Abstract
Programmed death receptor 1, PD1, modulates the function of immune cells by providing inhibitory signals and constitutes the marker of immune exhaustion. Monitoring the level of PD1 promises a useful diagnostic approach in autoimmune diseases and cancer. Here we describe the development of an ssDNA aptamer-based molecular probe capable of specific recognition of human PD1 receptor. The aptamer was selected using SELEX, its sequence was further optimized, and the affinity and specificity were determined in biochemical assays. The aptamer was converted into a fluorescent probe and its potential in molecular imaging was demonstrated in a culture of human cells overexpressing PD1 and murine pancreatic organoids / immune cells mixed co-culture model. We conclude that the provided aptamers are suitable probes for imaging of PD1 expressing immune cells even in complex cellular models and may find future utility as diagnostic tools.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-74891-7.
Subject terms: Cancer imaging, Imaging the immune system, Immunotherapy, Inflammation
Introduction
Modern imaging techniques allow tracking of multifactorial changes in complex biological systems but require specialized molecular probes in the first place. Immune checkpoint blocking antibodies targeted against the PD1 / PD-L1 pathway have revolutionized oncology in the last decade. Correspondingly, it was hypothesized that expression of PD-L1 on tumor cells may have predictive and/or prognostic value1–3, but the approach faced multiple limitations4,5 primarily because patients carrying tumors characterized by low PD-L1 expression can still benefit from anti-PD-L1 therapy.
PD1 has proved a more promising molecular marker. The receptor is expressed on the surface of immune cells and allows monitoring of changes related to exhaustion. As such, it promises useful diagnostic information in many pathological processes including autoimmune diseases6,7, acute and chronic infection8, sepsis9 and cancer. Interestingly, Xinyu Ren et al.10 have recently reported that in triple-negative breast cancer the PD1 expression in tumor-infiltrating lymphocytes (TILs), rather than PD-L1 in tumor cells, predicts survival. Tracking the expression of PD1 constitutes an interesting tool for imaging biological processes and an attractive candidate for a diagnostic marker in several diseases.
PD1 tumor imaging is unique because it is not the primary tumor, but rather the tumor environment, in particular the infiltrating immune cells11 which are visualized. Encouraging examples are available. Natarajan et al. developed an immune-PET tracer to image the tumor-infiltrating lymphocytes (TIL) expressing PD1. They labeled a murine anti–PD1 monoclonal antibody with 64Cu and successfully detected PD1-expressing murine TILs in a transgenic mouse model of melanoma using positron emission tomography (PET). The study demonstrated for the first time that PET may be used for the noninvasive imaging and determination of PD1 expression in vivo. In another study, Hettich et al.2 developed sensitive and high-resolution PET imaging radiotracers that enabled imaging of both the PD1 receptor and the PD-L1 ligand in malignant tumors in immunocompetent mice. Another study reported the evaluation of 89Zr-labeled antibody pembrolizumab in several mouse models, including mice implanted with human peripheral blood mononuclear cells3. These early proofs of concept demonstrated the utility of the approach, but successful implementation requires substantial further work.
Probes designed for molecular imaging in vivo must offer high sensitivity, low background noise, low toxicity and relative stability12. In this regard, oligonucleotide aptamers appear as excellent potential candidates13, exhibiting both low immunogenicity and high structural stability14. Initial comparative analysis of antibodies and aptamers (both in in vivo and in vitro imaging) indicates that aptamers offer consistently greater benefits compared to antibodies15,16 though only broader development and use will reveal the true potential. Aptamers are currently being considered for optical imaging (fluorescence and bioluminescence), magnetic resonance imaging (MRI), positron emission tomography (PET), single photon emission tomography (SPECT), computed tomography (CT) and ultrasound (USA)13. Moreover, several clinical trials are investigating the diagnostic potential13.
Several attempts have been made to develop aptamers directed against the human or mouse PD1 protein, which have been published to date17–21. These efforts mainly focused on developing a molecule with the potential to inhibit PD1/PDL1 interaction. Here we describe the development of an ssDNA aptamer-based molecular probe capable of specific recognition of human PD1 receptor. The aptamer was selected using SELEX and tested for specificity and affinity. The potential of the probe in molecular imaging was determined in cell and organoid cultures.
Results
Identification and characterization of ssDNA aptamers targeting human PD1
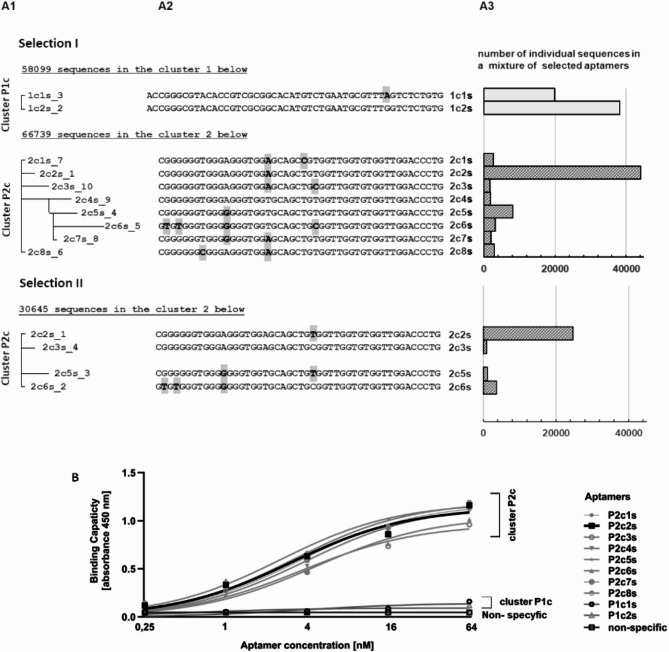
Aptamers were selected against two recombinant PD1 variants: (i) residues 21–168 and (ii) residues 25–167, both encompassing the major part of the extracellular domain of human PD1. Protein from two sources was used for selection to avoid selection of protein construct-specific aptamers. The selection was performed using the SELEX procedure with a gradual increase in the selective pressure22. Binding to the target protein was monitored after each selection cycle by ELISA. In each selection, the maximum saturation was observed after the 4th cycle (supplementary materials Fig. 1A). In subsequent cycles, a gradual decrease of the specific signal was observed for the reasons described below. Aptamer pools obtained after the 5th and subsequent selection cycles were analyzed by next-generation sequencing (NGS). The obtained sequences were grouped into clusters of similarity and the frequency of particular sequences within clusters was analyzed (Fig. 1A1–A3). Selection against PD1 extracellular domain (21–168) variant yielded two clusters of aptamers (P1c and P2c) (the nomenclature for individual aptamers was described in the supplementary data). Selection against PD1 extracellular domain (25–167) variant yielded a single cluster, corresponding to cluster P2c of the former selection (Fig. 1A1,A2) demonstrating that the selection converges at the PD1 core rather than picking up small differences in the protein construct design. Results obtained for cycles 5, 6 and 7 did not differ qualitatively (the same sequences were observed only with different relative frequencies) (see supplementary materials Fig. 2). The decreased binding capacity of aptamer pools after subsequent cycles starting from cycle 5 is explained by the increasing amount of low specific sequences and non-specific amplification products containing flanking sequences only (supplementary materials Fig. 2).
