Abstract
Interleukin-1 (IL-1) is a pro-inflammatory cytokine that exerts a wide range of neurological and immunological effects throughout the central nervous system (CNS) and is associated with the etiology of affective and cognitive disorders. The cognate receptor for IL-1, Interleukin-1 Receptor Type 1 (IL-1R1), is primarily expressed on non-neuronal cells (e.g., endothelial cells, choroidal cells, ventricular ependymal cells, astrocytes, etc.) throughout the brain. However, the presence and distribution of neuronal IL-1R1 (nIL-1R1) has been controversial. Here, for the first time, a novel genetic mouse line that allows for the visualization of IL-1R1 mRNA and protein expression (Il1r1GR/GR) was used to map all brain nuclei and determine the neurotransmitter systems which express nIL-1R1 in adult male mice. The direct responsiveness of nIL-1R1-expressing neurons to both inflammatory and physiological levels of IL-1β in vivo was tested. Neuronal IL-1R1 expression across the brain was found in discrete glutamatergic and serotonergic neuronal populations in the somatosensory cortex, piriform cortex, dentate gyrus, and dorsal raphe nucleus. Glutamatergic nIL-1R1 comprises most of the nIL-1R1 expression and, using Vglut2-Cre-Il1r1r/r mice, which restrict IL-1R1 expression to only glutamatergic neurons, an atlas of glutamatergic nIL-1R1 expression across the brain was generated. Analysis of functional outputs of these nIL-1R1-expressing nuclei, in both Il1r1GR/GR and Vglut2-Cre-Il1r1r/r mice, reveals IL-1R1+ nuclei primarily relate to sensory detection, processing, and relay pathways, mood regulation, and spatial/cognitive processing centers. Intracerebroventricular (i.c.v.) injections of IL-1 (20 ng) induces NFκB signaling in IL-1R1+ non-neuronal cells but not in IL-1R1+ neurons, and in Vglut2-Cre-Il1r1r/r mice IL-1 did not change gene expression in the dentate gyrus of the hippocampus (DG). GO pathway analysis of spatial RNA sequencing 1mo following restoration of nIL-1R1 in the DG neurons reveals IL-1R1 expression downregulates genes related to both synaptic function and mRNA binding while increasing select complement markers (C1ra, C1qb). Further, DG neurons exclusively express an alternatively spliced IL-1R Accessory protein isoform (IL-1RAcPb), a known synaptic adhesion molecule. Altogether, this study reveals a unique network of neurons that can respond directly to IL-1 via nIL-1R1 through non-autonomous transcriptional pathways; earmarking these circuits as potential neural substrates for immune signaling-triggered sensory, affective, and cognitive disorders.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12974-024-03287-1.
Introduction
Interleukin-1 (IL-1) is a critical pro-inflammatory cytokine that is involved in both health and disease. In disease, elevated levels of brain IL-1 are synonymous with neuroinflammation and are well known to alter HPA axis function, induce sickness behaviors, propagate inflammation through enhancing microglial function and peripheral immune cell trafficking into the CNS [33, 34], and contribute to neural damage via astrocyte ROS production [42]. Additionally, elevated IL-1 levels have also been associated with several affective and mood disorders along with decreased cognitive abilities [32]. However, in the absence of inflammation, basal IL-1 signaling is known to alter physiological brain functions, such as neuroendocrine activity [7], regulating normal sleep patterns [6, 28, 41], and aiding cognitive functioning [1, 56, 63] indicating the crucial role for IL-1 in normal CNS functions.
IL-1 signals through its sole receptor, the IL-1 Receptor Type 1 (IL-1R1), which we have previously found to be expressed in non-neuronal cells including brain endothelial cells, choroid plexus ependymal cells, tanycytes [8], and astrocytes [33]. These non-neuronal IL-1R1 expressing cells dictate most inflammatory processes during neuroinflammation mainly though canonical signaling pathways, such as p38α-MAPK and NFκB. These processes require IL-1R Accessory Protein (IL-1RAcP) to initiate intracellular signaling dependent on adaptor protein, MYD88, and induces the aforementioned signaling pathways. In situations like infection, injury, and stress, elevation of IL-1 levels can influence neural function and behavior through non-neuronal-derived inflammatory mediators, like ROS from astrocytes and prostaglandins from endothelial cells, leading to changes in behavior, such as sickness [20, 33], anxiety, and depressive-like behaviors [67]. On the other hand, although direct stimulation of neuronal IL-1R1 has been implicated in various neurophysiological activities, understanding how IL-1 directly affects neuronal activity and behavior through nIL-1R1, while avoiding the overt actions on non-neuronal cells, has been challenging [7, 40, 64]. An additional conceptual challenge is that neurons are thought to not express the canonical accessory protein, IL-1RAcP, and instead utilize an alternatively spliced isoform, IL-1RAcPb, which prevents MyD88-dependent signaling [53]. Therefore, nIL-1R1 can be utilized in an uncharacterized fashion compared to non-neuronal IL-1R1.
Previous investigations on nIL-1R1 distribution were incomplete and yielded controversial findings. Early studies utilizing in situ hybridization (ISH) or protein ligand binding (IL-1a, IL-1b, and IL-1ra) radiolabeling suggest IL-1 receptors exist only in choroid plexus and certain neurons of the brain. While these studies failed to detect most of the vascular IL-1R1, neuronal IL-1R1 was detected in the dentate gyrus (DG) and dorsal raphe nucleus [12, 18, 55]. Further ISH studies suggested that the primary IL-1R1 expressers are endothelial cells, choroid plexus cells, ependymal cells, and arcuate nucleus neurons. However, weak labeling in the dentate gyrus was discounted as an artifact of stuck riboprobe [17]. Follow up studies using immunohistochemical labeling produced discrepant results. For example, IL-1R1 was located on all hippocampal neurons [19], only in astrocytes and CA1 neurons but not dentate neurons [47], or in proliferating microglia [9], without detection of endothelial IL-1R1. These discrepant results demonstrate an urgent need to use better tools to accurately describe the expression patterns of IL-1R1.
In addition to clarifying non-neuronal IL-1R1 expressing cell types, we have previously identified and confirmed that IL-1R1 is expressed in neurons (nIL-1R1)—the highest levels being granule neurons of the DG [33]. Using conditional restoration or deletion of IL-1R1, we have shown that these nIL-1R1-expressing DG neurons are necessary and sufficient for stress-induced social behavior abnormalities, working memory dysfunctions, and stress-sensitization [14, 15, 22], which highlights that nIL-1R1 on these DG neurons is crucial for the induction of pathological behavioral responses in the presence of stress-induced neuroinflammation. IL-1R1 in dorsal raphe nucleus neurons is known to increase serotonin reuptake through enhanced SERT activity via a P38-MAPK dependent mechanisms [2, 67]. It is still not clear, however, what other neural circuits and neurotransmitter systems in the brain express nIL-1R1.
The lack of a comprehensive mapping of nIL-1R1 is a major gap in our knowledge, because nIL-1R1+ neurons could be vulnerable targets during neuroinflammation and become dysregulated. In addition, the mechanism by which nIL-1R1 affects neurons could involve non-conventional IL-1R1 signaling. By addressing these outstanding issues, we can contribute towards an enhanced understanding of IL-1-mediated pathogenesis and, in the future, identify circuit-specific behaviors that should be studied to examine IL-1-induced CNS effects. Here, global and cell-type specific IL-1R1 reporter mice were used to generate the most detailed map of brain regions, neural circuits, and neurotransmitter systems that express nIL-1R1. Furthermore, we demonstrate that nIL-1R1 does not affect neurocircuits through conventional intracellular IL-1R1 signaling pathways.
Materials and methods
Experimental models and subject details
Mice
Male C57BL/6 (6–8 weeks old) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and maintained in-house until desired ages. All other mouse lines were bred in-house. The IL-1R1-deficient Il1r1r/r mice (C57BL/6N-Il1r1tm1Quan/J, JAX 024101), IL-1R1 reporter mice (Il1r1GR/GR) and multiple cell-type-specific mouse lines, including Tie2-Cre-Il1r1r/r (JAX 004128), Camk2a-Cre-Il1r1r/r (JAX 005359) and Vglut2-Cre-Il1r1r/r (JAX 016963) mice were generated by our lab previously [33]. Gad2-Cre (Gad2tm2(cre)Zjh/J, JAX 010802), ePet1-Cre (B6.Cg-Tg(Fev-cre)1Esd/J, JAX 012712), NFκBGFP (FVB.Cg-Tg(HIV-EGFP, luc)8Tsb/J, JAX 027529) transgenic mice were purchased from The Jackson Laboratory and crossed to either Il1r1r/r or Il1r1GR/GR mice. IL-1 receptor AcPb knockout (Acpb−/−) mice were provided by Amgen (Seattle, WA). All mice were genotyped from tail snips by Transnetyx (Cordova, TN). Mice were housed under standard 12 h light/dark cycle conditions in rooms with controlled temperature and humidity. They were given standard rodent chow and sterilized tap water ad libitum. Male mice at 8 to 12 weeks of age were used for all experiments. All experiments were approved by the Florida Atlantic University Institutional Animal Care and Use Committee.
Surgical procedures
Mice were anesthetized with 5% isoflurane induction, placed in the stereotaxic frame (David Kopf Instruments), and maintained at 1.0 − 1.5% isoflurane during surgery. Coordinates for craniotomy were precisely located by a motorized stereotaxic instrument (Neurostar GmbH). The following coordinates were used: for lateral ventricle injections, AP, -0.6; ML, -1.0; DV, -2.5; for intra-hippocampus injections, AP, +/-1.8; ML, +/-1.8; DV, -1.7. 20 ng recombinant mouse IL-1β (R&D System, Cat 401-ML-005) in 2 µl of saline, 1 × 109 GC AAV2-CMV-Cre-eGFP (Cat: #105530-AAV2, RRID: Addgene_105545) or 1 × 109 GC AAV2-CMV-eGFP (Cat# 105545-AAV2, RRID: Addgene_105530) in 1 µl of saline was injected at a rate of 0.1 µl/min with a pulled glass capillary affixed to a nanoinjector. The glass pipette was left in place for 5 additional minutes and then slowly withdrawn (1 mm/min). Wounds were closed with surgical glue (Vetbond, 3 M). For post-operative pain management, mice received a single dose of intraperitoneal injection of buprenorphine-SR (0.36 mg/kg) after the surgery.
Tissue Preparation
Mice were euthanized with carbon dioxide or by overdose of isoflurane and perfused transcardially with ice cold 1 x PBS (pH 7.4), followed by ice cold 4% paraformaldehyde (Sigma-Aldrich, Cat # P6148) in phosphate buffer or ice cold glyoxal (Sigma-Aldrich, Cat# 128465) fixative (9% glyoxal/8% Acetic acid, pH 4) for labeling of IL-1R1-HA-tag [29]. Brains were removed, postfixed in their respective fixative for 24 h and then equilibrated in a cryoprotective solution of 30% sucrose at 4 °C for 24 h. Brains were frozen over dry ice and 40-µm-thick coronal or sagittal sections were generated with a cryostat according to standardized collection procedures [66] and stored in cryoprotectant (30% v/v Polyethylene glycol, 30% v/v ethylene glycol in 0.1 M PB) until use. Every 12th section collected was used for anatomical analysis.
Immunofluorescence labeling, processing, and imaging
Sections were washed in PBS, blocked with 5% normal donkey serum (1% BSA, 0.1% TritonX in PBS), and incubated with primary antibodies: rabbit-anti-Iba1 (Wako Chemicals, Cat# 019-19741, 1:1000), goat-anti-GFAP (Abcam, Cat# ab53554, 1:500), rabbit-anti-NeuN (MilliporeSigma, Cat# ABN78, 1:1000), rabbit-anti-RFP (Abcam, Cat# ab124754, 1:1000), goat-anti-tdTomato (OriGene, Cat# AB8181, 1:1000), goat-anti-ChAT (Abcam, Cat# ab53554, 1:300), rabbit-anti-TH (Millipore Sigma, Cat# AB152, 1:500), guinea pig-anti-Vglut2 (Millipore Sigma, Cat# AB2251-I, 1:500), chicken-anti-GFP ( Abcam, Cat#Ab13970, 1:500) or rabbit-anti-Tph2 antibody (Millipore Sigma, Cat# ABN60, 1:400). Primary antibody incubations were completed overnight at 4 °C. For HA labeling, glyoxal fixed sections were washed in 0.1% Triton-X in PBS (PBST) and then incubated in rabbit-anti-HA (Cell Signaling Technology, Cat# 3724, 1:500) and goat-anti-tdTomato (Origene, 1:1000) primary antibody solution overnight at RT. Sections were then washed in PBST and incubated with a fluorochrome-conjugated secondary antibody (Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 647, Thermofisher Scientific). Sections were cover-slipped with Vectashield with DAPI (#H-1500-10, Vector Laboratories) or Prolong Gold Antifade with DAPI (Invitrogen, Cat# P3697). Representative images were captured using a Nikon A1R confocal microscope. Multiple-channel images were overlaid and each stack was z-projected using ImageJ software (NIH, Bethesda, MD). For large area imaging, series of images were acquired by tile scans using a Leica TCS SP8 confocal microscope and stitched by the Leica Application Suite X software to produce a collage of brain sections. Colocalization was assessed using ImageJ by a trained experimenter.
