Abstract
Introduction
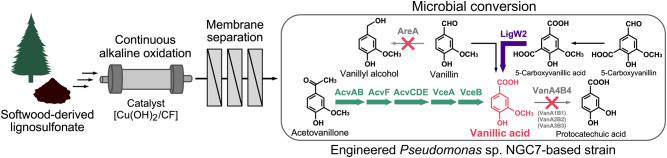
Lignin is a promising resource for obtaining aromatic materials, however, its heterogeneous structure poses a challenge for effective utilization. One approach to produce homogeneous aromatic materials from lignin involves the application of microbial catabolism, which is gaining attention. This current study focused on constructing a catabolic pathway in Pseudomonas sp. NGC7 to produce vanillate (VA) from aromatic compounds derived from the chemical depolymerization of sulfite lignin.
Results
Alkaline oxidation of sulfite lignin was performed using a hydroxide nanorod copper foam [Cu(OH)2/CF]-equipped flow reactor. The flow reactor operated continuously for 50 h without clogging and it yielded a sulfite lignin stream containing acetovanillone (AV), vanillin (VN), and VA as the major aromatic monomers. The catabolic pathway of Pseudomonas sp. NGC7 was optimized to maximize VA production from aromatic monomers in the sulfite lignin stream derived from this oxidation process. Pseudomonas sp. NGC7 possesses four gene sets for vanillate O-demethylase, comprising the oxygenase component (vanA) and its oxidoreductase component (vanB). Among these, the vanA4B4 gene set was identified as the key contributor to VA catabolism. To facilitate the conversion of AV to VA, AV-converting enzyme genes from Sphingobium lignivorans SYK-6 were introduced. The ΔvanA4B4 strain, harboring these AV-converting genes, produced VA from the sulfite lignin stream with 91 mol%. Further disruption of vanA1B1, vanA2B2, vanA3B3, and a vanillin reductase gene, in addition to vanA4B4, and introduction of a 5-carboxyvanillate decarboxylase gene from S. lignivorans SYK-6 to utilize 5-carboxyvanillin and 5-carboxyvanillate from the sulfite lignin stream for VA production achieved a VA yield of 103 mol%.
Conclusion
Developing methods to overcome lignin heterogeneity is essential for its use as a raw material. Consolidating continuous alkaline oxidation of lignin in a Cu(OH)2/CF-packed flow reactor and biological funneling using an engineered catabolic pathway of Pseudomonas sp. NGC7 is a promising approach to produce VA for aromatic materials synthesis. NGC7 possesses a higher adaptability to various aromatic compounds generated from the alkaline oxidation of lignin and its natural ability to grow on p-hydroxyphenyl, guaiacyl, and syringyl compounds underscores its potential as a bacterial chassis for VA production from a wide range of lignin-derived aromatic compounds.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-024-02590-z.
Keywords: Pseudomonas, Vanillate, Sulfite lignin, Continuous alkaline depolymerization, Biological funneling
Background
Lignin is the most abundant aromatic macromolecule in nature and a promising source for producing aromatic materials. However, its heterogeneous structure poses challenges for its use as a raw material. To produce homogeneous aromatic materials from lignin, it is desirable to first depolymerize lignin and then separate or fractionate a preferred aromatic compound from the mixture generated by the lignin depolymerization. Various physicochemical approaches have been proposed for separation and fractionation, including solid extraction using ion exchange or non-polar resins, centrifugal partition chromatography, solvent extraction, and membrane separation [1–5]. In addition to these methods, application of bacterial catabolism, known as biological funneling, is gaining attention as a mean to overcome lignin heterogeneity for material production [6, 7]. By applying the biological funneling concept, the production of aliphatic compounds, such as muconate, beta-ketoadipate, itaconate, and polyhydroxyalkanoate [8–13], and several aromatic monomers, such as vanillate (VA), protocatechuate, and gallate, from lignin-derived aromatic mixture obtained through chemical decomposition [14–16] has been reported. Among these aromatic compounds, VA is particularly promising, as it has been proposed for the synthesis of functionalized polymers [17–19]. Previously, we reported VA production from depolymerized sulfite lignin using an engineered Sphingobium lignivorans SYK-6-based strain [15]. In that study, by disrupting the genes responsible for tetrahydrofolate-dependent degradation of VA (ligM and desA) [20, 21], the strain produced VA from an aromatic mixture primarily composed of acetovanillone (AV), vanillin (VN), and VA, obtained through the copper hydroxide-catalyzed alkaline oxidation of sulfite lignin. Moreover, to the best of our knowledge, it was the first time that the lignin-derived VA was applicable to synthesize polyethylene vanillate via methyl esterification, hydroxyethylation, distillation, and catalytic polymerization [15]. In this study, we used Pseudomonas sp. NGC7 as a chassis to construct VA-producing strain. NGC7 can utilize various compounds having a guaiacyl nucleus [e.g., ferulate (FA), VA, and VN], a p-hydroxyphenyl nucleus [e.g., p-coumarate (CA), 4-hydroxybenzaldehyde (HBN), and 4-hydroxybenzoate (HBA)], and a syringyl nucleus [e.g., syringaldehyde (SN) and syringate (SA)] [22], and we have reported that NGC7 is a potent bacterial chassis for developing strains capable of producing value-added chemicals from lignin-derived aromatics mixture [9]. The specific growth rate measurements of NGC7 and SYK-6 in lysogeny broth (LB) in the presence of various concentrations of lignin-related aromatics (AV, VN, VA, HBN, HBA, SN, and SA) revealed that NGC7 exhibited higher growth rates than SYK-6 under all conditions examined (Figure S1). Phylogenetic and biochemical characterization strongly suggests that NGC7 belongs to Pseudomonas putida [9]. P. putida is well-known for its versatility as a biocatalyst [23], with KT2440 being the most extensively studied strain for engineering the production of chemicals from lignin-derived compounds [24, 25]. KT2440 can also utilize various compounds having a guaiacyl (e.g., FA, VA, and VN) and a p-hydroxyphenyl nucleus (e.g., CA, HBN, and HBA). However, it requires the overexpression of the genes responsible for VA O-demethylase, which consists of the oxygenase component (vanA) and oxidoreductase component (vanB), and the addition of glucose to utilize SA [26]. In contrast, NGC7 can originally utilize SA and SN as well as guaiacyl and p-hydroxyphenyl compounds without the need for additional carbon sources or gene overexpression. Moreover, it exhibits higher tolerance to AV, HBA, HBN, SA, SN, VA, and VN than KT2440 [9]. Therefore, in this study, we engineered the catabolic pathway of NGC7 to enable VA production from a mixture of aromatics generated by the copper hydroxide-catalyzed alkaline oxidative depolymerization of sulfite lignin.
