Abstract
To identify circRNAs associated with Parkinson’s disease (PD) we leveraged two of the largest publicly available studies with longitudinal clinical and blood transcriptomic data. We performed a cross-sectional study utilizing the last visit of each participant (N = 1848), and a longitudinal analysis that included 1166 participants with at least two time points. We identified 192 differentially expressed circRNAs, with effects that were sustained during disease, in mutation carriers, and diverse ancestry. The 192 circRNAs were leveraged to distinguish between PD and healthy participants with a ROC AUC of 0.797. Further, 71 circRNAs were sufficient to distinguish between genetic PD (AUC71 = 0.954) and, at-risk participants (AUC71 = 0.929) and healthy controls, supporting that circRNAs have the potential to aid the diagnosis of PD. Finally, we identified five circRNAs highly correlated with symptom severity. Overall, we demonstrated that circRNAs play an important role in PD and can be clinically relevant to improve diagnostic and monitoring.
Subject terms: Parkinson's disease, Diagnostic markers, High-throughput screening
Introduction
Parkinson’s disease (PD) affects more than six million people worldwide, with a prevalence projected to double in the next decades1. It is a neurodegenerative disease clinically defined by resting tremor, rigidity, bradykinesia, and postural instability2. Pathologically, it is characterized by Lewy bodies (LB) and neurites that are composed by aggregated and phosphorylated alpha-synuclein (α-Syn) and the degeneration of substantia nigra. Its cause is not fully understood yet. It is known that there are both genetic and environmental risk factors, but the complete causal pathway, or pathways, still remain elusive.
Circular RNAs (circRNAs) are non-coding RNAs that are the result of backsplicing events that take place during the maturation of linear RNA and lead to the creation of a covalently closed loop and an increase in their stability3–6. They are highly expressed in the nervous system, especially in synapses7 and have a role in neuronal development, aging5, and disease3,4,7–13. Among their many functions, they act as miRNA sponges to regulate gene expression, interact with proteins, and to generate protein products among many others3,4,7–12. Studies in PD have demonstrated that the substantia nigra accumulates circRNA in an age-related manner, event that is lost in PD, with a reduction in the overall number of circRNAs11. In blood, a study comparing four PD cases and four controls described 129 circRNAs up-regulated, and 282 down-regulated14 in PD participants compared to controls with an enrichment of PD terms. A study analyzing targeted circRNAs from PBMCs identified six circRNAs downregulated in PD10. They used four of them to build a classifier that showed an Area Under the ROC curve (AUC) of 0.86, evidencing the potential of circRNAs as biomarkers. To date, there is one high-throughput screening of circRNAs in blood of PD individuals, which focused on early-stage PD15. While they found three circRNAs downregulated in PD blood they were unable to replicate the results in an independent cohort15. Outside of the high-throughput realm, there are targeted analyses using Peripheral Blood Mononuclear Cells (PBMCs) with promising results10.
To better understand and describe the landscape of circRNAs in blood of PD participants compared to controls and evaluate their value as diagnostic and prognostic biomarkers we have quantified circRNAs in blood from the two largest longitudinal studies, the national Institute of Neurological Disorders and Parkinson’s Disease Biomarkers Program (PDBP)16, and the Parkinson’s Progression Markers Initiative (PPMI)17.
Material & methods
Study design
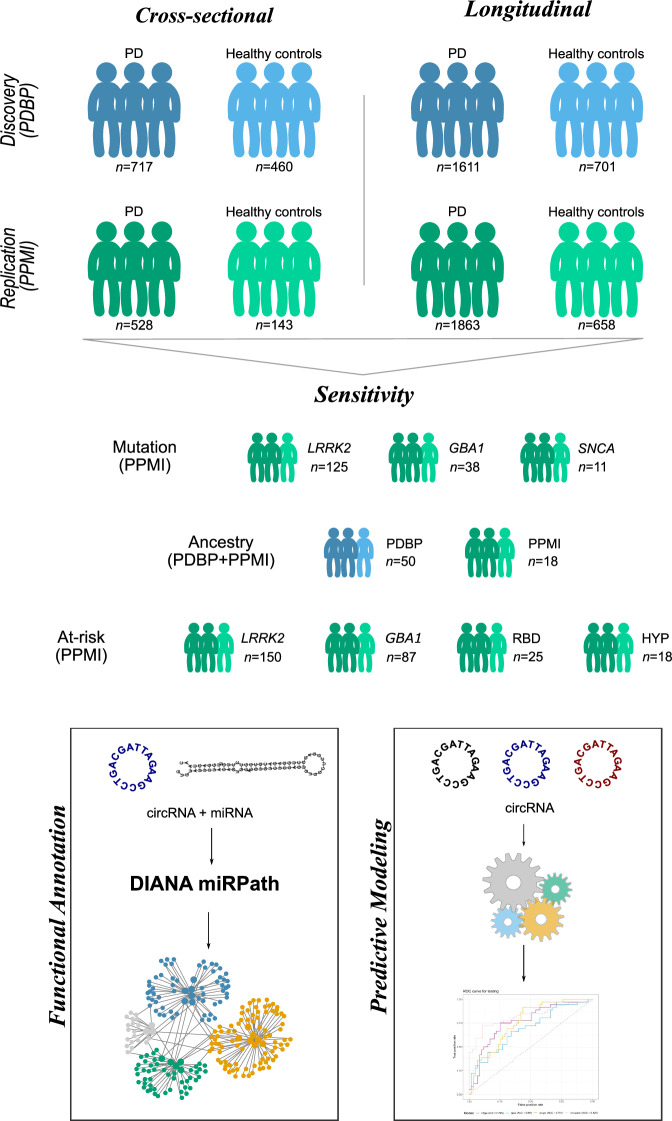
We analyzed the largest PD blood RNAseq datasets publicly available to date by combining the PDBP and PPMI studies. After raw data processing and stringent quality control, we compared the circular transcriptome between European Ancestry PD participants and controls to identify differentially abundant circRNAs. We leveraged the PDBP dataset (N = 1177) for discovery and PPMI (N = 671) for replication, followed by meta-analyses. We used gene-collapsed circRNA counts in the analyses and corrected all p-values using Benjamini-Hochberg (FDR) correction. Then, we investigated if the identified circRNAs were also associated with disease progression by performing longitudinal analyses of the RNAseq data collected across five visits. To confirm that the findings were not due to differences in cell proportions, or originated from the linear transcriptome, analyses were adjusted by digitally deconvoluted cell proportion and linear RNA quantification. Given the extensive information collected in PDBP and PPMI, we were able to perform several sensitivity analyses. We investigated if (i) the same circRNAs were differentially abundant in the African Ancestry individuals available in PDBP and PPMI, (ii) the changes in circRNA accumulation can be observed prior to symptom onset (PD-related mutation carriers and participants with REM sleep Behavior Disorder (RBD) or hyposmia), (iii) medication had effect on the circRNA landscape; and (iv) circRNA abundance associated with disease severity measured by the Unified Parkinson’s Disease Rating Scale (UPDRS) Part III or motor examination (UPDRS-III), and cognitive status measured by The Montreal Cognitive Assessment (MoCA). We then sought to understand the biology of the identified circRNAs and performed data integration with microRNA quantification (miRNA) and in-silico functional analyses. Finally, we assessed the diagnostic capacity of cirRNAs using machine learning approaches (Fig. 1). This research was conducted in accordance with the recommended protocols and written informed consent was obtained from all participants or their family members. The study was approved by the Washington University in Saint Louis Institutional Review Board (IRB ID 201701124 and 202004010).
Fig. 1. Study design summary.
We have followed a two-stage cross-sectional analysis with discovery (PDBP) and replication (PPMI) phases. Then we have performed sensitivity analysis stratifying by mutation, exploring individuals from African Ancestry, and those at risk due to being a carrier of a PD-causing mutation, or suffering from RBD or hyposmia. We also repeated the same analyses in a longitudinal manner using mixed models and leveraged the repeated measures available for each individual. Finally, we have functionally annotated the findings via circRNA-miRNA integration and leveraged circRNAs to build predictive models. The sample size (n) for each subgroup correspond to number of unique individuals except for the longitudinal comparison that correspond to total number of samples per group.
Dataset description
The present study includes two independent and publicly available datasets with longitudinal clinical and blood transcriptomic data available: PDBP16 and PPMI17. Both are observational multi-center studies aimed at identifying biomarkers of PD progression and improving the understanding of PD pathobiology. Participants (N = 1848; Table 1) are followed longitudinally with clinical assessments and biospecimen collection every six months, while blood RNAseq data is generated every six months in PDBP, and every six months during the first year and every 12 months thereafter in PPMI. To maximize the clinical differences between cases and controls when assessing the accumulation of circRNAs, we selected the last assessment of each participant instead of using baseline as previous publications15,18 for the cross-sectional analysis. We included a total of 717 cases and 460 control participants of European descent from the PDBP study (Table 1) as discovery, and 528 cases and 143 control participants of European descent form the PPMI study (Table 1) as replication. The two populations are comparable in age (p = 0.37), with mean age of 64.67 (±10.14) in PDBP and 64.24 (±9.59) in PPMI. Given the higher prevalence of PD in males19,20, both datasets had slightly higher proportion of male participants (>60%), with the exception of healthy control participants in PDBP, with more than 50% of females. Further, there was no significant difference between symptomatic PDBP and PPMI participants in terms of symptom severity as measured by UPDRS-III and MoCA (p = 0.22 and p = 0.73, respectively) (Table 1). UPDRS-III scores of PD participants were on average 25.88 (±13.35) and 24.95 (±13.06) in PDBP and PPMI, respectively, while healthy control participants presented mean UPDRS-III scores of 2.13 ( ± 5.07) and 1.46 (±2.61) in PDBP and PPMI, respectively. Regarding MoCA scores, they present narrow ranges in all groups, with means between 25 and 27 (Table 1). This might be explained by the enrolment of recently diagnosed individuals in both studies, in other words, the PD participants were in the early stages of the diseases and no cases have developed severe dementia. We have also included participants with African descent (N = 68) from the PDBP (N = 50) and PPMI (N = 18) studies to investigate if the findings are ancestry independent (Supplementary Table 1). There was no significant difference between mean ages of PDBP and PPMI (59.60 ± 9.48 vs 55.83 ± 14.35; p = 0.14) participants of African ancestry. PDBP African ancestry participants were evenly distributed by sex, with 53.33% of female participants, while PPMI had 33.33% female participants (p = 0.42). Similar to participants of European ancestry, no significant difference was observed in UPDRS-III and MoCA measured symptom severity (p = 0.73 and p = 0.21, respectively) between PDBP and PPMI participants of African ancestry. On average, PDBP participants scored 14.60 (±16.10) and PPMI participants 12.67 (±16.06) on UPDRSIII scale, while average values on MoCA scale were 23.13 (±4.29) and 25.18 (±4.85) in PDBP and PPMI, respectively.
