Abstract
Organismal communities associated with coral reefs, particularly invertebrates and microbes, play crucial roles in ecosystem maintenance and coral health. Here, we characterized the organismal composition of a healthy, non-urbanized reef (Site A) and a degraded, urbanized reef (Site B) in the Gulf of Eilat/Aqaba, Red Sea to assess its impact on coral health and physiology. Biomimetically designed terracotta tiles were conditioned for 6 months at both sites, then reciprocally transplanted, and scleractinian coral species, Acropora eurystoma and Stylophora pistillata, were attached for an additional 6 months. After 12 months, tiles from Site A transplanted to Site B exhibited greater invertebrate richness and diversity than Site B’s original tiles (via Cytochrome c. Oxidase subunit I metabarcoding). Key bacteria from the healthy reef were more prevalent on Site A tiles and on the tiles transplanted to Site B (via 16S rRNA gene sequencing). Corals originally from Site B attached to transplanted healthy tiles (Site A) showed higher photochemical capacity, increased endosymbionts, and reduced physiological stress, measured by total antioxidant capacity and an integrated biomarker response. Our findings demonstrate the successful transfer of organismal communities between reefs, highlighting the potential benefits of healthy reef-associated invertebrates and microbes on coral physiology and their implications for reef restoration strategies.
Subject terms: Microbial ecology, Molecular ecology, Biodiversity, Ecosystem ecology, Marine biology
Invertebrate and microbe communities support reef ecosystems and coral health. Here, the authors characterize these communities from degraded and healthy reefs, showing that transplanting these healthy communities improved coral health at degraded reefs.
Introduction
The degrading health of coral reefs caused by ecosystem changes can lead to a shift in available nutrients, chemicals, microbes, vertebrate and invertebrate communities, and coral health1–3. This degradation can be characterized by a loss of critical reef-associated organisms that can lead to the demise of ecosystem function and sustainability4. Benthic reef invertebrates and bacteria play key roles in mediating the early succession of the reef benthos1–3, the health of adult corals, and other physiological properties of the reef microhabitat5. These benthic communities are important for the growth, maintenance, and function of coral reef ecosystems2,3,5. Previous studies have demonstrated that the composition of bacteria associated with corals is highly dependent upon the structure of the surrounding benthic community, which can influence coral health and holobiont response to stress6–9. These observations suggest that invertebrates and microbes surrounding corals could play a role in their health, implicating the potential that organismal communities from healthy reefs may influence coral health differently than communities at degrading reefs.
Recently, a terrestrial study showed the resilience benefits of transplanting thermally tolerant microbes to tree seedlings and found that the seedlings exhibited a higher survival rate even when subjected to heat, or cold stress10. Previous research on coral reefs has demonstrated the critical role of microorganisms to help manage the coral microbiome response to various perturbations5,6,11–14. Rosado et al.15 exhibited the potential to engineer a community of beneficial microorganisms for corals (BMCs), which was used as a probiotic to inoculate corals exposed to bleaching temperatures and discovered that coral bleaching was partially mitigated as a result of the probiotic. Several coral reef studies have shown that BMCs16 and engineered coral-associated bacteria (probiotics)17, could be transferred to or used to inoculate stressed corals15–19 and therefore, suggested as a potential tool for coral reef restoration19,20. When the architectural complexity of a coral reef is diminished, fundamental microhabitats are also lost21–24, which can negatively impact the composition and diversity of the species relying on them25,26. Therefore, both structural and surface complexities have a significant role in influencing the benthic reef community27, though manipulating the influence of these communities on coral health and physiology, has yet to be explored.
In this work, we explored this concept further by determining if there was an inherent difference in the organismal composition between a healthy and degraded coral reef28,29 and if it is an important component in the maintenance of coral physiology and health. Biomimetic [ceramic] terracotta tiles were designed to emulate the natural topographic complexity of coral reef surfaces with various niches, holes, and crevices28,30, and used as substrates for accumulating reef organisms at a non-urbanized, healthy coral reef (Site A) and highly urbanized, degraded coral reef (Site B) in the Gulf of Eilat/Aqaba (GoE/A), Red Sea. Sites were determined by previous studies that detailed the nutrient, chemical, and biological parameters of each site29–31. Biomimetic tiles were deployed for 6 months, allowing the benthic community of each respective site to develop on tiles, a reciprocal transplant occurred where tiles containing the organismal community of a healthy coral reef (Site A) were transplanted to a degraded coral reef (Site B) and vice versa. After the reciprocal transplant, two species of branching corals were attached to tiles at Site A and B for an additional 6 months. At the end of the experiment (12 months), the community on the biomimetic tiles were assessed, observing a greater invertebrate community richness and diversity on the tiles from Site A that were transplanted to Site B, compared to Site B original tiles. We identified key bacteria from the healthy reef that were more prevalent on the Site A tiles that were transplanted to Site B. The corals that were attached to tiles from either site underwent several physiological tests that demonstrated that corals originally from Site B attached to the healthy tiles that were transplanted from Site A, had higher photochemical efficiency, increased endosymbionts, and reduced physiological stress, measured by total antioxidant capacity (TAC). Here, we harnessed a holistic approach to showcase the ability to transfer the organismal community from one coral reef to another. This method presents a potential restoration strategy as a stand-alone method or to boost current approaches, by using a coral reef ecosystem transplant (aCRET) to boost coral health and future reef resilience.
Results
Coral physiology - Electron transport rate
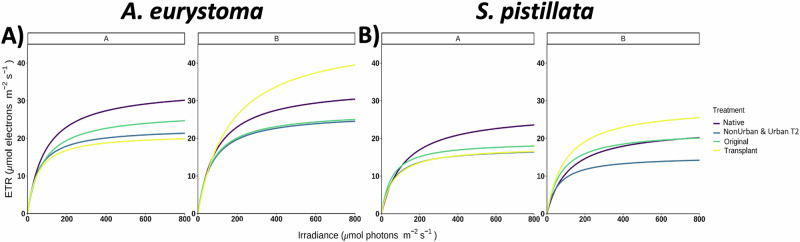
Pulse amplitude modulation (PAM) is commonly used to give an indication of endosymbiont and coral health32, declining photochemical efficiency and electron transport rates (ETR) are commonly linked with stress and indirectly associated with a reduction in photosynthetic production leading to early warnings of bleaching susceptibility. Here, we show a clear site and treatment difference between ETR curves, demonstrating that higher ETRs were correlated with a potential increase in photochemical efficiency in both Site A corals attached to the tiles at Site A, and corals from Site B that were attached to tiles transplanted from Site A to Site B.
There was a noticeable difference between site and treatments in ETRmax (p < 0.0001) and PAR50 (p < 0.0001) values for the coral Acropora eurystoma. In general, Site A A. eurystoma corals from the Native colonies at Site A and the corals attached to the Original tiles experienced a higher photochemical capacity for ETRmax (p < 0.001; Fig. 1A; Supplementary Table 2). In comparison to what was observed at Site B, the A. eurystoma corals that were attached to the Site A tiles transplanted to Site B (Transplant A) had a significantly higher photochemical capacity, compared to the A. eurystoma corals attached to all other tile treatments at Site B (p < 0.0001; Fig. 1A, B). Leading to a clear difference in the ETRmax values of the A. eurystoma corals affixed to the Transplant tiles across Site A and B (p < 0.0001; Fig. 1A; Supplementary Table 2). We observed similar patterns, when comparing PAR50 in A. eurystoma corals that were attached to Transplant tiles between Site A and B (p < 0.0001; Fig. 1; Supplementary Table 2). At Site B, A. eurystoma that were affixed to the Transplant tiles that came from Site A (Transplant A), experienced significant differences in light attenuation (p < 0.0001; Fig. 1A; Supplementary Table 2).
Fig. 1. Photochemical measurements of electron transport rate (ETR).
A ETR of A. eurystoma between Site A (healthy/NonUrban) and Site B (degraded/Urban) and each treatment. B ETR of S. pistillata between Site A and B and all treatments. Data is expressed as averages (n = 5). See Supplementary Fig. 2 for data ±SE and Supplementary Table 2.
It was observed that both coral species demonstrated a consistent response pattern, as Stylophora pistillata corals experienced differences between treatments at each site in ETRmax (p < 0.01) as well as site and treatment differences observed in measurements of PAR50 (p < 0.001; Fig. 1B). For example, at Site A, S. pistillata from the Native coral colonies at Site A and S. pistillata attached to the Original Site A tiles experienced a higher photochemical capacity than S. pistillata corals attached to the other tile treatments (Fig. 1B). Most notably, ETRmax values in S. pistillata corals were significantly different between the transplanted tiles, as S. pistillata corals attached to the tiles transplanted from Site A to Site B (Transplant A) had a higher photochemical capacity (p < 0.01; Fig. 1B; Supplementary Table 3). Similarly, measurements of PAR50 in S. pistillata corals that were attached to the Transplant tiles were significant across Site A and B (p < 0.01; Fig. 1B; Supplementary Table 3). As S. pistillata corals affixed to the Transplant A tiles at Site B had the highest PAR50 levels, compared to S. pistillata from the other treatments (p < 0.01; Fig. 1B; Supplementary Table 3).
