Abstract
Background
Premature infants are more prone to brain injuries owing to incomplete nervous system development and poor adaptation outside the mother’s body. Without timely intervention, premature infants with brain injuries often develop intellectual disabilities, causing significant burdens on families and the society. Multiple studies have shown that gut dysbiosis can affect the nervous system, and vice versa. This study aimed to explore the changes in gut microbiota of typical premature infants and those with brain injuries on the third and seventh days after birth using 16 S rRNA technology.
Methods
Fecal samples from typical premature infants (non-brain injury group, n = 17) and those with brain injuries (brain injury group, n = 21) were collected on days 1, 3, and 7 after birth for 16 S rRNA sequencing. Alpha diversity analysis was used to evaluate the diversity of gut microbiome. LEfSe and DESeq2 were used to analyze of the microorganisms’ characteristics and differentiate the microorganisms between the two groups.
Results
At the phylum level, Firmicutes, Proteobacteria, and Actinobacteria were the dominant flora in both groups. At the genus level, the proportion of Enterococcus in fecal samples of the brain injury group was higher than that of the non-brain injury group on day three after birth; however, the opposite was observed on day seven. Rothia and Lactobacillales were characteristic bacteria of the non-brain injury group on days three and seven after birth, whereas Enterococcus and Bifidobacteria were characteristic bacteria of the brain injury group on days three and seven after birth, respectively. Three days after birth, the Shannon and Simpson indices of the non-brain injury group were significantly higher than those of the brain injury group.
Conclusion
Premature infants with brain injuries have a unique gut microbiota that is different from that of typical premature infants, indicating correlation between brain injuries and gut microbiota.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03643-4.
Keywords: Brain injury, Premature infants, Gut microbiome, 16S rRNA sequencing, Gut microbiota-brain axis
Background
As a global event, preterm birth can increase the risk of infant death and various diseases. Premature babies are at a higher risk of developing metabolic syndrome and cognitive impairment [1]. The survival rate of premature babies has increased with the improvement of medical technology and perinatal care,; however, brain injury in premature infants (BIPI) remains a serious public health issue [2]. A previous study reported that 25–50% of infants with BIPI will have cognitive, behavioral, attention, or social deficits, and 5–10% of children with BIPI will have cerebral palsy and other diseases. Many disease occurrences caused heavy burdens on the families of children and society [3]. A study on complications in preterm infants indicated that fetal asphyxia can lead to hypoxic-ischemic brain damage, with its severity closely associated with gestational age. Specifically, among preterm infants, a low gestational age and birth weight increase the incidence of asphyxia [4]. In a follow-up study involving 100 premature infants, domestic researchers found that 46% did not meet the standard for the mental development index by the age of one year, while 49% failed to meet the standard for the psychomotor development index [5]. Furthermore, zhang et al. also found that the rate of mental and motor development delays in premature infants increased as gestational age decreased [5]. Currently, the diagnosis of brain injury in preterm infants is difficult and the effect of drug treatment is relatively poor. Therefore, it is urgent to find a new diagnosis and treatment plan.
The intestine is a complex organ composed of 1014 bacteria, totaling more than ten times the number of human cells [6]. Recent studies have shown that various human diseases, such as obesity and diabetes, may be related to microbial disorders in the intestines [7, 8]. In addition, multiple studies have shown that the gut microbiome can communicate with the brain through at least three parallel and interacting channels, using nervous, endocrine, and immune signals [9]. Animal experiments have revealed that the gut microbiota regulates anxiety, mood, cognition, and pain [10]. The gut microbiome–brain axis is a complex regulatory axis containing many metabolites and encapsulates the nervous system. Recent studies have suggested that this regulatory axis may include intestinal microbes and their metabolites, intestinal nervous system, central nervous system, and other nervous systems [11].
Abnormalities in the microbiota can result in an inflammatory response, leading to cytokine release and neuroinflammation [12]. Furthermore, the vagus nerve connects the gut nervous system to central nervous system by sending signals from microbes in the gut to the brain [13]. A previous study demonstrated that germ-free mice showed an abnormally strong response to restraint stress. In the absence of microbiota, the corticosterone concentration in mice was significantly increased. The stress performance of the germ-free mice could be partially reversed by colonization with different microbiota in adulthood [14].
A previous study suggested that the uterus is a sterile environment, and the colonization of microorganisms in the fetus occurs after birth. However, a recent study found that the fetus in pregnant women can indirectly contact the microorganisms present in the mother by swallowing amniotic fluid, indicating that the establishment of the gut microbiome starts from the fetal period [15]. For premature infants, factors such as birth by cesarean section, artificial feeding, infection, perinatal and postnatal antibiotics delay the colonization of the gut microbiome; thus, intestinal microecological disorders are more likely to occur, which in turn may disturb their nervous systems. The pattern of bacterial tailoring in preterm infants differs significantly from that of full-term infants owing to factors such as gestational age and antibiotic exposure. More than half of preterm infants have bacteria in their feces, and this percentage decreases with increasing gestational age [16]. In addition, studies have found that prematurity is associated with a significant risk of multiple neurological complications, including cerebral palsy.
Furthermore, the microbiota of preterm infants is associated with poor growth phenotype [17], and preterm infants have less intestinal microbial diversity and more potentially pathogenic bacteria [18]. The gut-brain axis is a complex communication network between the gut and brain. In this communication network, the central nervous system coordinates the digestive system whereas signals generated by the gut may also influence central nervous system non-brain injury [19]. However, few studies have reported whether there are differences in gut microbiome between preterm infants who developed brain injury and those who did not.
Based on the aforementioned research background, we hypothesized that the incidence of brain injury in preterm infants is associated with disturbances in gut microbiome. Therefore, we performed 16 S rRNA sequencing to compare the characteristics and differences in the gut microbiome of premature infants with brain injury to that of typical premature infants. Aiming to provide evidence for a potential link between brain injury and gut microbiome in preterm infants, as well as to propose new ideas for diagnosing and treating brain injuries in premature infants.