Table 1.
Demographic characteristics corresponding to the European descent individuals included in the cross-sectional analyses
| Parkinson’s Disease Biomarker Program (PDBP) | Parkinson’s Progression Markers Initiative (PPMI) | |||
|---|---|---|---|---|
| Healthy controls | Parkinson’s disease | Healthy controls | Parkinson’s disease | |
| Participants (N) | 460 | 717 | 143 | 528 |
| Mean age (IQR) | 62.83 (56.00–71.00) | 65.85 (60.00–72.00) | 63.93 (59.00–71.00) | 64.33 (57.75–71.00) |
| Female (N, %) | 251 (54.56%) | 269 (37.52%) | 52 (36.36%) | 203 (38.45%) |
| Mean UPDRS-III (IQR) | 2.13 (0.00–2.00) | 25.88 (16.00–33.00) | 1.46 (0.00–2.00) | 24.95 (15.75–33.25) |
| Mean MOCA (IQR) | 26.61 (25.00–28.50) | 25.15 (23.00–28.00) | 27.34 (26.00–29.00) | 26.32 (25.00–29.00) |
N Sample Size, IQR Interquartile Range, UPDRS-III Unified Parkinson’s Disease Rating Scale Part III, MoCA Montreal Cognitive Assessment.
PPMI is a comprehensive study that includes PD participants carriers of PD-causal mutations, and participants without diagnosis of PD, but at risk of developing it. More specifically, we included symptomatic PD participants with LRRK2 (N = 125), GBA (N = 38) and SNCA (N = 11) mutations, as well as non-symptomatic at-risk participants with LRRK2 (N = 150), GBA (N = 87), and SNCA (N = 3) mutations, and PD-associated syndromes RBD (N = 25) and hyposmia (N = 18; Supplementary Table 2). Detailed mutation information was available for majority of the participants. The most common mutation in the symptomatic LRRK2 participants was G2019S (n = 92), followed by R1441G (n = 14). Most of the symptomatic GBA1 participants were carriers of N409S (n = 30), while all of the symptomatic SNCA participants (n = 11) carried A53T mutation. The same mutations were the most common among the at-risk mutation carriers. Specifically, our study included at-risk LRRK2 participants with G2019S (n = 119) and R1441G (n = 14) mutations, GBA1 participants with N409S (n = 65) mutation and SNCA participants with A53T (n = 3) mutation. Mean age of symptomatic mutation carriers was significantly higher than that of at-risk mutation carriers (64.89 ± 9.13 vs 61.89 ± 9.07; p = 1.00 ×10−03). This was also true when broken down by gene, except in SNCA mutation carriers where there was no significant difference in mean age of symptomatic compared to at-risk participants (5.45 ± 11.10 vs 49.33 ± 7.50; p = 0.49) (Supplementary Table 2). Overall, symptomatic and at-risk mutation carriers had comparable proportion of females (50.57% and 56.43%, respectively, p = 0.23), but when broken down by mutation GBA+ symptomatic carriers had significantly lower proportion of female participants than at-risk GBA+ carriers (39.47% vs 62.07%, p = 0.02) (Supplementary Table 2).
Longitudinal blood transcriptomic data was available for 1,166 unique participants across five visits in both PDBP and PPMI studies (Table 2). Visits in PDBP were six months apart, while PPMI participants were followed every 6 months during the first year, and every twelve months thereafter. PDBP participants with more than one visit were significantly older than PPMI participants at first visit (64.58 ± 9.67 vs 61.59 ± 9.65, p = 1.52 ×10−7). Similar to the cross-sectional demographics, PDBP cohort is more evenly distributed between sexes, with 40–50% of female participant across visits, compared to PPMI that consists of 30-40% female participants (p = 0.11).
Table 2.
Demographic characteristics of the European descent participants included in the longitudinal analysis by time of sample collection
| Parkinson’s Disease Biomarker Program (PDBP) | Parkinson’s Progression Markers Initiative (PPMI) | ||||||
|---|---|---|---|---|---|---|---|
| Sampling time | Group | Samples (N) | Mean age | Female | Samples (N) | Mean age | Female |
| (IQR) | (N, %) | (IQR) | (N, %) | ||||
| Baseline | HC | 166 | 63.95 (56.00–71.00) | 80 (48.19) | 143 | 61.12 (56.00–68.00) | 52 (36.36) |
| PD | 362 | 64.77 (59.00–71.00) | 147 (40.60) | 474 | 61.54 (55.00–69.00) | 183 (38.60) | |
| Month 6 | HC | 139 | 64.59 (56.50–71.00) | 66 (47.48) | 129 | 61.64 (57.00–69.00) | 48 (37.21) |
| PD | 337 | 65.45 (59.50–71.50) | 140 (41.54) | 341 | 62.41 (55.00–69.00) | 139 (40.76) | |
| Month 12 | HC | 146 | 64.89 (57.00–71.75) | 68 (46.57) | 130 | 62.19 (57.00–69.00) | 45 (34.61) |
| PD | 333 | 65.94 (60.00–72.00) | 135 (40.54) | 388 | 62.34 (56.00–69.00) | 150 (38.66) | |
| Month 18 | HC | 128 | 64.48 (57.50–70.50) | 64 (50.00) | – | – | – |
| PD | 299 | 66.43 (60.50–72.50) | 124 (41.47) | – | – | – | |
| Month 24 | HC | 122 | 66.01 (59.00–72.00) | 60 (49.18) | 129 | 62.88 (58.00–70.00) | 49 (37.98) |
| PD | 280 | 67.17 (61.00–73.00) | 124 (44.28) | 381 | 63.74 (57.00–71.00) | 142 (37.27) | |
| Month 36 | PD | – | – | – | 127 | 63.88 (59.00–71.50) | 48 (37.79) |
| HC | – | – | – | 279 | 64.54 (58.00–72.00) | 94 (33.69) | |
Each participant must have available data in at least two time-points.
N Sample Size, IQR Interquartile Range, HC Healthy Control, PD Parkinson’s Disease.
Data processing and quality control
We accessed raw transcriptomic data (fastq files) from a total of 6362 ribodepleted blood RNA samples from 1848 unique participants from the PDBP and PPMI studies. Data generation, processing, and quality control for these datasets have been described elsewhere18,21,22. Due to the lack of availability of circular counts in the AMP-PD database, we re-processed the raw files using our in-house pipelines to obtain linear and circular counts7,23. For linear counts, we followed the TOPMed pipeline using the GRCh38 genome reference and the GENCODE 33 annotation7,24 (https://github.com/broadinstitute/gtex-pipeline/blob/master/TOPMed_RNAseq_pipeline.md). Briefly, the raw reads were aligned to the human reference genome using STAR (v.2.7.1a)25 and alignment quality was evaluated using sequencing metrics calculated using Picard tools (v.2.8.2)26. Gene expression was quantified using Salmon (v.1.2.0)27. All transcripts were collapsed to gene level prior to analysis. All samples with less than 50% of mapped reads in STAR or Salmon, samples that were outliers in Principal Component Analysis (PCA), or that had failed FastQC check in more than four categories were removed from downstream analyses. Linear transcripts with less than ten reads in more than 90% of the individuals (N = 42,092) were removed from the analyses. Counts were normalized using DESeq228 to adjust for library complexity and variance stabilizing transformation (vst) to obtain the final count matrix.
Circular RNA detection, annotation and quantification was performed using Detect CircRNA from Chimeric Reads (DCC v.0.4.8)29 using the software developer guidelines and as described previously23. Shortly, the raw reads were aligned a second time to the human reference genome (GRCh38) using STAR25 in chimeric alignment mode. Similar to linear RNA, circRNAs were collapsed by gene of origin prior to analyses. CircRNAs with missing counts in more than 75% of samples were removed from the analyses. To optimize the data processing time, we processed the samples from each visit separately, followed by integration of all visits for each study separately (Supplementary Fig. 1A, B). Then we calculated PCA using the 500 most variable circular transcripts for each dataset independently and removed samples that were more than three-standard deviations from the mean of PC1 or PC2 (Supplementary Fig. 1C, D). To normalize the counts, we used DESeq228 to adjust for library complexity and vst to obtain the final count matrix. As an additional QC step, we calculated count ratios between circRNAs and linear RNAs and kept only those circRNAs that had circRNA:linear RNA ratio of at least 0.1 in three or more samples7. Only circular transcripts that passed QC in both PDBP and PPMI datasets were included in the analyses. A total of 1789 circRNAs from five visits corresponding to 1848 individual participants were included in the downstream analyses.
Cross-sectional differential abundance analysis
We performed cross-sectional differential abundance analyses using DESeq228. To maximize the detection of differentially abundant circular transcripts due to PD and taking into consideration the longitudinal nature of PDBP and PPMI, we included one sample per individual, corresponding to the most recent visit. Only individuals with a diagnosis of PD (regardless of mutation status) and healthy controls were included in this analysis. A minimally adjusted model (sex and age at draw) was applied for discovery (PDBP) and replication (PPMI) followed by a meta-analysis using the R package metaRNAseq30. Only circRNAs with same direction of effect in both PDBP and PPMI datasets were considered for meta-analyses. We didn’t employ a log2 fold change cutoff, as it is not known how large of how small a change in circRNA abundance would be biologically or clinically relevant. Multiple test correction was performed using the FDR correction. FDR p-values below 0.05 in the meta-analysis were considered significant. Additionally, and to account for differences between PDBP and PPMI and ensure that the results from the meta-analyses were not driven by one population only, we considered two more stringent p-value thresholds: (i) circRNA that that were nominally significant in both datasets and FDR significant in the meta-analyses, and (ii) circRNA that passed multiple test correction in the discovery (PDBP), and the replication (PPMI).
