Abstract
Cat allergy affects ∼15% of the US population and can cause severe symptoms, including asthma. The major cat allergen, Fel d 1, drives IgE antibody responses. We conducted a comparative analysis of Fel d 1 genes, CH1 and CH2, and investigated structural features of Fel d 1 homologs across the family Felidae. The CH1 and CH2 domestic cat DNA references were used to identify homologous sequences in domestic and exotic cat genomes. Variability of these sequences within or across cat species was analyzed. Comprehensive alignments of Fel d 1 sequences and homologs from 276 domestic or exotic cats identified >100 unique, dissimilar substitutions in the protein sequences across Felidae. Selective pressure analyses of 37 exotic cat species revealed that Fel d 1 experienced positive selection, or greater variability over time, in CH1 and CH2. Linear regression of the mean pairwise identities of Fel d 1 DNA or protein sequences indicated that the genes largely reflected the evolution of Felidae. The Fel d 1 genes are highly variable (41 and 58% of the amino acid residues encoded by CH1 and CH2, respectively), suggesting that the biological function of Fel d 1, which is currently unknown, may vary among cat species and/or that Fel d 1 may be nonessential for cats. This is supported by Fel d 1 homology to nonessential proteins and recent evidence of healthy cats with CRISPR-edited CH2 genes. Fel d 1 variability could confer an evolutionary advantage for cats by allowing the allergen to bind different physiological ligands.
Keywords: cat allergen, Fel d 1, evolution, comparative genomics, gene editing
Significance Statement.
Domestic cat allergen frequently causes allergic disease, yet little is known about the diversity of cat allergen or how the allergen evolved. This study compared cat allergen genes among exotic and domestic cat species across millions of years of evolution. Genes encoding chain 1 and chain 2 of Fel d 1, the sentinel cat allergen, were variable along each evolutionary lineage. The diversity was mapped to specific residues in the protein structure at high resolution. The results suggest that Fel d 1 is a viable target for gene deletion to develop Fel d 1-free cats. Deleting or rendering the Fel d 1 genes nonfunctional could have therapeutic implications for cat allergy sufferers by removing the allergen from the source.
Introduction
Though domestic cat (Felis domesticus or Felis catus) is a common household pet, up to 15% of adults and children in the United States are allergic to cats (1). Symptoms of cat allergy present with varying degrees of severity ranging from rhinoconjunctivitis to severe asthma (2–4). Approximately 500,000 asthma attacks per year and 350,000 emergency care visits have been attributed to cat allergic disease (5). While both the rate of sensitization to cat allergen and the prevalence of cats as household pets remain high, current treatment options for cat allergy, such as cat allergen immunotherapy, have demonstrated varying degrees of success (6–8).
Several proteins produced by cats have been shown to elicit an allergic response (9). The major cat allergen, Fel d 1, causes IgE antibody-mediated sensitization in nearly 95% of cat allergic patients and accounts for 60–90% of the total anticat IgE (10–14). Fel d 1 is a heterodimer consisting of two chains, namely chain 1 (70 amino acids [AAs], 8kD protein) and chain 2 (92 AA, 10 kD) (7, 15). CH1 and CH2, the genes that encode chains 1 and 2, respectively, are located within a < 10,000-bp region in the cat genome (chromosome E2, assembly Felis_catus_8.0). The crystal structure of recombinant Fel d 1 indicates the presence of ligand-binding cavities, which may contribute to the allergenic function of the protein (16, 17). Fel d 1 is a secretoglobin produced in the sebaceous, salivary, perianal, and lachrymal glands of cats, with expression levels of the protein varying considerably across cats and even within the same cat over time (16, 18–21).
The biological function of Fel d 1 is currently unknown; however, studies of homologous proteins in other mammals suggest that the allergen may contribute to immune regulation, protection of the epithelium, or chemical communication among cats (7, 22–25). Several homologous protein sequences with ∼25–50% similarity to Fel d 1 have been identified in other species, including rabbit uteroglobin and lipophilin proteins, human Clara cell secretory protein, mouse androgen-binding protein (ABP), and a brachial gland secretory protein of the slow loris primate (7, 23, 26, 27). To our knowledge, the Fel d 1 genes are only expressed in species from the Felidae family. The first evidence of the existence of Fel d 1-like proteins in “big cats” and allergenic cross-reactivity between Fel d 1 and homologous proteins from other Felidae species was reported in 1990 by De Groot et al. (28). Another study reported that Fel d 1-specific monoclonal antibodies directed against one out of four identified epitopes, recognized a cross-reactive determinant in seven Felidae species (29). The preliminary analysis of the allergen sequences and orthologs (i.e. homologs derived from speciation) from domestic and exotic (nondomestic) cat species, respectively, indicated that the genes have evolved across Felidae (30).
Comparative genomic analyses may provide insight into the evolution and potential function of Fel d 1. Two working theories used to describe intra- and inter-species genome evolution include genetic drift (neutral evolution) and natural selection (31). The neutral model of evolution specifies that nondeleterious genetic mutations are the result of random chance rather than selective pressures from the environment. Furthermore, neutral evolution claims that deleterious mutations ultimately impact the fitness of an organism such that the mutations do not become fixed within the genome (32, 33). Two aspects of natural selection acting on populations of organisms are negative (purifying) selection and positive (adaptive or diversifying) selection. Natural selection describes the elimination of harmful mutations or the accumulation of beneficial ones to increase fitness for the preservation of a population that is well adapted to the environment (34).
By analyzing the Fel d 1 sequences and orthologs across domestic and exotic cats, this study highlights the evolutionary changes that have shaped the Fel d 1 gene sequences over millions of years. The goals are to elucidate details regarding the biological function or essential nature of the Fel d 1 protein in cats to evaluate whether targeting the allergen with gene editing technology is a rational approach for treating cat allergy.
