Abstract
The chloride channel transmembrane protein 16A (TMEM16A) possesses a calcium-activated property linked to tumor-promoting malignant phenotype and electrophysiological stability. Numerous studies have shown that TMEM16A exhibits aberrant amplification in various squamous cell carcinomas such as esophageal squamous cell carcinoma (ESCC) and is correlated with unfavorable outcomes of ESCC patients. Therefore, TMEM16A is considered as a promising therapeutic target for ESCC. Because of its intricate structure, the development of therapeutic antibodies directed against TMEM16A has not been documented. In this study, we produced a series of novel monoclonal antibodies targeting TMEM16A and identified mT16#5 as an antibody capable of inhibiting ESCC cells migration, invasion and TMEM16A ion channel activity. Additionally, based on the validation that TMEM16A was positively correlated with expression of EGFR and the interaction between them, the mT16#5 exhibited a synergistic inhibitory effect on ESCC metastasis and growth when administered in combination with Cetuximab in vivo. In terms of mechanism, we found that mT16A#5 inhibited the phosphorylation of PI3K, AKT and JNK. These results highlight the anti-growth and anti-metastasis capacity of the combination of mT16A#5 and Cetuximab in the treatment of ESCC by targeting TMEM16A and EGFR, and provide a reference for combinational antibody treatment in ESCC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-05830-3.
Keywords: Esophageal squamous cell carcinoma (ESCC), TMEM16A, Monoclonal antibodies (mAb), Cetuximab, Anti-metastasis
Introduction
Enhanced quality of life has led to alterations in human dietary patterns and behaviors, subsequently increasing the susceptibility to digestive tract malignancies. China is a significant contributor to the global burden of esophageal cancer, accounting for over half of reported cases. The incidence rate in esophageal squamous cell carcinoma (ESCC) of China stands at 5.6 cases per 100,000 individuals, with regional variations in mortality rates, peaking at 12.7 case per 100,000 individuals in certain rural areas [1, 2]. ESCC is characterized by the accumulation of epigenetic modifications, somatic mutations, and the dysregulation of a variety of proto-oncogenes, oncogenes, and cell adhesion molecules. Ultimately, it exhibits interactive superimposed effects, leading to its complex genesis and rapid metastatic properties [3, 4]. The utilization of molecularly targeted agents, including Cetuximab, Bevacizumab, Nivolumab, and other antibodies that have demonstrated notable efficacy in other cancer treatments, has shown reduction in the rate of ESCC metastasis, albeit falling short of anticipated outcomes [5–7]. The investigation of novel targeted agents for metastatic ESCC holds promise for enhancing treatment efficacy [8, 9]. A recent clinical trial reported a disease control rate of 100% for minimally invasive esophagectomy with Karelizumab in conjunction with chemotherapy and lapatinib, suggesting potential advancements in antibody therapy for ESCC, but longer follow-up is still needed to assess survival outcomes [10].
Numerous markers have been identified as prognostic indicators and potential therapeutic targets for ESCC, including NOTCH1, SPP1and CTSL [11–13]. However, there is currently a lack of reported antibody drugs targeting these markers. Ion channels are specialized membrane proteins that facilitate the passage of ions across lipid bilayers, thereby regulating cellular homeostasis through selective ion recognition and conduction. These channels play crucial roles in various cellular processes, including protein secretion, gene expression, and cell division. Tumors are characterized by aberrant expression and activity of ion channels, and these alterations have an equally driving role in the regulation of the malignant phenotype of tumors [14]. As a calcium-dependent chloride channel, TMEM16A has an eight-transmembrane structure and forms a half-inserted membrane-type structure between TMD6 and TMD7 of the transmembrane structural domains. The ion-conducting pore of TMEM16A consists of an hourglass-shaped pore-like structure from α3-α7 for ion penetration [15, 16]. In addition, TMEM16A is able to function as a homodimer that combines to form a pore-like structure in a double barrel [17]. The human TMEM16A gene, situated in the chromosomal region 11q13.3 known to harbor numerous pathogenic genes, exhibits aberrant amplification in oral cancer, head and neck squamous cell carcinoma, and gastrointestinal mesenchymal tumors, thereby correlating significantly with unfavorable patient prognosis [18–20]. TMEM16A can be activated by multiple signaling pathways within tumors, leading to the regulation of intracellular changes in chloride concentration and membrane potential through channel opening and closing. This activation subsequently triggers signaling pathways associated with the malignant phenotype of cancer cells [21, 22]. The primary signaling pathways responsible for activating TMEM16A are the EGFR and CAMKII pathways, with the former being particularly implicated in cancer metastasis [23–25]. Research has demonstrated that TMEM16A can enhance EGF-induced EGFR signaling and, when overexpressed, further amplifies EGFR/STAT3 signaling in cells [26, 27]. In addition, TMEM16A may also activate the EGFR signal pathway by increasing autocrine signaling of EGFR ligands [28]. Hence, TMEM16A exhibits qualities that make it a promising therapeutic target.
Presently, small molecule inhibitors such as synthetic T16Ainh-A01 and CaCCinh-A01 are the primary drugs targeting TMEM16A, effectively suppressing calcium-activated chloride currents and inhibiting cell proliferation and migration in certain cancer types [29, 30]. Furthermore, natural compounds like luteolin and silymarin have been identified as inhibitors of TMEM16A activity, also capable of inhibiting chloride currents and hindering tumor growth in animal models [31–34]. Despite these advancements, no inhibitor has progressed to the clinical trial phase as of yet on account of small molecule inhibitors are susceptible to interference from changes in the intracellular environment and their efficacy cannot be stabilized. Moreover, therapeutic antibodies targeting TMEM16A have not yet been reported.
In this study, we validated the negative correlation between TMEM16A high expression and lymph node metastasis as well as prognosis in patients with ESCC. Through bioinformatics analysis combined with examination of clinical specimens from ESCC patients, we found the interaction and correlations between TMEM16A and EGFR in ESCC. Subsequently, six monoclonal antibodies targeting TMEM16A were generated, leading to the identification and selection of mT16A#5, a functional antibody that demonstrated inhibitory effects on cell migration and invasion in vitro. The inhibitory impact of mT16A#5 on the ion channel activity of TMEM16A was further confirmed using a constructed ion channel detection system. Subsequent in vivo studies demonstrated that co-administration of mT16A#5 and Cetuximab resulted in a synergistic inhibitory effect on the metastasis of ESCC. Mechanistically, mT16A#5 inhibited the phosphorylation of PI3K, AKT and JNK. These pre-clinical results revealed anti-growth and anti-metastasis capacity of mT16A#5 for the treatment of ESCC by targeting TMEM16A.
Materials and methods
Cells and cell culture
The human ESCC cell lines KYSE-140, KYSE-410, KYSE-510 were obtained from ATCC. YES-2, TE12, KYSE30lm3, KYSE30luc, KYSE450lm2, and KYSE450luc were gifted by Prof. Zhihua Liu (Cancer Institute and Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical Collage). The KYSE30luc and KYSE450luc cell lines were obtained by labeling the KYSE30 and KYSE450 cells with luciferase reporter genes, which were injected into immunodeficient mice (SCID/Beige) via the tail vein. After 2–3 months, the lungs were taken out under aseptic conditions, minced and digested, and the primary culture of ESCC cells in lung metastases was carried out. These cells were injected secondly into SCID/Beige via the tail vein. KYSE30luc cells were screened for three rounds, while KYSE450luc cells were screened for two rounds, and finally a subset of cells with strong lung metastatic ability (KYSE30lm3 and KYSE450lm2) were obtained and cultured in RPMI-1640 medium supplemented with 15% FBS and 100 μg/mL G418 [35]. KYSE-140, KYSE-410, KYSE-510, YES-2, and TE12 were maintained in RPMI-1640 supplemented with 10% FBS. SP2/0 myeloma cells were maintained in RPMI-1640 supplemented with 20% FBS. All cells were grown at 37 °C with 5% CO2, 95% air atmosphere.
Small interfering RNA (siRNA) synthesis, vector construction, and transfection
The full-length sequence of human TMEM16A eukaryotic plasmid (pLVX-PURO-ANO1) and prokaryotic plasmids (pGEX-4 T-1-ANO1-T1-T4) were synthesized by Sangon Biotech (Suzhou, China). The vector pLVX-puro-MOCK and pGEX-4T1-GST were purchased from Public Protein/Plasmid Library (Nanjing, China). The genetically encodable biosensor of intracellular perchlorate concentration monitored by pCDNA3.1-Hygro EYFP H148Q/I152L was purchased from addgene (USA). The siRNAs, including si-EGFR1#1 (CUCCAGAGGAU GUUCAAUATT), siEGFR-#2 (GCCUUUGAGAACCUAGAAATT) and control siRNA (UUCU CCGAACGUGUCACGUTT) were synthesized by GenePharma (Shanghai, China). The shRNA of lentiviral knockout plasmids, including PLVX-TMEM16A-shRNA#1 (GCCGACGAAGAAG ATGTACCA), PLVX-TMEM16A-shRNA#2 (CAGCATCTATTTGACTTGTC) and PLVX-Non- silencing-shRNA (TTCTCCGAACGTGTCACGT) were provided by Beijing Syngentech. (Beijing, China).
Plasmids were transfected with Lipofectamine2000 (Invitrogen) for KYSE30luc and KYSE450luc cells. siRNA was transfected with Buffer Mate (GenePharma) for tumor cells. All transfections were performed according to the manufacturer’s instructions. The cells were collected and validated by Western blot 48 h after transfection.
Antibodies and proteins
Anti-TMEM16A (ab190803, WB 1:500, CO-IP 1:200), Anti-JNK1/JNK2/JNK3 (ab179461, WB 1:1000) and anti-JNK1/JNK2/JNK3 (phospho T183/T183/T221) (ab124956, WB 1:500) were purchased from Abcam. Anti-pEGFR-Tyr 992 (2235, WB 1:1000), anti-pEGFR-Tyr 1068 (#3777, WB 1:1000), anti-EGFR (#4267, WB 1:2000, CO-IP 1:500), anti-Akt (#14,702, WB 1:2000), anti-phospho-Akt (Ser473) (#4060, WB 1:1000), anti-phospho-PI3 Kinase p85 (Tyr458)/p55 (Tyr199) (#4228, WB 1:1000), and anti-PI3K p85 (#4257, WB 1:1000) were purchased from CST. Anti-GAPDH (10,494–1-AP, WB 1:2000) was purchased from Proteintech. Recombinant human EGFR protein (aa 668–1210, His & GST Tag) was purchased from Sino Biological. mIgG protein was purchased from Sigma.