Coral physiology - Chlorophyll and endosymbiont density
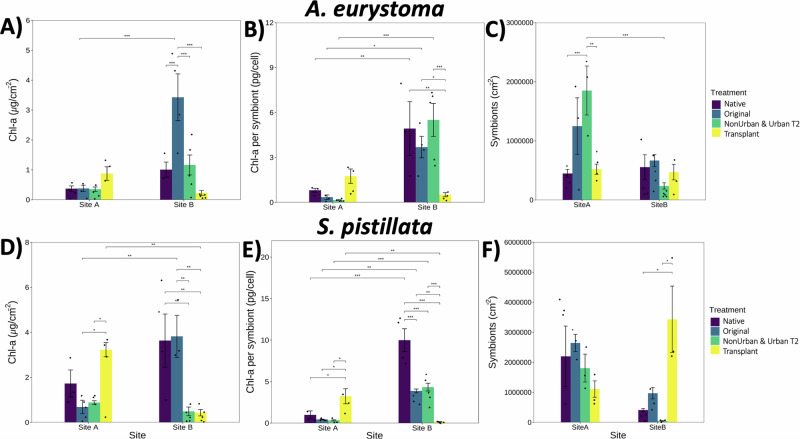
Variation in endosymbiont algae density and chlorophyll concentration in the tissues of corals is also broadly related to stress adaption, bleaching prediction, and symbiont stability. We observed a clear difference in Chl-a (µg/cm−2) in A. eurystoma between the sites and different treatments (p < 0.0001) (Fig. 2). Chl-a (µg/cm−2) in A. eurystoma corals that were attached to Original tiles at Site B was significantly higher (p < 0.0001) than at Site A (Fig. 2A; Supplementary Table 4). At Site B, the Chl-a (µg/cm−2) concentration in A. eurystoma corals on Original tiles experienced a significant increase compared to the Transplant A (p < 0.0001) tiles (Fig. 2A; Supplementary Table 4). Chl-a per endosymbiont (pg/cell) in A. eurystoma was significantly different among coral treatments across both sites (p < 0.001) (Fig. 2B; Supplementary Table 5). In particular, A. eurystoma corals that were affixed to Transplant tiles from Site A that moved to Site B, had less Chl-a per endosymbiont densities than the other tile treatments at Site B (p < 0.001 Native, p < 0.01 Original, and Urban T2 p < 0.0001; Fig. 2B). Endosymbiont density in A. eurystoma corals was significantly different, further demonstrating the consistent site and treatment effect (p < 0.001) (Fig. 2C). A. eurystoma corals, in general, had higher densities of endosymbionts per cell on the NonUrban T2 tiles compared to the Site A, Native A. eurystoma (p < 0.0001) and Transplant tiles (p < 0.001) at both sites (Fig. 2C; Supplementary Table 5).
Fig. 2. Chlorophyll-a and endosymbiotic algae measurements.
A A. eurystoma - chlorophyll-a per µg/cm−2 in each coral, (B) Chlorophyll-a per endosymbiont per cell, and (C) Density of endosymbionts per coral. D S. pistillata – chlorophyll-a per µg/cm−2 in each coral, (E) Chlorophyll-a per endosymbiont per cell, and (F) Density of endosymbionts per coral. Data is expressed as averages ±SE and black dots represent replicates (n = 5). Significance was determined by 2-way ANOVA and post-hoc tests across and within treatments, where p < 0.05 (*), p < 0.001 (**), and p < 0.0001 (***). Figure correlates to Supplementary Tables 4–7.
Site and treatment differences in Chl-a (µg/cm−2) concentrations in S. pistillata corals were consistent with the patterns we observed in A. eurystoma (p < 0.0001) (Fig. 2D). For example, S. pistillata corals from Site B that were affixed to the Original tiles at Site B contained a high density of Chl-a (µg/cm−2) (p < 0.001), compared to much lower concentrations in Chl-a (µg/cm−2) maintained in S. pistillata corals that were attached to the Transplant A tiles at Site B (p < 0.001; Fig. 2D and Supplementary Table 6). Whereas S. pistillata corals from Site A that were on the Site A Original (p < 0.01) and the NonUrban T2 tiles (p < 0.01), had considerably less Chl-a (µg/cm−2) concentrations than the Site A corals on the Transplant tiles that came from Site B (Fig. 2D; Supplementary Table 6). Chl-a (µg/cm−2) densities in Site B S. pistillata corals attached to the Site B Original tiles, were the highest across sites and significantly greater than in the S. pistillata corals that were on the Transplant A tiles (p < 0.001) (Fig. 2D; Supplementary Table 6). Chl-a (pg/cell) in S. pistillata corals remained highly significant when comparing across treatments and sites (p < 0.0001) (Fig. 2E). Site A S. pistillata corals attached to the Transplant tiles from Site B, had a greater density of Chl-a per endosymbiont (pg/cell) than the Native (p < 0.01), Original (p < 0.01), and NonUrban T2 treatments (Fig. 2E; Supplementary Table 7). At Site B, there was considerable variation among all tile treatments, as Site B S. pistillata corals contained high levels of Chl-a per endosymbionts when attached to tiles of all treatments, except for the Transplant A tiles at Site B (p < 0.0001; Fig. 2E and Supplementary Table 7). Similarly, to what was observed at Site A in endosymbiont densities in A. eurystoma, S. pistillata corals from Site A as well as the corals affixed to the Original Site A tiles, also experienced a higher abundance in endosymbionts compared to the other treatments (Fig. 2F). In contrast to the patterns of endosymbiont densities observed in A. eurystoma, S. pistillata corals at Site B attached to the Transplant A tiles, contained the highest density of endosymbionts, especially in comparison to the Native S. pistillata corals (p < 0.01) and S. pistillata attached to the Original tiles (p < 0.01) (Fig. 2F; Supplementary Table 7).
Coral physiology - Parameters of oxidative stress
Oxidative stress parameters are classic biomarkers of environmental stress in both algae and aquatic animals33. Higher TAC levels enable corals to counteract and mitigate oxidative stress conditions and is generally related to higher stress resistance34. Oxidative damage to lipids, such as lipid peroxidation (LPO) is a common biochemical outcome of oxidative stress in aquatic animals, and increased levels of LPO were commonly observed in corals under stress by light pollution32 and heat anomaly34. In general, increased activity of antioxidant defenses is a strategy to alleviate LPO, depending on the frequency and intensity of the stressor, increased antioxidant defense does not always translate to decreased oxidative damage to macromolecules35.
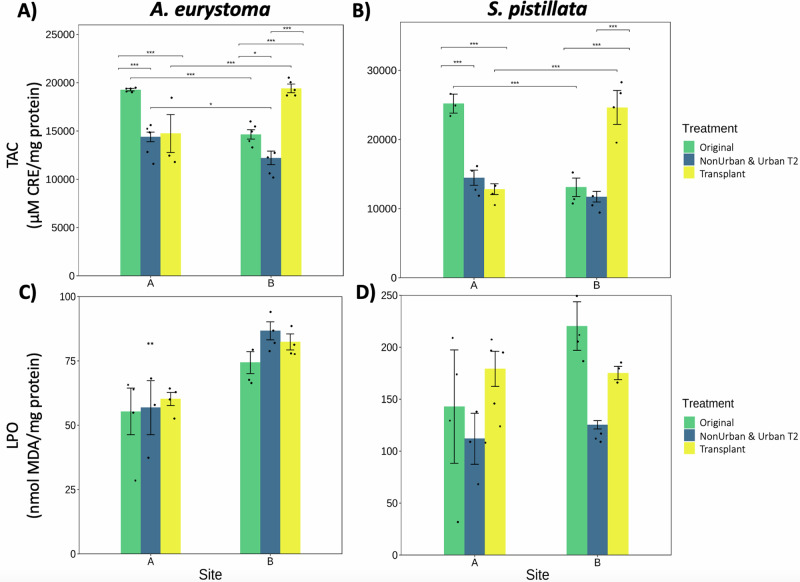
We observed a significant difference in TAC levels across site and treatments in the coral A. eurystoma (p < 0.0001; Fig. 3A). This was further pronounced in the elevated levels of TAC in A. eurystoma corals affixed to the Original Site A tiles, compared to the other tile treatments at Site A (p < 0.0001; Fig. 3A; Supplementary Table 8). A. eurystoma corals attached to both the Original tiles from Site A and the Transplant A tiles, also experienced elevated levels of TAC, where both treatments had the highest levels across sites (p < 0.0001; Fig. 3A). At Site B, we observed a similar pattern, where A. eurystoma corals that were attached to the Transplant A tiles had higher counteractive TAC levels (p < 0.0001), followed by the A. eurystoma corals that were attached to the Original Site B tiles (p < 0.0001; Fig. 3A).
Fig. 3. Oxidative stress parameters in corals.
A, B Total antioxidant capacity (TAC; µM CRE/mg protein) expressed in A. eurystoma and S. pistillata across sites and within each treatment, respectively. C, D Lipid peroxidation (LPO; measured as nmol MDA/mg protein) in A. eurystoma and S. pistillata across sites and within each treatment, respectively. Data is expressed as means, black dots are replicates, and ±SE (n = 5). Replicates with protein levels below a minimal standard were not included (minimum number of replicates n = 3). Significance was determined by 2-way ANOVA and post-hoc tests comparing across and within treatments, where p < 0.05 (*), p < 0.001 (**), and p < 0.0001 (***). Figure correlates to Supplementary Table 8.
There was a significant difference observed for TAC levels, across both sites and treatments in S. pistillata corals (p < 0.01; Fig. 3B). This interaction was further demonstrated at Site A, where S. pistillata corals affixed to the Original tiles contained the highest elevated levels of TAC compared to the other treatments (p < 0.0001; Fig. 3B; Supplementary Table 8). Similarly to A. eurystoma, S. pistillata corals attached to the Transplant A tiles, contained the highest levels of TAC at Site B compared to the other treatments (p < 0.0001; Fig. 3B).
There was a significant difference between the LPO levels of A. eurystoma between sites (p < 0.001), as lower levels of LPO were found in A. eurystoma corals at Site A in comparison to Site B (Fig. 3C). A. eurystoma corals attached to the Original Site A tiles had the lowest levels of LPO. In general, LPO was lower in S. pistillata corals at Site A compared to the corals attached to Site B tiles (Fig. 3D), however there was no significance for S. pistillata across sites and treatments.
