Abstract
The use of combinatorial siRNAs shows great promise for drug discovery, but the identification of safe and effective siRNA combinations remains challenging. Here, we develop a massively multiplexed technology for systematic screening of siRNA-based cocktail therapeutics. We employ composite micro-carriers that are responsive to near infrared light and magnetic field to achieve photoporation-facilitated siRNA transfection to individual cells. Thus, randomized gene silencing by different siRNA formulations can be performed with high-throughput single-cell-based analyses. For screening anti-cancer siRNA cocktails, we test more than 1300 siRNA combinations for knocking down multiple genes related to tumor growth, discovering effective 3-siRNA formulations with an emphasis on the critical role of inhibiting Cyclin D1 and survivin, along with their complementary targets for synergic efficacy. This approach enables orders of magnitude reduction in time and cost associated with largescale siRNA screening, and resolves key insights to siRNA pharmacology that are not permissive to existing methods.
Subject terms: High-throughput screening, Drug delivery, Biomedical engineering
Combined use of multiple siRNAs promises a new path for drug development. Here, the authors demonstrate a multiplexed combinatorial screening of thousands of siRNA formulations through highthroughput single-cell gene silencing, discovering the most effective anti-cancer siRNA cocktail therapeutics.
Introduction
Small interfering ribonucleic acids (siRNAs) are short double-stranded RNA sequences that bind to targeted messenger RNAs (mRNAs) after being introduced into cell cytoplasm, which action through a process known as RNA interference (RNAi), causing the targeted mRNA to break/degrade to stop its translation, thus suppressing gene expression. Since the discovery of RNAi1 and the demonstration of siRNA in post-transcriptional gene silencing2,3, there has been tremendous interest and effort to harness siRNAs for biomedical research and drug development4. However, for more than two decades, the success of using siRNA to treat human diseases has been very limited; thus far, US Food and Drug Administration (FDA) has approved only five siRNA-based therapeutics: patisiran, givosiran, lumasiran, inclisiran, and vutrisiran5. Not only is the delivery of siRNAs to the target cells a challenge, effective gene silencing by siRNA is also hindered by the presence of redundant or complementary signal transduction pathways6, which may diminish the action of a single siRNA and substantially offset the silencing effects.
To address these issues, researchers have devised strategies that target multiple genes or modulate both the upstream and downstream signaling pathways. For example, simultaneous silencing of Bcl-xl, Bcl-2, and Mcl-1 can potentially be forged as a treatment for breast cancer7; efficient and durable combinatorial RNAi in vivo was achieved by targeting redundant RAF signaling node in a mouse cancer model8; a nanovaccine carrying multiple siRNA elements exhibited a synergistic antitumor effect compared with single-target therapy9. These studies promise siRNA combinations for superior therapeutic efficacy in comparison to single gene silencing approaches. In fact, cocktail therapy, where three or more anti-retroviral medicines are simultaneously prescribed, has been practiced clinically for a long time and been the most effective anti-HIV treatment10. Likewise, the use of cocktail siRNAs for multiplexed gene silencing has been explored in the development of next-generation therapy. Recently, Tieu et al. showed that combinatorial knockdown screen identifies paired regulators of CAR T cell function, and multi-gene disruption broadly enhances the anti-tumor activity of CAR T cells11. Commonly, the selection of combinatorial siRNAs to form the most effective cocktail primarily relies on examining the silencing efficacy and safety of individual siRNAs, which is time-consuming and lacks the information regarding the synergic or counteractive function among them. If numerous siRNAs are concerned, it is challenging to screen different siRNA combinations and to determine the optimal cocktails. A quantitative, massively multiplexed, and systematic high-throughput platform technology would be greatly beneficial to enhance the clinical adoption and speed up the development of siRNA-based therapeutics.
This work aims to address the unmet needs by developing a high-throughput technology for screening siRNA-based cocktail therapeutics in a massively multiplexed, quantitative, and systematic manner. Technically, as one of the key innovations, we employ a multilayer functionalization of hierarchically assembled nano-/micro composite carriers (Knock-beads), rendering them with responsiveness to near infrared light and magnetic field to facilitate photoporation-induced cellular transfection of RNAs with single cell encoding capability. In a large population of cultured cells, randomized gene silencing by different siRNA variants is performed, recorded, and analyzed in a highly parallel manner. Our technology is capable of scaling to screening hundreds or even thousands of siRNA cocktail formulations in a single well, accelerating the testing of siRNA cocktails. Such benefits are not just limited to reducing the time and cost associated with siRNA screening by powers of magnitude, but also answer fundamental siRNA questions that are not addressable by existing methods. In addition to the technical advancement, as a proof-of-concept, we first perform a small screen with six cell apoptosis or cell cycle related siRNAs to test 54 siRNA combinations to seek the most effective cocktail for inhibiting cancer cell growth. With a customized multiplexed encoding strategy, we further demonstrate an expanded screening involving 20 siRNA target genes and test more than 1300 cocktail formulations composed of 1, 2, or 3 siRNAs for combinatorial knockdown of genes related to tumor growth, leading to the identification of the most effective 3-siRNA combinations as potential anti-cancer therapeutics with an emphasis on the critical role of inhibiting Cyclin D1 and survivin, as well as their complementary gene targets for synergic co-operation. We believe that this study demonstrates the path to a systematic and high-throughput platform technology that is greatly useful to accelerate further development of siRNA-based therapeutics and enhance their clinical adoption.
Results
System design
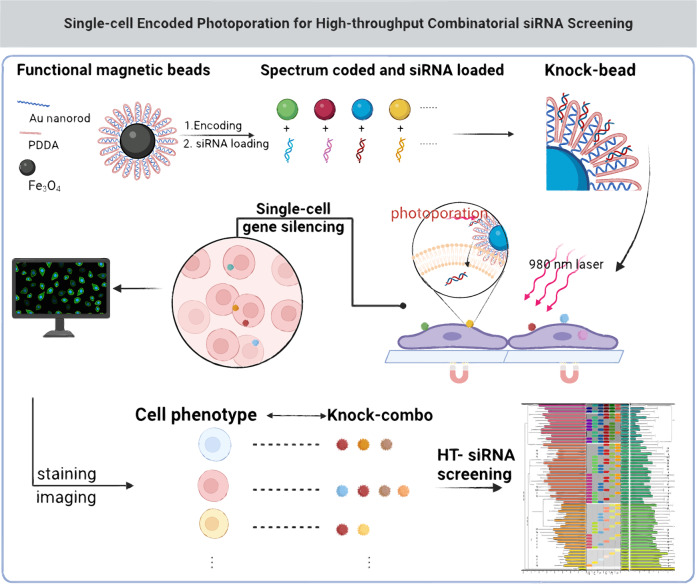
The system combined nanomaterial sensitized photoporation and single-cell encoded intracellular delivery of RNAs (scCode-fection), and was capable of tracing the gene silencing effect in a large number of single cells by a one or multiple siRNAs (Knock-combo) with a one-shot assay, thus to determine the best combination by single-cell based analysis. All the siRNA Knock-combos were applied to the same batch of cells subject to the same culture and treatment condition, thus avoiding any biased results. A multilayer functionalization was implemented for the hierarchically assembled nano-/micro composite carriers, Knock-beads. By using the multi-functional Knock-beads, photoporation facilitated intracellular delivery of RNAs could be tracked with single-cell resolution, and the associated gene silencing results could also be recorded and analyzed (Fig. 1).
Fig. 1. Illustration of the scCode-fection technique by using hierarchically assembled Knock-beads.
Fe3O4 beads are combined with Au nanorods and coated with PDDA, which binds siRNA, to create Knock-beads. The Au layer provides Knock-beads with photothermal responsiveness, facilitating intracellular delivery of siRNA through photoporation by 980 nm NIR laser. By utilizing a magnetic field, Knock-beads are immobilized on top of cells, enabling efficient cellular transfection of siRNAs by photoporation and single-cell encoded gene silencing in a large number of cells. Through only one-shot assay in a cell culture, more than 1300 siRNA combinations can be simultaneously analyzed to reveal the optimal siRNA therapeutic cocktail. Created in BioRender. Guo, F. (2024) BioRender.com/l65e561.
Specifically, a multiscale assembly of micro- and nano- structures along with a functionalization scheme was developed to fabricate the Knock-bead with multiple functions, including magnetic controllability for tight cell-bead interfacing, appropriate surface coating for siRNA loading/off-loading, and responsiveness to near infrared light (NIR) for photoporation. The microscale core, iron oxide (Fe3O4) beads, performs 3 functions: (1) a vehicle for siRNA delivery; (2) a spatially resolvable cellular interface for immobilization on the cell membrane by a magnetic field; (3) the means for fluorescence encoding of single-cell gene silencing by different siRNA combinations. For N siRNAs, the total number of possible gene silencing combinations would be:
| 1 |
If 6 siRNAs are considered, there are 64 possible cocktail combinations, all of which are tested in a one-shot assay. The tight interface between the Knock-beads and cells by magnetic force facilitates NIR-controlled photoporation and intracellular delivery of siRNAs without the Knock-bead entering cells, which remain on the cell membrane for later decoding purpose (Fig. 1).