Methods
Genomic data acquisition
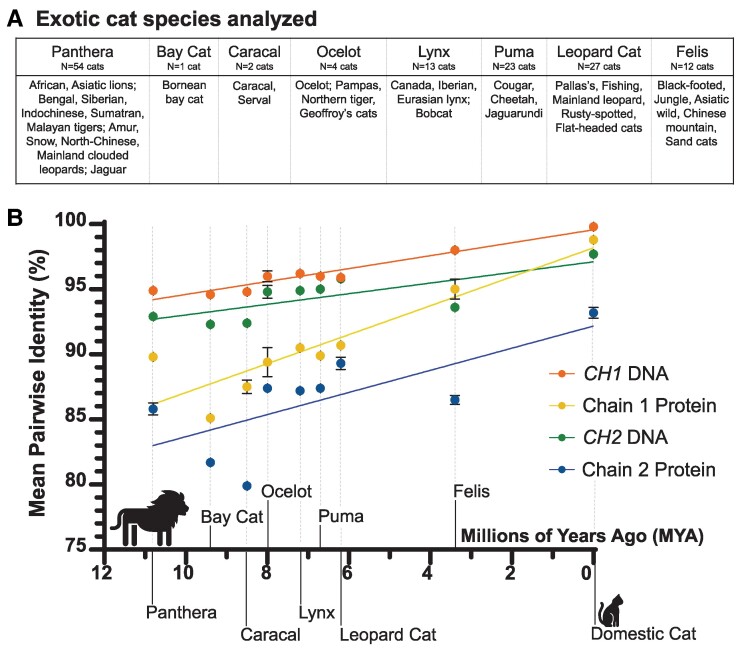
The full-length genomic DNA reference sequences for Fel d 1 chains 1 and 2 (National Center for Biotechnology Information [NCBI] accession X62477 and X62478, respectively) were used to perform a basic local alignment search (basic local alignment search tool [BLAST]) of the domestic and exotic (nondomestic) cat genomes in NCBI's sequence read archive (SRA) database. The whole-genome sequence fragments for each of the domestic or exotic cats were assembled to the Fel d 1 references to generate DNA consensus sequences (Geneious Prime, Boston, MA, USA). The coding DNA sequences for Fel d 1 CH1 (GenBank reference M74952.1) and CH2 (GenBank reference M77341.1) were extracted from the full-length DNA consensus sequences for each cat. Coding DNA sequences with gaps (no coverage) or regions of two or more consecutive, unknown nucleic acid residues (NN) were excluded from the analysis. The remaining coding DNA sequences were then translated and represented the open reading frames encoding for Fel d 1 protein and homologs, including the signal peptides (22 out of 92 AAs for chain 1 and 17 out of 109 AAs for chain 2). The signal peptide was included in the analysis to fully assess Fel d 1 DNA and protein sequence variability, inclusive of signal peptide mutations that could potentially affect the targeting or processing of the mature Fel d 1 protein. The final dataset included 90 domestic and 136 exotic cats (N = 1–21 cats per exotic species) from NCBI's SRA database (Tables S1 and S2, respectively). In total, 37 species of exotic (big or wild) cats that span the eight lineages (35) of the family Felidae were analyzed (Fig. 1A).
Fig. 1.
Regression analysis of Fel d 1 across the Felidae lineages. The Fel d 1 sequences and orthologs from domestic and exotic cats were aligned with the GenBank DNA or protein references for Fel d 1. A) The exotic cat species analyzed, representing the eight lineages of Felidae. B) Simple linear regression of the mean pairwise identities (mean ± SEM; %) of the Felidae lineages (N = 1–54 cats per lineage) and modern domestic cats (N = 140 cats) over the approximate evolutionary timeline of the cat family (36). R2 values: 0.93, 0.82, 0.57, and 0.51 for CH1 DNA (orange), chain 1 protein (yellow), CH2 DNA (green), and chain 2 protein (blue), respectively.
Domestic cat DNA extraction
Discarded tissue samples (feline testes, ovaries, or uteri) from the routine sterilization procedures of 50 short- or long-haired domestic cats of mixed ancestry were provided for the study (Charlottesville Albemarle Society for the Prevention of Cruelty to Animals). Genomic DNA was extracted from the tissue samples (DNeasy Blood and Tissue Kit, Qiagen), and Fel d 1 CH1 and CH2 were polymerase chain reaction (PCR) amplified, purified, Sanger sequenced, and translated as previously described (30). The DNA and protein sequences of these 50 cats (labeled “Domestic Cat 91”—“Domestic Cat 140”) were analyzed with the domestic cat Fel d 1 sequences obtained from NCBI's SRA database, for a total of 140 domestic cats.
Sequence alignment and identity analysis
Pairwise alignments of the domestic or exotic cat coding DNA or protein sequences with the GenBank references for Fel d 1 (coding DNA: M74952.1 and M77341.1; protein: AAC37318.1 and AAC41616.1) were performed, and percent identity was determined (CLUSTAL Omega, Geneious Prime) (Tables S3 and S4). The mean pairwise identity was calculated for each exotic cat species (N = 1–21 cats per species; Table S5) as well as for each of the eight lineages of the family Felidae (exotic cats only; N = 1–54 cats per lineage; Table S6). The average pairwise identity of the 140 domestic cats was determined. A linear regression of the mean pairwise identities of the domestic and exotic cats (exotic cats grouped by Felidae lineage) versus the estimated time (millions of years ago [MYA]) since diverging from domestic cats was performed (36).
Multisequence alignments of the domestic (Figs. S1–S4) or exotic (Figs. S5–S8) cat DNA or protein sequences were performed, and percent identity was determined (CLUSTAL Omega, Geneious Prime). Unrooted, maximum likelihood phylogenies of Fel d 1 chains 1 and 2 were generated from the multisequence alignments of all 140 domestic and 136 exotic cat sequences and orthologs to assess the conservancy and/or diversity of the sequences (37–40).
Fel d 1 sequence variation
The AA substitutions relative to the GenBank Fel d 1 references were identified in all domestic and exotic cats to evaluate Fel d 1 protein sequence variability (Tables S7 and S8, respectively). Changes in AA residue charge or polarity, as well as the addition or deletion of AA residues, were classified as dissimilar changes, while all other AA variations were identified as similar changes. The total number of AA changes as well as the number of unique changes at each AA residue was determined (Table S9). The variable AA residues identified in domestic and exotic cats were highlighted on the tetrameric crystal structure of recombinant Fel d 1 (17).
Potential structural effects of the variable AA residues on the Fel d 1 heterodimer were predicted using AlphaFold3 (41–43). AA substitutions encoded by > 50% of domestic or exotic cats were modeled on the heterodimeric reference structure for Fel d 1, protein database (PDB) 2EJN (17). The reference 2EJN Fel d 1 structure and the two AlphaFold models for the domestic or exotic cat substitutions were superimposed to predict and identify Fel d 1 structural changes due to the sequence variability of Fel d 1.
Selective pressure analysis
Selective pressure was estimated for each exotic cat species by comparing the DNA orthologs with the GenBank DNA references for Fel d 1 CH1 or CH2. One representative cat with pairwise identities similar to the species average identities was analyzed per exotic species (Table S10). The numbers of synonymous sites (S), synonymous substitutions (DS), nonsynonymous sites (N), and nonsynonymous substitutions (DN) in the exotic cat sequences were determined, and the resulting synonymous or nonsynonymous substitution rates (dS = DS/S and dN = DN/N, respectively) were calculated (44). The ratio of nonsynonymous and synonymous substitution rates (ω = dN/dS) was evaluated for each of the exotic species (Table S11). If there were no synonymous substitutions present in the exotic cat orthologs (DS = 0), then ω = dN/dS was estimated given: ω = dN/((DS + 1)/S).
Results
Comparative sequence analysis of Fel d 1
The Fel d 1 sequences of 90 domestic cats were identified by BLAST search of NCBI's SRA database, while the gene sequences of an additional 50 cats were determined by DNA extraction followed by Sanger sequencing (Table S1) (30). Multisequence alignments of all 140 domestic cat DNA sequences yielded the mean identities of 99 and 97% for CH1 and CH2, respectively (Figs. S1 and S2). Pairwise alignments of the domestic cat DNA sequences with the CH1 and CH2 reference sequences resulted in identities ranging from 93 to 100%, while pairwise alignments of the corresponding protein sequences with the GenBank references for Fel d 1 chains 1 and 2 produced identities of 84–100% (Table S3). Multisequence alignments of the domestic cat protein sequences resulted in the mean identities of 98 and 90% for chain 1 and chain 2, respectively (Figs. S3 and S4).