To ensure the robustness of the results, we performed several comprehensive analyses to verify that the identified statistical differences were not driven by confounding factors. One of the main confounders in blood transcriptomics are differences in cellular composition. EPIC31 was used to obtain the cell proportions, and those included in the model. Similarly, to evaluate if the findings could be driven by changes on the linear forms of the host genes, we performed the same analyses described above including the linear counts as a covariate in the model. Medication can be another confounding factor. We wanted to assess if the presence of PD medication (L-Dopa or Dopamine Agonist) had any impact on the differential accumulation. In consequence, we evaluated if the identified circRNAs were significant when comparing the PD participants that were being medicated at the time of blood draw compared to those that were not. Medication information is challenging to gather and harmonize due to its complexity and sparsity. Fortunately, we had access to individual data for the PPMI study, which we manually curated for each participant and visit prior to inclusion in the analysis. Medication status was simplified to yes (N = 443) or no (N = 85) to maximize the sample size of this sensitivity analysis.
Cross-sectional sensitivity analyses
To better understand the role of the differentially accumulated circRNAs in the context of PD, we assessed whether the counts of the circular transcripts identified in the cross-sectional analyses were correlated with UPDRS-III or MoCA for those participants (regardless of disease status) with the data available. Given the sample size and the limited time of follow-up, we considered correlations significant if p-value was lower than 0.05. Finally, to investigate if there was any difference between sporadic PD and mutation carriers in regard to circRNA abundance, we performed sensitivity analyses by comparing LRRK2, GBA, or SNCA mutation carriers to those of healthy controls or idiopathic PD. Additionally, we leveraged data from at risk participants (PPMI participants that carry a known PD mutation or suffer from RBD or hyposmia but have not been diagnosed with PD) to test if any of the differentially accumulated circRNAs could be used as early biomarker of PD. We compared if the counts in at-risk participants were different from those observed in healthy controls.
Finally, we took advantage of the diversity that these datasets provide to perform circRNA differential expression analyses in participants from African ancestry. Given the limited sample size, we combined participants from PDBM and PPMI studies (N = 26 PD participants and N = 42 healthy controls) and limited our analyses to the significant findings from the cross-sectional differential abundance analysis.
Longitudinal differential abundance analysis
To harness the longitudinal characteristics of the two studies included in this manuscript we used mixed models to perform differential abundance analyses. We included participants from PDBP (N = 547) and PPMI (N = 619) datasets with at least two clinical visits and blood transcriptomic data available after QC regardless of mutation status. All analyses were adjusted by circular transcript counts at first visit, sex, and age at draw. Participant ID was used as random effect. We modeled the trajectories using linear mixed model with status×time as the interaction term. The overall approach was similar to the one described for the cross-sectional analyses, but focusing on the circular transcripts that were found significant in the meta-analyses. Briefly, PDBP and PPMI were tested separately to perform a subsequent meta-analysis using only European ancestry cases and controls. Then, we explored if the linear transcripts, the cell proportion, or medication had an influence on the results. We investigated if the longitudinal circRNA counts for each of the transcripts was correlated with disease severity progression measured by UPDRS III and MoCA. The number of African ancestry individuals with at least two visits was very limited (N = 16), thus no testing on diverse ancestry was performed.
In-silico functional study
We explored the biological implications of the high confident circRNA identified in the cross-sectional analyses to minimize the number of predicted miRNAs and maximize the biological significance. We leveraged the Circular RNA Interactome website32 (accessed July 2023, last database update January 30th 2020) to predict which miRNAs have the potential to be targeted by each of the circRNA species identified in the present study. To reduce the number of miRNAs, and since miRNA sequence data is available for the PPMI dataset, we explored if the counts of any of the predicted miRNAs were correlated with the circRNA counts to identify the ones that are more likely to have biological consequences. After accessing the available miRNA count data from the PPMI study and filtering the low count miRNA data (standard quality control parameter of at least five counts in 90% of the sample). The miRNA counts were normalized using DESeq228 and the vst function (similar to what was described for the circular and the linear RNAs). We used Pearson correlation to assess which of the predictions hold true at the biological level. With the list of all miRNAs nominally significant (p < 0.05) for their correlation with each circRNA, we performed pathway analyses, grouped by circRNA, using the microT-CDS algorithm and the Kyoto Encyclopedia of Genes and Genomes (KEGG) form the DIANA mirPath software version 333, to identify pathways regulated by the predicted miRNAs and, consequently, the circRNAs targeting them.
Predictive model development and evaluation
To assess the predictive power of the identified circRNAs, we evaluated the ability to use circRNAs to differentiate between symptomatic participants and healthy controls, as well as at-risk individuals and healthy controls. First, we scaled the transcript counts between the PDBP and the PPMI datasets by computing the z-score. Then, we fit three generalized linear model using different combinations of the normalized circRNA counts and the glm function from the R stats package, adjusting by participants’ age and sex. We tested the predictive capabilities of the circRNAs that were significantly differentially accumulated in the cross-sectional meta-analyses, those circRNA that were nominally significant in both populations, and those that were significant after multiple test correction in both populations. Model performance was further evaluated in publicly available data from 28 dementia with Lewy bodies (DLB), 16 frontotemporal dementia (FTD) and 567 Alzheimer’s disease (AD) participants34. Data was processed the same way as described for PDBP and PPMI datasets and transcript counts were scaled using z-score. Risk score for each participant was calculated using the models previously defined using the combined PDBP and PPMI data. Predicted values above 0.50 were considered cases. We computed the ROC curves by comparing the predicted to the true disease status.
Additionally, we also examined whether circRNA levels can be used to differentiate between PD and healthy controls using supervised statistical and machine learning (ML) approaches. Since the goal is to detect PD associated signatures as soon as possible, we have used samples from the first visit in PDBP (N = 528) and PPMI (N = 617) datasets, and the 2849 circRNAs shared in this first visit (Supplementary Fig. 1). We scaled the vst-normalized counts from both datasets using z-scores. Then, PDBP was used as training data, and PPMI for testing. We adopted a hypothesis free approach by using all the common circRNAs into the modeling problem, resulting in a high dimensional modeling task. In consequence, the selected linear and non-linear approaches are well-suited to resolve this type of modeling scenarios and successfully select the most relevant features to build the best possible model. We used L1 LASSO and L2 ridge regularization statistical linear models as implemented in the glmnet R package35,36. The lambda hyperparameter was optimized with 10-fold cross-validation. Then prediction models were evaluated by 10-fold cross-validation and repeated ten times. We employed two ML non-linear bagged decision trees based approaches: random forests (RF) from the Ranger R package37, and the MiniPatch learning algorithm38. Model hyperparameter optimization was performed with a grid based search. Particularly for RF, we tested 30 values equally spaced from five up to three times the squared root of the number of circRNAs used as input by the number of predictors to use in each tree split. As feature selection criteria for each split, we tested at the hyperparameter Gini, Extratrees and Hellinger criteria. All forests have the same size, 300 trees and the tree branches are grown until the corresponding leave node for that branch has under 64 examples in size. Regarding the MiniPatch’s model selection, we tested five values equally spaced from 3% and 49% for the percentage of features to select for each patch. The percentage of samples to selected for each patch was configured across five values equally spaced from 60 and 100%. The selection criteria within each split that we tested were Gini and Entropy. The union of all circRNAs selected by LASSO, RF and MiniPatch were used as features to build the ridge regularization logistic regression model. Particularly, the most relevant features as determined by RF were those with an importance greater than a cut-off obtained by looking for a plateau at the curve formed all features used within the forest, ordered by importance at the y-axis. The regularization lambda parameter for the LASSO and ridge models was optimized by 10-fold cross-validation. We used balanced accuracy from 10-fold cross-validation of experiments repeated 10 times to select the best model for each technique. The circRNAs identified by the best model out of all four techniques were then functionally annotated with terms from the Gene Ontology as identified by the clusterProfiler R Package.
Results
CircRNAs are differentially accumulated in blood of PD cases compared to controls
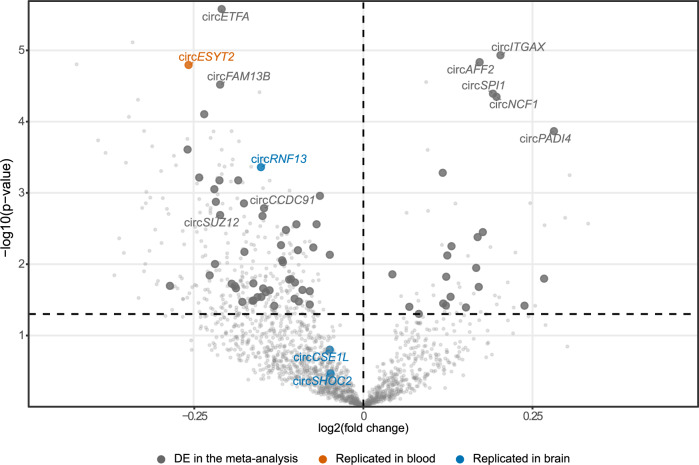
To identify circRNAs differentially accumulated in the blood of PD cases compared to controls, we used a cross-sectional approach, selecting the most recent blood sample available per individual. We included 1177 PDBP participants as discovery (Ncases = 717; Ncontrols = 460) and 671 from PPMI as replication (Ncases = 528; Ncontrols = 143; Table 1, Fig. 1). Meta-analyses revealed 192 circRNAs to be differentially accumulated when comparing cases and controls after multiple test correction, 141 of those downregulated and 51 upregulated (Fig. 2A, Supplementary Table 3). CircRHBDD1 was downregulated in blood from PD cases consistent with what has been previously reported15. Additionally, we identified three circRNA dysregulated in blood but previously described to be dysregulated in brain (circCSE1L, circRNF13, and circSHOC2) (Fig. 2, Supplementary Table 3)13,15. Of the 192 circRNAs, 71 were nominally significant in both the discovery and the replication dataset and nine after multiple test correction in both PDBP (circAFF2, log2FC = 0.172, p = 1.46× 10−5; circETFA, log2FC = −0.210, p = 2.63 ×10−6; circFAM13B, log2FC = −0.212, p = 3.02 ×10−5; circSPI1, log2FC = 0.192, p = 4.05 ×10−5; circSUZ12, log2FC = −0.212, p = 0.002) and PPMI (circAFF2, log2FC = 0.326, p = 1.28 ×10−7; circETFA, log2FC = −0.160, p = 0.006; circFAM13B, log2FC = −0.267, p = 3.97 ×10−4; circSPI1, log2FC = 0.333, p = 4.88 ×10−7; circSUZ12, log2FC = −0.280, p = 0.003) (Table 3).
