Abstract
Cysteine cathepsins are a family of lysosomal proteases that are often overexpressed in several human malignancies and haves been linked to cellular genomic alterations, disturbances in genomic stability, and the onset and spread of cancer. Recent studies have shown alterations in cysteine cathepsins in malignant ovarian tumors. However, it remains unclear whether there is a causal relationship between ovarian cancer, and its subtypes, and the cathepsin family. This study utilized two-sample Mendelian randomization (MR) analysis to examine this potential causal relationship. Genetic instruments derived from publicly available genetic summary data were used for the analyses. For MR analysis, the inverse-variance weighted method, weighted median method, and MR-Egger regression were employed. Multivariate MR analysis was performed concurrently. Univariate MR analysis indicated a strong correlation between decreased incidence of low-grade serous ovarian cancer and elevated levels of cathepsin L2 (odds ratio = 0.803, 95% confidence interval = 0.685–0.942, P = .007), whereas clear cell ovarian cancer showed a strong correlation with elevated levels of cathepsin H (odds ratio = 1.149, 95% confidence interval = 1.036–1.274, P = .008). Multivariate analysis, adjusted for 9 different cathepsins as covariates, confirmed the genetic relationships between cathepsin L2 and low-grade serous ovarian cancer and between cathepsin H and clear cell ovarian cancer. Our results suggest a causal relationship between cathepsins and ovarian malignancy and its subtypes. Cathepsin L2 has a protective effect on low-grade serous ovarian cancer, whereas cathepsin H is an adverse risk factor for clear cell ovarian cancer.
Keywords: cathepsins, Mendelian randomization, ovarian cancer
1. Introduction
Ovarian malignancies are a major contributor to cancer-related deaths worldwide and cause significant morbidity and mortality among women, ranking eighth in incidence and mortality among all cancers.[1] Pathologically, ovarian cancer is classified into low-grade serous, high-grade serous, clear cell, mucinous, and endometrioid types.[2] Multiple risk factors have been associated with the development and progression of ovarian cancer,[3] and maintaining homeostasis in the intracellular and tumor microenvironments is critical for ovarian cancer cells.[4,5] To preserve cellular integrity, cancer cells must regulate protein metabolism.[6] Therefore, increased levels of proteolytic enzymes are indispensable for tumor growth and proliferation.[7]
The human cysteine cathepsin family primarily consists of 9 members (cathepsins B, E, F, G, H, O, S, L2, and Z),[8] all containing cysteine and histidine residues that form a conserved active site.[9] The primary function of cathepsins is protein degradation within lysosomes, and is shared across the family. In normal cells, cathepsins predominantly act as endopeptidases within lysosomal vesicles and participate in physiological processes such as protein turnover, differentiation, and apoptosis.[10] Cathepsins also play roles in cancer development, progression, proliferation, and metastasis, and are considered relevant to the initiation and progression of diseases, including ovarian malignancies.[11,12]
Abnormal regulation of cathepsins at 1 or several levels can result in certain cathepsins being upregulated, membrane-bound, and secreted by tumor cells.[13] Studies on the expression of cathepsins in several types of cancer, including melanoma, lung, colon, prostate, and breast cancer, have revealed increased expression in tumor tissues.[14] It has been reported that cathepsins are intimately connected with the initiation and progression of cancer, including the attenuation of tumor development. The levels of cathepsins change with increasing grade of malignancy in tumors. Immunohistochemical studies have demonstrated that mature and/or precursor forms of cathepsins may be secreted near tumor boundaries and associated with plasma membrane proteins to facilitate invasion and metastasis.[15] In malignancies, cathepsins and their inhibitors are of potential therapeutic importance.[16] However, it is essential to determine which of the 9 cathepsins is most significant for the initiation of ovarian cancer. Moreover, the role of these proteases in the fundamental processes of ovarian cancer genesis and progression remains uncertain.
Mendelian randomization (MR) reduces the interference of confounding factors and is particularly useful when randomized controlled trials are impractical for investigating causal links and observational studies yield biased results due to confounders or reverse causality.[17] We aimed to identify the relationship of cathepsins with ovarian cancer and its various pathological subtypes to provide valuable evidence for managing ovarian malignancies. While the relationship between cathepsins and partial solid tumor malignancies has been elucidated, controversy remains regarding the relationship between cathepsins and ovarian malignancies.[18] Hence, we employed MR to establish a causal relationship between cathepsins and ovarian cancers. We first assessed this relationship using univariate MR analysis. Considering the potential interactions among cathepsins, we also conducted multivariate MR analysis to enhance the analytical strength.
1.1. Study methods
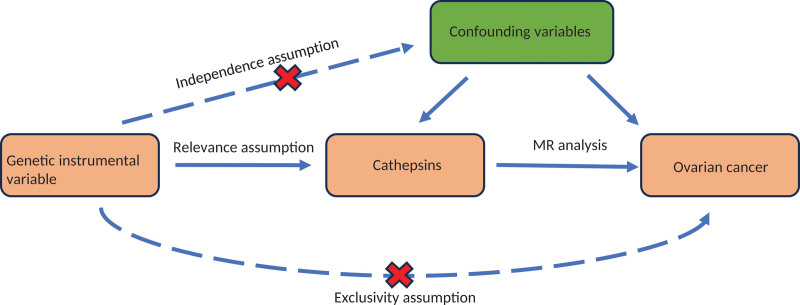
MR utilizes instrumental variables (IVs) to evaluate causality.[19] Genetic variants serve as natural IVs because they are assigned randomly at conception, creating genetic variation in exposure levels. Our research approach is depicted in Figure 1. The study drew on genome-wide association studies (GWAS) restricted to individuals of European ancestry. Ethical approval was obtained from institutional review boards for this study, and all participants provided informed consent.
Figure 1.
Flow chart of the Mendelian randomization analysis.
