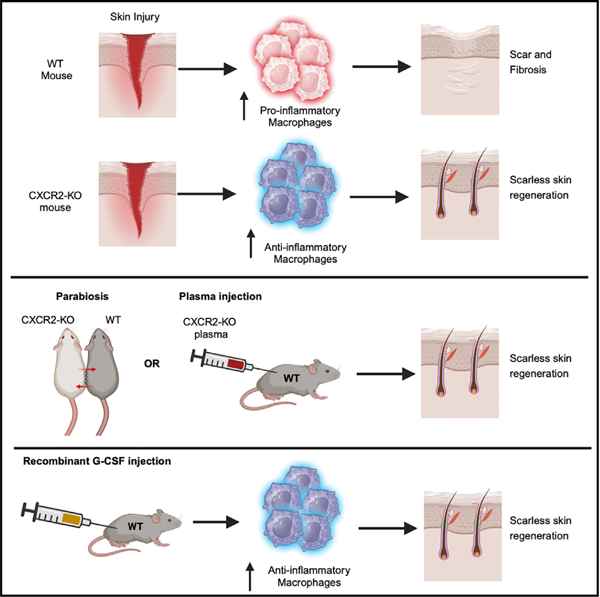
SUMMARY
Mammals typically heal with fibrotic scars, and treatments to regenerate human skin and hair without a scar remain elusive. We discovered that mice lacking C-X-C motif chemokine receptor 2 (CXCR2 knockout [KO]) displayed robust and complete tissue regeneration across three different injury models: skin, hair follicle, and cartilage. Remarkably, wild-type mice receiving plasma from CXCR2 KO mice through parabiosis or injections healed wounds scarlessly. A comparison of circulating proteins using multiplex ELISA revealed a 24-fold higher plasma level of granulocyte colony stimulating factor (G-CSF) in CXCR2 KO blood. Local injections of G-CSF into wild-type (WT) mouse wound beds reduced scar formation and increased scarless tissue regeneration. G-CSF directly polarized macrophages into an anti-inflammatory phenotype, and both CXCR2 KO and G-CSF-treated mice recruited more anti-inflammatory macrophages into injured areas. Modulating macrophage activation states at early time points after injury promotes scarless tissue regeneration and may offer a therapeutic approach to improve healing of human skin wounds.
Graphical abstract
In brief
Huang et al. demonstrate in mice that treatment of injured skin with exogenous G-CSF reduces scar formation and promotes full-thickness skin regeneration.
INTRODUCTION
Mice and humans generally heal skin wounds with a fibrotic scar within several weeks.1–5 Scar formation obliterates the native issue architecture, and repaired skin lacks accessory organs, including hair and sebaceous glands. Under specific circumstances, mice and humans may heal cutaneous injuries with scarless tissue regeneration, which heals tissues to their original architecture, with return of accessory skin organs.3–6 These findings suggest that the molecular mechanisms driving tissue regeneration remain conserved. Regenerative medicine is charged to find methods to promote scarless tissue regeneration.
We were interested in how immune cells may contribute to scarless tissue regeneration. Mice lacking the chemokine receptor CXCR2 (hereafter called CXCR2 knockout [KO]) exhibit defective neutrophil chemotaxis, which results in paradoxically increased circulating neutrophils due to defective retention of neutrophils within the bone marrow.7–9 One study showed that non-stented back wounds on CXCR2 KO mice closed with slower speed compared to control mice but did not assess scar formation.10 Since that study, non-stented mouse skin wounds have been shown to heal primarily by “wound contraction” due to an additional muscle layer rather than “wound healing.”11 Thus, the role of CXCR2 in wound healing remains largely unexplored.
Here, we show that loss of CXCR2 promotes scarless tissue regeneration in three different mouse models of skin injury. Elevated granulocyte colony stimulating factor (G-CSF) levels in CXCR2 KO blood is necessary and sufficient to induce scarless tissue regeneration.
RESULTS
Genetic deletion of Cxcr2 promotes scarless wound healing
The ear hole closure model involves through-and-through 2-mm ear wounds made through the ear pinnae that may heal with complete closure.12 We found that CXCR2 KO mice closed their ear holes faster and to a significantly smaller size compared to littermate wild-type (WT) or heterozygous controls (Figure 1A). Complete ear hole closure occurred in 50% of CXCR2 KO mice and 0% in the littermate control mice. Notably, this is the highest frequency of complete ear hole closure in our experience using any regeneration-competent mouse strain.12–14 H&E and trichrome staining of wound edge tissues from WT control littermates revealed horizontally oriented fibroblasts and glassy thickened collagen, indicating tissue fibrosis and scar formation (Figures 1B and S1A). Opposing cartilage endplates remained approximately 1.9 mm apart, confirming the absence of cartilage regeneration. By contrast, healed tissue from CXCR2 KO mice revealed normal tissue architecture with return of hair follicles and sebaceous glands (Figure 1B, arrows denote regenerated structures). Cartilage regeneration was demonstrated by new islands of proliferating chondrocytes and a shortened distance (~0.7 mm) between opposing cartilage endplates (Figure 1B). Immunostaining for keratin 14 and keratin 6 highlighted new hair follicle structures in healed areas of skin in CXCR2 KO mice. We measured fibrosis in the wounded area with picrosirius red staining and found significantly less fibrosis in CXCR2 KO healed tissue (Figure 1C). Finally, Ki-67 and phosphorylated histone H3 immunostaining at day 3 post injury revealed an ~3-fold increase in cell proliferation in CXCR2 KO skin compared to WT controls (Figure 1D). Furthermore, CD31 and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining also revealed an increase in angiogenesis and decreased apoptosis on CXCR2 KO skin, respectively (Figure 1D).
Figure 1. CXCR2-deficient mice promote ear tissue regeneration.
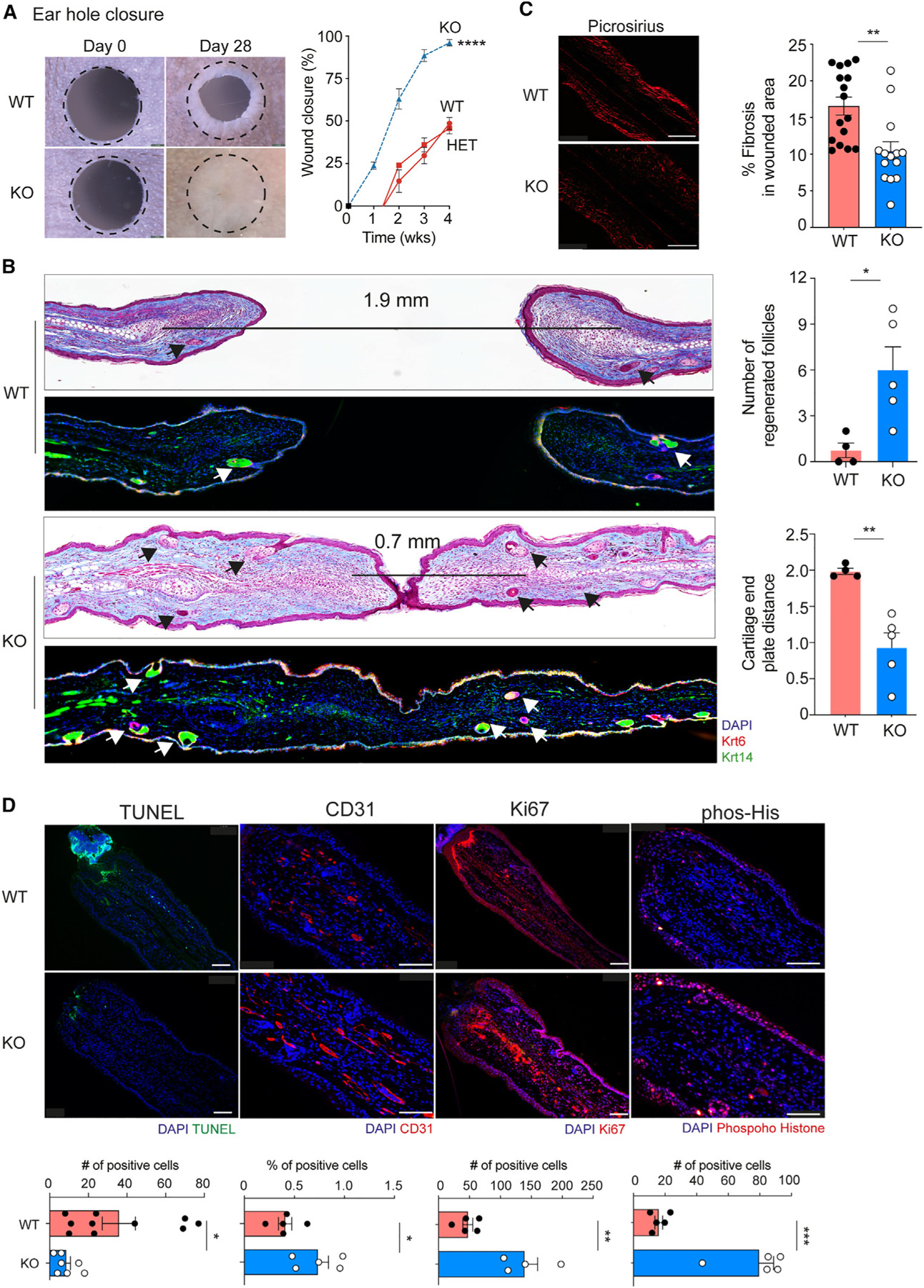
(A) Representative photographs and percentage of wound closure in wild-type (WT, n= 5), heterozygous (HET, n= 12), and CXCR2 KO homozygous (KO, dashed blue line, n = 6) mouse ears. A dotted circle represents the original 2-mm hole. 2-way ANOVA with KO compared to WT or heterozygous.
(B) Representative trichome stain and immunofluorescence of wounded ear tissue sections depicting neogenic hair follicles (Krt14+, Krt6+) from WT (n = 5–6) and CXCR2 KO (n = 5) mice. The distance between cartilage endplates is denoted by a black bar. Arrows indicated regenerated skin appendages. Right: quantitation of hair follicles in healed areas and distance between cartilage endplates. Unpaired two-tailed Student’s t test.
(C) Representative photographs and percentage of fibrosis assessed by picrosirius red staining in WT (n = 16) and KO (n = 14) wounds. Scale bars, 100 μM. Unpaired two-tailed Student’s t test.
(D) Representative images and quantification of immunofluorescence of tissue sections from wounded WT and CXCR2-KO mouse ears for apoptosis (TUNEL, n = 7), angiogenesis (CD31, n = 5) and cell proliferation (phosphorylated histone H3, n = 5 and Ki67, n=5 for WT, and n=4 for KO). Scale bars, 100 μM. Unpaired two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.01. Mean ± SEM are plotted.