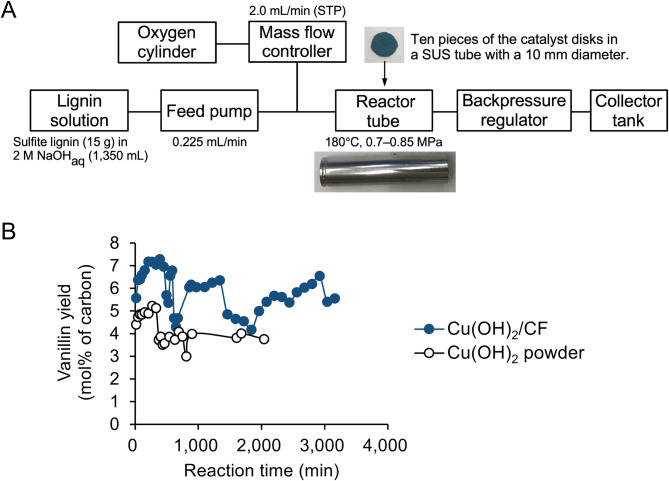
In our previous work, we optimized the alkaline oxidation conditions for sulfite lignin using oxygen as an oxidant and copper hydroxide as a catalyst to obtain a mixture of aromatic compounds suitable for subsequent biological conversion to VA. We performed repeated alkaline oxidations of sulfite lignin in batch mode, producing sufficient quantities of depolymerized products for biological conversion to VA [15]. In the present study, we advanced this process by alkaline oxidation of sulfite lignin in a continuous flow reactor equipped with an immobilized copper hydroxide-packed column (Fig. 1). We developed a hydroxide nanorod-modified copper foam [Cu(OH)2/CF], which demonstrated catalytic activity comparable to that of powdered copper hydroxide in the alkaline oxidation of sulfite lignin [27]. Therefore, in this study, we conducted continuous alkaline oxidation of sulfite lignin in a Cu(OH)2/CF-packed flow reactor. And, we optimized the catabolic pathway of Pseudomonas sp. NGC7 to produce VA from the mixture of aromatic compounds derived from the oxidation of sulfite lignin.
Fig. 1.
Outline of the Cu(OH)2/CF-packed column-equipped continuous flow reactor (A) and VN yield profiles (B). A single stainless column (10 mm in diameter, 50 mm in length) packed with 12 mmol of Cu2+ carrying powdered Cu(OH)2 (dot line, Tokyo Chemical Industry, Co. Ltd.) or Cu(OH)2/CF (solid line)
Materials and methods
Continuous flow catalytic alkaline oxidation of sulfite lignin
Sulfite lignin and sodium hydroxide were purchased from Tokyo Chemical Industry Co., Ltd., Japan, and FUJIFILM Wako Pure Chemical Corporation, Japan, respectively. Copper foam, with a thickness of 3 mm, was acquired from Xiamen TOB New Energy Technology Co. Ltd., China. Alkaline oxidation of sulfite lignin was conducted in an MCR-1000 continuous flow reactor system (TOKYO RIKAKIKAI CO., LTD., Japan), equipped with a reactor tube, an aluminum block heater for heating the tube, an inlet connector for liquid and gas supply, and an outlet connector. Cu(OH)2/CF was prepared from 3 mm-thick copper foam sheets, as previously reported [27]. Ten circular disks, each with a diameter of 10 mm, were fabricated from the prepared Cu(OH)2/CF and loaded into a stainless-steel (SUS316L) reactor tube with dimensions of 10 mm in diameter and 50 mm in length. The loaded catalyst contained ca. 12 mmol of Cu2+ on its surface. The reactor tube containing Cu(OH)2/CF was then mounted on a block heater of the MCR-1000 system. An alkaline sulfite lignin solution (15 g of sulfite lignin dissolved in 1350 mL of 2 M aqueous NaOH solution) was continuously supplied into the reactor tube at a flow rate of 0.225 mL/min using a high-pressure feed pump (EUI-22-110P, TOKYO RIKAKIKAI CO., LTD.). The outlet of the reactor tube was connected to a back-pressure regulator (P-787, IDEX Health & Science, LLC, WA) to maintain the reactor pressure between 0.75 and 0.85 MPa. Once the reactor tube was fully filled with the lignin solution, the temperature was increased to 180 °C. After the reactor temperature stabilized, gaseous oxygen was introduced at a flow rate of 2.0 mL/min using a mass-flow controller (SEC-E40MK3, HORIBA STEC, Co., Ltd., Japan) to initiate the reaction. The solution from the outlet was periodically sampled and analyzed using a high-performance liquid chromatography equipped with InertSustain C18 column (1.9 μm, 2.1 × 75 mm, GL Science, Inc., Japan) as previously reported [27] to monitor the yield changes of vanillin and other products over time. For comparison, a reactor tube loaded with 1.20 g of Cu(OH)2 powder (12.3 mmol Cu2+; FUJIFILM Wako Pure Chemical Corp.) was used.
The filtrated alkaline liquid fraction obtained from the alkaline oxidation reaction of sulfite lignin was adjusted to pH 10 using HCl (12 M or 6 M) and then filtrated through a vacuum filter system (0.22 μm pore size, Corning, Inc., NY). The permeate was further filtrated using an HP4750 high-pressure stirred cell (Sterlitech Corporation, WA) equipped with a 5,000 MWCO UF membrane (MT, Synder Filtration, CA). Subsequently, the 5-kDa UF permeate was filtrated using a 1,000 MWCO UF membrane (GE, Veolia Water Technologies & Solutions, PA) in the same cell. The 1-kDa UF permeate was then acidified to pH 3 with HCl and extracted three times with an equal volume of ethyl acetate. Ethyl acetate was evaporated, and the resulting extracts were dissolved in water with NaOH (pH = 8, designated the sulfite lignin stream).
Genome sequencing, assembly, and annotation
Genome sequencing was performed with an Illumina Hiseq-2500 sequencer using 100 bp paired-end mode by Hokkaido System Science Co., Ltd (Japan). Raw reads were filtered based on purity, retaining only those with chastity values of > 0.6. The resulting reads were pre-processed with the Cutadapt program (ver. 1.1) [28] and Trimmomatic (ver. 0.32) [29] to remove adapter sequences and low-quality reads. De novo genome assembly was performed using Velvet (ver. 1.2.10) [30] and Platanus (ver. 1.2.4) [31]. Whole genome comparison of the assembled genome was conducted using the Microbial Genome Atlas [32], which indicated that the NGC7 genome was most similar to P. putida KT2440 genome (average nucleotide identity, 90%). Consequently, the assembled contigs were sorted and combined based on the KT2440 genome using the ContigAligner program in GenomeMatcher (ver. 3.0) [33]. ORF prediction and annotation were performed via the RAST server (http://rast.nmpdr.org/) [34]. The annotated genome has been deposited in the DDBJ/EMBL/GenBank databases (accession number, SAMD00818483).
