Abstract
In non-small cell lung cancer (NSCLC), Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations are found in up to 30% of all cases, with the most prevalent mutations occurring in codons 12 and 13. The development of KRAS-targeted drugs like sotorasib and adagrasib has generated significant excitement in the clinical arena, offering new therapeutic options. Their potential for combination with other treatments broadens the scope for clinical exploration. Acquired resistance to KRAS exon 2 p.G12C inhibitors is a significant challenge, with several reported mechanisms. In this scenario, combination therapy strategies that include targeting Src Homology Region 2 Domain-Containing Phosphatase-2 (SHP2), Son of Sevenless Homolog 1 (SOS1), or downstream effectors of KRAS exon 2 p.G12C are showing promise in overcoming such resistance. However, the efficacy of immune checkpoint inhibitors in this context still requires comprehensive evaluation. The response to anti-Programmed Cell Death Protein 1/Programmed Cell Death Protein 1 Ligand (anti-PD-1/PD-L1) drugs in NSCLC may be significantly influenced by co-occurring mutations, underscoring the need for a personalized approach to treatment based on the specific genetic profile of each tumor.
Keywords: targeted therapies, sotorasib, adagrasib, resistance
Introduction
Lung cancer remains the leading cause of cancer-related deaths globally, with nearly 1.8 million fatalities annually. Among these, NSCLC represents the majority, accounting for about 85% of all lung cancer diagnoses, with adenocarcinoma being the most prevalent histological subtype. In recent years, advancements in molecular profiling and the development of novel targeted therapies have significantly transformed the treatment landscape for NSCLC. Simultaneously, the rise of immune checkpoint inhibitors has broadened the spectrum of therapeutic strategies available for lung cancer. Notably, combination therapies that incorporate immune checkpoint inhibitors alongside targeted drugs have shown superior efficacy compared to monotherapy approaches. Previously, certain molecular alterations like KRAS mutations were deemed untreatable, but the advent of new targeted drugs has opened up promising treatment possibilities for these challenging cases. This shift marks a pivotal moment in the ongoing efforts to effectively treat and manage NSCLC.
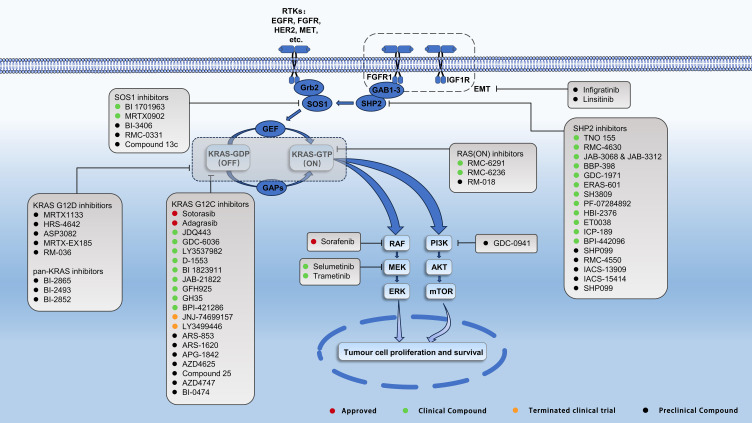
The KRAS protein, a small membrane-bound Guanosine Triphosphatase (GTPase), plays a critical role as a molecular switch regulating various cellular signaling functions. Its activity within the cell is determined by the balance between nucleotide hydrolysis and exchange, which governs the amount of active KRAS. In its GDP-bound form, KRAS remains in an “off” state. However, upon exposure to growth factors, KRAS transitions to the “on” state through the exchange of GDP for GTP. This activation process is facilitated by guanine nucleotide exchange factors (GEFs) like SOS1 and SOS2. Once activated, KRAS triggers downstream effector pathways, notably the Mitogen-Activated Protein Kinase (MAPK) and phosphoinositide 3-kinase (PI3K) pathways, which are essential for cell proliferation and survival. The return of KRAS to the “off” state occurs when GTP is hydrolyzed back to GDP, a reaction catalyzed by GTPase-activating proteins (GAPs) such as neurofibromatosis type 1 (NF1).1 This dynamic cycle of activation and deactivation is crucial for the precise regulation of cellular processes influenced by KRAS. This article comprehensively explores various aspects of KRAS mutations in NSCLC, with a focus on targeted therapies and combination treatment strategies for KRAS-mutant NSCLC.
Epidemiology of KRAS Mutations
KRAS mutations represent the most prevalent carcinogenic driver in NSCLC, with their incidence varying significantly based on the lung cancer’s pathological type, ethnicity, smoking history, and gender. In Caucasian populations, KRAS mutations are found in approximately 30% of lung adenocarcinomas, whereas they occur less frequently in lung squamous cell carcinoma, with an incidence ranging from 1.6% to 7.1%.2 In large cell lung cancer and adenosquamous carcinoma, the incidence rates are 12.8% and 10%, respectively. Beyond ethnic differences, KRAS mutations tend to be more common among women and individuals with a history of smoking.3
A strong correlation has been established between PD-L1 expression and KRAS mutations.4,5 NSCLC patients who are PD-L1 positive (TPS ≥ 1%) are more likely to have KRAS mutations, particularly the KRAS exon 2 p.G12C mutation, compared to those who are PD-L1 negative.5 The mutation rate in patients with high PD-L1 expression levels (≥50%) is significantly higher than in those with low levels (1–49%).4 This phenomenon may be associated with KRAS mutations that induce PD-L1, as confirmed in human NSCLC cell lines.6–8 Notably, the p.G12C mutation, characterized by the substitution of glycine with cysteine, is one of the most prevalent types of KRAS mutations, accounting for 40% of all such mutations in NSCLC.9 Nassar et al recently reported that specifically in NSCLC patients, the incidence of the KRAS exon 2 p.G12C mutation is 13.8%.10 In contrast, other significant mutations, such as p. G12V and p. G12D, represent 21% and 17% of KRAS mutations in NSCLC, respectively.9 Additionally, research by Dogan et al and Osta et al has highlighted that KRAS exon 2 p.G12C mutations are particularly prevalent among female patients and smokers.11 It has also been noted that there is no statistically significant difference in age among different KRAS mutation subtypes.12
Biological Characteristics of KRAS: Structure, Function, Downstream Pathway
KRAS is a gene that encodes a membrane-bound GTPase. This protein plays a pivotal role in cellular signaling and functions in an inactive state when bound to guanosine diphosphate (GDP) and in an active state when bound to guanosine triphosphate (GTP).13 The transition of RAS proteins to their active, GTP-bound state is facilitated by GEFs, such as the effector protein SOS1.14 Upon activation, KRAS can bind to downstream effector molecules either as monomers or dimers, initiating a variety of signaling pathways (Figure 1). Conversely, the inactivation of KRAS is regulated by GAPs, which increase the GTPase activity of KRAS, leading to a greater affinity for GDP and thus maintaining KRAS in its inactive state. This dynamic interplay between the active and inactive states of KRAS is fundamental to the regulation of various cellular processes and is a key focus in understanding the molecular mechanisms driving cancer and other diseases.
Figure 1.
[KRAS signaling pathway and targeted therapeutic agents.].
KRAS Mutation Subtypes and Genetic Heterogeneity
Incidence of Different Mutant Subtypes
KRAS mutations predominantly involve chromosome 12p12.1, with the most common alterations occurring at codons 12 and 13, and less frequently at codon 16. Remarkably, over 80% of oncogenic KRAS mutations are found at codon 12, which is strategically located near the nucleotide binding pocket and the adjacent effector protein switch of the KRAS protein.15 These mutations often result in the substitution of glycine at codon 12 with other amino acids, except proline. This alteration can lead to genomic heterogeneity in tumors harboring KRAS mutations, thus impeding GAP binding and promoting GTP hydrolysis. Consequently, this facilitates the accumulation of active KRAS-GTP, driving carcinogenic cell signaling.16 Among these, the most frequent mutations at KRAS codon 12 are the glycine-cysteine (KRAS exon 2 p.G12C), glycine-valine (KRAS exon 2 p.G12CV), and glycine-aspartic acid (KRAS exon 2 p.G12D) mutations.
Different Mutant Subtypes Have Different Prognoses
The prognostic implications of various KRAS mutation subtypes are currently unclear and subject to debate. While some studies indicate no significant differences in prognosis among the KRAS mutation subtypes,17 conflicting evidence exists. For example, several studies suggest that patients with the p. G12D mutation experience worse progression-free survival (PFS) and overall survival (OS),18 in contrast to those with the o, G12C mutation, who demonstrate better objective response rates (ORR), PFS, and OS.19 Conversely, other research has reported that the p. G12D mutation may be associated with better PFS and OS.20
Effects of Different Mutant Subtypes on Downstream Signaling Pathways
Different KRAS mutation subtypes have distinct impacts on downstream signaling pathways. For instance, in cell lines harboring KRAS exon 2 p.G12C or KRAS exon 2 p.G12CV mutations, there is a notable increase in RAS-related protein (RAL) A/B signaling,21 accompanied by a decrease in phosphorylated Protein Kinase B (AKT) levels. This pattern differs significantly compared to cell lines with other types of KRAS mutations or those with wild-type KRAS. In contrast, cell lines featuring KRAS exon 2 p.G12CD mutations show more effective activation of the PI3K-AKT pathway.22,23 The identification of these differing effects highlights the importance of developing treatment strategies for KRAS-mutated NSCLC that are specifically tailored to the unique characteristics of each mutation subtype. Such targeted approaches are likely to be more effective due to their precise alignment with the underlying molecular alterations.