Immunohistochemistry labeling, processing, and imaging
Sections were washed in PBS, incubated in 0.6% H2O2 solution in PBS for 30 min, washed in PBS, then blocked with 5% normal donkey serum (1% BSA, 0.1%TritonX in PBS) for 1 h and incubated with goat-anti-tdTomato (Origene, 1:1000). Primary antibody incubations were completed overnight at 4 °C. Sections were then washed in PBS and incubated with a biotinylated donkey-anti-rabbit secondary antibody (Jackson Immunoresearch, Cat# 711-035-152, 1:500). Sections were washed and then incubated in VECTASTAIN® Elite® ABC-HRP Kit, Peroxidase (Vector Labs, Cat# PK-6100) for 2 h according to manufacturer’s specification. Sections were washed and incubated in SIGMAFAST™ 3,3′-Diaminobenzidine solution (Sigma-Aldrich, Cat# D4418) for 7 min for visualization. Sections were plated on SuperFrost Plus slides (Fisher Scientific, Cat # 22-037-246) and cover-slipped with DPX (Sigma-Aldrich, Cat# 06522). 10x bright field tilescan images were acquired with BZ-X710 Keyence Microscope (Keyence, Itasca, IL) and stitched using Keyence Image analysis software.
Linear and non-linear registration
A modified QUINT workflow was used for determining Il1r1+ brain regions [62]. For linear registration, both fluorescent and DAB-stained sections were loaded into DeepSlice (https://www.deepslice.com.au/, RRID: SCR_023854,) for first round linear registration [10]. Results from DeepSlice linear registration were corrected by hand in QuickNII (QuickNII, RRID: SCR_016854). Non-linear registration was conducted in Visualign (Visualign, RRID: SCR_017978) using large anatomical structures to correct for warping or size of tissue samples [45]. An experienced experimenter then used the non-linear registration and Allen Atlas (v2017) overlay to identify tdTomato expressing brain regions. All sections were annotated using Visualign and confirmed with the Allen Brain Reference Atlas (Allen Brain Institute).
Golgi stain
Following the protocol outlined by FDNeurotech (FDNeurotechnologies, 2019), brains were extracted from male 12wk old WT and Il1r1r/r mice and incubated in a 1:1 ratio of impregnation solutions at RT for two weeks in the dark. After incubation, the brains were frozen in -80 °C isopentane. Subsequently, 100 μm sections were collected onto gelatin-coated slides (FDNeurotech, Cat #PO101) using a cryostat. The sections were stained using FDNeurotech solutions as recommended by the manufacturer. Following staining, the sections were dehydrated through successive rinses in increasing concentrations of ethanol and cleared with xylene. Finally, the slides were coverslipped using Eukitt mounting medium (Sigma Aldrich, Cat#03989).
Arborization and spine density
The neurons within the granule cell layer of the dentate gyrus in the hippocampus were imaged utilizing a brightfield camera attached to a Nikon A1 confocal microscope. Imaging was performed using a 60X magnification oil-immersion objective to capture z-stack images of individual neurons. Five neurons per animal were imaged. Each neuron was traced using Neurolucida 360 (MBF Bioscience, Williston, VT), and dendritic arborization as indicated by increasing branch order was analyzed and quantified with Neurolucida Explorer software. Second order branches were then cropped, and spine density analysis (spines/µm) was performed with Neurolucida 360.
RNA Isolation and Real-Time PCR
Mouse hippocampus was dissected from the brains on ice. Tissues were transferred to sterile Eppendorf tubes, suspended in the TRIzol reagent (Thermofisher Scientific, Cat# 15596026), and homogenized with a sonicator. Total RNA was isolated following the manufacture’s instruction and purity was determined with a Nanodrop spectrophotometer (Denville Scientific). RNAs were reverse transcribed into cDNAs using a Reverse Transcription Kit (Promega, Cat# A3500). To detect the restored IL-1R1 mRNA, SYBR qPCR was performed using the following primers, which detect RNA sequence between IL-1R1 Exon IX and Exon X: EX-F, 5′-CCTCACGGCTACAATTGTATGC-3′ and EX-R, 5′-CAAACTGTCCCTCCAAGACC-3′. qPCR was performed using the ABI PRISM 7500 sequence detection system (Applied Biosystems) and results were normalized based on reference cDNA levels (GAPDH). Relative gene expression was analyzed using the ΔΔCT method and results are expressed as fold difference from GAPDH.
Brain tissue dissection for RNA sequencing
For dentate gyrus RNA sequencing, RNA was extracted from microdissected regions of dentate gyrus in hippocampal slices from i.c.v IL-1 injected Vglut2-Cre-Il1r1r/r. At time of euthanasia, mouse brains were rapidly removed and snap-frozen in dry-ice cooled isopentane. Fresh-frozen tissue was cryosectioned at a thickness of 10 μm onto charged slides. Under a dissecting microscope, dentate gyrus granule cell layers were microdissected based on their optical density, immediately lysed, and RNA was extracted (PicoPure RNA extraction kit Cat# KIT0204, Thermofisher Scientific). RNA integrity was confirmed by Agilent BioAnalyzer and preamplification cycles were determined using expression of housekeeping genes (Gapdh, Actb).
For other brain regions dissection, each mouse brain was quickly removed and transferred to 35 mm petri dish containing 3 ml PBS at the time of euthanasia. Prefrontal cortex, hippocampus, midbrain and cerebellum were dissected out and collected in separate Eppendorf tubes under 5 min. All brain tissues were snap-frozen in dry-ice cooled isopentane. Total RNA was extracted from fresh frozen tissue samples using Qiagen RNeasy Plus Universal mini kit following manufacturer’s instructions (Qiagen, Hilden, Germany). Extracted RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and RNA integrity was checked using Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA).
RNA sequencing and data analysis
For microdissected dentate gyrus granule cells, RNA-seq was performed at the Hussman Institute for Human Genomics Sequencing Core Facility (University of Miami, Miami, FL). In brief, RNA-Seq libraries were prepared using the Ovation SoLo RNA-Seq System with AnyDeplete rRNA to remove rRNA and other abundant transcripts according to the manufacturer’s recommendation (Nugen). RNA-Seq libraries were run on an Illumina NextSeq 500 sequencing instrument according to the protocols described by the manufacturer. FASTQ files were aligned to the mouse mm10 genome using STAR Aligner [16]. Raw counts were normalized and differentially expressed genes were determined using the DESeq2 Bioconductor package in R [35].
For whole hippocampus and other brain regions from WT mice, RNA sequencing libraries were prepared using the NEB Next Ultra II RNA Library Prep Kit for Illumina with manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNAs were first enriched with Oligo (dT) beads. Enriched mRNAs were fragmented for 15 min at 94 °C. First strand and second strand cDNAs were subsequently synthesized. cDNA fragments were end repaired and adenylated at 3’ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by limited-cycle PCR. The sequencing libraries were validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA), and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA).
For alternative splicing analysis, aligned bam files (mm10) from bulk RNA-seq experiments were used to generate Sashimi plots with Integrative Genomics Viewer (IGV) [48].
Bioinformatic analysis of scRNA-seq data
The bioinformatics analysis was performed using the publicly available scRNA-seq database generated from different mouse and human brain regions in three previous studies [24, 26, 50], using counting matrix and cell clustering information provided by the authors deposited at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE71585, GSE134163 and GSE116470. Briefly, cells with Il1r1 transcripts were tagged, classified based on cell clustering information provided in the corresponding previous studies, counted and presented as the percentage of IL-1R1+ cells in all cells or subclustered cell population.
Spatial Transcriptomics
To perform Nanostring Digital Spatial Profiling (DSP)®, brains from Il1r1r/r mice 1mo following AAV-injections were fixed in 4% paraformaldehyde, postfixed in 4% paraformaldehyde for 24 h at 4 °C, and subsequently cryoprotected in a 10%, 20%, then 30% sucrose solution each for 24 h. The brains were embedded in optimal cutting temperature (OCT) compound and frozen on dry ice. Cryosections of 10 μm thickness were prepared using a cryostat and mounted on Superfrost Plus slides. For section staining, the tissue sections were permeabilized with 0.1% Triton X-100 in PBS for 10 min and then blocked with 5% normal goat serum in PBS for 1 h at room temperature. The sections were incubated overnight at 4 °C with primary antibodies anti-NeuN (1:1000, Millipore ABN78) and anti-GFP (1:100, Invitrogen A21311) in blocking solution. Following primary antibody incubation, the sections were washed with PBS and incubated with fluorophore-conjugated secondary antibodies (Alexa Fluor 488 and Alexa Fluor 594, Invitrogen) for 1 h at room temperature. After additional washing, DSP probes were applied according to the manufacturer’s protocol. The prepared slides were then loaded onto the Nanostring GeoMx DSP instrument, and regions of interest (ROIs) were selected based on segmentation of fluorescence imaging to identify GFP+NeuN+ neurons of the dentate gyrus. UV light was used to photocleave oligonucleotide tags from the probes within these ROIs. The oligonucleotide tags were collected, sequenced, and aligned using Mouse Whole Transcriptome Atlas (mWTA). Data was normalized to third quartile (Q3) and assay quality control with a criterion of limit of quantitation (LOQ) in at least 10% of the ROI using the negative probes as background. No samples were detected below the 50% LOQ warning limit. The normalized data were analyzed using Nanostring’s analysis software. Left vs. right DGs of the same animal were compared using a paired t-test with Benjamin-Hochberg correction to identify differentially expressed genes which were enriched in GFP+NeuN+ cells in either Cre+ or Cre neurons. A p-value less than 0.05 was set as criteria for genes to be used in Gene Ontology enrichment analysis. Genes were entered into the GO software and biological process, cellular component, and molecular functions pathways were identified for both left (GFP) vs. right sides (Cre-GFP) of the DG neurons (https://geneontology.org/).
Basescope in situ hybridization
For analysis of Il1RAcP isoform distribution, Basescope in situ hybridization (ISH) was utilized (ACDBio, Newark, CA). 10 μm thick hippocampal sections from paraformaldehyde fixed WT and Il1rAcPbKO mice were generated via cryostat. Antigen retrieval for 5 min at 90 °C in antigen retrieval buffer (ACDBio, Cat#322000) was conducted prior to ISH. Sections were incubated with custom 1 ZZ probes targeting 1519–1560 bp of NM_008364.2 (BA-Mm-Il1rap-tv1-E10E11) to detect Il1rAcP (blue-green) and a 1 ZZ probe targeting 1520–1563 bp of NM_001159318.1 (BA-Mm-Il1rap-tv4-E10E11) to detect Il1rAcPb (red). The AcPb signal was detected following a 10 min incubation with Fast Red and detection of the AcP signal was detected following 15 min incubation with Fast Green.
Statistical analysis
All data are expressed as treatment means ± standard error of the mean (SEM). Standard one-way or two-way ANOVA was used to analyze data. When appropriate, significant main and interaction effects were subjected to F-protected LSD test for post hoc analyses for further comparison. Observations of more than three interquartile ranges from the first and third quartile were excluded from analyses. An alpha level of p < 0.05 was used as the criterion for the rejection of the null hypothesis. Data were analyzed using GraphPad Prism statistical software (GraphPad Prism version 8.0.1 for Windows, GraphPad Software, Boston, Massachusetts USA, www.graphpad.com) or Nanostring DSP platform (Nanostring, Seattle, WA).
Contact for reagents and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ning Quan (nquan@health.fau.edu).
Results
Potential neuronal IL-1R1 expression throughout the brain
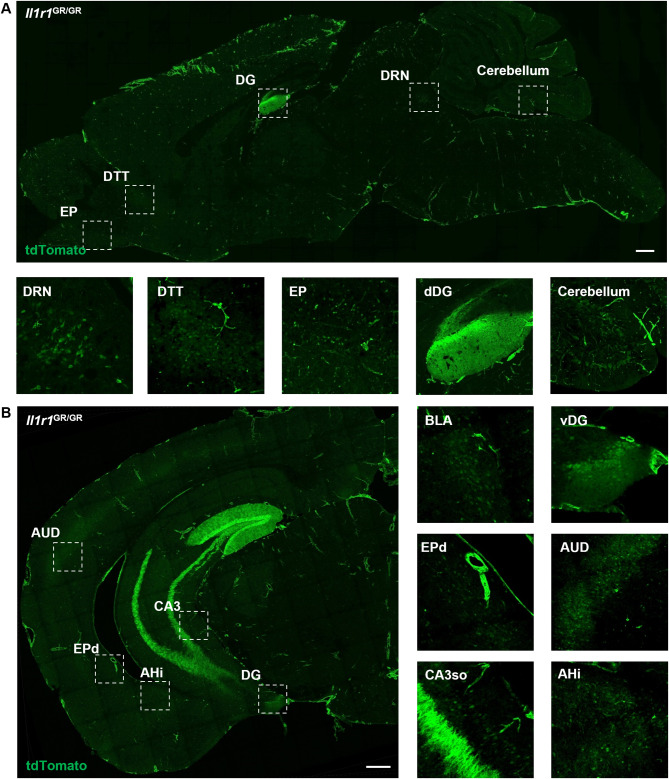
First, we determined the general expression patterns of potential neuronal IL-1R1 in the brain of adult male mice. The Il1r1 reporter mouse (Il1r1GR/GR) co-expresses Il1r1 mRNA and tdTomato [34] and was previously used to identify Il1r1 expressing cell types in the brain [33]. In regard to potential neuronal IL-1R1 (nIL-1R1) expression, nIL-1R1 was found to be highly expressed in the dentate gyrus (DG) of the hippocampus. Beyond the hippocampus, we identified and annotated potential Il1r1 expressing nuclei throughout the entire brain of the Il1r1GR/GR mice. An anti-RFP antibody, which is cross-reactive to endogenous tdTomato, was used to amplify the tdTomato signal that is co-expressed with Il1r1 mRNA and was pseudo-colored green for visualization purposes. Sagittal sections of Il1r1GR/GR brains reveal distinct neuronal populations which express nIl1r1 (Fig. 1A). White boxes indicate regions where nIl1r1 was annotated, such as the dorsal raphe nucleus (DRN), dorsal tenia tecta (DTT), external plexiform layer of the olfactory bulb (EP), dentate gyrus (DG), and cerebellum (Fig. 1A, below). Further annotation of nIl1r1-expressing regions throughout the brain was performed using the QUINT pathway [62] and the Allen Brain Atlas (Allen Reference Atlas – Mouse Brain [brain atlas]. Available from atlas.brain-map.org). Linear and non-linear registrations of coronal sections were performed on tilescan images of tdTomato immunofluorescent labeling in Il1r1GR/GR mice, revealing additional nIl1r1 expressing nuclei (Fig. 1B). White dashed boxes indicate regions with prominent Il1r1 expression; for example, the auditory cortex region (AUD), granule layer of the dorsal DG (dDG), ventral DG (vDG), dorsal endopiriform nucleus (EPd), the amygdalohippocampal area (AHi ) and cornu ammonis field 3 strata oriens (CA3so) but not the CA3 strata lacunosum-moleculare, radiatum, lucidum, or pyramidale (CA3slm, ra, sp, luc, py) (Fig. 1B, below). Table 1 lists all regions of the brain in which potential nIL-1R1-expressing nuclei were detected, ordered from anterior to posterior. Annotated coronal brain sections from Il1r1GR/GR mice following non-linear registration were used to generate Table 1.
