Abstract
Background
T cell-based immunotherapies including immune checkpoint blockade and chimeric antigen receptor T cells can induce durable responses in patients with cancer. However, clinical efficacy is limited due to the ability of cancer cells to evade immune surveillance. While T cells have been the primary focus of immunotherapy, recent research has highlighted the importance of natural killer (NK) cells in directly recognizing and eliminating tumor cells and playing a key role in the set-up of an effective adaptive immune response. The remarkable potential of NK cells for cancer immunotherapy is demonstrated by their ability to broadly identify stressed cells, irrespective of the presence of neoantigens, and their ability to fight tumors that have lost their major histocompatibility complex class I (MHC I) expression due to acquired resistance mechanisms.
However, like T cells, NK cells can become dysfunctional within the tumor microenvironment. Strategies to enhance and reinvigorate NK cell activity hold potential for bolstering cancer immunotherapy.
Methods
In this study, we conducted a high-throughput screen to identify molecules that could enhance primary human NK cell function. After compound validation, we investigated the effect of the top performing compounds on dysfunctional NK cells that were generated by a newly developed in vitro platform. Functional activity of NK cells was investigated using compounds alone and in combination with checkpoint inhibitor blockade. The findings were validated on patient-derived intratumoral dysfunctional NK cells from different cancer types.
Results
The screening approach led to the identification of a Casitas B-lineage lymphoma (Cbl-b) inhibitor enhancing the activity of primary human NK cells. Furthermore, the Cbl-b inhibitor was able to reinvigorate the activity of in vitro generated and patient-derived dysfunctional NK cells. Finally, Cbl-b inhibition combined with T-cell immunoreceptor with Ig and ITIM domains (TIGIT) blockade further increased the cytotoxic potential and reinvigoration of both in vitro generated and patient-derived intratumoral dysfunctional NK cells.
Conclusions
These findings underscore the relevance of Cbl-b inhibition in overcoming NK cell dysfunctionality with the potential to complement existing immunotherapies and improve outcomes for patients with cancer.
Keywords: Immunotherapy, Natural Killer - NK, Immune Checkpoint Inhibitor, Intratumoral
WHAT IS ALREADY KNOWN ON THIS TOPIC
Natural killer (NK) cells are emerging as new players in the cancer immunity cycle and are especially important effectors in the context of major histocompatibility complex class I (MHC I) loss tumors. Of clinical relevance, NK cell number and infiltration have been shown to be associated with improved patient survival. However, NK cells, similarly to T cells, can be characterized by a dysfunctional phenotype in the tumor. Therefore, it becomes critical to identify new ways to enhance NK cell activity but also strategies to overcome the dysfunctional status characterizing NK cells in the tumor.
WHAT THIS STUDY ADDS
In this study, we show that a Casitas B-lineage lymphoma (Cbl-b) inhibitor compound, which was identified through a small molecule library screening approach on human NK cells, enhances primary human NK cell activity. Furthermore, we show for the first time that Cbl-b inhibition overcomes NK dysfunctionality and reinvigorates NK cell activity of in vitro generated and patient-derived intratumoral NK cells. In addition, we provide a rationale to further investigate Cbl-b inhibitors in combination with T-cell immunoreceptor with Ig and ITIM domains (TIGIT) blockade to further increase NK cell activity for cancer immunotherapy.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
This study underscores the promise of Cbl-b inhibitors in the context of cancer immunotherapy, particularly within the sphere of MHC I loss tumors. While the impact of Cbl-b inhibition on T-cell function is well-documented, our findings extend this understanding by providing further evidence of its efficacy in enhancing NK cell functionality. Significantly, we present novel insights demonstrating that Cbl-b inhibition overcomes NK cell dysfunctionality of both in vitro and intratumoral NK cells from cancer patients, marking a significant advancement in the field.
Introduction
Over the last decade, immunotherapy has fundamentally transformed the approach to treating patients with cancer. This shift has been driven by the development of immune checkpoint blockade (ICB), which focuses on disrupting key inhibitory signaling pathways within T cells, such as programmed cell death protein 1 (PD-1)/programmed death ligand 1 (PD-L1), cytotoxic T lymphocyte-associated protein 4 (CTLA-4), and, more recently lymphocyte activation gene-3 (LAG-3) and T-cell immunoreceptor with Ig and ITIM domains (TIGIT).1,3 However, the efficacy of these therapies remains constrained by various mechanisms of resistance, resulting in a limited number of responding patients.4,6 One notable challenge is posed by the intrinsic ability of cancer cells to evade adaptive immunity through the loss of major histocompatibility complex class I (MHC I) and antigen presentation. MHC I loss can be the result of different molecular mechanisms including transcriptional downregulation, somatic mutation, deletion of MHC I structural genes, and epigenetic silencing.7 Furthermore, MHC I loss has been correlated to mechanisms of acquired resistance to ICB treatments.8 9
Unlike T cells and B cells of the adaptive immune system, which require antigen presentation and clonal expansion to respond to a pathogen, natural killer (NK) cells can recognize and kill compromised cells without prior sensitization due to their ability to recognize stressed cells through a delicate balance of activating and inhibitory receptors signals. Activating receptors recognize stress-induced ligands on target cells, while inhibitory receptors engage with MHC I molecules, which are typically expressed on all healthy cells.10,12 The lack of “self” signals unleash the NK cells to attack and kill these cancer cells. Thus, NK cells represent a valuable asset to overcome resistance and complement T-cell response in cancer therapy.11,13
Of clinical relevance, in the last few years NK cells have been described as having both a prognostic and predictive role across cancer types. NK cell number and infiltration was associated with increased overall survival in several cancer types such as melanoma, lung and breast cancer.14,17 Similarly to T cells, NK cells can also become dysfunctional when infiltrating the tumor.18 The main causes of this phenotype have been associated with an imbalanced expression of receptor ligands promoting inhibitory signals, the presence of immunosuppressive cytokines and the hypoxic tumorous environment.18
Therefore, it becomes relevant to identify strategies to increase their activity but also to reinvigorate their functionality, to effectively support and complement T-cell responses for an efficient antitumor response.
In this work, we aimed to identify molecules to enhance NK cell activity for cancer immunotherapy. At this scope, we developed a high throughput small molecule library screen on primary human NK cells in co-culture with an MHC I knock-out (KO) A549 non-small lung cancer (NSCLC) cell line and identified a Casitas B-lineage lymphoma (Cbl-b) inhibitor that increases NK cell effector functions. Cbl-b is an E3 ubiquitin ligase that has been shown to regulate the stability of proteins involved in the signaling pathway of lymphocytes’ effector functions.19 20 Here, we show for the first time that Cbl-b inhibition can reinvigorate the activity of intratumoral dysfunctional NK cells purified from patients with cancer and potentiate the activity of TIGIT checkpoint blockade.
Results
A small molecule library screening approach identifies Cbl-b inhibitor enhancing primary human NK cell activity
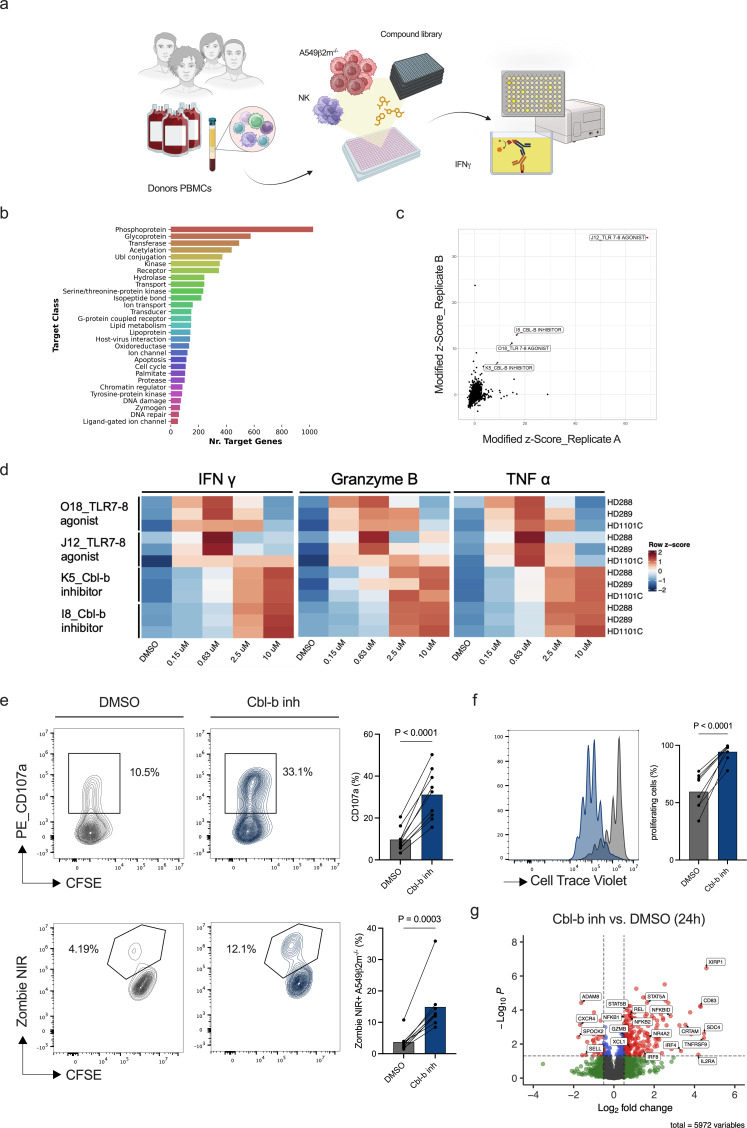
To identify compounds enhancing human NK cell antitumoral activity, we designed and optimized a small molecule library screening approach using primary human NK cells in co-culture with target cancer cells (figure 1a). CD56+ NK cells were isolated from healthy donors peripheral blood mononuclear cells (PBMCs) and primed with interleukin (IL)-15-enriched medium. IL-15 is an essential mediator of peripheral NK cell homeostasis and a multifunctional cytokine that supports the endurance, growth, and cytotoxic capabilities of NK cells.21,23 To establish an NK activation window suitable for detecting compound activity, we titrated IL-15 concentrations and identified both suboptimal and optimal concentrations for the screening (online supplemental figure S1a,b). As a cancer cell target, we used a ß2m gene-KO A549 NSCLC cell line (A549 ß2m−/−) obtained by CRISPR/Cas9 mediated KO (online supplemental figure S1c). Among the potential markers to monitor NK cell activation, interferon gamma (IFN-γ) was selected for its scalability for a high-throughput screen and measured by a 384-well optimized ELISA approach.