Prognostic Impact of KRAS Mutations
The prognostic impact of KRAS mutations in NSCLC patients at both early and advanced stages is increasingly recognized. According to the 2022 National Comprehensive Cancer Network (NCCN) guidelines, KRAS mutations are considered an adverse prognostic factor relative to tumors without these mutations.24 These mutations are associated with poor prognosis across all stages of NSCLC and present a higher risk of recurrence post-early complete resection.3,11,25 Tumors with KRAS exon 2 p.G12C mutations are particularly noted for their higher rates of lymphatic invasion and elevated tumor mutation burdens.26 Research involving surgically resected lung adenocarcinoma patients demonstrates that those with KRAS-mutant tumors experience worse disease-free survival (DFS) and overall survival (OS) compared to those with wild-type tumors, a difference that persists even when Epidermal Growth Factor Receptor (EGFR) mutations are excluded.3,27
It is crucial to recognize that prognosis varies significantly with different KRAS mutation subtypes and co-mutations. For instance, a single-center retrospective observational study of advanced NSCLC patients treated with at least one therapy reported that those with KRAS exon 2 p.G12CD mutations had a notably better response rate than those without this subtype (50% vs 29.4%; Odds Ratio: 2.4; 95% Confidence Interval: 0.70–8.18; P=0.162).20 Furthermore, in multivariate analysis, the KRAS exon 2 p.G12CD mutation was significantly associated with improved PFS (HR: 0.37; 95% CI: 0.12–0.84; P=0.021) and OS (HR: 0.22; 95% CI: 0.07–0.66; P=0.007).
The predictive significance of KRAS mutations as prognostic biomarkers for immune-oncology drugs, cytotoxic chemotherapy, and targeted therapy is of great importance to clinicians treating metastatic NSCLC.28 With the advent of immune checkpoint inhibitors, the presence of Serine/Threonine Kinase 11 (STK11) co-mutations is increasingly significant for prognosis.
Co-mutations
The most frequent co-mutations in NSCLC with KRAS exon 2 p.G12C mutations are STK11, Tumor Protein p53 (TP53), and Kelch-like ECH-associated protein 1 (KEAP1). These mutations occur in 10.3–28.0% of cases for STK11, 17.8–50.0% for TP53, and 6.3–23.0% for KEAP1.29,30 Co-mutations can significantly influence the effectiveness of chemotherapy, immunotherapy, and molecular targeted therapies in treating tumors. Inactivation of the STK11 gene is linked to a tumor immune microenvironment that excludes T-cells, reducing the efficacy of PD-L1 inhibition. This resistance is observed in KRAS-mutant lung adenocarcinoma, which often results in poor clinical outcomes when treated with PD-L1 inhibitors and chemo-immunotherapy.31–33 Furthermore, co-alterations in STK11 and KEAP1 are associated with unfavorable clinical outcomes in KRAS-mutant NSCLC treated with platinum-based chemotherapy,31,34 as well as in advanced NSCLC treated with docetaxel.34,35 Co-mutations of TP53 and KRAS correlate with genomic instability, a higher tumor mutational burden, and increased PD-L1 expression, leading to variable responses.36 While patients with TP53/KRAS mutations may show enhanced responses to PD-L1 inhibitors, these mutations also predict a shortened overall survival when patients receive platinum-based adjuvant chemotherapy.37
The CodeBreak 100 and KRYSTAL-1 clinical trials explored the impact of co-occurring genomic alterations in STK11, KEAP1, and TP53 on the clinical outcomes of NSCLC patients with KRAS exon 2 p.G12C mutations treated with sotorasib and adagrasib.38,39 In these studies, the response rates to sotorasib and adagrasib across key co-mutated subgroups were as follows: STK11 (40–41%), TP53 (39–51%), with KEAP1 co-mutations resulting in numerically lower (ORR) of 20–29% for both inhibitors. Additionally, the exploratory data from the CodeBreak 200 trial showed that sotorasib, when compared to docetaxel in advanced NSCLC patients with prior platinum treatment and KRAS exon 2 p.G12C mutations, consistently benefited patients across these key co-mutated subgroups.40 This research underscores the importance of understanding how genomic alterations co-occur in KRAS G12C-mutated NSCLC, providing a valuable framework for stratifying patients by co-mutation status for targeted therapy with KRAS exon 2 p.G12C inhibitors, thereby potentially enhancing patient outcomes.
Inhibitors Targeted on KRAS Exon 2 P. G12C Mutation
For over forty years, targeting KRAS mutations was deemed a formidable challenge, with limited progress in developing effective chemotherapy or targeted treatments specifically for these mutations, leading to few clinically approved options. However, recent advancements in understanding the KRAS signaling pathway have paved the way for the development of targeted therapies (Figure 1). A significant milestone was reached with Sotorasib, which became the first direct KRAS inhibitor to enter clinical trials in 2018. It was subsequently approved in 2021 by the Food and Drug Administration (FDA) and European Medicines Agency (EMA) for treating second-line KRAS exon 2 p.G12C mutated locally advanced or metastatic NSCLC. In clinical trials, Sotorasib demonstrated a disease remission rate of 32.2% and a control rate of 88.1%, with an objective response rate of 37.1% in 124 patients with KRAS exon 2 p.G12C mutated NSCLC.38,41 Similarly, Adagrasib has also gained FDA approval, showing promising results in both preclinical and clinical trials, with a recent study reporting a remission rate of 45% and a disease control rate of 96.1%.42,43 Both drugs achieve their therapeutic effect by covalently and irreversibly binding to cysteine in the mutated protein, thereby inhibiting its function and biological activity. This approach, targeting the switch II pocket structure of the mutated KRAS protein, marks a significant breakthrough in the field of oncology, offering new treatment avenues for what were once considered “undruggable” oncogenic proteins (Figure 1).
Sotorasib
The results from the Phase 2 CodeBreak100 trial demonstrate that sotorasib, when used as a monotherapy in patients with advanced NSCLC harboring the KRAS exon 2 p.G12C mutation and who had previously undergone other anticancer treatments, offered sustained clinical benefits. The trial reported an overall response rate of 37.1%, with a median PFS of 6.8 months and a median OS of 12.5 months. Notably, the incidence of treatment-related adverse events was generally mild and manageable.38,44 Further, the Phase 3 CodeBreak200 trial data revealed that sotorasib, compared to docetaxel, significantly improved PFS in patients with advanced NSCLC with the KRAS exon 2 p.G12C mutation and prior antitumor drug treatments, while also exhibiting a more favorable safety profile.45 Although the primary endpoint PFS for CodeBreak 200 reached statistical significance the median PFS difference was low, raising concerns about the actual clinical benefit in the Sotorasib arm.
The FDA’s denial of sotorasib (Lumakras) supplemental new drug application (sNDA) was primarily due to limited clinical efficacy and the inability to reliably interpret the primary endpoint. One key issue was that the actual PFS benefit of Lumakras was smaller than reported by Amgen, with the PFS difference between Lumakras and chemotherapy possibly as brief as 5 days. Additionally, the FDA attributed the limited clinical benefits to potential biases and trial execution flaws. Several indicators suggested systematic bias in the confirmatory trial, such as dropout, discordance, crossover, and a independence. These factors may have led to an overestimation of sotorasib’s PFS benefit.
In the trial, only 2 patients in the sotorasib group were untreated, while 23 patients (13%) in the control group withdrew consent within the first 5 weeks, with no post-baseline imaging. This severe early dropout imbalance introduces significant uncertainty, as control group patients who withdrew consent may have been in better health and more motivated for treatment, leading to a potential underestimation of the control group’s outcomes. Although imaging discordance is common in clinical studies, the discordance pattern in the CodeBreak 200 trial raised concerns. Early discordance was higher in the control group (58% vs 69%), while late discordance was more prevalent in the experimental group (42% vs 31%). These discrepancies suggest a potential bias in favor of the experimental group, especially given that the PFS gain was only 5 weeks—shorter than the study’s imaging assessment interval.
Despite allowing crossover in the control group to minimize dropout, the FDA found that early crossover patients who were censored for PFS had better prognoses. Specifically, the 19 patients who crossed over early had better OS outcomes than the 27 patients who crossed over after BICR confirmation, with median OS of 18.9 months versus 11.8 months. This raises the question: would the trial outcomes have been different if early crossover patients had better prognoses? Hypothetical analysis suggests that if the 20 early dropout control patients and the 19 early crossover patients had a 50% or greater lower risk of disease progression compared to the trial group, the PFS hazard ratio (HR) would have lost its statistical significance.