Effect of each vanillate O-demethylase gene disruption on ability of NGC7 to grow on VA and SA
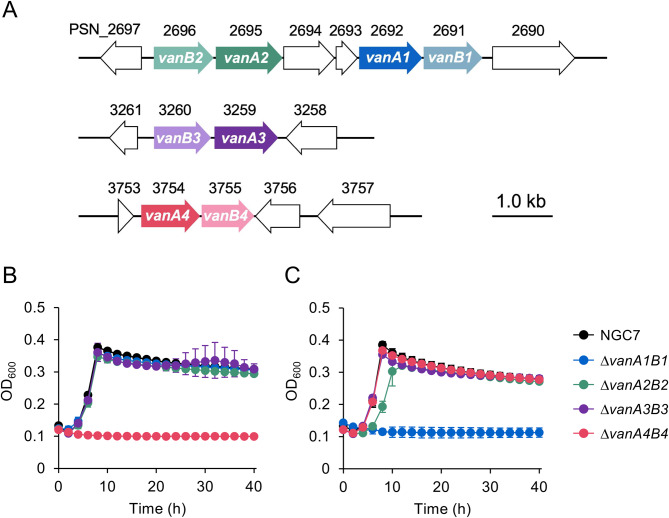
All strains (Table S1), plasmids (Table S1), and oligonucleotide sequences (Table S3) used in this study are listed in the supporting materials. To disrupt each region containing vanA1 and vanB1, vanA2 and vanB2, vanA3 and vanB3, or vanA4 and vanB4, the 5′- (ca. 1 kb) and 3′-stream (ca. 1 kb) regions were amplified by PCR and cloned into BamHI-digested pK18mobsacB [35] using NEBuilder HiFi DNA assembly (New England Biolabs, MA) to generate pK18ΔvanA1B1, pK18ΔvanA2B2, pK18ΔvanA3B3, or pK18ΔvanA4B4, respectively (Table S2). For disrupting the vanA1B1-containing region, NGC7 was transformed with pK18ΔvanA1B1 by electroporation (15 kV/cm), and the transformants were incubated on LB agar plates containing 25 mg/L kanamycin (Km) at 30 °C. The Km-resistant clones were shake-cultured in LB containing 25%(w/v) sucrose at 30 °C, followed by streaking a portion of the culture on LB agar plates containing 25% (w/v) sucrose and incubated at 30 °C. Several colonies sensitive to Km were selected, and the deletion of vanA1B1 was confirmed by PCR (designated NGC711; Figure S2, Table S1). Similarly, the regions containing vanA2 and vanB2, vanA3 and vanB3, or vanA4 and vanB4, were disrupted using pK18ΔvanA2B2, pK18ΔvanA3B3, or pK18ΔvanA4B4, respectively, resulting in strains designated NGC709, NGC710, or NGC711.
The proliferation abilities of the strains NGC7, NGC708, NGC709, NGC710, and NGC711 on VA and SA were evaluated by measuring the optical density at 600 nm (OD600) using a microplate spectrophotometer (567 cpm, 30 °C; BioTek Epoch2, Agilent Technologies, Inc., CA) in triplicate. Each strain was shake-cultured in 10 mL of LB at 30 °C for 16 h. The cells were collected by centrifugation, washed twice with Wx medium consisting of 9.8 g/L Na2HPO4·12H2O, 1.7 g/L KH2PO4, 1.0 g/L (NH4)2SO4, and the trace metal solution [20], and then suspended in Wx medium to be used as an inoculum. A portion of the inoculum was added to adjust OD600 of 0.2 in 0.2 mL of Wx medium containing 5 mM VA or SA.
Effect of VA O-demethylase genes disruption on VA yield from sulfite lignin-derived aromatics
NGC711 was transformed with pSEVAacv and pTS093vce, and the resulting strain, NGC711[pSEVAacv, pTS093vce], was shake-cultured in 10 mL of LB containing 25 mg/L Km and 15 mg/L tetracycline (Tc) at 30 °C for 16 h. A portion of the culture was centrifuged, harvested cells were washed twice with MMx-3 buffer [34.2 g/L Na2HPO4·12H2O, 6.0 g/L KH2PO4, 2.5 g/L (NH4)2SO4, and 1.0 g/L NaCl], and then resuspended in the same buffer to an OD600 of 10 to use an inoculum. Ten milliliters of MMx-3 [MMx-3 buffer, 493 mg/L MgSO4·7H2O, 15 mg/L CaCl2·2H2O, 5 mg/L FeSO4·7H2O] containing 0.1 mL of inoculum, 15 g/L glucose, 0.2 mL of the sulfite lignin stream, 25 mg/L Km, and 15 mg/L Tc was shake-incubated at 30 °C.
During the deletion of the four vanAB gene sets of NGC7, it was discovered that NGC7 harbors the aminoglycoside 3′-phosphotransferase gene (aph) and exhibits weak Km resistance. Then, the 5′- (ca. 1.2 kb) and 3′-stream regions (ca. 1.2 kb) of aph were amplified by PCR and cloned into pK18mobsacB using NEBuilder HiFi DNA assembly (pK18Δaph, Table S2). aph was then deleted from NGC712, a mutant deficient in vanA1B1 and vanA4B4, which had been constructed by transforming NGC708 with pK18ΔvanA4B4_2. Subsequently, vanA2B2 and vanA3B3 were deleted in order using pK18ΔvanA2B2_2 and pK18ΔvanA3B3, respectively. The resulting strain NGC720, in which all vanAB gene sets were disrupted, was transformed with pSEVAacv and pTS093vce. The VA-producing ability of NGC720[pSEVAacv, pTS093vce] was then evaluated in the same manner as described above. VA yield was calculated as [VA (mol) at 68 h/sum of AV, VN, and VA (mol) at the start] × 100%.
Effects of VN reductase disruption and 5CVA decarboxylase introduction on VA yield from the aromatics in sulfite lignin stream
The VN reductase gene areA was deleted from the genomic DNA of NGC720 via homologous recombination using a pK18mobsacB-derived plasmid. This plasmid carried a 3-kb fragment composed of the 5′- (ca. 1.5 kb) and 3′-stream (ca. 1.5 kb) regions of areA amplified by PCR and cloned into pK18mobsacB using NEBuilder HiFi DNA assembly (pK18ΔareA, Table S2). The effect of areA deletion on VN conversion was evaluated using the resting cell reaction method as follows: the ΔareA strain NGC729 was shake-cultured in 10 mL of LB containing 10 mM VN at 30 °C for 16 h, a portion of the culture was centrifuged, collected cells were washed twice with MMx-3 buffer, and then resuspended in the same buffer to an OD600 of 10 for use as an inoculum. The MMx-3 buffer (1.2 mL) containing 0.12 mL of the inoculum and 10 mM VN was shake-incubated using a Bioshaker (30 °C, 1,500 rpm, DWMax M·BR-034, TAITEC CORPORATION, Japan). Two hundred portions of the suspension were periodically collected, and the concentrations of VA, VN, and vanillyl alcohol were measured.
The Δaph strain NGC715 was evaluated for its ability to oxidize 5-carboxyvanillin (5CVN) using the resting cell reaction method. The strain was shake-cultured in 10 mL of LB at 30 °C for 16 h. A portion of the culture was centrifuged, collected cells were washed twice with MMx-3 buffer, and then resuspended in the same buffer to an OD600 of 10 for use as an inoculum. The MMx-3 buffer (1.2 mL) containing 0.12 mL of the inoculum and 1.0 mM 5CVN was shake-incubated with a Bioshaker (30 °C, 1,500 rpm, DWMax M·BR-034). Two hundred portions of the suspension were periodically collected, and the concentrations of 5CVN and 5-carboxyvanillate (5CVA) were measured.
