Abstract
The kidney plays a crucial role in maintaining the body’s microenvironment homeostasis. However, current treatment options and therapeutic agents for chronic kidney disease (CKD) are limited. Fortunately, the advent of kidney organoids has introduced a novel in vitro model for studying kidney diseases and drug screening. Despite significant efforts has been leveraged to mimic the spatial-temporal dynamics of fetal renal development in various types of kidney organoids, there is still a discrepancy in cell types and maturity compared to native kidney tissue. The extracellular matrix (ECM) plays a crucial role in regulating cellular signaling, which ultimately affects cell fate decision. As a result, ECM can refine the microenvironment of organoids, promoting their efficient differentiation and maturation. This review examines the existing techniques for culturing kidney organoids, evaluates the strengths and weaknesses of various types of kidney organoids, and assesses the advancements and limitations associated with the utilization of the ECM in kidney organoid culture. Additionally, it presents a discussion on constructing specific physiological and pathological microenvironments using decellularized extracellular matrix during certain developmental stages or disease occurrences, aiding the development of kidney organoids and disease models.
Keywords: extracellular matrix, kidney organoids, decellularized extracellular matrix, microenvironment, disease models
1 Introduction
The kidney, is a vital organ that plays an important role in maintaining the homeostasis of human body. Chronic kidney disease (CKD), a progressive and irreversible loss of kidney function, is becoming increasingly prevalent due to rising comorbidities such as diabetes, hypertension, obesity, and an aging population (Kishi et al., 2024; GBD Chronic Kidney Disease Collaboration, 2020). Current therapeutic approaches for CKD are limited, relying predominantly on antihypertensive agents, antidiabetic medications, and pharmacological strategies aimed at controlling disease progression (Kishi et al., 2024). These treatments, required prolonged administration and exhibited only moderate efficacy, failing to halt the progression of kidney injury to end-stage kidney disease (ESKD), defined as an eGFR below 15 mL/min/1.73 m2 (Levey et al., 2005). Animal disease models has substantially enhanced our understanding of the CKD pathophysiology and the clinical pharmacodynamics (Schnell et al., 2022; Fu et al., 2024). However, the interspecies differences significantly hinder the accurate extrapolation of disease mechanisms and the therapeutic efficacy. This challenge emphasizes the necessity for improved disease models that accurately reflect renal pathogenesis and facilitate precise drug screening approaches (Musah et al., 2024).
Organoids, as self-assembled 3D cellular structures in vitro, retain key characteristics of their in vivo counterparts and have emerged as powerful tools for developmental biology and drug screening. Kidney organoids have been a subject of research for nearly a decade (Taguchi et al., 2014). Over this time, their maturation has steadily advanced, enabling their use in constructing kidney disease models and drug screening (Musah et al., 2024; Tabibzadeh and Morizane, 2024; Dilmen et al., 2024; Long et al., 2024; Oishi et al., 2024; Chambers et al., 2023). However, their functional maturation and structural organization remain a challenge, in part due to the complexity of the kidney microenvironment (Garreta et al., 2019). The integration of extracellular matrix (ECM) components into organoid cultures has emerged as a promising strategy to mimic the in vivo environment, supporting more accurate tissue development and improving the functionality of kidney organoids (Kim J. W et al., 2022; Lacueva-Aparicio et al., 2022).
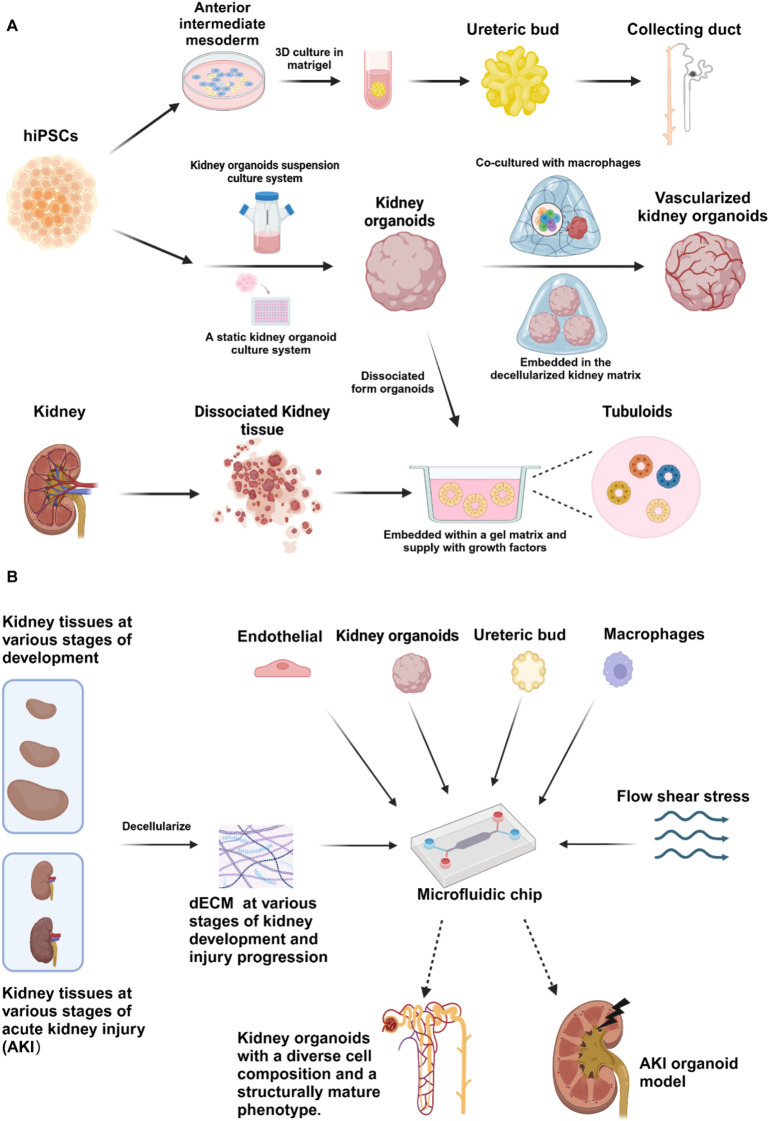
This article provides a comprehensive review of the current methods for the cultivation of kidney organoids (Figure 1A), with a particular focus on discussing the advantages and disadvantages of utilizing ECM for the culture of kidney organoids. It also proposes corresponding improvement strategies and outlines the future directions for the cultivation and application of kidney organoids.
FIGURE 1.
Various strategies are employed in the fabrication of kidney organoids (Created in BioRender.com). (A) Current procedures employed to construct ureteric bud (UB) organoids, kidney organoids and tubuloids using iPSCs and primary cells obtained from renal tissues. UB organoids are developed by differentiating hiPSCs into anterior intermediate mesoderm, which is subsequently embedded in Matrigel for 3D culture, leading to the formation of UB organoids that further differentiate into organoids containing collecting ducts. Kidney organoids, on the other hand, can be generated from hiPSCs using either suspension or static culture systems, with variations in the cultivation process resulting in vascularized kidney organoids. Finally, tubuloids are derived by embedding cells within a gel matrix and supplementing them with growth factors; their cellular origin may be either primary renal cells or dissociated organoids. Solid arrows indicate the processing steps, while dashed lines signify areas that have been magnified for clarity. (B) Schematic representation of further advancements in generation of sophisticated kidney organoids and acute kidney injury (AKI) organoid model achieved on organoids-on-chip system, incorporating the induction of decellularized renal extracellular matrix from specific developmental and pathological stages, co-culture with various exogenous cells, and introduction with fluidic shear stress, in order to simulate in vivo microenvironment. Solid arrows indicate the processing steps, while dashed arrows represent abstract model diagrams.
