Abstract
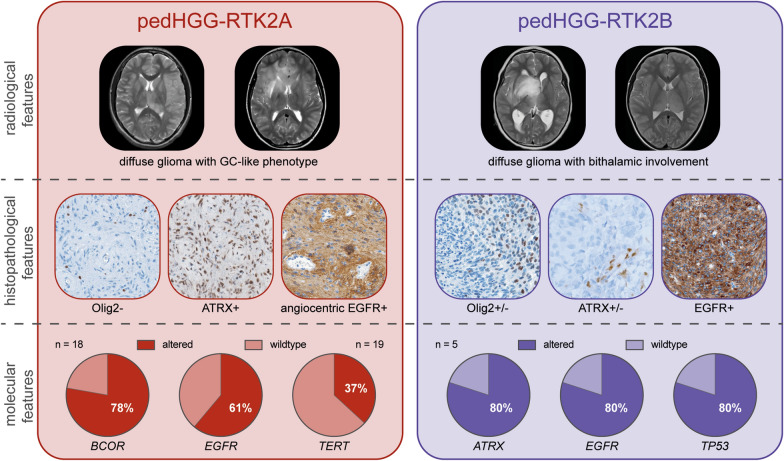
Diffuse pediatric-type high-grade gliomas (pedHGG), H3- and IDH-wildtype, encompass three main DNA-methylation-based subtypes: pedHGG-MYCN, pedHGG-RTK1A/B/C, and pedHGG-RTK2A/B. Since their first description in 2017 tumors of pedHGG-RTK2A/B have not been comprehensively characterized and clinical correlates remain elusive. In a recent series of pedHGG with a Gliomatosis cerebri (GC) growth pattern, an increased incidence of pedHGG-RTK2A/B (n = 18) was observed. We added 14 epigenetically defined pedHGG-RTK2A/B tumors to this GC series and provided centrally reviewed radiological, histological, and molecular characterization. The final cohort of 32 pedHGG-RTK2A/B tumors consisted of 25 pedHGG-RTK2A (78%) and seven pedHGG-RTK2B (22%) cases. The median age was 11.6 years (range, 4–17) with a median overall survival of 16.0 months (range 10.9–28.2). Seven of 11 of the newly added cases with imaging available showed a GC phenotype at diagnosis or follow-up. PedHGG-RTK2B tumors exhibited frequent bithalamic involvement (6/7, 86%). Central neuropathology review confirmed a diffuse glial neoplasm in all tumors with prominent angiocentric features in both subclasses. Most tumors (24/27 with available data, 89%) diffusely expressed EGFR with focal angiocentric enhancement. PedHGG-RTK2A tumors lacked OLIG2 expression, whereas 43% (3/7) of pedHGG-RTK2B expressed this glial transcription factor. ATRX loss occurred in 3/6 pedHGG-RTK2B samples with available data (50%). DNA sequencing (pedHGG-RTK2A: n = 18, pedHGG-RTK2B: n = 5) found EGFR alterations (15/23, 65%; predominantly point mutations) in both subclasses. Mutations in BCOR (14/18, 78%), SETD2 (7/18, 39%), and the hTERT promoter (7/19, 37%) occurred exclusively in pedHGG-RTK2A tumors, while pedHGG-RTK2B tumors were enriched for TP53 alterations (4/5, 80%). In conclusion, pedHGG-RTK2A/B tumors are characterized by highly diffuse-infiltrating growth patterns and specific radiological and histo-molecular features. By comprehensively characterizing methylation-based tumors, the chance to develop specific and effective therapy concepts for these detrimental tumors increases.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40478-024-01881-1.
Keywords: Receptor tyrosine kinase, RTK2A, RTK2B, Methylation, Gliomatosis cerebri, Pediatric high-grade glioma, pedHGG
Introduction
Molecular characterization of gliomas in childhood and adolescence has substantially increased the understanding of these tumors, including their different sites of origin in the central nervous system (CNS) [4–6]. For example, the discovery of the histone-3 alterations in pediatric high-grade gliomas (pedHGG) led to the characterization of diffuse midline glioma, H3 K27-altered [10, 15]. DNA-methylation profiling has evolved as a robust and reproducible diagnostic tool for classification of tumors of CNS [1, 14]. The brain tumor classifier version 12.5 defined twelve different diffuse pedHGG subclasses, some of which have been incorporated into the latest World Health Organization (WHO) Classification of Tumors of the CNS [10, 21]. However, due to the rarity of individual subclass, there is little information on the radiological or histopathological features of some of these tumors, and also on clinical characteristics of affected patients. Within the WHO CNS 2021 defined group of pedHGG, H3- and IDH-wildtype (pedHGG H3-/IDH-WT), there are the two epigenetically closely related, receptor-tyrosine-kinase-2-A/B subclasses (pedHGG-RTK2A/B). Recently, pediatric gliomas with gliomatosis cerebri (GC)—an extensively infiltrating growth pattern affecting at least three contiguous hemispheric lobes of the brain—were found frequently classified as pedHGG-RTK2A/B [12]. However, since the first description (n = 18) in 2017 [7], no study has comprehensively characterized the genotype–phenotype correlations of the two RTK2A/B subclasses.
We assembled a cohort of 14 pedHGG-RTK2A/B unselected cases and added them to the published European GC cohort (n = 18) [12]. Thereby, we radiologically, histopathologically, and genetically characterized 32 pedHGG-RTK2 tumors and identified differences between the RTK2A and RTK2B subclass.