Mouse ears are a unique anatomic location, and we wanted to determine whether these observations were generalizable to other areas of skin. Stented dorsal back skin wounds on mice typically heal with scar formation but may also exhibit regenerative phenotypes.15 CXCR2-KO mice closed stented 6 mm dorsal back skin wounds faster than WT littermate controls (Figure 2A). Histological analysis of wound edge tissue from WT control littermates revealed horizontally oriented fibroblasts, absence of secondary skin organs, and a scar diameter of ~3 mm (Figures 2B and S1B). In contrast, injured CXCR2 KO skin demonstrated normal tissue architecture and return of hair follicles and sebaceous glands. Healed skin from CXCR2 KO mice developed 8-fold smaller scars and less fibrosis compared to WT mice (Figures 2C and 2D).
Figure 2. CXCR2-deficient mice promote complete tissue regeneration.
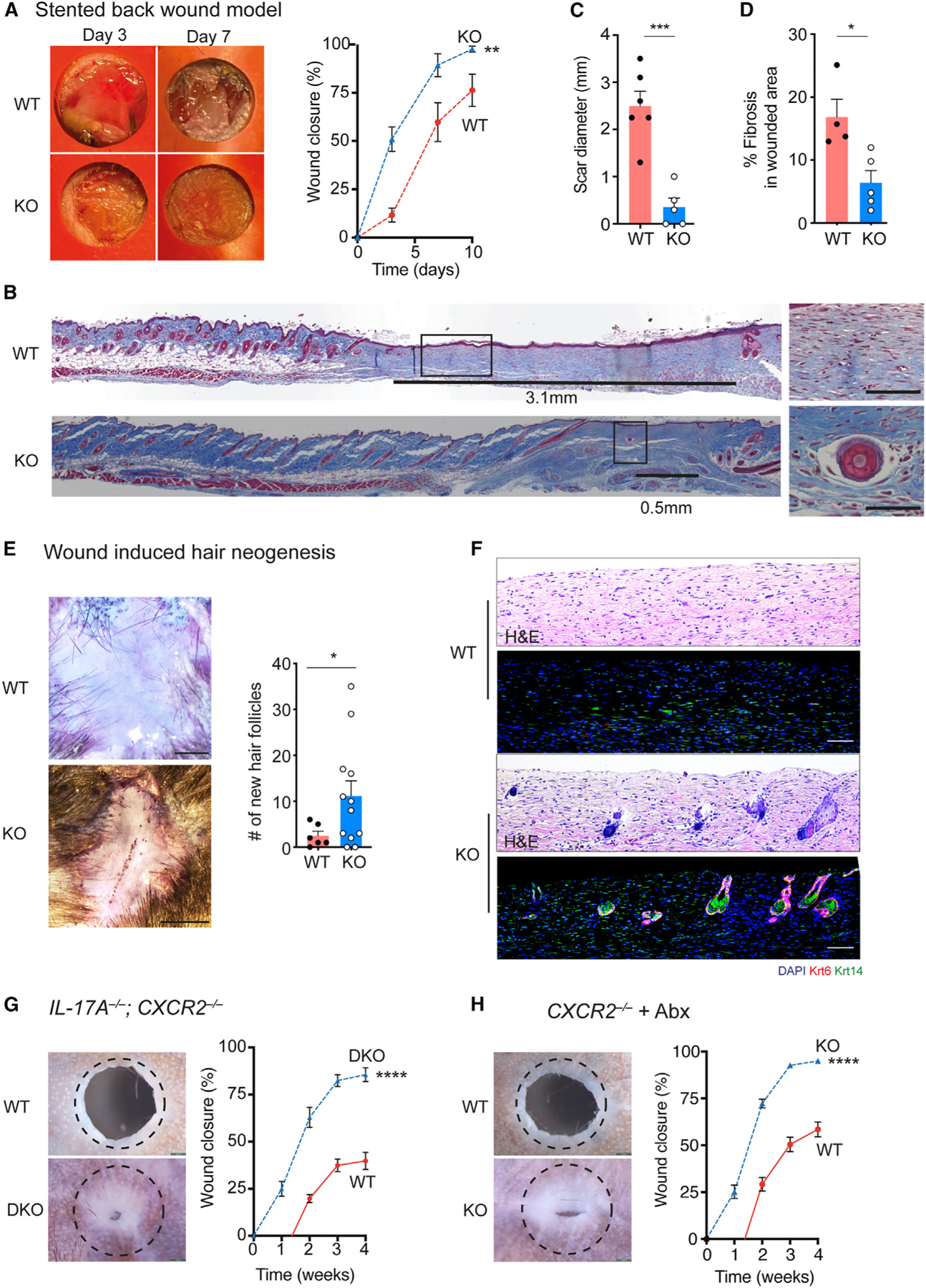
(A) Representative photographs and percentage of wound closure in WT (n = 8) and CXCR2 KO (n = 16) stented back wounds. 2-way ANOVA.
(B) Representative trichrome-stained tissue from WT and CXCR2 KO back wounds at 21 days after injury. Scar length denoted by black line. Higher magnification images from boxed areas. Scale bars, 100 μM.
(C) Scar size measured from histology sections in WT (n= 6) and CXCR2 KO wounds (n = 5). Unpaired two-tailed Student’s t test.
(D) Wound fibrosis assessed by picrosirius red staining in WT (n= 4) and CXCR2-KO wounds (n = 5). Unpaired two-tailed Student’s t test.
(E) Representative photographs and quantification of hair follicle regeneration in WIHN from WT (n = 6) and CXCR2-KO wounds (n = 12). Unpaired two-tailed Student’s t test.
(F) Representative H&E stain and immunofluorescence of healed WIHN skin, depicting the appearance of new hair follicle structures (Krt14+, Krt6+). Scale bars, 100 μM.
(G) Representative photographs and percentage of ear hole closure in littermate WT control (n = 8) and IL-17−/−; CXCR2−/− double-KO mice (n = 5). A dotted circle represents the original 2-mm hole. 2-way ANOVA.
(H) Representative photographs and percentage of ear hole closure in littermate WT control (n= 14) and CXCR2−/− (n= 11) mice treated with antibiotics. A dotted circle represents the original 2-mm hole. 2-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Mean ± SEM are plotted.
Wound-induced hair neogenesis (WIHN) model involves large excisional back wounds that may regenerate with new hair follicles.16 Compared to control littermates, CXCR2 KO mice exhibited a 6-fold increase in the number of regenerated hair follicles (Figure 2E). We serially sectioned healed skin from the center of the wounds at 5-week post injury, and immunostaining for keratin 14 and keratin 6 confirmed the appearance of new hair follicle structures specifically in healed skin from CXCR2 KO mice (Figure 2F). Taken together, three different skin injury models demonstrated that CXCR2 KO mice heal with faster wound closure, decreased scar formation, and improved tissue regeneration.
Improved healing in CXCR2 KO mice is not due to increased IL-17A or circulating neutrophils
CXCR2 KO mice exhibit baseline increased circulating levels of interleukin-17A (IL-17A) and neutrophils.7 We generated mice lacking both CXCR2 and IL-17A (CXCR2−/−; IL-17A−/−), and these mice continued to exhibit improved ear hole closure compared to littermate controls (Figure 2G). Prior work showed that the gut microbiome regulated circulating neutrophil levels in CXCR2 KO mice, and antibiotic treatment would resolve this neutrophilia, which we confirmed.7 Antibiotic-treated CXCR2 KO mice closed ear holes more efficiently than antibiotic-treated WT mice, similar to non-antibiotic-treated CXCR2 KO mice (Figure 2H). We concluded that the improved healing phenotype in CXCR2 KO mice was not dependent on IL-17A, commensal bacteria, or increased circulating neutrophils.
Cell-specific Cxcr2 KO mice exhibit partial tissue regeneration
We used single-cell RNA sequencing (scRNA-seq) on injured skin to identify cell types that express Cxcr2. We collected wound-edge tissue from WT and CXCR2 KO mice at days 0, 3, and 7 after injury. We generated 95,474 high-quality scRNA-seq profiles (Table S1). Unsupervised clustering of scRNA-seq profiles identified 43 cell clusters, which were annotated to 15 cell types based on marker gene identification, lineage marker genes, and mapping to single-cell databases (Figures 3A and S2A–S2C; Tables S2 and S3). The Cxcr2 transcript was predominantly expressed in neutrophils and was also found in keratinocytes, fibroblasts, and macrophages (Figures 3B and S2D). Flow cytometry and immunostaining (Ly6G+) on injured wound-edge skin confirmed increased recruitment of neutrophils in WT mice at days 3 and 7 after injury, which was markedly reduced in CXCR2 KO mice (Figures 3C, 3D, S2E, and S2F).
Figure 3. Cell-type-specific CXCR2 KO mice exhibit partial tissue regeneration.
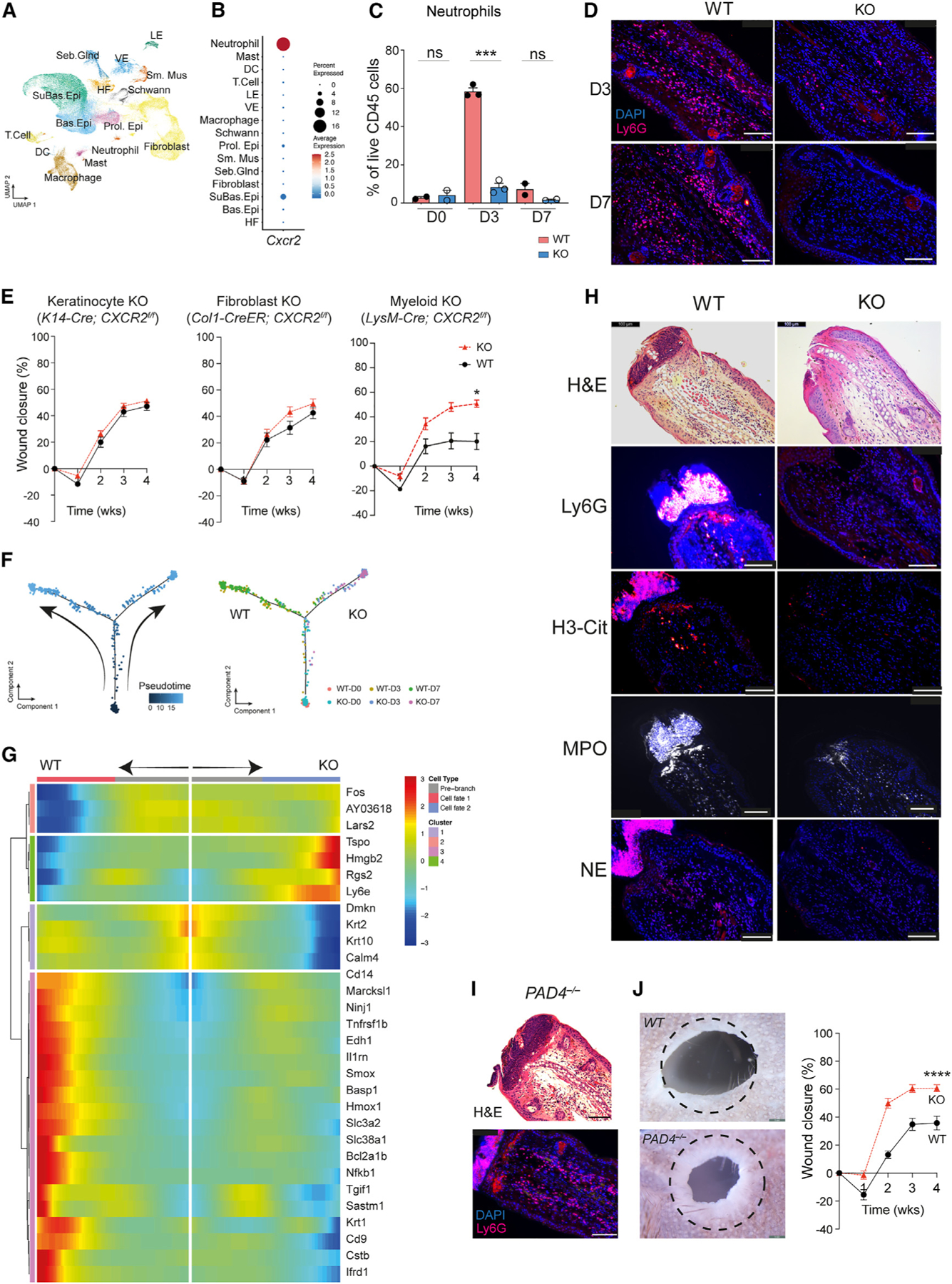
(A) Uniform manifold approximation and projection depicting cell clusters from WT (n= 2) and CXCR2 KO (n = 3) in wound-edge skin from the ear hole closure model on days 0, 3, and 7 after injury.