Fel d 1 orthologous sequences from 136 exotic (nondomestic) cat genomes representing 37 species of big or wild cats were identified by BLAST search (Table S2). The exotic cats span the eight lineages of the family Felidae, namely Panthera, Bay Cat, Caracal, Ocelot, Lynx, Puma, Leopard cat, and Domestic cat (labeled “Felis”) lineages, and include species that diverged from domestic cats up to ∼11 MYA (35, 36). Pairwise alignments of the exotic cat DNA orthologs with the GenBank Fel d 1 reference sequences for domestic cat resulted in identities ranging from 91 to 99%, whereas pairwise alignments of the corresponding protein orthologs with the domestic cat references produced identities of 80–98% (Table S4). Multisequence alignments of the exotic cat orthologs yielded the mean DNA identities of 96% for CH1 and 94% for CH2 and produced the mean protein sequence identities of 92 and 88% for chains 1 and 2, respectively (Figs. S5–S8). The multisequence alignments of all 276 domestic and exotic cats analyzed produced DNA identities of 97% for CH1 and 94% for CH2, with the corresponding protein identities of 93 and 86% for Fel d 1 chains 1 and 2, respectively. The pairwise and multisequence alignment data for the domestic and/or exotic cats are summarized in Table 1.
Table 1.
Average pairwise and multisequence identities of domestic and/or exotic cats.
| DNA | Protein | |||||
|---|---|---|---|---|---|---|
| CH1 | CH2 | Chain 1 | Chain 2 | |||
| Pairwise (aligned with domestic cat reference) | Domestic | Mean ± SEM | 99.8% ± 0.1 | 97.7% ± 0.2 | 98.8% ± 0.2 | 93.2% ± 0.4 |
| Range | 96.2–100% | 93.0–100% | 90.3–100% | 83.7–100% | ||
| Exotic | Mean ± SEM | 95.7% ± 0.1 | 94.2% ± 0.1 | 90.4% ± 0.2 | 86.9% ± 0.3 | |
| Range | 94.6–99.3% | 91.2–96.7% | 85.1–97.8% | 79.8–90.8% | ||
| Domestic and exotic | P (ANOVA) | 1.2E − 162 | 8.3E − 51 | 2.9E − 107 | 5.7E − 27 | |
| Multisequence | Domestic | Mean | 99.5% | 96.6% | 97.9% | 89.6% |
| Exotic | Mean | 95.9% | 94.2% | 92.3% | 87.9% | |
| Domestic and exotic | Mean | 96.8% | 94.3% | 92.7% | 86.4% | |
The mean pairwise identities of each exotic cat lineage as well as the average identity of 140 domestic cats were plotted against the estimated evolutionary timeline (MYA) of Felidae (Fig. 1A and B). The simple linear regression resulted in R2 values of 0.93, 0.82, 0.57, and 0.51 for CH1 DNA, chain 1 protein, CH2 DNA, and chain 2 protein, respectively (Fig. 1B). Single-factor ANOVA tests determined that for each dataset (CH1 DNA, chain 1 protein, CH2 DNA, and chain 2 protein), the mean pairwise identities were significantly different across lineages, with P < 0.01 (Table 1).
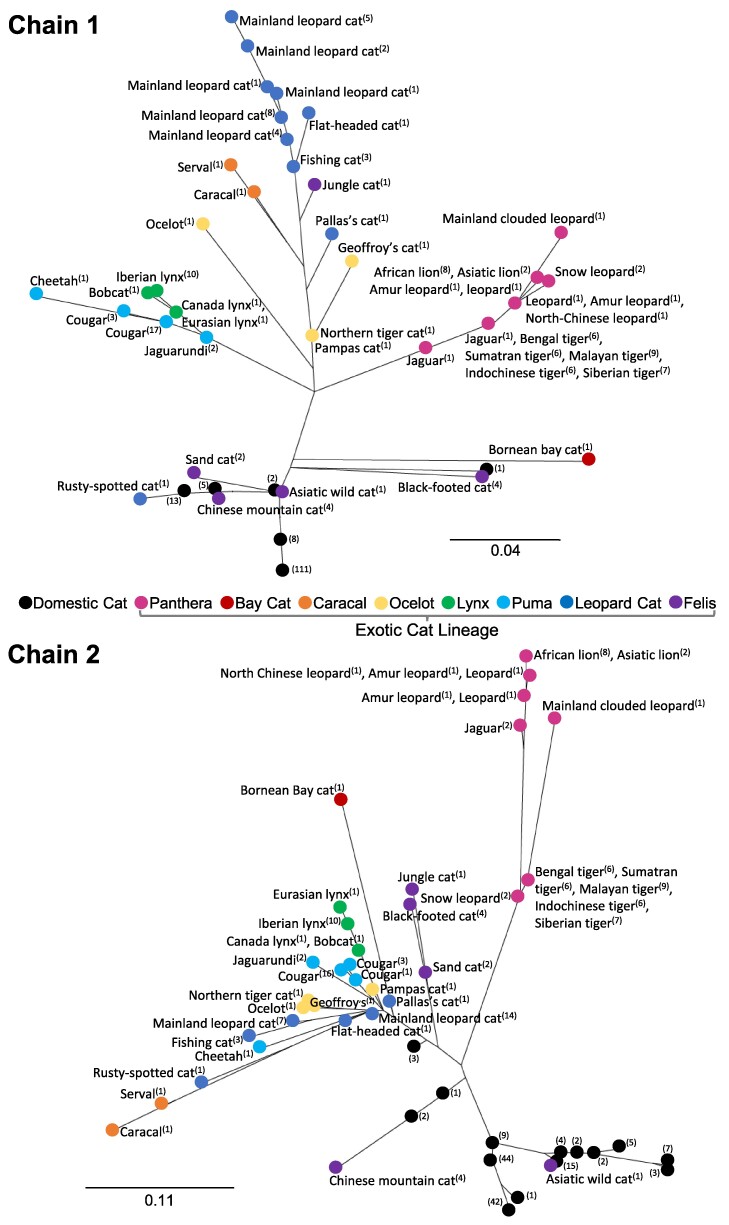
Unrooted, maximum likelihood phylogenies of the Fel d 1 chain 1 and chain 2 protein sequences were generated from the multisequence alignments of all domestic and exotic cats analyzed (Fig. 2). The data indicate that, on average, for both chains 1 and 2, the distributions of cats are predominantly clustered by lineage. The chain 1 phylogeny largely reflects the evolutionary timeline of Felidae. By contrast, chain 2 shows considerably more variability, particularly among the domestic cats (shown in black, Fig. 2).
Fig. 2.
Diversity of the Fel d 1 genes across Felidae. Unrooted, maximum likelihood phylogenies of Fel d 1 chain 1 (Top) and chain 2 (Bottom) assembled from the alignments of the domestic and exotic cat protein sequences and orthologs. Each datapoint indicates the species and number of cats represented for that species. Domestic cats (black), exotic cat lineages: Panthera (magenta), Bay Cat (red), Caracal (orange), Ocelot (yellow), Lynx (green), Puma (cyan), Leopard Cat (blue), and Felis (purple). Scale bars represent 0.04 and 0.11 AA substitutions per residue for chain 1 and chain 2, respectively.
Fel d 1 sequence and structure variation across Felidae
AA substitutions in the domestic and exotic cat Fel d 1 sequences and orthologs were identified to assess the variability and evolution of the Fel d 1 genes (Tables S7 and S8). The total and unique AA residue changes relative to the domestic cat references were determined for all cats analyzed and were further characterized by the fractions of similar or dissimilar AA changes (Table S9).
