Abstract
Background
Calcium-dependent protein kinases (CDPKs) phosphorylate downstream target proteins in response to signals transmitted by free calcium ions (Ca2+, one of the second messengers) and thus play important regulatory roles in many biological processes, such as plant growth, development, and stress response.
Results
A bioinformatic analysis, as well as thorough evolutionary and expression investigations, were conducted to confirm previous reports of functional evidence for plant CDPKs. Using the Phytozome database’s BLAST search engine and the HMM search tool in TBtools software, we discovered that CDPKs are well conserved from green algae to flowering angiosperms in various gene family sizes. Additional investigations of the obtained CDPKs revealed high conservation of domain and motif numbers, gene architectures, and patterns. However, this conservation differed among plant species. Phylogenetic analysis demonstrated that the CDPK gene family diverged from a common ancient gene. Similarly, investigations into plant interspecies evolutionary relationships revealed common ancestral plant species, suggesting speciation of plants and evolution based on plant adaptation and diversification. A search for the driving force of CDPK gene family expansion revealed that dispersed duplication events, among other duplication events, contributed largely to CDPK gene family expansion. Gene localization analysis in P. trichocarpa demonstrated that most CDPK genes are localized within several cell organelles and bind other kinases and proteins to perform their biological functions efficiently. Using RNA-seq data and qPCR analyses, we postulated that PtCDPKs play functional roles in abiotic stress responses by regulating cold, heat, drought and salt stress to varying extents.
Conclusion
The CDPK genes are well conserved in plants and are critical entities in abiotic stress regulation, and further exploration and manipulation of these genes in the future may provide solutions to some of the challenges in agriculture, forestry and food security.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10962-3.
Keywords: Calcium-dependent protein kinase, Evolution, Phylogenetics, Transcriptome expression, Abiotic stress responses, Populus
Background
Plants are susceptible to extreme environmental cues and depend on molecular interactions and regulations for adaptation, distribution, growth, and development. In order to acclimate to adverse stress conditions, plants have evolved complex signal transduction pathways governed by several biological entities, including transcription factors (TFs), genes, and other molecules [1]. Notably, calcium ions (Ca2+) act as crucial secondary messengers in signal transduction pathways, conveying stress impulses to four main key calcium-binding proteins - calcineurin B-like proteins (CBLs), calmodulin (CaM), calmodulin-like protein (CML), and calcium-dependent protein kinases (CDPKs) [2, 3]. Previous studies have demonstrated that each of the aforementioned calcium-binding proteins responds to abiotic stresses in plants and interacts with downstream stress response target genes to regulate and alleviate plant stresses [4–6]. Specifically, the CDPKs first reported in peas [7] and later confirmed in soybean [8]to belong to a large multigene family [9, 10]. Research has shown that plant CDPKs are monomeric proteins that have a conserved domain structure. Structural analysis revealed that these proteins are composed of an autoinhibitory domain (A), also referred to as the CDPK activation domain (CAD). CAD is formed from a pseudosubstrate region, and the N-terminal domain determines substrate specificity, providing engineering manipulation capacity for the variable N-terminal domain. Additionally, a regulatory calmodulin-like calcium-binding domain and a variable N-terminal domain (N) connect to a Ser/Thr kinase domain (K) [9]. The C-terminus of CDPKs contains calcium-binding regulatory domains that contain at most five elongation factor (EF)-hand motifs. These EF-hand motifs array into two lobes with diverse Ca2+ affinities, suggesting diverse functional roles of the CDPKs [10]. In addition, the pairing of the EF-hand motifs helps stabilize the protein for Ca2+ binding [11, 12].
Accumulating genomic sequencing and genome-wide analyses of plants have confirmed the importance of CDPK gene structure, gene conservation in plants, and divergence from common ancestral genes. Through selection pressure, some CDPKs have adapted and play essential roles in extant plants. Recent evidence has shown that CDPKs decode internal stimuli or environmental signal responses generated by transient cytosolic calcium signals to induce spatial and temporal expression of CDPKs. However, these expression levels are regulated depending on the extent of external stimuli [13, 14]. Furthermore, CDPK genes can form isoforms that have specific or overlapping functions that are mainly restricted to the N-terminal variable domain. These findings imply that CDPK isoforms or different CDPKs may play multiple roles or are involved in the same biological function. For instance, research in Arabidopsis has shown seven AtCDPKs that play a crucial role in drought and osmotic stresses, including three from Group I (AtCPK4/6/11), three from Group II (AtCPK3/21/23), and one from Group III (AtCPK10). Additional research has shown that these AtCDPKs regulate drought and osmotic stresses by controlling stomatal movements in response to developmental cues and environmental stimuli [15]. CDPKs also regulate other abiotic stresses, including heat, cold, and salt. In detail, at low temperatures, CDPK genes reduce cell damage by transmitting low-temperature stress signals in the form of phosphorylation activation of cold-responsive genes. Thereby activating their transcription and responses.CDPK17 from O. sativa were reportedto increase cold stress tolerance by improving the activity of plant membrane channels and sugar metabolism and interacting with downstream genes such as OsICE and OsCBF [16]. In C. glaucum, CDPKs were shown to phosphorylate CgbHLH001 via posttranslational regulation and to positively regulate cold stress and reduce cell damage [17].
Plant CDPKs also protect the cell membrane from oxidative damage induced by drought stress, consequently regulating drought stress. In Arabidopsis, CDPK3 enhances drought stress resistance by inhibiting the K+ inward channel (KAT1) and stomatal opening and enhancing reactive oxygen species (ROS) detoxification in the cell membrane [18]. These studies provide solid evidence that the CDPK gene family is crucial for regulating plant abiotic stress. In addition, genome-wide and extensively expressed sequence tag projects have identified multigene families of CDPKs from various plants, including wheat [19], rice [20], grape [21], and banana [22]. Previous publications have investigated the evolution and functions of CDPKs in response to external stimuli [6, 23–25]. In this study, we provided an update on the conservation and evolution of plant CDPKs and elucidated the possible abiotic functions of CDPK genes in populus using transcriptome data and qPCR analyses.
Results
CDPK genes in land plants and their conserved characteristics
To gain insight into the distribution of the CDPK genes in plants, we used the BLASTP search tool in the Phytozome v13 database [26] and the HMMER search tool in TBtools. After eliminating redundant and imperfect sequences without full open reading frames, we identified 1124 CDPK protein sequences from 51 land plant species (Table S1). 35 CDPK protein sequences were obtained from our recently acquired Populus deltoides x Populus euramericana ‘Nanlin 895’ transcriptome data. CDPK protein sequences were present in ancient vascular plant lineage members such as Selaginella moellendorffii and green algae members, ranging from eudicots to monocots in varying numbers. We also identified several CDPK proteins in aquatic plants, such as Volvox carteri. Generally, most CDPKs were identified in angiosperms. The full CDPK protein sequences ranged from 181 to 560 amino acids, except in C. reinhardtii, which had five of the protein sequences, CrCPK2/3/4/7/8, carrying 1800, 1979, 1041, 1829, and 1042 amino acids, respectively. This variation in protein length was related to differences in the total number of EF-hands, varying lengths of the N-terminal variable domain (N-VD), and the C-terminus.