Fig. 2. Volcano plot showing the cross-sectional differential expression analysis (PDBP).
Highlighted in dark gray are those circRNA that replicated in the PPMI cohort, in orange and blue those already described to be differentially accumulated in previous publications in blood and brain tissue respectively.
Table 3.
Analysis results for the nine high-confidence interval circRNA
| circAFF2 | circCCDC91 | circETFA | circFAM13B | circITGAX | circNCF1 | circPADI4 | circSPI1 | circSUZ12 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Analysis | Dataset | beta | p value | beta | p value | beta | p value | beta | p value | beta | p value | beta | p value | beta | p value | beta | p value | beta | p value |
| Cross-sectional | PDBP | 0.172 | 1.46 ×10−5 | −0.147 | 0.002 | −0.21 | 2.63 ×10−6 | −0.212 | 3.02 ×10−5 | 0.203 | 1.17 ×10−5 | 0.197 | 4.48 ×10−5 | 0.282 | 1.36 ×10−4 | 0.192 | 4.05 ×10−5 | −0.212 | 0.002 |
| PPMI | 0.326 | 1.28 ×10−7 | −0.186 | 0.006 | −0.16 | 0.006 | −0.267 | 3.97×10−4 | 0.207 | 0.001 | 0.293 | 1.08 ×10−4 | 0.295 | 0.004 | 0.333 | 4.88 ×10−7 | −0.28 | 0.003 | |
| Meta | – | 5.23 ×10−11 | – | 1.16 ×10−4 | – | 3.07 ×10−7 | – | 2.31 ×10−7 | – | 3.40 ×10−7 | – | 9.72 ×10−8 | – | 8.20 ×10−6 | – | 5.06 ×10−10 | – | 8.39 ×10−5 | |
| Ancestry | PDBP+PPMI | 0.203 | 0.484 | 0.101 | 0.678 | −0.021 | 0.943 | 0.352 | 0.186 | −0.111 | 0.687 | 0.124 | 0.618 | 0.012 | 0.979 | 0.03 | 0.922 | −0.954 | 0.016 |
| Longitudinal | PDBP | 0.023 | 0.409 | −0.037 | 0.332 | −0.022 | 0.529 | −0.046 | 0.247 | 0.079 | 0.011 | 0.014 | 0.679 | 0.059 | 0.127 | 0.048 | 0.122 | −0.036 | 0.404 |
| PPMI | 0.029 | 0.138 | −0.071 | 0.006 | −0.013 | 0.574 | −0.025 | 0.34 | 0.025 | 0.204 | 0.052 | 0.023 | −0.007 | 768 | 0.032 | 0.094 | −0.035 | 0.211 | |
| Meta | – | 0.219 | – | 0.014 | – | 0.666 | – | 0.292 | – | 0.016 | – | 0.081 | – | – | – | 0.063 | – | 0.295 | |
| Sensitivity (cross-sectional) | LRRK2vsHC | 0.464 | 6.66 ×10−9 | −0.251 | 0.004 | −0.109 | 0.159 | −0.573 | 6.51 ×10−9 | 0.252 | 0.003 | 0.461 | 1.23 ×10−6 | 0.012 | 0.929 | 0.526 | 2.43 ×10−9 | −0.374 | 0.003 |
| GBAvsHC | 0.599 | 2.13 ×10−7 | −0.44 | 1.80 ×10−4 | −0.123 | 0.274 | −0.749 | 4.37 ×10−8 | 0.415 | 0.002 | 0.713 | 1.37 ×10−6 | 0.338 | 0.078 | 0.584 | 6.08 ×10−6 | −0.795 | 5.26 ×10−5 | |
| SNCAvsHC | 0.776 | 1.47 ×10−4 | −0.038 | 0.857 | 0.176 | 0.376 | −0.103 | 0.667 | 0.199 | 0.399 | 0.523 | 0.046 | 0.884 | 0.013 | 0.437 | 0.066 | −0.248 | 0.448 | |
| At risk participants (cross-sectional) | LRRK2vsHC | 0.294 | 1.43 ×10−4 | −0.25 | 0.002 | −0.067 | 0.358 | −0.389 | 3.80 ×10−5 | 0.124 | 0.143 | 0.283 | 0.001 | −0.037 | 0.78 | 0.348 | 5.01 ×10−5 | −0.476 | 7.09 ×10−5 |
| GBAvsHC | 0.27 | 0.002 | −0.459 | 9.62 ×10−7 | −0.177 | 0.045 | −0.548 | 4.38 ×10−7 | 0.07 | 0.489 | 0.344 | 0.002 | −0.069 | 0.659 | 0.321 | 0.001 | −0.714 | 1.73 ×10−6 | |
| RBDvsHC | 0.507 | 4.03 ×10−4 | −0.381 | 0.012 | −0.174 | 0.235 | −0.477 | 0.006 | 0.149 | 0.357 | 0.273 | 0.133 | 0.078 | 0.75 | 0.562 | 5.26 ×10−4 | −0.469 | 0.048 | |
| HYPvsHC | 0.317 | 0.046 | −0.345 | 0.033 | −0.282 | 0.069 | −0.406 | 0.026 | 0.106 | 0.571 | 0.095 | 0.646 | 0.061 | 0.825 | 0.428 | 0.023 | −0.32 | 0.2 | |
Since we are repurposing linear RNA sequencing to call circRNAs, we verified that linear RNA counts were not driving our results by evaluating their differential accumulation using the same model. We found linear forms of 57 of the 192 and 25 of the 71 circRNAs to be differentially accumulated (Supplementary Table 4). When adjusting the model for circRNA differential abundance analyses by linear counts, 71 circRNAs remained significant (Supplementary Table 5). If we focus on the nine circRNAs that passed FDR correction in both datasets, none of their linear forms were significant, except for FAM13B (Table 3). We observed decreased levels of circFAM13B in PD (log2FC = −0.212, p = 3.97 ×10−4), as previously described14, along with increased levels of the linear form of FAM13B (log2FC = 0.036, p = 3.99 ×10−5). To understand if the signal was driven by the linear form of FAM13B, we included in the analyses both the linear and the circular RNA counts in addition to other covariates. Both forms of FAM13B, linear (log2FC = 0.043, p = 5.73 ×10−6) and circular (log2FC = −0.201, p = 4.35 ×10−7), remained significant with consistent effect sizes, suggesting that both participate in the association.
Cellular composition and medication affect circRNA accumulation
Since we are analyzing whole blood transcriptome, we repeated the analysis including cell proportions. We found 71 of the 192 (16 of the 71 if we only consider significance in both datasets) circRNAs identified in the cross-sectional DE analyses whose association seem to be driven by cellular composition (Supplementary Table 6). Of the nine circRNAs that remained significant after multiple test correction in both datasets, only circCCDC91 levels (log2FC = 0.263, p = 0.253; Table 3), were affected by cell proportions. CircCCDC91 was associated with neutrophils’ (p < 2 ×10−16), CD4 + T-cells’ (1.76 ×10−10) and monocytes’ (p = 9.54 ×10−7) cell proportions, a trend observed in most of the circRNA affected by cellular composition (Supplementary Table 7).
Medication is another key component that might be affecting the transcriptomic landscape. Even though medication is collected in both datasets, detailed and comprehensive medication information was only available for PPMI. We identified 15 of the 192 (5 of the 71) circRNAs that were associated with medication usage (Supplementary Table 6). Of the nine circRNAs, accumulation of two circRNAs, circITGAX (log2FC = 0.164, p = 0.042), and circCCDC91 (log2FC = 0.190, p = 0.025) seems to be affected by the presence of medication (Supplementary Fig. 2; Table 3).
CircRNA accumulation correlates with symptom severity
We evaluated if any of the nine high confidence circRNAs, were correlated with PD severity measured by UPDRS-III, or neurocognitive decline measured by MoCA (Supplementary Table 8). CircSPI1 (p = 1.45 ×10−7; r2 = 0.015), circAFF2 (p = 1.14 ×10−6; r2 = 0.013), circNCF1 (p = 3.53 ×10−9; r2 = 0.019) and circPADI4 (p = 1.14 ×10−8; r2 = 0.018) showed a significant correlation with UPDRS-III. Additionally, circITGAX (p = 2.81 ×10−8; r2 = 0.017) found to be associated with the presence of PD medication was also significantly correlated with UPDRS-III. Regarding cognitive measurements, we found that circAFF2 was significantly correlated with MoCA (r2 = 0.003; p = 0.031).
Genetic ancestry influences the circRNA landscape
We explored the effect that diverse ancestry has on the identified circRNAs. We observed that 99 circRNAs were in the same direction of effect when comparing the findings between European and African ancestry, ten of which were also nominally significant (Supplementary Figure 3, Supplementary Table 6). Further, we looked into the nine high confidence circRNAs, none of them were significant in the comparison (Table 3). Despite that, the direction of effect for seven of the nine circRNAs was consistent with that identified in the European ancestry individuals (circAFF2, log2FC = 0.203; circETFA, log2FC = −0.021; circFAM13B, log2FC = 0.352; circNCF1, log2FC = 0.124; circPADI4, log2FC = 0.012; circSPI1, log2FC = 0.030; circSUZ12, log2FC = −0.954).