Across all 276 cat sequences, fewer total or unique AA residue changes were identified in Fel d 1 chain 1 versus chain 2 and in domestic cats versus exotic cats (Table 2). In domestic cats alone, 76 and 60% of all AA changes in chains 1 and 2, respectively, were dissimilar owing to the addition or deletion of AA residues or changes in residue charge or polarity. Likewise in exotic cats, 77 and 49% of the total AA changes were dissimilar, respectively (Table 2). For unique changes, defined as the sum of distinct AA residue substitutions excluding repeats between cats, 74 and 58% of the unique AA changes in domestic cats were dissimilar in chains 1 and 2, respectively, compared with 54% of chain 1 and 57% of chain 2 in exotic cats (Table 2).
Table 2.
Summary of Fel d 1 amino acid (AA) residue changes in domestic and exotic cats.
| Chain 1 | Chain 2 | ||||
|---|---|---|---|---|---|
| Exotic | Domestic | Exotic | Domestic | ||
| Total changes | Sum | 1198 | 155 | 1951 | 1044 |
| Dissimilar | 924 (77%) | 117 (76%) | 964 (49%) | 627 (60%) | |
| Unique changes | Sum | 59 | 19 | 110 | 38 |
| Dissimilar | 32 (54%) | 14 (74%) | 63 (57%) | 22 (58%) | |
| Variable AA residues | 36 (39%) | 16 (17%) | 62 (57%) | 26 (24%) | |
| Variable AA residues (exotic or domestic cats) |
38 (41%) | 63 (58%) | |||
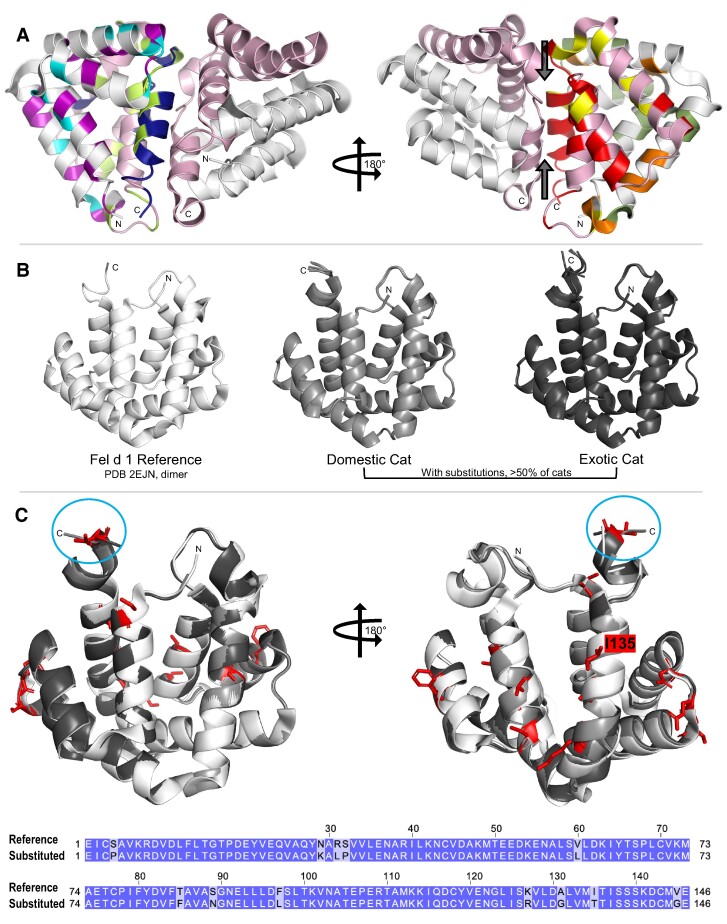
Of the 92 AA residues that comprise Fel d 1 chain 1, 17 and 39% of those chain 1 residues were determined to be variable, or to exhibit AA substitutions, in domestic and exotic cats, respectively (7). For the 109 AA residues of chain 2, 24% of the domestic cat and 57% of the exotic cat chain 2 residues were variable (Table 2) (7). Fifty-nine percent and 42% of the chain 1 and chain 2 AA residues, respectively, were conserved across all 276 domestic and exotic cats analyzed. In domestic cats alone, numerous spans of > 10 conserved AA residues were identified in chains 1 and 2. The variable and conserved AA residues in chain 1 and chain 2 were mapped onto the crystal structure of recombinant Fel d 1 (17) (Fig. 3A). For all cat species, the chain 2 dissimilar substitutions were primarily concentrated at the Fel d 1 dimer interface (Fig. 3A).
Fig. 3.
The sequence and structural variability of Fel d 1. The amino acid (AA) substitutions were mapped to the structure of recombinant Fel d 1 (PDB 2EJN (17)). A) Each molecule is chain 1 (white) linked to chain 2 (light pink) and represents the heterodimer in the natural allergen. Left: the AA substitutions identified in domestic cats alone (none), exotic cats alone (chain 1: purple; chain 2: light green), or both domestic and exotic cats (chain 1: cyan; chain 2: dark blue). Right: the similar (chain 1: orange; chain 2: yellow) and dissimilar (chain 1: green; chain 2: red) AA substitutions identified in domestic and/or exotic cats. The chain 2 dissimilar substitutions were concentrated at the Fel d 1 interface (arrows). B) Crystal structure and AlphaFold3 models of the Fel d 1 heterodimer: reference PDB: 2EJN (white, Left) and the AA substitutions identified in > 50% of domestic (light gray, Center) or exotic cats (dark gray, Right). C) Superposition of the AlphaFold3 models with the true Fel d 1 structure (PDB:2EJN). The RMSD between the true structure used as a reference and each of the two cat models is 0.34 (over 122 Cα atoms) for the domestic cat and 0.38 (over 119 Cα atoms) for the exotic cat. The low predicated local distance difference test (pLDDT) score for C terminus residues (Met144 to Glu146) of Fel d 1 suggests higher flexibility and probable conformational changes of these residues (blue circle). The AA substitutions in domestic (Ile135 only) or exotic cats are shown in red stick representation in the model structure. The sequence alignment of reference and substituted sequence shows AA substitutions (highlighted) in domestic (Ile135 only) or exotic cats.
The AA substitutions encoded by the majority (> 50%) of domestic or exotic cats were modeled on the heterodimeric reference structure for Fel d 1 using AlphaFold3 (41–43). The reference Fel d 1 structure (PDB 2EJN (17)) and the two AlphaFold3 models containing the domestic or exotic cat AA substitutions were superimposed to identify predicted changes in the Fel d 1 structure due to the variable AA residues (Figs. 3B and C). While 50% of the 12 AA substitutions included in the analysis were due to dissimilar AA residue changes, AlphaFold3 only predicted a shift in orientation of the C terminus of the Fel d 1 heterodimer (Fig. 3C).