Fig. 1.
Sequence analysis and binding capacity of aptamer pool after 7 rounds of selection. (A) Sequence analysis. (A1) (pseudo-)phylogenetic trees generated based on the multiple sequence alignment (MSA). (A2) Multiple sequence alignment (MSA) of particular aptamers within indicated clusters (flanking fragments, identical in all aptamers, were excluded from analysis). A3 Number of occurrences in the entire aptamer pool (based on NGS results). (B) Binding capacity of representative aptamers determined using ELISA.
The affinity of selected aptamers to PD1 was analyzed using ELISA. Cluster P1c specific to the selection against PD1 (residues 21–168) contained sequences of little or no affinity towards PD1. Cluster P2c, common to selections against both constructs, demonstrated readily detectable binding (Fig. 1B). Particular sequences within cluster P2c were not characterized by different binding capacities, consistent with minor differences in their nucleotide sequence. The dominant sequence within P2c cluster, the P2c2s aptamer was selected for the further analysis.
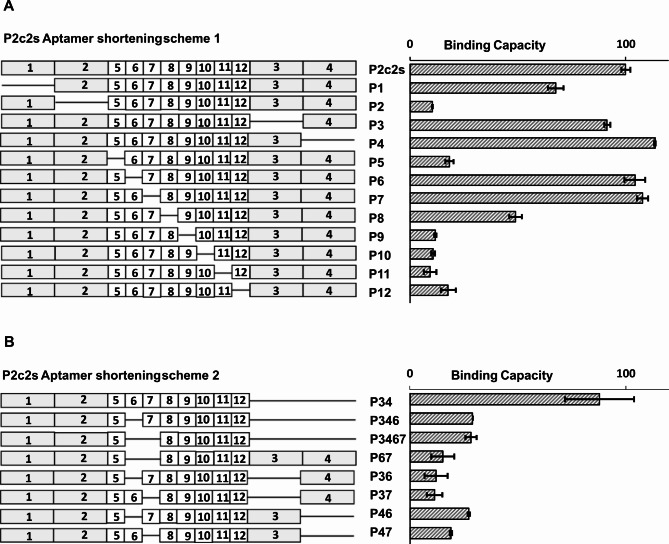
Aptamers in this study were derived from a library containing a relatively long randomized region of 50 consecutive nucleotides. Prior studies have indicated that often not all nucleotides within selected aptamers are necessary to maintain binding to the target protein. We, therefore, mapped P2c2s by positional scanning23 to determine the minimal sequence responsible for binding to the extracellular domain of PD1 (Fig. 2). P2c2s sequence was divided into 12 segments (6–10 nucleotides each) and variants were synthesized in each of which a single segment was deleted. The variants were tested by ELISA for their ability to interact with PD1. Removal of the flanking segments 3 and 4, and segments 6 and 7 within the variable region had no effect on the binding while the remaining variants were compromised (Fig. 2A). Based on the above results, P2c2s variants were designed lacking multiple segments identified as unnecessary for binding in positional scanning. Concomitant elimination of segments 3 and 4 have not affected the aptamer’s (P34) affinity for PD1 while all other tested combination variants resulted in the loss of activity (Fig. 2B).
Fig. 2.
Identification of minimal fragment of P2c2s responsible for PD1 interaction. (A) Positional scanning of P2c2s – deleted regions are marked. 1,2,3 and 4 encompass invariable primer interaction regions. (B) Tested combinations of regions identified as irrelevant for binding in positional scanning. Both panels: The binding capacity to PD1 as determined in ELISA is shown.
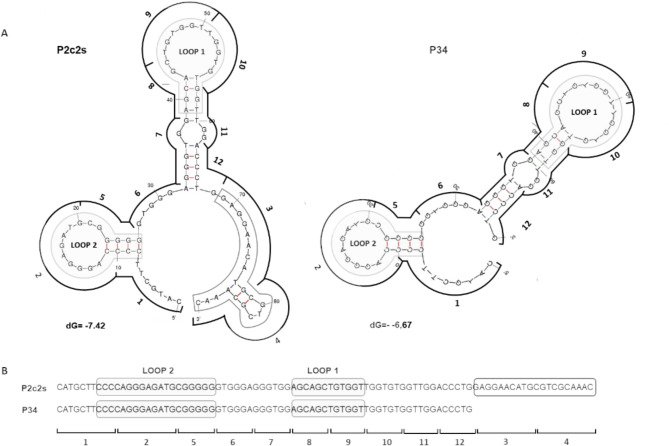
To rationalize the results of positional scanning we analyzed the computationally predicted secondary structures of P2c2s and P34 (https://rna.urmc.rochester.edu/RNAstructureWeb/index.html). The analysis suggests that each aptamer contains two hairpin motifs identical in P2c2s and P34 (Fig. 3A, Fig. S3). Segments 3 and 4 constitute an unstructured overhang in P2c2s and thereby could be removed (resulting in P34) without affecting the activity (Fig. 3A). Further elimination of segment 6 оr 7 reduced the affinity of the aptamer for PD1 (Fig. 3B). These mutations eliminated the segments of G-rich nucleotides probably supporting the aptamer structure (Fig. 3B). Elimination of the segments 2, 5, 8 and 9, each of which disrupts the predicted loops, led to an almost complete loss of affinity. Overall, the analysis revealed that the binding of P2c2s and P34 is primarily driven by two hairpin motifs and additionally stabilized by segments rich in G nucleotides. Further analysis was focused on the parent aptamer (P2c2s) and the P34 variant. In the supplementary data (Fig. S4), the proposed variants of the 3D structure of the P34 aptamer are provided. Additional analyses concerning surface charge or morphology were not carried out due to a lack of experimental data and inconsistent predictions generated by the software.
Fig. 3.
Analysis of the 2D structures of developed aptamers. (A) Prediction of secondary structure was carried out using the software available at: https://rna.urmc.rochester.edu/RNAstructureWeb/index.html (prediction of secondary structure common to more sequences) and confirmed by the software available at: http://www.unafold.org. (B) Putative loops (grey). Overhang found in P2c2s whose removal does not affect the affinity towards PD1 is highlighted by a black frame. Regions from positional scanning are marked as numbers corresponding to Fig. 2.