RNA extraction and quantitative RT-PCR (qPCR)
Cells were harvested in Trizol® reagent (Invitrogen), and total RNA was isolated according to the manufacturer’s instructions. Single-stranded cDNA was synthesized from 5 μg total RNA using M-MLV reverse transcriptase (Invitrogen), with an oligo(dT)18-mer and Random primers, in a final reaction volume of 20 μL. The resulting complementary DNA was subjected to Real-time PCR using SYBR Green qPCR Master Mix (Promega). GAPDH was used as an internal standard. Primers used for TMEM16A, forward: 5′-ACTACCACGAGGATGACAAGC-3′, reverse: 5′-TCTCTGCACAGCACGTTCC-3′; for EGFR, forward: 5′- AGGCACGAGTAA CAAGCTCACT-3’, reverse: 5’- ATGAGGACATAACCAGCCACC -3′; for GAPDH, forward: 5’-CATCAAGAAGGTGGTGAAGCAG-3′, reverse: 5′-CGTCAAAGGTGGAGGAGTGG-3′; for IFN-γ, forward: 5′-TCTTTGGGTCAGAGTTAAAGCCA-3′, reverse: 5′-TTCCATCTCGGCATACAGCAA-3′; for IL-1β, forward: 5′-ATGATGGCTTATTACAGTGGCAA-3′, reverse: 5′-GTCGGAGATTCGTAGCTGGA-3′;for IL-2, forward: 5′-TACAAGAACCCGAAACTGACTCG-3′, reverse: 5’-ACATGAAGGTAGTCTCACTGCC-3′; for IL-4, forward: 5’-CCAACTGCTTCCCCCTCTG-3′, reverse: 5′-TCTGTTACGGTCAACTCGGTG-3′; for IL-6, forward: 5′-ACTCACCTCTTCAGAACGAATTG-3′, reverse: 5′-CCATCTTTGGAAGGTTCAGGTTG-3′;for IL-8, forward: 5’-TTTTGCCAAGGAGTGCTAAAGA-3′, reverse: 5’-AACCCTCTGCACCCAGTTTTC-3′;for IL-10, forward: 5′-GACTTTAAGGGTTACCTGGGTTG-3′, reverse: 5’-TCACATGCGCCTTGATGTCTG-3′;for IL-12A, forward: 5’-CCTTGCACTTCTGAAGAGATTGA-3′, reverse: 5′-ACAGGGCCATCATAAAAGAGGT-3′; for IL-13, forward: 5′-CCTCATGGCGCTTTTGTTGAC-3′, reverse: 5’-TCTGGTTCTGGGTGATGTTGA-3′; for TNF-α, forward: 5’-CCTCTCTCTAATCAGCCCTCTG-3’, reverse: 5’-GAGGACCTGGGAGTAGATGAG-3’.
RNA transcriptome sequencing
The KYSE30lm3 and KYSE30luc cells were cultured for 48 h with complete medium. Then the cells were collected and the total RNA was extracted using TRIzol reagent (Invitrogen). The genomic DNA was removed using DNase I. RNA degradation and contamination was monitored on 1% agarose gels. Then RNA quality was determined by 2100 Bioanalyser (Agilent Technologies) and quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA sample (OD260/280 = 1.8 ~ 2.2, OD260/230 ≥ 2.0, RIN ≥ 8.0, 28S:18S ≥ 1.0, > 1 μg) was used to construct sequencing library. RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instruction (Illumian, San Diego, CA). The transcriptome library was prepared following TruSeq™ RNA sample preparation Kit from Illumian (San Diego, CA) using 1 μg of total RNA. Fastp (https://github.com/OpenGene/fastp) was used for quality control and read mapping. To identify DEGs between two different groups, the expression level of each gene was calculated according to the TPM method. RSEM (http://deweylab.biostat.wisc.edu/rsem/) was used to quantify gene abundances. GO (Gene Ontology, http://www.geneontology.org) and KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/) were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways at P-adjust ≤ 0.05compared with the whole-transcriptome background.
Migration and invasion assay
For migration assay, 5 × 104 KYSE30lm3/KYSE30luc and 2 × 105 KYSE450lm2/KYSE450luc cells were plated in 24-well Transwell plates with inserts (8-μm pore size, Corning) and were incubated at 37 °C for 24 h. Invasion assays were carried out in a 24-well Transwell unit on polycarbonate filter coated with Matrigel (Corning). Cell inserts were fixed with 4% PFA for 30 min, followed by PBS wash and 1% crystal violet staining to allow visualization. Nine random fields were captured per sample at 10 × magnification. The ratio of the average stained area to the field of view of each sample was calculated by Image J. All experiments were performed in triplicate.
Generation and purification of anti-TMEM16A monoclonal antibodies
BALB/C mouse were used for immunization. TMEM16A eukaryotic plasmid mixed with In vivo-jetPEI (Polyplus) transfection reagent was used as antigen. Ten days after the last boost the sera of the immunized mice were tested for the TMEM16A specific antibody by ELISA, and the spleen cells were isolated and fused with myeloma cells to obtain hybridomas. Limited dilution was performed as standard step to selected hybridoma cell clones. We used 293 T-TMEM16A cell as coating antigen of primary screening, and GST-T16A-T1-T4 prokaryotic protein and GST protein (400 ng/mL) recombinant protein as coating antigen in the next few rounds of screening. Antibodies were prepared by intra-peritoneal injection of hybridomas into 8-week-old BALB/C female mice. One week before the hybridoma injection, 500 μL of incomplete Freund's adjuvant was injected. After 10–15 days, the ascites was collected, and protein G agarose beads were used for antibody purification. After being concentrated by PEG20000, antibodies were dialyzed by PBS and identified with SDS-PAGE.
Immunoprecipitation and co-immunoprecipitation
KYSE30lm3 cells were lysed with IP buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl,1 mM EDTA and 0.5% NP-40) containing protease inhibitors (Roche, USA), and the lysates were used for IP with appropriate antibodies as well as protein G-Sepharose (GE Healthcare, USA). The precipitants were separated by SDS-PAGE and subjected to Western blotting with specific antibodies.
Western blot assay
For Western blot analysis, cells were collected and lysed with 40 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% (v/v) Triton X-100, 1 × cocktail of protease inhibitors and PMSF. Protein samples (20 μg per lane) were separated and transferred to a NC membrane. After blocking with 5% non-fat milk, the membranes were incubated with primary antibodies at 4 °C overnight. HRP-conjugated sheep anti-rabbit or anti-mouse IgG secondary antibodies (Vector, Burlingame, CA) were incubated for 1 h at room temperature. The protein bands were detected using the Super Enhanced chemiluminescence detection kit (Applygen Technologies Inc, Beijing, China).
Multiplex IHC
Immunofluorescence staining was performed using a Multiplex mIHC kit (TSA-RM-275, Panovue, China). Chips were heated at 60 °C for 20 min. After dehydrated, chips were incubated with formalin solution and 3% H2O2 for 10 min. Following antigen retrieval with pH 8.0 EDTA, the slides were blocked with 5% goat serum (ZLI-9056, ZSGB-BIO) for 1 h. Anti-EGFR antibody and anti-TMEM16 antibody as primary antibodies and then incubated with fluorescent antibodies. The chips were rinsed with washing buffer after each step. Following TSA deposition, the chips were again subjected to HIER to strip the tissue-bound primary/secondary antibody complexes and ready for labeling of the next marker. These steps were repeated until two markers were labeled and finally added with spectral 4′6-diamidno-2-phenylindole (DAPI) at 1:500 dilution. The chips were mounted in ProLong Diamond Anti-fade Mountant and cured in the dark at room temperature for 24 h. All images were acquired for each case using a Vectra 3 pathology imaging system and analyzed using inform software.
Measurement of TMEM16A chloride channel activity with MQAE
MQAE (S1082, Beyotime) is a fluorescently-labeled deoxyglucose analog that is used primarily to detect chloride channel activity in living cells. Briefly, the cells were cultured on glass-bottomed cell culture dishes (801,002, NEST, China). The cells were subsequently loaded with 1 mM MQAE in the dark for 2 h at 37 °C, and washed twice with the MQAE effluent to remove the unbound probe and re-added with 1 mL of MQAE effluent. Culture dishes were placed on the stage of the laser confocal microscope. The effluent was aspirated after selecting the field of view, and 500 μl of MQAE buffer with NaI (50 mM) was added. The focal length was adjusted at the excitation wavelength of 405 nm, and after 2 s of baseline fluorescence intensity in the field of view, 500 μl of ATP (200 μM) was added, then changes in the fluorescence intensity of cells under the field of view were recorded after continuous shooting after pipetting and mixing [36–38].
Measurement of chloride channel activity TMEM16A with EYFP-H148Q/I152L
The ESCC cells were transfected with the pCDNA3.0-YFP-H148Q/I152L plasmid for 24 h and were plated into a 96-well blackboard transparent bottom plate at 1 × 104/well after digested and resuspended. EYFP-H148Q/I152L fluorescence was monitored in cells with fluorescence microscopy after 24 h. After discarding the culture medium, the cells were washed in PBS for three times, then 50 μl/well of isotonic NaI Krebs-HEPES solution was added. Fluorescence was continuously measured at a rate of 5 s/point for 200 s, the first 10 s as the baseline, and after 10 s, the hypotonic Krebs-HEPES solution containing 200 μM ATP was injected into the well at a rate of 150 μl/well/s and the determination was continued. The parameters were set with excitation wavelength 514 nm and emission wavelength 550 nm [39, 40].
In vivo assay
Animal experiments were approved by the Biomedical Ethical Committee of Peking University Cancer Hospital & Institute and performed along institutional animal welfare protocols concordant with the NIH guidelines. Six-week-old male SCID/Beige mice were purchased from Charles River (Beijing, China). For footpad tumor model, KYSE30lm3 cells (2 × 105) were injected into the right footpad of mice which were killed 8 weeks later. For tumor growth model, KYSE30lm3 cells (2 × 106) were injected subcutaneously into the flanks of mice, which were subsequently killed 4 weeks later. All mice were randomized into four groups of 6 mice each and treated with mT16A#5 (10 mg/kg), Cetuximab (1 mg/kg), Combination (mT16A#5 (10 mg/kg) plus Cetuximab (1 mg/kg)), or equal volume of PBS twice a week.
