Abstract
Personality is influenced by both genetic and environmental factors and is associated with other psychiatric traits such as anxiety and depression. The ‘big five’ personality traits, which include neuroticism, extraversion, agreeableness, conscientiousness and openness, are a widely accepted and influential framework for understanding and describing human personality. Of the big five personality traits, neuroticism has most often been the focus of genetic studies and is linked to various mental illnesses, including depression, anxiety and schizophrenia. Our knowledge of the genetic architecture of the other four personality traits is more limited. Here, utilizing the Million Veteran Program cohort, we conducted a genome-wide association study in individuals of European and African ancestry. Adding other published data, we performed genome-wide association study meta-analysis for each of the five personality traits with sample sizes ranging from 237,390 to 682,688. We identified 208, 14, 3, 2 and 7 independent genome-wide significant loci associated with neuroticism, extraversion, agreeableness, conscientiousness and openness, respectively. These findings represent 62 novel loci for neuroticism, as well as the first genome-wide significant loci discovered for agreeableness. Gene-based association testing revealed 254 genes showing significant association with at least one of the five personality traits. Transcriptome-wide and proteome-wide analysis identified altered expression of genes and proteins such as CRHR1, SLC12A5, MAPT and STX4. Pathway enrichment and drug perturbation analyses identified complex biology underlying human personality traits. We also studied the inter-relationship of personality traits with 1,437 other traits in a phenome-wide genetic correlation analysis, identifying new associations. Mendelian randomization showed positive bidirectional effects between neuroticism and depression and anxiety, while a negative bidirectional effect was observed for agreeableness and these psychiatric traits. This study improves our comprehensive understanding of the genetic architecture underlying personality traits and their relationship to other complex human traits.
Subject terms: Genetic variation, Human behaviour
Using genome-wide association studies and meta-analyses on dimensions of personality from large existing datasets, Gupta et al. find novel genomic loci that refine our understanding of the genetic architecture of complex traits.
Main
Personality dimensions influence behaviour, thoughts, feelings and reactions to different situations. A valuable construct within the field of psychological research has converged on five different dimensions to characterize human personality: neuroticism, extraversion, agreeableness, conscientiousness and openness1,2. Personality dimensions could be playing an important role in the susceptibility and resilience to diagnosis of psychiatric disorders and their relationship with other health-related traits and responses to treatment.
The last decade has seen an increasing interest in understanding the dimensions of human personality through the lens of genetics. Depression is one mental disorder that has been studied with respect to its relationship to personality traits, with a large portion of genetic risk for depression being captured by neuroticism3. The same study found a modest negative association of genetic depression risk with conscientiousness, with small contributions from openness, agreeableness and extraversion. Neuroticism is one of the most studied dimensions of the ‘big five’ personality traits and numerous studies have found positive correlations with depression, anxiety and other mental illnesses3–5. Schizophrenia has also been associated with personality traits, especially neuroticism, which has been shown to increase risk for diagnosis6. A study using data from the Psychiatric Genomics Consortium (PGC) and personal genomics company 23andMe found two genomic loci to be common between neuroticism and schizophrenia. This study also reported six loci shared between schizophrenia and openness7.
The past 15 years have seen an explosion in the use of the genome-wide association study (GWAS). In 2010, Marleen de moor et al. from the Genetics of Personality Consortium (GPC) published a GWAS of the ‘big five’ personality traits conducted with 17,375 adults from 15 different samples of European ancestry (EUR)8. This study found two genome-wide significant (GWS) variants near the RASA1 gene on 5q14.3 for openness and one near KATNAL2 on 18q21.1 for conscientiousness but no significant associations for other personality traits. GPC then conducted studies on extraversion and neuroticism in their second phase and meta-analyses were performed. A GWAS of neuroticism that was conducted on approximately 73,000 subjects identified rs35855737 in the MAG1 gene as a GWS variant9. Although the sample size was increased substantially to 63,030 subjects in phase II, no GWS variants were detected for extraversion in that study10. In 2016, Lo et al. identified six loci associated with different personality traits, including loci for extraversion11. A paper that investigated neuroticism along with subjective well-being and depressive symptoms leveraging the UK Biobank (UKB) and other published data12 was published this same year. A more detailed picture of neuroticism genetics was presented by Nagel et al. 201813, where the authors collected neuroticism genotype data of 372,903 individuals from the UKB and performed a meta-analysis by combining the summary statistics from this UKB sample, 23andMe and GPC phase 1 samples, increasing the total sample size to 449,484. They identified a total of 136 loci and 599 genes showing GWS associations to neuroticism. In 2021, Becker et al. conducted a polygenic index study and created a resource with GWAS meta-analysis summary statistics combining different data cohorts for a large number of traits, including neuroticism, thus increasing the total sample size of neuroticism meta-analysis to 484,560 and increasing the number of novel GWS loci (although this was not the focus of this work)14. They also identified six genomic loci for extraversion.
In this work, we conducted GWAS of each of the ‘big five’ personality traits in a sample of ~224,000 individuals with genotype data available from the Million Veteran Program (MVP). Using linkage disequilibrium score regression (LDSC), we estimated the single-nucleotide polymorphism (SNP)-based heritability of each of the five personality traits. We then combined the MVP data with other sources of personality GWAS summary statistics from GPC and UKB and performed meta-analyses for each of the five personality traits, including as many as ~680,000 participants for the largest meta-analysis of neuroticism so far. To gain insights into the biology of these traits, we performed transcriptome-wide association studies (TWAS) and proteome-wide association studies (PWAS) followed by pathway and drug perturbation analyses and variant fine-mapping. We also studied the overlap of these personality traits with anxiety and other complex traits through phenome-wide genetic correlations and conditional analyses. We performed drug perturbation analyses with genes associated with neuroticism and found convergence on drugs for major depressive disorder (MDD). Finally, we conducted Mendelian randomization (MR) experiments to investigate the causal relationship of neuroticism and agreeableness, the two most genetically divergent traits, with depression and anxiety.
Results
MVP GWAS
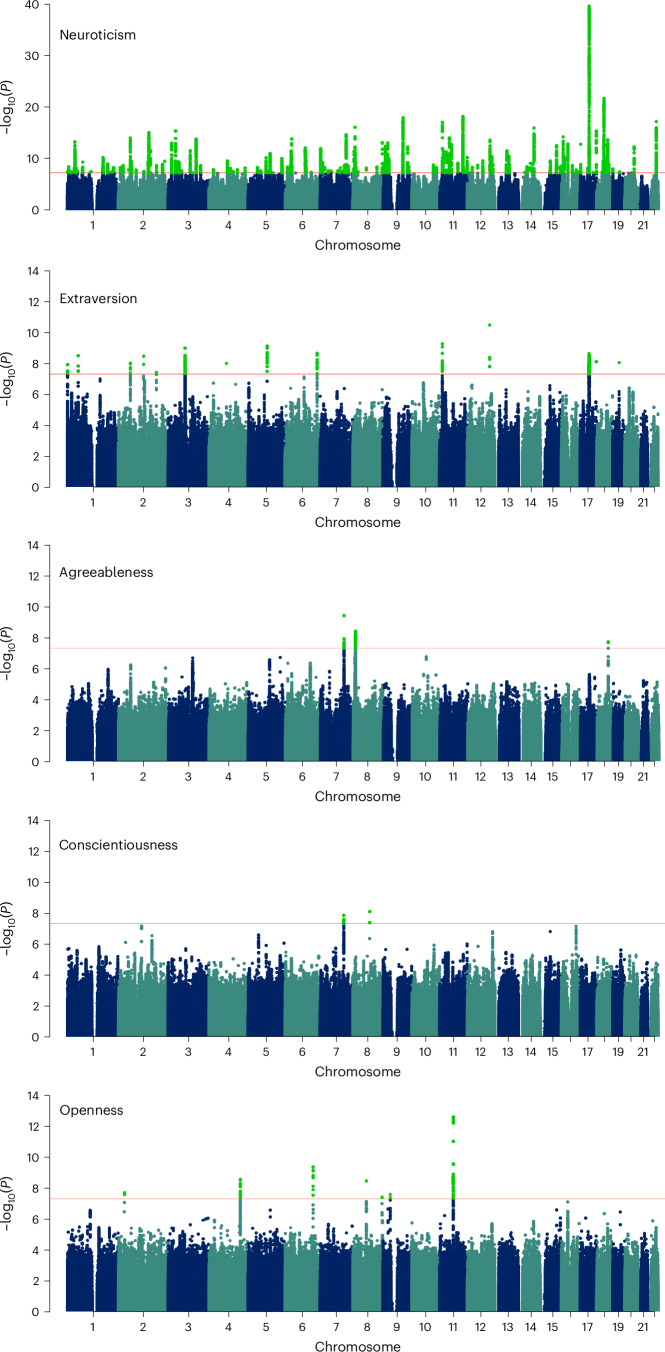
In the EUR GWAS in the MVP cohort, we identified in total 34 unique independent genomic loci significantly associated (P value <5 × 10−8) with at least one of the five personality traits (Table 1). The highest numbers of loci were found for extraversion and neuroticism (11 for each) while conscientiousness showed only two loci. In the MVP we identified 4,036 GWS variants (P < 5 × 10−8) for neuroticism across 7 independent genomic loci harbouring genes including MAD1L1, MAP3K14, CRHR1, CRHR1-IT1 and VK2 (P < 5 × 10−8). Of these seven loci, two (rs2717043 and rs4757136) were also reported to be GWS in Nagel et al.13. We identified 11 GWS loci for extraversion, the largest number of GWS loci to be identified for this trait. Associations for extraversion were found near several genes, including CRHR1, MAPT and METTL15 (total 90 genes). For the two conscientiousness loci, the first locus maps to a region near the genes FOXP2, PPP1R3A and MDFIC and the second locus maps to the ZNF704 gene, all of which are protein coding genes. For openness, 7 loci were identified spanning over 39 genes, including BRMS1, RIN1 and B3GNT1. For agreeableness, 3 loci were identified spanning 19 genes, including SOX7, PINX1 and FOXP2. The Manhattan plots for all five traits are shown in Supplementary Fig. 1.