2 Strategies for the construction of kidney organoids
Based on the single-cell sequencing data, 25 distinct cell types have been identified within the adult kidney tissue (Balzer et al., 2022). The development of the mammalian kidney initiates with the emergence of the nephrogenic cord, which is sequentially exposed to Wnt/β-catenin and BMP signaling to form the intermediate mesoderm (IM) (Schnell et al., 2022). This process provides two sources of progenitor cells for the differentiation of the collecting duct (CD) and the functional kidney units. Specifically, it involves the ureteric bud (UB), which originates from the anterior intermediate mesoderm (aIM), and the metanephric mesenchyme, which arises from the posterior intermediate mesoderm (pIM). However, currently kidney organoid culture strategies are unable to simultaneously provide appropriate culture conditions for both types of progenitor cells, which are requisite for replicating the reciprocal inductive signals observed in vivo. Specifically, GDNF secreted by MM initiates UB branching, while WNT9B secreted by UB initiates the mesenchymal-to-epithelial transition of the nephron (Oxburgh, 2018). Since the first report of nephron organoids induced from hiPSCs in 2014 (Taguchi et al., 2014), various protocols for constructing nephron organoids have endeavored to mimic the early kidney embryonic development. These protocols involved the induction of mesoderm formation in embryoid bodies through the activation of BMP4 and WNT signaling pathway using the GSK3 inhibitor (CHIR99021) (Lindsley et al., 2006; Magro-Lopez et al., 2024), followed by exposure to FGF9 to induce and maintain the niche of nephron progenitor cells (NPCs) (Muthukrishnan et al., 2015). The advancements in these methods have enabled the development of kidney organoids that provide a model that closely resembles human physiology, allowing the study of kidney biology at the organ level, and is superior to traditional two-dimensional culture systems or non-primate models.
Over the past decade, several laboratories have consistently improved the process of lineage reproduction of kidney organoids in vitro. Resulting in models that feature a broader spectrum of specialized cell types and increased structural complexity (Table 1). These advancements have proven to be crucial in understanding the pathogenesis of human kidney diseases and facilitating extensive drug screening (Taguchi and Nishinakamura, 2017; Takasato et al., 2014; Takasato et al., 2015; Kumar et al., 2019; Freedman et al., 2015; Huang et al., 2024; Li et al., 2016; Low et al., 2019; Morizane and Bonventre, 2017; Lawlor et al., 2021) (Table 1). However, the current established kidney organoids are characterized by an immature fetal state and transcriptionally similar to the first or second trimester of human fetal kidney, and lack an integrated vascular system, severely limiting their growth rate and long-term culture in vitro. A multimodal atlas of kidney organoid differentiation has delineated at least 15 highly specialized cell types, with off-target cell proportions varying from 6% (Combes et al., 2019) to 20% (Wu et al., 2018). Notably, the reproduction of distal cell types (mainly distal tubule and collecting duct cells) in organoids is comparatively less sophisticated than that of proximal cell types (mainly proximal tubule cells) (Yoshimura et al., 2023). Consequently, although kidney organoids demonstrate morphological similarity to the developing renal tissue, they encounter significant hurdles in attaining complete maturation and intricacy, especially with regard to replicating the in vivo filtration capabilities.
TABLE 1.
Induction strategies for kidney organoids.
| Ref. | Sources | Improvement | Method | Organoids |
|---|---|---|---|---|
| Krupa et al., 2024 (PMID: 38984433) | hiPSCs | Support the growth and maturation of kidney organoids | Cultured in Self-assembling polypeptide hydrogels and GelMA hydrogels | Kidney organoids |
| Garreta et al. (2019) (PMID:30778227) | hiPSCs | Accelerate the differentiation of kidney organoids | Cultured in 1kp polyacrylamide hydrogel | |
| Homan et al., 2019 (PMID:30742039) | hiPSCs | Generates vascularized kidney organoids with more mature podocyte, enhanced cellular polarity | 3D-printed millifluidic chips were embedded in gelatin-fibrin ECM and applied for low or high fluid shear stress | |
| Sun et al. (2020) (PMID:32698872) | hiPSCs | The usc organoids self-organized well with no significant cell death | Cultured in optimal kidney ECM | Usc organoids |
| Lee et al., 2021 (PMID:34748091) | hiPSCs | More mature podocytes and vascular structures | Microfluidic chip was coated with 1.5% Matrigel and 1.5% Matrigel containing 100 ng/mL VEGF | Kidney organoids |
| Kim J. W et al. (2022) (PMID:35322595) | hiPSCs | Increase the formation of blood vessel network and promote the maturation of kidney organoids | Kidney decellularized matrix hydrogel | |
| Nerger et al., 2024 (PMID:38180232) | hiPSCs | Affect the roundness of nephron segments, spatial localization and the ratio of glomerulus to tubules | Sodium alginate hydrogel | |
| Pecksen et al. (2024) (PMID:38702808) | hiPSCs | Decrease the apoptosis of iPSCs induced by CHIR, promote the differentiation of renal organoids and promote vascularization | A monocyte of human origin coculture with iPSCs by transwell cuture system | |
| Garreta et al. (2024) (PMID:38762768) | hiPSCs | Maintain organoid differentiation and promote vascularization | Kidney derived decellularized matrix hydrogel, hPSCs derived endothelial organoids were embedded for three-dimensional culture | |
| Maggiore et al. (2024) (PMID:38901605) | hiPSCs | Increase the vascularization of renal organoids and improve the maturity of nephron. Generate different endothelial cell subtypes | iETV2-iPSCs were integrated into the previously constructed renal organoid system, and ETV2 expression was induced at day5 after culture | |
| Takasato et al. (2015) (PMID:26444236) | hiPSCs | The first report for in vitro kidney organoids induction; Optimization the exposure period of Wnt, FGF, and RA in fate selection | 2D induced cell were spun down to form a pellet and transferred onto a Transwell under the sequential induction by Wnt, FGF | |
| Freedman et al. (2015) (PMID:26493500) | hiPSCs | Simplified induction procedure; validate the Genome-modified kidney organoids form PKD-specific cysts | Induction Procedure followed by the formation of cavitated spheroids, then through MET induction to form nephron-like organoids | |
| Morizane et al. (2015) (PMID: 26458176) | hiPSCs | First reported strategy to replated 2D NPCs into 3D suspension culture with distinct lumens mimics the nephron | Generation of SIX2+SALL1+WT1+PAX2+ NPCs with high efficiency followed by the formation of PAX8+LHX1+ renal vesicles that self-organized into 3D nephron structures | |
| Taguchi and Nishinakamura (2017) (PMID:29129523) | Mouse and hiPSCs | Higher-order structures with branching morphogenesis by co-culturing independently induced ureteric bud, nephron and stromal progenitor lineages | Identified mutually distinct inductive signals between the NP and UB lineages in every step of differentiation; Co-culture iNPs, iUB, and the Pdgfra+ SP population sorted from E11.5 embryonic kidneys. iUB: induced UB; iNPs: NP induction from the mESCs; SP: stromal progenitors | |
| Li et al. (2016), Huang et al. (2024) (PMID:27570066 PMID:38692273) | Primary mouse and human NPCs/hiPSCs | Develop systems for long-term expansion of induced NPCs generated nephron organoids with minimal off-target cell types and enhanced maturation of podocytes relative to other strategies; podocyte reprogramming to an NPC-like state | Manipulation of p38 and YAP activity allowed for long-term clonal expansion of primary mouse and human NPCs and induced NPCs from hiPSCs | |
| Vanslambrouck et al., 2022, 2023 (PMID:37770563 PMID:36209212) | hiPSCs | PT-enhanced organoids with distinct S1 - S3 proximal tubule cell types; improved albumin and organic cation uptake, improved expression of SARS-CoV-2 entry factors resulting in increased viral replication | Extended the monolayer differentiation of nephron progenitor to 12–14 days cultured in enhanced BMP7 condition to 10 ng/mL | |
| Przepiorski et al. (2018), Przepiorski et al. (2021) (PMID:30033089 PMID:33938892) | hiPSCs | Establish a fast, efficient and cost-effective suspension culture method, allows large-scale organoid production | Bioreactor-Based | |
