Abstract
The biogenesis of periplasmic and outer membrane proteins (OMPs) in Escherichia coli is assisted by a variety of processes that help with their folding and transport to their final destination in the cellular envelope. Chaperones are macromolecules, usually proteins, that facilitate the folding of proteins or prevent their aggregation without becoming part of the protein’s final structure. Because chaperones often bind to folding intermediates, they often (but not always) act to slow protein folding. Protein folding catalysts, on the other hand, act to accelerate specific steps in the protein folding pathway, including disulfide bond formation and peptidyl prolyl isomerization. This review is primarily concerned with E. coli and Salmonella periplasmic and cellular envelope chaperones; it also discusses periplasmic proline isomerization.
BIOGENESIS OF EXTRACYTOPLASMIC PROTEINS
In Gram-negative bacteria, proteins destined for the periplasm or outer membrane are, in the vast majority of cases, synthesized in the cytoplasm as precursors with a cleavable N-terminal signal sequence and then exported to their final destination in the cell envelope. Although various pathways exist for their transport across the inner membrane, most precursor proteins are translocated in unstructured states through an inner membrane channel formed by the heterotrimeric membrane complex SecYEG (1). After translocation and signal sequence cleavage, the newly exported extracytoplasmic proteins are sorted inside the periplasm via different folding pathways. While periplasmic localization is generally thought to be the default destination for proteins carrying cleavable signal sequences (2), folding intermediates of outer membrane proteins must transit through the periplasm in a conformational state compatible with their insertion into the lipid bilayer (3). Lipoproteins have their own dedicated secretion mechanism and chaperone, LolA. Lipoprotein secretion has recently been reviewed (4); it is not further described here.
The periplasm is separated from the extracellular milieu by only the porous outer membrane, which allows entry of anything below ∼600 Da, and is thus more susceptible to changes in the external environment than the cytoplasm. Periplasmic proteins must be able to cope with extreme changes in the environment. It is therefore not surprising that soluble periplasmic proteins are often highly stable, making them resistant to unfolding and aggregation (5). A disproportionate percentage of periplasmic proteins have had their structures determined by X-ray crystallography, which is understandable since highly stable proteins are often easier to crystallize (6). Given the high stability of periplasmic proteins, until fairly recently it was not even clear whether general periplasmic chaperones exist (7). However, as described below, it is now evident that there is an abundance of periplasmic chaperones, whose activity is particularly important under stress conditions.
The protein folding environment of the periplasm presents some unique differences from the protein folding challenges encountered in the cytosol. For example, the periplasm lacks ATP. Thus, it is clear that periplasmic chaperones must act using at least somewhat different mechanisms than those employed by cytosolic ATP-dependent chaperones. Another difference is that the cytosol is a reducing environment, whereas the periplasm is oxidizing. Disulfide bond formation and isomerization in periplasmic proteins is driven by the disulfide bond (DSB) family of enzymes. Null mutants in these DSB catalysts lead to steep declines in the abundance of a wide variety of proteins that contain disulfides necessary for their folding or stability (8, 9). The disulfide bond catalysts are not further discussed here, except to mention that DsbC and DsbG have both been shown to have chaperone activity in vitro (10, 11).
Proteins destined for the outer membrane or the exterior of the cell, such as pili, are not folded in the periplasm. They are instead kept in a partially unfolded, secretion-competent conformation as they transit the periplasm. The driving force for this transport and secretion may simply be the thermodynamic sink provided by outer membrane protein insertion into the outer membrane (12).
STRESS CHAPERONES
Although relatively few chaperones are involved in the folding of soluble periplasmic proteins under laboratory conditions, in nature, the E. coli periplasm is commonly faced with crises that can tax even the most stable proteins. Given the permeability of the outer membrane, the periplasm is especially vulnerable to the variety of different chemical stresses that Gram-negative bacteria are exposed to. These chemical agents can induce unfolding and/or aggregation of the periplasmic proteome. Thus far, several chaperones have been identified that combat periplasmic protein unfolding due to chemical agents. During transit through the stomach, for example, the E. coli periplasm is exposed to acid, which is a potent denaturant, or if swallowed by a herbivore, can be exposed to tannins, which are common in plants and are potent protein aggregation agents (13). As discussed below, periplasmic proteins require so much assistance remaining soluble under these conditions that E. coli induces chaperones to levels that approach 50% of the periplasmic proteome.
In addition to being required during stress and for outer membrane protein insertion, there are a number of elongated specialized structures present on the surface of bacteria, including pili, fimbriae, and curli, that require specialized chaperones for their assembly. Many of these structures appear to operate via a β-strand displacement mechanism, which we discuss further below.
REGULATORY MECHANISMS FOR STRESS-SPECIFIC PERIPLASMIC CHAPERONES
Organisms have evolved so that the expression level of periplasmic chaperones adapts to meet demand. The Cpx, sigmaE, Bae, Psp, and Rcs pathways in E. coli all sense alterations in the bacterial envelope and respond by inducing a variety of components involved in the formation and maintenance of the bacterial envelope, including many periplasmic chaperones and folding factors (14). The protease/chaperone DegP and the prolyl isomerase FkpA, for example, are induced by both the Cpx and sigmaE pathways. The chaperone Spy is induced by the Bae, Cpx, and RcsB pathways, whereas the outer membrane insertion chaperone SurA is only induced by the sigmaE pathway. These pathways are activated by a variety of stressors (15). For example, the potent protein denaturant, ethanol, activates all five pathways, suggesting that all of these pathways are responsive to protein unfolding (16). However, tannins only induce the Bae pathway. Bae induction by tannins is so strong that the Bae-controlled chaperone Spy becomes nearly half of the periplasmic mass after tannin treatment (13, 16). Acid treatment is also known to unfold proteins while also strongly inducing expression of the acid-responsive chaperones HdeA and HdeB (17).
Different protein denaturants act with varying degrees of effectiveness on different sets of proteins and induce distinct unfolded state ensembles (18). Thus, it is not surprising that different chaperones are needed to cope with these various denatured states. It seems likely that different chaperones have evolved to recognize not just different proteins but also different unfolded states of the same protein. Characterizing the presumably complex and overlapping substrate specificity of the various periplasmic chaperones is in its early stages.
The Cpx and Bae responses are classic two-component regulatory systems (19, 20). The Cpx response is one means of detecting and responding to periplasmic protein folding problems (21). Many of the Cpx inducers, including overexpression of misfolded proteins, disruption of periplasmic disulfide bond formation, and treatment with denaturing alcohols act to disturb envelope folding. About 50 transcriptional units are induced by the Cpx response, many of which are cell envelope proteins (14). Some of the proteins most strongly induced by the Cpx response encode periplasmic folding and degradation factors, including Spy, DsbA, DegP, and HtpX. The Cpx response also plays roles in virulence, pili formation (22), and drug resistance (23), and helps regulate inner membrane transport and energy metabolism (21). The Bae response, in contrast, triggers the induction of only about 7 transcriptional units. Since Spy, a periplasmic chaperone, is by far the most strongly induced protein, it seems likely that the Bae response is mainly also designed to deal with protein folding issues. However, the Bae response also induces multidrug exporters; thus, this regulon may also help deal with toxic small molecules (24).
Both the Cpx and Bae systems consist of a histidine sensor kinase, CpxA or BaeS, respectively, that acts to autophosphorylate its own transmitter domain (24). Activation is achieved after this phosphoryl group is transferred to the receiver domain of the response regulator CpxR or BaeR, respectively. These regulators then act as transcription factors to activate the Cpx and Bae responses (14). CpxP, the protein most strongly induced by the Cpx response, acts as an auxiliary regulator by sensing misfolded periplasmic proteins and modulating the activity of CpxA, likely by downregulating its autokinase activity (25). CpxP is homologous to the periplasmic chaperone Spy and the periplasmic zinc binding proteins, ZraP and CnrX (26, 27). CpxP has weak chaperone activity in vitro (13), but this is most likely a by-product of its need to bind and sense misfolded periplasmic proteins for regulating CpxA rather than its functioning directly as a general chaperone in vivo. Most chaperones are highly abundant proteins, but CpxP is only present at about 40 molecules per cell (28).
The Rcs system also acts through a phospho-relay system. For the sigmaE stress regulon, the accumulation of outer membrane porins and lipopolysaccharides act as inducing signals that cause the release of the transcription factor sigmaE through the regulated proteolysis of an inner membrane protein RseA, which normally acts to sequester sigmaE during nonstress periods. The Psp envelope stress-responsive system also acts by sequestering a transcription factor, in this case PspF, a sigma54 enhancer that is normally sequestered through binding by PspA. PspB and PspC bind to PspA after they themselves sense the activation signal, causing the release of PspA from PspF and thus activation of sigma54.
