Abstract
Mutations in creatine transporter SLC6A8 cause creatine transporter deficiency (CTD), which is responsible for 2% of all cases of X-linked intellectual disability. CTD has no current treatments and has a high unmet medical need. Inspired by the transformational therapeutic impact of small molecule “correctors” for the treatment of cystic fibrosis, which bind to mutated versions of the CFTR ion channel to promote its trafficking to the cell surface, we sought to identify small molecules that could stabilize SLC6A8 as a potential treatment for CTD. We leveraged a novel chemoproteomic technology for ligand discovery, reactive affinity probe interaction discovery, to identify small-molecule fragments with photoaffinity handles that bind to SLC6A8 in a cellular environment. We synthesized a library of irreversible covalent analogs of these molecules to characterize in functional assays, which revealed molecules that could promote the trafficking of mutant SLC6A8 variants to the cell surface. Further medicinal chemistry was able to identify reversible drug-like small molecules that both promoted trafficking of the transporter and also rescued creatine uptake. When profiled across the 27 most prevalent SLC6A8 missense variants, we found that 10–20% of patient mutations were amenable to correction by our molecules. These results were verified in an endogenous setting using the CRISPR knock-in of selected missense alleles. We established in vivo proof-of-mechanism for correctors in a novel CTD mouse model with the P544L patient-defined variant knocked in to the SLC6A8 locus, where treatment with our orally bioavailable and brain penetrant tool corrector increased brain creatine levels in heterozygous female mice, validating correctors as a potential therapeutic approach for CTD.
Introduction
Rare diseases are rooted in human genetics, where mutations are often mapped to a causal gene. Characterizing the functional consequences of these mutations on the resulting protein is critical to understanding the molecular defect causing the disease and aiding in the development of a therapeutic approach tailored to that defect. This has been the case with cystic fibrosis, where mutations in the CFTR channel cause a chloride transport defect, and a therapeutic approach was developed to correct these mutant transporters, which has led to the clinical approval for the triple combination of elexacaftor, tezacaftor, and ivacaftor,1−3 as well as others. We believe that a similar approach can be applied to creatine transporter deficiency (CTD), as considered here.
CTD is the second most common cause of X-linked intellectual disability, after Fragile X syndrome.4 It is caused by mutations in the creatine transporter gene, SLC6A8. CTD is one of three cerebral creatine deficiency syndromes, which arise from inborn errors in creatine metabolism. The other two inherited diseases result from mutations in the enzymes responsible for creatine synthesis, arginine/glycine amidinotransferase (AGAT) and guanidinoacetate methyltransferase (GAMT) deficiency.5,6
Creatine acts as a phosphate shuttle for ATP regeneration and is especially critical in organs with high energy demands, such as the brain. CTD patients therefore exhibit many cerebral phenotypes, oftentimes presenting with developmental and language delays, intellectual disability, and seizures. Although symptoms can present in infancy, CTD is typically diagnosed around two years of age. Because SLC6A8 is X-linked, more severe phenotypes emerge in male CTD patients. However, heterozygous female carriers have been reported to exhibit symptoms as well.7 While patients with AGAT or GAMT deficiency can be effectively treated with creatine supplementation, there is currently no treatment for CTD.5,6,8,9
In the CTD patient population, more than 80 variants in the SLC6A8 gene have been reported, which cause a partial or complete loss of its creatine transport function.10−14 The most common genetic aberrations are single amino acid variants, either missense or in-frame deletions, which together account for more than 70% of the CTD population. Frameshift, nonsense, large deletion, and splice variants account for the remainder of the CTD population and result in larger genetic aberrations that likely cause severe structural changes to the SLC6A8 protein. Although genetic aberrations occur throughout the SLC6A8 locus, many variants localize to the hydrophobic domain of the transporter, including transmembrane domains 7 and 8, which are likely responsible for the binding and transport of creatine directly.15
Understanding the nature of the deficit caused by each unique variant in SLC6A8 and its link to the phenotypic consequence in patients is crucial for developing effective treatments for CTD. The effects of some SLC6A8 variants on the function of the creatine transporter, including residual transport activity and trafficking defects due to retention in the endoplasmic reticulum, have been previously characterized across a variety of systems and methods of quantification, including patient-derived fibroblasts and exogenously expressed constructs.14,16−20 However, due to differences in experimental systems and measurements, it can be difficult to directly compare variants across studies.
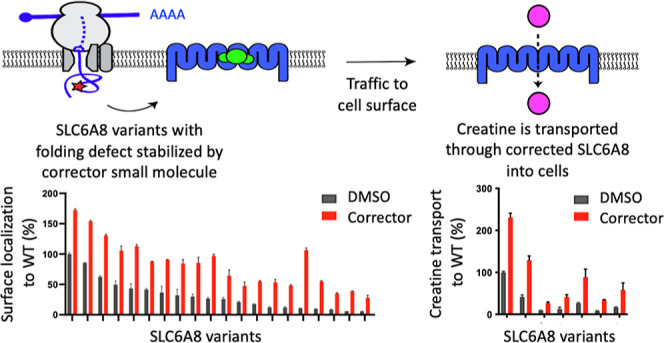
In this study, we developed assays to quantify both the trafficking of SLC6A8 to the cell surface and its residual creatine transport function, which enabled characterization of 27 different prevalent SLC6A8 variants found in CTD patients. We identified mutations as having either predominantly trafficking defects or transport defects. Because many SLC6A8 variants had a substantial defect in surface localization but retained residual transport function, we developed small molecule correctors using a novel chemoproteomic approach for ligand discovery, called reactive affinity probe interaction discovery (RAPID), as a strategy to augment surface localization to increase creatine transport, a mechanism that corrects the underlying molecular defect. We developed a translational pipeline that bridges an engineered cellular system to the endogenous SLC6A8 regulation and creatine transport and finally to a knock-in animal model of disease to probe correction of one SLC6A8 variant, P544L. This mouse model phenocopied the deficits in brain creatine observed in patients harboring the same SLC6A8 variant. Treatment with a tool SLC6A8 corrector, the discovery of which is described here, restored brain creatine levels following oral dosing of vehicle-treated SLC6A8 variant mice. Our results describe the functional deficits of SLC6A8 variants, introduce a translational path for the discovery of therapeutics for CTD, and establish proof of mechanism for a corrector approach to treat CTD.