(B) Dot plot demonstrating levels and percentages of cells expressing Cxcr2.
(C) Quantification of neutrophils in WT and CXCR2 KO wounded skin by flow cytometry. n=2 for day 0 and day 7 groups; n=3 for day 3 groups. Unpaired two-tailed Student’s t test.
(D) Representative image of immunofluorescence for neutrophils (Ly6G+) in WT and CXCR2 KO wound-edge skin at day 3. Scale bars, 100 μM.
(E) Ear hole closure in control and cell-specific CXCR2 KO mice: keratinocyte (K14-Cre; CXCR2f/f, n = 16 and 20 for control and KO, respectively), fibroblasts (Col1-Cre-ER; CXCR2f/f, n = 8 for each group), myeloid cells (neutrophils and macrophages, LysM-Cre; CXCR2f/f, n = 7 for each group). 2-way ANOVA.
(F) Pseudotime trajectory analysis of neutrophils from WT and CXCR2 KO mice. Each dot represents a cell. Left: kinetics. Right: sample origin.
(G) Differentially expressed genes identified in pseudotime branched expression analysis modeling analysis.
(H) Representative images of immunofluorescence detecting neutrophils (Ly6G) and NETs (citrullinated-H3 [H3-Cit], myeloperoxidase [MPO], and neutrophil elastase [NE]) in WT and CXCR2 KO mice. n = 5. Scale bars, 100 μM.
(I) Representative image of immunofluorescence detecting neutrophils (Ly6G) in PADI4 KO mice. n = 3. Scale bars, 100 μM.
(J) Representative photographs and percentages of ear hole closure in control and PADI4 KO mice. n = 10 for each group. A dotted circle represents the original 2-mm hole. 2-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Mean ± SEM are plotted.
To identify the responsible cell type(s), we generated mice lacking Cxcr2 specifically in keratinocytes (K14-Cre; CXCR2f/f), fibroblasts (COL1A1-CreER; CXCR2f/f), or myeloid cells (neutrophils and macrophages, LysM-Cre; CXCR2f/f). We confirmed functional deletion by real-time PCR (Figure S2G). In the ear hole closure model, keratinocyte- or fibroblast-specific CXCR2 KO mice did not exhibit improved ear hole closure compared to control littermates (Figure 3E). In contrast, myeloid-specific CXCR2 KO mice closed ear holes to a significantly smaller size (~40%) compared to littermate controls (~20%). However, none of the mice exhibited complete ear hole closure (Figure 3E). Therefore, myeloid-specific Cxcr2 KO mice partially recapitulated the improved healing phenotype of global CXCR2 KO.
CXCR2 regulates neutrophil activation
CXCR2 is well known to regulate neutrophil chemotaxis, but a role in neutrophil activation is less studied. We used pseudotime analysis to assess gene expression changes within skin-infiltrating WT and CXCR2 KO neutrophils. The analysis revealed two distinct branches in the pseudotime path, and neutrophils from WT and KO mice localized to individual branches (Figures 3F and S2H). Branch 1 contained WT day 3 and day 7 neutrophils, and many of the top genes, including Cd14, Marcks11, Smox, Il1rn, and Basp1, showed overlap with an inflammatory neutrophil gene signature reported previously in inflamed joint, lung, and peritoneum neutrophils.17 Gene Ontology analysis corroborated this interpretation, highlighting biological processes such as wound-involved inflammation, response to DNA damage, and regulation of apoptosis as key features (Figure S3). Branch 2 contained KO day 3 and day 7 neutrophils and did not exhibit induction of inflammatory genes. Recent work demonstrated that, similar to macrophages, neutrophils may exhibit different activation states.17,18 We conclude that CXCR2 is necessary for neutrophils to adopt an inflammatory neutrophil state to promote fibrosis and scar formation.
Activated neutrophils secrete neutrophil extracellular traps (NETs), and wound-edge tissue from WT mice collected on day 3 post injury exhibited exuberant NET formation (Figure 3H). Wound-edge tissue from CXCR2 KO mice exhibited minimal NET formation. Interestingly, wound scabs are thought to be composed of dried blood and cellular debris. In WT mice, neutrophils and NETs localized specifically to the wound scab and constituted a large portion of it (Figure 3H). CXCR2 KO mice did not develop wound scabs at early time points. Finally, we obtained mice lacking PAD4−/− (hereafter called PADI4 KO), which exhibit normal neutrophil homing but are specifically unable to generate NETs (Figure 3I). Prior work used WIHN to demonstrate that PADI4 KO mice modestly increased hair follicle regeneration 3-fold.19 In the ear hole closure assay, PADI4 KO mice exhibited improved ear hole closure compared to littermate controls (Figure 3J). However, none of the mice exhibited complete ear hole closure. Thus, mice lacking NETs exhibit a partial regenerative phenotype, similar to myeloid-specific CXCR2 KO mice.
A circulating factor in CXCR2 KO mice promotes scarless tissue regeneration
Since cell-specific CXCR2 KO mice did not fully phenocopy the global CXCR2 KO phenotype, we used parabiosis to assess whether a circulating factor in CXCR2 KO mice contributes to tissue regeneration. We generated parabiosis pairs of WT:CXCR2 KO mice and control pairs (WT:WT and CXCR2 KO:CXCR2 KO). We use the ear hole injury model to minimize further trauma to the back skin. As expected, CXCR2 KO:CXCR2 KO pairs closed ear holes faster and to a smaller size compared to WT:WT pairs (Figure 4A). The WT:CXCR2 KO parabiosis pairs closed ear holes with a slower speed compared to the CXCR2 KO:CXCR2 KO control pairs but achieved the same final ear hole size (Figure 4A, blue line vs. red line). Therefore, one or more circulating factor(s) in CXCR2 KO blood promote(s) scarless wound healing.
Figure 4. G-CSF is necessary and sufficient to reduce scarring and to promote scarless tissue regeneration.
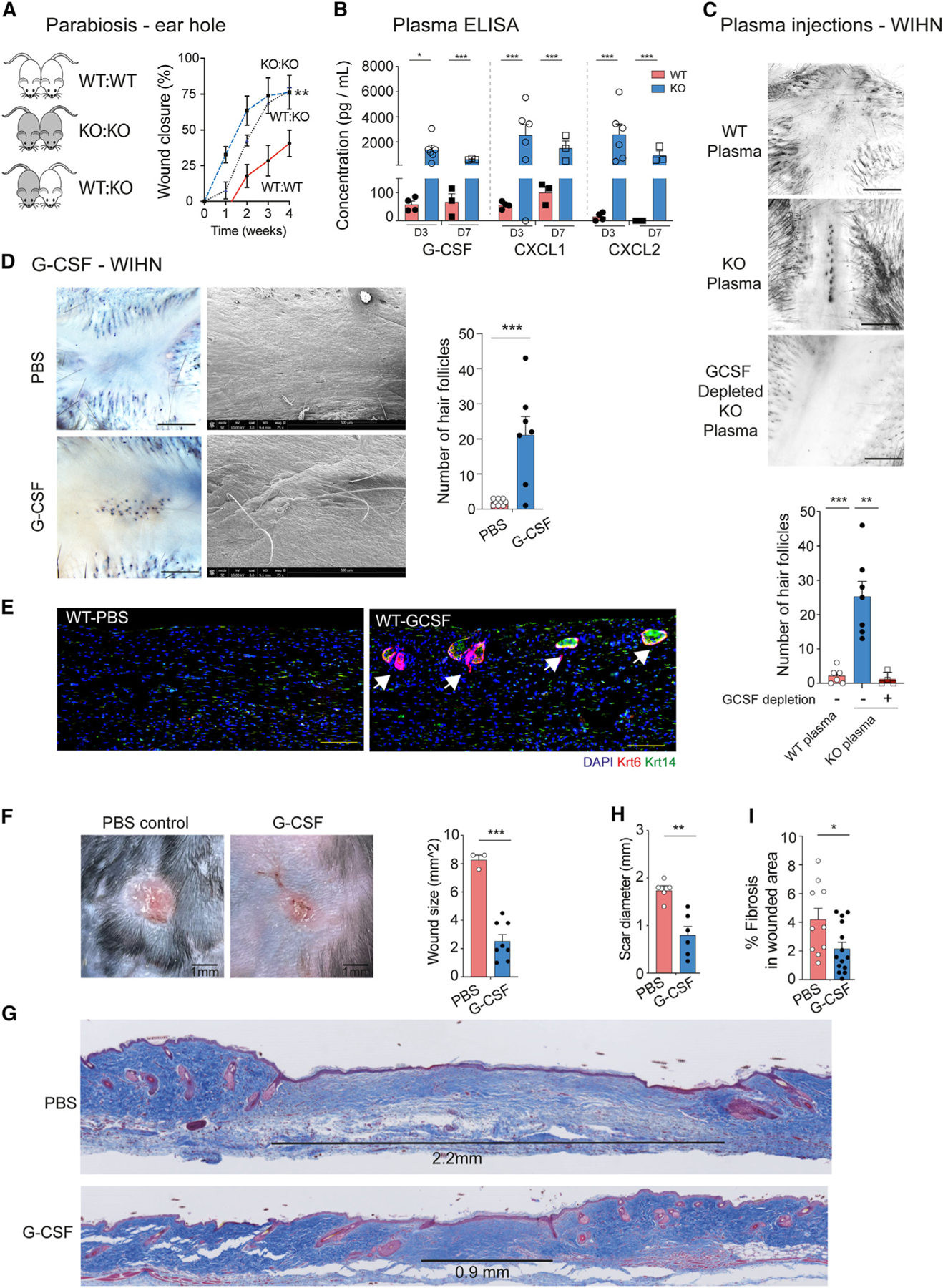
(A) Parabiosis between WT:WT (n = 4, red solid line), CXCR2 KO:CXCR2 KO (n = 3, blue dotted line), and WT:CXCR2 KO mice (n = 4, black dotted line). Shown is the percentage of ear hole closure. 2-way ANOVA comparing WT:KO pairs to WT:WT pairs.