STRESS-SPECIFIC CHAPERONES
HdeA and HdeB
E. coli must transit through the acidic stomach of warm-blooded animals to colonize the intestines. Whereas the cytosol is protected from the low pH environment because of the relative impermeability of the inner membrane to protons, the periplasmic proteome is unprotected from this acid stress (pH ∼2) because of the high permeability of the outer membrane to small molecules (<600 Da). Since the periplasm is devoid of ATP, the canonical ATP-dependent foldase chaperones of the cytosol are unavailable to protect periplasmic proteins from the acid stress.
However, E. coli contains two small (∼10 kDa) chaperones called HdeA and HdeB that protect the periplasmic proteome during acute acid stress (17, 29). Both the hdeA and hdeB genes share an operon and are located on an acid fitness island (30). HdeA is reported to be the 6th most abundant protein in the cell during stationary phase (31). This point is made more remarkable by the fact that HdeA is localized to the periplasm, a compartment that takes up only 10 to 20% of the total cell mass. HdeA’s abundance likely reflects the cell’s need to be prepared for two considerations that occur during acid stress: (1) nearly all proteins unfold in response to acid stress, and (2) the decline in pH that accompanies ingestion of bacteria into the mammalian gut is so fast that it precludes effective transcriptional or translational responses.
A number of reports suggest that HdeA functions at extreme acidic pH values (pH 1–3), whereas HdeB is active under more mild acid stress (pH 4–5) (32–34). Both proteins suppress the aggregation of proteins at their respective optimal pH values. Indeed, several in vivo cross-linking studies have revealed that HdeA and HdeB both bind a broad range of the periplasmic proteome, albeit at different pH conditions and with differing substrate specificity (35). Although HdeA and HdeB have only 17% sequence identity, they are structurally similar. Both proteins exist as well-folded α-helical dimers at neutral pH that bury large hydrophobic surfaces in their dimer interfaces (36). HdeB appears to be less important than HdeA for acid resistance; naturally occurring mutants that disrupt the ATG codon of HdeB even exist in some E. coli strains.
The chaperone activity of both HdeA and HdeB is regulated at the posttranslational level, which is likely necessary to respond to rapid changes in environmental pH. On exposure to low pH, HdeA undergoes a rapid, dramatic conformational change from a well-folded chaperone-inactive α-helical dimer (36) to partially disordered chaperone-active monomers (37–39). The large hydrophobic stretches buried in the dimer-dimer interface at neutral pH have been shown to be the substrate binding site of the HdeA monomers. Computational and mutagenesis studies revealed that the protonation of two aspartic acids in key salt bridges is necessary to monomerize HdeA (39–43). After activation, the chaperone-active monomers then tightly bind (nM Kd values) to substrate proteins unfolded by the acid stress to prevent them from aggregating. Interestingly, the HdeA monomers adopt different conformations when bound to different substrates, illustrating how HdeA can promiscuously bind to a variety of different client proteins. Return to neutral pH causes the release of bound substrates, allowing them to refold back into their native state, and HdeA returns to its chaperone-inactive dimer conformation.
In contrast, HdeB remains dimeric at its chaperone-active pH values. However, the HdeB dimer appears to undergo some structural rearrangements between pH 7 and pH 4, and the loops between helices 2 and 3 become less dynamic when HdeB is transitioned from pH 7 to pH 4 (44). The actual substrate binding site on HdeB has remained elusive.
Spy
Spy (Spheroplast Protein Y) (45) was discovered as a chaperone by a genetic selection that links protein stability to antibiotic resistance in the E. coli periplasm (13). The selection used a biosensor in which an unstable variant of immunity protein 7 (Im7) was inserted between two halves of β-lactamase, an enzyme that confers antibiotic resistance to penicillin. Spy was identified by its ability to enhance the stability of the Im7 insert and therefore enhance the antibiotic resistance of E. coli carrying the biosensor. Spy is a small (16 kDa) ATP-independent chaperone that can prevent aggregation of a variety of proteins and can promote the refolding of client proteins at substoichiometric concentrations (13). The crystal structure of Spy (13, 46) shows that its active form is a thin, cradle-shaped all α-helical dimer with a large hydrophobic surface in the middle of the cradle surrounded by basic residues on the edges of the cradle. It is normally not expressed under nonstress conditions but is massively upregulated upon protein unfolding stresses such as ethanol, butanol, or tannins, comprising a high percentage of the periplasmic proteome during these stress conditions (16, 47). Moreover, deletion of the spy gene makes E. coli more sensitive to tannin treatment. Its expression is controlled by the Cpx and Bae systems, both of which respond to protein unfolding stress in the cellular envelope. Spy mutants are sensitive to and accumulate zinc. It is not yet clear if this is due to an effect of Spy on zinc transport proteins or if Spy directly binds zinc (48). However, the Spy homologue, ZraP, is a zinc binding protein and also has chaperone activity in vitro (27).
The mechanism of chaperone action for Spy on its client, Im7, has been investigated in great detail (49–51). Unlike most chaperones, Spy uses complementary charge-charge interactions to bind to its unfolded substrates. Long-range electrostatic interactions between the positively charged binding site on Spy and negatively charged substrates accelerates Spy’s association rate constant for substrates so that client binding is diffusion limited. This allows Spy to rapidly associate with unfolded, aggregation-prone clients to prevent their aggregation. After recognition of clients through long-range electrostatics, hydrophobic interactions between the central Spy cradle and the unfolded substrate stabilize the complex (50). Remarkably, Spy then allows Im7 to fold from its unfolded conformation into its native structure while remaining continuously bound to the chaperone surface (51). By doing so, Spy minimizes the concentration of unbound protein folding intermediates that would otherwise be aggregation prone. Upon substrate folding, the hydrophobic contacts between Spy and its client are reduced, which increases the dissociation rate constant of the now-native client from Spy and causes release of the native client. Thus, client refolding triggers its own release from Spy, precluding the need for any cofactors or cochaperones to regulate Spy’s chaperone function (50). Recent crystallographic experiments using an iodine-labeled Im7 substrate have captured snapshots of Im7 as it refolds while continuously bound to the surface of Spy. In these snapshots, the Im7 bound to Spy adopts conformations ranging from an unfolded conformation to a native-like structure (49).
CHAPERONES INVOLVED IN OUTER MEMBRANE PROTEINS BIOGENESIS
The outer membrane of E. coli contains mainly two types of proteins, lipoproteins and β-barrel membrane proteins. Since the biogenesis of lipoproteins and outer membrane proteins is the subject of recent reviews (52), we will restrict our discussion to the role that the periplasmic chaperones SurA, Skp, and DegP play in maintaining the solubility of outer membrane proteins as they transit the periplasmic space.
Skp
Skp is a Seventeen Kilodalton Periplasmic protein (53). Initial evidence for Skp playing a role in the folding pathway of outer membrane proteins included its retention on an OmpF affinity column (54) and the observation that skp gene mutants can be isolated based on their increased sigma E activity (55). Skp is a nonessential periplasmic protein (54), and outer membrane protein levels are not significantly affected by deletion of skp (56). However, Skp-SurA double mutants are inviable, providing evidence that Skp and SurA function in parallel pathways (57). SurA depletion in a Skp null strain diminishes the levels of nearly all β-barrel proteins (56). Likewise, double mutants lacking both Skp and the periplasmic protease DegP are not viable at 37°C, possibly due to the accumulation of protein aggregates in the periplasm (58). SurA appears to participate in the primary pathway for outer membrane protein insertion, whereas Skp appears to provide a secondary pathway that is specifically necessary for some proteins such as LptD (59).
More than 30 proteins bind to Strep-tagged Skp, most of which are outer membrane proteins (56). Skp is involved in the handoff of substrates to the β-barrel assembly machine (BAM) complex, which helps to fold proteins and insert them into the outer membrane. Although Skp is primarily involved in the transport and folding of outer membrane proteins, it also interacts with some soluble periplasmic proteins including β-lactamase, OppA, and maltose binding protein (60), and its overexpression has been shown to be helpful for periplasmic recombinant expression of some proteins (61, 62).
Skp binds a variety of unfolded outer membrane proteins with binding affinities in the nM range, preventing their aggregation, and facilitates their insertion into negatively charged lipid membranes (63–65). Skp substrates include OmpA, LptD, and FhuA, and binding to these substrates occurs via aromatic residues and electrostatic interactions (66). Furthermore, biochemical experiments show that Skp selectively binds the newly synthesized outer membrane protein, PhoE immediately after its translocation across the inner membrane (67).