Benthic community characterization
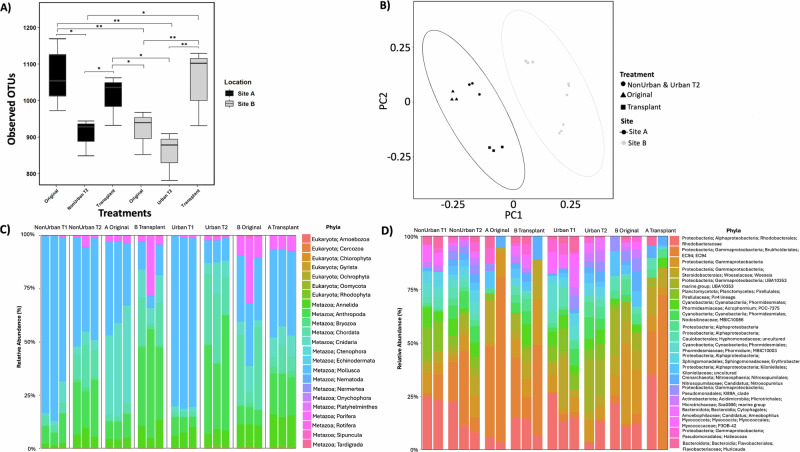
Twenty-eight main phyla were identified via COI belonging to metazoans (1623 OTUs), algae (146 OTUs), and eukaryotes (6 OTUs). Differences in invertebrate communities between site (A and B) and tile treatment (Original, NonUrban T1 & T2 and Urban T1 & T2, and Transplant) were highly significant (p < 0.0001), accounting for 19.7% of community variance (Supplementary Table 1). The strongest factors were the treatments across sites where most of the variance was observed (p < 0.0001; 37.7%), while tiles within sites accounted for 22.7% of variance (p < 0.0001) (Supplementary Table 1). Within Site A, tiles accounted for 79.3% of variance (p < 0.0001) and 72% at Site B (p < 0.0001) (Supplementary Table 1).
The Original A and Transplant A tiles (from Site A) harbored a significantly richer invertebrate community than the Original tiles at Site B (p < 0.001; Fig. 4A), suggesting a consistently higher species richness associated with Site A. The community composition within each site exhibited overall similarity; however a significant divergence was observed in the Transplant A tiles, which were markedly different from the other tiles at Site B and more closely resembled their origin at Site A (Fig. 4B).
Fig. 4. Taxonomic richness, community composition, and normalized abundances of invertebrate (COI) and bacteria (16S) communities.
A Observed invertebrate species richness between sites and treatments, using pairwise Wilcoxon Signed Rank tests; significance indicated by p < 0.05 (*) and p < 0.001 (**). The box represents the interquartile range with the bottom and top edges representing the 25th and 75th percentiles. The line inside the box indicates the median (50th percentile), while the whiskers indicate min and max values, excluding outliers (n = 18). B Observed community composition of invertebrate species on tiles from different sites and treatments. Ellipses show distinct clustering using the Bray-Curtis dissimilarities to test PERMANOVA differences between site and treatment (n = 18). C Percent relative abundance of the 23 most abundant invertebrate phyla within each site, treatment, and timepoint (n = 24). D Percent relative abundance of the 20 most abundant bacteria phyla (to family level) within each site, treatment, and timepoint (n = 20). Missing samples are due to QIIME’s quality filtering or a low read abundance. Figure correlates to Supplementary Figs. 3–6 and Tables 1, 9–10.
The top phyla from the organisms collected from tiles at Site A (all treatments; not including the Transplant from Site B) were Mollusca (49.2%), Cnidaria (16.9%), Arthropoda (11.5%), Chordata (6.3%), Annelida (3.7%), and Porifera (1.6%) (Fig. 4C; Supplementary Fig. 3). At Site A, the highest diversity of species was from Arthropoda (576 OTUs), followed by Cnidaria (318 OTUs), Mollusca (174 OTUs), and Annelida (155 OTUs). At Site B (all treatments; not including the Transplant from Site A), the following main phyla consisted of Mollusca (33.9%), Arthropoda (18.6%), Chordata (16.5%), Annelida (7.7%), Cnidaria (6.1%), and Porifera (5.8%) (Fig. 4C; Supplementary Fig. 3). The highest levels of OTU diversity at Site B were found among the phyla Arthropoda (580 OTUs), Cnidaria (318 OTUs), Mollusca (174 OTUs), and Annelida (155 OTUs), similarly to Site A. The Site A tiles that were transplanted to Site B (Transplant A) had a higher percentage of reads from Arthropoda (40%), Cnidaria (10.9%), Porifera (10.8%), and Annelida (9.5%) than tiles at Site B (Fig. 4C). Additionally, organismal abundance increased by 15% on NonUrban T2 tiles at Site A and by 22.5% on Urban T2 tiles at Site B compared to their respective T1 counterparts (Fig. 4C). A differential abundance analysis was conducted to compare Original A, Original B, and Transplant A tiles, determining seven significantly different phyla between the treatments: Cnidaria (e.g., corals, anemones, and hydroids), Bryozoa (colonial filter feeders), Annelida (segmented worms), Chordata (e.g., tunicates and ascidians), Rotifera (rotifers), Sipuncula (unsegmented worms), and Chlorophyta (green algae) (p-adj < 0.001; Supplementary Figs. 3–4).
We examined the potential differences in the microbial community on the biomimetic tiles across sites, finding differences in the bacteria community between treatments (p < 0.01) (Fig. 4D; Supplementary Fig. 5). Overall, the abundance of the top 20 bacteria families was relatively similar amongst treatments, with the most ubiquitous bacteria belonging to the phyla Proteobacteria. However, the most notable difference occurred between the Original A, Original B, and Transplant A tiles, with a Proteobacteria from the order Burkholderiales EC94, previously identified from sponge tissues and corals36–38. Site A Original tiles had a higher relative abundance of Burkholderiales EC94 (44.8%) than Site B Original tiles (1.6%). Transplant A tiles retained a higher relative abundance of EC94 (29.3%), compared to the other tiles at Site B (Fig. 4E; Supplementary Fig. 6). A differential abundance analysis determined that there were ten different bacteria families that had significantly different abundances between the Original A tiles and the Transplant A tiles (p-adj < 0.05; Supplementary Table 9). The same analysis was conducted to compare the Original B tiles to the Transplant A tiles, which identified 21 different bacteria families (p-adj < 0.05), which included Burkholderiales EC94 (p-adj<0.05; Supplementary Table 10). Suggesting that the community of bacteria was more closely associated with the dominant invertebrate phyla found on the tiles, such as Cnidaria and Porifera (Fig. 4C), which was also consistent with visual observations.
Effect of the organismal community on coral physiology and health
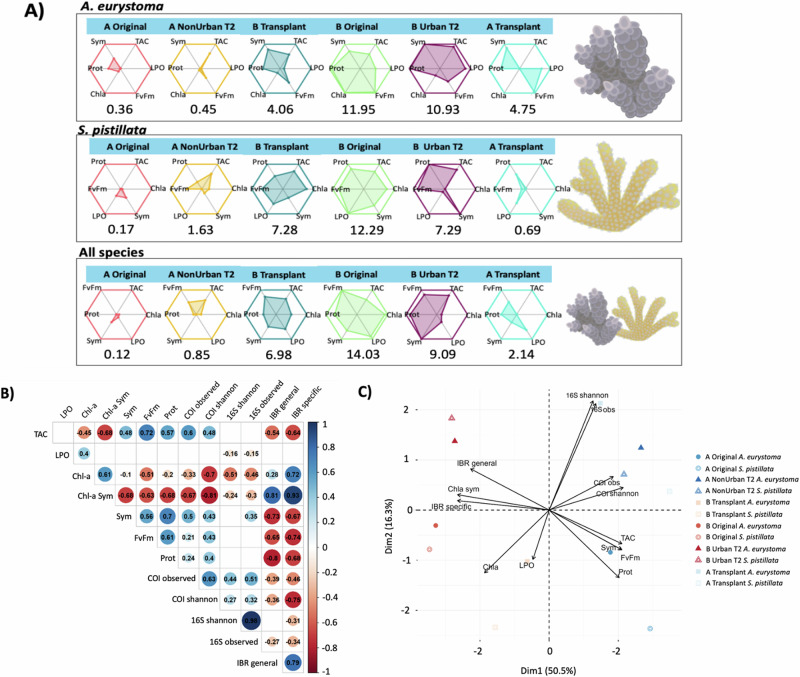
The relationship between the coral physiology parameters (biomarkers) were indicated by different levels of stress through an integrated biomarker response (IBR) for each site and treatment. A. eurystoma corals attached to Site A tiles maintained a higher level of health compared to corals from Site B treatments (Supplementary Table 11), including the A. eurystoma corals attached to Site A tiles that were transplanted to Site B (p < 0.001; Fig. 5A). S. pistillata exhibited a similar response to A. eurystoma (Supplementary Table 12), where corals attached to Site A tile treatments (including the Transplant A tiles that moved to Site B; p < 0.00001), performed significantly better than the S. pistillata corals attached to Site B tiles (Fig. 5A). Both coral species attached to tiles from Site A (including the Transplanted A tiles that moved to Site B) experienced elevated levels of health descriptors than the corals attached to the tiles at Site B (Fig. 5A; Supplementary Table 13).
Fig. 5. The relationship between the different organismal communities (COI and 16S) and coral physiology.
A Coral health was assessed by a stress index (IBR). Areas derived from star plots show the level of alteration in the physiological biomarkers in each site and treatment. The higher numerical values associated with each plot indicate increased physiological stress. B The correlation between the organismal community and coral physiology parameters denoted by colors representing positive and negative values. The size of the circle is proportional to the strength of the correlation (r2), also indicated by the numbers. Blank spaces indicate no significant correlation (p < 0.05). C The PCA plot visualizes the proximity and direction of the vectors and each physiological test, with the symbols referring to the different treatment and coral species. Figure correlates to Supplementary Figs. 7–9 and Tables 11–13.