Hierarchical assembly of micro-/nano-structures
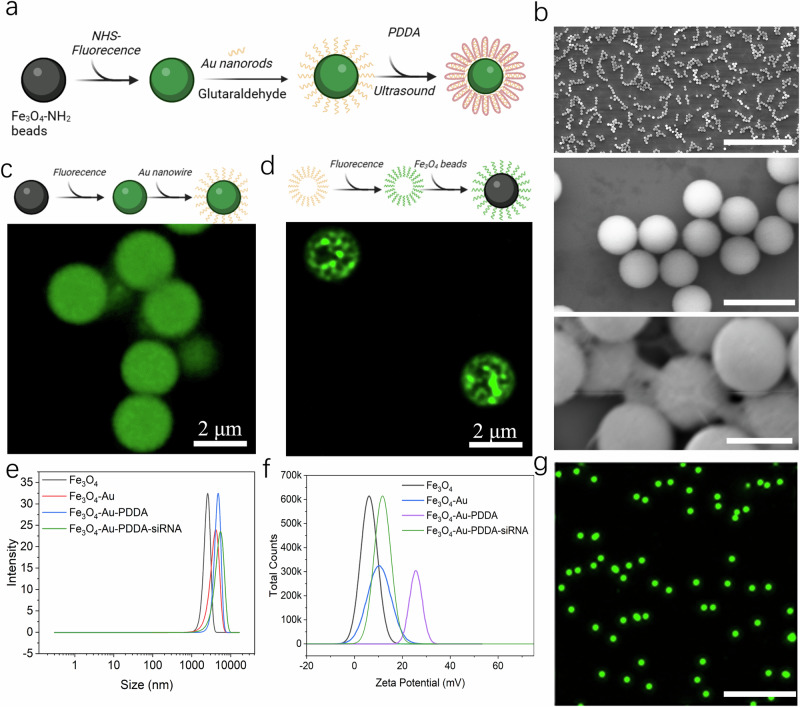
As the first key step of the scCode-fection, the establishment of Knock-bead will enable photoporation-facilitated intracellular delivery of siRNAs. A Knock-bead was produced by hierarchical assembly of microscale Fe3O4 core and nanoscale gold (Au) nanorod, which was achieved by covalent linkage of Au nanorods to the surface of Fe3O4 beads by glutaraldehyde crosslinking (Fig. 2a, b). By our design, the Au nanorods provide photo-thermal responsiveness to NIR to generate localized heating; temporarily disrupting the cell membrane and enable the transfer of exogenous molecules into the cells. The nanoscale size also allows for precise localization of the NIR energy transduction and better control over targeted intracellular delivery in individual single cells12–14. The Fe3O4-Au composite was spherical and homogeneously dispersed as shown by scanning electron microcopy (SEM); the Au nanorods were observed to form a rough texture on the Fe3O4 beads (Fig. 2b). To further prove the successful assembly of the Au nanorods on the Fe3O4 beads, prior to the Fe3O4-Au crosslinking, we stained either the Fe3O4 beads or the Au nanorods with a fluorescent dye (Alexa-647) for two batches of experiments, and obtained confocal microscopic images after the assembly procedure (Fig. 2c, d). The results showed different mode of fluorescent patterns as a result of the differential staining: solid sphere for dyes on Fe3O4 beads and sporadic puncta for dyes on Au nanorods, suggesting successful and stable linkage between the two components.
Fig. 2. Physical characterization of Knock-beads.
a The chemical synthesis route of Knock-beads. Created in BioRender. Guo, F. (2024) BioRender.com/y59k149. b SEM images showing the core part of Knock-beads at different scales: Fe3O4-Au (top, scale bar 70 μm), Fe3O4 beads (middle, scale bar 5 μm), and Fe3O4-Au (bottom, scale bar 2 μm). c Fluorescence image showing Fe3O4 magnetic beads that were first labeled with Alexa-647 dye and then conjugated to Au nanorods. Created in BioRender. Guo, F. (2023) BioRender.com/d92g406. d Fluorescence image showing the Au nanorods that were first labeled with Alexa-647 dye and then conjugated to Fe3O4 magnetic beads. Created in BioRender. Guo, F. (2023) BioRender.com/d92g406. e Dynamic light scattering results showing the size variation of Fe3O4, Fe3O4-Au, Fe3O4-Au-PDDA beads and Fe3O4-Au-PDDA-siRNA conjugates (8 × 105 beads, 20 pmol siRNA input). f Zeta potential analysis of Fe3O4, Fe3O4-Au, Fe3O4-Au-PDDA beads and Fe3O4-Au-PDDA-siRNA conjugates (8 × 105 beads, 20 pmol siRNA input). g Fluorescent image of Knock-beads in PBS solution, scale bar 20 μm. Three independent experiments were conducted (b, c, d, g). Source data are provided as a Source Data file.
To load siRNA on the Fe3O4-Au composite assembly to form the functional Knock-beads, the outer layer of the Fe3O4-Au composite is coated with poly-dimethyl diallyl ammonium chloride (PDDA) to facilitate the absorption of negatively charged nucleic acids (e.g. double stranded siRNA, Fig. 2a). Particle analysis clearly showed the incremental size change of the beads (Fe3O4, Fe3O4-Au, Fe3O4-Au-PDDA, and Fe3O4-Au-PDDA-siRNA) throughout the functionalization procedures (Fig. 2e). The addition of Au nanorods resulted in an increase in diameter from ~2.8 μm to ~3.8 μm and PDDA coating further increased the Knock-beads to ~4.4 μm in diameter. After loading siRNAs, the diameter slightly increased to ~4.5 μm. The surface charge was also changed, the zeta potential for Fe3O4, Fe3O4-Au, Fe3O4-Au-PDDA, and Fe3O4-Au-PDDA-siRNA was +6 mV, +11 mV, +24 mV and + 12 mV, respectively (Fig. 2f, Supplementary Note 1, Supplementary Fig. 1). The positively charged surface facilitated effective absorption of negatively charged siRNAs to form the final functional Knock-beads. The beads showed a good dispersion in solution to facilitate subsequent homogenous cellular interfacing (Fig. 2g).
Photothermal responsiveness
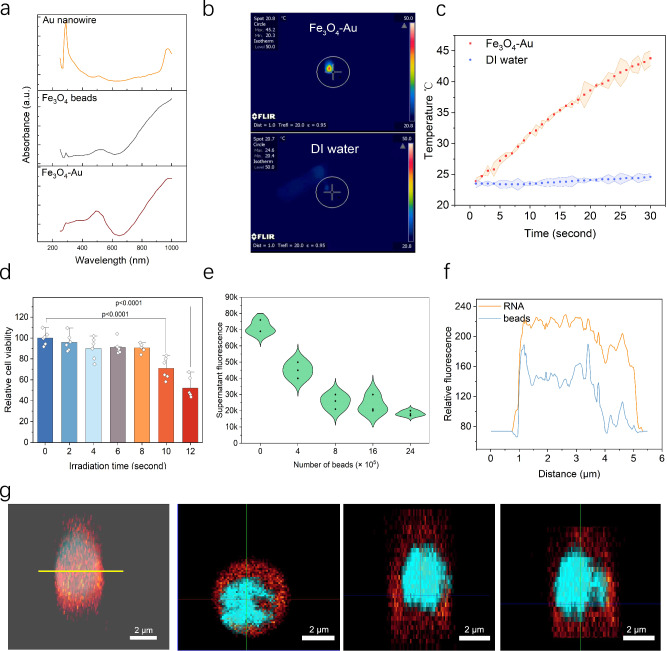
The wavelength of the laser irradiation plays a crucial role in the generation of photothermal effect for cellular transfection purpose. We measured the absorbance spectrum of the Fe3O4-Au-PDDA composite to assess its response to different wavelengths of light. For the Au nanorods used in our study, the absorbance peaked at approximately 980 nm (Fig. 3a), which could be attributed to the high aspect ratio of the gold nanorods and indicated a suitable wavelength for subsequent photoporation15. The additional absorption peak at approximately 510 nm was likely attributed to inter-band electronic transitions16,17. The tuned responsiveness to 980 nm NIR also ensures good biocompatibility and minimal photothermal damage to biological samples when compared to the use of laser in UV or blueish ranges. Under NIR irradiation at a power of 0.6 W/cm2 (pulse width 16 μs), the local temperature surrounding the Knock-beads reached a temperature around 45 °C, and prolonged irradiation induced accumulated thermal effects. The visualization of this photothermal effect was captured and analyzed using a NIR camera, as presented in Figs. 3b, c. Without the addition of Knock-beads, the temperature of DI water only exhibited a slight increase of approximately 3 °C following the same NIR irradiation. These results demonstrate the sensitive response of the Knock-beads to 980 nm NIR.
Fig. 3. Photothermal responsiveness and siRNA loading efficiency of Knock-beads.
a UV absorbance spectrum of Au nanorods, Fe3O4, and Fe3O4-Au beads. b Infrared imaging of the photothermal effect of Knock-beads (top) and DI water (control, bottom). A 980 nm laser (16 μs pulse width) with an intensity of 0.6 W/cm2 was applied for 30 s. c Time—temperature graph of Knock-beads (red) or DI water (blue) when stimulated by 980 nm laser. d Impact of different NIR-irradiation times (2 s, 4 s, 6 s, 8 s, 10 s, and 12 s) on cell viability 24 h after photoporation with Knock-beads. Data are presented as the mean ± SD, n = 6, analyzed using unpaired one-way ANOVA. e Characterization of siRNA loading efficiency of Knock-beads using prelabeled siRNAs pull-down assay. f The fluorescence distribution of a siRNA-loaded Knock-bead along the yellow line in panel (g). g 3D fluorescence images of Knock-beads (blue) loaded with siRNA (red) and the top, left, and front view. Source data are provided as a Source Data file.
