Abstract
Carya illinoinensis is an economically important nut tree, and its chloroplast (cp.) genome has been reported; however, its mitochondrial (mt) genome remains unknown. In the present study, we assembled the first mt genome of C. illinoinensis. The circular mt genome of C. illinoinensis is 495,205 bp long, with 37 protein-coding genes(PCGs), 24 tRNA genes, and 3 rRNA genes. All the tRNAs could be folded into typical cloverleaf secondary structures, with lengths of 58–88 bp. A conserved U-U-C-x-A-x2 consensus nucleotide sequence was discovered in the Ψ–loops of tRNA sequences. In addition, 447 dispersed repeats were detected, as well as found 482 RNA editing sites and 9,960 codons in the mt genome. Furthermore, a total of 27 DNA sequences with a length of 43,277 bp were transferred from the cp. to the mt genome, and eight integrated cp-derived genes (trnL-CAA, trnV-GAC, trnD-GUC, trnW-CCA, trnN-GUU, trnH-GUG, trnM-CAU, and rps7) were identified. We also obtained 1,086 hits, including 364.023 kp of nuclear genome sequences, that were transferred to the mt genome. To determine the evolutionary position of C. illinoinensis, we conducted a phylogenetic analysis of the mitogenomes of C. illinoinensis and 14 other taxa. The results strongly suggested that C. illinoinensis and Fagus sylvatica formed a single clade with 100% bootstrap support. This study sequenced comprehensive data on the C. illinoinensis mitochondrial genome and provided insights into the conserved sequences of tRNA genes, which could facilitate evolutionary research in other Carya trees in the future.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75324-1.
Keywords: Carya illinoinensis, Mitochondrial genome, TRNA, RNA editing, Phylogenetic analysis
Subject terms: Evolution, Genetics, Plant sciences
Introduction
The mitochondrion is a semiautonomous eukaryotic organelle that participates in energy conversion, biosynthesis, and signal transduction in living cells1. The animal mt genome is about 16 to 17 kb long and forms a single circular assembly molecule2. In contrast, the plant mitogenome is more highly complex and diverse in terms of size, structure, gene content, and gene order3. The size of plant mitogenomes varies greatly, ranging from 66 kb (Viscum scurruloideum)4 to 11,300 kb (Silene conica)5, with most genomes between 200 and 800 kb in length6. The gene content of plant mitogenomes also varies considerably, ranging from 32 to 67 genes, and some genes, such as those encoding NADH dehydrogenase, ATP synthase, ubiquinol cytochrome, and cytochrome c biogenesis, are highly conserved7. For most seed plants, nuclear genetic information is inherited from both parents, whereas the DNA of cp. and mt are derived from maternal genes8. This genetic mechanism makes it easier to study genetics because the genetic information comes from the maternal line9. In addition, recent studies have reported intergenomic gene transfer among the nuclear, plastid, and mt genomes, which is a common phenomenon in plant evolution10,11.
C. illinoinensis, commonly known as pecan, belongs to the Juglandaceae family and is the most valuable nut tree native to North America. It is widely distributed and can tolerate various environmental conditions (between 30° N and 42° N)12. The pecan is commercially produced in New Mexico, Georgia, Louisiana, and Texas, as well as Mexico13. It was introduced to China at the end of the 19th century. In recent years, pecan has been proven to be suitable for planting in southern areas of the Yangtze River valley, including Jiangsu, Anhui, and Zhejiang Provinces, and is widely grown in China14,15. In comparison with most other nuts, pecans contain high quantities of unsaturated fatty acids and high levels of antioxidants, as well as a series of phytochemicals such as phenolic compounds16. Pecans are also a rich source of dietary fibre, protein, minerals, and vitamins17. Recently, the cp. genomes of C. illinoinensis cv. pawnee18, C. illinoinensis cv. Wichita19, C. illinoinensis cv. 87MX3-2.11, and C. illinoinensis cv. Lakota20 were identified, and nuclear genome sequencing has been performed in C. illinoinensis21. The sequencing of more cp. genomes and genomes will facilitate the identification of genetic variations, and provide new insights into the study of interspecific relationships of pecans. However, to date, there have been no reports on the mt genomes of any species.
In this study, we aimed to assemble the full mt genome of C. illinoinensis via a combination of third-generation sequencing and second-generation sequencing techniques. After the mt genome was assembled, the secondary structure and conservation of tRNAs were identified. The repeat sequences, synonymous codon use, RNA editing, DNA transfer, and phylogenetic relationships were also analysed. These results may help to better elucidate the features of the C. illinoinensis mt genome.
Results
Genomic features of the C. illinoinensis mt genome
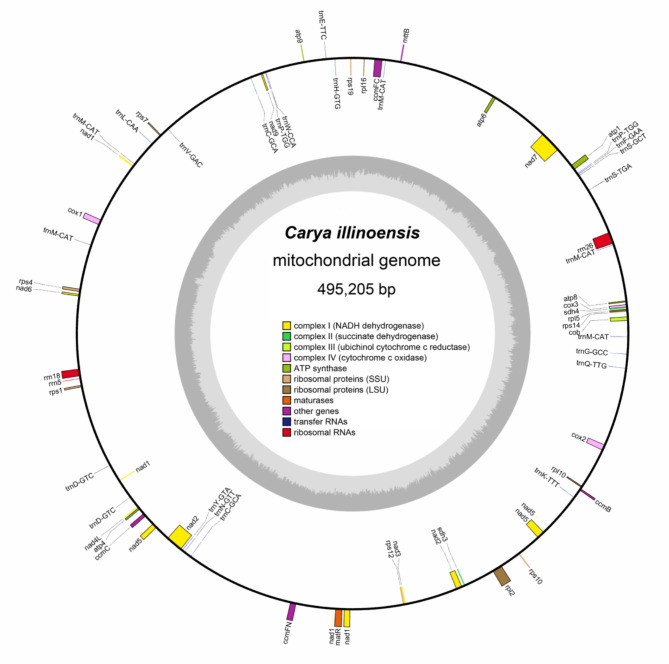
The C. illinoinensis genome sequence was submitted to the GenBank database (accession number PRJNA824975). We assembled and annotated a high-quality mitogenome for C. illinoinensis via second- and third-generation sequencing methods. The Illumina sequencing yielded 19,113,015 reads, with a minimum coverage depth of 52x and an average of 355.4x across the entire genome (Figure S1, and Tables S1). For the Nanopore sequencing, we obtained 1,629,169 reads, with an average read length of 6,007 bp and an N50 of 10,756 bp (Figure S2, and Tables S2). The C. illinoinensis mt genome is circular with a length of 495,205 bp (Fig. 1). The nucleotide composition of the mt genome was 27.32% A, 27.70% T, 22.52% G, and 22.46% C, and the GC content was 44.98% (Table S3). There were 64 genes annotated in the mt genome, including 37 PCGs, 24 tRNA genes, and 3 rRNA genes. PCGs, tRNAs, and rRNAs made up 6.11%, 0.36%, and 1.12% of the total mt genome, respectively.
Fig. 1.
The circular map of C. illinoinensis mt genome. Genes positioned on the exterior and interior of the circle are transcribed in a clockwise and counterclockwise direction, respectively. The dark gray area in the inner ring indicates the GC content. The different color blocks inside represent different functional gene groups.
The C. illinoinensis mt genome encodes 37 different proteins, which can be divided into 10 categories (Table 1). The start codon of all PCGs was ATG, and the use rates of the TAA, TGA, and TAG stop codons were quite different. The use rates of TAA, TGA, and TAG were 54.05% (20/37), 32.43% (12/37), and 13.51% (5/37), respectively, with the stop codon TAA being the most prevalent. In addition, 10 intron-containing genes were identified in the mt genome of C. illinoinensis, among which the ccmFC, cox2, rps2, rps19, and rps4 genes included one intron; nad4 contained three introns; and nad1, nad2, nad5, and nad7 included four introns.
Table 1.