P2c2s and P34 recognize PD1 with high affinity and specificity
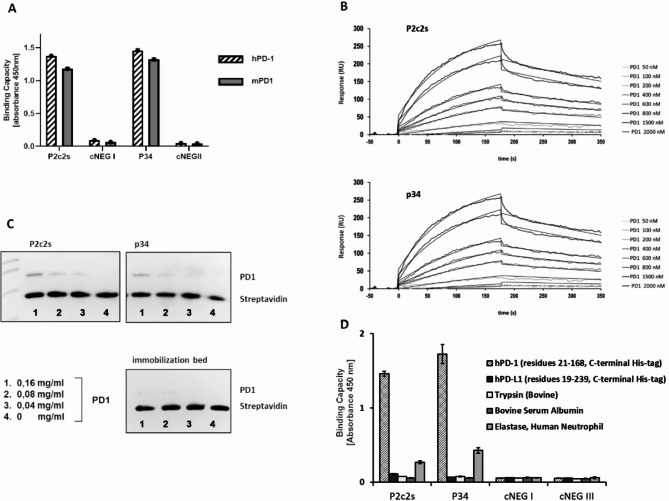
High affinity towards PD1 of aptamers belonging to cluster P2c including P2c2s compared to P1c suggested the specificity of P2c2s (Fig. 1B). To further assess the specificity of aptamers selected in this study we compared the affinity of P2c2s and P34 to the binding of non-specific sequences composed of flanking fragments and (AAAACCCCTTTTGGGG)n repeats (n corresponds to the length of the variable region). No efficient binding was detected for cNEG1 and cNEG2 (corresponding to P2c2s and P34, respectively) while the parent aptamers were characterized by high signal (Fig. 4A, Figure S5A).
Fig. 4.
Specificity and affinity of P2c2s and P34. (A) Selective binding of indicated aptamers to recombinant human PD1 (hPD1, residues 21–168, C-terminal His-tag) and to recombinant mouse PD1 (mPD1, residues 25–167, C-terminal His-tag) as determined using ELISA. cNEG1 and cNEG2 – non-specific controls. (B) Representative SPR sensograms characterizing binding of tested aptamers to immobilized PD1. (C) Target protein binding by aptamers immobilized on magnetic beads as analyzed by SDS-PAGE. (D) Specificity of P2c2s and P34. The affinity of tested aptamers to a set of proteins as determined by ELISA.
To further characterize the ability of P2c2s and P34 to recognize PD1, the aptamers were immobilized on magnetic beads and their ability to retain the purified extracellular domain of PD1 was tested. Both P2c2s and P34 were able to retain PD1 in such a setup. Empty beads have shown some unspecific binding, but less than any of the productive aptamers (Fig. 4C, Figure S6).
To evaluate the specificity of P2c2s and P34 we tested binding (if any) to human PD1 and arbitrarily selected proteins otherwise irrelevant to this study. Neither P2c2s, P34 nor any of the unspecific sequences tested have shown detectable affinity to mouse PD-L1 (residues 19–134), bovine serum albumin or trypsin demonstrating again that only P2c2s and P34, but not the unspecific sequences, specifically recognized PD1 (Fig. 4D). Certain unspecific binding to human elastase was observed, but with significantly lower affinity compared to PD1. This may be explained by the fact that elastase is known for its affinity to DNA24. Significantly, both aptamers relevant to this study recognized human extracellular domain of PD1 and mouse extracellular domain of PD1 (with high homology to the human domain) demonstrating a high level of specificity.
Having demonstrated the specificity of P2c2s and P34 we established the kinetic parameters of the binding events by SPR (Fig. 4B). Both aptamers interacted with the extracellular domain of human PD1 (21–168) with high nanomolar affinity (Table 1). Truncation of the region irrelevant for binding in P34 resulted in an almost 2x increase in affinity compared to P2c2s. The increase in affinity was reflected both in increased kon and decreased koff.
Table 1.
Characterisation of the binding kinetics of P2c2s and P34 with the extracellular domain of PD1 (residues 21-168;C-terminal His-tag).
| Aptamer | Dissociation constant | Association rate constant | Dissociation rate constant |
|---|---|---|---|
| P2c2s | Kd = 934 nM | kon = 603,4 M −1s −1 | koff = 0,00455 s −1, |
| P34 | Kd = 508 nM | kon = 3842,33 M−1 s−1 | koff = 0,001939 s−1 |
P2c2s and P34 target PD1 at the cell surface
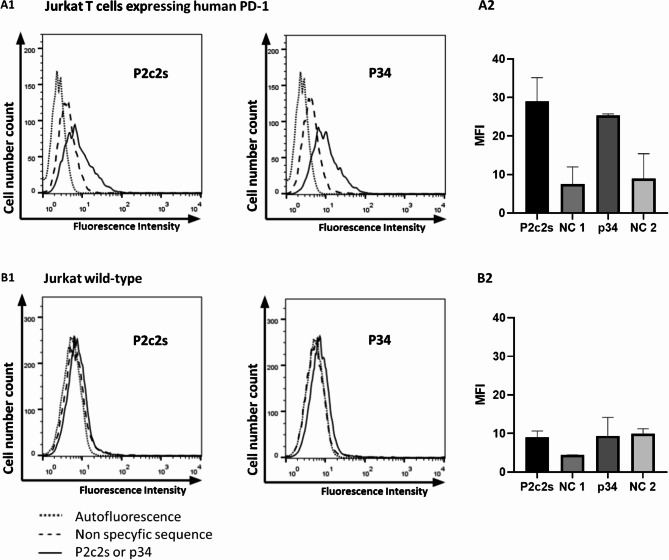
Encouraging biochemical results prompted us to evaluate the ability of obtained aptamers to selectively detect PD1 at the cell surface. To this end, staining of Jurkat-PD1 cells stably overexpressing the PD1 receptor and wild-type cells with negligible expression of PD1 was compared. Non-toxic concentrations of aptamers were selected based on LDH and MTT analyses (supplementary materials Fig. 8). FITC labeled P2c2s and P34 efficiently stained Jurkat cells overexpressing PD1 (Fig. 5A1, Fig. S5B) but not wild-type cells (Fig. 5B1,B2). Some unspecific binding of sequences non-specific for PD1 (NC1, NC2) was observed, but the signal was lower compared to the aptamers of interest (Fig. 5A2). The above studies were confirmed using cells characterized by a high endogenous level of PD1 expression such as: WM266 and WM115 cells (melanoma)25, HT29 (colon cancer)26, PANC-1 (pancreatic cancer)27, and A549 cells (lung cancer)28 (supplementary materials Figs. 7 and 9).
Fig. 5.