Methods
Patients and experimental design
Between March 2019 and January 2020, 21 premature infants diagnosed with brain injury admitted to the North Sichuan Medical College Hospital neonatal intensive care unit were enrolled in the brain injury group. The non-brain injury group included 17 premature babies without brain injury who were hospitalized during the same period. The inclusion criteria were as follows: (1) family members of the children voluntarily joined, cooperated with the study, and signed the informed consent form; (2) all newborns and premature babies born at a gestational age of 30–37 weeks [20]; and and hospitalization for more than 7 days; (3) infants had no history or family history of organic diseases. The exclusion criteria were children with (1) congenital malformations of the heart, brain, kidneys, or respiratory tract, inherited metabolic diseases, or chromosomal diseases; (2) use of hormones and probiotics after birth; and (3) a history of brain injury-related diseases (in the non-brain injury group only). All enrolled neonates were evaluated using electroencephalogram (EEG) and cranial ultrasound within 3 days of birth, followed by weekly follow-up and cranial MRI before discharge.
Diagnostic criteria for brain injury in premature infants: manifestations of premature infants non-specific symptoms such as Poor response and hypotonia, and satisfy any of the following criteria: (1) Cranial imaging shows white matter damage or dysplasia; (2) Ultrasound abnormalities, which manifested as a thickening of the ventricular wall, widening of the lateral ventricles, enlargement of the brain cavity, strong echogenicity, and visible positive lesions were evaluated. (3) Excluding intracranial hemorrhage and others in congenital cases, cranial MRI shows short T1, T2 signals, and diffusion-weighted imaging shows abnormally high signal intensity. This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Beichuan Medical College (No. 2020ER038-1).
Clinical information collection
The information collected from the mothers of the enrolled children included whether mother experienced pregnancy complications (hypertension, abnormal thyroid function, diabetes, etc.), and whether she experienced premature rupture of the membranes. We also recorded the children’s information: sex; gestational age; birth weight; birth method; feeding status (including the time of feeding, the time to reach total gastrointestinal tract feeding, breastfeeding or milk powder feeding, etc.); whether there was suffocation; sepsis; congenital heredity metabolic diseases; any chromosomal diseases, gastrointestinal malformations, etc.; whether any injuries were diagnosed as intracranial hemorrhage; neonatal hypoxic-ischemic encephalopathy; bilirubin encephalopathy; repeated hypoglycemia or other diseases that seriously affect the nervous system; whether microecological agents were used; whether antibiotics were used; and if mechanical ventilation were used.
Diagnostic criteria for asphyxia
The diagnosis of asphyxia is contingent upon the presence of several key indicators: (1) Prenatal risk factors (etiology) that may contribute to asphyxia; (2) 1-minute Apgar score ≤ 7, which must include evidence of respiratory depression (clinical manifestation); (3) Umbilical artery blood pH < 7.20, such as umbilical artery blood pH < 7.00 and 1-minute Apgar score ≤ 7 (pathophysiological nature); (4) The presence of hypoxic-ischemic organ damage; (5) The exclusion of other potential causes for a low Apgar score or no other combined factors sufficiently account for the findings in items 2 to 4. The last four indicators are deemed essential for diagnosis, while the first item serves as an auxiliary indicator. In this study, premature infants who met five or four (2–5) of above criteria were diagnosed as asphyxia.
Fecal collection
We collected fecal samples from the two groups of premature infants on the first, third, and seventh days after birth. Specifically, 4 g of feces from each premature infant was placed in four sterile tubes using sterile spoons. The tubes were sealed with sealing films and immediately frozen in liquid nitrogen and stored at -80 °C in a freezer for later analysis.
DNA extraction, PCR amplification, and Miseq library construction
Fecal microbial genomic DNA was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, USA), following the manufacturer’s instructions. DNA was run on 1% agarose gel to test its integrity. The purity and concentration of the DNA were determined using the NanoDrop2000 [21].
V3-V4 of the 16 S DNA were amplified; the primers used were 338 F-ACTCCTACGGGAGGCAGCAG and 806R-GGACTACHVGGGTWTCTAAT. The PCR system included a 4 µL 5× FastPfu Buffer, 2 µL dNTPs (2.5 mM), 0.4 µL FastPfuPolymerase, 10 ng TemplateDNA, 0.2 µL BSA, 0.8 µL Reverse Primer (5 µM), and 0.8 µL Forward Primer (5 µM). Finally, ddH2O was added to the system until the total volume reached 20 µL. The ABI GeneAmp system (ABI, USA) was used to perform the PCR.
A NEXTFLEX Rapid DNA-Seq Kit was used to construct the library. Sequencing was performed using a MiseqPE300 platform [22].
Operational taxonomic units (OTUs) analysis
First, the QIIME2 demux plug-in was used to split the original sequence, and the QIIME2 dada2 plug-in was used to perform quality non-brain injury, trimming, denoising, splicing, and removing the chimera of the split-sequence to obtain OTUs [23]. Subsequently, according to the 338 F/806R primers, the GREENGENES database, with 99% similarity to version 13_8, was pruned to the region of V3-V4 [24]. The QIIME2 feature-classifier plug-in was then used to align OTU sequences, and the alignment threshold was set to 70% to obtain the species classification information table. Finally, the QIIME2 feature-table plug-in was used to eliminate all contaminating mitochondrial and chloroplast sequences to obtain amplified feature sequences.
Alpha and beta diversity analysis
Bacteria diversity in the two sample groups was analyzed by observing the commonly used alpha diversity, Shannon, and Simpson indices were used to evaluate the microbiota richness and evenness of a single sample [25]. In addition, the principal co-ordinates analysis (PCoA) chart based on Bray-Curtis was used to present the results of beta diversity analysis. This method was used to rank and reduce the dimensionality of the flora, allowing for an analysis of the similarities and differences in the gut microbiome between the two groups. The Wilcoxon test rank-sum test was used to compare the alpha diversity index between groups. The difference was statistically significant at p < 0.05.