For effective gene silencing screening, cell viability is crucial for unbiased results, especially after the photoporation using the Knock-beads. 24 h following NIR irradiation, we assessed cell viability using the 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 3d). At specified laser power (e.g. 0.6 W/cm2) with varying irradiation times (2, 4, 6, 8, 10, and 12 s), the cell viability was evaluated to optimize the irradiation parameters. We found that more than 85% of the cells were viable after an 8 s NIR irradiation at 0.6 W/cm2; longer NIR irradiation of 10 s and 12 s reduced the cell viability to 70% and 55%, respectively. Therefore, NIR irradiation duration was kept brief ( < 8 s) to avoid any major damage or interference to regular cell function.
Knock-bead mediated loading and release of siRNAs
The efficiency of siRNA loading onto the Knock-beads was assessed by measuring the decrease in fluorescence of a solution containing prelabeled siRNAs following a pull-down assay by the Knock-beads. Our results suggest that approximately 64.3% of siRNA (2 μL, 10 μM) was absorbed by ~8 × 105 beads. Increasing Knock-beads (16 × 105, 24 × 105, and 32 × 105) tended to deplete the siRNAs from the solution by 66.0%, 73.8% and 80.3% respectively, confirming an efficient siRNA loading on the Knock-beads by electrostatic interaction (Fig. 3e). As a result of siRNA binding (up to 80 pmol for 8 × 105 beads), the Knock-beads zeta potential decreased from 22 ± 0.99 mV to 4.71 ± 1.08 mV. Further increasing the siRNA inputs (e.g. 160 pmol) caused the zeta potential to turn negative (Supplementary Fig. 2), suggesting an overloading status. In our study, the siRNA was loaded to Knock-beads at an input of 20 pmol, which is equivalent to 9.8 × 106 molecules per bead. The loading efficiency did not show any sequence specific variation (Supplementary Fig. 3). Fluorescence images were captured to further confirm the presence of RNA loading on the Knock-beads (Fig. 3f, g). In these images, the Fe3O4-Au core was stained with Alexa-568 (blue), while the RNA was stained with cy5 (red). The 3D sectioning images clearly showed that the RNAs were successfully absorbed onto the surface of the beads (Fig. 3g).
For releasing siRNA from the Knock-beads, it is expected that the NIR-induced local temperature increase in a Knock-bead disrupts the van der Waals interactions at the interface of the Fe3O4-Au surface and PDDA coating, causing the PDDA-siRNA to detach from the Knock-beads. For the loaded Knock-beads, NIR irradiation of 2–8 s induced a release of siRNA by 53.5–87% of the pre-loads (Supplementary Fig. 4). Such releasing profile also showed a good consistency across different siRNA sequences (Supplementary Fig. 5), thus ensuring the reliability for later use for high-throughput screening.
Photoporation facilitated intracellular delivery of siRNAs using Knock-beads
For a tight cellular interfacing between the Knock-beads and cells, a magnetic field (10 kilo-Gauss caused by Neodymium magnet) was utilized to strategically position the Knock-beads on individual cells. The Knock-beads were used to spatially encode the siRNA delivery into contacting cells via photoporation. Upon the off-loading of siRNA from the Knock-beads, beads remained on the cell for later decoding of gene silencing with single cell resolution. When applied to cultured A549 cells, we used pulsed NIR irradiation (0.6 W/cm2, 16 μs pulse width, 8 s) to induce photoporation for intracellular delivery of siRNAs. The success was evidenced by the presence of cy5-labeled siRNAs within the cytoplasmic region (Fig. 4a–g). The intracellular translocation of siRNA did not vary across different siRNAs (Supplementary Fig. 6). 5 h after photoporation, the siRNA signal was predominantly localized within the cytoplasm of the cells, indicating the successful internalization and intracellular delivery of the siRNAs. (Supplementary Fig. 7).
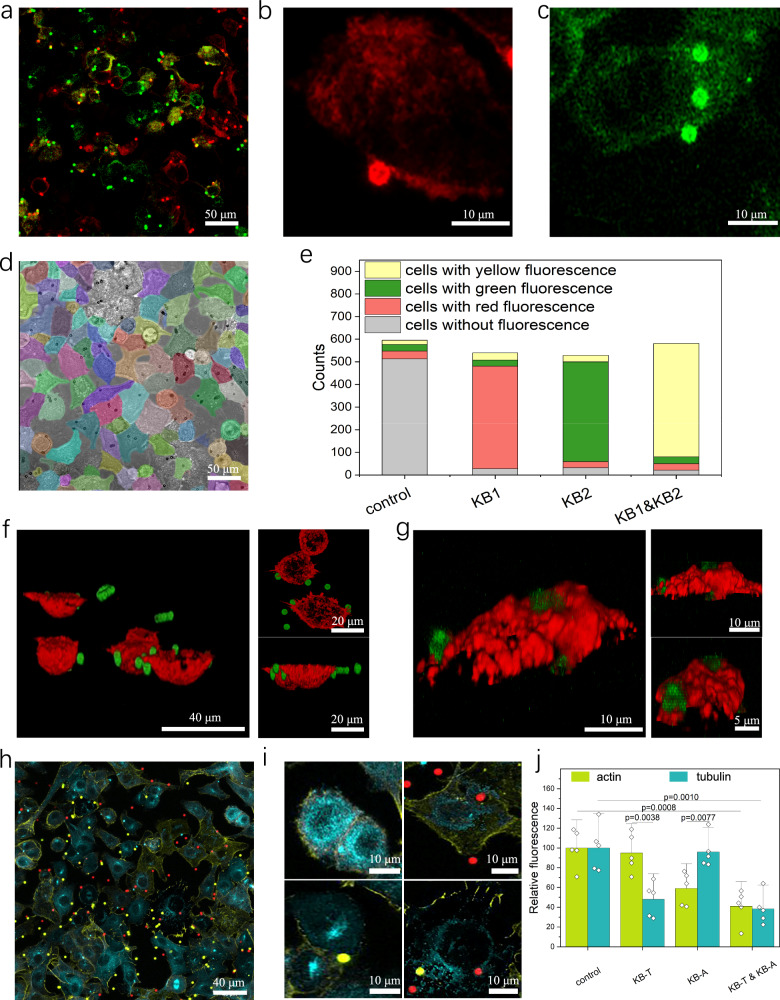
Fig. 4. Single-cell encoded siRNA delivery and gene silencing.
a–c Intracellular delivery with a magnetic field, where RNAs are labeled using cy5 (red) and Fam (green). Red beads are loaded with RNA-cy5, and green beads are loaded with RNA-Fam. d Cell segmentation visualized by a customize algorithm. The cell outline was visualized in different colors to help cell segmentation. e Number of cells categorized differently in (d): control refers cells without Knock-beads, KB1 refers cells with KB1 Knock-beads, KB2 refers cells with KB2 cells. f 3D images of Knock-beads(green) and diffused siRNA(red) and the front and top view. g 3D images of Knock-beads(green) and cell actin(red) and the front and top view. h Cells treated with two types of Knock-beads: anti-actin (KB-A, red) and anti-tubulin (KB-T, yellow). Actin is labeled in yellow, and tubulin is labeled in blue. i Magnified view of (h) cells that contacts with different Knock-beads: top-left is control, top-right contacts with KB-A, bottom-left contacts with KB-T, bottom-right contacts both KB-T and KB-A. j Quantification of fluorescence intensity for actin and tubulin: control refers cells without Knock-beads, KB-T refers cells with KB-T Knock-beads, KB-A refers cells with KB-A Knock-beads. Data are presented as the mean ± SD, n = 5, analyzed using two-tailed unpaired t test. For panel (a–c, f–h), three independent experiments were conducted. Source data are provided as a Source Data file.
To demonstrate Knock-bead encoded siRNA delivery into different cells, two types of siRNAs: siRNA-FAM and siRNA-cy5 were separately loaded to form two different composites of Knock-beads (KB1-cy5, KB2-FAM), which were then mixed and introduced to cultured A549 cells to perform NIR-photoporation with a magnetic field applied. The encoding of scCode-fection is achieved by discrete distribution of the Knock-beads over the cells, which are decoded by fluorescent microscopy of the cell-contacting Knock-beads. The magnetic force helped to create a tight interface between the Knock-beads and cells, which was critical for achieving single-cell specificity of the photoporation-facilitated intracellular delivery of siRNAs. After the siRNA was off-loaded from the Knock-beads, the beads remained on cell membrane for later decoding and analysis. For every sample, images were acquired in large areas to collect sufficient data for statistical analysis. The identification and segmentation of individual cells/Knock-beads was automated by using customize developed algorithms (Fig. 4d)18. Interestingly, for the cells in contact with only KB1-cy5 or KB2-FAM, the majority of the cell population (~90%) showed correct delivery of the designed cargo siRNA-cy5 (carried by KB1-cy5) or siRNA-FAM (carried by KB2-FAM), respectively (Fig. 4a–c). For cells that were interfaced with both KB1-cy5 and KB2-FAM, both siRNA-cy5 and siRNA-FAM were identified in their cytoplasm (Fig. 4e). 3D confocal images of the transfected cells further showed that the siRNA-cy5 was inside cytoplasm (Fig. 4f) and the Knock-bead remains in contact with the outer cell membrane (Fig. 4g). The tight interfacing between the cell and Knock-beads was critical for the highly specific single-cell-encoded siRNA delivery, avoiding any analytical error resulted from the diffused mobility of the Knock-beads. Without the help by the magnetic field, more than 55% of the cells received the wrong type of RNAs (Supplementary Fig. 8).