The presence of PD related mutations exacerbates circRNA differences
For the overall analyses, and to maximize statistical power, we included all PD cases regardless of mutation status. Thus, we assessed if there were differences between mutation carriers and idiopathic PD cases, in other words, if mutations were contributing to or driving the association. We observed that mutations in each the LRRK2, the GBA1 and the SNCA affected circRNA landscape in PD participants. We first investigated the overlap between the cross-sectional analysis and the comparison of mutation carriers to healthy controls. We observed a greater overlap with the cross-sectional analysis in the LRRK2+ (138/192) and the GBA1+ (131/192) carriers, than in the SNCA + (32/192) carriers (Supplementary Fig. 3, Supplementary Table 6). When we focused on the findings of the nine high confidence circRNAs identified in the cross-sectional analysis, we found that five of the nine circRNAs, circAFF2, circITGAX, circNCF1, circSUZ12, and circSPI1, were differentially accumulated in both idiopathic PD and mutation carriers with stronger associations in mutation carriers (circAFF2 log2FC = 0.286, p = 1.752 ×10−6; circNCF1 log2FC = 0.375, p = 1.518 ×10−7; circSUZ12 log2FC = −0.258, p = 0.007; circSPI1 log2FC = 0.309, p = 6.699 ×10−7; Supplementary Table 6). In contrast, circFAM13B and circCCDC91 were differentially accumulated in mutation carriers (circFAM13B log2FC = −0.577, p = 6.238 ×10−11; circCCDC91 log2FC = −0.278, p = 3.887 ×10−4) but not in idiopathic PD, suggesting that the signal identified in our previous analysis was driven by the genetic form of the disease. The opposite was true for circETFA and circPADI4 which were significant in idiopathic PD (circETFA log2FC = −0.190, p = 0.002; circPADI4 log2FC = 0.365, p = 0.001) but not in mutation carriers. When broken down by gene, circFAM13B, circCCDC91, circSPI1 and circSUZ12 were differentially accumulated in both the LRRK2+ (circFAM13B log2FC = 0.252, p = 0.003; circCCDC91 log2FC = −0.251, p = 0.004; circSPI1 log2FC = 0.526, p = 2.43 ×10−9; circSUZ12 log2FC = −0.374, p = 0.003) and the GBA1+ (circFAM13B log2FC = −0.749, p = 4.370 ×10–8 circCCDC91 log2FC = −0.44, p = 1.80 ×10−4; circSPI1 log2FC = 0.584, p = 6.08 ×10−6; circSUZ12 log2FC = −0.795, p = 5.26 ×10−5), whereas circPADI4 (log2FC = 0.884, p = 0.013) was differentially accumulated in the SNCA+ carriers only (Table 3). Altogether, suggesting the presence of heterogeneity in the circular transcriptome in relation to PD genetic background.
CircRNA accumulation reflects disease-associated changes over time in individuals with Parkinson’s Disease
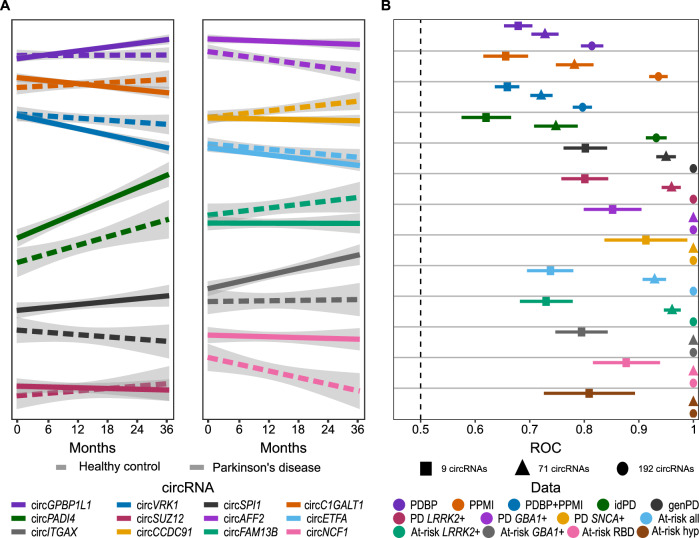
To leverage the longitudinal design of both the PDBP and PPMI studies, we also explored accumulation of circRNAs using mixed models to identify circular transcripts that change over time. We included 547 PDBP participants (Ncasses = 376, Ncontrols = 171) and 617 PPMI participants (Ncasses = 474, Ncontrols = 143) with at least two visits available for each participant (Table 2) and then performed the meta-analysis. When evaluating the intercepts of the mixed model, they were correlated with what we identified in the cross-sectional analysis (Supplementary Fig. 4, Supplementary Table 9), supporting our findings that there are changes in circRNA landscape associated with the presence of the disease. In the meta-analysis, we identified 114 circRNAs with significantly different trajectories between PD and healthy controls, 14 of which were nominally significant in both PDBP and PPMI populations. The three most significant findings for the interaction term were circGPBP1L1 (beta = 0.059, p = 1.351 ×10−4), circC1GALT1 (beta = −0.099, p = 6.859 ×10−4) and circVRK1 (beta = −0.111, p = 6.861 ×10−4) (Supplementary Table 9). When evaluating the nine high confidence circRNAs all directions of effect were consistent with those identified in the cross-sectional results (Table 3), except circPADI4 that had different directions of effect between PDBP and PPMI population in the longitudinal analysis (Table 3). We found that the rate of change in circRNA abundance (measured as slope) of two of the nine high confidence circRNAs differed significantly between PD cases and controls (circCCDC91, p = 0.018; circITGAX, p = 0.027; Fig. 3A).
Fig. 3. circRNAs change over the course of the disease and can be leveraged diagnostic biomarkers.
A Line plots showing trajectories of circRNA abundances for the top three significant circRNAs in longitudinal analysis, as well as the nine high-confidence circRNAs from cross-sectional analysis; B Whisker plot showing ROC AUC values with 90% confidence interval evaluating the ability of three combination of circRNAs to distinguish between PD participants and healthy controls or at-risk participants and healthy controls.
Similar to the approach followed for the cross-sectional analyses, several sensitivity analyses were performed (Supplementary Table 10). We found that trajectories of 91 of the 114 circRNAs were affected by cellular composition, and 41 of the 114 were impacted by medication usage. We further evaluated symptomatic participants with PD-associated genetic mutations and observed that 24/114 and 9/114 circRNAs have significantly different trajectories in the LRRK2+ and the GBA1+ carriers compared to healthy control participants, respectively. The top three circRNAs whose trajectories differed significantly between PD and healthy controls, circGPBP1L1, circC1GALT1 and circVRK1 were significant in idiopathic participants (circGPBP1L1 beta = 0.004, p = 0.007; circC1GALT1 beta = −0.007, p = 0.003; circVRK1 beta = −0.052, p = 0.037). Taking a closer look at the nine high-confidence circRNAs, we observed that cirAFF2 (beta = 0.085, p = 0.007) and circNCF1 (beta = 0.149, p = 1.273 ×10−4) were significant in mutation carriers, while circCCDC91 (beta = −0.007, p = 0.006) was significant in idiopathic participants only (Supplementary Table 10). Despite not reaching statistical significance, all but circETFA (beta = 0.030, p = 0.411), had direction of effect largely consistent with previous observations in both idiopathic and genetic cases. However, broken down by gene, three out of nine circRNAs, circAFF2 (beta = 0.072, p = 0.036), circFAM13B (beta = −0.106, p = 0.028), and circNCF1 (beta = 0.152, p = 3.097 ×10−4) had significantly different trajectories in LRRK2+ mutation carriers, and circAFF2 (beta = 0.341, p = 0.015) and circNCF1 (beta = 0.518, p = 0.003) in the GBA1+ carriers, suggesting that the circAFF2 association might be driven by the GBA1+ carriers. Longitudinal analyses were not possible for the SNCA+ carriers due to very limited sample size (N = 8).
CircRNA accumulation starts before symptom onset
We examined whether changes in circRNA accumulation can be observed in participants who are at high risk of developing PD, namely carriers of known PD-related mutations that have not been diagnosed, or those who show known the PD associated syndromes, RBD or hyposmia. None of the at-risk participants were included in the cross-sectional or longitudinal analyses described above. The analyses of individuals at-risk were done in the PPMI dataset exclusively due participant availability, and included 150 LRRK2+, 87 GBA1+, and three SNCA+ mutation carriers, along with 25 participants exhibiting RBD, and 18 participants with hyposmia. Given the reduced number of at-risk SNCA+ carriers, no analyses were performed in that sub-group.
Out of the 192 circRNAs that were significant after multiple test correction in the cross-sectional meta-analysis and 71 circRNA that were significant in the meta-analysis and also nominally significant in both PDBP and PPMI populations, most of them were already accumulating in individuals at-risk. The greatest overlap was observed in the LRRK2+ (123/192 and 52/71) and the GBA1+ (130/192 and 60/71) carriers, followed by participants with RBD (71/192 and 32/71) (Supplementary Table 11). However, a moderate number of circRNAs seem to accumulate before symptom onset in participants with hyposmia (35/192 and 12/71) (Supplementary Table 11). Similar to symptomatic participants, when focusing on the nine high confidence circRNAs (Table 3), we found that circAFF2 (log2FC = 0.294, p = 1.42 ×10−4), circCCDC91 (log2FC = −0.250, p = 0.004), circFAM13B (log2FC = −0.389, p = 3.80 ×10−5), circITGAX (log2FC = 0.251, p = 0.003), circNCF1 (log2FC = 0.461, p = 1.231 ×10−6), circSUZ12 (log2FC = −0.476, p = 7.09 ×10−5) and circSPI1 (log2FC = 0.348, p = 5.01 ×10−5), were differentially accumulated in the LRRK2+ and the GBA1+ (circAFF2, log2FC = 0.270, p = 0.002; circCCDC91 log2FC = −0.440, p = 1.802 ×10−4; circFAM13B, log2FC = −0.548, p = 4.38 ×10−7; circITGAX log2FC = 0.415, p = 0.002; circNCF1 (log2FC = 0.713, p = 1.373 ×10−6; circSUZ12, log2FC = −0.714, p = 1.73 ×10−6; circSPI1, log2FC = 0.321, p = 0.001) carriers compared to healthy controls. When comparing the LRRK2 to the GBA1 at-risk participants, we found 23 of the 192 DE circRNAs to also be differentially expressed between the two groups of at-risk participants (Supplementary Table 12). Additionally, we identified 185 circRNAs that were not DE in comparison of PD to healthy controls, that were DE in comparison of the LRRK2 to the GBA1 at-risk participants (Supplementary Table 12). Additionally, circETFA (log2FC = −0.177, p = 0.045) was associated with the GBA1+ carriers. Further, we found four circRNAs, circAFF2 (log2FC = 0.507, p = 4.030 ×10−4), circFAM13B (log2FC = −0.477, p = 0.006), circSPI1 (log2FC = 0.562, p = 5.260 ×10−4) and circSUZ12 (log2FC = −0.469, p = 0.048), differentially accumulated in participants with RBD, and three circRNAs, circAFF2 (log2FC = 0.317, p = 0.046), circFAM13B (log2FC = −0.406, p = 0.026) and circSPI1 (log2FC = 0.428, p = 0.023), differentially accumulated in participants with hyposmia.