1.2. Exposures chosen
Significant single-nucleotide polymorphisms (SNPs) associated with cathepsin levels were identified from relevant GWAS databases using a P-value threshold of <5E‐6 to select SNPs for use as IVs. Only independent SNPs with no linkage disequilibrium (R2 < 0.001 within a clumping window >10,000 kb) were retained. The strength of the associations between IVs and exposure factors was assessed using an F-statistic > 10, which indicates a robust correlation. The formula for calculating the F value is as follows: F = [R2 × (N ‐ 1 ‐ K)]/[K × (1 ‐ R2)]. The common formula for R2 is: R2 = 2 × MAF × (1-MAF) × β2, where MAF refers to the secondary allele frequency and β refers to the influence value of the exposure. At the same time, the statistical power was calculated.
1.3. Outcomes chosen
We accessed publicly available GWAS summary statistics for assessing the relationship between SNPs and ovarian cancer, including 25,509 cases and 40,941 controls, as well as with different pathological subtypes of ovarian cancer: low-grade serous (40,941 cases vs 43,907 controls), high-grade serous (13,037 cases vs 40,941 controls), clear cell (1366 cases vs 40,941 controls), mucinous (2566 cases vs 40,941 controls), and endometrioid ovarian cancer (2810 cases vs 40,941 controls) (Table 1).
Table 1.
GWAS summary datasets on ovarian cancer and its subtypes.
| Cancer | ncase | ncontrol | Dataset |
|---|---|---|---|
| Ovarian cancer | 25,509 | 40,941 | ieu-a-1120 |
| Low-grade serous ovarian cancer | 40,941 | 43,907 | ieu-a-1229 |
| High-grade serous ovarian cancer | 13,037 | 40,941 | ieu-a-1121 |
| Clear cell ovarian cancer | 1366 | 40,941 | ieu-a-1124 |
| Mucinous ovarian cancer | 2566 | 40,941 | ieu-a-1231 |
| Endometrioid ovarian cancer | 2810 | 40,941 | ieu-a-1125 |
GWAS = genome-wide association studies.
1.4. Statistics and reproducibility
To compute the causal effect, the primary statistical analysis method employed was the random-effects inverse-variance weighted (IVW) method. Additionally, we utilized techniques like the weighted median (WM) and MR-Egger to produce consistent outcomes.[20] The IVW Wald ratio, based on how each variant affects exposure, calculates the probability of the cancer under investigation. Subsequently, an overall summary estimate was produced by combining the distinct MR estimates. The weighted median and MR-Egger are 2 complementary techniques that serve as robust methods for confirming the presence of pleiotropy in MR data. MR-Egger regression acts both as an estimator and as a method to weight SNP-outcome associations linearly on SNP-exposure associations. The weighted median method can provide a sensitivity analysis when multiple genetic variants are involved.[21] Simple Mode (SM) and weighted mode (WM) are added, which can serve as a supplement to the IVW method. Horizontal pleiotropic effects were detected using the MR-Egger intercept.[22] To evaluate the validity of our hypothesis, we carried out numerous statistical tests and sensitivity analyses. Heterogeneity was assessed using Cochran Q statistic to analyze differences between estimates.[23] We implemented the leave-one-out method to locate SNPs that might disproportionately affect the outcomes and to further assess the amplitude of change in the findings. Multivariate MR expands upon standard univariate MR; multivariate IVW considers multiple exposures simultaneously to examine how the cathepsins interact with one another and their pathogenic effects on different ovarian cancer subtypes.[24] Reverse MR was conducted to investigate reverse causality and to support the existence of bidirectional causal relationships by treating ovarian cancer as the exposure and cathepsin levels as the outcome. All presented P-values are bilateral, with statistical significance set at the 5% level. In order to control the false-positive error rate, a conservative Bonferroni adjustment of the P-value threshold was adopted to determine causality. P < .05/9 = 0.0055 was regarded as having significant causality, and P < .05 was regarded as having potential causality. Statistical analyses were conducted using R software version 4.3.2, along with the packages “TwoSampleMR” and “MendelianRandomization.”