Linear discriminant analysis Effect size (LEfSe)
LEfSe analysis in multi-level species was used to analyze the bacteria with different abundances between the non-brain injury and the brain injury groups [26]. Linear discriminant analysis (LDA) of LEfSe was used to estimate the effect of each component (species). LDA > 2 and p < 0.05 were used as the screening threshold difference.
Statistical analysis
Statistical analysis was performed using the SPSS statistical software (IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp.). Measurement data are expressed as‾χ ± s, and t-tests were used to compare groups. Enumeration data are expressed as rates, and the comparison between groups was tested using the Fisher’s exact probability method. DESeq2 difference analysis [27] and LEfSe analysis were used for the abundance analysis at the genus and species levels. A rarefaction curve was constructed to evaluate the sequencing depth.
Results
Comparison of baseline data between groups
The brain injury group included 9 boys and 12 girls, with an average gestational age of 33.6 ± 1.79 weeks and an average birth weight of 2089.1 ± 543.5 g. The non-brain injury group consisted of 10 boys and 7 girls, with an average gestational age of 33.2 ± 1.85 weeks and an average birth weight of 1946.5 ± 456.5 g. Children in both groups were given milk (Zaoqi nengen, Nestle, Germany) within six hours after birth and achieved total gastrointestinal nutrition within one week. We further compared clinical information of the two groups. The results indicated that sex, gestational age, birth weight, production method, feeding method, time to reach the total gastrointestinal tract feeding, sepsis, premature rupture of membranes, mechanical ventilation, pregnancy complications, and incidence of antibiotic use in the two groups showed no significant differences (p > 0.05). However, a significant difference in the proportion of asphyxia during childbirth was observed between the two groups (p < 0.05) (Table 1). In addition, intracranial hemorrhage, bilirubin encephalopathy, and repeated hypoglycemia disease did not occur in both groups.
Table 1.
Clinical information
| Clinical factors | Brain injury | Non-brain injury | p |
|---|---|---|---|
| Information for premature infant | |||
| Sex (male) | 9(42.85%) | 10(58.82%) | 0.51 |
| Gestational age (week) | 33.6 ± 1.8 | 33.2 ± 1.9 | 0.47 |
| Birth weight (g) | 2089.1 ± 543.5 | 1946.5 ± 456.5 | 0.39 |
| Time of feeding (h) | 4.42 ± 1.16 | 4.71 ± 1.44 | 0.51 |
| Time to reach the total gastrointestinal tract feeding (day) | 5.09 ± 0.88 | 4.52 ± 0.94 | 0.06 |
| Cesarean section | 16(76.19%) | 11(64.71%) | 0.49 |
| Septicemia | 12(57.14%) | 5(29.41%) | 0.11 |
| Asphyxia during childbirth | 8(38.09%) | 0(0.00%) | < 0.001 |
| Mechanical ventilation | 8(38.09%) | 2(11.76%) | 0.13 |
| Breastfeeding | 16(76.19%) | 11(64.71%) | 0.49 |
| Antibiotic use | 12(57.14%) | 5(29.41%) | 0.11 |
| Information for mothers | |||
| Premature rupture of membranes | 9(42.85%) | 2(11.76%) | 0.07 |
| Complications during pregnancy | 6(28.57%) | 7(41.17%) | 0.50 |
Differences in the composition of the two groups of samples at the phylum level
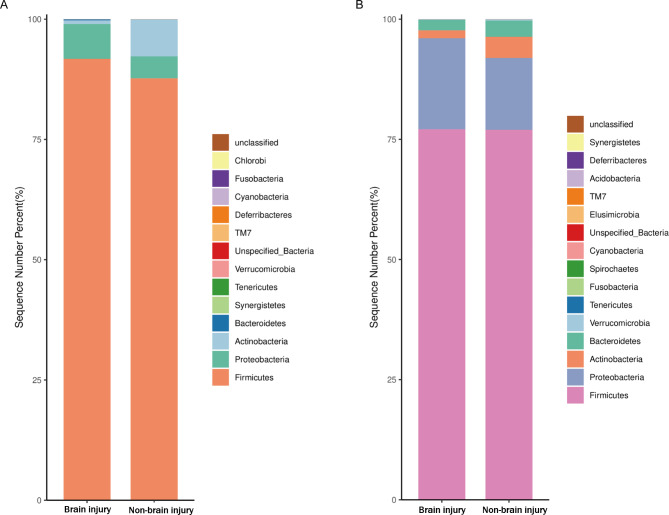
At the phylum level, the main components of samples from the brain injury and non-brain injury groups included Firmicutes, Proteobacteria, and Actinobacteria (Fig. 1). The results of the DESeq2 analysis revealed that the relative abundances of Firmicutes and Proteobacteria in the brain injury group on the third day after birth were significantly greater than those in the non-brain injury group (p < 0.05), whereas the content of Actinobacteria showed the opposite trend (p < 0.05). On the seventh day after birth, the changes in the contents of Firmicutes and Proteobacteria between samples of the two groups were not statistically significant (p > 0.05). The abundance of Actinobacteria in the brain injury group was significantly higher than that in the non-brain injury group on the third day after birth, whereas the abundance of Bacteroides on the seventh day after birth was significantly higher than that on the third day.
Fig. 1.