| Kumar et al. (2019) (PMID:30846463) | hiPSCs | Modified suspension culture method, a three- to fourfold increase in final cell yield and a 75% reduction in cost per million organoid-derived kidney cells compared with static culture | Low speed (60 rpm) swirling on an orbital shaker to form cell aggregates | |
| Lawlor et al. (2021) (PMID:33230326) | hiPSCs | Rapid and high-throughput; highly uniformly patterned; increasing nephron yield | 3D bioprinting | |
| Howden et al. (2021) (PMID:33378647) | Distal nephron epithelim from kidney organoids | Shift the identity of this GATA3+/EPCAM + epithelial population toward UE, including ureteric tips, cortical and medullary UE, by altering the in vitro culture conditions | GATA3+/EPCAM + epithelial population isolated by FASC and induced in the presence of GDNF, CHIR, FGF2, ATRA and Y-27632 | UB/Collecting duct organoids |
| Mae et al. (2020) (PMID:32726627) | hiPSCs | Induced UB organoids have tubular lumens and repeat branching and differentiated into collecting duct progenitors. | iPSC induced to AIM and then ND stage embedded in 2% Matrigel for 6 days to constitute induced UB organoids with epithelial polarity and tubular lumens | |
| Uchimura et al. (2020) (PMID:33326782) | hiPSCs | Combine independently differentiated MM-like and UB-like progenitors to generate human kidney organoids with a collecting system. Detect for the first time urothelial (Uro) cells | Aldosterone and AVP drive collecting duct maturation | |
| Zeng et al. (2021) (PMID:34131121) | Mouse and human fetal kidneys; hiPSCs | UB organoids generate collecting duct organoids, with differentiated principal and intercalated cells Develope a screen to establish conditions supporting the differentiation of CD organoids |
Sorting of KIT + cells were used to enrich the precursor population then induced in chemically-defined culture conditions | |
| Shi et al., 2023 (PMID:36038632) | hiPSCs | Exhibit authentic morphological behavior and responses to developmental stimuli; recapitulate the morphogenetic pattern in isolated UBs | Ensure at least 90% efficiency at the mesendodermal and pronephric intermediate mesoderm (IM) stages without using cell sorting or mechanical dissection | |
| Yousef Yengej et al. (2023) (PMID:36724260) | iPSC-derived organoids | Selectively expand the mature functional renal epithelium without off-target cells and provide easy apical access that enables evaluation of tubular transport | Tubular fragments and cells from D7+18 organoids resuspended in Basement Membrane Extract (BME) gel and plated on suspension culture wells plates | Tubuloid organoids |
| Lindoso et al., 2022 (PMID:35841001) | Tubuloid-derived cells + EVs | EVs from kidney tubular epithelial cells can phenotypically improve in vitro tubuloid maturation | Tubuloids cultured with EV | |
| Schutgens et al. (2019), Gijzen et al. (2021) (PMID:30833775 PMID:33674788) | Adult kidney tissue or urine | Long-term growth and can be expanded for at least 20 passages; Model infectious, malignant and hereditary kidney diseases; Adopt a tubular conformation and display active (trans-) epithelial transport function |
Establishing kidney tubuloids and characterization of tubuloid cell–derived 3D tubular structures in a perfused microfluidic multi-chip platform, the OrganoPlate 3-Lane |
Ref. for Reference, hiPSCs, for human indued pluripotent stem cells; USCs, for urine-derived stem cells, Extracellular vesicle (EVs), Ureteric bud (UB), Nephron progenitor cells (NPCs).
Furthermore, attempts have been dedicated to construct higher-order kidney organoids with higher lineage integrity and fully recapitulating in vivo renal developmental structures. To achieve this, some groups have developed ureteric bud (UB)/CD organoids derived from hiPSCs or UB progenitor cells extracted from mouse and human fetal kidneys, characterized by expandable, serially passaged and repeat branching morphogenesis (Howden et al., 2021; Mae et al., 2020; Shi et al., 2023; Uchimura et al., 2020; Zeng et al., 2021). Additionally, several proof-of-concept studies generated engineered kidney by aggregating 3D co-cultured NPCs with UB organoids derived from mice (Zeng et al., 2021), which preliminarily replicated the interconnected nephron and CD structures mimicking the reiterative inductive process of kidney development in vitro, shedding light on the developmental and regeneration mechanisms of the CD system (Zeng et al., 2021). However, akin to the limitations of nephron organoids, CD organoids derived from UB progenitors remain phenotypically immature compared to their in vivo counterparts, and functional evaluation demonstrating secretion and electrolyte reabsorption process is yet to be fully established (Mae et al., 2020).
To overcome the limitations of the extensive induction time and inadequate maturity of kidney organoids, researchers have turned to the induction and cultivation of tubuloids derived from primary renal tubular epithelial cells which were also found in urine (Gijzen et al., 2021; Schutgens et al., 2019). Intriguingly, tubuloid-derived cells can form polarized, leak-tight kidney tubules capable of performing trans-epithelial transporter activity (Yousef Yengej et al., 2023). Tubuloids serve as a highly physiologically relevant model for simulating infectious, malignant, and genetic kidney diseases, including tubulopathies such as Fanconi syndrome (Jamalpoor et al., 2021), re-invigorating the understanding of renal transport mechanisms, drug screening, and personalized medicine. Nonetheless, the capacity of tubuloids to accurately replicate diseases with complex multi-cellular interactions and intricate pathological mechanisms necessitates further investigation.
Although kidney organoid with similar degrees of differentiation have been established via cohort of procedures, most induction programs are time intensive and require the administration of costly exogenous growth factors, severely limiting the development of large-scale organoid culture strategies (Morizane and Bonventre, 2017). The complexity and high costs associated with these methods pose significant barriers to their widespread use in research and therapeutic applications. Therefore, establishing a controllable, and highly reproducible culture process, is essential for optimizing the lifespan, architecture complexity, homogeneity, and differentiation fidelity of organoids. This prospect is crucial for the standardization and large-scale generation of the next-generation organoids. Alan’s (Przepiorski et al., 2018; Przepiorski et al., 2021) and Little’s laboratory (Kumar et al., 2019) have developed suspension organoid culture systems resulted in a 3-4 fold increase in final cell yield compared to static culture, providing a highly promising platform for the automation and large-scale production of kidney organoids. Furthermore, the integration of bioprinting technologies can automate the production of organoids with highly homogeneity in conformation, improving the throughput of manufacture up to 9 fold (Lawlor et al., 2021). The large-scale production of kidney organoids via bioengineering strategies provides versatile platform for optimizing organoid technology towards revolutionizing the regenerative medicine and clinical applications. The integration of automated high-throughput imaging techniques enables the phenotypic analysis of kidney organoids, which can be utilized for drug screening related to nephrological disorders (Czerniecki et al., 2018; Wang et al., 2022).
In summary, the advancements of kidney organoids with the capability of recapitulating early temporal-spatial embryonic developmental trajectories are now used as faithful substitutes in studying kidney development in vitro. Organoid models enable the reproduction of tissue structures, providing opportunities to investigate the mechanism of kidney development and disease through functional screening (Huang et al., 2024). However, the efficacy of kidney organoid models depends on their developmental fidelity to primary tissue, and to what extent can they mimic embryonic organ development on cellular characteristics and architectural complexity levels remains challenging (Little and Combes, 2019). Consequently, modifying the in vivo biophysical microenvironment in spatiotemporal dimensions exhibits great potential for driving the determination of cell fate and commitment to lineage during organoid development.
