Abstract
Objective
The objective of the current study was to investigate the correlation between polymorphisms in extracellular matrix‐degrading enzymes and the risk of intervertebral disc degeneration (IDD) diseases.
Methods
The databases PubMed, Embase, and Cochrane Database were systematically queried from the inception until March 2023 to ascertain studies that meet the eligibility criteria. Utilizing a standardized data collection form to extract data from individual studies. The data were quantified using odds ratio (OR) along with its corresponding 95% confidence interval (95% CI), following an allelic model of inheritance.
Results
The study included a total of nine studies and indicated that the presence of rs17576 in the MMP9 gene was significantly associated with an increased risk of IDD diseases (GG: 1.30, 95% CI [1.09–1.55], p = 0.004). The presence of other polymorphisms in extracellular matrix‐degrading enzymes did not exhibit a significant association with the susceptibility to IDD.
Conclusion
The current study demonstrated a noteworthy correlation between the GG genotype of MMP‐9 rs17576 and susceptibility to IDD. The available evidence is insufficient to substantiate the correlation between other extracellular matrix‐degrading enzymes and susceptibility to IDD. The constraints of this analysis necessitate further research involving larger sample sizes across diverse ethnicities to provide a comprehensive understanding of the true impact of these polymorphisms on susceptibility to IDD.
Keywords: extracellular matrix‐degrading enzymes, intervertebral disc degeneration, polymorphisms
Understanding the genetic variations of genes can predict which individuals have a higher probability of developing IDD, enabling earlier prevention. Currently, there is a lack of systematic integration regarding the acellular matrix degradation enzyme polymorphism in IDD. A notable correlation between the MMP‐9 rs17576 mutation and an elevated susceptibility to disc degeneration.
![]()
1. INTRODUCTION
Intervertebral disc degeneration (IDD) is a disease characterized by a significant decrease in the elasticity and height of intervertebral discs, ultimately leading to impaired shock absorption function and loss of impact‐absorbing capacity. With the continuous evolution of the aging and degenerative process, there is an increased chance of fibrous annular wall rupture, leading to extrusion of the nucleus pulposus and compression of the neural structures. 1 , 2 This is a common factor contributing to the occurrence of lower back pain. It is reported that the total medical expenses paid for patients with lower back pain exceeded 9 billion dollars. 3 , 4 Therefore, the degeneration of intervertebral discs imposes a heavy burden on individuals and the economy.
There are many causes of lower back pain, including trunk posture, high BMI, age, local pressure load, genetic factors, and congenital conditions such as scoliosis and spinal deformities caused by traumatic lesions. In addition, some unhealthy habits such as smoking and drinking alcohol, as well as certain underlying diseases like diabetes, can exacerbate the progression of the disease to some extent. 1 , 4 , 5 Therefore, the development of IDD is a multifactorial process. However, some research indicates that factors related to the environment may not be the primary determinant of IDD, as genetic factors appear to play a decisive role. 6 , 7 Genes can regulate the physiology of intervertebral discs through their effects on the regeneration, structure, or homeostasis of the discs. 8 Therefore, understanding the genetic variations of genes can predict which individuals have a higher probability of developing IDD, enabling earlier prevention. Additionally, this is beneficial for developing more effective treatment methods.
In the past few years, there has been a notable rise in the quantity of research conducted on the mechanism underlying IDD. Understanding the genetic progress in this field can greatly facilitate the development of new diagnostic and treatment methods. Currently, there is a lack of systematic integration regarding the acellular matrix degradation enzyme polymorphism in IDD. Therefore, the objective of the current study was to analyze the existing body of literature concerning the polymorphism of extracellular matrix‐degrading enzymes and genetic mechanisms associated with the pathogenesis of IDD.
2. METHODS
2.1. Literature search
The literature search for the current study encompassed three databases (PubMed, Embase, and Web of Science Database), including studies published prior to March 2023. The keywords encompassed “ADAMTS protein,” “a disintegrin and metalloproteinase with thrombospondin motifs protein,” “matrix metalloproteinase,” “MMPs,” “polymorphisms,” “IDD,” or “intervertebral disc degeneration.” The article reference lists were also screened manually.