Histogram of the relative abundance of the samples at phylum level in the brain injury and non-brain injury groups on the third (A) and seventh (B) days after birth
Differences in the composition of the two groups of samples at the genus level
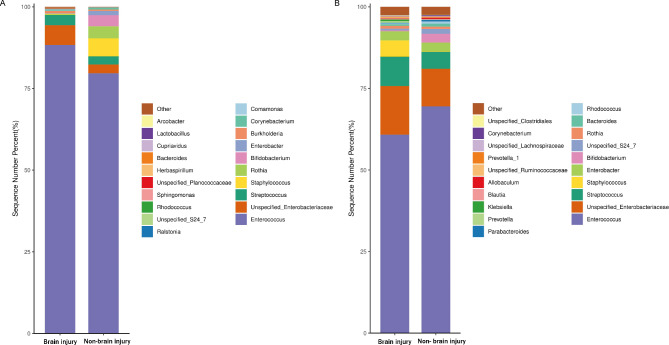
As shown in Fig. 2, at the genus level, the relative abundance of Enterococcus in each group was the highest. On the third day after birth, the relative abundance of Enterococcus in the brain injury group was significantly higher than in the non-brain injury group (p < 0.05), whereas the relative abundances of Bifidobacterium and Rothia were significantly lower (p < 0.05). On the seventh day after birth, the relative abundance of Staphylococcus in the brain injury group was significantly higher than that in the non-brain injury group (p < 0.05), whereas the relative abundance of Bifidobacterium was significantly lower (p < 0.05). On the seventh day after birth, no statistical difference in the relative abundance of Enterococcus was observed between the two groups.
Fig. 2.
Histogram of the relative abundance of the samples at genus level in the brain injury and non-brain injury groups on the third (A) and seventh (B) days after birth
Screening of differential bacteria between the brain injury group and the experimental group
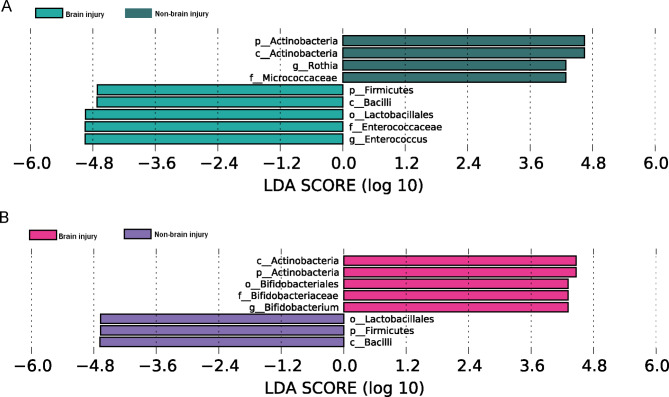
LEfSe analysis identified differentially abundant bacterial taxa between the two groups. The results revealed that 3 days after birth, Enterococcus and Rothia showed higher LDA score based on LEfSe in the brain injury and non-brain injury groups, respectively (Table 2; Fig. 3A). Our results also identified Lactobacillales with a higher LDA score in the brain injury group, reflecting a significant increase in frequency in the brain injury group compared with that in the non-brain injury group (Table 3; Fig. 3B).
Table 2.
Differential bacteria in the samples three days after birth between brain injury group and control group
| Different species | LogMax Mean | Group | LDA score | P |
|---|---|---|---|---|
| k__Bacteria.p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Enterococcaceae.g__Enterococcus | 5.97313 | Brain injury | 4.9495 | 0.0087 |
| k__Bacteria.p__Actinobacteria.c__Actinobacteria.o__Actinomycetales.f__Micrococcaceae.g__Rothia | 4.63037 | Non-brain injury | 4.2848 | 0.0319 |
Fig. 3.
Classification of the differential flora in the stool samples in the brain injury and non-brain injury groups 3 days (A) and 7 days (B) after birth
Table 3.
Differential bacteria in the samples seven days after birth between brain injury group and control group
| Different Species | LogMax Mean | Group | LDA score | P |
|---|---|---|---|---|
| p__Actinobacteria.c__Actinobacteria.o__Bifidobacteriales.f__Bifidobacteriaceae.g__Bifidobacterium | 4.72027 | Brain injury | 4.3108 | 0.0376 |
| p_Firmicutes.c_Bacilli.o_Lactobacillales | 5.95649 | Control | 4.6689 | 0.0364 |
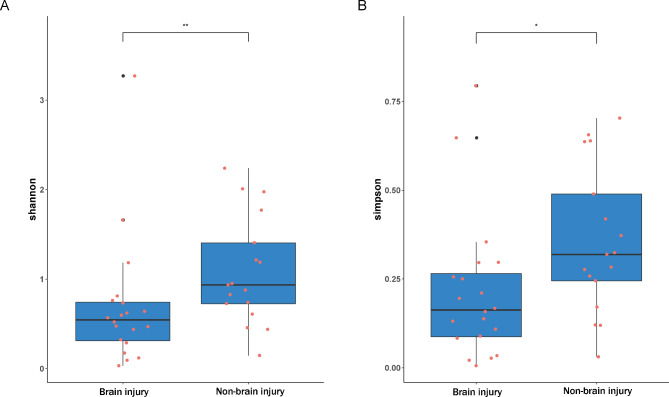
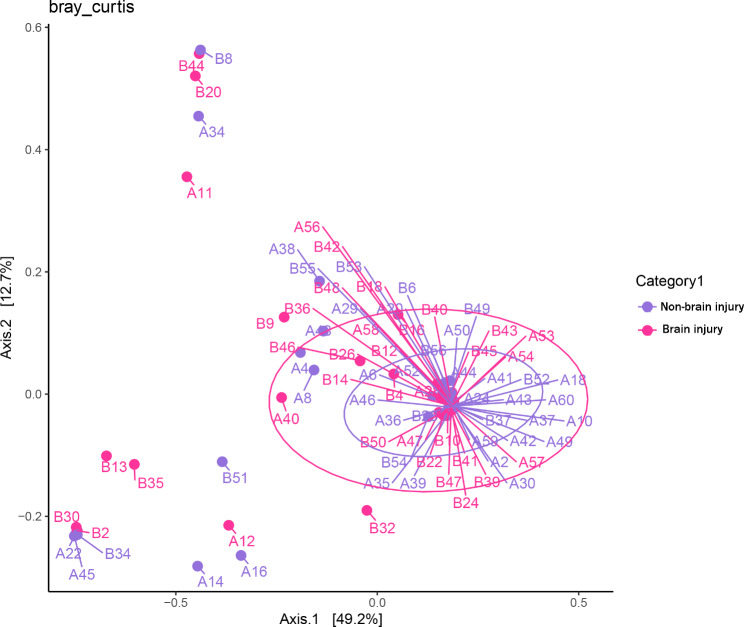
Alpha and beta diversity comparison
We compared the observed OTUs, Chao1, Shannon, Faith PD, and Simpson indices of the brain injury and non-brain injury groups (Table 4). The results of the samples comparison 3days after birth showed that the Shannon and Simpson indices of the non-brain injury group were significantly higher than those of the brain injury group (p < 0.05) (Fig. 4). The results of the sample comparison 7 days after birth further revealed no significant difference between the indices (Table 4). There was no significant difference in microbial beta diversity between brain injury and non-brain injury (Fig. 5).