P2c2s and p34 recognize PD1 at the cell surface. Cells were incubated with tested FITC-labeled aptamers and the labeling was evaluated using flow cytometry. (A1) Jurkat T cells stably expressing human PD1 (B1) Wild-type cells. (A2, B2) Quantitative representation of data shown in (A1) and (B1). NC1, NC2 were non-specific aptamers corresponding in length to P2c2s and p34, respectively.
P34 detects PD1 expression in the organoid tumor model
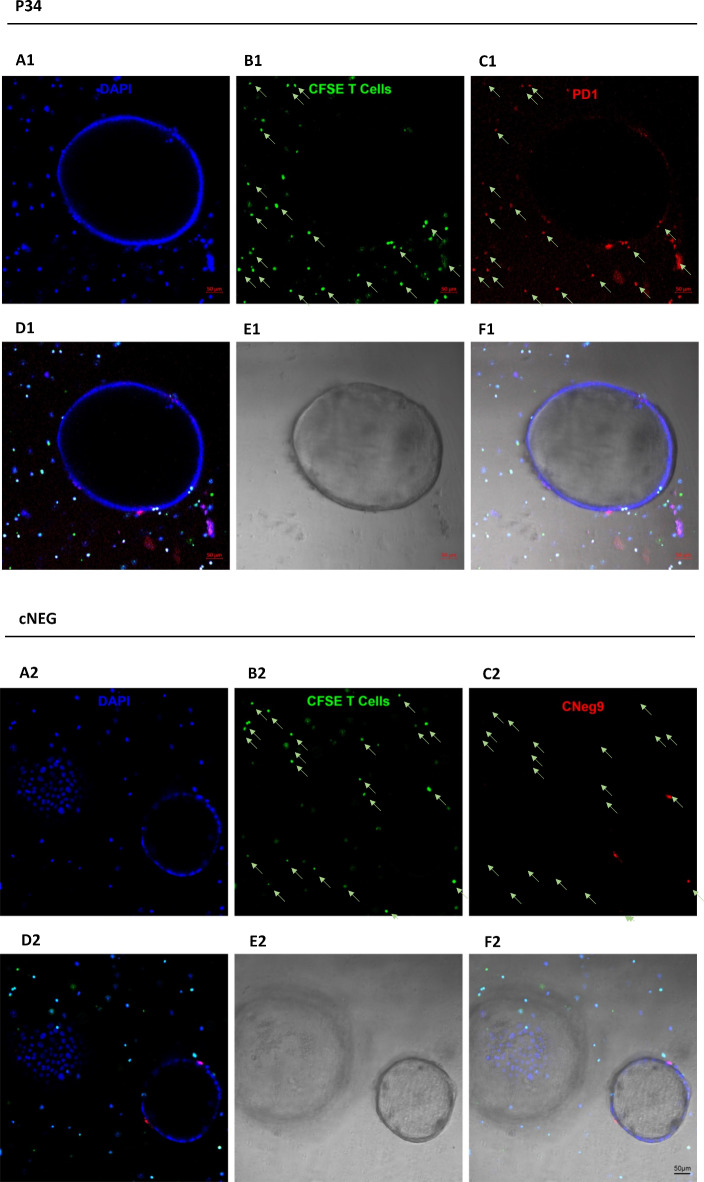
The PD1 overexpression model demonstrated that P2c2s and P34 specifically detect PD1 at the cell surface. Nonetheless, the overexpression model is characterized by physiologically irrelevant (high) expression of the receptor. To evaluate the ability of P34 to recognize PD1 in a more physiologically relevant setup, we established a three-dimensional organotypic co-culture system combining tumor-derived KPC murine pancreatic organoids and T-cells (Fig. 6A1,A2). Organoids were incubated with Texas Red (TxRd) labeled P34 (Fig. 6C1) or unspecific sequence (Fig. 6C2) and staining was analyzed using confocal microscopy. The TxRd-P34 labeling was characterized by a speckled pattern (Fig. 6C1) which colocalized with CFSE-labeled T-cells (Fig. 6B1,D1,F1) and not other organoid components (Fig. 6A1,D1,E1,F1). The non-specific sequence stained neither T cells (Fig. 6B2,C2) nor other organoid components (Fig. 6A2,D2,E2,F2). These results demonstrate that P34 selectively labeled PD1 at the surface of immune cells in a complicated environment of a three-dimensional organotypic co-culture system.
Fig. 6.
P34 specifically recognizes PD1 at the surface of T cells in KPC organoid / T-cell co-culture. Ex vivo live staining. (A1, A2) Nuclei staining with DAPI (blue), (B1, B2 ) T-cells labeled with CSFE (indicated by arrows) and PD1 expressed on T-cells: P34-TxRd staining (C1), staining with non-specific sequence cNEG-TxRd (C2); (D–F) the overlay of selected channels and the colocalization of signals.
Discussion
Diagnosis based on localized biopsy is often of limited value due to the heterogeneity of expression of the key markers. Non-invasive methods enabling in vivo assessment of heterogeneity throughout the tumor tissue are promising alternatives. Highly specific probes required for imaging additionally enable tracking of changes in marker expression over time, for example during treatment29. In this manuscript, we reported the development and initial evaluation of such a tool - an ssDNA probe suitable for molecular imaging of PD1 expression. PD1 is expressed, among others, on exhausted lymphocytes signifying the reduction in the capacity of the immune system to respond. Thereby, PD1 is considered as a marker in cancer and acute and chronic infections.
Antibody-mediated PD1 blockade proved efficient in clinics30,31, but the PD1 imaging strategy is unique in cancer because in most cases PD1 is expressed on infiltrating immune cells rather than the primary tumor as demonstrated in our organoid model and by others. A number of studies documented the utility of such an approach1,2,32–34 and the prognostic value has been demonstrated10. Therefore, assessment of tumor-infiltrating lymphocytes in histopathological samples may provide important prognostic information and may also be useful in predicting treatment response. For example, Hettich et al.2 developed PET imaging radiotracers enabling imaging of both PD1 and PD-L1 ligands in malignant tumors in immunocompetent mice. In these studies combination immunotherapy resulted in massive tumor infiltration by PD1+ lymphocytes, suggesting the possible utility of PD1 probes in monitoring the progress of the therapy in vivo. Moreover, the pattern of antigen expression in secondary lymphoid organs could have been monitored.
PD1 probes may also find utility in infectious and autoimmune diseases. PD1 axis negatively affects the activity of T cells preventing autoimmune reactions. Continuous exposure to the antigen in chronic infections leads to sustained expression of PD1 thus suppressing the response. This may limit the immune-mediated removal of pathogens. The important role of PD1 expression in acute and chronic infections has been documented both in vivo and ex vivo8. The pioneering research of Blackburn et al.35 demonstrated that antigen-specific T cells from mice infected with a chronic LCMV virus strain exhibit higher and more sustained level of PD1 expression compared to antigen-specific T cells from mice infected with acute LCMV strain. Further, these authors have demonstrated that blockage of inhibitory receptors (PD1 and LAG-3) improved T cell responses and diminished viral load in vivo. Later studies confirmed the utility of targeting PD1 in potentiating the response against pathogens8.