(B) ELISA measuring cytokine expression in injured WT (n = 4 for day 3, n = 3 for day 7) and CXCR2 KO (n = 6 for day 3, n = 3 for day 7) plasma. Unpaired two-tailed Student’s t test.
(C) WT (n = 6), CXCR2 KO (n = 7) and G-CSF depleted CXCR2 KO (n = 4) plasma was injected daily into the wound bed of WT mice undergoing WIHN for the first 3 days after injury. Shown are representative photographs and quantification of hair follicles. Scale bars, 100 μM. Unpaired two-tailed Student’s t test.
(D) G-CSF (n = 9) or PBS (control, n = 7) was injected daily into the wound bed of WT mice undergoing WIHN for the first 3 days after injury. Representative photographs of whole-mount and scanning electron microscopy demonstrating unpigmented hairs in the center of the healed areas. Right: quantification of hair follicles. Scale bars, 100 μM. Unpaired two-tailed Student’s t test.
(E) Representative immunofluorescence images of PBS and G-CSF-injected WT wound beds depicting hair follicle structures (Krt14+, Krt6+) Scale bars, 100 μM.
(F) Representative photographs and quantification of scar size of G-CSF-treated (n = 8) or PBS-treated (control, n = 3) stented back wounds at day 28 after injury. Scale bars, 1 mm. Unpaired two-tailed Student’s t test.
(G) Representative trichrome-stained tissue sections from G-CSF- or PBS-treated stented back wounds. A black line highlights scar size.
(H) Quantification of scar diameter for G-CSF (n = 6) or PBS-treated (n = 3) mice. Unpaired two-tailed Student’s t test.
(I) Wound fibrosis assessed by picrosirius red staining in G-CSF-treated (n = 14 sections) or PBS-treated (n = 10 sections). Unpaired two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Mean ± SEM are plotted.
To identify the circulating factor, we performed Luminex multianalyte cytokine analysis on CXCR2 KO and littermate control plasma collected at day 3 and day 7 after injury. We assessed 12 major cytokines, including interferon γ, IL-1β, tumor necrosis factor alpha (TNF-α), MCP-1, and IL-10. Consistent with prior studies, we found increased levels of CXCL1, CXCL2, and G-CSF in CXCR2 KO plasma (Figures 4B and S4A).7 Interestingly, wound-edge tissue samples in WT and CXCR2-KO mice did not exhibit a similar induction of these cytokines, which suggests that increased circulating levels were not triggered by local injury (Figure S4B). CXCL1 and CXCL2 are the canonical ligands of CXCR2, and we focused on G-CSF.
G-CSF is necessary and sufficient to reduce scar formation and promote scarless tissue regeneration
To test the necessity of G-CSF to promote scarless tissue regeneration, we collected plasma from injured WT and CXCR2 KO mice. Within CXCR2 KO plasma, we efficiently depleted G-CSF using antibody-conjugated beads (Figure S4C). We injected WT, CXCR2 KO, or G-CSF-depleted CXCR2 KO plasma in a circumferential pattern around the wound bed of WT mice undergoing WIHN for the first 3 days (Figure 4C). WT mice treated with CXCR2 KO plasma exhibited 5-fold increased hair follicle regeneration compared to WT mice treated with WT plasma (Figure 4C). Strikingly, WT mice treated with G-CSF depleted CXCR2 KO plasma did not exhibit increased hair follicle regeneration. Thus, G-CSF in CXCR2 KO plasma is necessary for tissue regeneration.
To test the sufficiency of G-CSF to promote tissue regeneration, we performed a similar WIHN experiment and substituted plasma injections with recombinant G-CSF or PBS (control) (Figure 4D). G-CSF-treated mice exhibited a 5-fold increase in hair follicle regeneration compared to control mice. Notably, whole-mount microscopy and scanning electron microscopy visualized newly unpigmented hairs in healed G-CSF-treated skin (Figure 4D, bottom). We serially sectioned healed skin from the center of the wounds 5 week post injury, and immunostaining for keratin 14 and keratin 6 confirmed the appearance of new hair follicle structures specifically in G-CSF-treated mice (Figure 4E). In addition, we repeated the G-CSF experiment in the stented back wound model. G-CSF-treated mice healed with >3-fold less scar formation compared to control mice (Figure 4F). Histological analysis of wound edge tissue from PBS-treated WT mice revealed horizontally oriented fibroblasts, absence of secondary hair organs, and an average scar diameter of ~1.8 mm (Figures 4F, 4G, and S3D). In contrast, G-CSF-treated WT mice exhibited an average scar diameter of ~0.5 mm and less fibrosis (Figures 4H and 4I). Taken together, exogenous G-CSF is necessary and sufficient to reduce scar formation and to promote complete tissue regeneration.
Injured CXCR2 KO skin recruits more anti-inflammatory macrophages
To understand how G-CSF may reduce scar formation and promote tissue regeneration, we subclustered immune cells in our scRNA-seq dataset and identified 11 immune cell types (Figures S5A–S5C; Table S2). Neutrophils and macrophages were the primary cell types in injured skin to express the G-CSF receptor (Csfr3) (Figure 5A). Since CXCR2 KO mice exhibited less neutrophil recruitment to injured tissue, we hypothesized that macrophages were the major receivers of G-CSF. Immunofluorescence confirmed that macrophages in WT and CXCR2 KO injured skin expressed the G-CSF receptor (Figure S5D).
Figure 5. G-CSF polarizes macrophages to an anti-inflammatory phenotype to promote tissue regeneration.
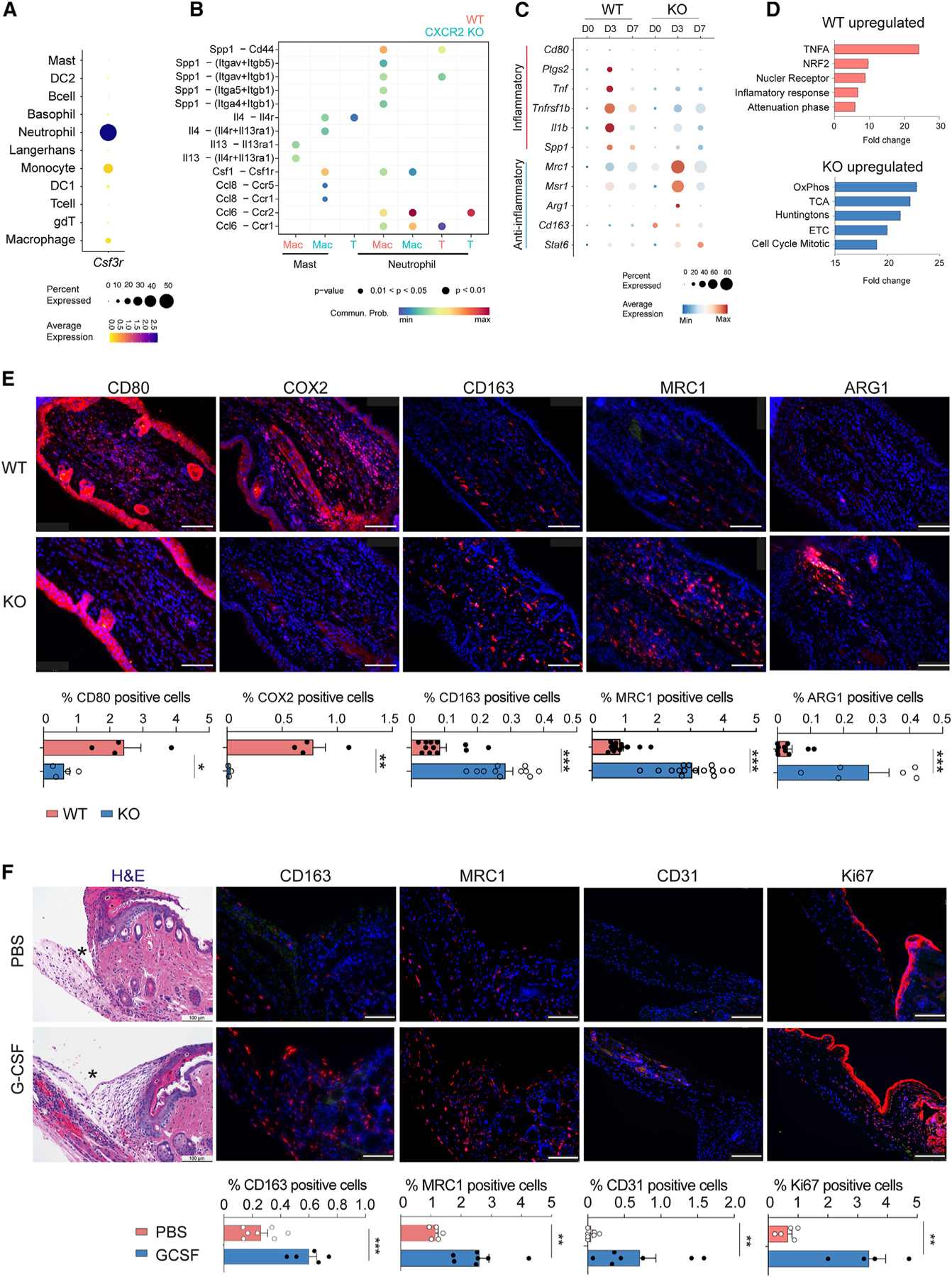
(A) Dot plot demonstrating average expression and percentage of immune cells expressing Csf3r.
(B) Analysis of key cell-to-cell interactions between immune cells in the ear skin of WT (salmon color) and CXCR2 KO (blue color) mice. Mac, macrophage; T, T cell.
(C) Dot plot demonstrating average gene expression of key genes between WT and CXCR2 KO macrophages.
(D) GSEA of macrophage populations in WT and CXCR2 KO wounded skin.
(E) Representative images and quantification of immunofluorescence of WT and CXCR2 KO wounded skin for CD80 (n = 4), COX2 (n = 4), CD163 (n = 12 for WT and n = 11 for KO), MRC1 (n = 10 for WT and n = 6 for KO), and ARG1 (n = 10 for WT and n = 6 for KO). Cell percentages are calculated with total DAPI+ cells as the denominator. Scale bars, 100 μM. Unpaired two-tailed Student’s t test.
(F) Representative H&E immunostaining images and quantification of PBS- and G-CSF-injected stented back wounds of WT mice for CD163 (n = 7 for PBS and n = 5 for G-CSF), MRC1 (n = 5 for PBS and n = 7 for G-CSF), CD31 (n = 8 for PBS and n = 7 for G-CSF) and Ki67 (n = 5 for PBS and n = 4 for G-CSF). Cell percentages are calculated with total DAPI+ cells as the denominator. Scale bars, 100 μM. Unpaired two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.01. Mean ± SEM are plotted.