Table 1.
Genomic loci identified in the MVP cohort for the five personality traits
| Lead SNP | Position | Effect size | s.e.m. | P | Gene |
|---|---|---|---|---|---|
| Neuroticism | |||||
| rs4129585 | 8:143312933:C:A | −0.05241 | 0.006589 | 1.80027 × 10−15 | TSNARE1 |
| rs574307253 | 17:43667635:G:A | 0.059817 | 0.007909 | 3.95325 × 10−14 | – |
| rs116956554 | 17:44699851:A:G | 0.057427 | 0.008282 | 4.09437 × 10−12 | NSF |
| rs2001433 | 8:10903475:A:T | 0.046409 | 0.006702 | 4.38959 × 10−12 | XKR6 |
| rs7396943 | 11:13328979:C:G | 0.046283 | 0.006822 | 1.16916 × 10−11 | BMAL1 |
| rs7825636 | 8:8578229:G:C | 0.04446 | 0.006712 | 3.49922 × 10−11 | – |
| rs6948912 | 7:2076701:T:C | −0.04524 | 0.007191 | 3.14987 × 10−10 | MAD1L1 |
| rs2139053 | 2:58156539:C:T | 0.042452 | 0.006905 | 7.86894 × 10−10 | – |
| rs615632 | 8:9796321:T:C | 0.040914 | 0.00673 | 1.20538 × 10−9 | – |
| rs117713019 | 8:143444050:T:C | 0.08952 | 0.015421 | 6.4366 × 10−9 | TSNARE1 |
| rs6498809 | 16:61833811:T:C | 0.03672 | 0.006642 | 3.23728 × 10−8 | CDH8 |
| Extraversion | |||||
| rs35424804 | 11:13248730:T:C | 0.037166 | 0.005994 | 5.6352 × 10−10 | – |
| rs17688916 | 17:43778680:A:T | −0.04406 | 0.007384 | 2.41754 × 10−9 | – |
| rs12971383 | 19:31876692:C:A | 0.054198 | 0.009231 | 4.33065 × 10−9 | – |
| rs3764002 | 12:108618630:T:C | −0.03928 | 0.006717 | 4.97048 × 10−9 | WSCD2 |
| rs1011501 | 5:93008722:C:T | −0.03858 | 0.006619 | 5.60839 × 10−9 | FAM172A |
| rs35918640 | 17:79452756:AT:A | 0.034989 | 0.006064 | 7.93604 × 10−9 | – |
| rs5831479 | 2:58167698:G:GA | −0.03516 | 0.006154 | 1.10698 × 10−8 | – |
| rs11209774 | 1:71834574:G:T | −0.03406 | 0.005964 | 1.12553 × 10−8 | – |
| rs7606514 | 2:185130889:G:A | −0.04297 | 0.007632 | 1.79308 × 10−8 | – |
| rs7739331 | 6:92315317:T:G | −0.03211 | 0.005872 | 4.5495 × 10−8 | − |
| rs1444978 | 4:85363489:C:T | 0.036666 | 0.0067175 | 4.81211 × 10−8 | − |
| Agreeableness | |||||
| rs17137124 | 7:114210814:C:T | −0.03273 | 0.005234 | 3.99629 × 10−10 | FOXP2 |
| rs7833945 | 8:10700266:G:T | −0.03161 | 0.005375 | 4.05674 × 10−9 | − |
| rs7240986 | 18:53195249:A:G | −0.0307 | 0.005464 | 1.92256 × 10−8 | TCF4 |
| Conscientiousness | |||||
| rs78446248 | 8:81443461:A:G | −0.08581 | 0.014907 | 8.61644 × 10−9 | − |
| rs936145 | 7:114297180:A:G | −0.0331 | 0.005848 | 1.50791 × 10−8 | FOXP2 |
| Openness | |||||
| rs7570 | 11:66610645:C:G | −0.04711 | 0.006439 | 2.54365 × 10−13 | C11orf80 |
| rs117890891 | 6:135928772:G:T | 0.145052 | 0.023233 | 4.28749 × 10−10 | − |
| rs919013 | 4:152945667:C:T | 0.033444 | 0.005625 | 2.7658 × 10−9 | − |
| rs6996198 | 8:65463442:T:C | −0.04602 | 0.007788 | 3.43929 × 10−9 | − |
| rs6725323 | 2:29377923:T:A | −0.03361 | 0.005991 | 2.03021 × 10−8 | CLIP4 |
| rs61689447 | 9:35777442:G:G:TT | −0.03312 | 0.005946 | 2.55878 × 10−8 | − |
| rs11996715 | 8:141647291:A:C | 0.030703 | 0.005588 | 3.91526 × 10−8 | − |
Two GWS variants were found for agreeableness in the African ancestry (AFR) sample. Variants rs2393573 (effect size, −0.106; standard error of the mean (s.e.m.), 0.018; 95% confidence interval (CI) −0.071, 0.141; P = 7.502 × 10−9) and rs112726823 (effect, −0.720; s.e.m., 0.130; 95% CI 0.465, 0.975; P 3.268 × 10−8) mapped near CCDC6 and ARHGAP24. We did not find any GWS variants for any of the other four personality traits in the AFR sample; the multiple subthreshold findings from this analysis may reach the GWS threshold in a larger sample. A list of lead independent SNPs found in the AFR sample for each trait is provided in Supplementary Tables 1–5.
Meta-analysis in EUR populations
The meta-analysis for neuroticism showed associations with 208 independent GWS loci. The increased power due to the inclusion of MVP data resulted in the identification of 79 additional GWS loci, which were not significant in the previous study13. Only five loci identified previously (rs1763839, rs2295094, rs11184985, rs579017 and rs76923064) were no longer significant in our meta-analysis. A total of 17 loci of these 79 have also been discovered in the polygenic index study (Supplementary Table 6). Thus, we found 62 novel loci associated with neuroticism in our meta-analysis. SNPs and loci were mapped to genes based on chromosomal position, expression quantitative trait loci (eQTL) and chromatic interaction15. A total of 231 genes were found significant in the MAGMA (Multi-marker Analysis of GenoMic Annotation) gene-based test16. NSF, KANSL1, FMNL1, PLEKHM1 and CRHR1 (P < 2.850 × 10−40) were among the top significant hits. The largest number of significant loci are located on chromosome 11, followed by chromosome 1. The GWS associations also include two loci with variants rs7818437 (effect, −0.021; s.e.m., 0.002; 95% CI −0.017, 0.025; P = 7.599 × 10−17) and rs76761706 (effect, −0.035; s.e.m., 0.002; 95% CI −0.031, 0.039; P = 2.850 × 10−40) located in inversion regions on chromosome 8 and 17, respectively. Variants in these two inversion regions were also previously reported to be significantly associated with neuroticism in the study by Okbay et al.12.
For extraversion, after meta-analysing the MVP and GPC data, the number of significant loci increased to 14. The lead signals were located on chromosomes 1–6,11,12, 17 and 19. The most significant locus harbours genes in/near WSCD2 (P < 3.449 × 10−11) located on chromosome 12.
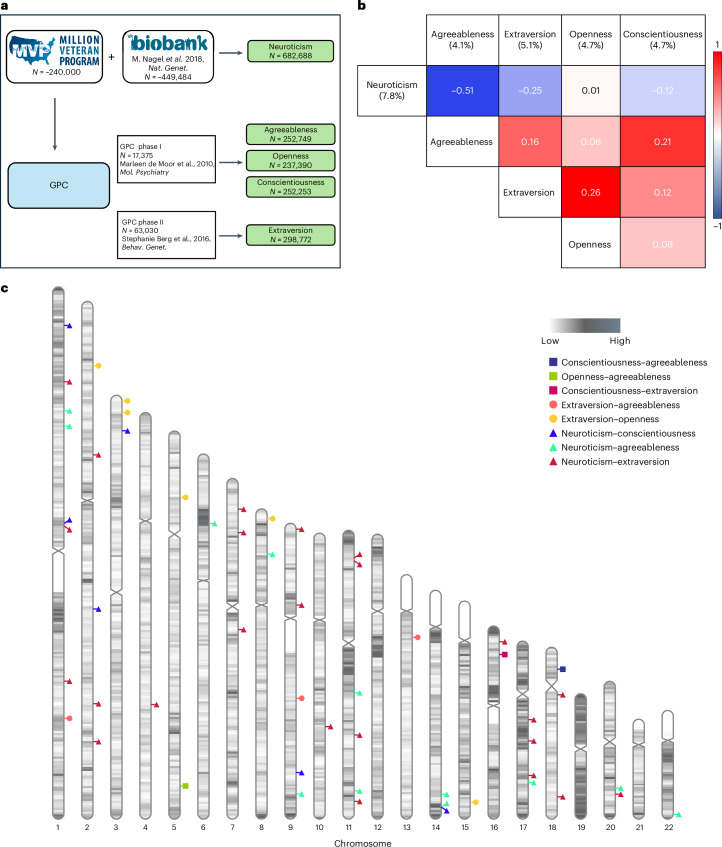
Chromosome 11 contains significant variant associations from three traits, namely neuroticism, extraversion and agreeableness, with neuroticism and extraversion both having findings near the ‘basic helix-loop-helix ARNT like 1’ (ARNTL1, also known as BMAL1) gene, with opposing and significant direction of effect at common variants. Complete information of all identified significant loci for each of the five traits with full statistics is provided in Supplementary Tables 6–10. The cohorts used in meta-analysis are depicted in Fig. 1a. Manhattan plots for meta-analyses of each of the five traits are depicted in Fig. 2.