Results
Functional Characterization of a Panel of Patient-Defined SLC6A8 Variants
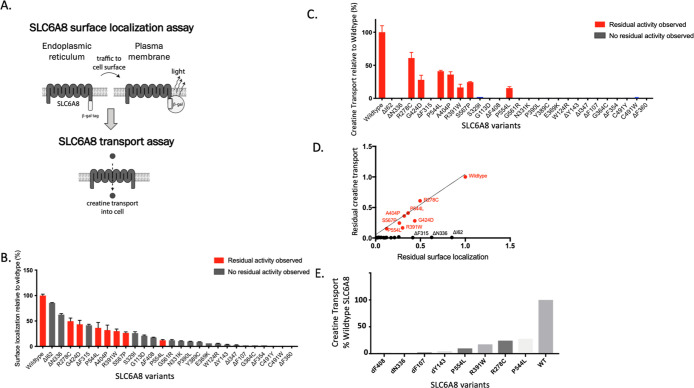
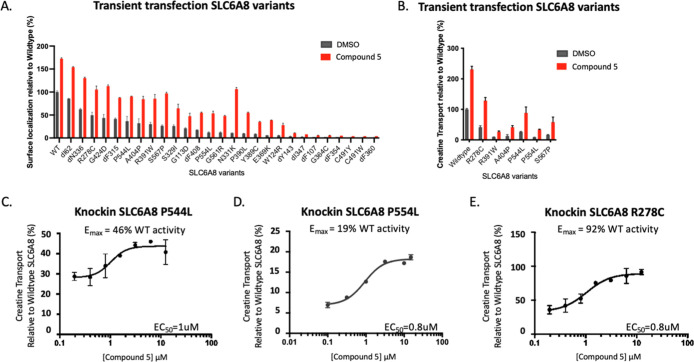
To quantify the surface localization of SLC6A8 variants, we established a trafficking assay using the PathHunter Assay technology21 (Figure 1A). This assay harnessed an enzyme fragment complementation system to generate a chemiluminescent signal only when SLC6A8 was successfully trafficked to the cell surface. We selected a panel of 27 pathogenic SLC6A8 variants based on a cross section of the reported single amino acid variants, selected to represent variants that were both missense mutations as well as single amino acid deletions13 (Figure S1A,B). Single amino acid variants were selected because they may result in subtle conformational changes more amenable to correction than larger genetic aberrations. Variants were prioritized based on prevalence, where multiple de novo occurrences were previously reported (Figure S1B). These SLC6A8 variants were expressed in the PathHunter MEM-EA U-2 OS cells lacking endogenous SLC6A8, where the wild-type locus was deleted using the CRISPR/cas9 system. We quantified surface localization of each SLC6A8 variant as a percentage of wildtype SLC6A8 (Figure 1B). All variants had less surface localization relative to the wild-type protein, indicating trafficking defects.
Figure 1.
Characterization of SLC6A8 variants by trafficking and transport activity. (A) Schematic of assays developed to identify correctors of SLC6A8 with a trafficking assay to measure surface localization of SLC6A8 and a transport assay to measure creatine transport through SLC6A8 into the cell. (B,C) Trafficking assay measuring surface localized SLC6A8 and intracellular creatine levels detected by RapidFire mass spectrometry of SLC6A8 variants following transient transfection in the PathHunter MEM-EA U-2 OS cells lacking endogenous SLC6A8 (values represent means normalized to wildtype SLC6A8 calculated from n = 2 technical replicates with standard deviation). (D) Correlation between surface localization and residual creatine transport of SLC6A8 variants profiled in (B,C). (E) Residual creatine transport in knock-in variants (values represent mean SLC6A8 knock-in variant as a percentage of wildtype SLC6A8 calculated from n = 3 technical replicates with standard deviation).
To quantify the residual function of SLC6A8 variants, we measured creatine transport into the cells from the surrounding media using the mass-spectrometry-based detection of D3-creatine. We utilized the same transiently transfected system as that described above to quantify surface localization. While most variants did not transport any detectable amount of creatine, there were 7 variants with residual creatine uptake, up to 50% of wildtype (labeled in red in Figure 1C). These variants with residual activity include P554L, R278C, R391W, A404P, G424D, S567P, and P544L, and represent an estimated 10–20% of the CTD patient population (Figure S1A,B).
When the defects in trafficking were correlated with defects in transport capacity, several categories of SLC6A8 variants emerged. Many variants resulted in a complete loss of creatine transport but maintained a significant presence at the cell surface, indicating a predominantly transport defect. Variants with transport defects included ΔN336 and ΔI62 variants, falling on the x-axis (Figure 1D). A second category of variants was defined by having seemingly proportional levels of surface localization and transport capacity, which was indicative of a functional transporter where lower levels of transport were likely attributed to trafficking defects, such as P544L and A404P variants. Variants R391W and G424D exhibited both a trafficking and transport defect, as they have less activity, as predicted from their surface localization. These variants fell just below the linear correlation. The remaining variants had both a severe trafficking defect and no detectable creatine transport. For these variants, we could not discern whether the inherent defect was attributed predominantly to a trafficking defect, a transport defect, or some other mechanism.
While transient overexpression of SLC6A8 variants enabled high-throughput characterization of their functional consequences, we wanted to ensure our results were not confounded by supraphysiological expression levels or dysregulation of creatine homeostasis. Therefore, we selected 8 variants to knock in at the endogenous SLC6A8 locus of U-2 OS cells using CRISPR/Cas9 genomic editing with homologous repair. Four pathogenic variants were selected with residual activity and four without, as defined by our previous characterization. Cell lines harboring these variants were clonally isolated, and their genotypes were confirmed by next-generation sequencing. The residual creatine transport activity of these knock-in cell lines corroborated the exogenous expression system, where P544L, R278C, P554L, and R391W retained creatine transport and ΔY143, ΔF107, ΔN336, and ΔF408 did not (Figure 1E). We hypothesized that variants with predominantly trafficking defects would be amenable to correction with a small molecule.
Discovery of a Novel Corrector of SLC6A8 Variants Using RAPID, a Novel Chemoproteomic Approach for Ligand Discovery
RAPID is a target agnostic in-cell assay that utilizes reactive fragments to identify binding sites on any target protein, where a small-molecule binder can be the starting point for a medicinal chemistry effort or can be used as a displacement probe against a reversible library in a high-throughput screen22 (Figure S2A,B). We employed both strategies to identify novel small molecule binders of SLC6A8, explore their pharmacology, and select the best starting point as the basis of a medicinal chemistry effort to develop correctors of SLC6A8.
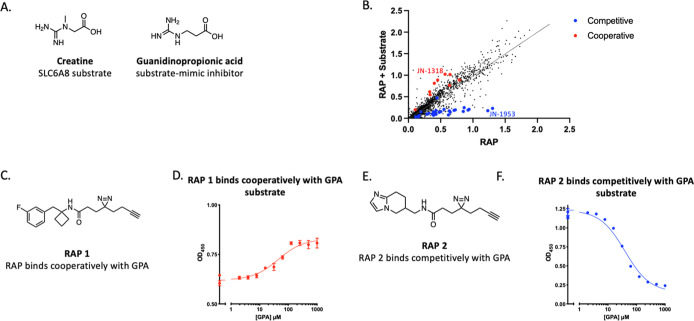
First, we conducted a high-throughput reactive affinity probe (RAP) binding assay (RAPID) to screen a library of approximately 2000 RAPs, bearing diazirine photo-cross-linking and click handle functionalities, and identify those that bind specifically to SLC6A8. To enrich for true RAP hits, we screened the library of RAPs in the presence and absence of a known ligand, guanidinopropionic acid (GPA). GPA is a substrate-mimetic inhibitor of SLC6A823 that shares structural similarities with the creatine substrate but cannot be transported (Figure 2A). Performing the screen in the presence and absence of GPA allowed for the identification of GPA-cooperative and GPA-competitive RAPs (Figure 2B). GPA competitive and cooperative RAPs were further profiled at a fixed dose with varying concentrations of GPA, which revealed that the labeling by all the RAPs was modulated with an EC50 value commensurate with the known binding affinity of GPA (∼20 μM) (Figure S2C,D). RAP 1 was identified as an example of a selective SLC6A8 RAP that only labeled SLC6A8 in the presence of the GPA substrate (Figure 2C,D). An example of a RAP that labeled SLC6A8 in a manner competitive with GPA was RAP 2 (Figure 2E,F).