2. Results
2.1. Univariate analysis
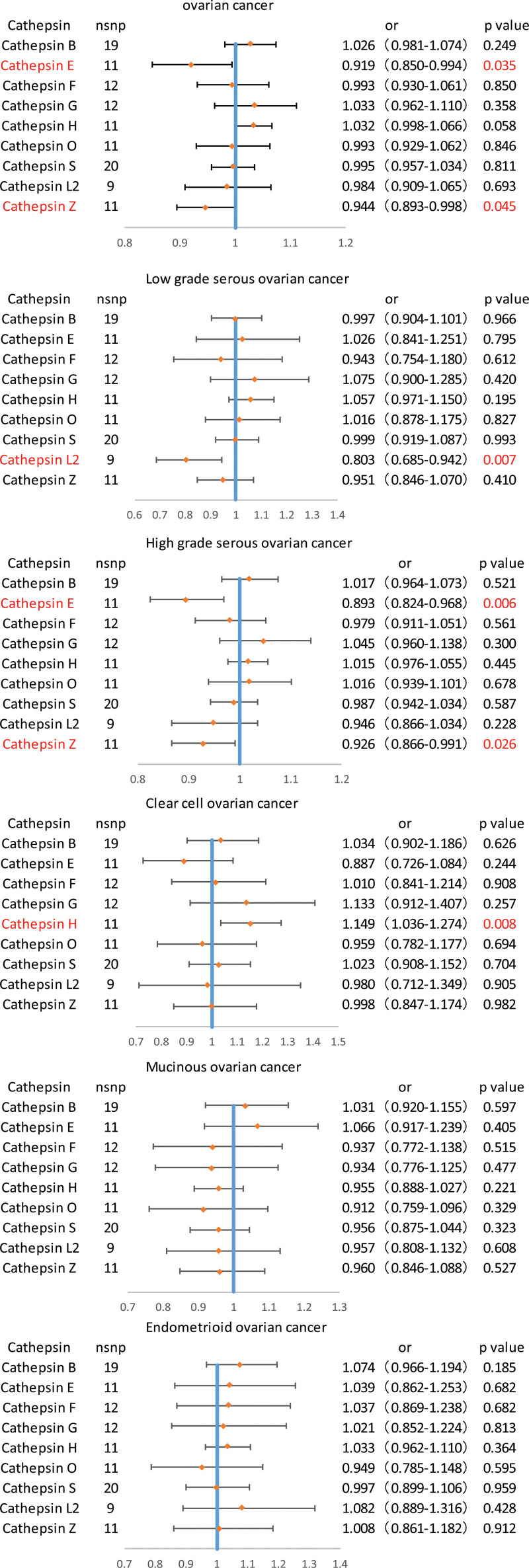
Causal relationships were assessed between various cathepsins and ovarian cancer, including its different pathological subtypes (high-grade serous ovarian cancer, low-grade serous ovarian cancer, clear cell ovarian cancer, mucinous ovarian cancer, and endometrioid ovarian cancer). There were 130 strong, independent, and replicated SNP exposures extracted from the GWAS statistical summary data (Table S1, Supplemental Digital Content, http://links.lww.com/MD/N779). Two-sample Mendelian randomization analysis involving 9 cathepsins was performed to evaluate the causal relationship between each cathepsin and ovarian cancer. The results of the univariate MR analysis indicated the associations of between cathepsin E (odds ratio [OR] = 0.919, 95% confidence interval [CI] = 0.850–0.994, P = .0357) and cathepsin Z (OR = 0.944, 95% CI = 0.893–0.998, P = .0458) with the predisposition to ovarian cancer. Subsequent MR analyses were conducted for the 9 cathepsins to determine their relationship with the different pathological subtypes of ovarian cancer. The incidence of low-grade serous ovarian cancer showed a significant correlation with elevated levels of cathepsin L2 (OR = 0.803, 95% CI = 0.685–0.942, P = .0071), while clear cell ovarian cancer was significantly correlated with increased levels of cathepsin H (OR = 1.149, 95% CI = 1.036–1.274, P = .0081). Additionally, our results suggests that cathepsin E (OR = 0.893, 95% CI = 0.824–0.968, P = .00634) and cathepsin Z (OR = 0.926, 95% CI = 0.866–0.991, P = .0267) are associated with high-grade serous cancer (Fig. 2). The weighted median, MR-Egger, simple mode, and weighted mode methods, which are 4 complementary methods, further verified the accuracy of these results. Although the MR-Egger, weighted median, simple mode, and weighted mode results do not fully support a causal relationship between cathepsin and ovarian cancer and its subtypes, the results of IVW method, MR-Egger, weighted median, simple mode, and weighted mode are in the same direction (B-value is in the same direction), indicating that the results are reliable (Table 2). No discernible heterogeneity was observed (Cochrane Q P-value > .05; Table S2, Supplemental Digital Content, http://links.lww.com/MD/N779), and the Leave-one-out analyses showed that the results were stable (Figures S1–S6, Supplemental Digital Content, http://links.lww.com/MD/N778). The MR-Egger intercept was utilized to assess the presence of horizontal pleiotropy; however, horizontal pleiotropy was not demonstrated in this study (Table S3, Supplemental Digital Content http://links.lww.com/MD/N779).
Figure 2.
Univariate Mendelian randomization (MR) analysis of inverse-variance weighted results of forest graphs.
Table 2.
Causal association of cathepsins on ovarian cancer and its histological subtypes estimated by univariable Mendelian randomization analysis.
| cancer | Cathepsin | nsnp | Inverse variance weighted | MR-Egger | Weighted median | Simple mode | Weighted mode | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P-value | OR(95%CI) | P-value | OR(95%CI) | P-value | OR(95%CI) | P-value | OR(95%CI) | P-value | |||
| Ovarian cancer | Cathepsin B | 19 | 1.026 (0.981–1.074) | .249 | 1.041 (0.936–1.158) | .462 | 1.008 (0.943–1.078) | .801 | 0.972 (0.867–1.090) | .636 | 1.000 (0.936–1.068) | .993 |
| Cathepsin E | 11 | 0.919 (0.850–0.994) | .036 | 0.974 (0.827–1.147) | .763 | 0.934 (0.848–1.027) | .162 | 0.969 (0.830–1.131) | .695 | 0.972 (0.845–1.118) | .698 | |
| Cathepsin F | 12 | 0.993 (0.930–1.061) | .851 | 1.002 (0.846–1.188) | .974 | 1.020 (0.941–1.107) | .617 | 1.024 (0.901–1.163) | .723 | 1.026 (0.929–1.133) | .627 | |
| Cathepsin G | 12 | 1.033 (0.962–1.110) | .359 | 1.002 (0.867–1.158) | .969 | 1.032 (0.938–1.136) | .510 | 1.004 (0.867–1.163) | .956 | 0.997 (0.860–1.155) | .969 | |
| Cathepsin H | 11 | 1.032 (0.998–1.066) | .058 | 1.049 (1.003–1.098) | .065 | 1.037 (1.001–1.076) | .043 | 1.028 (0.940–1.124) | .563 | 1.038 (1.002–1.075) | .067 | |
| Cathepsin O | 11 | 0.993 (0.929–1.062) | .847 | 0.915 (0.797–1.050) | .240 | 1.004 (0.919–1.098) | .915 | 1.031 (0.891–1.193) | .691 | 1.031 (0.904–1.176) | .659 | |