2.2. Selection criteria
The article selection was carried out based on the presence of a title and abstract. Subsequently, further exclusions were made after obtaining full texts. Among the selected articles, data extraction and assessment of study quality were performed according to the specified eligibility criteria. Eligible criteria included individuals with a confirmed diagnosis of IDD by imaging and/or clinical classification, along with gene polymorphism identification in both cohorts, based on present nomenclature. The analysis included results from multiple studies to minimize the potential bias that may arise when relying solely on findings reported in a single study. Additionally, the study required an adequate description of study demographics and a clearly defined methodology. Exclusions comprised languages other than English, animal studies, as well as incomplete data.
2.3. Data extraction
Two investigators independently conducted the reading and data extraction process. They utilized a standardized data extraction form for each study and jointly reviewed the articles in case of any discrepancies in their interpretation. Attempts were made to contact the corresponding author in instances where data were unavailable.
2.4. Methodological quality assessment
The utilization of the Newcastle–Ottawa Scale (NOS), which is widely recognized as a valuable instrument for assessing the methodological soundness of non‐randomized studies, was implemented to assess potential bias risks. The domains of “Selection” (consisting of 4 items), “Comparability” (comprising 1 item), and “Exposure” (including 3 items) were meticulously assessed, with each high‐quality criterion being awarded a star.
2.5. Statistical analysis
Examining the potential association between candidate genes and susceptibility to IDD by comparing the frequencies of alleles in individuals with IDDs and controls was the primary objective of this study. The frequencies of alleles were calculated for both individuals with IDDs and controls in each study that was incorporated into the analysis. The Q test was used to assess the statistical heterogeneity among the selected studies. The I 2 statistic was reported, with higher values indicating increased heterogeneity. If the I 2 exceeds 50%, analyses were conducted utilizing the Mantel–Haenszel (M–H) approach, which follows a fixed‐effect model. Otherwise, the random‐effect model was employed by incorporating inverse variance adjusted for inter‐study variability. The odds ratios (ORs) and their corresponding 95% confidence intervals (CI) were presented in forest plots. All statistical analyses were conducted using Revman 5.4 software.
3. RESULTS
3.1. Study selection
Three databases yielded a total of 3122 items. After evaluation of titles and abstracts, 171 papers remained for further analysis. Following the elimination of duplicates, applying exclusion criteria, and evaluation of full texts, nine studies 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 were ultimately considered eligible and incorporated into this analysis (Figure S1).
3.2. Characteristics and methodological quality assessment of studies
The included studies were case–control designs. Six studies 9 , 10 , 11 , 14 , 16 , 17 were from Asia, two studies 12 , 15 from Europe, and one study 13 from Africa. The mean age of the included studies was 40.4 years. All studies were diagnosed with LDD through magnetic resonance imaging or CT. The key features of the studies included were succinctly outlined in Table 1. The outcomes derived from the NOS evaluations can be found in Table 2. The NOS ratings ranged from 4 to 7 score and the average NOS score was 6.3 score, indicating a moderate to high level of methodological quality among the included studies.
TABLE 1.
Main characteristics of the included studies.
| Included studies | Country | Sample size | Sex (male/female) | Mean age | Variant | |
|---|---|---|---|---|---|---|
| LDD | Control | |||||
| Dong et al. 2007 9 | China | 162 | 318 | 240/240 | 24.5 | rs243865 (C/T) |
| Kitis et al. 2018 10 | Turkey | 199 | 197 | 198/198 | 43.3 | rs243865 (C/T) |
| YUAN et al. 2010 11 | China | 178 | 284 | 180/282 | 43.6 | rs731236 (5A/6A) |
| Noponen‐Hietala et al. 2003 12 | Finland | 29 | 56 | – | 48.5 | rs731236 (5A/6A) |
| Zawilla et al. 2014 13 | Egypt | 84 | 60 | 83/61 | 43.8 | rs731236 (5A/6A) |
| Hirose et al. 2008 14 | Japan | 847 | 896 | 1083/660 | 50.9 | rs17576 (G/A) |
| Kelempisioti et al. 2011 15 | Finland | 150 | 246 | 155/241 | 19.0 | rs17576 (G/A) |
| Jiang 2017 16 | China | 428 | 400 | 450/378 | 47.7 | rs162509 (C/T) |
| Wu et al. 2013 17 | China | 489 | 558 | 520/536 | 42.0 | rs162509 (C/T) |
TABLE 2.