Gene profile and organization of C. Illinoinensis mitogenome.
| Group of genes | Gene name | Length | Start codon | Stop codon | Amino acids |
|---|---|---|---|---|---|
| NADH dehydrogenase | nad1**** | 978 | ATG | TAA | 326 |
| nad2**** | 1467 | ATG | TAA | 489 | |
| nad3 | 357 | ATG | TAA | 119 | |
| nad4*** | 1488 | ATG | TGA | 496 | |
| nad4L | 273 | ATG | TAA | 91 | |
| nad5***** | 2013 | ATG | TAA | 671 | |
| nad6 | 618 | ATG | TAA | 206 | |
| nad7**** | 1185 | ATG | TAG | 395 | |
| nad9 | 573 | ATG | TAA | 191 | |
| ATP synthase | atp1 | 1530 | ATG | TGA | 510 |
| atp4 | 597 | ATG | TAG | 199 | |
| atp6 | 951 | ATG | TAA | 317 | |
| atp8 | 480 | ATG | TAA | 160 | |
| atp9 | 258 | ATG | TAA | 86 | |
| Cytohrome c biogenesis | ccmB | 615 | ATG | TGA | 205 |
| ccmC | 1047 | ATG | TGA | 349 | |
| ccmFC* | 1317 | ATG | TGA | 439 | |
| ccmFN | 1734 | ATG | TGA | 578 | |
| Cytochrome c oxidase | cox1 | 1584 | ATG | TAA | 528 |
| cox2* | 783 | ATG | TAA | 261 | |
| cox3 | 798 | ATG | TGA | 266 | |
| Ubichinol cytochrome c reductase | cob | 1182 | ATG | TGA | 394 |
| Maturases | matR | 1971 | ATG | TAG | 657 |
| Transport membrance protein | mttB | 348 | ATG | TAG | 116 |
| Ribosomal proteins (LSU) | rpl10 | 489 | ATG | TAA | 163 |
| rpl16 | 249 | ATG | TAA | 83 | |
| rpl2* | 999 | ATG | TAA | 333 | |
| rpl5 | 552 | ATG | TAA | 184 | |
| Ribosomal proteins (SSU) | rps1 | 606 | ATG | TAA | 202 |
| rps10 | 330 | ATG | TAA | 110 | |
| rps12 | 378 | ATG | TGA | 126 | |
| rps14 | 261 | ATG | TAG | 87 | |
| rps19* | 297 | ATG | TGA | 99 | |
| rps4 | 825 | ATG | TAA | 275 | |
| rps7* | 432 | ATG | TAA | 144 | |
| Succinate dehydrogenase | sdh3 | 318 | ATG | TGA | 106 |
| sdh4 | 387 | ATG | TGA | 129 | |
| Ribosomal RNAs | rrn18 | 2050 | |||
| rrn26 | 3396 | ||||
| rrn5 | 117 | ||||
| Transfer RNAs | trnC-GCA(2) | (73, 71) | |||
| trnD-GTC(2) | (74, 58) | ||||
| trnE-TTC | 72 | ||||
| trnF-GAA | 74 | ||||
| trnG-GCC | 72 | ||||
| trnH-GTG | 74 | ||||
| trnK-TTT | 73 | ||||
| trnL-CAA | 71 | ||||
| trnM-CAT(5) | (74, 74, 74, 73, 73) | ||||
| trnN-GTT | 72 | ||||
| trnP-TGG(2) | (75, 74) | ||||
| trnQ-TTG | 72 | ||||
| trnS-GCT | 88 | ||||
| trnS-TGA | 87 | ||||
| trnV-GAC | 72 | ||||
| trnW-CCA | 74 | ||||
| trnY-GTA | 83 |
Notes: The numbers after the gene names indicate the duplication number, and the superscripts *,***, and****represent one, three and four introns contained, respectively.
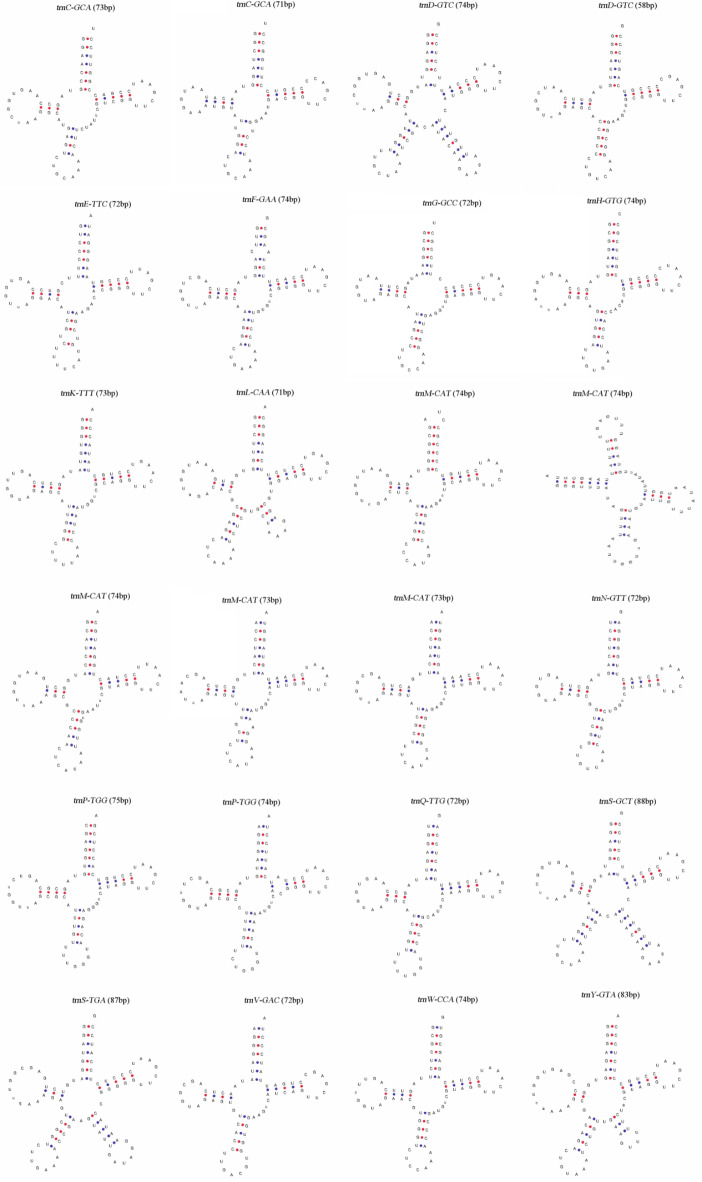
Conservation sequences of the tRNA gene secondary structures
Twenty-four distinct tRNA genes were found in the C. illinoinensis mt genome. All the tRNA genes were involved in the transport of the 20 amino acids, suggesting that two or more tRNAs might transport the same amino acid to different codons. For example, trnS-GCT and trnS-TGA are associated with the synonymous codons GCU and GCA, which are involved in the transportation of serine. All the tRNAs could be folded into typical cloverleaf secondary structures and possessed an acceptor arm, anticodon arm, anticodon loop, D-arm, D-loop, Ψ-arm, and Ψ-loop (Fig. 2). We observed that tRNAs trnD-GTC and trnM-CAT had different structures, and that five tRNAs (trnD-GTC, trnL-CAA, trnS-GCT, trnS-TGA, and trnY-CTA) possessed an additional variable region that formed a stem loop.
Fig. 2.
Structure of C. illinoinensis mt genome tRNAs.
The 24 tRNA sequences of the pecan mt genome were analysed for consensus bases (Table 2), and no conserved sequences were found in the acceptor arm, D-arm, or D-loop. However, the first positions of the acceptor arm and D-arm were both G nucleotides, accounting for 75% and 83.33%, respectively. The first and end positions of the D-loop were mostly A nucleotides, accounting for 75% and 95.83%, respectively. The number of base pairs in the acceptor arm was 6 and 7; most D-arms had 3 bp and 4 bp, and only trnY-GTA had 2 bp. The number of bases contained in the D-loop was 7–11 bp and 13 bp; the highest proportion of bases was 9 bp (25%), and the lowest was 13 bp (0.04%). The number of base pairs in the anticodon arm and anticodon loops was relatively stable. The anticodon arm was mainly 5 bp in length, accounting for 91.67%, and the remaining 4 bp, accounted for only 8.33%. The anticodon loops all had 7 bp, and the common sequence mainly appeared in the last two positions, which were A-A nucleotides, accounting for 70.83%. Each Ψ-arm had 5 bp, the last two positions of which were mainly G-G nucleotides, except for trnV-GAC, which ended in A-G nucleotides. Each Ψ-arm had 5 bp, and the last two positions were mainly G-G nucleotides, except for trnV-GAC nucleotides, which ended in A-G nucleotides. The position of the Ψ-loop was the most conserved, and all the Ψ-loops were 7 bp. The first three base pair combinations were U-U-C nucleotides, and the fifth position was an A nucleotide. The conserved region was U-U-C-x-A-x2 nucleotides in the Ψ-loops of the C. illinoinensis mt genome.
Table 2.