| Cathepsin S | 20 | 0.995 (0.957–1.034) | .811 | 0.976 (0.916–1.041) | .484 | 0.979 (0.929–1.032) | .445 | 0.995 (0.897–1.103) | .922 | 0.983 (0.929–1.040) | .553 | |
| Cathepsin L2 | 9 | 0.984 (0.909–1.065) | .694 | 1.077 (0.895–1.297) | .455 | 0.970 (0.880–1.069) | .545 | 0.933 (0.809–1.075) | .363 | 0.939 (0.818–1.078) | .399 | |
| Cathepsin Z | 11 | 0.944 (0.893–0.998) | .046 | 0.890 (0.819–0.967) | .022 | 0.931 (0.863–1.004) | .066 | 0.937 (0.825–1.064) | .338 | 0.926 (0.854–1.005) | .095 | |
| Low-grade serous ovarian cancer | ||||||||||||
| Cathepsin B | 19 | 0.997 (0.904–1.101) | .966 | 0.997 (0.790–1.257) | .982 | 1.013 (0.878–1.169) | .852 | 1.095 (0.861–1.392) | .470 | 1.035 (0.887–1.208) | .667 | |
| Cathepsin E | 11 | 1.026 (0.841–1.251) | .795 | 1.047 (0.682–1.609) | .835 | 1.081 (0.872–1.340) | .475 | 1.116 (0.814–1.530) | .511 | 1.106 (0.840–1.456) | .490 | |
| Cathepsin F | 12 | 0.943 (0.754–1.180) | .612 | 0.721 (0.416–1.249) | .271 | 1.039 (0.852–1.268) | .699 | 1.037 (0.748–1.438) | .832 | 1.043 (0.824–1.320) | .735 | |
| Cathepsin G | 12 | 1.075 (0.900–1.285) | .420 | 0.982 (0.673–1.433) | .930 | 1.043 (0.843–1.290) | .696 | 0.928 (0.657–1.311) | .680 | 1.048 (0.739–1.487) | .798 | |
| Cathepsin H | 11 | 1.057 (0.971–1.150) | .195 | 1.120 (1.004–1.251) | .073 | 1.070 (0.990–1.156) | .086 | 1.139 (0.946–1.370) | .199 | 1.070 (0.989–1.158) | .125 | |
| Cathepsin O | 11 | 1.016 (0.878–1.175) | .827 | 0.920 (0.682–1.241) | .600 | 1.074 (0.881–1.309) | .478 | 1.120 (0.837–1.500) | .463 | 1.120 (0.820–1.530) | .491 | |
| Cathepsin S | 20 | 0.999 (0.919–1.087) | .993 | 1.033 (0.900–1.184) | .646 | 0.984 (0.878–1.102) | .782 | 0.969 (0.786–1.194) | .769 | 0.977 (0.868–1.100) | .702 | |
| Cathepsin L2 | 9 | 0.803 (0.685–0.942) | .007 | 0.678 (0.468–0.981) | .078 | 0.843 (0.680–1.046) | .121 | 0.877 (0.634–1.212) | .449 | 0.867 (0.639–1.177) | .387 | |
| Cathepsin Z | 11 | 0.951 (0.846–1.070) | .410 | 0.869 (0.732–1.032) | .144 | 0.922 (0.785–1.083) | .325 | 1.056 (0.793–1.407) | .718 | 0.884 (0.743–1.051) | .194 | |
| High-grade serous ovarian cancer | ||||||||||||
| Cathepsin B | 19 | 1.017 (0.964–1.073) | .521 | 1.034 (0.911–1.174) | .603 | 1.000 (0.926–1.080) | .981 | 1.016 (0.885–1.167) | .823 | 1.001 (0.919–1.090) | .986 | |
| Cathepsin E | 11 | 0.893 (0.824–0.968) | .006 | 0.961 (0.813–1.136) | .656 | 0.952 (0.851–1.066) | .399 | 0.966 (0.825–1.132) | .680 | 0.966 (0.834–1.120) | .658 | |
| Cathepsin F | 12 | 0.979 (0.911–1.051) | .561 | 1.054 (0.885–1.257) | .563 | 1.011 (0.917–1.116) | .814 | 1.020 (0.865–1.202) | .822 | 1.017 (0.886–1.167) | .819 | |
| Cathepsin G | 12 | 1.045 (0.960–1.138) | .300 | 1.059 (0.891–1.258) | .529 | 1.016 (0.911–1.132) | .771 | 0.996 (0.835–1.189) | .968 | 0.992 (0.853–1.154) | .918 | |
| Cathepsin H | 11 | 1.015 (0.976–1.055) | .445 | 1.018 (0.965–1.074) | .520 | 1.011 (0.968–1.055) | .604 | 0.997 (0.892–1.114) | .956 | 1.011 (0.968–1.056) | .620 | |
| Cathepsin O | 11 | 1.016 (0.939–1.101) | .678 | 0.950 (0.806–1.120) | .562 | 1.012 (0.913–1.122) | .806 | 1.017 (0.880–1.176) | .822 | 1.021 (0.875–1.192) | .794 | |
| Cathepsin S | 20 | 0.987 (0.942–1.034) | .587 | 0.971 (0.900–1.047) | .459 | 0.956 (0.900–1.014) | .137 | 0.954 (0.828–1.098) | .517 | 0.957 (0.899–1.019) | .182 | |
| Cathepsin L2 | 9 | 0.946 (0.866–1.034) | .228 | 1.058 (0.861–1.299) | .607 | 0.942 (0.838–1.059) | .323 | 0.944 (0.782–1.140) | .566 | 0.943 (0.808–1.100) | .474 | |
| Cathepsin Z | 11 | 0.926 (0.866–0.991) | .027 | 0.882 (0.799–0.973) | .034 | 0.926 (0.846–1.014) | .097 | 0.948 (0.820–1.097) | .492 | 0.930 (0.845–1.023) | .167 | |
| Clear cell ovarian cancer | ||||||||||||
| Cathepsin B | 19 | 1.034 (0.902–1.186) | .626 | 1.158 (0.838–1.601) | .384 | 1.153 (0.961–1.385) | .124 | 1.095 (0.784–1.530) | .599 | 1.149 (0.912–1.449) | .254 | |
| Cathepsin E | 11 | 0.887 (0.726–1.084) | .244 | 1.079 (0.719–1.620) | .719 | 0.874 (0.671–1.138) | .320 | 0.779 (0.493–1.232) | .311 | 0.822 (0.527–1.280) | .405 | |
| Cathepsin F | 12 | 1.010 (0.841–1.214) | .908 | 0.704 (0.451–1.098) | .152 | 1.033 (0.805–1.326) | .795 | 0.952 (0.603–1.502) | .835 | 1.195 (0.864–1.653) | .305 | |