Newcastle–Ottawa Scale for risk of bias assessment of studies included in the analysis.
| Study | Selection | Comparability | Exposure | Overall | |||||
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of exposed cohort | Selection of nonexposed | Ascertainment of exposure | Outcome not present at start | Assessment of exposure | Same method of ascertainment for cases and controls | Non‐response rate | |||
| Yuan 2010 | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Zawilla 2014 | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Noponen‐Hietala 2003 | ★ | ★ | ★ | ★ | ☆☆ | ★ | ☆ | ☆ | 5 |
| Hirose 2008 | ☆ | ★ | ★ | ☆ | ★☆ | ☆ | ★ | ☆ | 4 |
| Kelempisioti 2011 | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Dong 2007 | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Kitis 2018 | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Jiang 2017 | ★ | ★ | ★ | ★ | ★☆ | ★ | ★ | ☆ | 7 |
| Wu 2013 | ★ | ★ | ☆ | ★ | ★☆ | ★ | ★ | ☆ | 6 |
Note: ☆, score of 0; ★, score of 1; ★★, score of 2.
3.3. MMP2 rs243865
The analysis of MMP2 rs243865 was performed using data from two separate studies. 9 , 10 The frequency of individuals carrying the CC/CT genotype was ~96.4% (348 out of 361) among IDD cases, compared to 96.9% (499/515) in controls (TT), as observed in the selected studies. The analysis yielded an OR of 1.30, with a 95% CI ranging from 0.22 to 7.68 and a p‐value of 0.77. There was significant level of heterogeneity observed among the studies included (I 2 = 62%) (Figure 1).
FIGURE 1.
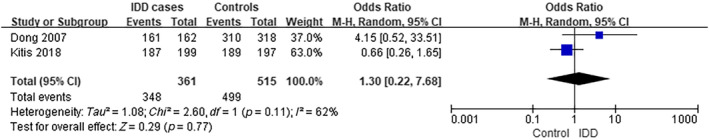
Forest plot of rs243865 in MMP2 gene and the risk of IDD (CC/CT vs. TT). CI, confidence interval; IDD, intervertebral disc degeneration; M–H, Mantel–Haenszel.
3.4. MMP3 rs731236
The analysis of MMP3 rs731236 was performed using data from three separate studies. 11 , 12 , 13 The frequency of individuals carrying the CC/CT genotype was ~39.2% (228/582) among IDD cases, compared to 29.3% (234/800) in controls (6A), as observed in the selected studies. The analysis yielded an OR of 1.57, with a 95% CI ranging from 0.93 to 2.68 and a p‐value of 0.09. There was a significant level of heterogeneity observed among the studies included (I 2 = 73%) (Figure 2). In addition, the frequency of 5A5A genotype carriers was ~11.0% (32/291) among IDD cases, compared to 7.25% (29/400) in controls (5A6A/6A6A), as observed in the selected studies. The analysis yielded an OR of 1.54, with a 95% CI ranging from 0.89 to 2.67 and a p‐value of 0.12. There was a low level of heterogeneity observed among the studies included (I 2 = 39%) (Figure 3).
FIGURE 2.
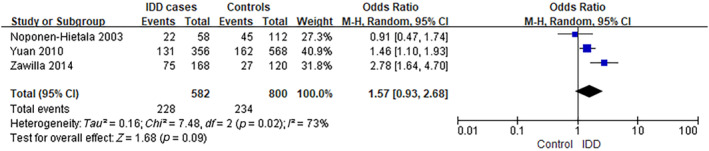
Forest plot of rs731236 in MMP3 gene and the risk of IDD (5A vs. 6A). CI, confidence interval; IDD, intervertebral disc degeneration; M–H, Mantel–Haenszel.
FIGURE 3.