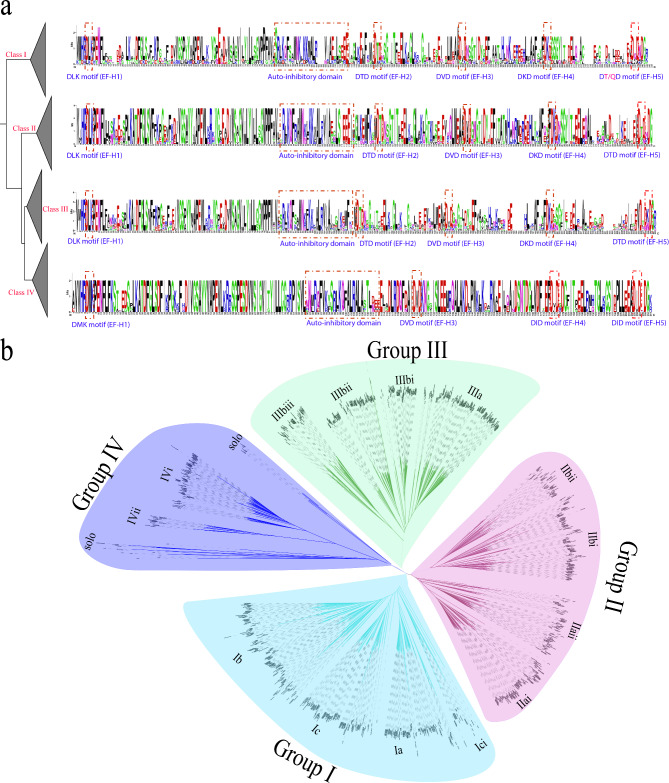
To understand the domain arrangements and patterns in the CDPK plants, we searched for conserved domains and motifs using multiple sequence alignment in Geneious Prime software and the InterPro online tool (Figs S1, S2). Consistent with previous research, we observed a conserved DLK motif in all the CDPK protein sequences toward the N-terminus. Furthermore, the CDPKs contained an autoinhibitory domain, which was absent in PtCDPK20 and PtCDPK21. However, the structure of autoinhibitory domain differed among the CDPK protein classes (Fig. 1a). For instance, the autoinhibitory domain of class IV proteins was prefixed with an NNM sequence differing from that of other classes that had an SRLK sequence. Additionally, four conserved EF hands were present in the C-terminus of all the CDPKs. EF-hand 2, which had a conserved DVD sequence, was imperfect in almost half of the investigated CDPKs, carrying only the last D or being replaced by a T in some sequences. Likewise, the EF-hand sequences differed with protein class. For example, in class IV, the DLK motif (EF-hand 1) was substituted with the DMK motif. The full details of the domain and motif structures are depicted in Fig. 1a. Taken together, these findings indicate that the CDPK genes are well conserved in land plants to varying degrees, suggesting a diverse conserved mode of function.
Fig. 1.
The CDPK conserved domain in land plants. a. EF-hand and autoinhibitory structures in different CDPK protein classes. The sequence logos were generated by the WebLogo online tool (https://weblogo.berkeley.edu/logo.cgi) based on the protein class alignments constructed using Mega11. The overall height of each stack letter indicates the sequence conservation at that position (measured in bits), whereas the height of the symbols within each stack reflects the relative frequency of the corresponding amino acid at that position. The red-marked region shows the conserved domains. The far-left tree was not drawn to scale and does not represent any phylogenetic distributions; b. Evolutionary relationships of the CDPK gene family in 52 land plants. An unrooted NJT tree was constructed with full protein sequences in MEGA X software at a bootstrap value of 1000, and the other parameters were held constant. Phylogenic classes were designated based on protein clustering and previous publications. Different color schemes show different CDPK protein classes. Different groups and subgroups are labeled in the figure
Additional conserved domain analysis using the NCBI-CDD and displayed through the TBtools software revealed protein clustering into evolutionary clades based on domain conservation (Fig S1). Proteins clustered together based on similar domain arrangement, conservation, and total number. The number of domains within each sequence varied from 2 to 5, with the majority of the CDPKs having at least two conserved domains. However, all the CDPKs contained the STKc_CAMK domain at the N-terminus and an EF-hand domain in the form of either the EF-hand_7 or FRQ1 superfamily. Nonetheless, some CDPK sequences carry additional conserved domains.
Evolutionary relationship of CDPK proteins in land plants
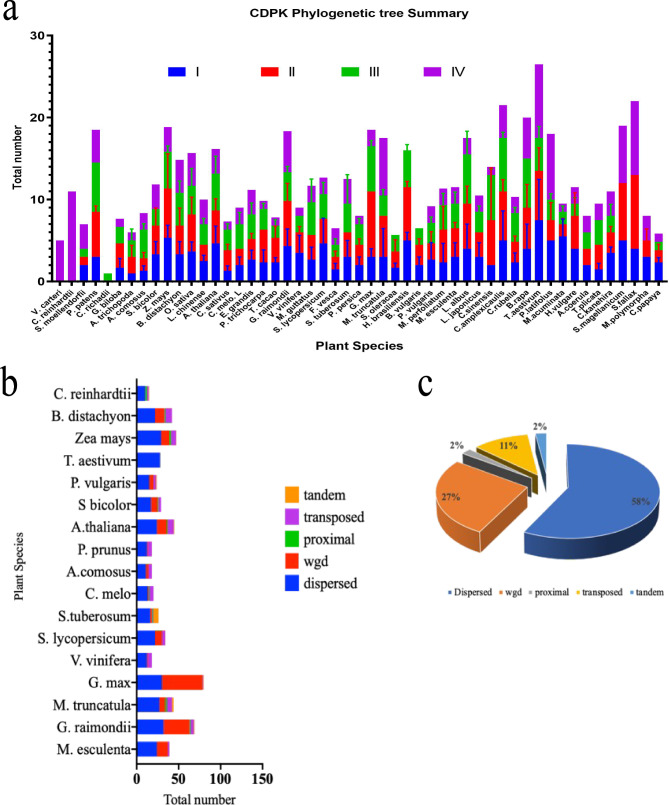
To understand the evolutionary relationships of CDPK proteins, we constructed a phylogenetic tree of CDPK protein sequences among the 52 plant species using the NJT and ML methods (Fig. 1b, Fig. S2, respectively). Based on the phylogenetic tree topology, the CDPK proteins were divided into four main groups and several subgroups (Fig. 1b, Fig S2). These findings were consistent with the previous CDPK evolution classification [6, 27]. CDPK proteins were unevenly distributed within each group, showing conservation during the evolution of land plants. Protein sequences from different plants clustered together based on protein arrangement, conserved domains, and possibly functions. CDPK proteins from higher plants even clustered with lower plant CDPK proteins; for example, M. polymorpha CDPK4 was in the same evolutionary cluster (class IV) as AtCDPKs. This finding suggested that these AtCDPKs retain conserved functional domains. Detailed analysis revealed that Group I was divided into three subgroups: Group I (A-C) and Group IC had an additional subdivision (Group IC-i) (Fig. 1b). Group II had two subgroups, which were further subclassified into two minor subgroups, Group II (ai-ii; bi-ii). Group III had two subgroups (IIIa-b); subgroup IIIb had three additional subgroups (IIIbi-iii). Group IV had three subgroups, subgroup IVbi-ii and an unclassified subgroup. Total number comparisons between groups which revealed that Classes III and II had the most and least number of protein sequences, 319 and 226, respectively (Fig. 2a).
Fig. 2.
Summary of the CDPK gene family phylogenetic tree and gene duplication events. a. A detailed bar graph of the distribution of the CDPK protein sequences within each class denoted with blue, red, green, and purple for classes I-IV, respectively. Plant species are labeled on the x-axis, and the y-axis shows the total numbers; b. The total number of duplication event types present in individual plant species generated using the (http://pdgd.njau.edu.cn) [85]; c. The total distribution of different duplication events in the investigated plant species. Different color backgrounds denote different duplication events, as shown in the key
To trace gene divergence, we computed the phylogenetic relationships between the plant species using Xshell ortho-Finder software (Fig S3). The findings showed that all the plants evolved from a common ancestral plant and diversified into different plant clades. However, there was little evolution in ferns and mosses compared to angiosperms. Speciation was greater in the angiosperms than in the later plants. Additionally, speciation was greater in the Eudicots than in the monocots, indicating that eudicots contribute more to the plant gene pool, including the CDPK gene family.