| Cathepsin G | 12 | 1.133 (0.912–1.407) | .257 | 1.119 (0.721–1.737) | .626 | 1.118 (0.849–1.473) | .423 | 1.129 (0.727–1.753) | .599 | 1.104 (0.726–1.680) | .652 | |
| Cathepsin H | 11 | 1.149 (1.036–1.274) | .008 | 1.134 (0.976–1.318) | .132 | 1.133 (1.010–1.271) | .033 | 1.219 (0.904–1.642) | .223 | 1.132 (1.011–1.268) | .057 | |
| Cathepsin O | 11 | 0.959 (0.782–1.177) | .694 | 0.830 (0.544–1.267) | .411 | 1.021 (0.766–1.362) | .883 | 1.011 (0.665–1.538) | .961 | 1.032 (0.692–1.539) | .880 | |
| Cathepsin S | 20 | 1.023 (0.908–1.152) | .704 | 1.037 (0.854–1.261) | .712 | 1.026 (0.875–1.202) | .750 | 1.021 (0.763–1.367) | .890 | 1.028 (0.873–1.211) | .745 | |
| Cathepsin L2 | 9 | 0.980 (0.712–1.349) | .905 | 0.837 (0.378–1.854) | .675 | 0.967 (0.703–1.329) | .837 | 1.464 (0.752–2.848) | .294 | 0.801 (0.483–1.327) | .413 | |
| Cathepsin Z | 11 | 0.998 (0.847–1.174) | .982 | 0.897 (0.708–1.136) | .392 | 0.923 (0.743–1.146) | .468 | 0.952 (0.668–1.357) | .791 | 0.889 (0.715–1.106) | .316 | |
| Mucinous ovarian cancer | ||||||||||||
| Cathepsin B | 19 | 1.031 (0.920–1.155) | .597 | 0.824 (0.642–1.057) | .146 | 1.041 (0.889–1.219) | .614 | 1.214 (0.914–1.612) | .198 | 1.041 (0.889–1.219) | .611 | |
| Cathepsin E | 11 | 1.066 (0.917–1.239) | .405 | 1.078 (0.794–1.464) | .639 | 1.063 (0.874–1.294) | .536 | 1.024 (0.767–1.367) | .875 | 1.063 (0.874–1.294) | .768 | |
| Cathepsin F | 12 | 0.937 (0.772–1.138) | .515 | 1.027 (0.626–1.684) | .917 | 0.877 (0.723–1.063) | .182 | 0.901 (0.655–1.240) | .536 | 0.877 (0.723–1.063) | .388 | |
| Cathepsin G | 12 | 0.934 (0.776–1.125) | .477 | 0.712 (0.505–1.005) | .083 | 1.014 (0.804–1.278) | .902 | 1.115 (0.715–1.738) | .641 | 1.014 (0.804–1.278) | .660 | |
| Cathepsin H | 11 | 0.955 (0.888–1.027) | .221 | 0.970 (0.877–1.072) | .569 | 0.959 (0.882–1.043) | .329 | 0.967 (0.812–1.151) | .714 | 0.959 (0.882–1.043) | .334 | |
| Cathepsin O | 11 | 0.912 (0.759–1.096) | .329 | 0.582 (0.422–0.803) | .009 | 1.003 (0.800–1.256) | .976 | 1.103 (0.712–1.708) | .669 | 1.003 (0.800–1.256) | .602 | |
| Cathepsin S | 20 | 0.956 (0.875–1.044) | .323 | 1.017 (0.879–1.175) | .821 | 0.999 (0.890–1.122) | .998 | 0.896 (0.690–1.164) | .422 | 0.999 (0.890–1.122) | .855 | |
| Cathepsin L2 | 9 | 0.957 (0.808–1.132) | .608 | 0.955 (0.636–1.434) | .833 | 0.928 (0.746–1.155) | .506 | 0.905 (0.646–1.269) | .579 | 0.928 (0.746–1.155) | .491 | |
| Cathepsin Z | 11 | 0.960 (0.846–1.088) | .527 | 0.938 (0.772–1.140) | .540 | 0.973 (0.824–1.148) | .746 | 0.805 (0.602–1.077) | .175 | 0.973 (0.824–1.148) | .794 | |
| Endometrioid ovarian cancer | ||||||||||||
| Cathepsin B | 19 | 1.074 (0.966–1.194) | .185 | 1.134 (0.878–1.465) | .345 | 1.125 (0.971–1.304) | .115 | 1.162 (0.885–1.527) | .294 | 1.148 (0.976–1.351) | .113 | |
| Cathepsin E | 11 | 1.039 (0.862–1.253) | .682 | 1.212 (0.829–1.773) | .345 | 1.112 (0.907–1.364) | .303 | 1.225 (0.890–1.686) | .241 | 1.217 (0.905–1.637) | .223 | |
| Cathepsin F | 12 | 1.037 (0.869–1.238) | .682 | 0.958 (0.609–1.506) | .857 | 0.952 (0.803–1.129) | .576 | 0.953 (0.744–1.220) | .707 | 0.931 (0.766–1.133) | .493 | |
| Cathepsin G | 12 | 1.021 (0.852–1.224) | .813 | 0.847 (0.591–1.213) | .386 | 0.993 (0.799–1.235) | .954 | 1.152 (0.783–1.695) | .488 | 0.906 (0.643–1.275) | .582 | |
| Cathepsin H | 11 | 1.033 (0.962–1.110) | .364 | 1.107 (1.003–1.221) | .073 | 1.063 (0.983–1.149) | .121 | 1.009 (0.802–1.271) | .938 | 1.065 (0.987–1.149) | .138 | |
| Cathepsin O | 11 | 0.949 (0.785–1.148) | .595 | 0.868 (0.580–1.298) | .509 | 1.060 (0.865–1.298) | .572 | 1.095 (0.829–1.444) | .537 | 1.080 (0.824–1.414) | .590 | |
| Cathepsin S | 20 | 0.997 (0.899–1.106) | .959 | 0.938 (0.791–1.113) | .476 | 0.988 (0.878–1.113) | .852 | 0.832 (0.627–1.106) | .221 | 0.971 (0.860–1.096) | .639 | |
| Cathepsin L2 | 9 | 1.082 (0.889–1.316) | .428 | 1.169 (0.716–1.909) | .549 | 0.945 (0.752–1.186) | .626 | 0.913 (0.652–1.278) | .608 | 0.894 (0.632–1.265) | .544 | |
| Cathepsin Z | 11 | 1.008 (0.861–1.182) | .912 | 0.893 (0.714–1.116) | .348 | 0.955 (0.812–1.122) | .577 | 1.051 (0.804–1.373) | .725 | 0.956 (0.803–1.139) | .628 | |
CI = confidence interval, OR = odds ratio, SE = standard error, SNP = single-nucleotide polymorphism.