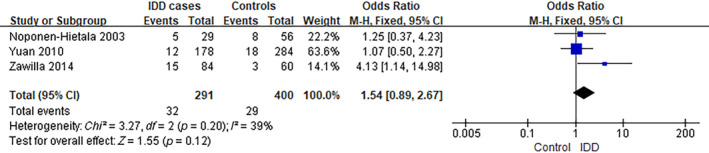
Forest plot of rs731236 in MMP3 gene and the risk of IDD (5A5A vs. 5A6A/6A6A). CI, confidence interval; IDD, intervertebral disc degeneration; M–H, Mantel–Haenszel.
3.5. MMP9 rs17576
The analysis of MMP9 rs17576 was performed using data from two separate studies. 14 , 15 The frequency of individuals carrying the G genotype was ~64.8% (1292/1994) among IDD cases, compared to 58.6% (1339/2284) in controls (A), as observed in the selected studies. The analysis yielded an OR of 1.15, with a 95% CI ranging from 0.88 to 1.51 and a p‐value of 0.30. There was a significant level of heterogeneity observed among the studies included (I 2 = 65%) (Figure 4). However, the frequency of GG genotype carriers was ~44.4% (443/997) among IDD cases, compared to 36.6% (418/1142) in controls (GA/AA), as observed in the selected studies. The analysis yielded an OR of 1.30, with a 95% CI ranging from 1.09 to 1.55 and a p‐value of 0.004. There was a low level of heterogeneity observed among the studies included (I 2 = 40%) (Figure 5).
FIGURE 4.
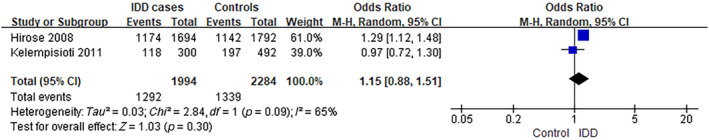
Forest plot of rs17576 in MMP9 gene (G vs. A) and the risk of IDD. CI, confidence interval; IDD, intervertebral disc degeneration; M–H, Mantel–Haenszel.
FIGURE 5.
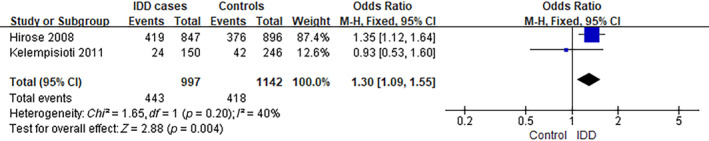
Forest plot of rs17576 in MMP9 gene (GG vs. GA/AA) and the risk of IDD. CI, confidence interval; IDD, intervertebral disc degeneration; M–H, Mantel–Haenszel.
3.6. ADAMTS5 rs162509
The analysis of ADAMTS5 rs162509 and G genotype was performed using data from two separate studies. 16 , 17 The frequency of individuals carrying the G genotype was ~61.9% (1093/1766) among IDD cases, compared to 59.2% (1073/1814) in controls, as observed in the selected studies. The analysis yielded an OR of 1.13, with a 95% CI ranging from 0.77 to 1.67 and a p‐value of 0.53. There was a significant level of heterogeneity observed among the studies included (I 2 = 88%) (Figure 6).
FIGURE 6.
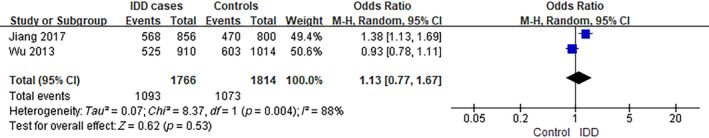
Forest plot of rs162509 in ADAMTS5 gene and the risk of IDD (G vs. C). CI, confidence interval; IDD, intervertebral disc degeneration; M–H, Mantel–Haenszel.