Fig. 1. Personality GWAS meta-analysis and genetic correlations among the five personality traits.
a, Data collection of the five personality traits. b, Genetic correlation matrix among the five personality traits (meta-data). The heritability value of the respective trait is written in parenthesis. c, A karyogram showing the regions with significant local genetic correlation (rG > 0.3) between different personality traits.
Fig. 2. Meta-analysis data GWAS Manhattan plots of the five personality traits.
The GWS variants in light-green colour. Reported P values are two-sided and not corrected for multiple testing. GWS threshold (P = 5 × 10−8) is used to define significant variants and depicted by red line.
Trans-ancestry analysis
We performed trans-ancestry meta-analysis of the five personality traits combining EUR and AFR GWAS for each of the five traits using inverse variance weighing in METAL17. For neuroticism, the trans-ancestry analysis identified a total of 216 GWS loci, of which 16 are novel, that is, they were not GWS in the EUR meta-analysis (Supplementary Tables 11–15). Of the 208 GWS loci for neuroticism in the EUR meta-analysis, 200 remained GWS in trans-ancestry analysis, while the remaining 8 showed a marginally higher P value and thus do not pass the threshold for being GWS in trans-ancestry. For agreeableness and conscientiousness, in addition to the loci that were shown to be GWS in their respective EUR meta-analysis, two more novel loci (rs140242735 located on chromosome 8 and rs10864876 located on chromosome 2 for agreeableness and conscientiousness, respectively) were identified as GWS in the trans-ancestry analysis. In case of openness, two loci out of the three that identified as GWS in EUR remained GWS in the trans-ancestry analysis. For extraversion, in total 13 were identified as GWS in the trans-ancestry analysis, of which 10 were also GWS in the EUR meta-analysis and 3 were newly identified.
TWAS
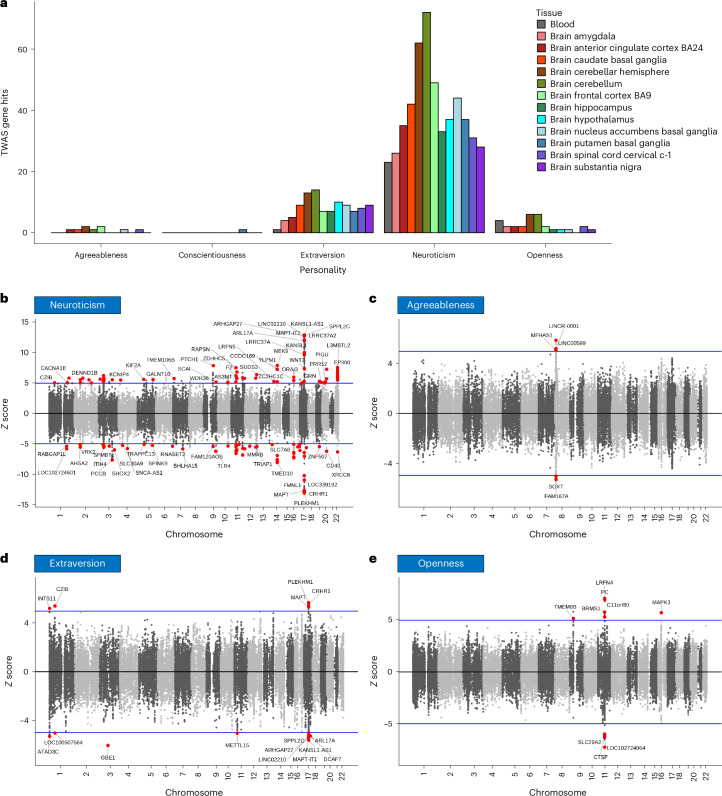
We performed TWAS for each of the ‘big five’ personality traits in EUR (meta-analysis) using FUSION18 and the GWAS summary statistics. We performed a multi-tissue TWAS in 13 different brain subtissues and blood using their respective expression profiles from Genotype Tissue-Expression project (GTEx v8)19. From a total 10,386 genes tested, we identified a total 175, 24, 5, 1 and 11 genes showing significant gene–trait associations across the 13 subtissues in neuroticism, extraversion, agreeableness, conscientiousness and openness, respectively, after Bonferroni correction for 135,018 tests (10,386 genes across 13 tissues) (Fig. 3a). Figure 3a shows the distribution of associations found across the 13 tissues for each trait. The highest number of gene–trait associations were found in brain caudate basal ganglia, cerebellum, cerebral hemisphere and frontal cortex regions for neuroticism and extraversion, while fewer TWAS gene–trait associations were identified for the other three personality traits, presumably owing to the comparatively lower power of their respective GWAS datasets.
Fig. 3. Transcriptome wide association study.
a, A bar chart showing the number of significant TWAS genes per transcripts found of four personality traits with significant findings in respective subtissues. Scatter plots of neuroticism (b), agreeableness (c), extraversion (d) and openness (e) with TWAS z-scores of each gene transcript plotted on the y axis and its respective chromosomal location plotted on the x axis. The significant hits are shown in red circles with mapped gene names as labels. The blue horizontal line indicates the significance threshold of the z-score corresponding to the Bonferroni-corrected, two-sided P value. Conscientiousness data is reported in Supplementary Table 22.
CRHR1, KANSLI1-AS1 and MAP-IT1 are among the top TWAS gene associations (P < 1.32 × 10−23) for neuroticism (Fig. 3b). The strong association of CRHR1 (encoding corticotropic-releasing hormone receptor), which in some prior work has been shown to be associated with treatment response to depression20, may suggest some common underlying elements regulating both neuroticism and depression. Extraversion also shows strong gene–trait associations with CRHR1, KANSL1-AS1 and MAPT-IT1 but with an opposite direction of effect to neuroticism. This may indicate some common genetic components whose differential behaviour regulates neuroticism and extraversion. There are nine such genes showing opposite direction of effect in neuroticism and extraversion (Supplementary Table 3).
LOC10271024064 and LRFN4 showed the strongest associations with openness and LINCR-0001 and FAM167A showed the strong associations with agreeableness, while only one gene, AP1G1, showed association with conscientiousness in the 13 tissues considered. The complete list of all GWS TWAS gene hits for the five personality traits is provided in Supplementary Table 22.
PWAS
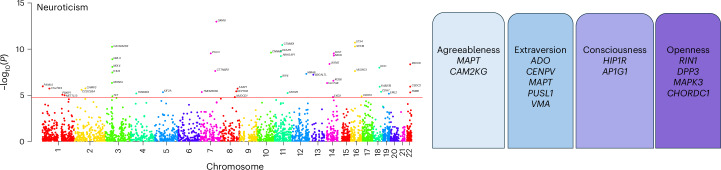
We investigated the association of personality traits with protein expression using PWAS. Based on the availability of protein profiles and the observed TWAS signal, dorsolateral prefrontal cortex brain protein profiles were chosen for the PWAS analysis. The PWAS identified 47 proteins to be significantly associated with neuroticism. Next, we checked the colocalization signal for these PWAS lead genes. Out of 47 PWAS lead genes, 35 genes showed a colocalization signal (H4 probability >0.5).
Five, two, two and four proteins were discovered for extraversion, agreeableness, conscientiousness and openness, respectively (Fig. 4). A complete list of all PWAS lead genes is provided in Supplementary Table 23.
Fig. 4. PWAS analysis.
A Manhattan plot is displayed showing the significant protein associations observed for neuroticism. The red line in the plot depicts the Bonferroni-corrected, two-sided P value threshold at 5% FDR. The boxes on the right show the significant proteins found for the respective four personality traits.
LDSC
We first used LDSC to calculate SNP-based heritability of each of the five personality traits within the MVP EUR cohort. The intercepts of the LDSC indicated no evidence for population stratification, with observed values of 1.01, 1.02, 0.99, 1.02 and 1.00 for neuroticism, extraversion, agreeableness, conscientiousness and openness, respectively. The SNP heritability ranges from 4% to 7% (Supplementary Fig. 2), with extraversion showing the highest heritability point estimate of all traits (neuroticism h2 = 0.0655; s.e.m., 0.004; 95% CI 0.058, 0.073; agreeableness h2 = 0.042; s.e.m., 0.003; 95% CI 0.036, 0.048; extraversion h2 = 0.071; s.e.m., 0.003; 95% CI 0.065, 0.077; openness h2 = 0.048; s.e.m., 0.003; 95% CI 0.042, 0.054; and conscientiousness h2 = 0.047; s.e.m., 0.003; 95% CI 0.041, 0.053).
For the MVP AFR cohort, cov-LDSC was utilized to estimate personality heritabilities (Methods)21. Relative to the MVP EUR cohort, neuroticism and extraversion showed lower heritability (4.47% and 3.30%, respectively) in the AFR cohort, while for agreeableness, the heritability was similar (4.24%) (Supplementary Table 1). The values were not significant for conscientiousness and openness in AFR.