5CVA decarboxylase genes (ligW and ligW2, accession no. AB033664 and AB089690, respectively) were amplified by PCR (Table S3) from the genomic DNA of Sphingobium lignivorans SYK-6 [36, 37]. These genes were then separately cloned into HindIII-digested pTS093vce (Table S2). NGC715 was transformed with either pTS093ligW-vce or pTS093ligW2-vce and subsequently evaluated for 5CVA decarboxylation activity using the resting cell reaction method, as described in previous sections, with the exception that 5CVA concentration was set at 200 µM. NGC729 was transformed with pSEVAacv and pTS093ligW2-vce, and the resulting strain NGC729[pSEVAacv, pTS093ligW2-vce] was assessed for its VA-producing ability under the same conditions. VA yield was calculated as [VA (mol) at 116 h/sum of AV, VN, VA, 5CVN, 5CVA (mol) at the start] × 100%.
Analytical methods
OD600 values were measured using a spectrophotometer (BIOmaster XB-10, TOMY CO., LTD., Japan), and glucose concentrations in the culture were determined with a Biosensor (BF-5, Oji Scientific Instruments). The concentrations of 5CVA, 5CVN, AV, VA, vanillyl alcohol, and VN in the culture were quantified with a high-performance liquid chromatography (HPLC, 1200 series, Agilent Technologies Inc.) equipped with a ZORBAX Eclipse Plus C18 column (reverse phase, 4.6 mm in diameter, 150 mm in length, 5 μm particle size) run at 40 °C with a mobile phase gradient [solvent A: 5% (v/v) CH3CN and 1% (v/v) CH3COOH in H2O; solvent B: 50% (v/v) CH3CN and 1% (v/v) CH3COOH in H2O]. The latter was introduced after the injection of samples and ramped from 0 to 20% in the first 8 min, and then to 100% in the next 5 min, and maintained for 5 min. The flow rate of the mobile phase was 1.0 mL/min, and the detection wavelengths were 254 nm (for VA and 5CVA), 280 nm (AV, VN, and vanillyl alcohol), and 330 nm (5CVN). Standards for 5CVN (Merck KGaA, Germany), AV (Tokyo Chemical Industry Co.), VA (FUJIFILM Wako Pure Chemical Corporation), vanillyl alcohol (Tokyo Chemical Industry Co.), and VN (Tokyo Chemical Industry, Co.) were acquired. 5CVA was prepared as previously described [37].
Results and discussion
Continuous alkaline oxidation of sulfite lignin
The sulfite lignin stream was prepared via alkaline oxidation in a flow reactor equipped with Cu(OH)2/CF-packed column (Fig. 1A). Our prior work required repeated batch reactions of the Cu(OH)2-catalyzed alkaline oxidation to continuously prepare aromatic monomers for biological processing. This study explored a flow reactor for continuous Cu(OH)2-catalyzed alkaline oxidation. The flow reactor loaded with Cu(OH)₂/CF effectively depolymerized lignin into monoaromatic phenolic compounds. The yield of vanillin, the most abundant product in the outlet solution, was monitored over time (Fig. 1B). Although the yield exhibited some fluctuations, it consistently ranged from 4 to 7% based on the carbon content in the lignin solution. For comparison, the lignin solution was also depolymerized using a flow reactor loaded with Cu(OH)2 powder. However, in this case, the vanillin yield over time was lower, ranging from 3 to 5%. In addition, after 2,000 min of operation, a sudden increase in reactor pressure indicated a blockage caused by aggregation of the catalyst powder. In contrast, the flow reactor loaded with Cu(OH)2/CF remained active even after 3,000 min of operation. The higher vanillin yield achieved with the Cu(OH)2/CF-loaded reactor is attributed to the improved contact between the lignin solution and catalyst, as the Cu(OH)2/CF catalyst resists aggregation and the mesh-shaped copper foam prevents the clogging issue experienced with powdered Cu(OH)2. Further detailed evaluations and optimizations of the Cu(OH)2/CF-catalyzed continuous alkaline oxidation process, including catalyst preparation and operating conditions, would contribute to developing further efficient lignin alkaline oxidation systems.
Revealing four gene sets homologous to VA O-demethylase oxygenase component (vanA) and VAO-demethylase oxidoreductase component (vanB)
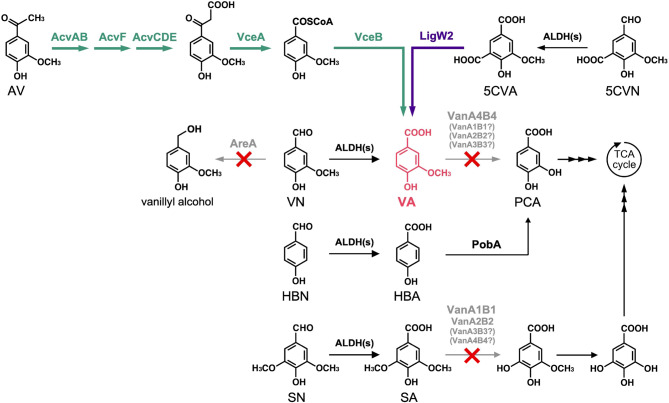
The draft genome sequence of the 6.34 Mb genome of Pseudomonas sp. NGC7 was analyzed to identify the genes responsible for VA degradation. The deduced amino acid sequences of VA O-demethylase oxygenase component (VanA, accession no. AAN69332.1) and VA O-demethylase oxidoreductase component (VanB, AAN69333.1) from P. putida KT2440 or tetrahydrofolate-dependent VA O-demethylase (LigM, BAK65949) from S. lignivorans SYK-6 were used as queries in a Basic Local Alignment Search Tool (TBLASTN) against the genome sequence of NGC7. This analysis revealed four coding sequences with amino acid sequence similarities with VanA and six with similarities to VanB (Table 1). In particular, vanA1 and vanB1, vanA2 and vanB2, vanA3 and vanB3, and vanA4 and vanB4 were adjacent to each other (Fig. 2A). The deduced amino acid sequences of VanA4 and VanB4 showed the highest identities (94% and 89%, respectively) with those of VanA and VanB from P. putida KT2440. No significant regions encoding LigM homologs were identified. Four mutants of NGC7, each deficient in vanA1B1, vanA2B2, vanA3B3, or vanA4B4, were constructed (Figure S2 and Table S1) and their availabilities to assimilate VA and its analog, SA, were evaluated (Fig. 2B) as NGC7 can naturally utilize not only VA but also SA as a sole source of carbon for biomass and energy (Fig. 3) [22] and it was reported that VanAB from P. putida KT2440 catalyzed O-demethylation of SA as well as VA [26]. Among the ΔvanAB mutants, NGC711, a ΔvanA4B4 strain, could not grow on VA, and the other three mutants did not show significant differences in growth compared to the wild-type strain NGC7 (Fig. 2B). Moreover, a hydroxybenzoate-derivative transporter gene (PSN_3757), which is part of the major facilitator superfamily, was found in the vicinity of vanA4B4 (Fig. 2A; Table 1). Although NGC711 did not exhibit any changes in growth on SA compared to NGC7, NGC708, a ΔvanA1B1 strain, lost its ability to grow on SA, and NGC709, a ΔvanA2B2 mutant, showed a slight decline in growth rate on SA compared to NGC7, NGC710 (a ΔvanA3B3 strain), and NGC711 (Fig. 2C). Between the vanA1B1 and vanA2B2 gene clusters, putative genes (PSN_2693 and PSN_2694) related to gallate metabolism were found (Fig. 2A; Table 1). This suggests that SA is metabolized through intermediates, such as 3-O-methylgallate and gallate, similar to the pathways in P. putida [38, 39]. Consequently, vanA1B1 and vanA2B2 are likely involved in SA catabolism in NGC7. These results suggest that vanA4B4 is primarily responsible for VA O-demethylation, vanA1B1 is involved in SA O-demethylation, and vanA2B2 also plays a role in SA catabolic step to some extent (Fig. 3). The functions of vanA3B3, vanB5, and vanB6 were not determined, as no genes associated with lignin-derived aromatics metabolism were identified in their vicinity.