The study also included a confirmation of progression (COP) process, separate from investigator and central imaging assessments. While the FDA typically views the COP process as minimally affecting the independence of results, concerns arose that irregular implementation may have biased the study’s final outcomes. Additionally, some critics questioned potential manipulation of the blinded independent central review (BICR) for PFS, raising concerns about the study’s integrity.
Adagrasib
Adagrasib, notable for its favorable pharmacokinetic properties, has a long half-life of 23 hours, exhibits dose-dependency, and is capable of penetrating the central nervous system.42,43,46 During the 2023 European Society for Medical Oncology (ESMO) meeting, the KRYSTAL-7 study presented promising results on the effectiveness and safety of adagrasib combined with pembrolizumab as a first-line treatment for patients with advanced NSCLC harboring the KRAS exon 2 p.G12C mutation.47 Specifically, for patients with a PD-L1 Tumor Proportion Score (TPS) of ≥ 50%, the regimen showed an ORR of 63% and a disease control rate (DCR) of 84%, offering preliminary evidence of its potential for long-lasting effects. This confirmed ORR of 63% notably surpasses the 39–45% ORR achieved with pembrolizumab monotherapy, underscoring the combination’s superior efficacy. Moreover, the median PFS has yet to be reached, even after a median follow-up of 10.1 months, indicating sustained benefits over time. In terms of safety, adagrasib and pembrolizumab together maintained the expected safety profiles seen when administered separately, with a lower incidence of treatment-related adverse events (TRAEs). Notably, only 4% of patients discontinued treatment due to TRAEs. Specifically, treatment-related liver events were reported in less than 10% of cases, with most being of minor severity, and no patients discontinued the treatment duo due to elevated Alanine Aminotransferase/Aspartate Aminotransferase (ALT/AST) levels or liver-associated TRAEs. Encouraged by these results, Mirati is preparing to launch a Phase III clinical trial to further investigate adagrasib with pembrolizumab as a first-line therapy in NSCLC patients with the KRAS exon 2 p.G12C mutation and a PD-L1 TPS of ≥50%. Complementing this, the ongoing confirmatory Phase III KRYSTAL-12 study is evaluating adagrasib against docetaxel in NSCLC patients with the KRAS exon 2 p.G12C mutation who have previously undergone treatment, aiming to validate the effectiveness of adagrasib in a broader treatment context.48
JDQ-443
JDQ-443 is another novel KRAS exon 2 p.G12C inhibitor reported in 2022, which exhibits superior structural features and enhanced therapeutic efficacy compared to existing KRAS inhibitors such as Sotorasib and Adagrasib. It binds covalently and irreversibly to the Switch II pocket of the KRAS exon 2 p.G12C protein. Uniquely, JDQ-443 also engages the C12 residue through a novel binding mode that avoids direct interaction with the H95 residue, which is associated with resistance.49,50 This interaction effectively locks the KRAS exon 2 p.G12C in an inactive state, inhibiting the excessive activation of the oncogenic pathway in tumors. In preclinical studies, JDQ-443 demonstrated potent inhibition of the KRAS exon 2 p.G12C signaling pathway and cellular proliferation, and exhibited dose-dependent anti-tumor activity.51 The ongoing KontRASt-01 study, an exploratory phase Ib/II trial, assesses the efficacy of JDQ-443 both as a monotherapy and in combination with TNO155 (an SHP2 inhibitor) and/or a PD-1 monoclonal antibody. The study reported an ORR of 45% according to Response Evaluation Criteria in Solid Tumors Version 1.1 (RECIST 1.1) criteria, which increased to 57% at the recommended 200mg BID dose level. This ORR surpasses those of Sotorasib and Adagrasib, marking it as the highest among current KRAS inhibitors. Regarding safety, most TRAEs were mild (grade 1–2). There were four instances of grade 3 TRAEs, but no severe adverse events (grade 4–5) were reported. These results affirm JDQ-443’s reliability and efficacy for treating NSCLC patients with KRAS exon 2 p.G12C mutations.52
Other KRAS Exon 2 P. G12C Inhibitors
The landscape of KRAS exon 2 p.G12C inhibitors has rapidly expanded following the successes of Sotorasib and Adagrasib. Currently, over 20 inhibitors targeting KRAS exon 2 p.G12C are in various stages of clinical research (Table 1). Notable among these are LY3537982, GFH925, JAB-21822, IBI351 (also known as GFH925), Garsorasib (D-1553), and others. In November 2023, the Center for Drug Evaluation (CDE) officially accepted the new drug application for IBI351,53 granting it priority review status. This decision was based on its potential to treat advanced NSCLC patients harboring the KRAS exon 2 p.G12C mutation, particularly those who have already received at least one systemic treatment. Furthermore, GDC-6036 is a promising KRAS exon 2 p.G12C inhibitor. According to the results of the Phase I trial,54 GDC-6036 treatment achieved durable clinical remission in patients with KRAS exon 2 p.G12C mutation-positive solid tumors, and the majority of adverse events were low-grade. In patients with NSCLC, the confirmed response rates was 53.4% (95CI 39.9% to 66.7%) and the median progression-free survival was 13.1 months (95% CI 8.8 months to unestimated). Of the 58 NSCLC patients with measurable disease, complete response rates were 2%, partial response rates were 59%, and disease stabilization was the best response in 29%. And Divarasib had a very rapid onset of action, with a median time from treatment initiation to achieving response of only 1.3 months and a median duration of response (DoR) of 14 months. In comparison to established drugs like Sotorasib (Lumakras) and Adagrasib (MRTX849), GDC-6036 demonstrates superior potency and selectivity in vitro, showing promise for advancing the treatment of KRAS mutations. The GO42144 study will subsequently explore the combination of Divarasib with a variety of other anticancer agents, such as PD-L1 inhibitors, PI3K alpha inhibitors, bevacizumab, and cetuximab, in an effort to delay the emergence of acquired resistance as much as possible, with the hope that Divarasib will bring greater therapeutic benefits.
Table 1.
Ongoing Trials with KRAS Exon 2 P. G12C Inhibitors in Lung Cancer
| Drug Inhibitor | Clinical Trial | Phase | Drug Combined | |
|---|---|---|---|---|
| 01 | GDC-6036 | NCT04449874 | I/II | Atezolizumab, cetuximab, bevacizumab, erlotinib, GDC1971, inavolisib |
| 02 | JDQ-443 | NCT04699188 | Ib/II | TNO155, tislelizumab |
| 03 | D-1553 |
NCT04585035 NCT05379946 |
I/II | Pembrolizumab, cetuximab, IN10018 |
| 04 | LY3537982, LY3499446 | NCT04956640 | I/II | Abemaciclib, erlotinib, pembrolizumab, temuterkib, LY3295668, cetuximab, TNO155 |
| 05 | JAB-21822 | NCT05002270 | I/II | Cetuximab |
| 06 | BI 1823911 | NCT04973163 | Ia/Ib | BI 1823911, BI 1701963, Midazolam |
| 07 | JAB-21822 | NCT05288205 | I/IIa | JAB-3312 |
| 08 | D-1553+IN10018 | NCT05379946 | I/II | D-1553+IN10018 |
| 09 | MK-1084 | NCT05067283 | Ia/Ib | Pembrolizumab, carboplatin, pemetrexed, cetuximab, oxaliplatin, leucovorin, 5-fluorouracil |
| 10 | JNJ-74699157 (ARS-3248) | NCT04006301 | I | NA |
| 11 | ARS-853 | Preclinical | NA | |
| 12 | ARS-1620 | Preclinical | NA | |
| 13 | ARS-3248 | NCT04006301 | I | NA |
| 14 | Compound1 | Preclinical | NA | |
| 15 | SML-10-70-1 | Preclinical | NA | |
| 16 | SML-8-73-1 | Preclinical | NA |
Inhibitors Targeted on KRAS Exon 2 P. G12D Mutation
KRAS exon 2 p.G12D mutations are more prevalent than KRAS exon 2 p.G12C mutations, yet, as of now, there are no approved targeted therapies for KRAS exon 2 p.G12CD worldwide. Drawing on the successful development strategies of KRAS exon 2 p.G12C inhibitors, several KRAS exon 2 p. G12D inhibitors have been developed (Table 2). Notably, MRTX1133 by Mirati Therapeutics is the first reported reversible inhibitor targeting KRAS exon 2 p. G12D.55 This compound selectively and reversibly inhibits KRAS exon 2 p. G12D, while sparing KRAS wild-type cells. In animal tumor models of pancreatic and colorectal cancer, MRTX1133 consistently shows dose-dependent inhibition of KRAS-dependent signaling. At a dosage of 3 mg/kg, it effectively inhibits the growth of G12D-mutant xenografts. However, MRTX1133 has not yet been reported to enter clinical trials. Other KRAS exon 2 p. G12D inhibitors are also predominantly in the early stages of clinical development. The research into KRAS exon 2 p. G12D inhibitors includes both small molecules and degraders, which vary in their methods of administration, including intravenous and oral dosing.
Table 2.