Before combining the MVP cohort-derived summary statistics with other data sources, we calculated the genetic correlation between the MVP personality summary statistics and other respective sources (Supplementary Table 2). A correlation coefficient value of 0.80 (s.e.m., 0.02) observed for the neuroticism summary statistics from the MVP cohort and Nagel et al. study13 suggests that there is limited heterogeneity between the two datasets and supports their use in a meta-analysis. As shown in Supplementary Table 2, the genetic correlations were high for all other four traits across data sources as well.
LDSC was used to estimate SNP-based heritability in the EUR participants for each personality trait in the meta-analysis. The SNP heritability values in the meta-analyses were similar to what was observed in the MVP-only cohort for the different traits in the EUR, with a decrease in heritability of extraversion from 7.1% to 5.1% (Fig. 1b).
Genetic correlation estimates were also obtained between the meta-analysis summary statistics for the five personality traits. We found a significant degree of varying genetic overlap among the five personality traits. The genetic correlations are presented in Fig. 1b. The highest correlation is observed between neuroticism and agreeableness with a rG = −0.51 (s.e.m., 0.030; P = 3.813 × 10−64).
Next, we estimated the genetic correlations of 1,437 traits listed in the Complex Traits Genetics Virtual Lab22 summary statistics record to find other traits related to the five personality traits (Supplementary Tables 16–20). A total of 325 traits showed significant genetic correlation following multiple testing correction to one or more personality traits. We found MDD and anxiety showed varying degrees of significant correlations to different personality traits as shown in Fig. 5. The highest genetic correlation is between neuroticism and anxiety (rG = 0.80). Neuroticism and agreeableness both show high genetic correlations to these traits, but in opposite directions with MDD (neuroticism rG = 0.68; s.em. 0.02; P < 5.00 × 10−100 and agreeableness rG = −0.35; s.e.m. 0.04; P = 1.53 × 10−22), manic behaviour (neuroticism rG = 0.44; s.e.m. 0.08; 95% CI 0.641, 0.719; P = 1.11 × 10−8 and agreeableness rG = −0.35; s.e.m. 0.11; 95% CI −0.134, 0.566; P = 1.556 × 10−3), anxiety (neuroticism rG = 0.80; s.e.m. 0.06; 95% CI 0.682, 0.918; P = 1.54×10−46 and agreeableness rG = −0.32; s.e.m. 0.08; 95% CI −0.163, 0.477; P = 7.28 × 10−5) and irritability (neuroticism rG = 0.70; s.e.m. 0.02; 95% CI 0.661; 0.739, P < 5.00 × 10−100 and agreeableness rG = −0.62; s.e.m. 0.04; 95% CI −0.542, 0.698; P = 9.76 × 10−61).
Fig. 5. Bar chart with SNP-based genetic correlation of each of the five personality traits with a different psychological disorder/trait or different behaviours plotted.
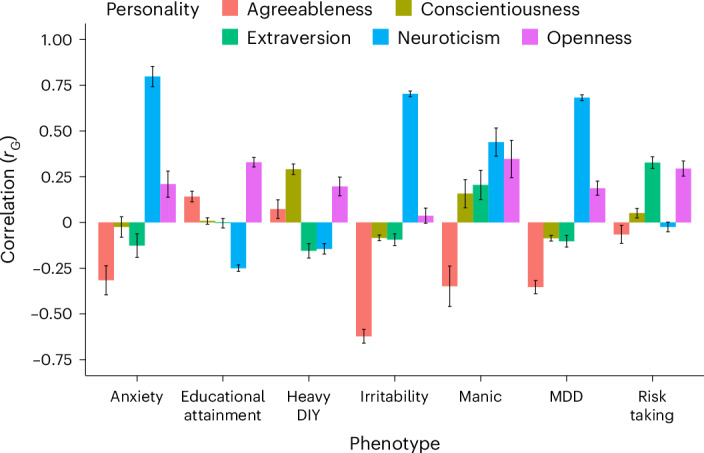
The y axis is the genetic correlation. Error bars (in black) indicate the 95% CIs of the estimated genetic correlation. Anxiety indicates substances taken for anxiety; medication is prescribed for at least 2 weeks. Heavy DIY activities describes the types of physical activity in last 4 weeks; for example, weeding, lawn mowing, carpentry and digging. Manic behaviour describes manic/hyper behaviour for 2 days. Detailed results for all traits, including the sample size of each of the traits, is presented in the Supplementary Tables 16–20.
Local genetic correlations
Global genetic correlations use the average squared signal over the entire genome, which may sometimes mask opposing local correlations in different genomic regions. To counter that, we also calculated the local genetic correlations among the five personality trait pairs using Local Analysis of [co]Variant Association (LAVA)23. All personality pairs showed varying degree of correlation in different genomic regions except for the neuroticism–openness pair, which showed negligible global (rG = −0.01) and no local genetic correlation between the two. The highest number of correlated genomic chunks were found for neuroticism–extraversion and neuroticism–openness pairs (Fig. 1c and Supplementary Table 21).
Variant fine-mapping
To identify well-supported possible causal variants from the large list of SNPs showing associations with the personality traits, we performed genome-wide variant fine-mapping using PolyFun24. In total, 166 unique variants were fine-mapped across the five personality traits. The number of variants fine-mapped for neuroticism, extraversion, agreeableness, conscientiousness and openness were 155, 8, 4, 7 and 3, respectively. The complete list of variants fine-mapped for each of the personality traits is provided in the Supplementary Tables 24–28.
Relationship between personality and psychiatric disorders
We performed additional analyses to help understand the significant differential genetic correlation observed between neuroticism and agreeableness with different psychiatric disorders such as MDD and anxiety.
Conditional analysis
Because the genetic correlation between anxiety and neuroticism was so high, we performed multi-trait-based conditional and joint analysis of neuroticism summary statistics conditioned on anxiety and MDD summary statistics individually. The anxiety and MDD summary statistic used is based on data from UKB, MVP and PGC with individuals of EUR ancestry (see Methods for details). We performed a similar analysis with agreeableness, which had a negative correlation with both MDD and anxiety, as a negative control.
After conditioning on MDD, the SNP heritability of the conditioned neuroticism summary statistic reduced significantly from 7.8% to 3% (Table 2). Out of the original 208 GWS leads, only 42 remained significant after conditioning, indicating there is substantial genetic overlap between neuroticism and MDD, which gets removed after conditioning. In case of conditioning on anxiety, again there is a decrease in neuroticism heritability, but to a lesser extent (Table 2). On conditioning agreeableness on MDD and anxiety, no significant reduction in heritability was observed. However, loss of one genomic locus, rs7240986 (18:53195249:A:G), was observed after conditioning on either anxiety or MDD for agreeableness.
Table 2.
Conditional analysis
| Primary trait | Trait conditioned on | h2 (s.e.m.) | No. of GWS loci before conditioning | h2 (s.e.m.) after conditioning | Z-difference | P value | No. of GWS loci after conditioning |
|---|---|---|---|---|---|---|---|
| Neuroticism | Anxiety | 0.078 (0.003) | 208 | 0.05 (0.001) | 8.85* | 8.41e-19 | 96 |
| Agreeableness | Anxiety | 0.041 (0.003) | 3 | 0.034 (0.003) | 1.65 | 0.01 | 2 |
| Neuroticism | MDD | 0.078 (0.003) | 208 | 0.03 (0.002) | 13.31* | 1.95e-40 | 42 |
| Agreeableness | MDD | 0.041 (0.003) | 3 | 0.036 (0.003) | 1.18 | 0.24 | 2 |
Drug perturbation analysis
We performed a drug perturbation analysis to find drug candidates for neuroticism-enriched genes using gene2drug software25. Gene2drug utilizes the Connectivity Map transcriptomics data of ~13,000 cell lines exposed to different drugs, and based on these gene expression profiles and then pathway expression profiles (PEPs), it first matches the query gene to its pathway and then to its potential candidate drug. This analysis predicted 298 unique drugs to correspond to the 231 significantly associated neuroticism genes. The top-scoring drug was found to be desipramine, which is a tricylic antidepressant. Some of the other drugs predicted are flupenthixol (anti-psychotic), tetryzoline (α-adrenergic agonist), doxorubicin (anthracycline/chemotherapy) and digitoxigenin (cardenolide). Based on these results, we repeated the drug perturbation analysis with depression-enriched genes. While there were only 51 genes common between neuroticism and depression gene sets, there was a convergence on drugs in the perturbation analysis. Out of 286 and 298 drugs predicted for depression and neuroticism, respectively, 167 drugs were common to both. The complete list of drugs is presented in Supplementary Tables 29 and 30.
MR
After establishing genetic overlap of neuroticism with MDD and anxiety, we carried out an MR analysis to explore the possibility of a causal relationship between genetic risk for neuroticism and MDD or anxiety. The results of the MR analysis using different methods are presented in Table 3. The results of MR indicate a bidirectional causal effect, with the exposure of MDD on neuroticism outcome showing an inverse variance weighting (IVW) effect value of 0.429 at a significant P value (2.072 × 10−85). The exposure of neuroticism on MDD shows a higher causal effect value of 0.834 with a significant P value (6.413 × 10−103). We performed sensitivity analysis of MR using MRlap, which corrects for different sources of bias, including sample overlap, because there are overlapping participants between the exposure and outcome datasets26. With MRlap, we observe similar results with positive significant corrected β values in MRlap performed between MDD and neuroticism in both directions (Supplementary Table 4).
Table 3.