Expansion of CDPK genes in land plants
Gene family sizes vary with plant species, and gene duplication events are responsible for the expansion of gene families. Thus, novel CDPK homologs have evolved and duplicated to attain functional specialization [28]. In this research, family sizes were inconsistent among the different plant species. The protein sequence numbers varied from 37 to 1 in G. max and C. richadii, respectively (Fig. 2a). Therefore, to gain insight into the mechanism of CDPK gene expansion in land plants, we investigated gene duplication events in several land plants (Fig. 2b/c; Table S3). Findings from 14 plants showed that dispersed duplication events contributed greatly to the expansion of the CDPK gene family. The tandem and proximal duplication events made the least contributions (Fig. 2c). In-depth analysis revealed that some duplication events were absent in other plants while present in other plants and that almost all duplication events occurred (Fig. 2c). For instance, in G. max, only three events were shown to have contributed to the expansion of the CDPK genes: dispersed, whole-genome duplication (WGD), and transposed duplication events. However, in S. tuberosum, all the investigated duplication events contributed to its gene family expansion. In total, different duplication events contributed to the expansion of the CDPK gene family in different plant species, and dispersed duplications made the greatest contribution.
Protein interaction networks
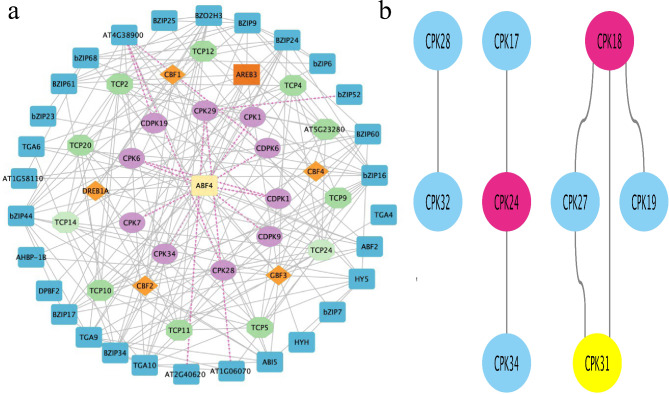
Protein-protein interaction (PPI) analysis plays a key role in predicting the functions of target proteins [29]. In previous research, we analyzed the PPIs of several proteins, including TCPs, bZIPs, and CBFs [30–33]. To predict the functions of CDPKs, we analyzed the PPIs of CDPKs with CDPKs and other protein families (bZIPs and CBFs) in Nanlin 895 using the String database (Fig. 3a). Generally, the results showed a densely interlinked network of proteins from four different protein families. Interestingly, the CDPK proteins had direct interactions with a few other protein families. For example, PtCDPK7/1/9/34/1/6 were directly linked to ABF4, a bZIP protein with specificity for abscisic acid-responsive elements (ABREs), to mediate ABA-dependent stress responses. On the other hand, PtCDPK19, PtCDPK6, and PtCDPK28 were linked to AT4G38900, a bZIP TF that is also essential for plant growth and development. Nonetheless, we observed that CDPKs had fewer interactions with each other; for instance, PtCPK18 was linked to PtCDPK27, PtCDPK19, and PtCDPK31 independently, and PtCDPK24 was linked to PtCDPK17 and PtCDPK34 (Fig. 3b). Therefore, we concluded that PtCDPKs preferably formed interactions with other gene families to facilitate their functional roles.
Fig. 3.
Protein-protein interactions. a. Network analysis of CDPK with TCP, BZIP, and CBF proteins generated by the String database (https://string-db.org/) using Arabidopsis plant species as a reference plant. The network was visualized with Cytoscape software. Different colors depict different proteins; each interaction is denoted by an interacting gray line, and colored lines show the interaction of the CDPKs. b. The protein-protein interaction network of PtCDPKs was analyzed using the STRING website (http://string-db.org, accessed on 25 June 2023) using the full-length protein sequences of the PtCDPK protein family. Arabidopsis thaliana was used as a reference plant species. Each PtCDPK protein is labeled at the node, and the gray line indicates interactions
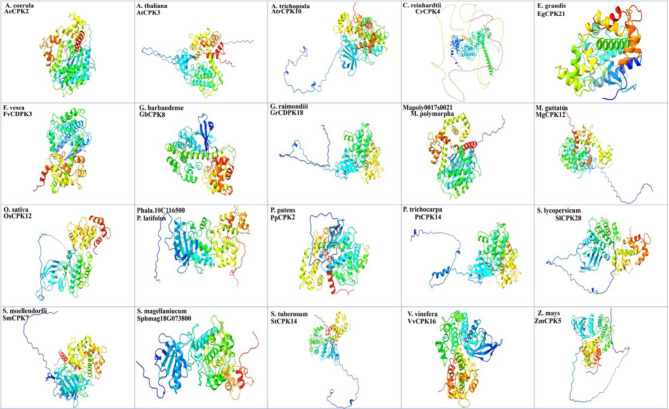
Three-dimensional (3D) protein structures provide knowledge related to structural functions. To gain insight into the possible functions of CDPKs, we selected 20 protein sequences from different plant species based on previous research. The selected protein sequences were modeled for 3D protein structures (Fig. 4). Generally, the protein tertiary structures composed of different subunits of both α-helix and β-pleated sheets that formed coils and stranded residues, aiding in hydrophobic interactions between protein-protein complexes.
Fig. 4.
The 3D protein structure prediction of 19 CDPK proteins from different plant species revealed potential strand, helical, and coil formation
Gene ontology analysis of the CDPK genes
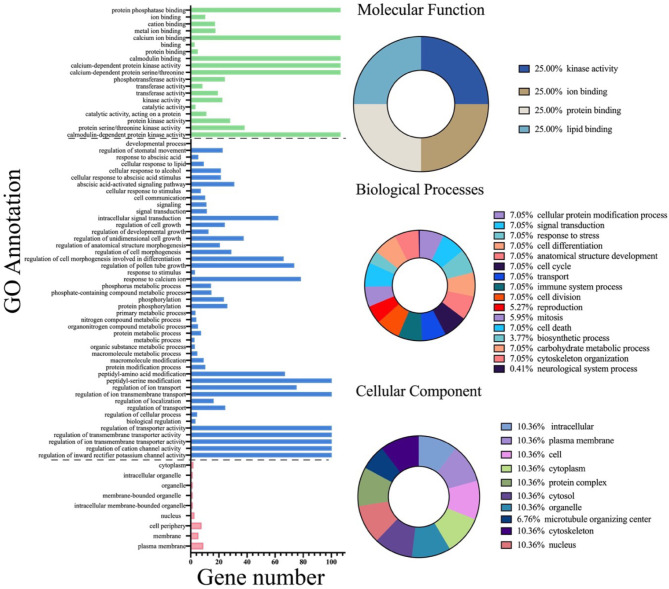
To predict the unified CDPK gene attributes in all the species investigated, we performed a Gene Ontology (GO) analysis (Fig. 5; Additional File 4). All the identified CDPK gene processes and functions were assigned to three independent GO terms: biological process (BP), molecular function (MF), and cellular component (CC). The MF category had comparably more processes and activities assigned than the BP and CC categories, constituting 45%, 35%, and 20%, respectively. In-depth analysis revealed that genes in the BP category were involved in various processes, including intracellular autophosphorylation, protein phosphorylation, and abscisic acid-activated signaling pathways. In contrast, the MF category had more genes involved in calmodulin-dependent protein kinase activity, ATP binding, and calcium-dependent protein serine/threonine kinase. The CC category showed that most of the CDPK genes were localized in the nucleus, while a few were localized in the plasmodesma. This result indicates that CDPKs are localized on the membrane and are involved in several signal transduction pathways.
Fig. 5.