Table 1.
Genes in the vicinity of vanA and vanB in NGC7
| Locus tag | Similar proteina | Organism | Accession No. | Identityb |
|---|---|---|---|---|
| vanA1B1, vanA2B2 | ||||
| PSN_2690 | Gallate transporter | P. putida | E8ZB61.1 | 29 |
| PSN_2691 (vanB1) | Carnitine monooxygenase reductase subunit | Acinetobacter baumannii ATCC 19606 | D0C9N8.1 | 28 |
| PSN_2692 (vanA1) | Vanillate O-demethylase oxygenase subunit | Pseudomonas sp. HR199 | O05616.1 | 42 |
| PSN_2693 | Gallate dioxygenase | P. putida KT2440 | Q88JX5.1 | 17 |
| PSN_2694 | Protocatechuate 4,5-dioxygenase beta chain | S. lignivorans SYK-6 | P22636.1 | 41 |
| PSN_2695 (vanA2) | Toluene-4-sulfonate monooxygenase system iron-sulfur subunit TsaM1 | Comamonas testosteroni | P94679.1 | 31 |
| PSN_2696 (vanB2) | Putative toluene-4-sulfonate monooxygenase system reductase subunit TsaB2 | C. testosteroni | Q9AHG2.1 | 38 |
| PSN_2697 | Uncharacterized HTH-type transcriptional regulator YdhC | Bacillus subtilis 168 | O05494.1 | 23 |
| vanA3B3 | ||||
| PSN_3258 | HTH-type transcriptional regulator PcaQ | Agrobacterium fabrum C58 | P0A4T6.1 | 28 |
| PSN_3259 (vanA3) | Putative toluene-4-sulfonate monooxygenase system iron-sulfur subunit TsaM2 | C. testosteroni | Q9AHG3.1 | 35 |
| PSN_3260 (vanB3) | Vanillate O-demethylase oxidoreductase | Pseudomonas sp. HR199 | O05617.1 | 56 |
| PSN_3261 | Not found | - | ||
| vanA4B4 | ||||
| PSN_3753 | Flagellar basal-body rod protein FlgG | Buchnera aphidicola Sg | Q8K9K4.1 | 14 |
| PSN_3754 (vanA4) | Vanillate O-demethylase oxygenase subunit | Pseudomonas sp. HR199 | O05616.1 | 78 |
| PSN_3755 (vanB4) | Vanillate O-demethylase oxidoreductase | P. putida | O54037.1 | 93 |
| PSN_3756 | Uncharacterized HTH-type transcriptional regulator YdhC | Bacillus subtilis 168 | O05494.1 | 27 |
| PSN_3757 | Gallate transporter | P. putida | E8ZB61.1 | 35 |
| vanB5 | ||||
| PSN_3718 | FMN reductase (NADH) RutF | Ancylobacter novellus DSM 506 | D7A989.1 | 44 |
| PSN_3719 (vanB5) | Vanillate O-demethylase oxidoreductase | Pseudomonas sp. HR199 | O05617.1 | 50 |
| PSN_3720 | 3-oxoacyl-[acyl-carrier-protein] reductase FabG | Staphylococcus epidermidis RP62A | Q5HPW0.1 | 34 |
| vanB6 | ||||
| PSN_5572 | Porin-like protein NicP | P. putida KT2440 | Q88FY7.1 | 34 |
| PSN_5573 (vanB6) | Vanillate O-demethylase oxidoreductase | P. putida | O54037.1 | 50 |
| PSN_5574 | Not found | - |
a Best-hit gene products from similarity search using BLAST program against UniProtKB/Swiss-Prot database are shown
b Each deduced amino acid sequence was compared using the EMBOSS Needle program
Fig. 2.
vanAB gene loci in the genome of Pseudomonassp. NGC7 (A) and the effect of each vanAB disruption on the assimilation of VA (B) and SA (C). (A) The functions of the gene products, identified through similarity searches using BLAST program are detailed in Table 1. (B) and (C) Each vanAB mutant was shake-cultured in 0.2 mL of Wx medium containing 5 mM VA or SA at 30 °C. Each value and error bar indicates the mean and standard deviation from triplicate experiments
Fig. 3.
Catabolic pathway of Pseudomonas sp. NGC7-based strain capable of producing VA from sulfite lignin stream. Four vanAB sets (vanA1B1, vanA2B2, vanA3B3, and vanA4B4) and the areA were disrupted using the homologous recombination methods. vanA4B4 is responsible for VA assimilation, whereas vanA1B1 and vanA2B2 are involved in SA assimilation. The role of vanA3B3 remains unclear. Two plasmids carrying acvA, acvB, acvC, acvD, acvE, and acvF, and vceA, vceB, and ligW2 from S. lignivorans SYK-6 were introduced. AcvAB, 4-acetyl-2-methoxyphenylphosphate/4-acetyl-2,6-dimethoxyphenylphosphate synthetase; AcvF, 4-acetyl-2-methoxyphenylphosphate/4-acetyl-2,6-dimethoxyphenylphosphate phosphatase; AcvCDE, biotin-dependent carboxylase; ALDHs, aldehyde dehydrogenases; AreA, VN reductase; LigW2, 5CVA decarboxylase; PobA, HBA monooxygenase; VanA1B1, VanA2B2, VanA3B3, and VanA4B4, isozymes of VA O-demethylase; VceA, vanilloyl acetate/3-(4-hydroxy-3,5-dimethoxyphenyl)-3-oxopropanoate-converting enzyme; VceB, vanilloyl-CoA/syringoyl-CoA thioesterase; Vdh, VN dehydrogenase. 5CVA, 5-carboxyvanillate; 5CVN, 5-carboxyvanillin; AV, acetovanillone; HBA, 4-hydroxybenzoate; HBN, 4-hydroxybenzaldehyde; PCA, protocatechuate; VA, vanillate; VN, vanillin; SA, syringate; SN, syringaldehyde
Modification of catabolic pathway of Pseudomonas sp. NGC7 to yield VA from sulfite lignin-derived aromatic monomers
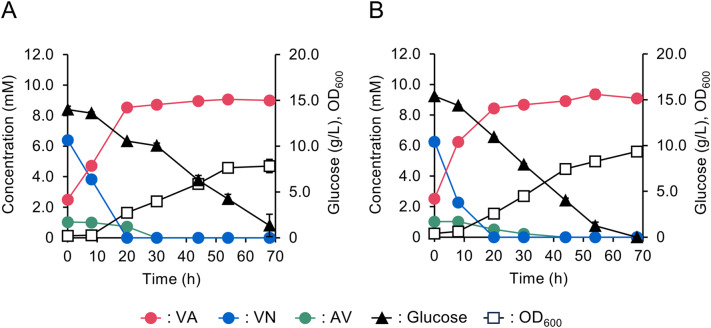