Cell proliferation assay
A Cell Counting Kit 8 (CCK-8, Dojindo Laboratories) was used to evaluate cell proliferation. At 24 h after transfection, the cells were seeded into 96-well plates with 1,000 cells per well and cultured for 24, 48, 72, 96, and 120 h. Then, 10 μl of CCK-8 solution was added to each well and incubated for 1 h. Optical density was examined at the wavelength of 450 nm.
ELISA
Detection of inflammatory factors in the KYSE30lm3 and KYSE30luc cell culture supernatant was performed by Univ-bio (Shanghai, China) using LabEx ELISA kit (MSD, K15049D-X).
Statically analysis
The SPSS (version 26) software package (SPSS Inc, Chicago, IL) was used for statistical analysis. The data were expressed as the mean ± SD, and the data from specific experiments were compared by one-way ANOVA, Student’s t-test, or χ2 test. GraphPad Prism 6 software (GraphPad Software, San Diego, CA, USA) was used for statistical figure or chart display. Chi-square analysis was used for differential expression of TMEM16A and EGFR in ESCC tissue microarrays. All experiments were repeated at least three times with consistent results. P-value < 0.05 was considered statistically significant.
Result
TMEM16A promotes ESCC cell motility and lymph node metastasis
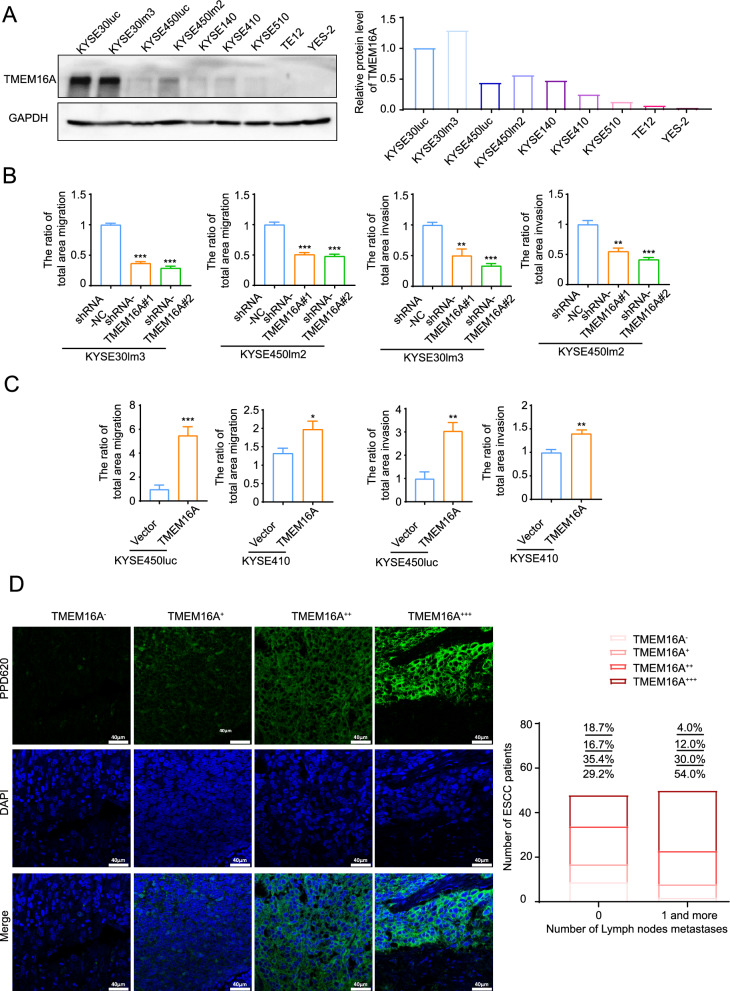
TMEM16A is overexpressed and promotes cell metastasis in a variety of squamous cell carcinomas including head and neck squamous carcinoma, oral squamous carcinoma, cervical squamous carcinoma and ESCC [19, 20, 41]. In this study, nine ESCC cell lines including KYSE30lm3, KYSE450lm2, KYSE30luc, KYSE450luc with different metastatic abilities and other five ESCC cell lines were selected for the detection of endogenous TMEM16A expression levels. The results showed that in the KYSE30lm3 and KYSE30luc cell lines with higher metastatic ability, the protein and transcript level of TMEM16A were significantly higher than those of other cell lines (Fig. 1A). We further constructed a TMEM16A knockdown model to validate the role of TMEM16A in cell motility (Supplement Fig S1A). The results of migration and invasion assays showed that knockdown of TMEM16A expression inhibited the migratory and invasive ability of cells (Fig. 1B, Supplement Fig S1B). Meanwhile, we examined the proliferative ability of these cell models, and the results of CCK8 assay showed that knockdown of TMEM16A diminished cell proliferation (Supplement Fig S1C). Correspondingly, we also performed transient overexpression of TMEM16A (Supplement Fig S1D), and the cells overexpressing TMEM16A showed stronger motility and proliferative ability than the control groups (Fig. 1C, Supplement Fig S1E, S1F). After verifying the motility-promoting effect of TMEM16A, we further clarified the correlation between TMEM16A level and lymph node metastasis by examining the tumor tissues of ESCC patients. As shown by fluorescencent immunohistochemistry, lymph node metastasis occurred in a significantly higher percentage of patients with high expression of TMEM16A in the tumor tissues of ESCC than in those with low expression of TMEM16A (Fig. 1D, Table 1). Thus, these data suggest that TMEM16A can promote ESCC cell motility and lymph mode metastasis.
Fig. 1.
TMEM16A promotes ESCC cell motility and lymph node metastasis. A TMEM16A was more highly expressed in ESCC cells with high metastatic ability. Western blot (left) and grayscale scanning analysis (right) of TMEM16A level in ESCC cell lines. GAPDH was used as a loading control. B, C TMEM16A promotes ESCC cell migration and invasion. B Transwell chamber assays and Matrigel invasion assays for KYSE30lm3 and KYSE450lm2 cells after TMEM16A knockdown. C Transwell chamber assays and Matrigel invasion assays for KYSE450luc and KYSE410 cells overexpressing TMEM16A. D Fluorescence immunohistochemistry staining showed that the level of TMEM16A in ESCC tissues was significantly correlated with the degree of lymph node metastasis. The right figure shows the statistical graph of TMEM16A level and the number of lymph node metastasis in patients. The fluorescence values (MFI) of all samples were sorted numerically from smallest to largest based on the relative values of fluorescence intensity. High/low TMEM16A expression was distinguished according to MFI quartiles. Lower quartile Q1 was 31.145, denoted as " + ". Median Q2 was 38.506, and upper quartile Q3 was 43.821, denoted as " + + ". Samples with MFI greater than 43.821 were labeled as " + + + ". No detectable fluorescence intensity was denoted as “-”
Table 1.
Correlation between clinical information and TMEM16A in tumor tissues of ESCC patients
| Variables | Number (n = 98) | TMEM16Alow (n = 25) | TMEM16Ahigh (n = 73) | χ2 | P |
|---|---|---|---|---|---|
| Gender | 1.545 | 0.487 | |||
| Male | 74 | 19 | 55 | ||
| Female | 24 | 6 | 18 | ||
| Age, years | 0.754 | 0.686 | |||
| < = 60 | 25 | 8 | 17 | ||
| > 60 | 73 | 17 | 56 | ||
| Number of Lymph nodes metastases | 4.859 | 0.037* | |||
| 0 | 48 | 17 | 31 | ||
| 1 or more | 50 | 8 | 42 | ||
| Pathology T stage | 5.063 | 0.536 | |||
| T1 | 1 | 1 | 0 | ||
| T2 | 18 | 5 | 13 | ||
| T3 | 77 | 19 | 58 | ||
| T4 | 1 | 0 | 1 | ||
| No detected | 1 | 0 | 1 |
* P < 0.05
Generation of TMEM16A mAbs and characterization of their specificities
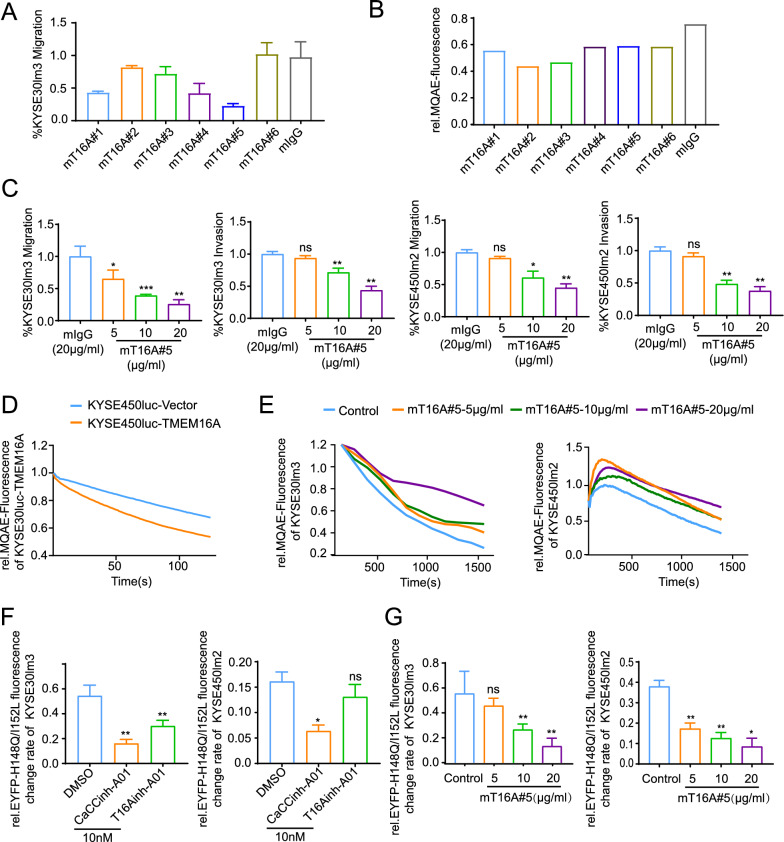
The experimental results outlined above provided evidence supporting TMEM16A as a promising target for anti-metastatic therapy in ESCC. Following immunization of mice with a TMEM16A eukaryotic plasmid, hybridoma fusion, and clonal screening, six strains of antibodies were successfully generated and purified (Supplement Fig S2A). To elucidate the specificity of these antibodies, four GST-tagged extracellular segments of TMEM16A were synthesized and extracted for comparison of binding specificity with the antibodies, which showed that all six mAbs could bind only to the fourth extracellular segment (Supplement Fig S2B, S2C). In order to assess the functional antibodies, we evaluated the inhibitory capacity of six antibodies on cell migration utilizing KYSE30lm3 with high endogenous levels of TMEM16A. Results indicated that mT16A#5 demonstrated the most potent inhibitory effect, with a 75% reduction in cell migration at an antibody concentration of 20 μg/ml (Fig. 2A). The impact of these antibodies on cellular chloride channel activity was determined through the utilization of the MQAE probe. In comparison to the control group, all six mAbs demonstrated inhibition of cellular chloride channel activity (Fig. 2B). Subsequently, a dose-dependent assay was conducted with the mT16A#5 antibody, which displayed the most potent inhibition capability on migration. The findings showed that inhibitory effects of this antibody on cell migration and invasion were dependent on the dosage administered (5 μg/ml to 20 μg/ml) (Fig. 2C, Supplement Fig S2D). The binding specificity of the antibody, along with its recognition specificity for endogenous TMEM16A protein, was validated through the examination of ESCC cell lines and ESCC tumor tissue sections exhibiting varying expression levels. The results clearly showed that mT16A#5 could recognize endogenous TMEM16A (Supplement Fig S2E, S2F). As previously discussed, TMEM16A has been identified as a key player in promoting tumorigenesis by modifying the cellular electrophysiological milieu through modulation of its ion