Fig. 1.
Representative images of neuronal IL-1R1 expression in the brain. Representative tilescan images of tdTomato labeling of a sagittal (A) and coronal (B) section from an Il1r1GR/GR mouse. Areas where high levels of neuronal IL-1R1 were found are marked with dashed squares and shown at higher magnification below (A) or to the right of the low-magnification image (B). Scale bar: 500 μm. DRN, dorsal raphe nucleus; DTT, dorsal tenia tecta; EP, endopiriform nucleus; dDG, dorsal dentate gyrus; vDG, ventral dentate gyrus; AUD, auditory cortex; CA3, Cornus ammonus 3; AHi, hippocampal-amygdalar transition area
Table 1.
Annotation of all nIl1r1-expressing nuclei in the Il1r1GR/GR mouse brain
| Abbreviations | Brain Regions (Anterior to Posterior) |
|---|---|
| MiA | Mitral cell layer of the main olfactory |
| AONd,1 | Anterior olfactory nucleus, dorsal part |
| MOs5,6a | Secondary motor area, layer 5 |
| PIR | Piriform area |
| MOp5 | Primary motor area, layer 5 |
| EPd | Endopiriform nucleus, dorsal part |
| PL5 | Prelimbic area, layer 6 |
| ILA6 | Infralimbic area, layer 6 |
| SSp5 | Primary somatosensory area, layer 5 |
| LSr | Lateral septal nucleus, rostral |
| ORBvl6a, l6a | Orbital area, ventrolateral part, layer 6a; lateral part, layer 6a |
| SSp4 | Primary somatosensory area, layer 4 |
| DTT/VTT | Dorsal taenia tecta/ Ventral taenia tecta |
| ACAd/v6a | Anterior cingulate area, dorsal part, layer 6a |
| GU2/3 | Gustatory areas, layer 2/3 |
| SSs5 | Supplemental somatosensory area, layer 5 |
| VISC5 | Visceral area, layer 5 |
| EPv | Endopiriform nucleus, ventral part |
| LSv | Lateral septal nucleus, rostral |
| PVH | Paraventricular hypothalamic nucleus |
| fi | Fimbria of the hippocampus |
| CLA | Claustrum |
| ArcL | Arcuate hypothalamic nucleus, lateral part |
| DG | Dentate gyrus, granule cell layer |
| AD | Anterodorsal nucleus of thalamus |
| MD | Mediodorsal nucleus of thalamus, lateral part |
| EPl | Endopiriform nucleus, lateral part |
| SSs4 | Supplemental somatosensory area, layer 4 |
| VISC2/3 | Visceral area, layer 2/3 |
| VMHdm | Ventromedial hypothalamic nucleus, dorsomedial part |
| BLAa | Basolateral amygdalar nucleus, anterior part |
| CA3so | CA3 stratum oriens |
| AUDp4, v4 | Primary/Ventral auditory area, layer 4 |
| ENTl5 | Entorhinal area, lateral part, layer 5 |
| BMAa | Basomedial amygdalar nucleus, anterior part |
| VISp4, rl4, al4 | Primary/Rostrolateral/Anterolateral visual area, layer 4 |
| MGd/v | Medial geniculate complex, dorsal/ventral |
| AHi | Hippocampal-amygdalar tranisiton area |
| RPa | raphe pallidus nucleus |
| AUDd4 | Dorsal auditory area, layer 4 |
| ENTm1,m2,m5 | Entorhinal area, medial part, dorsal zone, layer 2 |
| AUDpo4 | Posterior auditory area, layer 4 |
| DRN, DRD, DRVL, DRI | Dorsal raphe nucleus, dorsal/ventral/interfascicular part |
| PAG | Periaquaductal grey |
| CS | Superior central raphe nucleus (median raphe nucleus) |
| FLgr | Flocculus, Granular and Purkinje layers of cerebellum |
Due to the tdTomato reporter being under the control of endogenous Il1r1 promoters, some of the neuronal tdTomato expression was weak and could be misconstrued as background if not compared with a negative control. To prevent errors in our reporting, we confirmed that the nIl1r1 expression was not an artifact of our genetic mouse model or labeling methods by restricting IL-1R1 expression to endothelial cells with the Tie2-Cre promoter in our Il1r1r/r mouse line. Analysis of the whole brains of Tie2-Cre-Il1r1r/r mice show endothelial tdTomato expression is present throughout the entire brain. However, reported regions of nIl1r1 expression observed in Il1r1GR/GR were not present in the Tie2-Cre-Il1r1r/r sections such as the Pir, SSp, and DTT (Supplementary Fig. 1A-B).
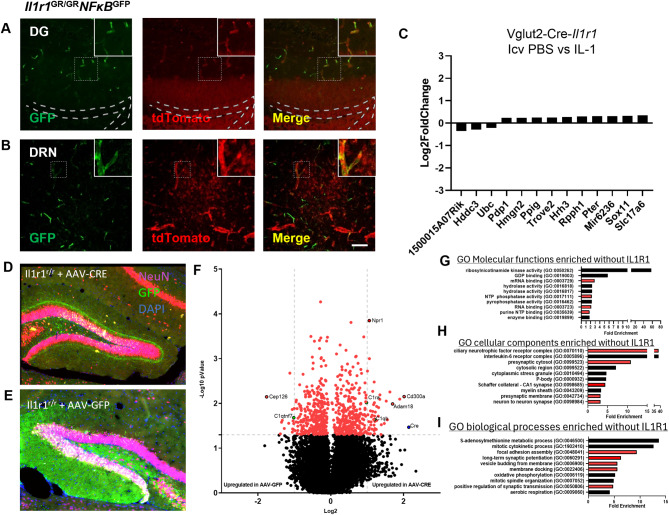
As proof of concept that IL-1R1 is typically expressed in specific sub-populations of neurons and can be selectively restored, we injected an adeno-associated virus co-expressing Cre and GFP (AAV2-Cre-GFP) into the major IL-1R1 expressing regions of the brain, the DG and DRN, of the Il1r1r/r mice. Following unilateral injection of AAV2-Cre-GFP, expression of IL-1R1 can be restored on one half of the DG as indicated by tdTomato fluorescence (Supplementary Fig. 1C, left side of image) and was absent in the non-injected contralateral side (right side of image). Furthermore, this targeted Cre expression in the DG or DRN restored IL-1R1 expression as indicated by tdTomato expression in those neurons (Supplemental Fig. 1D and E). However, not all GFP expressing neurons were tdTomato+, indicating that not all neurons which express Cre co-express nIL-1R1. Therefore, these data indicate that only neurons which typically use endogenous Il1r1 promoters in these discrete brain regions have the ability to express IL-1R1 and that viral-mediated nIL-1R1 restoration strategies can be implemented to decipher the function of nIL-1R1 in various brain regions. To gain an understanding of the specificity of the neuronal Il1r1-expressing circuits we analyzed the nIl1r1 distribution data from two perspectives: (1) functional neuroanatomy and (2) neurotransmitter-usage.
nIL-1R1 is primarily expressed on sensory and emotional processing circuits
All nIl1r1-expressing brain regions (n = 46) identified within the Il1r1GR/GR mice were classified as one of the following categories based on functional neuroanatomy: (1) sensory detection, relay, and processing; (2) emotional regulation, (3) spatial and cognitive Processes, (4) neuroendocrine response and (5) miscellaneous. Based on total nIl1r1 nuclei, 50% were related to sensory detection, relay, and processing; 26% were related to emotional regulation; 8.7% were related to spatial and cognitive processes, 5.5% were related to neuroendocrine responses, and 9% were determined miscellaneous (Supplementary Fig. 1F). These data point towards the sensory processing and emotional regulation nuclei as the key brain regions in which nIl1r1 can modify.
nIl1r1 is expressed in various, but limited, neurotransmitter systems
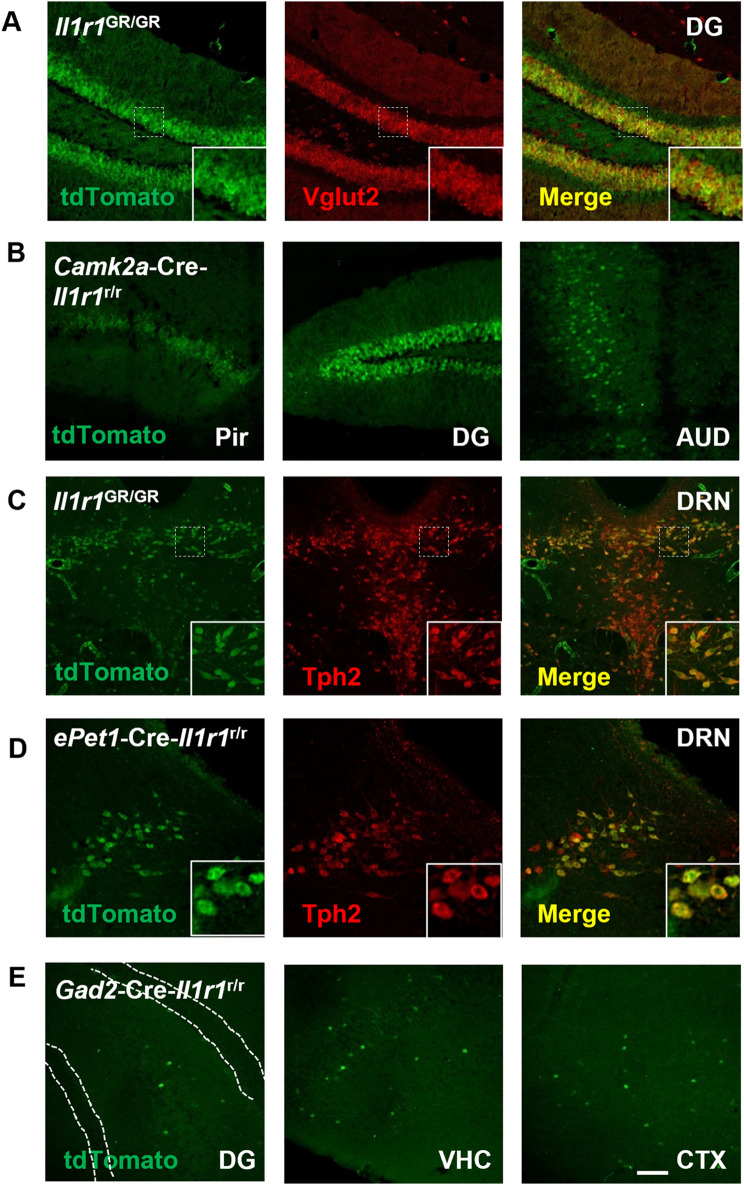
We next aimed to determine whether particular neurotransmitter systems express nIl1r1. To confirm the observed potential nIl1r1 expression are indeed neurons in specific neurotransmitter systems we (1) co-localized Il1r1 (tdTomato) and various neurotransmitter markers in the Il1r1GR/GR mice and (2) restored IL-1R1 expression with specific neurotransmitter-related Cre expression. In the dentate gyrus and basolateral amygdala of the Il1r1GR/GR mice, immunofluorescent labeling of tdTomato and vesicular glutamate transporter 2, Vglut2, is colocalized (Fig. 2A and Supplementary Fig. 2A, respectively). Restoration of IL-1R1 expression to excitatory forebrain neurons, using Camk2a-Cre-Il1r1r/r mice, revealed neuronal tdTomato expression in regions such as the Pir, DG, and AUD in the absence of endothelial Il1r1 (Fig. 2B). These data establish the nIl1r1 is primarily expressed in glutamatergic neurons.
Fig. 2.