2.2. Multivariable analysis
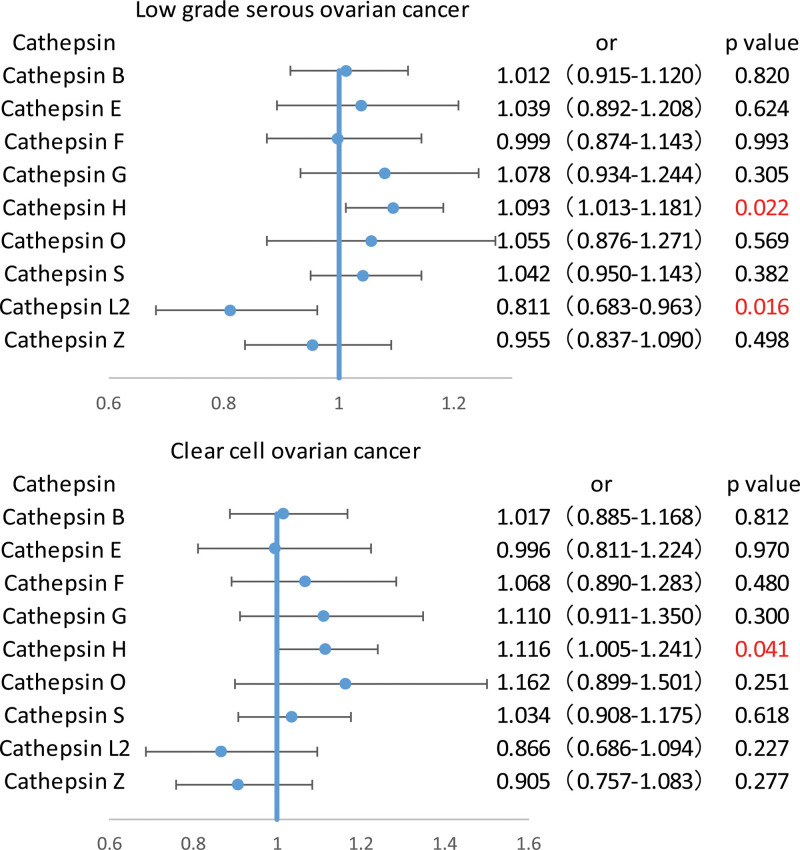
Furthermore, to exclude interference among cathepsins, multivariable MR assessments was conducted to assess the relationships of ovarian cancer and its different pathological subtypes with various cathepsins. The results indicated that after adjusting for other cathepsins using multivariable MR, cathepsin L2 levels remained strongly associated with the risk of low-grade serous ovarian cancer (OR = 0.811, 95% CI = 0.683–0.963, P = .016), and higher cathepsin H levels continued to be strongly related to the risk of clear cell ovarian cancer (OR = 1.116, 95% CI = 1.005–1.241, P = .041) as shown in Figure 3 and Table S4, Supplemental Digital Content, http://links.lww.com/MD/N779. However, no causal relationship was observed between cathepsins and other subtypes of ovarian cancer after adjustment for other cathepsins, with the results not reaching statistical significance. Furthermore, examination of the MR-Egger intercept did not indicate the presence of horizontal pleiotropy (Table S5, Supplemental Digital Content, http://links.lww.com/MD/N779).
Figure 3.
Part of forest graphs based on multivariate analysis.
2.3. Reverse MR analysis
Reverse MR analysis for identifying the potential for reverse causation showed that ovarian malignancies and their varying pathological subtypes had no significant association with cathepsin L2 and cathepsin H (P > .05), suggesting the absence of bidirectional causation (Table S6, Supplemental Digital Content, http://links.lww.com/MD/N779).
We determined that the efficacy and sample size of our study were sufficient through a power calculation, as shown in Table S7, Supplemental Digital Content, http://links.lww.com/MD/N779.
3. Discussion
Proteolytic events play a crucial role in the highly complex processes involved in the genesis and progression of malignant tumors.[25] Among the key players associated with these events, cathepsins have been identified as being particularly significant. In this study, we examined the potential causal relationship between cathepsins and ovarian cancer, including its pathological subtypes, using genetic instruments. To the best of our knowledge, this is the first study to demonstrate the causal relationship of ovarian cancer and its subtypes with various cathepsins. By integrating the results of from univariate, multivariate analyses, and reverse MR analyses, we concluded that cathepsin L2 has a protective effect, being negatively correlated with the risk of low-grade serous ovarian cancer. In contrast, cathepsin H levels showed a strong positive correlation with the risk of clear cell ovarian cancer, indicating that cathepsin H was a significant risk factor for this subtype.
The correlation between cathepsins and ovarian cancer has been documented in earlier observational studies.[26,27] A strong association has been reported between the emergence and development of ovarian cancer and cathepsins; however, reports on the link between cathepsin L2 and low-grade serous ovarian cancer, and between cathepsin H and clear cell ovarian cancer, are relatively scarce. Our IVW analysis showed that cathepsin H facilitates the occurrence and development of clear cell ovarian cancer, which aligns with the findings of previous studies regarding the role of cathepsins in the development of ovarian cancer. Interestingly, our results suggested that cathepsin L2 may reduce the risk of low-grade serous ovarian cancer, which differs from the results of most previous studies on the relationship between cathepsins and ovarian cancer. By employing multivariate and reverse MR analyses, we aimed to minimize confounding and reverse causal bias, thereby obtaining more reliable results.