Table 4.
Comparison of alpha diversity index between brain injury group and non-brain injury group
| Variable | p value | Method |
|---|---|---|
| Three days after birth | Wilcoxon | |
| Observed_OTUs | 0.239266 | Wilcoxon |
| Chao1 | 0.239266 | Wilcoxon |
| Shannon | 0.008012 | Wilcoxon |
| Faith_pd | 0.497465 | Wilcoxon |
| Simpson | 0.016868 | Wilcoxon |
| Seven days after birth | ||
| Observed_OTUs | 0.315563 | Wilcoxon |
| Chao1 | 0.315563 | Wilcoxon |
| Shannon | 0.8164933 | Wilcoxon |
| Faith_pd | 0.2203986 | Wilcoxon |
| Simpson | 0.7060043 | Wilcoxon |
Fig. 4.
Comparison of the Shannon index (A) and the Simpson index (B) of the brain injury and non-brain injury groups 3 days after birth.* P < 0.05, ** P < 0.01
Fig. 5.
Beta diversity between brain injury (purple) and non-brain injury (pink) groups 3 days (A) and 7 days (B) 3 after birth
Discussion
This study conducted a 16 S rRNA sequencing of fecal samples from premature infants with and without brain injury at 3 and 7 days after birth. The results revealed that the early dominant phyla in premature infants included Firmicutes, Proteobacteria, and Actinobacteria, whereas Enterococcus was the dominant flora at the genus level.This is consistent with a domestic study examining the relationship between neurodevelopment and gut microbiome in neonates, which included 66 preterm infants [28]. Furthermore, Arboleya et al. [29] reported that Firmicutes were predominant during the early stages of life in preterm infants, aligning with the results of this study, which observed the highest abundance of Firmicutes on the 3 and 7 days post-birth. In addition, the gut microbiome of premature infants is different from that of full-term infants. Although this study did not analyze the gut microbiome of full-term infants at an early stage, a better understanding of gut microbiome can provide deeper insights into the occurrence of brain injury in preterm infants. Compared with full-term infants, preterm infants show higher levels of facultative anaerobic microorganisms (such as Enterococcus and Staphylococcus) and lower levels of strict anaerobic microorganisms(such as Bifidobacterium and Bacteroides), it has also been demonstrated that the intestinal colonization of Bifidobacteria was delayed in preterm infants [30].
Many studies have confirmed that gut microbiome colonization of premature infants is related to many factors, including the mother’s diet, disease, drug use, birth methods, and feeding methods, and postnatal environment [31, 32]. No significant difference between the feeding methods or other factors between the two groups of preterm infants was observed in this study. However, more children in the brain injury group experienced asphyxia during childbirth. Several studies have confirmed that hypoxia severely affects neonatal cranial nerves [33]. Additionally, abnormal colonization of the gut microbiome may affect brain development and aggravate brain injury through the intestinal-brain axis [34]. Abnormal intestinal bacteria may cause intestinal inflammation, changes in blood-brain barrier permeability, decrease in neural substrate synthesis and secretion, or direct and indirect effects on the brain function by acting via the hypothalamic-pituitary-adrenal axis/parasympathetic nervous system [35, 36]. In contrast, brain injury in infants may also affect the establishment of normal gut microbiome through the bidirectional regulation pathway of the entero-brain axis [37].
Microbial invasion during the fetal period also affects gut microbiome colonization of premature infants; 25–40% of preterm births and 7–12% of preterm births with intact fetal membranes involve intrauterine microbial invasion, which is related to ascending vaginal microbes colonization and blood-borne transmission of maternal oral microbes [38–40]. Gestational age at delivery is significantly negatively correlated with bacterial abundance in the amniotic fluid, supporting the role of preterm intrauterine infection [41]. Bacterial infection of the amniotic fluid is also associated with cerebral palsy and intraventricular hemorrhage [42, 43]. Therefore, premature infants may be born with intestinal microflora disorder due to in utero infection. Early intestinal microflora disorder will not only cause immune, metabolic, and gastrointestinal dysfunctions but also cause central nervous system and subsequent brain damage [44].
This present study found that the Shannon and Simpson indices of the gut microbiome of the brain injury group were lower than those of the non-brain injury group on the third day after birth. On the seventh day, alpha diversity analysis result between the two groups was not statistically significant. Research has shown that patients with ischemic stroke exhibited reduced gut microbiome diversity compared to healthy controls during both the acute and recovery phases [45]. A recent meta-analysis study revealed a decreased diversity in the gut microbiome of individuals with anxiety disorders compared to healthy populations [46]. In addition, studies have found that a cocktail of antibiotics (ABX) depletes the gut microbes of a murine stroke model, worsening their outcomes [47]. ABX-treated APPSWE/PS1ΔE9 mice display lower alpha diversity, and the disturbance in intestinal microbial diversity induced by ABX also affects the neuroinflammatory response by reducing local glial hyperplasia and altering the morphology of the microglia [48]. Moreover, a study found a significant negative correlation between the 1-year-old children’s gut microbiome diversity measurement and connectivity between the left amygdala and mid/forebrain area overlapping with the thalamus. The higher diversity of the intestinal microbiome of 1-year-old infants may be related to lower fear/emotional processing efficiency [49]. However, another study showed a negative correlation between the alpha diversity of the gut microbes of 1-year-old children and the overall cognition and language functions of 2-year-old children [50]. The controversy may be due to the limitations of the method of microbial diversity calculation, such as bias of the extraction method and insufficient coverage. Despite ongoing controversies, most researchers agree that the disorder of gut microbiome is typically accompanied by a reduction in its diversity, and the diversity of gut microbiome is closely related to brain function.