Blocking the PD1 pathway in model studies improved control of the infection through an increased proliferation of T cells, the cytotoxic potential of CD8+ T cells and the production of cytokines8. In this context, tracking the changes in marker expression in tissues could be important in predicting responses to PD1 blocking therapy in chronic infectious conditions, improving the quality of treatment. Potential therapeutic intervention would require stringent monitoring of the response to preclude autoimmunity. PD1 imaging probes could potentially find utility in such instances.
In studies related to aptamers intended for cells or tissue imaging, researchers typically report Kd values in nanomolar range (ranging from several to several hundred nanomoles) obtained through flow cytometry. This applies to aptamers selected against entire cells36–39 as well as those chosen for specific molecular targets40–42. In our research, the Kd values obtained using Jurkat cells (which overexpress PD1) through flow cytometry were 237.1 nM and 264.9 nM, which is comparable to affinities of aptamers previously suggested as suitable for imaging. In line with the above, the utility of aptamers developed in this study to detect PD1 at the immune cell surface was demonstrated ex vivo using a pancreatic tumor model. We have shown that the aptameric probe specifically labels PD1-expressing T cells, allowing direct visualization of biodistribution. Further studies in living organisms are needed to determine the distribution of the probe, specificity and metabolism and finally the diagnostic/prognostic value. This study has set a solid ground for further (pre-) clinical development of aptameric PD1-directed probes for the diagnosis and monitoring of tumors and the progress of autoimmune disorders.
Methods
PD1 purification
Construct of the extracellular domain of human PD1 encompassing residues 21–168 (C-terminal His-tag) was expressed and purified as described previously43. Briefly, the protein was expressed in E. coli BL21 (DE3) as inclusion bodies. Cells were cultured at 37 °C in LB medium and induced with 1 mM Isopropyl-β-D-thiogalactoside (IPTG) at OD600 = 1.0. After further incubation at 37 °C for 3 h, the cells were collected and lysed by sonication in phosphate-buffered saline (PBS). Inclusion bodies were recovered by centrifugation, washed twice with 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.5% Triton X-100, 10 mM EDTA and 10 mM 2-mercaptoethanol followed by washing with the same buffer excluding the detergent. The inclusion bodies were dissolved in 50 mM Tris pH 8.0, 6 M GuHCl, 200 mM NaCl, and 10 mM 2-mercaptoethanol and refolded by drop-wise dilution into 0.1 M Tris pH 8.0, 0.4 M L-Arg hydrochloride, 2 mM EDTA, 5 mM cystamine and 0.5 mM cysteamine. After refolding, protein was dialyzed 3 times against 10 mM Tris pH 8.0 containing 20 mM NaCl, concentrated, and further purified by gel filtration on Superdex 75 (GE Healthcare, Chicago, Illinois, USA) in buffer containing; 25 mM Na2HPO4, 25 mM NaH2PO4 pH 6.4, 100 mM NaCl, 5 mM MgCl2 and 10 mM KCl or 10 mM Tris pH 8.0 containing 20 mM NaCl. The proteins were stored for further experiments as 0.2 mg/ml stocks at -80 °C. Recombinant human PD1 with C-terminal His tag (residues 25–167) (ab174035) and recombinant mouse PD1 (residues 21 to 167) - His tagged at C-Terminus (ab180051) were obtained from Abcam, Cambridge, UK.
In vitro selection of aptamers.
The single-stranded DNA (ssDNA) library (N50), composed of a random region of 50 nucleotides flanked by fixed 20 nucleotide primer sequences (5’-CATGCTTCCCCAGGGAGATG-N50-GAGGAACATGCGTCGCAAAC-3’) was synthesized at 0.2 µM scale and purified by HPLC (IBA, Göttingen, Germany). The ssDNA library and the ssDNA pools obtained after each subsequent selection cycle were renatured by incubation for 5 min at 92 ° C, 10 min 4 ° C and 15 min RT. Prior to productive selection, aptamers interacting with the carrier resin were removed by incubation with uncoated beads.
Aptamers were selected independently against PD1 extracellular domain constructs encompassing residues 21–168 (in house) and residues 25–167 (Abcam, UK, ab174035) both with C-terminal histidine tags. The target protein was immobilized (Dynabeads ™ His-Tag Isolation and Pulldown, Thermo Fisher Scientific, Waltham, USA) according to the manufacturer instructions. The beads were washed with PBS pH 7.4 supplemented with 5mM MgCl2, 10mM KCl and 0.01% Tween 20 (selection buffer) and resuspended in binding buffer (selection buffer supplemented with 40–120 µg/ml yeast tRNA (Invitrogen, Waltham, USA) and 125 µg/ml BSA (Bioshop, Canada Inc., Burlington, Canada). ssDNA library was added and incubated at 24 °C for 20 min with stirring. To increase the selective pressure, in consecutive cycles the amount of beads and concentration of ssDNA were gradually reduced while increasing the concentration of the competitor (yeast tRNA) (see supplementary materials Fig. 1). After incubation, the beads were rinsed, suspended in dH2O and ssDNA was amplified by PCR. 400 µl of PCR mix containing 1 µM primers (unmodified ss50_For: 5′-CATGCTTCCCCAGGGAGATG-3′ and 5’-phosphorylated ss50_Rev: 5′-GTTTGCGACGCATGTTCCTC-3′), 0.5 mM dNTPs, 2.5 mM MgCl2, 1.25U/100 µl of Taq polymerase (Thermo Fisher Scientific, Waltham, USA) was prepared and mixed with 3–0.3 µl of beads containing immobilized protein and bound aptamers. The PCR was performed for 30 cycles, consisting of denaturation at 95 °C for 2 min, annealing at 53 °C for 30 s, and extension at 72 °C for 30 s followed by the final polishing step of 72 °C for 5 min. The PCR products were extracted using phenol-chloroform-isoamyl alcohol (Sigma-Aldrich, St.Louis, USA) and precipitated overnight with ethanol at -20 °C. The DNA pellet was recovered by centrifugation, washed twice with 70% cold ethanol, dried, and dissolved in dH2O. The dsDNA was converted to relevant ssDNA by digestion with λ exonuclease (Thermo Fisher Scientific, Waltham, USA) in a 500 µl containing 100 U exonuclease at 37 °C, for 1 h with gentle shaking. ssDNA was extracted using phenol-chloroform-isoamyl alcohol (Sigma-Aldrich, St.Louis, USA) and dissolved in 100 µl dH2O. ssDNA samples were stored at -20 ° C for the next selection cycle. Enrichment of the pool in PD1 binding aptamers in subsequent selection steps was monitored by ELISA (see below). Aptamer pools were analyzed by NGS after 5-th, 6-th and 7th selection cycle (Genomed S. A., Warszawa, Poland).