Evidence suggests that Skp functions in the periplasm as a molecular chaperone for several β-barrel proteins by maintaining soluble conformations before their insertion and folding into the outer membrane bilayer. Skp also sequesters partially folded intermediates of soluble recombinant proteins including single-chain antibodies expressed in the periplasm, preventing their aggregation (68).
The crystal structure of this highly basic protein reveals three long α-helical hairpins protruding from a central β-barrel structure that defines a trimerization domain (69, 70). This homotrimeric complex has the overall appearance of a jellyfish that resembles the cytosolic holdase chaperone prefoldin, with three flexible tentacles defining a large internal cavity that constitutes its substrate binding site (68, 69). It has been shown that unfolded outer membrane proteins bind inside this central, predominantly hydrophobic cavity by surface-surface interactions. However, electrostatic interactions between Skp and the outer membrane proteins, PagP, are important for PagP folding and outer membrane insertion (71). It has been postulated that successive formation of increasing electrostatic interactions allows substrates to climb into Skp’s internal cavity (72).
The binding cavity of Skp has been estimated to accommodate substrate proteins up to ∼25 kDa in size. However, Skp is able to interact with unfolded outer membrane proteins (uOMPs) that are much larger than this. NMR experiments on the ∼35 kDa uOmpA bound to Skp showed that only the β-barrel domain of uOmpA is retained within the Skp binding cavity, providing an explanation for how Skp interacts with larger uOMPs. Very recent kinetic and ion-mobility mass spectrometry data showed that the central cavity of Skp can actually expand to accommodate larger substrates, and that multiple Skp molecules can bind a single uOMP for larger uOMP substrates. Indeed, in vitro experiments revealed that 2:1 or higher Skp:uOMP stoichiometries are necessary for Skp to suppress the aggregation of larger outer membrane protein clients (73). Both the uOMP and the Skp arms are flexible, and small angle neutron scattering (74) and modeling studies indicate that the mobility of Skp’s arms is important for allowing it to interact with a variety of conformations of the uOMP. After binding, however, Skp’s arms become more rigid (75). NMR studies indicate that many weak interactions occur between the substrate and the chaperone; this, combined with the high mobility and flexibility of both binding partners, can help explain the ability of Skp to interact promiscuously with various substrates and different conformations of proteins. These interactions have the end effect of shielding the aggregation-prone uOMP from the aqueous environment (52). In addition to preventing aggregation, it has been proposed that Skp traps folding intermediates, stabilizes dynamic unfolded states of its substrates, and funnels them toward their native conformations (76).
SurA
SurA is a 45-kDa protein that was originally identified as being required for survival of E. coli in stationary phase, but not during log phase (77). The physiological defects of surA mutants are indicative of outer membrane perturbations, but are more severe than those of skp mutants, suggesting that SurA plays a more dominant role as an outer membrane protein chaperone (55, 78, 79). These defects include hypersensitivity to bile salts, detergents, and hydrophobic antibiotics, and reduced levels of outer membrane porins and reduced virulence (80, 81). In surA mutants, unfolded monomeric species accumulate in the folding pathway of the maltoporin LamB (79). Deletion of SurA also leads to major decreases in the abundance of outer membrane proteins in Yersinia enterocolitica, which in turn substantially affects the virulence of this pathogen (82). Differential proteomics showed that a number of outer membrane proteins are less abundant in a surA mutant strain, although most of them are less abundant due to activation of the sigmaE stress response, which lowers the expression of outer membrane proteins in the surA mutant (83). The decrease in abundance correlates with decreased mRNA levels for all the outer membrane proteins except FhuA and LptD, suggesting that SurA is directly important for the folding of these two outer membrane proteins (83). The lptD gene actually shares an operon with surA, and overexpression of LptD in a strain lacking SurA fails to recover wild-type levels of LptD. Furthermore, these two outer membrane proteins show the most dramatic reduction in protein levels when SurA is depleted in a skp mutant strain. Although decreases at the RNA level appear sufficient to explain the decreases at the protein level for many outer membrane proteins, this does not necessarily eliminate the possibility that SurA plays an important role in the transport and folding of outer membrane proteins. Indeed, studies suggest that SurA does bind to the majority of outer membrane proteins during their transit through the periplasm, and SurA is essential for in vitro insertion of membrane proteins in a reconstituted system.
The surA gene is nonessential. However, like skp (84), a combination of surA and degP null mutations exhibits a synthetic lethal phenotype. This suggests that Skp and SurA may provide redundant chaperone functions in the periplasm (84, 85). This redundancy may explain why SurA mutants reveal relatively few obligate substrates. SurA cooperates with the disulfide isomerase DsbC in the folding of the outer membrane protein LptD (11). However, most models propose that SurA participates in the major pathway for outer membrane protein export and insertion, whereas Skp and DegP are involved in an auxiliary pathway (86). Further evidence for the existence of multiple parallel pathways for the folding and degradation of periplasmic proteins comes from an extensive genetic analysis conducted by the Ehrmann laboratory (57), where they individually, or in combination, mutated 15 different folding factors and proteases. All 7 different folding factors tested (SurA, DsbA, DsbC, FkpA, DegP, Skp, and PpiD) were synthetically lethal under at least one growth condition when combined either with each other or with a protease. A number of combinations also caused induction of the spy gene, which indicates activation of the periplasmic stress response, or leakage of β-galactosidase into the media, indicating a fragile cell envelope.
The primary structure of SurA is made up of four separate regions: an N-terminal region of ∼150 residues, two peptidyl prolyl isomerase (PPIase) domains of ∼100 residues each, and a short (∼40 residues) C-terminal extension. Although SurA exhibits low PPIase activity in vitro, the PPIase domains are nonessential: a variant of SurA in which the N-terminal region is directly fused to the C-terminal region without either of the two PPIase domains has in vitro chaperone activity and can almost entirely complement the in vivo phenotype of a surA deletion mutant, with the exception of novobiocin sensitivity (87). The two PPIase domains may, however, play a role in regulating the function of the chaperone domain (88, 89). The N-terminal structural domain has a deep elongated crevice that could accommodate unfolded polypeptides, suggesting a binding site for unfolded outer membrane proteins (90, 91). Consistent with this observation, SurA is known to bind several peptides with alternating aromatic amino acid motifs (Ar-X-Ar) frequently found in the sequences of outer membrane proteins (91, 92). Puzzlingly, crystallographic data showed that peptides containing these motifs actually bind to the first PPIase domain, not the chaperone domain (93); it is unclear if this binding to the first PPIase domain is relevant to SurA’s function as an outer membrane protein chaperone. It is also still unclear how SurA binds to unfolded outer membrane proteins to prevent their aggregation, because it seems to lack the ability to encapsulate substrates in the way that Skp does. Making SurA more rigid by introducing disulfide bonds does not negatively affect its function, implying that flexibility is not as important for SurA as it is for Skp (94). However, both Skp and SurA share an ability to interact with substrates in a flexible dynamic fashion (95). Recent single-molecule force microscopy experiments suggest that SurA facilitates the insertion of β-barrel outer membrane proteins into the lipid membrane one β-hairpin at a time until the entire outer membrane protein has been inserted (76). There is currently no evidence that SurA is involved in the folding of soluble periplasmic proteins.
PROLINE ISOMERASES
Peptidyl prolyl isomerases (PPIases) catalyze cis/trans proline isomerization, an important step in protein folding. Most proline residues are in the trans configuration, but some proteins require prolines in the cis configuration in order for them to adopt their native structure. Proline isomerases interconvert proline trans and cis isomers on a biologically meaningful timescale. Four enzymes with PPIase activity exist in the periplasm: PpiA, SurA, FkpA, and PpiD. Since isomerization of proline residues between the cis and trans configuration is often a rate-limiting step in protein folding, it is logical that the proline isomerization activity of these proteins should be important for protein folding. Perplexingly, this has been difficult to directly establish in vivo. For example, a quadruple mutant that lacks all four PPIases in E. coli shows a defect in pilus formation but is still viable (96).
The first proline isomerase, PpiA, is a cyclophilin homologue. Like many folding factors, PpiA is induced by the Cpx regulatory system (97). However, null mutants in ppiA have normal levels of periplasmic and outer membrane proteins and no detectable phenotype (98), raising questions about PpiA’s role in protein folding.