Randomized single-cell gene silencing
Next, we examined the localized gene-silencing among neighboring cells that received different species of Knock-beads. We tested two types of Knock-beads targeting actin (KB-A) and tubulin (KB-T) respectively, which are mixed and homogenously added to the population of culture A549 cells to randomize single-cell based gene silencing (Fig. 4h). Quantitative analysis showed that correct gene-silence was successfully achieved in the majority of the cells with either actin or tubulin expression effectively suppressed. Importantly, neighboring cells in contact with different Knock-beads (KB-A or KB-T, Fig. 4i, j) demonstrate significant suppression of the targeted genes, highlighting the specificity of the Knock-beads to only the cells in contact. However, we also observed that silencing actin alone may also influence the expression of tubulin, suggesting an interdependence between the actin and tubulin cytoskeletal networks19.
Additionally, we also performed quantitative PCR (qPCR) to confirm the gene-silencing efficacy of different Knock-beads loaded with siRNAs targeting different genes (actin, tubulin, Bcl-2, survivin, Mcl-1, c-Myc, Cyclin D1, PLK-1, Supplementary Fig. 9). At the protein level, we used western blot to show successful knockdown of actin in A549 cells by using the Knock-beads. For a direct observation with single cell resolution, we also used the Knock-beads to specifically Knock down enhanced green fluorescent protein (EGFP) in each individual EGFP-Hela cells that were in contact with the Knock-beads, and the cells showed a significant reduction in EGFP fluorescence (Supplementary Fig. 10, Supplementary Note 2). These results establish the Knock-beads as a legitimate tool for high-throughput gene silencing with single-cell addressability, and strongly support the development of more complex assays to enable screening of combinatorial siRNA cocktails.
Combinatorial screening of siRNA cocktails
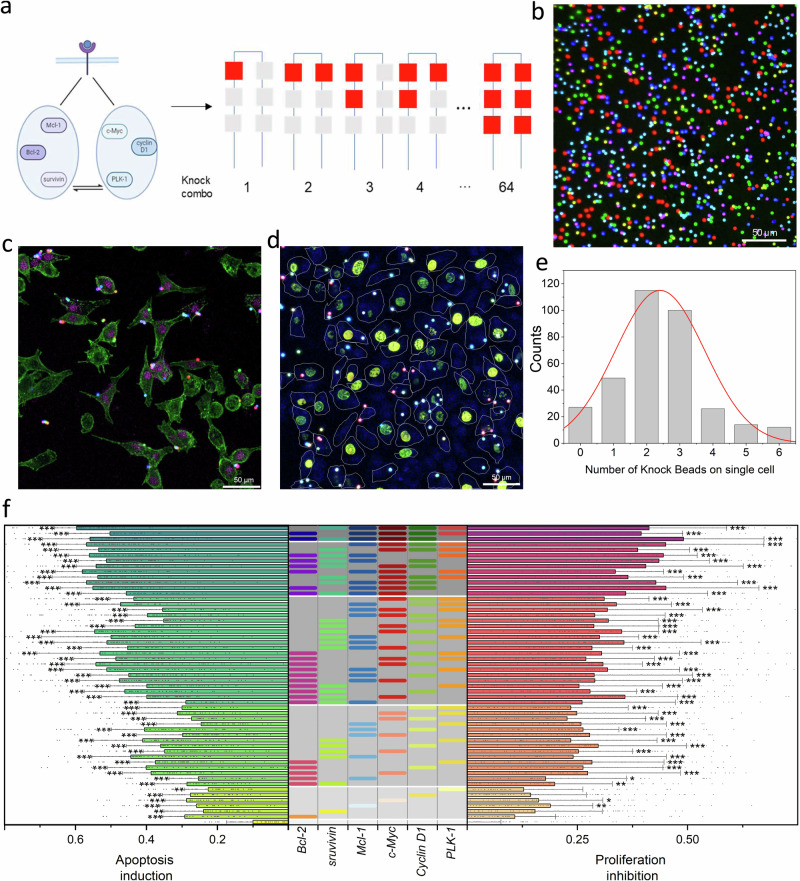
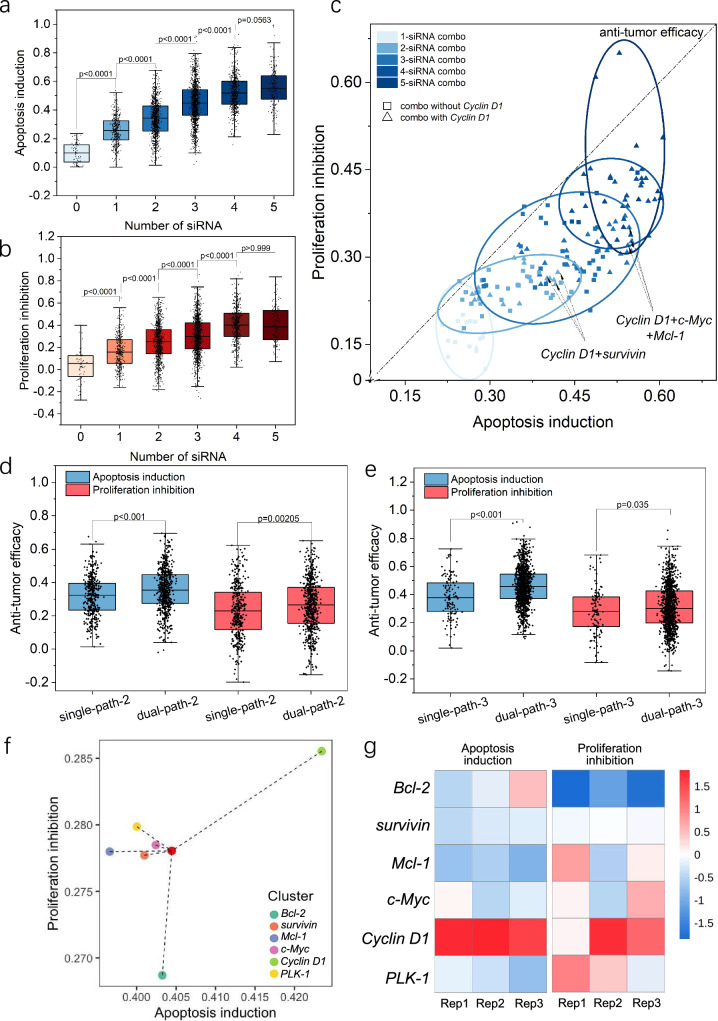
After the development of Knock-bead and the establishment scCode-fection technique, as a proof-of-concept demonstration, we performed a medium scale siRNA screening for tumor suppressing cocktails targeting cell cycle and apoptosis pathways. The use of siRNA in cancer therapy mainly concerns three crucial aspects: carcinogenesis, chemotherapeutic resistance, and tumor-host interactions20–24. In this study, the gene targets for siRNA interference included Bcl-2, survivin, and Mcl-1 on the apoptosis pathway25–27 and c-Myc, Cyclin D1, and PLK-1 on the cell-cycle pathway28–30. The sequences of the corresponding siRNAs are listed in Supplementary Table 1. Accordingly, 7 types of Knock-beads (6 siRNAs + a blank control without loading siRNA) were created (Supplementary Table 2, 3, and Supplementary Fig. 11), which theoretically gave 64 Knock-combos in total (Fig. 5a, b). For every sample of A549 cell culture, all the siRNA Knock-combo were tested in one-shot. The gene silencing efficacy was examined by TUNEL staining (for apoptosis, Fig. 5c) and Edu (for cell proliferation, Fig. 5d), and was quantitatively analyzed by confocal microscopy. Most of the cells were affected by multiple Knock-beads ranging from 1 to 6, following a Gaussian-like distribution (Fig. 5e, Supplementary Note 3, Supplementary Fig. 12). While a small proportion of cells were influenced by more than 4 Knock-beads, the image acquisition was performed to cover a large sample area for collecting sufficient data to carry out statistical analysis. Before a legitimate screening, the Knock-beads were tested for safe usage with mammalian cells to rule out the possibility of any photo-thermal damage that might affect cell viability. Upon NIR irradiation for photoporation of siRNAs, the Knock-bead induced temperature change does not harm the cells at the experimental conditions that recapitulate our screening parameters (Supplementary Fig. 13). The gene-silencing outcome for 54 Knock-combos was derived (Fig. 5f, Supplementary Note 4). In general, multiple siRNAs were significantly more effective at gene silencing than a single siRNA. As the number of genes silenced by siRNA transfection increased, from 1 to a maximum of 5 siRNAs, the cell apoptosis was increased by 25.80 ± 3.21%, 34.10 ± 1.64%, 45.12 ± 6.37%, 52.45 ± 5.97% and 55.35 ± 10.04%, respectively (mean±s.d., Fig. 6a). Similarly, cell proliferation was also inversely correlated with siRNA number, showing an inhibition of proliferation by 16.67 ± 1.41%, 24.71 ± 1.06%, 30.40 ± 5.16%, 40.34 ± 2.39% and 40.64 ± 4.61% as siRNA transfection increased from 1 to 5 (mean±s.d., Fig. 6b). These findings clearly indicated that multiple synergic siRNAs are significantly more efficient at inducing the desired gene inhibition and associated cellular responses. However, it is worth noting that there was only marginal inhibitory benefit when 4 or more siRNA were simultaneously transfected, suggesting the existence of an optimal siRNA number for a therapeutic cocktail if the 6 selected genes are to be used for cancer treatment.