Sequence analysis
FASTQ files containing demultiplexed sequences after barcode trimming were further trimmed of the adapters (primer attachment sites) and sequences were reversed if needed. Sequences shorter than 10 nucleotides were discarded. The remaining sequences were aligned into groups containing identical sequences. In further analysis, each group was represented by a common sequence (compressed_seq). The groups were identified by their cardinality. 10 most frequent compressed_seqs were clustered with the k-means algorithm. An optimal number of clusters was determined employing silhouette method and the silhouette score was computed for 2 < = k<=4. Then, the compressed_seqs in each cluster were aligned with ClustalW. The obtained alignments were subsequently processed with dnadist and neighbour programs from PHYLIP package to generate (pseudo-)phylogenetic trees representing relationships between compressed_seqs.
ELISA binding assay
The binding of selected aptamers to human PD1 was assessed by ELISA. 96-well microtiter plate (Nunc, Rochester, NY, USA) was coated with 100 µl of human PD1 (10 µg/ml in PBS) overnight at + 4 °C. All subsequent steps were performed at RT. Unbound PD1 was removed by washing in the selection buffer. Serial dilutions of biotinylated aptamers (100 µl) were added to the wells and incubated for 30 min. The control DNA sequences were used to assess non-specific binding. Unbound aptamers were removed by extensive washing in the selection buffer. 100 µl of horse-radish-peroxidase HRP-conjugated streptavidin (1:200 dilution; R&D Systems, Inc., Minneapolis, USA) was added to the wells. After 20 min of incubation, the unbound streptavidin was removed by washing in a selection buffer. Following, 100 µl of the HRP reagent substrate (R&D Systems, Minneapolis, USA) was added and incubated for 5–10 min. The reaction was stopped by adding 50 µl of 2 N H2SO4. Absorbance at 450 nm and 570 nm (correction for optical plate imperfections) was determined using the Infinite 200 PRO multimode reader (Tecan Group Ltd., Switzerland). Unspecific binding to proteins irrelevant to this study (mouse extracellular domain of PD-L1, human neutrophil elastase, bovine trypsin, and bovine serum albumin ) was analyzed in similar manner after coating the plates with each tested protein (10 µg / ml in PBS) separately.
Binding kinetics
Surface plasmon resonance (SPR) was used to characterize the binding kinetics of P2c2s and P34 to the extracellular domain of PD1(residues 21–168; C-terminal His-tag). The biotinylated aptamers were prepared as 5 nM stock in HBS-P buffer (0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% v/v Surfactant P20 (GE Healthcare, Chicago, Illinois, USA) and 5 mM MgCl2), boiled for 10 min at 95⁰ C, and cooled on ice for 10 min. Aptamers were immobilized at a density of approximately 700 RU (resonance units) on the surface of a SA chip (GE Healthcare, Chicago, Illinois, USA) according to the manufacturer’s protocol. The binding kinetics was analyzed in PD1 aptamer assay buffer (25 mM Na2HPO4, 25 mM NaH2PO4 pH 6.4, 100 mM NaCl, 5 mM MgCl2 and 10 mM KCl) with the use of Biacore X100 (GE Healthcare, Chicago, Illinois, USA). A single 20-sec pulse of 2 M KCl was used for chip regeneration in multi-cycle kinetics. The binding data were fitted to the Langmuir 1:1 binding model using Biacore X100 Evaluation software.
Immobilization assay
Biotinylated aptamers (P2c2s, P34) at 4 µM concentration were conjugated to Streptavidin Mag Sepharose (GE Healthcare, Chicago, USA) by incubation in PBS for 20 min at RT with constant shaking. The slurry was washed three times with PBS and blocked with 0,2% BSA (Bioshop) in SELEX buffer for 30 min in RT with constant shaking. After washing with SELEX buffer, the slurry was incubated with PD1 (165 µg/ml, 82,5 µg/ml or 40,7 µg/ml) in SELEX buffer supplemented with 40 µg/ml tRNA, for 20 min at RT with constant shaking. Samples were washed with SELEX buffer three times, eluted by boiling briefly in the loading buffer (3% SDS, 10% glycerol, 12,5 mM Tris-HCl, pH 6,8, 100 mM DTT, 0,05% bromophenol blue), and analyzed via SDS/PAGE. The Coomassie blue-stained gels were imaged with the use of ChemiDoc (Bio-Rad Laboratories, Inc. Hercules, USA).
Cell binding assay (flow cytometry)
Jurkat T-cells (wild type and stably overexpressing PD1) were maintained in RPMI 1640 medium (Gibco) supplemented with 10% FBS (InvivoGen, San Diego, CA, USA), at 37 °C with 5% CO2. Jurkat T-cells expressing hPD1 were maintained in a medium supplemented with selection antibiotics; hygromycin B (50 µg/ml; Sigma Aldrich, Darmstadt, Germany) and G418 (250 µg/ml; InvivoGen, San Diego, CA, USA). Additionally, the research used cells characterized by a high endogenous PD1 level: human melanoma cells (WM266 and WM115)25, colon cancer cells (HT29)26, pancreatic cancer cells (PANC-1)27, and lung cancer cells (A549)28. The WM266 and WM115 cells were cultured in RPMI medium, while HT29, PANC-1, and A549 cells were cultured in DMEM medium supplemented with 10% FBS (InvivoGen, San Diego, CA, USA), at 37 °C with 5% CO2.
Tested aptamers (P2c2s, P34 and nonspecific sequences) were obtained as conjugates with 5’-FAM, 5’-Texas Red or 5’-cyanine (5’-extremity of the oligonucleotide (5’-hydroxyle)). The cells were blocked in Hanks’ Balanced Salt solution (HBSS) supplemented with 1 mg/ml dextran (Merck Millipore, Darmstadt, Germany) and 1 mg/ml salmon sperm DNA (Merck Millipore, Darmstadt, Germany) for 30 min at 37 °C. Labeling with aptamers (0–250nM) was obtained by 30 min incubation of 0.25 × 106 cells in HBSS supplemented with MgCl2 and KCl, at 37 °C in the dark and with shaking (300 rpm). After incubation, cells were washed in HBSS and analysed by Flow Cytometry (BD FACSCalibur or BD Accuri c6). The binding of fluorophore-conjugated aptamers to the target protein was demonstrated as a percentage of fluorescent positive cells using CellQuestPro software. Antibodies against human PD1 (clone J105, Thermo Fisher Scientific, Waltham, MA, USA) were used as positive controls. Non-transfected Jurkat cells were used as negative controls.