It was confirmed that different vanAB genes are responsible for the assimilation VA and SA in NGC7, with vanA4B4 being identified as the major gene for VA catabolism. NGC711 was, thus, chosen to assess its potential for VA production from the sulfite lignin stream (Fig. 4). NGC711 was transformed with pSEVAacv and pTS093vce (Table S2). The resulting strain NGC711[pSEVAacv, pTS093vce] was cultured in a synthetic medium with glucose as a source of carbon and the sulfite lignin stream containing AV, VN, and VA (Fig. 4A). NGC711[pSEVAacv, pTS093vce] produced VA from the sulfite lignin stream with 91 ± 2 mol% yield. However, the sole disruption of vanA4B4 was insufficient to achieve the theoretical molar yield of VA (100 mol%, in which all AV and VN added to culture are converted to VA stoichiometrically). In addition, in a fed-batch culture using a mock solution, the yield decreased to 79 ± 7 mol% over time (Figure S3A). These results suggested that other vanAB genes may also be involved in VA degradation. In NGC7, vanA1B1 and vanA2B2 were considered to involve in SA degradation, but they may contribute to VA conversion, as VanABs from P. putida KT2440 [26] and Streptomyces sp. NL15-2 K [40] have been reported to demethylate VA analogs, such as syringate, 3-O-methylgallate, and veratrate. Therefore, strains NGC717[pSEVAacv, pTS093vce], NGC718[pSEVAacv, pTS093vce], and NGC720[pSEVAacv, pTS093vce] were cultured in fed-batch mode to evaluate their VA production yields. Disrupting vanA1B1 in addition to vanA4B4 resulted in a VA yield of 75 ± 2 mol% from the mock solution (Figure S3B). Further disruption of vanA2B2 increased the yield to 88 ± 5 mol% (Figure S3C). Disrupting vanA3B3 led to 84 ± 4 mol% yield (Figure S3D). NGC720[pSEVAacv, pTS093vce] was also evaluated for VA productivity from the sulfite lignin stream in a batch culture and demonstrated a VA yield of 93 ± 2 mol% (Fig. 4B). Despite this improvement, the yield was still below the theoretical molar yield.
Fig. 4.
Evaluation of VA-producing ability from lignin-derived aromatic compounds in the sulfite lignin stream. (A) NGC711[pSEVAacv, pTS093vce], (B) NGC720[pSEVAacv, pTS093vce].
During the incubation of NGC720[pSEVAacv, pTS093vce] in the synthetic medium containing glucose and the sulfite lignin stream, a specific peak was observed in the HPLC chromatogram (Figure S4). The UV spectrum and retention time were consistent with those of vanillyl alcohol. It has been reported that P. putida GN442, a KT2440-derived strain, deficient in aldehyde dehydrogenases involved in VN oxidation, produces vanillyl alcohol during the conversion of ferulate to VN via feruloyl-CoA synthetase and feruloyl-CoA hydratase/lyase reactions [41]. In addition, an aromatic aldehyde reductase (AreA) from P. putida KT2440 reduces coniferyl aldehyde to its alcohol form. The disruption of areA in GN442 prevented the strain from producing vanillyl alcohol during the conversion of ferulate to VN, suggesting that AreA is involved in VN reduction [42]. Analysis of the NGC7 genome identified a coding sequence at the PSN_2384 locus with a 97% of deduced amino acid sequence identity to vanillin reductase (areA) from KT2440 [42]. Then, the areA-disrupted strain NGC729 was evaluated for VN reduction activity in vivo (Figure S5). NGC720 produced vanillyl alcohol in addition to VA from VN. In contrast, NGC729 converted VN to VA stoichiometrically and was expected to produce VA from the sulfite lignin stream at theoretical yield. Consequently, the VA-producing ability of NGC729[pSEVAacv, pTS093vce] was assessed and found to produce VA from the mock solution with 96 ± 4 mol% yield in fed-batch culture (Figure S3E). This study clearly indicated that integrating disruptions of vanA4B4, vanA1B1, vanA2B2, vanA3B3, and areA are effective for achieving theoretical VA production from the major aromatic monomers in the sulfite lignin stream. However, it remains unclear whether the gene products of vanA1B1, vanA2B2, and vanA3B3 exhibit significant enzymatic activities on VA. Detailed characterization of VanA1B1, VanA2B2, VanA3B3, and VanA4B4 is currently underway and will be reported in future studies.
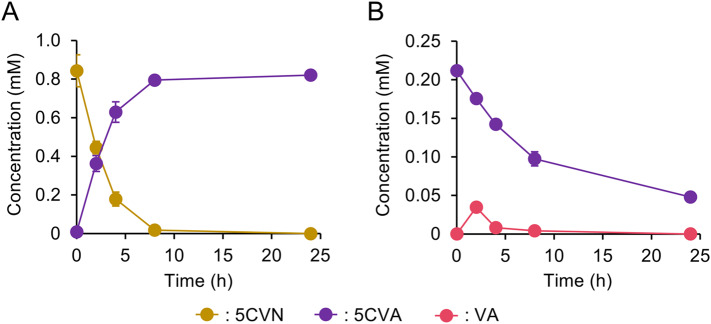
The sulfite lignin stream also contained lower amounts of HBA, HBN, 5CVA, and 5CVN (Table S4) and NGC7 utilized both HBN and HBA [22] (Fig. 3). To broaden the substrate range for VA production from the sulfite lignin stream, the conversion of 5CVN and 5CVA was engineered in NGC729 (Fig. 3). This was based on reports that 5CVA can be converted to VA via decarboxylation in S. lignivorans SYK-6 [36, 37]. In SYK-6, LigW and LigW2 were involved in the assimilation of 5,5′-dehydrodivanillic acid (DDVA), which is catabolized to VA through 5CVA. VA is then further metabolized via tetrahydrofolate-dependent VA O-demethylation and protocatechuate 4,5-ring cleavage [43]. Although NGC7 could oxidize 5CVN, it did not further convert 5CVA, as demonstrated in the in vivo assay (Fig. 5A). To confer 5CVA decarboxylation ability, a 5CVA decarboxylase gene from S. lignivorans SYK-6 (ligW2) was introduced into NGC715, a Δaph strain derivative of NGC7 (Table S1), as it was reported that a ligW2-disrupted mutant of SYK-6 utilizes DDVA slower than the ligW-disrupted one [36]. The NGC715 transformant carrying ligW2 successfully converted 5CVA (Fig. 5B). Therefore, ligW2 was further introduced into NGC729 (a ΔareA strain of NGC720) along with genes for AV conversion. The resulting strain NGC729[pSEVAacv, pTS093ligW2-vce] catabolized 5CVN, 5VA, AV, HBN, HBA, and VN in the sulfite lignin stream after 116 h of incubation and produced VA with 103 ± 1 mol% yield (Fig. 6). The slight increase in 5CVA observed during the incubation would suggest that unidentified compounds in the sulfite lignin stream may be converted to 5CVA. This conversion likely contributed to VA production exceeding the theoretical yield.