Materials and methods
Sample collection and clinical data
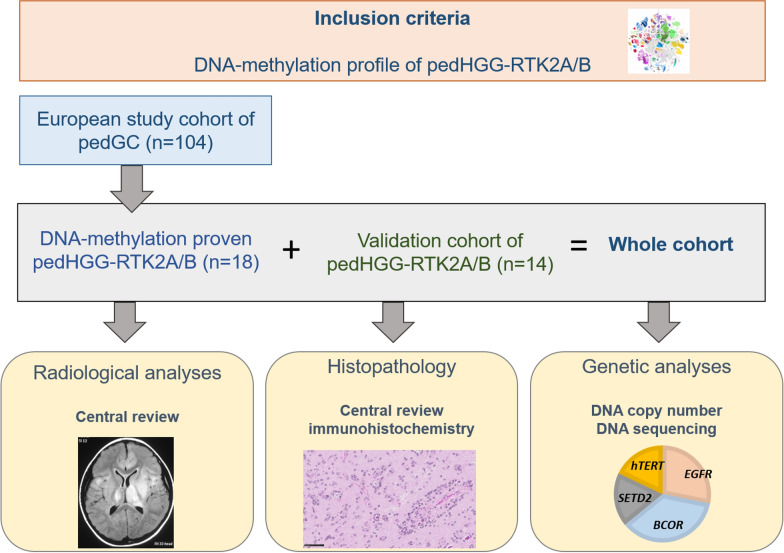
Our cohort comprises 32 tumors in total (the workflow of the study is summarized in Fig. 1). This series included 17/19 patients from the published European cohort of pediatric GC [12] classified as pedHGG-RTK2A/B). Two specimens were not included due to lack of tissue for histopathological characterization. One additional case, which had been classified as "Not elsewhere classified (NEC)" in the original publication [12], could be assigned to the pedHGG-RTK2A subclass, resulting in 18 cases (classified either as pedHGG-RTK2A (n = 15) or pedHGG-RTK2B (n = 3)). Fourteen pedHGG-RTK2 (RTK2A, n = 10; RTK2B, n = 4) were additionally identified and included. Patient characteristics and clinical data were retrieved from hospital records and included: sex, age at presentation, and medical history. Ethical approval was obtained from institutional review board (35–085 ex 22/23).
Fig. 1.
Design of the study. GC, gliomatosis cerebri; HGG, high-grade glioma; ped, pediatric
Central radiological review
The central radiological review was performed teleradiologically by two experienced neuroradiologists in consensus (VDR and BB). Preoperative magnetic resonance imaging (MRI) characterized the following: location, number of lobes involved, enhancement (intensity (mild/ moderate/ strong, with respect to the intensity of the choroid plexus), (extent (relative percentage of tumor volume)), presence of necrosis, and hydrocephalus. Tumors showing a diffuse infiltrative process involving at least three contiguous cerebral lobes of the brain were defined as GC in accordance with the 2007 WHO classification of CNS tumors [9], in which GC was defined as a distinct entity. The detailed characterization of GC was descripted previously [12]. If available, MRI of follow-up was analyzed regarding progression pattern.
Histopathological review and immunohistochemistry
The central pathology review was performed telepathologically and conjointly by six neuropathologists (ATE, LLF, PW, MA, TPi, GHG and PV). A representative paraffin block was selected for each sample. 3 μm-thick slides of formalin-fixed, paraffin-embedded (FFPE) tissues were used for immunostaining. The following primary antibodies were used: Glial fibrillary acidic protein (GFAP) (1:1000, clone 6F2, Dako, Glostrup, Denmark), OLIG2 (1:500, clone EP112, Roche Diagnostics GmbH, Mannheim, Deutschland), neurofilament protein (NF) (1:100, clone 2F11, Dako, Glostrup, Denmark), IDH1 R132H (1:40, clone H09, Clinisciences, Nanterre, France), ATRX (1:50, clone CL0537, Sigma-Aldrich, St. Louis, USA), EGFR (1:100, clone ZR16, NeoBiotech, Nanterre, France), H3K27me3 (1:2500, polyclonal, Diagenode, Liege, Belgium), EZHIP (1:75, polyclonal, Sigma-Aldrich, Bromma, Sweden), H3 G34R (1:1000, clone EPR23519, Abcam, Cambridge, United Kingdom), MSH2 (pre-diluted, clone FE11, Dako, Glostrup, Denmark), MSH6 (pre-diluted, clone 44, Dako, Glostrup, Denmark), MLH1 (pre-diluted, clone E505, Dako, Glostrup, Denmark), PMS2 (pre-diluted, clone EPR3947, Dako, Glostrup, Denmark), and Ki-67 (1:1000, clone Ki-67P, Dianova, Hamburg, Deutschland). External positive and negative controls were used for all antibodies.
DNA sequencing and DNA-methylation array analyses
Genomic DNA was extracted from fresh-frozen or FFPE tissue samples. Libraries were prepared for targeted or whole-exome sequencing using Illumina compatible protocols (Illumina, San Diego, CA). Variants were called with GATK/mutect2 using best practices and DNA copy number calculated from normalized coverage using Picard Tools CollectHsMetrics and segmented with DNAcopy [v1.72.3] as previously described [12].
DNA-methylation profiles were generated by hybridization to Infinium MethylationEPIC (850 k, v12.5) or Infinium HumanMethylation450 (450 k) BeadChip arrays (Illumina, San Diego, CA, USA) as previously described [16]. We only included pedHGG-RTK2A/B cases with high scores (> 0.9) with the Heidelberg Brain Tumor classifier (version v12.5), and/or consistent clustering with pedHGG-RTK2A/B by t-SNE in the previous GC study [12]. Integration of alterations and DNA copy number variants in the oncoprint was performed using Oncoprint function in package ComplexeHeatmap (v2.18.0) in R [19].
