error bars represent the standard error of the mean.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This project is supported by the National Natural Science Foundation of China (NSFC), 52273197 (B.Z.T.) and 52333007 (Z.Z.), National Key Research and Development Program of China, 2023YFB3810001 (Z.Z.), China Postdoctoral Science Foundation, 2023M733423 (W.-J.W.), Shenzhen Key Laboratory of Functional Aggregate Materials, ZDSYS20211021111400001 (B.Z.T.), the Science, Technology and Innovation Commission of Shenzhen Municipality, KQTD20210811090142053 (B.Z.T.), JCYJ20220818103007014 (B.Z.T.), GJHZ20210705141810031 (Z.Z.) and JCYJ2021324134613038 (Z.Z.), Shenzhen Medical Research Fund, A2403004 (W.-J.W.), and Longgang District Medical Key Discipline Construction Found (R.H.). The authors would like to thank the help of the materials characterization and preparation center of the Chinese University of Hong Kong, Shenzhen and the AIE Institute (www.aietech.org.cn) for providing some technical assistance.
Author contributions
W.-J.W., S.W., Z.Z., and B.Z.T. conceived the study and directed the project; W.-J.W., L.Z., and R.Z. designed and performed the experiments; L.H. performed the synthetic works; Q.W., X.-M.C., Z.Q., P.A., R.H. J.F., and G.Z. improved the manuscript. All authors have approved the final version of the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that all data generated or analyzed during this study are available in this published article and its supplementary information files or from the corresponding author upon request. The full image dataset is available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shaojuan Wang, Email: 609199354@qq.com.
Zheng Zhao, Email: zhaozheng@cuhk.edu.cn.
Ben Zhong Tang, Email: tangbenz@cuhk.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54291-1.
References
- 1.Grote, M. et al. The molecular vista: current perspectives on molecules and life in the twentieth century. Hist. Phil. Life Sci.43, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Regenmortel, M. H. V. Truth in science and in molecular recognition, post-truth in human affairs. J. Mol. Recognit.33, e2827 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Zhao, Z., Zhang, H., Lam, J. W. Y. & Tang, B. Z. Aggregation-induced emission: new vistas at the aggregate level. Angew. Chem. Int. Ed.59, 9888–9907 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Guo, T. et al. Towards bioresource-based aggregation-induced emission luminogens from lignin beta-O-4 motifs as renewable resources. Nat. Commun.14, 6076 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang, J., Fang, M. & Li, Z. Organic luminescent materials: the concentration on aggregates from aggregation‐induced emission. Aggregate1, 6–18 (2020). [Google Scholar]
- 6.Ma, J. Y. et al. Chemical–biological approaches for the direct regulation of cell–cell aggregation. Aggregate3, e166 (2022). [Google Scholar]
- 7.Lyon, A. S., Peeples, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol.22, 215–235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fassler, J. S., Skuodas, S., Weeks, D. L. & Phillips, B. T. Protein aggregation and disaggregation in cells and development. J. Mol. Biol.433, 167215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet.20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Yan, C. et al. Preparation of near-infrared AIEgen-active fluorescent probes for mapping amyloid-β plaques in brain tissues and living mice. Nat. Protoc.18, 1316–1336 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Cen, P. et al. Aggregation‐induced emission luminogens for in vivo molecular imaging and theranostics in cancer. Aggregate4, e352 (2023). [Google Scholar]
- 12.Su, H. et al. A brightly red emissive AIEgen and its antibody conjugated nanoparticles for cancer cell targeting imaging. Mater. Chem. Front.6, 1317–1323 (2022). [Google Scholar]
- 13.Li, D. et al. Photo-triggered cascade therapy: a NIR-II AIE luminogen collaborating with nitric oxide facilitates efficient collagen depletion for boosting pancreatic cancer phototheranostics. Adv. Mater. 36, e2306476 (2023). [DOI] [PubMed]
- 14.Kim, B. J. & Xu, B. Enzyme-instructed self-assembly for cancer therapy and imaging. Bioconjug. Chem.31, 492–500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Fabuel, I. et al. Aberrant upregulation of the glycolytic enzyme PFKFB3 in CLN7 neuronal ceroid lipofuscinosis. Nat. Commun.13, 536 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amer-Sarsour, F., Kordonsky, A., Berdichevsky, Y., Prag, G. & Ashkenazi, A. Deubiquitylating enzymes in neuronal health and disease. Cell Death Dis.12, 120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinha, D. et al. Cep55 overexpression promotes genomic instability and tumorigenesis in mice. Commun. Biol.3, 593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fentiman, I. S. Gamma-glutamyl transferase: risk and prognosis of cancer. Br. J. Cancer106, 1467–1468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao, X. et al. A tumor-targeting near-infrared heptamethine cyanine photosensitizer with twisted molecular structure for enhanced imaging-guided cancer phototherapy. J. Am. Chem. Soc.143, 20828–20836 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Cai, X.-M. et al. Natural acceptor of coumarin-isomerized red-emissive bioAIEgen for monitoring Cu2+ concentration in live cells via FLIM. Adv. Sci.11, 2307078 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, Z. et al. Aggregation-induced emission biomaterials for anti-pathogen medical applications: detecting, imaging and killing. Regen. Biomater.10, rbad044 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye, Z. et al. AIEgens for microorganism-related visualization and therapy. IMed1, e20220011 (2023). [Google Scholar]