4. DISCUSSION
In fact, extensive research has been carried out regarding the etiology and pathogenesis of IDD, but there are still many aspects that have not been fully elucidated. The development of intervertebral disc degenerative diseases seems to be influenced by a combination of genetic susceptibility and environmental factors. 6 , 18 , 19 However, research suggests that genetic factors have a predominant influence on the development of musculoskeletal degenerative conditions such as intervertebral disc degeneration and osteoarthritis, while environmental influences do not significantly contribute to their occurrence. 7 , 20 , 21 Therefore, identifying genetic variations linked to vulnerability towards intervertebral disc degeneration enables the prediction of the probability of developing intervertebral disc degenerative disease and its phenotypes, which is beneficial for the development of personalized prediction models in the future. 22 The current research offers a thorough investigation of the association between extracellular matrix‐degrading enzymes and vulnerability to IDD. The results of this research suggest a notable correlation between the MMP‐9 rs17576 mutation and an elevated susceptibility to disc degeneration.
The involvement of MMPs in the breakdown of the disc matrix is considered crucial in the development of IDD. MMP‐2 exhibits a preference for denatured collagen as its substrate. 23 Our analysis showed that MMP‐2 CC/CT genotype did not increase the risk of LDD. However, it is worth noting that Dong et al. identified the ‐1306C/T polymorphism of the MMP2 gene as a genetic predisposing factor for IDD in young Chinese individuals. This study revealed that individuals carrying the CC genotype exhibited a 3.08 times higher susceptibility to developing IDD compared to those harboring at least one T allele (TT or CT). 9 A subsequent research demonstrated a similar phenomenon in the ‐735C/T polymorphism of MMP2. The investigation revealed that individuals with TT or CT genotypes had a notably decreased likelihood (0.413‐fold) of developing IDD, whereas those with the CC genotype were at an increased risk (~2.5 times) compared to patients possessing the TT genotype. 24 Analogous to the ‐1306C/T SNP, it was observed that the presence of the T allele resulted in the disruption of a Sp1‐binding site, leading to a subsequent decrease in transcription. Conversely, the C allele, which is regarded as the wild‐type variant, exhibited an association with enhanced transcription. 24 These studies demonstrated the presence of multiple adjacent Sp1‐binding sites, the polymorphisms of which are associated with genetic susceptibility to IDD. 9 , 24 However, no significant correlation was found between the MMP2 polymorphism and the incidence of IDD in the Turkish population, leaving uncertainty regarding its contribution as a genetic susceptibility factor. 10 Thus, the current study indicated that there is a variation in the MMP2 polymorphism among different ethnic groups. Further investigations are warranted.
MMP‐3 belongs to the stromelysins subgroup of MMPs, 23 which primarily participate in the degradation process of laminas, proteoglycans, and other components within the extracellular matrix of intervertebral discs, as well as indirectly contributing to disc degradation by triggering the activation of other MMPs. 25 The levels of MMP‐3 have also been demonstrated to increase in reaction to inflammatory stimuli. 13 Additionally, the most extensively studied single nucleotide polymorphism (SNP) of MMP3 is the 5A variant allele situated in the gene's promoter region. The combined findings, however, do not demonstrate statistically significant correlation. In addition, in the study of MMP3 rs591058 C/T allele, Luo et al. 26 found a significant association between the T allele and an elevated risk of LDD (OR 1.37, p = 0.015), but Harshith et al. 27 did not find this association (p > 0.05). It is imperative to conduct an investigation into the correlation between MMP‐3 gene polymorphism and vulnerability to IDD in order to guarantee the credibility of forthcoming findings. Furthermore, our study found that the MMP9 GG allele significantly increased the risk of LDD compared with other genotypes. MMP‐9, a gelatinase that exhibits different levels of expression, has been associated with IDD. Although several reviews have indicated an increased risk of IDD associated with MMPs, the pooled results demonstrated that the majority of MMPs did not significantly elevate this risk, except for the GG allele of MMP9. However, given the limited research available, the polymorphism of MMPs still deserves further investigation and analysis.