The second proline isomerase, SurA, clearly plays an important role in outer membrane protein folding, as discussed above. In addition to its chaperone function, SurA also contains two PPIase domains that show PPIase activity in vitro (89). However, the role of SurA’s PPIase activity is unclear: mutants that disrupt SurA’s PPIase catalytic activity have little impact on its in vivo activity in outer membrane protein insertion (99). Instead, it is the peptide binding activity of this PPIase domain that seems to be important for SurA’s chaperone activity. Synthetic constructs made by inserting the chaperone domain of the unrelated chaperone, SlyD, into the second PPIase domain of SurA generates enzymes that are very effective in catalyzing proline isomerization, suggesting that the PPIase domains of SurA are not optimized for PPIase activity (100).
The third proline isomerase, FkpA, has a domain related to the FKBP family of prolyl isomerases as well as a chaperone domain (101). FkpA assists in the folding of recombinant proteins (102) and a MalF-LacZ fusion protein expressed in the periplasm (103). Furthermore, when overproduced, FkpA can rescue the lethal phenotype of a ΔsurA Δskp strain, suggesting that FkpA may be involved in outer membrane proteins biogenesis. It is not just the chaperone activity of FkpA that is important; the PPIase activity of FkpA has been shown to improve the proper periplasmic folding of colicin M (104). Although FkpA appears to only play a minor role in outer membrane protein biogenesis at 37°C, its importance increases with increasing growth temperature: FkpA function is as important as SurA for E. coli survival at 44°C (105). The binding rate and affinity of FkpA for outer membrane proteins increases under these conditions, suggesting that FkpA may be a temperature-responsive chaperone/PPIase for outer membrane protein biogenesis.
PpiD, like PpiA, is a periplasmic parvulin homologue. PpiD is anchored in the inner membrane by one transmembrane α-helix, and its parvulin-related PPIase domain extends into the periplasm. This PPIase domain, however, seems to be inactive (106–108). Overall, the PpiD protein does play a role in envelope biogenesis, because PpiD mutants are SDS/EDTA sensitive, indicating a cell wall defect. PpiD mutants, when combined with degP mutants, are, in addition, both temperature sensitive and salt sensitive (57). PpiD overproduction can rescue the lethality of a skp surA double mutant and the growth phenotype of a fkpA, ppiD, surA triple mutant in a way that is independent of its PPIase domain. Presumably, these effects are due to PpiD’s reported chaperone activity (109).
In summary, although E. coli possesses multiple periplasmic proteins with prolyl isomerase domains, the importance of these proteins appears to be more dependent on their chaperone activity than their PPIase activity.
DegP
degP (also known as htrA for high-temperature requirement) was first identified as a gene essential for growth at elevated temperatures where protein unfolding/misfolding is more pronounced. DegP is upregulated by the sigmaE and Cpx envelope stress response systems. As mentioned previously, deletion of degP shows a synthetically lethal phenotype when combined with either a surA or skp mutant, suggesting that DegP plays a role in outer membrane protein biogenesis. However, DegP is well known for its role as a serine protease in the periplasm towards unfolded proteins; degP null mutants accumulate misfolded proteins. However, DegP has also been reported to have chaperone activity, which is evident at temperatures below 28°C, whereas its protease activity dominates at higher temperatures (110). Structurally, DegP has an N-terminal chymotrypsin-related protease domain that contains an active site His-Asp-Ser motif and two PDZ domains at the C terminus. PDZ domains are often involved in protein-protein interactions. DegP forms large ring-shaped 12-mers or 24-mers that can encapsulate bound substrates. This encapsulation is thought to be important for preventing its substrates from interacting with solvent (111), similar to how Skp prevents aggregation of its substrates by sequestration. However, formation of 12-mers/24-mers is not required for protease function because a variant that is trimeric still shows proteolytic activity (112). Outer membrane proteins bound to DegP show a remarkable resistance to proteolysis compared to misfolded model substrates. (DegP-bound outer membrane proteins remain stable longer than the 30 minutes used in the assay.) Crystallographic and electron microscopy data revealed that outer membrane proteins encapsulated by DegP adopt a structured conformation within the DegP ring, perhaps explaining why they are not degraded by DegP. Although cage formation is not essential for proteolytic activity (112), mutations that interfere with the formation of cage-like structures result in rogue proteases, suggesting that cage formation provides a layer of protection against indiscriminate proteolysis (113). DegP has also been reported to form bowl-like structures on lipid membranes (114).
As a chaperone, DegP enhances the in vitro refolding yield of the E. coli periplasmic protein MalS at 28°C; refolding yields are similarly enhanced for the nonnative substrates citrate synthase and β-galactosidase. DegP has also been shown to suppress the aggregation of the nonnative substrate lysozyme. In addition, the temperature sensitivity of DegP deletions can be rescued by the overproduction of a protease-deficient DegP variant (115, 116), consistent with it having chaperone activity. Moreover, a protease-deficient DegP variant can rescue the lethality caused by the expression of the autotransporter virulence protein pertactin (117). Strikingly, heterologous overproduction of the sHSP chaperone CeHSP17 from Caenorhabditis elegans is able to overcome the temperature sensitivity of a DegP deletion, allowing growth of wild-type E. coli to temperatures as high as 50°C, which is about 3.5°C above the normal maximal growth temperature of E. coli (118). It is not immediately obvious if this eukaryotic protein compensates for the chaperone or for the protease activity of DegP. However, since CeHSP17 is a chaperone with no known protease activity, it is more likely that it compensates for DegP’s chaperone activity than its protease activity.
Results from the Fleming group suggest that DegP functions mainly as a protease during outer membrane protein biogenesis and that its chaperone activity does not significantly contribute to its role in OMP biogenesis (119). Although further work may eventually resolve the chaperone/protease DegP controversy, it is worth noting that the distinction between chaperones and proteases is ill defined and substrate dependent. Both chaperones and proteases need to interact transiently with substrate proteins to achieve their function. While cleavage of the substrate is a clear hallmark of proteases, if a protein lacks the cleavage recognition sequence for the protease, this resulting transient interaction may act to stabilize the substrate or affect its folding. The AAA+ family of proteins, for instance, plays roles in both folding and degradation, which depend on the presence of additional proteins and the substrate used (120). The distinction between chaperone and protease activities for DegP could depend on how well DegP binds a particular substrate and how quickly (or slowly) it proteolyzes that substrate under various environmental conditions. Furthermore, the dramatic changes in oligomerization state of DegP that occur could be used to regulate whether DegP acts as a protease or a chaperone, depending on the needs of the cell.
DegQ
There are three related members of the HtrA protein family in E. coli: DegP, DegQ, and DegS. All three contain both chymotrypsin-like protease domains and at least one PDZ domain and all form oligomers (121). Like DegP, DegQ is proposed to have both protease and chaperone activity. Evidence suggests that both DegP and DegQ switch between a hexameric resting state and higher-order oligomers in the presence of substrate (114, 122–124). A crystal structure of a DegQ-lysozyme complex has been obtained that consists of 12 DegQ molecules arranged in a cage surrounding 5 to 6 lysozyme molecules (125). Protease-deficient mutants of DegQ exhibit weak chaperone activity in refolding MalS (125). The structural similarities and differences between the DegQ structure and the GroEL/S double-donut structure have been described (122).
COMPARISON BETWEEN THE MECHANISMS OF PERIPLASMIC AND CYTOSOLIC CHAPERONES
It is interesting to more generally consider the similarities and differences between ATP-dependent cytosolic chaperones and ATP-independent periplasmic chaperones. Chaperones play important roles in the folding of proteins under both normal and stress conditions in both the periplasmic and cytosolic compartments. Chaperones in both compartments need to deal with similar issues, like the tendency of exposed hydrophobic residues to aggregate and the need to keep proteins in an at least partly unfolded secretion competent form. It is thus not surprising that some of the overall principles of chaperone action are shared between cytosolic and periplasmic chaperones. In the cytosol, hydrophobic residues generally end up buried within the hydrophobic core of the protein itself. In the periplasm, hydrophobic surfaces necessary for outer membrane proteins to be integrated into the outer membrane make them particularly aggregation prone while in an aqueous environment like the periplasmic space. To sequester hydrophobic residues, both cytosolic and periplasmic chaperones engage in encapsulation strategies, for example, within a closed chamber for the cytosolic chaperone GroEL or buried within the tentacles of the jellyfish-like periplasmic chaperone Skp.
One big difference is the lack of ATP in the periplasm. Many, but not all, cytosolic chaperones utilize ATP binding or hydrolysis to drive substrate binding and release. Those cytosolic “holdase” chaperones that do not use ATP generally work in concert with ATP-dependent chaperones. Without the driving force of ATP, periplasmic chaperones need to have much more finely balanced affinity constants for binding and release than cytosolic chaperones do. Otherwise, the chaperone either runs the risk of binding too tightly to its substrate, and never releasing it, or if it has a substantially higher affinity for the unfolded than the folded state, it will unfold the protein substrate by dragging the equilibrium in this direction. It has been hypothesized that the affinity of outer membrane proteins for the lipid environment provides the thermodynamic gradient that drives the transport of outer membrane proteins and substitutes for the need for ATP (12, 126).