3 The construction of a microenvironment specific to kidney tissue facilitates the differentiation of kidney organoids
Early embryonic development involves the formation of three germ layers, where cell fate is regulated by intracellular and extracellular signaling pathways. A multitude of studies have underscored the determinant role of specific transcription factors in lineage commitment (Wang et al., 2013; Takahashi and Yamanaka, 2006; Rigillo et al., 2021; Chen et al., 2024). However, research on how to specifically regulate cell fate through extrinsic signals is still limited (Walma and Yamada, 2020). The ECM significantly influences cell fate by activating various signaling pathways, which play a crucial role in cell fate decisions (Tang et al., 2022; Tang et al., 2013; Amran et al., 2024; Kersey et al., 2024; Li et al., 2024; Zhang et al., 2023; Sun et al., 2020). Therefore, elucidating the composition and dynamic changes of the ECM during the processes of cell development, aging, and disease progression is crucial for simulating and constructing the microenvironment of tissue at different developmental stages, injuries, and pathological processes.
The complexity of the ECM arises from its diverse constituents, including core structural proteins and regulatory factors that can initiate ECM remodeling and impact development and disease (Yamada et al., 2022; Rekad et al., 2022; Zhou et al., 2018; Damjanovski et al., 2001; Kaneko et al., 2024). Advances in tissue engineering allow for the simulation of ECM using biomaterials to achieve in vitro/in vivo cell fate regulation. However, disparities exist between commercial biomaterials and tissue ECM, which influence cell fate regulation and the efficacy of disease treatment (Zhang et al., 2023; Kim S. et al., 2022). Decellularized extracellular matrix (dECM) hydrogels are prepared through chemical or physical decellularization processes that remove immunogenic and pathogenic elements from natural tissues, followed by freeze-drying, grinding, and enzymatic digestion. These hydrogels retain the majority of bioactive proteins from the original tissue (Zhang W. et al., 2021). Therefore, the use of dECM derived from tissues to simulate the physiological microenvironment has attracted increasing attention for organoid studies. For example, Sun et al. demonstrated that the dECM hydrogels from spinal cord of neonatal rabbits can promote the axonal growth and functional maturation of spinal cord organoids (Sun et al., 2024). Similarly, in the aging process, the composition and mechanical properties of the ECM have also changed, thereby affecting tissue function. Culturing normal human mammary epithelial cells with the ECM from aged breast tissue reinforced the invasive capability of cells, and increased the expression of inflammatory cytokines and cancer-related genes and proteins (Bahcecioglu et al., 2021). Moreover, cervical squamous cell carcinoma (CSCC) patients’ adjacent cervical tissue can be used to prepare uterine cervical extracellular matrix (UCEM) hydrogels, which faithfully defined the microenvironment of cervical cancer tissue. CSCC organoids cultured with UCEM hydrogel exhibit superior characteristics compared to those cultured with Matrigel, as evidenced by increased expression of cervical cancer-related genes and signaling pathways, resulting in a closer resemblance to patient-derived CSCC tissues (Song et al., 2024). The above studies indicated that the preparation of dECM from tissues under different physiological/pathological conditions can help construct more mature organoids and disease models.
The influence of extracellular matrices (ECMs) on renal development and functionality has been extensively investigated, yielding insights into various aspects such as kidney morphogenesis, branching patterns, pathologies, and regenerative processes (Abdollahzadeh et al., 2022). Several research groups have employed proteomics to analyze the ECM composition in normally developing kidneys, aging kidneys, and kidney diseases (Diedrich et al., 2024; Rende et al., 2023; Randles et al., 2021; Eckersley et al., 2023; Li et al., 2023; Lipp et al., 2021; Lennon et al., 2014). Understanding the composition and dynamic changes of kidney ECM under different physiological and pathological conditions provides the basis for constructing microenvironments of renal tissues with diverse physiological and pathological characteristics. Furthermore, a series of studies has utilized kidney dECM for renal cell culture (Quinteira et al., 2024; Bongolan et al., 2022; Sobreiro-Almeida et al., 2020), renal injury repair (Kim et al., 2024), and organoid culture (Kim J. W et al., 2022; Garreta et al., 2024). In terms of renal cell culture, dECM-based hydrogels have been shown to effectively support renal progenitor cell survival, proliferation, and differentiation into tubular cells and podocytes, thereby providing a biocompatible platform conducive to renal regeneration (Quinteira et al., 2024). Furthermore, optimizing the decellularization process—such as using lower concentrations of SDS during the procedure—helps to preserve essential ECM components, enhancing renal cell survival and distribution, although challenges remain regarding mature cell migration (Bongolan et al., 2022). Additionally, dECM can serve as a substitute for the tubular basement membrane, simulating the physiological relevance of the in vivo environment. Co-culturing renal progenitors with endothelial cells has enabled the construction of a tubular bilayer model, which mimics the native tissue environment more closely (Sobreiro-Almeida et al., 2020). In the aspect of renal injury repair, an implantable decellularized extracellular matrix sponge has demonstrated not only rapid hemostasis during partial nephrectomy surgery but also superior wound healing, offering a promising solution for both managing renal hemorrhage and enhancing tissue regeneration at the lesion site (Kim et al., 2024). Collectively, these studies highlight the potential of dECM to advance renal research and therapeutic applications, including the enhancement of renal cell cultures and injury repair. Decellularized materials created in various laboratories have demonstrated the ability to promote differentiation, maturation, vascularization, and the development of tubular and glomerular-like structures in kidney organoids (Kim J. W et al., 2022; Garreta et al., 2024), reinforcing the promising role of dECM in advancing both basic research and clinical applications. Additionally, some laboratories have developed decellularized matrices from fibrotic kidneys to assess the impact of dECM on endothelial progenitor cells (Zhang R. et al., 2021). Although studies have not yet reported how these dECMs derived from pathological kidneys impact kidney organoid differentiation, they hold potential for constructing disease model organoids that may better simulate pathological conditions. However, there are still gaps in the maturity of kidney organoids (including the presence of precursor cells and cell cycle cells), the representation of cell types (lacking pericytes and distal tubular cells), and structural complexity (vascular wrapping and podocyte wrapping structures) compared to mature renal tissues (Kim J. W et al., 2022). In addition, the kidney organoids may contain off-target cell populations (Kim J. W et al., 2022). One potential explanation is that the current manufactured dECM primarily recapitulates the matrue renal-favor microenvironment. In contrast, kidney organoids are usually generated from hiPSCs, which contain numerous cells in the early stages of differentiation. As a result, the dECM derived from mature tissues may not be optimal for supporting the maturation of these early-stage differentiated cells in kidney organoids, leading to hindrances in their development. This mismatch between dECM derived from mature tissue and kidney organoids composed of early-stage differentiated cells highlights a crucial challenge in the field. Despite the absence of direct studies comparing early-stage and mature kidney dECM in renal organoid cultures, clues can be drawn from existing studies on the dECM in other organ systems. For instance, a study on rabbit spinal cord dECM found that neonatal dECM contained higher levels of proteins like pleiotrophin (PTN) and tenascin (TNC), which promote neural development, axonal growth, and regeneration, while mature dECM had more inhibitory components like chondroitin sulfate proteoglycans (CSPGs), limiting regenerative potential (Sun et al., 2024). This shift in ECM composition highlights a potential mismatch when applying mature tissue-derived ECM to support the maturation of progenitor cells in organoids. Early-stage ECM is optimized for promoting cell proliferation and differentiation, while mature ECM may lack these developmental cues, potentially hindering organoid maturation and limiting its functionality. By understanding and mimicking the developmental ECM environment, researchers may be able to better support the maturation and functional development of organoids, leading to more effective tissue models for both research and therapeutic applications.