Statistical analysis and reference cohorts
Data analysis was carried out with the software SPSS (v29) [3] and R (v4.0.3) [19] including a package for survival analysis (Therneau T. 2020 [v3.2–7]) [20]. Survival analyses were performed via Log-Rank test, Kaplan–Meier plots, and Cox-Regression (multivariate analyses). Comparisons between two groups regarding qualitative variables were conducted using Fisher's exact test. In all analyses, alpha was set at 0.05 as threshold for statistical significance. Since interpretations were explorative, adjustment of p-values for multiple testing was not performed.
Results
Clinical and radiological findings
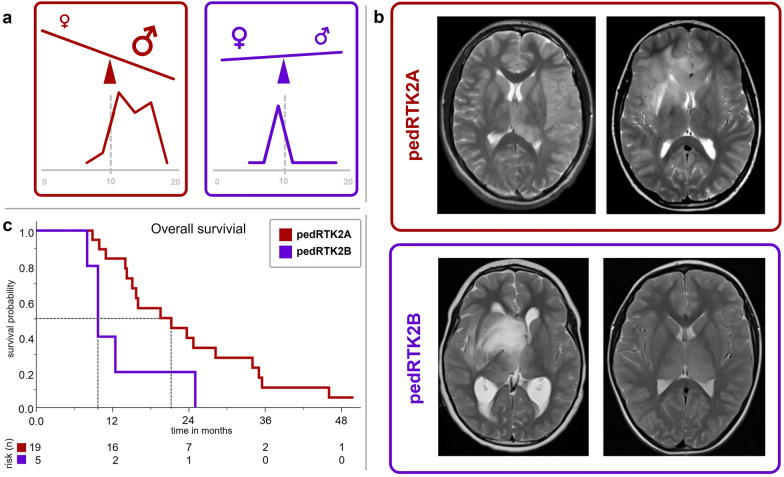
In total, we studied 32 pedHGG-RTK2A/B tumors (pedHGG-RTK2A: n = 25 [78%]; pedHGG-RTK2B: n = 7 [22%]) (Supplementary Tables 1 and 2). There was a significant difference of age at diagnosis between the two subclasses: 12.4 years in pedHGG-RTK2A (range 7–17 years) versus 8.3 years in pedHGG-RTK2B (range 4–17 years, p = 0.01, Supplementary Table 2). There were markedly more males than females in the pedHGG-RTK2A subclass (18 males, 7 females), whereas pedHGG-RTK2B seemed more balanced (3 males, 4 females). (Fig. 2a).
Fig. 2.
Comparison of main clinical parameters in pedHGG-RTK2A/B (pedHGG-RTK2A = red; pedHGG-RTK2B = purple) a Sex distribution and age according to subclass. Children with pedHGG-RTK2B were significantly younger (Graphic in the bottom row each ranging from 0 to 20 years of age; p = 0.01). b T2-weighted axial MR images of patients with pedHGG-RTK2A and pedHGG-RTK2B tumors, respectively. pedHGG-RTK2A (left); a diffuse infiltrating glioma mainly affecting the left parietal lobe at diagnosis is shown. This patient developed a gliomatosis cerebri (GC) phenotype (secondary GC) at progression; (right): a case with GC phenotype at diagnosis (primary GC). pedHGG-RTK2B (left); a glioma with diffuse infiltration of the midline (mesencephalon, basal ganglia and opticohypothalamic region) reaching the right temporal lobe. (right): a bithalamic tumor. c Kaplan–Meier plot displaying the overall survival (50% survival probability marked) of the two methylation-based subclasses
MRI at diagnosis was available in 29 patients, of whom 21 (72%) fulfilled the definition of GC. Of all unselected cases with radiologic characterization (11/14; pedHGG-RTK2A: n = 7, pedHGG-RTKB: n = 4), three patients showed a diffuse GC growing pattern at diagnosis and another four at progression (7/11; 64% in total). Of these seven cases, six tumors were of pedHGG-RTK2A subclass. In the entire radiologically characterized cohort, pedHGG-RTK2A tumors tended to be hemispheric, while additional bithalamic involvement was significantly more frequent in pedHGG-RTK2B tumors (pedHGG-RTK2A: 5/22 [23%] vs. pedHGG-RTK2B: 6/7 [86%]; p = 0.006). Two pedHGG-RTK2B tumors were located exclusively bithalamically (Fig. 2b). Tumors of pedHGG-RTK2A subclass with primary GC (n = 18) frequently involved both frontal lobes (11, 61%). Seven of all tumors (24%) showed additional infratentorial involvement. 60% (17/29) of the tumors showed no contrast enhancement at diagnosis; if enhancement was present (11/29, 38%), it was merely subtle in terms of extent and intensity. Signs of necrosis were rare (4/29, 14%) and only present to a small extent. Aside from bithalamic involvement and presence of a GC phenotype, no other radiological parameter including infratentorial involvement, necrosis, contrast enhancement, was associated with one subclass.