It is not absolutely necessary for each step in a chaperone cascade to be down a thermodynamic gradient, as long as the overall gradient is downhill. The use of ATP binding and hydrolysis in cytosolic chaperones allows for major shifts in binding affinity, allowing binding and release of a wide variety of substrate proteins. Periplasmic chaperones lack this resource and thus, in general, would need to more delicately balance substrate binding and release. This would appear to pose a problem for periplasmic chaperones that have broad substrate specificity, because one would expect that different substrates would bind with different affinities. This consideration may contribute to the higher prevalence of substrate-specific chaperones in the periplasm than in the cytosol. As previously mentioned, periplasmic chaperones can be present in astonishingly high concentrations. For example, Spy’s concentration following tannin or butanol stress approaches or exceeds 2 mM, and constitutes approximately 40 to 50% of the total protein content of the periplasm. This may be a reflection of simple mass action considerations. Spy’s mode of action involves very rapidly binding folding intermediates and allowing them to fold while bound to the chaperone. This rapid binding is dependent on the Kd of the interactions and the concentrations of the interacting partners. Since there is no possibility of ATP driving release, the affinity of the interaction cannot be too high, because this would prevent release of the substrate. Given this limitation, evolution has apparently chosen to ramp up the concentration of Spy after stress to very high levels likely to ensure rapid binding of folding intermediates so that Spy can effectively compete against aggregation. It may also reflect a high demand for chaperone activity following butanol or tannin stress that may induce widespread protein unfolding.
The stresses leading to induction of both periplasmic and cytosolic stress-induced chaperones overlap to some extent. Heat shock is the classical inducer of cytosolic chaperones, and extreme heat shock is an inducer of the sigmaE response, which in turn controls the chaperones, proteases, and outer membrane assembly factors BamA, BamC, BamD DegP, DegQ, DegS, DsbC, FkpA, GroL, and SurA (14). Ethanol and overproduction of unfolded proteins induce stress responses in both compartments (24, 127). Certain stresses, such as exposure to tannins or low pH, appear to specifically induce periplasmic stress responses (16, 31).
OTHER CHAPERONES
A number of other periplasmic proteins have been identified as having chaperone activity in vivo through a fold-or-die selection: OsmY, OppA, DppA, and Ivy (128). All four of these proteins can stabilize an unstable version of maltose binding protein in a fusion biosensor similar to the one that was used to discover Spy. These proteins were also shown to have in vitro chaperone activity in inhibiting the aggregation of a number of unrelated client proteins. OsmY is a highly soluble protein of unknown function that is upregulated by several different protein folding stress conditions, including osmotic stress. It is also very effective as a solubility-enhancing fusion tag, consistent with it having chaperone function. OppA and DppA are oligopeptide-binding proteins that facilitate the import of peptides into the cytosol. OppA has a preference for binding positively charged peptides (129), and DppA was reported to have chaperone activity in vitro (130). Their ability to bind peptides may allow them to bind unfolded proteins and therefore operate as chaperones. Ivy is a potent inhibitor of vertebrate lysozyme. However, deletion of ivy does not make E. coli more susceptible to hen egg white lysozyme, suggesting that Ivy may have other functions, consistent with its reported chaperone activity. Ivy, the sHSP chaperone IbpA, and the cytosolic chaperone GroEL are all induced in cultures that produce recombinant human growth hormone (131). It remains to be determined how the reported chaperone activity of OsmY, OppA, DppA, and Ivy (128) are related to their as yet poorly defined biological functions.
CHAPERONES INVOLVED IN THE FOLDING OF EXTRACELLULAR PROTEINS
A number of large filamentous structures exist on the surface of E. coli. These include pili, fimbriae, and curli. The polymerization of the major subunits in these structures is a facilitated process that, for pili and fimbriae, involves β-strand transfer. This is a structurally well-defined and elegant mechanism that neatly allows one protein to facilitate the folding of another without becoming part of its final structure. For type I fimbriae, the structural subunits of the fimbriae are called FimH, G, F, and A. All subunits contain an incomplete immunoglobulin type fold that are missing a single β-strand and thus expose a hydrophobic groove. Instead of promoting aggregation, this groove is filled by a β-strand donated by the dedicated chaperone/holdase FimC during transport through the periplasm. This process is called donor strand complementation. After transport through an outer membrane pore formed by the FimD usher, the β-strand of FimC is replaced by the β-strand of a previously transported structural subunit. This allows the structural subunits to polymerize in a linear series. Binding of the structural subunit-FimC heterodimer to the FimD usher opens the hydrophobic groove of the structural subunit and brings it into close proximity to another structural subunit, enabling polymerization through β-strand exchange between the two structural subunits. Other fimbriae and pili assemble by very similar mechanisms.
Curli are long extracellular polymers that are involved in biofilm formation, adhesion, and host invasion (132). They are composed of CsgA, the major structural subunit, and CsgB, a minor structural subunit, that together in a ∼20:1 ratio (133) form amyloid polymers that are functional (in contrast to most other amyloids, which are pathological). The curli fibrils form a tangled mess of thin threads that are the major constituents of the matrix in curli-associated biofilms (134). The chaperones involved in curli formation are CsgC, CsgE, and CsgF. CsgC performs a role somewhat analogous to that of the SecB cytosolic chaperone: namely, to keep the subunits in a secretion competent, partially unfolded configuration (132, 135, 136). In vitro, CsgC is extremely efficient at preventing CsgA polymerization, working in ratios as low as 1:500 (136). The chaperone Spy can also inhibit CsgA polymerization, but Spy is much less effective than CsgC (137). CsgE can also inhibit CsgA polymerization. It acts as a secretion adaptor to target CsgA to the outer membrane pore, which is composed of CsgG subunits. A nonamer of CsgE serves as a capping adaptor that transiently blocks the CsgG secretion channel (138). CsgF also forms a complex with the CsgG secretion channel. In contrast to the transient association of CsgE with the CsgG channel, CsgF is a constitutive component of the channel. In the absence of CsgF, CsgB can no longer function as a curli initiator and CsgA fails to polymerize. Thus, CsgF probably acts as a chaperone that couples CsgA secretion to polymerization (139). Following transit through this pore, CsgA and CsgB polymerize into cross-β-strand amyloid structures (see Fig. 1). Like fimbriae, these curli fibers are β-sheet-rich cross-β-strands. However, unlike fimbriae, polymerization of curli fibrils is not known to involve strand exchange between subunits. Fimbriae and curli also differ in the cellular location where they polymerize: fimbriae polymerize within the periplasm, whereas curli polymerize in the extracellular space (140). This is possibly to avoid the intracellular toxicity of amyloid assembly intermediates that are generated in curli formation (132).
Figure 1.
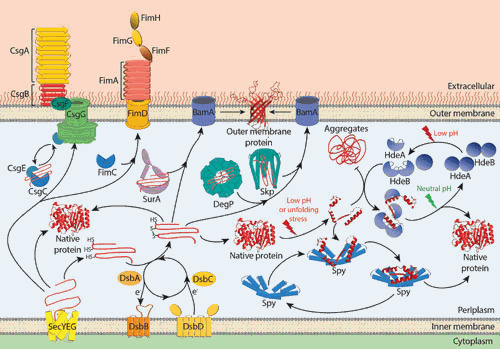
An overview of the chaperones and folding factors affecting protein folding in the periplasm of E. coli.
ROLE OF PERIPLASMIC CHAPERONES ON THE EXPRESSION OF RECOMBINANT PROTEINS
There have been a number of attempts at enhancing the expression of recombinant proteins in the periplasm by overexpression of periplasmic chaperones and folding catalysts. For example, a plasmid that simultaneously overexpresses DsbA, DsbC, FkpA, and SurA has been constructed and successfully used for this purpose (141, 142). However, this approach may not be a panacea for all expression issues. As discussed above, E. coli naturally responds to the presence of misfolded proteins in the cellular envelope by overexpressing periplasmic chaperones using the SigmaE, Cpx, and Bae regulatory systems. Given the likelihood that chaperones are naturally induced in response to recombinant protein overexpression and that our current grasp of chaperone-substrate specificity is poor, chaperone coinduction may not universally improve the folding of all overexpressed recombinant proteins. However, the folding environment of the periplasm can be optimized to improve the folding of specific proteins using folding biosensors. These biosensors link the in vivo stability of specific proteins to a genetically selectable marker, such as antibiotic resistance. Coupling chromosome mutations with a demand for high antibiotic resistance in strains containing these folding biosensors results in superfolder bacteria strains that are customized for promoting the expression and folding of specific unstable proteins. Applications of these folding biosensors have uncovered previously unrecognized periplasmic chaperones: Spy, OsmY, Ivy, DppA, and OppA. Given the wide range of potential protein folding problems faced by various proteins, it is not surprising that E. coli selects unique chaperone overproduction solutions for individual protein folding problems.