4 Conclusion and prospect
Amidst the rapid advancements in multidisciplinary technologies, despite significant advancements in cellular diversity, structural complexity, functional repertoire, and developmental maturity of kidney organoids, a discernible disparity remains when compared to mature renal tissues. To address this, one potential method is the construction of a tissue microenvironment based on tissue-specific dECM, which could facilitate the maturation of kidney organoids. Both human and porcine renal dECM have been found to promote the differentiation of kidney organoids (Garreta et al., 2024). This discovery not only paves the way for potential commercialization of renal dECM but also addresses ethical concerns related to the use of human dECM.
Although current dECM derived from mature renal tissues can partially promote the maturation and vascularization of organoids, there are limitations in terms of cell types and structures, with the presence of non-renal cell types. The continued differentiation and maturation of kidney organoids require an ECM that is distinct from mature renal tissues. To address this, single-cell sequencing can be utilized to analyze various stages of kidney development and aging, as well as different regions. Furthermore, the ECM can be identified using mass spectrometry. Through the integration and comparison of single-cell multi-omics data at different development stages of kidney and kidney organoid differentiation, it becomes possible to identify the ECM that best corresponds to the kidney organoids. Culturing organoids with the corresponding stage’s ECM and introducing exogenous cells such as macrophages (Liu et al., 2020; Pecksen et al., 2024) and endothelial cells (Maggiore et al., 2024), a complex cellular microenvironment can be constructed to simulate physiological conditions to the greatest extent (Figure 1B). Furthermore, microfluidic chips can be utilized to apply fluid shear stress to the three-dimensional co-cultured organoids, thereby mimicking the processes of kidney development, aging, and disease (Figure 1B). Developing organoids at these specific stages can help elucidate the mechanisms of development, aging, and disease occurrence, and also provide a promising direction for drug screening in nephropathy using kidney organoids.
There are numerous causes of kidney disease, including congenital genetic conditions such as polycystic kidney disease (Cornec-Le Gall et al., 2018), as well as a significant proportion of kidney diseases induced by nongenetic factors, such as obstructive nephropathy or nephrotoxic drugs leading to acute kidney injury (AKI) (Chávez-Iñiguez et al., 2020; Perazella and Rosner, 2022). Genetic factors, which induced kidney diseases can be modeled by gene editing of hiPSCs followed by the induction of kidney organoid to obtain the corresponding disease models. However, there is still limited research on how to construct kidney organoid disease models induced by nongenetic factors. Although several organoids models of AKI have been developed through the use of various inflammatory stimuli or nephrotoxic drugs (Morizane et al., 2015), there is still a certain gap between these organoid models and AKI due to the maturity of organoids (Bejoy et al., 2022). Moreover, due to the multitude of causes of AKI, various alterations in ECM proteins are also markers of AKI, such as nidogen-1 glycoprotein (Gui et al., 2024) and Metalloproteinase 1 and 3 (Klimm et al., 2024). However, our understanding of the dynamics of the overall ECM composition and cellular microenvironment during the occurrence and development of AKI is still limited. Therefore, how to use the ECM related to AKI diseases in combination with kidney organoids to construct a more physiologically relevant AKI model is also a direction for future research.
In summary, kidney organoids serve as crucial multicellular models for studying renal development, aging, and disease in vitro, and offer distinct advantages over traditional animal and cell models. Their greatest strength lies in the presence of multiple interacting cell types and a certain level of physiological structure, allowing them to simulate the microenvironment of kidney tissue in vitro. However, there remains a gap between current kidney organoids and mature renal tissues, both in terms of cell types and maturity. Furthermore, there is limited research on constructing organoids that precisely mimic specific stages of human kidney development, aging, and disease. One viable approach to address these challenges involves utilizing kidney dECM that correspond to the developmental stages of tissue. By co-culturing immune-related cells and creating a complex cellular microenvironment that closely resembles physiological conditions, it becomes possible to obtain more differentiated cell types and maturity in kidney organoids. Subsequently, these advanced models enable more accurate and reliable drug screening. Furthermore, the application of microfluidic chip technology enables the construction of micro-physiological models that replicate multi-organ interactions in disease states, facilitating the study of organ interactions under normal physiological and disease conditions and also drug screening. These avenues represent future directions for the advancement of kidney organoid research.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Guangzhou Basic and Applied Basic Research Project 2023A04J0731, Guangzhou Municipal Science and Technology Bureau- 202201011011, Guang Dong Basic and Applied Basic Research Foundation 2022A1515111041, the State Key Laboratory of Respiratory Disease (SKLRD) Open Project SKLRD-Z-202115, Guang Dong Basic and Applied Basic Research Foundation (Grant NO. 2021A1515110095), Guangzhou Basic and Applied Basic Research Project (Grant NO. 2024A04J3715), Young Scientists Fund of the National Natural Science Foundation of China (Grant NO. 32200678).
Author contributions
RW: Writing–original draft, Writing–review and editing. YS: Writing–original draft, Writing–review and editing. QL: Writing–review and editing. YX: Writing–review and editing. SL: Writing–review and editing. WG: Writing–review and editing. YX: Conceptualization, Writing–review and editing. SZ: Conceptualization, Supervision, Writing–original draft, Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abdollahzadeh F., Khoshdel-Rad N., Moghadasali R. (2022). Kidney development and function: ECM cannot be ignored. Differentiation 124, 28–42. 10.1016/j.diff.2022.02.001 [DOI] [PubMed] [Google Scholar]
- Amran A., Pigatto L., Farley J., Godini R., Pocock R., Gopal S. (2024). The matrisome landscape controlling in vivo germ cell fates. Nat. Commun. 15, 4200 10.1038/s41467-024-48283-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahcecioglu G., Yue X., Howe E., Guldner I., Stack M. S., Nakshatri H., et al. (2021). Aged breast extracellular matrix drives mammary epithelial cells to an invasive and cancer-like phenotype. Adv. Sci. (Weinh) 8, e2100128. 10.1002/advs.202100128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer M. S., Rohacs T., Susztak K. (2022). How many cell types are in the kidney and what do they do? Annu. Rev. Physiol. 84, 507–531. 10.1146/annurev-physiol-052521-121841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejoy J., Qian E. S., Woodard L. E. (2022). Tissue culture models of AKI: from tubule cells to human kidney organoids. J. Am. Soc. Nephrol. 33, 487–501. 10.1681/ASN.2021050693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongolan T., Whiteley J., Castillo-Prado J., Fantin A., Larsen B., Wong C. J., et al. (2022). Decellularization of porcine kidney with submicellar concentrations of SDS results in the retention of ECM proteins required for the adhesion and maintenance of human adult renal epithelial cells. Biomater. Sci. 10, 2972–2990. 10.1039/d1bm01017d [DOI] [PubMed] [Google Scholar]