Figure 2.
Identification of GPA-cooperative and competitive RAPs that label SLC6A8. (A) Chemical structures of creatine and GPA. (B) RAP quantification of labeling SLC6A8 in the presence or absence of GPA. Validated GPA-competitive RAPs are highlighted in blue, and validated GPA-cooperative RAPs are highlighted in red. (C) Chemical structure of RAP 1. (D) SLC6A8 labeling by 20 μM RAP 1 in the presence of indicated concentrations of GPA (values represent mean signal with n = 2 technical replicates with standard deviation). (E) Chemical structure of RAP 2. (F) As described in (D) with cells treated with 20 μM RAP 2 (values represent mean signal with n = 2 technical replicates with standard deviation).
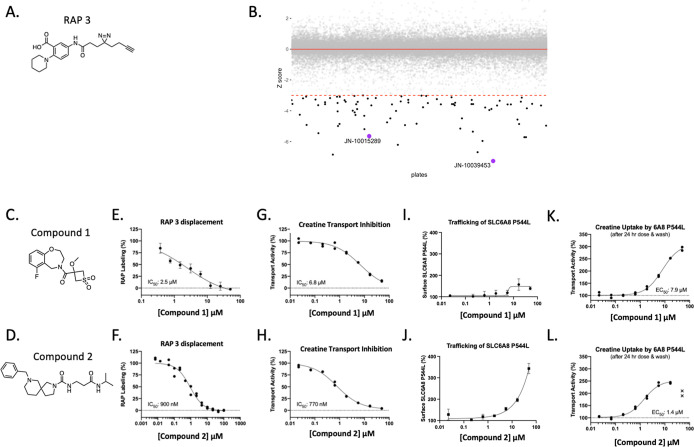
We next ran a RAPID high-throughput screen to identify reversible small molecules that prevented the binding of a selected RAP to SLC6A8 in cells overexpressing human SLC6A8. RAP 3 was selected from the validated RAPs due to its suitable window for the RAPID HTS (>2.5 fold). A library of 50,000 reversible compounds was screened at a single point at 50 μM concentration in competition with 20 μM RAP 3 (Figure 3A) and compounds that displaced the RAP were called hits based on a Z score cutoff of −3 (Figure 3B). These hits were validated in dose response against a fixed concentration of RAP 3. Compound 1 and compound 2 (Figure 3C,D) were two hits that validated the dose response (Figure 3E,F). All validated hits were subsequently profiled in functional assays and inhibited creatine transport of wildtype SLC6A8 after 30 min of pretreatment with compound (Figure 3G,H). After a 24 h incubation period, we observed that these two hit compounds increased the surface localization of a representative trafficking variant of SLC6A8, P544L (Figure 3I,J). After compound wash off, the hit compounds increased creatine transport, indicating that a functional SLC6A8 P544L transporter was indeed increased at the cell surface (Figure 3K,L). While there is precedent for using inhibitors as therapeutic chaperones of mutant proteins,24 we felt the challenge of balancing the pharmacology of correction with on-target inhibition was too complex to justify further medicinal chemistry investment in these HTS hits.
Figure 3.
RAPID HTS identified SLC6A8 inhibitors that correct the folding of the mutant transporter. (A) Structure of RAP 3. (B) A library of 50,000 compounds was screened against the RAP displacement probe RAP 3. Hits are highlighted in black with a z score of −3. (C,D) Chemical structures of RAPID HTS hit compound 1 and compound 2, respectively. (E,F) RAP 3 displacement with hits compound 1 and compound 2, respectively. (G,H) Creatine Transport Inhibition Assay: 30 min compound incubation followed by creatine uptake in HEK 293T cells overexpressing wildtype human SLC6A8. (I,J) Trafficking assay: 24 h of compound incubation in U-2 OS cells overexpressing SLC6A8 P544L variant. (K,L) creatine transport correction assay: 24 h compound incubation followed by compound wash off ahead of creatine transport in U-2 OS cells overexpressing SLC6A8 P544L variant. Values represent the mean signal with n = 2 technical replicates with standard deviation.
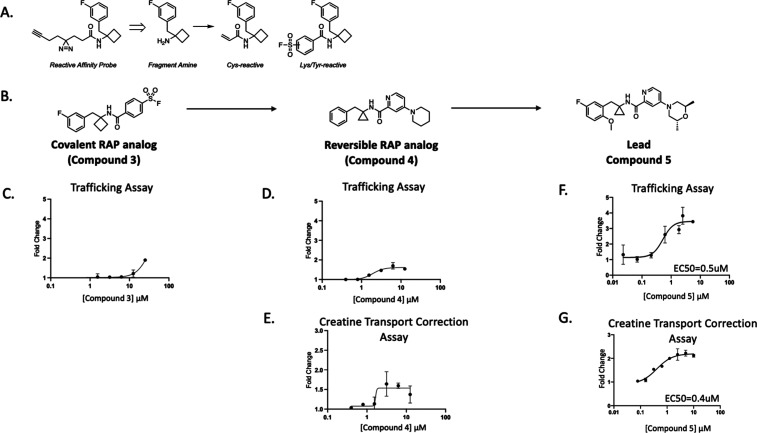
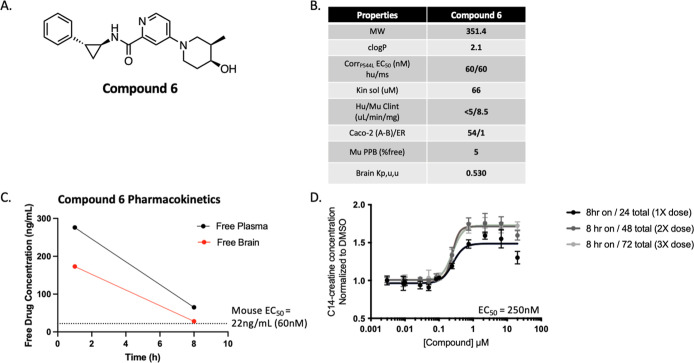
Without a suitable functional profile of SLC6A8 binders from hits emerging from the HTS, we turned to the RAP binders of SLC6A8 to see whether derivatization of these novel small molecule binders might yield a chemotype that enables trafficking and increased creatine transport without the liability of SLC6A8 inhibition. We elaborated a set of 40 validated competitive and cooperative RAPs into a small library of covalent probes by replacing their diazirine/alkyne linkers with a lysine/tyrosine reactive sulfonyl fluoride or a cysteine reactive acrylamide (Figure 4A). One such RAP analog, compound 3, was generated by replacing the alkyne linker of RAP 1 with a covalent sulfonyl fluoride motif (Figure 4B). Compound 3 revealed function at this binding site on SLC6A8, as seen by a dose-dependent increase in surface localization of SLC6A8 P544L by approximately 2-fold (Emax = > 50 μM) (Figure 4C). We further derived a reversible analog of compound 3 with a piperidine replacement of the sulfonyl fluoride, which resulted in compound 4. This compound showed improved potency by increasing the surface localization of P544L with an EC50 of 2 μM (Figure 4D). Compound 4 also enhanced the creatine transport of P544L with an EC50 of 2 μM (Figure 4E). Extensive medicinal chemistry and optimization led to compound 5 (Figure 4B). We observed improved potency for increasing the surface localization of P544L with a maximal effect of a 3.5-fold change and an EC50 of 0.5 μM (Figure 4F). Compound 5 improved the creatine transport of P544L with a maximal effect of a 2.2-fold change and an EC50 of 0.4 μM, without inhibiting creatine transport (Figures 4G and S3A).