Figure 1. A small molecule library screening approach identifies Cbl-b inhibitors enhancing the activity of primary human NK cells. (a) Experimental design of the small molecule library screening approach on primary human NK cells in co-culture with A549 β2m−/− (created with BioRender.com). NK cells were enriched from healthy donors’ PBMCs and primed with a suboptimal concentration of IL-15 (0.2 ng/mL). Following compound treatment, NK cells were co-cultured with A549 β2m−/− overnight. Screening hits were identified by measurement of IFN-γ secretion from supernatants. (b) Small molecule screening compounds target classes. (c) Volcano plot representing screening hits based on modified Z score value. Top four screening hits were identified based on a threshold of normalized modified Z score ≥3. (d) Heatmap of IFN-γ, granzyme B, and TNF-α concentrations for screening hits validation. Primary human NK cells from healthy donors PBMCs (n=3) were treated in dose response with the top four screening compounds and cytokines measured from the supernatants. (e) Representative flow cytometry plots for NK cell degranulation (CD107a) and NK cell cytotoxicity. NK cells from healthy donors PBMCs (n=9) were primed with a suboptimal concentration of IL-15 (0.2 ng/mL) and co-cultured with A549 β2m−/− (CFSE+) at an E:T ratio of 2:1 for 6 hours (p values are from paired t-test). (f) CellTrace Violet dilution and statistics of primary human NK cells after treatment with Cbl-b inhibitor (p values are from paired t-test). (g) Volcano plot showing differentially expressed proteins on primary human NK cells after overnight treatment with Cbl-b inhibitor in the presence of a suboptimal concentration of 0.2 ng/mL IL-15 (n=6; gray=NS, green=log2 FC, blue=adjusted p value, red=log2 FC and adjusted p value. For statistical analyses see “Material and methods”). Cbl-b, Casitas B-lineage lymphoma; CFSE, carboxyfluorescein succinimidyl ester; DMSO, dimethylsulfoxide; E:T, effector to target; IFN, interferon; IL, interleukin; NK, natural killer; PBMCs, peripheral mononuclear cells; TLR, toll-like receptor; TNF, tumor necrosis factor.
Purified NK cells and A549 ß2m−/− were plated in 384 wells in the presence of a suboptimal concentration of IL-15 and treated overnight with compounds (4,740) at a fixed concentration of 3 µM. The compound library consisted of commercial and proprietary small molecules annotated with assay results from public databases and publications targeting a broad range of pathways (figure 1b). The screening allowed the identification of top four hits (respectively O18, I8, K5 and J12) (figure 1c). These compounds were identified as two toll-like receptor (TLR) 7–8 agonists (O18 and J12) and two Cbl-b inhibitors (K5 and I8). The top four screening hits were validated in a dose-response assay across donors. IFN-γ, tumor necrosis factor alpha (TNF-α), and granzyme B (GrzB) were induced by TLR7-8 agonists and Cbl-b inhibitors (figure 1d). Cbl-b inhibitors induced NK cell effector functions in a dose-dependent manner, with the dose range between 0.15 and 10 µM, while TLR7-8 agonists exerted optimal effects at 0.63 µM.
Preclinical evidence showed that Cbl-b inhibitors are well tolerated in vivo and Cbl-b−/− mice are vital.24 Importantly, Cbl-b inhibition in T cells has been reported to increase T-cell activation and cytotoxic functions while reducing exhaustion.25 Here we decided to focus on Cbl-b inhibitors to investigate Cbl-b potential to revert dysfunctionality of intratumoral NK cells. Among the two inhibitors we followed-up on the compound inducing higher cytokine secretion (online supplemental figure S1e). Of note, the increased cytokine secretion was attributed to an effect of the compound on NK cells but not on cancer cells (online supplemental figure 1f).
Next, we examined the effect of Cbl-b inhibitor on NK cell-mediated cytotoxicity, the second effector function axis of NK cells. For this and the following experiments, the compound concentration of 3 µM was used, as it demonstrated higher activity while maintaining low toxicity (online supplemental figure S1g). Pretreatment of human NK cells with Cbl-b inhibitor resulted in increased NK degranulation (CD107a) and killing of A549 ß2m−/− cells (figure 1e). In addition, Cbl-b inhibition increased NK cell proliferation (figure 1f).
To gain more insights in the cellular pathways imposed by Cbl-b inhibition in NK cells, we performed a proteomic analysis of primary NK cells alone treated with Cbl-b inhibitor either for 3 hours or 24 hours (online supplemental figure S2a). While protein expression difference is minimal after 3 hours of treatment, most differences are observed after 24 hours of treatment (online supplemental figure S2a). Among the top upregulated proteins after 24 hours treatment, several belong to the IL-2/IL-15 signaling (STAT5A, STAT5B, IL-2RA) and nuclear factor kappa b (NF-kB) (NFKBID, NFKB2, NFKB1, REL) pathways (figure 1g), which might account for the effects of the Cbl-b inhibitor in increasing NK cell functionality.26 27 To obtain further information on protein interaction and associated pathways, the STRING database was used to perform a multiple protein analysis.28 Top upregulated proteins with the more stringent cut-off are shown in online supplemental figure S2b. Both the TNF/NF-kB and cytokine-related signaling pathways were enriched across data sets (Reactome pathways, Kyoto Encyclopedia of Genes and Genomes-KEGG pathways and, Gene Ontology-GO) with a false discovery rate <0.01 (online supplemental figure S2b and online supplemental table 1), thus substantiating previous observations on differentially expressed proteins. These results suggest a potential role of Cbl-b inhibition in enhancing IL-15 signaling and NF-kB activation.
In vitro generated dysfunctional NK cells are reinvigorated by Cbl-b inhibitor treatment
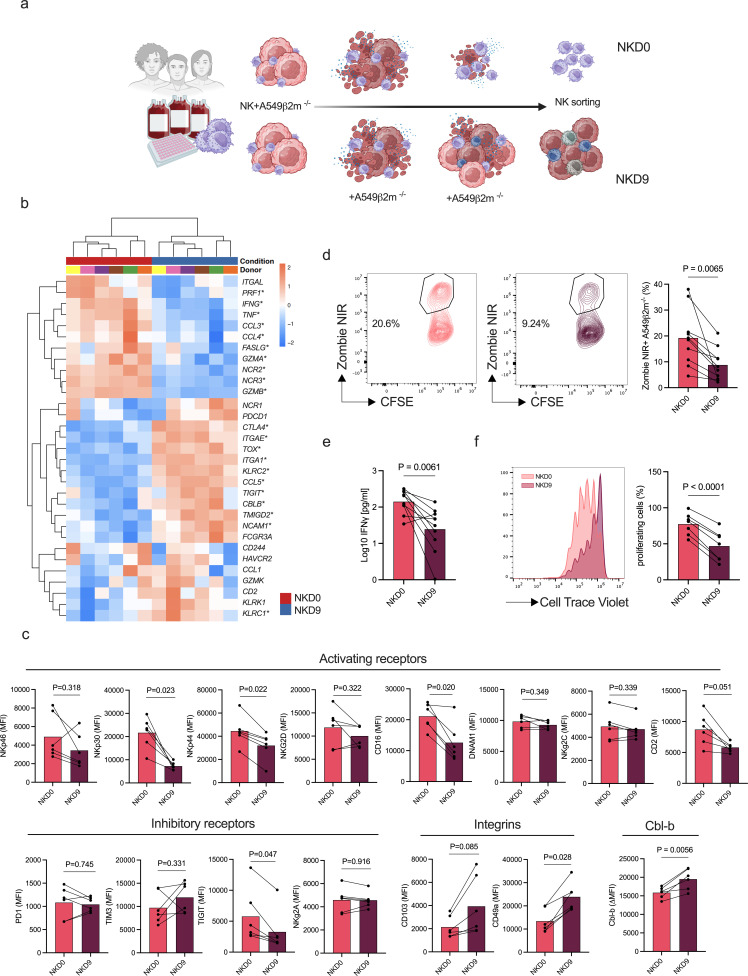
To gain more insights into the effect of Cbl-b inhibition in the context of NK cell dysfunction, we designed an in vitro model to generate dysfunctional NK cells. To this end, we co-cultured primary human NK cells with A549 ß2m−/− cancer cells continuously for 9 days (“NKD9”). As a control, NK cells from the same donor were exposed only once to the cancer cells at the beginning of the culture on day 0 (“NKD0”) (figure 2a).