We next performed comparative cell-cell communication analysis of immune cells between WT and KO mice, which revealed several significant alterations in signaling pathways (Figures 5B and S5E). First, WT mice exhibited induction of pro-inflammatory Spp1 from neutrophils at day 3 post injury, with macrophages and T cells serving as the primary recipients. Second, mast cells in KO mice induced Il4 and Csf1, factors known to polarize macrophages into an anti-inflammatory state. In contrast, WT mast cells predominantly induced Il13. Third, KO mice demonstrated a more robust Ccl6-Ccr2 interaction between neutrophils and macrophages/T cells. Previous studies have implicated Ccl6 and Ccr2 in promoting monocyte recruitment and anti-inflammatory macrophage polarization during cutaneous wound healing.20,21
Consistently, injury induced WT macrophages to express inflammatory marker genes, including Cd80, Ptgs2, Tnf, Il-1b, and Spp1 (Figure 5C). In contrast, injury induced CXCR2 KO macrophages to express anti-inflammatory macrophage marker genes, including Mrc1, Msr1, Arg1, Cd163, and Stat6 (Figure 5C). Gene Set Enrichment analysis (GSEA) in WT macrophages identified pathway changes consistent with an inflammatory phenotype, including TNF signaling, NRF2 activation, and inflammatory response (Figure 5D). CXCR2 KO macrophages exhibited pathway changes consistent with an anti-inflammatory phenotype, including oxidative phosphorylation, electron transport chain activation, and tricarboxylic acid (TCA) cycle activation (Figure 5D).
We validated macrophage gene expression changes by immunofluorescence on day 3 wound-edge tissues. WT skin exhibited increased expression of the pro-inflammatory proteins CD80 and COX2 (Ptgs2), whereas injured CXCR2 KO skin displayed increased expression of the anti-inflammatory proteins CD163, MRC1, and ARG1 (Figure 5E). These marker genes also co-stained with the macrophage marker F4/80 (Figure S6A). Taken together, injury induced macrophages in WT and CXCR2-KO mice to adopt a pro-inflammatory and anti-inflammatory phenotype, respectively.
G-CSF polarizes macrophages to adopt an anti-inflammatory phenotype to promote tissue regeneration
We wanted to test whether G-CSF may directly polarize macrophages into an anti-inflammatory phenotype. We treated bone marrow-derived monocytes with G-CSF or vehicle control and performed bulk RNA-seq. Consistent with results from prior studies, G-CSF induced expression of the same panel of anti-inflammatory marker genes, including Mrc1, Arg1, and Msr1 (Figure S6B).22–24 Next, we wanted to assess whether G-CSF may polarize infiltrating macrophages within injured skin. We collected wounded tissue at day 4 after injury in the stented back wound model from G-CSF-treated or PBS-treated WT mice. G-CSF-treated mice exhibited more anti-inflammatory macrophages (CD163 and MRC1) compared to PBS-treated mice (Figure 5F). Moreover, G-CSF-treated mice exhibited more angiogenesis (CD31) and proliferation (Ki67) (Figure 5F). Taken together, G-CSF reduces scar formation and promotes tissue regeneration through polarization of infiltrating macrophages into an anti-inflammatory phenotype.
DISCUSSION
CXCR2 KO mice exhibited robust scarless tissue regeneration across three different skin injury models. This regenerative ability is regulated by two distinct molecular mechanisms. (1) CXCR2 regulates neutrophil localization and activation. NETs secreted by activated neutrophils promote scar formation and fibrosis. (2) CXCR2 KO mice exhibit elevated levels of circulating G-CSF. G-CSF is necessary and sufficient to polarize macrophages into an anti-inflammatory state and promote complete tissue regeneration in WT mice.
Conventional wisdom says to leave wound scabs alone for optimal healing. We showed that wound scabs are predominantly composed of neutrophils and NETs. We speculate that myeloid-specific CXCR2 KO mice and PADI4 KO mice exhibited a partial regenerative phenotype during early time points because neutrophils were recruited only during the first few days after injury. NETs may be evolutionarily selected to help reduce wound infection. While pharmacologic PADI4 inhibitors for humans are still in early development, our results suggest that actively removing wound scabs may reduce scar formation.25,26
Prior work demonstrated that complete depletion of macrophages prevents wound healing, but depletion at different time points results in different outcomes.27 Depletion of early-stage macrophages reduced scar formation, and depletion of mid-stage macrophages prevented wound closure. Moreover, early- and late-stage wound macrophages exhibit distinct metabolic profiles.28 Early-stage inflammatory wound macrophages are more glycolytic, and late-stage anti-inflammatory wound macrophages use oxidative metabolism. We demonstrated that CXCR2 KO wound beds contained more early-stage macrophages that adopted an anti-inflammatory phenotype with activation of TCA cycle and oxidative metabolism genes. G-CSF also directly polarized macrophages into a similar anti-inflammatory phenotype. These results are consistent with prior work demonstrating that anti-inflammatory macrophages promote an anti-fibrotic response.29–33 Taken together, modulating macrophage activation states at early time points after injury promotes scarless tissue regeneration.
Our cell-cell communication analysis suggests that these two molecular mechanisms may be linked. Injury recruits and activates WT neutrophils to secrete inflammatory cytokines, including Spp1, that polarize infiltrating macrophages into an inflammatory subtype to drive scar formation and fibrosis. In contrast, injury induces less neutrophil recruitment in CXCR2-KO mice, and recruited CXCR2-KO neutrophils do not adopt an activated inflammatory state. The absence of neutrophil-secreted inflammatory cytokines and increased circulating G-CSF permit infiltrating macrophages to adopt an anti-inflammatory phenotype and to promote tissue regeneration. These results underscore the importance of the immune microenvironment in directing wound healing outcomes and are consistent with recent reports documenting that neutrophils may adopt different activation states, similar to macrophages.
Our G-CSF result is consistent with a prior study demonstrating that local injection of G-CSF increased the speed of wound closure in a rat skin excision model.34 It remains unclear how the global loss of CXCR2 results in increased circulating G-CSF. CXCR2 and G-CSF jointly regulate neutrophil homeostasis and exit from the bone marrow.35 CXCR2 signaling has been shown to reduce circulating G-CSF levels through commensal bacterium-stimulated IL-17A.7 However, our improved healing phenotype in CXCR2 KO mice was not dependent on IL-17A or commensal bacteria (Figures 2G and 2H). CXCR2 KO mice also exhibited neutrophil hyperplasia within the bone marrow, and more work is needed to study this potential link to circulating G-CSF levels.
Finally, a recent study demonstrated that exogenous complement factor H (CFH) reduced fibroblast-specific expression of Cxcl2, a CXCR2 ligand, to promote partial tissue regeneration.36 CFH led to reduced neutrophil recruitment and no changes in macrophage recruitment. This finding is consistent with our neutrophil-based mechanism of partial tissue regeneration. More work is needed to decipher epithelial-immune interactions during wound healing and to assess whether combination treatment of CFH and G-CSF will further augment wound healing compared to the individual components.
More than 100 million new acute skin wounds are created annually, and ~20 billion dollars are spent annually on treatment. Current treatment paradigms for acute wounds are lacking and represent a major unmet clinical need. Short courses of G-CSF are routinely and safely used in healthy volunteers who donate their stem cells for bone marrow transplantation.37 A pilot clinical trial tested systemic G-CSF in epidermolysis bullosa patients, a rare genetic skin disease with increased skin fragility, and found reduced wound sizes.38 Our results motivate initiating clinical trials to test G-CSF or scab modification to optimize human wound healing.
Limitations of the study
Our study uses mouse models of skin injury, which may not accurately replicate human wound healing biology. Second, we focused on early recruitment of neutrophils and macrophages. Third, while we identified G-CSF as a key factor, other circulating factors may also contribute to the regeneration process in CXCR2 KO mice.
RESOURCE AVAILABILITY
Lead contact
Requests for further information, resources, and reagents should be directed to Dr. Thomas Leung (thl@pennmedicine.upenn.edu).
Materials availability
This study did not generate new unique reagents. KO mice generated in this study are available upon request.
Data and code availability
All sequencing data have been deposited at the GEO and are publicly available as of the date of publication. (GEO: GSE245864; go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245864 and enter token gfuxokkajtybnmp into the box).
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
STAR★METHODS
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Study design
We predefined study components including rules for stopping data collection, data inclusion/exclusion criteria, and endpoint selection methods. Specific information is described in their relevant section. The overall objective of our study was to improve our understanding of tissue regeneration with respect to wound healing in different mice models. We used mice skin samples, in vivo mouse models, mouse genetics, and molecular biology to study this question. Presented data combines all experiments, and unless noted, all experiments were repeated at least 2 times independently. Animals used in this study were randomly assigned to experimental groups, and investigators were not blinded to allocation during experiments and outcome assessment unless noted in the text. Sample size justification for animal studies was based on preliminary experiments.
Mice
Mouse studies approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania (Protocol #805620 and performed in accordance with the NIH guidelines for the humane care of animals. Wild-type C57BL/6J (#000664), CXCR2−/− (#2724), CXCR2f/f (#24638), K14-Cre (#4782), COL1A1-CreER (#27751), LysM-Cre (#4781), and IL-17A−/− (#35717) mice were purchased from The Jackson Laboratory (JAX). All mice were group-housed in the animal facility of the University of Pennsylvania on a 12-h light/12-h dark cycle with ad libitum access to water and normal chow.
METHOD DETAILS
Injury models
Ear hole closure
For ear wounding, we used a standard 2mm mechanical punch (Roboz, Gaithersburg, MD) to create a hole in the center of each outer ear (pinna). Ear hole diameter was measured using a dissection microscope (Nikon) in the horizontal and vertical directions on a weekly basis. Ears were excluded if there were signs of wound infection, tearing of the ear, or abnormal geometric shape. These criteria were pre-established.
Wound induced hair neogenesis
1.5-cm2 full-thickness skin wounds were made as previously described.16 5-week later, de novo hair follicles were identified by whole-mount alkaline phosphatase staining of dermis preparations as previously described. Mouse plasma was freshly collected from injured WT or CXCR2 KO mice by cardiac punch under anesthetization. The collected plasma or recombinant G-CSF 50 μg/mL (or PBS, vehicle control) were injected intradermally in the mouse skin at 4 spots closely around the wound bed, in a total volume of 100 μL under anesthetization for three consecutive days.
Stented small back wounds
Briefly, a 6mm disposable biopsy punch (Acuderm) was used to make two circular full thickness wounds on the dorsal back skin of mice. Silicon wound splints (Grace Bio-Labs) were sutured with 4–0 Nylon to prevent skin contracture. Wounds were dressed with a sterile occlusive dressing and monitored daily. Borders were monitored by frequently application of permanent marker.
Parabiosis
Parabiosis surgery followed previously described procedures.43 Briefly, mirror-image incisions at the left and right flanks were made through the skin. Elbow and knee joints from each parabiont were sutured together with 3–0 Nylon, and the skin of each mouse was sutured with 4–0 Nylon to the skin of the adjacent parabiont. For overall health, several recovery characteristics were analyzed at various times after surgery, including weight and grooming responses, and animals were excluded if they failed overall health inspection. We waited 1 month after the parabiosis surgery to perform our standard ear punch assay.