- Chambers B. E., Weaver N. E., Wingert R. A. (2023). The “3Ds” of growing kidney organoids: advances in nephron development, disease modeling, and drug screening. Cells 12, 549. 10.3390/cells12040549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Iñiguez J. S., Navarro-Gallardo G. J., Medina-González R., Alcantar-Vallin L., García-García G. (2020). Acute kidney injury caused by obstructive nephropathy. Int. J. Nephrol. 2020, 8846622. 10.1155/2020/8846622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ye X., Zhong Y., Kang X., Tang Y., Zhu H., et al. (2024). SP6 controls human cytotrophoblast fate decisions and trophoblast stem cell establishment by targeting MSX2 regulatory elements. Dev. Cell 59, 1506–1522.e11. 10.1016/j.devcel.2024.03.025 [DOI] [PubMed] [Google Scholar]
- Combes A. N., Zappia L., Er P. X., Oshlack A., Little M. H. (2019). Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 11, 3. 10.1186/s13073-019-0615-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornec-Le Gall E., Torres V. E., Harris P. C. (2018). Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J. Am. Soc. Nephrol. 29, 13–23. 10.1681/ASN.2017050483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki S. M., Cruz N. M., Harder J. L., Menon R., Annis J., Otto E. A., et al. (2018). High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940. 10.1016/j.stem.2018.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanovski S., Amano T., Li Q., Pei D., Shi Y. B. (2001). Overexpression of matrix metalloproteinases leads to lethality in transgenic Xenopus laevis: implications for tissue-dependent functions of matrix metalloproteinases during late embryonic development. Dev. Dyn. 221, 37–47. 10.1002/dvdy.1123 [DOI] [PubMed] [Google Scholar]
- Diedrich A.-M., Daneshgar A., Tang P., Klein O., Mohr A., Onwuegbuchulam O. A., et al. (2024). Proteomic analysis of decellularized mice liver and kidney extracellular matrices. J. Biol. Eng. 18, 17. 10.1186/s13036-024-00413-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilmen E., Orhon I., Jansen J., Hoenderop J. G. (2024). Advancements in kidney organoids and tubuloids to study (dys)function. Trends Cell Biol. 34, 299–311. 10.1016/j.tcb.2023.09.005 [DOI] [PubMed] [Google Scholar]
- Eckersley A., Morais M. R., Ozols M., Lennon R. (2023). Peptide location fingerprinting identifies structural alterations within basement membrane components in ageing kidney. Matrix Biol. 121, 167–178. 10.1016/j.matbio.2023.07.001 [DOI] [PubMed] [Google Scholar]
- Freedman B. S., Brooks C. R., Lam A. Q., Fu H., Morizane R., Agrawal V., et al. (2015). Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715. 10.1038/ncomms9715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Xiang Yu, Wei Q., Ilatovskaya D., Dong Z. (2024). Rodent models of AKI and AKI-CKD transition: an update in 2024. Am. J. Physiol. Ren. Physiol. 326, F563–F583. 10.1152/ajprenal.00402.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreta E., Moya-Rull D., Marco A., Amato G., Ullate-Agote A., Tarantino C., et al. (2024). Natural hydrogels support kidney organoid generation and promote in vitro angiogenesis. Adv. Mater 36, e2400306. 10.1002/adma.202400306 [DOI] [PubMed] [Google Scholar]
- Garreta E., Prado P., Tarantino C., Oria R., Fanlo L., Martí E., et al. (2019). Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater 18, 397–405. 10.1038/s41563-019-0287-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD Chronic Kidney Disease Collaboration (2020). Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 395, 709–733. 10.1016/S0140-6736(20)30045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijzen L., Yousef Yengej F. A., Schutgens F., Vormann M. K., Ammerlaan C. M., Nicolas A., et al. (2021). Culture and analysis of kidney tubuloids and perfused tubuloid cells-on-a-chip. Nat. Protoc. 16, 2023–2050. 10.1038/s41596-020-00479-w [DOI] [PubMed] [Google Scholar]
- Gui Y., Fu H., Palanza Z., Tao J., Lin Y.-H., Min W., et al. (2024). Fibroblast expression of transmembrane protein smoothened governs microenvironment characteristics after acute kidney injury. J. Clin. Invest. 134, e165836. 10.1172/JCI165836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan K. A., Gupta N., Kroll K. T., Kolesky D. B., Skylar-Scott M., Miyoshi T. (2019). Flow-enhanced vascularization and maturation of kidney organoids in vitro . Nat Methods. 16 (3), 255–262. 10.1038/s41592-019-0325-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S. E., Wilson S. B., Groenewegen E., Starks L., Forbes T. A., Tan K. S., et al. (2021). Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk. Cell Stem Cell 28, 671–684.e6. 10.1016/j.stem.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Zeng Z., Kim S., Fausto C. C., Koppitch K., Li H., et al. (2024). Long-term expandable mouse and human-induced nephron progenitor cells enable kidney organoid maturation and modeling of plasticity and disease. Cell Stem Cell 31, 921–939.e17. 10.1016/j.stem.2024.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamalpoor A., van Gelder C. A., Yousef Yengej F. A., Zaal E. A., Berlingerio S. P., Veys K. R., et al. (2021). Cysteamine-bicalutamide combination therapy corrects proximal tubule phenotype in cystinosis. EMBO Mol. Med. 13, e13067. 10.15252/emmm.202013067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N., Hirai K., Oshima M., Yura K., Hattori M., Maeda N., et al. (2024). ADAMTS2 promotes radial migration by activating TGF-β signaling in the developing neocortex. EMBO Rep. 25, 3090–3115. 10.1038/s44319-024-00174-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey A. L., Cheng D. Y., Deo K. A., Dubell C. R., Wang T.-C., Jaiswal M. K., et al. (2024). Stiffness assisted cell-matrix remodeling trigger 3D mechanotransduction regulatory programs. Biomaterials 306, 122473. 10.1016/j.biomaterials.2024.122473 [DOI] [PubMed] [Google Scholar]
- Kim J. W., Nam S. A., Yi J., Kim J. Y., Lee J. Y., Park S.-Y., et al. (2022). Kidney decellularized extracellular matrix enhanced the vascularization and maturation of human kidney organoids. Adv. Sci. (Weinh) 9, e2103526. 10.1002/advs.202103526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Sen T., Lee J. Y., Cho D.-W. (2024). Degradation-controlled tissue extracellular sponge for rapid hemostasis and wound repair after kidney injury. Biomaterials 307, 122524. 10.1016/j.biomaterials.2024.122524 [DOI] [PubMed] [Google Scholar]
- Kim S., Min S., Choi Y. S., Jo S.-H., Jung J. H., Han K., et al. (2022). Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 13, 1692. 10.1038/s41467-022-29279-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S., Kadoya H., Kashihara N. (2024). Treatment of chronic kidney disease in older populations. Nat. Rev. Nephrol. 20, 586–602. 10.1038/s41581-024-00854-w [DOI] [PubMed] [Google Scholar]
- Klimm W., Szamotulska K., Karwański M., Bartoszewicz Z., Witkowski W., Rozmyslowicz T., et al. (2024). Tissue inhibitors of Metalloproteinase 1 (TIMP-1) and 3 (TIMP-3) as new markers of acute kidney injury after massive burns. Med. Sci. Monit. 30, e943500. 10.12659/MSM.943500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa I., Treacy N. J., Clerkin S., Davis J. L., Miller A. F., Saiani A., et al. (2024). Protocol for the growth and maturation of hipsc-derived kidney organoids using mechanically defined hydrogels. Curr Protoc. 4 (7), e1096. 10.1002/cpz1.1096 [DOI] [PubMed] [Google Scholar]