Figure 2. In vitro model for the generation of dysfunctional NK cells. (a) Schematic representation of the in vitro model (created with BioRender.com). Primary human NK cells were enriched from healthy donors PBMCs and co-cultured for 9 days after single (NKD0) or continuous (NKD9) exposure to A549 ß2m−/− NK cells were then sorted to be used in functional assays. (b) Heatmap of selected differentially expressed genes associated with NK cell functionality between NKD0 and NKD9 (n=6). Volcano plot with differentially expressed genes in online supplemental figure S4a. (c) Multicolor extracellular staining of in vitro generated NK cells from healthy donors PBMCs (n=6) and Cbl-b intracellular staining of in vitro generated NK cells (n=6). Cbl-b expression is represented as delta isotype MFI (p values are from paired t-test). (d) Representative flow cytometry plots of cytotoxicity of in vitro generated NK cells (n=10). NK-mediated cytotoxicity is represented as the percentage of dead (zombie NIR+) target cancer cells (CFSE+). Percentage of cytotoxicity was normalized by cancer cell viability (p values are from paired t-test). (e) Interferon-γ concentration of in vitro generated NK cells following restimulation with target cells (n=6) (p values are from paired t-test). (f) Flow cytometry histogram and statistics of CellTrace Violet dilution of in vitro generated NK cells (n=7, p values are from paired t-test with Benjamini-Hochberg correction). Cbl-b, Casitas B-lineage lymphoma; CFSE, carboxyfluorescein succinimidyl ester; MFI, mean fluorescence intensity; NK, natural killer; PBMCs, peripheral mononuclear cells; PD-1, programmed cell death protein; TIGIT, T-cell immunoreceptor with Ig and ITIM domains; TIM3, T-cell immunoglobulin and mucin domain 3.
We first transcriptionally characterized the in vitro generated NK cells using a bulk RNA sequencing approach (online supplemental figure S4a). The transcriptional profile of NKD9 cells exposed to cancer cells was characterized by lower expression of genes associated with NK functionality (GZMB, GZMA, IFNG, TNF, PRF1) when compared with NKD0 (figure 2b). Of note, the main activating receptors were downregulated (NCR2, NCR3) and the inhibitory receptors TIGIT, KLRC1 and CTLA-4 were upregulated. No modulation of additional inhibitory receptors was observed (PDCD1, HAVCR2). In addition, we noted an increase in markers associated with tissue and tumor residency (ITGA1 and ITGAE). The transcriptional signature identified in NKD9 compared with NKD0 was validated at the protein level for the activating receptors NKp30, NKp44, and CD49a (figure 2c). However, inhibitory receptor transcriptional modulation was not confirmed at the protein level (figure 2c). Interestingly, Cbl-b expression was upregulated on NKD9 (figure 2b,c), which could indicate a crosstalk between Cbl-b and NK cell functionality.
We then assessed the function of NK cells obtained from the in vitro co-culture. The NKD9 cells showed a decrease in IFN-γ production and cytotoxic potential compared with the control (NKD0) (figure 2d,e), in agreement with the transcriptional analysis (figure 2b). Furthermore, the NKD9 cells showed a lower proliferation potential when compared with the control (NKD0) (figure 2f). Overall, this data confirms the validity of our model in generating dysfunctional NK cells, to further assess the potential of compounds in reverting NK dysfunctionality.
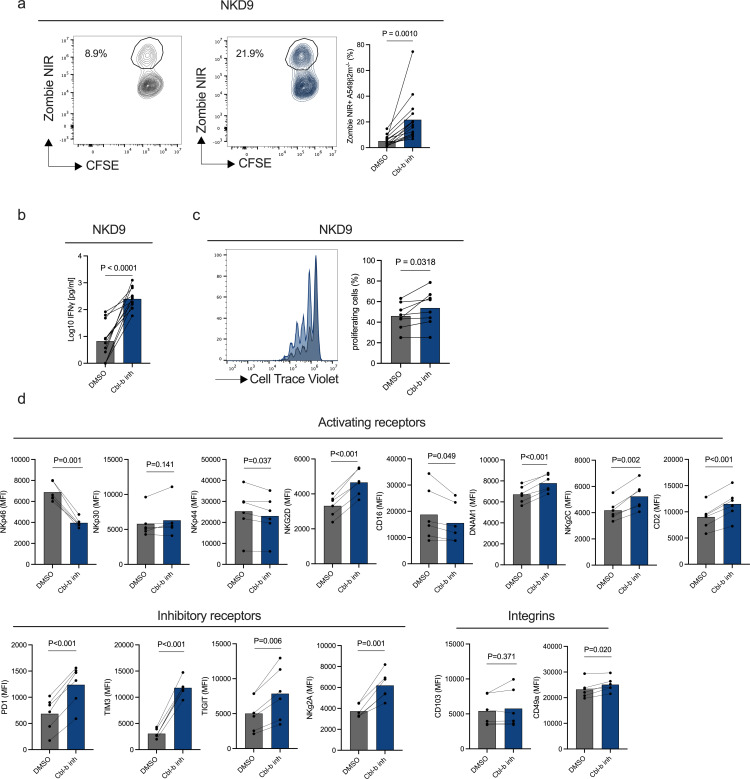
Next, we asked whether Cbl-b inhibitor treatment could revert the functional impairment of dysfunctional NKD9 cells. Cbl-b inhibitor treatment of NKD9 cells resulted in increased NK cell-mediated cytotoxicity, IFN-γ production and proliferation (figure 3a–c). Furthermore, Cbl-b inhibitor treatment resulted in modulation of main activating and inhibitory receptors (figure 3d). Taken together, Cbl-b inhibition reinvigorates NK cell cytotoxicity and proliferation potential in an in vitro system of dysfunctional NK cells continuously exposed to A549 ß2m−/− cancer cells.
Figure 3. Cbl-b inhibitor reinvigorates the activity of in vitro generated dysfunctional NK cells. In vitro generated dysfunctional NK cells (NKD9) from healthy donors PBMCs (n=6) were sorted and re-challenged with fresh A549 ß2m−/− (CFSE+) at a 2:1 E:T ratio overnight in the presence of DMSO or Cbl-b inhibitor (3 µM). (a) Cytotoxicity of in vitro generated dysfunctional NK cells (NKD9) following Cbl-b inhibitor treatment by flow cytometry. In vitro generated dysfunctional NK cells (NKD9) from healthy donors PBMCs (n=6) were sorted and re-challenged with fresh A549 ß2m−/− (CFSE+) in the presence of DMSO or Cbl-b inhibitor (3 µM) (p values are from paired t-test). (b) IFN-γ concentration of in vitro generated dysfunctional NK cells (NKD9) after Cbl-b inhibitor treatment (p values are from paired t-test). (c) Flow cytometry histogram and statistics of CellTrace Violet dilution of in vitro generated dysfunctional NK cells after Cbl-b inhibitor (3 µM) treatment (n=7). (P values are from paired t-test). (d) Multicolor extracellular staining of in vitro generated dysfunctional NK cells following Cbl-b inhibitor treatment (3 µM) (n=6, p values are from paired t-test with Benjamini-Hochberg correction). Cbl-b, Casitas B-lineage lymphoma; CFSE, carboxyfluorescein succinimidyl ester; E:T, effector to target; MFI, mean fluorescence intensity; NK, natural killer; PBMCs, peripheral mononuclear cells; PD-1, programmed cell death protein; TIGIT, T-cell immunoreceptor with Ig and ITIM domains; TIM3, T-cell immunoglobulin and mucin domain 3.