Fig. 5.
In vivo assay for 5CVN oxidation (A) and 5CVA decarboxylation (B). Resting cell reactions with NGC715 were performed to examine 5CVN (1 mM) oxidizing ability (A), and reactions with NGC715[pTS093ligW2-vce] (B) were performed to evaluate 5CVA (0.2 mM) decarboxylating activity. Each value and error bar indicates the mean and standard deviation from triplicate experiments
Fig. 6.
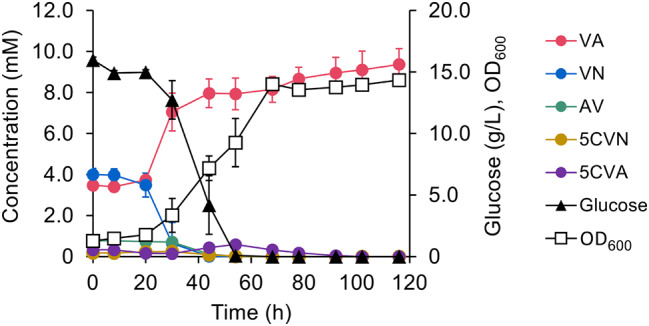
VA producing ability of NGC729[pSEVAacv, pTS093ligW2-vce] from the sulfite lignin stream. Each value and error bar indicates the mean and standard deviation from triplicate experiments
Conclusions
Developing methods and tools to manage the heterogeneity of lignin is essential for its use as a source for producing aromatic materials. Consolidating chemical depolymerization and biological funneling is a promising approach for this purpose. In the present work, the catabolic pathway of Pseudomonas sp. NGC7 was engineered to yield VA from the aromatic monomers present in the sulfite lignin stream, which is derived from alkaline oxidation through a flow reactor equipped with a Cu(OH)2/CF-packed column. The engineered strain incorporates the disruptions of four homologous gene sets for VA O-demethylase and VN reductase gene, and the introductions of 5CVA decarboxylase gene and AV-converting enzyme genes, all of which enable efficient VA production from the sulfite lignin stream. The NGC7 strain proliferates on lignin-derived p-hydroxyphenyl, guaiacyl, and syringyl compounds and exhibits tolerance to these compounds. Further efforts to engineer the catabolic pathway of NGC7 will facilitate the development of a strain capable of selectively producing VA from a mixture of aromatic compounds generated through the alkaline oxidation of lignin derived from hardwood and herbaceous sources.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by JST-MIRAI Program Grant Number JPMJMI19E2 and, in part, JST COI-NEXT Program Grant Number JPMJPF2104, and JSPS KAKENHI Grant Number 22K05399. We also thank the Gene Research Center and Shared Facility Center for Science and Technology of Hirosaki University for their technical support.
Abbreviations
- 5CVA
5-carboxyvanillate
- 5CVN
5-carboxyvanillin
- AV
Acetovanillone
- CF
Copper foam
- DDVA
5,5′-dehydrodivanillic acid
- HBA
4-hydroxybenzoate
- HBN
4-hydroxybenzaldehyde
- SA
Syringate
- SN
Syringaldehyde
- VA
Vanillate
- VN
Vanillin
- aph
Aminoglycoside 3′-phosphotransferase
- areA
Vanillin reductase
- desA
Tetrahydrofolate-dependent syringate O-demethylase
- ligM
Tetrahydrofolate-dependent vanillate O-demethylase
- ligW
5CVA decarboxylase
- ligW2
5CVA decarboxylase (isogene of ligW)
- vanA
Vanillate O-demethylase oxygenase component
- vanB
Vanillate O-demethylase oxidoreductase component
Author contributions
Mami Kamada: Investigation. Chieko Yasuda: Investigation. Yudai Higuchi: Investigation, Methodology, Visualization, Writing – review & editing. Akihiro Yoshida: Conceptualization, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft. Irwan Kurnia: Methodology, Investigation. Chiho Sakamoto: Investigation. Aya Takeuchi: Investigation. Yuta Osaka: Investigation. Kanami Muraki: Investigation. Naofumi Kamimura: Investigation, Methodology, Writing – original draft. Eiji Masai: Conceptualization, Funding acquisition, Supervision, Methodology, Writing – review & editing. Tomonori Sonoki: Conceptualization, Funding acquisition, Investigation, Supervision, Methodology, Visualization, Writing – original draft, Writing – review & editing. All authors reviewed the manuscript.
Data availability
All data are provided in the manuscript or in the supplementary information file. The genomic sequence data have been deposited in DDBJ/NCBI/EMBL under the accession no. SAMD00818483.
Declarations
Competing interests
Y.H, A.Y, N.K, E.M, and T.S. are the inventors on the patent related to this work (PCT/JP2022/13826). The authors declare no competing financial interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu Z, Jiang Y, Wang X, Xu J, Hu L. A review on the separation of lignin depolymerized products. Biomass Convers Biorefin. 2022;13(18):16667–83. [Google Scholar]
- 2.Gomes ED, Rodrigues AE. Lignin biorefinery: separation of vanillin, vanillic acid and acetovanillone by adsorption. Sep Purif Technol. 2019;216:92–101. [Google Scholar]
- 3.Moeller F, Klein J, Waldvogel SR. Selective Degradation of Technically Relevant Lignin to Vanillic Acid and Protocatechuic Acid. ChemSusChem. 2024:e202400759. [DOI] [PubMed]
- 4.Rocha ILD, da Costa Lopes AM, Ventura SPM, Coutinho JAP. Selective separation of vanillic acid from other lignin-derived monomers using centrifugal partition chromatography: the effect of pH. ACS Sustain Chem Eng. 2022;10(15):4913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, De Bruyn M, Barta K. Deriving high value products from depolymerized lignin oil, aided by (bio)catalytic funneling strategies. Chem Commun (Camb). 2023;59(66):9929–51. [DOI] [PubMed] [Google Scholar]
- 6.Linger JG, Vardon DR, Guarnieri MT, Karp EM, Hunsinger GB, Franden MA, et al. Lignin valorization through integrated biological funneling and chemical catalysis. PNAS. 2014;111(33):12013–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borchert AJ, Henson WR, Beckham GT. Challenges and opportunities in biological funneling of heterogeneous and toxic substrates beyond lignin. Curr Opin Biotechnol. 2022;73:1–13. [DOI] [PubMed] [Google Scholar]
- 8.Salvachúa D, Rydzak T, Auwae R, De Capite A, Black BA, Bouvier JT, et al. Metabolic engineering of Pseudomonas putida for increased polyhydroxyalkanoate production from lignin. Microb Biotechnol. 2020;13(1):290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akutsu M, Abe N, Sakamoto C, Kurimoto Y, Sugita H, Tanaka M, et al. Pseudomonas sp. NGC7 as a microbial chassis for glucose-free muconate production from a variety of lignin-derived aromatics and its application to the production from sugar cane bagasse alkaline extract. Bioresour Technol. 2022;359:127479. [DOI] [PubMed] [Google Scholar]
- 10.Choi S, Lee H-N, Park E, Lee S-J, Kim E-S. Recent advances in Microbial production of cis,cis-Muconic acid. Biomolecules. 2020;10(9):1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vardon DR, Franden MA, Johnson CW, Karp EM, Guarnieri MT, Linger JG, et al. Adipic acid production from lignin. Energy Environ Sci. 2015;8:617–28. [Google Scholar]