GO annotation of PtCDPK genes. The bar graph on the right shows a full summary of the gene allocation in different processes and responses. Pie charts on the right side show summaries
Transcriptome and qPCR analyses of PtCDPK gene expression
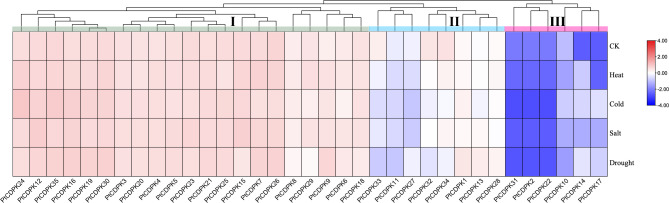
Transcriptome expression analysis provides insight into gene responses to various abiotic stresses. We performed transcriptome analysis in populus seedlings under four abiotic stresses (cold, heat, drought, and salt) (Fig. 6). The genes clustered together into three main groups based on similar gene expression values. To better analyze this result, we named the expression groups I-III. Compared with those in the control group, the genes in group I were highly upregulated in all treatment groups. Specifically, PtCDPK24/12/35/16/19/30 had the greatest upregulation under cold stress, while PtCDPK12/35/16/19/30 had the greatest upregulation under drought stress. This observation may suggest that PtCDPK genes in group I respond positively to abiotic stresses investigated. Group II comprised PtCDPK genes that were both fairly upregulated and downregulated. In detail, PtCDPK33/11/27 were fairly downregulated under all the stresses analyzed. PtCDPK1/34/13/28 were strongly upregulated. Overall, this observation suggested that some group II genes partially respond to abiotic stresses. Group III comprised significantly downregulated genes, implying that these genes do not respond to abiotic stresses and that they may negatively regulate the abiotic stresses analyzed. Overall, these findings support the hypothesis that CDPK genes respond to various abiotic stresses.
Fig. 6.
Transcriptome expression analysis of PtCDPK genes under four abiotic stresses (heat, cold, salt, and drought) was performed. The heatmap was generated using TBtools software
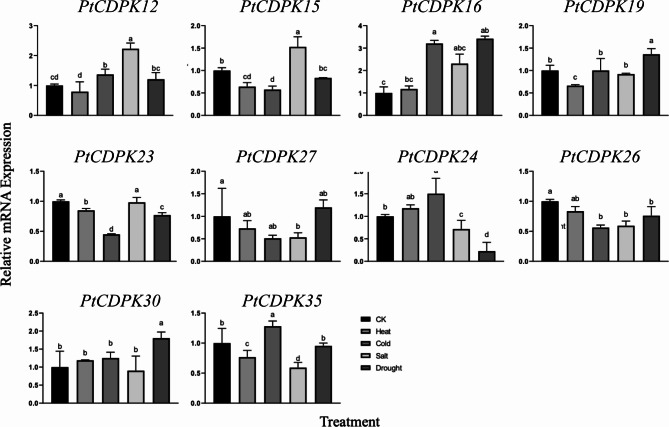
To validate expression results from transcriptome data under the four abiotic stresses, 10 PtCDPK genes were selected based on the transcriptome data for qPCR analysis (Fig. 7). The obtained results corresponded with the transcriptome data, and we concluded that PtCDPKs are to a greater extent involved in abiotic stress regulation.
Fig. 7.
qPCR expression analysis of PtCDPK genes. Relative expression values are shown in the bar graphs generated using GraphPad Prism. Significant differences are shown with error bars generated using the SEM of three replicates
Discussion
In the past decade, plant genomic analysis expertise has increased along with the characterization of several plant CDPKs and their respective functional regulations. Some plant CDPKs have been characterized in tomato [34, 35], grape [21], maize [36], B. distachyon [37], P. edulis [38], etc. To gain insight into the evolution and conservation of CDPK factors, we searched 52 plant species and identified their CDPK proteins, which had conserved DLK domains, autoinhibitory motifs, and EF-hand domains (Fig. 1; Fig S1/2). However, some sequences had incomplete or partially conserved domains, which we related to gene mutations, especially in the EF-hand region. This finding suggested that these sequences may lack sensitivity to Ca2+ concentration fluctuations [39]. To support these findings, we further analyzed the conserved domain arrangements in 41 different CDPK proteins. The results showed the conservation of 2 to 5 domains, characterized by the presence of STKc_CAMK, a calmodulin-like domain (CAM-LD), and EF-hands. This observation concurs with previous findings that the CDPK domain is a result of fusion between calmodulin and kinase domains, which facilitates the direct activation of CDPKs by calcium ions [40, 41]. Research has also shown that these conserved domains act as Ca2+-triggered switches that sense minute transient changes in the cellular Ca2+ concentration. Thus, they respond to signals by conformation changes upon Ca2+ binding. Specifically, the CAM-LD directly binds calcium, inducing intramolecular conformational changes that result in calcium-specific activation of the catalytic domain [42]. Additionally, CDPKs possess a bipartite nuclear localization signal sequence as a subdomain in their JD, which indicates the absence of consensus binding sites in their respective CAM-LD [2, 3, 43]. Nonetheless, a change in the CDPK globular structure may inhibit interactions with target proteins. Research in Arachis hypogea demonstrated that a deviation in the calcium-binding site confers a significantly lower affinity for calcium [43].
Gene duplication provides preliminary genetic materials for functional diversification through subfunctionalization or neofunctionalization [44]. Plants have exploited this mechanism through chromosomal or whole-genome duplications to increase their gene family size [45]. In this research, the gene family sizes varied among plant species between 5 and 40 protein sequences, which we related to gene duplication events. To understand the specific mechanisms involved in gene family expansion, we performed gene duplication event analysis (Fig. 2b). We observed that dispersed gene duplication events were responsible for the expansion of the CDPK gene family in plants, excluding G. max and G. raimondii. Similarly, several plant gene families, such as the TCP [33], PINIOD [46], and alpha prolamin [47] gene families, have been shown to have expanded through dispersed gene duplications. The WGD event was responsible for the expansion of the G. max and G. raimondii CDPK gene families. In support of these findings, previous research has shown that WGD in legumes has played a significant role in the expansion of their genome size and speciation [48]. Other gene families, such as the MADS-box [49] and Cycloidea-like [50] families, have also been shown to have expanded as a result of WGD events. Furthermore, we observed that WGD was prominent in other angiosperms to a certain extent. WGD was shown to be a key driver in angiosperms, contributing to both genome size and quality, hence the term WGD. However, not all paleopolyploids have undergone WGD events, and other mechanisms, such as segmental and tandem gene duplication, are involved in gene family expansions [51–53].
In addition, research has shown a high similarity of CDPK genes from distinguished gene pairs [54]. For instance, the amino acid sequences of CDPKs in (A) thaliana, AtCPK 4, and AtCPK11 exhibit 94% similarity, while those of 11 pairs of CDPKs in rice are closely related [20, 55]. These findings suggested that the gene family members were duplications of a single ancient gene. To support this phenomenon, research on wheat CDPKs has shown that TaCDPK7 and TaCDPK12 are identical and exist in other grass species, including rice, sorghum, maize, and (B) distachyon, indicating that these genes were strongly selected and conserved in the grass family [19].