REFERENCES
- 1.Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. 2004. X-ray structure of a protein-conducting channel. Nature 427:36–44. [PubMed] [DOI] [PubMed] [Google Scholar]
- 2.Danese PN, Silhavy TJ. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet 32:59–94. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Mogensen JE, Otzen DE. 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol 57:326–346. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Szewczyk J, Collet JF. 2016. The journey of lipoproteins through the cell: one birthplace, multiple destinations. Adv Microb Physiol 69:1–50. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Fu X, Shen J, Zhang H, Hong W, Chang Z. 2004. Periplasmic proteins of Escherichia coli are highly resistant to aggregation: reappraisal for roles of molecular chaperones in periplasm. Biochem Biophys Res Commun 316:795–801. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Park C, Zhou S, Gilmore J, Marqusee S. 2007. Energetics-based protein profiling on a proteomic scale: identification of proteins resistant to proteolysis. J Mol Biol 368:1426–1437. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wülfing C, Plückthun A. 1994. Protein folding in the periplasm of Escherichia coli. Mol Microbiol 12:685–692. [PubMed] [DOI] [PubMed] [Google Scholar]
- 8.Vertommen D, Depuydt M, Pan J, Leverrier P, Szikora J, Messens J, Bardwell JC, Collet JF. 2008. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol 67:336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardwell JC, McGovern K, Beckwith J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589. [PubMed] [DOI] [PubMed] [Google Scholar]
- 10.Shao F, Bader MW, Jakob U, Bardwell JC. 2000. DsbG, a protein disulfide isomerase with chaperone activity. J Biol Chem 275:13349–13352. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Song JL, Zhang S, Wang Y, Cui DF, Wang CC. 1999. Chaperone activity of DsbC. J Biol Chem 274:19601–19605. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Moon CP, Zaccai NR, Fleming PJ, Gessmann D, Fleming KG. 2013. Membrane protein thermodynamic stability may serve as the energy sink for sorting in the periplasm. Proc Natl Acad Sci USA 110:4285–4290. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JCA. 2011. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol 18:262–269. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet 5:e1000651. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leblanc SKD, Oates CW, Raivio TL. 2011. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol 193:3367–3375. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoetendal EG, Smith AH, Sundset MA, Mackie RI. 2008. The BaeSR two-component regulatory system mediates resistance to condensed tannins in Escherichia coli. Appl Environ Microbiol 74:535–539. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong W, Wu YE, Fu X, Chang Z. 2012. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol 20:328–335. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Freire E, Schön A, Hutchins BM, Brown RK. 2013. Chemical denaturation as a tool in the formulation optimization of biologics. Drug Discov Today 18:1007–1013. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raivio TL. 2005. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol 56:1119–1128. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Raffa RG, Raivio TL. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol 45:1599–1611. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Raivio TL. 2014. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta 1843:1529–1541. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. 2010. The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus. Mol Microbiol 76:1095–1110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weatherspoon-Griffin N, Yang D, Kong W, Hua Z, Shi Y. 2014. The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J Biol Chem 289:32571–32582. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macritchie DM, Raivio TL. 2009. Envelope Stress Responses. Ecosal Plus 3:3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Fleischer R, Heermann R, Jung K, Hunke S. 2007. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J Biol Chem 282:8583–8593. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Thede GL, Arthur DC, Edwards RA, Buelow DR, Wong JL, Raivio TL, Glover JNM. 2011. Structure of the periplasmic stress response protein CpxP. J Bacteriol 193:2149–2157. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Appia-Ayme C, Hall A, Patrick E, Rajadurai S, Clarke TA, Rowley G. 2012. ZraP is a periplasmic molecular chaperone and a repressor of the zinc-responsive two-component regulator ZraSR. Biochem J 442:85–93. [PubMed] [DOI] [PubMed] [Google Scholar]
- 28.Surmann K, Ćudić E, Hammer E, Hunke S. 2016. Molecular and proteome analyses highlight the importance of the Cpx envelope stress system for acid stress and cell wall stability in Escherichia coli. MicrobiologyOpen 5:582–596. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, Houry WA. 2010. Acid stress response in enteropathogenic gammaproteobacteria: an aptitude for survival. Biochem Cell Biol 88:301–314. [PubMed] [DOI] [PubMed] [Google Scholar]
- 30.Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol 189:2759–2768. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Link AJ, Robison K, Church GM. 1997. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18:1259–1313. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Malki A, Le HT, Milles S, Kern R, Caldas T, Abdallah J, Richarme G. 2008. Solubilization of protein aggregates by the acid stress chaperones HdeA and HdeB. J Biol Chem 283:13679–13687. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Ding J, Yang C, Niu X, Hu Y, Jin C. 2015. HdeB chaperone activity is coupled to its intrinsic dynamic properties. Sci Rep 5:16856. 10.1038/srep16856. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahl J-U, Koldewey P, Salmon L, Horowitz S, Bardwell JCA, Jakob U. 2015. HdeB functions as an acid-protective chaperone in bacteria. J Biol Chem 290:65–75. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, He D, Yang Y, Lin S, Zhang M, Dai S, Chen PR. 2016. Comparative proteomics reveal distinct chaperone-client interactions in supporting bacterial acid resistance. Proc Natl Acad Sci USA 113:10872–10877. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang F, Gustafson KR, Boyd MR, Wlodawer A. 1998. Crystal structure of Escherichia coli HdeA. Nat Struct Biol 5:763–764. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Salmon L, Stull F, Sayle S, Cato C, Akgül S, Foit L, Ahlstrom LS, Eisenmesser EZ, Al-Hashimi HM, Bardwell JCA, Horowitz S. 2018. The mechanism of HdeA unfolding and chaperone activation. J Mol Biol 430:33–40. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong W, Jiao W, Hu J, Zhang J, Liu C, Fu X, Shen D, Xia B, Chang Z. 2005. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J Biol Chem 280:27029–27034. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Foit L, George JS, Zhang BW, Brooks CL III, Bardwell JCA. 2013. Chaperone activation by unfolding. Proc Natl Acad Sci USA 110:E1254–E1262. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahlstrom LS, Law SM, Dickson A, Brooks CL III. 2015. Multiscale modeling of a conditionally disordered pH-sensing chaperone. J Mol Biol 427:1670–1680. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang BW, Brunetti L, Brooks CL III. 2011. Probing pH-dependent dissociation of HdeA dimers. J Am Chem Soc 133:19393–19398. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahlstrom LS, Dickson A, Brooks CL III. 2013. Binding and folding of the small bacterial chaperone HdeA. J Phys Chem B 117:13219–13225. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garrison MA, Crowhurst KA. 2014. NMR-monitored titration of acid-stress bacterial chaperone HdeA reveals that Asp and Glu charge neutralization produces a loosened dimer structure in preparation for protein unfolding and chaperone activation. Protein Sci 23:167–178. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, Rasmussen T, Harding AJ, Booth NA, Booth IR, Naismith JH. 2012. Salt bridges regulate both dimer formation and monomeric flexibility in HdeB and may have a role in periplasmic chaperone function. J Mol Biol 415:538–546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagenmaier S, Stierhof YD, Henning U. 1997. A new periplasmic protein of Escherichia coli which is synthesized in spheroplasts but not in intact cells. J Bacteriol 179:2073–2076. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon E, Kim DY, Gross CA, Gross JD, Kim KK. 2010. The crystal structure Escherichia coli Spy. Protein Sci 19:2252–2259. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srivastava SK, Lambadi PR, Ghosh T, Pathania R, Navani NK. 2014. Genetic regulation of spy gene expression in Escherichia coli in the presence of protein unfolding agent ethanol. Gene 548:142–148. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Wang D, Fierke CA. 2013. The BaeSR regulon is involved in defense against zinc toxicity in E. coli. Metallomics 5:372–383. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horowitz S, Salmon L, Koldewey P, Ahlstrom LS, Martin R, Quan S, Afonine PV, van den Bedem H, Wang L, Xu Q, Trievel RC, Brooks CL III, Bardwell JCA. 2016. Visualizing chaperone-assisted protein folding. Nat Struct Mol Biol 23:691–697. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koldewey P, Stull F, Horowitz S, Martin R, Bardwell JCA. 2016. Forces driving chaperone action. Cell 166:369–379. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stull F, Koldewey P, Humes JR, Radford SE, Bardwell JCA. 2016. Substrate protein folds while it is bound to the ATP-independent chaperone Spy. Nat Struct Mol Biol 23:53–58. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schiffrin B, Calabrese AN, Higgins AJ, Humes JR, Ashcroft AE, Kalli AC, Brockwell DJ, Radford SE. 2017. Effects of periplasmic chaperones and membrane thickness on BamA-catalyzed outer-membrane protein folding. J Mol Biol 429:3776–3792. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thome BM, Müller M. 1991. Skp is a periplasmic Escherichia coli protein requiring SecA and SecY for export. Mol Microbiol 5:2815–2821. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Chen R, Henning U. 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol Microbiol 19:1287–1294. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Missiakas D, Betton JM, Raina S. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol 21:871–884. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Denoncin K, Schwalm J, Vertommen D, Silhavy TJ, Collet JF. 2012. Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. Proteomics 12:1391–1401. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weski J, Ehrmann M. 2012. Genetic analysis of 15 protein folding factors and proteases of the Escherichia coli cell envelope. J Bacteriol 194:3225–3233. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schäfer U, Beck K, Müller M. 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem 274:24567–24574. [PubMed] [DOI] [PubMed] [Google Scholar]
- 59.Schwalm J, Mahoney TF, Soltes GR, Silhavy TJ. 2013. Role for Skp in LptD assembly in Escherichia coli. J Bacteriol 195:3734–3742. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jarchow S, Lück C, Görg A, Skerra A. 2008. Identification of potential substrate proteins for the periplasmic Escherichia coli chaperone Skp. Proteomics 8:4987–4994. [PubMed] [DOI] [PubMed] [Google Scholar]
- 61.Bothmann H, Plückthun A. 1998. Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat Biotechnol 16:376–380. [PubMed] [DOI] [PubMed] [Google Scholar]
- 62.Narayanan N, Chou CP. 2008. Physiological improvement to enhance Escherichia coli cell-surface display via reducing extracytoplasmic stress. Biotechnol Prog 24:293–301. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Patel GJ, Behrens-Kneip S, Holst O, Kleinschmidt JH. 2009. The periplasmic chaperone Skp facilitates targeting, insertion, and folding of OmpA into lipid membranes with a negative membrane surface potential. Biochemistry 48:10235–10245. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. 2003. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem 278:9092–9099. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.De Cock H, Schäfer U, Potgeter M, Demel R, Müller M, Tommassen J. 1999. Affinity of the periplasmic chaperone Skp of Escherichia coli for phospholipids, lipopolysaccharides and non-native outer membrane proteins. Role of Skp in the biogenesis of outer membrane protein. Eur J Biochem 259:96–103. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. 2007. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J Mol Biol 374:91–105. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Harms N, Koningstein G, Dontje W, Muller M, Oudega B, Luirink J, de Cock H. 2001. The early interaction of the outer membrane protein phoe with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J Biol Chem 276:18804–18811. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Entzminger KC, Chang C, Myhre RO, McCallum KC, Maynard JA. 2012. The Skp chaperone helps fold soluble proteins in vitro by inhibiting aggregation. Biochemistry 51:4822–4834. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walton TA, Sousa MC. 2004. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell 15:367–374. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Korndörfer IP, Dommel MK, Skerra A. 2004. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat Struct Mol Biol 11:1015–1020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.McMorran LM, Bartlett AI, Huysmans GHM, Radford SE, Brockwell DJ. 2013. Dissecting the effects of periplasmic chaperones on the in vitro folding of the outer membrane protein PagP. J Mol Biol 425:3178–3191. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lyu ZX, Shao Q, Gao YQ, Zhao XS. 2012. Direct observation of the uptake of outer membrane proteins by the periplasmic chaperone Skp. PLoS One 7:e46068. 10.1371/journal.pone.0046068. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schiffrin B, Calabrese AN, Devine PWA, Harris SA, Ashcroft AE, Brockwell DJ, Radford SE. 2016. Skp is a multivalent chaperone of outer-membrane proteins. Nat Struct Mol Biol 23:786–793. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaccai NR, Sandlin CW, Hoopes JT, Curtis JE, Fleming PJ, Fleming KG, Krueger S. 2016. Deuterium labeling together with contrast variation small-angle neutron scattering suggests how Skp captures and releases unfolded outer membrane proteins. Methods Enzymol 566:159–210. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burmann BM, Wang C, Hiller S. 2013. Conformation and dynamics of the periplasmic membrane-protein-chaperone complexes OmpX-Skp and tOmpA-Skp. Nat Struct Mol Biol 20:1265–1272. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Thoma J, Burmann BM, Hiller S, Müller DJ. 2015. Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer-membrane proteins. Nat Struct Mol Biol 22:795–802. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Tormo A, Almirón M, Kolter R. 1990. surA, an Escherichia coli gene essential for survival in stationary phase. J Bacteriol 172:4339–4347. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lazar SW, Kolter R. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol 178:1770–1773. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rouvière PE, Gross CA. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev 10:3170–3182. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Behrens-Kneip S. 2010. The role of SurA factor in outer membrane protein transport and virulence. Int J Med Microbiol 300:421–428. [PubMed] [DOI] [PubMed] [Google Scholar]
- 81.Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. 2006. Maturation of intracellular Escherichia coli communities requires SurA. Infect Immun 74:4793–4800. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weirich J, Bräutigam C, Mühlenkamp M, Franz-Wachtel M, Macek B, Meuskens I, Skurnik M, Leskinen K, Bohn E, Autenrieth I, Schütz M. 2017. Identifying components required for OMP biogenesis as novel targets for antiinfective drugs. Virulence 8:1170–1188. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9:2432–2443. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzitello AE, Harper JR, Silhavy TJ. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol 183:6794–6800. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 21:2473–2484. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goemans C, Denoncin K, Collet J-F. 2014. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta 1843:1517–1528. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Watts KM, Hunstad DA. 2008. Components of SurA required for outer membrane biogenesis in uropathogenic Escherichia coli. PLoS One 3:e3359. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Soltes GR, Schwalm J, Ricci DP, Silhavy TJ. 2016. The activity of Escherichia coli chaperone SurA is regulated by conformational changes involving a parvulin domain. J Bacteriol 198:921–929. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ricci DP, Schwalm J, Gonzales-Cope M, Silhavy TJ. 2013. The activity and specificity of the outer membrane protein chaperone SurA are modulated by a proline isomerase domain. MBio 4:1–9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bitto E, McKay DB. 2002. Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Structure 10:1489–1498. [DOI] [PubMed] [Google Scholar]
- 91.Bitto E, McKay DB. 2003. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem 278:49316–49322. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. 2005. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J Biol Chem 280:23540–23548. [PubMed] [DOI] [PubMed] [Google Scholar]
- 93.Xu X, Wang S, Hu YX, McKay DB. 2007. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J Mol Biol 373:367–381. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhong M, Ferrell B, Lu W, Chai Q, Wei Y. 2013. Insights into the function and structural flexibility of the periplasmic molecular chaperone SurA. J Bacteriol 195:1061–1067. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu S, Ge X, Lv Z, Zhi Z, Chang Z, Zhao XS. 2011. Interaction between bacterial outer membrane proteins and periplasmic quality control factors: a kinetic partitioning mechanism. Biochem J 438:505–511. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Justice SS, Hunstad DA, Harper JR, Duguay AR, Pinkner JS, Bann J, Frieden C, Silhavy TJ, Hultgren SJ. 2005. Periplasmic peptidyl prolyl cis-trans isomerases are not essential for viability, but SurA is required for pilus biogenesis in Escherichia coli. J Bacteriol 187:7680–7686. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11:1169–1182. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Kleerebezem M, Heutink M, Tommassen J. 1995. Characterization of an Escherichia coli rotA mutant, affected in periplasmic peptidyl-prolyl cis/trans isomerase. Mol Microbiol 18:313–320. [PubMed] [DOI] [PubMed] [Google Scholar]