Cbl-b inhibitor treatment combined with TIGIT checkpoint blockade increases reinvigoration of NK cell dysfunction
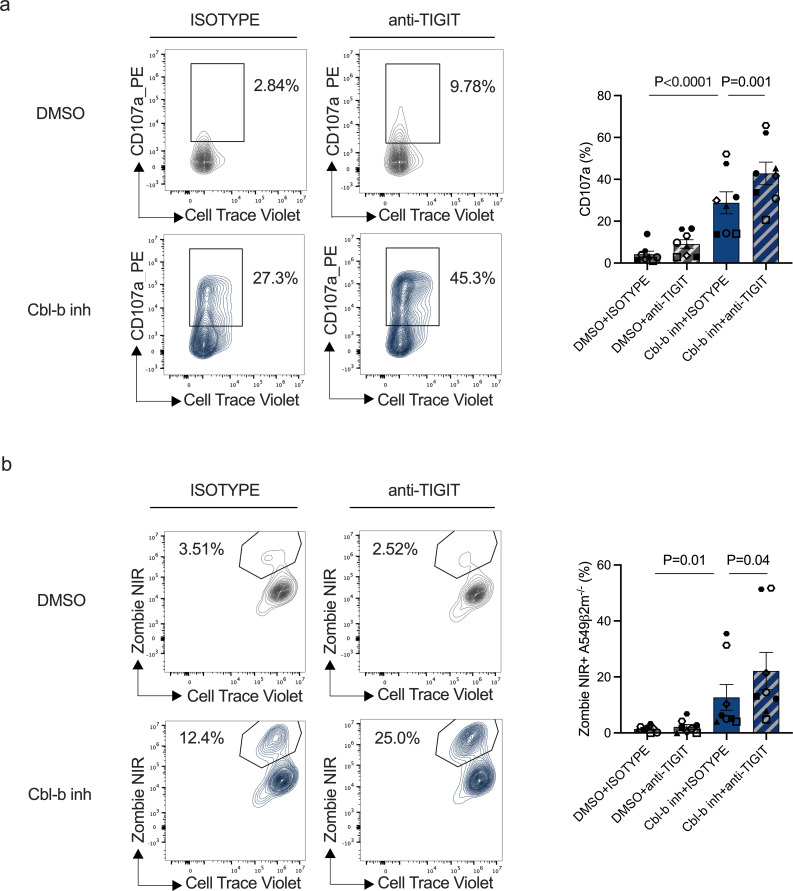
T-cell immunoglobulin and mucin domain 3 (TIM3), TIGIT and NKG2A inhibitory receptors play a key role in modulating NK cell activity in the tumor environment.29,31 As their expression was upregulated after Cbl-b inhibitor treatment (figure 3d), we asked whether a combination approach using Cbl-b inhibitor with inhibitory receptor blockade could further increase the activity of in vitro generated dysfunctional NK cells. A549 are known to express ligands for TIGIT and therefore be susceptible to TIGIT-mediated inhibition.32 To test Cbl-b and TIGIT combinatorial potential, we treated in vitro generated dysfunctional NK cells (NKD9) with Cbl-b inhibitor and a human anti-TIGIT antibody (tiragolumab). The combinations further increased NK cell activation compared with either Cbl-b inhibitor or anti-TIGIT antibody alone (figure 4a,b and online supplemental figure S5a). Our results suggest that the combination of Cbl-b inhibition and TIGIT blockade can further overcome NK cell dysfunctionality.
Figure 4. Combination of Cbl-b inhibitor with TIGIT checkpoint blockade results in increased reinvigoration of dysfunctional NK cells. (a) Degranulation (CD107a) of in vitro generated dysfunctional NK cells (NKD9) following Cbl-b inhibitor and TIGIT blockade (n=8, for p values see Material and methods). (b) Cytotoxicity of in vitro generated NK cells following Cbl-b inhibitor treatment and TIGIT blockade treatment (n=8, for p values see “Material and methods”). Cbl-b, Casitas B-lineage lymphoma; DMSO, dimethylsulfoxide; NK, natural killer; TIGIT, T-cell immunoreceptor with Ig and ITIM domains.
Cbl-b inhibitor reinvigorates patients tumor-infiltrating NK cells
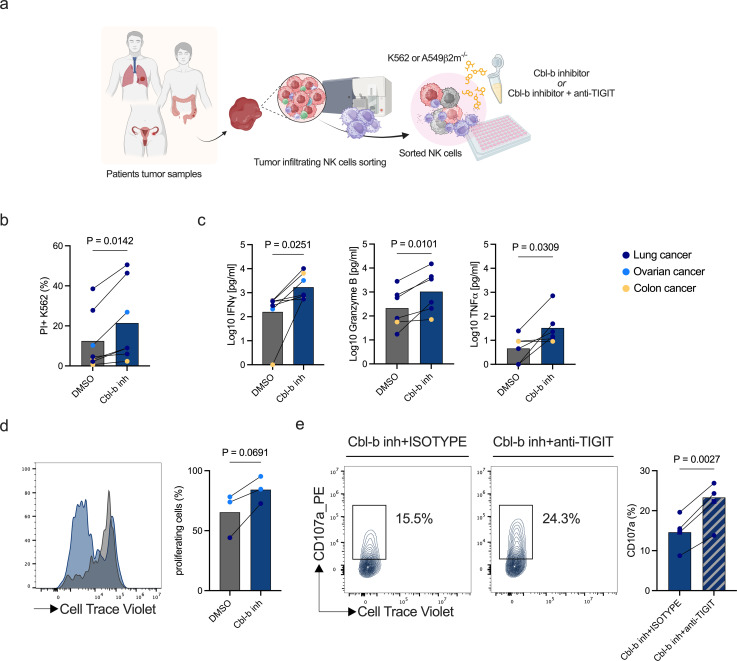
Tumor-infiltrating NK cells isolated from patients with cancer display a dysfunctional phenotype when compared with patients autologous peripheral PBMCs and those from healthy donors.33,36 We investigated whether Cbl-b inhibitors could reinvigorate and enhance the activity of patients’ tumor-infiltrating NK cells. Therefore, we sorted intratumoral NK cells from digested tumor samples obtained from patients with different cancer types (online supplemental table 2) and performed functional assays to check for NK activation after Cbl-b inhibitor treatment (figure 5a). Cbl-b inhibitor increased NK cell-mediated cytotoxicity and cytokine production across cancer types including lung, ovarian and colorectal cancer (figure 5b,c). Furthermore, treatment with Cbl-b inhibitor showed a trend of increased proliferation in tumor-infiltrating NK cells (figure 5d). This data shows that Cbl-b inhibition can reinvigorate the functionality and proliferation potential of tumor-infiltrating NK cells of patients from distinct cancer types.
Figure 5. Cbl-b inhibitor enhances the activity of intratumoral NK cells from cancer patient samples. (a) Schematic representing experimental design to assess the effect of Cbl-b inhibitor compound on tumor-infiltrating NK cells. Tumor-infiltrating NK cells were previously sorted from patients’ tumor samples and treated overnight with Cbl-b inhibitor in the presence of target cells (K562 or A549 ß2m−/−) (created with BioRender.com). (b) Flow cytometry-based cytotoxicity of Cbl-b inhibitor treated tumor-infiltrating NK cells (n=7 tumor digests, p values are from paired t-test). (c) IFN-γ, granzyme B and TNF-ɑ supernatant concentrations of Cbl-b inhibitor treated tumor-infiltrating NK cells from patients’ samples (n=6 tumor digests, p values are from paired t-test). (d) Histogram and statistics of flow cytometry CellTrace Violet dilution of patients tumor-infiltrating NK cells treated with Cbl-b inhibitor (n=3 tumor digests, p values are from paired t-test). (e) Degranulation (CD107a) of Cbl-b inhibitor treated intratumoral NK cells in combination with anti-TIGIT antibody (n=3 lung adenocarcinoma digests, n=1 lung adenocarcinoma pleural effusion, p values are from paired t-test). Cbl-b, Casitas B-lineage lymphoma; CFSE, carboxyfluorescein succinimidyl ester; DMSO, dimethylsulfoxide; IFN, interferon; NK, natural killer; TIGIT, T-cell immunoreceptor with Ig and ITIM domains; TNF, tumor necrosis factor.
In light of the findings on the effect of Cbl-b inhibition and TIGIT blockade on in vitro generated dysfunctional NK cells, we asked whether a combinatory treatment approach could increase the reinvigoration of dysfunctional patients intratumoral NK cells. While the treatment with Cbl-b inhibitor increased the expression of TIGIT (online supplemental figure S6a,b), the combination of Cbl-b inhibitor treatment and TIGIT blockade increased the activity and reinvigoration on patient-derived intratumoral NK cells (figure 5e).