Cathepsin L2 (CatL2), located on human chromosome 9q21–22, encodes a lysosomal proteinase.[28] The protective mechanism of cathepsin L2 against low-grade serous ovarian cancer is not fully understood, and further research is required for confirmation. Possible mechanisms include: (1) CatL2 may promote DNA repair through various molecular pathways to prevent cellular malignancy and promote, cell survival.[29] (2) CTSL2 could affect cellular responses to radiation and chemotherapeutic agents by influencing factors such as oxygenation, nutrient availability, and immune cell infiltration in the cellular microenvironment, thus maintaining cellular homeostasis and promoting drug resistance.[30,31] (3) CatL2 has the potential to hydrolyze cell surface proteins, exposing tumor antigens, which can enhance the antigen-presenting functions of immune cells and enable effective recognition and elimination of cancer cells by immune cells. Additionally, CatL2 may induce various inflammatory necrosis factors in the immune microenvironment to inhibit the growth of malignant cells.[32] (4) CatL2 may mediate apoptosis through proteolytic cascade reactions, revealing apoptotic inducers on the cell surface, thereby suppressing tumor growth.[33]
Cathepsin H (CatH), encoded by a gene on chromosome 15q24–25, has been implicated in tumorigenesis.[34] For example, Martínez suggested that CatH participates in modifications of the tumor microenvironment, facilitating cell-to-cell communication and formation of malignant cells. It may also accelerate the proliferation of cancer cells by processing transcription factors within the nucleus. Increased levels of CatH in early-stage tumors could assist in cancer diagnosis.[35] Similarly, Wang reported that CatH might facilitate tumor cell invasion of surrounding tissues through alterations in cell adhesion, anchoring to the matrix, and remodeling of the extracellular matrix. It may also promote the detachment of tumor cells from the primary site, degrade junctions between vascular endothelial cells, enable entry of tumor cells into the bloodstream, and hematogenous metastasis. In addition, CatH can cleave the extracellular matrix to aid metastatic cancer cell colonization at new sites and release growth factors trapped in the surrounding matrix to promote angiogenesis, invasion, and tumor development.[36] Kolwijck indicated that upregulation of CatH can lead to an imbalance between cathepsins and endogenous inhibitors; this altered enzyme/inhibitor balance can promote tumorigenesis. Cathepsin inhibitors have the potential to halt tumor progression, spread, and metastatic advancement, offering promising new therapeutic strategies against cancer progression and metastasis, in addition to providing novel insights for developing personalized treatment strategies for ovarian cancer.[37]
This study has some limitations. First, the results of this study are derived from statistical analysis, and there are few studies on the correlation between cathepsin and ovarian malignant tumors and their pathological subtypes; therefore, more basic and clinical studies are needed to support the results of this study. Second, due to the limited number of cases of ovarian cancer and its pathological subtypes, the power of MR Analysis was reduced in our study. Third, this study used abstract-level data on exposure and outcome, rather than individual-level data; consequently, subgroup analyses could not be performed. For example, our analysis was not stratified by tumor stage, nor did it consider the potential effect of cathepsin concentration on the outcome. Finally, because of the genetic diversity that exists between different populations, it is necessary to conduct a more comprehensive study of GWAS cases and gather more detailed information about the characteristics of the disease. The study participants were entirely of European descent, which limits the generalizability of our results to the wider population. Accordingly, these findings should be further validated in different populations.
In summary, our study results suggests that cathepsin L2 is negatively correlated with ovarian cancer risk and may function as a protective factor against low-grade serous ovarian cancer. Conversely, cathepsin H is positively correlated with an increased risk of clear cell ovarian cancer, indicating that it is a significant risk factor. These findings could aid in identifying new therapeutic targets for ovarian cancer subtypes and provide potential research directions for cathepsin-related treatments for these cancers.
Author contributions
Data curation: Jiaqi Ying, Fang Jie.
Formal analysis: Xia Chen.
Investigation: Fang Jie.
Methodology: Xia Chen.
Project administration: Huanyong Tian.
Resources: Tian Lv.
Software: Xia Chen.
Supervision: Jiaqi Ying, Fang Jie, Huanyong Tian.
Validation: Tian Lv.
Writing – original draft: Jiaqi Ying, Tian Lv.
Writing – review & editing: Huanyong Tian.
Supplementary Material
Abbreviations:
- CatH
- Cathepsin H
- CatL2
- Cathepsin L2
- CI
- confidence interval
- GWAS
- genome-wide association studies
- IVs
- instrument variants
- IVW
- inverse-variance weighting
- MR
- Mendelian randomization
- OR
- odds ratio
- SNP
- single-nucleotide polymorphism
This study did not require ethical approval due to the nature of the GWAS data, which was obtained and utilized in accordance with ethical standards and after obtaining written informed consent.
The authors have no funding and conflicts of interest. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request
Supplemental Digital Content is available for this article.
How to cite this article: Ying J, Chen X, Lv T, Jie F, Tian H. Mendelian randomization analysis to explore the relationship between cathepsins and malignant ovarian tumors. Medicine 2024;103:46(e40219).
JY and XC contributed equally to this work.
Contributor Information
Jiaqi Ying, Email: 470580052@qq.com.
Xia Chen, Email: 490102889@qq.com.
Tian Lv, Email: lt627756@163.com.
Fang Jie, Email: jiefang2030@163.com.