Figure 4.
RAP-derived noncovalent correctors of SLC6A8. (A) Schematic on library derivatization. (B) Chemical structures of compound 3, compound 4, and compound 5. (C) Trafficking of P544L SLC6A8 in the U-2 OS cell line following incubation with compound 3 for 24 h (values represent means normalized to DMSO calculated from n = 2 technical replicates with standard deviation). (D,E) Trafficking and transport assays following treatment of compound 4 for 24 h in the P544L SLC6A8 U-2 OS cell line with compound wash off before creatine transport (values represent means normalized to DMSO calculated from n = 2 technical replicates with standard deviation). (F,G) Trafficking and creatine transport assays following treatment of compound 5 for 24 h in the P544L SLC6A8 U-2 OS cell line with compound wash off before creatine transport (values represent means normalized to DMSO calculated from n = 2 technical replicates with standard deviation).
We next assessed the efficacy of compound 5 across our broader panel of patient-defined SLC6A8 variants. Compound 5 trafficked most variants of SLC6A8 and corrected the creatine transport of variants with residual activity (Figure 5A,B). The R278C variant reached cellular creatine levels above the wild-type protein, and R391W, A404P, P544L, P554L, and S567P substantially increased their level of creatine transport. Variants without residual creatine transport activity did not show any increase in creatine transport, although their surface localization was increased (Figures 5B and S3B). We measured the activity of compound 5 at the endogenous locus of SLC6A8 by treating several representative knock-in SLC6A8 variant cell lines with a dose titration of compound 5. After 24 h, compound 5 increased creatine transport in cell lines with variants P544L, P554L, and R278C to 46%, 19%, and 92% of wild-type activity, respectively, with an EC50 of approximately 1 μM (Figure 5C–E).
Figure 5.
Increased creatine transport function across SLC6A8 variants (A) Trafficking assay quantifying the surface localization of SLC6A8 variants normalized to wildtype in the presence of DMSO or compound 5 in U-2 OS cells (values represent means normalized to wildtype SLC6A8 from n = 2 technical replicates with standard deviation). (B) Creatine transport correction assay measuring creatine transport of SLC6A8 variants expressed in U-2 OS cells after a 24 h preincubation with either DMSO or compound 5 and wash off of compound ahead of transport (values represent means normalized to wildtype SLC6A8 from n = 2 technical replicates with standard deviation). (C–E) Creatine transport correction assay in the P544L, P554L, and R278C knock-in U-2 OS cell lines following preincubation with compound 5 for 24 h and wash off ahead of transport (values represent means normalized to wildtype SLC6A8 from n = 2 technical replicates with standard deviation).
To ensure that this chemical series of correctors followed the mechanism of trafficking SLC6A8 to the cell surface and correcting its function of transport, we characterized the selectivity of example correctors. Representative molecules from this chemical series did not enhance the transcription of the SLC6A8 locus (Figure S3C), nor enhance the surface localization of other proteins with trafficking defects, SLC6A3 G108Q and CFTR ΔF508 (Figures S3D and S3E), indicating that this mechanism appeared selective for specific SLC6A8 variants.
Characterization of an In Vivo Tool Corrector, Compound 6
To further understand the translation of SLC6A8 correctors in vivo, we developed an analog of compound 5, compound 6, with favorable pharmacokinetic properties in terms of permeability, solubility, and microsomal stability (Figure 6A,B). Compound 6 was dosed orally in wildtype C57BL/6J mice at 30 mg/kg and demonstrated high bioavailability (100%). Compound 6 reached in vivo brain exposure for 8 h above free-drug concentrations required for in vitro efficacy defined as continuous coverage above the in vitro mouse EC50 (Figures 6C and Figure S4A–C). The human and mouse potency for SLC6A8 P544L correction were both measured at 60 nM, demonstrating translation of the mechanism of Compound 6 to the mouse SLC6A8 P544L (Figure S4A,B).
Figure 6.
In vitro and in vivo characterization of compound 6 as a brain-penetrant and bioavailable tool compound. (A) Chemical structure of compound 6. (B) In vitro ADME profile of compound 6. (C) Free drug concentration in plasma and brain homogenate from wildtype C57BL/6J mice following treatment with compound 6 at 30mpk PO at the indicated time points (n = 1 animal per time point). (D) Pulse dose treatment paradigm of compound 6 in vitro to mimic 8 h in vivo brain coverage of EC50 after once daily dosing for 3 days. Creatine transport correction assay was performed in U-2 OS cells with P544L knock-in variant with C14-creatine.
Because compound 6 achieved 8 h of pharmacokinetic coverage of free drug levels in the brain followed by 16 h of washout, we replicated this dosing paradigm in vitro and assessed correction. We observed that this dosing paradigm of 8 h on/16 h wash off maintained correction as continuous coverage did, as shown in the P544L knock-in U-2 OS cells (Figure 6D). The overall maximal efficacy (Emax) was increased after the second dose to above 1.5-fold, and it was maintained with the third daily dose. To ensure conservation in the mechanism of action, the increase in SLC6A8 P544L surface localization was measured for compound 6 at a potency of 100 nM (Figure S4C). We selected compound 6 for in vivo proof-of-mechanism studies to be carried out in a mouse model of CTD.