Fig. 5. Combinatorial screening of siRNA cocktails for anti-tumor efficacy.
a Six siRNAs that related to cell apoptosis (survivin, Bcl-2, Mcl-1) and cell cycle (c-Myc, Cyclin D1, PLK-1) pathway form 64 combinations for relevant gene knock down. Created in BioRender. Guo, F. (2024) BioRender.com/v53d647. b Combined fluorescence image showing 7 types of spectrum-coded (Alexa-514, Alexa-568, and Alexa-647) Knock-beads. Confocal microscopic images showing the staining of apoptosis (c, TUNEL) or proliferation (d, Edu) markers for gene-silencing phenotype in cells treated with scCode-fection. The Knock-beads were overlayed with the cells. c Fluorescence image of actin (green) and TUNEL (magenta) staining to show cell apoptosis level. d Fluorescence image of Edu (cyan) staining to show cell proliferation level. The cell outlines (white) were derived from bright-field images. e Distribution of the number of Knock-beads on individual cells. f Combinatorial screening of 54 Knock-combos composed of 6 siRNAs for anti-tumor efficacy. The siRNA combinations are marked in the gray-shaded table (middle). The bar charts show the induction of cellular apoptosis (left) or inhibition of cell proliferation (right) as the result of gene-silencing by correspondent siRNA Knock-combos. Three rounds of screening were performed. For each knock combo, n > 20 per round, *P < 0.05, **P < 0.01, ***P < 0.001, analyzed using unpaired one-way ANOVA. More details about the statistical significance are provided in Supplementary Table 7. For panel (b–e), three independent experiments were conducted. Source data are provided as a Source Data file.
Fig. 6. Analysis of Knock-combo screening results to optimal siRNA cocktails.
a Effect of siRNA number on the induction of cellular apoptosis. b Effect of siRNA numbers on the inhibition of cellular proliferation. c Scattered plot showing the distribution of the knock-combo efficacy classified by the number of siRNAs. The dotted line (f(x) = x) refers to the total anti-tumor efficacy with equal weight of cellular apoptosis and proliferation in determining the final efficacy. Comparison between single or dual pathway gene silencing by different Knock-combos composed of 3 (d) or 2 (e) siRNAs. The black dots indicate the measurements from individual cells. f Centroid distribution illustrating the apoptosis induction and proliferation inhibition efficacy of Knock-combos containing specific siRNA. g Heatmap showing the apoptosis induction and proliferation inhibition efficacy for Knock-combos containing specific siRNA. The color index indicates the Z-score normalized phenotype quantification. Three rounds of screening were performed. For each knock combo, n > 20 per round. For panel (a, b), the data was analyzed using unpaired one-way ANOVA; for panel (d, e), the data was analyzed using non-parametric Mann–Whitney test. For the boxplots (a, b, d, e), the black dots indicate an overlay of individual data points, each box indicates the first quartile, median, third quartile of the data, the whisker limit is 1.5 times the interquartile range. Source data are provided as a Source Data file.
It is expected that siRNA interference of apoptosis or cell-cycle genes would cause mixed effects on both pathways given their close association in regulating cellular function31,32. Next, our analysis was oriented to answer whether a combinatory targeting of both pathways is more efficient than interfering a single pathway when using the same number of siRNAs. To compare the silencing efficacy, the six siRNAs (Bcl-2, survivin, Mcl-1 for apoptosis; c-Myc, Cyclin D1, PLK-1 for cell-cycle) were classified into four groups: single-path or dual-path using two (single-path-2 vs. dual-path-2) or three (single-path-3 vs. dual-path-3) siRNA conditions (Supplementary Table 4). When two siRNA were designed to target both pathways (9 Knock-combos), the overall silencing efficacy was higher compared to situations where both siRNAs targeted the same pathway (6 Knock-combos, 31.80 ± 11.84% vs. 35.63 ± 12.66% in apoptosis induction and 23.16 ± 15.02% vs. 26.25 ± 10.53% in proliferation inhibition). Similarly, targeting both pathways with 3 siRNA (dual-path-3, 9 Knock-combos) was more efficacious than targeting a single pathway (single-path-3, 2 Knock-combos) (38.13 ± 14.32% vs. 45.90 ± 13.44% in apoptosis induction and 28.50 ± 17.06% vs. 31.40 ± 16.14% in proliferation inhibition, Fig. 6d, e). If all the Knock-combos of 2 or 3 siRNAs were grouped together for analysis, dual pathway silencing still wins for better anti-tumor efficacy (Supplementary Fig. 14). These results are well aligned with previous research reporting that siRNA-based silencing of a single gene yields limited therapeutic effects8,20, due to the existence of complex signal transduction networks that are likely to be complementary or parallel and compensate for the single-gene silencing effects. Accordingly, the use of siRNA cocktails targeting multiple disease-related pathways represents an attractive and robust approach to develop siRNA-based therapeutics.
Based on the screening data, we performed further analysis to answer whether some siRNAs are more important in constructing an effective siRNA cocktail. Among the six siRNAs (targeting 6 different genes) examined here, we used a centroid analytical approach to assess the overall weights of a gene target in the two-dimensional coordinates of apoptosis induction and proliferation inhibition for all combinations containing a particular siRNA (Fig. 6f). Interestingly, while Cyclin D1 is usually categorized as a cell-cycle related gene33, siRNA inhibition of this target showed the greatest effects on induced cellular apoptosis (Fig. 6g). In comparison, siRNA inhibition of c-Myc showed almost equal efficacy on both apoptosis and cell proliferation related phenotypes. The heatmap of multiple biological replicates highlights the important role of Cyclin D1 in comparison to the other siRNA targets. Knock-combos that contain siRNA targeting Cyclin D1 showed higher anti-tumor efficacy, characterized by apoptosis induction and proliferation inhibition. A closer look at the screening results indicated that the most significant Knock-combo was siRNA-(Cyclin D1, Bcl-2) for 2-siRNA combos, siRNA-(Cyclin D1, survivin, c-Myc) for 3-siRNA combos (Fig. 6c), emphasizing the critical role of inhibiting Cyclin D1 in the construction of effective siRNA cocktails as potential anti-cancer therapeutics.
Expanded large-scale screening
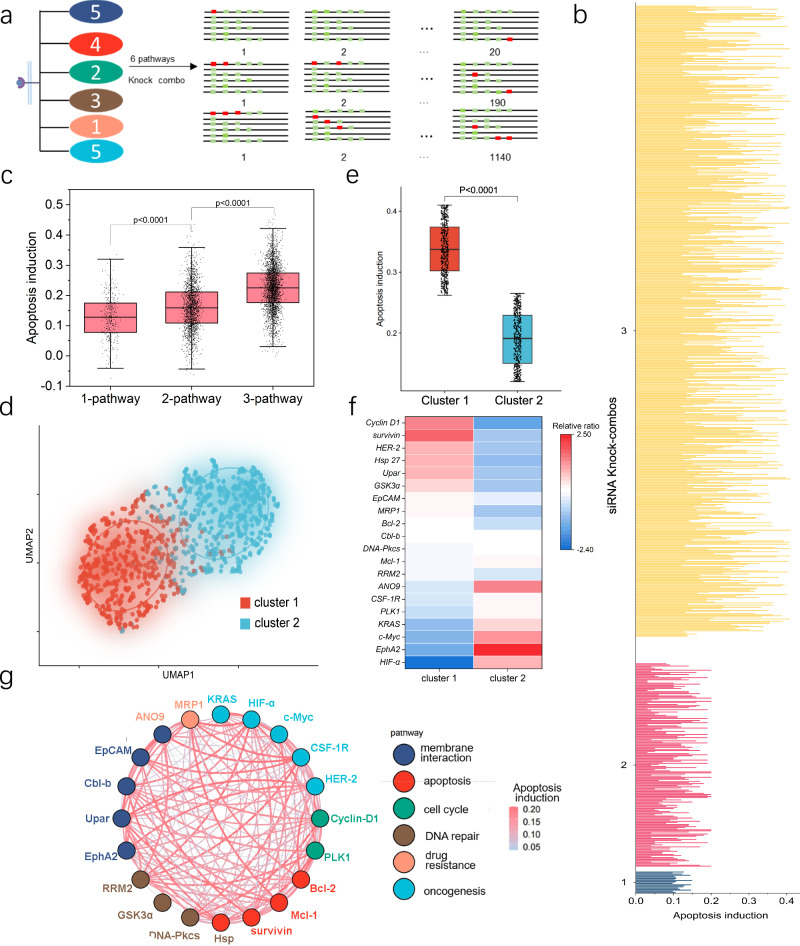
The translational potential of a high-throughput technology relies heavily on its extendibility for largescale screens. The challenging part is the phenotypic extraction of the massive number of Knock-combos in the overall cell population, which requires innovation in the following two aspects: (1) multiplexed encoding siRNA-loaded Knock-beads; (2) Phenotypic analysis and decoding of Knock-combos in a large number of cells. For multiplexed encoding, we used a combinatory fluorescence labeling of the Knock-beads to substantially increase the multiplexing throughput by using 5 fluorescent channels. Each Knock-bead was made with a programmed rainbow fluorescence composed of 5 different fluorophores (quantum dot 547, 572, 604, 641, and Alexa fluor 647). For spectrum digitization, we used 1 to denote the inclusion and 0 to denote the exclusion of a fluorophore and to compose a 5-digit spectral code for each Knock-bead variant. In this way, a 5-channel fluorescent system is expanded to encode 31 Knock-bead variants (25 – 1 = 31, Supplementary Table 5, Fig. 15), each of which can be assigned to a specific siRNA. At the analytical microscopy stage, the optical spectrum of the Knock-beads was decoded by using a machine learning algorithm34 (Supplementary Fig. 16). By using this system, we were able to demonstrate an expanded screening involving 20 siRNA targets and tested more than 1300 Knock-combos composed of 1–3 siRNAs for combinatorial knockdown (Fig. 7a). The 20 genes spread over 6 different cellular pathways: membrane interaction (5 siRNAs), apoptosis (4 siRNAs), cell cycle (2 siRNAs), DNA repair (3 siRNAs), drug resistance (1 siRNA), and oncogenesis (5 siRNAs). The phenotypic induction of cellular apoptosis was analyzed for more than 1300 Knock-combos involving 1, 2 or 3 siRNAs targeting a subset of the 20 genes (Fig. 7b).