The divergence of plant CDPKs has been estimated to occur between 270 and 340 MYA, closer to the period when land plants diversified into vascular and nonvascular plants [56, 57]. Using phylogenetic analysis, we showed that the CDPKs had a common ancestral gene and further diversified into four major evolutionary groups, implying that the most recent common ancestor had four CDPK genes. In this research, we speculated that Group IV CDPKs were the earliest lineage from the common ancestor of land plants, and we observed that they were more divergent. Other publications have shown that algal CDPKs have highly diverged due to the length of their evolution compared to that of other land plants [58]. Furthermore, the green algae (Chlorophyta) clade, i.e., C. reinhardtii and V. carteri, clustered in Group IV, consistent with previous research [6, 59], suggesting that this group contains highly conserved genes and can be used as a reference for genomic research [60]. However, other lower plant CDPKs, such as those of lycophytes and bryophytes, were also distributed within the remaining three major groups. For instance, Group IIb included CDPKs from lower plants, showing that CDPKs are ancient evolutionary factors that predate plant life and that their evolution played a major role in the adaptation and transition of plants to terrestrial life. Additional analysis showed a supplementary subdivision of the major groups into minor groups. Group I had three subgroups (Ia-c), and subgroup c had an additional subgroup (Ic-i). Group II had an additional two minor subgroups, Group IIa-b (i-ii). Group III had an additional minor classification in subgroup b, Group IIIb (i-iii). Group IV had four minor groups, which was consistent with previous findings [6]. Within these minor subgroups, monocot and eudicot CDPKs clustered into several evolutionary groups based on sequence similarity, possibly suggesting functional similarity. Overall, these findings on monocot and eudicot distributions suggest that the diversification of CDPK genes occurred in the ancestral CDPK genome. Similarly, the seedless plants formed monophyletic groups on separate branches; for instance, in Group IIIbii, P. patens, M. polymorpha, and S. phallax CDPKs clustered separately, suggesting that these genes are evolutionarily closer and diverged less from the common ancestor gene.
According to the interspecies evolutionary studies (Fig. S4), plants diverged from common ancestral plants into several species through speciation. However, speciation was greater in angiosperms, following the fact that angiosperms underwent two rounds of WGD. There is a complex pattern in both ancient and extant angiosperms that acts as a catalyst for plant diversification [61].
The above-discussed analyses showed that the CDPKs clustered based on sequence similarities, conserved domain arrangements, and evolutionary relationships. This conclusion can also suggest functional similarity of the clustered CDPKs, although there are functional variations that may be a result of several rounds of evolution and adaptations [62]. In retrospect, researchers have shown that CDPKs are involved in multiple functions, including growth, development, and stress response. Specifically, plant CDPKs perceive rapid intercellular changes in Ca2+ concentrations, which are then relayed further downstream through specific phosphorylation events to regulate various plant processes and external stresses (Fig. S5) [63]. In this study, we presented a comprehensive GO analysis of PtCDPKs provided by the RNA-seq data (Fig. 5). Generally, CDPKs are located in various organelles, with the majority localized in the plasma membrane, nucleus, and cytoplasm. This finding attests to the fact that CDPKs sense changes in the intracellular Ca2+ concentration and translate them into phosphorylation events that initiate downstream signaling processes [64]; thus, their localization and distribution are vital. In A. thaliana, AtCPK7, AtCPK8, AtCPK9, and AtCPK16, which are located in the cell membrane, function as Ca2+ direct transporters and important ion channels on the cell membrane [65]. In addition, AtCPK3 and AtCPK4 are located in the cytoplasm or nucleus, and AtCPK2 is located in the endoplasmic reticulum, suggesting that different PtCDPK family members distributed in different cell organelles may have different functions in cell-related abiotic stress regulation through the calcium-dependent pathway. Several studies have shown that abiotic stresses are regulated by CDPKs; for instance, CDPKs in potato were shown to initiate the regulatory response of ABF4 to drought and salt stress by regulating abscisic acid signaling pathways [66–68]. In this study, we also showed that several PtCDPKs interacted with ABF4 (Fig. 3), suggesting that PtCDPK29/6/1/9/28/34/7/19 may regulate drought stress by interacting with ABF4. We also revealed that the protein structures of CDPKs contribute to their overall function. Experiments in maize have also shown that the functions of most ZmCDPKs are largely determined by their structures. The ZmCDPKs carry four domains to which, in the absence of Ca2+, the junction region bound to the kinase domain acts as a pseudosubstrate that inhibits kinase activity. In the presence of Ca2+, the CAM domain changes the protein structure to activate ZmCDPK, after which the kinase domain is released to mediate phosphorylation and Ca2+ signaling [69–71].
Analyses of the expression patterns of the PtCDPKs using RNA-seq data and qPCR also revealed that the CDPKs respond to abiotic stresses, including drought, salt, heat, and cold (Figs. 6 and 7). Specifically, PtCDPKs from group I exhibited high expression patterns under all stresses at all time points investigated, and we assumed that these genes may regulate abiotic stresses by linking various genes, such as bZIPs, CBFs, TCPs, and/or even CDPKs. ZmCDPK35/37 localized on the plasma membrane and induced by an increase in Ca2+ concentrations during drought stress were shown to alleviate drought stress by activating ZmSLASC1-mediated CL and NO3 currents [72]. In other studies, AtCPK33 was shown to interact with thiamine thiazole synthase 1 (TH1) to regulate drought stress by regulating ABA-induced stomatal closure [73]. In G. hirsutum, GhCDPKs have been found to respond to salt stress through the ethylene signaling pathway [74], suggesting that PtCDPKs can also regulate salt stress by interacting with phytohormones, such as ethylene and ABA. AtCDPK12 was also shown to confer salt tolerance by regulating ion homeostasis and H2O2 production in the roots [74]. ZmCDPK1 was demonstrated to act as a negative regulator of cold stress by suppressing the expression of a cold-inducer marker gene, Zmfer3 (ZmCOI6.21) [75]. In this study, PtCDPK genes in groups II and III were downregulated, suggesting that these genes may act as negative regulators of the abiotic stresses investigated. Furthermore, the CDPKs were shown to interact with other plant transcription factors for full functionality in response to abiotic stresses (Fig. 3A). To support this finding, CDPK TFs have been shown to interact with and modulate the activity of components of the ICE-CBF-COR pathway [76]. These enzymes phosphorylate the ICE, thereby increasing its activity and stability, which activates CBF, leading to the modulation of downstream gene expression (Fig. S5). The activation of ICE and CBF by the CDPKs ultimately regulates cold-responsive genes, including CPR genes [77]. Overall, this analysis contributes to the understanding of the involvement of the CDPK gene in abiotic stress regulation and provides a firm foundation for additional functional characterization of CDPKs in populus.
Conclusion
CDPK genes are well conserved in the plant family. In this study, we have shown their evolution from an ancient common gene, which we concluded to have had four primary genes, which were duplicated through evolution and speciation into several CDPK plant gene families. Most of the analyzed duplication events were biased toward dispersed duplication events and WGD in legume flowering plants. However, there is a need for extensive studies on the estimation of the divergence times of duplicated gene pairs, and future research should focus on the history of CDPK genes in plants to understand their origins, diversification, and conservation across different plant lineages. Additionally, comparative genomics should provide insights into the functional significance of CDPK gene family expansion and contraction. We also established that CDPKs from different plant species exhibit similar functions in abiotic stress regulation due to domain and motif conservation and used PtCDPKs as a point of reference to emphasize that CDPKs regulate cold, heat, salt, and drought stresses. Therefore, we concluded that CDPK genes are actively involved in the regulation of abiotic stresses because they act as secondary messengers to sense intracellular Ca2+ concentration changes and relay signals downstream through phosphorylation (Fig S5). With the progression of research on the CDPK gene family, numerous plant CDPKs have been characterized, and their functions have been elucidated. There is still a need to bridge the gap in understanding the mode of action of CDPK in stress alleviation. The progression and integration of molecular biology studies have great potential to fill this gap. For instance, the advent of CRISPR/Cas-mediated plant genome editing techniques has led to plant genome knock-ins and knockouts, which are expected to answer many questions regarding CDPK gene responses and pathways. The current study provides a comprehensive overview of the evolution and functions of CDPK genes in plants, providing a foundation for further functional studies of this essential gene family in plants.
Methods
Mining ofCDPKgene families from various plant species.