Outcome data were available for 24/32 patients (Supplementary Table 1). Most patients (22/24, 92%) died because of disease progression. Median overall survival (OS) was 16.0 months (range 10.9–28.2) with a 2 year survival rate of 33%. There was no patient surviving longer than five years. Compared with pedHGG-RTK2A, pedHGG-RTK2B tumors were associated with significantly inferior survival (pedHGG-RTK2A: median OS 21.2 months [IQR 14.2–34.0], pedHGG-RTK2B: 9.7 months [9.7–12.4]; p = 0.022; Fig. 2c). Signs of necrosis on MRI at diagnosis were associated with inferior OS as well (8.8 vs. 21.2 months; p = 0.001). Of all other clinical and radiological parameters tested (age [± 10 yrs], sex, primary GC, bithalamic-, bihemispheric- or infratentorial involvement, contrast enhancement at diagnosis), none had a significant effect on overall survival in univariate analysis. In multivariate analysis (18 pedHGG-RTK2A and 5 pedHGG-RTK2B) including all above mentioned parameters, only necrosis on MRI at diagnosis remained associated with a significantly inferior survival (Supplementary Table 3).
Distinct histopathological features of pedHGG-RTK2A and pedHGG-RTK2B
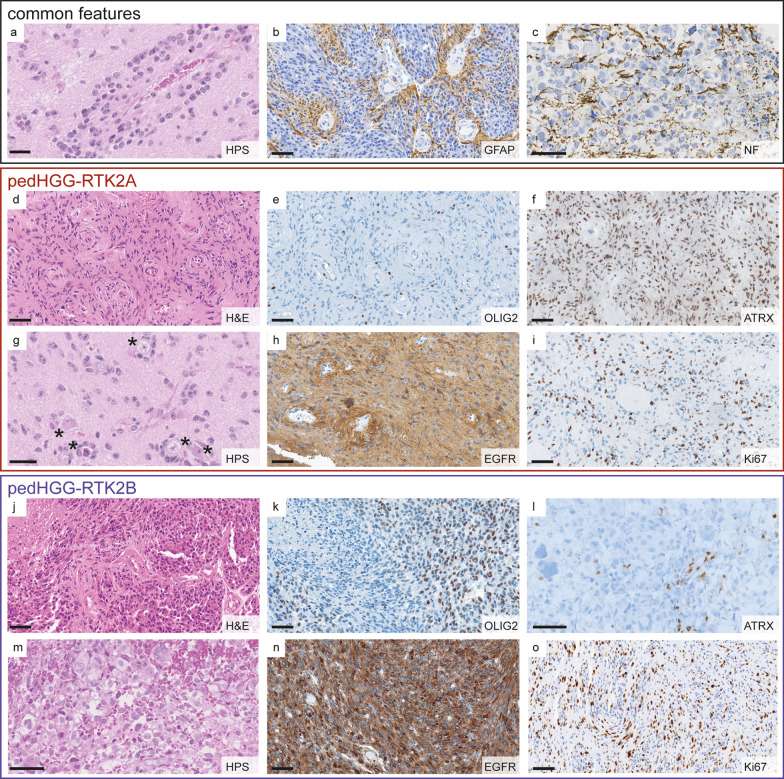
Both pedHGG-RTK2A and B subclasses presented as diffuse, moderately cellular gliomas, with frequent angiocentric growth patterns of tumor cells (pedHGG-RTK2A: 16/22 tumors, 73%, pedHGG-RTK2B: 4/6 tumors, 67%), easily identifiable on hematoxylin phloxin saffron stain (HPS) or hematoxylin and eosin stain (H&E) (Fig. 3a and Supplementary Table 4). All pedHGG-RTK2B (6/6) and most pedHGG-RTK2A (22/25, 88%) cases showed an astrocytic differentiation with expression of GFAP, in a diffuse pattern or with an angiocentric accentuation (Fig. 3b). The diffuse infiltrative behavior of the tumors was highlighted by neurofilament (NF) staining illustrating preexisting axons between the tumor cells (Fig. 3c). All tumors were negative for mutant IDH1-R132H protein. H3K27me3 expression was preserved (15/15 samples with available data) and, as expected, there was no expression of EZHIP (13/13 samples with available data) or mutant H3 G34R protein (16/16 samples with available data). There was no loss of expression of mismatch repair proteins in all tested pedHGG-RTK2A and B cases (14/14 samples).
Fig. 3.
Histopathological features in pedHGG-RTK2A/B. a–c Common angiocentric growth pattern of pedHGG-RTK2A and B a, astrocytic differentiation (GFAP expression). b diffuse infiltrating growth is highlighted by an axonal NF staining c. d–i Typical histological characteristics of pedHGG-RTK2A tumors d–i are OLIG2 negativity e, ATRX preservation f, perineuronal satellitosis (g; marked by stars), EGFR expression with angiocentric enhancement h and high Ki-67 index (20%). J–o pedHGG-RTK2B with heterogeneous OLIG2 expression (OLIG2 positive and OLIG2 negative tumor cell populations) k, loss of ARTX l, multinucleated giant cells m, diffuse EGFR positivity n and Ki-67 index of 15%. Scale bars; 20 µm a, g, 60 µm b–f, h–o
Some histopathological features appeared differently represented in pedHGG-RTK2A (Fig. 3d–i) versus pedHGG-RTK2B (Fig. 3j–o). Whereas pleomorphic xanthoastrocytoma (PXA)-like features were absent in pedHGG-RTK2B, three of 23 pedHGG-RTK2A cases (13%) exhibited pleomorphic cells with xanthomatous changes, reminiscent of PXA. However, no eosinophilic granular bodies, perivascular mononuclear inflammatory infiltrates or desmoplasia were observed. A subset of pedHGG-RTK2B cases presented giant multinucleated cells (2/6 tumors with available data; 33%) (Fig. 3m). Perineuronal satellitosis was frequently observed in pedHGG-RTK2A (16/19 tumors with available data, 84%) (Fig. 3g). Because most of the pedHGG-RTK2B samples did not contain cortical parenchyma, perineuronal satellitosis could not be evaluated in this subclass. Microcalcifications were less frequently observed in pedHGG-RTK2A (6/23 tumors with available data, 26%) than in pedHGG-RTK2B (3/7 tumors with available data, 43%). Immunohistochemical analysis revealed that all 22 cases of pedHGG-RTK2A with available data were negative for the transcription factor OLIG2 (Fig. 3e). In contrast, half of the pedHGG-RTK2B tumors expressed OLIG2 (3/6 tumors with available data, 50%) at least in a subset of tumor cells (Fig. 3k). In both subclasses, EGFR protein expression was commonly observed (Fig. 3h, n). Whereas all pedHGG-RTK2A but one (19/20 tumors with available data, 95%) diffusely expressed EGFR with focal stronger angiocentric staining, 71% (5/7) of pedHGG-RTK2B were positive for EGFR but angiocentric accentuation was absent. All cases of pedHGG-RTK2A with available data revealed preserved expression of ATRX (Fig. 3f). Interestingly, three of six (50%) pedHGG-RTK2B with available data showed a loss of ATRX expression (Fig. 3l). The proliferation index varied widely between tumors (pedHGG-RTK2A: 2 to 80% with a median of 10%, pedHGG-RTK2B: 5 to 40% with a median of 17.5%) (Fig. 3i, o).