Outcome of MR experiments performed using MR
| Trait | Two sample method | Exposure | Outcome | No. of instruments | β | P | Pleiotropy | Heterogeneity |
|---|---|---|---|---|---|---|---|---|
| Neuroticism | IVW | MDD | Neuroticism | 71 | 0.429 | 2.072 × 10−85 | 3.48 × 10−4 | 248.350 |
| MR Egger | 0.416 | 2.704 × 10−5 | 248.274 | |||||
| Weighed mean | 0.363 | 4.585 × 10−68 | ||||||
| Simple mode | 0.351 | 3.102 × 10−9 | ||||||
| Weighed mode | 0.336 | 3.427 × 10−12 | ||||||
| IVW | Neuroticism | MDD | 114 | 0.834 | 6.413 × 10−103 | 2.55 × 10−5 | 369.516 | |
| MR Egger | 0.791 | 7.795 × 10−5 | 369.340 | |||||
| Weighed mean | 0.734 | 3.418 × 10−66 | ||||||
| Simple mode | 0.748 | 1.729 × 10−6 | ||||||
| Weighed mode | 0.704 | 3.772 × 10−6 | ||||||
| IVW | Anxiety | Neuroticism | 75 | 0.179 | 1.248 × 10−15 | 0.007 | 389.419 | |
| MR Egger | −0.002 | 9.585 × 10−1 | 307.410 | |||||
| Weighed mean | 0.101 | 1.182 × 10−9 | ||||||
| Simple mode | 0.081 | 9.700 × 10−4 | ||||||
| Weighed mode | 0.081 | 3.227 × 10−3 | ||||||
| IVW | Neuroticism | Anxiety | 126 | 0.700 | 5.767 × 10−61 | −1.17 × 10−3 | 209.008 | |
| MR Egger | 0.766 | 3.174 × 10−5 | 208.767 | |||||
| Weighed mean | 0.706 | 8.209 × 10−40 | ||||||
| Simple mode | 0.821 | 2.764 × 10−6 | ||||||
| Weighed mode | 0.854 | 1.101 × 10−7 | ||||||
| Agreeableness | IVW | MDD | Agreeableness | 66 | −0.284 | 5.775 × 10−13 | 9.31 × 10−4 | 118.501 |
| MR Egger | −0.273 | 1.181 × 10−1 | 118.492 | |||||
| Weighed mean | −0.281 | 5.703 × 10−13 | ||||||
| Simple mode | −0.376 | 5.529 × 10−3 | ||||||
| Weighed mode | −0.376 | 3.823 × 10−3 | ||||||
| IVW | Agreeableness | MDD | 32 | −0.221 | 4.164 × 10−6 | −1.17 × 10−2 | 133.267 | |
| MR Egger | 0.127 | 3.521 × 10−1 | 106.341 | |||||
| Weighed mean | −0.172 | 6.621 × 10−5 | ||||||
| Simple mode | −0.261 | 2.316 × 10−2 | ||||||
| Weighed mode | −0.234 | 9.338 × 10−4 | ||||||
| IVW | Anxiety | Agreeableness | 68 | −0.241 | 7.734 × 10−16 | −5.40 × 10−3 | 102.166 | |
| MR Egger | −0.112 | 1.135 × 10−1 | 96.094 | |||||
| Weighed mean | −0.191 | 4.077 × 10−7 | ||||||
| Simple mode | −0.155 | 7.727 × 10−2 | ||||||
| Weighed mode | −0.172 | 4.346 × 10−2 | ||||||
| IVW | Agreeableness | Anxiety | 42 | −0.224 | 1.157 × 10−8 | −5.07 × 10−3 | 52.159 | |
| MR Egger | −0.068 | 6.059 × 10−1 | 50.235 | |||||
| Weighed mean | −0.198 | 1.436 × 10−4 | ||||||
| Simple mode | −0.188 | 1.395 × 10−1 | ||||||
| Weighed mode | −0.192 | 1.260 × 10−1 |
We also investigated the casual relationship of neuroticism with anxiety. On performing MR with anxiety exposure on neuroticism, we found a β value of 0.179 (P = 1.248 × 10−15) and a corrected β value with MRlap of 0.531 (P = 7.781 × 10−14) showing evidence of causality. On reversing the direction, the causality effect was stronger as seen by higher β value of 0.70 (P = 5.767 × 10−61) with MR and corrected β value of 0.548 (P = 1.129 × 10−40) with MRlap. This suggests that there is stronger evidence of causal effect of neuroticism on anxiety as compared with the reverse based on the genetic susceptibility. GWAS of anxiety and anxiety disorders are still relatively underpowered compared with neuroticism, limiting the number of available genetic instruments available for testing as exposures.
We investigated the causal effect of agreeableness on MDD and anxiety and vice versa. In the case of MR of MDD exposure on agreeableness outcome, a β value of −0.284 (P = 5.775 × 10−13) was observed indicating negative causal effect of MDD on agreeableness (Table 3 and Supplementary Table 4). The causal effect is bidirectional with similar values observed in the opposite direction as well. The results are consistent with genetic correlation findings where negative correlation was observed between agreeableness and MDD. MR analysis of agreeableness and anxiety also indicated bidirectional causal effect. However, here both the traits have limited instruments available.
Out-sample polygenic risk score prediction
We conducted polygenic prediction analysis to validate our findings using the Yale–Penn cohort27, which had NEO Personality Inventory (NEO PI-R) scores and genotype information available for 4,532 EUR individuals, and used those data to predict PRS for each of the big five personality traits (Methods). We found modest but significant r2 values in line with previous reports for all personality traits14: neuroticism of 2%, extraversion of 2%, openness of 2%, agreeableness of 3% and conscientiousness of 1%.
Discussion
We conducted a GWAS meta-analysis study of each of the ‘big five’ personality traits in a sample size of up to 682,688 participants. We combined original GWAS results from the MVP (available for all five traits) with summary statistics from the UKB (neuroticism only) and GPC (all traits except neuroticism) cohorts to perform a well-powered meta-analysis for EUR GWAS in each trait. We identified 468 independent significant SNPs associations mapping to 208 independent genomic loci, of which one-third are novel. We identified 231 significant gene associations with neuroticism in the gene-based analysis. The current study was also successful in identifying 23 significant genomic locus associations for the four other personality traits studied, for which prior knowledge in the literature was very limited. In AFR, we found lower heritabilities for neuroticism and extraversion and no significant results for conscientiousness and openness. We identified two GWS variants for agreeableness in AFR. This is probably a reflection of low power and underlines the critical need to increase recruitment in underrepresented groups. Our work provides new data to inform the underlying genetic architecture of personality traits.
Neuroticism, the trait with the largest available sample size in this study, is characterized by emotional instability, increased anxiousness and low resilience to stressful events. As such, it has been the focus of previous efforts in GWAS. As seen previously, neuroticism overlaps substantially with psychopathology, where it is usually viewed as a precursor or risk factor for depressive and anxiety symptoms. Extraversion had the second largest sample size and had the highest SNP-based heritability in the MVP. In our data, scoring high on extraversion was genetically correlated with risk-taking behaviours and had the second strongest negative genetic correlation with neuroticism. Agreeableness assays show how someone relates with other people, that is, how trusting one is or how likely to find fault in others. This trait was the most negatively correlated with neuroticism and irritability as well as MDD, anxiety and manic symptoms. Conscientiousness items relate to discipline and thoroughness, with specific questions being ‘are you lazy’ and ‘does a thorough job’. This trait was most closely associated with ‘types of physical activity in last 4 weeks: ‘heavy do-it-yourself (DIY)’. Finally, openness 10-item Big Five Inventory (BFI-10) items assay imagination and artistic interest. Openness was positively associated with extraversion and risk taking in our data. Educational attainment was positively correlated with openness and negatively associated with neuroticism, while the other three personality traits showed essentially no such overlap (Fig. 5). Since these are self-reported items, they naturally reflect one’s own assessment of one’s personality traits, which might filter actual traits and behaviour through a lens of how one wishes to appear or be perceived.
Using these GWAS summary statistics, with excellent power for neuroticism and moderate power for the other traits, we investigated the heritability of the different personality traits and studied genetic correlations among them using LDSC. SNP-based heritability for all five personality traits in EUR were statistically significant. Out of all the personality pairs studied, the strongest relationship was a negative genetic correlation observed between neuroticism and agreeableness (rG = −0.51, Fig. 1b). Examining the genetic correlations of the five personality traits with 1,437 external traits including depression (neuroticism rG = 0.68 and agreeableness rG = −0.35), manic behaviour (neuroticism rG = 0.44 and agreeableness rG = −0.35), anxiety (neuroticism rG = 080 and agreeableness rG = −0.33) and irritability (neuroticism rG = 0.70 and agreeableness rG = −0.62) further reflected a pattern of opposing relationships between these traits (Fig. 5 and Supplementary Tables 16–20). We also calculated local genetic correlations between personality pairs using LAVA, which helped in identifying the genomic regions playing roles in differential overlap in the genetic architecture of personality. This analysis identified several regions where the effect direction differed from the whole genome genetic correlation.
The MVP, our discovery dataset, is one of the world’s largest biobanks and is a valuable resource for genetic studies. Some previously published personality trait studies had significant contribution from UKB data. It is important to quantify the heterogeneity in these independent cohorts and the different definitions of personality phenotype within each. We investigated the genetic correlation between traits defined on the basis of different inventories (BFI-10, EPQ-RS and NEO-FFI) of personality ascertainment with different cohorts, namely MVP, UKB (part of Nagel et al. study) and GPC, respectively. For neuroticism, Nagel et al. and MVP studies showed a high rG value of 0.80 making these two independent cohorts suitable for meta-analysis (Supplementary Table 1). Similarly, for extraversion, NEO-FFI and two-item inventories showed high rG of 0.89 in the extraversion data of GPC and MVP studies. While for agreeableness, openness and conscientiousness, the rGs between MVP and GPC cohort were lower (0.63–0.72); this may be due to the small size of the GPC dataset for these traits and the correspondingly large standard errors around the point estimate. The point estimate is not necessarily biased in any particular direction, we only mean there is uncertainty. This limitation will be addressed by future GPC studies with larger sample sizes. No novel loci were identified in the meta-analysis with GPC for these traits.