Selective pressure analysis
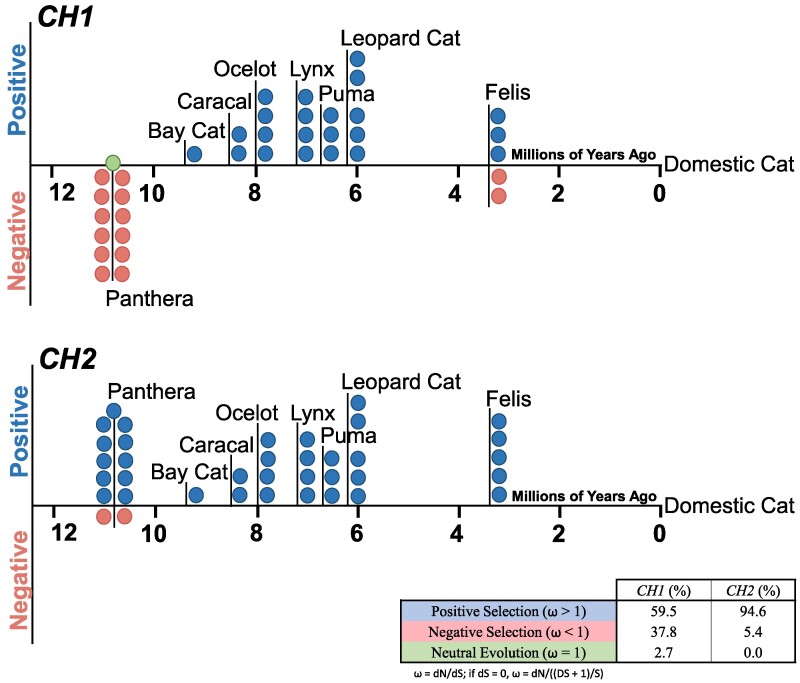
The exotic cat orthologs (one representative cat per species; Table S10) were compared with the GenBank DNA references for domestic cat Fel d 1 CH1 or CH2, and selective pressure was calculated for each exotic species (ω = dN/dS; Table S11). Positive or diversifying selection pressure was indicated by ω > 1, while ω < 1 or ω = 1 indicated negative selection or neutral evolution, respectively (44, 45). For Fel d 1 CH1, 59% of the exotic cats were under positive selection versus 38% negative selection or 3% neutral evolution. By contrast, 95 and 5% of the exotic cat CH2 orthologs reflected positive and negative selection, respectively. The majority of the species that exhibited negative or purifying selective pressure were members of the Panthera lineage that diverged from domestic cats ∼11 MYA (35) (Fig. 4).
Fig. 4:
Fel d 1 undergoes positive, diversifying selection across Felidae. DNA selective pressure analysis of Fel d 1 CH1 (Top) and CH2 (Bottom) in 37 exotic cat species, grouped by lineage. Each datapoint represents an exotic cat species, with positive selection (blue), negative selection (pink), and neutral evolution (green) indicated. The ratio of nonsynonymous and synonymous substitution rates (ω) was determined for each exotic species (Table S11).
Discussion
Genetic diversity
Fel d 1 identity roughly reflects the evolution of Felidae, with the more divergent exotic cats (e.g. Panthera, Bay Cat, and Caracal lineages) exhibiting relatively lower pairwise identities with Fel d 1 versus the exotic species more closely related to domestic cat (35). The coefficients of determination (R2 > 0.8) indicated that the lineage–average pairwise identities for CH1 DNA and chain 1 protein strongly correlated with time (MYA) since diverging from domestic cats. By contrast, time was only moderately predictive of the variance in the Fel d 1 chain 2 sequences (R2 > 0.5; CH2 DNA and chain 2 protein). The correlation between pairwise identity and the Felidae evolutionary timeline was also supported by the phylogenies for Fel d 1 chains 1 and 2. For example, the Felis lineage, which diverged from domestic cats ∼3 MYA and includes exotic species such as Chinese mountain, Asiatic wild, black-footed, sand, and jungle cats, had significantly shorter mean branch distances relative to domestic cats (mean ± SEM: chain 1: 0.08 ± 0.001; chain 2: 0.19 ± 0.001 AA substitutions per residue) versus the average distances between the more divergent Panthera lineage and domestic cats (chain 1: 0.15 ± 0.0001; chain 2: 0.28 ± 0.0008) (P < 0.001; Felis N = 1,680, Panthera N = 7,560).
Pairwise and multisequence alignments of the Fel d 1 genes across Felidae showed lower identities, or greater variability, among the chain 2 DNA and protein sequences versus chain 1. A higher proportion (58%) of the chain 2 AA residues in domestic or exotic cats were variable compared with chain 1 (41%), and ∼65% of the chain 1 or 2 variable residues stemmed from dissimilar AA changes. Across both domestic and exotic cats, the chain 2 DNA and protein sequences or orthologs were highly variable, partly due to the likely expression of several chain 2 isoforms (15). Exotic cats of the less divergent Felis lineage encoded a truncated isoform (similar to IAINEY, isoform CH2S (15)), while the more divergent exotic species preferentially encoded the standard CH2 isoform (135TTISSSKD142) (30). The domestic cats encoded either or a combination of the two chain 2 isoforms, which have previously been shown to be expressed in different feline tissues (15). This variable region of chain 2 comprises part of an α-helix (H8) that resides at the dimer interface of recombinant Fel d 1 and could significantly affect the dimeric assembly and the overall structural conformation of the allergen (17). This variable sequence is part of the dimeric interface that spans residues Ile125 to Met144 (according to PDBePISA analysis). The dimeric interface has a couple of additional structural features that could influence protein function. First, it includes the preceding stretch of residues 121–131, reported to have different conformation between chains 1 and 2 (17). This conformational change is driven by calcium coordination with residues Ile125 (chain 1) and Asp130 (chain 2) and determines the shape of water-filled cavities (17). However, recent strong evidence by the authors indicates that the calcium binding observed in the reference structure was most likely an artifact of crystallization. Second, the dimeric interface also contains residues Phe85, Gly131, and Leu132, which, together with Asn103 (outside the dimer interface), form a hydrophobic cavity on the interface which could influence function. These four residues are fairly conserved among the analyzed sequences. Therefore, among the residues that form the interface (Ile125–Met144), most sequence variability was observed for the stretch of (135TTISSSKD142), which may impact the physiological properties of the molecule as well as the tetrameric assembly (dimerization of the heterodimer). Since the function of Fel d 1 remains unknown, whether these sequence variabilities affect protein function cannot be ascertained. It can be speculated that AA changes may impact ligand (fatty acid and steroids) preference and overall physiological properties of the protein in the respective cats.
The alignments of the Fel d 1 sequences and orthologs determined that the chain 1 and chain 2 protein sequences exhibited greater variability than the corresponding DNA sequences. A substantial proportion (> 68%) of the CH1 or CH2 nucleotide substitutions occurred in the first or second codon positions of the AA residues, thus eliminating potential redundancy in the subsequent protein sequences. This is supported by the selective pressure analysis, which determined that most of the exotic cat species experienced positive or diversifying selection due to nucleotide substitutions that resulted in AA changes (i.e. greater nonsynonymous mutations). Though two Felis species (black-footed and Chinese mountain cats) showed negative selective pressure in CH1, the remaining exotic cats that deviated from the positive selection majority were more divergent species from the Panthera lineage. The largely positive selective pressure acting on Fel d 1 could indicate that CH1 and CH2 are evolving to increase fitness. Alternatively, given that negative selection of essential genes often manifests as lower rates of mutations and evolution, our predominantly positive selection data (59 and 95% positive selection for CH1 and CH2, respectively) coupled with the considerable sequence variability in the genes (41 and 58% variable AA residues for chains 1 and 2, respectively) may suggest that Fel d 1 function could be nonessential to cats (46). In particular for CH2, the large ratio of nonsynonymous to synonymous substitution rates (up to ω = 7.2) relative to CH1 (up to ω = 3.8) suggests that the corresponding chain 2 protein is mutating, or accumulating nonsynonymous substitutions, at a faster rate versus chain 1. Thus, there may be a stronger evolutionary drive for chain 2 sequence heterogeneity. The relatively high rate at which CH2 is mutating or evolving may indicate that chain 2, specifically, may be nonessential for cats, which is supported by recent evidence of healthy cats with CRISPR-edited CH2 (47).