Fig. 7. Expanded screening of 1315 Knock-combos composed of 1, 2, or 3 siRNAs targeting 20 genes.
a A set of 20 siRNAs targeting 6 different cellular pathways: membrane interaction (5 siRNAs), apoptosis (4 siRNAs), cell cycle (2 siRNAs), DNA repair (3 siRNAs), drug resistance (1 siRNA), and oncogenesis (5 siRNAs). The screening space (1350) is composed of 20 1-siRNA, 190 2-siRNA and 1140 3-siRNA Knock-combos. b Phenotypic analysis of apoptosis induction for all the tested 1-siRNA (blue), 2-siRNA (red) and 3-siRNA (yellow) Knock-combos. Data were collected from more than 20 cells for each of the 1315 Knock-combos. c Boxplots with scattered data points showing the comparison of the efficacy on apoptosis induction for combinatorial targeting of single, dual or multiple signaling pathways. d K-means clustering analysis of all the Knock-combos reveals two distinct clusters in the UMAP space. e Boxplots showing the comparison of apoptosis induction for the 2 identified Knock-combo clusters. f Enrichment analysis of the 20 siRNA targets in each identified Knock-combo clusters. g An interaction network showing the synergic co-operation in apoptosis induction for the 20 siRNAs to different gene targets, where the line color and thickness indicate the phenotypic efficacy of the corresponding combination. For panel (c, e), n = 7 for each Knock-combo, the data was analyzed using non-parametric Mann–Whitney test. The black dots indicate an overlay of individual data points, each box indicates the first quartile, median, third quartile of the data, the whisker limit is 1.5 times the interquartile range. Source data are provided as a Source Data file.
While the apoptosis level varied among different Knock-combos, it was clearly observed that multi-pathway knock-down was more effective than just targeting a single-pathway. Such synergic efficacy becomes even more evident for 3 siRNA combinations (Fig. 7c), which is consistent with our observations in previous smaller scale screening and emphasizes the importance and advantage of our innovation for high-throughput combinatorial siRNA screening. For all the 1315 siRNA Knock-combos, K-means clustering analysis was used to discover two distinct clusters, one of which showed significantly better efficacy for apoptosis induction (Fig. 7d, e, Supplementary Fig. 17). We then derived a relative ratio to show the enrichment of individual siRNA in each cluster (cluster 1, 2), showing the top-6 dominant role of Cyclin D1, survivin, HER2, Hsp 27, UPAR and GSK 3a for the apoptosis-pro cluster (cluster 1, Fig. 7f), which are well-represented in the top-2 apoptosis-pro Knock-combos (Supplementary Fig. 18). We then used a network interaction diagram to analyze the synergic efficacy from multiple siRNAs (Fig. 7g). As a hub gene, HER2 or Cyclin D1 showed the most connections to other genes, suggesting the important contributions from siRNAs targeting supplementary genes. On the other hand, uPAR gene appeared to be the best silencing partner for many leading siRNA targeting either survivin or HER2. These screening results provide valuable insights into the construction of combinatorial siRNAs therapeutics for anti-tumor purpose.
Discussion
Since the discovery of RNAi and the demonstration of siRNA in post-transcriptional gene silencing, there has been tremendous interest and effort in harnessing siRNAs for biomedical research and drug development. Thus far, the success has been limited. One of the major issues is that effective gene silencing by siRNA is often hindered by the presence of complementary or parallel signal transduction pathways, which may counteract the action of a single siRNA and substantially offset the therapeutic silencing effects. Alternatively, the use of cocktail siRNAs has been suggested as a feasible and promising drug discovery route. However, the identification of the most safe and efficient siRNA combination remains challenging and a crucial bottleneck. This study aims to address the problem by developing a high-throughput technology for screening siRNA-based cocktail therapeutics.
Technically, we employed a multilayer functionalization of hierarchical assembled nano-/micro composite carriers (Knock-beads) with responsiveness to NIR and magnetic field, to facilitate photoporation-induced cellular transfection of siRNAs with single cell encoding capability. Randomized gene silencing by different siRNA variants in many cells was performed, recorded, and analyzed in a highly parallel manner. As one of the key steps of the scCode-fection, photoporation allows for the temporary permeabilization of cell membranes by using NIR laser. We believe that the PDDA layer coated on the surface of the Fe3O4-Au beads plays a critical role for siRNA loading and release, which is controlled by NIR-induced photo-thermal effects via mediating the van der Waals interactions35–37. As the loading procedure, the PDDA layer was used to electrostatically capture siRNAs. A temperature increase of just 23 K can lead to a substantial decrease in van der Waals adhesion force from 106 nN to 28 nN38. In our system, the temperature of the Knock-beads was changed by gold nanorod-induced surface plasmon resonance35,39. The local temperature increase in each Knock-bead disrupts the van der Waals interactions, causing the PDDA-siRNA to detach from the Knock-beads (Supplementary Fig. 4). The nanoscale size of the gold nanorods also allows for precise localization of the laser energy and better control over the size and distribution of the created cell membrane disruption for intracellular translocation of the siRNAs13,40.
Meanwhile, the backbone of the Knock-beads, microscale Fe3O4 beads can be easily distinguishable in fluorescent imaging to facilitate later imaging-based decoding of the scCode-fection. The microscale Knock-beads are unlikely to be internalized by the cells via endocytosis due to the physical size41. It has been reported that micro-particles larger than 3 μm almost do not enter cells42,43. We believe that leaving the beads outside a cell would cause much less interference to cellular function than the intracellularly delivered ones. Apparently, the mobility of the Knock-beads from one cell to another could potentially cause diffused errors for the screening. Therefore, it is very important to make sure that the Knock-beads remain fixed on the contacting cells. The combination of magnetic fixation, chemical cross-linking, and size constraints ensures that the Knock-beads remain stably associated with the contacting cells throughout culturing, decoding and analysis processes, which is a critical aspect of the experimental design to allow single-cell-based encoding and decoding of the molecular information by the Knock-beads. Notably, while the static magnetic field exposure generally does not affect most gene expression or cellular responses, extra cautions should be taken for screening involving gene targets that are sensitive to magnetic field stimulation44.
Beyond the demonstrated combinatorial screening of siRNAs at different scales (e.g. 54 Knock-combos for 6 genes, >1300 Knock-combos for 20 genes), it is possible to further extend the screening throughput ( > 60 gene targets) to industrial scale ( > 10,000 Kock-combo) by using advanced multiplexing strategy to encode the Knock-beads. The spectrum encoding approach used in this study significantly boosts assay throughputs, which is achieved by a programmed rainbow fluorescence labeling using limited number of fluorophores. In this study, a 5-channel fluorescent system was implemented to encode various Knock-beads targeting 20 genes with some redundancy (Supplementary Table 5). An extension to a 10-channel system45 with binary labeling (0 or 1) would increase the multiplexing capacity to 1023 gene targets, which would further render millions of Knock-combos. Actually, in many drug combination therapies, such as the cocktail anti-viral therapy for HIV/AIDS management, 2 to 3 active components are typically used. Among more than 20 antiretroviral cocktails, 18 formulations are made of 2 to 3 components, and only 2 formulations consist of 4 components46. Recently, Tieu et al. developed a CRISPR-Cas9-based platform for combinatorial gene editing on CAR-T cells also focused on pairwise combinations of different signaling regulators11. Therefore, we believe that the most meaningful siRNA cocktail screening should be oriented towards Knock-combos consisting of 2 to 3 siRNAs. If so, within the gene space of 1023, the total possible Knock-combos would reach 178,433,024, which is already a scale inaccessible to any existing methods. But using the scCode-fection system, depending on the operation scale, hundreds or even thousands of siRNA cocktail formulations could be assayed in just a single well of cell culture, which would be tremendously useful for accelerating the development of siRNA-based therapeutics. Such benefits are not just limited to reducing the time and cost associated with siRNA screening by power of magnitude, but also rely on the capability to answer fundamental siRNA questions that are not addressable by existing methods, such as: (1) whether multiplexed gene silencing are more efficient than single target knockdown; (2) whether a combination of different signaling pathways related siRNAs are more efficient than multiple siRNAs targeting a single pathway; (3) which siRNAs are the most important elements in constructing an effective siRNA cocktail; (4) what is the optimal combination of siRNAs (precise and concise) to achieve the best synergic therapeutic gene-silence? The answers to these fundamental questions could be extensively explored by machine learning and bioinformatic analysis of the large number of single-cell gene silencing data enabled by the scCode-fection technology, as we have demonstrated in the screening of more than 1300 siRNA combinations concerning 20 genes in seeking the most effective cocktail for potential inhibition of cancer cell growth. We believe this study will lead to paths to a systematic and high-throughput platform that would be greatly useful to accelerate further development of siRNA-based therapeutics and enhance their clinical adoption.