TWAS revealed common genes with changes in gene expression but with opposite direction of effect for some personality traits. A study by Ward et al. in 2020 reported five of these genes (Supplementary Table 3) as eQTLs showing significant associations with mood instability28. This is further supported by the local genetic correlation studies (Supplementary Sheet 5) where we found genomic region 45883902-47516224 on chromosome 17, which harbours genes KANSL1-AS1, MAPT and MAPT-IT1, showing negative local genetic correlation between neuroticism and extraversion with a ρ value of −0.57 and r2 value of 0.32.
rs1876829, which maps to CRHR-Intronic Transcript 1, emerged as the lead SNP (P = 7.872 × 10−39) for neuroticism in the GWAS analysis. We also found multiple eQTL SNPs in this genomic region (rs8072451, rs17689471, rs173365 and rs11012) for the CRHR1 gene to be significantly associated (P value ranging from 1 × 10−5 to 1 × 10−37). The TWAS analysis showed significant association of this gene with neuroticism in nervous system tissues including caudate basal ganglia, frontal cortex, hippocampus and spinal cord cervical region. CRHR1 encodes the receptor of corticotropin-releasing hormone family, which are major regulators of the hypothalamic–pituitary–adrenal pathway29. Genetic variation in the corticotropin-releasing hormone system has been linked to several psychiatric illnesses30. Another study reported hypermethylation at corticotropin-releasing hormone-associated CpG site, cg19035496, in individuals with high general psychiatric risk score for disorders such as depression, anxiety, post-traumatic stress disorder and obsessive compulsive disorder31. Further, a study by Gelernter et al. found that CRHR1 significantly associated with re-experiencing post-traaumatic stress disorder symptoms32 and also maximum habitual alcohol intake33. This gene is also involved in hippocampal neurogenesis30, while reduced hippocampal activation is associated with elevated neuroticism34. This makes CRHR1 a good lead candidate to be followed in future studies to understand the molecular processes impacted by genetic variation underlying a range of psychiatric traits including neuroticism.
While gene expression associations give a wide array of information on the involvement of different genes regulating the different biological processes underlying the biology of traits, searching protein expression associations confers several advantages, as proteins are the final implementers in the functioning of all cells for many biological processes. Through PWAS studies, we found 47 proteins showing significant association with neuroticism in the dorsolateral prefrontal cortex. The PWAS analysis also identified leucine-rich repeat and fibronectin type III domain-containing 5 (LRFN5) protein association with neuroticism, and this protein is also involved in synapse formation. This protein has shown higher levels in patients with MDD and has been suggested as a potential MDD biomarker35.
Examples of genes for which we found converging evidence in neuroticism for transcript and protein-level associations with neuroticism include low-density lipoprotein receptor-related protein 4 (LRP4), syntaxin 4 (STX4) and metabolism of cobalamin associated B (MMAB) (Supplementary Table 31). LRP4 has diverse roles in neuromuscular junctions and in disorders of the nervous system, including Alzheimer’s disease and amyotrophic lateral sclerosis36, STX4 is implicated in synaptic growth and plasticity37, and MMAB, which catalyses the final step in the conversion of cobalamin (vitamin B12) into adenosylcobalamin (biologically active coenzyme B12), all of which have broad implications for brain function, including those in relation to methylmalonic acidaemia38. Low levels of plasma vitamin B12 have been found to be associated with higher depression cases in multiple studies39.
We investigated the relationship of these personality traits with other psychiatric traits, cognitive functions and disorders in a broad phenome-wide scan of genetic correlations with 1,437 traits. A total of 325 traits showed significant genetic correlations with at least one of the five personality traits following multiple testing correction. Two important traits that had some of the strongest associations were MDD and anxiety. Whereas the association of neuroticism with depression and anxiety has been previously considered4,13, our analysis revealed that another personality trait, agreeableness, is also strongly associated with both anxiety and depression but in the opposite direction to neuroticism, showing a potential protective relationship. MR indicated a strong bidirectional causal relationship between neuroticism with anxiety and depression, while showing a bidirectional protective relationship for agreeableness for both traits. Variance explained for neuroticism was attenuated upon conditioning for MDD but remained significant, indicating some independent genetic component for neuroticism despite the strong overlap. Similar, but with a less strong effect, was seen of anxiety on neuroticism, which may be partly due to lower power of available anxiety summary statistics. Larger studies of anxiety disorders are needed to better understand this relationship. Conversely, when we conditioned on agreeableness, for MDD and anxiety we observed a nominal but non-significant change in SNP-based heritability. We conducted MR to further discern these patterns and it showed bidirectional causal effects with neuroticism, confirming a high degree of inter-relatedness between the traits. Given the high degree of genetic overlap between trait neuroticism and the expectation of personality trait expression preceding age of onset for MDD, a high trait neuroticism may be considered an early risk factor for anxiety, depressive and related psychopathology. Indeed, studies have shown persistent elevated neuroticism through adolescence is a risk factor for later susceptibility to anxiety and MDD diagnosis40.
Personality phenotyping in The MVP sample were done using self-report for the short BFI-10 inventory. As such, data are relatively sparse compared with more robust instruments and do not have more in-depth features such as facets found in the NEO inventory. The nature of large biobank studies such as the MVP comes with a crucial advantage in recruitment and sample size, but comes with the sacrifice of deep phenotyping. Future studies that compare findings from more deeply phenotyped samples to more sparse phenotyping used by the MVP would be valuable to address this limitation. Additionally, while we greatly expand on the amount of data available for agreeableness, conscientiousness, openness and extraversion, they still lag behind what has been accomplished for neuroticism. This means genetic instruments defined for the other four traits may lack the precision available for neuroticism. Larger samples still need to be collected to better understand these other traits.
Personality traits are known to have complex interactions with other human behaviours. In this work we have conducted comprehensive genomic studies of personality traits. We performed a GWAS in the MVP sample, the largest and most diverse biobank in the world, in both EUR and AFR to better understand genetic factors underlying personality traits. We combined this information with previously published results in a large meta-analysis, identifying novel genetic associations with five personality traits studied. We identified interactions in a phenome-wide genetic correlation analysis, finding novel relationships between complex traits. We used in silico analysis techniques to identify genetic overlap and causal relationships with depression and anxiety disorders. We also characterized underlying biology using predicted changes in gene and protein expression, biological pathway enrichment and drug perturbation analysis. These results substantially enhance our knowledge of the genetic basis of personality traits and their relationship to psychopathology.
Methods
Inclusion and ethics statement
This research was not restricted or prohibited in the setting of any of the included researchers. All studies were approved by local institutional research boards and ethics review committees. MVP was approved by the Veterans Affairs central institutional research board. We do not believe our results will result in stigmatization, incrimination, discrimination or personal risk to participants.
Cohort and phenotype
We used data release version 4 of the MVP41. The BFI-10 was included as part of a self-report Lifestyle survey provided to MVP participants, with two items for each of the personality traits (Supplementary Fig. 3). For the MVP EUR participants, the mean age was ~65.5 years for each of the five traits and 8% of the sample was female. For MVP AFR, the mean age was ~60.6 years for each trait while 14.0% of the sample was female.
Genotyping and imputation
Genotyping and imputation of MVP subjects has been described previously41,42. A customized Affymetrix Axiom Array was used for genotyping. MVP genotype data for biallelic SNPs were imputed using Minimac443 and a reference panel from the African Genome Resources panel by the Sanger Institute. Indels and complex variants were imputed independently using the 1000 Genomes phase 3 panel44 and merged in an approach similar to that employed by the UKB. Ancestry group assignment within the MVP has been previously described45. Briefly, designation of broad ancestries was based on genetic assignment with comparison to 1000 Genomes reference panels44. Principal components to be used as covariates were generated within each assigned broad ancestral group.
GWAS and meta-analysis
We performed individual GWAS for each of the five personality traits in the MVP cohort41. The personality information along with genotype data were available for a total of 270,000 individuals with 240,000 EUR and 30,000 AFR. The GWAS was performed separately for each of the traits in the EUR and AFR datasets and the effect values were computed using linear regression.
MVP GWAS was conducted using linear regression in PLINK 2.0 using the first ten principal components, sex and age as covariates46. Variants were excluded if call missingness in the best-guess genotype exceeded 20%. Alleles with minor allele frequency (MAF) <0.1% were excluded. Additionally, only variants with an imputation accuracy of ≥0.6 were retained. After applying all filters, genotype data from 233,204, 235,742, 235,374, 234,880 and 220,015 participants were included for neuroticism, extraversion, agreeableness, conscientiousness and openness, respectively.
For meta-analysis, summary statistics generated in this study (referred to as MVP study) were combined using METAL17 with that from Nagel et al. and GPC phase I and II studies (Fig. 1a) based on the availability of data for respective traits. The z-scores of variants provided in the summary statistics were converted into β scores47. The inverse variance weighing scheme of METAL was applied to weight the effect sizes of SNPs from the different source studies. For neuroticism, summary statistics from MVP and Nagel et al. studies13 (excluding 23 and Me) were combined, increasing the total sample size to 682,688. For extraversion, summary statistics from MVP and GPC phase II study10 were combined, while summary statistics from MVP and GPC phase I study8 were combined for the respective meta-analysis of agreeableness, openness and conscientiousness. GPC data were already included in the neuroticism meta-analysis of Nagel et al.