- Kumar S. V., Er P. X., Lawlor K. T., Motazedian A., Scurr M., Ghobrial I., et al. (2019). Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Development 146, dev172361. 10.1242/dev.172361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacueva-Aparicio A., Lindoso R. S., Mihăilă S. M., Giménez I. (2022). Role of extracellular matrix components and structure in new renal models in vitro . Front. Physiol. 13, 1048738. 10.3389/fphys.2022.1048738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor K. T., Vanslambrouck J. M., Higgins J. W., Chambon A., Bishard K., Arndt D., et al. (2021). Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater 20, 260–271. 10.1038/s41563-020-00853-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. N., Choi Y. Y., Kim J. W., Lee Y. S., Choi J. W., Kang T., et al. (2021). Effect of biochemical and biomechanical factors on vascularization of kidney organoid-on-a-chip. Nano Converg. 8 (1), 35. 10.1186/s40580-021-00285-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon R., Byron A., Humphries J. D., Randles M. J., Carisey A., Murphy S., et al. (2014). Global analysis reveals the complexity of the human glomerular extracellular matrix. J. Am. Soc. Nephrol. 25, 939–951. 10.1681/ASN.2013030233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey A. S., Eckardt K.-U., Tsukamoto Y., Levin A., Coresh J., Rossert J., et al. (2005). Definition and classification of chronic kidney disease: a position statement from Kidney Disease: improving Global Outcomes (KDIGO). Kidney Int. 67, 2089–2100. 10.1111/j.1523-1755.2005.00365.x [DOI] [PubMed] [Google Scholar]
- Li L., He M., Tang X., Huang J., Li J., Hong X., et al. (2023). Proteomic landscape of the extracellular matrix in the fibrotic kidney. Kidney Int. 103, 1063–1076. 10.1016/j.kint.2023.01.021 [DOI] [PubMed] [Google Scholar]
- Li L., Jiao L., Feng D., Yuan Y., Yang X., Li J., et al. (2024). Human apical-out nasal organoids reveal an essential role of matrix metalloproteinases in airway epithelial differentiation. Nat. Commun. 15, 143. 10.1038/s41467-023-44488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Araoka T., Wu J., Liao H.-K., Li M., Lazo M., et al. (2016). 3D culture supports long-term expansion of mouse and human nephrogenic progenitors. Cell Stem Cell 19, 516–529. 10.1016/j.stem.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley R. C., Gill J. G., Kyba M., Murphy T. L., Murphy K. M. (2006). Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 133, 3787–3796. 10.1242/dev.02551 [DOI] [PubMed] [Google Scholar]
- Lindoso R. S., Yousef Yengej F. A., Voellmy F., Altelaar M., Mancheño Juncosa E., Tsikari T., et al. (2022). Differentiated kidney tubular cell-derived extracellular vesicles enhance maturation of tubuloids. J. Nanobiotechnology. 20 (1), 326. 10.1186/s12951-022-01506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp S. N., Jacobson K. R., Hains D. S., Schwarderer A. L., Calve S. (2021). 3D mapping reveals a complex and transient interstitial matrix during murine kidney development. J. Am. Soc. Nephrol. 32, 1649–1665. 10.1681/ASN.2020081204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. H., Combes A. N. (2019). Kidney organoids: accurate models or fortunate accidents. Genes Dev. 33, 1319–1345. 10.1101/gad.329573.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Dai S., Feng D., Qin Z., Peng X., Sakamuri S. S., et al. (2020). Distinct fate, dynamics and niches of renal macrophages of bone marrow or embryonic origins. Nat. Commun. 11, 2280. 10.1038/s41467-020-16158-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H.-Y., Qian Z.-P., Lan Q., Xu Y.-J., Da J.-J., Yu F.-X., et al. (2024). Human pluripotent stem cell-derived kidney organoids: current progress and challenges. World J. Stem Cells 16, 114–125. 10.4252/wjsc.v16.i2.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J. H., Li P., Chew E. G., Zhou B., Suzuki K., Zhang T., et al. (2019). Generation of human PSC-derived kidney organoids with patterned nephron segments and a de novo vascular network. Cell Stem Cell 25, 373–387. 10.1016/j.stem.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mae S.-I., Ryosaka M., Sakamoto S., Matsuse K., Nozaki A., Igami M., et al. (2020). Expansion of human iPSC-derived ureteric bud organoids with repeated branching potential. Cell Rep. 32, 107963. 10.1016/j.celrep.2020.107963 [DOI] [PubMed] [Google Scholar]
- Maggiore J. C., LeGraw R., Przepiorski A., Velazquez J., Chaney C., Vanichapol T., et al. (2024). A genetically inducible endothelial niche enables vascularization of human kidney organoids with multilineage maturation and emergence of renin expressing cells. Kidney Int. 10.1016/j.kint.2024.05.026 [DOI] [PubMed] [Google Scholar]
- Magro-Lopez E., Vazquez-Alejo E., La Espinar-Buitrago M. D., Muñoz-Fernández M. Á. (2024). Optimizing Nodal, Wnt and BMP signaling pathways for robust and efficient differentiation of human induced pluripotent stem cells to intermediate mesoderm cells. Front. Cell Dev. Biol. 12, 1395723. 10.3389/fcell.2024.1395723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R., Bonventre J. V. (2017). Kidney organoids: a translational journey. Trends Mol. Med. 23, 246–263. 10.1016/j.molmed.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R., Lam A. Q., Freedman B. S., Kishi S., Valerius M. T., Bonventre J. V. (2015). Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200. 10.1038/nbt.3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musah S., Bhattacharya R., Himmelfarb J. (2024). Kidney disease modeling with organoids and organs-on-chips. Annu. Rev. Biomed. Eng. 26, 383–414. 10.1146/annurev-bioeng-072623-044010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S. D., Yang X., Friesel R., Oxburgh L. (2015). Concurrent BMP7 and FGF9 signalling governs AP-1 function to promote self-renewal of nephron progenitor cells. Nat. Commun. 6, 10027. 10.1038/ncomms10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerger B. A., Sinha S., Lee N. N., Cheriyan M., Bertsch P., Johnson C. P., et al. (2024). 3D Hydrogel encapsulation regulates nephrogenesis in kidney organoids. Adv Mater. 36(14), e2308325. 10.1002/adma.202308325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi H., Tabibzadeh N., Morizane R. (2024). Advancing preclinical drug evaluation through automated 3D imaging for high-throughput screening with kidney organoids. Biofabrication 16, 035003. 10.1088/1758-5090/ad38df [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxburgh L. (2018). Kidney nephron determination. Annu. Rev. Cell Dev. Biol. 34, 427–450. 10.1146/annurev-cellbio-100616-060647 [DOI] [PubMed] [Google Scholar]
- Pecksen E., Tkachuk S., Schröder C., Vives Enrich M., Neog A., Johnson C. P., et al. (2024). Monocytes prevent apoptosis of iPSCs and promote differentiation of kidney organoids. Stem Cell Res. Ther. 15, 132. 10.1186/s13287-024-03739-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazella M. A., Rosner M. H. (2022). Drug-induced acute kidney injury. Clin. J. Am. Soc. Nephrol. 17, 1220–1233. 10.2215/CJN.11290821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przepiorski A., Crunk A. E., Holm T. M., Sander V., Davidson A. J., Hukriede N. A. (2021). A simplified method for generating kidney organoids from human pluripotent stem cells. J. Vis. Exp. 10.3791/62452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przepiorski A., Sander V., Tran T., Hollywood J. A., Sorrenson B., Shih J.-H., et al. (2018). A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Rep. 11, 470–484. 10.1016/j.stemcr.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinteira R., Gimondi S., Monteiro N. O., Sobreiro-Almeida R., Lasagni L., Romagnani P., et al. (2024). Decellularized kidney extracellular matrix-based hydrogels for renal tissue engineering. Acta Biomater. 180, 295–307. 10.1016/j.actbio.2024.04.026 [DOI] [PubMed] [Google Scholar]
- Randles M. J., Lausecker F., Kong Q., Suleiman H., Reid G., Kolatsi-Joannou M., et al. (2021). Identification of an altered matrix signature in kidney aging and disease. J. Am. Soc. Nephrol. 32, 1713–1732. 10.1681/ASN.2020101442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekad Z., Izzi V., Lamba R., Ciais D., van Obberghen-Schilling E. (2022). The alternative matrisome: alternative splicing of ECM proteins in development, homeostasis and tumor progression. Matrix Biol. 111, 26–52. 10.1016/j.matbio.2022.05.003 [DOI] [PubMed] [Google Scholar]