Neuronal IL-1R1 is expressed in diverse types of neurons classified by different neurotransmitters. (A) Representative images of tdTomato and Vglut2 labeling in Il1r1GR/GR hippocampal sections. Dashed squares mark the area shown at higher magnification on the bottom right. (B) Representative images of tdTomato from different CamK2a-Cre-Il1r1r/r brain sections. Pir, Piriform cortex; DG, dentate gyrus; AUD, primary auditory cortex. (C) Representative images of tdTomato and Tph2 labeling in Il1r1GR/GR dorsal raphe sections. A dashed square marks the area shown at higher magnification on the bottom right. DRN, dorsal raphe nucleus. (D) Representative images of tdTomato and Tph2 labeling in ePet1-Cre-Il1r1r/r dorsal raphe sections. Dashed squares mark the area shown at higher magnification on the bottom right. (E) Representative images of tdTomato from different Gad2-Cre-Il1r1r/r brain sections, white dotted lines denote granule cell layer of the DG. DG, Dentate gyrus, VHC, ventral hippocampus, CTX, cortex. Scale bar: 200 μm
IL-1 is known to signal within serotonergic neurons [2][67], therefore we examined whether IL-1R1 was expressed in serotonergic neurons. Specifically, the dorsal raphe nucleus (DRN) is known as the primary serotonergic nuclei of the brain and can respond to IL-1 stimulation. To confirm IL-1R1 is expressed in serotonergic neurons, Il1r1GR/GR coronal brain sections containing the DRN were immunofluorescently co-labeled for tdTomato and tryptophan hydroxylase 2 (Tph2), the rate limiting enzyme in serotonin production. Most, but not all, neurons of the DRN were both Tph2+ and tdTomato+ (Fig. 2C). Restricting IL-1R1 expression to serotonergic neurons using ePet1-Cre-Il1r1r/r mice, DRN neurons were found to co-express tdTomato and Tph2 via immunofluorescent labeling. Other serotonergic brain regions such as the raphe pallidus nucleus (RPa) and raphe nucleus magnus (RMG) also show colocalization of both tdTomato and Tph2 (Supplementary Fig. 2C); however, these regions had fewer Il1r1+ cells compared to the DRN. These data indicate that subsets of serotonergic neurons express nIl1r1.
Other neurotransmitter systems which were thought to be influenced directly by IL-1 are cholinergic [59], dopaminergic [38], and GABAergic neurons [3, 4, 68]. Colocalization of choline acetyltransferase (ChAT) with tdTomato in the horizontal limb of diagonal band (HDB) was not observed (Supplementary Fig. 2B). Further, in dopaminergic nuclei of the brain, the DRN, ventral tegmental area (VTA), substantia nigra (SN), colocalization of tyrosine hydroxylase (TH) and tdTomato expressing neurons was not found (Supplementary Fig. 2D-G). After restoring IL-1R1 expression to only GABAergic neurons, by crossing a Gad2-Cre with the Il1r1r/r mouse, tdTomato expression throughout the brain was sparse but identifiable. tdTomato+ GABA-ergic neurons were identified in the DG hilus, but not the granule cell layer, ventral hippocampus (VHC), and throughout the cortex (CTX, Fig. 2E). In Gad2-Cre-Il1r1r/r mice, qPCR of Il1r1 mRNA expression in the hippocampus show detectable but significantly lower levels of Il1r1 mRNA compared to WT controls (Supplementary Fig. 2H). This data shows IL-1R1 is not expressed in acetylcholine or dopaminergic neurons; however, IL-1R1 is sparsely expressed in subsets of GABAergic neurons.
Confirmation of neuronal IL-1R1 expression from scRNAseq databases
As an unbiased confirmation of nIl1r1 distribution, we analyzed publicly available single cell RNA sequencing (scRNAseq) of various brain regions in mice [50]. The predominate cellular expression of Il1r1 in most brain regions were endothelial and choroid plexus cells, however, astrocyte and mural cell Il1r1 expression was detected (Supplementary Fig. 3A). Out of all the cells analyzed, neuronal Il1r1 was detected throughout the brain in the hippocampus (2%), followed by the thalamus (0.5%), then frontal cortex (0.3%). These subsets of neurons were then classified by their neurotransmitter-related gene expression. Within the hippocampus, frontal and posterior cortex, and thalamus, Il1r1 expressing cells were predominately glutamatergic (Supplementary Fig. 3B, grey). Other regions show sparse GABAergic expression of Il1r1 such as the entoperduncular region, globus pallidus and substantia nigra (Supplementary Fig. 3B, orange). scRNAseq data from the mouse DRN conducted by Huang et al. was analyzed for Il1r1 expression patterns [26]. Il1r1 expressing cells were primarily vascular cells and astrocytes. Of all the total cells analyzed, only 1.5% of the cells expressed detectable transcripts of Il1r1. Of all the neurons isolated, 7% expressed Il1r1 mRNA and a majority (86%) of the Il1r1+ neurons were serotonergic. The DRN is comprised of a variety of serotonergic neurons subtypes and these subtypes are known to project to different brain regions, which could inform how IL-1 can alter neurocircuitry and behavior. Of the serotonergic neurons, Il1r1 expression is highest in 5-HT subtype II (7%, orange), followed by 5-HT subtype IV (6%, yellow), 5-HT subtype I (2%, blue), and then 5-HT subtype III (1%). Il1r1 expression was not identified in 5-HT subtype V (light blue) (Supplementary Fig. 3C). scRNAseq from human prefrontal cortex (PFC) and hippocampus (HC) confirm Il1r1 expression is indeed expressed in human excitatory neurons of the PFC and dentate gyrus (Supplementary Fig. 3D); however, one of the major differences in humans is that Il1r1 is expressed in inhibitory neurons of the PFC suggesting species-specific Il1r1 expression [24].
Detailed mapping of glutamatergic neuronal IL-1R1 expression throughout the brain
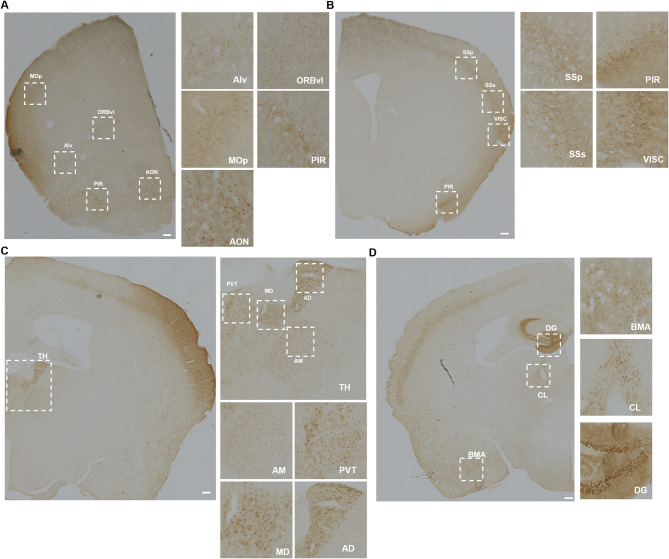
Our immunofluorescence and scRNAseq analysis of multiple brain regions strongly suggests that nIL-1R1 is primarily expressed on glutamatergic neurons. To verify and detail which of the brain nuclei express nIL-1R1 in glutamatergic neurons, Vglut2-Cre-Il1r1r/r mice were used to restore IL-1R1 only in glutamatergic neurons. Immunohistochemical labeling of endogenous tdTomato in sections of Vglut2-Cre-Il1r1r/r mice is required as tdTomato expression in neurons is weak (Supplemental Fig. 7G). QUINT pathway analysis was used to annotate glutamatergic nIL-1R1 expression throughout the brain. Without confounding non-neuronal IL-1R1 expression and using higher contrast IHC techniques, glutamatergic nIL-1R1 was detected in 49 individual brain regions. Glutamatergic nIL-1R1 was detected in many anterior brain regions such as the olfactory regions of the anterior olfactory nucleus (AON) and piriform cortex (PIR) along with other motor regions of the primary motor cortex (MOp) (Fig. 3A). Other IL-1R1 expressing regions that were detected in the Il1r1GR/GR mice were not found to express tdTomato in Vglut2-Cre-Il1r1r/r such as the dorsal raphe nucleus (DRN), lateral septum (LS), fimbria (fi) and central nucleus of the raphe (CS) (Supplementary Fig. 4A-D). These regions are well known to be comprised of serotonergic or GABAergic neurons. nIl1r1 expression in the PIR extends from anterior (+ 2.3 mm from bregma) to posterior (-0.5 mm from bregma) (Supplementary Fig. 4E). More posteriorly, glutamatergic nIl1r1 was found to be expressed in regions of the somatosensory cortex (SSp, SSs, VISC) but not in the adjacent motor cortex (Fig. 3B). More apparent labeling of thalamic Il1r1 expression was detected in the Vglut2-Cre-Il1r1r/r sections. Nuclei within the thalamus were detected to have clear cell bodies in the anterior dorsal nucleus (AD), medial dorsal (MD), and paraventricular nucleus of the thalamus (PVT) (Fig. 3C). In the PVT, glutamatergic neuronal tdTomato expression was found be highly expressed in the anterior (-0.25 mm relative to bregma) and posterior (-1.5 mm relative to bregma) PVT, whereas the medial (-0.5–1.0 mm relative to bregma) PVT shows fewer tdTomato + cells (Supplemental Fig. 4F). This suggests that some nuclei can have differential expression of nIl1r1 in subregions of a nuclei. Other glutamatergic nIl1r1-expressing regions identified in the Il1r1GR/GR mice, such as the dentate gyrus (DG), claustrum (CLA) and basomedial amygdala (BMA) were verified in the Vglut2-Cre-Il1r1r/r (Fig. 3D).
Fig. 3.
Representative images of IL-1R1 expression on glutamatergic neurons in different brain sections. (A) Representative images of tdTomato immunolabeling in Vglut2-Cre-Il1r1r/r mice, highlighting the cortex and olfactory areas. MOp, primary motor area; ORBvl, orbital area, ventrolateral part; Alv, agranular insular area, ventral part; PIR, piriform area; AON, anterior olfactory nucleus. The dashed squares indicate areas with high neuronal IL-1R1 expression and are magnified. Scale bar = 200 μm (B) Representative images of tdTomato immunolabeling in the Vglut2-Cre-Il1r1r/r somatosensory areas, visceral areas, and piriform area. SSp, primary somatosensory area; SSs, supplementary somatosensory area; VISC, visceral areas; PIR, piriform area. Scale bar: 200 μm (C) Representative images of DAB labeling in various thalamic nuclei of the Vglut2-Cre-Il1r1r/r mouse. AD, anterodorsal nucleus; MD, mediodorsal nucleus of thalamus; PVT, paraventricular nucleus of thalamus; AM, anteromedial nucleus. Scale bar: 200 μm (D) Representative images of tdTomato immunolabeling in Vglut2-Cre-Il1r1r/r mouse dentate gyrus (DG), central lateral nucleus of the thalamus (CL), and basomedial amygdalar nucleus (BMA). Scale bar = 200 μm
Similar to the analysis conducted in Il1r1GR/GR mice, functional neuroanatomical analysis was conducted in the Vglut2-Cre-Il1r1r/rmice. Brain regions were categorized as (1) sensory detection, relay, and processing; (2) emotional Regulation, (3) spatial and cognitive processes, (4) neuroendocrine response and (5) miscellaneous. Based on all total nIl1r1+ nuclei in Vglut-Cre-Il1r1r/r mice, 49% were related to sensory detection, relay, and processing; 20% were related to emotional regulation; 12% were related to spatial and cognitive processes, 8% were related to neuroendocrine responses, and 10% were determined as miscellaneous (Supplementary Fig. 1G). A fully annotated list containing all glutamatergic nIl1r1 expression is provided in Table 2.
Table 2.