Implications for biological function
Though the definitive biological function of Fel d 1 remains unknown, the substantial fraction of chemically dissimilar AA substitutions identified in chains 1 and 2 may have a measurable impact on the structure and, ultimately, the function of the protein. In chain 2, many of the variable AA residues, particularly those resulting from dissimilar AA substitutions, were found to reside at the dimer interface of the recombinant Fel d 1 tetramer. Alternatively, many of the dissimilar chain 1 residues appear to be concentrated on the outward facing surfaces of the chain 1 alpha helices (17). Dissimilar AA changes (e.g. the addition or loss of AA residues; changes in AA charge or polarity) in the Fel d 1 protein sequences may significantly alter the secondary, tertiary, or quaternary structures of the allergen. Several dissimilar chain 2 AA residues on the α-helices of the Fel d 1 dimer interface have been previously identified as potential binding sites for steroid or semiochemical ligands (30, 48). While AlphaFold3 predicted minimal structural changes to the Fel d 1 heterodimer due to recurring AA substitutions (substitutions observed in > 50% of domestic or exotic cats), the model may underestimate the effects of those protein sequence changes. A documented limitation of AlphaFold is an inability to predict the effects of point mutations on protein structure, stability, or functionality (49). Producing Fel d 1 mutants that incorporate the recurring AA substitutions may more definitively elucidate the structural and/or functional contribution of those variable residues.
Assuming that the structure of recombinant Fel d 1 resembles that of the natural protein, the considerable variability in the Fel d 1 AA residues could indicate that the allergen may have evolved to bind a variety of ligands. Previously, a docking study predicted critical residues for ligand or steroids binding on Fel d 1, among which chain 1 residues Val10, Phe13, Leu14, and Tyr21 and chain 2 residues Phe80, Phe84, Val87, Met112, Tyr119, Asp130, and Met134 are conserved across domestic and exotic cats (48). Among the chain 2 residues, variability is observed for residues Phe85, Gly131, Thr135, and Ser138. Phe85 is changed to Thr85 in all exotic cats, and Gly131 is changed to Ala131 in most exotic cats (six have Thr131 and one has Val131). Similarly, Thr135 is substituted by Ile135 in some domestic and exotic cats. Phe85 is predicted to interact with steroids hydrophobically, so substitution to a polar residue like Thr85 in exotic cats suggests a possible difference in ligand preference. Similarly, Thr135 and Ser138 are among the chain 2 residues (Thr135–Asp142) in which most variability is observed among sequenced cats. As such, addition/deletion or substitution of these residues may impact the ligand preference.
Alternatively, the data may suggest that the function of these variable residues, such as ligand-binding capabilities, may be nonessential. For chain 2, in particular, the abundance of dissimilar AA changes at the interface between the two Fel d 1 heterodimers may impair dimer assembly, thus impacting the quaternary structure and potential functionality of the protein. Efforts to determine the structure and relevant functional epitopes of the natural Fel d 1 allergen are in progress, which would help in resolving the biological function of the protein.
Across all domestic and exotic cats analyzed, ∼60 and 40% of the chain 1 and 2 AA residues, respectively, were determined to be conserved. In domestic cats alone, 84% of chain 1 and 74% of chain 2 AA residues were conserved, corresponding to multiple spans of 40–80 conserved DNA nucleotides in the Fel d 1 genes. While many conserved nucleotides and AA residues were identified in the Fel d 1 sequences, the average CH1 and CH2 identities determined across Felidae are relatively low and on par with the noncoding, genome-wide identity averages between cat species (50). This suggests that the Fel d 1 genes may not be conserved in evolution, which could indicate that the allergen does not serve an essential function in cats or that the function of the protein varies among cat species. Though the definitive biological function of Fel d 1 should be ascertained experimentally, the data may support removing the gene sequences from domestic cats as a treatment approach for cat allergy.
Of the nearly 300 cats analyzed, two cats encoded heterozygous mutations in CH1 or CH2 that could significantly impair Fel d 1 protein translation and subsequent expression. A cougar (Exotic Cat 2) had a heterozygous frameshift mutation in CH1 that would correspond with a mutated Leu34 and an early stop codon at AA position Leu36. A black-footed cat (Exotic Cat 30) encoded a heterozygous nonsense mutation in CH2, corresponding with a stop codon at Glu68 and a truncated Fel d 1 protein. While each of these mutations is presumably monoallelic and may have negligible phenotypic consequences in the affected cats, the prevalence of such potentially significant mutations in living cats (0.7% of 276 cats analyzed), which could result in the expression of a nonfunctional Fel d 1 protein, further supports the theory that Fel d 1 may not be essential for cats.
Significance
The conserved DNA spans of CH1 and CH2 identified in the analysis could serve as viable targets for editing the Fel d 1 genes using CRISPR technology (30). In particular, CRISPR–Cas9 would allow for targeted deletions with superior throughput, efficiency, and precision compared with preceding gene editing approaches (51). CRISPR editing poses a unique opportunity for significant disease improvement compared with existing treatment options for cat allergy and will undoubtedly benefit cat allergic patients by effectively removing the Fel d 1 genes from the source (30, 52). In fact, a recent study by Lee et al. (47) reports that Fel d 1 chain 2 was genome-edited using CRISPR, which resulted in disrupted Fel d 1 expression and secretions, and in the generation of cats that were “very healthy and active, and there was no special issue with their health”. Furthermore, mouse ABP which is reportedly involved in chemical communication among mice shows 35–60% sequence identity with Fel d 1, though an unequivocal function for mouse ABP has not been ascertained. Deletion of mouse ABP resulted in significantly lower levels of salivary ABP than in normal mice, but otherwise ABP deficient mice had normal growth curves, tissue histology, fecundity, and longevity (53). These studies provide further evidence that Fel d 1 may be nonessential in cats, although its precise function is not known.
To our knowledge, this is the most comprehensive sequence analysis of any allergen genes to date. Some allergens have a large number of reported isoallergens or isoforms, for example, 27 and 24 isoforms are listed in the WHO/International Union of Immunological Societies (IUIS) Allergen Nomenclature database for allergens Bet v 1 and Der p 1, respectively. A previous study of the major birch pollen allergen, Bet v 1, which included the alignments of 264 Bet v 1-like sequences, revealed that the homologous proteins varied considerably with evolution and showed a ubiquitous distribution of Bet v 1-related proteins among all taxonomic domains (eukaryote, bacteria, and archaea organisms) (54). Given the prevalence of Bet v 1 isoforms and homologous proteins, the comprehensive analysis supports the potential discovery of allergen cross-reactivity, conformational or functional epitopes, or possible natural hypoallergens for the treatment of birch allergic disease (55, 56). Similarly, an alignment of 12 variants of Amb a 1, an allergen from short ragweed pollen, determined the sequence identities of the five corresponding Amb a 1 isoallergens, while a study of Der p 2, a major house dust mite allergen, examined the effect of Der p 2 sequence variants on IgE antibody binding (57, 58).