Ex vivo aptamer analysis in KPC organoid/T-cell co-culture
The Pdx1-Cre; Kras+/LSL-G12D; Trp53+/LSL-R172H (KPC) mice with developed pancreatic tumors were used in the analysis. 6 to 8-week old mice were purchased from Charles River Laboratory. The study was carried out in compliance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. All experiments were carried out with approval of the Animal Care and Use Committee at SCUT. And all methods were carried out in accordance with relevant guidelines and regulations. Mouse pancreatic tumor cells were isolated from the tumor bulk of mice older than 8 weeks as previously described44. Briefly, mouse pancreatic tumors were minced and enzymatically dissociated with 5 mg/ml Collagenase type XI (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1 mg/ml Dispase (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 1% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) in DMEM medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C for a maximum of 16 h. The isolated material was incubated with TrypLE (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C for 10 min, embedded into growth factor-reduced Matrigel (Corning), and cultured in mouse complete medium (AdDMEM/F12 (Gibco) supplemented with 1% penicillin/streptomycin (Gibco), 1% GlutaMAX (Gibco), 10 mM HEPES (Gibco), 1:50 B27 supplement (Gibco), 1.25 mM N-Acetylcysteine (Sigma Aldrich, Darmstadt, Germany), 10% (vol/vol) Rspo1-conditioned media, 10 mM Nicotinamide (Sigma Aldrich, Darmstadt, Germany), 10 nM recombinant human-gastrin I (Tocris), 50 ng/ml recombinant mouse EGF (Gibco), 100 ng/ml recombinant human FGF10 (Peprotech, London, UK), 0.5 µM A83-01 (Tocris), and 100 ng/ml recombinant human Noggin (Peprotech, London, UK)). Mouse’s complete medium was changed twice a week. All animal experiments were approved by the Animal Care and Use Committee at SCUT, and every effort was made to minimize suffering from experiments.
KPC organoids were cultured with CFSE-labelled T cells. Prior to aptamer staining, organoids were incubated with blocking buffer (HBSS + 0.1 mg/mL dextran + 1 mg/mL salmon sperm DNA) for 30 min. 50 nM P34-TxR aptamer or TxR labeled negative control aptamer were added and incubated with organoids for 6 h. After this time, organoids were washed with HBSS by pipetting. Images were captured on a confocal fluorescence microscope (Carl Zeiss).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the Jagiellonian Centre of Innovation (Cracow, Poland) for performing the SPR measurements. The authors would like to thank the South China University of Technology (Guangzhou, China) for their cooperation in developing the KPC model.
Author contributions
S.M., Conceptualization; S.M., A.C., A.S.G., Methodology; S.M., A.C., E.Z., B.P., W.G., M.KA., A.S.G., K.M., B.Z., M.B.M., Investigation; W.G.: Software; S.M., A.C., E.Z., B.P., W.G., B.CH., A.S.G., J.W., M.W., M.B.M., J.K., G.D., P.M., Data analysis; B.CH., Formal analysis; J.W., J.K., G.D., P.M., Resources; S.M., G.D., P.M., Supervision; S.M., Project administration; S.M., M.B.M., G.D., PM, Funding, S.M., B.CH., M.KA., A.S.G., G.D., P.M., Writing – original draft; S.M., B.CH., J.K., G.D., P.M., Writing – review&editing. All authors read and approved the final manuscript.
Funding
This research was supported in part by the National Science Centre, Poland, grant number 2017/25/B/NZ1/00827 (to G.D.) and 2018/02/X/NZ1/02015 (to S.M). P.M. was supported by the Norwegian Research Council NFR 296129 and 311544. M.B.M. and S.M. were supported by the National Centre for Research and Development, Poland, grant number: NOR/SGS (0141/2020-00 Aptaquest). S.M. was supported by TEAM TECH CORE FACILITY/2017-4/6 grant from the Foundation for Polish Science. A.C. was supported by the by Polish National Agency for Academic Exchange, NAWA Polish Returns 2018 (PPN/PPO/2018/1/00046/U/00001). Some of the research was carried out with equipment purchased with the financial support of the European Regional Development Fund in the framework of the Polish Innovation Economy Operational Program (contract no. POIG.02.01.00-12-167/08, project Małopolska Centre of Biotechnology).
Data availability
The data presented in this study are available in the main text and the supplementary materials of this article.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal experiments were approved by the Animal Care and Use Committee at SCUT. The study was carried out in compliance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. And all methods were carried out in accordance with relevant guidelines and regulations.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stanisław Malicki, Email: stanislaw.malicki@uj.edu.pl.
Grzegorz Dubin, Email: grzegorz.dubin@uj.edu.pl.
Piotr Mydel, Email: piotr.mydel@uib.no.