The BLASTP search tool [26] was implemented in the Viridiplantae clade search in Phytozome. v13 database, using Arabidopsis thaliana CDPK proteins as queries obtained from the TAIR database (https://www.arabidopsis.org/). The threshold value was set at an E value < 10− 5. Additionally, the HMM models of PKinase (Pfam-ID: PF00069) and EF-hand (Pfam-ID: PF13499) of the CDPK gene family were obtained from the Pfam database v.28 [78]. The proteomes of the 51 plant species were searched via simple HMMER searches in TBtools [79], with an E < 10− 5. After screening, at least 50% of the identified CDPKs considered had a coverage of the Pfam domain model, and the CDD search tools from both the NCBI CDD (https://ncbi-nlm-nih-gov.brum.beds.ac.uk/Structure/bwrpsb/bwrpsb.cgi) and SMART (http://smart.embl-heidelberg.de/) were further used to authenticate the obtained putative CDPKs. Then, the EF-hand and N-myristoylation motifs were predicted by PROSITE (https://prosite.expasy.org/scanprosite/) [80]. Hits with significant similarity were classified as CDPKs based on three criteria: (I) a cutoff BLAST and HMMER score of at least 250 and at least an E-value of < 10− 5; (II) the presence of CDPK conserved domains, namely, the N_VD, CAM-like, PK, AJ, and CT variable domains; and (III) the presence of at most five functional EF-hands within the CAM-like domain. Additionally, any CDPK sequences with degenerate EF-hands, as determined by InterProScan, were excluded. Refer to the Additional File, Table S1, for all the included plant species and their information.
Multiple sequence alignments, phylogenetic and gene duplication event analyses
Multiple sequence alignments and phylogenetic analyses were carried out using Geneious Prime v. 2024.0. Full CDPK protein sequences were used for phylogenetic analysis via the NJT method, with 1000 bootstrap replicates, and the threshold value was set at 50%. To authenticate the results of the NJT method, a maximum likelihood (ML) method was used in MEGA X to generate a CDPK phylogenetic tree, with the bootstrap value set at 1000 and other options set as default. The consensus trees were beautified using the online iTOL tool [81]. To understand the gene duplication events of the CDPK genes in several plant species, we computed the duplication event types for 17 different plant species using the plant duplicate gene database PDGD (http://pdgd.njau.edu.cn8080; accessed 11 March 2023) based on their availability in the database.
Protein-to-protein interactions and protein 3D structure predictions
To gain insight into the functions and interactions of the identified CDPKs, we investigated protein-protein interactions in Arabidopsis CDPKs using the online tool String database based on orthogonal analysis. The network was visualized using Cytoscape software. Additionally, the TCP and bZIP TFs were included in the PPI network analysis to possibly outline interactions of the CDPKs with other abiotic stress-responsive proteins. The online SWISS-MODEL database (https://swissmodel.expasy.org/) was used to generate CDPK protein models, which were subsequently visualized using Chimera X software.
Plant material culture and abiotic stress treatment
Populus seedlings (Populus deltoides x Populus euramericana ‘Nanlin 895’) were grown in a growth chamber and potted in pots containing a mixture of vermiculite, perlite, and organic matter (3:1:1). The chamber conditions were set at 23 °C, 5000 lx light intensity, 16 h (h) light/8 h darkness, and relative humidity of 55% [32]. At five weeks, the plants were separated into five groups: control (22 °C), heat stress (40 °C), cold stress (-4 °C), drought stress (40% polyethylene glycol/PEG6000), and salt stress (200 mM NaCl). These conditions were set based on previous methods by Hwarari et al. [30, 82]. Three biological replicates were used for each stress treatment. Both the tender and mature leaves of the seedlings were extracted at three days after stress treatment in a 50 mL centrifuge tube and then placed in liquid nitrogen for quick freezing and stored at -80 °C for additional experimentation.
RNA-Seq and qPCR analyses of PtCDPKs in response to abiotic stresses
Transcriptome sequencing was performed on the samples mentioned above, and transcriptome data were obtained for the Pt CDPK gene family members. Based on the RNA-seq data, the transcript abundances of the CDPK genes were calculated using fragments per kilobase of exons per million mapped reads (FPKM) values. Microsoft Excel 2016 was used to convert (FPKM + 1) to log base 2, and TBtools (TBtools.v1.09854) was used to create a heatmap of CDPK gene expression.
qPCR expression analysis was used to validate the RNA-seq data expression analysis, and 10 Pt CDPKs were selected based on the RNA-seq expression patterns. The selected genes were expressed under cold, heat, drought, and salt stress conditions at 3 days. The primers were designed with Batch Primer Design [83], as shown in the Additional file (Table S2). Total plant RNA was extracted according to the instructions of the Plant Total RNA Extraction Kit, and cDNA was synthesized with a reverse transcription kit. RT‒qPCR was performed on a LightCycler 480 II (Roche, Switzerland) using the SYBR Green Premix Pro Taq HS qPCR Kit (AG11701, ACCURATE Biotechnology, HUNAN, Co., Ltd.). A 20 µL reaction mixture was prepared according to the instructions of the fluorescence quantitative kit. RT-qPCR was performed at 95 °C for 2 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Three technical replicates were used per treatment, and the gene expression values were averaged. PtActin and PtUCBQ10 were selected as internal reference genes, and gene expression changes were analyzed by the 2−△△CT method [84]. The obtained data were summarized and analyzed in MS Excel and displayed with GraphPad software.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to the funders of this research, the editors, and the reviewers for the effort and time spent giving helpful comments to improve our work.
Abbreviations
- CDPK
Calcium-Dependent Protein Kinase
- Ca2+
Calcium ion
- RNA-seq
RNA sequence
- CBL
Calcineurin B-like Protein
- CaM
Calmodulin
- CML
Calmodulin-Like Protein
- CAD
CDPK Activation Domain
- EF
Elongation Factor
- NJT
Neighbor-Joining Tree
- PPI
Protein-Protein Interaction
- 3D
Three-Dimensional
- GO
Gene Ontology
- BP
Biological Process
- MF
Molecular Function
- CC
Cellular Component
- WGD
Whole-Genome Duplication
Author contributions
H.D. and Z.M. conceived, planned, coordinated the project, and finalized the manuscript. Z.M., M.X., and H.D. performed the experiments and data analysis and wrote the draft. T.M. and F.Z. validated and contributed to the data analysis and curation and revised the manuscript. F.Z. and L.Y. contributed to data curation, and funded this research.
Funding
This research was supported by STI 2030- Major Projects (No. 2023ZD0405602) and the Research Startup Fund for High-Level and Highly Educated Talents of Nanjing Forestry University.
Data availability
Transcriptome datasets are also available on the NCBI website; the accession number is PRJNA1172555 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1172555, accessed on October 16, 2024).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhiying Mu and Mingyue Xu contributed equally to this work.
Contributor Information
Delight Hwarari, Email: tondehwarr@njfu.edu.cn.
Fu-Yuan Zhu, Email: fyzhu@njfu.edu.cn.