Il1r1-expressing brain regions in the Vglut2-Cre-Il1r1r/r mouse
| Abbreviations | Brain Regions (Anterior to Posterior) |
|---|---|
| ORBl2/3, 6a, vl5, vl6a, m5, m6a | Orbital area, lateral part, layer 2/3 |
| PL5 | Prelimbic area, layer 5 |
| ACAd5 | Anterior cingulate area, dorsal part, layer 5 |
| MOs2/3, s6a | Secondary motor area, layer 2/3 |
| MOp2/3, p5, p6a | Primary motor area, layer 2/3 |
| AId6a, v2/3 | Agranular insular area, dorsal, layer 6a; ventral, layer 2/3 |
| MOB | Main olfactory bulb |
| AONm, pv | Anterior olfactory nucleus, medial part; posteroventral part |
| PIR | Piriform area |
| VTT/DTT | Ventral taenia tecta; Dorsal taenia tecta |
| SSp2/3, 4, 5 | Primary somatosensory area, layer 2/3, 4, 5 |
| SSs2/3 | Supplemental somatosensory area, layer 2/3, 4, 5 |
| VISC2/3, 4, 5 | Visceral area, layer 2/3, 4, 5 |
| GU2/3, 4, 5, 6a | Gustatory areas, layer 2/3 |
| AIp2/3 | Agranular insular area, posterior, layer 2/3 |
| AUDd2/3, 4, p4, v2/3, v4 | Dorsal auditory area, layer 2/3, 4; posterior layer 4; ventral 2/3, 4 |
| TEa2/3, 4 | Temporal association areas, layer 2/3, 4 |
| ECT2/3 | Ectorhinal area, layer 2/3 |
| PERI2/3 | Perirhinal area, layer 2/3 |
| ENTl3 | Entorhinal area, lateral part, layer 3 |
| COAa | Cortical amygdalar area, anterior |
| Epv/d | Endopiriform nucleus, ventral and dorsal part |
| CLA | Claustrum |
| PAA | Piriform-amygdalar area |
| AAA | Anterior amygdalar area |
| BLAa | Basolateral amygdalar nucleus, anterior part |
| MEAav | Medial amygdalar nucleus, anteroventral part |
| TU | Tuberal nucleus |
| BMAa | Basomedial amygdalar nucleus, anterior/posterior part |
| DG | Dentate gyrus, granule cell layer |
| ARH | Arcuate hypothalamic nucleus |
| VMH | Ventromedial hypothalamic nucleus |
| PVH | Paraventricular hypothalamic nucleus |
| RE | Nucleus of reuniens |
| RH | Rhomboid nucleus |
| PVT | Paraventricular nucleus of thalamus |
| CM | Central medial nucleus of thalamus |
| MD | Mediodorsal nucleus of thalamus |
| AD, AM | Anterodorsal nucleus/Anteromedial nucleus of thalamus |
| SMT | Submedial nucleus of thalamus |
| PCN | Paracentral nucleus of thalamus |
| IMD | Intermediodorsal nucleus of thalamus |
| CL | Central lateral nucleus of thalamus |
| LD, LP | Lateral dorsal/ posterior nucleus of thalamus |
| PO | Posterior complex of thalamus |
| MGd, v, m | Medial geniculate complex, dorsal, ventral, medial |
| VISpm4 | Posteromedial visual area, layer 4 |
| VCO | Ventral/Dorsal cochlear nucleus |
| FLgr | Flocculus, granular layer of cerebellum |
Protein expression of IL-1R1 in glutamatergic neurons
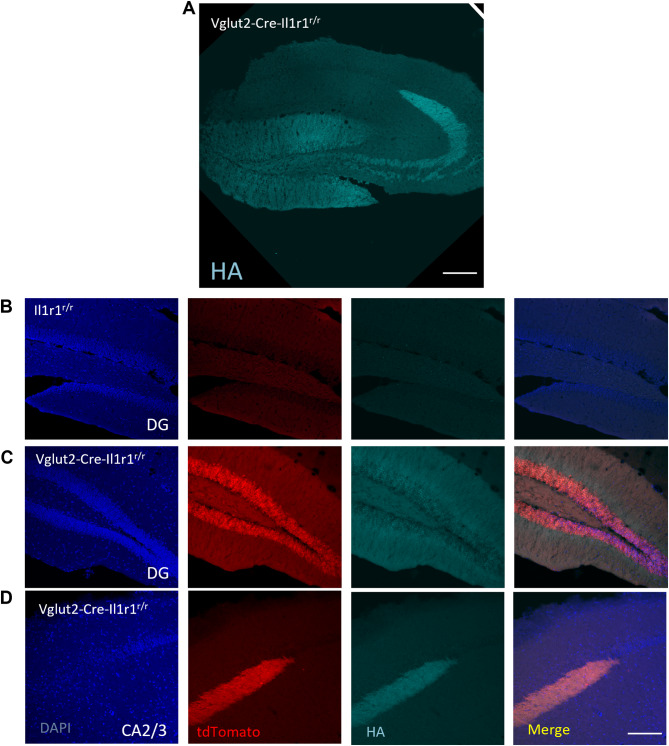
We next sought to determine the IL-1R1 protein expression patterns in brain regions with high nIl1r1 expression. Immunohistochemical labeling of IL-1R1 has been difficult in previous studies. Using the Vglut-Cre-Il1r1r/r mice where Il1r1 mRNA is tracked via cytosolic tdTomato expression and a 3xHA tag is fused to the intracellular region of the IL-1R1 protein, we have successfully labeled IL-1R1 protein via the HA tag in DG neurons using glyoxal-based fixatives (Fig. 4A). In an IL-1R1 knockout (Cre negative Il1r1r/r littermates) no tdTomato (Il1r1 mRNA) or HA tag (IL-1R1 protein) was detected (Fig. 4B). In the Vglut2-Cre-Il1r1r/r mice concordant tdtomato (red) and HA tag (cyan) was detected in the DG neurons (Fig. 4C). TdTomato filled the entire cellular processes of DG neurons (Fig. 4C, red) whereas HA tag was localized on the cellular membrane (Fig. 4C, cyan). Mossy fiber terminals from the DG and projecting to the CA2/3, express both tdTomato and HA tag, however, no IL-1R1 expression was found in the CA2/3 pyramidal cells (Fig. 4D). These data show, nIL-1R1 protein is concordant with Il1r1 mRNA expression and is expressed throughout the entire dentate gyrus neuron.
Fig. 4.
IL-1R1 protein expression is localized across the entire neuron of the dentate gyrus. (A) Representative tilescan image of immunofluorescent labeling of HA-tag in the hippocampus of glyoxal-fixed Vglut2-Cre-Il1r1r/r mice. (B-C) 20x micrograph of immunofluorescent labeling in the DG of DAPI, tdTomato, and HA-tag in Il1r1r/r (B) or Vglut2-Cre-Il1r1r/r (C) mice. (D) 20x micrograph of immunofluorescent labeling in the CA3 of DAPI, tdTomato, and HA-tag in Vglut2-Cre-Il1r1r/r (D) mice
IL-1R1 expression does not alter gross neuronal structure or excitatory synaptic density in the dentate gyrus
Since IL-1R1 is expressed by most, if not all, dentate gyrus neurons we examined whether the presence of nIL-1R1 can alter the structural anatomy of DG neurons. To address this, we used Golgi staining (FDNeuroTech) to measure neuron complexity and synaptic spine density in male 12wk old WT (n = 4) and Il1r1r/r (n = 3) mice. Individual Golgi-stained neurons within the dentate gyrus were imaged and then traced using Neurolucida360 (MBF Neuroscience). Branch order analysis of these neurons was conducted to analyze the dendritic complexity. Segments of secondary dendrites from each neuron were analyzed for spine density. There were no differences in branch order (Supplementary Fig. 5A-E) or synaptic spine density (Supplementary Fig. 5F-G) between WT and Il1r1r/r mice in the dentate gyrus. These data suggest that global elimination of IL-1R1 does not lead to altered dendritic or synaptic structure in the dentate gyrus of adult male mice.
IL-1R1 expressing neurons do not utilize canonical NFκB signaling pathways
Although nIL-1R1 is distributed throughout the brain in discrete glutamatergic neurons, it is unknown whether glutamatergic nIL-1R1 would respond to an IL-1β stimulus like that of other IL-1R1 expressing cells (e.g. endothelial). Typically, in non-neuronal cells, IL-1/IL-1R1 signaling induces the canonical NFκB pathway activation, however, in neurons the signaling pathways are disputed. To determine whether IL-1R1 expressing neurons initiate canonical NFκB signaling following IL-1 stimulation, we crossed the NFκB-GFP reporter mouse to our global IL-1R1 reporter (Il1r1GR/GR) to generate a double reporter mouse (Il1r1GR/GRNFκBGFP). NFκB expression was examined throughout the brain at 2, 4, 8, and 24 h following intracerebroventricular IL-1 (20 ng) or PBS administration via immunofluorescent labeling of GFP and tdTomato. The timepoint where NFκB-GFP was first detected, by a given cell type was reported. In the PBS sham surgeries, no NFκB-GFP was detected at any timepoint, whereas tdTomato was, as previously described, detected in DG and DRN neurons, endothelial cells, and ependymal cells (Supplementary Fig. 6B-D). Following i.c.v. IL-1β, NFκB-GFP was detected in the ependymal cells lining the 3rd ventricle at 2 h (Supplementary Fig. 6E), in infiltrating leukocytes near blood vessels at 4 h (Supplementary Fig. 6F), in choroid plexus epithelial cells at 8 h (Supplementary Fig. 6G) and in brain vasculature at 24 h post injection (Fig. 5A-B). No GFP expression was detected in Il1r1 expressing neurons of the DG or DRN across all timepoints; however, GFP was detected in the hippocampal and DRN endothelial cells at 24 h indicating IL-1β reached these nIL-1R1 expressing regions (Fig. 5A-B). This indicates that neuronal IL-1R1 does not utilize the canonical NFκB signaling pathway.
Fig. 5.
Neuronal IL-1R1-mediated signaling is independent of NF-κB pathway and chronic restoration of DG neuronal IL-1R1 reveals negative regulation of synaptic-related pathways. (A-B) Representative images of GFP and tdTomato labeling in Il1r1GR/GRNFκBGFP DG (A) and DRN (B) sections. The dashed square marks the area shown at higher magnification on the top right. Dashed lines mark the dentate gyrus granule cell layer. DG, dentate gyrus; DRN, dorsal raphe nucleus. Scale bar = 200 μm. (C) Log2 fold change of top 13 differentially expressed genes of microdissected dentate gyrus following i.c.v. IL-1 (20 ng) compared to PBS. (D-E) Immunofluorescent labeling of NeuN (Red), GFP (Green), and DAPI (Blue) in Il1r1r/r mice injected with AAV-CMV-CRE-GFP into the right dentate (D) and AAV-CMV-GFP injected into the left dentate (E). (F) Volcano plot of differentially expressed genes between AAV-Cre versus AAV-GFP injected dentate gyrus neurons within Il1r1r/r mice (n = 3). Vertical dotted lines indicate a log2 value of 1 and horizontal dotted lines indicate a -Log10 p value of 1.3. Red data points were used in GO pathway enrichment analysis in G-I. (G-I) GO Molecular function (G), Cellular component (H), and Biological Process (I) pathway analysis of differentially expressed genes in the AAV-CMV-GFP injected Il1r1r/r DG neurons
Direct stimulation of nIL-1R1 in Vglut2-Cre-Il1r1r/r mice does not lead to acute downstream gene expression
Transcriptional pathways in nIL-1R1-expressing neurons related to direct IL-1 stimulus have yet to be determined. To determine which signaling pathways nIL-1R1 may use, Vglut2-Cre-Il1r1r/r mice were injected i.c.v. with PBS or IL-1β (20 ng), 3 h later the DG was microdissected, RNA was isolated, and RNA sequencing of the tissue was performed. Gene expression was analyzed using the DESeq2 Bioconductor package in R as previously reported [14, 69]. The threshold of a log2Fold Change > 1 was set as a biologically significant gene. Of the top differentially expressed genes compared with PBS controls, none reached our criteria (Fig. 5C). However, some genes reached statistical significance below a Log2Fold change of 1: 1500015A07Rik, Hddc3, and Ubc were downregulated genes and Pdp1, Hmgn2, Ppig, Trove2, Hrh3, Rpph1, Pter, Mir6236, Sox 11, Slc17a6 were marginally upregulated genes. Following GO pathway analysis, these genes did not associate with any specific cellular, molecular, or biological pathways. At this acute timepoint, we conclude, stimulation of nIL-1R1 with IL-1β does not change transcriptional activity.
Next, we performed an exploratory study to determine which genes and pathways are influenced by the restoration of DG nIL-1R1 for an extended period of time. Bilateral AAV injections were performed in Il1r1r/r mice in which AAV-Cre-GFP was injected into the right DG (to restore IL-1R1 expression in neurons) and AAV-GFP vector control was injected into the left DG. Injected mice were allowed for nIL-1R1 expression to occur for 1 month (n = 3). To verify that AAV-Cre would indeed restore IL-1R1 protein to only one hemisphere of the brain, triple immunofluorescent labeling for GFP, tdTomato, and HA tag was conducted. GFP, indicating successful viral transfection, was detected in DG neurons on both sides of the hippocampus, whereas, tdTomato and HA tag were detected only on the AAV-Cre, but not in the AAV-GFP, side (Supplemental Fig. 7E-F). AAV-injected brains were collected and GeoMx spatial transcriptomics (Nanostring) was performed to isolate GFP+NeuN+ neurons from either the right and left hippocampus (Fig. 5D-E), respectively, in order to identify nIL-1R1-associated genes and pathways following restoration of IL-1R1. Volcano plot of gene expression revealed genes associated with AAV-CRE (i.e. the Il1r1 expressing DG) contained complement (C1ra, C1qa) and immune-related genes (ADAM18, Cd300a) (Fig. 5F). GO pathway analysis of all genes above a -Log10pvalue of 1.3 (p < 0.05) that were increased with the AAV-CRE injected DG showed no enrichment for any specific GO pathway. However, GO pathway analyses (molecular function, cellular components, and biological processes) identified significantly enriched pathways in the AAV-GFP injected DG (i.e. the Il1r1-null DG). These include RNA and mRNA binding pathways (Fig. 5G, red bars), synaptic regulation pathways (Fig. 5H, red bars) and synaptic transmission, formation, and adhesion pathways (Fig. 5I, red). Thus, while IL-1β did not induce acute gene expression in the DG neurons, the observed transcriptional effects from long term nIL-1R1 restoration indicates nIL-1R1, nonetheless, modulates neuronal gene expression over time.