Ongoing Trials with KRAS Exon 2 P. G12D Inhibitors
| Drug Inhibitor | Phase | Cancer Type | Clinical Trial | |
|---|---|---|---|---|
| 01 | MRTX-1133 | I/II | NSCLC, PDAC, CRC, Other Solid Tumors | NCT05737706 |
| 02 | HRS-4642 | I | With Advanced Solid Tumors Harboring KRAS exon 2 p. G12D Mutation | NCT05533463 |
| 03 | ASP3082 | I | Solid Tumors | NCT05382559 |
| 04 | INCB161734 | I | Solid Tumors | NCT06179160 |
| 05 | RMC-9805 | I | NSCLC, CRC, PDAC, Advanced Solid Tumors | NCT06040541 |
| 06 | AST2169 | I | With Advanced Solid Tumors Harboring KRAS exon 2 p. G12D Mutation | CXHL2301442 |
| 07 | QLC1101 | Clinical approval | With Advanced Solid Tumors Harboring KRAS exon 2 p. G12D Mutation | NCT06403735 |
| 08 | JAB-22000 | Preclinical | PDAC, NSCLC, CRC | NA |
| 09 | UA022 | Preclinical | Tumors | NA |
Abbreviations: NSCLC, non-small cell lung cancer; PDAC, pancreatic ductal adenocarcinoma; CRC, colorectal cancer.
Pan-KRAS Inhibitors
Beyond the drug therapies targeting KRAS exon 2 p. G12D, pan-KRAS targeted therapies, which aim at multiple or all KRAS mutation variants, have provided new therapeutic choices for numerous cancer types with KRAS mutations. Among the recent developments, the Memorial Sloan Kettering Cancer Center in the USA has reported promising results with an inhibitory molecule, BI-2865. This molecule is noted for its ability to deactivate 18 of the 24 most commonly observed KRAS mutant proteins in cancer.56 Additionally, the company Revolution is making strides with RMC-6236, a pan-RAS inhibitor. Specifically targeting advanced solid tumors with KRAS G12 mutations (except KRAS exon 2 p.G12C), RMC-6236 is currently undergoing Phase 1 clinical trials. Notably, in animal models of non-small cell lung and pancreatic cancers, which were implanted with tumors exhibiting various KRAS gene mutations, RMC-6236 showed potent anticancer effects.57
BI-2865
Recently, Boehringer Ingelheim (BI) and the Memorial Sloan Kettering Cancer Center (MSKCC) unveiled a new Pan-RAS inhibitor, BI-2865, in a publication in Nature.56 Originating from the KRAS G12C inhibitor BI-0474, BI-2865 emerged as an optimized version after researchers removed its covalent warhead. This modification reduced its potency against the p.G12C mutation alone but expanded its inhibitory effect to cover other KRAS mutations, including p.G12D and p. G12V. In vitro experiments demonstrated that BI-2865 effectively inhibits the growth of cancer cells with various KRAS mutations, including G12C, G12V, and G12D. Its inhibitory effect on G12C mutations is comparable to that of Sotorasib. Out of the 24 common KRAS mutations found in human cancers, BI-2865 can inhibit 18, showcasing its broad potential in targeting KRAS-driven malignancies.
RMC-6236
The revised text effectively presents RMC-6236, an innovative oral, non-covalent RASMULTI(ON) inhibitor that specifically targets mutations and variants within classical RAS subtypes, exhibiting selectivity in both GTP-bound and dissociated states.58 In preclinical studies, RMC-6236 has been shown to consistently and powerfully suppress the growth of diverse RAS mutant tumors, particularly in cases of pancreatic and NSCLC with KRAS G12X mutations, where X represents A, D, R, S, or V. The Phase I clinical trials involved patients who had progressed after standard treatments, predominantly those with NSCLC featuring KRAS exon 2 p. 12D and 12V mutations. For NSCLC treated solely with RMC-6236, the ORR reached 38%, with a DCR of 85%. The median response time recorded was 1.4 months, the average duration of treatment was 3.1 months, and the longest PFS spanned nearly one year.59,60
Acquired Resistance to KRAS Exon 2 P. G12C Mutation
Mechanisms of Acquired Resistance to KRAS Exon 2 P. G12C
The development of acquired resistance to KRAS inhibitors in cancer treatment can be understood through two primary categories: on-target and off-target mechanisms. On-target resistance mechanisms are specifically associated with mutations occurring in the switch-II binding pocket of the KRAS protein. These mutations can alter the binding affinity of the drug, thereby diminishing its efficacy.61 Acquired KRAS activating mutations and the amplification of the KRAS gene are key factors that mediate resistance via on-target mechanisms. They achieve this by reactivating the RAS signaling pathway, which the KRAS inhibitors initially aim to suppress. Understanding these mechanisms is crucial for developing more effective strategies to overcome or prevent resistance to KRAS inhibitors in cancer therapy (Figure 1).
Secondary mutations in KRAS play a significant role in the development of resistance to KRAS exon 2 p.G12C inhibitors. In cases of resistance to sotorasib, secondary mutations in KRAS are observed in 87.3% of cases. Among these, KRAS exon 2 p. G13D is the most frequent mutation in sotorasib-resistant clones, accounting for 23%, followed by R68M and A59S mutations, each constituting 21.2%.62 In the context of resistance to adagrasib, KRAS Q99L emerges as the most prevalent secondary mutation, found in 52.8% of cases, with Y96D and R68S mutations following at 15.3% and 13.9% respectively.
Off-target resistance mechanisms to KRAS inhibitors involve a range of biological processes including the activation of bypass signaling pathways, epithelial-mesenchymal transition (EMT) mechanisms, and shifts toward cellular senescence. These mechanisms enable cancer cells to maintain signal activation and continue proliferating, even under the stress of KRAS inhibitor treatment. A key discovery in this area was made by Suzuki et al, who identified substantial upregulation of Mesenchymal to epithelial transition factor (MET) and hepatocyte growth factor (HGF) in cells that had developed resistance to sotorasib. They found that amplification of the MET gene played a crucial role in the acquired resistance to this drug. Importantly, the deletion of MET in resistant cells restored their sensitivity to sotorasib in vitro. This re-sensitization to the drug was also accompanied by the resumption of sustained Extracellular Regulated Protein Kinases (ERK) phosphorylation, highlighting the significance of MET in the resistance mechanism and offering a potential therapeutic target to overcome resistance in KRAS-mutated cancers.63
EMT represents another key non-genomic transcriptional reprogramming mechanism that contributes to acquired resistance in targeted cancer therapies.64 In the context of KRAS exon 2 p.G12C mutated NSCLC, the adoption of EMT characteristics has been linked to the development of acquired resistance to sotorasib, both in vitro and in xenograft models.65 This phenomenon was observed in H358 and LU65 cell lines that developed resistance to sotorasib, as evidenced by a decrease in e-cadherin and an increase in vimentin expression, which are hallmarks of EMT.65
Overall, despite the recent development of sotorasib and adagrasib, biological mechanisms of acquired resistance have yet been described in both pre-clinical and clinical specimens. Numerous emerging resistance mechanisms are being identified, and the resistance mechanisms related to KRAS resemble a vast network that requires exploration.66,67
Targeting Acquired Resistance to KRAS Exon 2 P. G12C Inhibitors
As detailed by Canon et al sotorasib may augment the efficacy of targeted therapies, including those that inhibit both upstream and downstream effectors of KRAS exon 2 p.G12C.68 Their synergy scoring analysis with various targeted therapies supports the concept that sotorasib can enhance therapeutic efficacy and help overcome acquired resistance. In studies involving the H358 xenograft model, combining sotorasib with Mitogen-Activated Protein Kinase/Extracellular Signal-Regulated Kinase (MEK) inhibitors led to a substantial reduction in tumor volume, markedly greater than using either monotherapy.68
Additionally, the KRAS exon 2 p.G12C/Y96D mutation is known to cause cross-resistance to both sotorasib and adagrasib by modifying drug binding.62,69 In response, Tanaka et al identified structurally and functionally distinct KRAS exon 2 p.G12C inhibitors that are effective against the Y96D mutation.69 RM-018, a novel KRAS exon 2 p.G12C inhibitor, exemplifies this by using cyclophilin A to bind and inhibit the GTP-bound state of KRAS exon 2 p.G12C. Both in vitro and in vivo studies demonstrate RM-018’s capability to decrease cell viability and significantly reduce RAS-MAPK signaling in cell lines with the KRAS exon 2 p.G12C mutation.70,71 When administered to the H358 KRAS G12C-NSCLC xenograft model, RM-018 induced dose-dependent tumor regression and was well tolerated. Importantly, RM-018 also showed in vitro efficacy against acquired resistance.69
Combination therapies that target both upstream and downstream signaling pathways of KRAS offer another avenue to circumvent acquired resistance.63,65,72 Additionally, the simultaneous inhibition of SOS1 and SHP2 presents a novel strategy for overcoming resistance to KRAS exon 2 p.G12C inhibitors.42
Combined with Inhibitors of Other Signaling Pathways
The potential synergy between the PI3K/AKT/mTOR and RAS/RAF/MEK signaling pathways suggests that targeting both pathways simultaneously, or even just one, could be an effective approach in treatments centered on signal transduction pathways (Table 3). Notably, the combined blockade of PI3K/mTOR and MEK has demonstrated significant effectiveness in mouse models with KRAS-induced adenocarcinoma.73 The activation of the PI3K pathway is dependent on RAS and its catalytic subunit p110α. By inhibiting this subunit, the PI3K pathway can be effectively suppressed, leading to hindered tumor growth. This effect is further enhanced when MEK inhibition is applied concurrently, showcasing the potential of a dual inhibition strategy in managing KRAS-driven cancers.74,75
Table 3.