Many studies have affirmed the two-way communication mechanism between the gut microbiome and central nervous system regarding immunity, inflammation and metabolism. The gut microbiome can regulate the production of myelin sheath in the prefrontal cortex of mice [51]. Moreover, the gut microbiome can trigger the production of pro-inflammatory cytokines by activating various lymphocytes and innate immune cells, which act on afferent nerve receptors to transmit signals to the brain. They can also reach the brain through direct diffusion or transport of proteins through the blood-brain barrier [52]. Gut microbiome disturbance can cause abnormal activation of the immune system, affecting the development of neurons and oligodendrocytes and the formation of myelin, resulting in abnormal nervous system development [53]. Most of the pathogenic bacteria reported in the literature belong to the phylum Firmicutes and Proteobacteria. Firmicutes are more resistant to environmental changes due to their unique cell walls; The gut microbiome of preterm infants with brain injury undergoes significant alterations in both physical and chemical environments, leading to the dominance of Firmicutes such as Enterococcus and Staphylococcus [54]. The abnormal colonization of Proteobacteria in the human intestine will significantly reduce the resistance of the inherent flora to pathogenic microorganisms, promote the intestinal and systemic inflammatory response, and lead to central nervous system damage through the pathways mentioned above [55]. In addition, research has demonstrated that reduced expression levels of Bifidobacterium, Lactococcus, and Bacteroides, alongside elevated expression levels of Streptococcus, Salmonella, Clostridium, and Staphylococcus, could contribute to an increased incidence of brain injury in preterm infants [56]. This is consistent with the significant increase in the abundance of Firmicutes, Proteobacteria, Enterococcus, and Staphylococcus in premature infants with brain injury in this present study.
Short-chain fatty acids (SCFAs) produced by intestinal microbes can cross the blood-brain barrier and interact with microglia, and high SCFAs concentrations can reduce the inflammatory response of peripheral monocytes. A new study has also shown that SCFAs can regulate Alzheimer’s disease, disrupting the function of specific microglia [57]. Additionally, Bifidobacteria can increase SCFA content [58]. In this study, we found that the relative proportion of Bifidobacteria in the brain injury group was lower than that in the non-brain injury group. We also found that the non-brain injury group had a higher abundance of Rothia, however, no reports of brain development related to Rothia were available.
This study had some limitations. First, brain injury is a persistent pathological change that may require long-term monitoring. This study only tested stool samples at 3 and 7 days after the birth of premature infants. Second, as a single-center study, this study included a small sample size of only 38 premature infants. More research centers and larger sample sizes are needed in the future. Finally, this study did not measure bacterial metabolites and cytokines; thus, further analysis in future studies should also be considered.
Conclusion
In summary, the dominant gut microbiome of preterm infants one week after birth consists of Firmicutes, Proteobacteria, and Actinobacteria at the phylum level, with Enterococcus being prominent at the genus level. Furthermore, compared with preterm infants without brain injury, the gut microbiome diversity of premature infants with brain injury was decreased; concurrently, there was a reduction in the relative abundance of Actinobacteria, while the relative abundance of Firmicutes and Proteobacteria increased. At the genus level, a decline in the relative abundance of Bifidobacterium was observed, alongside an increase in the relative abundance of Enterococcus and Staphylococcus. Indicating a high correlation between preterm infants with brain injury and the abundance and diversity of gut microbiome. Therefore, focusing on alteration in the gut microbiome of preterm infants is beneficial for the diagnosis and treatment of brain injury.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Sup Figure 1. Annotated diagram for each classification level of the sample
Supplementary Material 2: Sup Figure 2. 16S rRNA sequencing rarefaction curve of the stool samples of premature infants 3 (A) and 7 (B) days after birth
Acknowledgements
Not applicable.
Abbreviations
- BIPI
Brain injury in premature infants
- OTUs
Operational taxonomic units
- SCFAs
Short-chain fatty acids
Author contributions
Conception and design of the research: Jing Zhao and Li Hou; acquisition of data: Li Hou and Linlin Yin; analysis and interpretation of data: Hong Deng and Lin Jiang; statistical analysis: Li Hou, Lu Dai, and Linlin Yin; obtaining funding: Jing Zhao; drafting the manuscript: Li Hou and Linlin Yin; manuscript revision for important intellectual content: Jing Zhao. All authors have read and approved the final manuscript.
Funding
This work was supported by the School Cooperation Research Fund of Nanchong City (No. 20SXZRKX003), Research and Development Program of Affiliated Hospital of North Sichuan Medical College (No. 2021ZK004 and No. 2021LC010), and Scientific Research Projects of North Sichuan Medical College (No. CBY22-ZDA04). The funders had no involvement in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; nor in the decision to submit the article for publication.
Data availability
The datasets generated during the current study are available in the NCBI BioProject (PRJNA933769) database repository [http://www.ncbi.nlm.nih.gov/bioproject/933769].