DNA sequencing
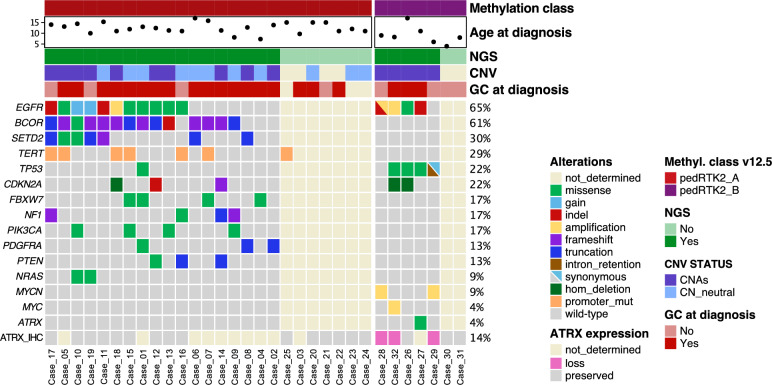
DNA-sequencing data was available for 23/32 (72%) tumors (pedHGG-RTK2A: 18, pedHGG-RTK2B: 5) (Fig. 4). In both epigenetically defined subclasses, alterations in EGFR—especially mutations—were frequently detected (pedHGG-RTK2A: 11/18, 61%; pedHGG-RTK2B 4/5, 80%). In other aspects, the subclasses differed substantially: recurrent altered genes in pedHGG-RTK2A comprised BCOR (14/18, 78%, especially frameshift and truncation mutations), SETD2 (7/18, 39%) and the promoter region of hTERT (7/19, 37%). In pedHGG-RTK2B, four of five (80%) tumors showed TP53 alterations and two tumors presented MYCN amplification (without germline alterations). In one sample an ATRX mutation was detected. Copy number variation (CNV) analyses derived from DNA-methylation profiling revealed that pedHGG-RTK2B presented consistently with CN alterations, mainly gains and amplifications (5/5 tumors with available data) whereas in pedHGG-RTK2A the CN variations were less common (10/21 tumors with available data). There was no difference in molecular features between tumors with a GC phenotype at diagnosis and those without.
Fig. 4.
Overview of the main clinical and molecular features in the pedHGG-RTK2A/B cohort. Epigenetically defined pedHGG-RTK2A (n = 25) and pedHGG-RTK2B (n = 7) cases are presented in columns and mutational status or clinical characteristics in rows. Molecular information was derived from DNA sequencing and DNA methylation array analyses, except for immunohistological data of ATRX expression. Fulfilled primary GC criteria on MRI (i.e., involvement of at least three contiguous cerebral lobes at diagnosis) is indicated as ‘GC at diagnosis’. Other abbreviations are as follow. CNV: copy-number variations; CNAs: copy-number alterations; CN_neutral: copy-number neutral; hom_deletion homozygous deletion; NGS: next-generation DNA sequencing; promoter_mut: promoter mutation; ATRX_IHC: ATRX expression assessed by immunohistochemistry
Discussion
Very few data are available in the literature concerning the clinical, radiological, and detailed histomolecular features of diffuse pedHGG H3-/IDH-WT. The current WHO classification states that three subtypes exist, characterized by PDGFRA (RTK1 subtype), EGFR (RTK2 subtype), and MYCN amplifications (MYCN subtype) [7]. However, these alterations are not constantly observed in these three subtypes and are not specific to each subgroup [7]. The DNA methylation-based CNS tumor classification (random forest classifier, version 12.5) identified different subclasses of pedHGG-RTK1 (A, B, and C) and -RTK2 (A and B) without definition of biological features or possible clinical implications. In this study, we showed that diffuse pedHGG-RTK2, subclasses A and B, presented distinct radiological and histomolecular features. The combination of the histomorphological, immunophenotypical and molecular features of pedHGG-RTK2B tumors including the presence of multinucleated tumor cells, loss of ATRX expression and frequent detection of TP53 alterations may be initially mistaken for diffuse hemispheric gliomas, H3 G34-mutant, but G34 mutations can be excluded by either immunohistochemistry or sequencing. Furthermore, the infiltration of midline structures is exceptional in diffuse hemispheric gliomas, H3 G34-mutant [8, 13]. Major histopathological features of pedHGG-RTK2A tumors such as their angiocentric growth pattern and OLIG2 immunonegativity are shared by angiocentric gliomas, which unlike pedHGG-RTK2A tumors are commonly restricted to one lobe of the brain (particularly the temporal or the frontal lobe) and show an indolent biology [10]. Still, angiocentric glioma may be a potential differential diagnosis in case of not fulfilled GC criteria (4/22 pedHGG-RTK2A in the current series with available radiological data). (Epi) genetic analyses should be performed for an accurate diagnosis.