Discussion
In this study, we developed a small molecule library screening approach to identify compounds enhancing human NK cell activity for cancer immunotherapy. To mimic the situation where NK cells encounter tumor cells characterized by loss of MHC I expression, a common immune evasion strategy in tumors, primary isolated human NK cells were co-cultured with A549 ß2m−/− NSCLC as target cells. The screening approach led to the identification and validation of four hits which included two TLR 7–8 agonists and two Cbl-b inhibitors. The identified TLR agonists target the two endosomal TLRs, TLR7 and TLR8, which biologically recognize single-stranded RNA and trigger pro-inflammatory responses.37 However, when compared with the myeloid compartment the expression of both TLR7-8 is relatively low in NK cells. Previous literature has indeed shown that despite a direct effect on NK cells, the main efficacy is largely mediated by dendritic cells (DCs).38,41
The second group of identified screening hits are Cbl-b inhibitors which target the E3 ubiquitin ligase Cbl-b. Cbl-b has been described as a main regulator of the activation threshold in lymphocytes.19 20 24 42 43 Using an in vitro set-up with suboptimal IL-15 concentration, we show that Cbl-b inhibitor increases effector functions and proliferation of human NK cells. This data is in line with recent findings showing the enhancement in NK cell activation in response to stimuli following genetic knock-down of Cbl-b in primary human NK cells.44 Previous literature described this mechanism as the result of a regulation of phosphorylation or stability of signaling proteins. Cbl-b activity has been broadly described in the context of T-cell activation by regulating TCR and CD28 signals via the non-degradative-ubiquitination of ZAP70, VAV and p85 (PI3K).45 46 In the context of NK cells, a Cbl-b−/− model has been shown to increase NK tumor surveillance and control metastatic spread by mediating the ubiquitination of TAM (TYRO3, AXL and MER) receptors and their internalization.24 A more recent study has highlighted the role of Cbl-b in regulating the degradation of LAT downstream of TAM receptors.47 Current research on TAM receptor expression in human NK cells has produced conflicting results, indicating a need for further investigation into the mechanisms downstream of Cbl-b inhibition in humans.44 48 On the other hand, LAT is known to be a key mediator of NK-activating signaling in human NK cells.49 To gain further insights into the signaling pathways associated with enhanced NK cell functionality, we performed a proteomic analysis of human primary NK cells after treatment with Cbl-b inhibitor. We did not detect any of the TAM receptors, further supporting previous data on TAM receptor expression in primary human NK cells.44 48 Among the proteomic results, we observed an enrichment in the proteins of the IL-2/IL-15 pathway (STAT5A, STAT5B, IL-2R). Among these proteins, the STAT5 proteins were previously shown to be directly ubiquitinated in DCs by both c-Cbl and Cbl-b.50 This data suggests a potential role of Cbl-b in regulating response to IL-15 signaling probably through STAT5 protein stability in NK cells.
Extending the current knowledge on Cbl-b inhibition on NK cell functions from healthy donors, we investigated whether Cbl-b inhibition could reinvigorate dysfunctional NK cells obtained from patients with cancer. Of note, Cbl-b inhibition was shown to restore the effector functions of exhausted PD-1+TIM-3+CD8+ tumor infiltrating lymphocytes (TILs) in an in vivo mouse tumor model.25 Similarly to T cells, NK cells can become dysfunctional induced by imbalanced receptor expression and suppressive signals in the tumor microenvironment as recently reviewed by Cozar and coworkers.18 To this end, we set-up an in vitro model characterized by a co-culture of primary human NK cells and target cancer cells for 9 days (NKD9). This induced a dysfunctional NK cell phenotype, characterized by reduced killing capacity, cytokine production and downregulation of main activating receptors. NKp30 and CD16 receptors were shown to be downregulated in tumor-associated NK cells across cancer types.51,53 In addition, tissue-resident markers (CD103 and CD49a) were induced. Using a mouse model to track newly arrived NK cells in the tumor, Dean et al demonstrated an early upregulation of CD49a on tumor entry that is associated with reduced effector functions.54 Of note, we observed a discrepancy between the increased gene expression of TIGIT and the unchanged protein level on in vitro-generated NKD9 cells. This discrepancy may be attributed to post-transcriptional mechanisms or kinetics of TIGIT protein regulation within the in vitro model. Therefore, in regards to the functional capability, activating receptor expression, and tissue-resident markers, our platform recapitulates the main features of intratumoral NK cells.
In this in vitro generated dysfunctional context, Cbl-b treatment increased functions and proliferation of dysfunctional NK cells (NKD9). We characterized the receptor-expression changes, highlighting the upregulation of inhibitory receptors, including PD-1, TIM-3, TIGIT, NKG2A. These receptors have been previously shown to regulate NK cell functionality in the tumor.12 Therefore, we investigated the effects of a combination treatment with TIGIT blockade and Cbl-b inhibition and discovered a further functional improvement of the in vitro generated NK cells.
To further validate our findings on Cbl-b inhibition potential to reinvigorate NK cell dysfunctionality, we tested Cbl-b inhibition on patient-derived tumor-infiltrating NK cells from lung, ovarian and colon cancer. Our results demonstrate that inhibition of Cbl-b rejuvenates the effector functions of tumor-infiltrating human NK cells, thereby reinvigorating their dysfunctional state. A combinatory treatment with TIGIT blockade further increased the reinvigoration of dysfunctional tumor-infiltrating NK cells. Given that Cbl-b inhibitors are presently being evaluated in clinical trials for various conditions, including NSCLC, our results advocate for additional in vivo investigations into the therapeutic efficacy of these inhibitors in conjunction with checkpoint blockade immunotherapy.55 56 Furthermore, the recent clinical trials on TIGIT blockade have shown promising results yet to be validated.2 57 In this scenario, the combination of Cbl-b inhibition with TIGIT blockade could be a potential strategy to be considered and further investigated to improve TIGIT blockade therapeutic efficacy.
Materials and methods
Cell culture
A549 (ATCC-CCL185) and K562 (ATCC-CCL243) were cultured in RPMI-1640 medium (Sigma, R8758) supplemented with 10% heat-inactivated (56°C, 30 min) fetal bovine serum (PAN Biotech, P30-5500) and 1% penicillin-streptomycin (Sigma, P4333). Cell lines were passaged every 2–3 days. For adherent cell lines, cells were treated with Trypsin-EDTA 0.05% (Thermo Fisher, 25300–054). Primary human NK cells were cultured in DMEM-F12 (Thermo Fisher, 11320033) supplemented with 10% heat inactivated (56°C, 30 min) human serum (University Hospital Basel), 1 mM Sodium Pyruvate (Sigma, S8636), 1% MEM non-essential amino acids (Sigma, M7145), 10 mM HEPES (Thermo Fisher, 15630–056) and 50 nmol/L beta-mercaptoethanol (Thermo Fisher, 31 350–010). NK cell culture medium was supplemented with recombinant human IL-15 (PeproTech, 200-15-100UG). Cytokine concentration was adapted to the experimental requirements as described.
Generation of ß2M KO cell lines
Plasmid CRISPR vector, containing CAS9 nuclease, gRNA cassette for ß2m and puromycin resistance were purchased from ATUM (plasmid “pD1431-Apuro:378738”). Before transfection, cell lines were cultured for at least 1 week, in antibiotic-free medium. A549 cells were transfected using Lipofectamine 3000 (Invitrogen, L3000-015) according to the manufacturer’s instructions. ß2m KO validation was performed by immunofluorescence flow cytometry staining of HLA-ABC, as described below. To obtain higher purity, ß2m KO cells were further sorted using FACSAria III or FACS SCORPAria (BD) based on HLA-ABC expression.
Isolation of primary human NK cells
Human PBMCs were isolated from healthy donors’ buffy coats (Blood Bank, University Hospital Basel) by density gradient centrifugation using Lymphoprep (STEMCELL, 07851) and SepMate PBMCs isolation tubes (STEMCELL, 85460). If required, an additional step of red blood cell lysis was performed using 1× RBC Lysis Buffer (Invitrogen, 00-4333-57).
After isolation, PBMCs were frozen in fetal bovine serum supplemented with 10% dimethyl sulfoxide (Sigma, D2650) at −80°C. For long storage, PBMCs frozen vials were kept in liquid nitrogen until use. Primary human NK cells were isolated from PBMCs using the “NK isolation Kit, human” (Miltenyi, 130-092-657) or “EasySep Human NK Cell Isolation Kit” (STEMCELL, 17955) according to manufacturer’s instructions.
Isolation of human intratumoral NK cells
Tumor samples were obtained from patients undergoing surgery at the University Hospital Basel. Tumor samples were mechanically dissociated and digested using accutase (Innovative Cell Technologies, AT-104), collagenase IV (Worthington, LS004188), hyaluronidase (Sigma-Aldrich, H6254) and DNAse type IV (Sigma-Aldrich, D5025). After digestion, cell suspensions were used immediately or in case of longer storage, kept in liquid nitrogen until use. Intratumoral CD56+ NK cells were sorted from tumor digests through a flow cytometry-based sorting using FACSAria III or FACS SORP Aria (BD).
In vitro NK dysfunction model
Primary human NK cells were negatively isolated from healthy donors PBMCs as previously described. After isolation, NK cells were co-cultured with A549 ß2m−/− at an effector to target (E:T) ratio of 2:1 in tissue culture treated 96-well plates (FALCON, 353077) using supplemented DMEM with 10 ng/mL IL-15. NKD0 were co-cultured only once with A549 ß2m−/− on day 0. NKD9 were co-cultured with A549 ß2m−/− for 9 days. Every 3 days, half of the medium was refreshed and for the NKD9 condition fresh A549 ß2m−/− were added. After 9 days of culture, cells were sorted (Propidium iodide−, CD56+) using FACSAria III or FACS SORPAria (BD) to be used for experiments.