channel activity, as well as facilitating cellular motility via various signaling pathways [16, 18]. Studies have shown that both CaCCinh-A01 and T16Ainh-A01, specific inhibitors of TMEM16A, are capable of suppressing the function of TMEM16A ion channels. Furthermore, it has been observed that inhibition of chloride channel activity with T16Ainh-A01 can lead to the attenuation of cellular metastatic potential in certain cancer types [28, 29]. To further investigate the dose-dependent inhibitory effect of antibody mT16A#5 on chloride channel activity, KYSE30luc with exogenous overexpression of TMEM16A and its control counterpart (Supplement Fig S2G) were assessed using an MQAE probe. It was observed that the overexpression of TMEM16A in cell lines treated with antibody mT16A#5 exhibited a consistent duration of detection following the addition of ATP. Additionally, the rapid quenching of blue fluorescence on the membrane surface of the TMEM16A-overexpressing cell line compared to the control cell line was noted at the same time period after the addition of ATP, suggesting that overexpression of TMEM16A could promote the increase of the cellular chloride channel activity (Fig. 2D, Supplement S2H). This finding further confirmed the stability of the assay system for detecting TMEM16A chloride channel activity. The concentration gradient treatment experiments with the mT16A#5 antibody revealed a concentration-dependent inhibitory effect on cellular chloride channel activity at concentrations of 10–20 μg/ml (Fig. 2E). To validate this inhibitory effect, we introduced the halide-sensitive cytoplasmic fluorescent sensor EYFP-H148Q/I152L into ESCC cell lines and established an assay system to assess the impact of CaCCinh-A01 and T16Ainh-A01 on TMEM16A channel activity. We successfully achieved stable overexpression of the halide ion sensor protein EYFP-H148Q/I152L in ESCC cell lines (Supplement Fig S2I) and established an assay system to assess the inhibitory impact of CaCCinh-A01 and T16Ainh-A01 on TMEM16A chloride channel function (Fig. 2F). Subsequently, we utilized this system to investigate the influence of the antibody mT16A#5 on cellular chloride channel activity. Our findings were in line with those obtained using the MQAE probe, indicating that the inhibitory effect of antibody mT16A#5 on cellular chloride channel activity is contingent upon concentration (Fig. 2G). These results suggested that mT16A#5 exhibits dose-dependent inhibition on ESCC cell migration, invasion, and chloride channel activity.
Fig. 2.
Inhibition of cell migration, invasion and chloride channel activity by TMEM16A-specific antibody mT16A#5. A Transwell chamber assays were employed to compared the ability of 6 anti-TMEM16A monoclonal antibodies to inhibit migration of ESCC cell lines. B MQAE probe fluorescence quenching assay were compared the inhibitory ability of 6 anti-TMEM16A monoclonal antibodies on chloride channel activity of ESCC cell lines. C Transwell chamber assays and Matrigel invasion assay demonstrated the mT16A#5 inhibited the cell motility in a concentration-dependent manner. D MQAE probe detected significant up-regulation of chloride channel activity with KYSE30luc cells overexpressing TMEM16A. E MQAE probe detected the concentration-dependent inhibitory effect of mT16A#5 on chloride channel activity in KYSE30lm3 and KYSE450lm2 cells. F The halide-sensitive cytoplasmic fluorescent sensor EYFP-H148Q/I152L detected the inhibitory effect of TMEM16A inhibitors CaCCinh-A01 and T16Ainh-A01 on cellular chloride ion activity at a concentration of 10 nM. G EYFP-H148Q/I152L system detected the concentration-dependent inhibitory effect of mT16A#5 on cellular chloride channel activity of KYSE30lm3 and KYSE450lm2 cells. *P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05
The reciprocal regulatory effects between TMEM16A and EGFR
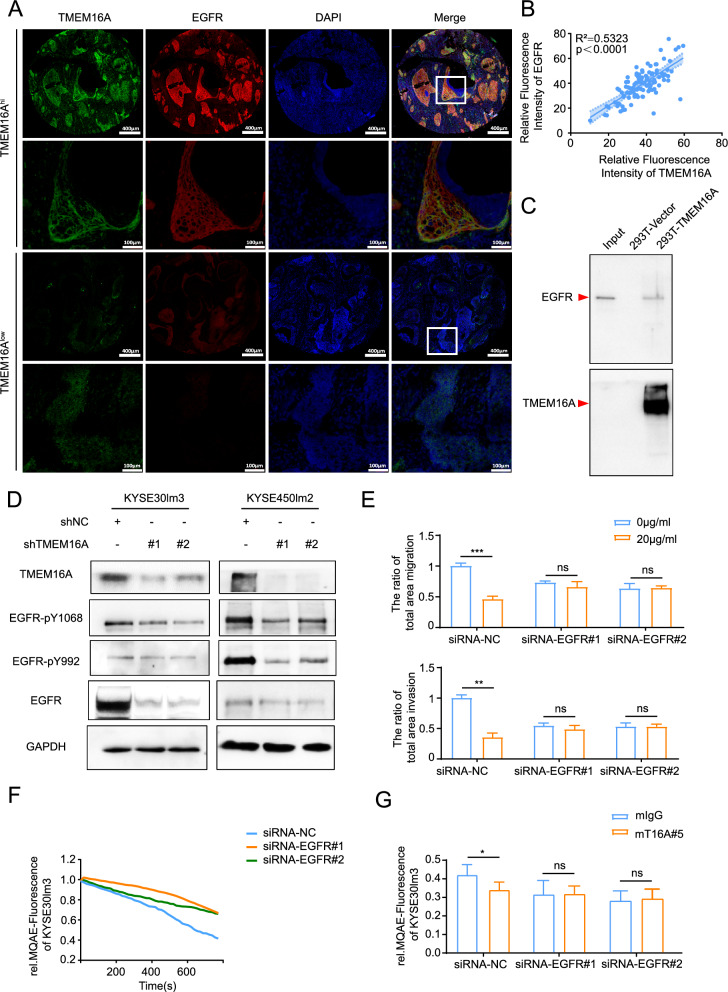
By carrying out cytological experiments, we have determined that the monoclonal antibody mT16#5 exhibits inhibitory effects on cell motility and chloride channel activity. This prompts that targeting TMEM16A with this antibody may hold promise for treating metastasis in ESCC. In contrast, the efficacy of targeted antibody therapy for ESCC metastasis, such as targeting EGFR, HER2, and PD-1, has shown limited effectiveness in clinical trials [1, 3, 5]. It is well established that TMEM16A plays a role in promoting tumor growth, metastasis, and angiogenesis through the activation of various signaling pathways, including EGFR, CAMKII, P38, and Ras [21, 23, 24]. Of these pathways, the activation of the EGFR is closely associated with the metastasis of ESCC. The interaction between EGFR and TMEM16A has been observed in multiple cancer types, including breast cancer [25]. To explore the potential correlation and regulatory relationship between EGFR and TMEM16A in ESCC, a preliminary analysis was conducted to confirm the transcriptional level expression correlation using data from the TCGA database (Supplement Fig S3A). Subsequently, the protein levels of EGFR and TMEM16A were examined in tumor tissues obtained from 98 ESCC patients, revealing a significant positive correlation as anticipated (Fig. 3A, B). Further investigation was carried out to explore the regulatory interaction between EGFR and TMEM16A in ESCC, as well as the potential synergistic effects of targeting both proteins with specific antibodies. Co-localization of EGFR and TMEM16A was demonstrated using confocal laser scanning microscopy (Supplement Fig S3B). Pull-down and CO-IP assays further validated the interaction between EGFR and TMEM16A (Fig. 3C, Supplement Fig S3C). The knockdown of TMEM16A in these cells resulted in a decrease in total EGFR protein levels, with differential down-regulation of phosphorylated EGFR proteins pY1068 and pY992, albeit to a lesser extent than the decrease of total protein (Fig. 3D, Supplement Fig S3D). This suggested that TMEM16A plays a role in maintaining EGFR protein levels. Additionally, depletion of TMEM16A resulted in a significant reduction in EGFR at the transcriptional level (Supplement Fig S3E). Treatment with the anti-TMEM16A antibody mT16A#5 in EGFR knockdown and control cells revealed further insights. Upon treatment with the mT16A#5, it was observed that the inhibitory effects on the migration and invasion of EGFR knockdown cells were minimal at a concentration of 20 μg/ml compared to control cells (Fig. 3E). These results suggested that the impact of mT16A#5 on cell motility is dependent on EGFR expression. Next, we detected the migratory and invasive ability of cells under double knockdown of EGFR and TMEM16A. The inhibitory effect of knocking down EGFR on cell migration and invasion was weaker than that of knocking down TMEM16A on cell migration and invasion, while double knockdown of EGFR and TMEM16A had the strongest inhibitory effect on cell migration invasion (Supplement Fig S3F, S3G). This may be due to the differences in the downstream signaling pathways that the two act on, thus exerting a combined effect on the inhibition of cell movement. Interestingly, EGFR knockdown led to reduced chloride channel activity in KYSE30lm3 cells, and mT16A#5 failed to inhibit chloride channel activity upon EGFR knockdown, suggesting a potential regulatory function of EGFR in ion channel activity of TMEM16A (Fig. 3F, G).