Nucleotide sequence in mitogenome tRNA of C. Illinoinensis.
| TRNA | Acceptor arm | D-arm | D-loop | Anti-codon arm | Anti-codon loops | Ψ-arm w | Ψ-loop | Variable region |
|---|---|---|---|---|---|---|---|---|
| trnC-GCA | GGAACCG | GCC | AAGUGGCUAA | GAGU | CUGCAAA | GUCGG | UUCGAAU | |
| trnC-GCA | GGCUAGG | ACAU | AAUGGAA | UUGGA | CUGCAAA | GACGG | UUCGACC | |
| trnD-GTC | GGAGGUA | GCU | GAGUGGCUUAA | UUGGU | UUGCUAA | AUGGG | UUCGAAU | AUACAA/GAAGA |
| trnD-GTC | GGGAUUG | GUUC | AAUCGGUCA | CCGCC | CUGUCAA | GCGGG | UUCGAGC | |
| trnE-TTC | GUCCCUU | GUCC | AGUGGGUUA | UCGUC | UUUUCAU | ACGGG | UUCGAUU | |
| trnF-GAA | GUUCAGG | GCUC | AGCUGGUUA | AAGGA | CUGAAAA | AGUGG | UUCGAAU | |
| trnG-GCC | GCGGAA | GCUU | AAUGGUA | UAGCC | UUGCCAA | GAGGG | UUCAAGU | |
| trnG-GTG | GCGGAUG | GCC | AAGUGGAUCAA | GUGGA | UUCUGAA | GCGGG | UUCAAUC | |
| trnK-TTT | GGGUGUA | GCUC | AGUUGGUA | UUGGG | CUUUUAA | GCAGG | UUCGAGU | |
| trnL-CAA | GCCUUGG | GUG | AAAUGGUAGA | CGAGA | CUCAAAA | GGAGG | UUCGAGU | GCU/AAAG |
| trnM-CAT | GCGGGG | GAG | GAAUUGGUCGA | UCAGG | CCCAUGA | GCAGG | UUCGAAU | |
| trnM-CAT | GGGCUUA | GUUU | AAUUGGUUG | ACCG | CUCAUAA | GUAGG | UUCGAGC | |
| trnM-CAT | GCAUCCA | GCU | GAUGGUUAA | CCCAA | CUCAUAA | GUAGG | UUCAAUU | |
| trnM-CAT | ACCUACU | GCUC | AGCAAUUA | UUGCU | CUCAUAA | AUUGG | UUCAAAU | |
| trnM-CAT | ACCUACU | ACUC | AGCGGUUA | UCGCU | UUCAUAC | AUUGG | UUCAAAU | |
| trnN-GTT | UCCUCAG | GCUC | AGUGGUA | GUCGG | CUGUUAA | GUAGG | UUCAAAU | |
| trnP-TGG | CGAGGUG | GCGC | AGUCUGGUCA | UCUGU | UUUGGGU | AUAGG | UUCGAAU | |
| trnP-TGG | AGGGAUG | GCGC | AGCUUGGUA | UUUGU | UCUGGGU | ACGGG | UUCCAAU | |
| trnQ-TTG | UGGAGUA | GCC | AAGUGGUAA | UCGGU | UUUUGGU | AAAGG | UUCGAAU | |
| trnS-GCT | GGAGGUA | GCU | GAGUGGCUUAA | UUGGU | UUGCUAA | AUGGG | UUCGAAU | AUACAA/GAAGA |
| trnS-TGA | GGAUGGA | UCU | GAGCGGUUGAA | UCGGU | CUUGAAA | GGGGG | UUCGAAU | GUAUU/GAUAGG |
| trnV-GAC | AGGGAUA | ACUC | AGCGGUA | UCACC | UUGACGU | AUCAG | UUCGAGC | |
| trnW-CCA | GCGCUCU | GUUC | AGUUCGGUA | UGGGU | CUCCAAA | GUAGG | UUCAAAU | |
| trnY-GTA | GGGAGAG | GC | CGAGUGGUCAAAA | ACAGA | CUGUAAA | GUAGG | UUCGAAU | GAA/CUUU |
A total of 6 bp mismatches were observed in the 24 tRNAs. Three mismatches occurred in the anticodon, specifically, U-U (twice) and G-A mismatches, and the other three occurred in the codon, U-U (twice) and C-A mismatches.
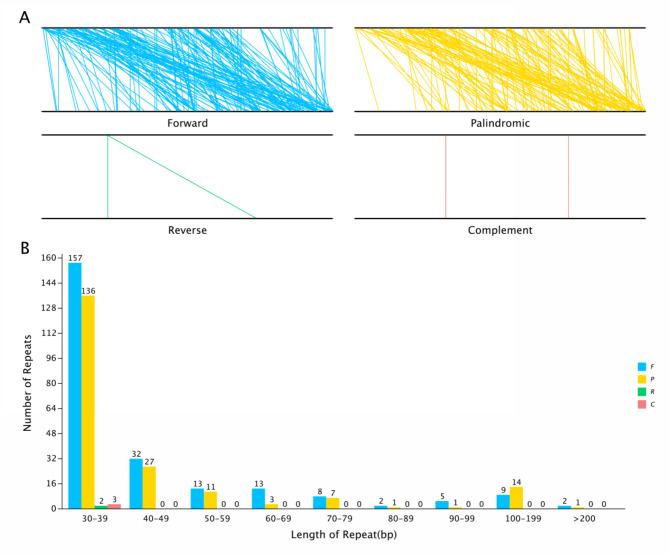
Repeat sequence analysis
Dispersed sequences are repetitive sequences scattered throughout the genome. In the present study, a total of 447 dispersed repeats were identified in the C. illinoinensis mt genome, including 241 forward (53.91%), 201 palindromic (44.97%), 2 reverse (0.45%), and 3 complementary (0.67%) repeats. The distribution of the dispersed repeats is shown in Fig. 3. Most repeats were 30–39 bp long (298 repeats, 64.88%); however, three sequences were longer than 200 bp. Most of the repeats were concentrated in intergenic spacers (IGSs), and the remaining repeats were located in coding regions.
Fig. 3.
The dispersed repeat sequences in the C. illinoinensis mt genome. (A) The four different types of repeats are distributed in the genome; the mt genome is represented by the two black lines, and the line segments are linked to the same repeats. (B) Distribution of lengths of dispersed repeats in the mt genome. The X-axis shows the type of dispersed repeats; the Y-axis shows the number of scattered repeats.
SSRs are DNA fragments with lengths of 1–6 bp. Unique SSR markers are excellent tools for intraspecific population genetic variation research, interspecific evolutionary studies, and identification studies22,23. In this study, a total of 432 SSRs were detected in the C. illinoinensis mitogenome, including 162 (37.50%) monomers, 189 (43.75%) dimers, 22 (5.09%) trimers, 54 (12.50%) tetramers, and 18 (1.16%) pentamer repeats (Table S4). Among all the SSRs, more than 81% were monomeric or dimeric repeats. Further analysis of SSR repeat units revealed that 87.65% of the monomers had A/T contents and that 66.48% of the dinucleotide repeats were AT/TA or AG/TC.
Codon preference analysis
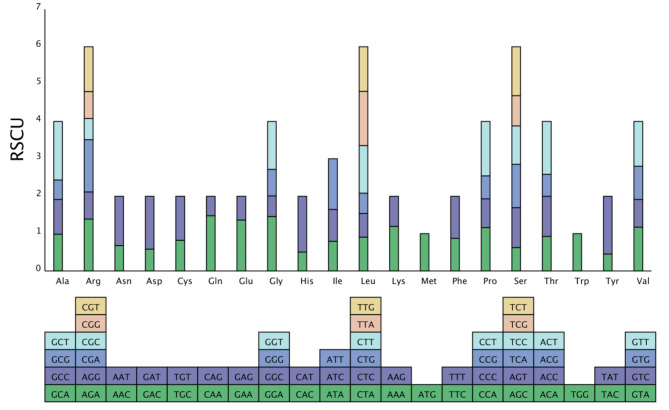
In C. illinoinensis, 9,960 amino acids are encoded. The most frequently used amino acids were Ser (905, 9.09%), Leu (834, 8.37%), and Ile (750, 7.3%), and the least common amino acids were Trp (1.50%) and Cys (1.36%) (Fig. 4). Owing to the degeneracy of codons, each amino acid is encoded by more than one codon (synonymous codon) in organisms24. The utilization rate of codons varies greatly among different species; this inequality of codons is called relative synonymous codon usage (RSCU). The RSCU is thought to be the result of natural selection in organisms, and amino acids are thought to preferentially use codons whose RSCU higher than 125. Codon preference analysis was performed on 37 unique PCGs of C. illinoinensis mt, and the codon usage of individual amino acids is shown in Table S5. The results revealed that all the genes were encoded by 9,960 codons, and 64 different codons encoded the 20 amino acids. The most frequently used codons were UUU (Phe), AUU (Ile), and UUC (Phe), which were used 373 (3.74%), 338 (3.39%), and 289 (2.90%) times, respectively. There were 31 codons (one stop codon) with an RSCU > 1, indicating that the usage frequency of these codons was greater than that of other synonymous codons. Among these codons, 27 ended with the A/U base, accounting for 87.10% (27/31), suggesting that high-frequency codons tend to end in A/U bases.