Beyond our analysis of Fel d 1, a number of genetic studies have been carried out in cats with the aims of elucidating the origins of cat domestication, cataloging the genetic and phenotypic diversity of domestic cats, and aiding the conservation of threatened Felidae species (59–61). In particular, a recent study of > 11,000 domestic cat genomes evaluated the prevalence and distribution of genetic variants associated with heritable diseases and physical traits across 90 breeds of cats, highlighting the value of large-scale comparative genomic analyses in supporting the health and wellness of cats (60). Additionally, cats have served as model organisms for human genetic disease including heritable conditions such as polycystic kidney disease, dwarfism, and a form of retinal blindness, pointing to the value of cats and cat genomic analyses in translational research and human healthcare (62).
Conclusion
Across all cat species analyzed, our data indicate that the Fel d 1 gene sequences vary considerably, which could have meaningful implications regarding the evolution, structure, or function of the allergen. While the study relies on the assumption that the natural protein resembles the structure of recombinant Fel d 1, the data highlight the abundant, dissimilar sequence changes that will presumably affect Fel d 1 structural conformation and likely, in turn, impact Fel d 1 binding to relevant ligands and variant-specific antibodies. The substantial variability revealed by our comparative genomics analysis suggests that Fel d 1 is not well conserved over the evolution of Felidae, which hints that the protein may not be essential for cats. A lack of an essential biological function for the allergen would suggest that Fel d 1 is a rational target for gene deletion, which may have clinical benefits for individuals suffering from cat allergy.
Supplementary Material
Contributor Information
Clifford W Cleveland, III, InBio, Charlottesville, 700 Harris St, VA 22903, USA.
Brian W Davis, Department of Veterinary Integrative Biosciences, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, TX 77843, USA.
Kriti Khatri, Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.
Anna Pomés, InBio, Charlottesville, 700 Harris St, VA 22903, USA.
Martin D Chapman, InBio, Charlottesville, 700 Harris St, VA 22903, USA.
Nicole F Brackett, InBio, Charlottesville, 700 Harris St, VA 22903, USA.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
Research reported in this publication was funded by internal funds provided by the InBio company. The research was also supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01A1077653 (A.P. and M.D.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Contributions
C.W.C.: Investigation, Data curation; B.W.D.: Conceptualization, Methodology, Formal analysis, Writing review and editing; K.K.: Methodology, Visualization, Data curation, Writing review and editing; A.P.: Validation, Visualization, Data curation, Writing review and editing; M.D.C.: Conceptualization, Project administration, Writing review and editing; N.F.B.: Conceptualization, Supervision, Visualization, Formal analysis, Writing original draft.
Data Availability
The data supporting the findings of this study are available within the article and its supplementary materials. Cat raw genomic data are publicly available in NCBI's sequence read archive (SRA) database.
References
- 1. Salo PM, et al. 2014. Prevalence of allergic sensitization in the United States: results from the national health and nutrition examination survey (NHANES) 2005–2006. J Allergy Clin Immunol. 134:350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingram JM, et al. 1995. Quantitative assessment of exposure to dog (Can f 1) and cat (Fel d 1) allergens: relation to sensitization and asthma among children living in Los Alamos, New Mexico. J Allergy Clin Immunol. 96:449–456. [DOI] [PubMed] [Google Scholar]
- 3. Chan SK, Leung DYM. 2018. Dog and cat allergies: current state of diagnostic approaches and challenges. Allergy Asthma Immunol Res. 10:97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suzuki S, et al. 2019. Characterization of sensitization to furry animal allergen components in an adult population. Clin Exp Allergy. 49:495–505. [DOI] [PubMed] [Google Scholar]
- 5. Gergen PJ, et al. 2018. Sensitization and exposure to pets: the effect on asthma morbidity in the US population. J Allergy Clin Immunol Pract. 6:101–107.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel D, et al. 2013. Fel d 1-derived peptide antigen desensitization shows a persistent treatment effect 1 year after the start of dosing: a randomized, placebo-controlled study. J Allergy Clin Immunol. 131:103–109.e1-e7. [DOI] [PubMed] [Google Scholar]
- 7. Morgenstern JP, et al. 1991. Amino acid sequence of Fel dI, the major allergen of the domestic cat: protein sequence analysis and cDNA cloning. Proc Natl Acad Sci U S A. 88:9690–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvarez-Cuesta E, et al. 2007. Sublingual immunotherapy with a standardized cat dander extract: evaluation of efficacy in a double blind placebo controlled study. Allergy. 62:810–817. [DOI] [PubMed] [Google Scholar]
- 9. Matricardi PM, et al. 2016. EAACI molecular allergology User's Guide. Pediatr Allergy Immunol. 27(Suppl 23):1–250. [DOI] [PubMed] [Google Scholar]
- 10. Ohman JL Jr, Marsh DG, Goldman M. 1982. Antibody responses following immunotherapy with cat pelt extract. J Allergy Clin Immunol. 69:320–326. [PubMed] [Google Scholar]
- 11. Lowenstein H, Lind P, Weeke B. 1985. Identification and clinical significance of allergenic molecules of cat origin. Part of the DAS 76 study. Allergy. 40:430–441. [DOI] [PubMed] [Google Scholar]
- 12. de Groot H, van Swieten P, van Leeuwen J, Lind P, Aalberse RC. 1988. Monoclonal antibodies to the major feline allergen Fel d I. I. Serologic and biologic activity of affinity-purified Fel d I and of Fel d I-depleted extract. J Allergy Clin Immunol. 82:778–786. [DOI] [PubMed] [Google Scholar]
- 13. Chapman MD, Aalberse RC, Brown MJ, Platts-Mills TA. 1988. Monoclonal antibodies to the major feline allergen Fel d I. II. Single step affinity purification of Fel d I, N-terminal sequence analysis, and development of a sensitive two-site immunoassay to assess Fel d I exposure. J Immunol. 140:812–818. [PubMed] [Google Scholar]
- 14. van Ree R, van Leeuwen WA, Bulder I, Bond J, Aalberse RC. 1999. Purified natural and recombinant Fel d 1 and cat albumin in in vitro diagnostics for cat allergy. J Allergy Clin Immunol. 104:1223–1230. [DOI] [PubMed] [Google Scholar]
- 15. Griffith IJ, et al. 1992. Expression and genomic structure of the genes encoding FdI, the major allergen from the domestic cat. Gene. 113:263–268. [DOI] [PubMed] [Google Scholar]
- 16. Kaiser L, et al. 2003. The crystal structure of the major cat allergen Fel d 1, a member of the secretoglobin family. J Biol Chem. 278:37730–37735. [DOI] [PubMed] [Google Scholar]
- 17. Kaiser L, et al. 2007. Structural characterization of the tetrameric form of the major cat allergen Fel d 1. J Mol Biol. 370:714–727. [DOI] [PubMed] [Google Scholar]
- 18. Charpin C, et al. 1991. Fel d I allergen distribution in cat fur and skin. J Allergy Clin Immunol. 88:77–82. [DOI] [PubMed] [Google Scholar]
- 19. van Milligen FJ, Vroom TM, Aalberse RC. 1990. Presence of Felis domesticus allergen I in the cat's salivary and lacrimal glands. Int Arch Allergy Appl Immunol. 92:375–378. [DOI] [PubMed] [Google Scholar]