Methods
Fabrication of knock-beads
The fabrication is based on two processes: Connection of Fe3O4-NH2 with gold nanorods through glutaraldehyde and positive coating of PDDA. To prepare Fe3O4-Au assemblies, 20 μL stock solution of Fe3O4-NH2 (Invitrogen Thermo Fisher, 2.8 μm in diameter, 2 × 109 beads/mL) were incubated in 15% Glutaraldehyde aqueous solution (Sigma-Aldrich) at room temperature on a lab rotator (DLAB, MX-RD-Pro) at 30 rpm overnight. After three washes with deionized water, the beads were incubated with 100 μL of Au-NH2 solution (Sigma-Aldrich, 70 nm in width, 1μm in length) while rotating on a lab rotator (DLAB, MX-RD-Pro) at 30 rpm for 2 h. Finally, the assemblies were collected with a magnetic field and blocked with 3% bovine serum albumin (BSA) for 2 h.
For spectrum coding, the Fe3O4-Au assemblies were washed three times with deionized water. Then the Fe3O4-Au assemblies were incubated with specific fluorescence dyes according to their spectrum codes (Supplementary Table 3, Table 5). For example, to prepare code 100 (Alexa-514 positive, Alexa-568 negative, and Alexa-647 negative), 1 μL NHS-Alexa-514 (Thermo Fisher) was added to Fe3O4-Au assemblies and rotated at 30 rpm (DLAB, MX-RD-Pro) for 2 h followed by three rinses with deionized water. Other codes were prepared in a similar way using a different mixture of the three fluorescent dyes.
PDDA was coated for loading negative charged RNAs. Briefly, Fe3O4-Au assemblies were washed twice with deionized water and then resuspended in 200 μL of 15% (v/v) PDDA solution (Sigma Aldrich). The suspension was sonicated in a bath for 10 min. The Knock-beads were then washed three times with deionized water and resuspended in 100 μL of PBS for storage.
Dynamic light scattering for size characterization
The size of Fe3O4, Fe3O4-Au, and Fe3O4-Au-PDDA was determined using a Particle Size Analyzer (Zeta sizer, Malvern Panalytical). In this experiment, a 20 μL solution of each particle type (Fe3O4, Fe3O4-Au, and Fe3O4-Au-PDDA) was added to 500 μL of deionized water. The Particle Size Analyzer was set to detect the size by selecting water as the detection solution and the zeta size mode. For zeta potential measurements, the same process was followed, with the only difference being the selection of the zeta potential mode.
Evaluation of RNA loading efficiency on Knock-beads
To determine the RNA loading efficiency of Knock-beads, we utilized fluorescence detection of unbound RNA-cy5. Briefly, the Knock-beads were incubated with RNA-cy5 in deionized water rotating at 30 rpm at room temperature. The mixture was then subjected to a magnetic field, ensuring that the beads were attracted to the side of the tube. The supernatant was then carefully collected. The fluorescence intensity of the supernatant was measured using MD SpectraMax M5e microplate reader. The RNA loading efficiency was calculated based on the fluorescence intensity of the supernatant, as well as the initial concentration of RNA-cy5. This procedure was repeated three times to ensure reproducibility and accuracy of the results.
Scanning electron microscopy
Scanning electron microscopy (SEM; Philips FEG SEM XL30) was employed to characterize the morphology of Fe3O4-Au. Briefly, the Fe3O4-Au assemblies were deposited onto a silicon wafer and allowed to dry overnight. The dried samples were imaged using a scanning electron microscope at a voltage of 10 kV or 20 kV. SEM images were analyzed using ImageJ software to determine the size distribution of the Fe3O4-Au assemblies.
Fluorescence microscopy
The fluorescence images were captured with confocal laser scanning microscope (Leica SP8, 40× water immersion objective, NA 1.1, Leica Microsystems). Briefly, after washing with PBS, the Knock-beads were mounted with an antifade mounting medium and imaged using a confocal laser scanning microscope. For quantitative analysis, all the imaging parameters, including the laser power, gain, and scanning speed were fixed across the microscopy process. Scanning was performed with a 1 μm step size along the Z-axis spanning the whole cell. 3D reconstruction was then performed to visualize the relative position of Knock-beads, cells, and diffused fluorescence dyed RNA. Confocal microscopic images were analyzed using ImageJ software to determine the fluorescence labeling and distribution of Knock-beads.
For data collection from a screening, imaging-based decoding was performed according to the spectrum encoding (Supplementary Table 5). The excitation of the 4 quantum dots (emission peak at 547 nm, 572 nm, 604 nm, 641 nm) was 405 nm laser. The emission window was set to be 10 nm wide. To acquire a sufficient number of cells, we employed the navigator model of Leica Microsystems to capture over 200 tiles from a single culture dish for large area coverage. The resulting large-scale tiles were merged using Leica software to ensure smooth calibration along each edge of the view arrays.
Cell culture
A549 (CRM-CCL-185 from American Type Culture Collection, ATCC) and Hela:EGFP (gift from Prof. Linfeng Huang’s lab, Duke Kunshan University) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The medium was changed every two days, and cells were subcultured at 80–90% confluency using Trypsin-EDTA.
Evaluation of photothermal effect and cell viability
The solution of Fe3O4-Au beads, containing 1 × 107 beads/mL in 2 mL of PBS, was subjected to laser irradiation using a 980 nm laser with a power density of 0.6 W/cm2 and pulse width of 16 μs. Infrared photography was recorded using an infrared camera (Nikon). Cell viability was tested by adding 5 × 104 beads to A549 cells in a 24-well plate (1 × 105 cells per well), followed by irradiation with a 0.6 W/cm2 in 16 μs pulse width laser for 2, 4, 6, 8, 10, and 12 s. 24 h after NIR irradiation, 100 μL of MTT solution (Sigma-Aldrich, 1 mg/mL) was added and incubated for further 4 h. The MTT-containing medium was then removed, and the formazan crystals were dissolved in dimethyl sulfoxide (DMSO). The absorbance of each well was measured at 570 nm using Molecular Devices SpectraMax M5e Microplate Reader. The viability of the cells was expressed as the percentage of the absorbance relative to the control group.
Photoporation-based intracellular delivery
The Knock-beads were used to deliver fluorescence RNA into A549 using photoporation under a magnetic field. Briefly, cells were seeded in glass bottom plates at a density of 1 × 106 cells/plate and allowed to adhere overnight. The cells were then incubated with the Knock-beads at a concentration of 4 × 105 for 30 s to allow for absorption of the beads after discharged medium. The cells were then irradiated with a 980-nm laser (16 μs pulse width) for 8 s at a power of 0.6 W/cm2 within a magnetic field. After 2 h of photoporation, the cells were washed 2 times with PBS within a magnetic field. The efficiency of RNA delivery was determined by visualizing the cells under a confocal microscope and analyzing the uptake of the fluorescence dye with ImageJ.
Immunocytochemistry
Actin and tubulin were stained for evaluation of the single cell gene knockdown efficiency. TUNEL and Edu were stained to evaluate the anti-tumor efficiency of related Knock-combo. For cell fixation, cells were washed with PBS and fixed with 4% paraformaldehyde (Thermo) at room temperature for 30 min. For permeabilization, cells were incubated in triton X-100 (0.1% in PBS) at room temperature for 10 min and then rinsed with PBS. For actin and tubulin staining, 20 h after photoporation, 2 μL of Actin-Tracker (Actin-tracker kit from Beyotime) were added to each dish and incubated in the dark at room temperature for 60 min. Then, 2 μL of Tubulin-tracker (Tubulin-tracker kit from Beyotime) were added to each dish and incubated in the dark for 60 min. To detect apoptosis, One Step TUNEL Apoptosis Assay Kit from Beyotime was used 20 h after photoporation. Terminal Deoxynucleotidyl Transferase (TdT) and fluorescein-dUTP were added to each dish, and cells were incubated at 37 °C for 1 h in the dark allowing for TdT-mediated dUTP Nick-End labeling. To detect proliferation, BeyoClick™ EdU Cell Proliferation Kit from Beyotime was used. Briefly, 5-ethynyl-2’-deoxyuridine (EDU) at 10 mM was added to the cells before fixation and permeabilization, then incubated for 3 h. To perform the click reaction, 430 μL of click reaction buffer, 20 μL of CuSO4, 1 μL of Azide, and 50 μL of click additive solution were added to each dish, followed by a 30-minute incubation in dark at room temperature.
Cell segmentation
For cell segmentation, the images were first loaded into the Python-based CellPose software. The brightfield channel was used to derive an evaluation of the expected cell diameter in pixels. The grayscale fluorescent channels were then selected for the segmentation processing. The CellPose algorithm was then executed to generate a mask outlining the boundary of individual cells. Finally, the cell outlines were exported as an ImageJ ZIP file, which was imported to ImageJ using ROI Manager for further analysis.