Positive numbers were attributed to significant values for both A. eurystoma and S. pistillata corals attached to all tiles at Site A. Increased values of health for TAC, Protein, Fv/Fm, and Symbionts were also related to increased richness and diversity of the organismal community (i.e., invertebrates, microorganisms, and bacteria) at Site A (Fig. 5B; Supplementary Figs. 6–8). Site B tiles (except for the corals on the Transplant tiles that went from Site A to Site B) were associated with negative values for the IBR – stress index, Chlorophyll, and Symbionts (Fig. 5B). These comparisons are further visualized in Site A tiles (including corals attached to Transplant tiles that moved from Site A to Site B), which are clustered on the positive side of PC1 (Fig. 5C). Higher richness (COI) was significantly correlated with elevated levels of TAC, Fv/Fm, proteins, and a lower IBR, while an increased diversity of bacteria (16S rRNA) was associated with less LPO (Fig. 5B).
Discussion
This study constitutes an exploration of the invertebrate and bacteria communities of coral reefs from healthy and degraded reefs in the GoE/A, and the effect that it has on coral physiology. We demonstrate that the organismal communities of a healthy coral reef are associated with a greater richness of reef invertebrates and microorganisms than that of a degraded reef. Furthermore, we validate that transferring benthic organismal communities from a healthy reef to a nearby degraded reef on substrates could be used as a mechanism to boost the health of corals at degraded reefs. Therefore, we propose aCRET as a restoration strategy to holistically support degraded reefs. It is important to note that the aCRET method may not be suitable for all global coral reefs and coral species, as some organisms within these communities may not always be conducive to boosting coral health. A minimum amount of time may also be required to maintain the improved health conditions of corals at a degraded site. Nevertheless, this research highlights the potential of harnessing the invertebrate and bacteria communities of healthy reefs as a support system for deteriorated corals, in conjunction with biomimetically-designed tiles or substrates as a scalable, transferable ecosystem to aid adjacent degraded reefs in the interim or to compliment current reef restoration initiatives.
Key physiological functions of corals from the degraded site were improved after performing aCRET from the healthy to degraded reef. Overall, S. pistillata responded slightly stronger to aCRET from the healthy site than A. eurystoma, however it is evident that both species were significantly boosted (Fig. 5). For example, corals from the degraded site attached to healthy reef tiles experienced an increase in photochemical capacity after 6 months (Fig. 1). Additionally, S. pistillata secured to the tiles from the healthy reef transferred to the degraded site, had a much higher symbiont count, than the other treatments, analogous to the corals native to the healthy reef (Fig. 2). In other studies, from the GoE/A, branching corals located at healthy coral reefs compared to highly urbanized degraded reefs, also experienced a similar occurrence, where corals exhibited a higher photochemical capacity, reduced levels of chlorophyll, and a higher symbiont count32,39. For example, the elevated chlorophyll levels observed in the corals at Site B may be attributed to the excess nutrients and pollution reported there, with the notable exception of corals affixed to the transplanted tiles. Key nutrients and microbes associated with the benthic invertebrate communities of the healthy reef, could play a role in helping corals cope with the increased urbanization at the site32.
This boost in coral physiology could also be explained by an increased antioxidant capacity (TAC) displayed in corals from the healthy reef and the corals attached to the tiles that were transplanted from the healthy reef. TAC can mitigate potential detrimental effects caused by stress39, such as oxidative stress and cellular damage. Decreased values of TAC can indicate more antioxidants are being used to counteract the overproduction of reactive oxygen species (ROS), which signifies oxidative damage32. We observed higher TAC levels in corals from the healthy reef and the corals on the transplanted healthy reef tiles that were at the degraded site, signifying low stress and high tolerance (Fig. 3). In contrast to other studies32,39, we did not observe a linear inverse relationship, with higher levels of TAC on treatments with lower levels of LPO. However, we did see a significant decrease in LPO of corals at the healthy coral reef in contrast to the degraded site. This could suggest that when corals attached to the tiles that came from the degraded reef (Site B) moved to the healthy reef (Site A), they were immersed in the healthy reef environment and were able to cope better with potential stress and conduct cellular repairs. Adversely, the corals attached to the tiles with the healthy reef community (Site A) that were transplanted to the degraded reef (Site B) did not experience the same benefit, despite TAC already being significantly boosted. The pool of non-enzymatic antioxidants (assessed by TAC levels) was boosted in corals attached to tiles containing the healthy reef organisms, but this was not reflected in a reduction of LPO at Site B (Fig. 3). The increased TAC was not enough to counteract the oxidative stress generated at Site B, at least for A. eurystoma. Despite the higher level of health descriptors, the transplanted healthy reef tiles from Site A to B could not mitigate the LPO and increased chlorophyll (that could be a compensatory mechanism) that the Site B corals experienced (Fig. 5). Nevertheless, IBR values show decreased overall physiological stress in the corals attached to healthy Site A tiles at Site B, compared to corals attached to tiles kept for >6 months at the degraded site (Site B). Using this integrative approach, we consider the combined effects of biomarkers, providing a more holistic view of the corals’ condition rather than individual biomarkers alone40,41. We acknowledge that metrics of coral health while taken individually, may not conclusively determine coral health, but when combined represent a more comprehensive insight into the shift in coral health, contributing to our understanding of the physiological state of the transplanted corals. Our findings indicate significant signs in these physiological measurements, which are well-established proxies for assessing coral health. For instance, enhancements in antioxidant capacity, although not directly indicative of overall health, suggest an adaptive response to stress, which is crucial for survival and acclimatization under new environmental conditions. Similarly, changes in photopigmentation and symbiont densities are indicative of photoacclimation processes, which play a vital role in the corals’ ability to adapt to varying light conditions—an important factor in their potential use for reef restoration.
Differences among free-living microorganisms and invertebrate communities between healthy and degraded coral reefs are known to be highly distinct and often location-specific13,42–45. Across reef habitats, the variation among these communities could be related to different nutrient regimes29,46, fluctuations in environmental conditions2, and various biotic patterns13,45, whereas site-specificity could also be connected to the structural complexity and benthic diversity of the reef. The degraded, highly urbanized reef (Site B) in this study has historically been characterized by more detrimental changes, such as spikes in salinity, nitrates, phosphates, and ammonia, in comparison to the healthy, non-urban reef31,32, which has been known to support an abundance of diverse marine species (Supplementary Data 1)29,31. The pH and temperature have remained some of the only consistent parameters between the two reef sites in the last several years31 (Supplementary Data 1). The environmental condition of the urbanized site was most likely caused by local eutrophication (e.g., excess nutrients) and human-induced perturbations, such as pollution and artificial light at night (ALAN), which is highly prevalent in the GoE/A, causing disturbances in coral physiology and spawning29,31,32,39. Promoting resilience and enforcing the persistence of coral-dominance at declining coral reefs can be challenging47. Although some corals can survive in degraded conditions, there could still be room for improvement, by introducing beneficial communities to boost their current health state and encourage long-term resilience.
In this study, we identified differences in the invertebrate and bacteria communities between a degraded and healthy reef, as the healthy reef sustained a more diverse and richer community of organisms (Fig. 4). We visually noticed a higher presence of fouling organisms, such as ascidians and annelids, as well as an increase in turf algae on tiles that never left Site B (i.e., photography and visual observations), which may have suppressed the habitation of other important reef-building invertebrates. It was observed that the healthy reef (Site A) contained a much higher abundance of species from the phyla, Cnidaria and Mollusca, as well as bacteria directly linked to sponges38 and some corals36–38, such as the Gammaproteobacteria, Burkholderiales EC9448,49. Burkholderiales EC94 is one of the more abundant bacteria identified with Red Sea coral reefs and had a higher abundance on Site A tiles compared to all tiles across sites, including the Site A tiles that were transplanted to B37. In particular, EC94 bacteria has been found in abundance within sponge tissues constituting more than 70% of the microbial profile38. This prevalence likely facilitates the frequent discovery of sponge-associated bacteria within corals, which may be attributed to shared environmental niches (i.e., biofilms) and potential symbiont transmission between sponge and coral holobionts38, as there was a visual presence and moderate abundance of sponges identified on the tiles at both sites. Notably, there was a higher diversity and richness of Cnidarians - corals and hydrozoa, from the tiles of the healthy reef, including the transplanted healthy tiles (Site A) that went to the degraded site (Site B), which could also be consistent with the dominance of Burkholderiales EC94 bacteria. While these key organisms may illustrate broader patterns, it is crucial to emphasize the collective role of the invertebrate and microbial communities at the different sites in influencing coral physiology. Numerous studies underscore the importance of a diverse reef community, highlighting how a variety of species contributes to reef vitality5,24,26,27. Recognizing that although some species may have a greater impact on coral health, the dynamics of the reef community as whole and their combined interactions contribute to improvements in coral physiology.
The benthic communities of a coral reef are often correlated with the reef’s surface, structural, and cryptic complexity24–27,30,50. Nelson et al. 24, observed that structural and micro-complexity were more of a determining factor in regulating assemblages of invertebrates, rather than live coral. Previous studies cited micro-niche partitioning as a way for corals and reef-associated communities to sustain richness and abundance to effectively maintain resources30,50, similarly to the improvements in coral physiology we observed. The reef community composition was found to be dependent on the rate of increasing complexity, and that abundance and richness could be just as important as diversity26. Moreover, we are mindful of the potential to introduce invasive species and pathogens on transplanted communities, however, to mitigate this we advocate for using local and nearby communities in proximity to the degrading reef. Future studies should indeed include broader ecological and biosecurity assessments to mitigate the risks associated with transplantation and ensure sustainable restoration practices. We recommend the continued exploration and utilization of aCRET method in conjunction with optimized 3D printed biomimetic substrates30 and large-scale artificial hybrid reefs28,51,52 as part of upscaling measures, to bring structures that have been pre-inoculated with a healthy reef community to help support nearby reefs. Light-based 3D printing could also be used as an effective way to create living coatings from bioink or microstructures53,54 of beneficial organismal communities19, which are encapsulated in a hydrogel matrix (e.g., bacteria and algae)55. These living materials could cover large-scale 3D printed ceramic structures or directly 3D bioprinted as a biomimetic structure56,57 to upscale restoration for global coral reefs28. As this method holds numerous promising benefits, it is vital to continue understanding the potential of boosting the health of corals, by utilizing key reef communities of bacteria and invertebrates to support inexpensive, scalable, and holistic restoration efforts.