Targeted miRNAs analysis suggests involvement of circRNA in known PD related pathways
To better understand the biological implications of the nine high confidence circRNAs identified in the previous analysis, we listed their predicted miRNA targets using CircInteractome web tool32. By doing so, we obtained a list of 282 miRNA targets for circAFF2, 271 for circCCDC91, 96 for circETFA, 178 for circFAM13B, 285 for circITGAX, 32 for circNCF1, 26 for circSPI1, 313 for circSUZ12 and none for circPADI4. To ensure the biological relevance of the analyses and reduce the number of miRNAs, we leveraged the miRNA counts available in the PPMI dataset. We performed correlation analyses between normalized circRNA and normalized miRNA counts to retain the miRNA binding sites with biological evidence (p < 0.05 for the correlation between circRNA and miRNA) for downstream analyses. We reduced the miRNA targets to 27 miRNAs for circAFF2, 37 miRNAs for circCCDC91, 7 miRNAs for circETFA, 20 miRNAs for circFAM13B, 32 miRNAs for circITGAX, two miRNAs for circNCF1, two miRNAs for circSPI1, and 30 miRNAs for circSUZ12 (Supplementary Table 13). All miRNAs significantly correlated with circAFF2, circETFA, circFAM13B, and circSPI1 overlapped with the 30 miRNAs significantly correlated with circSUZ12. We then performed pathway analyses with DIANA mirPath using the 61 miRNAs as input. We observed enrichment in several KEGG terms such as dopaminergic synapse39–41 (p = 5.411 ×10−4) and long-term depression42 (p = 6.20 ×10−5), both of them previously associated with PD, and enriched with miRNAs associated with seven of the nine circRNAs (Supplementary Table 14). Another term that we found enriched in miRNAs correlated with all nine circRNAs is the Hippo signaling pathway (p = 7.680 ×10−8) (Supplementary Table 14), which has previously been described to play a role in ischemia-associated CNS diseases and PD43,44. Ubiquitin mediated proteolysis, suspected to be impaired in PD and contributing to Lewy body formation45–47, was also found enriched (p = 0.006) for miRNAs targeted by five of the nine circRNAs (Supplementary Table 14). Together, these findings add evidence to the potentially role of circRNAs in PD pathogenesis.
CircRNAs are informative as biomarkers of Parkinson’s disease to potentially aid diagnosis
We investigated the predictive power of our findings to differentiate between PD and healthy controls, as well as at-risk and healthy control participants, in the most recent samples from PDBP and PPMI datasets. We analyzed three circRNA sets: 1) with 192 circRNAs which were significant in the cross-sectional meta-analysis after multiple test correction, 2) with 71 circRNAs that were significant in the meta-analysis and nominally significant in both PBDP and PPMI populations, and 3) nine circRNAs that passed multiple test correction in meta-analysis, as well as in PDBP and PPMI datasets separately, and adjusted all analyses by age and sex. We tested the performance in PDBP and PPMI separately and also combined the two datasets. We found that the set with 192 circRNAs had the best predictive value of the three, with a ROC AUC value of 0.797 in the combined dataset and 0.814 in PDBP and 0.936 in PPMI datasets (Supplementary Table 15, Fig. 3B). The two smaller circRNA sets had a moderate ability to predict PD with AUC values of 0.721 (AUCPDBP = 0.728; AUCPPMI = 0.782) and 0.659 (AUCPDBP = 0.679; AUCPPMI = 0.656) in the sets with 71 and nine circRNAs, respectively (Supplementary Table 15, Fig. 3B). When we split the PPMI participants into those with idiopathic and genetic forms of PD, we observed that the set with 192 circRNAs had a high predictive value for both idiopathic and genetic PD (0.932 and 1.00, respectively) (Supplementary Table 15, Fig. 3B). However, the two smaller sets maintained high predictive value only in genetic PD (AUC9 = 0.807, AUC71 = 0.954), with only moderate ability to differentiate between idiopathic PD and healthy controls (AUC9 = 0.620, AUC71 = 0.748) (Supplementary Table 15, Fig. 3B). This is also reflected in at-risk individuals, where the 192 circRNAs set was able to differentiate between at-risk and healthy individuals with an AUC of 1.000, while some power lost in the sets with 71 (AUC71 = 0.929) and nine circRNAs (AUC9 = 0.738) (Supplementary Table 15, Fig. 3B).
Next, we evaluated the power of the three circRNA sets to differentiate between healthy controls and dementia with Lewy bodies (DLB, n = 28), frontotemporal dementia (FTD, n = 16) and Alzheimer’s disease (AD, n = 567). As counts were not available for all 192 DE circRNAs, the three circRNA sets had to be sized down to 1) 169/192 circRNAs which were significant in the cross-sectional meta-analysis after multiple test correction, 2) 63/71 circRNAs that were significant in the meta-analysis and nominally significant in both PBDP and PPMI populations, and 3) eight/nine circRNAs that passed multiple test correction in meta-analysis, as well as in PDBP and PPMI datasets separately. We observed that the smallest set of eight circRNAs had a moderate ability to differentiate controls from DLB (AUC8 = 0.741) and AD (AUC8 = 667), but not FTD (AUC8 = 0.595), (Supplementary Fig. 5, Supplementary Table 16). Further, the predictive ability of the models progressively decreased with the addition of circRNAs. As such, models with 63 and 169 circRNAs were not able to distinguish FTD (AUC63 = 0.554, AUC169 = 0.418) or AD (AUC63 = 0.581, AUC169 = 0.563) from healthy controls, while they still had a moderate ability to predict DLB (AUC63 = 0.692, AUC169 = 0.686) (Supplementary Fig. 5, Supplementary Table 16).
We have also attempted an unbiased and hypothesis free approach to select the most informative circRNAs to predict PD participants at first visit. We evaluated two statistical linear approaches, logistic regression with LASSO and ridge regularization, and two nonlinear ML approaches: random forests and MiniPatch, for binary classification models. We obtained a 149 circRNA model with MiniPatch, which was the best performing (AUC149 = 0.825; Supplementary Fig. 6A, B), followed by the ridge model with 227 circRNA (AUC227 = 0.784; Supplementary Fig. 6A, B). Functional annotation on the linear gene versions of the 149 circRNAs detected by MiniPatch generated no significant GO annotation after multiple test correction but some nominally identified suggestive terms like organic acid transmembrane transport (p = 0.001), chromatin organization (p = 0.002) and neuron to neuron synapse (p = 0.007) (Supplementary Fig. 6C, Supplementary Table 17). MiniPatch is a nonlinear approach, which suggest that the software is going beyond the simple but restrictive paradigm of transcripts acting independently in an additive manner. Non-assumption of this paradigm might help discover new patterns of transcript cluster interactions. We found 78% overlap between the two non-linear algorithms, RF and MiniPatch, but only 10% overlap between MiniPatch and LASSO linear approach. This evidence contributes to this idea that non-linear ML models might be more informative to predict complex diseases.
Discussion
In this study we leveraged the largest to date publicly available longitudinal blood RNAseq data from two of the largest active PD studies, PDBP and PPMI, to identify circRNAs that were differentially accumulated in relation to PD. Its design attempted to maximize differences between cases and controls by including the last visit of each individual instead of baseline as previous reports48. By including the last visit, the disease is more advanced, and thus the differences are potentially more pronounced. Finally, and for the first time in PD, we have taken into consideration the longitudinal design of the study and included multiple observations per participant to not only identify circRNA that are potentially differentially accumulated but also find those that change with time and have the potential to be leveraged to follow the progression of the disease.
Overall, we identified 192 circRNA transcripts that were associated with PD at the time of the last visit, and 114 whose trajectories differed significantly between cases and controls. Even though the exact function of circRNA is still to be deciphered, many functions have been already attributed. They have been found to act as miRNA sponges to regulate gene expression, interact with proteins, and to generate protein products among many others3,4,7–12. When biologically contextualizing our findings predicting miRNA binding sites, we observed an enrichment in dopaminergic synapse term. It is well established that PD is caused by the death of dopaminergic neurons39–41, thus, it is plausible to think that circRNAs might be participating as regulatory molecules. Strikingly, we seem to be capturing neuronal death in blood, suggesting that those changes are being captured as the result of blood-brain barrier disruption49. Additionally, we found Hippo pathway43,44, which has already been linked with PD, to be enriched in conjunction to all our circRNAs. Together with enrichment in other PD-associated pathways such as long-term depression and ubiquitin-mediated proteolysis, these results show that blood circRNAs may reflect changes related to PD pathobiology, potentially leaking from the brain. As further support to the relevance of circRNAs in PD, we found that genes giving yield to 18 of the identified 192 DE circRNAs, including ITGAX, have previously been nominated as potentially PD-causal genes, with varying levels of support50. Several of the genes have been recorded in association with neurodegenerative diseases such as AD (ZGRF151), amyotrophic lateral sclerosis (NUP5452, NEK153) and frontotemporal dementia (NUP5452), and other nervous system and movement disorders (MED1354, CDKN355, TLK256,57, REV3L58).
We further evaluated our findings in diverse genetic backgrounds, finding that both ancestry and mutation carrier status contributed to PD circRNA landscape. Our results point to some differences in circRNA expression patterns between participants of African and those of European ancestry, supporting the urgency to increase the diversity of the cohorts to not only validate and universalize scientific findings, but also to understand better the pathobiology underlying PD and provide personalized medicine.