References
- 1.Waadt R, Seller CA, Hsu P-K, Takahashi Y, Munemasa S, Schroeder JI. Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol. 2022:1–15. [DOI] [PMC free article] [PubMed]
- 2.Christodoulou J, Malmendal A, Harper JF, Chazin WJ. Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J Biol Chem. 2004;279(28):29092–100. [DOI] [PubMed] [Google Scholar]
- 3.Bender KW, Blackburn RK, Monaghan J, Derbyshire P, Menke FL, Zipfel C, et al. Autophosphorylation-based calcium (ca(2+)) sensitivity priming and ca(2+)/Calmodulin inhibition of Arabidopsis thaliana ca(2+)-dependent protein kinase 28 (CPK28). J Biol Chem. 2017;292(10):3988–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao S, Zhang Q, Liu M, Zhou H, Ma C, Wang P. Regulation of plant responses to salt stress. Int J Mol Sci. 2021;22(9):4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keshan R, Patra A, Mehta S, Abdelmotelb K, Lavale SA, Chaudhary M, et al. Expression and regulation of stress-responsive genes in plants under harsh environmental conditions. Harsh environment and plant resilience: Springer; 2021. pp. 25–44. [Google Scholar]
- 6.Valmonte GR, Arthur K, Higgins CM, MacDiarmid RM. Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 2014;55(3):551–69. [DOI] [PubMed] [Google Scholar]
- 7.Trewavas AJ, Malho R. Signal Perception and Transduction: the origin of the phenotype. Plant Cell. 1997;9(7):1181–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putnam-Evans CL, Harmon AC, Cormier MJ. Purification and characterization of a novel calcium-dependent protein kinase from soybean. Biochemistry. 1990;29(10):2488–95. [DOI] [PubMed] [Google Scholar]
- 9.Parvathy ST. Versatile roles of ubiquitous calcium-dependent protein kinases (CDPKs) in plants. Indian Soc Oilseeds Res. 2018;35:1–11. [Google Scholar]
- 10.Zeng H, Zhang Y, Zhang X, Pi E, Zhu Y. Analysis of EF-hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front Plant Sci. 2017;8:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohanta TK, Yadav D, Khan AL, Hashem A, Abd_Allah EF, Al-Harrasi A. Molecular players of EF-hand containing calcium signaling event in plants. Int J Mol Sci. 2019;20(6):1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaur A, Upadhyay SK. EF-hand domain-containing proteins: diversity and role in plants. Cation Transporters in Plants: Elsevier; 2022. pp. 185–203.
- 13.Mathivanan S. Abiotic stress-Induced Molecular and physiological changes and adaptive mechanisms in plants. Abiotic Stress Plants. 2021:315.
- 14.Köster P, DeFalco TA, Zipfel C. Ca2 + signals in plant immunity. EMBO J. 2022:e110741. [DOI] [PMC free article] [PubMed]
- 15.Chandran V, Stollar EJ, Lindorff-Larsen K, Harper JF, Chazin WJ, Dobson CM, et al. Structure of the Regulatory Apparatus of a calcium-dependent protein kinase (CDPK): a Novel Mode of calmodulin-target recognition. J Mol Biol. 2006;357(2):400–10. [DOI] [PubMed] [Google Scholar]
- 16.Almadanim MC, Alexandre BM, Rosa MT, Sapeta H, Leitão AE, Ramalho JC, et al. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose‐phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017;40(7):1197–213. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Wang J, Zhang S, Yu Q, Lan H. Investigation of the nature of CgCDPK and CgbHLH001 interaction and the function of bHLH transcription factor in stress tolerance in Chenopodium glaucum. Front Plant Sci. 2021;11:603298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bundó M, Coca M. Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J Exp Bot. 2017;68(11):2963–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A-L, Zhu Y-F, Tan X-M, Wang X, Wei B, Guo H-Z, et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L). Plant Mol Biol. 2008;66(4):429–43. [DOI] [PubMed] [Google Scholar]
- 20.Asano T, Tanaka N, Yang G, Hayashi N, Komatsu S. Genome-wide identification of the Rice Calcium-dependent Protein Kinase and its closely related kinase gene families: Comprehensive Analysis of the CDPKs Gene Family in Rice. Plant Cell Physiol. 2005;46(2):356–66. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K, Han Y-T, Zhao F-L, Hu Y, Gao Y-R, Ma Y-F, et al. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 2015;15(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Hu W, Ren L, Jia C, Liu J, Miao H, et al. Identification, expression, and Interaction Network analyses of the CDPK Gene Family Reveal their involvement in the Development, Ripening, and abiotic stress response in Banana. Biochem Genet. 2020;58(1):40–62. [DOI] [PubMed] [Google Scholar]
- 23.Klimecka M, Muszyńska G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim Pol. 2007;54(2):219–33. [PubMed] [Google Scholar]
- 24.Schulz P, Herde M, Romeis T. Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 2013;163(2):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao X, Cox KL Jr, He P. Functions of calcium-dependent protein kinases in plant innate immunity. Plants. 2014;3(1):160–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A, Sagar S, Biswas DK. Calcium dependent protein kinase, a versatile player in plant stress management and development. CRC Crit Rev Plant Sci. 2017;36(5–6):336–52. [Google Scholar]
- 28.Tong T, Li Q, Jiang W, Chen G, Xue D, Deng F, et al. Molecular evolution of calcium signaling and transport in plant adaptation to abiotic stress. Int J Mol Sci. 2021;22(22):12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchand A, Van Hall-Beauvais AK, Correia BE. Computational design of novel protein–protein interactions–An overview on methodological approaches and applications. Curr Opin Struct Biol. 2022;74:102370. [DOI] [PubMed] [Google Scholar]
- 30.Hwarari D, Guan Y, Li R, Movahedi A, Chen J, Yang L. Comprehensive Bioinformatics and expression analysis of TCP Transcription Factors in Liriodendron chinense Reveals Putative Abiotic Stress Regulatory Roles. Forests. 2022;13(9):1401. [Google Scholar]
- 31.Li M, Hwarari D, Li Y, Ahmad B, Min T, Zhang W et al. The bZIP transcription factors in Liriodendron chinense: genome-wide recognition, characteristics and cold stress response. Front Plant Sci. 2022;13. [DOI] [PMC free article] [PubMed]
- 32.Guan Y, Liu S, Wu W, Hong K, Li R, Zhu L, et al. Genome-wide identification and cold stress-induced expression analysis of the CBF gene family in Liriodendron chinense. J Forestry Res. 2021;32(6):2531–43. [Google Scholar]
- 33.Zhou H, Hwarari D, Ma H, Xu H, Yang L, Luo Y. Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology. Front Genet. 2022;13. [DOI] [PMC free article] [PubMed]
- 34.Hu Z, Lv X, Xia X, Zhou J, Shi K, Yu J, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front Plant Sci. 2016;7:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J, Wang B, Yang T, Li N, Yang H, Yu Q, et al. A calcium-dependent protein kinase gene SpCPK33 from Solanum pennellii associated with increased cold tolerance in tomato. J Plant Physiol. 2022;279:153834. [DOI] [PubMed] [Google Scholar]
- 36.Khalid MHB, Raza MA, Yu HQ, Khan I, Sun FA, Feng LY, et al. Expression, subcellular localization, and interactions of CPK family genes in maize. Int J Mol Sci. 2019;20(24):6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen F, Ye F, Xiao Z, Liao L, Li T, Jia M, et al. Genome-wide survey and expression analysis of calcium-dependent protein kinase (CDPK) in grass Brachypodium distachyon. BMC Genomics. 2020;21(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M, Liu H, Wang L, Zhang X, He W, Xiang Y. Comparative genomic analysis of the CPK gene family in Moso bamboo (Phyllostachys edulis) and the functions of PheCPK1 in drought stress. Protoplasma. 2022:1–17. [DOI] [PubMed]
- 39.Heizmann CW. Ca(2+)-Binding proteins of the EF-Hand Superfamily: diagnostic and prognostic biomarkers and novel therapeutic targets. Methods Mol Biol. 2019;1929:157–86. [DOI] [PubMed] [Google Scholar]
- 40.Harper JF, Sussman MR, Schaller GE, Putnam-Evans C, Charbonneau H, Harmon AC. A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science. 1991;252(5008):951–4. [DOI] [PubMed] [Google Scholar]
- 41.Das Gupta M, Chaudhuri S. CDPKs in Plant Signaling Networks. In: Sopory SK, Oelmüller R, Maheshwari SC, editors. Signal Transduction in plants: current advances. Boston, MA: Springer US; 2001. pp. 145–55. [Google Scholar]