Compound 6 Increased Cerebral Creatine Levels in a Novel CTD Mouse Model with Knock-in P549L (Homologous Mutation to the Human P544L)
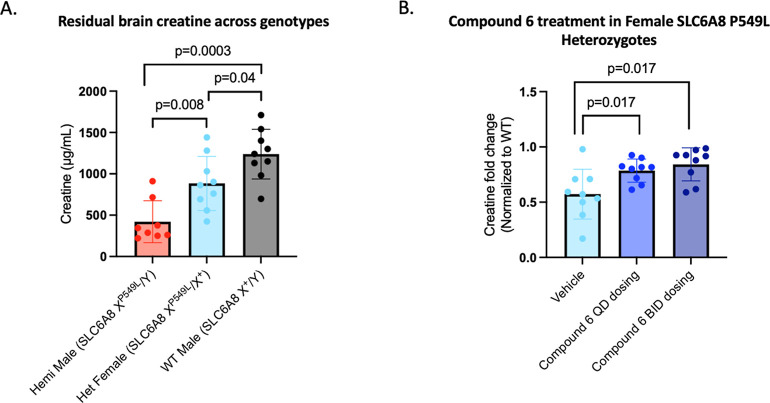
All CTD mouse models previously characterized harbor a deletion of the SLC6A8 locus. We generated a novel mouse model by introducing the P549L variant at the endogenous SLC6A8 locus to generate a knock-in model, as the presence of a misfolded protein is critical to evaluating the corrector approach. Sanger sequencing confirmed the introduction of the P549L variant into the germline (Figure S5A). Brain creatine levels from three brain regions per mouse were measured by using mass spectrometry in brain homogenates from male and female mice harboring the P549L mutation. P549L male hemizygous mice had approximately 33% creatine levels of wildtype littermates, while heterozygous females had levels approximately double that of the males (71% of wildtype), consistent with the X-linked inheritance pattern of the SLC6A8 gene and concordant with patients bearing this mutation (Figure 7A, Supporting Information Table). Heterozygous female mice exhibited decreased levels of creatine in the brain relative to wildtype mice, consistent with reports of female carriers exhibiting clinical symptoms, albeit milder than male patients.7,25
Figure 7.
P549L mouse model characterization and compound 6 efficacy. (A) Residual brain creatine for each genotype of SLC6A8 P549L knock-in C57/BL6J mice. Significance was first tested with a Kruskal–Wallis rank sum test (p value of 0.0008) with post hoc analysis using Wilcoxon tests, giving significant p values indicated on the graph (samples were analyzed from three brain regions per animal, n = 3 animals/group). (B) Creatine measured from brain homogenates from heterozygous female mice treated with compound 6 normalized to wildtype female mice treated with vehicle. The heterozygous female mice in the QD group received 4 daily doses of 30 mg/kg, and the heterozygous female mice in the BID group received 2 doses at 0 and 8 h of 30 mg/kg BID. Creatine was measured from three brain regions per mouse (left hemisphere, right cortex, and right hippocampus) and analyzed together for significance tested with a Kruskal–Wallis rank sum test (p value of 0.013) with post hoc analysis using Wilcoxon tests, giving significant p values indicated on the graph (n = 3 animals/group).
Given the reduced creatine levels observed in the brains of these heterozygous SLC6A8 P549L female mice and the availability of animals early in model development, we designed a small-scale acute study aimed to understand whether this reduction could be rescued using our corrector molecule approach. Guided by exposures from a single 30 mg/kg dose that achieved desired levels of compound 6 for 8 h in the mouse brain, we evaluated once-daily (QD) and twice-daily (BID) PO dosing regimens of compound 6 in heterozygous female mice, with total creatine measurements from brain homogenates from the left hemisphere, right cortex, and right hippocampus taken from three mice per treatment group following treatment with compound 6. The QD group received 4 daily doses of compound 6, with brain tissue harvested at day 4. The BID group received twice daily dosing of compound 6 for 1 day, with brain tissue harvested at day 3. In both the BID and QD groups compared to vehicle control, compound 6 significantly increased brain creatine levels in the P549L heterozygous females toward wildtype levels (Figure 7B, Supporting Information Table). The translation of these results into hemizygous male mice will be critical to determining the suitability of the corrector approach in vivo as a therapeutic approach for select variants of SLC6A8. Since male hemizygous mouse brains have a lower residual level of creatine, we would expect the correct approach to rescue the creatine deficit; however, the magnitude toward wildtype of the increase remains to be determined in further studies.
Conclusions
The study of molecular defects of patient-defined variants in rare diseases can guide the development of novel therapeutics tailored toward correcting the underlying cause of the disease. Here, we characterized a subset of patient-defined mutations in SLC6A8 by quantifying the residual surface localization and creatine transport capacity. We developed a novel corrector for variants of SLC6A8 based on a substrate-cooperative RAP binder and evaluated its activity across a number of SLC6A8 variants to assess which variants were amenable to correction. Finally, we developed a CTD mouse model with the P544L patient-defined variant (homologous to mouse P549L) edited directly into the SLC6A8 locus. Using this model in a pilot study, we showed that our corrector significantly increased creatine levels in the brain of heterozygous female mice, validating correctors as a potential therapeutic approach for CTD.
In this study, we directly compared the surface localization and creatine transport function of 27 patient-described SLC6A8 variants, nearly doubling the number of variants assessed within a single study previously reported. The high-throughput functional characterization of patient variants delineates the spectrum of deficits and allows the categorization of variants that behave similarly. This characterization can aid the development of therapeutic approaches for specific profiles of variants and rapidly bridge translation to variants with a similar profile. We compared the residual creatine levels by generating 8 representative knock-in variants to both validate our high-throughput system and ensure the conservation of physiologically relevant mechanisms. As hypothesized, SLC6A8 variants with trafficking defects and some residual transport function proved to be correctable, with creatine levels of select knock-in variants reaching up to wildtype levels. This category of variants with residual activity represents 10–20% of the CTD patient population. We believe that this profile predicts the response to the corrector approach in vitro. Variants with both trafficking and transport defects may require a second molecule with a distinct mechanism to restore the transport function.
The in vitro and in vivo models of CTD developed in this study showed an increase of creatine levels with corrector treatment, with some variants showing partial restoration and others where creatine levels reached up to wildtype. In order to test how much creatine restoration is necessary to impact key disease phenotypes, in vivo studies correlating creatine restoration in the brain leading to improved disease phenotypes are needed. However, there is a gene dosage effect for SLC6A8, where disease-causing variants that retain some creatine transport function are present with less extreme disease phenotypes compared with variants that abolish all creatine transport function.7 Additionally, female heterozygotes that are carriers of SLC6A8 loss of function variants usually present with milder, if any, disease phenotypes compared with hemizygous males. This suggests a gene dosage effect exists for SLC6A8, and any increase in creatine would likely be beneficial, with closer to wildtype levels more desirable. In vivo experiments will confirm this hypothesis and define the threshold for curative brain levels of creatine. It is also possible that modest correction could be combined with a high dose of creatine to address the deficit. Perhaps creatine supplementation alone would be of benefit for patients with variants of SLC6A8 with some residual activity.