Informatic analysis of the screening data
For the centroid approach in Fig. 6f, the centroid of a specific type of siRNA Knock-combo, denoted as , refers to the average position of a group of two-dimensional points. For each siRNA combo , where represents the efficacy of apoptosis induction and represents the efficacy of proliferation inhibition, the centroid for n Knock-combos is obtained by:
| 2 |
For clustering analysis in Fig. 7, K-Means clustering was used. The optimal number of clusters (k) was determined using silhouette statistics. The K-Means algorithm was then applied to the preprocessed data with multiple random initializations to find the global optimum for unsupervised derivation of optimal number clusters of the Knock-combos based on the phenotypic features related to cellular apoptosis or proliferation.
To prioritize the enrichment of a gene targeted siRNA in the identified Knock-combo clusters (Fig. 7f), the relative enrichment ratio of each siRNA was calculated using the Z-score normalization defined as:
| 3 |
where X is the appearance frequency of a siRNA for all the Knock-combos in a cluster, μ is the mean appearance frequency of all siRNAs in that cluster, and σ is the standard deviation of the appearance frequency for all siRNAs in the same cluster. The appearance frequency of a siRNA in a cluster is calculated as: counts of Knock-combos containing the siRNA divided by the total number of siRNAs appeared in all Knock-combos of that cluster.
To analyze the interaction network and to show the synergic co-operation in apoptosis induction for the siRNAs to their 20 gene targets (Fig. 7g), a network diagram was used. The siRNAs were represented as the nodes connected by lines, where the line color and thickness indicate the phenotypic efficacy of the corresponding combination siRNA silencing of the node genes. The line color and thickness were indexed by calculating the mean apoptosis induction among all the Knock-combos containing the two node gene siRNAs (connection score). A hub score of each node was also derived by taking the sum of all the connections scores for the lines originated from one node.
Statistics & reproducibility
At least three independent biological replicates were used for all experiments. Statistical significance for comparing two experimental conditions is calculated by two-tailed unpaired t test; for comparing one control with multiple experimental conditions, the statistical significance is calculated using the unpaired one-way ANOVA test. For screening data analysis, unpaired nonparametric Mann–Whitney test is used to compare different groups. A p value less than 0.05 is considered to be statistically significant, and all the p values are indicated in the respective figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was supported by National Natural Science Foundation of China (U20A20194, P.S.), by General Research Fund (11215920, 11220024, 11218522, 11218523, P.S.) from the Research Grants Council of Hong Kong SAR, Shenzhen-Hong Kong-Macau Science and Technology Program (Category C, SGDX2020110309300502, P.S.), by a Kunshan Shuangchuang Talent Grant (KSSC202102069, L.H.). Support from Innovation and Technology Commission of Hong Kong through the Center for Cerebro-Cardiovascular Health Engineering and funds from City University of Hong Kong (7005084, 7005206, 7005642, 7020003, 7020077, 9680233, 9240060, P.S.) are also acknowledged.
Author contributions
P.S. supervised the research. P.S., X.J. and F.Guo conceived the project. F.Guo and X.J. designed the experiments and carried out the investigation. C.X. contributed to the experimental works. H.S. contributed to the data analysis. ZH.L. and BY.K. contributed to the RNA synthesis. L.H. contributed to the validation of siRNA gene silencing. B.G. and F.Gao contributed to the informatic analysis of the large-scale screening data. F.Guo and P.S. wrote the manuscript. R.Y. and ZP.L. contributed to the writing of the manuscript.
Peer review
Peer review information
Nature Communications thanks Guanyou Lin, Zhuxian Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data generated in this study are provided in the Supplementary Information/Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Feng Guo, Xianglin Ji.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53419-7.
References
- 1.Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in. Nature391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Hamilton, A. J. et al. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science286, 950–952 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature411, 494–498 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Friedrich, M. et al. Therapeutic siRNA: state-of-the-art and future perspectives. BioDrugs36, 549–571 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padda, I. S., Mahtani, A. & Parmar, M. Small Interfering RNA (siRNA) Therapy. In StatPearls [Internet]. (StatPearls Publishing, Treasure Island (FL), 2024) https://www.ncbi.nlm.nih.gov/books/NBK580472/. [PubMed]
- 6.Nussinov, R. et al. Are parallel proliferation pathways redundant? Trends Biochem Sci.45, 554–563 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Jain, A. et al. Dendrimer mediated targeting of siRNA against polo-like kinase for the treatment of triple negative breast cancer. J. Biomed. Mater. Res A107, 1933–1944 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Amen, A. M. et al. Endogenous spacing enables co-processing of microRNAs and efficient combinatorial RNAi. Cell Rep. Methods2, 100239 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, M. et al. In situ cocktail nanovaccine for cancer immunotherapy. Adv. Sci.10, 2207697 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch, D. H. et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature503, 224–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tieu, V. et al. A versatile CRISPR-Cas13d platform for multiplexed transcriptomic regulation and metabolic engineering in primary human T cells. Cell187, 1278–1295 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasanzadeh Kafshgari, M. et al. Antibody-functionalized gold nanostar-mediated on-resonance picosecond laser optoporation for targeted delivery of RNA therapeutics. Small17, 2007577 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Chow, T. H. et al. Gold nanobipyramids: an emerging and versatile type of plasmonic nanoparticles. Acc. Chem. Res.52, 2136–2146 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Morshedi Rad, D. et al. A comprehensive review on intracellular delivery. Adv. Mater.33, e2005363 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Khalavka, Y. et al. Synthesis of rod-shaped gold nanorattles with improved plasmon sensitivity and catalytic activity. J. Am. Chem. Soc.131, 1871–1875 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Liu, L. et al. Gold photosensitized SrTiO3 for visible-light water oxidation induced by Au interband transitions. J. Mater. Chem. A2, 9875–9882 (2014). [Google Scholar]
- 17.Guerrisi, M. et al. Splitting of the interband absorption edge in Au. Phys. Rev. B12, 557–563 (1975). [Google Scholar]
- 18.Stringer, C. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Po’uha, S. T. et al. Partial depletion of gamma-actin suppresses microtubule dynamics. Cytoskeleton70, 148–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Z. et al. Unravelling the enigma of siRNA and aptamer mediated therapies against pancreatic cancer. Mol. Cancer22, 8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Almeida, L. C. et al. DNA damaging agents and DNA repair: From carcinogenesis to cancer therapy. Cancer Genet252-253, 6–24 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Kara, G. et al. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev.182, 114113 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Holohan, C. et al. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer13, 714–726 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Pogge von Strandmann, E. et al. Tumor–host cell interactions in ovarian cancer: pathways to therapy failure. Trends Cancer3, 137–148 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Diepstraten, S. T. et al. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat. Rev. Cancer22, 45–64 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Altieri, D. C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer8, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Wang, H. et al. Targeting MCL-1 in cancer: current status and perspectives. J. Hematol. Oncol.14, 67 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madden, S. K. et al. Taking the Myc out of cancer: toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer20, 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lourenco, C. et al. MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer21, 579–591 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Peng, F. et al. Regulated cell death (RCD) in cancer: Key pathways and targeted therapies. Signal Transduct. Tar.7, 286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X. et al. Involvement of TRPC1 and Cyclin D1 in human pulmonary artery smooth muscle cells proliferation induced by cigarette smoke extract. Curr. Med Sci.40, 1085–1091 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Ma, C. et al. USP13 deubiquitinates and stabilizes Cyclin D1 to promote gastric cancer cell cycle progression and cell proliferation. Oncogene42, 2249–2262 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montalto, F. I. et al. Cyclin D1 in cancer: a molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells9, 2648 (2020). [DOI] [PMC free article] [PubMed]
- 34.Breiman, L. Random forests. Mach. Learn.45, 5–32 (2001). [Google Scholar]
- 35.Amendola, V. et al. Surface plasmon resonance in gold nanoparticles: A review. J. Phys.-Condens Mat.29, 203002 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Liu, X. et al. Understanding the effect of different polymeric surfactants on enhancing the silicon/reduced graphene oxide anode performance. J. Phys. Chem. C.119, 5848–5854 (2015). [Google Scholar]
- 37.Luo, D. et al. Interparticle forces underlying nanoparticle self-assemblies. Small11, 5984–6008 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Pinon, A. V. et al. Thermal effects on van der Waals adhesive forces. Phys. Rev. B93, 035424 (2016).
- 39.You, M. et al. Quantifying and adjusting plasmon-driven nano-localized temperature field around gold nanorods for nucleic acids amplification. Small Methods5, e2001254 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Lee, J. K. et al. Spontaneous formation of gold nanostructures in aqueous microdroplets. Nat. Commun.9, 1562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gratton, S. E. et al. The effect of particle design on cellular internalization pathways. Proc. Natl Acad. Sci. USA105, 11613–11618 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, Y. et al. Size-dependent cytotoxicity of gold nanoparticles. Small3, 1941–1949 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Kus-Liskiewicz, M. et al. Biocompatibility and cytotoxicity of gold nanoparticles: recent advances in methodologies and regulations. Int. J. Mol. Sci.22, 10952 (2021). [DOI] [PMC free article] [PubMed]
- 44.Ongaro, A. et al. Pulsed electromagnetic fields stimulate osteogenic differentiation in human bone marrow and adipose tissue derived mesenchymal stem cells. Bioelectromagnetics35, 426–436 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Shi, H. et al. Highly multiplexed spatial mapping of microbial communities. Nature588, 676–681 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, G. et al. Approved HIV reverse transcriptase inhibitors in the past decade. Acta Pharm. Sin. B12, 1567–1590 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are provided in the Supplementary Information/Source Data file. Source data are provided with this paper.