- 99.Behrens S, Maier R, de Cock H, Schmid FX, Gross CA. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J 20:285–294. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Geitner AJ, Varga E, Wehmer M, Schmid FX. 2013. Generation of a highly active folding enzyme by combining a parvulin-type prolyl isomerase from SurA with an unrelated chaperone domain. J Mol Biol 425:4089–4098. [PubMed] [DOI] [PubMed] [Google Scholar]
- 101.Ramm K, Plückthun A. 2000. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. II. Isomerase-independent chaperone activity in vitro. J Biol Chem 275:17106–17113. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Bothmann H, Plückthun A. 2000. The periplasmic Escherichia coli peptidylprolyl cis,trans-isomerase FkpA. I. Increased functional expression of antibody fragments with and without cis-prolines. J Biol Chem 275:17100–17105. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Dwyer RS, Malinverni JC, Boyd D, Beckwith J, Silhavy TJ. 2014. Folding LacZ in the periplasm of Escherichia coli. J Bacteriol 196:3343–3350. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Helbig S, Patzer SI, Schiene-Fischer C, Zeth K, Braun V. 2011. Activation of colicin M by the FkpA prolyl cis-trans isomerase/chaperone. J Biol Chem 286:6280–6290. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ge X, Lyu ZX, Liu Y, Wang R, Zhao XS, Fu X, Chang Z. 2014. Identification of FkpA as a key quality control factor for the biogenesis of outer membrane proteins under heat shock conditions. J Bacteriol 196:672–680. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weininger U, Jakob RP, Kovermann M, Balbach J, Schmid FX. 2010. The prolyl isomerase domain of PpiD from Escherichia coli shows a parvulin fold but is devoid of catalytic activity. Protein Sci 19:6–18. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sachelaru I, Petriman NA, Kudva R, Koch HG. 2014. Dynamic interaction of the sec translocon with the chaperone PpiD. J Biol Chem 289:21706–21715. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Antonoaea R, Fürst M, Nishiyama K, Müller M. 2008. The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47:5649–5656. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Matern Y, Barion B, Behrens-Kneip S. 2010. PpiD is a player in the network of periplasmic chaperones in Escherichia coli. BMC Microbiol 10:251. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Spiess C, Beil A, Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339–347. [DOI] [PubMed] [Google Scholar]
- 111.Krojer T, Sawa J, Schäfer E, Saibil HR, Ehrmann M, Clausen T. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453:885–890. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Kim S, Sauer RT. 2012. Cage assembly of DegP protease is not required for substrate-dependent regulation of proteolytic activity or high-temperature cell survival. Proc Natl Acad Sci USA 109:7263–7268. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim S, Sauer RT. 2014. Distinct regulatory mechanisms balance DegP proteolysis to maintain cellular fitness during heat stress. Genes Dev 28:902–911. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shen Q-T, Bai X-C, Chang L-F, Wu Y, Wang H-W, Sui S-F. 2009. Bowl-shaped oligomeric structures on membranes as DegP’s new functional forms in protein quality control. Proc Natl Acad Sci USA 106:4858–4863. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Misra R, CastilloKeller M, Deng M. 2000. Overexpression of protease-deficient DegP(S210A) rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J Bacteriol 182:4882–4888. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.CastilloKeller M, Misra R. 2003. Protease-deficient DegP suppresses lethal effects of a mutant OmpC protein by its capture. J Bacteriol 185:148–154. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Braselmann E, Chaney JL, Champion MM, Clark PL. 2016. DegP chaperone suppresses toxic inner membrane translocation intermediates. PLoS One 11:e0162922. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ezemaduka AN, Yu J, Shi X, Zhang K, Yin CC, Fu X, Chang Z. 2014. A small heat shock protein enables Escherichia coli to grow at a lethal temperature of 50°C conceivably by maintaining cell envelope integrity. J Bacteriol 196:2004–2011. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Costello SM, Plummer AM, Fleming PJ, Fleming KG. 2016. Dynamic periplasmic chaperone reservoir facilitates biogenesis of outer membrane proteins. Proc Natl Acad Sci USA 113:E4794–E4800. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elsholz AKW, Birk MS, Charpentier E, Turgay K. 2017. Functional diversity of AAA+ protease complexes in Bacillus subtilis. Front Mol Biosci 4:44. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim DY, Kim KK. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol 38:266–274. [DOI] [PubMed] [Google Scholar]
- 122.Jiang J, Zhang X, Chen Y, Wu Y, Zhou ZH, Chang Z, Sui S-F. 2008. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc Natl Acad Sci USA 105:11939–11944. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim S, Grant RA, Sauer RT. 2011. Covalent linkage of distinct substrate degrons controls assembly and disassembly of DegP proteolytic cages. Cell 145:67–78. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455–459. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Malet H, Canellas F, Sawa J, Yan J, Thalassinos K, Ehrmann M, Clausen T, Saibil HR. 2012. Newly folded substrates inside the molecular cage of the HtrA chaperone DegQ. Nat Struct Mol Biol 19:152–157. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Plummer AM, Fleming KG. 2016. From chaperones to the membrane with a BAM! Trends Biochem Sci 41:872–882. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jeong SM, Lee HJ, Park YM, Kim JS, Lee SD, Bang IS. 2017. Inducible spy transcription acts as a sensor for envelope stress of Salmonella typhimurium. Korean J Food Sci Anim Resour 37:134–138. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lennon CW, Thamsen M, Friman ET, Cacciaglia A, Sachsenhauser V, Sorgenfrei FA, Wasik MA, Bardwell JCA. 2015. Folding optimization in vivo uncovers new chaperones. J Mol Biol 427:2983–2994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Klepsch MM, Kovermann M, Löw C, Balbach J, Permentier HP, Fusetti F, de Gier JW, Slotboom DJ, Berntsson RPA. 2011. Escherichia coli peptide binding protein OppA has a preference for positively charged peptides. J Mol Biol 414:75–85. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Matsuzaki M, Kiso Y, Yamamoto I, Satoh T. 1998. Isolation of a periplasmic molecular chaperone-like protein of Rhodobacter sphaeroides f. sp. denitrificans that is homologous to the dipeptide transport protein DppA of Escherichia coli. J Bacteriol 180:2718–2722. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Champion KM, Nishihara JC, Aldor IS, Moreno GT, Andersen D, Stults KL, Vanderlaan M. 2003. Comparison of the Escherichia coli proteomes for recombinant human growth hormone producing and nonproducing fermentations. Proteomics 3:1365–1373. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Van Gerven N, Klein RD, Hultgren SJ, Remaut H. 2015. Bacterial amyloid formation: structural insights into curli biogensis. Trends Microbiol 23:693–706. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bian Z, Normark S. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J 16:5827–5836. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. 2013. Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 4:e00645-13. 10.1128/mBio.00645-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Taylor JD, Hawthorne WJ, Lo J, Dear A, Jain N, Meisl G, Andreasen M, Fletcher C, Koch M, Darvill N, Scull N, Escalera-Maurer A, Sefer L, Wenman R, Lambert S, Jean J, Xu Y, Turner B, Kazarian SG, Chapman MR, Bubeck D, de Simone A, Knowles TPJ, Matthews SJ. 2016. Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones. Sci Rep 6:24656. 10.1038/srep24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Evans ML, Chorell E, Taylor JD, Åden J, Götheson A, Li F, Koch M, Sefer L, Matthews SJ, Wittung- Stafshede P, Almqvist F, Chapman MR. 2015. The bacterial curli system possesses a potent and selective inhibitor of amyloid formation. Mol Cell 57:445–455. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Evans ML, Schmidt JC, Ilbert M, Doyle SM, Quan S, Bardwell JCA, Jakob U, Wickner S, Chapman MR. 2011. E. coli chaperones DnaK, Hsp33 and Spy inhibit bacterial functional amyloid assembly. Prion 5:323–334. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shu Q, Krezel AM, Cusumano ZT, Pinkner JS, Klein R, Hultgren SJ, Frieden C. 2016. Solution NMR structure of CsgE: structural insights into a chaperone and regulator protein important for functional amyloid formation. Proc Natl Acad Sci USA 113:7130–7135. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nenninger AA, Robinson LS, Hultgren SJ. 2009. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci USA 106:900–905. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shriver-Lake LC, Goldman ER, Zabetakis D, Anderson GP. 2017. Improved production of single domain antibodies with two disulfide bonds by co-expression of chaperone proteins in the Escherichia coli periplasm. J Immunol Methods 443:64–67. [PubMed] [DOI] [PubMed] [Google Scholar]
- 142.Sun P, Tropea JE, Waugh DS. 2011. Enhancing the solubility of recombinant proteins in Escherichia coli by using hexahistidine-tagged maltose-binding protein as a fusion partner. Methods Mol Biol 705:259–274. [PubMed] [DOI] [PubMed] [Google Scholar]