In this work, we evidenced that diffuse pedHGG-RTK2, sublass A harbor EGFR amplifications and hTERT mutations less frequently than reported in a previous article (EGFR amplification and hTERT mutations were observed in 3% and 37% of our cohort, respectively, vs. 50% and 64% in pedHGG-RTK2 in [7]). There may be two reasons for these differences. First, our current series (n = 32) was partly selected based on their GC-like growth pattern. Secondly, in 2017, the description of the three distinct subclasses was based on the first version of the methylation classifier (v11b4). It is possible that the annotation to specific methylation subclasses of diffuse pedHGG have changed in the novel classifier 12.5. The current study used the v12.5 of the classifier which do no formally differ from v12.8. Moreover, the current classifier identified eight subclasses of these three subtypes of pedHGG, thus highlighting the increasingly complex epigenetic annotation of these pediatric gliomas. Our study demonstrated that no specific alterations define the pedHGG-RTK2, subclasses A and B, but recurrent alterations (EGFR, BCOR, hTERT and SETD2 alterations for pedHGG-RTK2A, and TP53, ATRX and EGFR alterations for pedHGG-RTK2B) were found in different frequencies. Because there are no defining alterations for the subclasses, specific variants cannot be used as surrogate markers for the epigenetically defined subclasses. Furthermore, EGFR alterations were reported in other types of diffuse pedHGG, including the novel subclass of diffuse midline gliomas (DMG), H3 K27-altered, EGFR-mutant [11, 16]. Whereas this tumor type seems to present frequently as thalamic or bithalamic tumors, some “supratentorial” cases have been reported [16]. Conversely, some tumors of the current series were solely bithalamic at diagnosis and a subset of previously reported bithalamic gliomas, EGFR-mutant reported by Mondal et al. were reclassified as pedHGG-RTK2 [16]. Fortunately, DMG, H3 K27-altered without H3 K27M mutation may be easily identified using immunohistochemistry (EZHIP immunopositivity and loss of H3K27me3 expression, these findings being absent in diffuse pedHGG-RTK2) [16, 18]. Interestingly, the particular radiological and histomolecular pattern of pedHGG-RTK2A (“GC”-like, angiocentric pattern, OLIG2 immunonegativity, EGFR and hTERT mutations) seems to be the same as in two previously reported cases [17]. In the presence of a GC-like diffuse glioma with aforementioned histopathologic and molecular markers there is a high probability of a pedHGG-RTK2A/B tumor after exclusion of diffuse hemispheric glioma, H3 G34-mutant and DMG, H3 K27-altered, EGFR-mutant.
Previously, patients with diffuse pedHGG-RTK2 tumors have been described with a significantly longer median OS (44 months, compared to RTK1 and MYCN subtypes, the latter being the most pejorative subtype) [7]. In our study, 22/24 patients died having a median survival period of 16 months (varying from 8 to 49 months after initial diagnosis). This may be partly due to the fact that the current series mainly comprised primary GC-like tumors (21/29 cases) which represent a disseminated form of glioma, contributing to a more grim prognosis [12]. Due to the large number of GC presentation in this series, we were not able to determine the prognostic relevance of a GC phenotype per se within the two subclasses. Overall, our study confirms the highly infiltrative potential of these tumors—especially pedHGG-RTK2A tumors seem to exhibit tendency to develop a frequent GC-like phenotype. Despite a potential inclusion bias (18/32 cases of the current study were selected from the European GC cohort), this GC-like phenotype was confirmed as frequent in the additional 14 other unselected cases. Whereas the subtypes of pedHGG-RTK1 or pedHGG-MYCN are associated with Constitutional Mismatch Repair Deficiency syndrome or Li-Fraumeni syndrome, no genetic tumor syndromes involving the CNS have been described to date in pedHGG-RTK2A/B [2, 10].
Our study also highlights that, in addition to DNA-methylation based classification, histopathological and radiological assessments are necessary to gain a comprehensive knowledge of tumors, and we assume that only this multimodal approach enables the development of specific therapeutic concepts. The significance of EGFR for the biological behavior of pedHGG-RTK2A/B tumors cannot be reasonably assessed based on our immunohistopathological and molecular data only. However, until preclinical models for these tumors are available, it might be worth investigating recurrent altered genes like EGFR as potential therapeutic targets. As our study cohort was small and consequently susceptible to biases, certain prognostic parameters such as methylation subclass or the presence of necrosis on MRI need to be confirmed by larger, prospective clinical studies.
To conclude, herein we performed a systematic histopathological, genetic and clinical analysis of diffuse pedHGG-RTK2A/B cases identifying recurrent radiological, histopathological and molecular features (summarized in Fig. 5). Further investigations are required, in particular concerning the clinical behavior and therapeutic implications of these rare tumors.
Fig. 5.