Fig. 3.
Mutual regulation between TMEM16A and EGFR. A, B Detection of TMEM16A and EGFR levels in tumors and adjacent normal tissues of 98 ESCC patients. A mIHC staining of tissues from patients with ESCC patients with fluorescent dye coupled to excitation light of 520 nm for the EGFR antibody, fluorescent dye coupled to excitation light of 650 nm for the TMEM16A antibody, and DAPI was used for staining nuclei. B The expression of TMEM16A and EGFR in ESCC tissues was positively correlated with each other. The staining results were statistically analyzed, and the spectral splitting and fluorescence semi-quantification were carried out after removing the background value of autofluorescence in the tissues. C Pull-down assay detected the interaction of TMEM16A with EGFR extracellular segment protein (aa 668–1210) in 293 T cells overexpressing TMEM16A. EGFR extracellular segment was used as Input. D Knockdown of TMEM16A down-regulated EGFR protein levels. E The inhibitory effect of mT16A#5 on migration and invasion of KYSE30lm3 cells was dependent on EGFR expression. F MQAE system assay showed that knockdown of EGFR was able to down-regulate the chloride channel activity of KYSE30lm3 cells. G. Effect of antibodies mIgG (20 μg/ml) and mT16A#5 (20 μg/ml) on the chloride channel activity in EGFR knockdown on KYSE30lm3 by the MQAE probe. **P < 0.01, ***P < 0.001, ns P > 0.05
Synergistic effect of mT16A#5 and Cetuximab on the inhibition of ESCC metastasis and growth
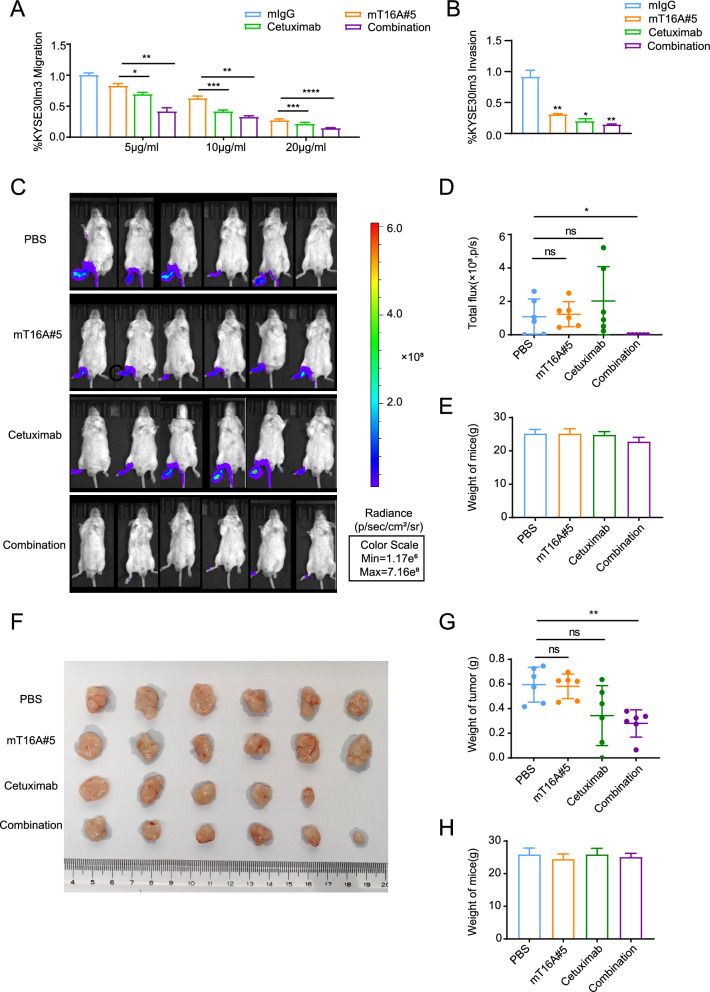
After establishing the correlations and interactions between EGFR and TMEM16A in ESCC, as well as the dependence of mT16#5-inhibited motility on EGFR, we postulated that the combination of mT16#5 and Cetuximab, an EGFR-targeting antibody, would exhibit a synergistic effect in inhibiting metastasis in ESCC. We found that co-administration of equal concentrations of mT16#5 and Cetuximab yielded a more pronounced inhibitory effect on cell migration and invasion compared to either antibody alone, confirming a synergistic effect in these assays (Fig. 4A, B, Supplement Fig S4A, 4B, 4C). Subsequently, a mouse model of ESCC was utilized to confirm this synergistic effect. A footpad metastasis model was established in mice, followed by treatment with mT16A#5 antibody and Cetuximab. After four weeks of consecutive treatment, almost no footpad tumors were detected in mice in the combination treatment group, whereas almost all tumors in mice in the other groups had footpad metastases, revealing that the combination treatment had a synergistic effect on inhibiting ESCC metastasis (Fig. 4C, D, Supplement Fig S4D). Although we did not observe a significant inhibitory effect of mT16A#5 on cell proliferation in the in vitro experiments, the effect of the combination of Cetuximab and mT16A#5 on cell proliferation showed a tendency to suppress the proliferation (Supplement Fig S4E). This may be due to the homogeneous cellular environment present in in vitro experiments, in contrast to the tumor microenvironment where antibodies exert tumor-inhibitory effects through multiple signaling pathways. Consequently, we conducted combination therapy on an animal model of esophageal squamous carcinoma. In the treatment of subcutaneous hormonal tumors in mice, only one mouse in the cetuximab treatment group did not develop a tumor, while the average weight of subcutaneous tumors in the combination treatment group was lower than in each of the single treatment group, therefore the combination of antibodies had a synergistic effect on the inhibition of ESCC growth (Fig. 4F, G). Similar to the results of metastasis model, there was no significant difference in body weight was detected between the four groups, suggesting treatment with these antibodies was well tolerated (Fig. 4E, H).
Fig. 4.
Synergistic effects of mT16A#5 and Cetuximab on the inhibition of ESCC metastasis and growth. A, B Combination of mT16A#5 and Cetuximab inhibited migration and invasion of KYSE30lm3 cells. A Transwell chamber assays. B Matrigel invasion assays. C–E mT16A#5 and Cetuximab have synergistic effect on inhibiting footpad metastasis of ESCC in mice. C KYSE30lm3 cells were injected subcutaneously into the footpad of the right hindfoot of mice, and whole-body imaging was performed on each group of mice after four weeks of consecutive treatment to observe the metastasis of the footpad tumor. D Statistical graph of fluorescence intensity of each mouse foot. E Statistical graph of body weight of mice in each group at the end of treatment. F–H The mT16A#5 and Cetuximab had a synergistic effect on the inhibition of subcutaneous ESCC tumor growth in mice. F KYSE30lm3 cells were injected into the subcutaneous part of the right coeliac region of mice. The mice were executed after four weeks of treatment, and the tumors were stripped and observed for growth. G Statistical graph of subcutaneous tumor mass of mice in each group at the end of treatment. H Statistical graph of body weight of mice in each group at the end of treatment *P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05
Antibody mT16A#5 inhibits phosphorylation of PI3K-AKT/JNK signaling pathway
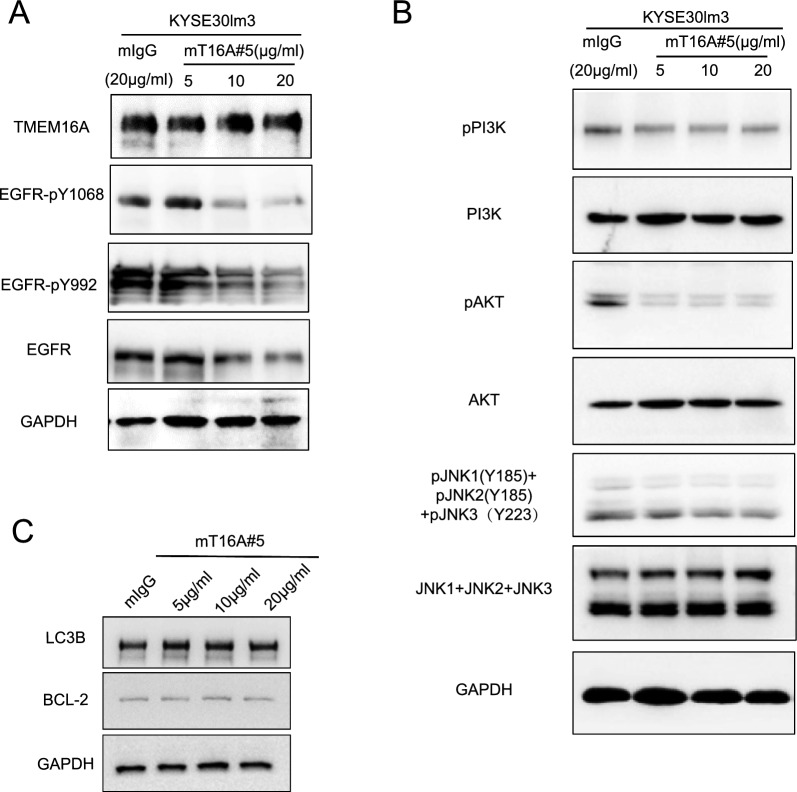
Given the interplay between TMEM16A and EGFR, we postulated that mT16A#5 may affect EGFR-related signaling pathway. Western blot analysis revealed changes in total EGFR cellular protein and phosphorylated protein levels in cells treated with mT16#5 compared to mIgG. Notably, a decrease in total EGFR protein levels of cells treated by mT16#5 was observed (Fig. 5A, Supplement Fig S5A). However, there was no significant alteration in the transcriptional levels of both TMEM16A and EGFR (Supplement Fig S5B). It was hypothesized that mT16#5 may reduce the overall EGFR protein content by facilitating EGFR degradation. That is, mT16#5 inhibited the receptor activation induced by EGF to a certain extent. Through RNA transcriptome sequencing of KYSE30lm3 and KYSE30luc cells, we identified target molecules with high expression in the downstream signaling pathway of EGFR, such as TNF signaling pathway and MAPK signaling pathway (Supplement Fig S5C). Based on the relevance of these signaling pathways to inflammation, we detected expression of a subset of inflammatory factors in KYSE30lm3 and KYSE30luc cells. Transcript levels of target genes which belong to these signaling pathways were quantified by qRT-PCR (Supplement Fig S5D, S5E). Additionally, protein levels in the cell culture media were quantified by ELISA (Supplement Fig S5F). Results of qRT-PCR and ELISA verified the difference in the inflammatory factors between these two cell lines. Subsequent analysis of the related proteins and their phosphorylation in mT16#5-treated cells revealed a down-regulation in the phosphorylation levels of PI3K, AKT, and JNK1,2,3 proteins (Fig. 5B). Hence, it was hypothesized that mT16#5 may exert its inhibitory effect on tumor cell motility by modulating the PI3K-AKT/JNK signaling pathway. Based on the role of the PI3K-AKT and MAPK signaling pathways in regulating autophagy, we investigated the impact of mT16A#5 on autophagy in KYSE30lm3 cells. The analysis of autophagy markers LC3B and Beclin2 revealed that mT16A#5 did not significantly inhibit cellular autophagy (Fig. 5C).