- 12.Werner AZ, Cordell WT, Lahive CW, Klein BC, Singer CA, Tan EC, et al. Lignin conversion to β-ketoadipic acid by Pseudomonas putida via metabolic engineering and bioprocess development. Sci Adv. 2023;9(36):eadj0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmore JR, Dexter GN, Salvachua D, Martinez-Baird J, Hatmaker EA, Huenemann JD, et al. Production of itaconic acid from alkali pretreated lignin by dynamic two stage bioconversion. Nat Commun. 2021;12(1):2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai C, Xu Z, Zhou H, Chen S, Jin M. Valorization of lignin components into gallate by integrated biological hydroxylation, O-demethylation, and aryl side-chain oxidation. Sci Adv. 2021;7:eabg4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higuchi Y, Ishimaru H, Yoshikawa T, Masuda T, Sakamoto C, Kamimura N, et al. Successful selective production of vanillic acid from depolymerized sulfite lignin and its application to poly(ethylene vanillate) synthesis. Bioresour Technol. 2023;385:129450. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yue C, Wei W, Shang Y, Zhang P, Ye BC. Construction of a p-coumaric and ferulic acid auto-regulatory system in Pseudomonas putida KT2440 for protocatechuate production from lignin-derived aromatics. Bioresour Technol. 2021;344(Pt B):126221. [DOI] [PubMed]
- 17.Righetti MC, Marchese P, Cavallo D, Celli A, Marega C. Coexistence of two lamellar populations in poly(ethylene vanillate) reorganized upon heating. Polymer. 2024;293:126659. [Google Scholar]
- 18.Balla ED, Papadopoulos L, Ainali NM, Kourtidou D, Grigora M-E, Tzetzis D, et al. Poly(ethylene furanoate-co-ethylene vanillate) biobased copolymers: impact of the incorporation of vanillic acid units in poly(ethylene furanoate). Eur Polym J. 2022;176:111429. [Google Scholar]
- 19.Xanthopoulou E, Terzopoulou Z, Zamboulis A, Koltsakidis S, Tzetzis D, Peponaki K, et al. Poly(hexylene vanillate): synthetic pathway and remarkable properties of a novel alipharomatic lignin-based polyester. ACS Sustain Chem Eng. 2023;11(4):1569–80. [Google Scholar]
- 20.Kasai D, Kamimura N, Tani K, Umeda S, Abe T, Fukuda M, et al. Characterization of FerC, a MarR-type transcriptional regulator, involved in transcriptional regulation of the ferulate catabolic operon in Sphingobium sp. strain SYK-6. FEMS Microbiol Lett. 2012;332(1):68–75. [DOI] [PubMed] [Google Scholar]
- 21.Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, et al. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol. 2004;186(9):2757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinoda E, Takahashi K, Abe N, Kamimura N, Sonoki T, Masai E. Isolation of a novel platform bacterium for lignin valorization and its application in glucose-free cis,cis-muconate production. J Ind Microbiol Biotechnol. 2019;46(8):1071–80. [DOI] [PubMed] [Google Scholar]
- 23.Wackett LP. Pseudomonas putida-a versatile biocatalyst. Nat Biotechnol. 2003;21(2):136–8. [DOI] [PubMed] [Google Scholar]
- 24.Johnson CW, Salvachúa D, Rorrer NA, Black BA, Vardon DR, John PCS, et al. Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule. 2019;3(6):1523–37. [Google Scholar]
- 25.Kumar M, You S, Beiyuan J, Luo G, Gupta J, Kumar S, et al. Lignin valorization by bacterial genus Pseudomonas: state-of-the-art review and prospects. Bioresour Technol. 2021;320(Pt B):124412. [DOI] [PubMed] [Google Scholar]
- 26.Notonier S, Werner AZ, Kuatsjah E, Dumalo L, Abraham PE, Hatmaker EA, et al. Metabolism of syringyl lignin-derived compounds in Pseudomonas putida enables convergent production of 2-pyrone-4,6-dicarboxylic acid. Metab Eng. 2021;65:111–22. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida A, Kurinia I, Higuchi Y, Osaka Y, Yasuta C, Sakamoto C, et al. Direct catalytic oxidation of rice husk lignin with hydroxide nanorod-modified copper foam and muconate production by engineered Pseudomonas sp. NGC7. J Biosci Bioeng. 2024;138(5):431–8. [DOI] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–2. [Google Scholar]
- 29.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008;18(5):821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24(8):1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez -RLM, Gunturu S, Harvey WT, Rosselló-Mora R, Tiedje JM, Cole JR, et al. The Microbial genomes Atlas (MiGA) webserver: taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acid Res. 2018;46(W1):W282–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145(1):69–73. [DOI] [PubMed] [Google Scholar]
- 36.Peng X, Masai E, Kasai D, Miyauchi K, Katayama Y, Fukuda M. A second 5-carboxyvanillate decarboxylase gene, ligW2, is important for lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl Environ Microbiol. 2005;71(9):5014–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng X, Masai E, Kitayama H, Harada K, Katayama Y, Fukuda M. Characterization of the 5-carboxyvanillate decarboxylase gene and its role in lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl Environ Microbiol. 2002;68(9):4407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donnelly M, Dagley S. Production of methanol from aromatic acids by Pseudomonas putida. J Bacteriol. 1980;142(3):916–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tack BF, Chapman PJ, Dagley S. Metabolism of gallic acid and syringic acid by Pseudomonas putida. J Biol Chem. 1972;247(20):6438–43. [PubMed] [Google Scholar]
- 40.Nishimura M, Nishimura Y, Abe C, Kohhata M. Expression and substrate range of Streptomyces Vanillate demethylase. Biol Pharm Bull. 2014;37(9):1564–8. [DOI] [PubMed] [Google Scholar]
- 41.Graf N, Altenbuchner J. Genetic engineering of Pseudomonas putida KT2440 for rapid and high-yield production of vanillin from ferulic acid. Appl Microbiol Biotechnol. 2014;98(1):137–49. [DOI] [PubMed] [Google Scholar]
- 42.García-Hidalgo J, Brink DP, Ravi K, Paul CJ, Lidén G, Gorwa-Grauslund MF. Vanillin Production in Pseudomonas: whole-genome sequencing of Pseudomonas sp. Strain 9.1 and reannotation of Pseudomonas putida CalA as a Vanillin Reductase. Appl Environ Microbiol. 2020;86(6):e02442–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamimura N, Takahashi K, Mori K, Araki T, Fujita M, Higuchi Y, et al. Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ Microbiol Rep. 2017;9(6):679–705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are provided in the manuscript or in the supplementary information file. The genomic sequence data have been deposited in DDBJ/NCBI/EMBL under the accession no. SAMD00818483.