Among the nine strongest candidates from our analyses, some of them have already been associated with PD, especially the linear form of those transcripts. FAM13B was found to be differentially expressed in PD brains but not in blood59. Interestingly, another study including eight participants did report the dysregulation of the FAM13B in blood14. In the present study with more than 1,800 subjects, we observed changes in the linear forms of FAM13B in blood of PD participants, confirming that FAM13B is indeed differentially accumulated in the blood. Additionally, we also report that the circular form of the FAM13B is differentially accumulated in blood of PD participants compared to controls. Overall, these findings suggest that FAM13B might be a key player in PD pathogenesis. The presence of circular forms suggests that there are tightly regulated processes associated with this gene. Looking at longitudinal data, circFAM13B levels change over time in the LRRK2+ carriers, but not in other participants with the genetic forms of PD. This could indicate that while circFAM13B does correlate with PD diagnosis in all mutation carriers, it does so via a different process in the LRRK2+ which more closely reflects the advancement of the disease. SPI1, a transcription factor involved in myeloid cell development and function, is known for its role in Alzheimer’s disease (AD)60. Given its association with AD, and our findings, circSPI1 was found to be associated with PD independently of medication and cell counts, we investigated if the association with PD status might be driven by cognitive function measured by MoCA. Unfortunately, the correlation was not significant. Despite the negative results and given the close to normal values of MoCA in the population included in this study, it is plausible to think that this correlation might become significant once the disease advances. Alternatively, circSPI1 might influence PD differently than AD, or even have a different function than the linear form, thus explaining the lack of association.
Among the novel circRNA associations described in this paper, several of the host genes have not been previously linked to PD. For example, the AFF2 has not been previously reported in PD, but it has been reported to contribute to axonal degeneration and TDP-43 pathology in frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS)61. Given the involvement of the AFF2 in neurodevelopment62,63 and neurodegeneration61, and the presence of co-pathology and overlap across neurodegenerative diseases, it would be reasonable to postulate that circAFF2 is integral to proper functioning of the nervous system as well. This is further supported by our finding of circAFF2 being significantly differentially expressed, including across different genetic backgrounds, with consistent direction of effect.
When diving into the mutation carriers only, this study included participants with mutations in the LRRK2, the GBA1 and the SNCA genes, with highly heterogenous results. It is known that the most common LRRK2 mutation, Gly2019Ser, leads to constitutive activation of the LRRK2, which leads to activation of neuronal death pathway, and possibly upregulation of the SNCA64,65. On the other hand, the most common SNCA mutations are copy-number mutation, more specifically duplications66. Altogether, it is not surprising to observe such variability in circRNA landscape, as regulatory pathways that are triggered by the LRRK2 mutation to bring about PD, might not be affected in the SNCA mutation carriers, given that the SNCA is downstream from the LRRK2. Our findings regarding circFAM13B, circSPI1, and circSUZ12 that are significantly associated with the LRRK2 mutation carriers, but not the SNCA seem to support this hypothesis. Differences in pathway regulation between the LRRK2 and the SNCA mutation carriers are further emphasized by opposing directions of effect of circETFA in the two. Mutations in the GBA1 gene lead to decrease in GCase enzyme activity, resulting in lysosome malfunctioning67,68. GCAse impairment has been suggested to promote alpha-synuclein accumulation in PD. This points to possible overlap between regulatory pathways, that involve circFAM13B, circSPI1, and circSUZ12, that are differentially accumulated in the GBA1 compared to the LRRK2 mutation carriers, potentially leading to increased alpha-synuclein pathology in both. Further analyses in a larger sample sizes are required to verify these findings given the limited sample size and variability of the results.
We demonstrated that circRNA not only capture relevant biological events but have some ability to differentiate between PD cases and controls, despite having moderate accuracy. Currently, PD is diagnosed based on clinical and neuroimaging criteria, and then monitored using clinical tests that assess the motor and non-motor symptoms of the disease. However, there are no molecular diagnostic or prognostic biomarkers available for PD, or in general, for most of the neurodegenerative diseases69. Circular RNA have already been demonstrated to be informative for PD, with intrinsic qualities that are highly desired in a biomarker. They are highly stable and very abundant in the brain, which means they can potentially leak to the CSF or blood via blood brain barrier breakdown and can be measured in biofluids49, they can be measured by real-time PCR, and are stable due to being circular6,69. We tested different sets of circRNA, showing that 71 circRNA are sufficient to distinguish participants at-risk of PD from healthy participants with a high accuracy, pointing to their potential utility for PD diagnosis. On top of that, we identified five circRNA that were correlated with UPDRS-III, suggesting their potential clinical use to monitor drug response. Given the lack of medically useful molecular tools to monitor or diagnose disease, these findings have the potential to be relevant clinically, if validated. The identified circRNA are promising measurements to expedite diagnosis and facilitate motor symptom monitoring by providing much needed non-invasive molecular tests for PD care.
This study had several limitations. While we had access to the two largest longitudinal datasets (4833 samples, 1789 circRNAs), longitudinal analyses are underpowered, and larger sample sizes are needed to appropriately power these analyses. Likewise, the number of African American participants, at-risk individuals, or mutation carriers was very limited, thus we did not perform de novo discovery in these groups, but rather leveraged them to validate the main findings. Further efforts are needed to actively recruit participants from diverse backgrounds. Furthermore, mutation and medication data were absent from the PDBP dataset, along with the recruitment of “at risk” individuals, which did not allow for a straight comparison of the two populations. Finally, we are repurposing traditional RNAseq data to identify and quantify circRNAs, rather than purifying and subsequently sequencing circRNAs. Despite the need for some additional analysis and validation in the future, we have successfully replicated findings from other groups, supporting the validity of this approach.
In conclusion, this is the largest study to date describing and biologically contextualizing the circRNA landscape in blood in relation to PD. We identified and replicated nine circRNAs differentially accumulated in PD compared to healthy controls and linked to biologically pathways relevant for the disease. More importantly, we have demonstrated that circRNA not only have a biological role in PD but can also be leveraged as biomarkers with the potentially aid the diagnosis and facilitate the monitoring of PD with minimally invasive molecular measurements.
Supplementary information
Acknowledgements
We thank all the participants and their families along with the institutions and all the staff who provided plasma tissue, without whom this study would not have been possible. We thank Dr. Abdallah Eteleeb for providing an outline upon which we built our data processing pipeline and for his hands-on help processing part of the data. Next, we would like to thank Drs. Joe Boktor and Sarkis Mazmanian, for kindly providing the deconvoluted blood cell counts using Immunedeconv tool that was leveraged to add cellular composition in the present manuscript. This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, the Neurogenomics and Informatics Center (NGI: https://neurogenomics.wustl.edu/) and the Departments of Neurology and Psychiatry at Washington University School of Medicine. Data used in the preparation of this article were obtained from the Accelerating Medicine Partnership® (AMP®) Parkinson’s Disease (AMP PD) Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. The AMP® PD program is a public-private partnership managed by the Foundation for the National Institutes of Health and funded by the National Institute of Neurological Disorders and Stroke (NINDS) in partnership with the Aligning Science Across Parkinson’s (ASAP) initiative; Celgene Corporation, a subsidiary of Bristol-Myers Squibb Company; GlaxoSmithKline plc (GSK); The Michael J. Fox Foundation for Parkinson’s Research ; Pfizer Inc.; AbbVie Inc.; Sanofi US Services Inc.; and Verily Life Sciences. ACCELERATING MEDICINES PARTNERSHIP and AMP are registered service marks of the U.S. Department of Health and Human Services. Clinical data and biosamples used in preparation of this article were obtained from the Michael J. Fox Foundation for Parkinson’s Research (MJFF) and National Institutes of Neurological Disorders and Stroke (NINDS) Parkinson’s Disease Biomarkers Program (PDBP) and MJFF Parkinson’s Progression Markers Initiative (PPMI) studies. This work was supported by grants from the Department of Defense (W81XWH2010849), Bright Focus Foundation (A2021033S), Michael J. Fox Foundation (MJFF-021599), National Institute of Health (P30AG066444, R00AG062723, U19AG03243812, R01AG053267), Alzheimer’s Association (DIAN-TU-PP-22-872356 and DIANTUOLE21725093) and an NGI Pilot Grant. PPMI is sponsored by The Michael J. Fox Foundation for Parkinson’s Research and supported by a consortium of scientific partners: 4D Pharma, Abbvie, AcureX, Allergan, Amathus Therapeutics, Aligning Science Across Parkinson’s, AskBio, Avid Radiopharmaceuticals, BIAL, BioArctic, Biogen, Biohaven, BioLegend, BlueRock Therapeutics, Bristol-Myers Squibb, Calico Labs, Capsida Biotherapeutics, Celgene, Cerevel Therapeutics, Coave Therapeutics, DaCapo Brainscience, Denali, Edmond J. Safra Foundation, Eli Lilly, Gain Therapeutics, GE HealthCare, Genentech, GSK, Golub Capital, Handl Therapeutics, Insitro, Jazz Pharmaceuticals, Johnson & Johnson Innovative Medicine, Lundbeck, Merck, Meso Scale Discovery, Mission Therapeutics, Neurocrine Biosciences, Neuron23, Neuropore, Pfizer, Piramal, Prevail Therapeutics, Roche, Sanofi, Servier, Sun Pharma Advanced Research Company, Takeda, Teva, UCB, Vanqua Bio, Verily, Voyager v. 25MAR2024 Therapeutics, the Weston Family Foundation and Yumanity Therapeutics. The PPMI investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit www.ppmi-info.org. The Parkinson’s Disease Biomarker Program (PDBP) consortium is supported by the National Institute of Neurological Disorders and Stroke (NINDS) at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. The PDBP investigators have not participated in reviewing the data analysis or content of the manuscript.
Author contributions
L.I., A.B. and Y.S. conceived and wrote this article. L.I. and C.C. conceptualized the research plan. L.I., A.B., and Y.S. designed the analysis plan. Y.S., A.B., S.S., T.P., R.K., J.S., D.D., B.P. and C.M. performed the analyses. T.P., R.K. and C.M. developed the RShiny browser associate with the manuscript. J.A.P. and J.B. performed predictive modeling. A.B., Y.S., S.S., R.K., C.M., J.A.P., J.B., C.C., and L.I. discussed the project, revised the manuscript, and provided critical feedback.
Data availability
CircRNA count matrices are available from AMP-PD (https://amp-pd.org/). Summary results for all circRNAs and linear RNAs quantified in both PDBP and PPMI can be found in our transcript level browser (pdbloodtranscriptomics.wustl.edu).
Code availability
All original code has been deposited at GitHub (https://github.com/Ibanez-Lab/BloodCircularRNA-ParkinsonsDisease) and is publicly available as of the date of publication.
Competing interests
The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. C.C. is a member of the advisory board of Vivid genetics, Halia Therapeutics and ADx Healthcare and has received research support from Biogen, EISAI, Alector and Parabon. The rest of the authors report no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-024-00839-3.