Methods
Study sites and experimental design
Two coral reefs were chosen as study sites based on their location, health, and biodiversity status in the GoE/A. Permits to conduct this research were granted by the Israel Nature Parks Authority for deploying and retrieving tiles (Permit #2020/42549) and collection and sampling of corals (Permit#2021/42831). The healthy reef (Site A) is characterized by accreting patch reefs and a coral reef biorock structure in the southernmost, non-urbanized portion of the Gulf, away from human disturbances (Supplementary Fig.1). This site was chosen based on its high diversity, abundance, and richness of coral reef species30 and its stable environmental condition (i.e., temperature, water quality, nutrient levels) (Supplementary Data 1)29,31. The degraded reef (Site B) is in the northernmost area of the Gulf, roughly 6 km from Site A, in a heavily urbanized region due to human impacts and characterized by fringing reefs interspersed with sand and seagrass58. Site B is known to experience poor water quality, exposure to salinity fluctuations, pollution, and enriched nutrients (e.g., nitrates and phosphates) (Supplementary Data 1)29,31. Both sites contain small and large patch reefs in addition to long-standing artificial structures such as a biorock reef and nursery tables sitting at roughly 8–12 m.
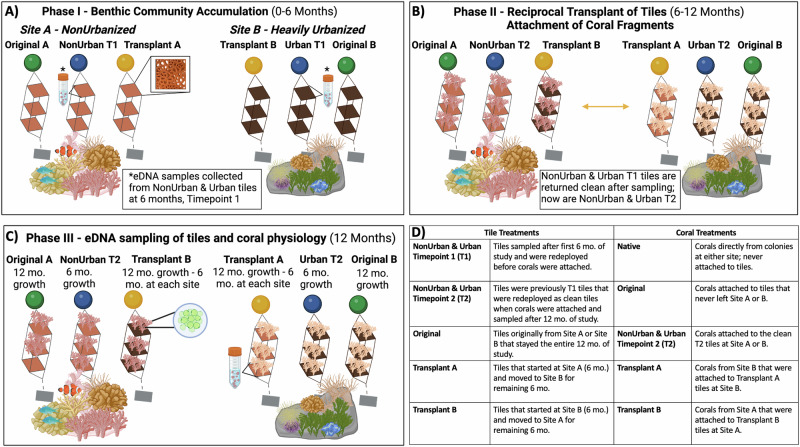
We deployed 18 biomimetic tiles (25 cm × 25 cm) fabricated with 3D design, printing, and molding to replicate the rugosity of natural reef surfaces (Supplementary Fig. 1)30. Three biomimetic tiles were held together in a stainless-steel frame (one complete structure; n = 3, per site) and were spaced apart by 45 cm to allow light access and water flow30. Biomimetic tiles within each frame were attached to the coral reef at roughly 8 m at Site A (n = 9) and Site B (n = 9) from February 2021 to February 2022. The tile treatments at either site were described by the time and status of the organismal community accumulated on each tile (Fig. 6A–D). Site A and B each contained a set of Original tiles (Original A and Original B) that never left the site and stayed for the total 12-month duration of the study. The next tile treatment was sampled twice during the study for eDNA (i.e., marine invertebrates and bacteria); the first time was after the first 6 months of community development to mark timepoint 1, before corals were attached (Phase I). Site A tiles were referred to as NonUrban T1 and Site B tiles were referred to as Urban T1 tiles (Phase I). After sampling, these tiles were scrapped clean, washed, and dried visibly of any organisms or biofilms, and redeployed as clean tiles for an additional 6 months at Site A as NonUrban T2, and at Site B as Urban T2 tiles (Phase II), where they remained at either site until they were sampled for the second time at the end of the study (Phase III). The last tile treatment were the Transplant tiles that were reciprocally transferred from either site, Transplant A tiles (conditioned at Site A) went to Site B and Transplant B tiles (conditioned at Site B) went to Site A (6 months of growth at both sites) (Phase II; Fig. 6B). Tiles were monitored visually every 2 months using photogrammetry.
Fig. 6. Experimental design demonstrating the three main phases of the 12-month study.
A Phase I (Month 0-6), nine biomimetic tiles were deployed at Site A and B. After 6 months, only the NonUrban T1 and Urban T1 tiles were collected and sampled from each site and redeployed as clean tiles in the water as NonUrban T2 and Urban T2. B Phase II (Month 6), the Transplant tiles were reciprocally transferred from either site. Fragments of A. eurystoma and S. pistillata were attached to all tiles. C After another 6 months, in Phase III (12 months total) all corals and tiles were collected for analysis. D Glossary of terms related to tile and coral treatments. Figure correlates to Supplementary Fig. 1. Created in BioRender. Levy, O.30 BioRender.com/t97k483.
After the reciprocal transplant of biomimetic tiles between sites, fragments were collected from mature, native colonies of two common branching corals at each site to attach to tiles. This study utilized two prevalent scleractinian coral species in the GoE/A: Acropora eurystoma, a broadcast spawner from the Complexa clade, and Stylophora pistillata, a brooder from the Robusta clade58. Corals were collected (n = 5, per species) per site at a depth of 8 m, fragmented into smaller colonies and secured to each tile treatment for an additional 6 months. Coral treatments were based on the specific tile treatment that they were attached to during the study (Fig. 6B, C). Native corals were directly collected from colonies at either site and were never attached to tiles, serving as the site baseline. Original corals remained on the original Site A or Site B tiles that stayed at one site for all 12-months. NonUrban T2 and Urban T2 corals were attached to clean tiles that stayed at either site until the end of the study. Transplant corals were on the tiles that came from the opposite site (Transplant A - conditioned at A went to Site B and Transplant B – conditioned at B went to Site A) (Fig. 6B, C). All remaining corals not used in the experiment were replanted back on their original reef. Tiles and corals were monitored every 2 months using photogrammetry. After the second 6-month period (Phase III; 12 months), the organisms on all tiles were assessed using eDNA (COI and 16S rRNA) to determine species diversity, richness, and relative abundance. Coral health was evaluated based on photochemical efficiency (i.e., ETR curves and Fv/Fm), protein content of coral tissues, endosymbiont algae density and chlorophyll concentration, and TAC and LPO as a measure of oxidative stress and cellular damage.
Sample collection and processing
Organismal community samples were collected from biomimetic tiles at each site at time point 1 (end of Phase I) after first 6 months (n = 8, 2 controls) and again at 12 months (Phase III) (n = 18, 2 controls) via SCUBA diving (Fig. 6). Biomimetic tiles were photographed inside and outside of the water before sampling. A clean control tile was brought into the water at each site during each tile retrieval timepoint. All tiles were secured in a sterile bag in the field, before arriving to a wet lab at the Interuniversity Institute for Marine Sciences in Eilat (IUI). The seawater that was collected in the bag with the tile was not included in the processing. Sessile reef organisms from both sides of the tile were scraped into a sterile tray, collected, and homogenized as a whole community30,59. Samples were preserved and frozen at −20 °C for DNA extractions. To avoid potential contamination between samples all tools and equipment were sterilized with three disinfectant baths of 10% bleach, 90% ethanol, and fresh water, which were replaced regularly.
Corals that were never attached to tiles and served as the Native (baseline) treatment for each site were snap-frozen in liquid nitrogen at the time of fragmentation at the end of the first 6 months (Phase II). At the end of the 12-month study, coral fragments that were attached to tiles for 6 months (Phase III) were collected. Corals were carefully removed from each tile, labeled, and sealed in sterile bags in the field at both sites. Corals were separated by treatment and site (n = 5) in outdoor, open system water tables at IUI, before measuring quantum yield with PAM (Imaging PAM) fluorometry. Water tables were maintained with a consistent pH, salinity, and temperature with a direct flow from the GoE/A. Subsequently, all corals were wrapped in aluminum foil, tagged, and snap-frozen in liquid nitrogen before being stored at −80 °C until future processing.
Coral physiology
Coral fragments of A. eurystoma (n = 5) and S. pistillata (n = 5) from each site and treatment were assessed for several physiological parameters as a proxy for health and stress: pulse amplitude modulation (Imaging PAM), total protein concentration, algae density, chlorophyll concentration, TAC, and LPO.
Coral photochemical measurements were conducted using an Imaging PAM (Maxi-PAM, Heinz Walz GmbH, Germany). All coral fragments were adapted in total darkness for 20 min before measurements occurred. Fluorescence was measured as rapid light curves (RLCs) and were interpreted as ETR as illuminations increased at 120-s intervals (0, 20, 55, 110, 185, 280, 335, 395, 460, 530, 610, 700 µmol quanta m−2 s−1)32. Images of coral fluorescence were analyzed in the Imaging-Win software program (v2.56p; Walz Gmbh) by selecting two areas in the top and bottom of branches. The software generated the following measurements which were used to calculate maximum ETR through photosystem II (PSII) (ETRmax) obtained through fitted RLCs; Photosynthetically active radiation (PAR) was used to measure the intensity of useable light for photosynthesis across a spectrum; Fo and Fm were used to calculate the Fv/Fm, which is the difference between maximum fluorescence and minimum fluorescence; and Y(NPQ), which is the irradiance at the point that the absorbed quanta is dissipated through a process of nonphotochemical quenching60.