Summary of pedHGG-RTK2A/B main characteristics. Most frequent radiological, histolopathological and molecular features detected in the RTK2A (left) and RTK2B (right) subclasses are displayed. Abbreviations: pedHGG: pediatric high-grade glioma. T1 CE: T1-weighted image after application of a gadolinium based contrast agent
Supplementary Information
Acknowledgements
We would like to thank the laboratory technicians at GHU Paris Neuro Sainte-Anne Hospital and at the Institute of Neuropathology, University of Bonn Medical Center, for their assistance, and the SIOPE GC consortium. We thank the German and Styrian Childhood Cancer Foundation (‘Deutsche Kinderkrebsstiftung’, ‘Steirische Kinderkrebshilfe’) for financial support. DC and JG acknowledge the Necker Imagine Tumor and DNA biobank (BB-033-00065) for tumor provision. We thank Theresa Hammerl (Graz University of Technology) for technical support.
Author contributions
ATE, JG, GN, VDR, BB, DS, MB, TPe, DV, DJ, AMLM, MLG, JG, AOvB, MK, MH, CMK compiled the MRI and clinical records; ATE, LLF, FG, MA, AMe, TPi, PW, GHG and PV conducted the neuropathological examinations; JZ, AMa, YG, CJ, YA and DC conducted the molecular studies; GN and RK performed the statistical analyses; ATE, LLF, GN, VDR, BB, GHG, and PV drafted the manuscript; all authors reviewed the manuscript.
Funding
This study including the contribution of the Neuroradiological Reference Center in Augsburg (BB), the Neuropathological Reference Center in Bonn (TPi), and the HIT-HGG/GBM Studies (GN, TPe, MB, and CMK,) were supported by the Styrian- and German Childhood Cancer Foundation (‘Steirische Kinderkrebshilfe’, ‘Deutsche Kinderkrebsstiftung’), respectively. DC and JG acknowledge funding from ‘Etoile de Martin’ and ‘Les Boucles du Cœur de la Fondation Carrefour’ for the project RARE. C.J. acknowledges funding from the ‘Rudy A Menon Foundation’, ‘CRIS Cancer Foundation’, the ‘Ollie Young Foundation’, ‘Cancer Research UK’ (C13468/A23536, DRCRPG-Nov21\100002) and the Joshua Bembo Project, as well as NHS funding to the ICR/RMH Biomedical Research Centre, and is recipient of joint grant support from Izas la princesa guisante, L’union des rayons de soleils and the AYJ Fund. The Pediatric Cancer Center Barcelona (AMLM) acknowledges funding from the ‘Izas, la Princesa Guisante Foundation’. LLF acknowledges funding by the BONFOR program of the Medical Faculty of the University of Bonn (grant ID 2022–1A-09) and the Neuro-aCSis program, supported by the German Research Foundation (DFG) (grant ID 2024–12-03). DS acknowledges funding from PRIMUS/19/MED/06, Charles University Grant Agency, Czech Republic. MLG acknowledges support by the Italian Ministry of Health and by Regione Liguria (Ricerca Finalizzata di Rete NET-2019–12371188, GLI-HOPE). M.L.G. acknowledges support by the Italian Ministry of Health and by Regione Liguria (Ricerca Finalizzata di Rete NET-2019–12371188, GLI-HOPE). We remember and thank Prof. Felice Giangaspero for his all-life dedication to childhood brain tumors.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the Medical University of Graz (35–085 ex 22/23).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest directly related to the topic of this article.
Footnotes
Felice Giangaspero: Deceased.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Arnault Tauziède-Espariat, Lea L. Friker and Gunther Nussbaumer: Shared first authorship.
David Castel, Gerrit H. Gielen and Pascale Varlet: Shared last authorship.
References
- 1.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Hölsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Brück W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hänggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Mühleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Müller HL, Rutkowski S, von Hoff K, Frühwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu C-M, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blümcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schüller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A, Pfister SM (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerrini-Rousseau L, Tauziède-Espariat A, Castel D, Rouleau E, Sievers P, Saffroy R, Beccaria K, Blauwblomme T, Hasty L, Bourdeaut F, Grill J, Varlet P, Debily M-A (2023) Pediatric high-grade glioma MYCN is frequently associated with Li-Fraumeni syndrome. Acta Neuropathol Commun 11:3. 10.1186/s40478-022-01490-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.IBM Corp. Released (2020) IBM SPSS Statistics for Windows, Version 29.0. Armonk, NY: IBM Corp
- 4.International Agency for Research on Cancer, editor. WHO Classification of Tumours Editorial Board. Central nervous system tumours. 5th ed. Lyon (France); 2021
- 5.Jones C, Baker SJ (2014) Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 10.1038/nrc3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones C, Karajannis MA, Jones DTW, Kieran MW, Monje M, Baker SJ, Becher OJ, Cho Y-J, Gupta N, Hawkins C, Hargrave D, Haas-Kogan DA, Jabado N, Li X-N, Mueller S, Nicolaides T, Packer RJ, Persson AI, Phillips JJ, Simonds EF, Stafford JM, Tang Y, Pfister SM, Weiss WA (2017) Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro-Oncol 19:153–161. 10.1093/neuonc/now101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korshunov A, Schrimpf D, Ryzhova M, Sturm D, Chavez L, Hovestadt V, Sharma T, Habel A, Burford A, Jones C, Zheludkova O, Kumirova E, Kramm CM, Golanov A, Capper D, von Deimling A, Pfister SM, Jones DTW (2017) H3-/IDH-wild type pediatric glioblastoma is comprised of molecularly and prognostically distinct subtypes with associated oncogenic drivers. Acta Neuropathol 134:507–516. 10.1007/s00401-017-1710-1 [DOI] [PubMed] [Google Scholar]