Clinical Trials Assessing the Safety and Efficacy of Combining KRAS Exon 2 P. G12C Inhibitors with Other Cancer Therapies
| Combination Target | Combination Options | NCT Number | Phase |
|---|---|---|---|
| PD1/PD-L1 | Sotorasib + Pembrolizumab |
NCT04185883 NCT03600883 |
I/II |
| Adagrasib + Pembrolizumab |
NCT04613596 NCT03785249 |
II | |
| Sotorasib + AMG404 | NCT04185883 | I/II | |
| GDC-6036 + Atezolizumab | NCT04449874 | I | |
| LY3537982 + Sintilimab | NCT04956640 | I/II | |
| JDQ443 + Spartalizumab ±TNO155 | NCT04699188 | Ib/II | |
| SHP2 inhibitors | Sotorasib + RMC4630 |
NCT04185883 NCT05054725 |
II |
| Sotorasib + TNO155 | NCT04449874 | I | |
| Adagrasib + TNO155 | NCT04330664 | I/II | |
| GDC-6036 + GDC-1971 | NCT04185883 | I/II | |
| JDQ443 + TNO155 ±Spartalizumab | NCT04699188 | Ib/II | |
| SOS1 inhibitors | Adagrasib + BI1701963 | NCT04975256 | I/Ib |
| EGFR and pan ERBB inhibitors | Adagrasib + Afatinib | NCT03785249 | I/II |
| Sotorasib + Afatinib | NCT04185883 | I/II | |
| Sotorasib + MVASI | NCT05180422 | I/II | |
| JDQ443 + Cetuximab | NCT05358249 | Ib/II | |
| LY3537982 + Cetuximab | NCT04956640 | I/II | |
| LY3537982 + erlotinib | NCT04956640 | I/II | |
| GDC-6036 + Cetuximab | NCT04449874 | I | |
| GDC-6036 + Erlotinib | NCT04449874 | I | |
| MEK inhibitor | Sotorasib + Trametinib | NCT04185883 | I/II |
| Sotorasib + Avutometinib | NCT05074810 | I/II | |
| JDQ443 + Trametinib | NCT05358249 | Ib/II | |
| ERK inhibitors | LY3537982 + Temuterkib | NCT04956640 | I/II |
| mTOR inhibitor | Sotorasib + Everolimus | NCT04185883 | I/II |
| CDK4/6 inhibitors | LY3537982 + Abemaciclib | NCT04956640 | I/II |
| Sotorasib + Palbociclib | NCT04185883 | I/II | |
| Adagrasib + Palbociclib | NCT05178888 | I | |
| JDQ443 + Ribociclib | NCT05358249 | Ib/II |
Abbreviations: PD-1/PD-L1, programmed cell death protein 1/programmed cell death protein 1 ligand; SHP2, src homology region 2 domain-containing phosphatase-2; SOS1, son of sevenless homolog 1; EGFR, epidermal growth factor receptor; ERBB, The family of ERBB or epidermal growth factor (EGF) receptors includes four members: EGFR/ERBB1, ERBB2, ERBB3 and ERBB4. EGFR and ERBB2; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase; ERK, extracellular regulated protein kinases; mTOR, mammalian target of rapamycin; CDK 4/6, cyclin-dependent kinase 4/6.
Targeting RTKs Involved in by-Pass Signaling Pathways and Epithelial-to-Mesenchymal Transition
The strategy of dual inhibition, targeting both MET and KRAS exon 2 p.G12C, has proven effective in reducing tumor growth in sotorasib-resistant xenograft mouse models. This approach suggests that combining MET and KRAS exon 2 p.G12C inhibitors could be a viable treatment option for NSCLC patients who have both MET amplification and KRAS exon 2 p.G12C mutations.63 The efficacy of such dual blockade underscores the potential of combination therapies in overcoming resistance mechanisms and improving treatment outcomes for patients with complex genetic profiles in NSCLC.
In the context of KRAS exon 2 p.G12C inhibitor therapy, the activation of EMT and the PI3K pathway are recognized as contributing factors to both intrinsic and acquired resistance. While SHP2 plays a relatively minor role in activating the PI3K pathway, it is crucial in activating the MAPK pathway, particularly through SOS1 activation. SHP2 facilitates this process by interacting with SRC kinase,76 leading to the subsequent activation of the RAS signaling pathway. The use of combination therapy, such as sotorasib or adagrasib with RMC-4550, has shown enhanced efficacy compared to monotherapy. In the first human clinical trial phase 1 results, RMC-4630, used as a monotherapy, achieved a 71% disease control rate in NSCLC patients (n=19) with KRAS exon 2 p.G12C mutations.77
Targeting SHP2 and SOS1
Sotorasib Combined with TNO155
TNO-155, an allosteric SHP2 inhibitor, when combined with sotorasib, has been found to effectively inhibit ERK phosphorylation and reduce RAS-GTP levels in met-amplified H23-sotorasib resistant cells, more so than with monotherapy.63 The synergistic effect of combining KRAS exon 2 p.G12C inhibitors with PI3K and SHP2 inhibitors has been demonstrated to cause tumor regression in mouse models that acquired resistance to AMG510.65
JDQ443 Combined with TNO155
SHP2 inhibitors have long been the subject of debate, primarily because they are challenging to develop as standalone drugs. However, since SHP2 is situated upstream of the KRAS pathway, there is intense interest in developing SHP2 inhibitors in combination with KRAS inhibitors.78,79 In preclinical models, the combination of JDQ443 and the SHP2 inhibitor TNO155 has shown to enhance the antitumor efficacy beyond what is achieved with JDQ443 alone as a KRAS exon 2 p.G12C inhibitor. At the ESMO conference, researchers presented preliminary results from a dose-escalation study involving JDQ443+TNO155 in patients with advanced KRAS G12C-mutant solid tumors, including NSCLC.79 As of February 1, 2023, 50 patients had participated in this dose-escalation treatment. The patient cohort consisted of individuals diagnosed with NSCLC (24 patients, 48%), colorectal cancer (19 patients, 38%), pancreatic cancer (3 patients, 6%), cholangiocarcinoma (2 patients, 4%), duodenal cancer (1 patient, 2%), and ovarian cancer (1 patient, 2%). Notably, 17 of these patients had previously been treated with KRAS exon 2 p.G12C inhibitors.
Throughout the trial, six different dosing regimens of JDQ443+TNO155 were explored. Forty-four patients experienced TRAEs, with 18 reporting grade 3–4 TRAEs. Fortunately, no grade 5 TRAEs occurred. Efficacy was confirmed in patients with NSCLC, cholangiocarcinoma, duodenal cancer, and ovarian cancer. Of particular note, among the 12 evaluable NSCLC patients who had previously received KRAS exon 2 p.G12C inhibitor treatment, 4 (33.3%) displayed a confirmed response, achieving a disease control rate of 66.7% (8/12). The maximum tolerated dose (MTD) was not reached, leading to the selection of JDQ443 at 200 mg BID combined with TNO155 at 10 mg BID 2/1 as the recommended dose for further evaluation. This combination has shown strong tolerability and potential to address resistance to KRAS inhibitors while also providing synergistic efficacy. Future updates will include detailed biomarker data.
BI-3406 Combined with Adagrasib
SOS1 has emerged as a promising therapeutic target in cancers with KRAS mutations, owing to its pivotal role in feedback activation of the Mitogen-activated Protein Kinase (MAPK) pathway. Recently developed BI3406, a selective SOS1 inhibitor, specifically binds to the catalytic site of SOS1, blocking its interaction with GDP-bound KRAS.80 Preclinical studies of BI-3406 have shown its efficacy extends beyond KRAS exon 2 p.G12C mutation models, inducing tumor growth inhibition in xenograft models of various KRAS mutations including KRAS exon 2 p.G12C (MIA PaCa-2 cells), G12V (SW620 cells), G13D (LoVo cells), and G12S (A549 cells). In NSCLC and CRC (Colorectal Cancer) models, compared to administering KRAS exon 2 p.G12C inhibitors alone, combination therapy with SOS1 inhibitors enhances drug efficacy and delays the onset of acquired resistance.81,82
Combined Chemotherapy
KRAS exon 2 p.G12C inhibitors AMG-510 and MRTX 849 are being explored in combination with carboplatin and the CDK 4/6 (Cyclin-dependent Kinase) inhibitor palbociclib for the treatment of mutant lung cancer.42,68 Individual treatments with AMG-510 or carboplatin can inhibit tumor growth in models resistant to MRTX 849, yet their combined use significantly amplifies antitumor activity in mouse xenografts.68 Similarly, in MRTX 849-resistant models with a homozygous deletion of Cyclin Dependent Kinase Inhibitor 2A (CDKN 2A), the concurrent administration of MRTX 849 and palbociclib markedly induces tumor regression by targeting the Retinoblastoma (Rb)/E2F transcription factor pathway.42 Additionally, the combinations of AMG-510 with erlotinib or afatinib, and MRTX 849 with afatinib, show high efficacy in tumor models that depend significantly on RTK-mediated feedback resistance mechanisms. However, these combinations are less effective in other KRAS exon 2 p.G12C mutation models.83,84
Moreover, the KRAS exon 2 p.G12C inhibitor ARS-1620 and the cMET-targeted crizotinib demonstrate synergistic effects.85 In preclinical settings, AMG-510 also exhibits efficacy when used with the chemotherapeutic agent carboplatin.68 These findings highlight the diverse biological responses of KRAS mutant cells to combinations of KRAS inhibitors with chemotherapeutic agents. However, the broad applicability of these specific combinations to different tumor types and KRAS mutations still requires further validation.