Dentate gyrus neurons utilize a unique splice variant of the IL-1R1 accessory protein (IL-1RAcP)
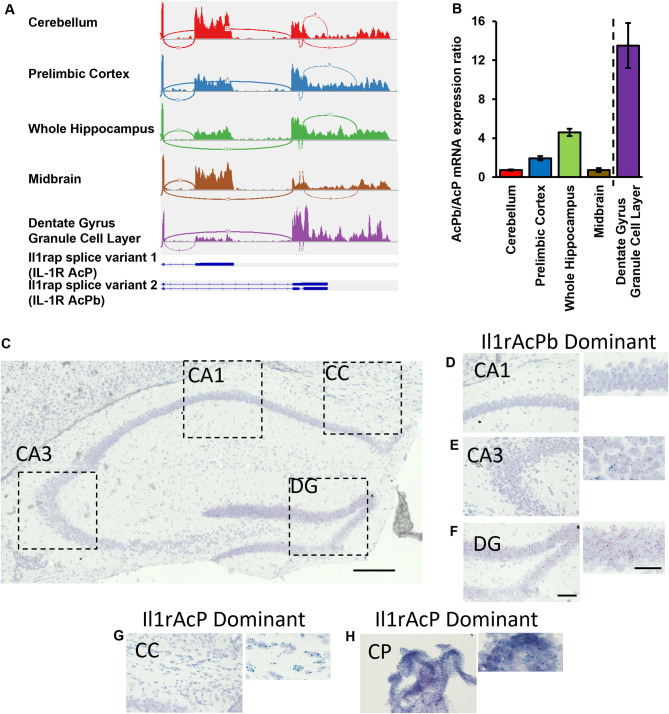
Due to the lack of acute neuronal transcriptional activity, following direct IL-1β stimulation, alternative pathways for IL-1R1 signaling were hypothesized. Typically, the IL-1R1 signaling cascade requires an accessory protein (AcP), named IL-1RAcP, to initiate intracellular signaling. In the central nervous system, neurons use a CNS-restricted isoform, called IL-1RAcPb, that blocks canonical MyD88 binding and can possibly signal though other transcription factors [27, 53] or act as trans-synaptic adhesion molecules [61, 65]. It is unclear whether IL-1RAcPb is expressed in the same regions which express nIL-1R1 and whether neurons solely express IL-1RAcP variant 1 (AcP, exon 10–11), IL-1RAcPb variant 2 (AcPb, exon 10–12), or a combination of the two isoforms. IL-1RAcP and IL-1RAcPb gene fragments were quantified from bulk RNA sequencing data throughout various brain regions such as the cerebellum, prelimbic cortex, hippocampus, midbrain, and isolated dentate gyrus granule cells. The Sashimi plot reports the frequency of exonal usage and connection and mapped exonal usage is reported in Fig. 6A. The ratio of AcPb: AcP expression by brain region was analyzed. Within the cerebellum and midbrain, AcPb: AcP ratio was near 1, suggesting an even distribution pattern. However, in other regions such as the prelimbic cortex and whole hippocampus, the ratio of AcPb: AcP increased 2–4 times indicating that the primary expressed isoform is the alternative spliced AcPb. Isolated dentate gyrus granule cells show high specificity for only AcPb, as the AcPb: AcP ratio was near 12 (Fig. 6B). Confirmation of this expression pattern was conducted with Basescope in situ hybridization (ACDBio). Probes were generated that selectively bind to either the exon junction between exon 10 and exon 11 which encodes AcP or between exon 10 and 12 which encodes AcPb. Puncta of AcP (Blue-Green) and AcPb (Red) mRNA were detected in brain sections of the hippocampus (Fig. 6C). AcPb dominant regions reside in the neuronal layers of the CA1 (Fig. 6D), CA3 (Fig. 6E), and strongest expression in the DG (Fig. 6F). AcP puncta were detected sparsely throughout the CA3 region but not DG or CA1 cell layers. AcP-dominant expression was identified in the corpus callosum (CC, Fig. 6G) and choroid plexus (CP, Fig. 6H). AcPb KO mice (Amgen) were used to validate specificity of the probes and determine whether the expression pattern of AcP changes when AcPb is eliminated. AcPb KO mice showed complete elimination of AcPb mRNA expression in the DG, CA1 and CA3 neurons (Supplementary Fig. 7A-C) while AcP was still detected in the CC and ChPlx (data not shown). All samples were run in tandem with positive and negative control probes (Supplementary Fig. 7D). Interestingly, elimination of AcPb did not permit AcP expression in the dentate, suggesting restricted AcPb expression in neurons.
Fig. 6.
IL-1R1 expressing neurons utilize alternatively spliced accessory protein, IL-1RAcPb. (A) Sashimi plots from RNAseq data analyses of different brain areas and schematics showing the last two exons of Il1rap. (D) Quantification of AcPb/AcP RNA transcripts ratio in different brain areas. Dashed line separates two different RNAseq experiments. Error bars represent the mean ± SEM. (C) Photomicrograph of the WT hippocampus following BaseScope in situ hybridization of exon junction regions for IL-1RAcP (Blue-Green) and IL-1RAcPb (Red). Scale bar = 300 μm Dotted lines indicate magnified regions presented in D-G. (D-F) Photomicrographs of predominantly IL-1RAcPb expression regions, such as the CA1 (D), CA3 (E), and DG (F), which are magnified images to the right of each corresponding image. (G-H) Photomicrographs of predominantly IL-1RAcP expressing regions of the corpus callosum (G) and the choroid plexus (H), which are magnified images to the right of each corresponding image. DG, dentate gyrus; CC, corpus callosum; CP, choroid plexus. Scale bar = 100 μm
Discussion
Our data clarifies a decades-long controversy of which brain nuclei and neurotransmitter systems express neuronal IL-1R1 in mice. A thorough brain-wide mapping of nIl1r1 using both global IL-1R1 and glutamatergic-restored nIL-1R1 reporter mice was conducted, adding many new IL-1R1 expressing neuron populations while verifying previously established regions. This analysis revealed the somatosensory and glutamatergic systems as the primary expressors of nIl1r1, which have been overlooked and understudied. Furthermore, direct stimulation of neurons with IL-1 reveals a lack of transcriptional activity to acute IL-1. Restoration of IL-1R1 to DG neurons shows genes associated with synaptic function are downregulated in the presence of nIL-1R1. Neurons identified as nIL-1R1 expressers exclusively utilize an isoform of the IL-1R accessory protein (AcPb). Taken together, specific nIL-1R1 modulated neural circuits were identified and are likely to influence neuronal function through non-conventional signaling pathways.
Mapping of IL-1R1 expression has been previously conducted many times that resulted in contradictory conclusions. Radiolabeled ligand binding studies was the first attempt to describe the distribution of IL-1R in the brain. Farrar et al. showed low-magnification autoradiographs of radiolabeled IL-1α bound to many neural structures of the rat brain such as the dentate gyrus, olfactory bulb, and piriform cortex, cingulate cortex, and cerebellar granule cell layers while discounting vascular labeling entirely [18]. Follow-up studies in mice using radiolabeled IL-1β and IL-1α show these ligands highly bind to the hippocampus and unilateral ablation of hippocampal neurons prevents binding of IL-1α, solidifying the hippocampus as one of the brain regions that binds to IL-1 [5, 55]. These ligand binding assays did not permit specification of which type of IL-1R is expressed as IL-1β, IL-1α, and IL-1ra all bind to both IL-1R1 and IL-1R type 2 (IL-1RII), a non-functional decoy receptor; however, IL-1RII is not known to be expressed in high enough levels in the hippocampus to be considered a confound. More attempts to accurately describe IL-1R1 distribution patterns used in situ hybridization (ISH) or immunohistochemistry (IHC). Initial ISH studies verified the DG as the strongest neuronal Il1r1 mRNA expresser and CA3, DRN, and thalamic nuclei exhibiting lower levels of expression in mice. Other discrepant, but species-specific, results contest the DG as being an Il1r1-expressing region and identify only non-neuronal (endothelial and choroid plexus), basolateral amygdala, paraventricular nucleus of the hypothalamus, and trigeminal nerve in rats [12]. IHC-based studies, using unconventional fixation methods, show that while IL-1R1 protein of CFW/D mice was detected in the choroid plexus ependymal cells and DG, it was also detected throughout the CA1-4 neurons but not in blood vessels [19]. These discordant results demanded a thorough follow up using more sophisticated techniques, such as genetic reporters, conditional expression of IL-1R1, and standardized mapping procedures to definitively describe nIl1r1 expression which was completed in this study.
One major advantage of the current study is that sensitive genetic techniques can be leveraged to detail all IL-1R1-expressing neuronal populations. Previously these genetic mouse lines were used to identify that the DG is the primary expressor of neuronal IL-1R1 [14, 33, 37] which was verified here. Additionally, the usage of the Vglut2-Cre-Il1r1r/r, CamKii-Cre-Il1r1r/r, ePet1-Cre-Il1r1r/r, and Gad2-Cre-Il1r1r/r mice allowed for neuronal/neurotransmitter specific restoration of IL-1R1 expression without confounds of non-neuronal cells or other populations of neurons. For instance, ePet1-Cre-Il1r1r/r (data not shown) and Gad2-Cre-Il1r1r/r mice do not have tdTomato within the dentate gyrus neurons or other excitatory neurons. All IL-1R1+ neurons identified in the global reporter mice (Il1r1GR/GR) were also identified in each of the conditional neuronal restoration models (i.e. Vglut2-Cre-Il1r1r/r, CamKii-Cre-Il1r1r/r, ePet1-Cre-Il1r1r/r, and Gad2-Cre-Il1r1r/r) indicating there were no gross unintentional changes to nIl1r1 distribution between mouse lines. Using Tie2-Cre-Il1r1r/r mice as a negative control prevents artifacts contributed to technical issues during IHC and imaging since endothelial cells will exclusively be labeled and any neuronal tdTomato expression can be determined as artifact. In this study, none of the listed nuclei in Tables 1 and 2 were present in sections from Tie2-Cre-Il1r1r/r mice. The results from this study were corroborated by analysis of publicly available single-cell sequencing data from other groups.
DRN nIl1r1 distribution is not well described in the literature. Analysis of single cell RNAseq data in the DRN shows that nIl1r1 is predominantly expressed in subtype II and subtype IV serotonergic neurons. Subtype II occupies the dorsal regions of the DRN, are enriched for Tph, and are thought to project to the sensory cortical regions compared to motor regions. Subtype IV is known to be enriched in the medial parts of the DRN and express higher levels of vesicular glutamate transporter, Slc1a8. These subtype IV neurons project to the striatum and throughout broad cortical regions [26]. Further characterization of the DRN neuronal projections is crucial to dissect individual behaviors that nIL-1R1 will modify.
Due to the lack of the full distribution patterns of nIL-1R1 in the mouse, the understanding of potential functional outputs of nIL-1R1 is incomplete. In the literature, the primary outcomes of central IL-1 are sickness behaviors, anxiety-like behavior, changes to sleep, depressive-like behaviors, and alterations to memory/cognition. Sickness behaviors induced by IL-1, such as food and water intake, locomotor activity, anhedonia, and social avoidance, can be blocked by eliminating endothelial-produced prostaglandin synthesis and/or inhibiting canonical non-neuronal IL-1R1 signaling [20, 33]. Anxiolytic behaviors were described in IL-1R1KO mice [30, 39] and the anxiogenic effect of chronic, but not acute, stress required IL-1R1 signaling [36]. Furthermore, studies in IL-1R1 KO mice show IL-1R1 is important for cognition and memory, however, this is controversial and refuted [1, 30, 39]. Administration of low levels of IL-1 or blockade of physiological IL-1 in the brain was shown to alter slow-wave sleep [6, 41] and non-specific neuronal IL-1R1 overexpression altered sleep patterns suggesting neuronal IL-1 signaling can modify sleep [28]. However, these studies do not specify which neural circuits are influenced by IL-1 and the overexpression of IL-1R1 in neurons is not restricted to only circuits which should express IL-1R1. Other nIL-1R1 mediated functions have been shown in the DRN in response to LPS. Low levels of LPS induce a serotonergic nIL-1R1-dependent depressive-like phenotype in the tail suspension tests [67]. Recently, genetic mouse lines were used to restrict nIL-1R1 to glutamatergic neurons and revealed that chronic stress requires neuronal IL-1R1 to alter cognitive and social-avoidance behaviors, demonstrating the functional role of glutamatergic nIL-1R1 in the context of psychosocial-related neuroinflammatory conditions [14, 15, 22].
Direct IL-1β signaling impacting neuronal activity has also been a topic of contentious debate. As reviewed by Nemeth and Quan, the impact of direct IL-1β administration is highly dependent on neuronal cell type, preparation, and brain nuclei. Many studies suggest that IL-1 can suppress neuronal excitability in the amygdala, anterior hypothalamus, dorsal motor nucleus, and hippocampus, whereas IL-1β enhances activity in the hypothalamus, subfornical organ, and trigeminal ganglion. Due to these discrepancies, we hypothesize that multiple factors may modify whether IL-1β is an activator or inhibitor of neuronal activity such as the working concentration of IL-1, neuron type, or whether the observed results were MyD88-dependent [40]. The current study verified that nIL-1R1 is highly expressed in the dentate, sparsely in the CA3, and not CA1. Because our results show stimulation of nIL-1R1 did not induce downstream MyD88 signaling, we hypothesize that any effect of neuronal activity in the CA3 and CA1 neurons is the result of indirect IL-1R1 signaling, most likely though astrocytes or endothelium. Areas such as the amygdala, hypothalamic nuclei, and dentate gyrus require further electrophysiological studies with IL-1R1 restricted only to neurons to avoid confounding indirect effects.
New neural systems where IL-1 may act was identified in this study. Our data shows that the primary functional groups identified to express nIl1r1 are related to somatosensory detection, relay, and processing. The expression of glutamatergic IL-1R1 in nearly the entire somatosensory cortex (e.g., gustatory, visceral, visual, and auditory) and the thalamic relay centers (e.g., AD, AM, SMT, and PCN) suggests that IL-1 may modify sensory perception and detection. Indeed, studies in the rat suggest that central IL-1 administration may alter sensory perception such as taste reactivity [31, 54]. Whether nIL-1R1 is responsible for modulating sensory processing has yet to be studied.