Primary human NK small molecule library screening
NK cells from PBMCs were isolated using the human negative “NK isolation Kit” (Miltenyi, 130-092-657). Following isolation, primary NK cells were plated in 384 well plates (Corning, 3701) using an electronic 384 channel pipette (VIAFLO 384, INTEGRA Biosciences). 20,000 cells per well were plated in a final volume of 40 µL NK medium supplemented with 0.2 ng/mL of recombinant human IL-15. Control wells were seeded in the presence of 10 ng/mL recombinant human IL-15. IL-15 screening suboptimal concentration was defined by performing a titration and measuring the respective NK response via ELISA IFN-γ readout (online supplemental figure S1a). NK cells were rested overnight before compound treatment. Primary NK cells were treated with a single 3 µM dose of small molecules compounds with a final dimethylsulfoxide (DMSO) concentration of 0.1% per well. Compound treatment was performed using an electronic 384 channel pipette (VIAFLO 384, INTEGRA Biosciences). After compound treatment, NK cells were rested for 2 hours at 37°C. A549ß2m−/− cells were kept in culture for 1 week before the screening. After NK cells compound treatment, A549ß2m−/− cells were detached, counted and co-cultured with treated primary NK cells in an E:T ratio of 2:1. After overnight co-culture, supernatants were collected using an electronic 384 multichannel pipette (VIAFLO 384, INTEGRA Biosciences) and transferred to MAXISORP NUNC 384 plates (Thermo Fisher, 464718).
Screening readout was assessed by measuring IFN-γ concentration from the supernatants using the OPTEIA Human IFN-γ ELISA Kit (BD Bioscience, 555142) according to manufacturer’s instructions. Washing steps were performed using a 384 well plate washer (BioteK405ST, Agilent). Finally, screening results were acquired using an absorbance microplate reader (SPECTROstar Nano, BMG LabTech). The screening was performed in two technical replicates.
The results from the drug screen were analyzed using R (V.) 4.3.2 in RStudio (V.2023.12.0+369). The analysis has been deposited on GitHub and released on Zenodo at https://doi.org/10.5281/zenodo.10577977. Briefly, the mean IFN-γ ELISA results in pg/mL for the negative control (0.2 ng/mL IL-15) and positive control (10 ng/mL IL-15) were calculated for each plate replicate. The percentage of effect for each drug and control was calculated based on these mean values per plate as . The z-prime quality metric was calculated for each plate (online supplemental figure S1d). Two plates were excluded due z-prime values below 0.5. Due to well location bias on the plate (wells on outer edges showed higher values likely due to evaporation), the percentage effect values were normalized by the median for this location (eg, well A1) over all plates. From these normalized values, the robust hit identification metric was calculated as . Screening hits were selected by having a modified Z score value higher than 2.5 across replicates.
Flow cytometry
For surface staining, cells were previously stained with a fixable live/dead Zombie dye for 20 min at 4°C. Cells were then treated with an Fc receptor binding inhibitor (Invitrogen, 14-9161-73) 1:100 diluted. After 20 min of incubation at 4°C cells were washed with FACS buffer (phosphate-buffered saline (PBS), 0.5 mM EDTA, 2% fetal calf serum, 0.1% sodium azide (NaN3) and resuspended in the antibody staining mix and incubated for 30 min at 4°C.
For intracellular staining, after surface staining, cells were washed with FACS buffer and fixed for 20 min at room temperature with IC fix buffer (eBioscience, 00-8222-49). Following fixation cells were washed in perm buffer (eBioscience, 00-8333-56) and resuspended in the staining mix for 30 min at room temperature. After staining, samples were resuspended in FACS buffer and acquired at a flow cytometer (CytoFLEX, Beckmann Coulter or Aurora, Cytek).
The multicolor flow cytometry panel used to characterize NK cells can be found in online supplemental table 3. The complete list of antibodies, cell tracers and viability dyes used in all the experiments can be found in online supplemental table 4.
NK degranulation
NK cell degranulation was assessed by co-culturing NK cells (primary human NK cells or in vitro generated dysfunctional NK cells or human intratumoral NK cells) with target cancer cells (K562 or A549 ß2m−/−) in U-bottom 96-well tissue culture plates (Falcon, 353077). Cells were cultured in supplemented DMEM in the presence of a suboptimal concentration of recombinant human IL-15. To measure degranulation, CD107a antibody was added at the beginning of the co-culture. Cells were co-cultured at a 2:1 E:T ratio for 4 hours at 37°C. To assess NK degranulation after compound treatment, NK cells were pretreated with Cbl-b inhibitor (3 µM) overnight before co-culture. For TIGIT blockade following Cbl-b inhibitor treatment, anti-TIGIT antibody (Tiragolumab, Selleckchem A2028) or its isotype control Human IgG1 (Selleckchem, A2051) were used at a concentration of 10 µg/mL for 1 hour at 37°C before co-culture.
At the end of the co-culture, cells were stained with a viability dye (PI or Zombie NIR) and samples readout was performed by flow cytometry (CytoFLEX, Beckmann Coulter). An example of the gating strategy is provided in online supplemental figure S3a.
NK killing assays and cytokines quantification
NK cell mediated killing was assessed by co-culturing NK cells (primary human NK cells or in vitro generated dysfunctional NK cells or human intratumoral NK cells) with target cancer cells (K562 or A549 ß2m−/−) in U-bottom 96-well tissue culture plates (Falcon, 353077). Cells were cultured in supplemented DMEM in the presence of a suboptimal concentration of recombinant human IL-15. Target cells were stained with cell tracers (CellTrace Violet or carboxyfluorescein succinimidyl ester-CFSE) before co-culture. The co-culture was performed at an E:T ratio of 2:1 overnight. To measure NK-mediated killing, the frequency of viability dye (Zombie NIR−/− or PI) positive target cells was calculated. This value was then adjusted by subtracting the frequency of viability dye positive target cells in the absence of NK cells.
An example of the gating strategy is provided in online supplemental figure S3b.
To assess NK-mediated killing with compound treatment, cells were treated with Cbl-b inhibitor (3 µM) overnight. For combination treatment with TIGIT blockade, NK cells were pretreated overnight with Cbl-b inhibitor (3 µM) and then treated with anti-TIGIT antibody (Tiragolumab, Selleckchem A2028) or its isotype control Human IgG1 (Selleckchem, A2051) at a concentration of 10 µg/mL at 37°C for 1 hour before co-culture with target cells.
Supernatants were collected after overnight co-culture for cytokines quantification. IFN-γ measurement was performed using the OptEIA Human IFN-γ ELISA Kit (BD Bioscience, 555142) according to manufacturer’s instructions. For multiplex cytokines (IFN-γ, GrzB and TNF-α) measurements the following kits were used: BD Cytometric Bead Array (CBA) Human IFN-γ Flex Set, BD Cytometric Bead Array (CBA) Human Granzyme B Flex Set (BD Bioscience, 560304) and the BD Cytometric Bead Array (CBA) Human TNF Flex Set (BD Bioscience, 558273). Multiplex cytokine measurement was performed by flow cytometry (iQue, Sartorius).
NK proliferation assay
To assess NK cell proliferation, NK cells were stained with a cell tracer dye (CellTrace Violet) and cultured in DMEM supplemented medium in the presence of IL-15. NK cells were left in culture for 3–5 days. After culture, cells were stained with a viability dye and proliferation assessed by flow cytometry measuring CellTrace Violet dilution. An example of the gating strategy is provided in online supplemental figure S3c.
Proteomics
Primary human NK cells were negatively isolated from healthy donors’ buffy coats (n=6). After NK cell isolation, cells were plated in 0.2 ng/mL IL-15 supplemented NK medium and treated for 3 hours or 24 hours with Cbl-b inhibitor (3 µM). After treatment, cells were harvested and rinsed with cold PBS. Cell pellets were prepared and immediately stored at −80°C until use.
Cells pellets were lysed in 50 µL of lysis buffer (1% sodium deoxycholate, 10 mM TCEP, 100 mM Tris, pH=8.5) using 10 cycles of sonication (30 s on, 30 s off per cycle) on a Bioruptor Sonicator (Diagenode). Following sonication, protein lysates were reduced by incubation at 95°C for 10 min. Samples were then alkylated using 15 mM chloroacetamide at 37°C for 30 min and further digested using sequencing-grade modified trypsin (1/50 w/w, ratio trypsin/protein; Promega, USA) at 37°C for 12 hours. After digestion, the samples were acidified using TFA (final concentration 1%). Peptide desalting was performed using iST cartridges (PreOmics, Germany) following the manufacturer’s instructions. After drying the samples under vacuum, peptides were stored at −20°C and dissolved in 0.1% aqueous formic acid solution at a concentration of 0.5 mg/mL on use.
Dried peptides were resuspended in 0.1% aqueous formic acid and subjected to liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis using a Q Exactive HF mass spectrometer fitted with an UltiMate 3000 nano-LC (both Thermo Fisher Scientific) and a custom-made column heater set to 60°C. Peptides were resolved using an RP-HPLC column (75 µm × 30 cm) packed in-house with C18 resin (ReproSil-Pur C18–AQ, 1.9 µm resin; Dr Maisch GmbH) at a flow rate of 0.3 μL min 1. The following gradient was used for peptide separation: from 2% B to 7% B over 5 min, to 30% B over 75 min, to 50% B over 10 min, to 95% B over to 2 min followed by 10 min at 95% B. Buffer A was 0.1% formic acid in water and buffer B was 80% acetonitrile, 0.1% formic acid in water.