Coral tissues were removed from previously frozen fragments using an airbrush and ice-cold filtered seawater (FSW) (0.22 µM), followed by homogenization for 30 s (Polytron™ PT2100 Benchtop Homogenizer, Kinematica, Switzerland). Aliquots of 1 ml were divided into Eppendorf tubes, sonicated in an ice water bath, and vortexed. Surface area was determined using a wax [single] dipping technique on coral skeletons61,62. Total protein (holobiont) was processed using a bovine serum albumin as a standard (Quick Start Bradford Protein Assay Kit 1, Bio-Rad, Israel) and a standard curve was generated (Bradford 1976) in a 96-well plate. Total protein content was determined with a Biotek Synergy HT microplate reader (Marshall Scientific, USA) (595 nm and 450 nm). Normalizations were conducted according to the volume (ml) of tissue and the skeletal surface area (cm−2). Algal density was determined from a 100 µl sub-sample and counted on a hemocytometer under a microscope under x100 magnification. Five squares were counted (10 µl per square at depth of 0.1 mm) and the average counts were multiplied by 10,000 (number of cells/ml). Chlorophyll concentrations were achieved after samples were centrifuged for 5 min at 5000 g at 4 °C, decanted and resuspended in 1 ml FSW. One ml of 90% acetone was added to the remaining pellet and incubated for approximately 24 h at 4 °C in total darkness. Chlorophyll-a concentrations were measured with a UV-1900i UV-VIS Spectrophotometer (Shimadzu Scientific Instruments, Japan) at 630, 663, and 750 nm. Chlorophyll concentrations were calculated and normalized to algae counts and surface area as previously described63.
TAC and LPO assays were conducted as proxies for oxidative stress. The OxiSelectTM TAC Assay Kit (Cell Biolabs Inc., USA) was used to measure TAC of low molecular scavengers according to the manufacturer’s protocol in a 96-well plate. This method is widely used as a proxy of the non-enzymatic antioxidant defense system40. A Biotek Synergy HT microplate reader (Marshall Scientific, USA) was used to read absorbance (490 nm), data was normalized according to the total protein content in each well and conveyed as µM copper reducing equivalents (CRE)/mg protein. Oxidative lipid damage was measured according to a protocol described by Federici et al. 64 in 96-well plates, based on the 2-thiobarboturic acid reactive substances (TBARS)64. Lipids are prevalent targets of oxidative damage, and TBARS quantification is a frequent technique to evaluate cellular damage in aquatic organisms exposed to environmental stressor. Lipid damage was measured by the reaction between malondialdehyde (MDA) (a by-product of LPO) and thiobarbituric acid. Readings of autofluorescence were calculated at 535 and 549 nm using a Biotek Synergy HT microplate reader (Marshall Scientific, USA). Normalization of the results was conducted according to the total protein content in each well and conveyed as nmol MDA/mg protein. Homogenates with protein levels below a minimal standard point curve were not included (minimum number of replicates n = 3).
DNA extraction, amplification, and sequencing
eDNA of bacteria and invertebrates from the biomimetic tiles were extracted using the DNeasy PowerMax Soil Kit (Qiagen, Hilden, Germany) from bulk sessile samples according to Levy et al.30. The DNeasy PowerClean Cleanup Kit (Qiagen, Hilden, Germany) was used to purify genomic DNA following the manufacturer’s protocol. Amplification of the 313 bp fragment of the mitochondrial Cytochrome c. Oxidase subunit I (COI) region occurred using a two-step PCR process. Primers targeting metazoans (i.e., mlCOIintF and jgHCO2198)65 were synthetized with locus-specific overhangs30, and index specific primers (Illumina Metagenomic Sequencing Library Prep, pp. 6–14) were attached. PCRs and purification steps were executed according to Levy et al.30. Final PCR products were measured with Qubit dsDNA HS Assay (Invitrogen, USA) and average fragment size was estimated with a 4200 TapeStation System and a D1000 ScreenTape (Agilent, USA). Equimolar concentrations of PCR products were pooled together. Libraries were sequenced using an Illumina MiSeq v2 500-cycle Kit (Illumina, San Diego, USA) on an Illumina MiSeq platform. Marine bacteria were targeted using the prokaryote 16S ribosomal RNA (rRNA) gene from the V4 region, using an updated version of the primer set 515 F (GTGYCAGCMGCCGCGGTAA)66 and 806 R (GGACTACNVGGGTWTCTAAT)67. PCRs, amplicon purification, library prep, and 16S sequencing with the Illumina Miniseq MO 2 × 150 (Illumina, San Diego, USA) occurred at the Genome Research Center at the University of Illinois-Chicago.
Raw reads of marine invertebrates (COI) were processed from FASTQ files with the DADA2 package version 1.26.068 and R software v.4.1.069. Primers were trimmed from reads with the filterAndTrim function, quality-filtered, merged, and chimeras were eliminated to create amplicon sequence variants (ASVs) (DADA2 parameters: maxN = 0, maxEE = c (2,2), runcQ = 10, trimLeft = 26, with pseudo-pooling)30. After quality control steps, 6,264,202 sequences remained (53%), averaging 223,722 (SD = 50,205) reads per sample. All samples from Site A, including the first 6-month time point, but excluding the Transplant tiles accounted for 1,928,128 reads (SD = 67,702) and 2,071150 reads (SD = 54,284) at Site B. The Transplant tiles that came from Site B totaled 602,607 reads (SD = 24,096), while the Transplant tiles from Site A totaled 672,491 (SD = 24,455). The ASVs were then further curated using LULU v0.170, with 28,406 (81%) remaining ASVs. In VSEARCH48, the remaining ASVs were clustered into OTUs (3704 OTUs) at 97% similarity, and an OTU table was generated. We employed a multifaceted approach to assign taxonomy for OTUs using QIIME (v. 1.9)71, implementing uclust against Midori (version GB239)72 and MetaCOXI73 nucleotide databases with Bayesian Least Common Ancestor analysis74. The Ribosomal Database Project75 classifier alongside a CO1 classifier76 was utilized for further classifications, before utilizing blastx against the Midori amino acid database, followed by BASTA77 to identify the Last Common Ancestor of 70% of the top hits. This multi-step approach ensured comprehensive taxonomic annotation of OTU sequences, harnessing both nucleotide and amino acid databases to optimize classification accuracy. Following the automated assignment for COI OTU taxonomic classifications, manual curation was conducted. In cases where taxonomic consensus was achieved across methodologies, we retained the most detailed taxonomic information. Any discrepancies were resolved by adopting the taxonomy of the lowest common ancestor.
Demultiplexed raw sequence data were imported into QIIME 2 v2022.2 for marine bacteria 16S sequence data processing78. We first performed rarefaction, calculated alpha diversity metrics, and then assigned taxonomy to all sequences using QIIME2. Reads were trimmed, low quality sequences were filtered, and the remaining data was denoised, merged, and chimeras were removed, using the DADA268 workflow implemented in QIIME2. Sequence counts were normalized to a depth of 5051 per sample to estimate alpha diversity metrics (species richness, Shannon index, and Faith’s phylogenetic diversity). A taxonomy classifier was then trained on reference 16S sequences (trimmed to V4 region) obtained from the SILVA 16S database version 13879. This classifier was subsequently used to infer taxonomy of representative sequences produced by DADA2. The final non-normalized ASV counts, and taxonomic assignments were exported from QIIME2 for statistical analysis in R.
Statistical analysis
The nonlinear relationship between ETRs and PAR, under different sites and treatments, was modeled with a two-parameter rectangular hyperbola (Michaelis-Menten) regression. Each coral sample from both species (n = 5, per species, per treatment) was fit to a curve defined by the equation:
| 1 |
Where X is PAR and Y is ETR, a is the maximum ETR level reached (asymptote value) and b is the half-saturation constant - the PAR value at which the curve reaches half of the asymptote ETR value, known as PAR50. The regression fitting was performed using the drm function and MM.2 self-starting function from the drc R package80. Each parameter (a and b) was then compared between sites and treatments with a two-way ANOVA. Post-hoc analysis was performed as pairwise comparisons defined by linear contrasts. Finally, p-values were adjusted for multiple comparisons with the Benjamini-Hochberg (FDR) procedure.
Effects of site and treatment on oxidative stress parameters (TAC and LPO) in corals were assessed using two-way ANOVA tests. We compared TAC and LPO at the end of the experiment as it is a highly sensitive parameter to environmental changes (e.g., water temperature, endosymbiont density) and were not able to compare to native corals from either site. Chlorophyll-a and endosymbiont algae (Symbiodiniaceae) of both coral species were also evaluated using two-way ANOVA tests. Post-hoc analysis was performed as pairwise comparisons defined by linear contrasts. P-values were then adjusted for multiple comparisons with the FDR procedure. Homogeneity of variances and data normality assumptions were checked by graphical inspection of residuals. In all cases, the significance level adopted was 95% (α = 0.05). Results were expressed as mean ± SE and statistical tests and graphical visualizations were conducted in R69.
The coral physiology parameters were integrated into a stress index using the IBR approach. The IBR was modeled to assess and compare the level of physiological stress in each location/treatment condition, for each species, and for both species combined. Calculations followed the method developed by Devin et al.81, which is based on the calculation initially described in Beliaeff and Burgeot49, using R scripts integrated into the CALculate IBR Interface82. Briefly, biomarker values were standardized and multiplied by a coefficient matrix that represents a priori stress responses for each biomarker. The coefficient is set to −1 when inhibition is the stress response for the specific biomarker (e.g., decreased photochemical efficiency), and +1 when the expected stress response is upregulation (e.g., increased oxidative damage). Final values were computed in a radar diagram. The area of the diagram corresponds to the stress index, which is calculated for every possible order of the biomarkers along the radar diagram using a permutation procedure. The final IBR value is an average of all permutations, with the highest values indicating the highest biological alteration. Overall relationships between the physiological responses, IBR, benthic community descriptors (COI and 16S), and treatments were evaluated with principal component analyses (PCA) and Pearson correlations83. Variables were centered and scaled prior to analysis. All analyses and graphs were conducted in R69.