A series of secreted ADAMTS proteases is encoded by the ADAMTS genes, which are recognized for their regulatory roles in coagulation, development, angiogenesis, and ECM homeostasis. 18 The expression of several ADAMTSs has been identified in connective tissues. 28 Research had demonstrated an association between variants within several ADAMTS genes and the occurrence of degenerative disorders affecting the musculoskeletal system, such as LDD, 29 Achilles tendon pathology, 30 and osteoarthritis. 31 , 32 Currently, ADAMTS4 and ADAMTS5 are the most extensively studied genes in the ADAMTSs family, playing a vital part in the development and advancement of cartilage injury. 33 , 34 The research on ADAMTS5 in IDD disease has been more extensive among them. Therefore, our current research is focused on investigating the variations within ADAMTS5 and their correlation with the susceptibility to IDD disease. The recognition of ADAMTS5 as biomarkers for assessing the extent of initial‐stage damage in cartilage affected by osteoarthritis has been proven through many experimental studies conducted both in vivo and in vitro. However, there are few studies on ADAMTS5 in IDD. ADAMTS5 (rs162509) did not appear to increase the risk of IDD based on the current analysis, but there was high heterogeneity between the two studies. Another study reported that ADAMTS5 (rs162509) increased the risk of IDD (OR 1.281, p = 0.04), but it was not included in the analysis because it only reported the OR value and p value. 29
It is noteworthy that researchers have found that the synergistic effect of genetic and environmental factors increases the risk of developing IDD. 11 Therefore, future endeavors ought to vigorously intensify their focus on unraveling the intricate interplay between genetic and environmental factors, thereby shedding light on the multifaceted and complex mechanisms that underlie the development of IDD. Furthermore, delving into the upstream mechanisms that regulate the expression of extracellular matrix degrading enzymes holds significant research importance. By targeting upstream regulatory genes that orchestrate the synthesis and catabolism of genes crucial for matrix production and degradation, we can gain valuable insights into how genetic variations modulate the activity of these enzymes and their precise roles in the pathogenesis of IDD. This endeavor not only sheds new light on the etiology of IDD but also presents promising molecular targets for the development of targeted therapeutic interventions.
Despite strictly adhering to the methods of analysis, there are still certain limitations. Firstly, there is a high degree of heterogeneity in some of the results obtained from this study. Although all the studies analyzed followed a case–control design, propensity matching was not employed, which may introduce confounding factors and could be one of the reasons for significant heterogeneity observed in this analysis. In addition, the pathogenesis of IDD is a complex interplay between genetic factors at various polymorphic loci and the cumulative impact of gene–environment interactions, which can either enhance or obscure the functional roles of involved genes.
5. CONCLUSION
The current analysis demonstrated a noteworthy correlation between the GG genotype of MMP‐9 rs17576 and susceptibility to IDD. The available evidence is insufficient to substantiate the correlation between other extracellular matrix‐degrading enzymes and susceptibility to IDD. In consideration of the limitations inherent in the current study, it is strongly recommended that further studies be conducted encompassing larger sample sizes across diverse ethnicities to provide a more comprehensive understanding of the true impact of these polymorphisms on susceptibility to IDD.
AUTHOR CONTRIBUTIONS
Di Zhao: Conceptualization; formal analysis; investigation; validation; writing—original daft. Yao‐xing Dou: Methodology; data curation; software; formal analysis; writing—original daft. Ling‐feng Zeng: Formal analysis; software; methodology. Yan‐hong Han: Data curation; software; formal analysis. Fang‐zheng Lin: Data curation; formal analysis; visualization. Nan‐jun Xu: Formal analysis; visualization. Jun Liu: Conceptualization; supervision; writing—review and editing. Yu‐ping Zeng: Conceptualization; investigation; writing—review and editing. All authors have approved the submitted manuscript.
FUNDING INFORMATION
This study was supported by the National Key Research and Development Program (2021YFC1712804).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
FIGURE S1. PRISMA flow diagram.
ACKNOWLEDGMENT
We are deeply grateful to the authors who have provided the original research materials.
Zhao D, Dou Y, Zeng L, et al. The effects of extracellular matrix‐degrading enzymes polymorphisms on intervertebral disc degeneration. JOR Spine. 2024;7(4):e70012. doi: 10.1002/jsp2.70012
Di Zhao and Yao‐xing Dou contributed equally to this study.
Contributor Information
Jun Liu, Email: liujun3040@126.com.
Yu‐ping Zeng, Email: zyp8365@163.com.