Fig. 4.
Relative synonymous codon usage (RSCU) in the C. illinoinensis mt genome. The X-axis shows the various amino acids and codon families; the Y-axis shows RSCU values. The boxes below represent all the codons that encode each amino acid, and the height of the top column represents the sum of all the codon RSCU values.
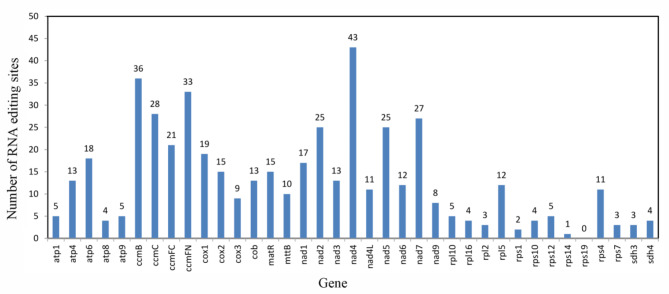
Prediction of RNA editing sites
RNA editing is a posttranscriptional process that converts specific cytidines to uridines in the cp. and mt genomes of land plants26. This process is necessary for gene expression, as it increases protein conservation among plants by modifying codons. In this study, 482 RNA editing sites in 37 PCGs were predicted in the mt genome of C. illinoinensis (Table 3). Among those PCGs, only 1 gene (rps19) encoded none of the RNA editing sites, and 36 genes had RNA editing sites. The nad4 encoded the most RNA editing sites (43 sites, 8.92%), followed by the ccmB gene, which had 36 RNA editing sites. The rps14 and rps1 genes had the lowest number of RNA editing events, with only one and two editing sites, respectively (Fig. 5). Among those sites, 69.29% (334 sites) were located at the second position of the triplet codon, 30.71% (148 sites) occurred in the first position of the codon, and none were locate at the predicted third base position.
Table 3.
Prediction of RNA editing sites.
| Type | RNA-editing | Number | Percentage |
|---|---|---|---|
| Hydrophobic | CTC(L) = > TTC(F) | 7 | 33.20% |
| CTT(L) = > TTT(F) | 10 | ||
| CCA(P) = > CTA(L) | 50 | ||
| CCC(P) = > CTC(L) | 8 | ||
| CCC (P) = > TTC (F) | 6 | ||
| CCG (P) = > CTG (L) | 33 | ||
| CCT (P) = > TTT (F) | 10 | ||
| CCT(P) = > CTT(L) | 25 | ||
| GCA (A) = > GTA (V) | 1 | ||
| GCG (A) = > GTG (V) | 7 | ||
| GCT (A) = > GTT (V) | 3 | ||
| Hydrophilic | CGC (R) = > TGC (C) | 9 | 12.86% |
| CGT (R) = > TGT (C) | 28 | ||
| CAC (H) = > TAC (Y) | 9 | ||
| CAT (H) = > TAT (Y) | 16 | ||
| Hydrophobic-hydrophilic | CCA(P) = > TCA(S) | 9 | 7.68% |
| CCT (P) = > TCT (S) | 16 | ||
| CCC (P) = > TCC (S) | 9 | ||
| CCG(P) = > TCG(S) | 3 | ||
| Hydrophilic-hydrophobic | TCA (S) = > TTA (L) | 70 | 45.85% |
| TCC (S) = > TTC (F) | 27 | ||
| TCG (S) = > TTG (L) | 41 | ||
| TCT (S) = > TTT (F) | 36 | ||
| ACA (T) = > ATA (I) | 4 | ||
| ACC (T) = > ATC (I) | 2 | ||
| ACG (T) = > ATG (M) | 6 | ||
| ACT (T) = > ATT (I) | 5 | ||
| CGG(R) = > TGG(W) | 30 | ||
| Hydrophilic- stop | CGA (R) = > TGA (X) | 1 | 0.41% |
| CAA (Q) = > TAA (X) | 1 |
Fig. 5.
Distribution of RNA-editing sites in the C. illinoinensis mt PCGs. The X-axis shows the name of genes; the Y-axis shows the number of RNA-editing sites.
RNA editing results in diverse start and stop codons, which might lead to the premature termination of PCGs27. Additionally, further analysis revealed that 45.85% (221 sites) of the RNA edited amino acids were converted from a hydrophilic to a hydrophobic amino acid, 33.20% (160 sites) from a hydrophobic to another hydrophobic amino acid, 12.86% (62 sites) from a hydrophilic to another hydrophilic amino acid, and 7.68% (37 sites) from a hydrophobic to a hydrophilic. Only two amino acids, glutamine and arginine, were converted to a stop codon. Among these amino acids, most tended to be converted from proline to leucine (24.27%, 117 sites), serine to leucine (213.03%, 111 sites), or serine to phenylalanine (13.07%, 63 sites). The remaining 191 RNA editing sites were distributed in other RNA editing types, including arginine to cysteine, proline to serine, arginine to tryptophan, histidine to tyrosine, leucine to phenylalanine, proline to phenylalanine, alanine to valine, threonine to isoleucine, threonine to methionine, and glutamine and arginine to X, where X represents a stop codon. The results revealed that amino acids tended to be leucine after RNA editing, which was supported by the fact that 47.30% (228 sites) of the edits were converted to leucine.
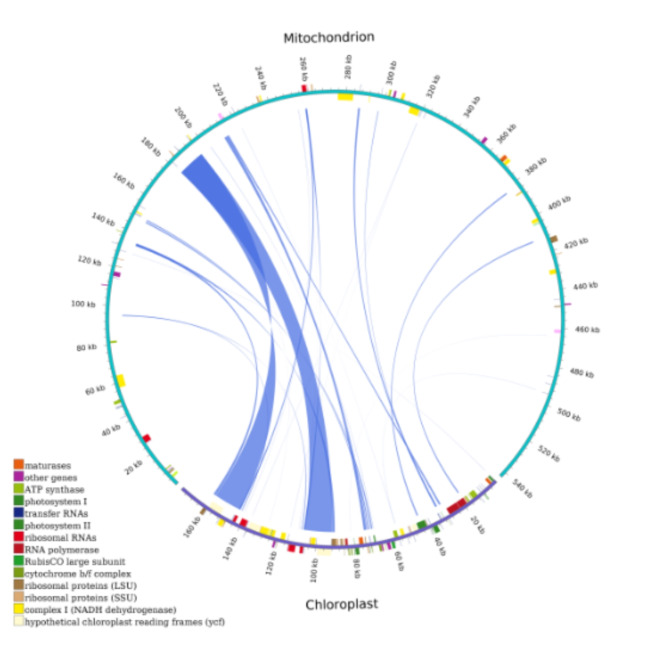
DNA migration among cp., mt, and nuclear
The C. illinoinensis mitogenome sequence (495,205 bp) is approximately 3.08 times longer than its cp. genome (160,819 bp)28. A total of 27 fragments with a length of 43,277 bp, accounting for 8.74% of the mitogenome, migrated from the cp. to the mt genome of C. illinoinensis (Fig. 6). The homologous fragments varied widely, with the shortest being 39 bp and the longest being 15,012 bp. Eight integrated cp-derived genes were located on these fragments, including seven tRNA genes and one PCG gene, namely, trnL-CAA, trnV-GAC, trnD-GUC, trnW-CCA, trnN-GUU, trnH-GUG, trnM-CAU, and rps7. The data also revealed that some PCGs, such as rpl23, rpl2, psaB, rpoC2, and psbE, migrated from the cp. to the mitogenome. However, most of these genes lost their integrity during evolution, and only partial sequences of these genes can be found in the mitogenome (Table 4). The different completeness levels of the transferred PCGs and tRNA genes suggested that tRNA genes were much more conserved in the mt genome than PCGs, indicating that tRNAs play an indispensable role in mitochondria.