- 20. De Andrade AD, et al. 1996. Fel d I levels in cat anal glands. Clin Exp Allergy. 26:178–180. [DOI] [PubMed] [Google Scholar]
- 21. Bastien BC, Gardner C, Satyaraj E. 2019. Influence of time and phenotype on salivary Fel d1 in domestic shorthair cats. J Feline Med Surg. 21:867–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Durairaj R, Pageat P, Bienboire-Frosini C. 2018. Another cat and mouse game: deciphering the evolution of the SCGB superfamily and exploring the molecular similarity of major cat allergen Fel d 1 and mouse ABP using computational approaches. PLoS One. 13:e0197618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scheib H, et al. 2020. The toxicological intersection between allergen and toxin: a structural comparison of the cat dander allergenic protein Fel d1 and the slow loris brachial gland secretion protein. Toxins (Basel). 12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emara M, et al. 2011. Recognition of the major cat allergen Fel d 1 through the cysteine-rich domain of the mannose receptor determines its allergenicity. J Biol Chem. 286:13033–13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herre J, et al. 2013. Allergens as immunomodulatory proteins: the cat dander protein Fel d 1 enhances TLR activation by lipid ligands. J Immunol. 191:1529–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hilger C, et al. 2014. Identification and isolation of a Fel d 1-like molecule as a major rabbit allergen. J Allergy Clin Immunol. 133:759–766. [DOI] [PubMed] [Google Scholar]
- 27. Karn RC. 1994. The mouse salivary androgen-binding protein (ABP) alpha subunit closely resembles chain 1 of the cat allergen Fel dI. Biochem Genet. 32:271–277. [DOI] [PubMed] [Google Scholar]
- 28. de Groot H, van Swieten P, Aalberse RC. 1990. Evidence for a Fel d I-like molecule in the “big cats” (Felidae species). J Allergy Clin Immunol. 86:107–116. [DOI] [PubMed] [Google Scholar]
- 29. Vailes LD, et al. 1994. Fine specificity of B-cell epitopes on Felis domesticus allergen I (Fel d I): effect of reduction and alkylation or deglycosylation on Fel d I structure and antibody binding. J Allergy Clin Immunol. 93:22–33. [DOI] [PubMed] [Google Scholar]
- 30. Brackett NF, Davis BW, Adli M, Pomés A, Chapman MD. 2022. Evolutionary biology and gene editing of cat allergen, Fel d 1. CRISPR J. 5:213–223. [DOI] [PubMed] [Google Scholar]
- 31. Jensen JD, et al. 2019. The importance of the neutral theory in 1968 and 50 years on: a response to Kern and Hahn 2018. Evolution. 73:111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kimura M. 1968. Evolutionary rate at the molecular level. Nature. 217:624–626. [DOI] [PubMed] [Google Scholar]
- 33. Kimura M. 1979. The neutral theory of molecular evolution. Sci Am. 241:98–100, 102, 108 passim. [DOI] [PubMed] [Google Scholar]
- 34. Darwin C. 1859. On the origin of species by means of natural selection, or, the preservation of favoured races in the struggle for life. London: J. Murray. [PMC free article] [PubMed] [Google Scholar]
- 35. O'Brien SJ, Johnson WE. 2007. The evolution of cats. Genomic paw prints in the DNA of the world's wild cats have clarified the cat family tree and uncovered several remarkable migrations in their past. Sci Am. 297:68–75. [PubMed] [Google Scholar]
- 36. O'Brien SJ, et al. 2008. State of cat genomics. Trends Genet. 24:268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bianchini G, Sanchez-Baracaldo P. 2024. TreeViewer: flexible, modular software to visualise and manipulate phylogenetic trees. Ecol Evol. 14:e10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jumper J, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Varadi M, et al. 2022. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abramson J, et al. 2024. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 630:493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kryazhimskiy S, Plotkin JB. 2008. The population genetics of dN/dS. PLoS Genet. 4:e1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kimura M. 1977. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 267:275–276. [DOI] [PubMed] [Google Scholar]
- 46. Hurst LD, Smith NG. 1999. Do essential genes evolve slowly? Curr Biol. 9:747–750. [DOI] [PubMed] [Google Scholar]
- 47. Lee SR, et al. 2024. Generation of Fel d 1 chain 2 genome-edited cats by CRISPR-Cas9 system. Sci Rep. 14:4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bienboire-Frosini C, Durairaj R, Pelosi P, Pageat P. 2020. The major cat allergen Fel d 1 binds steroid and fatty acid semiochemicals: a combined in silico and in vitro study. Int J Mol Sci. 21:1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yang Z, Zeng X, Zhao Y, Chen R. 2023. AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct Target Ther. 8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cho YS, et al. 2013. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun. 4:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 32:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brackett NF, Pomés A, Chapman MD. 2021. New frontiers: precise editing of allergen genes using CRISPR. Front Allergy. 2:821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chung AG, Belone PM, Bimova BV, Karn RC, Laukaitis CM. 2017. Studies of an androgen-binding protein knockout corroborate a role for salivary ABP in mouse communication. Genetics. 205:1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Radauer C, Lackner P, Breiteneder H. 2008. The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol. 8:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gieras A, et al. 2011. Mapping of conformational IgE epitopes with peptide-specific monoclonal antibodies reveals simultaneous binding of different IgE antibodies to a surface patch on the major birch pollen allergen, Bet v 1. J Immunol. 186:5333–5344. [DOI] [PubMed] [Google Scholar]
- 56. Wagner S, et al. 2008. Naturally occurring hypoallergenic Bet v 1 isoforms fail to induce IgE responses in individuals with birch pollen allergy. J Allergy Clin Immunol. 121:246–252. [DOI] [PubMed] [Google Scholar]
- 57. Radauer C, et al. 2014. Update of the WHO/IUIS allergen Nomenclature database based on analysis of allergen sequences. Allergy. 69:413–419. [DOI] [PubMed] [Google Scholar]
- 58. Tanyaratsrisakul S, et al. 2016. Effect of amino acid polymorphisms of house dust Mite der p 2 variants on allergic sensitization. Allergy Asthma Immunol Res. 8:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nilson SM, et al. 2022. Genetics of randomly bred cats support the cradle of cat domestication being in the near east. Heredity (Edinb). 129:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anderson H, et al. 2022. Genetic epidemiology of blood type, disease and trait variants, and genome-wide genetic diversity in over 11,000 domestic cats. PLoS Genet. 18:e1009804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ito H, Nakajima N, Onuma M, Murayama M. 2020. Genetic diversity and genetic structure of the wild tsushima leopard cat from genome-wide analysis. Animals (Basel). 10:1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lyons LA. 2021. Cats—telomere to telomere and nose to tail. Trends Genet. 37:865–867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and its supplementary materials. Cat raw genomic data are publicly available in NCBI's sequence read archive (SRA) database.