Our novel P549L (homologous to human P544L) knock-in mouse model recapitulated a deficit in brain creatine levels, consistent with CTD patients harboring the P544L mutation. As the first knock-in mouse model for CTD, we anticipate that the P549L mouse will be a critical model to study physiological and behavioral phenotypes associated with this variant, as have been seen with SLC6A8 knockout models.26 Additionally, this model can be used to test new therapeutic approaches and assess improvements in brain creatine levels as well as behavioral phenotypes. To aid in the translational assessment of this corrector approach and other therapeutic approaches to treating CTD, we have made the P549L mouse model available to the scientific community through the Jackson Laboratories (036022—C57BL/6J-Slc6a8em1Jnana/J).
In summary, we developed a novel corrector for variants of SLC6A8, with creatine levels normalizing to wild-type levels in a subset of knock-in variants. These studies propose the corrector approach as a novel therapeutic opportunity for CTD, a rare disease with a lack of treatment options and a high unmet medical need. We anticipate that the in vitro and in vivo models developed and characterized here will aid in the further investigation of the corrector approach as a therapeutic route as well as other novel treatment options for CTD.
Methods
Mammalian Cell Culture
U-2 OS and HEK 293T cells were purchased from ATCC and were grown in RPMI medium 1640 (Thermo Fisher Scientific, A10491-01) or DMEM (Thermo Fisher Scientific, 10566016) medium, respectively, supplemented with 10% fetal bovine serum (FBS) (Gibco, 10438026) and 1% penicillin–streptomycin (Gibco 15140122). Cells were grown at 37 °C in a humidified CO2 incubator.
Cell Line Generation for Trafficking and Creatine Transport Assays
U-2 OS MEM-EA cells were purchased from Eurofins (catalog no. 93-1101C3). CRISPR knockout of wild-type SLC6A8 was performed using the CRISPR methods detailed below using SLC6A8 knockout guides ordered from Synthego. From these parental cells, transiently transfected or stable cell lines expressing SLC6A8 CTD mutants were made using standard cell culture protocols involving transfections of plasmids followed by antibiotic selection for the stable lines. These plasmids encoded CTD mutant SLC6A8 proteins with a C-terminal ProLink2 tag. U-2 OS MEM-EA cells and derived stable cell lines were grown in RPMI medium 1640 (Thermo Fisher Scientific, A10491-01) supplemented with 10% FBS (Gibco, 10438026), 250 μg/mL hygromycin B (Thermo Fisher Scientific, 10687010), 5 μg/mL Blasticidin (Thermo Fisher Scientific, A1113903), and 1% penicillin–streptomycin (Gibco 15140122). HEK 293T cells were transfected with human wild-type SLC6A8 and selected with 5 μg/mL Blasticidin (Thermo Fisher Scientific, A1113903). Cells were grown at 37 °C in a humidified CO2 incubator.
Generation of Knock-in Cell Lines and Next-Generation Sequencing
Guide RNAs and homology repair templates were purchased from Synthego and IDT. These SLC6A8 variants include P544L, R391W, R278C, P554L, ΔF408, ΔN336, ΔF107, and ΔY107. Nucleofection of guides and HDR templates were performed using the Lonza 4D-Nucleofector following manufacturer protocols specific to the U-2 OS cell line. Clonal expansion of single cell clones was validated by Illumina MiSeq 2500 paired-end next-generation sequencing.
SLC6A8 Trafficking Assay
U-2 OS MEM-EA cells stably expressing the SLC6A8 CTD mutant P544L with a C-terminal Prolink tag were plated into white-walled 96-well plates (Corning, catalog no. 3903) at a density of 20,000 cells per well. After 24 h, compounds were dispensed directly into the plated cells using the Tecan D300e Digital Dispenser. After an additional 24 h, the media with compound was removed. Luminescence indicative of SLC6A8 CTD mutant cell surface localization was measured according to the manufacturer’s protocol using the PathHunter Detection kit (Eurofins catalog no. 93-0001L) and an EnVision plate reader (PerkinElmer, 2104 multilabel reader). Briefly, after the media was removed, 90uLs of the PathHunter reagent solution were added to each well using a multichannel pipette. The plates were then placed in the dark for 1 h and then measured with the EnVision plate reader. Data were analyzed in Excel. Fold changes were computed with respect to DMSO.
SLC6A8 Creatine Transport Assay
Transport Correction
Stable cell lines expressing human SLC6A8 variants (U-2 OS and HEK 293T), U-2 OS wildtype, and U-2 OS knock-in SLC6A8 variant cell lines were plated into 96-well plates (Corning, catalog no. 3595) at a density of 40,000 cells per well. After 24 h, compounds were dispensed directly into the plated cells using the Tecan D300e Digital Dispenser. After an additional 24 h, the media with compound was removed. Cells were then incubated with a solution of D3-creatine (SIGMA, 616249–1G) in medium (without FBS). The concentration of D3-creatine was prepared at 100 μM for the HEK 293T and U-2 OS cells expressing a stable variant and at 25 μM for the U-2 OS cells (both wildtype and with knock-in SLC6A8 mutations). This solution was incubated with the cells at 37 °C for 30 min. After the incubation, the medium was removed, and the cells were washed once with 180 μL of phosphate buffered solution (PBS). To extract metabolites, water was added to the cells for 1 h with vigorous shaking at 700 rpm. Cell extracts were analyzed on an ABSciex-4000 triple quad mass spectrometer coupled with a RapidFire sample desalting/injection system with a graphitic carbon desalting column and a basic buffer system in reverse phase. Abundances of D3-creatine were analyzed in Excel, and then fold-changes were computed with respect to DMSO. For radiometric experiments, the same creatine transport correction protocol was followed as described above using 25 μM C14-creatine (Moravek, MC 204). Cells were lysed using scintillation fluid and radioactivity was detected using the Microbeta2 plate reader (PerkinElmer).
Transport Inhibition
For transport inhibition, the same protocol was followed as above, except that transport inhibition assays were performed with a 30 min compound incubation and without compound wash off before creatine substrate addition.
RAPID
RAPID methods are summarized with schematics and details in Figure S2 with further details in ref (22). Briefly, cells overexpressing epitope-tagged human SLC6A8 were incubated with a specified RAP that binds to the target of interest via its binding element. The RAP was covalently bound to the target following UV cross-linking of its reactive moiety. Cells were lysed, and the target of interest was captured on antibody-coated plates via the epitope tag. Biotin was conjugated to the RAP’s reporter group in a click reaction, and the biotinylated RAP was detected by an HRP-tagged streptavidin conjugate in a chemiluminescent reaction. In the RAPID high-throughput screen, cells expressing the human SLC6A8 were coincubated with a specified RAP and a library of small molecules. Small molecules with an affinity for the target of interest may compete or cooperate with RAP binding.