It is unknown whether a modification to sensory processing through neuronal IL-1R1 contributed to the behaviors reported following stress, neuroinflammation, or required for homeostasis. Indeed, infants which scored higher on sensory processing assessments also showed higher signs of stress [49], enhanced sensory processing in humans predicted a link between stress and the prevalence of depression [60], and patients with PTSD showed increased severity of sensory and executive dysfunctions [11]. Taken together, it is important to follow up with our findings to probe somatosensory-related nIL-1R1-expressing brain regions to verify (1) whether nIL-1R1 is required for behavioral outcomes and (2) whether somatosensory processing is affected by IL-1 and is the underlying determinant for the alteration of cognitive and affective behavior.
Intracellular nIL-1R1 signaling has been controversial. Some previous studies suggest that nIL-1R1 on hippocampal [43] and dorsal raphe neurons [67] utilize P38α-MAPK pathways. Others suggest hippocampal nIL-1R1 signals through MYD88-dependent SrC [13, 21, 58]. Yet another study suggested nIL-1R1 initiates AKT pathways in cultured neurons co-expressing IL-1R1 and IL-1RAcPb [46]. It should be noted that many of these studies are conducted in vitro and in primary cultures. These preparations may be contaminated with non-neuronal IL-1R1 expressing cells which use canonical AcP-dependent pathways, leading to attributing AcP-dependent pathways to nIL-1R1 activation. Current results show AcPb is the splice variant that is exclusively expressed in neurons. AcPb is known to block MYD88 binding to IL-1R1 [27, 53]; therefore, any MYD88-dependent mechanisms can be excluded as a downstream pathway of nIL-1R1. Alternatively, nIL-1R1 could modify neuronal activity via its extracellular domain. This is because AcPb extracellular domains are known to regulate synaptic formation in vitro via trans-synaptic adhesion. In vivo AcPbKO mice show decreased spine density in cortex and CA1 neurons [65]. Considering our sequencing data, this mechanism is highly plausible as acute IL-1 stimulation did not alter any gene expression, whereas long term restoration of nIL-1R1 decreased many synapse-related pathways, potentially by sequestering AcPb into an IL-1/IL-1R1/AcPb complex, removing the ability for AcPb to function as a transsynaptic adhesion molecule. Further, increased complement gene (C1qb and C1ra) expression and downregulation of mRNA binding was associated with nIL-1R1 expression which could indicate formation of weak synapses tagged for pruning [25, 57] or, as recent studies shows, C1q production can negatively regulate translation through ribonucleoprotein complexes [51] resulting in reduced synaptic proteins. Interestingly, Il1r1-deficient mice (Il1r1r/r) did not show any changes to neuronal morphology or spine density in the DG neurons. Whether neuronal morphology and spine density in other IL-1R1− neurons synaptically connected to an IL-1R1+ neuron is dependent on nIL-1R1 remains to be determined.
While this study provided the most detailed analysis of neuronal Il1r1 expression in the brain, this study has a few limitations that should be addressed. Firstly, while systematic, our section sampling procedure is limited in spatial resolution due to using adjacent sections that are 440 μm between samplings. Therefore, there may be nuclei that were not detected in this study. Additionally, while using tdTomato for identifying Il1r1 positive neurons is currently the most sensitive methodology, this still may miss very low-level expression of nIL-1R1. It is well known that functional responses to IL-1 can be detected with as few as six proteins of IL-1R1 [52]. Secondly, we understand that this study uses only male mice in a limited age range and that neuroinflammatory processes can be both sexually dimorphic and age dependent which limits the generalizability of the results. Future studies are required to identify both age and sex specific expression of nIL-1R1 throughout the brain. Thirdly, the sequencing results yielded no changes in gene expression following i.c.v. IL-1 and we suggest a non-transcriptional activity of nIL-1R1; however, this does not exclude that IL-1 activity may take longer than 3 h. Follow-up studies of slower-acting transcriptional responses should be analyzed. Results from long-term expression of nIL-1R1 in this study suggests that nIL-1R1-expression may downregulate synaptic pathways in the absence of pathologically elevated IL-1 levels. Requiring chronic, compared to acute, IL-1 signaling to see the effects of IL-1-mediated negative regulation has been demonstrated previously [44] and should be considered in future studies. Fourthly, using invasive intracerebral injection procedures and AAVs to administer Cre recombinase to the DG may alter gene pathways. Indeed, one limitation of this study is the lack of naïve control samples. Further we did detect multiple interferon-related genes were elevated with the addition of AAV-CRE but not AAV-GFP which could indicate nIL-1R1 has other actions and could confound the analysis. However, the gene expression described in this study is unlikely due to inflammatory processes, as the tissues were selected 1 month following surgery. Another concern is AAV administration could cause leakage of the blood brain barrier (BBB). Guo et al. showed at 1 × 108 GC of AAV9, an even more immunogenic serotype compared to the AAV2 used in the current study, had no effect on BBB permeability, tight junction expression, or cytokine expression. Additionally, a three to ten times higher titer of AAV than what is used in this study is needed to cause BBB permeability changes [23]. Therefore, it is unlikely that the surgical or viral responses influenced our data. Overall, this study identifies discrete nIL-1R1 neural networks, details the neurotransmitter systems usage by nIL-1R1 neurons, and provides novel mechanistic insights to how nIL-1R1 may function, thus leading to a more comprehensive understanding of the brain IL-1R1 system.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Dr. James Krueger for generously providing the IL-1RAcPbKO mice for our studies.
Author contributions
D.P.N, X.L., and N.Q. conceptualized and designed all experiments. D.P.N wrote the main manuscript text and generated main and supplemental figures. D.P.N and X.L. analyzed all data. D.P.N., X.L., M.C.M., H.N., G.M., M.S.S., M.I.S., S.J.M., A.H., D.J.D., T.F., R.R.A., A.C.N., B.O., K.G.W., J.S.J.E., J.Y., A.D.C., D.B.M., P.A.G.-K. conducted the data collection. G.M. prepared Fig. 4. M.I.S prepared Supp. Figure 5. Q.Z provided RNA sequencing dataset. N.Q., J.P.G., R.D.B., J.F.S. provided supervision and conceptual insights.
Funding
This study was supported by the National Institute of Mental Health (NIMH) grant MH-109165 and National Institute of Neurological Disorders and Stroke (NINDS) NS116914 to NQ. DPN and DJD were supported by NIDCR Training Grant 2T32DE01432016. MCM and MIS were supported by IMPRIS.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethical approval
All experiments were approved by the Florida Atlantic University Institutional Animal Care and Use Committee under protocol A22-39 and A22-14.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoyu Liu is a co-first author.
Daniel P. Nemeth, Xiaoyu Liu and Ning Quan contributed equally to this work.
Contributor Information
Daniel P. Nemeth, Email: nemethd@fau.edu
Ning Quan, Email: nquan@health.fau.edu.
References
- 1.Avital A, Goshen I, Kamsler A, Segal M, Iverfeldt K, Richter-Levin G, Yirmiya R. Impaired interleukin-1 signaling is associated with deficits in hippocampal memory processes and neural plasticity. Hippocampus. 2003;13(7):826–34. 10.1002/hipo.10135. [DOI] [PubMed] [Google Scholar]
- 2.Baganz NL, Lindler KM, Zhu CB, Smith JT, Robson MJ, Iwamoto H, Deneris ES, Hewlett WA, Blakely RD. A requirement of serotonergic p38α mitogen-activated protein kinase for peripheral immune system activation of CNS serotonin uptake and serotonin-linked behaviors. Translational Psychiatry. 2015;5(11):e671. 10.1038/tp.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajo M, Patel RR, Hedges DM, Varodayan FP, Vlkolinsky R, Davis TD, Burkart MD, Blednov YA, Roberto M. Role of MyD88 in IL-1β and ethanol modulation of GABAergic Transmission in the Central Amygdala. Brain Sci. 2019;9(12). 10.3390/brainsci9120361. Article 12. [DOI] [PMC free article] [PubMed]
- 4.Bajo M, Varodayan FP, Madamba SG, Robert AJ, Casal LM, Oleata CS, Siggins GR, Roberto M. IL-1 interacts with ethanol effects on GABAergic transmission in the mouse central amygdala. Front Pharmacol. 2015;6:49. 10.3389/fphar.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ban E, Milon G, Prudhomme N, Fillion G, Haour F. Receptors for interleukin-1 (α and β) in mouse brain: mapping and neuronal localization in hippocampus. Neuroscience. 1991;43(1):21–30. 10.1016/0306-4522(91)90412-h. [DOI] [PubMed] [Google Scholar]
- 6.Baracchi F, Opp MR. Sleep-wake behavior and responses to sleep deprivation of mice lacking both interleukin-1β receptor 1 and tumor necrosis factor-α receptor 1. Brain Behav Immun. 2008;22(6):982–93. 10.1016/j.bbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besedovsky HO. The immune system as a sensorial system that can modulate brain functions and reset homeostasis: the immune system as a sensorial system. Ann N Y Acad Sci. 2019;1437(1):5–14. 10.1111/nyas.13935. [DOI] [PubMed] [Google Scholar]
- 8.Böttcher M, Müller-Fielitz H, Sundaram SM, Gallet S, Neve V, Shionoya K, Zager A, Quan N, Liu X, Schmidt-Ullrich R, Haenold R, Wenzel J, Blomqvist A, Engblom D, Prevot V, Schwaninger M. NF-κB signaling in tanycytes mediates inflammation-induced anorexia. Mol Metabolism. 2020;39:101022. 10.1016/j.molmet.2020.101022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruttger J, Karram K, Wörtge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, Mack M, Pinteaux E, Müller W, Zipp F, Binder H, Bopp T, Prinz M, Jung S, Waisman A. Genetic cell ablation reveals clusters of local self-renewing Microglia in the mammalian Central Nervous System. Immunity. 2015;43(1):92–106. 10.1016/j.immuni.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Carey H, Pegios M, Martin L, Saleeba C, Turner AJ, Everett NA, Bjerke IE, Puchades MA, Bjaalie JG, McMullan S. DeepSlice: Rapid fully automatic registration of mouse brain imaging to a volumetric atlas. Nat Commun. 2023;14(1):5884. 10.1038/s41467-023-41645-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clancy K, Ding M, Bernat E, Schmidt NB, Li W. Restless rest: intrinsic sensory hyperactivity and disinhibition in post-traumatic stress disorder. Brain. 2017;140(7):2041–50. 10.1093/brain/awx116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham ET, Wada E, Carter DB, Tracey D, Battey J, De Souza EB, De Souza EB. In situ histochemical localization of type I interleukin-1 receptor messenger RNA in the central nervous system, pituitary, and adrenal gland of the mouse. J Neurosci. 1992;12(3):1101–14. 10.1523/jneurosci.12-03-01101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis CN, Tabarean I, Gaidarova S, Behrens MM, Bartfai T. IL-1β induces a MyD88-dependent and ceramide-mediated activation of src in anterior hypothalamic neurons. J Neurochem. 2006;98(5):1379–89. 10.1111/j.1471-4159.2006.03951.x. [DOI] [PubMed] [Google Scholar]
- 14.DiSabato DJ, Nemeth DP, Liu X, Witcher KG, O’Neil SM, Oliver B, Bray CE, Sheridan JF, Godbout JP, Quan N. Interleukin-1 receptor on hippocampal neurons drives social withdrawal and cognitive deficits after chronic social stress. Mol Psychiatry. 2020;1–13. 10.1038/s41380-020-0788-3. [DOI] [PMC free article] [PubMed]
- 15.DiSabato DJ, Yin W, Biltz RG, Gallagher NR, Oliver B, Nemeth DP, Liu X, Sheridan JF, Quan N, Godbout JP. (2022). IL-1 Receptor-1 on Vglut2 + neurons in the hippocampus is critical for neuronal and behavioral sensitization after repeated social stress. Brain, Behavior, & Immunity - Health, 26, 100547. 10.1016/j.bbih.2022.100547 [DOI] [PMC free article] [PubMed]
- 16.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinf (Oxford England). 2013;29(1):15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ericsson A, Liu C, Hart RP, Sawchenko PE. Type 1 interleukin-1 receptor in the rat brain: distribution, regulation, and relationship to sites of IL-1-induced cellular activation. J Comp Neurol. 1995;361(4):681–98. 10.1002/cne.903610410. [DOI] [PubMed] [Google Scholar]
- 18.Farrar WL, Kilian PL, Ruff MR, Hill JM, Pert CB. Visualization and characterization of interleukin 1 receptors in brain. J Immunol (Baltimore Md: 1950). 1987;139(2):459–63. [PubMed] [Google Scholar]
- 19.French RA, VanHoy RW, Chizzonite R, Zachary JF, Dantzer R, Parnet P, Bluthé R-M, Kelley KW. Expression and localization of p80 and p68 interleukin-1 receptor proteins in the brain of adult mice. J Neuroimmunol. 1999;93(1):194–202. 10.1016/S0165-5728(98)00224-0. [DOI] [PubMed] [Google Scholar]