Marine invertebrate community analyses were conducted using the vegan package in R software v4.1.069,78. OTU tables were created from QIIME v1.9 using the core_diversity_analyses.py function71. Pairwise dissimilarity distance matrices were calculated with the Bray-Curtis metric from abundances30. Bray-Curtis ranges between 0 and 1: 0 refers to samples with identical OTU composition and abundance and 1 indicates samples with no shared species. A principal coordinate analysis (PCoA) was used to visualize Bray-Curtis dissimilarities in community composition between sites and treatments and was tested using permutational multivariate analysis of variance (PERMANOVA)84. The PERMANOVA was conducted to estimate partitioning of potential differences across relevant factors. The PERMANOVA was run using the full dataset to test for variation between the samples across both sites and within sites (i.e., factor Location, factor Treatments, factor Location*Treatment, and factor Site A and factor Site B). The PERMANOVA analyses were calculated with the adonis function using the vegan R package v2.5-7 with 9999 permutations78,84. Observed species richness of OTUs for marine invertebrates was conducted between sites and treatments using pairwise Wilcoxon Signed Rank tests. Alpha diversity was estimated with the specnumber function in the vegan R package 2.5-730. Bar plots were created with the 23 most abundant taxa. Replicates that were not included were due to QIIME’s quality filtering. Significance levels across certain groups were conducted and illustrated with the ggpubr package85. Graphical illustrations were produced with R packages ggplot286 and treemapify87.
The ASV counts table from QIIME v2 and metadata were imported into R v4.2.1 to conduct 16S amplicon-based microbial community composition analyses. A centered log-ratio transformation was applied to ASV counts bacterial abundances were summarized as barplots using the ampvis 2 R package88. Association between microbial community composition with treatments and sites were assessed using a PERMANOVA. Bar plots were created from the 20 most abundant taxa. Replicates that were not included were due to QIIME’s quality filtering or because of too few reads. Statistical differences in alpha diversity across site and treatment were tested using one-way ANOVA and Tukey’s test for post-hoc pairwise comparisons.
Differential abundance analyses were performed for both invertebrate COI and bacteria 16S, using DESeq289 to identify taxa associated with the different conditions. The approximate posterior estimation for general linear model coefficients (apeglm)90 was applied to shrink the log2 fold changes, improving the interpretability of the results. Phyla or OTUs/ASVs with an adjusted p-value (p-adj) < 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank the staff and students at the Interuniversity Institute for Marine Sciences in Eilat for their hospitality and assistance. We appreciate everyone who helped in this study, including Ryan Sirota and all members of the Levy Lab for Molecular Marine Ecology, especially Bar Feldman, Shachaf Ben-Ezra, Inbal Ayalon, and Leviah Zand. Thank you to the members of the Design-Tech Lab at the Technion Institute of Technology, especially Ofer Berman. A big thank you to Matthieu Leray and David Kline for their guidance and expertise. Thank you to Orly Yaron, Sarit Karako-Lampert, and the Genome Research Center at the University of Illinois-Chicago for the sequencing work. Thank you to Isabel Flores for illustrating the beautiful corals in Fig. 5A. N.L. was supported by the Bar-Ilan University President’s Scholarship, the Israeli Ministry of Integration and Aliyah Scholarship, and the Red Sea Reef Foundation. E.T. was supported by the Polack Fund and Stem Fund. This manuscript represents partial fulfillment of the requirements for a Ph.D. thesis for N.L. at the Mina and Everard Goodman Faculty of Life Sciences at Bar-Ilan University, Israel.
Author contributions
Conception and study design by: N.L. and O.L. Data collection by: N.L., J.A.M., N.SB., E.T., and O.L. Data analysis and interpretation by: N.L., J.A.M., N.SB., D.G.B., T.D., J.I.C.B., J.Y.L., and O.L. The manuscript was written by N.L. with critical contributions by: J.A.M., D.G.B., T.D., J.Y.L., and O.L. All authors contributed to the drafts and final revisions of the manuscript.
Peer review
Peer review information
Nature Communications thanks Linda Wegley Kelly and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The sequencing data generated from this study has been deposited in the NCBI public database under accession code PRJNA1114437. The supporting figures and tables of sequencing and coral physiology data generated in this study are provided in the Supplementary Information file. Water quality information to inform this study can be found in the Supplementary Data 1 file. The coral physiology data generated in this study is publicly available at SEANOE (sea scientific open data publication) [10.17882/102809]. All data accession codes can be found in the Supplementary Data 2 file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Natalie Levy, Email: nrlevy@ucsd.edu.
Oren Levy, Email: oren.levy@biu.ac.il.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54149-6.
References
- 1.Nyström, M. et al. Confronting feedbacks of degraded marine ecosystems. Ecosystems15, 695–710 (2012). [Google Scholar]
- 2.Glasl, B., Webster, N. S. & Bourne, D. G. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol.164, 91 (2017). [Google Scholar]
- 3.Nelson, C. E., Wegley Kelly, L. & Haas, A. F. Microbial interactions with dissolved organic matter are central to coral reef ecosystem function and resilience. Ann. Rev. Mar. Sci.15, 431–460 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Kennedy, E. V. et al. Avoiding coral reef functional collapse requires local and global action. Curr. Biol.23, 912–918 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Remple, K. L. et al. Coral reef biofilm bacterial diversity and successional trajectories are structured by reef benthic organisms and shift under chronic nutrient enrichment. NPJ Biofilms Microbiomes7, 84 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourne, D. G., Morrow, K. M. & Webster, N. S. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol.70, 317–340 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Marangon, E., Laffy, P. W., Bourne, D. G. & Webster, N. S. Microbiome-mediated mechanisms contributing to the environmental tolerance of reef invertebrate species. Mar. Biol.168, 89 (2021). [Google Scholar]
- 8.Zaneveld, J. R. et al. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat. Commun.7, 11833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guibert, I., Lecellier, G., Torda, G., Pochon, X. & Berteaux-Lecellier, V. Metabarcoding reveals distinct microbiotypes in the giant clam Tridacna maxima. Microbiome8, 57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allsup, C. M., George, I. & Lankau, R. A. Shifting microbial communities can enhance tree tolerance to changing climates. Science380, 835–840 (2023). [DOI] [PubMed] [Google Scholar]
- 11.Reshef, L., Koren, O., Loya, Y., Zilber-Rosenberg, I. & Rosenberg, E. The coral probiotic hypothesis. Environ. Microbiol.8, 2068–2073 (2006). [DOI] [PubMed] [Google Scholar]
- 12.West, A. G. et al. The microbiome in threatened species conservation. Biol. Conserv.229, 85–98 (2019). [Google Scholar]
- 13.Glasl, B., Smith, C. E., Bourne, D. G. & Webster, N. S. Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. Peer J.7, e6377 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Oppen, M. J. H. & Blackall, L. L. Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol.17, 557–567 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Rosado, P. M. et al. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J.13, 921–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peixoto, R. S., et al Beneficial Microorganisms for Corals (BMC): proposed Mechanisms for Coral Health and Resilience. Front. Microbiol.8, 341 (2017). [DOI] [PMC free article] [PubMed]
- 17.Blackall, L. L., Dungan, A. M., Hartman, L. M. & van Oppen, M. J. Probiotics for corals. Microbiol. Aust.41, 100 (2020). [Google Scholar]
- 18.Damjanovic, K., van Oppen, M. J. H., Menéndez, P. & Blackall, L. L. Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in acropora tenuis and platygyra daedalea. Front. Microbiol.10, 1702 (2019). [DOI] [PMC free article] [PubMed]
- 19.Peixoto, R. S. & Voolstra, C. R. The baseline is already shifted: marine microbiome restoration and rehabilitation as essential tools to mitigate ecosystem decline. Front. Mar. Sci.10, 1218531 (2023).
- 20.Vanwonterghem, I. & Webster, N. S. Coral reef microorganisms in a changing climate. iScience23, 100972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santoro, E. P. et al. Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Sci. Adv.7, eabg3088 (2021). [DOI] [PMC free article] [PubMed]
- 22.Damjanovic, K., Blackall, L. L., Webster, N. S. & van Oppen, M. J. H. The contribution of microbial biotechnology to mitigating coral reef degradation. Micro. Biotechnol.10, 1236–1243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doering, T. et al. Towards enhancing coral heat tolerance: a “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome9, 102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, H. R., Kuempel, C. D. & Altieri, A. H. The resilience of reef invertebrate biodiversity to coral mortality. Ecosphere7, e01399 (2016).
- 25.Alvarez-Filip, L., Dulvy, N. K., Gill, J. A., Côté, I. M. & Watkinson, A. R. Flattening of Caribbean coral reefs: region-wide declines in architectural complexity. Proc. R. Soc. B Biol. Sci.276, 3019–3025 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Filip, L., Dulvy, N. K., Côté, I. M., Watkinson, A. R. & Gill, J. A. Coral identity underpins architectural complexity on Caribbean reefs. Ecol. Appl.21, 2223–2231 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Torres-Pulliza, D. et al. A geometric basis for surface habitat complexity and biodiversity. Nat. Ecol. Evol.4, 1495–1501 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Berman, O., Levy, N., Parnas, H., Levy, O. & Tarazi, E. Exploring new frontiers in coral nurseries: leveraging 3D printing technology to benefit coral growth and survival. J. Mar. Sci. Eng.11, 1695 (2023). [Google Scholar]
- 29.Rosenberg, Y. et al. Urbanization comprehensively impairs biological rhythms in coral holobionts. Glob. Chang. Biol.28, 3349–3364 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy, N. et al. Evaluating biodiversity for coral reef reformation and monitoring on complex 3D structures using environmental DNA (eDNA) metabarcoding. Sci. Total Environ.856, 159051 (2023). [DOI] [PubMed]
- 31.Shaked, Y. & Genin, A. Annual Scientific Report 2021. https://iui-eilat.huji.ac.il/uploaded/NMP/reports/NMP%20Report%202021.pdf (2022).