Fig. 6.
DNA transfer between mt and mcp genomes in C. illinoinensis. The graph displays the entire genomes of mt and cp. in cyan and purple, respectively. The different color blocks represent different functional gene groups.
Table 4.
Fragment transferred from cp. To mt in C. Illinoinensis.
| Fragments | Length (bp) | start | end | Cp genes | Mt genes |
|---|---|---|---|---|---|
| 1 | 15,012 | 91,918 | 106,928 | rpl23 (partical: 6.38%), trnI-CAU, ycf2, ycf15, trnL-CAA, ndhb, rps7, trnA-UGC, trnV-GAC, rrn16S (partical: 42.32%) | trnV-GAC, rps7, trnL-CAA, trnM-CAU |
| 2 | 15,012 | 14,3934 | 158,944 | rrn16S (partical: 42.32%), trnV-GAC, trnA-UGC, rps7, ndhb, trnL-CAA, ycf15, ycf2, trnI-CAU, rpl23 (partical: 6.38%) | trnV-GAC, rps7, trnL-CAA, trnM-CAU |
| 3 | 1,379 | 159,052 | 160,430 | rpl23 (partical: 55.67%), rpl2 (partical: 79.05%) | ORF |
| 4 | 1,379 | 90,432 | 91,810 | rpl2 (partical:79.05%); rpl23 (partical: 55.67%) | ORF |
| 5 | 1,470 | 73,194 | 74,643 | rpl33, rps18, rpl20 | ORF |
| 6 | 999 | 32,610 | 33,607 | trnD-GUC | trnD-GUC |
| 7 | 936 | 30,335 | 31,270 | petN | ORF |
| 8 | 684 | 18,580 | 19,263 | rpoC2 (partical: 16.29%) | ORF |
| 9 | 726 | 44,030 | 44,755 | psaB (partical: 6.53%); psaA (partical: 24.72%) | ORF |
| 10 | 425 | 142,348 | 142,772 | ORF | ORF |
| 11 | 425 | 108,090 | 108,514 | ORF | ORF |
| 12 | 458 | 72,531 | 72,988 | psaJ | ORF |
| 13 | 777 | 69,916 | 70,670 | psbE (partical: 51.59%) | ORF |
| 14 | 640 | 71,428 | 72,059 | petG, petL, trnW-CCA | trnW-CCA |
| 15 | 889 | 143,488 | 144,351 | rrn16S (partical: 57.95%) | rrn18 (partical: 41.90%) |
| 16 | 889 | 106,511 | 107,374 | rrn16S (partical: 57.95%) | rrn18 (partical: 41.90%) |
| 17 | 147 | 69,242 | 69,388 | psbJ | ORF |
| 18 | 83 | 136,237 | 136,319 | trnN-GUU | trnN-GUU |
| 19 | 83 | 114,543 | 114,625 | trnN-GUU | trnN-GUU |
| 20 | 368 | 33,198 | 33,556 | trnD-GUC (partical: 89.19%) | trnD-GUC |
| 21 | 86 | 20 | 105 | trnH-GUG | trnH-GUG |
| 22 | 78 | 57,284 | 57,361 | trnM-CAU | trnM-CAU |
| 23 | 88 | 140,924 | 141,011 | ORF | ORF |
| 24 | 88 | 109,851 | 109,938 | ORF | ORF |
| 25 | 77 | 57,284 | 57,360 | trnM-CAU | trnM-CAU |
| 26 | 40 | 81,667 | 81,706 | ORF | ORF |
| 27 | 39 | 126,557 | 126,595 | ndhA (partical: 1.71%) | ORF |
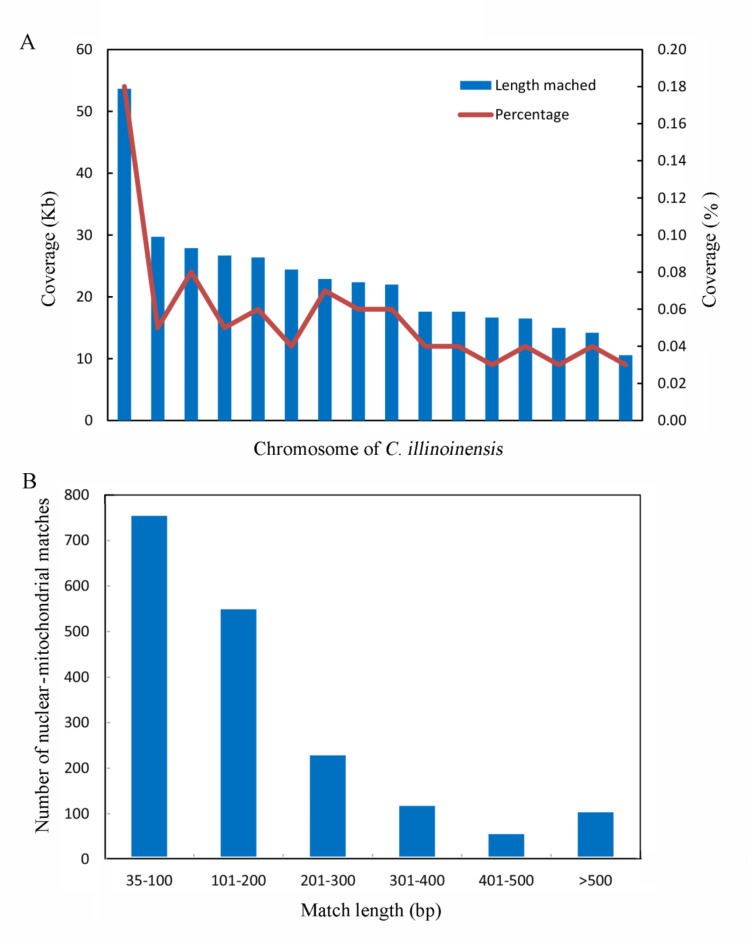
The C. illinoinensis mt genome was searched against its available nuclear genome, and 1,086 hits were obtained, including 364.023 kp of nuclear genome sequences that were transferred to the mt genome. The mitochondrial–nuclear alignment showed that hits occurred on every chromosome (Fig. 7A). However, the total length of the hits and the percent coverage on every chromosome were different. Chromosome 16 had the maximum total length of hits (53.643 kb) and the highest coverage (0.18%). In contrast, chromosomes 5, 7, and 8 had the lowest coverage (0.03%). In addition, the fragment lengths were mainly between 35 bp and 200 bp (Fig. 7B), and the largest fragment was 15.012 kb in length on chromosome 16, with a homology of 98%. Interestingly, we found that most of the homologous sequences (19/27) of the cp. and mt genes were located on chromosome 16 (Table S6), indicating that the exchange of genetic material between the organelles and the nuclear genome of C. illinoinensis occurred mainly on chromosome 16.
Fig. 7.
Characteristics of mt and nuclear homologous sequences in C. illinoinensis. A. The percentage distribution between shared mt and nuclear matches. Blue boxes show the number of matches. The red lines represent the coverage of matches on mt and nuclear genomes. B The length distribution between shared mt and nuclear matches .
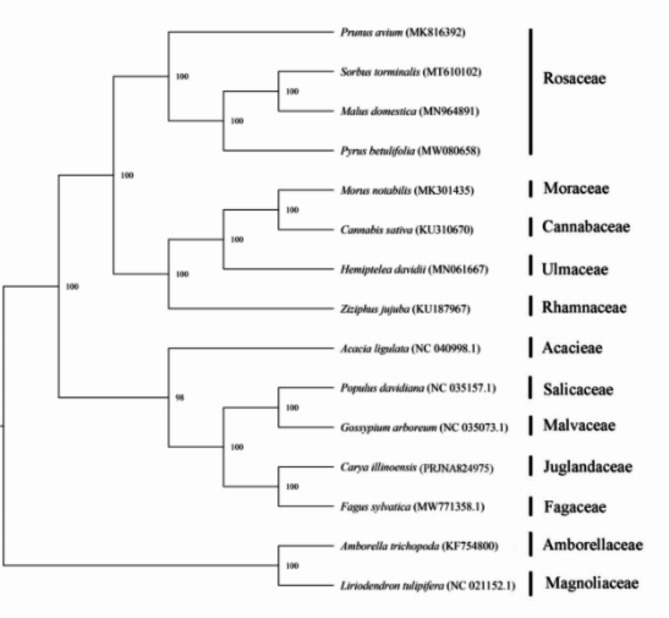
Phylogenetic analysis
To understand the evolution of C. illinoinensis, we downloaded 14 plant mitogenomes from GenBank (https://www.ncbi.nlm.nih.gov/genome/browse/) and constructed a phylogenetic tree based on 36 conserved mitochondrial PCGs. As shown in Fig. 8, all the nodes in the generated tree had bootstrap support values greater than 98%, including 12 nodes with 100% support. The phylogenetic tree strongly suggested (100% bootstrap support) the close phylogenetic relationship between C. illinoinensis and Fagus sylvatica, and these two plants belong to the order Magnoliales, and the family Lauraceae. Overall, the results of our analysis of the mitogenomes provide a valuable foundation for future analyses of the phylogenetic affinities of Carya species.