Declarations
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Beichuan Medical College (No. 2020ER038-1). All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from each guardian.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cerasani J, Ceroni F, De Cosmi V. Human Milk Feeding and Preterm Infants’ Growth and Body Composition: A Literature Review. 2020, 12(4). [DOI] [PMC free article] [PubMed]
- 2.Volpe JJ. Brain injury in premature infants: a complex amalgam of destructive and developmental disturbances. Lancet Neurol. 2009;8(1):110–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allin M, Walshe M, Fern A, Nosarti C, Cuddy M, Rifkin L, Murray R, Rushe T, Wyatt J. Cognitive maturation in preterm and term born adolescents. J Neurol Neurosurg Psychiatry. 2008;79(4):381–6. [DOI] [PubMed] [Google Scholar]
- 4.TIAN Z H NYZ. Research progress of the interaction between gut microbes and brain gut axis. Infect Disease Inform. 2016;29(05):302–7. [Google Scholar]
- 5.ZHANG M KXN, QIAN H Y, et al. Case-control study on risk factors and complications of premature. Chin J Child Health Care. 2015;11(23):1181–4. [Google Scholar]
- 6.Ma Q, Li Y, Li P, Wang M, Wang J, Tang Z, Wang T, Luo L, Wang C, Wang T, et al. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed Pharmacotherapy = Biomedecine Pharmacotherapie. 2019;117:109138. [DOI] [PubMed] [Google Scholar]
- 7.Pushpanathan P, Mathew GS, Selvarajan S, Seshadri KG, Srikanth P. Gut microbiota and its mysteries. Ind J Med Microbiol. 2019;37(2):268–77. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Wang K, Wang X, Pang Y, Jiang C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell. 2021;12(5):360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA. 2011;108(38):16050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–12. [DOI] [PubMed] [Google Scholar]
- 11.Wang HX, Wang YP. Gut microbiota-brain Axis. Chin Med J. 2016;129(19):2373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quigley EMM. Microbiota-Brain-Gut Axis and neurodegenerative diseases. Curr Neurol Neurosci Rep. 2017;17(12):94. [DOI] [PubMed] [Google Scholar]
- 13.Forsythe P, Bienenstock J, Kunze WA. Vagal pathways for microbiome-brain-gut axis communication. Adv Exp Med Biol. 2014;817:115–33. [DOI] [PubMed] [Google Scholar]
- 14.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558(Pt 1):263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Sci (New York NY). 2014;345(6198):760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JK, Hern Tan LT. Exploring the role of gut Bacteria in Health and Disease in Preterm neonates. 2020, 17(19). [DOI] [PMC free article] [PubMed]
- 17.Lu J, Claud EC. Connection between gut microbiome and brain development in preterm infants. Dev Psychobiol. 2019;61(5):739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bresesti I, Salvatore S, Valetti G, Baj A. The Microbiota-Gut Axis in Premature Infants: Physio-Pathological Implications. 2022, 11(3). [DOI] [PMC free article] [PubMed]
- 19.Bonaz B, Bazin T, Pellissier S. The Vagus nerve at the interface of the Microbiota-Gut-Brain Axis. Front NeuroSci. 2018;12:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L J: Obstetrics and gynecology. Beijing: People’s Medical Publishing House; 2016.
- 21.Yu S, Wang Y, Li X, Yu F, Li W. The factors affecting the reproducibility of micro-volume DNA mass quantification in Nanodrop 2000 spectrophotometer. Optik - Int J Light Electron Opt. 2017;145:555–60. [Google Scholar]
- 22.Yeh YC, Needham DM, Sieradzki ET, Fuhrman JA. Taxon Disappearance from Microbiome Analysis Reinforces the Value of Mock Communities as a Standard in Every Sequencing Run. mSystems 2018, 3(3). [DOI] [PMC free article] [PubMed]
- 23.Hall M, Beiko RG. 16S rRNA Gene Analysis with QIIME2. Methods in molecular biology (Clifton, NJ) 2018, 1849:113–129. [DOI] [PubMed]
- 24.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichler M, Coskun ÖK, Ortega-Arbulú AS, Conci N, Wörheide G. A 16S rRNA gene sequencing and analysis protocol for the Illumina MiniSeq platform. 2018, 7(6):e00611. [DOI] [PMC free article] [PubMed]
- 26.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP. Bioconductor Workflow for Microbiome Data Analysis: from raw reads to community analyses. F1000Research. 2016;5:1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y M. Preliminary study on the relationship between neonatal meurodevelopment and intestinal flora. University of South China; 2020.