Fig. 5.
mT16A#5 inhibits phosphorylation of PI3K-AKT/JNK signaling pathway. A Western Blot detected the effect of antibody mT16A#5 on the levels of EGFR and its phosphorylated protein level in ESCC cells. B Western Blot detected the effect of antibody mT16A#5 on the total and phosphorylation protein level of PI3K-AKT/JNK signaling pathway related proteins in ESCC cells. C Western Blot detected the effect of antibody mT16A#5 on the LC3B and BCL-2 protein in KYSE30lm3
Discussion
Prior to its identification as a chloride channel, TMEM16A was utilized as a prominent biomarker for gastrointestinal gliomas in clinical diagnostics [42]. Through extensive research on various tumor types, it was discovered that TMEM16A exhibited elevated expression levels due to copy number abnormalities in a range of tumors, with a significant correlation of the low survival rate. The elevated expression of TMEM16A in ESCC tumor tissues has been found to be strongly associated with a negative prognosis, prompting researchers to consider it as a promising target for diagnostic and therapeutic interventions [43, 44]. Despite ongoing efforts to develop novel drugs targeting TMEM16A for inhibiting tumor metastasis, progress in this area remains at an early stage. While small molecule inhibitors CaCCinh-A01 and T16Ainh-A01 have been extensively investigated for their potential in tumor therapy, they have not yet progressed to clinical trials [1, 2, 45]. In this study, we for the first time successfully generated a functional antibody targeting TMEM16A that has the ability to inhibit cellular chloride currents and can inhibit ESCC metastasis after being combined with Cetuximab.
As a calcium-dependent chloride channel, TMEM16A has garnered significant interest in the context of diseases characterized by current dysregulation. The development of inhibitors for this channel was initially motivated by the need to suppress abnormal cellular discharges and treat associated conditions. Consequently, numerous studies have been conducted to identify TMEM16A inhibitors through screening for compounds that inhibit cellular chloride ions. Notable examples include natural compounds such as tannins, lignans, and eugenol, as well as the synthetic compound ZAF, which has received approval for clinical use in treating asthma [31, 32, 46]. As oncology research advances and targeted therapies become more prevalent, researchers have revised the screening criteria for TMEM16A inhibitors to focus on their inhibitory effects on tumor cell phenotypes. This has led to the identification of specific inhibitors capable of inhibiting tumor cell proliferation, migration, invasion, and inducing apoptosis. For example, compound CaCCinh-A01, known for its effectiveness as a TMEM16A blocker, not only inhibits chloride channel activity but also promotes the degradation of TMEM16A proteins. Another inhibitor, compound T16Ainh-A01, has demonstrated the ability to inhibit the proliferation of gastric cancer cells and pancreatic cancer cell lines [28, 29]. It is noteworthy that CaCCinh-A01, despite exhibiting a more potent inhibitory effect on TMEM16A chloride channel activity at equimolar concentrations, demonstrated minimal inhibitory effect on tumor growth [47, 48]. Consequently, there appears to be no direct correlation between the suppression of chloride channel activity and the extent of inhibition of malignant tumor phenotypes for inhibitors. This phenomenon parallels the attributes of the specific antibodies generated in this study, particularly antibody mT16A#5. That is, while mT16A#5 exhibits the most potent capacity to inhibit the malignant phenotype of the tumor, it does not demonstrate the highest inhibitory effect on TMEM16A chloride channel activity. This discrepancy can be attributed to the fact that the phosphorylation level of EGFR and the activity of the PI3K-AKT/JNK signaling pathway represent only a subset of the numerous factors influencing cellular metastasis and ion channel activity. The activity of the TMEM16A ion channel is influenced not only by signaling pathways implicated in cellular metastasis but also by the exposure of its calcium binding site and the activation of various receptors. The differential inhibition of TMEM16A ion channel activity by distinct antibodies may be attributed to their varying capacities to obscure the number of TMEM16A calcium binding sites and receptors.
EGFR is a tyrosine kinase receptor situated on the cell membrane, and its mediated signaling plays a pivotal role in tumor development [49, 50]. Upon binding to ligands such as EGF, EGFR undergoes dimerization and subsequently activates intracellular domain tyrosine kinase activity, initiating downstream cell signaling pathways to facilitate transmembrane signal transduction. Previous studies have documented the direct or indirect interaction between EGFR and TMEM16A, which plays a crucial role in mediating the activation of the MAPK signaling pathway. Animal model research on lung cancer has demonstrated that TMEM16A can enhance tumor growth and metastasis in lung cancer through the EGFR/MAPK-dependent signaling pathway [22, 24]. Furthermore, the EGF-initiated EGFR/STAT3 signaling pathway has been shown to induce the upregulation of TMEM16A, thereby activating STAT3 signaling in breast cancer cells [27]. Nevertheless, the potential interaction between TMEM16A and EGFR in ESCC remains unexplored. In the study, the relationship between transcript levels of TMEM16A and EGFR content in ESCC has been investigated using the TCGA database. Additionally, we have conducted mIHC analysis to confirm the positive correlation between TMEM16A and EGFR expression in tumor tissues from patients with ESCC for the first time. We validated the role of TMEM16A in regulating EGFR transcription and overall protein levels using an ESCC cell model. During our investigation into the mechanism of action of the antibody mT16A#5, we discovered that mT16A#5 could inhibit the PI3K-AKT/JNK signaling pathway. The critical role of the PI3K-AKT signaling pathway in tumor metastasis has been well-documented in the literature. Specifically, TMEM16A has been shown to enhance MAPK/AKT signaling by activating the EGF receptor and other pathways, thereby creating a positive feedback loop and promoting the further expression of EGFR [23, 24, 51]. This result suggested that TMEM16A may participate in the regulation of EGFR expression by activating PI3K-AKT/JNK, and enhance its interaction with EGFR.
Based on the interaction of TMEM16A with EGFR and the high expression of both in ESCC, as well as the phenomenon that inhibition of TMEM16A improves the response to antibody-mediated EGFR-targeted therapy as indicated by Sucheta et al., TMEM16A may play a crucial role in the response to antibody-mediated EGFR-targeted therapy [50]. The present study was carried out with the combination of mT16A#5 and Cetuximab. Inhibition of TMEM16A has been shown to enhance the therapeutic efficacy of Cetuximab treatment, as combined inhibition of migratory invasion of ESCC cells to a greater extent than either component alone at the same concentrations. Subsequent animal studies have corroborated these observations. This suggests that simultaneous inhibition of EGFR and TMEM16A has a better synergistic anti-tumor effect. Cetuximab has been shown to bind to EGFR, forming an antibody-receptor complex that leads to intracellular degradation and downregulation of EGFR. In this study, it was observed that knockdown of EGFR resulted in decreased TMEM16A protein levels. The combination of Cetuximab and mT16#5 exhibited a synergistic effect, likely attributed to mT16#5 reducing cellular TMEM16A chloride channel activity and exerting a tumor-suppressing effect through the PI3K-AKT/JNK signaling pathway. Additionally, the utilization of Cetuximab not only demonstrated binding inhibition of the EGFR ligand receptor, but also potentiating the suppression of cellular chloride channel activity and PI3K-AKT/JNK signaling pathway by mT16#5.
In conclusion, our study not only validated the potential of TMEM16A as a therapeutic target for metastatic ESCC through analysis of clinicopathological samples, but also demonstrated the efficacy of a functional monoclonal antibody against TMEM16A in inhibiting metastatic ESCC. Additionally, we conducted preliminary investigations into the mechanism of action of this antibody. Therapeutic antibodies targeting metastatic cancer through inhibiting TMEM16A have not been previously documented, thus this study introduces a novel idea for antibody therapy targeting TMEM16A in ESCC.