Quantitative Polymerase Chain Reaction (qPCR)
Cells were plated at a density of 120,000 cells per well in 24-well plates in 500 μL of RPMI medium (Gibco, A1049101) supplemented with 10% FBS (Gibco, 16140071) and 1% penicillin–streptomycin (P/S) (Gibco, 15070063). The following day, cells were treated with compound and incubated for 24 h. Cells were removed from the incubator and washed once with DPBS (Gibco, 14190144) before freezing in the plates at −80 °C until collection. The RNeasy Plus Mini Kit (Qiagen, 74134) was used to perform cell lysis and RNA extraction. Concentrations of total RNA were measured using the Nanodrop (Thermo Scientific, ND-ONE-W) and normalized to the lowest concentration before generating complementary DNA (cDNA) from each normalized sample of RNA using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems, 4387406). RT-qPCR was performed in triplicate reactions on the QuantStudio 5 (Applied Biosystems, A31674) utilizing probes specific for SLC6A8 and housekeeping genes: beta-actin and 18s. Relative quantification of the expression of SLC6A8 was normalized to the average expression of the housekeeping genes.
Immunoblotting
Cell pellets were lysed for 15 min on ice using RIPA buffer with an added protease inhibitor cocktail (Roche, 4693132001). The lysate was spun at 16,000 g for 15 min at 4 °C. The protein concentration of the lysate was quantified using the BCA assay (Pierce, 23225). The primary antibodies used were V5 (Cell Signaling Technologies, 13202) and GAPDH (97166). Fluorescence-labeled secondary antibodies (LI-COR) and the OdysseyCLxImager (LI-COR) were used to image and visualize the immunoblot.
Generation of SLC6A8 Variant Mouse
The generation of the SLC6A8 P549L (homologous to the P544L mutation in human) mice was generated and maintained on a C57BL/6J background through the Jackson laboratories using the CRISPR/cas9 system to insert the variant into the genomic locus of SLC6A8. The guide sequence was TAGGTGTTGTTGTAGACCAG, and the HDR template was AAGTGGTGTGGTGTAGTGAGGCACCAGTGACCGCAGGCATCTGTGTCCACAGGGCATCTTCATCTTCAACATTGTGTACTACGAGCTGCTGGTCTACAACAACACCTACGTGTACCCATGGTGGGGT. Genomic editing was verified using Sanger sequencing.
In Vivo Pharmacokinetics in Wildtype Mouse
Compound 6 was formulated in 10% DMSO and 20% (w/v) HP-b-CD in water. Male wildtype C57/6J mice were administered a single oral dose by gavage, and samples were collected at 1 and 8 h (brain, n = 1/TP) and 0.083, 0.25, 0.5, 1, 2, 4, 8, 24 h (plasma, n = 3/TP) post dose. Brain tissue was collected following cardiac puncture and whole-body perfusion with PBS, stored on ice, and then dissected. Brain tissue samples were homogenized (1:3 ratio) in PBS, and then, the homogenate and plasma samples were measured for drug concentrations by LC/MS/MS.
In Vivo Pharmacodynamics
Compound 6 was formulated in 10% DMSO in 20% (w/v) HP-b-CD in water. Female wildtype and P549L heterozygous C57/6J mice (N = 3/group) were dosed at 30 mg/kg BID and QD PO. At each time point, brain tissue was collected following cardiac puncture and whole-body perfusion with PBS, dissected (left hemisphere, right cortex, and right hippocampus), then snap frozen, and stored at −70 °C until analysis. Endogenous brain creatine levels were measured from the brain homogenate (1:3 ratio in PBS). Brain homogenates and plasma samples were analyzed for drug and creatine concentrations by LC/MS/MS.
Acknowledgments
We’d like to thank all authors for their contributions to these studies. We’d also like to thank the Association of Creatine Deficiencies, Dr. Nicola Longo, and Dr. Sonja Sucic for valuable discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.4c00571.
Novel corrector for variants of SLC6A8 (XLSX)
Chemical synthesis and compound characterization (PDF)
Pathogenic genetic aberrations reported in SLC6A8, validated GPA-competitive and cooperative RAPs that label SLC6A8, characterization of the activity and selectivity of compound 5, in vitro characterization of compound 6 as a corrector for SLC6A8 P544L, and genetic confirmation of SLC6A8 P549L knock-in mice (PDF)
Author Contributions
§§ L.N.G. and G.M. contributed equally to this work.
The authors declare the following competing financial interest(s): Some authors are current or former employees of Jnana Therapeutics; J.C.B. is a Venture Partner with RA Ventures (part of RA Capital).
Supplementary Material
References
- Middleton P. G.; Mall M. A.; Dřevínek P.; Lands L. C.; McKone E. F.; Polineni D.; Ramsey B. W.; Taylor-Cousar J. L.; Tullis E.; Vermeulen F.; et al. Elexacaftor–tezacaftor–ivacaftor for cystic fibrosis with a single Phe508del allele. N. Engl. J. Med. 2019, 381 (19), 1809–1819. 10.1056/nejmoa1908639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goor F.; Straley K. S.; Cao D.; González J.; Hadida S.; Hazlewood A.; Joubran J.; Knapp T.; Makings L. R.; Miller M.; et al. Rescue of ΔF508-CFTR trafficking and gating in human cystic fibrosis airway primary cultures by small molecules. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2006, 290 (6), L1117–L1130. 10.1152/ajplung.00169.2005. [DOI] [PubMed] [Google Scholar]
- Van Goor F.; Hadida S.; Grootenhuis P. D.; Burton B.; Stack J. H.; Straley K. S.; Decker C. J.; Miller M.; McCartney J.; Olson E. R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. U.S.A. 2011, 108 (46), 18843–18848. 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J.; Rosenberg E. H.; Almeida L. S.; Wood T. C.; Jakobs C.; Stevenson R. E.; Schwartz C. E.; Salomons G. S. X-linked creatine transporter (SLC6A8) mutations in about 1% of males with mental retardation of unknown etiology. Hum. Genet. 2006, 119, 604–610. 10.1007/s00439-006-0162-9. [DOI] [PubMed] [Google Scholar]
- Item C. B.; Stöckler-Ipsiroglu S.; Stromberger C.; Mühl A.; Alessandrì M. G.; Bianchi M. C.; Tosetti M.; Fornai F.; Cioni G. Arginine: glycine amidinotransferase deficiency: the third inborn error of creatine metabolism in humans. Am. J. Hum. Genet. 2001, 69 (5), 1127–1133. 10.1086/323765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Figura K., Hanefeld F., Isbrandt D., Stöckler-Ipsiroglu S.. Guanidino- acetate methyltransferase deficiency. The Metabolic and Molecular Basis of Inherited Disease, 8th ed.: Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Eds.; McGraw-Hill: New York, 2001; 1897–1908. [Google Scholar]