REFERENCES
- 1. Rodrigues‐Pinto R, Richardson SM, Hoyland JA. An understanding of intervertebral disc development, maturation and cell phenotype provides clues to direct cell‐based tissue regeneration therapies for disc degeneration. Eur Spine J. 2014;23(9):1803‐1814. [DOI] [PubMed] [Google Scholar]
- 2. Battie MC, Lazary A, Fairbank J, et al. Disc degeneration‐related clinical phenotypes. Eur Spine J. 2014;23(Suppl 3):S305‐S314. [DOI] [PubMed] [Google Scholar]
- 3. Luo X, Pietrobon R, Sun SX, Liu GG, Hey L. Estimates and patterns of direct health care expenditures among individuals with back pain in the United States. Spine (Phila Pa 1976). 2004;29(1):79‐86. [DOI] [PubMed] [Google Scholar]
- 4. Kadow T, Sowa G, Vo N, Kang JD. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthop Relat Res. 2015;473(6):1903‐1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hadjipavlou AG, Tzermiadianos MN, Bogduk N, Zindrick MR. The pathophysiology of disc degeneration: a critical review. J Bone Joint Surg Br. 2008;90(10):1261‐1270. [DOI] [PubMed] [Google Scholar]
- 6. Battie MC, Videman T, Levalahti E, Gill K, Kaprio J. Genetic and environmental effects on disc degeneration by phenotype and spinal level: a multivariate twin study. Spine (Phila Pa 1976). 2008;33(25):2801‐2808. [DOI] [PubMed] [Google Scholar]
- 7. Battie MC, Videman T, Gibbons LE, Fisher LD, Manninen H, Gill K. Volvo award in clinical sciences. Determinants of lumbar disc degeneration. A study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine (Phila Pa 1976). 1995;20(24):2601‐2612. [PubMed] [Google Scholar]
- 8. Walker CT, Bonney PA, Martirosyan NL, Theodore N. Genetics underlying an individualized approach to adult spinal disorders. Front Surg. 2016;3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong DM, Yao M, Liu B, Sun CY, Jiang YQ, Wang YS. Association between the ‐1306C/T polymorphism of matrix metalloproteinase‐2 gene and lumbar disc disease in Chinese young adults. Eur Spine J. 2007;16(11):1958‐1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitis S, Coskun ZM, Tasdemir P, Tuncez E, Zamani AG, Acar A. Analysis of genetic polymorphisms associated with intervertebral disc degeneration. Cell Mol Biol (Noisy‐le‐Grand). 2018;64(10):61‐65. [PubMed] [Google Scholar]
- 11. Yuan HY, Tang Y, Liang YX, et al. Matrix metalloproteinase‐3 and vitamin d receptor genetic polymorphisms, and their interactions with occupational exposure in lumbar disc degeneration. J Occup Health. 2010;52(1):23‐30. [DOI] [PubMed] [Google Scholar]
- 12. Noponen‐Hietala N, Kyllonen E, Mannikko M, et al. Sequence variations in the collagen IX and XI genes are associated with degenerative lumbar spinal stenosis. Ann Rheum Dis. 2003;62(12):1208‐1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zawilla NH, Darweesh H, Mansour N, et al. Matrix metalloproteinase‐3, vitamin D receptor gene polymorphisms, and occupational risk factors in lumbar disc degeneration. J Occup Rehabil. 2014;24(2):370‐381. [DOI] [PubMed] [Google Scholar]
- 14. Hirose Y, Chiba K, Karasugi T, et al. A functional polymorphism in THBS2 that affects alternative splicing and MMP binding is associated with lumbar‐disc herniation. Am J Hum Genet. 2008;82(5):1122‐1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelempisioti A, Eskola PJ, Okuloff A, et al. Genetic susceptibility of intervertebral disc degeneration among young Finnish adults. BMC Med Genet. 2011;12:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang H, Yang Q, Jiang J, Zhan X, Xiao Z. Association between COL11A1 (rs1337185) and ADAMTS5 (rs162509) gene polymorphisms and lumbar spine pathologies in Chinese Han population: an observational study. BMJ Open. 2017;7(5):e15644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu N, Chen J, Liu H, et al. The involvement of ADAMTS‐5 genetic polymorphisms in predisposition and diffusion tensor imaging alterations of lumbar disc degeneration. J Orthop Res. 2014;32(5):686‐694. [DOI] [PubMed] [Google Scholar]