Supplementary Information
Supplementary material 1: Fig S1 Effects of TMEM16A on cell proliferation and Migration. A. TMEM16A protein level in KYSE30lm3 and KYSE450lm2 cells withTMEM16A knockdown. GAPDH was used as a loading control. B. Transwell chamber assays and Matrigel invasion assays for KYSE30lm3 and KYSE450lm2 cells after TMEM16A knockdown. C. Cell proliferation assay kit detects proliferation ability of KYSE30lm3 and KYSE450lm2 cells withTMEM16A knockdown. D. TMEM16A protein level in KYSE450luc and KYSE410 cells withTMEM16A overexpression. GAPDH was used as a loading control. E. Transwell chamber assays and Matrigel invasion assays for KYSE450luc and KYSE410 cells overexpressing TMEM16A. F. Cell proliferation assay kit detects proliferation ability of KYSE450luc and KYSE410 cells withTMEM16A overexpression. *P<0.05, **P<0.01, ***P<0.001. Fig S2 Preparation and functional assay of TMEM16A antibody. A. Anti-TMEM16A monoclonal antibody mT16A#1-mT16A#16 stained with Coomassie Brilliant Blue. B. Purification and quantification of four GST-tagged extracellular segments of TMEM16A proteins. C. ELISA was performed to detect the specificity of six anti-TMEM16A monoclonal antibodies recognizing the four extracellular segments of TMEM16A. D. Antibody mT16A#5 concentration-dependently inhibited the migration and invasion of KYSE30luc overexpressing TMEM16A. E. IHC detected the mT16A#5’s binding specificity to endogenous TMEM16A in ESCC cells. mIgG was used as control. F. IHC detected the antibody mT16A#5’s binding specificity to endogenous TMEM16A in ESCC tumor tissues. mIgG was used as control. G. Western Blot detected the TMEM16A protein level after overexpression of TMEM16A in KYSE30luc cells. GAPDH was used as a loading control. H. MQAE probe was used to detect the inhibitory effect of TMEM16A inhibitor CaCCinh-A01 on the chloride activity of KYSE30lm3 cells at a concentration of 10 nM. I. Expression efficiency assay of the halide-sensitive cytoplasmic fluorescent sensor EYFP-H148Q/I152L stably transfected in KYSE30lm3 cells. *P<0.05, **P<0.01, ***P<0.001. Fig S3 TMEM16A has interaction with EGFR. A. Correlation prediction of TMEM16A and EGFR transcript levels in tumor tissues of ESCC patients in TCGA database. B. Cell membrane co-localization of endogenous TMEM16A and EGFR in KYSE30lm3 cells. C. TMEM16A and EGFR interaction detected by CO-IP assay. D. Grey scale values of total protein and phosphorylated protein content of EGFR after knockdown of TMEM16A detected by Western Blot. in Fig 3D. E. TMEM16A and EGFR transcript levels after knockdown of TMEM16A. F. Western Blot detected the TMEM16A and EGFR protein level after dual silencing of EGFR and TMEM16A in KYSE30luc cells. GAPDH was used as a loading control. G. Transwell chamber assays and Matrigel invasion assays for KYSE30lm3 and KYSE450lm2 cells after dual silencing of EGFR and TMEM16A. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Fig S4 Synergistic effects of mT16A#5 and Cetuximab on the inhibition of ESCC metastasis. A. Migration assay to detect the inhibitory effect of the combination of mT16A#5 and Cetuximab on esophageal squamous carcinoma cells KYSE30lm3. B. Inhibitory effect of combination of mT16A#5 and Cetuximab on esophageal squamous carcinoma cells KYSE30lm3 detected by invasion assay. C. Inhibition of KYSE450lm2 in ESCC cells by the combination of mT16A#5 and Cetuximab as detected by migration assay and invasion assay. D. KYSE30lm3 cells were injected subcutaneously into the foot pads of the right hind feet of mice, and foot pad tumor metastasis was observed after four weeks of continuous treatment. E. Inhibition of KYSE30lm3 in ESCC cells by the combination of mT16A#5 and Cetuximab as detected by CCK8 assay. The antibody mT16A#5 had no effect on cell proliferation in the range of tested concentration, and the combination of mT16A#5 and Cetuximab have showed trend of proliferation inhibition on KYSE30lm3. mIgG, mT16A#5, Cetuximab, combination. *P<0.05, **P<0.01, ns P>0.05. Fig S5 mT16A#5 inhibits phosphorylation of PI3K-AKT/JNK signaling pathway. A. Grayscale scanning of protein levels in Fig 5A. B. Detection of changes of TMEM16A and EGFR transcript levels in cells treated with the antibody mT16A#5. C. KEGG enrichment analysis of DE genes in the KYSE30lm3VS KYSE30luc. D. Statistical graph of number of DEGs in the KYSE30lm3 VS KYSE30luc. E. Transcript levels of inflammatory factors in the KYSE30luc and KYSE30lm3 cells were quantified by qRT-PCR. F. Protein levels of inflammatory factors in the KYSE30luc and KYSE30lm3 cell culture media were quantified by ELISA. *P<0.05, **P<0.01,***P<0.001,ns P>0.05.
Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFC2501001) and funding from Peking University Cancer Hospital & Institute (A002207). We are very grateful to Professor Zhihua Liu (Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing) for gifting KYSE30luc/KYSE30lm3 and KYSE450luc/KYSE450lm2 cells.
Author contributions
YZ performed the research, data analysis, visualization and wrote the manuscript. LM performed the project administration, resources and methodology. LQ performed the conceptualization, methodology and manuscript review and revision. CZ performed the supervision. LW performed the resources. JM performed the validation. CL and CS performed the funding acquisition. CS developed the idea for the study.
Data availability
All relevant data are within the manuscript and its supplementary material files.
Declarations
Competing interests
The authors have declared that no competing interest exists.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Caiyun Liu, Email: liucaiyun23@163.com.
Chengchao Shou, Email: cshou@vip.sina.com.
References
- 1.Zhu H, Ma X, Ye T, et al. Esophageal cancer in China: practice and research in the new era. Int J Cancer. 2023;152(9):1741–51. [DOI] [PubMed] [Google Scholar]
- 2.He J, Chen WQ, Li ZS, et al. Expert group of china guideline for the screening, early detection and early treatment of esophageal cancer; work group of china guideline for the screening, early detection and early treatment of esophageal cancer. [China guideline for the screening, early detection and early treatment of esophageal cancer (2022, Beijing)]. Zhonghua Zhong Liu Za Zhi. 2022 Jun 23;44(6):491–522 [DOI] [PubMed]
- 3.Li SW, Zhang LH, Cai Y, et al. Deep learning assists detection of esophageal cancer and precursor lesions in a prospective, randomized controlled study. Sci Transl Med. 2024;16(743):5395. [DOI] [PubMed] [Google Scholar]
- 4.Guo D, Sheng K, Zhang Q, et al. Single-cell transcriptomic analysis reveals the landscape of epithelial-mesenchymal transition molecular heterogeneity in esophageal squamous cell carcinoma. Cancer Lett. 2024;10(587): 216723. [DOI] [PubMed] [Google Scholar]
- 5.Kato K, Doki Y, Chau I, et al. Nivolumab plus chemotherapy or ipilimumab versus chemotherapy in patients with advanced esophageal squamous cell carcinoma (CheckMate 648): 29-month follow-up from a randomized, open-label, phase III trial. Cancer Med. 2024;13(9): e7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Z, Zhang Y, Fan Q, et al. Paclitaxel and cisplatin with or without cetuximab in metastatic esophageal squamous cell carcinoma: a randomized, multicenter phase II trial. Innovation. 2022;3(3): 100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang YM, Hong P, Xu WW, et al. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. 2020;5(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbaum MW, Gonzalez RS. Targeted therapy for upper gastrointestinal tract cancer: current and future prospects. Histopathology. 2021;78(1):148–61. [DOI] [PubMed] [Google Scholar]
- 9.Zhao D, Guo Y, Wei H, et al. Multi-omics characterization of esophageal squamous cell carcinoma identifies molecular subtypes and therapeutic targets. JCI Insight. 2024;23: 171916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, Wu C, Zhao J, et al. Camrelizumab, chemotherapy and apatinib in the neoadjuvant treatment of resectable oesophageal squamous cell carcinoma: a single-arm phase 2 trial. EClinical Medicine. 2024;6(71): 102579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Bao W, Zhu X, et al. Molecular characteristics and multivariate survival analysis of 43 patients with locally advanced or metastatic esophageal squamous cell carcinoma. J Thorac Dis. 2024;16(3):1843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Li Y, Wang L, et al. SPP1 represents a therapeutic target that promotes the progression of oesophageal squamous cell carcinoma by driving M2 macrophage infiltration. Br J Cancer. 2024. 10.1038/s41416-024-02683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Wang J, Shi Y, et al. Cathepsin L promotes oesophageal squamous cell carcinoma development and may be associated with tumour-associated macrophages. Heliyon. 2024;10(7):29273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prevarskaya N, Skryma R, Shuba Y. Ion channels in cancer: are cancer hallmarks oncochannel opathies? Physiol Rev. 2018;98(2):559–621. [DOI] [PubMed] [Google Scholar]
- 15.Shi S, Pang C, Guo S, et al. Recent progress in structural studies on TMEM16A channel. Comput Struct Biotechnol J. 2020;21(18):714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Q, Guo S, Wang X, et al. Recent advances in TMEM16A: Structure, function, and disease. J Cell Physiol. 2019;234(6):7856–73. [DOI] [PubMed] [Google Scholar]
- 17.Lim NK, Lam AK, Dutzler R. Independent activation of ion conduction pores in the double-barreled calcium-activated chloride channel TMEM16A. J Gen Physiol. 2016;148(5):375–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Yu Z, Wang H, et al. Role of ANO1 in tumors and tumor immunity. J Cancer Res Clin Oncol. 2022;26:12–8. [DOI] [PubMed] [Google Scholar]
- 19.Duran C, Hartzell HC. Physiological roles and diseases of TMEM16/Anoctamin proteins: are they all chloride channels? Acta Pharmacol Sin. 2011;32(6):685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Zou L, Ma K, et al. Cell-specific mechanisms of TMEM16A Ca2+-activated chloride channel in cancer. Mol Cancer. 2017;16(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta AK, Boggs K, Khimji AK, et al. Signaling through the interleukin-4 and interleukin-13 receptor complexes regulates cholangiocyte TMEM16A expression and biliary secretion. Am J Physiol Gastrointest Liver Physiol. 2020;318(4):G763–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo S, Zhang L, Li N. ANO1: more than just calcium-activated chloride channel in cancer. Front Oncol. 2022;6(12): 922838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britschgi A, Bill A, Brinkhaus H, et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci USA. 2013;110(11):e1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bill A, Gutierrez A, Kulkarni S, et al. ANO1/TMEM16A interacts with EGFR and correlates with sensitivity to EGFR-targeting therapy in head and neck cancer. Oncotarget. 2015;6(11):9173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lev S. Targeted therapy and drug resistance in triple-negative breast cancer: the EGFR axis. Biochem Soc Trans. 2020;48(2):657–65. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Zhao D, Spring DJ, et al. Genetics and biology of prostate cancer. Genes Dev. 2018;32(17–18):1105–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Yao F, Luo S, et al. A mutual activation loop between the Ca2+-activated chloride channel TMEM16A and EGFR/STAT3 signaling promotes breast cancer tumorigenesis. Cancer Lett. 2019;455:48–59. [DOI] [PubMed] [Google Scholar]