Histology and immunohistochemistry
Standard histology and immunostaining protocols were followed. Briefly, immunohistochemical analysis was performed on 6 μm-thick sections of mouse skin. TUNEL assay was conducted using DeadEnd Flurometric TUNERL System (G3250, Promega according to manufacturer’s protocol. The following primary antibodies were used: rabbit monoclonal anti-Arginase-1 (93668, Cell Signaling Technology), rabbit monoclonal anti-Ki-67 (9129, Cell Signaling), rabbit polyclonal anti-CD80 (8679, ProSci), rabbit polyclonal anti-Histone H3 (ab5103, Abcam), goat polyclonal anti-MPO (AF3667, R&D Systems), rabbit monoclonal anti-MRC1 (24595, Cell Signaling), rat monoclonal anti-F4/80 (ab6640, Abcam), rabbit polyclonal anti-Neutrophil Elastase (ab68672, Accam), mouse monoclonal anti-cytokeratin 14 (ab7800, Abscam), rabbit monoclonal anti keratin 6 (SAB5500131, Sigma), rabbit monoclonal anti-Phospho-Histone H3 (53348, Cell Signaling), rabbit monoclonal anti-CD31 (77699, Cell Signaling), rabbit monoclonal anti-F4/80 (70076, Cell Signaling), rabbit polyclonal anti-COX2 (ab15191, Abcam), rabbit anti-iNOS (2982, Cell Signaling), rabbit polyclonal anti-GCSFR (bs-2574R, Bioss) rabbit monoclonal anti-CD163 (68922, Cell Signaling) and rat anti-Ly6G (551459, BD Pharmingen). The following secondary antibodies were used: goat polyclonal anti-Rabbit IgG (H + L) Alexa Fluor 488(A-11008 Thermo-Fisher), goat polyclonal anti-Rat IgG (H + L) Alexa Fluor 647 (A-21247 Thermo-Fisher) and donkey polyclonal anti-rabbit IgG (H + L) Alexa Fluor 555(A-31572 Thermo-Fisher). Trichome staining was performed as per standard histology procedure. After staining, images were analyzed using a Leica Microsystems DM6 B microscope equipped with a DFC9000 Camera or Keyence imaging system. A minimum of 4–6 sections were stained per sample. Secondary antibody control was included for every experiment. Unwounded skin was included as a control for each antibody. Representative images were selected for figure panels. Immunofluorescent images were analyzed using FIJI.44 For immunostaining quantification, positive cells were counted and normalized to the total number of DAPI+ cells within the same section. This was reported as ‘cells per section’ in the figures. Additionally, we performed co-staining experiments where inflammatory (COX2) or anti-inflammatory (MRC1) markers were co-stained with a macrophage marker (F4/80). The percentage of macrophages expressing each marker was calculated by dividing the number of double-positive cells (F4/80+ and marker+) by the total number of F4/80+ cells in the same section.
Scanning electron microscope
Scanning electron microscope experiments were carried out at CDB Microscopy Core (Dept. of Cell and Developmental Biology at the Perelman School of Medicine, University of Pennsylvania). WIHN skin samples were washed three times with 50mM Na-cacodylate buffer, fixed for 2 h with 2.5% glutaraldehyde in 50mM Na-cacodylate buffer (pH 7.3), and dehydrated in a graded series of ethanol concentrations through 100% over a period of 1.5 h. Dehydration in 100% ethanol was done three times. After 100% ethanol step dehydrated samples were incubated for 20min in 50% HMDS in ethanol followed by three changes of 100% HMDS (Sigma-Aldrich Co.) and followed by overnight air-drying as described previously*. Then samples were mounted on stubs and sputter coated with gold palladium. Specimens were observed and photographed using a Quanta 250 FEG scanning electron microscope (FEI, Hillsboro, OR, USA) at 10 kV accelerating voltage.
Picrosirius red (PSR) staining
The paraffin sections were de-waxed, hydrated and the nuclei were stained with hematoxylin. Picrosirius red (Sigma-Aldrich, 365548) was then added for 1 h. The slides were washed twice with acidified water (0.05% glacial acetic acid). The slides underwent dehydration in three changes of 100% ethanol, followed by clearing in xylene, and finally, mounting in a resinous medium. The PSR images were acquired on a Leica DM6B-Z microscope using a light polarizer equipped with a 32 mm quarter-wave plate and an ICT/P analyzer module. The acquired images were analyzed for fibrosis by quantifying the percent of collagen 1 (PSR) signal within the wounded tissue region. Representative images were selected for figure panels.
Flow cytometry
Freshly dissected tissue from the rim of healing wounds were dissociated with Liberase TL (Roche) for 90 min at 37°C. Single cell suspensions were washed in PBS and resuspended in FACS buffer (PBS +0.05% NaN3 + 2% FBS). Neutrophils were quantified using a modified version of a previously described flow cytometry gating strategy.45 Cells were stained with Zombie GreenTM cell viability dye (423111, Zombie Green Biolegend) for 15 min in the dark at room temperature. Cells were pretreated with Fc-blocking agent TruStain FcX anti-mouse CD16/32 (101320, clone 93) and subsequently stained with the following monoclonal antibodies: CD45 (103130, clone 30-F11, BioLegend), F4/80 (123149, clone BM8, BioLegend), CD11b (101218, clone M1/70, BioLegend), GR-1 (127617, clone 1A8, BioLegend). Samples were acquired on a four-laser BD LSRII flow cytometer and all sample data was analyzed using FloJo software version 10.8.1 (BD).
Luminex ELISA assay
The quantification of chemokine/cytokines from mouse plasma and tissue protein extracts was performed using a Luminex 100 system at the University of Maryland SOM Cytokine Core Laboratory (CCL). For each assay, at least two technical replicates were used, and the number of biological replicates is mentioned in the individual data figure legends. The following chemokine/cytokines used were in the assay: IL-1beta, IL-10, TNF-alpha, G-CSF, IFN-gamma, IL-15, MCP-1, MIP-1 alpha, MIP-2, KC and MIP-1 beta.
Real-time RT–PCR
Freshly dissected tissues were collected in TRI Reagent (Zymo) and then mechanically disrupted (Fastprep 24, Lysing Matrix D, MP Bio). Total RNA was isolated by Direct-Zol RNA MicroPrep (Zymo). RNA concentration was measured by Nanodrop 1000 (Thermo Scientific). cDNA synthesis was performed with Maxima Reverse Transcriptase or Superscript IV VILO (Thermo Scientific) per manufacturer’s instructions. One-step quantitative RT–PCR was performed and analyzed using an ABI ViiA7 Real-Time PCR detection system (Applied Biosystems) with TaqMan one-step RT–PCR Master Mix Reagents.
Single-cell gene RNA-Seq sequencing and analysis
Freshly dissected tissue from the rim of healing wounds was dissociated with scissors in a serum free RPMI 1640 media with DNase I (0.2 mg/mL, 12633012, Thermo Fisher Scientific), 20 mM HEPES and 0.25 mg/mL Liberase TL (5401020001, Roche) and incubated for 90 min at 37°C. The digestion was stopped by adding 100 μL FBS and 3 μL of 0.5 M EDTA and filtered through a 70-mm cell strainer (22–363–548, Fisher Scientific). The cells were washed twice with PBS containing 1% BSA and resuspended in PBS containing 0.04% BSA and counted. The scRNA-seq was performed using 10X Chromium 3 v3.1 kit (1000268, 10X Genomics). The sequencing libraries were prepared per manufacturer’s protocol and sequenced 2×100bp paired-end run on the Illumina HiSeq2000/HiSeq2500 platforms at the BGI America. The raw and processed sequencing data details are given in Table S1.
scRNA-seq data analysis
The scRNA sequencing data was mapped to the GRCm38 reference genome to generate gene count and cell barcode matrices using the “cellranger count” function from the cellranger pipeline (version 5.0.1, 10X Genomics). All downstream analysis steps were performed using the R package Seurat46 (ver. 4.3.0, https://github.com/satijalab/seurat) unless otherwise noted. In brief, seurat functions ‘Read10X’ and ‘CreateSeuratObject’ were used to import and created a merged Seurat object from all filtered feature barcode matrices generated by the cellranger pipeline. Cells with less <250 genes, <500 UMI, <0.80 log10 Genes per UMI, and more than 10% mitochondrial reads were excluded from the merged Seurat object for further analysis. Genes that were detected in less than 10 cells were also discarded. DoubletFinder was used to identify potential cell doublets as a final quality control.42 To determine and regress out the effect of cell cycle, each cell was given a cell cycle phase score using the Seurat function ‘CellCycleScoring’.47 The data was then log-normalized and scaled by linear regression against the number of reads. The FindVariableFeatures function followed by SelectIntegrationFeatures function (nfeatures = 3000) were used to identify variable genes from merged Seurat object. For cross-tissue data integration and batch correction, ‘FindIntegrationAnchors’ and ‘IntegrateData’ were applied to the merged Seurat object. Dimensionality reduction was performed using the RunPCA and RunUMAP function generated UMAP plots. Next, Louvain clustering was performed with the ‘FindClusters’ function using the first 40 PCs and at resolution 1.4. We used the ElbowPlot function in Seurat, visual inspection of DimHeatmap plots at different dimensions and R package clustree to choose an optimum number of dimensions and resolution.
Cell type annotation
We used two complementary approaches to annotate the identities of different cell clusters: (1) we checked the expression of lineage-specific marker genes identified from previously published single-cell RNA-Seq studies in our query cluster marker genes list and in differentially expressed genes of the query cluster. (2) We applied an unbiased cell type recognition method named SingleR (R package),48 which leverages mapping of the genes from the query cluster to the reference transcriptomic datasets. We first applied SingleR to determine if the predicted annotations were consistent with our findings and then assigned the identity to the cluster. The sample statistics and marker gene dot plots were made by using dittoSeq (v 1.4.1). The uniform manifold approximation and projection (UMAP) were applied to visualize the single cell transcriptional profile in 2D space based on the SNN graph described above.49 Other bar plots, boxplots, violin plots and heatmaps were generated by customized R code through ggplot2 (v3.2.1, R package).40
Pseudotime trajectory analysis
The single cell pseudotime trajectory analysis was performed using the Monocle2 R package (v 2.18.0). The mice neutrophil subclusters were used as an input for pseudotime analysis, and genes expressed in at least 5% of the cells were selected to construct the pseudotime trajectory. Following dimensionality reduction using PCA and tSNE method, we ran the densityPeak algorithm to cluster cells based on each cell’s local density (P) and the nearest distance (Δ). Default values were chosen for parameters of the DDRTree method and visualization of dynamically expressed genes along the pseudotime was performed using the ‘plot_genes_in_pseudotime’ function with the default parameters.