Immunotherapy in KRAS Mutation NSCLC
Beyond chemotherapy and targeted therapies, the role of KRAS mutations in determining the efficacy of immune checkpoint inhibitors (ICIs) in NSCLC patients has gained attention. At the 2023 ESMO conference, researchers presented the Phase II KRYSTAL-7 study results, which involved combining adagrasib with pembrolizumab as a first-line treatment for patients with advanced NSCLC that harbors KRAS exon 2 p.G12C mutations.47 For patients exhibiting a PD-L1 expression of ≥50%, the ORR was 63%, and the DCR reached 84%. The median duration of response (DOR) has not yet been reached, with a 95% confidence interval (CI) of 12.6 months to not estimable (NE); similarly, the median PFS was also not reached, with a 95% CI of 8.2 months to NE. These promising results support the advancement to Phase III clinical trials to compare the efficacy of adagrasib combined with pembrolizumab against pembrolizumab monotherapy in treating first-line NSCLC patients with PD-L1 expression of ≥50% and KRAS exon 2 p.G12C mutations.86
A single-center retrospective study yielded inconclusive results regarding the correlation between immunotherapy and KRAS mutations, finding no significant association.87 In contrast, another study reported contradictory findings, suggesting that KRAS mutations may serve as predictive markers for extended responses to ICIs in NSCLC patients, particularly those with high PD-1 expression, in future clinical trials.88 It is typically observed that patients with KRAS mutations have higher PD-L1 expression and a greater tumor mutational burden (TMB) compared to those with wild-type KRAS. Tumors characterized by high TMB are known to be highly immunogenic, often described as “hot” or inflammatory tumors. These tumors are distinguished by the infiltration of immune cells, especially cytotoxic T lymphocytes, which can potentially make them more responsive to immunotherapy.89
The Choice of Treatment Options
Currently, treatments for NSCLC primarily include surgery, chemotherapy, immunotherapy, and targeted therapies.90 Sotorasib is available for patients with locally advanced or metastatic NSCLC harboring KRAS G12C mutations who have previously undergone at least one systemic therapy. However, The first-line clinical treatment regimen for KRAS-mutant NSCLC patients remains similar to that for patients without driver genes, primarily involving immunotherapy combined with chemotherapy, where bevacizumab plays a significant role. Targeted therapy is typically reserved as a second-line treatment. Additionally, local treatment methods such as surgery and radiotherapy are crucial options. Sequential chemoradiotherapy following chemo-immunotherapy is also one of the available regimens.
For metastatic NSCLC with KRAS mutations, the first-line treatment typically involves PD-L1-guided combined chemotherapy and immunotherapy. Although recent data suggest that the effectiveness of this approach requires further validation, it is considered superior to immunotherapy alone for certain patients. Treatment decisions for metastatic KRAS-mutated NSCLC, similar to those for NSCLC without driver mutations, are largely dependent on PD-L1 expression levels. Patients with PD-L1 levels ≥50% usually receive monotherapy with immunotherapy, while those with PD-L1 <50% are typically treated with a combination of immunotherapy and chemotherapy.
In addition to PD-L1 status, other biomarkers like KRAS mutation subtypes, co-mutation status, and tumor mutation burden should also be considered to stratify patients and tailor the most effective treatment strategy. Properly evaluating the status of RAS gene mutations and interpreting the biological and clinical significance of different KRAS and NRAS mutations is essential for the comprehensive management of patients with advanced NSCLC and colorectal cancer (CRC). To support healthcare professionals in precision oncology, Malapelle et al have developed an Italian knowledge database (www.rasatlas.com) that summarizes real-world data on RAS gene mutations in lung and colorectal cancers.91
Conclusion
The realm of KRAS-targeted drug development is still in its nascent stages, with the advent of KRAS exon 2 p.G12C inhibitors marking a breakthrough in the longstanding challenge of targeting KRAS mutations. This development represents just the beginning, the first chapter in unraveling the complexities of the KRAS enigma. Currently, the field is evolving rapidly, entering a new phase of exploration beyond the confines of KRAS exon 2 p.G12C inhibitors. Like other targeted therapies, the mechanisms of acquired resistance to KRAS exon 2 p.G12C inhibitors are diverse and complex. In this context, the formulation of combination therapies, particularly those incorporating SHP2 and SOS1 inhibitors, is emerging as a promising approach to tackle both intrinsic and acquired resistance.
Preclinical studies indicate that combining KRAS exon 2 p.G12C inhibitors with agents targeting upstream and downstream pathways is often essential for effective treatment. The efficacy of ICIs in KRAS-mutant NSCLC, however, remains an area of ongoing research. The response to ICIs appears to be heavily influenced by specific co-existing mutations and the type of KRAS mutation variant present. As such, there is a pressing need for continued research and development efforts to expand the therapeutic options available for effectively targeting KRAS mutations. Only through such concerted efforts will we be able to build a comprehensive arsenal against KRAS-driven cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15(5):290–301. doi: 10.1038/nrc3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrendt SA, Decker PA, Alawi EA, et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92(6):1525–1530. doi: [DOI] [PubMed] [Google Scholar]
- 3.Izar B, Zhou H, Heist RS, et al. The prognostic impact of KRAS, its codon and amino acid specific mutations, on survival in resected stage I lung adenocarcinoma. J Thorac Oncol. 2014;9(9):1363–1369. [DOI] [PubMed] [Google Scholar]
- 4.Pisapia P, Iaccarino A, De Luca C, et al. Evaluation of the molecular landscape in PD-L1 positive metastatic NSCLC: data from Campania, Italy. Int J Mol Sci. 2022;23(15) 8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karatrasoglou EA, Chatziandreou I, Sakellariou S, et al. Association between PD-L1 expression and driver gene mutations in non-small cell lung cancer patients: correlation with clinical data. Virchows Arch. 2020;477(2):207–217. doi: 10.1007/s00428-020-02756-1 [DOI] [PubMed] [Google Scholar]
- 6.Sumimoto H, Takano A, Teramoto K, Daigo Y. RAS-mitogen-activated protein kinase signal is required for enhanced PD-L1 expression in human lung cancers. PLoS One. 2016;11(11):e0166626. doi: 10.1371/journal.pone.0166626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N, Fang W, Lin Z, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017;66(9):1175–1187. doi: 10.1007/s00262-017-2005-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura Y, Sunaga N. Role of immunotherapy for oncogene-driven non-small cell lung cancer. Cancers. 2018;10(8):245. doi: 10.3390/cancers10080245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169–6177. doi: 10.1158/1078-0432.CCR-11-3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRAS (G12C) somatic mutations across race, sex, and cancer type. N Engl J Med. 2021;384(2):185–187. doi: 10.1056/NEJMc2030638 [DOI] [PubMed] [Google Scholar]
- 11.El Osta B, Behera M, Kim S, et al. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: the lung cancer mutation consortium experience. J Thorac Oncol. 2019;14(5):876–889. doi: 10.1016/j.jtho.2019.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Judd J, Abdel Karim N, Khan H, et al. Characterization of KRAS mutation subtypes in non-small cell lung cancer. Mol Cancer Ther. 2021;20(12):2577–2584. doi: 10.1158/1535-7163.MCT-21-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10(12):842–857. doi: 10.1038/nrc2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–225. doi: 10.1016/S0092-8674(00)00114-8 [DOI] [PubMed] [Google Scholar]
- 15.Wiesweg M, Kasper S, Worm K, et al. Impact of RAS mutation subtype on clinical outcome-a cross-entity comparison of patients with advanced non-small cell lung cancer and colorectal cancer. Oncogene. 2019;38(16):2953–2966. doi: 10.1038/s41388-018-0634-0 [DOI] [PubMed] [Google Scholar]