- 29.Arboleya S, Sánchez B, Solís G, Fernández N, Suárez M, Hernández-Barranco AM, Milani C, Margolles A, de Reyes-Gavilán L, Ventura CG. M : Impact of Prematurity and Perinatal antibiotics on the developing intestinal microbiota: a functional inference study. Int J Mol Sci 2016, 17(5). [DOI] [PMC free article] [PubMed]
- 30.Arboleya S, Binetti A, Salazar N, Fernández N, Solís G, Hernández-Barranco A, de Margolles A Los Reyes-Gavilán CG, Gueimonde M. Establishment and development of intestinal microbiota in preterm neonates. FEMS microbiology ecology 2012, 79(3):763–772. [DOI] [PubMed]
- 31.Underwood MA, Sohn K. The microbiota of the extremely Preterm Infant. Clin Perinatol. 2017;44(2):407–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baranowski JR, Claud EC. Necrotizing enterocolitis and the Preterm infant Microbiome. Adv Exp Med Biol. 2019;1125:25–36. [DOI] [PubMed] [Google Scholar]
- 33.Silveira RC, Procianoy RS. Hypothermia therapy for newborns with hypoxic ischemic encephalopathy. Jornal De Pediatria. 2015;91(6 Suppl 1):S78–83. [DOI] [PubMed] [Google Scholar]
- 34.Bao W, Sun Y, Lin Y, Yang X, Chen Z. An integrated analysis of gut microbiota and the brain transcriptome reveals host-gut microbiota interactions following traumatic brain injury. Brain Res. 2023;1799:148149. [DOI] [PubMed] [Google Scholar]
- 35.Tang W, Zhu H, Feng Y, Guo R, Wan D. The impact of gut microbiota disorders on the blood–brain barrier. Infect Drug Resist 2020:3351–63. [DOI] [PMC free article] [PubMed]
- 36.Hattay P, Prusator D, Tran L, Greenwood-Van Meerveld B. Psychological stress‐induced colonic barrier dysfunction: role of immune‐mediated mechanisms. Neurogastroenterology Motil. 2017;29(7):e13043. [DOI] [PubMed] [Google Scholar]
- 37.Rogers MB, Simon D, Firek B, Silfies L, Fabio A, Bell MJ, Yeh A, Azar J, Cheek R, Kochanek PM. Temporal and spatial changes in the microbiome following pediatric severe traumatic brain injury. Pediatric critical care medicine 2022. [DOI] [PMC free article] [PubMed]
- 38.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, et al. A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reproductive Immunol (New York NY: 1989). 2014;71(4):330–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.León R, Silva N, Ovalle A, Chaparro A, Ahumada A, Gajardo M, Martinez M, Gamonal J. Detection of Porphyromonas gingivalis in the amniotic fluid in pregnant women with a diagnosis of threatened premature labor. J Periodontol. 2007;78(7):1249–55. [DOI] [PubMed] [Google Scholar]
- 41.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE. 2008;3(8):e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Z, Ma L, Luo K, Bajaj M, Chawla S, Natarajan G, Hagberg H, Tan S. Chorioamnionitis in the development of cerebral palsy: a Meta-analysis and systematic review. Pediatrics 2017, 139(6). [DOI] [PMC free article] [PubMed]
- 43.Jung EY, Park KH. Amniotic Fluid Infection, Cytokine Levels, and Mortality and Adverse Pulmonary, Intestinal, and Neurologic Outcomes in Infants at 32 Weeks’ Gestation or Less. 2017, 32(3):480–7. [DOI] [PMC free article] [PubMed]
- 44.O’Mahony SM, Clarke G, Dinan TG, Cryan JF. Early-life adversity and brain development: is the microbiome a missing piece of the puzzle? Neuroscience 2017, 342:37–54. [DOI] [PubMed]
- 45.NAN Y LZ, GUO G H. Diversity analysis of intestinal flora in patients with ischemic stroke at different stages. Chongqing Med J. 2019;48(09):1587–9. [Google Scholar]
- 46.Sublette ME, Cheung S, Lieberman E, Hu S, Mann JJ, Uhlemann AC, Miller JM. Bipolar disorder and the gut microbiome: a systematic review. Bipolar Disord. 2021;23(6):544–64. [DOI] [PubMed] [Google Scholar]
- 47.Winek K, Engel O, Koduah P, Heimesaat MM, Fischer A, Bereswill S, Dames C, Kershaw O, Gruber AD, Curato C, et al. Depletion of cultivatable gut microbiota by broad-spectrum antibiotic pretreatment worsens Outcome after Murine Stroke. Stroke. 2016;47(5):1354–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minter MR, Zhang C, Leone V, Ringus DL, Zhang X, Oyler-Castrillo P, Musch MW, Liao F, Ward JF, Holtzman DM, et al. Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci Rep. 2016;6:30028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao W, Salzwedel AP, Carlson AL, Xia K, Azcarate-Peril MA, Styner MA, Thompson AL, Geng X, Goldman BD, Gilmore JH, et al. Gut microbiome and brain functional connectivity in infants-a preliminary study focusing on the amygdala. Psychopharmacology. 2019;236(5):1641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loughman A, Ponsonby AL, O’Hely M, Symeonides C, Collier F, Tang MLK, Carlin J, Ranganathan S, Allen K, Pezic A, et al. Gut microbiota composition during infancy and subsequent behavioural outcomes. EBioMedicine. 2020;52:102640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. 2017, 114(40):10713–8. [DOI] [PMC free article] [PubMed]
- 52.Farzi A, Fröhlich EE, Holzer P. Gut microbiota and the neuroendocrine system. Neurotherapeutics: J Am Soc Experimental Neurother. 2018;15(1):5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu J, Lu L, Yu Y, Cluette-Brown J, Martin CR, Claud EC. Effects of Intestinal Microbiota on Brain Development in Humanized Gnotobiotic mice. Sci Rep. 2018;8(1):5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caparrós-Martín JA, Lareu RR, Ramsay JP, Peplies J, Reen FJ, Headlam HA, Ward NC, Croft KD, Newsholme P, Hughes JD, et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin N-R, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. [DOI] [PubMed] [Google Scholar]
- 56.Murthy P, Zein H, Thomas S, Scott JN, Abou Mehrem A, Esser MJ, Lodha A, Metcalfe C, Kowal D, Irvine L, et al. Neuroprotection Care Bundle implementation to decrease Acute Brain Injury in Preterm infants. Pediatr Neurol. 2020;110:42–8. [DOI] [PubMed] [Google Scholar]
- 57.Wenzel TJ, Gates EJ, Ranger AL, Klegeris A. Short-chain fatty acids (SCFAs) alone or in combination regulate select immune functions of microglia-like cells. Mol Cell Neurosci. 2020;105:103493. [DOI] [PubMed] [Google Scholar]
- 58.Libbey JE, Sanchez JM, Doty DJ, Sim JT, Cusick MF, Cox JE, Fischer KF, Round JL, Fujinami RS. Variations in diet cause alterations in microbiota and metabolites that follow changes in disease severity in a multiple sclerosis model. Beneficial Microbes. 2018;9(3):495–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Sup Figure 1. Annotated diagram for each classification level of the sample
Supplementary Material 2: Sup Figure 2. 16S rRNA sequencing rarefaction curve of the stool samples of premature infants 3 (A) and 7 (B) days after birth
Data Availability Statement
The datasets generated during the current study are available in the NCBI BioProject (PRJNA933769) database repository [http://www.ncbi.nlm.nih.gov/bioproject/933769].