Fig. 8.
The phylogenetic relationships of C. illinoinensis.
Discussion
Compared with those of animals, Plant mitochondria have more complex structures because they have variable genome sizes, multiple types of repeated sequences, and a large number of noncoding regions29,30. The rapid development of next-generation sequencing technology has accelerated the release of complex mt genomes, including Acer truncatum (2022)31, Salix wilsonii (2022)32, Momordica charantia (2023)33, and Apostasia shenzhenica (2023)34. In this study, for the first time, we described the basic characteristics of the C. illinoinensis mt genome. These findings provide important information for understanding the function, inheritance, and evolutionary relationships of the mt genome. The C. illinoinensis mt genome was a circular sequence with a length of 495,205 bp and a 44.98% GC content. The GC content was comparable to that of other sequenced plant mitogenomes, for example, A. truncatum (45.68%)31, S. glauca (44.07%)9, and Beta vulgaris (45.68%)35, but higher than that of the C. illinoinensis cp. genome (36.15%)18. The GC content is an the important component of different genomic regions, and variation in the GC content can be used to understand the evolution of genomes36. In addition, most sequences are noncoding in the C. illinoinensis mitogenome, and PCGs account for only 6.11%, which is probably due to the frequent recombination of repeated sequences and the integration of foreign sequences in the mitogenome during evolution.
Usually, tRNAs are composed of 70–100 nucleotides and are commonly found in all organisms. The nucleotide sequence of a tRNA forms a cloverleaf secondary structure through hydrogen bonds and then folds into an L-shaped tertiary structure37. This study predicted that all the pecan mitochondrial tRNA genes had a typical cloverleaf structure, and the results showed that trnD-GTC and trnM-CAT possessed different structures, and trnD-GTC, trnL-CAA, trnS-GCT, trnS-TGA, and trnY-CTA had an additional variable region. Owing to the particularity of the plant mitochondrial genome, analysing its tRNA genes can help in understanding its molecular composition, conservation, evolutionary history, and other information38. Previous studies reported that there was a conserved nucleotide sequence in tRNA, which was limited to the Ψ-loop39. This study revealed that the Ψ-loops of the tRNAs in pecan mitochondria was also the most conserved, with 7 nucleotides conserved. The conserved sequence could be summarized as U-U-C-x-A-x2, with a common sequence U-U-C in the Ψ-loop. In other regions of the tRNA, no completely conserved sequences were found, only some more conserved nucleotides. The first nucleotide of the tRNA in the acceptor arm was mostly the G nucleotide, and the first nucleotide with the highest frequency in the D-arm was also the G nucleotide. The first position in the D-loop was usually the A nucleotide, and the last position is usually the A nucleotide. In the anticodon loop, the last two positions were dominated by A-A nucleotides. The last two positions of the Ψ-arm were mainly G-G nucleotides.
The repeat sequences are potentially important markers for population and evolutionary analyses40. Repeats in mt are essential for intermolecular recombination, which can contribute to extreme mitogenome sizes and structural variations6,41. In this study, dispersed repeats and SSR loci were intensively investigated. A total of 447 dispersed repeats were identified in the C. illinoinensis mitogenome, and 357 repeats were less than 50 bp long, accounting for 2.47% of its genome. The maximum length of the repeats was 335 bp, and the repeats were not of medium or large size. This finding suggests that intermolecular recombination is less frequent in the mitogenome34. We also detected 432 SSRs in the C. illinoinensis mitogenome; among these SSRs, the number of monomeric and dimeric repeats was the greatest. SSRs containing AT/TA repeat motifs are more likely to appear in the cp. genome of Carya18,42, as well as in the mt genome.
RNA editing is a posttranscriptional process that can alter genetic information at the mRNA level in the mt genomes of higher plants, resulting in more efficient protein folding7. In this study, 482 RNA editing sites were identified in 37 PCGs of the C. illinoinensis mt genome. Among the codon transfer types, TCA = > TTA was the most common, with 70 editing sites, followed by CCA = > CTA, with 50 editing sites. After RNA editing, 7.68% of the hydrophobic amino acids became hydrophilic, and 45.85% of the hydrophilic amino acids became hydrophobic. Consistent results were found in the genomes of both Bupleurum chinense35 and Diospyros oleifera43, where the most abundant transfer type in this plant was TCA = > TTA, which had been edited to change the hydrophobicity of more than half of the amino acids. In previous studies, RNA edits that occurred at the second position of a codon accounted for more than half of the total edits9. In the C. illinoinensis mt genome, 69.29% of the editing sites were also located at the second-position base of the triplet codon. In addition, the selection of mt genome editing sites in C. illinoinensis showed a strong bias, with C-T editing being the most common type of editing, except for one T-C editing, which is the most popular editing type in plant mt genomes according to several studies35,44. After RNA editing, some of the encoded amino acids became stop codons (TAA, TAG, and TGA). In the C. illinoinensis mt genome, two codons (CGA = > TGA, CAA = > TAA) were edited to generate a stop codon, which resulted in the coding process being stopped prematurely, thereby altering the function of the related gene.
Information on DNA transfer events between different genomes (mt, cp., and nuclear) has been obtained by sequencing analysis in many plants45,46. Previous studies revealed that DNA transfer events occur primarily from organelle genomes to the nuclear genome in angiosperms, followed by transfer from the nuclear genome and plastid genome to the mitogenome31,47–49. The transfer of DNA sequences among the cp. and mt genomes has been frequently observed in the mitogenome50. In many cases, the cp. DNA content in the mt genomes of most plants is 3–6%, sometimes as high as approximately 10% 51. In this study, a total length of 43,277 bp, was found to be transferred from the cp. genome to the mt genome, accounting for 8.74% of the mitogenome, which was greater than the mt genome lengths of A. truncatum (2.36%)31, Liriodendron tulipifera (3%)52, and Suaeda glauca (5.18% )9, which is comparable to those of Vitis vinifera (8.8%)53 and less than those of Cucurbita pepo (11.5%)41. The transfer of tRNA genes is most commonly observed in the transfer of DNA fragments from the cp. genome to the mt genome54. A total of 27 homologous fragments transferred from the cp. genome to the mt genome were identified, and these homologous fragments contained 8 integrated genes, 7 of which were tRNA genes. The different levels of integrity of the transferred PCGs and tRNA genes indicated that the tRNA genes were much more conserved in the mt genome, suggesting that they played an indispensable role in the mt genome. The metastases of tRNAs can be traced back to the memory of early horizontal gene transfer events. According to previous studies, cp-derived trnM-CAU first appeared in gymnosperms55; cp-derived trnD-GUC mainly appeared in dicotyledons44; and both trnM-CAU and trnD-GUC were found in the C. illinoinensis mt genome. However, the lack of cp-derived trnA-UGC, which is commonly detected in angiosperms, was lost during the early evolution of terrestrial plants45,51, suggesting that special evolutionary events may have occurred during C. illinoinensis formation.
In high plants, the total length of transferred DNA varies depending on the plant species, ranging from 50 kb (Arabidopsis thaliana) to 1.1 Mb (Oryza sativa subsp. Japonica)56. According to our study, a total of 1,086 hits covering 364.023 kp of nuclear DNA have been transferred into the mitogenome of C. illinoinensis. Although the nuclear–mt transfer process has occurred in every pecan chromosome, the total length of hits and the percent coverage differ. Cheng et al.9 and Ma et al.31 reported similar results in Acer truncatum and Glycine max, respectively. Chromosome 16 had the longest total length (53.643 kb), whereas chromosome 8 had the shortest total length (10.559 kb). The transferred fragment lengths were mainly between 35 bp and 200 bp, and the largest fragment length was 15.012 kb. The transfer from the nucleus to the mt can be ambiguous because of the difficulty in determining the orientation of the transfers9.