- 16.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheffler M, Ihle MA, Hein R, et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol. 2019;14(4):606–616. doi: 10.1016/j.jtho.2018.12.013 [DOI] [PubMed] [Google Scholar]
- 18.Cai D, Hu C, Li L, et al. The prevalence and prognostic value of KRAS co-mutation subtypes in Chinese advanced non-small cell lung cancer patients. Cancer Med. 2020;9(1):84–93. doi: 10.1002/cam4.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amanam I, Mambetsariev I, Gupta R, et al. Role of immunotherapy and co-mutations on KRAS-mutant non-small cell lung cancer survival. J Thorac Dis. 2020;12(9):5086–5095. doi: 10.21037/jtd.2020.04.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caballé-Perez E, Hernández-Pedro N, Ramos-Ramírez M, et al. Impact of KRAS(G12D) subtype and concurrent pathogenic mutations on advanced non-small cell lung cancer outcomes. Clin Transl Oncol. 2024;26(4):836–850. doi: 10.1007/s12094-023-03279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadal E, Beer DG, Ramnath N. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2015;10(2):e9–10. doi: 10.1097/JTO.0000000000000438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S, Long BN, Boris GH, et al. Structural insight into the rearrangement of the switch I region in GTP-bound G12A K-Ras. Acta Crystallogr D Struct Biol. 2017;73(12):970–984. doi: 10.1107/S2059798317015418 [DOI] [PubMed] [Google Scholar]
- 23.Cruz-Migoni A, Canning P, Quevedo CE, et al. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc Natl Acad Sci U S A. 2019;116(7):2545–2550. doi: 10.1073/pnas.1811360116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(5):497–530. doi: 10.6004/jnccn.2022.0025 [DOI] [PubMed] [Google Scholar]
- 25.Guan JL, Zhong WZ, An SJ, et al. KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol. 2013;20(4):1381–1388. doi: 10.1245/s10434-012-2754-z [DOI] [PubMed] [Google Scholar]
- 26.Jones GD, Caso R, Tan KS, et al. KRAS (G12C) mutation is associated with increased risk of recurrence in surgically resected lung adenocarcinoma. Clin Cancer Res. 2021;27(9):2604–2612. doi: 10.1158/1078-0432.CCR-20-4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadal E, Chen G, Prensner JR, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2014;9(10):1513–1522. doi: 10.1097/JTO.0000000000000305 [DOI] [PubMed] [Google Scholar]
- 28.Manolakos P, Ward LD. A critical review of the prognostic and predictive implications of KRAS and STK11 mutations and co-mutations in metastatic non-small lung cancer. J Pers Med. 2023;13(6):1010. doi: 10.3390/jpm13061010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salem ME, El-Refai SM, Sha W, et al. Landscape of KRAS G12C, associated genomic alterations, and interrelation with immuno-oncology biomarkers in KRAS -mutated cancers. JCO Precis Oncol. 2022;6(6):e2100245. doi: 10.1200/PO.21.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spira AI, Tu H, Aggarwal S, et al. A retrospective observational study of the natural history of advanced non-small-cell lung cancer in patients with KRAS p.G12C mutated or wild-type disease. Lung Cancer. 2021;159:1–9. doi: 10.1016/j.lungcan.2021.05.026 [DOI] [PubMed] [Google Scholar]
- 31.Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24(2):334–340. doi: 10.1158/1078-0432.CCR-17-1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8(7):822–835. doi: 10.1158/2159-8290.CD-18-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negrao MV, Araujo HA, Lamberti G, et al. Comutations and KRASG12C inhibitor efficacy in advanced NSCLC. Cancer Discov. 2023;13(7):1556–1571. doi: 10.1158/2159-8290.CD-22-1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricciuti B, Arbour KC, Lin JJ, et al. Diminished efficacy of programmed death-(Ligand)1 inhibition in STK11- and KEAP1-mutant lung adenocarcinoma is affected by KRAS mutation status. J Thorac Oncol. 2022;17(3):399–410. doi: 10.1016/j.jtho.2021.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh A, Daemen A, Nickles D, et al. NRF2 activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin Cancer Res. 2021;27(3):877–888. doi: 10.1158/1078-0432.CCR-20-1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi: 10.1158/1078-0432.CCR-16-2554 [DOI] [PubMed] [Google Scholar]
- 37.Shepherd FA, Lacas B, Le Teuff G, et al. Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2017;35(18):2018–2027. doi: 10.1200/JCO.2016.71.2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skoulidis F, Li BT, Dy GK, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med. 2021;384(25):2371–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jänne PA, Riely GJ, Gadgeel SM, et al. Adagrasib in non-small-cell lung cancer harboring a KRAS(G12C) mutation. N Engl J Med. 2022;387(2):120–131. doi: 10.1056/NEJMoa2204619 [DOI] [PubMed] [Google Scholar]
- 40.Skoulidis F, Langen AD, Paz-Ares LG, et al. Biomarker subgroup analyses of CodeBreaK 200, a phase 3 trial of sotorasib versus (vs) docetaxel in patients (pts) with pretreated KRAS G12C-mutated advanced non-small cell lung cancer (NSCLC). J clin oncol. 2023;41(16_suppl):9008. doi: 10.1200/JCO.2023.41.16_suppl.9008 [DOI] [Google Scholar]
- 41.Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383(13):1207–1217. doi: 10.1056/NEJMoa1917239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hallin J, Engstrom LD, Hargis L, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10(1):54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jänne PA, Rybkin I, Spira A, et al. KRYSTAL-1: updated safety and efficacy data with adagrasib (MRTX849) in NSCLC with KRASG12C mutation from a phase 1/2 study. 2020.
- 44.Spira AI, Wilson FH, Shapiro G, et al. Patient-reported outcomes (PRO) from the phase 2 CodeBreaK 100 trial evaluating sotorasib in KRAS p.G12C mutated non-small cell lung cancer (NSCLC). J clin oncol. 2021;39(15_suppl):9057. doi: 10.1200/JCO.2021.39.15_suppl.9057 [DOI] [Google Scholar]
- 45.De Langen AJ, Johnson ML, Mazieres J, et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRAS(G12C) mutation: a randomised, open-label, phase 3 trial. Lancet. 2023;401(10378):733–746. doi: 10.1016/S0140-6736(23)00221-0 [DOI] [PubMed] [Google Scholar]
- 46.Ou SI, Jänne PA, Leal TA, et al. First-in-human phase I/IB dose-finding study of adagrasib (MRTX849) in patients with advanced KRAS(G12C) solid tumors (KRYSTAL-1). J Clin Oncol. 2022;40(23):2530–2538. doi: 10.1200/JCO.21.02752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garassino MC, Theelen WSME, Jotte R, et al. LBA65 KRYSTAL-7: efficacy and safety of adagrasib with pembrolizumab in patients with treatment-naïve, advanced non-small cell lung cancer (NSCLC) harboring a KRASG12C mutation. Ann Oncol. 2023;34:S1309–S1310. doi: 10.1016/j.annonc.2023.10.066 [DOI] [Google Scholar]
- 48.Mok TSK, Lawler WE, Shum MK, et al. KRYSTAL-12: a randomized phase 3 study of adagrasib (MRTX849) versus docetaxel in patients (pts) with previously treated non-small-cell lung cancer (NSCLC) with KRASG12C mutation. J clin oncol. 2021;39(15_suppl):TPS9129–TPS9129. doi: 10.1200/JCO.2021.39.15_suppl.TPS9129 [DOI] [Google Scholar]
- 49.Weiss A, Lorthiois E, Barys L, et al. Discovery, preclinical characterization, and early clinical activity of JDQ443, a structurally novel, potent, and selective covalent oral inhibitor of KRASG12C. Cancer Discov. 2022;12(6):1500–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorthiois E, Gerspacher M, Beyer KS, et al. JDQ443, a structurally novel, pyrazole-based, covalent inhibitor of KRAS(G12C) for the treatment of solid tumors. J Med Chem. 2022;65(24):16173–16203. [DOI] [PubMed] [Google Scholar]
- 51.Li L, Zhao H, Peng X, et al. Discovery of novel Quinazoline-based KRAS G12C inhibitors as potential anticancer agents. Bioorg Med Chem. 2022;71:116962. doi: 10.1016/j.bmc.2022.116962 [DOI] [PubMed] [Google Scholar]
- 52.Tan DS, Shimizu T, Solomon B, et al. Abstract CT033: KontRASt-01: a phase Ib/II, dose-escalation study of JDQ443 in patients (pts) with advanced, KRAS G12C-mutated solid tumors. Cancer Res. 2022;82(12_Supplement):CT033–CT033. [Google Scholar]
- 53.Yuan Y, Jin Y, Jin Y, et al. Efficacy and safety of IBI351 (GFH925) monotherapy in metastatic colorectal cancer harboring KRAS G12C mutation: preliminary results from a pooled analysis of two phase I studies. J clin oncol. 2023;41(16_suppl):3586. doi: 10.1200/JCO.2023.41.16_suppl.3586 [DOI] [Google Scholar]