- 42.Gao Q, Xiong T, Li X, Chen W, Zhu X. Calcium and calcium sensors in fruit development and ripening. Sci Hort. 2019;253:412–21. [Google Scholar]
- 43.Raichaudhuri A, Bhattacharyya R, Chaudhuri S, Chakrabarti P, Dasgupta M. Domain analysis of a groundnut calcium-dependent protein kinase: nuclear localization sequence in the junction domain is coupled with nonconsensus calcium binding domains. J Biol Chem. 2006;281(15):10399–409. [DOI] [PubMed] [Google Scholar]
- 44.Ray S, Agarwal P, Arora R, Kapoor S, Tyagi AK. Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol Genet Genomics. 2007;278(5):493–505. [DOI] [PubMed] [Google Scholar]
- 45.Magadum S, Banerjee U, Murugan P, Gangapur D, Ravikesavan R. Gene duplication as a major force in evolution. J Genet. 2013;92(1):155–61. [DOI] [PubMed] [Google Scholar]
- 46.Bai J, Song MJ, Gao J, Li G. Whole genome duplication and dispersed duplication characterize the evolution of the plant PINOID gene family across plant species. Gene. 2022;829:146494. [DOI] [PubMed] [Google Scholar]
- 47.Xu J-H, Messing J. Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proceedings of the National Academy of Sciences. 2008;105(38):14330-5. [DOI] [PMC free article] [PubMed]
- 48.Shoemaker RC, Schlueter J, Doyle JJ. Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol. 2006;9(2):104–9. [DOI] [PubMed] [Google Scholar]
- 49.Dong Z-c, Zhao Z, Liu C-w, Luo J-h, Yang J, Huang W-h, et al. Floral patterning in Lotus japonicus. Plant Physiol. 2005;137(4):1272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukuda T, Yokoyama J, Maki M. Molecular evolution of cycloidea-like genes in Fabaceae. J Mol Evol. 2003;57(5). [DOI] [PubMed]
- 51.Sémon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17(6):505–12. [DOI] [PubMed] [Google Scholar]
- 52.Veitia RA, Birchler JA. Gene-dosage issues: a recurrent theme in whole genome duplication events. Trends Genet. 2022;38(1):1–3. [DOI] [PubMed] [Google Scholar]
- 53.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geng S, Zhao Y, Tang L, Zhang R, Sun M, Guo H, et al. Molecular evolution of two duplicated CDPK genes CPK7 and CPK12 in grass species: a case study in wheat (Triticum aestivum L). Gene. 2011;475(2):94–103. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez Milla MA, Uno Y, Chang I-F, Townsend J, Maher EA, Quilici D, et al. A novel yeast two-hybrid approach to identify CDPK substrates: characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein1. FEBS Lett. 2006;580(3):904–11. [DOI] [PubMed] [Google Scholar]
- 56.Raven JA, Edwards D. Roots: evolutionary origins and biogeochemical significance. J Exp Bot. 2001;52(suppl1):381–401. [DOI] [PubMed] [Google Scholar]
- 57.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hazra A, Dasgupta N, Sengupta C, Das S. MIPS: functional dynamics in evolutionary pathways of plant kingdom. Genomics. 2019;111(6):1929–45. [DOI] [PubMed] [Google Scholar]
- 59.Chen F, Fasoli M, Tornielli GB, Dal Santo S, Pezzotti M, Zhang L, et al. The evolutionary history and diverse physiological roles of the grapevine calcium-dependent protein kinase gene family. PLoS ONE. 2013;8(12):e80818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salomé PA, Merchant SS. A series of fortunate events: introducing Chlamydomonas as a reference organism. Plant Cell. 2019;31(8):1682–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soltis PS, Soltis DE. Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol. 2016;30:159–65. [DOI] [PubMed] [Google Scholar]
- 62.Panchy N, Lehti-Shiu M, Shiu S-H. Evolution of gene duplication in plants. Plant Physiol. 2016;171(4):2294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asano T, Hayashi N, Kikuchi S, Ohsugi R. CDPK-mediated abiotic stress signaling. Plant Signal Behav. 2012;7(7):817–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao X-H, Yang M, Sui J-L, Qi J-Y, Fang Y-J, Hu S-N, et al. The calcium-dependent protein kinase (CDPK) and CDPK-related kinase gene families in Hevea brasiliensis—comparison with five other plant species in structure, evolution, and expression. FEBS Open Bio. 2017;7(1):4–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, et al. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 2003;132(4):1840–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muñiz García MN, Cortelezzi JI, Fumagalli M, Capiati DA. Expression of the Arabidopsis ABF4 gene in potato increases tuber yield, improves tuber quality and enhances salt and drought tolerance. Plant Mol Biol. 2018;98(1–2):137–52. [DOI] [PubMed] [Google Scholar]
- 67.Pan W, Zheng P, Zhang C, Wang W, Li Y, Fan T, et al. The effect of ABRE BINDING FACTOR 4-mediated FYVE1 on salt stress tolerance in Arabidopsis. Plant Sci. 2020;296:110489. [DOI] [PubMed] [Google Scholar]
- 68.Hwang K, Susila H, Nasim Z, Jung JY, Ahn JH. Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate Drought-Accelerated Flowering. Mol Plant. 2019;12(4):489–505. [DOI] [PubMed] [Google Scholar]
- 69.Reddy AS. Calcium: silver bullet in signaling. Plant Sci. 2001;160(3):381–404. [DOI] [PubMed] [Google Scholar]
- 70.Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol. 2005;6(7):555–66. [DOI] [PubMed] [Google Scholar]
- 71.Du H, Chen J, Zhan H, Li S, Wang Y, Wang W, et al. The roles of CDPKs as a convergence point of different signaling pathways in Maize Adaptation to Abiotic Stress. Int J Mol Sci. 2023;24(3):2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li XD, Gao YQ, Wu WH, Chen LM, Wang Y. Two calcium-dependent protein kinases enhance maize drought tolerance by activating anion channel ZmSLAC1 in guard cells. Plant Biotechnol J. 2022;20(1):143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C-L, Wang M, Wu X-M, Chen D-H, Lv H-J, Shen J-L, et al. THI1, a Thiamine Thiazole synthase, interacts with Ca2+-Dependent protein kinase CPK33 and modulates the S-Type Anion Channels and Stomatal Closure in Arabidopsis. Plant Physiol. 2015;170(2):1090–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pires IS, Negrão S, Pentony MM, Abreu IA, Oliveira MM, Purugganan MD. Different evolutionary histories of two cation/proton exchanger gene families in plants. BMC Plant Biol. 2013;13:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.WECKWERTH P, EHLERT B. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015;38(3):544–58. [DOI] [PubMed] [Google Scholar]
- 76.Hwarari D, Guan Y, Ahmad B, Movahedi A, Min T, Hao Z, et al. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int J Mol Sci. 2022;23(3):1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guan Y, Hwarari D, Korboe HM, Ahmad B, Cao Y, Movahedi A, et al. Low temperature stress-induced perception and molecular signaling pathways in plants. Environ Exp Bot. 2023;207:105190. [Google Scholar]
- 78.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar Gustavo A, Sonnhammer ELL, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2020;49(D1):D412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. [DOI] [PubMed] [Google Scholar]
- 80.Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, et al. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30(1):235–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hwarari D, Radani Y, Guan Y, Chen J, Liming Y. Systematic characterization of GATA Transcription Factors in Liriodendron chinense and Functional Validation in Abiotic Stresses. Plants. 2023;12(12):2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pandey GK, Kanwar P, Singh A, Steinhorst L, Pandey A, Yadav AK, et al. Calcineurin B-Like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 2015;169(1):780–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Z, Halford NG, Liu C, Real-Time Quantitative PCR. Primer design, reference gene selection, calculations and statistics. Metabolites. 2023;13(7):806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee T-H, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2012;41(D1):D1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome datasets are also available on the NCBI website; the accession number is PRJNA1172555 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1172555, accessed on October 16, 2024).