- 32.Ayalon, I., de Barros Marangoni, L. F., Benichou, J. I. C., Avisar, D. & Levy, O. Red Sea corals under Artificial Light Pollution at Night (ALAN) undergo oxidative stress and photosynthetic impairment. Glob. Chang. Biol.25, 4194–4207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lushchak, V. I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol.101, 13–30 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Marangoni, L. FdeB. et al. Oxidative stress biomarkers as potential tools in reef degradation monitoring: a study case in a South Atlantic reef under influence of the 2015–2016 El Niño/Southern Oscillation (ENSO). Ecol. Indic.106, 105533 (2019). [Google Scholar]
- 35.Marangoni, L. FdeB. et al. Copper effects on biomarkers associated with photosynthesis, oxidative status and calcification in the Brazilian coral Mussismilia harttii (Scleractinia, Mussidae). Mar. Environ. Res.130, 248–257 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Zapata, F. L. et al. Holobiont diversity in a reef-building coral over its entire depth range in the mesophotic zone. Front. Mar. Sci.5, 29 (2018).
- 37.Osman, E. O. et al. Coral microbiome composition along the northern Red Sea suggests high plasticity of bacterial and specificity of endosymbiotic dinoflagellate communities. Microbiome8, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, J. A. et al. Phylogeny resolved, metabolism revealed: functional radiation within a widespread and divergent clade of sponge symbionts. ISME J.15, 503–519 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg, Y., Doniger, T. & Levy, O. Sustainability of coral reefs are affected by ecological light pollution in the Gulf of Aqaba/Eilat. Commun. Biol.2, 289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marques, J. A., Abrantes, D. P., Marangoni, L. F. B. & Bianchini, A. Ecotoxicological responses of a reef calcifier exposed to copper, acidification and warming: a multiple biomarker approach. Environ. Pollut.257, 113572 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Dias, M. et al. Integrative indices for health assessment in reef corals under thermal stress. Ecol. Indic. 113, 106230 (2020).
- 42.Sweet, M. J., Croquer, A. & Bythell, J. C. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs30, 39–52 (2011). [Google Scholar]
- 43.Tout, J. et al. Variability in microbial community composition and function between different niches within a coral reef. Micro. Ecol.67, 540–552 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Glasl, B., Bourne, D. G., Frade, P. R. & Webster, N. S. Establishing microbial baselines to identify indicators of coral reef health. Microbiol. Aust.39, 42 (2018). [Google Scholar]
- 45.Glasl, B. et al. Comparative genome-centric analysis reveals seasonal variation in the function of coral reef microbiomes. ISME J.14, 1435–1450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martiny, J. B. H., Jones, S. E., Lennon, J. T. & Martiny, A. C. Microbiomes in light of traits: a phylogenetic perspective. Science350, aac9323 (2015). [DOI] [PubMed]
- 47.Graham, N. A. J. & Nash, K. L. The importance of structural complexity in coral reef ecosystems. Coral Reefs32, 315–326 (2013). [Google Scholar]
- 48.Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beliaeff, B. & Burgeot, T. Integrated biomarker response: A useful tool for ecological risk assessment. Environ. Toxicol. Chem. 21, 1316–1322 (2002). [PubMed]
- 50.Hooper, D. U. et al. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature486, 105–108 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Levy, N. et al. Emerging 3D technologies for future reformation of coral reefs: enhancing biodiversity using biomimetic structures based on designs by nature. Sci. Total Environ.830, 154749 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Berman, O. et al. Design and application of a novel 3D printing method for bio-inspired artificial reefs. Ecol. Eng.188, 106892 (2023). [Google Scholar]
- 53.Wangpraseurt, D., You, S., Sun, Y. & Chen, S. Biomimetic 3D living materials powered by microorganisms. Trends Biotechnol.40, 843–857 (2022). [DOI] [PubMed] [Google Scholar]
- 54.Wangpraseurt, D. et al. Bioprinted living coral microenvironments mimicking coral‐algal symbiosis. Adv. Funct. Mater.32, 2202273 (2022).
- 55.Datta, D. et al. Phenotypically complex living materials containing engineered cyanobacteria. Nat. Commun.14, 4742 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roger, L. M. et al. Nanobiotech engineering for future coral reefs. One Earth6, 778–789 (2023). [Google Scholar]
- 57.Roger, L. et al. Nanotechnology for coral reef conservation, restoration and rehabilitation. Nat. Nanotechnol.18, 831–833 (2023). [DOI] [PubMed] [Google Scholar]
- 58.Shlesinger, Y. & Loya, Y. Coral community reproductive patterns: red sea versus the great barrier reef. Science228, 1333–1335 (1985). [DOI] [PubMed] [Google Scholar]
- 59.Leray, M. & Knowlton, N. DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proc. Natl. Acad. Sci. USA112, 2076–2081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang, W., Zhang, S.-B. & Cao, K.-F. Evidence for leaf fold to remedy the deficiency of physiological photoprotection for photosystem II. Photosynth. Res.110, 185–191 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Veal, C. J., Carmi, M., Fine, M. & Hoegh-Guldberg, O. Increasing the accuracy of surface area estimation using single wax dipping of coral fragments. Coral Reefs29, 893–897 (2010). [Google Scholar]
- 62.Lavy, A. et al. A quick, easy and non‐intrusive method for underwater volume and surface area evaluation of benthic organisms by 3D computer modelling. Methods Ecol. Evol.6, 521–531 (2015). [Google Scholar]
- 63.Jeffrey, S. W. & Humphrey, G. F. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochemie und Physiologie der Pflanz.167, 191–194 (1975). [Google Scholar]
- 64.Federici, G., Shaw, B. & Handy, R. Toxicity of titanium dioxide nanoparticles to rainbow trout (Oncorhynchus mykiss): gill injury, oxidative stress, and other physiological effects. Aquat. Toxicol.84, 415–430 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Leray, M. et al. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool.10, 34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parada, A. E., Needham, D. M. & Fuhrman, J. A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol18, 1403–1414 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Apprill, A., Mcnally, S., Parsons, R. & Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol.75, 129–137 (2015). [Google Scholar]
- 68.Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing Vienna, Austria, (2019).
- 70.Frøslev, T. G. et al. Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun.8, 1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Machida, R. J., Leray, M., Ho, S.-L. & Knowlton, N. Metazoan mitochondrial gene sequence reference datasets for taxonomic assignment of environmental samples. Sci. Data4, 170027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balech, B., Sandionigi, A., Marzano, M., Pesole, G. & Santamaria, M. MetaCOXI: an integrated collection of metazoan mitochondrial cytochrome oxidase subunit-I DNA sequences. Database2022, baab084 (2022). [DOI] [PMC free article] [PubMed]
- 74.Gao, X., Lin, H., Revanna, K. & Dong, Q. A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinforma.18, 247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cole, J. R. et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res.42, D633–D642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Porter, T. M. & Hajibabaei, M. Automated high throughput animal CO1 metabarcode classification. Sci. Rep.8, 4226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kahlke, T. & Ralph, P. J. BASTA – taxonomic classification of sequences and sequence bins using last common ancestor estimations. Methods Ecol. Evol.10, 100–103 (2019). [Google Scholar]
- 78.Oksanen, J. & Simpson, G. L. The Vegan Package Designing Efficient Sampling Schemes for Monitoring Boreal Forest Bird Communities View Project Vegan: Community Ecology Package View Project. http://cran.r-project.org/ (2009).
- 79.Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ritz, C., Baty, F., Streibig, J. C. & Gerhard, D. Dose-response analysis using R. PLoS ONE10, e0146021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Devin, S., Burgeot, T., Giambérini, L., Minguez, L. & Pain-Devin, S. The integrated biomarker response revisited: optimization to avoid misuse. Environ. Sci. Pollut. Res.21, 2448–2454 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Devin, S., Arnould, P. Y., Minguez, L., Pain-Devin, S. & Jouffret, V. Correction to: CalIBRi: a web interface to calculate Integrated Biomarker Index (Environmental Science and Pollution Research, (2014), 21, 4, (2448−2454), 10.1007/s11356-013-2169-9). Environ. Sci. Pollut. Res.30, 67912–67913 (2023). [DOI] [PubMed] [Google Scholar]
- 83.Borcard, D., Gillet, F. & Legendre, P. Numerical Ecology with R. (Springer New York, New York, NY, 2011). 10.1007/978-1-4419-7976-6.
- 84.Anderson, M. J. Permutational Multivariate Analysis of Variance (=PERMANOVA=). in Wiley StatsRef: Statistics Reference Online 1–15 (Wiley, 2017). 10.1002/9781118445112.stat07841.
- 85.Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.4.0 (2020).
- 86.Wickham, H. Data Analysis. In ggplot2 (Springer, Cham, 2016). 10.1007/978-3-319-24277-4_9.
- 87.Wilkins, D. treemapify: Draw Treemaps in ‘ggplot2’. R package version 2.5.5 (2021).
- 88.Andersen, K. S., K. R. H., K. S. M., & A. M. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. 362 10.1101/299537.
- 89.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequencing data generated from this study has been deposited in the NCBI public database under accession code PRJNA1114437. The supporting figures and tables of sequencing and coral physiology data generated in this study are provided in the Supplementary Information file. Water quality information to inform this study can be found in the Supplementary Data 1 file. The coral physiology data generated in this study is publicly available at SEANOE (sea scientific open data publication) [10.17882/102809]. All data accession codes can be found in the Supplementary Data 2 file.