- 8.Kurokawa R, Baba A, Kurokawa M, Pinarbasi ES, Makise N, Ota Y, Kim J, Srinivasan A, Moritani T (2022) Neuroimaging features of diffuse hemispheric glioma, H3 G34-mutant: a case series and systematic review. J Neuroim Off J Am Soc Neuroim 32:17–27. 10.1111/jon.12939 [DOI] [PubMed] [Google Scholar]
- 9.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109. 10.1007/s00401-007-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncol 23:1231–1251. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mondal G, Lee JC, Ravindranathan A, Villanueva-Meyer JE, Tran QT, Allen SJ, Barreto J, Gupta R, Doo P, Van Ziffle J, Onodera C, Devine P, Grenert JP, Samuel D, Li R, Metrock LK, Jin L-W, Antony R, Alashari M, Cheshier S, Whipple NS, Bruggers C, Raffel C, Gupta N, Kline CN, Reddy A, Banerjee A, Hall MD, Mehta MP, Khatib Z, Maher OM, Brathwaite C, Pekmezci M, Phillips JJ, Bollen AW, Tihan T, Lucas JT, Broniscer A, Berger MS, Perry A, Orr BA, Solomon DA (2020) Pediatric bithalamic gliomas have a distinct epigenetic signature and frequent EGFR exon 20 insertions resulting in potential sensitivity to targeted kinase inhibition. Acta Neuropathol 139:1071–1088. 10.1007/s00401-020-02155-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nussbaumer G, Benesch M, Grabovska Y, Mackay A, Castel D, Grill J, Alonso MM, Antonelli M, Bailey S, Baugh JN, Biassoni V, Blattner Johnson M, Broniscer A, Carai A, Colafati GS, Colditz N, Corbacioglu S, Crampsie S, Entz-Werle N, Eyrich M, Friker LL, Frühwald MC, Garrè ML, Gerber NU, Giangaspero F, Gil-da-Costa MJ, Graf N, Hargrave D, Hauser P, Herrlinger U, Hoffmann M, Hulleman E, Izquierdo E, Jacobs S, Karremann M, Kattamis A, Kebudi R, Kortmann R-D, Kwiecien R, Massimino M, Mastronuzzi A, Miele E, Morana G, Noack CM, Pentikainen V, Perwein T, Pfister SM, Pietsch T, Roka K, Rossi S, Rutkowski S, Schiavello E, Seidel C, Štěrba J, Sturm D, Sumerauer D, Tacke A, Temelso S, Valentini C, van Vuurden D, Varlet P, Veldhuijzen van Zanten SEM, Vinci M, von Bueren AO, Warmuth-Metz M, Wesseling P, Wiese M, Wolff JEA, Zamecnik J, Morales La Madrid A, Bison B, Gielen GH, Jones DTW, Jones C, Kramm CM (2024) Gliomatosis cerebri in children: a poor prognostic phenotype of diffuse gliomas with a distinct molecular profile. Neuro-Oncol. 10.1093/neuonc/noae080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picart T, Barritault M, Poncet D, Berner L-P, Izquierdo C, Tabouret E, Figarella-Branger D, Idbaïh A, Bielle F, Bourg V, Vandenbos FB, Moyal EC-J, Uro-Coste E, Guyotat J, Honnorat J, Gabut M, Meyronet D, Ducray F (2021) Characteristics of diffuse hemispheric gliomas, H3 G34-mutant in adults. Neuro-Oncol. 10.1093/noajnl/vdab061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priesterbach-Ackley LP, Boldt HB, Petersen JK, Bervoets N, Scheie D, Ulhøi BP, Gardberg M, Brännström T, Torp SH, Aronica E, Küsters B, den Dunnen WFA, de Vos FYFL, Wesseling P, de Leng WWJ, Kristensen BW (2020) Brain tumour diagnostics using a DNA methylation-based classifier as a diagnostic support tool. Neuropathol Appl Neurobiol 46:478–492. 10.1111/nan.12610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D-AK, Tönjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jäger N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Frühwald MC, Roggendorf W, Kramm C, Dürken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482:226–231. 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- 16.Sievers P, Sill M, Schrimpf D, Stichel D, Reuss DE, Sturm D, Hench J, Frank S, Krskova L, Vicha A, Zapotocky M, Bison B, Castel D, Grill J, Debily M-A, Harter PN, Snuderl M, Kramm CM, Reifenberger G, Korshunov A, Jabado N, Wesseling P, Wick W, Solomon DA, Perry A, Jacques TS, Jones C, Witt O, Pfister SM, von Deimling A, Jones DTW, Sahm F (2021) A subset of pediatric-type thalamic gliomas share a distinct DNA methylation profile, H3K27me3 loss and frequent alteration of EGFR. Neuro-Oncol 23:34–43. 10.1093/neuonc/noaa251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith HL, Collins J, Park D, Darlington W, Quezado M, Aldape K, Warnke P, Pytel P (2021) Pediatric gliomas presenting with gliomatosis-like spread, lack of contrast enhancement, EGFR mutation, and TERT promoter variants. J Neuropathol Exp Neurol 80:1134–1136. 10.1093/jnen/nlab093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tauziède-Espariat A, Debily M-A, Castel D, Grill J, Puget S, Roux A, Saffroy R, Gauchotte G, Wahler E, Hasty L, Chrétien F, Lechapt E, Dangouloff-Ros V, Boddaert N, Sievers P, Varlet P (2022) Deciphering the genetic and epigenetic landscape of pediatric bithalamic tumors. Brain Pathol Zurich Switz 32:e13039. 10.1111/bpa.13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team RC A language and environment for statistical computing
- 20.Therneau T. (2020) A Package for Survival Analysis in R. https://CRAN.R-project.org/package=survival
- 21.WHO Classification of Tumours Editorial Board. Central Nervous System tumours. Lyon (France): International Agency for Research on cancer; 2021
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.