In conclusion, we presented the first mt genome assembly of a juglandaceae plant, C. illinoinensis. The C. illinoinensis mitogenome was circular, with a length of 495,205 bp. The conserved U-U-C-x-A-x2 consensus nucleotide sequence was found in the Ψ-loop of the tRNA. Furthermore, sequence repeats, codon preference, and RNA editing were analysed in the mitogenome, and DNA transfer events were detected among the cp., mt, and nuclear genomes. Finally, the evolutionary status of C. illinoinensis was verified by phylogenetic analysis. This study provides insights into the conserved sequences of tRNA genes and the evolution of the C. illinoinensis mitogenome.
Materials and methods
Plant materials and DNA sequencing
The plant materials of the pecan cultivar Xinxuan-4, which were collected from the seedling57 in Jintan District, Changzhou, Jiangsu Province, China (31° 42’ N, 119° 21’ E), were planted at Nanjing Botanical Garden, Jiangsu Province, China, (32° 03’ N, 118° 49’ E). Fresh leaves of Xinxuan-4 were collected and rapidly stored at -80 °C. Total genomic DNA was extracted via the modified CTAB method58.
The samples with good purity were retained for sequencing, following the standard sequencing protocol according to the manufacturer’s instructions (Illumina Inc., San Diego, CA, USA). Library construction was performed via the Truseq Nano DNA HT Sample Preparation Kit (Illumina USA). DNA underwent sonication and fragmentation to achieve a target size of 350 bp, which was subsequently amplified through PCR. Purified PCR products were obtained via the AMPure XP purification kit, and size distribution was assessed with the Agilent 2100 Bioanalyzer. Quantification of the library was performed via real-time PCR. Sequencing was done with paired-end PE-150 bp on the Illumina HiSeq 2500 platform, while the same DNA sample also underwent single-molecule real-time sequencing via Nanopore-based ONT (Oxford Nanopore Technologies). After sequencing, Trimmomatic v0.36 was used to remove low-quality bases and adaptor sequences from the raw Illumina reads59.
After the samples successfully underwent quality control, the third-generation sequencing experiment was conducted. Genomic DNA was randomly fragmented, and large DNA fragments were enriched and purified via magnetic beads. These large fragments were then cut and recovered, with any damage to the fragmented DNA repaired. After purification, end repair was performed on both ends of the DNA fragments, and an A tail was added. The connection reaction utilized the joints from the SQK-LSK109 kit. The constructed DNA library was then quantitatively assessed. Once a library of appropriate concentration was prepared, it was loaded into the flow cell and transferred to the Oxford Nanopore PromethION sequencer for real-time single molecule sequencing. The third-generation sequencing data were filtered via Filtlong v0.2.1 software and analyzed with Perl scripts.
Assembly and annotation of the mt genome
To obtain a high-quality C. illinoinensis mt genome, second-generation data were used fastp v0.20.0 (https://github.com/OpenGene/fastp) software to obtain high-quality reads. The original third-generation data were spliced via Canu assembly software to obtain the contigs60, the parameters setting were “ genome size = 5 m, and correctedErrorRate = 0.03, then the contigs were compared to the plant mt gene database via BLAST v2.6 (https://blast.ncbi. nlm nih gov/ Blast.cgi). The contigs that aligned with mt genes were used as the seed sequences, which were extended and cyclized using the original data to determine the master structure (or subloop) of the ring; then the assembly was performed by NextPolish v1.3.1 61 (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using second- and third-generation data sequencing for correction. The specific parameters were “rerun = 3, -max_depth = 100 ”.
The annotation of the draft mt genome of C. illinoinensis was performed as previously described35. The encoded proteins and rRNAs were annotated via BLASTn searches of the published plant mt sequences at the National Center for Biotechnology Information (NCBI), and further adjustments were made on the basis of closely related species. The tRNAs were annotated via tRNAscanSE62 (http://lowelab.ucsc.edu/tRNAs can-SE/). ORFfinder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to examine open reading frames (ORFs), and OrganellarGenomeDRAW1 (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) was used to construct the mt genome. The software of tRNAScan-SE 2.0 was used to predict the tRNA gene structure63 (http://lowelab.ucsc.edu/tRNAscan-SE).
Analysis of repeat sequences
Repeat structures including the forward (F), reverse (R), complement(C), and palindromic (P) repeats were analysed by vmatch v2.3.0 (http://www.vmatch.de/) software and Perl scripts. The minimum length was set to 30 bp, and the hamming distance was set to 3. A simple repeat sequence (SSR) is a type of tandem repeat sequence with a dozen nucleotides consisting of several nucleotides (usually 1 to 6) as repeat units. The software of the MicroSAtellite identification tool (Misa, https://webblast.ipk-gatersleben.de/misa/)64 was used to analyse the mt SSRs. The parameters used were as follows: mono-nucleotides repeated 8 times; di-nucleotides repeated 5 times; trinucleotides repeated 4 times; and tetra-, penta-, and hexa-nucleotides repeated 3 times.
Synonymous Codon usage analysis
Relative synonymous codon usage (RSCU) was used to characterize the synonymous codon usage with CodonW1.4.4 (http://codonw.sourceforge.net/) of the mt genome, and the R package ggplot2 was used for plotting.
RNA editing analyses
The editing sites in the mt RNA of C. illinoinensis were identified via the mt gene-encoding proteins of plants as reference proteins. Site analysis was conducted via the Plant Predictive RNA Editor (PREP) suite (http://prep.unl.edu/), with a cut-off value of 0.2.
DNA transformation
The genomes (cp. and nuclear) of C. illinoinensis28,65 were downloaded from the NCBI Organelle Genome Resources Database. The homologous fragments in the mt and cp. genomes were identified via BLAST v2.10.1 software. The screening criteria were as follows: ≥ 70%, E-value ≤ 1e−10, and length ≥ 40.
Phylogenetic analysis
The conserved PCGs from the mt genomes of C. illinoinensis and 14 other taxa were used for phylogenetic tree analysis. The 15 mt genomes were downloaded from NCBI, and the conserved PCGs were extracted via TBtool software. The acquired sequences were subsequently aligned via Muscle software with default parameters.Bayesian analysis was conducted via the MrBayesv3.2.7 software, with Markov chain Monte Carlo (MCMC) iterative calculations performed. A total of 1 million iterations were conducted, with sampling every 100 iterations. The results showed that the first 25% of the system tree (burn-in) was removed, and a consensus tree was obtained with the majority of rules agreeing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are very thankful to the editor-in-chief and the reviewer for their suggestions for improvement on this article.
Author contributions
Y.C. designed the experiments. Y.C. and C.F. wrote the manuscript. Y.C. and S. Z. prepared samples and generated the experiments. Y.Z. and W. W. collected and analyzed the data. W.W. and C.Z. provided suggestions and revised the paper. All authors have read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (32001344), the Natural Science Foundation of Jiangsu Province, China (BK20200290)
Data availability
The data were deposited under the NCBI SRA accession PRINA824975 (submission numberSRR18718033) (https://www.ncbi.nlm.nih.gov/search/all/?term=%20SRR18718033).
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The sampling of pecan cultivar Xinxuan-4 is not endangered in China, and no specific permission was required for the collection. The materials in this study were collected in the germplasm resource nursery of the Nanjing Botanical Garden with permission. Our experimental study complied with relevant institutional, national, and international guidelines and legislation. This article does not contain any studies with human participants or animals performed by any of the author.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liuchun Feng, Email: fenglc2021@163.com.
Cancan Zhu, Email: zcc@cnbg.net.
References
- 1.Greiner, S., Lehwark, P. & Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res.47, W59–W64 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picard, M. & Shirihai, O. S. Mitochondrial signal transduction. Cell Metabol.34, 1620–1653 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mower, J. P., Sloan, D. B. & Alverson, A. J. Plant mitochondrial genome diversity: The genomics revolution. in Plant Genome diversity volume 1: Plant Genomes, Their Residents, and Their Evolutionary Dynamics 123–144 (2012).