The mass spectrometer was operated in data-independent(DIA) acquisition mode. For the MS1, the following parameters were set: Resolution: 120,000 FWHM (at 200 m/z), AGC target: 3e6, maximum injection time: 100 ms, scan range: 350–1,600 m/z. MS2 scans were acquired using the following parameters: Resolution: 30,000 FWHM (at 200 m/z), AGC target 3e6, maximum injection time: auto, isolation window: 17 m/z, normalized collision energy 28%. Each DIA cycle consisted of 30 equal SWATH windows covering m/z range of 400–900. All spectra were acquired in centroid mode.
The acquired raw-files were searched by direct DIA against the human UniProt protein database (version February 2022) and commonly observed contaminants (in total 20,752 sequences) by the SpectroNaut software (Biognosys, V.16.3.221108.5300) using default settings. The search criteria were set as follows: full tryptic specificity was required (cleavage after lysine or arginine residues unless followed by proline), two missed cleavages were allowed, carbamidomethylation (C) was set as fixed modification and oxidation (M) and N-terminal acetylation as a variable modification. The false identification rate was set to 1%. The search results were exported from SpectroNaut and protein abundances were statistically analyzed using MSstats (V.4.4.1).
Bulk RNA sequencing
In vitro generated dysfunctional cells from six donors at day 9 of culture were isolated as described above and washed with PBS. After washing, cell pellets of each condition were stored at −80 until use.
RNA extraction was performed using the Qiagen RNeasy Plus Mini Kit (250) (Qiagen, 74136) according to manufacturer’s instructions.
Sequencing was performed on two lanes of the Illumina NovaSeq 6000 instrument resulting in 51nt-long paired-end reads.
The data set was analyzed by the Bioinformatics Core Facility, Department of Biomedicine, University of Basel. Complementary DNA reads were aligned to “hg38” genome using Ensembl 104 gene models with the STAR tool (V.2.7.10a) with default parameter values except the following parameters: outFilterMultimapNmax=10, outSAMmultNmax=1, outSAMtype=BAM SortedByCoordinate, outSAMunmapped=Within. At least 20 M read pairs were mapped per sample.
The software R (V.4.1.1) and the tool featureCounts from Subread (V.2.0.1) package from Bioconductor (V.3.14) were used to count aligned reads per gene with default parameters except: -O, -M, --read2pos=5, --primary, -s 2, -p, -B.
Further analysis steps were performed using R (V.4.2.0) and multiple packages from Bioconductor (V.3.15). The package edgeR (V.3.38.4) was used to perform differential expression analysis. Genes included in the analysis were filtered by the function “filterByExpr” with default parameters.
Statistical analysis
The statistical analysis was performed using the software Prism V.10.0.3 (GraphPad Software, La Jolla, California, USA). Data was considered significant with p values<0.05. Statistical testing for figure 4a,b was performed in R (V.4.2.0) by constructing a linear model based on measured experimental values (function lm) and subsequent testing linear hypotheses (paired t-test) using the function “glht” from the multcomp package (V.1.4.20).
supplementary material
Acknowledgements
We thank the flow cytometry facility of the University of Basel in particular Stella Stefanova, Dr. Irene Calvo, Dr. Jelena Markovic, Dr. Morgane Hilpert, Dr. Cecile Cumin and Mihaela Barbu-Stevanovic for help with cell sorting and advice for flow cytometry panel optimization. We thank the Roche pRED Flow Cytometry Center of Excellence for help with cell sorting and advice for flow cytometry panel optimization. We thank the Genomics Facility Basel of the University of Basel and the Department of Biosystems Science and Engineering, ETH Zurich for the library preparation and sequencing. We thank Dr. Robert Ivanek and Dr. Julien Roux from the DBM Bioinformatics Core Facility for support and feedback with bioinformatics and statistical analysis. We thank Dr. Karen Olivia Dixon and Dr. Stephanie Kueng for the valuable feedback and discussions. We thank Rosa Sousa for her precious support with material ordering and procurement logistics, especially during COVID times. We thank Tina Zimmermann, Anne Marie Mueller and Debora Paduraru for technical feedback and support. We thank all the donors and patients for allowing the use of their samples and making possible these findings. Calculations for this project were performed at sciCORE (http://scicore.unibas.ch/) scientific computing center at the University of Basel.
Footnotes
Funding: This project was supported by Roche pRED Oncology and a grant from Monique Dornonville Foundation.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Consent obtained directly from patient(s).
Ethics approval: This study was approved by Ethikkommission Nordwestschweiz and University Hospital Basel (reference EKNZ 2018-01990 and EKNZ 2019-02106). Participants gave informed consent to participate in the study before taking part.
Data availability free text: The sequencing and proteomics data are available in public, open access repositories. The sequencing data has been deposited in the European Genome-Phenom Archive (EGA) under the ID: EGAD50000000819. Proteomics data has been deposited and is available in the “massIVE” database (MASSIVE ID: MSV000095572, ProtemeXchange ID: PXD054820). All other data are available in the main text or in the supplementary materials.
Map disclaimer: The inclusion of any map (including the depiction of any boundaries therein), or of any geographical or locational reference, does not imply the expression of any opinion whatsoever on the part of BMJ concerning the legal status of any country, territory, jurisdiction or area or of its authorities. Any such expression remains solely that of the relevant source and is not endorsed by BMJ. Maps are provided without any warranty of any kind, either express or implied.
Contributor Information
Sofia Tundo, Email: sofia.tundo@unibas.ch.
Marcel Trefny, Email: marcel.trefny@med.uni-muenchen.de.
Andrijana Rodić, Email: andrijana.rodic@unibas.ch.
Olivia Grueninger, Email: olivia.grueninger@roche.com.
Nicole Brodmann, Email: nicole.brodmann@bluewin.ch.
Anastasiya Börsch, Email: anastasiya.boersch@unibas.ch.
Clara Serger, Email: c.serger@unibas.ch.
Jonas Fürst, Email: jonas.fuerst@unibas.ch.
Melanie Buchi, Email: melanie.buchi@unibas.ch.
Katarzyna Buczak, Email: katarzyna.buczak@unibas.ch.
Alex T Müller, Email: alex.mueller@roche.com.
Lisa Sach-Peltason, Email: lisa.sach-peltason@roche.com.
Leyla Don, Email: don.leyla@gmx.de.
Petra Herzig, Email: petra.herzig@unibas.ch.
Didier Lardinois, Email: didier.lardinois@usb.ch.
Viola Heinzelmann-Schwarz, Email: Viola.Heinzelmann@usb.ch.
Kirsten D Mertz, Email: Kirsten.Mertz@ksbl.ch.
Aljaž Hojski, Email: aljaz.hojski@unibas.ch.
Karin Schaeuble, Email: karin.schaeuble@unibas.ch.
Heinz Laubli, Email: heinz.laeubli@unibas.ch.
Marina Natoli, Email: marina.natoli@hotmail.it.
Alberto Toso, Email: tosoa1980@gmail.com.
Thuy T Luu, Email: luuthanhthuyvn88@gmail.com.
Alfred Zippelius, Email: alfred.zippelius@usb.ch.
Andrea Romagnani, Email: andrea.romagnani@roche.com.
Data availability statement
Data are available upon reasonable request.