The independent GWS loci for each of the personality traits were identified by clumping all SNPs using PLINK v1.9 software48. P value cut-off of 5 × 10−8, MAF >0.0001, distance cut-off of 1 MB and r2 < 0.1 were used to define the lead SNPs using the 1000 Genomes phase 3 European reference panel44. The genes are mapped for the identified lead SNPs using biomaRt package in R49. The same parameters were used to define novel independent loci for comparison from the Nagel et al.13 and Becker et al.14 summary statistics (excluding 23 and Me).
Trans-ancestry analysis
Trans-ancestry analysis for each of the five personality phenotypes was performed by combining their respective summary statistics from AFR and EUR analyses using METAL17. As with the EUR-only meta-analysis, the inverse variance weighing scheme of METAL was applied to weight the effect sizes of SNPs from the two ancestries. We identified independent SNPs in the same manner as described above for the ancestry-stratified GWAS.
LDSC and SNP heritability
LDSC was performed based on the linkage disequilibrium reference from the 1000 Genomes data for all EUR cohorts and SNP heritability for each of the five personality traits was calculated50. To investigate the relation among the different personality traits, the LDSC-based correlation was also calculated between each pair of traits51. LDSC was also used to calculate genetic correlation of the personality traits with multiple other phenotypes (1,437 traits) with the Complex Traits Genetics Virtual Lab webtool22. A P value cut-off of 6.9 × 10−6 (0.05/(1437 × 5)) was applied to filter the significant correlating pair of traits after multiple test correction.
For MVP AFR, linkage disequilibrium scores were computed from the approximately 123,000 AFR individuals’ genotype data in the MVP cohort using covariant LDSC software21. This linkage disequilibrium reference panel was then utilized to calculate SNP heritability in the MVP AFR cohort using LDSC.
Local genetic correlations
We used LAVA23 to calculate local heritability for the five personality traits and local genetic correlations for each pair. The genome was divided into 2,495 genomic chunks/loci to attain minimum linkage disequilibrium between them and maintain an approximate equal size of around 1 MB. The local heritability of each of the five personality traits was calculated for each of the 2,495 loci. For a given personality trait pair, local genetic correlations were calculated only for pairs that had significant local heritability (Bonferroni-corrected P value at 5% false discovery rate (FDR)) for both traits of the pair. Bonferroni multiple testing correction was also applied to genetic correlated P value to consider significant correlated pairs.
TWAS
FUSION software18 was used to perform TWAS. FUSION first estimates the SNP heritability of steady-state gene and uses the nominally significant (P < 0.05) genes for training the predictive models. The predictive model with significant out-of-sample R2 (>0.01) and nominal P < 0.05 in the five-fold cross-validation was then used for the predictions in the GWAS data. The process is performed for all five personality EUR GWAS data with 10,386 unique genes spanning over the 13 selected tissues. The expression weight panels for 13 a priori selected tissues were taken from GTEx v819. We selected the different available brain tissues and whole blood as the tissues of interest, where Bonferroni corrections at FDR <0.05 were applied with the 10,386 genes test for the 13 tissues to find the genes with significant hits (P < 3.703 × 10−7).
PWAS
We performed PWAS to test the association between genetically regulated protein expression and different personality traits individually using FUSION software18. The weights for genetic effect on protein expression for the PWAS were from the Wingo et al. study52. In the PWAS, we integrated the protein weights with the summary statistics from the GWAS of each of the personality traits, respectively. Next, to decrease the probability of linkage contributing to the significant association in the PWAS, we performed colocalization analysis using COLOC53. In COLOC, we determined if the genetic variants that regulate protein expression colocalize with the GWAS variants for the personality trait. Significant proteins in the PWAS that also have COLOC posterior probability of hypothesis 4 (PP4) >50% have a higher probability of being consistent with a causal role in the personality trait of interest.
Fine-mapping
To identify likely causal variants, we performed variant fine-mapping using Polyfun software24. Since the fine-mapping was performed on the same EUR data, SNP-specific prior causal probabilities were taken directly from the pre-computed causal probabilities of 19 million imputed UKB SNPs with MAF >0.01 based on 15 UKB traits analysis. The fine-mapping was performed on the GWAS sumstats for each of the five personality traits. SuSiE54 was used to map the posterior causal probabilities of the SNPs. The SNPs with posterior inclusion probability (PIP) value >0.95 were considered as significant for neuroticism, while a more relaxed cut-off of PIP >0.80 was used for other four personality traits to avoid loss of causal variant information due to the relatively less power in their respective datasets.
Conditional analysis
Conditional analysis was performed to investigate the possible mediating effects between depression or anxiety and neuroticism or agreeableness. Neuroticism meta-data GWAS summary statistics were used and conditioned on MDD and anxiety in individual runs. The MDD summary statistics were from Levey et al. study55 and include a meta-analysis from the MVP, UKB, PGC and FinnGen. The anxiety summary statistics were taken from Levey et al. study42. With depression/anxiety studies as covariate traits, the conditional analysis of neuroticism (target trait) was carried out using multi-trait-based conditional and joint analysis utility of genome-wide complex trait analysis56. Similarly, the same method was used to perform conditional analysis of agreeableness on MDD and anxiety.
Drug perturbation analysis
FUMA was used to carry out the MAGMA-based gene-association tests to find significantly associated genes for a trait from its GWAS summary data15. Drugs were searched for both neuroticism and MDD individually using their respective significantly associated genes derived from neuroticism meta-analysis summary statistics and MDD GWAS summary statistics from the Levey et al. summary statistics. To predict drug candidates for a given trait, significant genes associated with neuroticism/depression were given as input to gene2drug R-package25. Pre-computed Pathway Expression Profiles of the Connectivity Map data were taken from Drug Set Enrichment Analysis (DSEA) website. For each query gene, a maximum of five predicted drugs were predicted. Further, the drugs showing an E score >0.5 and a P value less than 1 × 10−6 were considered significant. The process was repeated for MDD.
MR
MR was performed to study the causal relationship between four pairs of traits: neuroticism and MDD, neuroticism and anxiety, agreeableness and MDD, and agreeableness and anxiety. These traits had the highest genetic correlation. The summary statistics described previously for conditional analysis for all four traits were used for carrying out MR analysis as well. TwoSample MR package was used to perform the MR57. For each pair of traits, the TwoSample MR was run twice to see the effect of exposure of each of the two traits on the outcome of the other. After harmonizing the exposure and outcome instruments sets, clumping of SNPs (distance of 500 kb, r2 = 0.05) was performed before conducting the MR analysis. Because some of our samples included in the analysis of personality overlap with our outcomes and exposures of interest, and a TwoSample MR is not robust to sample overlap, we also performed a sensitivity analysis for each trait pair using the MRlap package26. MRlap is specifically designed to account for many assumptions of MR, including sample overlap. It first calculates observed MR-based effect values and then a corrected effect value by using the genetic covariance calculated by LDSC.
Out-sample polygenic risk score prediction
The Yale–Penn cohort includes participants recruited from sites in the eastern United States58. A total of 11,705 participants completed the 240-item revised NEO PI-R, which assesses the domains of the five-factor model of personality: neuroticism, extraversion, openness to experience, agreeableness and conscientiousness59. Each domain has six facets. For example, the facets of neuroticism are anxiety, angry hostility, depression, self-consciousness, impulsiveness and vulnerability. Each item is rated on a five-point scale. Of the Yale–Penn participants with a NEO score, 4,582 were assigned to the broadly defined EUR group using the same methods as in the MVP sample and were unrelated. We used PRS-CS, Python software that uses Bayesian regression and continuous shrinkage priors, to calculate posterior effect sizes per SNP60. The 1000 Genomes linkage disequilibrium reference panel was used. The training datasets were summary statistics from the EUR meta-analysis for each of the five personality factors. The target dataset was a PLINK-formatted binary file set containing genotype information from the Yale–Penn participants48. Once score per SNP was generated by PRS-CS and PLINK was used to generate a score for each individual by summing SNP effect48. The lm (linear model) function in R was used to regress NEO PI-R scores on PRS, using age, sex and the first ten within-ancestry principal components as covariates61.
Ethics oversight
Research involving MVP in general is approved by the Veterans Affairs Central institutional research board; the current project was also approved by institutional research boards in West Haven, CT.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Figs. 1–3, Tables 1–4 and VA Million Veteran Program core acknowledgement.
Thirty-one supplementary tables.
Acknowledgements
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award no. 5IK2BX005058. This publication does not represent the views of the Department of Veteran Affairs or the United States Government. A.W. was supported by a BLRD CDT award from the US Department of Veterans Affairs no. 1IK4BX005219 and grant I01 BX005686. A.W. and T.W. were supported by R01 grant no. AG072120. J.G. was supported by US Department of Veterans Affairs grant 5I01CX001849-04 and NIH grants R01DA037974 and R01DA058862. H.K. was supported by US Department of Veterans Affairs grant I01 BX004820 and the VISN4 Mental Illness Research, Education and Clinical Center of the Crescenz VAMC. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Detailed MVP Core team acknowledgements are included in the supplement.