References
- 1.Natarajan, A. et al. Novel radiotracer for immunoPET imaging of PD-1 checkpoint expression on tumor infiltrating lymphocytes. Bioconjug. Chem.26, 2062–2069 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hettich, M., Braun, F., Bartholomä, M. D., Schirmbeck, R. & Niedermann, G. High-resolution PET imaging with therapeutic antibody-based PD-1/PD-L1 checkpoint tracers. Theranostics6, 1629–1640 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.England, C. G. et al. Preclinical pharmacokinetics and Biodistribution studies of 89Zr-Labeled pembrolizumab. J. Nucl. Med.: Off. Publ. Soc. Nucl. Med.58, 162–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maleki Vareki, S., Garrigós, C. & Duran, I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit. Rev. Oncol. Hematol.116, 116–124 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Davis, A. A. & Patel, V. G. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer7, 52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Álvarez-Sierra, D. et al. Analysis of the PD-1/PD-L1 axis in human autoimmune thyroid disease: Insights into pathogenesis and clues to immunotherapy associated thyroid autoimmunity. J. Autoimmun.103, 102285 (2019). [DOI] [PubMed] [Google Scholar]
- 7.MR, J. et al. Downregulation of immunosuppressive molecules, PD-1 and PD-L1 but not PD-L2, in the patients with multiple sclerosis. Iran. J. Allergy Asthma Immunol.15, 296–302 (2016). [PubMed] [Google Scholar]
- 8.Jubel, J. M., Barbati, Z. R., Burger, C., Wirtz, D. C. & Schildberg, F. A. The role of PD-1 in acute and chronic infection. Front. Immunol.11, 487 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson, J. K., Zhao, Y., Singer, M., Spencer, J. & Shankar-Hari, M. Lymphocyte subset expression and serum concentrations of PD-1/PD-L1 in sepsis - pilot study. Crit. Care. (London, England)22, (2018). [DOI] [PMC free article] [PubMed]
- 10.Ren, X. et al. PD1 protein expression in tumor infiltrated lymphocytes rather than PDL1 in tumor cells predicts survival in triple-negative breast cancer. Cancer Biol. Ther.19, 373–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehlerding, E. B., England, C. G., McNeel, D. G. & Cai, W. Molecular imaging of immunotherapy targets in Cancer. J. Nuclear Medicine: Official Publication Soc. Nuclear Med.57, 1487–1492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarty, R., Goel, S. & Cai, W. Nanobody: the ‘magic bullet’ for molecular imaging? Theranostics 4, 386–398 (2014). [DOI] [PMC free article] [PubMed]
- 13.Yoon, S. & Rossi, J. J. Targeted Molecular Imaging using aptamers in Cancer. Pharmaceuticals (Basel Switzerland)11, (2018). [DOI] [PMC free article] [PubMed]
- 14.Que-Gewirth, N. S. & Sullenger, B. A. Gene therapy progress and prospects: RNA aptamers. Gene Ther.14, 283–291 (2007). [DOI] [PubMed] [Google Scholar]
- 15.De Castro, M. A. G., Hobartner, C. & Opazo, F. Aptamers provide superior stainings of cellular receptors studied under super-resolution microscopy. PloS One12, (2017). [DOI] [PMC free article] [PubMed]
- 16.Melancon, M. P. et al. Selective uptake and imaging of aptamer- and antibody-conjugated hollow nanospheres targeted to epidermal growth factor receptors overexpressed in head and neck cancer. ACS nano. 8, 4530–4538 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prodeus, A., Abdul-Wahid, A., Fischer, N. W., Huang, E. H. & Cydzik, M. Targeting the PD-1/PD-L1 immune evasion axis with DNA aptamers as a novel therapeutic strategy for the treatment of disseminated cancers. Mol. Therapy - Nucleic Acids. 4, e237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, H., Lam, C. H., Li, X., West, D. L. & Yang, X. Selection of PD1/PD-L1 X-Aptamers. Biochimie. 145, 125–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khedri, M. et al. Development and evaluation of novel aptamers specific for human PD1 using hybrid systematic evolution of ligands by exponential enrichment approach. Immunol. Investig.49, 535–554 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Sun, Y. et al. Bispecific aptamer-based recognition-then-conjugation strategy for PD1/PDL1 axis blockade and enhanced immunotherapy. ACS Nano16, 21129–21138 (2022). [DOI] [PubMed] [Google Scholar]
- 21.CA2956718A1. - An antagonistic pd-1 aptamer and its applications in cancer therapy related applications - Google Patents. June (2024). https://patents.google.com/patent/CA2956718A1/en. (Accessed: 24th.
- 22.Malicki, S. et al. Imaging of clear cell renal carcinoma with immune checkpoint targeting aptamer-based probe. Pharmaceuticals (Basel Switzerland)15, (2022). [DOI] [PMC free article] [PubMed]
- 23.Gao, S., Zheng, X., Jiao, B. & Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem.408, 4567–4573 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Belorgey, D. & Bieth, J. G. DNA binds neutrophil elastase and mucus proteinase inhibitor and impairs their functional activity. FEBS Lett.361, 265–268 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Kleffel, S. et al. Melanoma Cell-intrinsic PD-1 receptor functions promote Tumor Growth. Cell. 162, 1242–1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ieranò, C. et al. In PD-1 + human colon cancer cells NIVOLUMAB promotes survival and could protect tumor cells from conventional therapies. J. Immunother. Cancer10, (2022). [DOI] [PMC free article] [PubMed]
- 27.Gao, M. et al. Direct therapeutic targeting of immune checkpoint PD-1 in pancreatic cancer. Br. J. Cancer. 120, 88–96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan, F. et al. Elevated Cellular PD1/PD-L1 expression confers Acquired Resistance to Cisplatin in Small Cell Lung Cancer cells. PloS One11, (2016). [DOI] [PMC free article] [PubMed]
- 29.AN, N. et al. Whole body PD-1 and PD-L1 positron emission tomography in patients with non-small-cell lung cancer. Nat. Commun.9, (2018). [DOI] [PMC free article] [PubMed]
- 30.SL, T. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med.366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science. 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.X, G. et al. Development of 99m Tc-conjugated JS001 antibody for in vivo mapping of PD-1 distribution in murine. Bioorg. Med. Chem. Lett.29, 2178–2181 (2019). [DOI] [PubMed] [Google Scholar]
- 33.H, H. et al. Evaluation of 124 I-JS001 for hPD1 immuno-PET imaging using sarcoma cell homografts in humanized mice. Acta Pharm. Sinica B. 10, 1321–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broos, K. et al. Noninvasive imaging of the PD-1:PD-L1 immune checkpoint: embracing nuclear medicine for the benefit of personalized immunotherapy. Theranostics. 8, 3559–3570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blackburn, S. D. et al. Coregulation of CD8 + T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol.10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon, S. et al. Targeted delivery of C/EBPα -saRNA by pancreatic ductal adenocarcinoma-specific RNA aptamers inhibits Tumor Growth in vivo. Mol. Therapy: J. Am. Soc. Gene Therapy. 24, 1106–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan, M. et al. Selection and characterization of DNA aptamer for metastatic prostate cancer recognition and tissue imaging. Oncotarget. 7, 36436–36446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, M., Yang, T., Chen, Z., Wang, Z. & He, N. Differentiating breast cancer molecular subtypes using a DNA aptamer selected against MCF-7 cells. Biomaterials Sci.6, 3152–3159 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Liu, Y. et al. DNA aptamer S11e recognizes fibrosarcoma and acts as a tumor suppressor. Bioactive Mater.12, 278–291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu, Y., Li, X., An, Y., Duan, J. & Yang, X. Da. Selection of a novel CD19 aptamer for targeted delivery of doxorubicin to lymphoma cells. Oncotarget. 9, 26605–26615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, G. et al. Selection and characterization of DNA aptamer against glucagon receptor by cell-SELEX. Sci. Rep.7, (2017). [DOI] [PMC free article] [PubMed]
- 42.Wang, T., Rahimizadeh, K. & Veedu, R. N. Development of a novel DNA oligonucleotide targeting low-density lipoprotein receptor. Mol. Therapy Nucleic Acids. 19, 190–198 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zak, M. K. et al. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget. 24, 7, 30323–30335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartfeld, S. & Clevers, H. Organoids as model for infectious diseases: culture of human and murine stomach organoids and microinjection of Helicobacter pylori. J. Vis. Exp.: JoVE (2015). (2015). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the main text and the supplementary materials of this article.