- 28.Song Y, Gao J, Guan L, et al. Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-α signaling. Cell Death Dis. 2018;9(6):703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JY, Youn HY, Choi J, et al. Anoctamin-1 affects the migration and invasion of anaplastic thyroid carcinoma cells. Anim Cells Syst. 2019;23(4):294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi S, Guo S, Chen Y, et al. Molecular mechanism of CaCCinh-A01 inhibiting TMEM16A channel. Arch Biochem Biophys. 2020;695:108650. [DOI] [PubMed] [Google Scholar]
- 31.Shi S, Ma B, Sun F, et al. Zafirlukast inhibits the growth of lung adenocarcinoma via inhibiting TMEM16A channel activity. J Biol Chem. 2022;298(3):101731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo Y, Jeong SB, Woo JH, Kwon OB, Lee S, Oh HI, Jo S, Park SJ, Namkung W, Moon UY, Lee S. Diethylstilbestrol, a novel ANO1 inhibitor, exerts an anticancer effect on non-small cell lung cancer via inhibition of ANO1. Int J Mol Sci. 2021;22(13):7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imran M, Rauf A, Abu-Izneid T, et al. Luteolin, a flavonoid, as an anticancer agent: a review. Biomed Pharmacother. 2019;112:108612. [DOI] [PubMed] [Google Scholar]
- 34.Wadhwa K, Pahwa R, Kumar M, et al. Mechanistic insights into the pharmacological significance of silymarin. Molecules. 2022;27(16):5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X, Huang F, Ma G, et al. Dysregulated ceramides metabolism by fatty acid 2-hydroxylase exposes a metabolic vulnerability to target cancer metastasis. Signal Transduct Target Ther. 2022;7(1):370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson C, Roomans GM. Determination of chloride efflux by X-ray microanalysis versus MQAE-fluorescence. Microsc Res Tech. 2002;59(6):531–5. [DOI] [PubMed] [Google Scholar]
- 37.Kovalchuk Y, Garaschuk O. Two-photon chloride imaging using MQAE in vitro and in vivo. Cold Spring Harb Protoc. 2012;7:778–85. [DOI] [PubMed] [Google Scholar]
- 38.Weilinger NL, Wicki-Stordeur LE, Groten CJ, LeDue JM, Kahle KT, MacVicar BA. KCC2 drives chloride microdomain formation in dendritic blebbing. Cell Rep. 2022;41(4): 111556. [DOI] [PubMed] [Google Scholar]
- 39.Rhoden KJ, Cianchetta S, Stivani V, et al. Cell-based imaging of sodium iodide symporter activity with the yellow fluorescent protein variant YFP-H148Q/I152L. Am J Physiol Cell Physiol. 2007;292(2):814–23. [DOI] [PubMed] [Google Scholar]
- 40.Rhoden KJ, Cianchetta S, Duchi S, et al. Fluorescence quantitation of thyocyte iodide accumulation with the yellow fluorescent protein variant YFP-H148Q/I152L. Anal Biochem. 2008;373:239–46. [DOI] [PubMed] [Google Scholar]
- 41.Jo S, Yang E, Lee Y, Jeon D, Namkung W. Cinobufagin exerts anticancer activity in oral squamous cell carcinoma cells through downregulation of ANO1. Int J Mol Sci. 2021;22(21):12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CH, Liang CW, Espinosa I. The utility of discovered on gastrointestinal stromal tumor 1 (DOG1) antibody in surgical pathology-the GIST of it. Adv Anat Pathol. 2010;17(3):222–32. [DOI] [PubMed] [Google Scholar]
- 43.Yu Y, Cao J, Wu W, et al. Genome-wide copy number variation analysis identified ANO1 as a novel oncogene and prognostic biomarker in esophageal squamous cell cancer. Carcinogenesis. 2019;40(10):1198–208. [DOI] [PubMed] [Google Scholar]
- 44.Li S, Wang Z, Geng R, Zhang W, Wan H, Kang X, Guo S. TMEM16A ion channel: a novel target for cancer treatment. Life Sci. 2023;15(331): 122034. [DOI] [PubMed] [Google Scholar]
- 45.Petrillo A, Smyth EC. Immunotherapy for squamous esophageal cancer: a review. J Pers Med. 2022;12(6):862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Namkung W, Thiagarajah JR, Phuan PW, et al. Inhibition of Ca2+-activated Cl- channels by gallotannins as a possible molecular basis for health benefits of red wine and green tea. FASEB J. 2010;24(11):4178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauter DRP, Novak I, Pedersen SF, Larsen EH, Hoffmann EK. ANO1 (TMEM16A) in pancreatic ductal adenocarcinoma (PDAC). Pflugers Arch. 2015;467(7):1495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fröbom R, Sellberg F, Xu C, Zhao A, Larsson C, Lui WO, Nilsson IL, Berglund E, Bränström R. Biochemical Inhibition of DOG1/TMEM16A achieves antitumoral effects in human gastrointestinal stromal tumor cells In vitro. Anticancer Res. 2019;39(7):3433–42. [DOI] [PubMed] [Google Scholar]
- 49.Levantini E, Maroni G, Del Re M, Tenen DG. EGFR signaling pathway as therapeutic target in human cancers. Semin Cancer Biol. 2022;85:253–75. [DOI] [PubMed] [Google Scholar]
- 50.Kulkarni S, Bill A, Godse NR, Khan NI, Kass JI, Steehler K, Kemp C, Davis K, Bertrand CA, Vyas AR, Holt DE, Grandis JR, Gaither LA, Duvvuri U. TMEM16A/ANO1 suppression improves response to antibody-mediated targeted therapy of EGFR and HER2/ERBB2. Genes Chromosomes Cancer. 2017;56(6):460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crottès D, Lin YT, Peters CJ, Gilchrist JM, Wiita AP, Jan YN, Jan LY. TMEM16A controls EGF-induced calcium signaling implicated in pancreatic cancer prognosis. Proc Natl Acad Sci USA. 2019;116(26):13026–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1: Fig S1 Effects of TMEM16A on cell proliferation and Migration. A. TMEM16A protein level in KYSE30lm3 and KYSE450lm2 cells withTMEM16A knockdown. GAPDH was used as a loading control. B. Transwell chamber assays and Matrigel invasion assays for KYSE30lm3 and KYSE450lm2 cells after TMEM16A knockdown. C. Cell proliferation assay kit detects proliferation ability of KYSE30lm3 and KYSE450lm2 cells withTMEM16A knockdown. D. TMEM16A protein level in KYSE450luc and KYSE410 cells withTMEM16A overexpression. GAPDH was used as a loading control. E. Transwell chamber assays and Matrigel invasion assays for KYSE450luc and KYSE410 cells overexpressing TMEM16A. F. Cell proliferation assay kit detects proliferation ability of KYSE450luc and KYSE410 cells withTMEM16A overexpression. *P<0.05, **P<0.01, ***P<0.001. Fig S2 Preparation and functional assay of TMEM16A antibody. A. Anti-TMEM16A monoclonal antibody mT16A#1-mT16A#16 stained with Coomassie Brilliant Blue. B. Purification and quantification of four GST-tagged extracellular segments of TMEM16A proteins. C. ELISA was performed to detect the specificity of six anti-TMEM16A monoclonal antibodies recognizing the four extracellular segments of TMEM16A. D. Antibody mT16A#5 concentration-dependently inhibited the migration and invasion of KYSE30luc overexpressing TMEM16A. E. IHC detected the mT16A#5’s binding specificity to endogenous TMEM16A in ESCC cells. mIgG was used as control. F. IHC detected the antibody mT16A#5’s binding specificity to endogenous TMEM16A in ESCC tumor tissues. mIgG was used as control. G. Western Blot detected the TMEM16A protein level after overexpression of TMEM16A in KYSE30luc cells. GAPDH was used as a loading control. H. MQAE probe was used to detect the inhibitory effect of TMEM16A inhibitor CaCCinh-A01 on the chloride activity of KYSE30lm3 cells at a concentration of 10 nM. I. Expression efficiency assay of the halide-sensitive cytoplasmic fluorescent sensor EYFP-H148Q/I152L stably transfected in KYSE30lm3 cells. *P<0.05, **P<0.01, ***P<0.001. Fig S3 TMEM16A has interaction with EGFR. A. Correlation prediction of TMEM16A and EGFR transcript levels in tumor tissues of ESCC patients in TCGA database. B. Cell membrane co-localization of endogenous TMEM16A and EGFR in KYSE30lm3 cells. C. TMEM16A and EGFR interaction detected by CO-IP assay. D. Grey scale values of total protein and phosphorylated protein content of EGFR after knockdown of TMEM16A detected by Western Blot. in Fig 3D. E. TMEM16A and EGFR transcript levels after knockdown of TMEM16A. F. Western Blot detected the TMEM16A and EGFR protein level after dual silencing of EGFR and TMEM16A in KYSE30luc cells. GAPDH was used as a loading control. G. Transwell chamber assays and Matrigel invasion assays for KYSE30lm3 and KYSE450lm2 cells after dual silencing of EGFR and TMEM16A. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Fig S4 Synergistic effects of mT16A#5 and Cetuximab on the inhibition of ESCC metastasis. A. Migration assay to detect the inhibitory effect of the combination of mT16A#5 and Cetuximab on esophageal squamous carcinoma cells KYSE30lm3. B. Inhibitory effect of combination of mT16A#5 and Cetuximab on esophageal squamous carcinoma cells KYSE30lm3 detected by invasion assay. C. Inhibition of KYSE450lm2 in ESCC cells by the combination of mT16A#5 and Cetuximab as detected by migration assay and invasion assay. D. KYSE30lm3 cells were injected subcutaneously into the foot pads of the right hind feet of mice, and foot pad tumor metastasis was observed after four weeks of continuous treatment. E. Inhibition of KYSE30lm3 in ESCC cells by the combination of mT16A#5 and Cetuximab as detected by CCK8 assay. The antibody mT16A#5 had no effect on cell proliferation in the range of tested concentration, and the combination of mT16A#5 and Cetuximab have showed trend of proliferation inhibition on KYSE30lm3. mIgG, mT16A#5, Cetuximab, combination. *P<0.05, **P<0.01, ns P>0.05. Fig S5 mT16A#5 inhibits phosphorylation of PI3K-AKT/JNK signaling pathway. A. Grayscale scanning of protein levels in Fig 5A. B. Detection of changes of TMEM16A and EGFR transcript levels in cells treated with the antibody mT16A#5. C. KEGG enrichment analysis of DE genes in the KYSE30lm3VS KYSE30luc. D. Statistical graph of number of DEGs in the KYSE30lm3 VS KYSE30luc. E. Transcript levels of inflammatory factors in the KYSE30luc and KYSE30lm3 cells were quantified by qRT-PCR. F. Protein levels of inflammatory factors in the KYSE30luc and KYSE30lm3 cell culture media were quantified by ELISA. *P<0.05, **P<0.01,***P<0.001,ns P>0.05.
Data Availability Statement
All relevant data are within the manuscript and its supplementary material files.