Author contributions
D.F.L. and P.G. designed the study. P.G. and D.F.L. drafted the manuscript. J.G. and M.B.S. provided ongoing feedback and refinement of the analytical plan, as well as early feedback on the drafted manuscript. D.F.L. and P.G. conducted GWAS on included cohorts. D.F.L. and P.G. discussed, created and refined the phenotype in the MVP. P.G. and M.G. discussed and refined MVP analytic plans. P.G. and Y.L. conducted TWAS and PWAS analysis with guidance from A.W., T.W. and D.F.L. S.B conducted out-sample PRS into the Yale–Penn cohorts with guidance from J.G. and H.R.K. P.G. D.F.L., M.G., S.B., Y.L., A.W., T.W. and K.A. conducted original analyses. D.F.L., T.W. and A.W. supervised original analyses. All authors critically evaluated and revised the manuscript.
Peer review
Peer review information
Nature Human Behaviour thanks Robert Krueger, Aysu Okbay and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All MVP summary statistics are made available through dbGAP request at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v11.p1. Meta-analysis summary statistics are available through the Levey lab website at https://medicine.yale.edu/lab/leveylab/data/. Meta-analysis data will also be made available via the Complex Trait Genetics Virtual Lab at https://vl.genoma.io/.
Code availability
No custom code was developed for analyses in this manuscript. All code used is cited and described in the methods. Software versions are accessible via PLINK v1.9 at https://www.cog-genomics.org/plink/1.9/, PLINK v2.0 at https://www.cog-genomics.org/plink/2.0/ and Polyfun: version 1.0.0 SuSiE package version: 0.11.92.
Competing interests
H.R.K. is a member of advisory boards for Dicerna Pharmaceuticals, Sophrosyne Pharmaceuticals, Clearmind Medicine and Enthion Pharmaceuticals; a consultant to Sobrera Pharmaceuticals; the recipient of research funding and medication supplies for an investigator-initiated study from Alkermes; and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the past 3 years by Alkermes, Dicerna, Ethypharm, Lundbeck, Mitsubishi and Otsuka. J.G. and H.R.K. are holders of US patent 10,900,082 titled: ‘Genotype-guided dosing of opioid agonists’, issued 26 January 2021. J.G. is paid for editorial work on the journal Complex Psychiatry. The remaining authors declare no competing interests. J.G. is named as an inventor on PCT patent application no. 15/878,640 entitled ‘Genotype-guided dosing of opioid agonists’, filed 24 January 2018, and issued on 26 January 2021, as US patent no. 10900082. M.B.S. has stock options in Oxeia Biopharmaceuticals and EpiVario. He has been paid for his editorial work on Depression and Anxiety (Editor-in-Chief), Biological Psychiatry (Deputy Editor) and UpToDate (Co-Editor-in-Chief for Psychiatry). No other authors report competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Daniel F. Levey, Email: daniel.levey@yale.edu
VA Million Veteran Program:
Priya Gupta, Marco Galimberti, Sarah Beck, Henry R. Kranzler, Murray B. Stein, Joel Gelernter, and Daniel F. Levey
Supplementary information
The online version contains supplementary material available at 10.1038/s41562-024-01951-3.
References
- 1.John, O. P. & Srivastava, S. in Handbook of Personality: Theory and Research (eds Pervin, L. A. and John, O. P.) (Guilford Press, 1999).
- 2.McCrae, R. R. & Costa, P. T. Personality in Adulthood: A Five-factor Theory Perspective (Guilford Press, 2003).
- 3.Kendler, K. S. & Myers, J. The genetic and environmental relationship between major depression and the five-factor model of personality. Psychol. Med.40, 801–806 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Hettema, J. M. et al. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am. J. Psychiatry163, 857–864 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Hettema, J. M., Prescott, C. A. & Kendler, K. S. Genetic and environmental sources of covariation between generalized anxiety disorder and neuroticism. Am. J. Psychiatry161, 1581–1587 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Van Os, J. & Jones, P. B. Neuroticism as a risk factor for schizophrenia. Psychol. Med31, 1129–1134 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Smeland, O. B. et al. Identification of genetic loci shared between schizophrenia and the Big Five personality traits. Sci. Rep.7, 2222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Moor, M. H. et al. Meta-analysis of genome-wide association studies for personality. Mol. Psychiatry17, 337–349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Moor, M. H. et al. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry72, 642–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Berg, S. M. et al. Meta-analysis of genome-wide association studies for extraversion: findings from the genetics of personality consortium. Behav. Genet.46, 170–182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo, M. T. et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat. Genet.49, 152–156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet.48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagel, M. et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet.50, 920–927 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Becker, J. et al. Resource profile and user guide of the Polygenic Index Repository. Nat. Hum. Behav.5, 1744–1758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe, K. et al. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Leeuw, C. A. et al. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol.11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gusev, A. et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet.48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium, G. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramoz, N. et al. Corticotropin releasing hormone receptor CRHR1 gene is associated with tianeptine antidepressant response in a large sample of outpatients from real-life settings. Transl. Psychiatry10, 378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo, Y. et al. Estimating heritability and its enrichment in tissue-specific gene sets in admixed populations. Hum. Mol. Genet.30, 1521–1534 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuéllar-Partida, G. et al. Complex-Traits Genetics Virtual Lab: a community-driven web platform for post-GWAS analyses. Preprint at bioRxiv10.1101/518027 (2019).
- 23.Werme, J. et al. An integrated framework for local genetic correlation analysis. Nat. Genet.54, 274–282 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Weissbrod, O. et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet.52, 1355–1363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Napolitano, F. et al. gene2drug: a computational tool for pathway-based rational drug repositioning. Bioinformatics34, 1498–1505 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Mounier, N. & Kutalik, Z. Bias correction for inverse variance weighting Mendelian randomization. Genet. Epidemiol.47, 314–331 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Gelernter, J. et al. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry19, 41–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward, J. et al. The genomic basis of mood instability: identification of 46 loci in 363,705 UK Biobank participants, genetic correlation with psychiatric disorders, and association with gene expression and function. Mol. Psychiatry25, 3091–3099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman, J. P. et al. Regulation of the hypothalamic–pituitary–adrenocortical stress response. Compr. Physiol.6, 603–621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koutmani, Y. et al. CRH promotes the neurogenic activity of neural stem cells in the adult hippocampus. Cell Rep.29, 932–945 e7 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Jokinen, J. et al. Epigenetic changes in the CRH gene are related to severity of suicide attempt and a general psychiatric risk score in adolescents. EBioMedicine27, 123–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelernter, J. et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat. Neurosci.22, 1394–1401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelernter, J. et al. Genome-wide association study of maximum habitual alcohol intake in >140,000 U.S. European and African American veterans yields novel risk loci. Biol. Psychiatry86, 365–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magal, N., Hendler, T. & Admon, R. Is neuroticism really bad for you? Dynamics in personality and limbic reactivity prior to, during and following real-life combat stress. Neurobiol. Stress15, 100361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, K. et al. LRFN5 and OLFM4 as novel potential biomarkers for major depressive disorder: a pilot study. Transl. Psychiatry13, 188 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePew, A. T. & Mosca, T. J. Conservation and innovation: versatile roles for LRP4 in nervous system development. J. Dev. Biol.9, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris, K. P. et al. The postsynaptic t-SNARE Syntaxin 4 controls traffic of Neuroligin 1 and Synaptotagmin 4 to regulate retrograde signaling. eLife5, e13881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, T. et al. Methylmalonic acidemia: neurodevelopment and neuroimaging. Front. Neurosci.17, 1110942 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sangle, P. et al. Vitamin B12 supplementation: preventing onset and improving prognosis of depression. Cureus12, e11169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aldinger, M. et al. Neuroticism developmental courses—implications for depression, anxiety and everyday emotional experience; a prospective study from adolescence to young adulthood. BMC Psychiatry14, 210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaziano, J. M. et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol.70, 214–223 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Levey, D. F. et al. Reproducible genetic risk loci for anxiety: results from approximately 200,000 participants in the Million Veteran Program. Am. J. Psychiatry177, 223–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das, S. et al. Next-generation genotype imputation service and methods. Nat. Genet.48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genomes Project, C. et al. A global reference for human genetic variation. Nature526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, H. et al. Multi-ancestry study of the genetics of problematic alcohol use in over 1 million individuals. Nat. Med.29, 3184–3192 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, Z. et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet.48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durinck, S. et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics21, 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet.47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wingo, T. S. et al. Shared mechanisms across the major psychiatric and neurodegenerative diseases. Nat. Commun.13, 4314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet.10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, G. et al. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B82, 1273–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levey, D. F. et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat. Neurosci.24, 954–963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, Z. et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun.9, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherva, R. et al. Genome-wide association study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry73, 472–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa, P. T. Jr. & McCrae, R. R. in The SAGE Handbook of Personality Theory and Assessment Vol. 2 (eds Boyle, G. M. G. J. & Saklofske, D. H.) 179–198 (Sage Publications, 2008).
- 60.Ge, T. et al. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun.10, 1776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R: A Language and Environment for Statistical Computing (R Core Team, 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–3, Tables 1–4 and VA Million Veteran Program core acknowledgement.
Thirty-one supplementary tables.
Data Availability Statement
All MVP summary statistics are made available through dbGAP request at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001672.v11.p1. Meta-analysis summary statistics are available through the Levey lab website at https://medicine.yale.edu/lab/leveylab/data/. Meta-analysis data will also be made available via the Complex Trait Genetics Virtual Lab at https://vl.genoma.io/.
No custom code was developed for analyses in this manuscript. All code used is cited and described in the methods. Software versions are accessible via PLINK v1.9 at https://www.cog-genomics.org/plink/1.9/, PLINK v2.0 at https://www.cog-genomics.org/plink/2.0/ and Polyfun: version 1.0.0 SuSiE package version: 0.11.92.