- 20.Gabriel Knoll J, Krasnow SM, Marks DL. (2017). Interleukin-1β signaling in fenestrated capillaries is sufficient to trigger sickness responses in mice. Journal of Neuroinflammation, 14. 10.1186/s12974-017-0990-7 [DOI] [PMC free article] [PubMed]
- 21.Ghosh B, Green MV, Krogh KA, Thayer SA. Interleukin-1β activates an src family kinase to stimulate the plasma membrane Ca2 + pump in hippocampal neurons. J Neurophysiol. 2016;115(4):1875–85. 10.1152/jn.00541.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman EJ, Biltz RG, Packer JM, DiSabato DJ, Swanson SP, Oliver B, Quan N, Sheridan JF, Godbout JP. Enhanced fear memory after social defeat in mice is dependent on interleukin-1 receptor signaling in glutamatergic neurons. Mol Psychiatry. 2024;1–14. 10.1038/s41380-024-02456-1. [DOI] [PMC free article] [PubMed]
- 23.Guo Y, Chen J, Ji W, Xu L, Xie Y, He S, Lai C, Hou K, Li Z, Chen G, Wu Z. High-titer AAV disrupts cerebrovascular integrity and induces lymphocyte infiltration in adult mouse brain. Mol Therapy - Methods Clin Dev. 2023;31:101102. 10.1016/j.omtm.2023.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, Weitz DA, Rozenblatt-Rosen O, Zhang F, Regev A. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14(10):955–8. 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, Lemere CA, Selkoe DJ, Stevens B. (2016). Complement and Microglia Mediate Early Synapse Loss in Alzheimer Mouse Models. Science (New York, N.Y.), 352(6286), 712–716. 10.1126/science.aad8373 [DOI] [PMC free article] [PubMed]
- 26.Huang KW, Ochandarena NE, Philson AC, Hyun M, Birnbaum JE, Cicconet M, Sabatini BL. Molecular and anatomical organization of the dorsal raphe nucleus. eLife. 2019;8:e46464. 10.7554/eLife.46464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Smith DE, Ibáñez-Sandoval O, Sims JE, Friedman WJ. Neuron-specific effects of Interleukin-1β are mediated by a Novel Isoform of the IL-1 receptor accessory protein. J Neurosci. 2011;31(49):18048–59. 10.1523/JNEUROSCI.4067-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingiosi AM, Raymond RM, Pavlova MN, Opp MR. Selective contributions of neuronal and astroglial interleukin-1 receptor 1 to the regulation of sleep. Brain Behav Immun. 2015;48:244–57. 10.1016/j.bbi.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Konno K, Yamasaki M, Miyazaki T, Watanabe M. Glyoxal fixation: an approach to solve immunohistochemical problem in neuroscience research. Sci Adv. 2023;9(28):eadf7084. 10.1126/sciadv.adf7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo JW, Duman RS. Interleukin-1 receptor null mutant mice show decreased anxiety-like behavior and enhanced fear memory. Neurosci Lett. 2009;456(1):39–43. 10.1016/j.neulet.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.László BR, Hormay E, Szabó I, Mintál K, Nagy B, László K, Péczely L, Ollmann T, Lénárd L, Karádi Z. Disturbance of taste reactivity and other behavioral alterations after bilateral interleukin-1β microinjection into the cingulate cortex of the rat. Behav Brain Res. 2020;383:112537. 10.1016/j.bbr.2020.112537. [DOI] [PubMed] [Google Scholar]
- 32.Levine J, Barak Y, Chengappa KNR, Rapoport A, Rebey M, Barak V. Cerebrospinal cytokine levels in patients with Acute Depression. Neuropsychobiology. 1999;40(4):171–6. 10.1159/000026615. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Nemeth DP, McKim DB, Zhu L, DiSabato DJ, Berdysz O, Gorantla G, Oliver B, Witcher KG, Wang Y, Negray CE, Vegesna RS, Sheridan JF, Godbout JP, Robson MJ, Blakely RD, Popovich PG, Bilbo SD, Quan N. Cell-type-specific interleukin 1 receptor 1 signaling in the Brain regulates distinct neuroimmune activities. Immunity. 2019;50(2):317–e3336. 10.1016/j.immuni.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Yamashita T, Chen Q, Belevych N, Mckim DB, Tarr AJ, Coppola V, Nath N, Nemeth DP, Syed ZW, Sheridan JF, Godbout JP, Zuo J, Quan N. Interleukin 1 type 1 receptor restore: a genetic mouse model for studying interleukin 1 receptor-mediated effects in specific cell types. J Neurosci. 2015;35(7):2860–70. 10.1523/JNEUROSCI.3199-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKim DB, Niraula A, Tarr AJ, Wohleb ES, Sheridan JF, Godbout JP. Neuroinflammatory Dynamics Underlie Memory Impairments after repeated Social Defeat. J Neurosci. 2016;36(9):2590–604. 10.1523/JNEUROSCI.2394-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurray KMJ, Winter A, Ahlbrand R, Wilson A, Shukla S, Sah R. Subfornical organ interleukin 1 receptor: a novel regulator of spontaneous and conditioned fear associated behaviors in mice. Brain Behav Immun. 2022;101:304–17. 10.1016/j.bbi.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohankumar PS, Thygarajan S, Kaleem Quadri S. Interleukin-1 stimulates the release of dopamine and dihydroxyphenylacetic acid from the hypothalamus in vivo. Life Sci. 1991;48(9):925–30. 10.1016/0024-3205(91)90040-I. [DOI] [PubMed] [Google Scholar]
- 39.Murray CL, Obiang P, Bannerman D, Cunningham C. Endogenous IL-1 in cognitive function and anxiety: a study in IL-1RI–/– mice. PLoS ONE. 2013;8(10):e78385. 10.1371/journal.pone.0078385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemeth DP, Quan N. Modulation of neural networks by Interleukin-1. Brain Plast. 2021;7(1):17–32. 10.3233/BPL-200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Opp MR, Krueger JM. Anti-interleukin-1 beta reduces sleep and sleep rebound after sleep deprivation in rats. Am J Physiology-Regulatory Integr Comp Physiol. 1994;266(3):R688–95. 10.1152/ajpregu.1994.266.3.R688. [DOI] [PubMed] [Google Scholar]
- 42.Pinteaux E, Rothwell NJ, Boutin H. Neuroprotective actions of endogenous interleukin-1 receptor antagonist (IL-1ra) are mediated by glia. Glia. 2006;53(5):551–6. 10.1002/glia.20308. [DOI] [PubMed] [Google Scholar]
- 43.Prieto GA, Smith ED, Tong L, Nguyen M, Cotman CW. Inhibition of LTP-Induced translation by IL-1β reduces the level of newly synthesized proteins in hippocampal dendrites. ACS Chem Neurosci. 2019;10(3):1197–203. 10.1021/acschemneuro.8b00511. [DOI] [PubMed] [Google Scholar]
- 44.Prieto GA, Snigdha S, Baglietto-Vargas D, Smith ED, Berchtold NC, Tong L, Ajami D, LaFerla FM, Rebek J, Cotman CW. Synapse-specific IL-1 receptor subunit reconfiguration augments vulnerability to IL-1β in the aged hippocampus. Proc Natl Acad Sci. 2015;112(36):E5078–87. 10.1073/pnas.1514486112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puchades MA, Csucs G, Ledergerber D, Leergaard TB, Bjaalie JG. Spatial registration of serial microscopic brain images to three-dimensional reference atlases with the QuickNII tool. PLoS ONE. 2019;14(5):e0216796. 10.1371/journal.pone.0216796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian J, Zhu L, Li Q, Belevych N, Chen Q, Zhao F, Herness S, Quan N. Interleukin-1R3 mediates interleukin-1–induced potassium current increase through fast activation of akt kinase. Proc Natl Acad Sci. 2012;109(30):12189–94. 10.1073/pnas.1205207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravizza T, Vezzani A. Status epilepticus induces time-dependent neuronal and astrocytic expression of interleukin-1 receptor type I in the rat limbic system. Neuroscience. 2006;137(1):301–8. 10.1016/j.neuroscience.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 48.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryckman J, Hilton C, Rogers C, Pineda R. Sensory processing disorder in preterm infants during early childhood and relationships to early neurobehavior. Early Hum Dev. 2017;113:18–22. 10.1016/j.earlhumdev.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, Goeva A, Nemesh J, Kamitaki N, Brumbaugh S, Kulp D, McCarroll SA. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174(4):1015–e103016. 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott-Hewitt N, Mahoney M, Huang Y, Korte N, de Soysa TY, Wilton DK, Knorr E, Mastro K, Chang A, Zhang A, Melville D, Schenone M, Hartigan C, Stevens B. Microglial-derived C1q integrates into neuronal ribonucleoprotein complexes and impacts protein homeostasis in the aging brain. Cell. 2024;0(0). 10.1016/j.cell.2024.05.058. [DOI] [PubMed]
- 52.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10(2):89–102. 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 53.Smith DE, Lipsky BP, Russell C, Ketchem RR, Kirchner J, Hensley K, Boissonneault V, Plante M-M, Rivest S, Huang Y, Friedman W, Sims JE. A novel CNS-Restricted isoform of the IL-1R accessory protein modulates neuronal responses to IL-1. Immunity. 2009;30(6):817–31. 10.1016/j.immuni.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takács G, Lukáts B, Papp S, Szalay C, Karádi Z. Taste reactivity alterations after IL-1β microinjection into the ventromedial hypothalamic nucleus of the rat. Neurosci Res. 2008;62(2):118–22. 10.1016/j.neures.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 55.Takao T, Tracey DE, Mitchell WM, De EBS. Interleukin-1 receptors in mouse brain:characterization and neuronal localization. Endocrinology. 1990;127(6):3070–8. 10.1210/endo-127-6-3070. [DOI] [PubMed] [Google Scholar]
- 56.Takemiya T, Fumizawa K, Yamagata K, Iwakura Y, Kawakami M. Brain Interleukin-1 facilitates learning of a water maze spatial memory Task in Young mice. Front Behav Neurosci. 2017;11:202. 10.3389/fnbeh.2017.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, Yu J, Perez-Torres C, Frouin A, Wilton DK, Funk K, DeMasters BK, Jiang X, Bowen JR, Mennerick S, Robinson JK, Garbow JR, Tyler KL, Suthar MS, Klein RS. A complement–microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534(7608). 10.1038/nature18283. Article 7608. [DOI] [PMC free article] [PubMed]
- 58.Viviani B, Bartesaghi S, Gardoni F, Vezzani A, Behrens MM, Bartfai T, Binaglia M, Corsini E, Di Luca M, Galli CL, Marinovich M. Interleukin-1β enhances NMDA receptor-mediated intracellular calcium increase through activation of the src family of Kinases. J Neurosci. 2003;23(25):8692–700. 10.1523/JNEUROSCI.23-25-08692.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong R, Lénárt N, Hill L, Toms L, Coutts G, Martinecz B, Császár E, Nyiri G, Papaemmanouil A, Waisman A, Müller W, Schwaninger M, Rothwell N, Francis S, Pinteaux E, Denés A, Allan SM. Interleukin-1 mediates ischaemic brain injury via distinct actions on endothelial cells and cholinergic neurons. Brain Behav Immun. 2019;76:126–38. 10.1016/j.bbi.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, Zhang R, Li X, Feng T, Yan N. The moderating role of sensory processing sensitivity in the link between stress and depression: a VBM study. Neuropsychologia. 2021;150:107704. 10.1016/j.neuropsychologia.2020.107704. [DOI] [PubMed] [Google Scholar]
- 61.Yamagata A, Yoshida T, Sato Y, Goto-Ito S, Uemura T, Maeda A, Shiroshima T, Iwasawa-Okamoto S, Mori H, Mishina M, Fukai S. Mechanisms of splicing-dependent trans-synaptic adhesion by PTPδ–IL1RAPL1/IL-1RAcP for synaptic differentiation. Nat Commun. 2015;6(1):6926. 10.1038/ncomms7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yates SC, Groeneboom NE, Coello C, Lichtenthaler SF, Kuhn P-H, Demuth H-U, Hartlage-Rübsamen M, Roßner S, Leergaard T, Kreshuk A, Puchades MA, Bjaalie JG. QUINT: Workflow for Quantification and Spatial Analysis of Features in histological images from Rodent Brain. Front Neuroinformatics. 2019;13:75. 10.3389/fninf.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yirmiya R. Brain Interleukin-1 is involved in spatial memory and Passive Avoidance Conditioning. Neurobiol Learn Mem. 2002;78(2):379–89. 10.1006/nlme.2002.4072. [DOI] [PubMed] [Google Scholar]
- 64.Yirmiya R, Goshen I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain Behav Immun. 2011;25(2):181–213. 10.1016/j.bbi.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 65.Yoshida T, Shiroshima T, Lee S-J, Yasumura M, Uemura T, Chen X, Iwakura Y, Mishina M. Interleukin-1 receptor accessory protein organizes neuronal synaptogenesis as a cell adhesion molecule. J Neurosci. 2012;32(8):2588–600. 10.1523/JNEUROSCI.4637-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao X, van Praag H. Steps towards standardized quantification of adult neurogenesis. Nat Commun. 2020;11(1). 10.1038/s41467-020-18046-y. [DOI] [PMC free article] [PubMed]
- 67.Zhu C-B, Lindler KM, Owens AW, Daws LC, Blakely RD, Hewlett WA. Interleukin-1 receptor activation by systemic lipopolysaccharide induces behavioral despair linked to MAPK regulation of CNS serotonin transporters. Neuropsychopharmacology. 2010;35(13):2510–20. 10.1038/npp.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu G, Okada M, Yoshida S, Mori F, Ueno S, Wakabayashi K, Kaneko S. Effects of interleukin-1β on hippocampal glutamate and GABA releases associated with Ca2+-induced Ca2 + releasing systems. Epilepsy Res. 2006;71(2):107–16. 10.1016/j.eplepsyres.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Zhu L, Liu X, Nemeth DP, DiSabato DJ, Witcher KG, Mckim DB, Oliver B, Le X, Gorantla G, Berdysz O, Li J, Ramani AD, Chen Z, Wu D, Godbout JP, Quan N. Interleukin-1 causes CNS inflammatory cytokine expression via endothelia-microglia bi-cellular signaling. Brain Behav Immun. 2019;81:292–304. 10.1016/j.bbi.2019.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.