References
- [1].Guo JZ, Xiao Q, Gao S, Li XQ, Wu QJ, Gong TT. Review of Mendelian randomization studies on ovarian cancer. Front Oncol. 2021;11:681396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McGregor SM. Pathologic classification of ovarian cancer. Methods Mol Biol. 2022;2424:11–40. [DOI] [PubMed] [Google Scholar]
- [3].Kroeger PT, Jr, Drapkin R. Pathogenesis and heterogeneity of ovarian cancer. Curr Opin Obstet Gynecol. 2017;29:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li X, Zhang HS. Amino acid metabolism, redox balance and epigenetic regulation in cancer. FEBS J. 2024;291:412–29. [DOI] [PubMed] [Google Scholar]
- [5].Liu J, Zhang W, Wang Z, et al. Cathepsin V is correlated with the prognosis and tumor microenvironment in liver cancer. Mol Carcinog. 2024;63:400–16. [DOI] [PubMed] [Google Scholar]
- [6].Liang R, Tan H, Jin H, Wang J, Tang Z, Lu X. The tumour-promoting role of protein homeostasis: implications for cancer immunotherapy. Cancer Lett. 2023;573:216354. [DOI] [PubMed] [Google Scholar]
- [7].Kim EK, Song MJ, Jang HH, Chung YS. Clinicopathologic analysis of cathepsin B as a prognostic marker of thyroid cancer. Int J Mol Sci. 2020;21:9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brix K, Dunkhorst A, Mayer K, Jordans S. Cysteine cathepsins: cellular roadmap to different functions. Biochimie. 2008;90:194–207. [DOI] [PubMed] [Google Scholar]
- [9].Ranade H, Paliwal P, Chaudhary AA, et al. Predicting diagnostic potential of cathepsin in epithelial ovarian cancer: a design validated by computational, biophysical and electrochemical data. Biomolecules. 2021;12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Patel S, Homaei A, El-Seedi HR, Akhtar N. Cathepsins: Proteases that are vital for survival but can also be fatal. Biomed Pharmacother. 2018;105:526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–4. [DOI] [PubMed] [Google Scholar]
- [12].Zhang L, Wei L, Shen G, et al. Cathepsin L is involved in proliferation and invasion of ovarian cancer cells. Mol Med Rep. 2015;11:468–74. [DOI] [PubMed] [Google Scholar]
- [13].Kuester D, Lippert H, Roessner A, Krueger S. The cathepsin family and their role in colorectal cancer. Pathol Res Pract. 2008;204:491–500. [DOI] [PubMed] [Google Scholar]
- [14].Khaket TP, Kwon TK, Kang SC. Cathepsins: potent regulators in carcinogenesis. Pharmacol Ther. 2019;198:1–19. [DOI] [PubMed] [Google Scholar]
- [15].Yuzhalin AE, Lim SY, Kutikhin AG, Gordon-Weeks AN. Dynamic matrisome: ECM remodeling factors licensing cancer progression and metastasis. Biochim Biophys Acta Rev Cancer. 2018;1870:207–28. [DOI] [PubMed] [Google Scholar]
- [16].Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr Pharm Des. 2007;13:387–403. [DOI] [PubMed] [Google Scholar]
- [17].Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: a review. Res Synth Methods. 2019;10:486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li J, Tang M, Gao X, Tian S, Liu W. Mendelian randomization analyses explore the relationship between cathepsins and lung cancer. Commun Biol. 2023;6:1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27:3253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sanderson E, Spiller W, Bowden J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat Med. 2021;40:5434–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol. 2019;48:713–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rudzińska M, Parodi A, Soond SM, et al. The role of cysteine cathepsins in cancer progression and drug resistance. Int J Mol Sci. 2019;20:3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pranjol MZ, Gutowski N, Hannemann M, Whatmore J. The potential role of the proteases cathepsin D and cathepsin L in the progression and metastasis of epithelial ovarian cancer. Biomolecules. 2015;5:3260–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fan X, Wang C, Song X, Liu H, Li X, Zhang Y. Elevated Cathepsin K potentiates metastasis of epithelial ovarian cancer. Histol Histopathol. 2018;33:673–80. [DOI] [PubMed] [Google Scholar]
- [28].Itoh R, Kawamoto S, Adachi W, Kinoshita S, Okubo K. Genomic organization and chromosomal localization of the human cathepsin L2 gene. DNA Res. 1999;6:137–40. [DOI] [PubMed] [Google Scholar]
- [29].Zhang D, Tang B, Xie X, Xiao YF, Yang SM, Zhang JW. The interplay between DNA repair and autophagy in cancer therapy. Cancer Biol Ther. 2015;16:1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang X, Wang J, Li X, Wang D. Lysosomes contribute to radioresistance in cancer. Cancer Lett. 2018;439:39–46. [DOI] [PubMed] [Google Scholar]
- [31].Zhao Y, Shen X, Zhu Y, et al. Cathepsin L-mediated resistance of paclitaxel and cisplatin is mediated by distinct regulatory mechanisms. J Exp Clin Cancer Res. 2019;38:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Conus S, Simon HU. Cathepsins and their involvement in immune responses. Swiss Med Wkly. 2010;140:w13042. [DOI] [PubMed] [Google Scholar]
- [33].Hua T, Robitaille M, Roberts-Thomson SJ, Monteith GR. The intersection between cysteine proteases, Ca2+ signalling and cancer cell apoptosis. Biochim Biophys Acta Mol Cell Res. 2023;1870:119532. [DOI] [PubMed] [Google Scholar]
- [34].Deussing J, Roth W, Rommerskirch W, Wiederanders B, von Figura K, Peters C. The genes of the lysosomal cysteine proteinases cathepsin B, H, L, and S map to different mouse chromosomes. Mamm Genome. 1997;8:241–5. [DOI] [PubMed] [Google Scholar]
- [35].Martínez JF, Aparicio JR, Peiró G, et al. Study of the expression of cathepsins in histological material from pancreatic lesions. Rev Esp Enferm Dig. 2016;108:780–4. [DOI] [PubMed] [Google Scholar]
- [36].Wang Y, Zhao J, Gu Y, et al. Cathepsin H: Molecular characteristics and clues to function and mechanism. Biochem Pharmacol. 2023;212:115585. [DOI] [PubMed] [Google Scholar]
- [37].Kolwijck E, Kos J, Obermajer N, et al. The balance between extracellular cathepsins and cystatin C is of importance for ovarian cancer. Eur J Clin Invest. 2010;40:591–9. [DOI] [PubMed] [Google Scholar]