- 18. Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ikegawa S. The genetics of common degenerative skeletal disorders: osteoarthritis and degenerative disc disease. Annu Rev Genomics Hum Genet. 2013;14:245‐256. [DOI] [PubMed] [Google Scholar]
- 20. Zengini E, Finan C, Wilkinson JM. The genetic epidemiological landscape of hip and knee osteoarthritis: where are we now and where are we going? J Rheumatol. 2016;43(2):260‐266. [DOI] [PubMed] [Google Scholar]
- 21. Loughlin J. The genetic epidemiology of human primary osteoarthritis: current status. Expert Rev Mol Med. 2005;7(9):1‐12. [DOI] [PubMed] [Google Scholar]
- 22. Lv ZT, Gao ST, Cheng P, et al. Association between polymorphism rs12722 in COL5A1 and musculoskeletal soft tissue injuries: a systematic review and meta‐analysis. Oncotarget. 2018;9(20):15365‐15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ricard‐Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3(1):a4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Gu Z, Qiu G. Association of the polymorphism of MMP2 with the risk and severity of lumbar disc degeneration in the Chinese Han population. Eur Rev Med Pharmacol Sci. 2013;17(13):1830‐1834. [PubMed] [Google Scholar]
- 25. Takahashi M, Haro H, Wakabayashi Y, Kawa‐uchi T, Komori H, Shinomiya K. The association of degeneration of the intervertebral disc with 5a/6a polymorphism in the promoter of the human matrix metalloproteinase‐3 gene. J Bone Joint Surg Br. 2001;83(4):491‐495. [DOI] [PubMed] [Google Scholar]
- 26. Luo Y, Wang J, Pei J, et al. Interactions between the MMP‐3 gene rs591058 polymorphism and occupational risk factors contribute to the increased risk for lumbar disk herniation: a case‐control study. J Clin Lab Anal. 2020;34(7):e23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harshitha SM, Sibin MK, Chetan GK, Dhananjaya IB. Association of CILP, COL9A2 and MMP3 gene polymorphisms with lumbar disc degeneration in an Indian population. J Mol Neurosci. 2018;66(3):378‐382. [DOI] [PubMed] [Google Scholar]
- 28. Kevorkian L, Young DA, Darrah C, et al. Expression profiling of metalloproteinases and their inhibitors in cartilage. Arthritis Rheum. 2004;50(1):131‐141. [DOI] [PubMed] [Google Scholar]
- 29. Rajasekaran S, Kanna RM, Senthil N, et al. Genetic susceptibility of lumbar degenerative disc disease in young Indian adults. Eur Spine J. 2015;24(9):1969‐1975. [DOI] [PubMed] [Google Scholar]
- 30. El KL, Posthumus M, Collins M, Handley CJ, Cook J, Raleigh SM. Polymorphic variation within the ADAMTS2, ADAMTS14, ADAMTS5, ADAM12 and TIMP2 genes and the risk of Achilles tendon pathology: a genetic association study. J Sci Med Sport. 2013;16(6):493‐498. [DOI] [PubMed] [Google Scholar]
- 31. Poonpet T, Honsawek S, Tammachote N, Kanitnate S, Tammachote R. ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet Mol Res. 2013;12(4):5301‐5309. [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez‐Lopez J, Pombo‐Suarez M, Loughlin J, et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthr Cartil. 2009;17(3):321‐327. [DOI] [PubMed] [Google Scholar]
- 33. Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD. Inhibition of ADAM‐TS4 and ADAM‐TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem. 2002;277(25):22201‐22208. [DOI] [PubMed] [Google Scholar]
- 34. Verma P, Dalal K. ADAMTS‐4 and ADAMTS‐5: key enzymes in osteoarthritis. J Cell Biochem. 2011;112(12):3507‐3514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. PRISMA flow diagram.