References
- 1.Dorsey, E. R., Sherer, T., Okun, M. S. & Bloem, B. R. The Emerging Evidence of the Parkinson Pandemic. J. Parkinsons Dis.8, S3–S8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizek, P., Kumar, N. & Jog, M. S. An update on the diagnosis and treatment of Parkinson disease. CMAJ Can. Med. Assoc. J.188, 1157–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett, S. P. & Salzman, J. Circular RNAs: analysis, expression and potential functions. Development143, 1838–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, X., Yang, L. & Chen, L. L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell71, 428–442 (2018). [DOI] [PubMed] [Google Scholar]
- 5.D’Ambra, E., Capauto, D. & Morlando, M. Exploring the Regulatory Role of Circular RNAs in Neurodegenerative Disorders. Int. J. Mol. Sci.20, 5477 (2019). [DOI] [PMC free article] [PubMed]
- 6.Doxakis, E. Insights into the multifaceted role of circular RNAs: implications for Parkinson’s disease pathogenesis and diagnosis. NPJ Parkinsons Dis.8, 7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dube, U. et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci.22, 1903–1912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H. H. et al. Circular RNA detection identifies circPSEN1 alterations in brain specific to autosomal dominant Alzheimer’s disease. Acta Neuropathol. Commun.10, 29 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta, S. L., Dempsey, R. J. & Vemuganti, R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol.186, 101746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravanidis, S. et al. Differentially Expressed Circular RNAs in Peripheral Blood Mononuclear Cells of Patients with Parkinson’s Disease. Mov. Disord.36, 1170–1179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanan, M. et al. A Parkinson’s disease CircRNAs Resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol. Med.12, e13551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, M. & Bian, Z. The Emerging Role of Circular RNAs in Alzheimer’s Disease and Parkinson’s Disease. Front Aging Neurosci.13, 691512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, X. et al. Circular RNAs in the human brain are tailored to neuron identity and neuropsychiatric disease. Nat. Commun.14, 5327 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong, F. et al. RNA-sequencing of peripheral blood circular RNAs in Parkinson disease. Med. (Baltim.)100, e25888 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittle, B. J. et al. Early-stage idiopathic Parkinson’s disease is associated with reduced circular RNA expression. npj Parkinsons Dis.10, 1–14 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ofori, E., Du, G., Babcock, D., Huang, X. & Vaillancourt, D. E. Parkinson’s disease biomarkers program brain imaging repository. Neuroimage124, 1120–1124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkinson Progression Marker, I. The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol.95, 629–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig, D. W. et al. RNA sequencing of whole blood reveals early alterations in immune cells and gene expression in Parkinson’s disease. Nat. Aging1, 734–747 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Cerri, S., Mus, L. & Blandini, F. Parkinson’s Disease in Women and Men: What’s the Difference? J Parkinsons Dis. 9, 501–515 (2019). [DOI] [PMC free article] [PubMed]
- 20.Ou, Z. et al. Global Trends in the Incidence, Prevalence, and Years Lived With Disability of Parkinson’s Disease in 204 Countries/Territories From 1990 to 2019. Front Public Health9, 776847 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenthal, L. S. et al. The NINDS Parkinson’s Disease Biomarkers Program. Mov. Disord.31, 915–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gwinn, K. et al. Parkinson’s disease biomarkers: perspective from the NINDS Parkinson’s Disease Biomarkers Program. Biomark. Med.11, 451–473 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, H.-H. et al. Circular RNA detection identifies circPSEN1 alterations in brain specific to autosomal dominant Alzheimer’s disease. Acta Neuropathologica Commun.10, 29 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Z. et al. Genetic variants associated with Alzheimer’s disease confer different cerebral cortex cell-type population structure. Genome Med. 10, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard Toolkit (Broad Institute, 2019).
- 27.Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. methods14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng, J., Metge, F. & Dieterich, C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics32, 1094–6 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Rau, A., Marot, G. & Jaffrezic, F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinforma.15, 91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salas, L. A. et al. Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling. Nat. Commun.13, 761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dudekula, D. B. et al. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol.13, 34–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vlachos, I. S. et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 43, W460–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez, M. V. et al. Genetic and multi-omic resources for Alzheimer disease and related dementia from the Knight Alzheimer Disease Research Center. Sci. Data11, 768 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman, J. H., Hastie, T. & Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw.33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 36.Tay, J. K., Narasimhan, B. & Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Softw.106, 1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright, M. N. & Ziegler, A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw.77, 1–17 (2017). [Google Scholar]
- 38.Gan, L. & Allen, G. I. Fast and interpretable consensus clustering via minipatch learning. PLOS Computational Biol.18, e1010577 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, J.-S. et al. Cellular and synaptic mechanisms for Parkinson’s disease-related chronic pain. Mol. Pain.17, 1744806921999025 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latif, S. et al. Dopamine in Parkinson’s disease. Clin. Chim. Acta522, 114–126 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Stern, S. et al. Reduced synaptic activity and dysregulated extracellular matrix pathways in midbrain neurons from Parkinson’s disease patients. npj Parkinsons Dis.8, 1–16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prange, S., Klinger, H., Laurencin, C., Danaila, T. & Thobois, S. Depression in Patients with Parkinson’s Disease: Current Understanding of its Neurobiology and Implications for Treatment. Drugs Aging39, 417–439 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, X. et al. An update on the role of Hippo signaling pathway in ischemia-associated central nervous system diseases. Biomedicine Pharmacother.162, 114619 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Ahn, E. H., Kang, S. S., Qi, Q., Liu, X. & Ye, K. Netrin1 deficiency activates MST1 via UNC5B receptor, promoting dopaminergic apoptosis in Parkinson’s disease. Proc. Natl Acad. Sci. USA117, 24503–24513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNaught, K. S. & Jenner, P. Proteasomal function is impaired in substantia nigra in Parkinson’s disease. Neurosci. Lett.297, 191–194 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Buneeva, O. & Medvedev, A. Atypical Ubiquitination and Parkinson’s Disease. Int J. Mol. Sci.23, 3705 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi, M., Du, X., Jiao, Q., Chen, X. & Jiang, H. Expanding the role of proteasome homeostasis in Parkinson’s disease: beyond protein breakdown. Cell Death Dis.12, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parkinson’s disease is associated with an imbalance in circular RNA expression. https://www.researchsquare.com10.21203/rs.3.rs-3202518/v1 (2023).
- 49.Al-Bachari, S., Naish, J. H., Parker, G. J. M., Emsley, H. C. A. & Parkes, L. M. Blood-Brain Barrier Leakage Is Increased in Parkinson’s Disease. Front Physiol.11, 593026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grenn, F. P. et al. The Parkinson’s Disease Genome-Wide Association Study Locus Browser. Mov. Disord.35, 2056–2067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rochín-Hernández, L. J. et al. The Proteome Profile of Olfactory Ecto-Mesenchymal Stem Cells-Derived from Patients with Familial Alzheimer’s Disease Reveals New Insights for AD Study. Int J. Mol. Sci.24, 12606 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khalil, B., Linsenmeier, M., Smith, C. L., Shorter, J. & Rossoll, W. Nuclear-import receptors as gatekeepers of pathological phase transitions in ALS/FTD. Mol. Neurodegener.19, 8 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brenner, D. et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain139, e28 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Snijders Blok, L. et al. De novo mutations in MED13, a component of the Mediator complex, are associated with a novel neurodevelopmental disorder. Hum. Genet137, 375–388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaletzky, J. et al. Phosphatidylinositol-5-Phosphate Activation and Conserved Substrate Specificity of the Myotubularin Phosphatidylinositol 3-Phosphatases. Curr. Biol.13, 504–509 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Lelieveld, S. H. et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci.19, 1194–1196 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Reijnders, M. R. F. et al. De Novo and Inherited Loss-of-Function Variants in TLK2: Clinical and Genotype-Phenotype Evaluation of a Distinct Neurodevelopmental Disorder. Am. J. Hum. Genet102, 1195–1203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomas-Roca, L. et al. De novo mutations in PLXND1 and REV3L cause Möbius syndrome. Nat. Commun.6, 7199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benoit, S. M. et al. Expanding the search for genetic biomarkers of Parkinson’s disease into the living brain. Neurobiol. Dis.140, 104872 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Huang, K. et al. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci.20, 1052–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuva-Aydemir, Y., Almeida, S., Krishnan, G., Gendron, T. F. & Gao, F.-B. Transcription elongation factor AFF2/FMR2 regulates expression of expanded GGGGCC repeat-containing C9ORF72 allele in ALS/FTD. Nat. Commun.10, 5466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eigenhuis, K. N., Somsen, H. B. & van den Berg, D. L. C. Transcription Pause and Escape in Neurodevelopmental Disorders. Front Neurosci.16, 846272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poeta, L., Drongitis, D., Verrillo, L. & Miano, M. G. DNA Hypermethylation and Unstable Repeat Diseases: A Paradigm of Transcriptional Silencing to Decipher the Basis of Pathogenic Mechanisms. Genes11, 684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rui, Q., Ni, H., Li, D., Gao, R. & Chen, G. The Role of LRRK2 in Neurodegeneration of Parkinson Disease. Curr. Neuropharmacol.16, 1348–1357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tolosa, E., Vila, M., Klein, C. & Rascol, O. LRRK2 in Parkinson disease: challenges of clinical trials. Nat. Rev. Neurol.16, 97–107 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Cherian, A. & Divya, K. P. Genetics of Parkinson’s disease. Acta Neurol. Belg.120, 1297–1305 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Vidyadhara, D. J., Lee, J. E. & Chandra, S. S. Role of the endolysosomal system in Parkinson’s disease. J. Neurochem150, 487–506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muñoz-Delgado, L. et al. Peripheral inflammatory immune response differs among sporadic and familial Parkinson’s disease. npj Parkinsons Dis.9, 1–9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parnetti, L. et al. CSF and blood biomarkers for Parkinson’s disease. Lancet Neurol.18, 573–586 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CircRNA count matrices are available from AMP-PD (https://amp-pd.org/). Summary results for all circRNAs and linear RNAs quantified in both PDBP and PPMI can be found in our transcript level browser (pdbloodtranscriptomics.wustl.edu).
All original code has been deposited at GitHub (https://github.com/Ibanez-Lab/BloodCircularRNA-ParkinsonsDisease) and is publicly available as of the date of publication.