- 54.Sacher A, LoRusso P, Patel MR, et al. Single-agent divarasib (GDC-6036) in solid tumors with a KRAS G12C mutation. N Engl J Med. 2023;389(8):710–721. doi: 10.1056/NEJMoa2303810 [DOI] [PubMed] [Google Scholar]
- 55.Hallin J, Bowcut V, Calinisan A, et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat Med. 2022;28(10):2171–2182. doi: 10.1038/s41591-022-02007-7 [DOI] [PubMed] [Google Scholar]
- 56.Kim D, Herdeis L, Rudolph D, et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature. 2023;619(7968):160–166. doi: 10.1038/s41586-023-06123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mullard A. Glue-based KRAS inhibitors make their debut cancer trial mark. Nat Rev Drug Discov. 2023;22(12):942. doi: 10.1038/d41573-023-00169-8 [DOI] [PubMed] [Google Scholar]
- 58.Gustafson WC, Wildes D, Rice MA, et al. Direct targeting of RAS in pancreatic ductal adenocarcinoma with RMC-6236, a first-in-class, RAS-selective, orally bioavailable, tri-complex RASMULTI(ON) inhibitor. J clin oncol. 2022;40(4_suppl):591. doi: 10.1200/JCO.2022.40.4_suppl.591 [DOI] [Google Scholar]
- 59.Jiang J, Jiang L, Maldonato BJ, et al. Translational and therapeutic evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-driven cancers. Cancer Discov. 2024, 14, Of1–of24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arbour KC, Punekar S, Garrido-Laguna I, et al. 652O preliminary clinical activity of RMC-6236, a first-in-class, RAS-selective, tri-complex RAS-MULTI(ON) inhibitor in patients with KRAS mutant pancreatic ductal adenocarcinoma (PDAC) and non-small cell lung cancer (NSCLC). Ann Oncol. 2023;34:S458. doi: 10.1016/j.annonc.2023.09.1838 [DOI] [Google Scholar]
- 61.Romero D. Uncovering adagrasib resistance. Nat Rev Clin Oncol. 2021;18(9):541. doi: 10.1038/s41571-021-00545-6 [DOI] [PubMed] [Google Scholar]
- 62.Koga T, Suda K, Fujino T, et al. KRAS secondary mutations that confer acquired resistance to KRAS G12C inhibitors, sotorasib and adagrasib, and overcoming strategies: insights from in vitro experiments. J Thorac Oncol. 2021;16(8):1321–1332. doi: 10.1016/j.jtho.2021.04.015 [DOI] [PubMed] [Google Scholar]
- 63.Suzuki S, Yonesaka K, Teramura T, et al. KRAS inhibitor resistance in MET-amplified KRAS (G12C) non-small cell lung cancer induced by RAS- and non-RAS-mediated cell signaling mechanisms. Clin Cancer Res. 2021;27(20):5697–5707. doi: 10.1158/1078-0432.CCR-21-0856 [DOI] [PubMed] [Google Scholar]
- 64.Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer. 2020;20(12):743–756. doi: 10.1038/s41568-020-00302-4 [DOI] [PubMed] [Google Scholar]
- 65.Adachi Y, Ito K, Hayashi Y, et al. Epithelial-to-mesenchymal transition is a cause of both intrinsic and acquired resistance to KRAS G12C inhibitor in KRAS G12C-mutant non-small cell lung cancer. Clin Cancer Res. 2020;26(22):5962–5973. doi: 10.1158/1078-0432.CCR-20-2077 [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Liu L, Pei J, et al. Tissue factor overexpression promotes resistance to KRAS-G12C inhibition in non-small cell lung cancer. Oncogene. 2024;43(9):668–681. doi: 10.1038/s41388-023-02924-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lv X, Lu X, Cao J, et al. Modulation of the proteostasis network promotes tumor resistance to oncogenic KRAS inhibitors. Science. 2023;381(6662):p.eabn4180. doi: 10.1126/science.abn4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575(7781):217–223. doi: 10.1038/s41586-019-1694-1 [DOI] [PubMed] [Google Scholar]
- 69.Tanaka N, Lin JJ, Li C, et al. Clinical acquired resistance to KRAS(G12C) Inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11(8):1913–1922. doi: 10.1158/2159-8290.CD-21-0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nichols RJ, Schulze CJ, Bermingham A, et al. A06 tri-complex inhibitors of the oncogenic, GTP-bound form of KRASG12C overcome RTK-mediated escape mechanisms and drive tumor regressions in preclinical models of NSCLC. J Thorac Oncol. 2020;15(2):S13–S14. doi: 10.1016/j.jtho.2019.12.035 [DOI] [Google Scholar]
- 71.Schulze CJ, Bermingham A, Choy TJ, et al. Abstract PR10: tri-complex inhibitors of the oncogenic, GTP-bound form of KRASG12C overcome RTK-mediated escape mechanisms and drive tumor regressions in vivo. Mol Cancer Ther. 2019;18(12_Supplement):PR10–PR10. doi: 10.1158/1535-7163.TARG-19-PR10 [DOI] [Google Scholar]
- 72.Dunnett-Kane V, Nicola P, Blackhall F, Lindsay C. Mechanisms of resistance to KRAS(G12C) Inhibitors. Cancers. 2021;13(1):151. doi: 10.3390/cancers13010151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Castellano E, Sheridan C, Thin MZ, et al. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell. 2013;24(5):617–630. doi: 10.1016/j.ccr.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan TL, McCormick F. Killing tumors by keeping ras and PI3’ kinase apart. Cancer Cell. 2013;24(5):562–563. doi: 10.1016/j.ccr.2013.10.015 [DOI] [PubMed] [Google Scholar]
- 76.Zhang SQ, Yang W, Kontaridis MI, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13(3):341–355. doi: 10.1016/S1097-2765(04)00050-4 [DOI] [PubMed] [Google Scholar]
- 77.Ou SI, Koczywas M, Ulahannan S, et al. A12 the SHP2 inhibitor RMC-4630 in patients with KRAS-mutant non-small cell lung cancer: preliminary evaluation of a first-in-man phase 1 clinical trial. J Thorac Oncol. 2020;15(2):S15–S16. doi: 10.1016/j.jtho.2019.12.041 [DOI] [Google Scholar]
- 78.Falchook G, Li BT, Marrone KA, et al. OA03.03 sotorasib in combination with RMC-4630, a SHP2 inhibitor, in KRAS p.G12C-mutated NSCLC and other solid tumors. J Thorac Oncol. 2022;17(9, Supplement):S8. doi: 10.1016/j.jtho.2022.07.022 [DOI] [Google Scholar]
- 79.Negrao MV, Cassier PA, Solomon B, et al. MA06.03 KontRASt-01: preliminary safety and efficacy of JDQ443 + TNO155 in patients with advanced, KRAS G12C-mutated solid tumors. J Thorac Oncol. 2023;18(11, Supplement):S117–S118. doi: 10.1016/j.jtho.2023.09.151 [DOI] [Google Scholar]
- 80.Hofmann MH, Gmachl M, Ramharter J, et al. BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2021;11(1):142–157. doi: 10.1158/2159-8290.CD-20-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Daley BR, Sealover NE, Sheffels E, et al. SOS1 inhibition enhances the efficacy of and delays resistance to G12C inhibitors in lung adenocarcinoma. bioRxiv. 2023. [DOI] [PubMed] [Google Scholar]
- 82.Thatikonda V, Lu H, Jurado S, et al. Combined KRAS(G12C) and SOS1 inhibition enhances and extends the anti-tumor response in KRAS(G12C)-driven cancers by addressing intrinsic and acquired resistance. bioRxiv. 2023. doi: 10.1101/2023.01.23.525210 [DOI] [Google Scholar]
- 83.Ryan MB, Fece de la Cruz F, Phat S, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRAS(G12C) inhibition. Clin Cancer Res. 2020;26(7):1633–1643. doi: 10.1158/1078-0432.CCR-19-3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Misale S, Fatherree JP, Cortez E, et al. KRAS G12C NSCLC models are sensitive to direct targeting of KRAS in combination with PI3K inhibition. Clin Cancer Res. 2019;25(2):796–807. doi: 10.1158/1078-0432.CCR-18-0368 [DOI] [PubMed] [Google Scholar]
- 85.Wang H, Wang L, Zhang Y, et al. Inhibition of glycolytic enzyme hexokinase II (HK2) suppresses lung tumor growth. Cancer Cell Int. 2016;16(1):9. doi: 10.1186/s12935-016-0280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37(7):537–546. doi: 10.1200/JCO.18.00149 [DOI] [PubMed] [Google Scholar]
- 87.Sciortino C, Viglialoro V, Nucci M, et al. Response to immunotherapy in KRAS G12C mutated NSCLC: a single-centre retrospective observational study. Oncotarget. 2022;13(1):686–693. doi: 10.18632/oncotarget.28230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cefalì M, Epistolio S, Ramelli G, et al. Correlation of KRAS G12C mutation and high PD-L1 expression with clinical outcome in NSCLC patients treated with anti-PD1 immunotherapy. J Clin Med. 2022;11(6):1627. doi: 10.3390/jcm11061627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu C, Zheng S, Jin R, et al. The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett. 2020;470:95–105. doi: 10.1016/j.canlet.2019.10.027 [DOI] [PubMed] [Google Scholar]
- 90.NCCN. NCCN clinical practice guidelines in oncology-non-small cell lung cancer [version 4.2024 — April 10, 2024]. Available from: https://www.nccn.org/guidelines/category_12024. Accessed November 9, 2024. [DOI] [PubMed]
- 91.Malapelle U, Passiglia F, Cremolini C, et al. RAS as a positive predictive biomarker: focus on lung and colorectal cancer patients. Eur J Cancer. 2021;146:74–83. doi: 10.1016/j.ejca.2021.01.015 [DOI] [PubMed] [Google Scholar]