Ligand receptor analysis
We used R package CellChat (1.5.0) to study the ligand-receptor interaction networks between different immune cell subclusters. We performed the ligand receptor interaction analysis on the immune subcluster from the scRNA-seq dataset. The analysis was performed on the paracrine signaling network. For our analysis we considered ligand-receptor interactions that were expressed in at least 10 cells. The CellChat algorithm calculates an aggregated ligand-receptor interaction score base on a method called ‘trimean’. The CellChat algorithm has the added advantage of comparing two or more single-cell datasets and gives a comparative score for the given cell types. These scores represent the probability of interaction among the ligand-receptor pairs. The probability was then visualized using functions such as netAnalysis_signalingRole_scatter, which visualizes the major sender and receiver across all cell types, and netAnalysis_signalingChanges_scatter, which identifies the major signaling networks acting within a given cell type.
Functional enrichment analysis
We used SCPA to assess gene set enrichment in both wild-type and knockout conditions, leveraging the Molecular Signatures Database (MSigDB), KEGG pathway, and Reactome pathway database.50 From this analysis, pathways with a Qval >4, where Qval signifies statistical significance (defined as sqrt(–log10(Bonferroni-adjusted p-value))), were chosen. Subsequently, we visualized the pathways with the most significant fold change (FC) enrichment scores.
Depletion of G-CSF in CXCR2 KO plasma
For GCSF depletion we used anti-G-CSF antibody bound magnetic beads using a magnetic conjugation kit (Abcam ab269890). Briefly, 20 μg of Rat monoclonal anti-G-CSF (MAB414, R&D Systems) was conjugated to 20 μg of magnetic beads as per manufacturer’s protocol. Following conjugation, the beads (1 μg/μL) were stored in the storage buffer provided in the kit. For GCSF depletion, we incubated 10 μL of G-CSF antibody conjugated beads with 100 μL of plasma, at room temperature for 30 min. The magnetic beads were then removed from plasma using a magnetic column and GCSF-depleted CXCR2-KO plasma was tested for GCSF depletion efficiency using Mouse G-CSF Quantikine ELISA kit (MCS00, R&D Systems).
QUANTIFICATION AND STATISTICAL ANALYSIS
For in vivo time courses comparing hole size, data were each analyzed using 2-way analysis of variance (ANOVA) using a temporal main effect, a main effect comparing treatment, and an interaction of the two main effects. For tests such that the two-way ANOVA indicates significant time-treatment interactions, additional 2-tailed Student’s t test was used, with p values of less than 0.05 considered significant. The conclusions for the test between treatments using the two methods are identical. 2-tailed Student’s t test was used to determine significance, with p values of less than 0.05 considered significant. Higher levels of significance are indicated by the following: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 in the text.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Arginase-1 | Cell Signaling Technology | Cat# 93668; RRID:AB_2800207 |
| CD80 | ProSci | Cat#8679 |
| Histone H3 | Abcam | Cat# ab5103; RRID:AB_304752 |
| MPO | R and D Systems | Cat# AF3667; RRID:AB_2250866 |
| MRC1 | Cell Signaling Technology | Cat# 24595; RRID:AB_2892682 |
| F4/80 | Cell Signaling Technology | Cat# 70076; RRID:AB_2799771 |
| COX2 | Abcam | Cat# ab15191; RRID:AB_2085144 |
| iNOS | Cell Signaling Technology | Cat# 2982; RRID:AB_1078202 |
| GCSFR | Bioss | Cat# bs-2574R; RRID:AB_10857313 |
| CD163 | Cell Signaling Technology | Cat# 68922; RRID:AB_3105898 |
| Ly6G | BD Pharmingen | Cat# 551459; RRID:AB_394206 |
| IgG (H + L) Alexa Fluor™ 488 | Thermo-Fisher | Cat# A-11008 (also A11008); RRID:AB_143165 |
| IgG (H + L) Alexa Fluor™ 647 | Thermo-Fisher | Cat# A-21247; RRID:AB_141778 |
| IgG (H + L) Alexa Fluor™ 555 | Thermo-Fisher | Cat# A-31572 (also A31572); RRID:AB_162543 |
| CD16/32 | BioLegend | Cat# 101320 (also 101319); RRID:AB_1574975 |
| CD45 | BioLegend | Cat# 103130 (also 103129); RRID:AB_893339 |
| F4/80 | BioLegend | Cat# 123149; RRID:AB_2564589 |
| CD11b | BioLegend | Cat# 101220 (also 101218); RRID:AB_493546 |
| CD31 | Cell Signaling Technology | Cat#77699; RRID:AB_2722705 |
| G-CSF | R and D Systems | Cat# MAB414; RRID:AB_2085954 |
| Ki-67 | Cell Signaling | Cat# 9129; RRID: AB_2687446 |
| Elastase | Abcam | Cat# ab68672; RRID:AB_1658868 |
| Cytokeratin 14 | Abcam | Cat# ab7800; RRID:AB_306091 |
| keratin 6 | Sigma | Cat# SAB5500131 |
| Phospho-Histone H3 | Cell Signaling Technology | Cat# 53348; RRID: AB_2799431 |
| CD45 | BioLegend | Cat# 103130; RRID:AB_893339 |
| F4/80 | BioLegend | Cat# 123149; RRID: AB_2564589 |
| CD11b | BioLegend | Cat# 101218; RRID: AB_389327 |
| GR-1 | BioLegend | Cat# 127617; RRID: AB_1877262 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Gibco RPMI media | Thermo Fisher Scientific | 12633012 |
| Dnase I Recombinant, Rnase-free | Millipore Sigma | 4716728001 |
| 1x HBSS | Corning | 21–023-CV |
| HEPES-Buffer 1M, pH 7.5 | Boston BioProducts | BBH-75-K-500 |
| Liberase TL Research Grade | Thermo Fisher Scientific | NC1328423 |
| FBS | Corning | 35–010-CV |
| Bovine Serum Albumin | Sigma-Aldrich | A7906–50G |
| 70 μm Strainer | Fisher Scientific | 22–363-548 |
| 40 μm Strainer | Corning | 431750 |
| 1x DPBS | Gibco | 14190–136 |
| TRIzol | Thermo Fisher Scientific | 15596026 |
| UltraPure Phenol:Chloroform:Isoamyl Alcohol | Thermo Fisher Scientific | 15593031 |
| 0.5 M EDTA, pH 8.0 | Invitrogen | 15575–038 |
| Paraformaldehyde Solution, 4% in PBS | Thermo Scientific | J19943-K2 |
| DMSO | Fisher Scientific | 200–664-3 |
| OCT | Fisher Health Care | 4585 |
| Agencourt RNA Clean XP Beads | Beckman Coulter | A63987 |
| SPRI Beads | Beckman Coulter | B23318 |
| G-CSF | Med Chem Express | HY-P70608 |
| Magnetic Conjugation kit | Abcam | ab269890 |
| Mouse G-CSF Quantikine ELISA kit | R and D Systems | MCS00 |
| DeadEnd Flurometric TUNERL System | Promega | G3250 |
| Picrosirius red | Sigma-Aldrich | 365548 |
| Zombie GreenTM cell viability dye | Biolegend | 423111 |
| TruStain FcX anti-mouse CD16/32 | Biolegend | 101320, clone 93 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Chromium Next GEM Single Cell 3′ Kit v3.1 | 10x Genomics | 1000268 |
| Chromium Next GEM Chip G Single Cell Kit | 10x Genomics | 1000120 |
| Dual Index Kit TT Set A | 10x Genomics | 1000215 |
| Chromium i7 Sample Index Plate | 10x Genomics | 120262 |
| NEBNext Ultra DNA Library Prep Kit for Illumina | NEB | E7370 |
| Nextseq 500/550 Mid-Output v2.5 Kit | Illumina | 200024904 |
| NEBNext Low Input Prep Kit | NEB | E7630L |
|
| ||
| Deposited data | ||
|
| ||
| Raw scRNA-seq data | This Study | GEO: GSE245864 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Wild-type C57BL/6J | The Jackson Laboratory (JAX) | #000664 |
| Mouse: CXCR2−/− | The Jackson Laboratory (JAX) | #2724 |
| Mouse: CXCR2f/f | The Jackson Laboratory (JAX) | #24638 |
| Mouse: K14-Cre | The Jackson Laboratory (JAX) | #4782 |
| Mouse: COL1A1-CreER | The Jackson Laboratory (JAX) | #27751 |
| Mouse: LysM-Cre | The Jackson Laboratory (JAX) | #4781 |
| Mouse: IL-17A−/− | The Jackson Laboratory (JAX) | #35717 |
| Mouse: Pad4−/− | The Jackson Laboratory (JAX) | #030315 |
|
| ||
| Software and algorithms | ||
|
| ||
| cellranger pipeline 5.0.1 | 10x Genomics | https://support.10xgenomics.com/single-cell/software/overview/welcome |
| Seurat 4.3.0 | Hao et al., 202139 | https://github.com/satijalab/seurat/ |
| FACSDiva 7 | BD Biosciences | https://www.bdbiosciences.com/en-us |
| FlowJo 10.4.2 | VD Biosciences | NA |
| Prism8 | GraphPad Inc. | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH, USA | https://imagej.nih.gov/ij/ |
| DESeq2 v | Bioconductor project, USA | https://bioconductor.org/packages/DESeq2/ |
| ggplot2 v3.2.1 | Wickham40 | https://CRAN.R-project.org/package=ggplot2 |
| ComplexHeatmap | Bioconductor project, USA | https://bioconductor.org/packages/ComplexHeatmap/ |
| fgsea | Bioconductor project, USA | https://bioconductor.org/packages/fgsea/ |
| R | The R Foundation | https://www.r-project.org |
| UMAP | https://github.com/lmcinnes/umap | |
| Adobe Illustrator 2022 | Adobe | N/A |
| CellChat | Jin et al., 202141 | http://www.cellchat.org/ |
| DoubletFinder | McGinnis et al.42 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| clustree | CRAN | https://cran.r-project.org/web/packages/clustree/index.html |
| Enrichplot | Bioconductor project, USA | https://bioconductor.org/packages/release/bioc/html/enrichplot.html |
Highlights.
Mice lacking CXCR2 heal injured skin without a scar
CXCR2-KO mice exhibit increased circulating G-CSF
G-CSF is necessary and sufficient to drive scarless skin regeneration
G-CSF polarizes wound macrophages to adopt an anti-inflammatory state
ACKNOWLEDGMENTS
We thank Sixia Huang at the Penn Skin Biology Disease Resource Center (SBDRC) for support and George Cotsarelis, Mitchell Lazar, Todd Ridky, and Ronadip Banerjee for careful reading of this manuscript and critical comments. We thank Yuri Veklich at the University of Pennsylvania Cell & Developmental Biology (CDB) Microscopy Core for support with SEM data acquisition.
T.H.L. receives support from the NIH (R01AR079483), VA (I01RX002701), Moseley Foundation, Edwin and Fanny Gray Hall Center for Human Appearance, and H.T. Leung Foundation. S.S. and A.K. received support from the Dermatology Foundation.
Footnotes
DECLARATION OF INTERESTS
A provisional patent has been filed with the US Patent and Trademark Office regarding CXCR2, G-CSF, and NETosis on reducing scar formation and promoting tissue regeneration.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114742.