- 23.Obara, R. et al. Gamma-Glutamyltranspeptidase (GGT)-activatable fluorescence probe for durable tumor imaging. Angew. Chem. Int. Ed.60, 2125–2129 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Zhai, W. et al. Universal scaffold for an activatable photosensitizer with completely inhibited photosensitivity. Angew. Chem. Int. Ed.58, 16601–16609 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Ebo, J. S. et al. An in vivo platform to select and evolve aggregation-resistant proteins. Nat. Commun.11, 1816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, Q. et al. Enzyme-activatable polymer-drug conjugate augments tumour penetration and treatment efficacy. Nat. Nanotechnol.14, 799–809 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Bakthavatsalam, S. et al. Leveraging gamma-glutamyl transferase to direct cytotoxicity of copper dithiocarbamates against prostate cancer cells. Angew. Chem. Int. Ed.57, 12780–12784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, X., He, S., Cheng, P. & Pu, K. A dual-locked tandem fluorescent probe for imaging of pyroptosis in cancer chemo-immunotherapy. Adv. Mater.35, e2206510 (2023). [DOI] [PubMed] [Google Scholar]
- 29.Lin, J. et al. Stimuli-responsive macrocyclization scaffold allows in situ self-assembly of radioactive tracers for positron emission tomography imaging of enzyme activity. J. Am. Chem. Soc.144, 7667–7675 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Luo, Z. et al. Targeted delivery of a gamma-glutamyl transpeptidase activatable near-infrared-fluorescent probe for selective cancer imaging. Anal. Chem.90, 2875–2883 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Li, H. et al. Imaging gamma-glutamyltranspeptidase for tumor identification and resection guidance via enzyme-triggered fluorescent probe. Biomaterials179, 1–14 (2018). [DOI] [PubMed] [Google Scholar]
- 32.An, R., Wei, S., Huang, Z., Liu, F. & Ye, D. An activatable chemiluminescent probe for sensitive detection of gamma-glutamyl transpeptidase activity in vivo. Anal. Chem.91, 13639–13646 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Yang, M., Li, X. & Yoon, J. Activatable supramolecular photosensitizers: advanced design strategies. Mater. Chem. Front.5, 1683–1693 (2021). [Google Scholar]
- 34.Zhang, H., Forman, H. J. & Choi, J. Gamma-glutamyl transpeptidase in glutathione biosynthesis. Methods Enzymol.401, 468–483 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Batsios, G. et al. In vivo detection of gamma-glutamyl-transferase up-regulation in glioma using hyperpolarized gamma-glutamyl-[1-(13)C] glycine. Sci. Rep. 10, 6244 (2020). [DOI] [PMC free article] [PubMed]
- 36.Cho, E. J. et al. Gamma-glutamyl transferase and risk of all-cause and disease-specific mortality: a nationwide cohort study. Sci. Rep.13, 1751 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai, C. et al. Gamma-glutamyltransferase activity (GGT) is a long-sought biomarker of redox status in blood circulation: a retrospective clinical study of 44 types of human diseases. Oxid. Med. Cell Longev.2022, 8494076 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pompella, A., De Tata, V., Paolicchi, A. & Zunino, F. Expression of gamma-glutamyltransferase in cancer cells and its significance in drug resistance. Biochem. Pharmacol.71, 231–238 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Zeng, Q., Li, X., Xie, S., Xing, D. & Zhang, T. Specific disruption of glutathione-defense system with activatable single molecule-assembled nanoprodrug for boosted photodynamic/chemotherapy eradication of drug-resistant tumors. Biomaterials290, 121867 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Pompella, A., Corti, A., Paolicchi, A., Giommarelli, C. & Zunino, F. Gamma-glutamyltransferase, redox regulation and cancer drug resistance. Curr. Opin. Pharmacol.7, 360–366 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Liu, Y., Wan, Y., Jiang, Y., Zhang, L. & Cheng, W. GPX4: The hub of lipid oxidation, ferroptosis, disease and treatment. BBA-Rev. Cancer1878, 188890 (2023). [DOI] [PubMed] [Google Scholar]
- 42.Yang, W. S. & Stockwell, B. R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol.26, 165–176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doll, S. et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol.13, 91–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown, Z. J., Heinrich, B. & Greten, T. F. Mouse models of hepatocellular carcinoma: an overview and highlights for immunotherapy research. Nat. Rev. Gastro. Hepat.15, 536–554 (2018). [DOI] [PubMed] [Google Scholar]
- 45.He, L., Tian, D.-A., Li, P.-Y. & He, X.-X. Mouse models of liver cancer: Progress and recommendations. Oncotarget6, 23306–23322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, W. J. et al. Induction and monitoring of DNA phase separation in living cells by a light-switching ruthenium complex. J. Am. Chem. Soc.143, 11370–11381 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Frisch, M. J. et al. Gaussian 16 Rev. B.01. (2016).
- 48.Hariharan, P. C. & Pople, J. A. Accuracy of AHnequilibrium geometries by single determinant molecular orbital theory. Mol. Phys.27, 209–214 (2006). [Google Scholar]
- 49.Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem.30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West, M. B. et al. Novel insights into eukaryotic gamma-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme. J. Biol. Chem.288, 31902–31913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, T. et al. Semi-quantitative activity assays for high-throughput screening of higher activity gamma glutamyl transferase and enzyme immobilization to efficiently synthesize L-theanine. J. Biotechnol.330, 9–16 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data generated or analyzed during this study are available in this published article and its supplementary information files or from the corresponding author upon request. The full image dataset is available from the corresponding author upon request. Source data are provided with this paper.