- Rende U., Ahn S. B., Adhikari S., Moh E. S., Pollock C. A., Saad S., et al. (2023). Deciphering the kidney matrisome: identification and quantification of renal extracellular matrix proteins in healthy mice. Int. J. Mol. Sci. 24, 2827. 10.3390/ijms24032827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigillo G., Basile V., Belluti S., Ronzio M., Sauta E., Ciarrocchi A., et al. (2021). The transcription factor NF-Y participates to stem cell fate decision and regeneration in adult skeletal muscle. Nat. Commun. 12, 6013. 10.1038/s41467-021-26293-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell J., Achieng M., Lindström N. O. (2022). Principles of human and mouse nephron development. Nat. Rev. Nephrol. 18, 628–642. 10.1038/s41581-022-00598-5 [DOI] [PubMed] [Google Scholar]
- Schutgens F., Rookmaaker M. B., Margaritis T., Rios A., Ammerlaan C., Jansen J., et al. (2019). Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 37, 303–313. 10.1038/s41587-019-0048-8 [DOI] [PubMed] [Google Scholar]
- Shi M., McCracken K. W., Patel A. B., Zhang W., Ester L., Valerius M. T., et al. (2023). Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types. Nat. Biotechnol. 41, 252–261. 10.1038/s41587-022-01429-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreiro-Almeida R., Melica M. E., Lasagni L., Romagnani P., Neves N. M. (2020). Co-cultures of renal progenitors and endothelial cells on kidney decellularized matrices replicate the renal tubular environment in vitro . Acta Physiol. (Oxf) 230, e13491. 10.1111/apha.13491 [DOI] [PubMed] [Google Scholar]
- Song H., Jiang H., Hu W., Hai Y., Cai Y., Li H., et al. (2024). Cervical extracellular matrix hydrogel optimizes tumor heterogeneity of cervical squamous cell carcinoma organoids. Sci. Adv. 10, eadl3511. 10.1126/sciadv.adl3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Zhang S., Zhou T., Shan Y., Gao F., Zhang Y., et al. (2020). Human urinal cell reprogramming: synthetic 3D peptide hydrogels enhance induced pluripotent stem cell population homogeneity. ACS Biomater. Sci. Eng. 6, 6263–6275. 10.1021/acsbiomaterials.0c00667 [DOI] [PubMed] [Google Scholar]
- Sun Z., Chen Z., Yin M., Wu X., Guo B., Cheng X., et al. (2024). Harnessing developmental dynamics of spinal cord extracellular matrix improves regenerative potential of spinal cord organoids. Cell Stem Cell 31, 772–787.e11. 10.1016/j.stem.2024.03.007 [DOI] [PubMed] [Google Scholar]
- Tabibzadeh N., Morizane R. (2024). Advancements in therapeutic development: kidney organoids and organs on a chip. Kidney Int. 105, 702–708. 10.1016/j.kint.2023.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., et al. (2014). Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67. 10.1016/j.stem.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Taguchi A., Nishinakamura R. (2017). Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell 21, 730–746. 10.1016/j.stem.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P. X., Becroft M., Vanslambrouck J. M., Stanley E. G., Elefanty A. G., et al. (2014). Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 16, 118–126. 10.1038/ncb2894 [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P. X., Chiu H. S., Maier B., Baillie G. J., Ferguson C., et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. 10.1038/nature15695 [DOI] [PubMed] [Google Scholar]
- Tang Y., Rowe R. G., Botvinick E. L., Kurup A., Putnam A. J., Seiki M., et al. (2013). MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev. Cell 25, 402–416. 10.1016/j.devcel.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Zhu L., Cho J.-S., Li X.-Y., Weiss S. J. (2022). Matrix remodeling controls a nuclear lamin A/C-emerin network that directs Wnt-regulated stem cell fate. Dev. Cell 57, 480–495.e6. 10.1016/j.devcel.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura K., Wu H., Yoshimura Y., Humphreys B. D. (2020). Human pluripotent stem cell-derived kidney organoids with improved collecting duct maturation and injury modeling. Cell Rep. 33, 108514. 10.1016/j.celrep.2020.108514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanslambrouck J. M., Wilson S. B., Tan K. S., Groenewegen E., Rudraraju R., Neil J., et al. (2022). Enhanced metanephric specification to functional proximal tubule enables toxicity screening and infectious disease modelling in kidney organoids. Nat Commun. 13(1), 5943 10.1038/s41467-022-33623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanslambrouck J. M., Tan K. S., Mah S., Little M. H. (2023). Generation of proximal tubule-enhanced kidney organoids from human pluripotent stem cells. Nat Protoc. 18 (11), 3229–3252. 10.1038/s41596-023-00880-1 [DOI] [PubMed] [Google Scholar]
- Walma D. A., Yamada K. M. (2020). The extracellular matrix in development. Development 147, dev175596. 10.1242/dev.175596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang L., Huang W., Su H., Xue Y., Su Z., et al. (2013). Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods 10, 84–89. 10.1038/nmeth.2283 [DOI] [PubMed] [Google Scholar]
- Wang Q., Lu J., Fan K., Xu Y., Xiong Y., Sun Z., et al. (2022). High-throughput “read-on-ski” automated imaging and label-free detection system for toxicity screening of compounds using personalised human kidney organoids. J. Zhejiang Univ. Sci. B 23, 564–577. 10.1631/jzus.B2100701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Uchimura K., Donnelly E. L., Kirita Y., Morris S. A., Humphreys B. D. (2018). Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23, 869–881. 10.1016/j.stem.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. M., Doyle A. D., Lu J. (2022). Cell-3D matrix interactions: recent advances and opportunities. Trends Cell Biol. 32, 883–895. 10.1016/j.tcb.2022.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura Y., Muto Y., Ledru N., Wu H., Omachi K., Miner J. H., et al. (2023). A single-cell multiomic analysis of kidney organoid differentiation. Proc. Natl. Acad. Sci. U. S. A. 120, e2219699120. 10.1073/pnas.2219699120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef Yengej F. A., Jansen J., Ammerlaan C. M., Dilmen E., Pou Casellas C., Masereeuw R., et al. (2023). Tubuloid culture enables long-term expansion of functional human kidney tubule epithelium from iPSC-derived organoids. Proc. Natl. Acad. Sci. U. S. A. 120, e2216836120. 10.1073/pnas.2216836120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Huang B., Parvez R. K., Li Y., Chen J., Vonk A. C., et al. (2021). Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat. Commun. 12, 3641. 10.1038/s41467-021-23911-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Jiang J., Yu Y., Wang F., Gao N., Zhou Y., et al. (2021). Analysis of structural components of decellularized scaffolds in renal fibrosis. Bioact. Mater. 6, 2187–2197. 10.1016/j.bioactmat.2020.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhai M., Xu Y., Han J., Chen J., Xiong Y., et al. (2023). Decellularised spinal cord matrix manipulates glial niche into repairing phase via serglycin-mediated signalling pathway. Cell Prolif. 56, e13429. 10.1111/cpr.13429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du A., Liu S., Lv M., Chen S. (2021). Research progress in decellularized extracellular matrix-derived hydrogels. Regen. Ther. 18, 88–96. 10.1016/j.reth.2021.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Horowitz J. C., Naba A., Ambalavanan N., Atabai K., Balestrini J., et al. (2018). Extracellular matrix in lung development, homeostasis and disease. Matrix Biol. 73, 77–104. 10.1016/j.matbio.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]