REFERENCES
- 1.Eming SA, Martin P, and Tomic-Canic M (2014). Wound repair and regeneration: mechanisms, signaling, and translation. Sci. Transl. Med 6, 265sr6. 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guenin-Mace L, Konieczny P, and Naik S (2023). Immune-Epithelial Cross Talk in Regeneration and Repair. Annu. Rev. Immunol 41, 207–228. 10.1146/annurev-immunol-101721-062818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurtner GC, Werner S, Barrandon Y, and Longaker MT (2008). Wound repair and regeneration. Nature 453, 314–321. 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 4.Murawala P, Tanaka EM, and Currie JD (2012). Regeneration: the ultimate example of wound healing. Semin. Cell Dev. Biol 23, 954–962. 10.1016/j.semcdb.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Seifert AW, Monaghan JR, Voss SR, and Maden M (2012). Skin regeneration in adult axolotls: a blueprint for scar-free healing in vertebrates. PLoS One 7, e32875. 10.1371/journal.pone.0032875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godwin JW, Pinto AR, and Rosenthal NA (2013). Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 110, 9415–9420. 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, Guttentag SH, Kolls JK, Oliver PM, Bushman FD, and Worthen GS (2012). Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest 122, 974–986. 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tateda K, Moore TA, Newstead MW, Tsai WC, Zeng X, Deng JC, Chen G, Reddy R, Yamaguchi K, and Standiford TJ (2001). Chemokine-dependent neutrophil recruitment in a murine model of Legionella pneumonia: potential role of neutrophils as immunoregulatory cells. Infect. Immun 69, 2017–2024. 10.1128/IAI.69.4.2017-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai WC, Strieter RM, Mehrad B, Newstead MW, Zeng X, and Standiford TJ (2000). CXC chemokine receptor CXCR2 is essential for protective innate host response in murine Pseudomonas aeruginosa pneumonia. Infect. Immun 68, 4289–4296. 10.1128/IAI.68.7.4289-4296.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devalaraja RM, Nanney LB, Du J, Qian Q, Yu Y, Devalaraja MN, and Richmond A (2000). Delayed wound healing in CXCR2 knockout mice. J. Invest. Dermatol 115, 234–244. 10.1046/j.1523-1747.2000.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galiano RD, Michaels J 5th, Dobryansky M, Levine JP, and Gurtner GC (2004). Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen 12, 485–492. 10.1111/j.1067-1927.2004.12404.x. [DOI] [PubMed] [Google Scholar]
- 12.Bedelbaeva K, Snyder A, Gourevitch D, Clark L, Zhang XM, Leferovich J, Cheverud JM, Lieberman P, and Heber-Katz E (2010). Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc. Natl. Acad. Sci. USA 107, 5845–5850. 10.1073/pnas.1000830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung TH, Snyder ER, Liu Y, Wang J, and Kim SK (2015). A cellular, molecular, and pharmacological basis for appendage regeneration in mice. Genes Dev 29, 2097–2107. 10.1101/gad.267724.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiguchi MA, Spencer CA, Leung DH, and Leung TH (2018). Aging Suppresses Skin-Derived Circulating SDF1 to Promote Full-Thickness Tissue Regeneration. Cell Rep 24, 3383–3392.e5. 10.1016/j.celrep.2018.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mascharak S, Talbott HE, Januszyk M, Griffin M, Chen K, Davitt MF, Demeter J, Henn D, Bonham CA, Foster DS, et al. (2022). Multi-omic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell 29, 315–327.e6. 10.1016/j.stem.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, and Cotsarelis G (2007). Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320. 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 17.Grieshaber-Bouyer R, Radtke FA, Cunin P, Stifano G, Levescot A, Vijaykumar B, Nelson-Maney N, Blaustein RB, Monach PA, and Nigrovic PA; ImmGen Consortium (2021). The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat. Commun 12, 2856. 10.1038/s41467-021-22973-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jerome AD, Sas AR, Wang Y, Hammond LA, Wen J, Atkinson JR, Webb A, Liu T, and Segal BM (2024). Cytokine polarized, alternatively activated bone marrow neutrophils drive axon regeneration. Nat. Immunol 25, 957–968. 10.1038/s41590-024-01836-7. [DOI] [PubMed] [Google Scholar]
- 19.Wier E, Asada M, Wang G, Alphonse MP, Li A, Hintelmann C, Sweren E, Youn C, Pielstick B, Ortines R, et al. (2021). Neutrophil extracellular traps impair regeneration. J. Cell Mol. Med 25, 10008–10019. 10.1111/jcmm.16896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boniakowski AE, Kimball AS, Joshi A, Schaller M, Davis FM, den-Dekker A, Obi AT, Moore BB, Kunkel SL, and Gallagher KA (2018). Murine macrophage chemokine receptor CCR2 plays a crucial role in macrophage recruitment and regulated inflammation in wound healing. Eur. J. Immunol 48, 1445–1455. 10.1002/eji.201747400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng X, Ji Y, Zhang C, Jin T, Li J, and Guo J (2023). CCL6 promotes M2 polarization and inhibits macrophage autophagy by activating PI3-kinase/Akt signalling pathway during skin wound healing. Exp. Dermatol 32, 403–412. 10.1111/exd.14718. [DOI] [PubMed] [Google Scholar]
- 22.Hollmen M, Karaman S, Schwager S, Lisibach A, Christiansen AJ, Maksimow M, Varga Z, Jalkanen S, and Detmar M (2016). G-CSF regulates macrophage phenotype and associates with poor overall survival in human triple-negative breast cancer. OncoImmunology 5, e1115177. 10.1080/2162402X.2015.1115177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagiannidis I, de Santana Van Vilet E, Said Abu Egal E, Phinney B, Jacenik D, Prossnitz ER, and Beswick EJ (2020). G-CSF and G-CSFR Induce a Pro-Tumorigenic Macrophage Phenotype to Promote Colon and Pancreas Tumor Growth. Cancers 12, 2868. 10.3390/cancers12102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen Q, Kong Y, Zhao HY, Zhang YY, Han TT, Wang Y, Xu LP, Zhang XH, and Huang XJ (2019). G-CSF-induced macrophage polarization and mobilization may prevent acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 54, 1419–1433. 10.1038/s41409-019-0449-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee W, Ko SY, Mohamed MS, Kenny HA, Lengyel E, and Naora H (2019). Neutrophils facilitate ovarian cancer premetastatic niche formation in the omentum. J. Exp. Med 216, 176–194. 10.1084/jem.20181170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steele CW, Karim SA, Leach JDG, Bailey P, Upstill-Goddard R, Rishi L, Foth M, Bryson S, McDaid K, Wilson Z, et al. (2016). CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell 29, 832–845. 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Müller W, Roers A, and Eming SA (2010). Differential roles of macrophages in diverse phases of skin repair. J. Immunol 184, 3964–3977. 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- 28.Willenborg S, Injarabian L, and Eming SA (2022). Role of Macrophages in Wound Healing. Cold Spring Harb. Perspect. Biol 14, a041216. 10.1101/cshperspect.a041216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasuya A, Ito T, and Tokura Y (2018). M2 macrophages promote wound-induced hair neogenesis. J. Dermatol. Sci 91, 250–255. 10.1016/j.jdermsci.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Cao Q, Harris DCH, and Wang Y (2015). Macrophages in kidney injury, inflammation, and fibrosis. Physiology 30, 183–194. 10.1152/physiol.00046.2014. [DOI] [PubMed] [Google Scholar]
- 31.Wynn TA, and Vannella KM (2016). Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 44, 450–462. 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klinkert K, Whelan D, Clover AJP, Leblond AL, Kumar AHS, and Caplice NM (2017). Selective M2 Macrophage Depletion Leads to Prolonged Inflammation in Surgical Wounds. Eur. Surg. Res 58, 109–120. 10.1159/000451078. [DOI] [PubMed] [Google Scholar]
- 33.Rohani MG, McMahan RS, Razumova MV, Hertz AL, Cieslewicz M, Pun SH, Regnier M, Wang Y, Birkland TP, and Parks WC (2015). MMP-10 Regulates Collagenolytic Activity of Alternatively Activated Resident Macrophages. J. Invest. Dermatol 135, 2377–2384. 10.1038/jid.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen GY, Park IH, Song YS, Joo HW, Lee Y, Shin JH, Kim KS, and Kim H (2016). Local injection of granulocyte-colony stimulating factor accelerates wound healing in a rat excisional wound model. Tissue Eng. Regen. Med 13, 297–303. 10.1007/s13770-016-9054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Day RB, and Link DC (2012). Regulation of neutrophil trafficking from the bone marrow. Cell. Mol. Life Sci 69, 1415–1423. 10.1007/s00018-011-0870-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mack KL, Talbott HE, Griffin MF, Parker JBL, Guardino NJ, Spielman AF, Davitt MF, Mascharak S, Downer M, Morgan A, et al. (2023). Allele-specific expression reveals genetic drivers of tissue regeneration in mice. Cell Stem Cell 30, 1368–1381.e6. 10.1016/j.stem.2023.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grupp SA, Frangoul H, Wall D, Pulsipher MA, Levine JE, and Schultz KR (2006). Use of G-CSF in matched sibling donor pediatric allogeneic transplantation: a consensus statement from the Children’s Oncology Group (COG) Transplant Discipline Committee and Pediatric Blood and Marrow Transplant Consortium (PBMTC) Executive Committee. Pediatr. Blood Cancer 46, 414–421. 10.1002/pbc.20800. [DOI] [PubMed] [Google Scholar]
- 38.Fine JD, Manes B, and Frangoul H (2015). Systemic granulocyte colony-stimulating factor (G-CSF) enhances wound healing in dystrophic epidermolysis bullosa (DEB): Results of a pilot trial. J. Am. Acad. Dermatol 73, 56–61. 10.1016/j.jaad.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickham H (2016). ggplot2 : Elegant Graphics for Data Analysis. Use R!, 2nd ed. (Springer International Publishing: Imprint: Springer; ), pp. 33–186. [Google Scholar]
- 41.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using. CellChat. Nat. Commun 12, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 8, 329–337.e4. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer CA, and Leung TH (2019). Research Techniques Made Simple: Parabiosis to Elucidate Humoral Factors in Skin Biology. J. Invest. Dermatol 139, 1208–1213.e1. 10.1016/j.jid.2019.03.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ubags NDJ, and Suratt BT (2018). Isolation and Characterization of Mouse Neutrophils. Methods Mol. Biol 1809, 45–57. 10.1007/978-1-4939-8570-8_4. [DOI] [PubMed] [Google Scholar]
- 46.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. (2016). Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313. 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol 20, 163–172. 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 50.Bibby JA, Agarwal D, Freiwald T, Kunz N, Merle NS, West EE, Singh P, Larochelle A, Chinian F, Mukherjee S, et al. (2022). Systematic single-cell pathway analysis to characterize early T cell activation. Cell Rep 41, 111697. 10.1016/j.celrep.2022.111697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited at the GEO and are publicly available as of the date of publication. (GEO: GSE245864; go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE245864 and enter token gfuxokkajtybnmp into the box).
This paper does not report the original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