- Kamp J. M. v. d.; Mancini G. M. S.; Pouwels P. J.; Betsalel O. T.; Van Dooren S. J. M.; de Koning I.; Steenweg M.; Jakobs C.; van der Knaap M.; Salomons G. Clinical features and X-inactivation in females heterozygous for creatine transporter defect. Clin. Genet. 2011, 79 (3), 264–272. 10.1111/j.1399-0004.2010.01460.x. [DOI] [PubMed] [Google Scholar]
- Cecil K. M.; Salomons G. S.; Ball W. S. Jr; Wong B.; Chuck G.; Verhoeven N. M.; Jakobs C.; DeGrauw T. J. Irreversible brain creatine deficiency with elevated serum and urine creatine: a creatine transporter defect?. Ann. Neurol. 2001, 49 (3), 401–404. 10.1002/ana.79. [DOI] [PubMed] [Google Scholar]
- Stöckler S.; Hanefeld F.; Frahm J. Creatine replacement therapy in guanidineoacetate methyltransferase deficiency, a novel inborn error of metabolism. Lancet 1996, 348 (9030), 789–790. 10.1016/s0140-6736(96)04116-5. [DOI] [PubMed] [Google Scholar]
- van de Kamp J. M.; Betsalel O. T.; Mercimek-Mahmutoglu S.; Abulhoul L.; Grünewald S.; Anselm I.; Azzouz H.; Bratkovic D.; de Brouwer A.; Hamel B.; et al. Phenotype and genotype in 101 males with X-linked creatine transporter deficiency. J. Med. Genet. 2013, 50 (7), 463–472. 10.1136/jmedgenet-2013-101658. [DOI] [PubMed] [Google Scholar]
- Mencarelli M. A.; Tassini M.; Pollazzon M.; Vivi A.; Calderisi M.; Falco M.; Fichera M.; Monti L.; Buoni S.; Mari F.; et al. Creatine transporter defect diagnosed by proton NMR spectroscopy in males with intellectual disability. Am. J. Med. Genet., Part A 2011, 155 (10), 2446–2452. 10.1002/ajmg.a.34208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T.; Ito S.; Ohta Y.; Tachikawa M.; Wada T.; Terasaki T.; Ohtsuki S. Abnormal N-glycosylation of a novel missense creatine transporter mutant, G561R, associated with cerebral creatine deficiency syndromes alters transporter activity and localization. Biol. Pharm. Bull. 2017, 40 (1), 49–55. 10.1248/bpb.b16-00582. [DOI] [PubMed] [Google Scholar]
- Fokkema I. F.; Taschner P. E.; Schaafsma G. C.; Celli J.; Laros J. F.; den Dunnen J. T. LOVD v. 2.0: the next generation in gene variant databases. Hum. Mutat. 2011, 32 (5), 557–563. 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Salomons G. S.; van Dooren S. J.; Verhoeven N. M.; Cecil K. M.; Ball W. S.; Degrauw T. J.; Jakobs C. X-linked creatine-transporter gene (SLC6A8) defect: a new creatine-deficiency syndrome. Am. J. Hum. Genet. 2001, 68 (6), 1497–1500. 10.1086/320595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freissmuth M.; Stockner T.; Sucic S.. SLC6 transporter folding diseases and pharmacochaperoning. In Targeting Trafficking in Drug Development; Springer, 2018; pp 249–270. [DOI] [PubMed] [Google Scholar]
- Betsalel O. T.; Pop A.; Rosenberg E. H.; Fernandez-Ojeda M.; Jakobs C.; Salomons G. S.; Salomons G. S. Detection of variants in SLC6A8 and functional analysis of unclassified missense variants. Mol. Genet. Metab. 2012, 105 (4), 596–601. 10.1016/j.ymgme.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Rosenberg E. H.; Muñoz C. M.; Degrauw T. J.; Jakobs C.; Salomons G. S. Overexpression of wild-type creatine transporter (SLC6A8) restores creatine uptake in primary SLC6A8-deficient fibroblasts. J. Inherited Metab. Dis. 2006, 29 (2–3), 345–346. 10.1007/s10545-006-0271-6. [DOI] [PubMed] [Google Scholar]
- Salazar M. D.; Zelt N. B.; Saldivar R.; Kuntz C. P.; Chen S.; Penn W. D.; Bonneau R.; Koehler Leman J.; Schlebach J. P. Classification of the molecular defects associated with pathogenic variants of the SLC6A8 creatine transporter. Biochemistry 2020, 59 (13), 1367–1377. 10.1021/acs.biochem.9b00956. [DOI] [PubMed] [Google Scholar]
- Rosenberg E. H.; Muñoz C. M.; Betsalel O. T.; van Dooren S. J.; Fernandez M.; Jakobs C.; deGrauw T. J.; Kleefstra T.; Schwartz C. E.; Salomons G. S. Functional characterization of missense variants in the creatine transporter gene (SLC6A8): improved diagnostic application. Hum. Mutat. 2007, 28 (9), 890–896. 10.1002/humu.20532. [DOI] [PubMed] [Google Scholar]
- El-Kasaby A.; Kasture A.; Koban F.; Hotka M.; Asjad H. M.; Kubista H.; Freissmuth M.; Sucic S. Rescue by 4-phenylbutyrate of several misfolded creatine transporter-1 variants linked to the creatine transporter deficiency syndrome. Neuropharmacology 2019, 161, 107572. 10.1016/j.neuropharm.2019.03.015. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Jones A.; Olson K. R.; Peng K.; Wehrman T.; Park A.; Mallari R.; Nebalasca D.; Young S. W.; Xiao S. H. A Homogeneous Enzyme Fragment Complementation-Based β-Arrestin Translocation Assay for High-Throughput Screening of G-Protein-Coupled Receptors. SLAS Discovery 2008, 13 (8), 737–747. 10.1177/1087057108321531. [DOI] [PubMed] [Google Scholar]
- Labenski M. T., Rettenmaier T., Jones L., Muncipinto G. . Reactive affinity probe-interaction discovery platform. WO 2021076680 A1, 2021. https://patents.google.com/patent/WO2021076680A1/en.
- Saltarelli M. D.; Bauman A. L.; Moore K. R.; Bradley C. C.; Blakely R. D. Expression of the rat brain creatine transporter in situ and in transfected HeLa cells. Dev. Neurosci. 1996, 18 (5–6), 524–534. 10.1159/000111450. [DOI] [PubMed] [Google Scholar]
- Fan J. Q.; Ishii S.; Asano N.; Suzuki Y. Accelerated transport and maturation of lysosomal α–galactosidase A in Fabry lymphoblasts by an enzyme inhibitor. Nat. Med. 1999, 5, 112–115. 10.1038/4801. [DOI] [PubMed] [Google Scholar]
- Hautman E. R.; Kokenge A. N.; Udobi K. C.; Williams M. T.; Vorhees C. V.; Skelton M. R. Female mice heterozygous for creatine transporter deficiency show moderate cognitive deficits. J. Inherited Metab. Dis. 2014, 37, 63–68. 10.1007/s10545-013-9619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton M. R.; Schaefer T. L.; Graham D. L.; Degrauw T. J.; Clark J. F.; Williams M. T.; Vorhees C. V. Creatine transporter (CrT; Slc6a8) knockout mice as a model of human CrT deficiency. PLoS One 2011, 6 (1), e16187 10.1371/journal.pone.0016187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.