References
- 1.Tawbi HA, Schadendorf D, Lipson EJ, et al. Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N Engl J Med. 2022;386:24–34. doi: 10.1056/NEJMoa2109970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho BC, Abreu DR, Hussein M, et al. Tiragolumab plus atezolizumab versus placebo plus atezolizumab as a first-line treatment for PD-L1-selected non-small-cell lung cancer (CITYSCAPE): primary and follow-up analyses of a randomised, double-blind, phase 2 study. Lancet Oncol. 2022;23:781–92. doi: 10.1016/S1470-2045(22)00226-1. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Goswami S, Raychaudhuri D, et al. Immune checkpoint therapy—current perspectives and future directions. Cell. 2023;186:1652–69. doi: 10.1016/j.cell.2023.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Postow MA, Chesney J, Pavlick AC, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006–17. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med. 2019;381:1535–46. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 6.Hellmann MD, Paz-Ares L, Bernabe Caro R, et al. Nivolumab plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2019;381:2020–31. doi: 10.1056/NEJMoa1910231. [DOI] [PubMed] [Google Scholar]
- 7.Dhatchinamoorthy K, Colbert JD, Rock KL. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front Immunol. 2021;12:636568. doi: 10.3389/fimmu.2021.636568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gettinger S, Choi J, Hastings K, et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov. 2017;7:1420–35. doi: 10.1158/2159-8290.CD-17-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenfeld AJ, Hellmann MD. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell. 2020;37:443–55. doi: 10.1016/j.ccell.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivori S, Vacca P, Del Zotto G, et al. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol . 2019;16:430–41. doi: 10.1038/s41423-019-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf NK, Kissiov DU, Raulet DH. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat Rev Immunol. 2023;23:90–105. doi: 10.1038/s41577-022-00732-1. [DOI] [PubMed] [Google Scholar]
- 12.Vivier E, Rebuffet L, Narni-Mancinelli E, et al. Natural killer cell therapies. Nature New Biol. 2024;626:727–36. doi: 10.1038/s41586-023-06945-1. [DOI] [PubMed] [Google Scholar]
- 13.Kärre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002;55:221–8. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- 14.Barry KC, Hsu J, Broz ML, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24:1178–91. doi: 10.1038/s41591-018-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Concha-Benavente F, Kansy B, Moskovitz J, et al. PD-L1 Mediates Dysfunction in Activated PD-1+ NK Cells in Head and Neck Cancer Patients. Cancer Immunol Res. 2018;6:1548–60. doi: 10.1158/2326-6066.CIR-18-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, et al. A Gene Signature Predicting Natural Killer Cell Infiltration and Improved Survival in Melanoma Patients. Cancer Immunol Res. 2019;7:1162–74. doi: 10.1158/2326-6066.CIR-18-0500. [DOI] [PubMed] [Google Scholar]
- 17.Youn J-I, Park S-M, Park S, et al. Peripheral natural killer cells and myeloid-derived suppressor cells correlate with anti-PD-1 responses in non-small cell lung cancer. Sci Rep. 2020;10:9050. doi: 10.1038/s41598-020-65666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cózar B, Greppi M, Carpentier S, et al. Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 2021;11:34–44. doi: 10.1158/2159-8290.CD-20-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachmaier K, Krawczyk C, Kozieradzki I, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature New Biol. 2000;403:211–6. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Sun SC. Targeting ubiquitin signaling for cancer immunotherapy. Signal Transduct Target Ther. 2021;6:16. doi: 10.1038/s41392-020-00421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carson WE, Giri JG, Lindemann MJ, et al. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J Exp Med. 1994;180:1395–403. doi: 10.1084/jem.180.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntington ND, Puthalakath H, Gunn P, et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nat Immunol. 2007;8:856–63. doi: 10.1038/ni1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luu TT, Ganesan S, Wagner AK, et al. Independent control of natural killer cell responsiveness and homeostasis at steady-state by CD11c+ dendritic cells. Sci Rep. 2016;6:37996. doi: 10.1038/srep37996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paolino M, Choidas A, Wallner S, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature New Biol. 2014;507:508–12. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar J, Kumar R, Kumar Singh A, et al. Deletion of Cbl-b inhibits CD8+ T-cell exhaustion and promotes CAR T-cell function. J Immunother Cancer. 2021;9:e001688. doi: 10.1136/jitc-2020-001688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston JA, Bacon CM, Finbloom DS, et al. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc Natl Acad Sci USA. 1995;92:8705–9. doi: 10.1073/pnas.92.19.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra A, Sullivan L, Caligiuri MA. Molecular Pathways: Interleukin-15 Signaling in Health and in Cancer. Clin Cancer Res. 2014;20:2044–50. doi: 10.1158/1078-0432.CCR-12-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Bi J, Zheng X, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol. 2018;19:723–32. doi: 10.1038/s41590-018-0132-0. [DOI] [PubMed] [Google Scholar]
- 30.André P, Denis C, Soulas C, et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells Graphical Abstract Highlights d Blocking NKG2A unleashes both T and NK cell effector functions d Combined blocking of the NKG2A and the PD-1 axis promotes anti-tumor immunity d Blocking NKG2A and triggering CD16 illustrates the efficacy of dual checkpoint therapy. Cell. 2018;175:1731–43. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Huang Y, Tan L, et al. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int Immunopharmacol. 2015;29:635–41. doi: 10.1016/j.intimp.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Luo J, Chen Y, et al. Combined evaluation of the expression status of CD155 and TIGIT plays an important role in the prognosis of LUAD (lung adenocarcinoma) Int Immunopharmacol. 2020;80:106198. doi: 10.1016/j.intimp.2020.106198. [DOI] [PubMed] [Google Scholar]
- 33.Trefny MP, Kaiser M, Stanczak MA, et al. PD-1+ natural killer cells in human non-small cell lung cancer can be activated by PD-1/PD-L1 blockade. Cancer Immunol Immunother . 2020;69:1505–17. doi: 10.1007/s00262-020-02558-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platonova S, Cherfils-Vicini J, Damotte D, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71:5412–22. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- 35.Russick J, Joubert P-E, Gillard-Bocquet M, et al. Natural killer cells in the human lung tumor microenvironment display immune inhibitory functions. J Immunother Cancer. 2020;8:e001054. doi: 10.1136/jitc-2020-001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mele D, Pessino G, Trisolini G, et al. Impaired intratumoral natural killer cell function in head and neck carcinoma. Front Immunol. 2022;13:997806. doi: 10.3389/fimmu.2022.997806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitzgerald KA, Kagan JC. Toll-like Receptors and the Control of Immunity. Cell. 2020;180:1044–66. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornung V, Rothenfusser S, Britsch S, et al. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol . 2002;168:4531–7. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 39.Hart OM, Athie-Morales V, O’Connor GM, et al. TLR7/8-mediated activation of human NK cells results in accessory cell-dependent IFN-gamma production. J Immunol . 2005;175:1636–42. doi: 10.4049/jimmunol.175.3.1636. [DOI] [PubMed] [Google Scholar]
- 40.Gorski KS, Waller EL, Bjornton-Severson J, et al. Distinct indirect pathways govern human NK-cell activation by TLR-7 and TLR-8 agonists. Int Immunol. 2006;18:1115–26. doi: 10.1093/intimm/dxl046. [DOI] [PubMed] [Google Scholar]
- 41.Veneziani I, Alicata C, Pelosi A, et al. Toll-like receptor 8 agonists improve NK-cell function primarily targeting CD56brightCD16− subset. J Immunother Cancer. 2022;10:e003385. doi: 10.1136/jitc-2021-003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yasuda T, Tezuka T, Maeda A, et al. Cbl-b positively regulates Btk-mediated activation of phospholipase C-gamma2 in B cells. J Exp Med. 2002;196:51–63. doi: 10.1084/jem.20020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sohn HW, Gu H, Pierce SK. Cbl-b negatively regulates B cell antigen receptor signaling in mature B cells through ubiquitination of the tyrosine kinase Syk. J Exp Med. 2003;197:1511–24. doi: 10.1084/jem.20021686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu T, Chen L, Mansour AG, et al. Cbl-b Is Upregulated and Plays a Negative Role in Activated Human NK Cells. J Immunol . 2021;206:677–85. doi: 10.4049/jimmunol.2000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiang YJ, Kole HK, Brown K, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nat New Biol. 2000;403:216–20. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 46.Fang D, Liu YC. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nat Immunol. 2001;2:870–5. doi: 10.1038/ni0901-870. [DOI] [PubMed] [Google Scholar]
- 47.Chirino LM, Kumar S, Okumura M, et al. TAM receptors attenuate murine NK-cell responses via E3 ubiquitin ligase Cbl-b. Eur J Immunol. 2020;50:48–55. doi: 10.1002/eji.201948204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giroud P, Renaudineau S, Gudefin L, et al. Expression of TAM-R in Human Immune Cells and Unique Regulatory Function of MerTK in IL-10 Production by Tolerogenic DC. Front Immunol. 2020;11:564133. doi: 10.3389/fimmu.2020.564133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vivier E, Nunès JA, Vély F. Natural Killer Cell Signaling Pathways. Science. 2004;306:1517–9. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- 50.Xu F, Liu C, Dong Y, et al. Ablation of Cbl-b and c-Cbl in dendritic cells causes spontaneous liver cirrhosis via altering multiple properties of CD103+ cDC1s. Cell Death Discov. 2022;8:142. doi: 10.1038/s41420-022-00953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vujanovic L, Chuckran C, Lin Y, et al. CD56dim CD16− Natural Killer Cell Profiling in Melanoma Patients Receiving a Cancer Vaccine and Interferon-α. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pesce S, Tabellini G, Cantoni C, et al. B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. Oncoimmunology. 2015;4:e1001224. doi: 10.1080/2162402X.2014.1001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petricevic B, Laengle J, Singer J, et al. Trastuzumab mediates antibody-dependent cell-mediated cytotoxicity and phagocytosis to the same extent in both adjuvant and metastatic HER2/neu breast cancer patients. J Transl Med. 2013;11:1–11. doi: 10.1186/1479-5876-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dean I, Lee CYC, Tuong ZK, et al. Rapid functional impairment of natural killer cells following tumor entry limits anti-tumor immunity. Nat Commun. 2024;15:683.:683. doi: 10.1038/s41467-024-44789-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharp A, Williams A, Blagden SP, et al. A first-in-human phase 1 trial of nx-1607, a first-in-class oral CBL-B inhibitor, in patients with advanced solid tumor malignancies. J C O. 2022;40:TPS2691. doi: 10.1200/JCO.2022.40.16_suppl.TPS2691. [DOI] [Google Scholar]
- 56.Triozzi P, Kooshki M, Alistar A, et al. Phase I clinical trial of adoptive cellular immunotherapy with APN401 in patients with solid tumors. J Immunotherapy Cancer. 2015;3 doi: 10.1186/2051-1426-3-S2-P175. [DOI] [Google Scholar]
- 57.Genentech Press releases. 2022. https://www.gene.com/media/press-releases/14951/2022-05-10/genentech-reports-interim-results-for-ph Available.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request.