- 4.Skippington, E., Barkman, T. J., Rice, D. W. & Palmer, J. D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci.112, E3515–E3524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sloan, D. B. et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol.10, e1001241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo, W. et al. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol.33, 1448–1460 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Bi, C., Lu, N., Xu, Y., He, C. & Lu, Z. Characterization and analysis of the mitochondrial genome of common bean (Phaseolus vulgaris) by comparative genomic approaches. Int. J. Mol. Sci.21, 3778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birky, C. W. Jr. Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc. Natl. Acad. Sci.92, 11331–11338 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, Y. et al. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda Glauca. BMC Genom.22, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bock, R. Witnessing genome evolution: Experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu. Rev. Genet.51, 1–22 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Zhao, N., Grover, C. E., Chen, Z., Wendel, J. F. & Hua, J. Intergenomic gene transfer in diploid and allopolyploid Gossypium. BMC Plant Biol.19, 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grauke, L., Wood, B. W. & Harris, M. K. Crop vulnerability: Carya. HortScience51, 653–663 (2016). [Google Scholar]
- 13.Sagaram, M., Lombardini, L. & Grauke, L. Variation in leaf anatomy of pecan cultivars from three ecogeographic locations. J. Am. Soc. Hortic. Sci.132, 592–596 (2007). [Google Scholar]
- 14.Chen, Y. et al. Field investigation of resistance against black spot of different pecan varieties in Jintan, Changzhou. J. Jiangsu Forestry Sci. Technol.45, 26–29 (2018). [Google Scholar]
- 15.Chen, X., Zhu, C., Zhang, S., Lu, X. & Chen, Y. Study on photosynthesis of the pecan under the stress of black spot disease. North. Hortic.7, 40–45 (2023). [Google Scholar]
- 16.Bolling, B. W., Chen, C. Y. O., McKay, D. L. & Blumberg, J. B. Tree nut phytochemicals: composition, antioxidant capacity, bioactivity, impact factors. A systematic review of almonds, brazils, cashews, hazelnuts, macadamias, pecans, pine nuts, pistachios and walnuts. Nutr. Res. Rev.24, 244–275 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Zhu, C., Deng, X. & Shi, F. Evaluation of the antioxidant activity of Chinese Hickory (Carya cathayensis) kernel ethanol extraction. Afr. J. Biotechnol.7, 13 (2008).
- 18.Mo, Z. et al. The chloroplast genome of Carya illinoinensis: Genome structure, adaptive evolution, and phylogenetic analysis. Forests11, 207 (2020). [Google Scholar]
- 19.Feng, G., Mo, Z. H. & Peng, F. R. The complete chloroplast genome sequence of Carya illinoinensis Cv. Wichita and its phylogenetic analysis. Mitochondrial DNA Part B5, 2235–2236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, X. et al. Chloroplast genome sequences of Carya illinoinensis from two distinct geographic populations. Tree. Genet. Genomes16, 48 (2020). [Google Scholar]
- 21.Huang, C. Y., Ayliffe, M. A. & Timmis, J. N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature422, 72–76 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Liu, Q. et al. Comparative chloroplast genome analyses of Avena: Insights into evolutionary dynamics and phylogeny. BMC Plant. Biol.20, 406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh, N., Pal, A. K., Roy, R. K., Tamta, S. & TS, R. Development of cpSSR markers for analysis of genetic diversity in Gladiolus cultivars ScienceDirect. Plant. Gene10, 31–36 (2017). [Google Scholar]
- 24.Wald, N., Alroy, M., Botzman, M. & Margalit, H. Codon usage bias in prokaryotic pyrimidine-ending codons is associated with the degeneracy of the encoded amino acids. Nucleic Acids Res.40, 7074–7083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo, L. H. et al. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 12, e0171264 (2017). [DOI] [PMC free article] [PubMed]
- 26.Raman, G. & Park, S. Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. Longicalyncinus, from a comparative genomics perspective. PLoS ONE10, e0141329 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hia, F. & Takeuchi, O. The effects of codon bias and optimality on mRNA and protein regulation. Cell. Mol. Life Sci.78, 1909–1928 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, Y. et al. Chloroplast genome sequencing of Carya Illinoinensis Cv. Xinxuan-4, a new pecan pollinated cultivar. Fruit Res.4, 1–11 (2024).
- 29.Chevigny, N., Schatz-Daas, D., Lotfi, F. & Gualberto, J. M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci.21, 328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wynn, E. L. & Christensen, A. C. Repeats of unusual size in plant mitochondrial genomes: Identification, incidence and evolution . G3 Genes Genomes Genet.9, 549–559 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, Q. et al. Assembly and comparative analysis of the first complete mitochondrial genome of Acer Truncatum Bunge: a woody oil-tree species producing nervonic acid. BMC Plant Biol.22, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han, F., Qu, Y., Chen, Y., Xu, L. & Bi, C. Assembly and comparative analysis of the complete mitochondrial genome of Salix Wilsonii using PacBio HiFi sequencing. Front. Plant Sci.13, 1031769 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niu, Y. et al. Analysis of the complete mitochondrial genome of the bitter Gourd (Momordica charantia). Plants12, 1686 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke, S. J. et al. Apostasia mitochondrial genome analysis and monocot mitochondria phylogenomics. Int. J. Mol. Sci.24, 7837 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao, Y., Zhang, X., Li, Z., Song, Y. & Sun, Z. Assembly and comparative analysis of the complete mitochondrial genome of Bupleurum chinense DC. BMC Genom.23, 1–17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh, R., Ming, R. & Yu, Q. Comparative analysis of GC content variations in plant genomes. Trop. Plant. Biol.9, 136–149 (2016). [Google Scholar]
- 37.Tamura, K. Origins and early evolution of the tRNA molecule. Life5, 1687–1699 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren, J. M. & Sloan, D. B. Interchangeable parts: The evolutionarily dynamic tRNA population in plant mitochondria. Mitochondrion52, 144–156 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Roovers, M., Droogmans, L. & Grosjean, H. Post-transcriptional modifications of conserved nucleotides in the T-loop of tRNA: A tale of functional convergent evolution. Genes12, 140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, L. et al. The development of SSR markers based on RNA-sequencing and its validation between and within Carex L. species. BMC Plant Biol.21, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alverson, A. J. et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol.27, 1436–1448 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, J., Li, X., Chen, X., Huang, X. & Jin, S. The complete chloroplast genome of Carya cathayensis and phylogenetic analysis. Genes13, 369 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, Y. et al. Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Diospyros Oleifera, the first representative from the family Ebenaceae. Heliyon 8 (2022). [DOI] [PMC free article] [PubMed]
- 44.Edera, A. A. & Sanchez-Puerta, M. V. Computational detection of plant RNA editing events. RNA Edit. Methods Protoc. 13–34 (2021). [DOI] [PubMed]
- 45.Bergthorsson, U., Adams, K. L., Thomason, B. & Palmer, J. D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature424, 197–201 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet.5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Martin, W. et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature393, 162–165 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Zhao, N., Wang, Y. & Hua, J. The roles of mitochondrion in intergenomic gene transfer in plants: A source and a pool. Int. J. Mol. Sci.19, 547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice, D. W. et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science342, 1468–1473 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Straub, S. C., Cronn, R. C., Edwards, C., Fishbein, M. & Liston, A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol. Evol.5, 1872–1885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams, K. L., Qiu, Y. L., Stoutemyer, M. & Palmer, J. D. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci.99, 9905–9912 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong, S. et al. The draft mitochondrial genome of Magnolia biondii and mitochondrial phylogenomics of angiosperms. PLoS ONE15, e0231020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin, X., Gao, Y., Song, S., Hassani, D. & Lu, J. Identification, characterization and functional analysis of grape (Vitis vinifera L.) mitochondrial transcription termination factor (mTERF) genes in responding to biotic stress and exogenous phytohormone. BMC Genom.22, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bi, C. et al. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Res. Int. (2016). [DOI] [PMC free article] [PubMed]
- 55.Filip, E. & Skuza, L. Horizontal gene transfer involving chloroplasts. Int. J. Mol. Sci.22, 4484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, D. R., Crosby, K. & Lee, R. W. Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol. Evol.3, 365–371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, Y. et al. Transcriptomic analysis to unravel potential pathways and genes involved in Pecan (Carya illinoinensis) resistance to Pestalotiopsis microspora. Int. J. Mol. Sci.23, 11621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doyle, J. J. & Doyle, J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. (1987).
- 59.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res.27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu, J., Fan, J., Sun, Z. & Liu, S. NextPolish: A fast and efficient genome polishing tool for long-read assembly. Bioinformatics36, 2253–2255 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Chan, P. P. & Lowe, T. M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Gene Predict. Methods Protoc. 1–14 (2019). [DOI] [PMC free article] [PubMed]
- 63.Lowe, T. M. & Chan, P. P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res.44, 54–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics33, 2583–2585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang, Y. et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. GigaScience8, giz036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data were deposited under the NCBI SRA accession PRINA824975 (submission numberSRR18718033) (https://www.ncbi.nlm.nih.gov/search/all/?term=%20SRR18718033).